Summary
Background
Studies on the fusion-inhibitory peptides derived from the heptad repeat 1 and 2 (HR1 and HR2) regions of the HIV-1 envelope glycoprotein gp41 provided crucial information on the viral fusogenic mechanism. We used a similar approach to study the fusogenic mechanism of severe-acute-respiratory-syndrome-associated coronavirus (SARS-CoV).
Methods
We tested the inhibitory activity against infection of two sets of peptides corresponding to sequences of SARS-CoV spike protein HR1 and HR2 regions and investigated the interactions between the HR1 and HR2 peptides by surface plasmon resonance, sedimentation equilibration analysis, circular dichroism, native polyacrylamide-gel electrophoresis, size exclusion high-performance liquid chromatography, and computer-aided homology modelling and molecule docking analysis.
Findings
One peptide, CP-1, derived from the HR2 region, inhibited SARS-CoV infection in the micromolar range. CP-1 bound with high affinity to a peptide from the HR1 region, NP-1. CP-1 alone had low -helicity and self-associated to form a trimer in phosphate buffer (pH 7·2). CP-1 and NP-1 mixed in equimolar concentrations formed a six-helix bundle, similar to the fusogenic core structure of HIV-1 gp41.
Interpretation
After binding to the target cell, the transmembrane spike protein might change conformation by association between the HR1 and HR2 regions to form an oligomeric structure, leading to fusion between the viral and target-cell membranes. At the prefusion intermediate state, CP-1 could bind to the HR1 region and interfere with the conformational changes, resulting in inhibition of SARS-CoV fusion with the target cells. CP-1 might be modifiable to increase its anti-SARS-CoV activity and could be further developed as an antiviral agent for treatment or prophylaxis of SARS-CoV infection.
Introduction
The global outbreak of severe acute respiratory syndrome (SARS)1 has seriously threatened public health and socioeconomic stability. The disorder is caused by a novel coronavirus, SARS-associated coronavirus (SARS-CoV).2, 3, 4, 5, 6 Angiotensin-converting enzyme 2 is a functional receptor for the virus,7 but the mechanism by which SARS-CoV enters target cells is unclear. Elucidation of the fusogenic mechanism will provide important information for development of antiviral drugs and vaccines for treatment and prevention of SARS.
SARS-CoV could have originated from wild animals because a very similar coronavirus has been isolated from animals such as Himalayan palm civets.8 This coronavirus shares genetic and structural properties with others, including murine hepatitis virus and bovine coronavirus.9, 10, 11, 12 For example, SARS-CoV also expresses abundant spike glycoprotein on its surface in a trimeric or dimeric structure, forming spikes about 20 nm long and 10 nm wide at the bulbous end.2, 3, 4, 5, 6 The spike protein of SARS-CoV, which is involved in virus binding, fusion, and entry, is a typical class I viral fusion protein, similar to the transmembrane glycoproteins of many enveloped viruses.13 Therefore, information obtained from studies of other enveloped viruses such as HIV-1 could be useful for investigation of the fusogenic mechanism mediated by the SARS-CoV spike protein.
In the early 1990s, Wild and co-workers and our group discovered potent HIV-1 fusion-inhibitory peptides derived from the gp41 heptad repeat 1 and 2 (HR1 and HR2) regions, designated N-peptides and C-peptides, respectively.14, 15, 16 One of the C-peptides, T-20 (Trimeris, Research Triangle Park, NC, USA), was recently approved by the US Food and Drug Administration as the first of a new class of anti-HIV drugs—fusion inhibitors. Studies on the mechanism of action of these anti-HIV-1 peptides provided important information to elucidate the fusogenic mechanism mediated by HIV-1 gp41.17, 18, 19 After binding of gp120 to CD4 and a coreceptor, gp41 changes conformation via HR1 and HR2 interaction to form a fusogenic hexameric core structure, which brings the viral and target-cell membranes into close proximity, resulting in fusion between them. At the intermediate state, a C-peptide can bind the viral gp41 HR1 region and block formation of the six-helix bundle, thus inhibiting virus fusion with the target cell.20 The discovery of these HIV-1 fusion-inhibitory peptides launched a new line of investigation of viral fusogenic mechanisms and identification of antiviral peptides against infection by many other enveloped viruses—simian21 and feline22 immunodefi-ciency viruses, Ebola virus,23 Newcastle disease virus,24 and respiratory syncytial virus.25
Similarly to HIV-1 gp41, the S2 domain of SARS-CoV spike protein contains HR1 and HR2 sequences, which tend to form a coiled-coil structure (figure 1 ). The aminoacid sequences of peptides derived from the HR1 and HR2 regions of SARS-CoV spike protein are similar to those from the HIV-1 gp41 HR1 and HR2 regions (figure 1); the similarity suggests that these two transmembrane envelope glycoproteins share a common mechanism mediating fusion between the viral and target-cell membranes. With a similar approach to that used for studying HIV-1 gp41, we designed and synthesised a series of peptides corresponding to the sequences of SARS-CoV spike protein HR1 and HR2 regions and used these peptides as probes to study the fusogenic mechanism of the virus and to identify antiviral peptides.
Figure 1.
Schematic representation of SARS-CoV (strain TOR2) spike protein, sequence similarities between SARS-CoV and HIV-1 N-peptides and C-peptides, and helical wheels of HR1 and HR2
SP=signal peptide; TM=transmembrane domain; CP=cytoplasmic domain. Residue numbers of each region correspond to their positions in spike protein of SARS-CoV. Six peptides corresponding to the sequences of HR1 and HR2 regions are also shown in the upper diagram.
Methods
Peptides were synthesised on an Applied Biosystems Pioneer peptide synthesiser (ABI, Framingham, MA, USA) by standard Fmoc/2-(1H-benzo-triazole-1-yl)-1,1,3,3-tetramethyluro-nium hexafluorophosphate (HBTU) methods, purified to homogeneity by high-performance liquid chromatography (HPLC: Agilent Technologies, Palo Alto, CA, USA), and characterised with a MALDI TOF mass spectrometer (ABI Voyager DE, Foster City, CA). The peptide sequences are shown in table 1 . Biotinylated CP-1 and NP-1 were prepared by addition of biotin with HBTU to the N-termini of the peptides during their synthesis.
Table 1.
Sequences of synthetic peptides derived from HR1 and HR2 regions of SARS-CoV spike protein
| Peptide | Residue region | Residue number | Sequence |
|---|---|---|---|
| NP-1 | 892–931 | 40 | GVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQ |
| NP-2 | 920–953 | 34 | LTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFG |
| NP-3 | 946–981 | 36 | KQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG |
| NP-4 | 981–1014 | 34 | GRLQSLQTYVTQQLIRAAEIRASANLAATKMSEC |
| CP-1 | 1153–1189 | 37 | GINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYE |
| CP-2 | 1163–1199 | 37 | KEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVW |
The inhibitory activity of the synthetic peptides on SARS-CoV infection of Vero E6 cells was assessed as previously described.26 100 TCID50 (50% tissue-culture infectious dose) of SARS-CoV WHU strain (accession number AY394850) was mixed with an equal volume of a peptide at graded concentrations and incubated at 37°C for 30 min. The mixture was added to monolayers of Vero E6 cells in 96-well tissue-culture plates. After incubation at 37°C for 1 h, the supernatants were removed and fresh medium was added. On day 3 after infection, the cytopathic effect was recorded and the concentrations of the peptides resulting in 50% inhibition (IC50) were calculated.
To measure the binding affinity or kinetic dissociation constant of the SARS-CoV related peptides, we used surface plasmon resonance with an optical biosensor (model BIA 2000, BIAcore Inc, Piscataway, NJ, USA). Biotinylated CP-1 (2 μmol/L) was immobilised on streptavidin sensor chips (BIAcore). A surface reference to which no biotinylated CP-1 was bound was included in the analysis. The derivatised sensor chips were washed and equilibrated in phosphate-buffered saline (pH 7·4, 20 μL/min), and the peptides to be tested were introduced at five serial dilutions in running buffer (phosphate-buffered saline, pH 7·4, and 0·005% P20) in 200 μL injection volumes and at a flow rate of 20 μL/min. The measurement was done at 25°C for 10 min of association, followed by 15 min of dissociation. The sensogram response data were analysed by commercial software (BIA-Evaluation version 3.1), and the kinetic dissociation constant was calculated. Each experiment was done at least three times.
The secondary structure of the peptides was determined by circular-dichroism spectroscopy as previously described.27 The isolated peptides or peptide mixtures were diluted in phosphate buffer (50 mmol/L sodium phosphate, pH 7·2) at the final concentration of 10 μ mol/L, and the circular-dichroism spectra were acquired on a spectropolarimeter (Model J-715, Jasco Inc, Japan) at 4°C with a bandwidth of 5·0 nm, resolution 0·1 nm, path length 0·1 cm, response time 4·0 s, and scanning speed 50 nm/min. Thermal denaturation was monitored at 222 nm by application of a thermal gradient of 5°C/min. The spectra were corrected by the subtraction of a blank corresponding to the solvent. The melting curve was smoothed and the midpoint of the thermal unfolding transition (Tm) values was calculated with Jasco software utilities as previously described.28
The apparent molecular masses of the peptides and peptide mixtures were measured by sedimentation equilibrium analysis on a Beckman Coulter Optima XL-I Analytical Ultracentrifuge equipped with a real-time video-based data-acquisition system and Rayleigh optics. The peptides were run at three initial concentrations (250 μ mol/L, 500 μ mol/L, and 1000 μ mol/L) in 10 mmol/L sodium phosphate, pH 7·2, containing 150 mmol/L sodium chloride and 5 mmol/L tris(2-carboxyethyl)phosphine hydrochloride. Sample volumes of 110 μ L were used in six-sector cells equipped with quartz windows. All experiments were done at 25°C, and sedimentation equilibrium data were collected at 48 000 rpm (167731× g) for NP-1, 35 000 rpm (89 180× g) for CP-1, and 25 000 rpm (45 500×g) for CP-1/NP-1 complex. Sedimentation equilibrium profiles were edited by the program REEDIT (Jeff Lary, National Analytical Ultracentrifuge Center, Storrs, CT, USA) and analysed by a non-linear, least-squares method with the program WINNONLIN.29 The values of the partial specific volumes for the peptides were calculated from the aminoacid composition with the program SEDNTERP(David Hayes, Magdalen College, Oxford, UK; Tom Laue, University of New Hampshire, Durham, NH, USA; John Philo, Amgen, Thousand Oaks, CA, USA).
Native polyacrylamide-gel electrophoresis (N-PAGE) was done as previously described.30 The isolated peptides and peptide mixtures at indicated concentrations were mixed with Tris-glycine native sample buffer (Invitrogen, Carlsbad, CA, USA) at a ratio of one to one and were then loaded (25 μL each well) on a 10×1 cm Tris-glycine precast gel (18%). Gel electrophoresis was carried out with constant 125 V at room temperature for 2 h. The gel was then stained with Coomassie blue. The image was taken by a FluorChem 8800 Imaging System (Alpha Innotech Corp, San Leandro, CA, USA). In another set of experiments, mixtures of NP-1 and biotinylated CP-1 were analysed by N-PAGE as described above. The peptides in the gel were transferred to a nitrocellulose membrane (Amersham Pharmacia Biotech Inc, Piscataway, NJ, USA), which was blocked with 3% non-fat milk at 4°C overnight, followed by incubation with streptavidin-conjugated horseradish peroxidase at room temperature for 1 h. After extensive washing, the membrane was stained with 3,3 -diaminobenzidine dye and the image was taken as described above.
A Bio-Sil SEC 125-5 column (300×7·8 mm, Bio-Rad, Hercules, CA, USA) was used for size-exclusion HPLC (SE-HPLC) to assess oligomer formation.30 The HR1-derived peptide was incubated with HR2-derived peptide (final concentration of peptide 50 μmol/L) at 37°C for 30 min, and then a 20 μL sample was injected into the column connected to an HPLC system (Waters Delta 600, Milford, MA, USA). Phosphate-buffered saline (pH 7·2) was used as the mobile phase with a flow rate of 1·0 mL/min. The ultraviolet absorbance at 195–320 nm was recorded with a photodiode array detector (Waters 996). A Bio-Rad gel-filtration standard, consisting of thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1·35 kDa) was used to define the molecular masses of the peptides and their complex.
SARS-CoV spike protein was analysed for the probability of forming α-helical coiled-coil structures by use of the program MultiCoil;31 sequences are submitted to (http://jura.wi.mit.edu/cgi-bin/multicoil/multicoil.pl).
To predict the conformation of the NP-1/CP-1 complex by homology modelling and docking techniques, we first looked at the sequence alignment of the NP-1 and CP-1 peptides with the corresponding N-peptides and C-peptides of HIV-1 gp41. This analysis showed about 45–50% sequence similarity (figure 1). We used the X-ray crystal structure of the gp41 core structure (pdb code 1env) to build a model of interaction between the SARS-CoV NP-1 and CP-1 peptides as described below. We aligned HIV-1 N-peptide N36 and SARS-CoV N-peptide NP-1, and HIV-1 C-peptide C43 and SARS-CoV C-peptide CP-1 by use of the program ClustalX.32 Any insertions and deletions were manually adjusted to avoid gaps in the helix regions. Homology models of each peptide were built by the automated software Modeller 4.033 within Quanta 2000 (Accelrys, San Diego, CA, USA) running on a Silicon Graphics Octane with a dual R12000 processor (sgi, Mountain View, CA, USA). The models were optimised with the “Refine 3” option in Modeller, which uses conjugated gradient together with molecular dynamics by simulated annealing techniques. Five models for each run were built, and the model with the lowest objective function was selected. The protein geometry was verified by Quanta Protein Health Check, and the resulting model was evaluated further for its stereochemical properties by Procheck software.34 We used the docking program GRAMM35 to model the interactions between NP-1 and CP-1 peptides. This docking algorithm predicts the structure of a protein (peptide) complex by maximising the geometric match of the images of two protein (peptide) molecules. The details of the docking approach have been described elsewhere.36
Role of the funding source
The funding source had no role in study design; collection, analysis, or interpretation of data; the writing of the report; or the decision to submit it for publication.
Results
We analysed the spike protein of SARS-CoV strain Tor25 by a PredictProtein program.37 The protein consists of an extracellular domain (aminoacid residues 1–1199), a transmembrane domain (residues 1200–1215), and an intracellular domain (residues 1216–1255). A signal peptide (residues 1–13) at the amino-terminus was observed, but no obvious fusogenic peptide sequence was identified. The spike proteins of most coronaviruses (except porcine transmissible gastroenteritis virus and feline infectious peritonitis virus) are cleavable by cellular proteases to yield S1 (receptor binding) and S2 (virus fusion) subunits.9, 10, 12, 38 SARS-CoV spike protein does not contain a typical cleavage site (a basic aminoacid sequence—eg, RRFRR, GRHRR, KRRSRR).9 Therefore, the boundary between the S1 and S2 domains cannot be accurately identified at present. As in other coronavirus spike proteins, the S2 domain has a more conserved sequence than the S1 domain. The latter is a compact globular structure as predicted by the PredictProtein program Globe for prediction of globularity. The S2 domain contains HR1 (residues 892–1013) and HR2 (residues 1145–1194), similar to other class I viral fusion proteins.9, 13, 39 Analysis by the MultiCoil program31 showed that the sequences in the HR1 and HR2 regions have a tendency to form trimeric or dimeric helix bundles, consistent with the observation that coronavirus spike proteins are homotrimers or homodimers.9 Thus, these regions may take part in the oligomerisation and conformational changes of spike protein during virus–cell fusion events. The spike protein of SARS-CoV has only about 24% pairwise aminoacid identity with the corresponding proteins of other coronaviruses, consistent with the observation by Rota and colleagues.6 However, the identity of their HR1 and HR2 sequences is much higher (table 2 ), which suggests that the sequences in the HR1 and HR2 regions are more conserved than other sequences in spike protein. Alignments of the peptides derived from the HR1 and HR2 regions of SARS-CoV spike protein and the HIV-1 gp41 revealed high sequence similarity between HIV-1 N-peptide N36 and SARS-CoV N-peptide NP-1 and between HIV-1 C-peptide C43 and SARS-CoV C-peptide CP-1 (45% and 51%, respectively; figure 1). Thus, the HR1 and HR2 regions in SARS-CoV spike protein and HIV-1 gp41 may share certain structural and functional features.
Table 2.
Comparison of pairwise aminoacid identity and similarity of spike glycoproteins of SARS-CoV and other coronaviruses
| Group and virus | Host | Main disease |
Identity (%) |
Similarity (%) |
||||
|---|---|---|---|---|---|---|---|---|
| S protein | HR1 | HR2 | S protein | HR1 | HR2 | |||
| G1 | ||||||||
| HCoV-229E | Human being | Respiratory infection | 22·7 | 44·1 | 24·6 | 43·3 | 66·2 | 54·1 |
| PRCoV | Pig | Respiratory infection | 22·4 | 50·8 | 21·3 | 42·0 | 68·9 | 50·8 |
| FIPV | Cat | Infectious peritonitis | 20·7 | 43·4 | 23·8 | 41·6 | 64·0 | 47·6 |
| PEDV | Pig | Enteritis | 20·5 | 44·1 | 24·6 | 39·8 | 64·0 | 52·5 |
| CCoV |
Dog |
Enteritis |
20·4 |
44·9 |
21·3 |
40·2 |
62·5 |
50·8 |
| G2 | ||||||||
| MHV | Mouse | Hepatitis, enteritis, encephalitis | 30·2 | 54·9 | 40·0 | 51·7 | 76·2 | 74·0 |
| HCoV-OC43 | Human being | Respiratory infection | 27·2 | 55·7 | 42·0 | 45·9 | 76·2 | 70·0 |
| BCoV | Cattle | Enteritis | 27·0 | 56·6 | 42·0 | 46·0 | 75·4 | 70·0 |
| HEV |
Pig |
Encephalomyelitis, respiratory infection |
26·7 |
54·9 |
42·0 |
45·8 |
75·4 |
70·0 |
| G3 | ||||||||
| IBV |
Chicken |
Respiratory infection |
23·2 |
43·4 |
40·0 |
40·9 |
68·9 |
62·0 |
| Average | .. | .. | 24·1 | 49·3 | 32·2 | 43·7 | 69·8 | 60·2 |
HCoV=human coronavirus; PRCoV=porcine respiratory coronavirus; FIPV=feline infectious peritonitis virus; PEDV=porcine epidemic diarrhoea virus; CCoV=canine coronavirus; MHV=murine hepatitis virus; BCoV=bovine coronavirus; HEV=hepatitis E virus; IBV=infectious bronchitis virus.
We have also noticed that most of the residues located at the “a” and “d” positions of the helical wheels are hydrophobic (figure 1; grey boxes) and more conserved than those in the “b”, “c”, and “f” positions, which are hydrophilic (figure 1; white boxes). This feature suggests that these helices form homologous oligomers by hydrophobic association through “knobs-into-holes” packing interactions at the “a” and “d” positions, in a way similar to interactions between the N-helices in the HIV-1 gp41 core.18 The residues at the “b”, “c”, and “f” positions form a hydrophilic face towards the solution, making the oligomeric bundles water soluble.
Since the HR1 and HR2 sequences of SARS-CoV spike protein are highly similar to those of HIV-1 gp41, we hypothesised that the HR1 and HR2 domains have similar functions to those of HIV-1 gp41—ie, participating in the viral fusion process.17, 40 Therefore, we designed and synthesised six peptides overlapping the sequences of the HR1 and HR2 regions (table 1 figure 1). The peptides range from 34 to 40 aminoacid residues in length.
Only the peptide CP-1 had inhibitory activity on SARS-CoV-induced cytopathic effect in Vero E6 cells, with IC50 values of about 19 μmol/L (figure 2 ); NP-1 had a marginal anti-SARS-CoV activity at 50 μmol/L. Other peptides did not show inhibitory activity at 50 μmol/L (data not shown), which suggests that CP-1, perhaps like the HIV-1 gp41 C-peptides, interacts with a site in the HR1 region and interferes with the conformational change of the spike protein during the viral fusion process. To identify the CP-1 binding site in HR1, we immobilised biotinylated CP-1 on streptavidin sensor chips and searched for the CP-1-binding peptides from HR1 region, as determined by surface plasmon resonance with BIACORE. CP-1 strongly bound to NP-1 with a high binding affinity or kinetic dissociation constant (1·62×10−8 mol/L; figure 2), whereas other peptides derived from the HR1 region had very low or no binding activity to CP-1 (data not shown). CP-2, which has a ten-residue shift towards the C-terminus of CP-1 (figure 1; table 1), did not bind to NP-1, which suggests that the N-terminal residues of CP-1 are important for the interaction between NP-1 and CP-1 or that the C-terminal residues in CP-2 interfere with binding between NP-1 and CP-1.
Figure 2.
Inhibitory activity of CP-1 on SARS-CoV infection and interaction between CP-1 and NP-1
Note log scale in upper diagram.
Circular-dichroism spectroscopic analysis showed that the peptide CP-1 had a low content of α-helicity (with minima at 208 nm and 222 nm) in phosphate buffer (pH 7·2); NP-1 and other N-peptides had random coil structure in the same solution. When NP-1 was mixed with CP-1 in equimolar concentrations, the α-helicity increased (figure 3 ). However, the α-helicity did not change when NP-1 was mixed with CP-2, or when CP-1 was mixed with other peptides from HR1. These findings suggest that NP-1 and CP-1 associate to form a complex with α-helical conformation. This helical complex is relatively stable in phosphate buffer with a Tm of about 59°C (figure 3).
Figure 3.
Secondary structures of CP-1, NP-1, and CP-1/NP complex in phosphate buffer
Upper panel: circular-dichroism spectra for CP-1 (10 μmol/L), NP-1 (10 μmol/L), and their complex in phosphate buffer (pH 7·2) at 4°C. Lower panel: circular-dichroism signal at 222 nm for the NP-1/CP-1 complex as a function of temperature. Insert: curve of the first derivative (d[θ]/dT) against temperature (T), which was used to determine the Tm value.
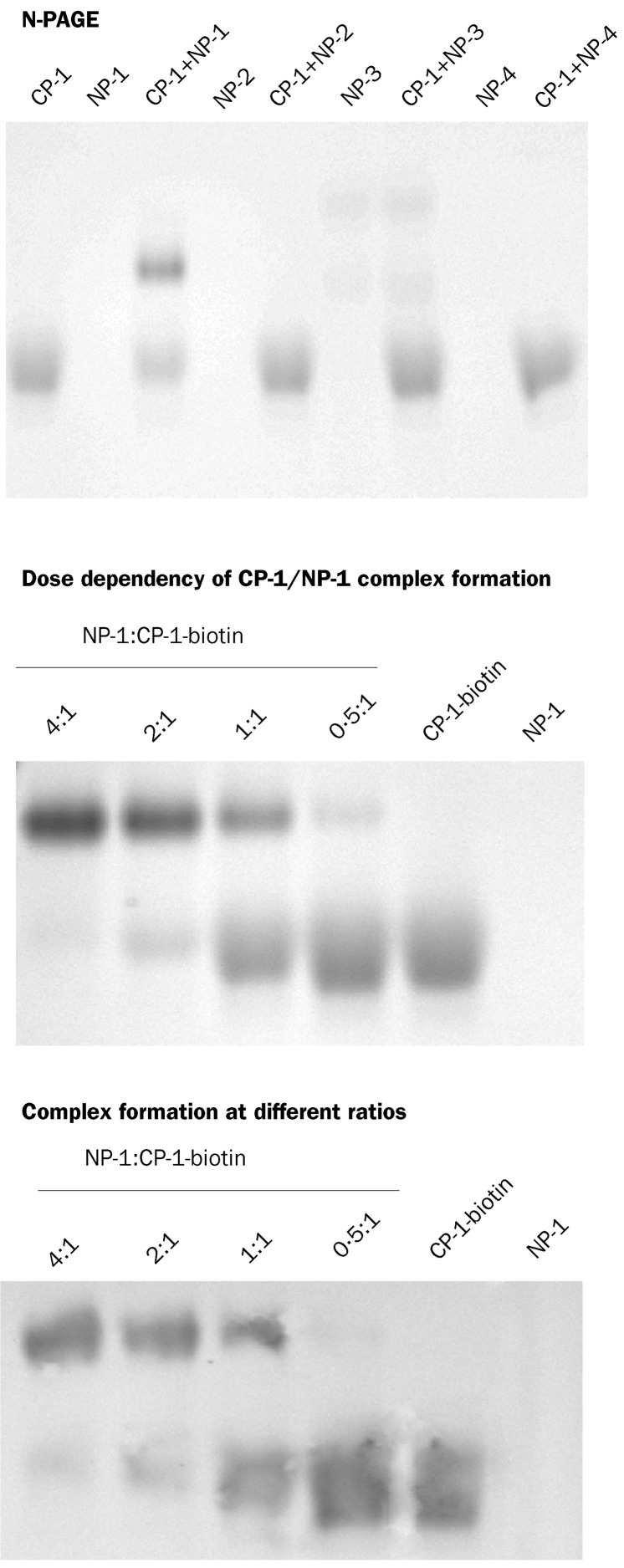
We have previously shown by N-PAGE that the HIV-1 gp41 peptides N36 and C34 can interact with each other to form a six-helix bundle.30 By a similar approach, we investigated whether CP-1 also forms an oligomeric complex with peptides derived from the HR1 region. With the exception of NP-3, N-peptides showed no bands in the gel (figure 4 ) because they carried net positive charges under the native electrophoresis conditions (table 1) and could therefore have migrated up and off the gel. NP-3 did migrate down in the gel because it has net zero charge, but it showed two bands, perhaps because it has differing conformations. Peptide CP-1 showed a band in the lower part of the gel. The mixture of NP-1 and CP-1 showed two bands: the lower one had the same position as isolated CP-1, and the upper one is expected to be a higher order of oligomeric complex formed by NP-1 and CP-1. To confirm whether this band indeed represents a complex formed between NP-1 and CP-1, we analysed the mixture of NP-1 and biotinylated CP-1 at different ratios by N-PAGE in two gels. One was stained with Coomassie blue (figure 4, middle gel). The peptides and their complex in another gel were transferred to a nitrocellulose membrane, which was stained with streptavidin-conjugated horseradish peroxidase and its substrate (figure 4, lower gel). With increasing concentrations of NP-1, the intensity of upper bands increased and that of the lower bands decreased, which suggests that formation of the upper band depends on the concentration of NP-1 and confirms that the band does not represent aggregates of CP-1.
Figure 4.
Association of CP-1 with N-peptides derived from SARS-CoV spike protein HR1 region, as determined by N-PAGE
For N-PAGE, the final concentration of the individual peptides in each preparation was 40 μmol/L.
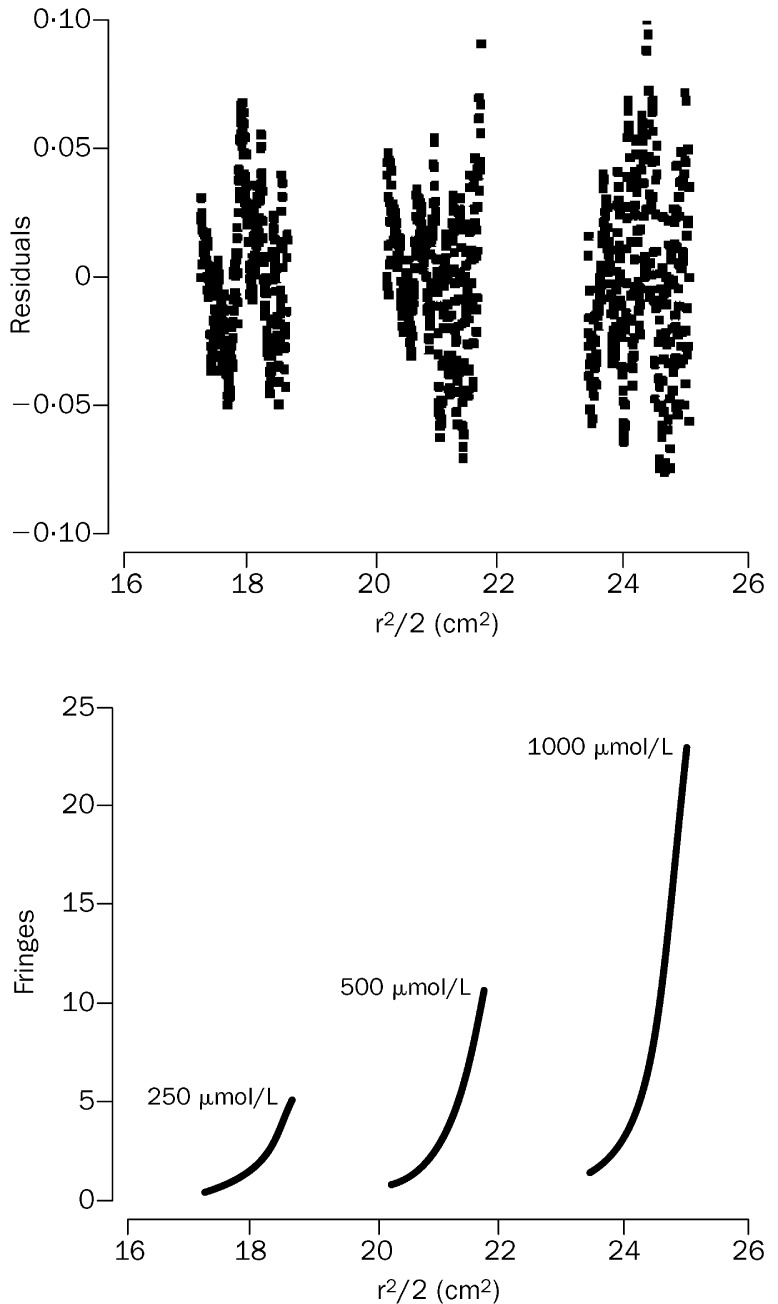
Since the migration speed of a molecule or a complex in N-PAGE depends on both the size and the net charge of the molecule or complex, this method cannot be used to measure exactly the molecular mass of NP-1/CP-1 complex in the gel or to find out whether the lower band of CP-1 is monomer or oligomer. Therefore, we analysed the apparent molecular masses of the peptides NP-1, CP-1, and NP-1/CP-1 complex by sedimentation equilibration, which was successfully used to find out the mass of the six-helix bundle formed by the HIV-1 gp41 N-peptides and C-peptides.17 The isolated NP-1 was a monomer and the molecular mass obtained for CP-1 corresponded to a trimer in phosphate-buffered saline (table 3 ). The HPLC-purified mixture of NP-1 and CP-1 showed a complex with a molecular mass of about 24 781 Da (figure 5 ), which was close to the calculated molecular mass of a trimer of dimers. These results were confirmed by SE-HPLC; NP-1 showed a single peak of monomer and CP-1 a major peak corresponding to a trimer, whereas the mixture of NP-1 and CP-1 showed three peaks corresponding to NP-1 monomer, CP-1 trimer, and NP-1/CP-1 trimer of heterodimer (hexamer) (figure 6 ). These findings suggest that NP-1 is a monomeric structure and that CP-1 has a trimeric conformation in phosphate-buffered saline. Once they are mixed at equimolar concentration, they interact to form a six-helix bundle consisting of three molecules of NP-1 and three of CP-1. This notion is consistent with the biophysical properties of the HIV-1 gp41 N-peptides and C-peptides as shown by sedimentation equilibration and SE-HPLC.17, 30, 41
Table 3.
Molecular masses of the isolated peptides NP-1 and CP-1 and the NP-1/CP-1 complex measured by sedimentation equilibrium
|
Peptide |
Mass of monomer (Da) |
Centrifugation speed |
Molecular mass (Da) | |
|---|---|---|---|---|
| rpm | ×g | |||
| CP-1 | 4171 | 35 000 | 89180 | 12616 |
| NP-1 | 4409 | 48 000 | 167731 | 4700 |
| CP-1/NP-1 | 8580 | 25 000 | 45500 | 24781 |
Figure 5.
Sedimentation-equilibration profiles of CP-1/NP-1 complex
Plots show concentration gradients detected as fringes (Y axis) as a function of the squared radial distance in cm2 (X axis). Samples were run at 25 000 rpm (45 500× g) at 25°C with three initial concentrations (lower panel). The global fit from the three sets of data and with a model of a single ideal solute is shown as the single line going through the data points. The fit gives a molecular mass of 24 781 Da, in close agreement with the theoretical molecular mass for a trimer of dimers (25 740 Da). Residuals plot shows random distribution indicating the goodness of the fit (upper panel).
Figure 6.
SE-HPLC profiles of NP-1, CP-1, and the mixture of NP-1 and CP-1
The final concentration of peptides was 50 μmol/L. The bottom panel shows Bio-Rad gel filtration standard and the plot of logarithmic molecular mass (MW) against retention time (insert). Peaks 1–5 and points 1–5 in the insert correspond to thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1·35 kDa).
Alignments of the NP-1 and CP-1 peptides on the corresponding HIV-1 gp41 N-peptides and C-peptides revealed reasonable sequence similarities and gave us confidence that a homology model of NP-1 and CP-1 association could be built on the basis of the X-ray crystal structure of the HIV-1 gp41 core formed by the gp41 N-peptides and C-peptides.18, 19 NP-1 showed a typical coiled-coil structure (figure 7 ). Most of CP-1 had an -helical conformation with the exception of a kink in the middle of the structure. The structure of the possible complex between these two peptides was then investigated by use of specialised protein-docking software.35 The residues at the “a” and “d” positions in the NP-1 and CP-1 are hydrophobic. The complex structure formed by the docking technique also shows that there are hydrophobic interactions between these two peptides: eg, L898, I905, F909, I913, and I916 of NP-1 interact very well with L1182, V1171, L1168, I1165, and I1161 of CP-1, respectively. Owing to the lack of crystal coordinates of the sequence beyond the N-terminus of the HIV-1 gp41 peptide C34,18 the corresponding N-terminal sequence (residues 1153–1159) of CP-1 could not be modelled and its interaction with the corresponding region in the C-terminus of NP-1 could not be predicted.
Figure 7.
Interaction between NP-1 and CP-1 as predicted by molecular modelling
Discussion
Discovery of the peptidic HIV-1 fusion inhibitors14, 15, 16 and elucidation of their mechanism of action17, 18, 19 have provided important information to explain the fusogenic mechanism of HIV-1 and other enveloped viruses.21, 22, 23, 24, 25 By a similar approach, we identified an anti-SARS-CoV peptide from the HR2 region of the SARS-CoV spike protein. This finding conforms with the prediction by Gallaher and Garry that peptides overlapping the sequences of HR regions of SARS-CoV spike protein might have inhibitory activity on SARS-CoV infection (http://www.virology.net/articles/sars/s2model.html).
The anti-SARS-CoV peptide CP-1 and other peptides derived from the HR regions of the spike protein can be used as probes to investigate the roles of HR1 and HR2 regions in the process of SARS-CoV entry into target cells. The biophysical studies showed that the peptides derived from these regions of SARS-CoV spike proteins have similar properties to those derived from the HIV-1 gp41 HR1 and HR2 regions, respectively. For example, like the HIV-1 gp41 N-peptides and C-peptides, NP-1 and CP-1 from SARS-CoV can interact to form high-order oligomeric helices, which may have important roles in the fusion of SARS-CoV with target cells. On the basis of results from this study and knowledge about HIV-1 gp41 and other viral transmembrane proteins,14, 15, 16, 17, 18, 19, 21, 22, 23, 24, 25 we propose a model to elucidate the fusogenic mechanism mediated by SARS-CoV spike protein and the mechanism by which CP-1 inhibits SARS-CoV infection (figure 8 ). In the native state, spike proteins on the virus surface could be in oligomeric form through the self-association of their partly helical HR2 regions to form the stems of the spikes on SARS-CoV; HR1 regions may be in random coil conformation covered by S1 domain. After the binding of the S1 domain to a receptor (eg, angiotensin-converting enzyme 2)7 on the target cell (receptor-binding state), the S2 domain changes conformation by forming α-helices, extending and inserting its inert fusion peptide into the target-cell membrane, and exposing HR1 and HR2 regions (intermediate pre-hairpin state). Then the HR1s form a trimer and the HR2 trimer dissociates to monomers. Subsequently, the HR2 monomers pack onto the grooves of the HR1 trimer to form the six-helix bundle. This fusion-active core structure brings the viral and target-cell membranes into close proximity, resulting in fusion between the membranes and formation of fusion pores, which allow the virus genome to enter the target cell. If a C-peptide, such as CP-1, is present in the intermediate state, it could bind to the HR1 region and block hexamer formation, thereby inhibiting fusion. However, more direct evidence is needed to validate this model. Nevertheless, the information obtained from other coronaviruses can provide indirect evidence to support the hypothesis.
Figure 8.
Illustration of the conformational changes of SARS-CoV spike protein during the process of fusion between the virus and target-cell membranes
Luo and colleagues42 have shown that the spike protein of murine hepatitis virus, like HIV-1 gp41,13, 18 contains HR1 and HR2 regions that may have important roles in spike-protein-mediated cell-cell fusion and virus entry. Double alanine substitutions of the four middle heptadic leucine and isoleucine residues within the HR2 region lead to significant reduction in fusion activity and defects in spike oligomerisation. Bosch and co-workers43 reported the structural and functional characterisation of the fusion core complex formed by the HR1 and HR2 regions of the murine hepatitis virus spike protein. Although our group and theirs have worked independently on distinct coronaviruses by different approaches, we have obtained surprisingly similar results. First, only peptides derived from the HR2 region, not the HR1 region, have antiviral activity. Second, HR2 peptides derived from both murine hepatitis virus and SARS-CoV spike proteins have antiviral activity in the micromolar range. Third, the fragments involving the interaction between the HR1 and HR2 in spike proteins of both viruses are located in the same regions. These findings suggest that murine hepatitis virus and SARS-CoV have a common fusogenic mechanism, which may be shared by other coronaviruses. The peptides derived from the HR2 regions of the SARS-CoV and murine hepatitis virus spike proteins have much less potent inhibitory activity than HIV-1 gp41 C-peptides (eg, C34 and T-20), which have activity in the nanomolar range.16, 44 SARS-CoV and murine hepatitis virus may therefore have a fusogenic mechanism different from that of HIV-1. However, the peptides derived from the HR2 regions of many other enveloped viruses, including Ebola virus,23 Newcastle disease virus,24 parainfluenza virus,45 and respiratory syncytial virus,25 inhibit the corresponding virus infection also in the micromolar range, and these viruses have similar fusogenic mechanisms to HIV-1.23 Two major factors can affect the potency of a fusion-inhibitory peptide: the sensitivity of a virus to the corresponding antiviral peptides; and the sequence and conformation of the inhibitory peptide. The sensitivity of HIV-1 to the fusion-inhibitory peptide T-20 correlates with fusion kinetics, envelope/coreceptor affinity, and receptor density.46 Different viruses might have differing fusion rates or receptor-binding affinities. Therefore, these viruses might have different sensitivity to the corresponding inhibitory peptides. The antiviral activity of the peptidic fusion inhibitors depends on their optimum peptide sequences and conformations. Changes in their sequences or conformations could substantially affect their antiviral activity and the stability of the complexes they form.17, 47, 48 However, optimisation of the peptide sequence and conformation could improve antiviral activity.
RELEVANCE TO PRACTICE.
BACKGROUND
Insights into the viral fusogenic mechanism have been gained from studies on the peptidic HIV-1 fusion inhibitors. The SARS-CoV spike glycoprotein contains heptad repeat 1 and 2 (HR1 and HR2) sequences that are similar to those from HIV-1 envelope glycoprotein, gp41. This similarity might help to elucidate the mechanism by which SARS-CoV enters target cells, and thus could have implications for treatment.
This paper assesses the ability of synthesised peptides derived from the SARS-CoV HR1 and HR2 regions to inhibit a cytopathic effect in tissue culture, and it investigates the interactions between HR1 and HR2 peptides. Only the CP-1 peptide (derived from the HR2 region) had strong inhibitory activity in tissue culture. CP-1 and the NP-1 peptide (derived from the HR1 region) interacted to form a six-helix structure similar to the fusogenic core structure of HIV-1 gp41.
IMPLICATIONS
These findings confirm that the carboxyl terminal portion of the spike glycoprotein of SARS-CoV is a type 1 fusion glycoprotein. A possible fusogenic mechanism involves CP-1 binding to the HR1 region, thereby interfering with conformational changes that lead to fusion between the viral and target-cell membranes.
Therefore, CP-1 can be used as a lead in designing more potent anti-SARS-CoV peptides for drug development. Use of such peptides for treatment of SARS is better than application of anti-HIV peptides for chemotherapy of AIDS because only a few doses should be needed to treat an individual with acute SARS-CoV infection (no chronic SARS has been reported), whereas an HIV-infected individual might have to use T-20, one of the most expensive anti-HIV drugs, for life. Furthermore, the anti-SARS-CoV peptides might also be used prophylactically as nasal spray or inhalation formulations to protect at-risk populations working and travelling in SARS-endemic areas. The peptides derived from the HR1 and HR2 regions of SARS-CoV spike protein could be used to establish high-throughput assays for screening non-peptidic SARS-CoV fusion inhibitors; we have successfully developed several such assays with the HIV-1 gp41 N-peptides and C-peptides for identification of small-molecule HIV-1 fusion inhibitors.20, 49, 50 Any compounds that block the HR1–HR2 interaction could be used as leads for development of virus fusion inhibitors.
Contributors
S Jiang planned, directed, and executed this project. S Liu carried out N-PAGE, circular dichroism, and SE-HPLC analysis. G Xiao did the virus inhibition assay. S Liu and G Xiao made equal contributions.
Y Chen and H Xiong carried out the surface plasmon resonance analysis.
Y He coordinated the study. J Niu and J Farmar synthesised the peptides.
C R Escalante carried out sedimentation equilibration analysis.
A K Debnath did the molecular modelling and docking study. P Tien supervised the virus inhibition assay.
Conflict of interest statement
None declared.
GLOSSARY
- heptad repeat
Occurrence of a group of seven aminoacids many times in a protein sequence. The seven aminoacids are labelled a to g; those at a and d are generally hydrophobic and the others are hydrophilic.
- sedimentation equilibrium
A method for measuring protein molecular masses in solution and for studying interactions between proteins, including associative states, heterogeneity, and some thermodynamic properties.
- six-helix bundle
A complex formed by six rod-like structures. Each rod consists of a peptide with -helical conformation, like a spring. Three peptides form the central trimeric core, and the other three peptides pack into the grooves on the surface of the central trimeric core.
References
- 1.WHO Summary of probable SARS cases with onset of illness from Nov 1, 2002, to July 31, 2003. www.who.int (accessed Dec 19, 2003)
- 2.Ksiazek TG, Erdman D, Goldsmith CS. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Drosten C, Gunther S, Preiser W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. NEnglJ Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Peiris JS, Lai ST, Poon LL. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marra MA, Jones SJM, Astell CR. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 6.Rota PA, Oberste MS, Monroe SS. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 7.Li WH, Moore MJ, Vasilieva NY. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan Y, Zheng BJ, He YQ. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 9.Cavanagh D. The coronavirus surface glycoprotein. In: Siddell SG, editor. The coronaviridae. Plenum Press; New York: 1995. pp. 73–114. [Google Scholar]
- 10.Lai MM, Cavanagh D. The molecular biology of coronaviruses. AdvVirus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallagher TM. A role for naturally occurring variation of the murine coronavirus spike protein in stabilizing association with the cellular receptor. J Virol. 1997;71:3129–3137. doi: 10.1128/jvi.71.4.3129-3137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes KV. SARS-associated coronavirus. N Engl J Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- 13.Gallaher WR, Ball JM, Garry RF. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 14.Jiang S, Lin K, Strick N. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 15.Wild C, Oas T, McDanal C. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wild CT, Shugars DC, Greenwell TK. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 18.Chan DC, Fass D, Berger JM. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 19.Weissenhorn W, Dessen A, Harrison SC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–428. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 20.Jiang S, Zhao Q, Debnath AK. Peptide and non-peptide HIV fusion inhibitors. Curr Pharm Des. 2002;8:563–580. doi: 10.2174/1381612024607180. [DOI] [PubMed] [Google Scholar]
- 21.Blacklow SC, Lu M, Kim PS. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry. 1995;34:14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- 22.Lombardi S, Massi C, Indino E. Inhibition of feline immunodeficiency virus infection in vitro by envelope glycoprotein synthetic peptides. Virology. 1996;220:274–284. doi: 10.1006/viro.1996.0315. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe S, Takada A, Watanabe T. Functional importance of the coiled-coil of the Ebola virus glycoprotein. J Virol. 2000;74:10194–10201. doi: 10.1128/jvi.74.21.10194-10201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu M, Wang E, Liu Y. Six-helix bundle assembly and characterization of heptad repeat regions from the F protein of Newcastle disease virus. J Gen Virol. 2002;83:623–629. doi: 10.1099/0022-1317-83-3-623. [DOI] [PubMed] [Google Scholar]
- 25.Wang E, Sun X, Qian Y. Both heptad repeats of human respiratory syncytial virus fusion protein are potent inhibitors of viral fusion. Biochem Biophys Res Commun. 2003;302:469–475. doi: 10.1016/s0006-291x(03)00197-9. [DOI] [PubMed] [Google Scholar]
- 26.Qin E, Zhu Q, Yu M. A complete sequence and comparative analysis of a SARS-associated virus (isolate BJ01) Chin Sci Bull. 2003;48:941–948. doi: 10.1007/BF03184203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Q, Ernst JT, Hamilton AD. XTT formazan widely used to detect cell viability inhibits HIV type 1 infection in vitro by targeting gp41. AIDS Res Hum Retroviruses. 2002;18:989–997. doi: 10.1089/08892220260235353. [DOI] [PubMed] [Google Scholar]
- 28.Jones DK, Badii R, Rosell FI. Bacterial expression and spectroscopic characterization of soybean leghaemoglobin a. Biochem J. 1998;330:983–988. doi: 10.1042/bj3300983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson ML, Correia JJ, Yphantis DA. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Zhao Q, Jiang S. Determination of the HIV-1 gp41 postfusion conformation modeled by synthetic peptides: applicable for identification of the HIV-1 fusion inhibitors. Peptide. 2003;24:1303–1313. doi: 10.1016/j.peptides.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Wolf E, Kim PS, Berger B. MultiCoil: a program for predicting two-and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Gibson TJ, Plewniak F. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 35.Vakser IA, Matar OG, Lam CF. A systematic study of low-resolution recognition in protein-protein complexes. Proc Natl Acad Sci USA. 1999;96:8477–8482. doi: 10.1073/pnas.96.15.8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katchalski-Katzir E, Shariv I, Eisenstein M. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci USA. 1992;89:2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rost B. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 38.Krueger DK, Kelly SM, Lewicki DN. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion. J Virol. 2001;75:2792–2802. doi: 10.1128/JVI.75.6.2792-2802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White JM. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 40.Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 41.Dwyer JJ, Hasan A, Wilson KL. The hydrophobic pocket contributes to the structural stability of the N-terminal coiled coil of HIV gp41 but is not required for six-helix bundle formation. Biochemistry. 2003;42:4945–4953. doi: 10.1021/bi027283n. [DOI] [PubMed] [Google Scholar]
- 42.Luo Z, Matthews AM, Weiss SR. Amino acid substitutions within the leucine zipper domain of the murine coronavirus spike protein cause defects in oligomerization and the ability to induce cell-to-cell fusion. J Virol. 1999;73:8152–8159. doi: 10.1128/jvi.73.10.8152-8159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosch BJ, van der ZR, de Haan CA. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang S, Lin K, Strick N. Inhibition of HIV-1 infection by a fusion domain binding peptide from HIV-1 envelope glycoprotein gp41. Biochem Biophys Res Commun. 1993;195:533–538. doi: 10.1006/bbrc.1993.2078. [DOI] [PubMed] [Google Scholar]
- 45.Yao Q, Compans RW. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology. 1996;223:103–112. doi: 10.1006/viro.1996.0459. [DOI] [PubMed] [Google Scholar]
- 46.Reeves JD, Gallo SA, Ahmad N. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci USA. 2002;99:16249–16254. doi: 10.1073/pnas.252469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang S, Lin K. Effect of amino acid replacements, additions and deletions on the antiviral activity of a peptide derived from the HIV-1 gp41 sequence. Peptide Res. 1995;8:345–348. [PubMed] [Google Scholar]
- 48.Chan DC, Chutkowski CT, Kim PS. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. ProcNatl Acad Sci USA. 1998;95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang S, Lin K, Zhang L. A screening assay for antiviral compounds targeted to the HIV-1 gp41 core structure using a conformation-specific monoclonal antibody. J Virol Methods. 1999;80:85–96. doi: 10.1016/s0166-0934(99)00041-5. [DOI] [PubMed] [Google Scholar]
- 50.Debnath AK, Radigan L, Jiang S. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J Med Chem. 1999;42:3203–3209. doi: 10.1021/jm990154t. [DOI] [PubMed] [Google Scholar]