Abstract
Here we identified a hydrophobic 6.4 kDa protein, Cox26, as a novel component of yeast mitochondrial supercomplex comprising respiratory complexes III and IV. Multi-dimensional native and denaturing electrophoretic techniques were used to identify proteins interacting with Cox26. The majority of the Cox26 protein was found non-covalently bound to the complex IV moiety of the III–IV supercomplexes. A population of Cox26 was observed to exist in a disulfide bond partnership with the Cox2 subunit of complex IV. No pronounced growth phenotype for Cox26 deficiency was observed, indicating that Cox26 may not play a critical role in the COX enzymology, and we speculate that Cox26 may serve to regulate or support the Cox2 protein. Respiratory supercomplexes are assembled in the absence of the Cox26 protein, however their pattern slightly differs to the wild type III–IV supercomplex appearance. The catalytic activities of complexes III and IV were observed to be normal and respiration was comparable to wild type as long as cells were cultivated under normal growth conditions. Stress conditions, such as elevated temperatures resulted in mild decrease of respiration in non-fermentative media when the Cox26 protein was absent.
Keywords: Blue native electrophoresis, Cytochrome c oxidase, Protein composition, Respiratory supercomplexes, III–IV supercomplexes, Saccharomyces cerevisiae
1. Introduction
Under aerobic conditions, mitochondria are the main producers of ATP by the oxidative phosphorylation system (OXPHOS). The OXPHOS enzymes are multi-subunit complexes in the inner mitochondrial membrane [1] that have been isolated as detergent-resistant individual complexes and, more recently, also as higher order structures, such as the dimeric and oligomeric ATP synthase and the respiratory supercomplex enzymes composed of the cytochrome bc1 complex (complex III) and the cytochrome c oxidase (COX, complex IV), also termed the III–IV supercomplex or “respirasomes” [2–8]. In eukaryotic organisms which contain the traditional complex I-NADH dehydrogenase (e.g. mammals), the supercomplex arrangement of I-III–IV has been shown to exist [4–6, 8]. Blue-native electrophoresis (BNE, [9,10]), clear-native electrophoresis (CNE, [8,11]), and high resolution clear-native electrophoresis (hrCNE, [12]) have been commonly used for the separation and characterization of these higher ordered structures. Respiratory supercomplexes have also been analysed by gel filtration and separation by sucrose density gradient centrifugation, especially for electron microscopic single particle analysis of the large protein complexes [13–21].
Purification of the mitochondrial supercomplexes under mild detergent solubilization conditions has resulted in the identification of previously unidentified supernumerary subunits. For example, the homodimeric yeast F1Fo-ATP synthase complex was found to contain membrane embedded subunits e and g, which are non-essential for catalysis, but serve to stabilize the dimeric form of the enzyme complex [2]. These proteins are also important to generate cristae morphology [21] and to impose a strong curvature to the membranes [22,23]. Bovine ATP synthase contains two further accessory subunits, AGP and MLQ, with so far unknown function [24]. We asked here whether the yeast III–IV supercomplex enzymes contain additional, previously unknown protein components that are potentially important to stabilize supercomplexes or to support functional roles of supercomplex enzymes [25–28]. To date the respiratory III–IV supercomplex from the yeast Saccharomyces cerevisiae (S. cerevisiae) has been reported to contain at least 21 subunits, i.e. ten subunits of complex III [29] and eleven subunits of complex IV [30].
Evidence to indicate that the yeast III–IV supercomplex may contain additional, accessory proteins, has been suggested by a recent three-way proteomics strategy that identified additional candidate proteins potentially associated with complexes III and IV [31]. The proteins, originally termed Aim31 and Aim38, were identified in this study. These proteins were later functionally analysed and confirmed as components of the III–IV supercomplex and were subsequently renamed as respiratory supercomplex factors 1 and 2, Rcf1 and Rcf2 [33–35]. A further protein, encoded by the YDR119W-A gene was also suggested from the proteomic analysis to co-purify with the III–IV supercomplexes [31], but further verification and analysis of this protein had not been undertaken so far.
In this present study, we have employed a high-resolution 2D-native electrophoresis system to further characterize the composition of the yeast III–IV supercomplex and to identify novel protein subunits. Through this analysis we have confirmed that the protein encoded by the YDR119w-a gene is indeed a subunit of the III–IV supercomplex. Moreover our analysis demonstrates that this protein is a subunit of complex IV with a direct interaction to the catalytically important subunit Cox2. We propose here to rename it Cox26.
2. Materials and methods
2.1. Materials, media, strains, culture conditions
Imidazole, 6-aminohexanoic acid, digitonin (purity >50%) were obtained from Fluka. Digitonin was used directly without recrystallization. Acrylamide and bisacrylamide (the commercial twice crystallized products), Coomassie blue G-250 (Serva Blue G) and Digitonin used for experiments in Figs. 2 and 3 were purchased from Serva. All other chemicals were from Sigma. Yeast strains used in this study were wild type (WT) W303–1A (Mat a, leu2, trp1, ura3, his3, ade2) and the cox26 null mutant, Δcox26 (W303–1A, leu2, trp1, ura3, ade2, COX26::KAN). The Cox26 gene deletion yeast strain, Δcox26 was constructed in the W303–1A genetic background, as follows: Introducing the kanamycin resistance gene (KANr) into the COX26 (YDR119w-a) gene locus of wild type cells, resulted in the complete deletion of the open reading frame. The KANr gene was amplified from the plasmid pFA6a-KANMX6 [32].
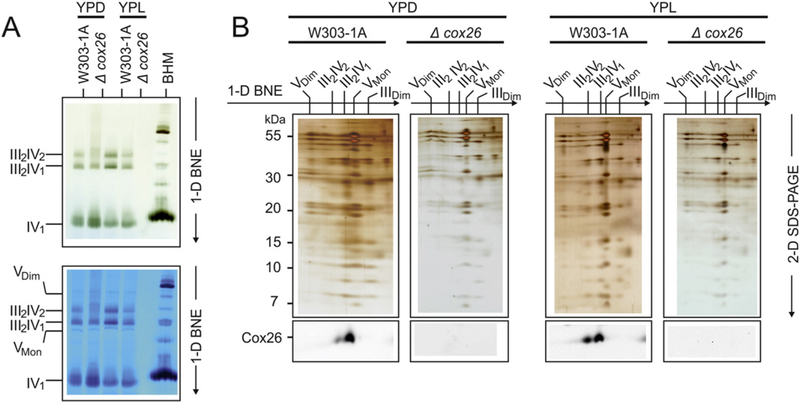
Fig. 2.
Cox26 stabilizes III–IV supercomplexes. (A) BNE of digitonin-solubilized mitochondrial complexes from wild type (W303–1A) and Cox26 null mutant strain (Δcox26) grown under identical conditions in YPD and YPL. Individual or supercomplex associated complex IV was detected by a specific in-gel complex IV activity stain [12]. Decreased amounts of supercomplexes were detected in the mutant strain in both conditions with even less stability in cells grown in glucose (upper panel). The same gel was stained with Coomassie to show complex V as loading control (lower panel). (B) 2-D Tricine-SDS-PAGE of complexes from wild type and Δcox26 strain using 13% acrylamide gels revealed identical subunit pattern for complexes III and IV. Decoration with an antibody against Cox26 shows presence of Cox26 in III–IV supercomplexes in wild type mitochondria. Assignment of protein complexes: according to Fig. 1. BHM, bovine heart mitochondria as native mass ladder.
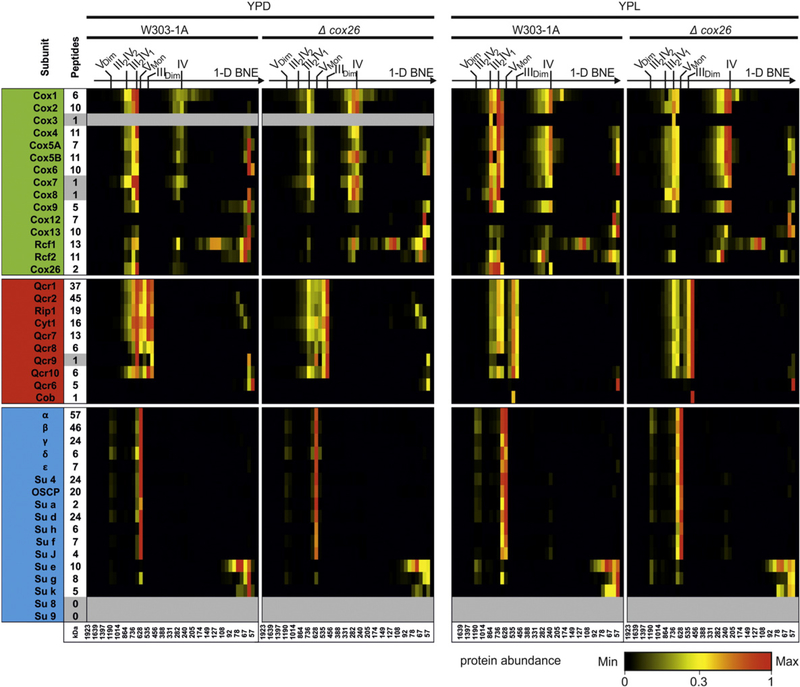
Fig. 3.
Composition of III–IV supercomplexes. 1-D BNE gels were fractionated and proteins were identified and quantified by mass spectrometry. Identified proteins were quantified by intensity-based absolute quantification values (IBAQ) using only unique and razor peptides (column indicate number of peptides used for quantification). Heatmaps show protein profiles normalized to the maximum abundance of a protein within BN lanes of wild type and Δcox26 mutant for each condition, respectively. The majority of Cox12 and Cox13 was identified on the electrophoretic front indicating either higher abundance as free protein or dissociation of proteins from the complex under experimental conditions used here. Cytochrome b (Cob) was quantified only by one peptide with a modified side (methionine oxidation).Very small proteins like Cox7, Cox8, Qcr9 and the hydrophobic protein Cox3 were identified with one peptide within the complete dataset that could not give full quantitative data for the entire lane. ATP synthase subunits 8 and 9 escaped from identification as they are very hydrophobic and/or small and difficult to identify in complex samples.
Strains were grown in the following media: YPGal (yeast extract and bactopeptone containing 2% galactose), YPD (YP + 3% glucose), YPL (YP + 2% lactate), YPEG (YP + 2% ethanol, 3% glycerol), and YPGalLac (YP + 2% galactose and 0.5% lactate) at temperatures of 30 °C and 37 °C.
2.2. Isolation of mitochondria
Yeast cells were harvested by centrifugation at 1800 g and washed with sucrose buffer (250 mM sucrose, 5 mM 6-aminohexanoic acid, 1 mM EDTA, and 10 mM sodium phosphate, pH 7.0). Five grammes of sedimented cells, 5 ml of glass beads (0.25–0.5 mm) and 5 ml of sucrose buffer were vortexed in a 50 ml tube 10 times for 1 min with 1 min cooling intervals on ice. Following dilution with 10 ml sucrose buffer, the sedimented glass beads were removed and the supernatant was centrifuged for 20 min at 1250 g. Mitochondrial membranes were collected by 30 min centrifugation at 18,000 g, and stored in sucrose buffer at − 80 °C.
2.3. Electrophoretic and associated techniques
Mitochondrial membranes were thawed shortly before blue-native electrophoresis (BNE). Aliquots containing 400 μg protein or multiples thereof were then sedimented by 15 min centrifugation at 20,000 g, and solubilized by digitonin for the first dimension BNE using a digitonin/protein ratio of 3 (g/g) as described [10,36]. Two-dimensional native electrophoresis (2-D BNE/BNE), or 2-D BNE/SDS-PAGE [10], doubled SDS-PAGE (dSDS-PAGE) and silver staining [37] were performed as described. A novel technique for semidry electroblotting of hydrophobic proteins from high percentage acrylamide gels was used [36]. An antibody against the C-terminal 13 amino acids of the Cox26 protein was generated in rabbits. The antibody was used for immunological detection on PVDF membranes as described [24].
In order to isolate sufficient Cox26 protein for Edman sequencing, Cox26 was prepared by a 3-D electrophoretic protocol on a preparative scale. Three 14 cm wide gels were used for first dimension BNE to separate the mitochondrial complexes from 15 mg total mitochondrial protein. The band of the larger supercomplex was excised from each of the gels and cut into two halves. Stacks of these 6 gel strips (height around 1.5 cm; width around 7 cm) were then placed on top of another native gel and resolved by second dimensional modified BNE. Dodecylmaltoside (DDM, 0.02% w/v) that was added to the cathode buffer for modified BNE served to dissociate supercomplexes into the individual constituent complexes and to pull individual, loosely attached proteins to the running front of BNE. Beginning with the upper edge of the dark blue Coomassie-dye front, several horizontal 2–3 mm strips were then excised. The gel strips were cut into smaller pieces (width around 2 cm) and placed as a stack of gel strips (height around 1–1.5 cm) on top of a 1.5 mm thick gel for SDS-PAGE. The stack was wetted for 2 h with 1% SDS, 1% mercaptoethanol, then resolved using 16% acrylamide gels containing 6 M urea for Tricine-SDS-PAGE [34], and finally electroblotted onto PVDF-membranes. The bands were Coomassie-stained on the PVDF membrane and used for Edman degradation in a 473A protein sequencer (Applied Biosystems) as described [2]. Complex IV was stained in 1-D BN-gels as described [12].
2.4. High-resolution respirometry
Mitochondrial routine respiration of yeast cells was measured using high-resolution respirometry (Oxygraph-2 k, Oroboros Instruments, Innsbruck, Austria) with DatLab software 5.2.1.51 (Oroboros Instruments). Measurements were performed at 30 °C or 37 °C in yeast growth medium YPD or in YPL. Routine respiration (R) was measured for 20 min followed by stepwise titration of uncoupler FCCP (Sigma Aldrich, Munich, Germany) until maximal uncoupled respiration (E) was reached. Residual oxygen consumption (ROX) was determined after addition of KCN (2 mM final concentration). Absolute respiration rates were normalized for the total number of cells per chamber. As the mutant differs in size of the cells compared to wild type cells at 37 °C, flux control ratios (FCR; R/E; routine respiration over maximal uncoupler respiration) were used to evaluate the respiration independent of cell size and/or mitochondrial content [38].
2.5. Quantitative mass spectrometry
Protein abundance profiles of subunits within complexes and supercomplexes were analysed by a combination of 1-D BNE and quantitative mass spectrometry (MS) [39]. Briefly, 1-D BNE (linear 3–18% acrylamide gradient) was cut into 46 even slices and digested with trypsin as described [39].
Liquid chromatography/mass spectrometry (LC/MS) was performed on Thermo Scientific™ Q Exactive Plus equipped with an ultra-high performance liquid chromatography unit (Thermo Scientific Dionex Ultimate 3000) and a Nanospray Flex Ion-Source (Thermo Scientific). Peptides were loaded on a C18 reversed-phase precolumn (Thermo Scientific) followed by separation on a with 2.4 μm Reprosil C18 resin (Dr. Maisch GmbH) in-house packed picotip emitter tip (diameter 100 μm, 15 cm long from New Objectives) using an gradient from mobile phase A (4% acetonitrile, 0.1% formic acid) to 30% mobile phase B (80% acetonitrile, 0.1% formic acid) for 20 min followed by a second gradient until 60% B for 10 min with a flow rate 400 nl/min and washout with 99% B for 5 min.
MS data were recorded by data dependent acquisition Top10 method selecting the most abundant precursor ions in positive mode for HCD fragmentation. The full MS scan range was 300 to 2000 m/z with resolution of 70,000, and an automatic gain control (AGC) value of3*106 total ion counts with a maximal ion injection time of 160 ms. Only higher charged ions (2 + ) were selected for MS/MS scans with a resolution of 17,500, an isolation window of 2 m/z and an automatic gain control value set to 105 ions with a maximal ion injection time of 150 ms. Selected ions were excluded in a time frame of 30s following fragmentation event. Data were acquired in profile mode.
MS Data were analysed by MaxQuant (v1.5.2.8) [40]. Proteins were identified using yeast reference proteome database UniProtKB with 6630 entries, released in 11/2013. The enzyme specificity was set to Trypsin, missed cleavages were limited to 2 and the minimum peptide length was seven amino acids. Acetylation ( + 42.01) at N-terminus and oxidation of methionine ( + 15.99) were selected as variable modifications and carbamidomethylation ( + 57.02) as fixed modification on cysteines. False discovery rate (FDR) for the identification protein and peptides was 1%.
Intensity-based absolute quantification (IBAQ) values were recorded. We calculated the sum of all IBAQ values of each protein group and sample. The median of IBAQ sums were computed. For normalization we multiplied all IBAQ values of the Δcox26 mutant dataset with a factor F. F is the ratio between the median of the wildtype (Mwt) and the median of the mutant (Mmut) for one condition: F = Mwt/Mmut. To visualize data in a heat map, values were normalized to maximum appearance within two native lanes comparing wild type and Δcox26. Slice number of mitochondrial complex III dimer (472 kDa), Complex IV (215 kDa), respiratory supercomplex III2IV1 (687 kDa), supercomplex III2IV2 (902 kDa), complex V monomer (573 kDa) and dimer (1209 kDa) were used for native mass calibration. The equation [f(x) = 36.062 × e^(0.0875×), R2 = 0.9938] obtained by exponential regression was used to calculate the native masses of each slice.
3. Results
Yeast mitochondrial supercomplexes containing respiratory complexes III and IV can be solubilized by the mild detergent digitonin and isolated by BNE. Here we used an improved 3-D electrophoretic separation technique to further characterize the proteome of yeast III–IV supercomplex and to search for novel protein components of these supercomplexes.
3.1. Identification of the supercomplex-associated Cox26 protein
The yeast S. cerevisiae requires functional mitochondrial oxidative phosphorylation complexes for ATP generation when growing on lactate or glycerol as the sole carbon source. Therefore in wild type mitochondria, cytochrome c-oxidase (complex IV) levels are high under these non-fermentative conditions. BNE analysis indicated that most of the complex IV was assembled to a supercomplex (III2-IV2) containing dimeric complex III and two copies of monomeric complex IV (Fig. 1A [4,41]).
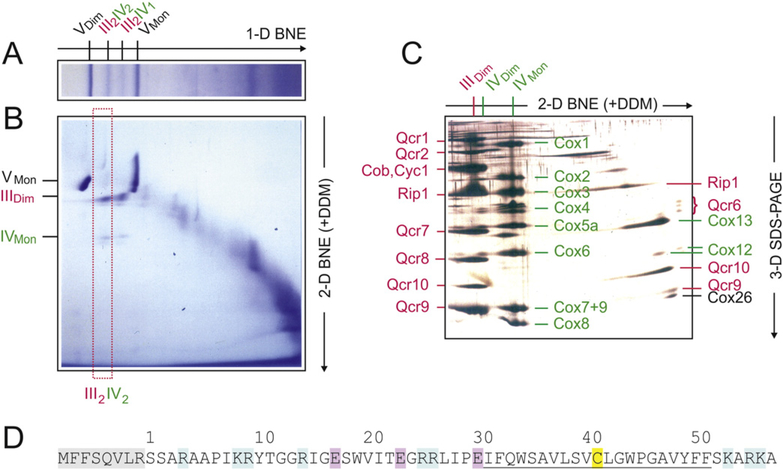
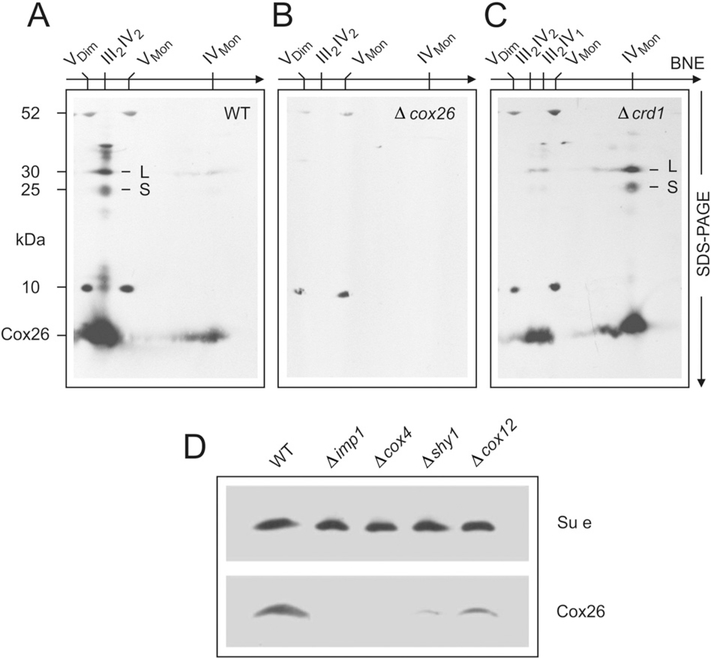
Fig. 1.
Three-dimensional electrophoresis of yeast mitochondria to identify Cox26, a novel protein co-migrating with respiratory supercomplexes. III2IV2, III2IV1, respiratory supercomplexes containing dimeric complex III and two or one copy of complex IV, respectively. VDim. VMon, IVDim. IVMon. IIIDim, dimeric and monomeric complexes V, IV, and III. (A) Yeast mitochondria were solubilized by digitonin and the mitochondrial (super)complexes were separated by BNE. (B) A gel strip from 1-D BNE was used for 2-D resolution by modified BNE, i.e. DDM was added to the cathode buffer to release individual complexes from the supercomplexes. (C) A gel strip from 2-D BNE was used for 3-D SDS-PAGE. Subunits of dimeric complex III (IIIDim) and the detergent-labile subunits (assigned on the right side) were marked red. Subunits of dimeric and monomeric complex IV (IVDim and IVMon) and the detergent-labile subunits were marked green. The subunits were assigned according to previous Edman sequencing [29] and/or detection by specific antibodies. One single protein, Cox26, was identified as a novel protein component of supercomplexes. It was identified as the smallest protein in the group of detergent-labile subunits (assigned on the right side of Fig. 1C). (D) Sequence and structural features of Cox26 protein. The preprotein contains a presequence of eight amino acids (shaded grey). Basic and acidic residues are marked blue and red, respectively. A cysteine residue (C41 marked yellow) is located within a predicted transmembrane helix (underlined).
The supercomplex containing only one copy of complex IV (III2-IV1) was also observed. Under these electrophoresis conditions, both the III–IV supercomplex species were flanked by monomeric and dimeric ATP synthase (complex V) with native masses of 573 kDa and 1.209 MDa, respectively [42] (Fig. 1A). Gel strips from 1-D BNE were then resolved by second dimension modified BNE, i.e. 0.02% DDM was added to the Coomassie-dye containing cathode buffer (Fig. 1B). The mixed micelles of DDM and the anionic Coomassie-dye mimic some properties of anionic detergents. They can release individual complexes III and IV from the supercomplexes and monomeric complex V from the dimeric form. We expected that the mixed micelles would also dissociate detergent-labile subunits of complexes III and IV and potentially release some novel previously unidentified protein components of supercomplexes. The released components were expected to migrate close to the electrophoretic front because of their low mass. In fact, when resolving the 2-D gel strip (boxed in Fig. 1B) by 3-D SDS-PAGE (Fig. 1C), a number of detergent-labile subunits of complex III (marked red) and of complex IV (marked green) were identified by immunodetection on Western blots and/or by Edman degradation (data not shown). One protein spot just below Qcr9 (denoted Cox26; right side of Fig. 1C) did not correspond to any known subunit of complexes III and IV and thus was identified as a novel protein subunit of the III–IV supercomplex and referred to here as Cox26. Several lines of evidence suggested that this Cox26 protein was associated and not just co-migrating with the III–IV supercomplexes. Accidental comigration of this small 6.4 kDa membrane protein with huge III–IV supercomplex (≤ 1 MDa) can be excluded because of their size differences and since no other protein complexes potentially binding Cox26 and co-migrating with the III–IV supercomplexes were identified. Unusual migration behaviour, as often observed with water-soluble proteins and pI values in the weekly acidic to neutral range, can also be excluded for Cox26, since Cox26 alone, once dissociated from the III–IV supercomplex with the DDM, migrates to the running front (Fig. 1C) like all previously analysed small membrane proteins that bind the anionic Coomassie-dye. We therefore conclude that Cox26 protein is a physical component of the III–IV supercomplex.
In order to increase the protein amount about 30 fold for Edman sequencing, the 3-D electrophoretic approach of Fig. 1 was repeated on a preparative scale. The amino-terminal sequence of the Cox26 protein was determined to be SSARAAPIKR. TFASTA search of the S. cerevisiae genome identified a previously annotated protein YDR119w-a on the S. cerevisiae chromosome IV. The analysis indicated that eight amino acids (shaded grey in Fig. 1D) had been removed in the mature Cox26 protein, suggesting that these residues may have constituted a mitochondrial targeting signal in the Cox26 precursor protein. The predicted mature protein of 58 amino acids had an isoelectric point in the basic range (pI = 10.52), and a mass of 6442.5 Da. The hydrophobic protein (Gravy score 0.048) contained a single cysteine residue (C41, marked yellow in Fig. 1D) in the center of a putative transmembrane helix (underlined) predicted by the TMpred [43]. No significant similarity to other proteins could be identified except with non-annotated homologues in other yeast species.
3.2. Cox26 protein stabilizes III–IV supercomplexes
To determine the functional relevance of the Cox26 protein, we constructed a null mutant yeast strain, deficient in the COX26 gene. We first analysed whether Cox26 was required for the assembly/stability of respiratory III–IV supercomplexes. The Cox26 null mutant (Δcox26) and wild type (W303–1A) strains were grown under identical conditions (YPD and YPL, 30 °C), mitochondria were isolated and prepared for the analysis of supercomplexes by BNE.
The supercomplexes were visualized by complex IV stain. Both supercomplexes III2–IV2 and III2–IV1 were assembled but appeared reduced in the Δcox26 mutant mitochondria relative to the wild type control (Fig. 2A). The decreased levels of supercomplexes and increased amount of individual complex IV was even more pronounced in cells grown with glucose as carbon source (Fig. 2A left lanes). A Cox26-specific antibody was used to verify the presence of the Cox26 protein in the III–IV supercomplexes from wild type mitochondria and its absence from the Cox26 null mutant in 2-D SDS-PAGE (Fig. 2B, lower panels).
We further analysed subunit composition in complexes and supercomplexes by mass spectrometry. The 1-D BNE lanes of wild type and Δcox26 mutant were fractionated into 46 even slices and measured by mass spectrometry.
Protein abundance profiles of subunits showed that in absence of Cox26 all identified complex III and complex IV subunits assemble into supercomplexes including the recently reported supercomplex associated proteins Rcf1 and Rcf2 (Fig. 3). Again the levels of supercomplexes appear reduced in both growth conditions. Increased amounts of complex IV were detected in cells grown under glucose as carbon source (Supplementary Fig. 1 ). Based on these data, we concluded that the presence of Cox26 is not essential for supercomplex formation and composition, but has an impact on the level and stability of the III–IV arrangement. The absence of Cox26 resulted in a clear difference in fermentative compared to non-fermentative carbon sources, suggesting that Cox26 might be involved in the regulation of III–IV supercomplexes under fermentative conditions.
3.3. Functional analysis of role of cox26
Next we analysed if the absence of Cox26 alters mitochondrial function, in particular respiratory capacity. Cytochrome c reductase (complex III) and cytochrome c oxidase (complex IV) activities from wild type and Cox26 deletion strains (Δcox26) were measured and found to be comparable (Supplementary Table 1), suggesting that Cox26 is not required for optimal catalytic activity of complexes III and complexes IV.
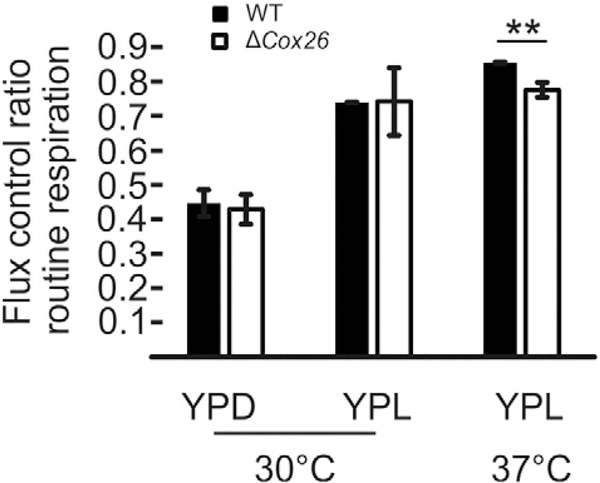
To analyse the impact of the Cox26 deletion on mitochondrial OXPHOS function, cells were analysed by high-resolution respirometry (Fig. 4). As cell size differs between wild type and mutant especially when grown at 37 °C (see below), the routine respiration was normalized to the maximal uncoupled respiration (flux control ratio; FCR) to evaluate respiration independent of cell size. Yeast strains grown in YPD or YPL media showed no differences in their routine FCR when grown at 30 °C. In contrast when cells were grown in YPL media a temperature induced decrease was detected in mutant cells (Fig. 4). We conclude that although Cox26 is not essential for supercomplex function, stress conditions seemed to reduce the respiration capacity under non-fermentable conditions when Cox26 was absent.
Fig. 4.
Flux control ratio routine respiration ofW303–1Aand Δcox26. Cells grown in YPD or YPL and were analysed by high resolution respirometry at 30 °C and 37 °C. Error bars indicate standard deviation of three independent experiments. **, mark significant difference from studentSs t-test p < 0.01.
To ascertain if the presence of Cox26 was required to support the respiratory activity or growth of the yeast strain, growth analysis of the Δcox26 strain was performed. Typical growth curves of wild type and Δcox26 strains are presented in Supplementary Fig. 2A–D. No obvious changes of the initial growth rates at 30 °C were observed under all tested fermentative and non-fermentative conditions. However, the optical density reached after 24 h tended to be higher with the Δcox26 strain. We asked whether differences in cell size or cell number could explain the unexpected high absorbance of Δcox26 at times greater than 24 h when the wild type strain had reached stationary phase. When grown at 30 °C for 24 h, cell size (Supplementary Fig. 3A) and cell number (data not shown) were comparable for wild type and Δcox26 strains on fermentative and non-fermentative media. Comparing cell size of wild type and Δcox26 strains that were grown on fermentative media at 37 °C for 24 h revealed that cells of the Δcox26 strain were almost doubled in size (Supplementary Fig. 3BC).
3.4. Cox26 protein is bound to complex IV
Immunodecoration of Western blots of 2-D BNE/SDS gels from wild type and Δcox26 mitochondria (Fig. 5A–B) with the Cox26-specific antisera indicated that the Cox26 protein was predominantly found co-migrating with the III–IV supercomplex in the wild type sample.
Fig. 5.
Cox26 protein is associated with complex IV. Assignment of complexes as in Fig. 1. Digitonin-solubilized mitochondrial complexes from (A) wild type yeast, (B) Δcox26 strain, and (C) Δcrd1 mutant with defective cardiolipin synthase were separated by BNE, followed by Tricine-SDS-PAGE in the second dimension using 13% acrylamide gels for Tricine-SDS-PAGE and electroblotted onto PVDF membranes. Anti-Cox26 antibody identified Cox26 (6.4 kDa) and two larger bands, L and S, with apparent masses around 25 kDa and 30 kDa, in the column of subunits of supercomplexes (e.g. in Figure partA). It also identified monomeric complex IV, e.g. in the Δcrd1 strain (figure partC). Detection of10 kDa and 52 kDa subunits of complex V seemed non-specific cross-reactions by the Cox26 antibody that mark the position of the monomeric and dimeric ATP synthase complexes. (D) Estimation of Cox26 protein amounts in null mutant strains not containing assembled complex IV (Δimp1 and Δcox4) or containing reduced amounts of complex IV (Δshy1 and Δcox12). Complex V amounts were estimated by a specific anti-subunit e antibody as loading control.
In addition, Cox26 was detected in the region of the gel where the free monomeric complex IV species migrated, suggesting that Cox26 is associated with the complex IV (COX) species (Fig. 5). The level of Cox26 co-migrating with the free complex IV species was significantly enhanced when mitochondria isolated from the Δcrd1 strain were analysed (Fig. 5C). The Δcrd1 strain is deficient in the cardiolipin synthase (Crd1 ) enzyme responsible for the synthesis of the mitochondrial lipid cardiolipin. Cardiolipin stabilizes the III–IV supercomplex [44–47] and thus under the solubilization and electrophoresis conditions used here, a significant increase in the level of free complex IV (and a corresponding reduction in III–IV levels) was obtained. The level of Cox26 protein found co-migrating with the free complex IV species was significantly increased in the Δcrd1 sample, indicating that a correlation existed between the levels and migration behaviour of the Cox26 protein and the assembly state complex IV (Fig. 5C). This observation supports the conclusion that the Cox26 protein partitions with complex IV and thus its association with the III–IV supercomplex is because it is a subunit of the complex IV enzyme.
To further explore the association of Cox26 with the III–IV supercomplex and also with the free complex IV species, 2-D BNE/SDS analysis followed by Western blot and immunodecoration with the Cox26 antibody were also performed with mitochondria isolated from a variety of null mutants of subunits of complex III (Δqcr6, 8, 9, 10) and of complex IV (Δcox8, 12, 13). In all cases where III–IV supercomplexes were detectable (Δqcr6, 10 and Δcox8, 12, 13), Cox26 was observed to comigrate with the supercomplexes (data not shown). In mutants where complex III was not assembled and thus no III–IV supercomplexes were formed (e.g. Δqcr8, 9), Cox26 was found to co-migrate with the free complex IV species (data not shown). In mutants with partial or complete loss of complex IV, like Δcox12, Δshy1, Δcox4, and Δimp1, the steady state levels of Cox26 was also reduced or undetectable in total protein extracts, indicating that Cox26 depends on the presence of complex IV for its stability within the mitochondria (Fig. 5D). Taking these results together, we conclude the observed comigration of Cox26 with the III–IV supercomplex species is thus due to the association of Cox26 with the complex IV aspect of the III–IV supercomplex. Furthermore, in our analysis Cox26 was always bound to complex IV and/or respiratory supercomplexes and was not detected as free protein in relevant amounts. This was also supported by gel filtration analyses (Supplementary Fig. 4) showing that Cox26 co-eluted only with complexes III and IV in the supercomplex fractions. We conclude that essentially all Cox26 in the mitochondrial membrane was physically associated with COX enzyme (complex IV).
3.5. Hydrophobic and covalent interactions between Cox26 and complex IV
The immunodecoration of the Western blotted native gels in Fig. 5A&C with Cox26-specific antisera, revealed also the additional presence of two larger Cox26-containing species, a larger and a smaller one (termed L and S in Fig. 5A&C) and located in the 25–30 kDa range. The absence of these protein spots in the Δcox26 sample analysed in parallel, confirmed that these L- and S-protein species, specifically contain the Cox26 protein. In the wild type mitochondrial sample, both the Cox26-containing L- and S-species co-migrated in the first dimension native electrophoresis with Cox26 in the III–IV supercomplex species (Fig. 5A). From their size (25–30 kDa), we suggest the L- and S- containing Cox26 species each represent the tight physical association of the Cox26 protein with another subunit of complex IV, which is not disrupted during the electrophoresis conditions used here. Consistently, in the Δcrd1 sample, these larger Cox26-containing L- and S-forms co-migrated with Cox26 in the free complex IV species.
In order to further characterize the larger Cox26-containing L and S- species, and to identify the partner proteins of the Cox26, we used a 3-D electrophoretic system. 1-D BNE and 2-D modified BNE were performed under non-reducing conditions to initially isolate the III–IV supercomplexes and then to separate into the individual complexes III and IV. The 3-D SDS-PAGE step was then finally used to separate the protein subunits under reducing and non-reducing conditions (Supplementary Fig. 5). This assay allowed dissociation of Cox26 from supercomplexes and individual complex IV in three steps (Supplementary Fig. 5C). The presence of the L- and S-Cox26 containing species was observed only under the non-reducing 3-D SDS PAGE analysis, and their absence was notable in the parallel reducing gel analysis. This suggests that the Cox26-population present in the L- and S-containing species, may represent a Cox26 protein in disulfide bond partnership with another complex IV subunit. As the apparent masses of bands L and S were around 25–30 kDa, subunits Cox4, Cox3, and Cox2 of complex IV (around 14–25 kDa) are potential partners covalently bound to the 6.4 kDa Cox26 protein. It is important to note that this 3-D PAGE analysis indicates that only a small fraction of the total Cox26 species existed in the larger L- and S-arrangement. The majority of Cox26 was released as a monomeric protein during 2-D modified BNE, i.e. under native and non-reducing conditions, indicating that most of the Cox26 protein was in a non-covalent relationship with complex IV under these experimental conditions used.
An even more complex 4-D electrophoretic technique was next employed to identify the Cox26 binding partners in the Cox26 L and S-arrangement. Again, 1-D BNE was applied first to isolate the III–IV supercomplexes. Complex IV was then released and isolated from the III–IV supercomplexes by the addition of DDM in the 2-D modified BNE. Gel pieces from the 2-D BN gel containing complex IV and associated L and S-Cox26 containing species were finally analysed by two orthogonal SDS gels (3-D and 4-D SDS separations, shown in Fig. 6).
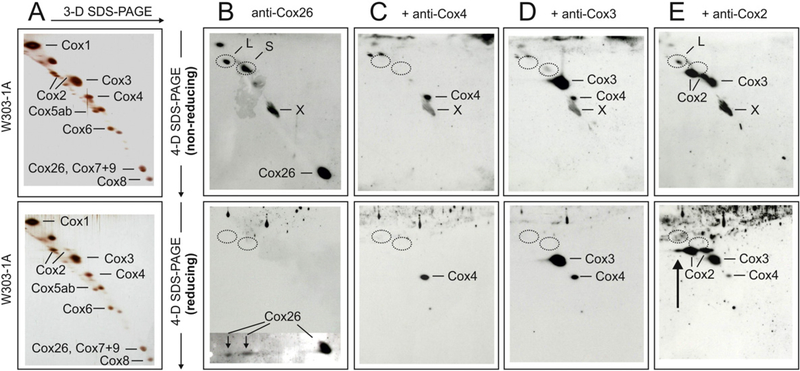
Fig. 6.
Evidence for covalent interaction of Cox26 and Cox2 proteins in isolated complex IV. Gel pieces from 2-D BNE/modified BNE containing complex IV were resolved under non-reducing conditions by 3-D Tricine-SDS-PAGE using 9% acrylamide gels. 3-D gel strips were then incubated under non-reducing (upper panels) or reducing conditions (lower panels) followed by 4-D Tricine-SDS-PAGE using 16% acrylamide gels containing 6 M urea. The 4-D gels were silver-stained (A) or blotted onto PVDF membranes (B-E). Polyclonal anti-Cox26 antibody identified individual Cox26 protein and two bands (L and S) under non-reducing conditions (B, upper panel), and two Cox26 protein spots that were dissociated from bands L and S under reducing conditions (B, lower panel). A local defect of the PVDF membrane (marked X) was also recognized by the antibody. Circles mark the actual or expected positions of bands L and S. (C and D) Anti-Cox4 and anti-Cox3 antibodies that were added consecutively to the same blot membranes (without using stripping protocols) identified individual Cox4 and Cox3 proteins but no bands L and S. Reusing the same blots, an anti-Cox2 antibody finally recognized two forms of individual Cox2 protein and also band L under non-reducing conditions (E, upper panel) in addition to the protein spots recognized before by the anti-Cox4 and anti-Cox3 antibodies. Performing 4-D SDS-PAGE under reducing conditions (E, lower panel), band L was no longer detected but a third Cox2 spot immediately below the expected position ofband L appeared (marked by an arrow).
All 1-D, 2-D and 3-D gels were performed under non-reducing conditions. Both non-reducing (Fig. 6, upper panels) and reducing conditions (Fig. 6, lower panels) were used in parallel for the final 4-D SDS- PAGE. Silver-stained 3-D/4-D separations of complex IV from the W303–1A strain are shown in Fig. 6A. Subunits were assigned according to previous identification by amino-terminal protein sequencing and/or immunological detection. A few unmarked spots are regarded as contaminations. Corresponding Western blots are presented in figure parts B-E. A polyclonal anti-Cox26 rabbit antibody was applied first, followed consecutively by monoclonal mouse anti-Cox4, anti-Cox3, and anti-Cox2, antibodies on the same blot membrane (Fig. 6B–E). Under non-reducing conditions for 4-D SDS-PAGE, the anti-Cox26 antibody recognized individual Cox26 protein and bands L and S, and weakly also Cox1 (Fig. 6B, upper panel).
A similar blot using reducing conditions for 4-D SDS-PAGE revealed individual Cox26 straight below the expected positions of non-reduced bands Land S (circles in Fig. 6B, lower panel). The upper and lower blot membranes were then reused immediately for detection by the anti- Cox4 antibody, i.e. without using an antibody stripping protocol. This shortened protocol could be applied, since an anti-mouse antibody followed the previous anti-rabbit antibody and all signals from the previous detection were lost. Anti-Cox4 antibody recognized Cox4 at the expected position for individual Cox4 protein but did not detect bands L and S (Fig. 6C, upper panel) and no Cox4 protein was found released from bands L and S under reducing conditions (Fig. 6C, lower panel). This suggested that Cox4 is not a component of bands L and S. Using a similar approach, Cox3 was also excluded (Fig. 6D). In contrast, anti- Cox2 antibody always detected band L in Western blot approaches like those shown in Fig. 4E, upper panel, suggesting that band L comprises the Cox2 and Cox26 proteins. The corresponding control experiment to demonstrate the release of Cox2 under reducing conditions, revealed some extra Cox2 protein as an extension of the normal Cox2 spots immediately below band L, as expected (arrow in Fig. 6E, lower panel). Although this extra signal presumably corresponded partially or largely to Cox2 that was released from band L under reducing conditions, it should be noted that a minor signal intensity was also detected in the Δcox26 control lacking band L (Supplementary Fig. 6J, lower panel). For an overview of the complete set of Western blots including Δcox26 mutant see Supplementary Fig. 6. Together, all evidence points to association of Cox26 and Cox2 via a disulfide-bridge in band L. The identity of the second component in band S remains obscure. However Cox2, Cox3, and Cox4 which were not detected by specific antibodies in band S can be excluded as potential binding partners.
3.6. Model for the association of complexes III and ¡V and the binding of Cox26 protein
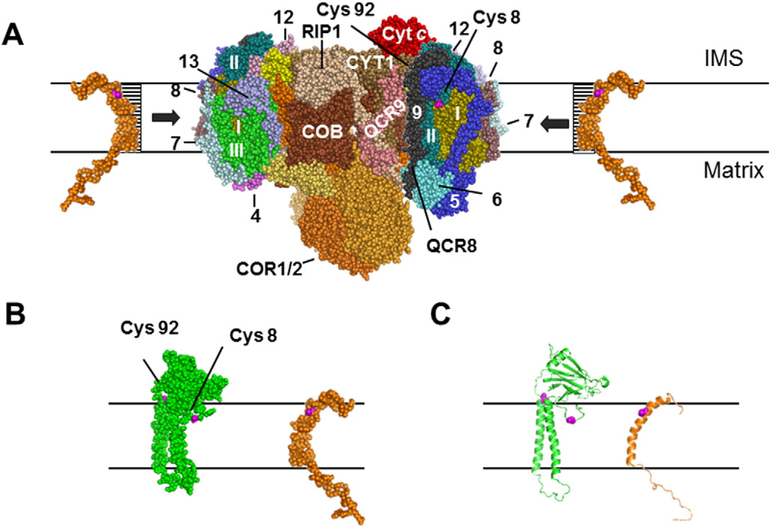
The model for the association of complexes III and IV in respirasomes is based on previous evidence suggesting that the interface between the two complexes comprises transmembrane helices of cytochromes b and c1 of complex III, and presumably at least one of the hydrophobic subunits of complex IV, Cox1, Cox2, and Cox3 [48]. According to the identified covalent interaction of Cox26 with Cox2 in the L-species, the putative binding site of Cox26 protein within the complex IV would be in close proximity of the Cox2 protein, as suggested in Fig. 7A.
Fig. 7.
Model for the position of Cox26 in the yeast supercomplex. (A) Model comprises cytochrome c1 (CYT1), cytochrome b (COB), RIP1,Rieske iron-sulphur protein, Core proteins 1 and 2 (COR1/2) and QCR6–9 of complex III, and all three hydrophobic subunits of complex IV, Cox1 (I),Cox2 (II), and Cox3 (III) and the subunits Cox5 (5), Cox7–9 (7–9),Cox12 (12), and Cox13 (13) of complex IV. The model was generated based on structural information of yeast complex III [49] and bovine complex IV [48] using the software PyMOL Molecular Graphics System, Version 1.2, Schrodinger, LLC. Cysteine-92 (Cys 92) which is located at the end of a transmembrane helix of Cox2 was only visible when the central complex IV subunits were inspected. After adding all other subunits Cys 92 seems to be hidden. Cysteine-8 (Cys 8) is located in the centre of a hydrophilic loop structure of Cox2. Cys 8 seems freely accessible to Cox26 protein even after complementation of the model structure with all accessory subunits of complex IV. (B, C) Cox26 was modelled using HHpred and Modellerwith the pdb files 3arc-L, 2kad_A and 2yus_A. The Cys 41 of Cox26 is located within a predicted transmembrane helix which could have direct access to Cys 8 of Cox2 at the hydrophilic boundary part of the membrane facing to the intermembrane space (IMS).
As our results suggested that cysteine-41 in the end of the predicted transmembrane helix of Cox26 forms a disulfide bridge with Cox2 we asked which one of the four cysteine residues of yeast Cox2 most likely interacted with cysteine-41 of Cox26. Since cysteines-206 and −210 of the mature Cox2 protein (cysteines-221 and −225 also counting the import sequence; UniProt P00410) are involved in CuA-binding, only cysteines-8 and −92 of the mature Cox2 protein seem relevant for potential disulfide bonding to Cox26. The positions of cysteines-8 (visible) and −92 (hidden) are indicated in Fig. 7A, as derived from the 3-D crystal structure of the bovine complex IV [43] and the sequences of the homologous yeast subunits. In the lower panel of Fig. 7, Cox2 and a modelled Cox26 are shown. Cysteine-92, which is located at the end of a transmembrane helix of Cox2, is only visible on the monomer of complex IV (small red spot on the surface of Cox2 Fig. 7BC). Cysteine-8 is located at the centre of a amphipathic loop region of Cox2. In contrast to cysteine-92, cysteine-8 seems accessible to the Cox26 protein even after all accessory subunits of complex IV were added to the model structure (Fig. 7A). We conclude that cysteine-8 of Cox2 is the most likely interaction partner of cysteine-41 in Cox26.
4. Discussion
A major technical aspect of this work was to demonstrate how multidimensional electrophoretic techniques can be used to identify noncovalent and covalent protein-protein interactions. Two native dimensions (1-D BNE and 2-D modified BNE) were used here to isolate native membrane protein complexes directly from biological membranes. This simple two-step native electrophoresis, performed on a micro-scale, can replace multi-step chromatographic isolation protocols that otherwise must be established in time consuming optimization trials and must be carried out on a much larger preparative scale. We also show that three-dimensional BNE/BNE/SDS-PAGE offers great advantages for the component analysis of multiprotein complexes and facilitates the identification of novel components (Fig. 1). Furthermore, 3-D BNE/BNE/SDS gels can give valuable qualitative and quantitative information on covalent and non-covalent interactions of protein components (Supplementary Fig. 5). Four-dimensional BNE/BNE/SDS/SDS-PAGE applying reducing and non-reducing conditions for the final 4-D SDS-PAGE was found a valuable means to identify inter-protein disulfide bridges (Fig. 6 and Supplementary Fig. 6).
The role of Cox26 on mitochondrial function is still not clear. Complex IV activity and respiratory function was comparable to wild type cells, but under stress conditions the respiratory capacity of the Δcox26 mutant was compromised. The observation of an increased cell volume of the Cox26 null mutant in fermentative media at 37 °C remains unclear. The systematic study from Jorgensen et al. [50] identified in more than 500 deletion mutants abnormal cell volume under standard growth conditions (30 °C, Glucose). Cox26 and other OXPHOS proteins were not reported under these conditions. We observed increased cell size only under elevated temperature. If this phenotype is related or not to the function of Cox26 as complex IV subunit remains to be elucidated.
The Cox26 protein was identified here as a protein component of yeast respirasomes containing dimeric complex III and one or two copies of monomeric complex IV [4,41]. Tight but detergent-sensitive binding to complex IV, and partial disulfide bonding to the Cox2 subunit initially seemed to point to a complex IV specific role. Our data here would indicate that the presence of Cox26 is not essential for, but may play a supporting role in the III–IV supercomplex formation and stability. The level and the appearance of supercomplexes seemed to be much more affected when cells were grown under fermentative conditions. Recently, independent studies characterized the III–IV supernumerary proteins Rcf1 and Rcf2 [33–35] and the suggestion that different forms of the III–IV supercomplexes may exist, for example a Rcf1-III–IV, a Rcf2-III–IV and III–IV population (i.e. without either Rcf1 or Rcf2), was proposed [34,35]. It is tempting to speculate therefore that different populations of the III–IV supercomplex may exist within a given mitochondria to serve specialized roles or optimized to function under different conditions. For example, these may reflect specialized III–IV populations operating in different mitochondrial sublocations (cristae membrane vs. inner boundary membrane, or arranged to optimally function under different metabolic or bioenergetics conditions). One study discussed the role of alternative supercomplexes to be protection for generation of reactive oxygen species (ROS) [34]. We report here that Cox26 was found in two arrangements within the III–IV supercomplex populations, one loosely attached and a smaller fraction covalently bound to Cox2 (the Cox26-L species) and another to a currently unknown protein (the Cox26 S-species). It is possible that the complex IV populations containing the different arrangements of Cox26 may represent complex IV species displaying for different enzymatic properties, however this requires further investigation.
In this study we analysed fermentative and non-fermentative growth conditions, both under atmospheric oxygen pressure. To deeper understand the function of this covalent interaction would be interesting for example to investigate supercomplex remodelling in hypoxia and conditions with elevated ROS levels. We found that Cox26 covalently interacts with Cox2 suggesting a regulatory role for this subunit ofthe catalytical core of complex IV. Cox2 interacts directly with cytochrome c and contains the dinuclear copper centre CuA. In this respect the binding of Cox26 with this central subunit could play a role in modulating the function of Cox2. In general complex IV is highly regulated [reviewed in 51]. The Cys-8 of Cox2 locates at the membrane boundary to the intermembrane space, a more oxidizing compartment with several redox pathways and functionally important disulfide bonds [51–55]. It is conceivable that the cellular redox state can have an impact on the thiol-disulfide balance of Cox2 and Cox26. Indeed many targets for redox-import Mia40/Erv1 system belong to factors of COX biosynthesis [56]. This pathway targets proteins with twin CX9C [57] or CX3C motifs. Therefore the single cysteine of Cox26 is unlikely to be a target of this system. However these redox pathways have been connected to complex IV assembly but a function of ROS generation systems that modulate the function of cytochrome c oxidase have not yet considered. In mammals complex III is supposed to generate ROS into the IMS and to target protein thiols more specific as “signalling ROS” [58]. It remains to be elucidated if condition of elevated ROS generation, hypoxia, redox pathways or carbon sources switches Cox26 from a mostly noncovalent bound subunit to a covalent COX-modulator.
5. Conclusion
In conclusion we identified the new complex IV subunit Cox26 that exists in close proximity to the catalytically important Cox2 subunit, where it can form an intersubunit disulfide bond. Our functional characterization of the Cox26 null mutant strain would suggest that Cox26 is not critically important for the catalysis or assembly of COX complex into the III–IV supercomplex but with impact on stability and organization of the higher molecular supercomplex with III2IV2 stoichiometry. We speculate here that the Cox26 protein may serve to regulate the COX activity possibly by modulating or supporting the Cox2 protein subunit.
Supplementary Material
Acknowledgments
We are grateful to Hermann Schägger, who discovered Cox26 and scientifically supported the work. We thank Martina Ding and Ulrich Brandt for helpful scientific discussions. We thank Ilka Siebels for support in oxygraph measurements. This work was supported by the Cluster of Excellence “Macromolecular Complexes” at the Goethe University Frankfurt (DFG Project EXC 115), the SFB815 project Z1 (to IW) and by Bundesministerium für Bildung und Forschung Grant BMBF 01GMIII3B mitoNET-Deutsches Netzwerk für mitochondriale Erkrankungen (to I.W.) and by the National Science Foundation (NSF) grants MCB0744067 and MCB 1157722 and the National Institutes of Health (NIH) NIH-GM101594 (to R.A.S.).
Abbreviations
- BN
blue native
- Coomassie dye
Coomassie blue G-250
- Complex III
ubiquinol:cytochrome c reductase
- Complex IV
cytochrome c oxidase
- Complex V
ATP synthase
- DDM
dodecyl-β-D-maltoside
- PVDF
polyvinylidenedifluoride
Footnotes
Conflict of interest
All authors have declared no conflicts of interest concerning the contents of the article.
Transparency document
The Transparency document associated with this article can be found, in online version.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbamcr.2016.04.012.
References
- [1].Hatefi Y, The mitochondrial electron transport and oxidative phosphorylation system, Annu. Rev. Biochem 54 (1985) 1015–1069. [DOI] [PubMed] [Google Scholar]
- [2].Arnold I, Pfeiffer K, Neupert W, A Stuart R, Schägger H, Yeast F1FO-ATP synthase exists as a dimer: identification of three dimer specific subunits, EMBO J. 17 (1998) 7170–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA, The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria, J. Biol. Chem 275 (2000) 18093–18098. [DOI] [PubMed] [Google Scholar]
- [4].Schägger H, Pfeiffer K, Supercomplexes in the respiratory chains of yeast and mammalian mitochondria, EMBO J. 19 (2000) 1777–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Eubel H, Heinemeyer J, Braun HP, Identification and characterization of respirasomes in potato mitochondria, Plant Physiol. 134 (2004) 1450–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Perales M, Eubel H, Heinemeyer J, Colaneri A, Zabaleta E, Braun HP, Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I + III2 levels and alters mitochondrial physiology in Arabidopsis, J. Mol. Biol 350 (2005) 263–277. [DOI] [PubMed] [Google Scholar]
- [7].Krause F, Reifschneider NH, Goto S, Dencher NA, Active oligomeric ATP synthases in mammalian mitochondria, Biochem. Biophys. Res. Commun 329 (2005) 583–590. [DOI] [PubMed] [Google Scholar]
- [8].Wittig I, Schägger H, Advantages and limitations of clear native polyacrylamide gel electrophoresis, Proteomics 5 (2005) 4338–4346. [DOI] [PubMed] [Google Scholar]
- [9].Schägger H, von Jagow G, Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form, Anal. Biochem 199 (1991) 223–231. [DOI] [PubMed] [Google Scholar]
- [10].Wittig I, Braun HP, Schägger H, Blue-native PAGE, Nat. Protoc 1 (2006) 416–428. [DOI] [PubMed] [Google Scholar]
- [11].Schägger H, Cramer WA, von Jagow G, Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis, Anal. Biochem 217 (1994) 220–230. [DOI] [PubMed] [Google Scholar]
- [12].Wittig I, Karas M, Schägger H, High resolution clear-native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes, Mol. Cell. Proteomics 6 (2007) 1215–1225. [DOI] [PubMed] [Google Scholar]
- [13].Minauro-Sanmiguel F, Wilkens S, Garcia JJ, Structure of dimeric mitochondrial ATP synthase: novel Fo bridging features and the structural basis of mitochondrial cristae biogenesis, Proc. Natl. Acad. Sci 102 (2005) 12356–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dudkina NV, Heinemeyer J, Keegstra W, Boekema EJ, Braun HP, Structure of dimeric ATP synthase from mitochondria: an angular association of monomers induces the strong curvature of the inner membrane, FEBS Lett. 579 (2005) 5769–5772. [DOI] [PubMed] [Google Scholar]
- [15].Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP, Structure of a mitochondrial supercomplex formed by respiratory chain complexes I and III, Proc. Natl. Acad. Sci. U. S. A 102 (2005) 3225–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dudkina NV, Sunderhaus S, Braun HP, Boekema EJ, Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria, FEBS Lett. 580 (2006) 3427–3432. [DOI] [PubMed] [Google Scholar]
- [17].Sunderhaus S, Dudkina NV, Jànsch L, Klodmann J, Heinemeyer J, Perales M, Zabaleta E, Boekema EJ, Braun HP, Carbonic anhydrase subunits form a matrix- exposed domain attached to the membrane arm of mitochondrial complex I in plants, J. Biol. Chem 281 (2006) 6482–6488. [DOI] [PubMed] [Google Scholar]
- [18].Schàfer E, Seelert H, Reifschneider NH, Krause F, Architecture of active mammalian respiratory chain supercomplexes, J. Biol. Chem 281 (2006) 15370–15375. [DOI] [PubMed] [Google Scholar]
- [19].Schàfer E, Dencher NA, Vonck J, Parcej DN, Three-dimensional structure of the respiratory chain supercomplex I(1)III(2)IV(1) from bovine heart mitochondria, Biochemistry 46 (2007) 12579–12585. [DOI] [PubMed] [Google Scholar]
- [20].Wittig I, Velours J, RA Stuart H Schägger, Characterization of domain-interfaces in monomeric and dimeric ATP synthase, Mol. Cell. Proteomics 7 (2008) 995–1004. [DOI] [PubMed] [Google Scholar]
- [21].Paumard J, Vaillier B, Coulary J, Schaeffer V, Soubannier V, Mueller DM, Brèthes D, di Rago JP, Velours J, The ATP synthase is involved in generating mitochondrial cristae morphology, EMBOJ. 21 (21) (2002) 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Davies KM, Anselmi C, Wittig I, Faraldo-Gómez JD, Kühlbrandt W, Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae, Proc. Natl. Acad. Sci. U. S. A 109 (2012) 13602–13607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Strauss M, Hofhaus G, Schroder RR, Kühlbrandt W, Dimer ribbons of ATP synthase shape the inner mitochondrial membrane, EMBOJ. 27 (2008) 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Meyer B, Wittig I, Trifilieff E, Karas M, Schägger H, Identification of two proteins associated with mammalian ATP synthase, Mol. Cell. Proteomics 6 (2007) 1690–1699. [DOI] [PubMed] [Google Scholar]
- [25].Bianchi C, Genova ML, Parenti Castelli G, Lenaz G, The mitochondrial respiratory chain is partially organized in a supercomplex assembly: kinetic evidence using flux control analysis, J. Biol. Chem 279 (2004) 36562–36569. [DOI] [PubMed] [Google Scholar]
- [26].Schägger H, De Coo R, Bauer MF, Hofmann S, Godinot C, Brandt U, Significance of respirasomes for the assembly/stability of human respiratory chain complex I, J. Biol. Chem 279 (2004) 36349–36353. [DOI] [PubMed] [Google Scholar]
- [27].Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Pérez-Martos A, Bruno C, Moraes CT,JA Enriquez, Respiratory complex III is required to maintain complex I in mammalian mitochondria, Mol. Cell 13 (2004) 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].De Coo IFM, Renier WO, Ruitenbeek W, Ter Laak HJM, Schägger H, van Oost BA, Smeets HJ, A 4 bp deletion in mitochondrial cytochrome b gene associated with Parkinsonism/MELAS overlap syndrome, Ann. Neurol 45 (1999) 130–133. [DOI] [PubMed] [Google Scholar]
- [29].Brandt U, Uribe S, Schägger H, Trumpower BL, Isolation and characterization of QCR10, the nuclear gene encoding the 8.5-kDa subunit 10 of the Saccharomyces cerevisiae cytochrome bc1 complex, J. Biol. Chem 269 (1994) 12947–12953. [PubMed] [Google Scholar]
- [30].Geier BM, Schägger H, Ortwein C, Link TA, Hagen WR, Brandt U, von Jagow G, Kinetic properties and ligand binding of the eleven subunit cytochrome c oxidase from Saccharomyces cerevisiae isolated with a novel large scale purification method, Eur. J. Biochem 227 (1995) 296–302. [DOI] [PubMed] [Google Scholar]
- [31].Helbig AO, de Groot MJ, A van Gestel R, Mohammed S, de Hulster EA, Luttik MA, Daran-Lapujade P, Pronk JT, Heck AJ, Slijper M, A three-way proteomics strategy allows differential analysis of yeast mitochondrial membrane protein complexes under anaerobic and aerobic conditions, Proteomics 9 (2009) 4787–4798. [DOI] [PubMed] [Google Scholar]
- [32].Wach A, Brachat A, R Pohlmann P Philippsen, New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae, Yeast 10 (1994) 1793–1808. [DOI] [PubMed] [Google Scholar]
- [33].Chen YC, Taylor EB, Dephoure N, Heo JM, Tonhato A, Papandreou I, Nath N, Denko NC, Gygi SP, Rutter J, Identification of a protein mediating respiratory supercomplex stability, Cell Metab. 15 (2012) 348–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Vukotic M, Oeljeklaus S, Wiese S, Vogtle FN, Meisinger C, Meyer HE, Zieseniss A, Katschinski DM, Jans DC, Jakobs S, Warscheid B, Rehling P, Deckers M, Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex, Cell Metab. 15 (2012) 336–347. [DOI] [PubMed] [Google Scholar]
- [35].Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart RA, Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex, Mol. Cell. Biol 32 (2012) 1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Schägger H, Tricine-SDS-PAGE, Nat. Protoc 1 (2006) 16–22. [DOI] [PubMed] [Google Scholar]
- [37].Rais I, Karas M, Schägger H, Two-dimensional electrophoresis for the isolation of integral membrane proteins and mass spectrometric identification, Proteomics 4 (2004) 2567–2571. [DOI] [PubMed] [Google Scholar]
- [38].Gnaiger E, Mitochondrial pathways and respiratory control, An Introduction to OXPHOS Analysis, fourth ed.OROBOROS MiPNet Publications; 2014, Innsbruck, Austria, 2014. [Google Scholar]
- [39].Heide H, Bleier L, Steger M, Ackermann J, Drose S, Schwamb B, Zornig M, Reichert AS, Koch I, Wittig I, Brandt U, Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex, Cell Metab. 16 (2012) 538–549. [DOI] [PubMed] [Google Scholar]
- [40].Cox J, Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification, Nat. Biotechnol 26 (2008) 1367–1372. [DOI] [PubMed] [Google Scholar]
- [41].Heinemeyer J, Braun HP, Boekema EJ, Kouril RA, A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria, J. Biol. Chem 282 (2007) 12240–12248. [DOI] [PubMed] [Google Scholar]
- [42].Wittig I, Schägger H, Structural organization of mitochondrial ATP synthase, Biochim. Biophys. Acta 1777 (2008) 592–598. [DOI] [PubMed] [Google Scholar]
- [43].Hofmann K, Stoffel W, TMbase — a database of membrane spanning proteins segments, Biol. Chem. Hoppe Seyler 374 (1993) 166. [Google Scholar]
- [44].Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, Pfanner N, Greenberg ML, Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function, J. Biol. Chem 275 (2000) 22387–22394. [DOI] [PubMed] [Google Scholar]
- [45].Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schägger H, Cardiolipin stabilizes respiratory chain supercomplexes, J. Biol. Chem 278 (2003) 52873–52880. [DOI] [PubMed] [Google Scholar]
- [46].Brandner K, Mick DU, Frazier AE, Taylor RD, Meisinger C, Rehling P, Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth syndrome, Mol. Biol. Cell 16 (2005) 5202–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].McKenzie M, Lazarou M, Thorburn DR, Ryan MT, Mitochondrial respiratory chain supercomplexes are destabilized in Barth syndrome patients, J. Mol. Biol 361 (2006) 462–469. [DOI] [PubMed] [Google Scholar]
- [48].Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S, The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A, Science 272 (1996) 1136–1144. [DOI] [PubMed] [Google Scholar]
- [49].Solmaz SR, Hunte C, Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer, J. Biol. Chem 283 (2008) 17542–17549. [DOI] [PubMed] [Google Scholar]
- [50].Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M, Systematic identification of pathways that couple cell growth and division in yeast, Science 297 (2002) 395–400. [DOI] [PubMed] [Google Scholar]
- [51].Bourens M, Fontanesi F, Soto IC, Liu J, Barrientos A, Redox and reactive oxygen species regulation of mitochondrial cytochrome c oxidase biogenesis, Antioxid. Redox Signal 19 (2013) 1940–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hu J, Dong L, Outten CE, The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix, J. Biol. Chem 283 (2008) 29126–29134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hell K, The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria, Biochim. Biophys. Acta 1783 (2008) 601–609. [DOI] [PubMed] [Google Scholar]
- [54].Gabriel K, Milenkovic D, Chacinska A, Müller J, Guiard B, Pfanner N, Meisinger C, Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway, J. Mol. Biol 365 (2007) 612–620. [DOI] [PubMed] [Google Scholar]
- [55].Bleier L, Drose S, Superoxide generation by complex III: from mechanistic rationales to functional consequences, Biochim. Biophys. Acta 1827 (2013) 1320–1331. [DOI] [PubMed] [Google Scholar]
- [56].Herrmann JM, Riemer J, Oxidation and reduction of cysteines in the intermembrane space of mitochondria: multiple facets of redox control, Antioxid. Redox Signal 13 (2010) 1323–1326. [DOI] [PubMed] [Google Scholar]
- [57].Longen S, Bien M, Bihlmaier K, Kloeppel C, Kauff F, Hammermeister M, Westermann B, Herrmann JM, Riemer J, Systematic analysis of the twin cx(9)c protein family, J. Mol. Biol 393 (2009) 356–368. [DOI] [PubMed] [Google Scholar]
- [58].Bleier L, Wittig I, Heide H, Steger M, Brandt U, Drose S, Generator-specific targets of mitochondrial reactive oxygen species, Free Radic. Biol. Med 78 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.