Abstract
Background
Drought stress is one of the major factors limiting wheat production globally. Improving drought tolerance is important for agriculture sustainability. Although various morphological, physiological and biochemical responses associated with drought tolerance have been documented, the molecular mechanisms and regulatory genes that are needed to improve drought tolerance in crops require further investigation. We have used a novel 4-component version (for overexpression) and a 3-component version (for underexpression) of a barley stripe mosaic virus-based (BSMV) system for functional characterization of the C2H2-type zinc finger protein TaZFP1B in wheat. These expression systems avoid the need to produce transgenic plant lines and greatly speed up functional gene characterization.
Results
We show that overexpression of TaZFP1B stimulates plant growth and up-regulates different oxidative stress-responsive genes under well-watered conditions. Plants that overexpress TaZFP1B are more drought tolerant at critical periods of the plant’s life cycle. Furthermore, RNA-Seq analysis revealed that plants overexpressing TaZFP1B reprogram their transcriptome, resulting in physiological and physical modifications that help wheat to grow and survive under drought stress. In contrast, plants transformed to underexpress TaZFP1B are significantly less tolerant to drought and growth is negatively affected.
Conclusions
This study clearly shows that the two versions of the BSMV system can be used for fast and efficient functional characterization of genes in crops. The extent of transcriptome reprogramming in plants that overexpress TaZFP1B indicates that the encoded transcription factor is a key regulator of drought tolerance in wheat.
Keywords: Barley stripe mosaic virus, C2H2 zinc finger proteins, Drought, Functional characterization, Gene overexpression, Plant transformation, RNA-Seq, siRNA, Transcriptome, Triticum aestivum
Background
Bread wheat (Triticum aestivum L.) is one of the most important crops worldwide and global demand is increasing. It was estimated that cereal production needs to increase by at least 50% between 2005 and 2050 [1, 2]. However, achieving this goal is uncertain due to limited land resources and the impact of various abiotic and biotic stresses. Drought stress is one of the major environmental stresses limiting crop productivity worldwide [3], and the frequency of drought spells is expected to increase with global climate change [4]. In order to improve crop yield, we must increase our understanding of the genetic and molecular mechanisms underlying the responses and tolerance mechanisms to various abiotic stresses in crops.
Most genomic studies have focused on plant models or on crops with diploid genomes (e.g. Arabidopsis thaliana and Oryza sativa) [5, 6]. These studies have provided valuable insights into different biological processes associated with various abiotic stresses in plants. While many conserved pathways are shared between models and crops, divergent functions sometimes arise between homologous genes during the course of evolution [7]. This limits direct translation of functional characterization results from model species to crops, and suggests that the identification of functional gene orthologues in crop plants such as wheat requires species-specific studies. Wheat is a hexaploid organism that originated from hybridization events between ancestral genomes. These events provided genetic diversity and plasticity [8] which is key to the success of this crop under different ecological conditions [9]. The diverse gene pools of cultivated or ancestral wheat provide a great opportunity to identify stress-associated genes and improve our knowledge of gene networks that may contribute to increase wheat performance under diverse abiotic stress conditions.
Plants, as sessile organisms, need to evolve different strategies to cope with and adapt to environmental changes. Exposure to abiotic stress induces physiological and metabolic responses that are mediated through complex signal transduction networks involving a great number of molecules and stress-responsive genes [10–13]. Drought stress is initiated by water deficit in soil, resulting in osmotic stress. Moreover, inhibition of CO2 fixation during drought leads to disturbances in the electron transport chain and photosystem activities in chloroplasts, resulting in increased ROS production and accumulation [14], which could be harmful to plants. In the course of evolution, plants have adapted dynamic responses at the morphological, physiological, and biochemical levels, allowing them to survive under rapidly changing environmental conditions. Adaptive responses associated with tolerance traits include cuticular wax biosynthesis on leaf surfaces, improved osmotic adjustment ability and increased cell wall elasticity to maintain tissue turgidity [15, 16] via the synthesis and accumulation of xyloglucan endotransglucosylase/hydrolase (XTH), cellulose synthase, pectin esterase, expansin, soluble carbohydrates and osmoprotectants like proline and glycine betaine [16, 17]. The manifestation of these morphological or physiological responses involves processes starting from perception of stress to the expression of large numbers of genes that increase the chances of survival. Increasing evidence supports that transduction of the stress signal and plant responses are mediated by calcium and the activation of several Ca2+ sensors [18–20]. In Arabidopsis, a study showed that overexpression of the Calmodulin 1 (CaM1) gene positively regulates NADPH oxidase RbohF, leading to abscisic acid (ABA)-triggered ROS production and stomatal closure [21].
ABA is a key drought-induced signal modulating physiological responses that eventually lead to acclimation and stress tolerance. Accumulation of ABA in leaves directly regulates stomatal movement [22, 23] and reduces water loss, resulting in drought avoidance [24]. The relationship between ABA sensitivity and drought tolerance has been demonstrated by a study in the wild wheat Aegilops tauschii in which drought-tolerant accessions of A. tauschii show significantly higher ABA sensitivity than drought-sensitive lines, and tend to accumulate more stress-responsive gene transcripts [23]. This suggests that ABA sensitivity is regulated by the expression of different genes involved in ABA perception/signaling. In studies using transgenic lines, overexpression of genes involved in ABA signaling such as the aspartic protease ASPG1, the NADP-malic enzyme or an E3 ubiquitin ligase enhanced tolerance to drought stress [25–27]. In another study, the loss of function through antisense regulation or by mutation of the receptor-like kinase1 (RPK1) in Arabidopsis decreased ABA sensitivity, stomatal closure and the expression of several stress-inducible genes such as LEA-like proteins, peroxidase, RD26, DnaJ-like protein, cytochrome P450 and SOD [28]. Enzymes belonging to the SnRK2 protein kinase subfamily are major regulators of plant response to ABA by direct phosphorylation of various downstream targets including transcription factors, the NADPH oxidase RbohF, LEA-like proteins, DREB (Dehydration-Responsive Element-Binding protein), slow anion channel (SLAC)-associated genes and antioxidant enzyme genes [29–31]. These studies show that drought tolerance is governed by a complex gene regulatory network which is still poorly understood in wheat.
Transcriptional factors are the most important regulatory proteins that modulate the expression of specific sets of genes [32–35]. They have major roles in plant responses to abiotic stresses, where they convert stress-induced signals to cellular responses. Drought stress up-regulated gene expression is driven by transcription factors belonging to families of drought response element binding protein/C-repeat binding factors (DREB/CBF), basic leucine zipper (bZIP), myeloblastosis oncogene (MYB), NAM, ATAF1/2 and CUC (NAC), nuclear factor Y (NF-Y), zinc finger proteins (ZFP), and proteins containing the highly conserved amino acid sequence WRKYGQK (WRKY) [36–41] via ABA-dependent or ABA-independent pathways. Functional analysis of stress-inducible transcription factors should provide more information on the complex regulatory gene networks and their involvement in abiotic stresses. In soy, overexpression studies of DREB1-type transcription factors showed that they induce a number of target genes belonging to dehydrins/LEA families, chaperones, and enzymes involved in detoxification and synthesis of secondary metabolites [42]. Several WRKY transcription factors have been implicated as regulators of stress responses and senescence in different plants [43]. Overexpression of GmWRKY27 reduces ROS levels and improves salt and drought tolerance in transgenic soy plants [44]. In transgenic rice plants, overexpression of OsWRKY89 leads to growth retardation, increased wax deposition on leaf surfaces, and ultraviolet B tolerance [45]. Another rice transcription factor, OsMYB2, confers tolerance to multiple stresses such as salinity, cold and drought by stimulating the accumulation of soluble sugars and proline [46], while overexpression of Arabidopsis MYB96 enhances drought tolerance via cuticular wax accumulation [47]. Similarly, the rice SERF1 transcription factor has been demonstrated to regulate the expression of different genes associated with salt tolerance including three Cys2/His2-type (C2H2) zinc finger proteins (ZFP179, ZFP182, ZFP252) (Schmidt et al., 2013). Overexpression of these C2H2 ZFP transcription factors in rice was shown to increase tolerance to salt and/or drought [48, 49]. Additional studies suggest that C2H2 ZFP transcription factors are involved in responses and tolerance to drought, cold, salt, high light and oxidative stresses in Arabidopsis thaliana [50–54] and rice [48, 55–57]. Genetic analysis revealed that ZAT10 and ZAT12, two widely studied members of the C2H2 ZFPs family in Arabidopsis, are required for the expression of genes encoding ROS-scavenging enzymes [52, 58, 59]. These results suggest that C2H2 ZFPs could play important roles in regulation of ROS signaling under abiotic stress. In wheat, at least 53 members of a C2H2 TaZFP subfamily (C12i) have been identified [60]. The latter study revealed that 37 TaZFP members are up-regulated by drought stress and by at least one other abiotic stress. However, the mechanisms by which this TaZFP subfamily coordinates stress responses in wheat is poorly understood. Among these 37 members, TaZFP1B (TaZFP1 from the B genome) showed strong expression under all stresses studied (high light, flooding, drought, H2O2), and was previously associated with Al tolerance [61]. This indicates that this gene could govern expression of stress-inducible genes and may play a significant role in various abiotic stresses with an oxidative stress component in wheat. Another wheat ZFP gene named TaZFP1 was recently shown to improve salt stress tolerance in tobacco [62]. However, this TaZFP1 has 8 C2H2 domains (compared to two C2H2 domains in TaZFP1B) and has no significant homology with TaZFP1B or any other member of the TaZFP subfamily that we previously identified [60].
Here, we focused on the functional characterization of the C2H2 zinc finger transcription factor TaZFP1B in response to drought stress in wheat using a novel four-component BSMV overexpression system and the well-characterized three-component BSMV system for gene down-regulation [63]. Our results show that TaZFP1B improves tolerance to drought stress by stimulating scavenging ROS systems and by up-regulating numerous genes which were shown to improve drought and ROS tolerance in transgenic studies using different plant species [64]. Evidence show that TaZFP1B is a key regulator of drought tolerance in wheat.
Results
TaZFP1B expression levels are positively associated with increased drought stress tolerance
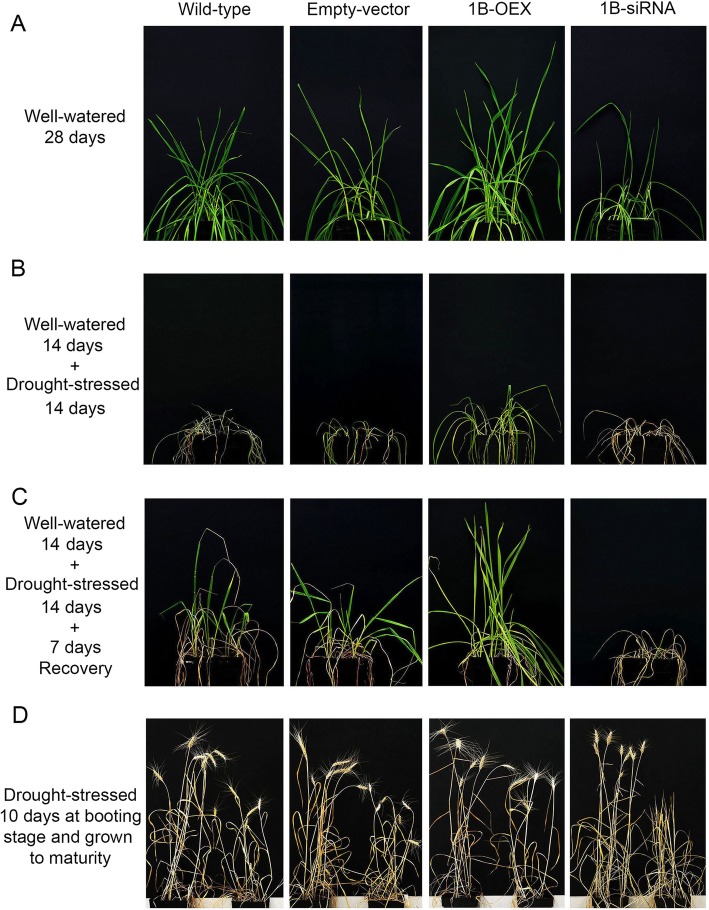
To investigate the effects of TaZFP1B on drought stress tolerance, the three or four-component system of the barley stripe mosaic virus was used to generate wheat plants with lower or higher TaZFP1B expression levels, respectively (Fig. 1a and b). These transformants were named empty vector (no cDNA insert), 1B-OEX (TaZFP1B overexpression) and 1B-siRNA (TaZFP1B silencing). To confirm that the new BSMV virus-mediated overexpression (VOX) system can be used to modify TaZFP1B expression, this transcript was analyzed by qRT-PCR (Fig. 1c). As expected, the TaZFP1B transcript level in 1B-OEX plants was about 12-fold higher than the level observed in wild-type plants under well-watered conditions. Also as expected, TaZFP1B expression in 1B-siRNA plants is low in well-watered plants, confirming an efficient targeting of the TaZFP1B transcripts by the siRNA. The siRNA specificity was verified by analyzing the expression profiles of the closest TaZFP relatives (TaZFP1A, 1D, 2B, 2D, 3B and 3D) (Additional file 1: Fig. S1). This analysis revealed that the 1B-siRNA also affected the expression of the homoeologous copy TaZFP1A (from the A genome) but did not target TaZFP2 or TaZFP3 transcripts [60]. Note that TaZFP2A and TaZFP3A have not been identified in wheat. Following a 7-day drought treatment, the TaZFP1B transcript level in wild-type plants was up-regulated by about 5-fold compared to well-watered plants. A similar result was observed in drought-stressed empty vector plants. On the other hand, the drought-induced up-regulation of TaZFP1B in 1B-OEX plants was about 6-fold higher than the up-regulation observed after drought stress in wild-type plants. We then sought to determine if an increased expression in TaZFP1B causes phenotypic changes (Fig. 2). Interestingly, overexpression of TaZFP1B improved plant growth under well-watered conditions (Fig. 2a). In 1B-OEX plants, the growth rate was more vigorous compared to the three other plants (wild-type, empty-vector and 1B-siRNA). On the other hand, silencing of TaZFP1B caused a slight loss of turgor. A 14-day drought stress significantly impaired the turgor of the four plant types, but the effect was less severe in 1B-OEX plants (Fig. 2b). Upon rewatering for 7 days, 1B-OEX plants recovered faster than the other plants (Fig. 2c). The fact that there is no visible phenotypic difference between the empty vector and wild-type plants indicates that the phenotypes under well-watered and drought conditions depend on TaZFP1B expression levels and not on the viral components used for infection. To verify the effect of drought on seed yield, water was withheld for 10 days after plants reached the booting stage (Fig. 2d). Our results show that drought stress significantly reduced the spike length, the total weight of grains per spike in all plants, the number of grains per spike and the average grain weight (weight of 10 grains). Interestingly, the total weight of grains per spike was higher in 1B-OEX plants while it was lower in 1B-siRNA plants compared to wild-type and empty vector plants (Table 1). Again, TaZFP1 expression levels likely explain the results obtained since the data are similar between empty-vector and wild-type plants, which show similar levels of TaZFP1B expression.
Fig. 1.
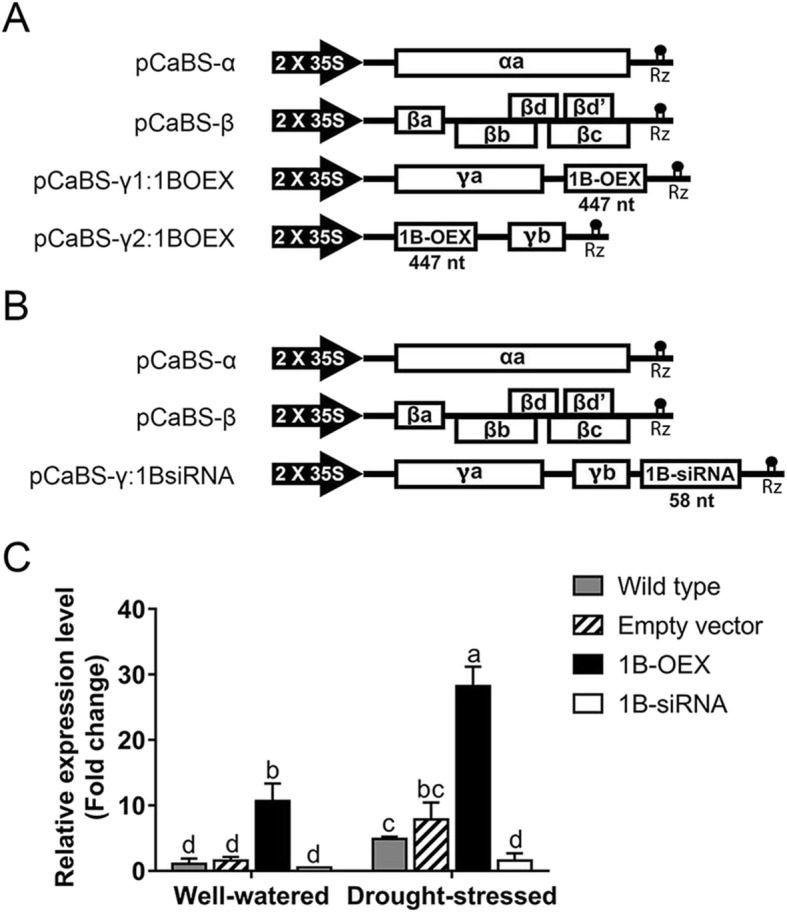
Schematic representations of the vectors used to modify TaZFP1B expression in wheat. a Vectors of the four-component BSMV system used for overexpression. b Vectors of the three-component BSMV system used for silencing. c Quantification of TaZFP1B transcripts in wheat. Wild-type, uninfected Atlas 66 plants; Empty-vector, Atlas 66 plants infected with the four basic (“empty”) plasmids: pCaBS-α, pCaBS-β, pCaBS-γ1:00 and pCaBS-γ2:00; 1B-OEX, Atlas 66 plants infected with the four plasmids described in (a); 1B-siRNA, Atlas 66 plants infected with the three plasmids described in (b). Values are means ±SD of four biological replicates. Different letters indicate statistically significant differences between samples (P < 0.05 by Tukey’s test)
Fig. 2.
TaZFP1B enhances the tolerance of wheat plants to drought stress. The different types of wheat plants (see Fig. 1) were grown for 14 days then were either well-watered for an additional 7 days (a) or drought-stressed by withholding water for 14 days (b). For recovery, control plants and drought-stressed plants were watered every day for an additional 7 days (total of 35 days of growth) (c). Photograph of wild-type, empty vector, 1B-OEX and 1B-siRNA plants (cv. Dakosta) subjected to drought stress for 10 days at the booting stage and then grown to maturity (d)
Table 1.
TaZFP1B transcript levels affect the wheat inflorescence parameters. Plants (cv. Dakosta) were grown and treated as described in Fig. 2
| Wild-type | Empty-vector | 1B-OEX | 1B-siRNA | |||||
|---|---|---|---|---|---|---|---|---|
| Well-watered | Drought | Well-watered | Drought | Well-watered | Drought | Well-watered | Drought | |
| Spike length (cm) | 7.10 ± 1.00a | 4.35 ± 0.58c | 6.64 ± 0.63a | 4.40 ± 0.65c | 6.89 ± 0.74a | 5.54 ± 0.58b | 5.57 ± 0.44b | 3.35 ± 0.48d |
|
Total weight of grains per spike (g) |
0.53 ± 0.17 b | 0.19 ± 0.04 d | 0.61 ± 0.11ab | 0.17 ± 0.01 d | 0.71 ± 0.08a | 0.30 ± 0.07c | 0.41 ± 0.10c | 0.08 ± 0.03 e |
| Number of grains per spike | 17.50 ± 5.68ab | 9.90 ± 2.23c | 19.17 ± 1.11ab | 9.29 ± 1.11c | 21.50 ± 4.50a | 12.36 ± 3.11bc | 16.14 ± 4.22b | 7.14 ± 2.12d |
| Weight of 10 grains (g) | 0.30 ± 0.03a | 0.19 ± 0.02c | 0.32 ± 0.04a | 0.18 ± 0.02c | 0.33 ± 0.02a | 0.24 ± 0.02b | 0.26 ± 0.03b | 0.11 ± 0.01d |
Values represent mean ± SD (N = 8). Different letters indicate significant differences between groups (P < 0.05)
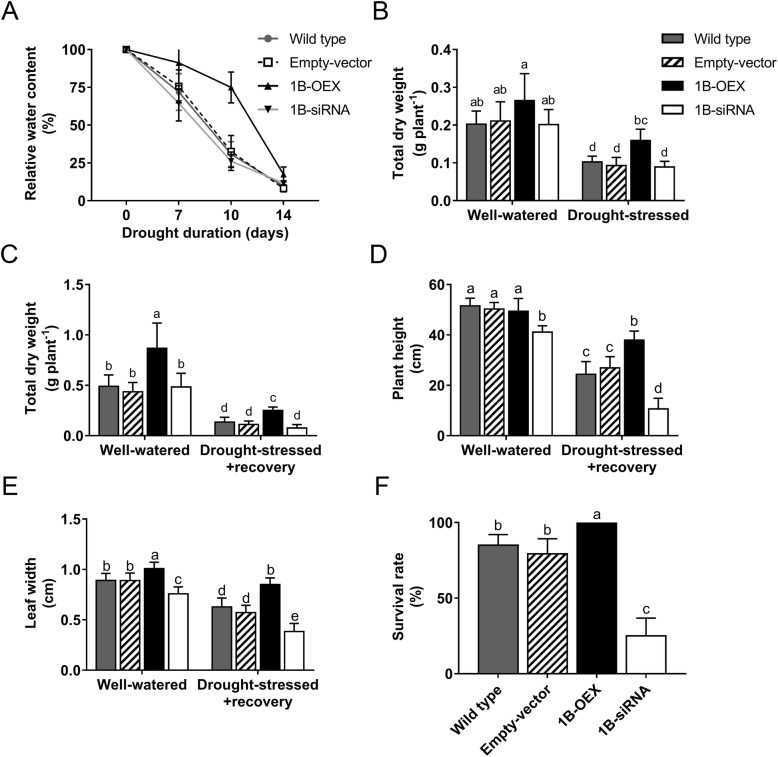
The effect of drought stress on other growth parameters was investigated. The relative water content (RWC) is an important indicator of water status. Plants were drought-stressed for 14 days and the RWC was determined at different time points. Results in Fig. 3a show that the RWC decreases quickly in the wild-type, empty vector and 1B-siRNA plants compared to 1B-OEX plants. A drought stress of 10 days resulted in a significantly higher RWC in 1B-OEX plants compared to the other plants. Results in Fig. 3b show that dry weight accumulation was not significantly different between the four types of plants under well-watered conditions. However, exposure of plants to drought stress for 14 days resulted in a reduction in dry weight compared to the well-watered plants, albeit this reduction was significantly less in 1B-OEX plants. After 35 days of growth under well-watered conditions, the dry weight was higher 1B-OEX plants than in the other plants, and this was also the case after 14 days of drought stress and 7 days of recovery (Fig. 3c). Despite the superior performance of 1B-OEX plants, there was a significant reduction in dry weight in drought-stressed + recovery plants compared to 1B-OEX well-watered plants that continue to grow under optimal conditions (Fig. 3c). There was a significant reduction in plant height in all plants after drought stress and recovery compared to well-watered plants. However, the plant height of the 1B-siRNA plants was lower compared to the three other types of plants under well-watered conditions or after drought-stress and recovery (Fig. 3d). After drought stress and recovery, 1B-OEX plants were the tallest, while the 1B-siRNA plants were the shortest. Under well-watered conditions, the 1B-OEX plants also developed wider leaves while the opposite effect occurred in the 1B-siRNA plants (Fig. 3e). After drought stress and recovery, the leaf width was reduced in all plants compared to well-watered plants. However, leaves of the 1B-OEX plants were the widest while they were the narrowest in the 1B-siRNA plants. After drought stress and recovery, all the 1B-OEX plants survived while the survival rate for the 1B-siRNA was only at 25% compared to 80–85% in wild-type and empty vector plants (Fig. 3f). Moreover, chlorophyll fluorescence imaging showed that a 10-day drought stress decreases chlorophyll content in the leaves of wild-type and empty vector plants, and even more sharply in 1B-siRNA plants (Additional file 2: Fig. S2). In contrast, chlorophyll fluorescence remains high in 1B-OEX plants even after 14 days of drought. These results suggest that TaZFP1B participates in chlorophyll stability under stress.
Fig. 3.
TaZFP1B overexpression improves the plants’ physiological parameters. Plants were grown and treated as described in Fig. 2. The following physiological parameters were measured: a relative water content; b total dry weight in well-watered and drought-stressed plants; c total dry weight in well watered plants and after drought-stress recovery; d plant height; e 2nd leaf width; and f survival rate after drought treatment and recovery. Values are means ±SD of four biological replicates. Different letters indicate statistically significant differences between samples (P < 0.05 by Tukey’s test)
TaZFP1B improves tolerance to drought-induced oxidative stress
The effect of TaZFP1B on ROS accumulation was investigated by analyzing the accumulation of 2′,7′-dichlorofluorescein (DCF) (Fig. 4a). Well-watered wild-type and empty vector plants show similar levels of DCF, while lower and higher levels of DCF were observed in 1B-OEX and 1B-siRNA plants, respectively. A 7-day drought stress led to a significant accumulation of DCF in wild-type and empty vector plants compared to well-watered plants. The DCF level was much higher in 1B-siRNA plants, while in contrast it was significantly lower in 1B-OEX plants.
Fig. 4.
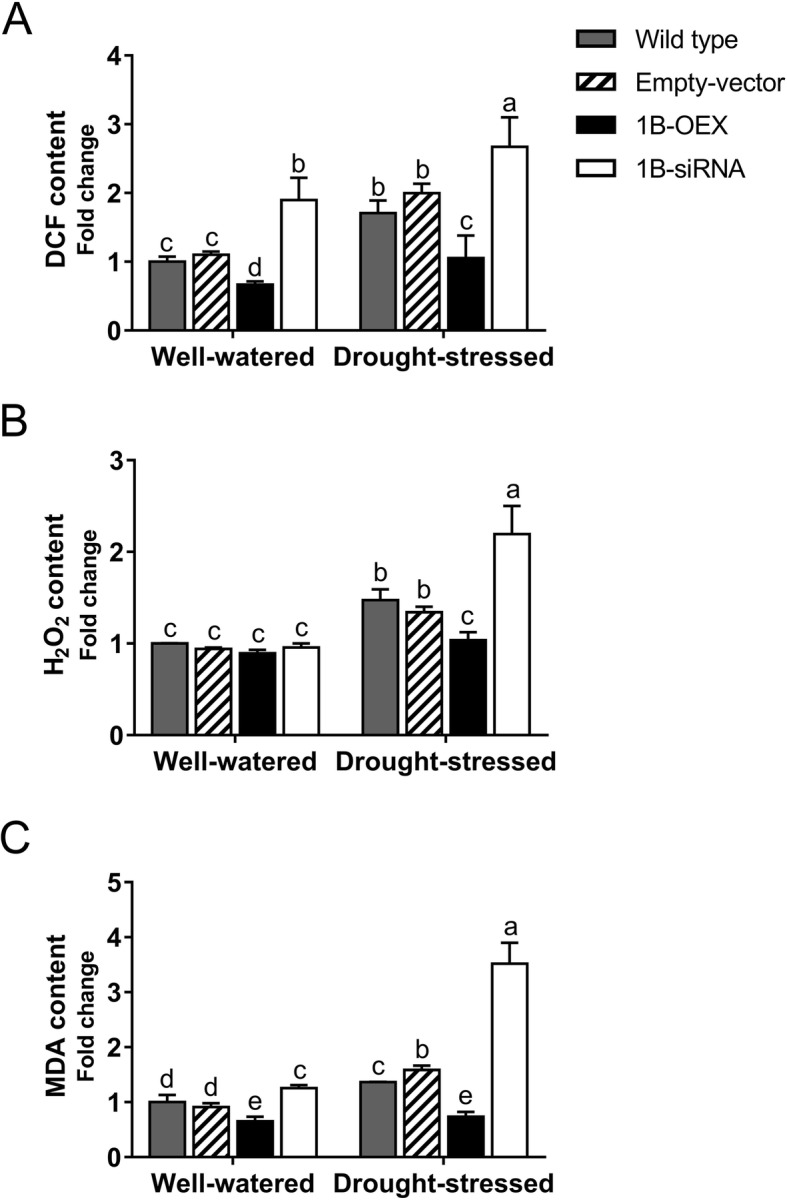
TaZFP1B reduces ROS accumulation under drought stress. Plants were grown and treated as described in Fig. 2 except that drought stress was applied for 7 days. Soluble extracts were prepared and contents in DCF (a), H2O2 (b) and malondialdehyde (c) were determined, as well as the protein content and the results are expressed as fold-change relative to well watered wild-type plants. Values are means ±SD of four biological replicates. Different letters indicate statistically significant differences between samples (P < 0.05 by Tukey’s test)
Since H2O2 is the most stable of the major ROS species produced under drought stress [65, 66], its production in leaf tissues was also examined (Fig. 4b). There are no significant changes in H2O2 content between the different well-watered plants. After 7 days of drought stress, there was a significant increase in H2O2 in wild-type and empty vector plants, and the highest level of H2O2 was observed in the 1B-siRNA plants. In contrast, there was no significant increase in H2O2 in the 1B-OEX plants.
The accumulation of various free radicals results in lipid peroxidation, which itself causes the formation of by-products such as malondialdehyde (MDA) [67]. Oxidative damage to lipids was estimated by measuring the MDA content in leaf tissues. As shown in Fig. 4c, well-watered 1B-OEX plants show reduced MDA accumulation and 1B-siRNA plants show increased accumulation. After drought stress, higher levels of MDA were observed in wild-type and empty vector plants, and even more so in the 1B-siRNA plants. In contrast, overexpression of TaZFP1B prevented drought-induced MDA accumulation.
Gene expression and activities of ROS scavenging systems
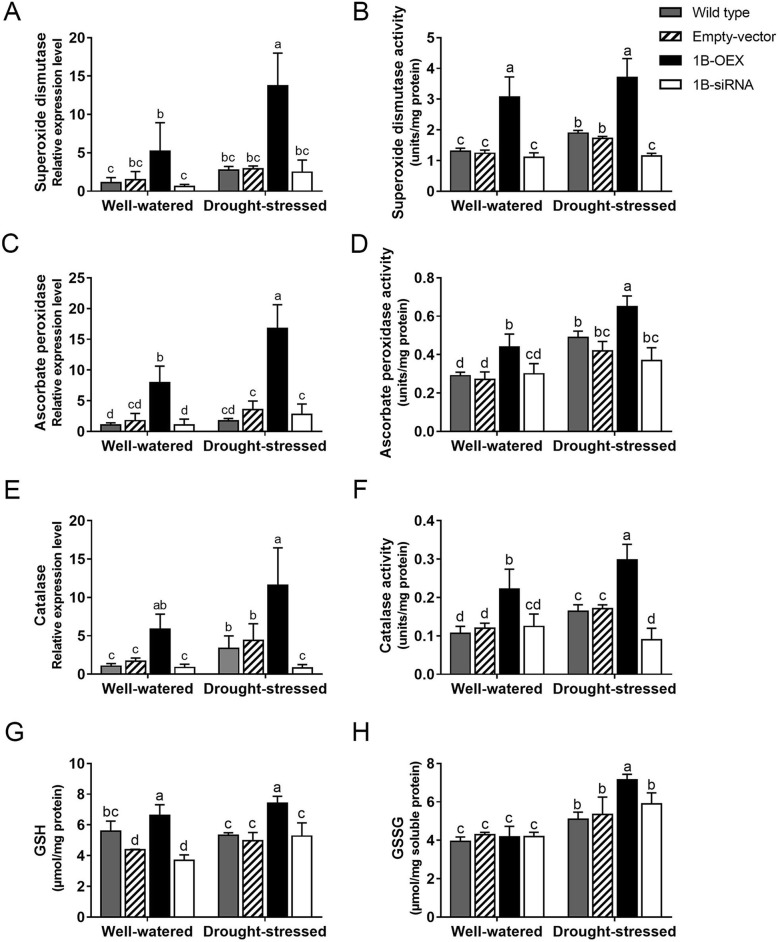
The presence of antioxidant enzymes and compounds is needed to maintain cellular ROS homeostasis under stress conditions [11]. Major ROS-scavenging enzymes of plants include superoxide dismutase (SOD), ascorbate peroxidase (APX) and catalase (CAT). To investigate whether TaZFP1B is required for ROS scavenging, genes encoding SOD (TRIAE_CS42_2BL_TGACv1_131439_AA0427700), APX (TRIAE_CS42_U_TGACv1_642188_AA2112960.5) and CAT (TRIAE_CS42_7DL_TGACv1_602975_AA1973160.1), which showed increased expression in RNA-Seq data, were selected for analysis by qRT-PCR (Fig. 5a, c and e respectively). Enzymatic activities of SOD, APX and CAT were also determined (Fig. 5b, d and f respectively). In well-watered conditions, a significant up-regulation of SOD expression and total SOD activity was observed in 1B-OEX plants while there is no significant difference in the three other types of plants (Fig. 5a and b). A 7-day drought stress up-regulated SOD RNA expression and SOD activity only slightly in wild-type and empty vector plants, but more strongly in 1B-OEX plants. SOD activity in 1B-siRNA plants was not induced by drought stress suggesting that TaZFP1B is needed for SOD up-regulation. Similar results were observed for the two other antioxidant enzymes (APX and CAT) (Fig. 5c to f).
Fig. 5.
TaZFP1B enhances the gene expressions and activities of ROS scavenging systems. Plants were grown and treated as described in Fig. 2 except that drought stress was applied for 7 days. Expression levels of SOD (a), APX (c) and CAT (e) were determined by qPCR and and the results are expressed as fold-change relative to well watered wild-type plants. The enzyme activity corresponding to these genes was also assayed (b, d and f). The contents in reduced and oxidized glutathione (GSH and GSSG) were also determined (g and h). Values are means ±SD of four biological replicates. Different letters indicate statistically significant differences between samples (P < 0.05 by Tukey’s test)
Non-enzymatic antioxidants such as reduced glutathione (GSH) have been reported to play a significant role in the management of oxidative stress [68]. We assayed the total glutathione content by measuring reduced (GSH) and oxidized (GSSG) glutathione in leaf tissues. As shown in Fig. 5g, a higher amount of GSH was observed in 1B-OEX plants compared to the wild-type, empty vector and 1B-siRNA plants under well-watered conditions. After 7 days of drought stress, similar amounts of GSH were observed in wild-type, empty vector and 1B-siRNA plants, while 1B-OEX plants showed a higher amount of GSH. Under control conditions, the four plant types had similar GSSG levels. Drought stress caused a significant accumulation of GSSG in the four plants and this accumulation was highest in 1B-OEX plants (Fig. 5h). These results suggest that the antioxidant capacity is increased by TaZFP1B.
Transcriptome modifications under well-watered conditions
To better understand the function of TaZFP1B at the molecular level, eight mRNA-Seq libraries were sequenced to analyze the transcript profiles. To determine how the TaZFP1B transcript level affects gene expression, the mRNA profiles of 1B-OEX and 1B-siRNA plants were compared to that of wild-type plants, under well-watered or drought conditions. The global expression pattern was visualized by generating a heat map of the differentially regulated transcripts between the different types of plants (Fig. 6). We found that 27 transcripts were up-regulated by at least 5-fold in 1B-OEX well-watered plants (Table 2). These up-regulated genes encode proteins involved in transcription, calcium binding, stress response, oxidation-reduction, cell wall and membrane structure, transport, cell cycle and carbohydrate metabolism. Some of these proteins have been associated with abiotic stress responses.
Fig. 6.
Heat map showing the differential gene expression. Plants were grown and treated as described in Fig. 2 except that drought stress was applied for 7 days, then RNA-Seq libraries were prepared and sequenced. The hierarchical clustering was generated using Spearman correlation coefficients of log2-transformed TPM expression values. The color scale indicates the expression levels (red, low expression; green, high expression). The 187 genes up-regulated at least five-fold in 1B-OEX plants under drought stress are listed in Table 4. The 96 genes down-regulated at least two-fold by TaZFP1B overexpression under drought stress are listed in Table 5
Table 2.
Genes up-regulated at least five-fold by TaZFP1B overexpression under well-watered conditions
| Gene number # | Gene | Annotation | Fold change |
|---|---|---|---|
| Transcription factor | |||
| 1 | TRIAE_CS42_3DL_TGACv1_249236_AA0842580.1 | Zinc finger homeodomain protein 5 | 55,9 |
| 2 | TRIAE_CS42_2AS_TGACv1_112557_AA0340950.6 | Eukaryotic translation initiation factor | 53,9 |
| 3 | TRIAE_CS42_3AL_TGACv1_196955_AA0663930.1 | Transcription factor PCF7 | 5,3 |
| Calcium binding proteins | |||
| 4 | TRIAE_CS42_5BL_TGACv1_405470_AA1328020.1 | Calreticulin | 5,3 |
| Stress-related proteins | |||
| 5 | TRIAE_CS42_5AL_TGACv1_376571_AA1238590.1 | DEAD-box ATP-dependent RNA helicase | > 100 |
| 6 | TRIAE_CS42_4AL_TGACv1_293117_AA1000210.1 | Aspartic protease | 11,7 |
| 7 | TRIAE_CS42_6AL_TGACv1_474009_AA1533790.1 | Lysine-specific demethylase JMJ706-like | 9,4 |
| 8 | TRIAE_CS42_7AS_TGACv1_569033_AA1805230.1 | ACT domain repeat protein | 7,3 |
| 9 | TRIAE_CS42_3B_TGACv1_222967_AA0774260.2 | Chitinase 10 | 6,6 |
| 10 | TRIAE_CS42_3DS_TGACv1_272269_AA0918140.2 | Disease resistance protein RPM1 | 5,2 |
| 11 | TRIAE_CS42_4AS_TGACv1_307024_AA1016270.3 | BTB/POZ domain-containing protein NPY4 | 5,1 |
| 12 | TRIAE_CS42_5AL_TGACv1_376986_AA1242950.1 | Glutathione S-transferase | 5,0 |
| Oxidation-reduction process | |||
| 13 | TRIAE_CS42_3AL_TGACv1_194817_AA0640010.2 | Aldo-keto reductase | 39,0 |
| 14 | TRIAE_CS42_5AL_TGACv1_376972_AA1242830.1 | Alcohol dehydrogenase ADH2H | 10,8 |
| 15 | TRIAE_CS42_2BL_TGACv1_129634_AA0391150.1 | DMR6-like oxygenase | 7,1 |
| Cell wall and membrane structure | |||
| 16 | TRIAE_CS42_7BS_TGACv1_592358_AA1936370.2 | Profilin actin binding protein | 38,5 |
| 17 | TRIAE_CS42_U_TGACv1_694116_AA2161830.1 | Xyloglucan endotransglucosylase/hydrolase | 6,5 |
| Transporters | |||
| 18 | TRIAE_CS42_6BL_TGACv1_500270_AA1602490.3 | TIC 20 protein | 6,8 |
| 19 | TRIAE_CS42_7DS_TGACv1_621736_AA2024850.1 | WAT1-related protein | 6,2 |
| 20 | TRIAE_CS42_3AL_TGACv1_195744_AA0653320.1 | Bidirectional sugar transporter SWEET | 5,9 |
| Cell cycle | |||
| 21 | TRIAE_CS42_4DS_TGACv1_362060_AA1176230.3 | Structural maintenance of chromosomes (SMC) protein | > 100 |
| 22 | TRIAE_CS42_2BL_TGACv1_130589_AA0414320.3 | Protein laz1 | > 100 |
| Carbohydrate metabolism-related proteins | |||
| 23 | TRIAE_CS42_7BL_TGACv1_578713_AA1899080.1 | Glucan endo-1,3-beta-glucosidase | > 100 |
| Others | |||
| 24 | TRIAE_CS42_U_TGACv1_641461_AA2095670.2 | Glutamate receptor interacting protein | > 100 |
| 25 | TRIAE_CS42_5BL_TGACv1_406408_AA1345220.1 | Ankyrin repeat containing protein | > 100 |
| 26 | TRIAE_CS42_6DL_TGACv1_526505_AA1685690.2 | F-box domain, cyclin-like domain containing protein | 31,0 |
| 27 | TRIAE_CS42_5DL_TGACv1_434415_AA1435460.1 | TolB-like domain containing protein | 7,5 |
Fold change, 1B-OEX to wild-type ratio. The gene numbers (#) are used in the text
Although a 5-fold induction in gene expression provides confidence that the genes are regulated by TaZFP1B, genes up-regulated between one and five-fold in 1B-OEX plants may nevertheless play a significant role in stress tolerance. For example, genes encoding enzymes known for their role in ROS scavenging (Additional file 3: Table S1) are of particular interest: SOD increases 2.5-fold, APX increases 2.1-fold and CAT increases 1.8-fold. Furthermore, overexpression of TaZFP1B also down-regulates 11 transcripts by at least 2-fold (Table 3).
Table 3.
Genes down-regulated at least two-fold by TaZFP1B overexpression under well-watered conditions
| Gene number # | Gene | Annotation | Fold change |
|---|---|---|---|
| Transcription factor | |||
| 28 | TRIAE_CS42_1AL_TGACv1_001758_AA0034810.2 | G-box binding factor | 2,8 |
| Transporter | |||
| 29 | TRIAE_CS42_3B_TGACv1_223624_AA0784820.1 | Protein DETOXIFICATION | 10,7 |
| 30 | TRIAE_CS42_U_TGACv1_642488_AA2118360.7 | Copper transporter CT1 | 10,4 |
| 31 | TRIAE_CS42_5BL_TGACv1_404850_AA1312820.1 | ZINC INDUCED FACILITATOR | 2,8 |
| Carbohydrate metabolism-related proteins | |||
| 32 | TRIAE_CS42_1DS_TGACv1_081598_AA0261790.1 | Malonyl-coenzyme A:anthocyanin 3-O-glucoside-6″-O-malonyltransferase-like | 47,9 |
| Others | |||
| 33 | TRIAE_CS42_3B_TGACv1_226496_AA0817320.2 | Unknown protein | 4,5 |
| 34 | TRIAE_CS42_7BS_TGACv1_593046_AA1947680.1 | Unknown protein | 4,3 |
| 35 | TRIAE_CS42_3B_TGACv1_221867_AA0752050.2 | Bark storage protein A-like | 2,9 |
| 36 | TRIAE_CS42_7DS_TGACv1_622168_AA2034370.3 | Protein REVEILLE | 2,4 |
| 37 | TRIAE_CS42_6BL_TGACv1_501185_AA1614740.1 | Unknown protein | 2,3 |
| 38 | TRIAE_CS42_3B_TGACv1_221388_AA0739090.1 | Unknown protein | 2,0 |
Fold change, wild-type to 1B-OEX ratio. The gene numbers (#) are used in the text
Transcriptome modifications under drought stress
To identify molecular pathways by which TaZFP1B confers drought tolerance in wheat, transcript levels were compared between 1B-OEX and wild-type plants under drought stress. Our analyses revealed that overexpression of TaZFP1B modifies the expression of many new genes during drought stress compared to the other three types of plants (Fig. 6). We found 187 transcripts that were up-regulated by at least 5-fold (Table 4) in drought-treated 1B-OEX plants compared to wild-type plants. Among these are genes encoding proteins involved in transcription, signal transduction, stress responses, oxidation-reduction processes, cell wall and membrane structure, cell cycle, transport, protein post-translational modifications, carbohydrate, fatty acid and nitrogen metabolisms, and other metabolisms. Of interest are the data on genes associated with ROS. Genes encoding enzymes that stimulate ROS production, for example NADPH oxidase (#143) and galactose oxidase (#149), are up-regulated concomitantly with genes encoding ROS-scavenging enzymes such as peroxidases (#137, #141, #142) and thioredoxin (#153). A host of genes known for their role in stress response and tolerance are also up-regulated (Table 4): genes encoding dehydrins (#91, #95, #100, #114), cold-responsive proteins COR14a (#69), glucan endo-1,3-beta-glucosidase (#182), repeat domain proteins (#92, #102, #125–127), NADP-dependent malic enzyme (#183), E3 ubiquitin-protein ligase (#178, #179), glycine or hydroxyproline-rich proteins (#113, #159) and genes involved in programmed cell death (#148, #160, #164, #165). Moreover, TaZFP1B up-regulates several genes encoding enzymes involved in cell wall modifications such as xyloglucan endotransglucosylase/hydrolase (#154) and pectinesterase (#155), and genes encoding other proteins associated with cell wall remodeling such as expansin (#156), remorin (#157) and WAX2 (#158). Several genes involved in carbohydrate and fatty acid metabolisms are also up-regulated (#182–195). On the other hand, 96 transcripts are down-regulated by at least 2-fold in drought-treated 1B-OEX plants compared to wild-type plants (Table 5). Many of these are genes involved in photosynthesis metabolism (#244–264). Together, these observations emphasize the role of TaZFP1B in transcriptional regulation under drought stress and suggest that TaZFP1B is a key regulator of stress-related genes which are important for drought stress and oxidative stress tolerance.
Table 4.
Genes up-regulated at least five-fold by TaZFP1B overexpression under drought stress
| Gene number # | Gene | Annotation | Fold change |
|---|---|---|---|
| Transcription-related proteins | |||
| 39 | TRIAE_CS42_3B_TGACv1_223209_AA0778000.3 | LSD1 zinc finger | > 100 |
| 40 | TRIAE_CS42_5DL_TGACv1_434621_AA1438900.6 | Scarecrow-like protein | > 100 |
| 41 | TRIAE_CS42_7BS_TGACv1_592226_AA1933690.4 | Transcriptional corepressor LEUNIG | > 100 |
| 42 | TRIAE_CS42_3AS_TGACv1_212728_AA0702890.1 | Transcriptional regulator RABBIT EARS | 42,2 |
| 43 | TRIAE_CS42_3AL_TGACv1_195838_AA0654650.3 | Double-stranded RNA-binding protein 1 | 28,9 |
| 44 | TRIAE_CS42_4DL_TGACv1_343075_AA1129010.3 | Trihelix transcription factor GT-2 | 24,1 |
| 45 | TRIAE_CS42_3AL_TGACv1_196955_AA0663930.1 | Transcription factor PCF7 | 22,5 |
| 46 | TRIAE_CS42_1DL_TGACv1_061611_AA0199920.1 | DnaJ homolog subfamily B | 20,5 |
| 47 | TRIAE_CS42_6BS_TGACv1_514227_AA1657240.1 | B-box zinc finger protein | 19,7 |
| 48 | TRIAE_CS42_2DL_TGACv1_159186_AA0534130.1 | WRKY transcription factor 12 | 18,4 |
| 49 | TRIAE_CS42_5BL_TGACv1_406061_AA1340040.1 | CBFIVd-B4 | 18,4 |
| 50 | TRIAE_CS42_2DL_TGACv1_159186_AA0534130.1 | WRKY transcription factor | 18,4 |
| 51 | TRIAE_CS42_2AL_TGACv1_095147_AA0307480.1 | Lateral organ boundaries transcription factor | 16,4 |
| 52 | TRIAE_CS42_5DL_TGACv1_435031_AA1445050.1 | Ocs element-binding factor | 14.2 |
| 53 | TRIAE_CS42_2BS_TGACv1_147417_AA0482900.1 | Zinc finger protein CONSTANS 15 | 15,8 |
| 54 | TRIAE_CS42_1DL_TGACv1_062044_AA0207980.1 | Transcription factor bHLH 112 | 15,1 |
| 55 | TRIAE_CS42_7AL_TGACv1_558337_AA1792520.4 | Zinc finger CCCH domain-containing protein | 14,2 |
| 56 | TRIAE_CS42_4DS_TGACv1_361293_AA1165150.6 | BTB/POZ domain-containing protein | 12,6 |
| 57 | TRIAE_CS42_1DL_TGACv1_064769_AA0235650.2 | DnaJ homolog subfamily C member | 11,9 |
| 58 | TRIAE_CS42_4AS_TGACv1_306719_AA1012420.1 | Homeobox-leucine zipper protein HOX12 | 11,9 |
| 59 | TRIAE_CS42_2AS_TGACv1_113089_AA0351070.5 | Heterogeneous nuclear ribonucleoprotein 1 | 10,7 |
| 60 | TRIAE_CS42_3DS_TGACv1_271642_AA0904520.1 | Ethylene-responsive transcription factor CRF1 | 10,5 |
| 61 | TRIAE_CS42_7DS_TGACv1_624521_AA2061810.2 | Transcription factor bHLH 78 | 9,7 |
| 62 | TRIAE_CS42_1DS_TGACv1_080414_AA0247520.3 | DnaJ homolog subfamily B member | 9,4 |
| 63 | TRIAE_CS42_5DS_TGACv1_457055_AA1481570.1 | Zinc finger protein C2H2 type | 9,3 |
| 64 | TRIAE_CS42_5BS_TGACv1_424687_AA1391330.5 | Transcription factor bZIP | 8,2 |
| 65 | TRIAE_CS42_5BL_TGACv1_406061_AA1340050.1 | Dehydration-responsive element-binding protein | 7,4 |
| 66 | TRIAE_CS42_1DS_TGACv1_080938_AA0255890.1 | C3HC4 type zinc-finger (RING finger) | 6,9 |
| 67 | TRIAE_CS42_3B_TGACv1_224651_AA0799380.1 | Lateral organ boundaries transcription factor | 6,9 |
| 68 | TRIAE_CS42_5DL_TGACv1_437778_AA1467310.5 | Two-component response regulator | 6,7 |
| 69 | TRIAE_CS42_2DL_TGACv1_162669_AA0563320.1 | Cold-responsive protein COR14a | 6,2 |
| 70 | TRIAE_CS42_5BS_TGACv1_424687_AA1391330.3 | Transcription factor bZIP | 5,9 |
| 71 | TRIAE_CS42_4DL_TGACv1_345275_AA1152550.1 | Homeobox-leucine zipper protein HOX13 | 5,7 |
| 72 | TRIAE_CS42_7BS_TGACv1_592977_AA1946950.1 | Transcription factor bHLH HEC2 | 5,6 |
| 73 | TRIAE_CS42_7AS_TGACv1_569468_AA1816690.1 | C2-C2 zinc finger | 5,6 |
| 74 | TRIAE_CS42_7BL_TGACv1_577432_AA1875300.1 | VQ domain containing protein | 5,5 |
| 75 | TRIAE_CS42_5AL_TGACv1_375766_AA1226930.1 | CBF IVd-A22 | 5,2 |
| 76 | TRIAE_CS42_6DS_TGACv1_544607_AA1748880.1 | B-box zinc finger protein | 5,2 |
| 77 | TRIAE_CS42_3AL_TGACv1_195166_AA0645630.3 | Transcription factor bHLH87 | 5,1 |
| Calcium binding protein, kinase or phosphatase | |||
| 78 | TRIAE_CS42_7BS_TGACv1_594052_AA1955780.1 | Phytosulfokine receptor 1 | 84,1 |
| 79 | TRIAE_CS42_5DL_TGACv1_434773_AA1441380.1 | LRR receptor-like serine/threonine-protein kinase | 35,9 |
| 80 | TRIAE_CS42_4DS_TGACv1_362249_AA1178220.1 | Calmodulin CML2 | 14,8 |
| 81 | TRIAE_CS42_7BL_TGACv1_577549_AA1878500.1 | Serine/threonine protein kinase | 14,0 |
| 82 | TRIAE_CS42_2DS_TGACv1_177289_AA0572410.1 | EF-Hand type domain containing protein | 9,2 |
| 83 | TRIAE_CS42_6DL_TGACv1_527174_AA1700020.3 | Mitogen-activated protein kinase kinase kinase | 7,6 |
| 84 | TRIAE_CS42_7AL_TGACv1_559296_AA1799010.1 | Receptor-like protein kinase | 6,9 |
| Stress-related proteins | |||
| 85 | TRIAE_CS42_4AS_TGACv1_307024_AA1016270.3 | BTB/POZ domain-containing protein NPY4 | > 100 |
| 86 | TRIAE_CS42_2AL_TGACv1_093753_AA0286070.3 | Protein DJ-1 homolog D | > 100 |
| 87 | TRIAE_CS42_U_TGACv1_641289_AA2090920.5 | Topless-related protein 1 | > 100 |
| 88 | TRIAE_CS42_3DS_TGACv1_272269_AA0918140.2 | Disease resistance protein RPM1 | > 100 |
| 89 | TRIAE_CS42_4AS_TGACv1_308773_AA1029400.2 | Pathogenesis-related protein | > 100 |
| 90 | TRIAE_CS42_7AS_TGACv1_569748_AA1823280.2 | Cysteine synthase | 86,2 |
| 91 | TRIAE_CS42_5DL_TGACv1_433513_AA1415280.1 | Dehydrin DHN2 | 73,3 |
| 92 | TRIAE_CS42_4BS_TGACv1_328898_AA1095240.1 | Pentatricopeptide repeat containing protein | 67,2 |
| 93 | TRIAE_CS42_2DL_TGACv1_163849_AA0564640.1 | Wound induced protein | 57,4 |
| 94 | TRIAE_CS42_2AS_TGACv1_115694_AA0372890.2 | Cytochrome P450 family protein | 50,4 |
| 95 | TRIAE_CS42_5BL_TGACv1_406032_AA1339460.1 | Dehydrin DHN2 | 50,4 |
| 96 | TRIAE_CS42_2AL_TGACv1_095873_AA0315400.1 | Mediator of ABA-regulated dormancy 1 | 48,4 |
| 97 | TRIAE_CS42_2DL_TGACv1_160571_AA0551860.1 | Mediator of ABA-regulated dormancy 1 | 31,6 |
| 98 | TRIAE_CS42_7BS_TGACv1_592018_AA1928290.1 | Glutathione S-transferase | 27,3 |
| 99 | TRIAE_CS42_2DL_TGACv1_160490_AA0551070.1 | Wound-responsive family protein | 26,3 |
| 100 | TRIAE_CS42_5AL_TGACv1_376309_AA1235150.1 | Dehydrin DHN2 | 22,9 |
| 101 | TRIAE_CS42_5AL_TGACv1_374112_AA1190770.1 | Aspartic protease | 22,7 |
| 102 | TRIAE_CS42_4AL_TGACv1_290111_AA0981810.2 | Tetratricopeptide repeat protein | 17,4 |
| 103 | TRIAE_CS42_7DL_TGACv1_602552_AA1960450.1 | Tubby-like protein | 17,0 |
| 104 | TRIAE_CS42_6AS_TGACv1_486459_AA1561390.1 | Auxin-responsive protein SAUR | 16,9 |
| 105 | TRIAE_CS42_2DL_TGACv1_158690_AA0524470.1 | Late embryogenesis abundant protein | 14,7 |
| 106 | TRIAE_CS42_2DL_TGACv1_158690_AA0524470.1 | Late embryogenesis abundant protein | 14,6 |
| 107 | TRIAE_CS42_4DL_TGACv1_343378_AA1133790.1 | Defensin | 14,1 |
| 108 | TRIAE_CS42_5BL_TGACv1_404954_AA1316190.1 | Cinnamoyl-CoA reductase | 13,1 |
| 109 | TRIAE_CS42_7AS_TGACv1_569113_AA1807500.2 | BTB/POZ and MATH domain-containing protein | 13,1 |
| 110 | TRIAE_CS42_5DL_TGACv1_433130_AA1403020.1 | Stress responsive A/B barrel domain-containing protein | 12,9 |
| 111 | TRIAE_CS42_U_TGACv1_640759_AA2072760.14 | Cysteine proteinase superfamily protein | 12,5 |
| 112 | TRIAE_CS42_7AS_TGACv1_569033_AA1805230.1 | ACT domain repeat protein | 11,6 |
| 113 | TRIAE_CS42_4BL_TGACv1_320342_AA1036020.1 | Glycine-rich protein | 11,3 |
| 114 | TRIAE_CS42_5AL_TGACv1_376309_AA1235140.1 | Dehydrin DHN1 | 11,2 |
| 115 | TRIAE_CS42_4AL_TGACv1_293117_AA1000210.1 | Aspartic protease | 10,9 |
| 116 | TRIAE_CS42_6BS_TGACv1_513372_AA1639240.1 | F-box/kelch-repeat protein SKIP4 | 10,5 |
| 117 | TRIAE_CS42_1BL_TGACv1_030562_AA0094080.1 | Cytochrome P450 85A1 | 10,5 |
| 118 | TRIAE_CS42_6DS_TGACv1_542552_AA1724150.1 | Auxin-responsive protein SAUR71 | 10,4 |
| 119 | TRIAE_CS42_5BL_TGACv1_406277_AA1343590.3 | Cysteine proteinase superfamily protein | 9,1 |
| 120 | TRIAE_CS42_7DS_TGACv1_621863_AA2028000.1 | Cytochrome c oxidase subunit 5B mitochondrial | 8,9 |
| 121 | TRIAE_CS42_5DL_TGACv1_434334_AA1434100.2 | F-box/kelch-repeat protein | 8,8 |
| 122 | TRIAE_CS42_5DL_TGACv1_435810_AA1455180.2 | Rhodanese-like domain-containing protein | 8,1 |
| 123 | TRIAE_CS42_5AL_TGACv1_376986_AA1242950.1 | Glutathione S-transferase | 7,8 |
| 124 | TRIAE_CS42_7BS_TGACv1_591856_AA1923720.1 | Aspartic protease | 7,5 |
| 125 | TRIAE_CS42_5DL_TGACv1_434499_AA1437000.1 | Pentatricopeptide repeat-containing protein | 7,2 |
| 126 | TRIAE_CS42_6AL_TGACv1_471352_AA1507530.3 | WD repeat-containing protein | 6,1 |
| 127 | TRIAE_CS42_6AL_TGACv1_471352_AA1507530.6 | WD repeat-containing protein | 6,1 |
| 128 | TRIAE_CS42_3B_TGACv1_223361_AA0780920.2 | Glutathione transferase GST | 5,8 |
| 129 | TRIAE_CS42_3AL_TGACv1_197123_AA0664910.3 | Universal stress protein PHOS32 | 5,7 |
| 130 | TRIAE_CS42_1BL_TGACv1_030249_AA0083800.1 | Universal stress protein PHOS32 | 5,7 |
| 131 | TRIAE_CS42_3AS_TGACv1_210945_AA0681930.1 | AAA-protein family | 5,5 |
| 132 | TRIAE_CS42_1AL_TGACv1_001038_AA0023900.1 | Universal stress protein PHOS | 5,5 |
| 133 | TRIAE_CS42_2AL_TGACv1_093314_AA0277190.2 | Heat shock protein | 5,3 |
| 134 | TRIAE_CS42_3DL_TGACv1_249203_AA0841110.1 | Abscisic stress-ripening protein | 5,3 |
| 135 | TRIAE_CS42_2AL_TGACv1_094123_AA0292860.2 | Elicitor responsive gene | 5,2 |
| 136 | TRIAE_CS42_6AS_TGACv1_487227_AA1569190.1 | Auxin-induced protein | 5,0 |
| Oxidation-reduction process | |||
| 137 | TRIAE_CS42_2DS_TGACv1_178485_AA0596270.1 | Peroxidase | 44,7 |
| 138 | TRIAE_CS42_4AL_TGACv1_290177_AA0982590.1 | Alcohol dehydrogenase ADH1A | 23,1 |
| 139 | TRIAE_CS42_7BL_TGACv1_576933_AA1860390.1 | Defensin | 19,0 |
| 140 | TRIAE_CS42_2AS_TGACv1_112531_AA0340030.1 | Flavin-containing monooxygenase | 18,9 |
| 141 | TRIAE_CS42_2DS_TGACv1_177840_AA0585640.1 | Peroxidase | 18,6 |
| 142 | TRIAE_CS42_2BS_TGACv1_146806_AA0473250.1 | Peroxidase | 15,9 |
| 143 | TRIAE_CS42_5AL_TGACv1_377290_AA1245640.4 | NADPH oxidase | 9,9 |
| 144 | TRIAE_CS42_2DL_TGACv1_160970_AA0555660.1 | Protochlorophyllide reductase A | 7,7 |
| 145 | TRIAE_CS42_2AL_TGACv1_096139_AA0317380.1 | Hyoscyamine 6-dioxygenase | 7,4 |
| 146 | TRIAE_CS42_5AL_TGACv1_376972_AA1242830.1 | Alcohol dehydrogenase ADH2H | 6,9 |
| 147 | TRIAE_CS42_2DL_TGACv1_158391_AA0517380.1 | FAD-dependent urate hydroxylase-like | 6,4 |
| 148 | TRIAE_CS42_2AL_TGACv1_096158_AA0317510.1 | Polyamine oxidase 4 | 6,0 |
| 149 | TRIAE_CS42_2AL_TGACv1_096643_AA0320430.1 | Galactose oxidase | 5,9 |
| 150 | TRIAE_CS42_7AS_TGACv1_570735_AA1840120.1 | Thiosulfate sulfurtransferase | 5,7 |
| 151 | TRIAE_CS42_1AL_TGACv1_002876_AA0045790.1 | Gibberellin 20 oxidase 2 | 5,5 |
| 152 | TRIAE_CS42_3AL_TGACv1_194843_AA0640510.2 | Selenium-binding protein | 5,4 |
| 153 | TRIAE_CS42_1BL_TGACv1_031952_AA0123300.2 | Thioredoxin protein | 5,0 |
| Cell wall and membrane structure | |||
| 154 | TRIAE_CS42_U_TGACv1_694116_AA2161830.1 | Xyloglucan endotransglucosylase/hydrolase | > 100 |
| 155 | TRIAE_CS42_1AL_TGACv1_000357_AA0009840.1 | Pectinesterase | 26,8 |
| 156 | TRIAE_CS42_5AS_TGACv1_393897_AA1277020.1 | Expansin-B3 | 19,0 |
| 157 | TRIAE_CS42_5DL_TGACv1_433202_AA1405370.1 | Remorin | 8,2 |
| 158 | TRIAE_CS42_6DL_TGACv1_527336_AA1702400.1 | Protein WAX2 | 7,3 |
| 159 | TRIAE_CS42_5AS_TGACv1_394403_AA1280080.3 | Hydroxyproline-rich glycoprotein family protein | 7,2 |
| Cell cycle | |||
| 160 | TRIAE_CS42_2BL_TGACv1_130589_AA0414320.3 | Protein laz1 | > 100 |
| 161 | TRIAE_CS42_U_TGACv1_640759_AA2072760.14 | Cysteine proteinases superfamily protein | 12,5 |
| 162 | TRIAE_CS42_5BL_TGACv1_406277_AA1343590.3 | Cysteine proteinases superfamily protein | 9,1 |
| 163 | TRIAE_CS42_3DL_TGACv1_249314_AA0845060.3 | Mitotic spindle checkpoint protein MAD1 | 7,4 |
| 164 | TRIAE_CS42_1BL_TGACv1_030567_AA0094200.2 | Protein LOL1 | 7,3 |
| 165 | TRIAE_CS42_5AL_TGACv1_375766_AA1226930.1 | Protein LOL1 | 6,6 |
| 166 | TRIAE_CS42_2DS_TGACv1_177975_AA0588450.1 | Cyclin-D2–2 | 5,1 |
| 167 | TRIAE_CS42_3DS_TGACv1_271852_AA0909480.1 | Cell division cycle-associated protein | 5,0 |
| Transporters | |||
| 168 | TRIAE_CS42_3DL_TGACv1_250015_AA0860390.2 | GABA transporter 1 | > 100 |
| 169 | TRIAE_CS42_3DL_TGACv1_249597_AA0852170.3 | Bidirectional sugar transporter SWEET | 86,3 |
| 170 | TRIAE_CS42_2AL_TGACv1_093880_AA0288670.1 | Non-specific lipid-transfer protein | 16,2 |
| 171 | TRIAE_CS42_5DL_TGACv1_432929_AA1394800.5 | Heavy metal-associated isoprenylated protein 32 | 10,4 |
| 172 | TRIAE_CS42_1DL_TGACv1_061859_AA0204490.1 | Non-specific lipid-transfer protein | 8,1 |
| 173 | TRIAE_CS42_3AL_TGACv1_195744_AA0653320.1 | Bidirectional sugar transporter SWEET | 6,5 |
| 174 | TRIAE_CS42_6DL_TGACv1_526901_AA1694680.2 | Bidirectional sugar transporter SWEET | 6,1 |
| 175 | TRIAE_CS42_1AL_TGACv1_003689_AA0051080.1 | Non-specific lipid-transfer protein | 6,0 |
| 176 | TRIAE_CS42_1DL_TGACv1_061102_AA0185280.1 | Non-specific lipid-transfer protein | 5,8 |
| 177 | TRIAE_CS42_7BS_TGACv1_593410_AA1951130.1 | Non-specific lipid-transfer protein | 5,6 |
| Post-translational protein modification | |||
| 178 | TRIAE_CS42_4DS_TGACv1_361658_AA1171150.4 | E3 ubiquitin-protein ligase ARI1 | 9,1 |
| 179 | TRIAE_CS42_3DS_TGACv1_272908_AA0926480.3 | E3 ubiquitin-protein ligase | 8,4 |
| 180 | TRIAE_CS42_1AL_TGACv1_000329_AA0009100.2 | Ubiquitin conjugation factor E4 protein | 7,9 |
| 181 | TRIAE_CS42_7BL_TGACv1_578537_AA1896750.2 | Dolichyl-diphosphooligosaccharide protein glycosyltransferase subunit DAD1 | 7,1 |
| Carbohydrate metabolism-related proteins | |||
| 182 | TRIAE_CS42_7BL_TGACv1_578713_AA1899080.1 | Glucan endo-1,3-beta-glucosidase 3 | > 100 |
| 183 | TRIAE_CS42_3AL_TGACv1_194492_AA0634060.2 | NADP-dependent malic enzyme | > 100 |
| 184 | TRIAE_CS42_3DL_TGACv1_249576_AA0851790.4 | Beta-galactosidase | 79,1 |
| 185 | TRIAE_CS42_5DL_TGACv1_436125_AA1458320.1 | Beta-glucosidase | 53,9 |
| 186 | TRIAE_CS42_2AS_TGACv1_112777_AA0345080.1 | Transketolase | 32,4 |
| 187 | TRIAE_CS42_5BL_TGACv1_405377_AA1326060.1 | Phosphoglycerate mutase-like protein | 12,8 |
| 188 | TRIAE_CS42_2BL_TGACv1_131851_AA0432880.2 | Aldose 1-epimerase | 10,2 |
| 189 | TRIAE_CS42_3B_TGACv1_222532_AA0766530.4 | ATP synthase subunit beta | 6,6 |
| 190 | TRIAE_CS42_2DL_TGACv1_158102_AA0509180.1 | Phenolic glucoside malonyltransferase | 6,2 |
| 191 | TRIAE_CS42_5AS_TGACv1_392538_AA1260340.7 | GDP-L-galactose phosphorylase | 6,1 |
| 192 | TRIAE_CS42_1DL_TGACv1_061598_AA0199620.1 | Beta-galactosidase 7 | 4,7 |
| Fatty acid metabolism-related proteins | |||
| 193 | TRIAE_CS42_3AS_TGACv1_210886_AA0680800.3 | Sphingosine-1-phosphate lyase | > 100 |
| 194 | TRIAE_CS42_1AS_TGACv1_019138_AA0061500.2 | GDSL esterase/lipase | 8,8 |
| 195 | TRIAE_CS42_1BL_TGACv1_030780_AA0100660.2 | Phospholipase D | 5,4 |
| Nitrogen metabolism-related proteins | |||
| 196 | TRIAE_CS42_4AS_TGACv1_307728_AA1023060.1 | Glutamine synthetase | 6,4 |
| Others | |||
| 197 | TRIAE_CS42_4DS_TGACv1_362426_AA1179710.6 | Cyclin-like F-box domain containing protein | > 100 |
| 198 | TRIAE_CS42_1BS_TGACv1_050119_AA0167640.2 | Unknown protein | > 100 |
| 199 | TRIAE_CS42_3AS_TGACv1_211314_AA0688450.2 | WPP domain-interacting tail-anchored protein | > 100 |
| 200 | TRIAE_CS42_5DL_TGACv1_434415_AA1435460.1 | TolB-like domain containing protein | 94,1 |
| 201 | TRIAE_CS42_6DL_TGACv1_528348_AA1713440.1 | Unknown protein | 84,4 |
| 202 | TRIAE_CS42_5BL_TGACv1_406843_AA1350500.1 | Unknown protein | 58,7 |
| 203 | TRIAE_CS42_5DL_TGACv1_433372_AA1411340.1 | Tryptophan synthase alpha chain-like | 52,7 |
| 204 | TRIAE_CS42_1AS_TGACv1_019383_AA0065910.3 | Isovaleryl-CoA dehydrogenase | 39,6 |
| 205 | TRIAE_CS42_4DL_TGACv1_343143_AA1130470.1 | Unknown protein | 27,7 |
| 206 | TRIAE_CS42_5DL_TGACv1_435042_AA1445250.2 | Obg-like ATPase 1 | 27,1 |
| 207 | TRIAE_CS42_1AS_TGACv1_019803_AA0071620.1 | Cyclin-like F-box domain containing protein | 23,5 |
| 208 | TRIAE_CS42_2BL_TGACv1_130998_AA0421050.2 | Peptidylprolyl isomerase | 17,2 |
| 209 | TRIAE_CS42_3DL_TGACv1_250372_AA0867150.1 | Unknown protein | 15,9 |
| 210 | TRIAE_CS42_3AL_TGACv1_195382_AA0648850.1 | Unknown protein | 13,9 |
| 211 | TRIAE_CS42_4BL_TGACv1_322190_AA1069510.1 | Anthocyanidin 3-O-glucosyltransferase | 12,2 |
| 212 | TRIAE_CS42_4BL_TGACv1_321444_AA1060550.2 | Elongator complex protein 6 | 10,8 |
| 213 | TRIAE_CS42_5DL_TGACv1_433709_AA1420080.1 | Nitrile-specifier protein 1 | 10,1 |
| 214 | TRIAE_CS42_5AL_TGACv1_375015_AA1213930.1 | Actin-depolymerizing factor 10 | 9,7 |
| 215 | TRIAE_CS42_7DL_TGACv1_602612_AA1962750.1 | Arogenate dehydrogenase 2 | 9,4 |
| 216 | TRIAE_CS42_3DL_TGACv1_249368_AA0846860.1 | Peptidase C1A, papain family protein | 7,5 |
| 217 | TRIAE_CS42_5AL_TGACv1_375655_AA1225200.2 | deSI-like protein | 7,3 |
| 218 | TRIAE_CS42_3B_TGACv1_224606_AA0798680.1 | Aromatic-ring hydroxylase domain containing protein | 7,1 |
| 219 | TRIAE_CS42_5BL_TGACv1_404868_AA1313220.1 | Nitrile-specifier protein 1 | 5,9 |
| 220 | TRIAE_CS42_4DL_TGACv1_343042_AA1128220.1 | Golgin subfamily A member | 5,8 |
| 221 | TRIAE_CS42_1DL_TGACv1_061281_AA0191130.2 | Plant UBX domain-containing protein 10 | 5,6 |
| 222 | TRIAE_CS42_1AL_TGACv1_000217_AA0006430.1 | Putative protein of unknown function (DUF640) | 5,4 |
| 223 | TRIAE_CS42_2AL_TGACv1_092997_AA0269140.3 | Imidazoleglycerol-phosphate dehydratase | 5,4 |
| 224 | TRIAE_CS42_5BL_TGACv1_404429_AA1299570.1 | Protein WVD2 | 5,2 |
| 225 | TRIAE_CS42_5AL_TGACv1_376738_AA1240450.1 | F-box domain, cyclin-like domain containing protein | 5,0 |
Fold change, 1B-OEX to wild-type ratio. The gene numbers (#) are used in the text
Table 5.
Genes down-regulated at least two-fold by TaZFP1B overexpression under drought stress
| Gene number # | Gene | Annotation | Fold change |
|---|---|---|---|
| Transcription factor | |||
| 226 | TRIAE_CS42_1AL_TGACv1_001758_AA0034810.2 | G-box binding factor | 5,0 |
| 227 | TRIAE_CS42_4DS_TGACv1_361864_AA1173710.2 | CCR4-NOT transcription complex subunit 3 | 3,0 |
| 228 | TRIAE_CS42_4AS_TGACv1_307339_AA1019660.8 | Scarecrow-like protein | 2,4 |
| Calcium binding protein, kinase or phosphatase | |||
| 229 | TRIAE_CS42_5BS_TGACv1_423377_AA1375480.3 | Dual specificity protein kinase shkD-like | 8,5 |
| 230 | TRIAE_CS42_4BL_TGACv1_320912_AA1051490.5 | Phosphoinositide phosphatase SAC2 | 4,5 |
| 231 | TRIAE_CS42_4DS_TGACv1_361621_AA1170770.1 | Type IV inositol polyphosphate 5-phosphatase 11 | 3,0 |
| 232 | TRIAE_CS42_7DL_TGACv1_603289_AA1980210.3 | Haloacid dehalogenase-like hydrolase | 2,9 |
| 233 | TRIAE_CS42_6DL_TGACv1_526671_AA1689500.1 | Calcium sensing receptor | 2,6 |
| Stress-related proteins | |||
| 234 | TRIAE_CS42_U_TGACv1_645365_AA2144110.3 | Protein argonaute | 15,4 |
| 235 | TRIAE_CS42_2AL_TGACv1_093339_AA0277990.3 | Pentatricopeptide repeat (PPR-like) superfamily protein | 6,2 |
| 236 | TRIAE_CS42_5DL_TGACv1_434794_AA1441820.1 | Disease resistance protein RPM1 | 3,5 |
| 237 | TRIAE_CS42_7BL_TGACv1_579887_AA1910750.10 | Spermidine synthase | 2,9 |
| 238 | TRIAE_CS42_5DS_TGACv1_456750_AA1477470.2 | Chloroplast stem-loop binding protein of 41 kDa | 2,9 |
| 239 | TRIAE_CS42_5AS_TGACv1_393102_AA1268440.1 | Chloroplast stem-loop binding protein of 41 kDa | 2,8 |
| 240 | TRIAE_CS42_2AS_TGACv1_112253_AA0334170.1 | Tetratricopeptide repeat containing protein | 2,6 |
| 241 | TRIAE_CS42_2DL_TGACv1_158673_AA0524320.3 | Zeaxanthin epoxidase | 2,0 |
| Oxidation-reduction process | |||
| 242 | TRIAE_CS42_7DL_TGACv1_603859_AA1990270.1 | NAD(P) H dehydrogenase (quinone) FQR1-like 1 | 14,6 |
| 243 | TRIAE_CS42_6AL_TGACv1_472100_AA1517740.2 | (+)-neomenthol dehydrogenase | 3,3 |
| Photosynthesis related proteins | |||
| 244 | TRIAE_CS42_4BS_TGACv1_327886_AA1077790.6 | Ribulose bisphosphate carboxylase/oxygenase activase A | 18,2 |
| 245 | TRIAE_CS42_1AS_TGACv1_019302_AA0064620.1 | PGR5-like protein 1A | 5,0 |
| 246 | TRIAE_CS42_2AL_TGACv1_094760_AA0302740.1 | Photosystem I subunit O | 3,8 |
| 247 | TRIAE_CS42_3DL_TGACv1_253211_AA0893780.1 | Chlorophyllide a oxygenase, chloroplastic | 2,9 |
| 248 | TRIAE_CS42_2DS_TGACv1_177171_AA0567560.1 | Ribulose bisphosphate carboxylase small chain | 2,8 |
| 249 | TRIAE_CS42_3DL_TGACv1_250162_AA0863350.3 | Carbonic anhydrase | 2,8 |
| 250 | TRIAE_CS42_2BL_TGACv1_130248_AA0407140.1 | Protein STAY-GREEN LIKE | 2,7 |
| 251 | TRIAE_CS42_U_TGACv1_642994_AA2125780.1 | Photosystem II 5 kD protein | 2,7 |
| 252 | TRIAE_CS42_3B_TGACv1_222152_AA0758310.2 | Carbonic anhydrase | 2,6 |
| 253 | TRIAE_CS42_4DL_TGACv1_342533_AA1116020.1 | Photosystem II subunit X | 2,6 |
| 254 | TRIAE_CS42_4AL_TGACv1_290053_AA0980930.6 | Ribulose bisphosphate carboxylase/oxygenase activase A | 2,5 |
| 255 | TRIAE_CS42_4DS_TGACv1_361664_AA1171210.2 | Ribulose bisphosphate carboxylase/oxygenase activase A, | 2,5 |
| 256 | TRIAE_CS42_6AS_TGACv1_486261_AA1559010.1 | Photosystem II 5 kD protein | 2,3 |
| 257 | TRIAE_CS42_2BL_TGACv1_129762_AA0395100.1 | Photosystem I reaction center subunit | 2,2 |
| 258 | TRIAE_CS42_5BL_TGACv1_404244_AA1291940.1 | Photosystem I reaction center subunit | 2,2 |
| 259 | TRIAE_CS42_2AL_TGACv1_093154_AA0273500.1 | Ribulose bisphosphate carboxylase/oxygenase activase | 2,2 |
| 260 | TRIAE_CS42_5AL_TGACv1_375138_AA1216740.1 | Chlorophyll a-b binding protein | 2,1 |
| 261 | TRIAE_CS42_2DL_TGACv1_162716_AA0563420.2 | Probable plastid-lipid-associated protein 7 | 2,0 |
| 262 | TRIAE_CS42_5BL_TGACv1_405399_AA1326490.1 | Chlorophyll a-b binding protein | 2,0 |
| 263 | TRIAE_CS42_3AS_TGACv1_210772_AA0678660.5 | SCAR-like protein 2 | 2,0 |
| 264 | TRIAE_CS42_4BS_TGACv1_329104_AA1097840.1 | Serine transhydroxymethyltransferase | 2,0 |
| Transporter | |||
| 265 | TRIAE_CS42_U_TGACv1_642488_AA2118360.7 | Copper transporter CT1 | 10,4 |
| 266 | TRIAE_CS42_1DL_TGACv1_061382_AA0193640.2 | Protein YIPF | 5,7 |
| 267 | TRIAE_CS42_3B_TGACv1_223624_AA0784820.1 | Protein DETOXIFICATION | 3,5 |
| 268 | TRIAE_CS42_5DL_TGACv1_435599_AA1452500.1 | Mitochondrial substrate carrier family protein C | 2,6 |
| 269 | TRIAE_CS42_5BS_TGACv1_423469_AA1377600.2 | CSC1-like protein | 2,4 |
| 270 | TRIAE_CS42_5BL_TGACv1_404850_AA1312820.1 | ZINC INDUCED FACILITATOR | 2,2 |
| 271 | TRIAE_CS42_7AL_TGACv1_557283_AA1779210.1 | ABC transporter B family | 2,2 |
| Cell cycle | |||
| 272 | TRIAE_CS42_2DL_TGACv1_160031_AA0545750.1 | Cyclin-P1–1 | 15,8 |
| Carbohydrate metabolism-related proteins | |||
| 273 | TRIAE_CS42_6DS_TGACv1_542696_AA1728130.1 | Glycosyltransferase | 9,3 |
| 274 | TRIAE_CS42_3DL_TGACv1_249217_AA0841740.2 | Sedoheptulose-1,7-bisphosphatase | 2,9 |
| 275 | TRIAE_CS42_1DS_TGACv1_081598_AA0261790.1 | Malonyl-coenzyme A:anthocyanin 3-O-glucoside-6″-O-malonyltransferase | 2,5 |
| 276 | TRIAE_CS42_2AL_TGACv1_093283_AA0276660.3 | Glyceraldehyde-3-phosphate dehydrogenase | 2,4 |
| 277 | TRIAE_CS42_2DL_TGACv1_158386_AA0517110.1 | Glyceraldehyde-3-phosphate dehydrogenase | 2,0 |
| Nitrogen metabolism-related proteins | |||
| 278 | TRIAE_CS42_2DL_TGACv1_161369_AA0558280.2 | Glutamine synthetase | 5,2 |
| 279 | TRIAE_CS42_U_TGACv1_640900_AA2078630.2 | Glutamine synthetase | 3,0 |
| Others | |||
| 280 | TRIAE_CS42_1BS_TGACv1_049885_AA0163610.1 | Unknown protein | 26,1 |
| 281 | TRIAE_CS42_4BS_TGACv1_330248_AA1106770.1 | Root phototropism protein 2 | 25,0 |
| 282 | TRIAE_CS42_7DL_TGACv1_605944_AA2008620.1 | Unknown protein | 14,8 |
| 283 | TRIAE_CS42_7BL_TGACv1_580679_AA1915040.1 | Unknown protein | 10,4 |
| 284 | TRIAE_CS42_6BL_TGACv1_501185_AA1614740.1 | Unknown protein | 9,2 |
| 285 | TRIAE_CS42_2AL_TGACv1_094031_AA0291380.1 | S-norcoclaurine synthase | 7,6 |
| 286 | TRIAE_CS42_1AL_TGACv1_003073_AA0047460.1 | Unknown protein | 6,9 |
| 287 | TRIAE_CS42_2BL_TGACv1_130918_AA0419860.1 | Unknown protein | 5,6 |
| 288 | TRIAE_CS42_7DS_TGACv1_622168_AA2034370.3 | Protein REVEILLE | 5,2 |
| 289 | TRIAE_CS42_3B_TGACv1_226496_AA0817320.2 | Unknown protein | 5,2 |
| 290 | TRIAE_CS42_5DS_TGACv1_457673_AA1488760.1 | Unknown protein | 5,2 |
| 291 | TRIAE_CS42_3B_TGACv1_221867_AA0752050.2 | Bark storage protein A-like | 4,9 |
| 292 | TRIAE_CS42_6AS_TGACv1_485239_AA1541330.2 | Unknown protein | 4,5 |
| 293 | TRIAE_CS42_7DS_TGACv1_625467_AA2065230.1 | Unknown protein | 4,1 |
| 294 | TRIAE_CS42_3B_TGACv1_223220_AA0778090.1 | Unknown protein | 3,7 |
| 295 | TRIAE_CS42_3AS_TGACv1_212545_AA0701630.2 | Unknown function | 3,5 |
| 296 | TRIAE_CS42_3B_TGACv1_224639_AA0799250.6 | Phosphatidate cytidylyltransferase | 3,3 |
| 297 | TRIAE_CS42_6AL_TGACv1_472100_AA1517740.2 | (+)-neomenthol dehydrogenase-like | 3,3 |
| 298 | TRIAE_CS42_3B_TGACv1_224639_AA0799250.6 | Phosphatidate cytidylyltransferase | 3,3 |
| 299 | TRIAE_CS42_5DL_TGACv1_434212_AA1431710.1 | Unknown protein | 3,3 |
| 300 | TRIAE_CS42_3B_TGACv1_220931_AA0724270.1 | Carboxyl-terminal-processing peptidase 1 | 3,2 |
| 301 | TRIAE_CS42_3AL_TGACv1_194142_AA0627380.1 | Unknown protein | 3,2 |
| 302 | TRIAE_CS42_6DL_TGACv1_527961_AA1710260.1 | Unknown protein | 2,9 |
| 303 | TRIAE_CS42_3B_TGACv1_224332_AA0795340.1 | Peptidase family M48 family protein | 2,7 |
| 304 | TRIAE_CS42_7BL_TGACv1_578255_AA1892020.2 | (S)-coclaurine N-methyltransferase-like | 2,7 |
| 305 | TRIAE_CS42_7DL_TGACv1_604164_AA1994640.2 | Unknown protein | 2,7 |
| 306 | TRIAE_CS42_3B_TGACv1_221388_AA0739090.1 | Unknown protein | 2,7 |
| 307 | TRIAE_CS42_1AL_TGACv1_000467_AA0012810.1 | Unknown protein | 2,6 |
| 308 | TRIAE_CS42_U_TGACv1_641221_AA2088730.1 | Unknown protein | 2,5 |
| 309 | TRIAE_CS42_5BL_TGACv1_405011_AA1317790.1 | Unknown protein | 2,4 |
| 310 | TRIAE_CS42_2BL_TGACv1_132610_AA0438610.1 | Aminomethyltransferase | 2,3 |
| 311 | TRIAE_CS42_7BS_TGACv1_593046_AA1947680.1 | Unknown protein | 2,3 |
| 312 | TRIAE_CS42_1DS_TGACv1_080446_AA0248140.1 | Unknown protein | 2,3 |
| 313 | TRIAE_CS42_4DL_TGACv1_342977_AA1126610.1 | Unknown protein | 2,2 |
| 314 | TRIAE_CS42_1AS_TGACv1_019551_AA0068040.1 | Unknown protein | 2,2 |
| 315 | TRIAE_CS42_6BL_TGACv1_499775_AA1591420.1 | Unknown protein | 2,2 |
| 316 | TRIAE_CS42_6BL_TGACv1_499688_AA1589190.1 | Unknown protein | 2,1 |
| 317 | TRIAE_CS42_6BS_TGACv1_514327_AA1658410.1 | Unknown protein | 2,1 |
| 318 | TRIAE_CS42_2DL_TGACv1_158386_AA0517100.1 | Unknown protein | 2,1 |
| 319 | TRIAE_CS42_2BL_TGACv1_129404_AA0382260.1 | Farnesyl pyrophosphate synthetase | 2,1 |
| 320 | TRIAE_CS42_4BS_TGACv1_329104_AA1097840.1 | Serine transhydroxymethyltransferase 1 | 2,0 |
| 321 | TRIAE_CS42_5DL_TGACv1_433432_AA1413030.1 | Unknown protein | 2,0 |
Fold change, wild-type to 1B-OEX ratio. The gene numbers (#) are used in the text
Discussion
The novel BSMV expression system allows functional gene characterization in wheat
Functional characterization in wheat is well-known as being more difficult to achieve than in model systems such as Arabidopsis thaliana. Furthermore, characterizing a crop gene in a heterologous system brings a host of questions that cannot be readily answered, and data interpretation cannot always be translated to the crop species. In this study, we demonstrate that the BSMV system allows for easy, fast and efficient gene characterization directly in wheat, an important crop species. Using the 4-component BSMV system that we have developed for VOX and the existing 3-component BSMV system for VIGS, we here show that the TaZFP1B transcription factor is required for oxidative and drought stress tolerance in wheat.
Drought is a major abiotic factor limiting growth and crop productivity worldwide. Improving drought tolerance in crops is an important consideration for agriculture sustainability, especially since climate change is expected to exacerbate the occurrence and severity of drought periods. From previous studies, we identified the C2H2-type zinc finger member TaZFP1B (previously named TaZFP2) as the gene most strongly up-regulated by various abiotic stresses (aluminum, high light, anoxia, H2O2 and drought) [60, 61]. Our current study demonstrates that overexpression of TaZFP1B does not cause growth reduction under normal growth conditions compared to other drought-associated transcription factors such as CBFs [69, 70]. Plants overexpressing TaZFP1B are more tolerant to drought while plants underexpressing TaZFP1B are more sensitive. The 1B-OEX plants have improved phenotypic parameters such as relative water content, dry matter production, shoot length, leaf width, survival rate and seed yield per spike compared to wild-type plants. This positive effect on growth might be mediated via a phytosulfokine receptor (#78) since overexpression AtPSKR1 improves growth in Arabidopsis [71].
The transcriptome profiling experiments performed in this study revealed that TaZFP1B regulates a collection of transcripts involved in stress response and tolerance. Most are stress-responsive genes involved in signaling, transcription, oxidation-reduction process, cell wall and membrane structure, transport, cell cycle and carbohydrate metabolism. Several genes that are strongly up-regulated have not been previously associated with drought or oxidative stress tolerance and will not be discussed here. However, their strong up-regulation suggest that further characterization of these genes may be of interest. Our analysis revealed that in response to drought stress, 188 genes are up-regulated by at least 5-fold in 1B-OEX plants compared to wild-type. It is difficult at this point to determine whether the genes identified in this study are true orthologs of genes whose function in stress tolerance has been demonstrated in transgenic studies. However, they could play similar roles and explain the improvement of drought tolerance in the 1B-OEX plants [72]. A model that summarizes changes in gene expression and their potential relationship to drought tolerance is presented in Fig. 7 to support the discussion.
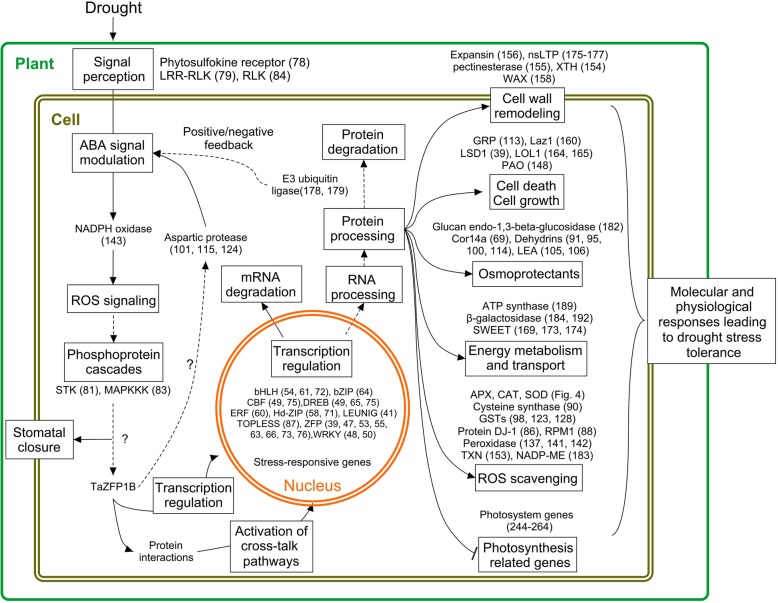
Fig. 7.
Summary of major changes in 1B-OEX plants and putative signaling pathways involved in drought tolerance. Solid lines indicate single-step reactions, and dashed lines indicate multi-step reactions. ABA, abscisic acid; APX, ascorbate peroxidase; bHLH, basic helix-loop-helix,; bZIP, basic leucine-zipper; CAT, catalase; CBF, core binding factor; COR, cold-regulated genes; DJ-1, protein deglycase DJ-1, DREB, dehydration-responsive element binding; ERF, ethylene response factor; GST, glutathione S-transferase; GRP, glycine-rich protein; Hd-ZIP, homeodomain-leucine zipper; HSP, heat shock proteins; Laz1, Lazarus 1; LEA, late embryogenesis abundant; LRR-RLK, leucine-rich repeats receptor-like kinase; LOL1, lsd one like 1; LSD1, lesion simulating disease 1; MAPKKK, mitogen-activated protein kinase kinase kinase; NADP-ME, NADP-dependent malic enzyme; nsLTP, non-specific lipid transfer protein; PAO, polyamine oxidase; SOD, superoxide dismutase; STK, serine/threonine kinase; ROS, reactive oxygen species; RUBISCO, ribulose bisphosphate carboxylase/oxygenase; TXN, thioredoxin protein; WAX; WRKY, transcription factor containing a highly conserved WRKY domain; and XTH, xyloglucan endotransglucosylase/hydrolase and ZFP, zinc finger protein;. Numbers refer to the corresponding genes in Tables 2, 3, 4 and 5
Signal perception and modulation
Drought triggers the production of the phytohormone ABA which in turn induces the expression of stress-related genes required for environmental adaptation. ABA-related responses are thus intricately related to drought responses. Leucine-Rich Repeat Receptor-Like Kinases (LRR-RLKs) (#79) belong to the large subfamily of Receptor-Like Kinases (RLKs) which are important mediators of environmental stimuli (Fig. 7). It has been proposed that LRR-RLKs might be involved in early responses to drought and ABA perception [28, 73]. Because of their roles in development and stress responses, LRR-RLKs are new potential targets for abiotic stress tolerance [74]. In Arabidopsis, the repression of the LRR-RLK RPK1 down-regulates many ABA-inducible genes, resulting in a decrease in ABA sensitivity and stomatal closure [28]. This suggests that RLKs function as important regulators in ABA signal transduction pathways. In addition, a LEUNIG and a TOPLESS-related proteins (#41, #87), which are known to act as transcriptional repressors, are induced by drought in 1B-OEX plants [75]. This suggests that transcriptional repression of genes negatively associated with drought tolerance might be a mechanism to study further.
Earlier studies showed that ABA induces ROS production through activation of NADPH oxidases [25]. The increased expression of NADPH oxidase (#143 and Additional file 4: Fig. S3A) in 1B-OEX plants may participate in modulating ABA signaling. ROS such as superoxide radicals and hydrogen peroxide are considered essential molecules in ABA signaling while excessive accumulation can be very toxic during drought stress. Therefore, regulatory mechanisms modulating ROS signal transduction and ROS detoxification are required to orchestrate the responses to ABA. ROS act as intracellular signals to trigger responses to drought stress [76, 77]. They induce phosphorylation and dephosphorylation events through the activation of protein kinases and phosphatases [78]. In this study, several protein kinases and phosphatases up-regulated 2 to 5-fold by overexpression of TaZFP1B were identified (Additional file 3: Table S1). Phosphoprotein cascades function as crucial regulators to mediate abiotic stress response and tolerance. Serine/threonine kinases (#81) and MAP kinases (#83) are able to phosphorylate a wide range of substrates and are associated with many different stress responses [79]. Our previous bioinformatic analysis has identified several putative phosphorylation sites for different kinases in the TaZFP1B amino acid sequence [60]. This suggests that phosphorylation may be required to fully activate TaZFP1B under drought stress (Fig. 7). Furthermore, studies have reported that the C2H2 zinc finger proteins ZAT10 and ZAT6 in Arabidopsis, and ZFP36 in rice, require kinase activation for their positive regulation of stress tolerance [57, 80, 81].
To control the level of ROS accumulation under stress, plants activate the expression of genes involved in antioxidant functions and production of stress proteins. In Arabidopsis, overexpression of the ASPG1 aspartic protease resulted in lower H2O2 levels with the parallel activation of detoxification enzymes (SOD and CAT), enhanced sensitivity to ABA and improved drought tolerance [24]. In accordance, our study revealed that genes encoding aspartic proteases are strongly up-regulated by TaZFP1B in 1B-OEX wheat plants under well-watered conditions (#6) and drought stress (#101, #115 (see also Additional file 4: Fig. S3B), #124). This result indicates that aspartic proteases could play an important role in enhancing drought tolerance and ROS detoxification mechanisms in wheat through the modulation of ABA sensitivity (Fig. 7).
Additionally, ROS accumulation is lower in 1B-OEX plants which is in accordance with the up-regulation of genes encoding SOD, APX, and CAT and the significant increase in SOD, APX and CAT activities. Furthermore, overexpression of TaZFP1B increases the expression of a gene encoding an ankyrin repeat containing protein (#25). It was shown that the AKR2A protein acts as a chaperone for APX3 in Arabidopsis [82], providing additional support to the observed increased APX activity. The induction of the Protein DJ-1 homolog D (#86) may contribute to the overall oxidative stress tolerance of 1B-OEX plants since Arabidopsis plants overexpressing AtDJ-1A show increased tolerance against various abiotic stresses, possibly by the interaction with SODs [83]. This suggests that the increased drought tolerance observed in wheat might be also mediated by the interaction with SODs, further linking drought and oxidative stresses. The increase in RPM1 (#88) can also contribute to the oxidative stress improvement, as suggested by the observation that SOD and CAT activities increase when AtRPM1 is overexpressed in Arabidopsis [84]. TaZFP1B OEX also increases GSH and GSSG contents, which are important metabolites for ROS detoxification in plants. It is possible that the increase in cysteine synthase (#90) contributes more Cys for the synthesis of GSH [85]. Together, these results suggest that overexpression of TaZFP1B triggers a greater capacity to maintain ROS homeostasis and improves both drought tolerance and productivity compared to wild-type wheat.
An increasing number of studies have reported key roles of ubiquitin-protein ligases (E3s) (#178 and #179 and Additional file 4: Fig. S3C) in plant developmental processes including responses to abiotic stresses [86]. In rice, the U-box E3 ligase OsPUB15 induced by H2O2 and drought stress plays an important role in plant tolerance. Its overexpression promoted growth under drought stress in transgenic plants [87]. In contrast, overexpression of AtPUB19 negatively regulated ABA signaling and decreased tolerance to drought stress [88]. Based on these studies, we hypothesize that TaZFP1B function might involve downstream E3 ligases (Fig. 7).
Physical interactions between transcription factors or with other protein complexes have emerged as important mechanisms allowing cross-talk between different pathways that lead to enhanced adaptability to environmental conditions [89]. This suggests that transcription factor members found to be up-regulated in 1B-OEX plants could be involved in specific or shared pathways. As regulators of many stress-responsive genes, transcription factors constitute one of the largest groups of genes differentially expressed in drought-treated 1B-OEX plants. Based on gene annotation, transcription factors belonging to large families are strongly up-regulated: bHLH (#54, #61, #72 and #77), bZIP (#64), CBF/DREB (#49 and #75), ERF (#60), HD-ZIP (#58 and #71), WRKY (#48 and #50) and ZFP (#39, #47, #53, #55, #63, #66, #73 and #76). These transcription factors could induce the expression of additional genes associated with the response to different hormonal signaling pathways involving abscisic acid (#101, #115, #124, #134 and #185), jasmonic acid (JA) (#89 and #107), auxin (#104, #118 and #136), gibberellic acid (GA) (#151), brassinosteroid (BR) (#117) and ethylene (#60) [38]. Many transcription factors within each family are known to enhance drought, salt, cold and osmotic stress tolerance [90, 91] and participate in the regulation of stress responses. Together, these observations suggest that TaZFP1B may be a molecular mediator engaged in a complex network involved in the response to drought stress.
ROS scavenging and energy supply
Overexpression of TaZFP1B resulted in the up-regulation of a number of genes encoding proteins involved in stress tolerance. Overexpression of TaZFP1B resulted in lower ROS accumulation and strong induction of ROS scavenging enzymes activity (SOD, APX and CAT). We also found that the NADP-malic enzyme (#183 and Additional file 4: Fig. S3D) is strongly up-regulated in 1B-OEX plants. This enzyme participates in CO2 fixation in plants. Additionally, in C3 plants, it is thought to be involved in the conversion of NADH to NADPH which improves cellular antioxidant defense [92]. In tobacco, the NADP-malic enzyme has been associated with drought stress acclimation [93]. Its overexpression resulted in a decrease in stomatal conductance, which improved water use efficiency [94]. This may be useful for the maintenance of growth during drought stress. Other genes involved in ROS scavenging have also been identified in our study. Different classes of glutathione S-transferase (GSTs) up-regulated in 1B-OEX plants (#12, #98 #123, #128, and Additional file 4: Fig. S3E and S3F) are known to promote detoxification of xenobiotics and to participate in the response to various abiotic stresses including oxidative stresses [95, 96]. Some theta, phy and tau GSTs have been shown to have glutathione peroxidase activity to reduce organic hydroperoxides of fatty acids, preventing oxidative damage [97].
Delaying leaf senescence was previously suggested to represent a strategy to enhance drought stress tolerance [98]. Slowing down photosynthesis is one of the main strategies to limit ROS production and propagation through down-regulation of components of the photosynthetic machinery [99]. We found that many genes involved in photosynthesis metabolism are down-regulated in 1B-OEX plants and, contrarily, overexpressed in 1B-siRNA plants during drought stress (Table 5). These genes include RUBISCO activase, RUBISCO small chain, and several other chloroplastic proteins (#244–264). Interestingly, the down-regulation of these genes in 1B-OEX plants is associated with the maintenance of a high chlorophyll content and delayed senescence compared to wild-type and 1B-siRNA plants (Fig. 2 and Additional file 2: Fig. S2). Maintaining the chlorophyll content while uncoupling photosynthesis may help the plant to survive the stress period while allowing it to recover more rapidly after stress.
During water deprivation, energy supply is severely limited. ATP synthase is a key enzyme involved in ATP synthesis during electron transport. Increased expression of ATP synthase (#189) was shown to improve drought tolerance in Arabidopsis [100]. Under water shortage, plant energy allocation strategy is crucial to increase survival, therefore the up-regulation of genes encoding bidirectional sugar transporters SWEET (#169, #173 and #174) could contribute to the allocation of energy in proper compartments to support stress tolerance mechanisms [101]. Interestingly, a reduced photosynthesis activity was previously observed in Arabidopsis with a concomitant increase in β-galactosidase activity (#184). The galactosidase activity possibly participates in the catabolic network of cell wall polysaccharides to produce sugars needed as energy source when photosynthate production is lower [102].
Osmoprotection and structural reinforcement
Osmotic adjustment is one of the most important mechanism used by plants to tolerate drought stress. In our study, we found that genes encoding CBF/DREB (#49, #65 and #75) are up-regulated in 1B-OEX plants under drought stress. The importance of DREB proteins in plant stress signaling and abiotic and biotic stress tolerance was previously reported [103]. Overexpression of CBF/DREB proteins enhances the expression of downstream target genes including Cor14a (#69), dehydrins (#91, #95, #100 and #114) and late embryogenesis abundant proteins (#105, #106) which are known to protect macromolecules from aggregation due to dehydration [104–106].
Additional genes encoding proteins involved in cell structure, elongation and maintenance, and cell wall or membrane metabolism such as xyloglucan endotransglucosylase/hydrolase (#154 and Additional file 4: Fig. S3G), WAX (#158), pectinesterase (#155) and expansin (#156) were also up-regulated by TaZFP1B overexpression during drought stress. These proteins play a major role in controlling cell wall extensibility and plasticity, two characteristics required to cope with a progressive decrease in water content. The latter results in considerable mechanical stress on plant cell architecture, therefore increasing elasticity and cellulose synthesis in cell wall would contribute to the maintenance of cell integrity and cell turgor in response to dehydration [17, 107, 108]. Moreover, changes in cell wall polysaccharides and proteins was observed in resurrection plants during drought stress and rehydration [109]. The majority of water loss occurs via transpiration through stomata, but studies have reported that under drought conditions, the synthesis of epicuticular wax on the leaf surface contributes to reducing water loss [110, 111]. WAX (#158) and non-specific lipid transfer protein genes (#172, #175–177 and Additional file 4: Fig. S3H) were reported to promote wax synthesis and to participate in cuticular wax deposition, respectively [110, 112]. Therefore, these results suggest that TaZFP1B regulates stress-responsive genes known for their capacity to enhance adaptation to drought stress (Fig. 7).
Programmed cell death and growth under stress
Ubiquitins are also involved in the regulation of programmed cell death (PCD) [113]. Abiotic stress-induced PCD is of considerable interest to ensure plant survival since it is a highly regulated process that facilitates the removal of unwanted and damaged cells, thus maintaining tissue and biological process homeostasis. Cell death is an integral part of plant growth and development, and also occurs as part of the plant response to abiotic stresses [114, 115]. In this study, genes regulating PCD and autophagy such as polyamine oxidase (#148), Laz1 (#160), LOL1 (#164, #165) and LSD1 (#39) were up-regulated in 1B-OEX plants [116–119]. Intriguingly, LOL1 (#164 and #165) and LSD1 (#39) have antagonistic effects on PCD. In Arabidopsis, overexpression of LOL1 enhances pathogen-driven hypersensitive response and oxidative stress-induced cell death [118] while LSD1 encodes a negative regulator of PCD [119]. Further studies have also reported that LSD1 contributes to plant vegetative and reproductive growth during drought stress by regulating PSII maximum efficiency, water use, cell wall structure and composition, and H2O2 concentration. This indicates that LSD1 could be involved in the regulation of photosynthesis, transpiration, cell signaling homeostasis, plant biomass production and seed yield [120, 121]. Similarly, emerging evidence suggest that glycine-rich proteins (GRPs) (#113) are involved in the regulation of plant cell growth and drought stress [122]. Overexpression of Arabidopsis glycine-rich RNA-binding proteins AtGRP2 or AtGRP7 in rice improves grain yield during drought stress [123], while AtGRP5 promotes cell elongation [124]. These findings suggest that a delicate balance of cell growth and cell death is needed during drought stress acclimation.
Conclusions
In conclusion, the functional characterization of TaZFP1B at the physiological and molecular levels allowed a better understanding of the importance of this transcription factor in wheat as well as its role in the response and tolerance to drought stress. The use of a novel VOX and VIGS system allowed the rapid functional gene characterization without the need to generate transgenic plant lines, which is a lengthy process in hexaploid wheat. The data presented here indicate that TaZFP1B is a key transcription factor orchestrating multiple molecular mechanisms involving the regulation of a collection of genes associated with stress tolerance, through the activation of multiple signaling pathways or via crosstalks between stress response pathways. The enhanced tolerance to drought or oxidative stress by the overexpression of TaZFP1B suggests that this transcription factor could be used as a marker for the selection of drought-tolerant wheat in breeding programs to improve productivity under climate change. Alternatively, a gene editing approach of the promoter region to enhance its expression is an interesting possibility for the improvement of wheat crop productivity under normal growth and drought conditions.
Methods
Plant materials and growth conditions
The Triticum aestivum cv. Atlas 66 winter wheat cultivar was obtained from Carver and colleagues [125] and was grown in our greenhouse facilities. This cultivar was selected for functional characterization of TaZFP1B (GenBank accession number MN577972) since this gene was identified from a previous study in this cultivar and was shown to respond to different abiotic stresses [60, 61]. In addition, to avoid the vernalization step required in winter cultivars, the commercial spring wheat T. aestivum cv. Dakosta (https://www.inspection.gc.ca/english/plaveg/pbrpov/cropreport/whe/app00009711e.shtml) was purchased from La Coop Fédérée (Saint-Hyacinthe, Québec Canada) and used to facilitate analysis of drought tolerance during booting. T. aestivum and Nicotiana benthamiana plants were grown in growth chambers under controlled conditions at 22 °C, a 14 h photoperiod, 100 μmol m− 2 s− 1 irradiance (fluorescent and incandescent lighting), and 70% relative humidity. Seeds were sown in a mixture of black earth, peat moss and perlite (2:1:1 ratio), and pots were watered every day for 14 days. Drought stress was then initiated by withholding water for different periods, as indicated in the Figure and Table legends. To determine the effect of drought stress on seed yield, water was withheld for 10 days when the developing head within the sheath of the flag leaf became visibly enlarged (booting stage Z45). Control samples were watered daily. Tissue sampling was performed at the same time of day to avoid circadian cycle influence.
Constructs for TaZFP1B silencing and overexpression
The four-component BSMV system developed previously was used for TaZFP1B virus-mediated overexpression (VOX) [63]. Compared to the three-component system, this modified system is based on four different plasmids (pCaBS-α, pCaBS-β, pCaBS-γ1 and pCaBS-γ2) where the DNA encoding the γ RNA has been split in two parts to allow for cloning of larger cDNA fragments or to increase expression by cloning genes in the two plasmids: pCaBS-γ1 and pCaBS-γ2. To obtain the TaZFP1B coding sequence (447 bp), total RNA was extracted from wheat root tips (cv. Atlas 66) exposed to aluminum, the mRNAs were reverse-transcribed to cDNAs using the SensiFAST™ cDNA Synthesis Kit (Bioline), and the TaZFP1B cDNA was PCR-amplified using the Q5 high fidelity DNA polymerase (New England Biolabs). This TaZFP1B coding sequence was cloned into pCaBS-γ1 and pCaBS-γ2 vectors using the primer pairs 1B-OEX_LIC1 and 1B-OEX_LIC2 (Additional file 5: Table S2). The resulting plasmids were used with the pCaBS-α and pCaBS-β vectors to maximize TaZFP1B overexpression (Fig. 1a) since the expression level is moderate with this system compared to strong promoters generally used in transgenic plants studies.
The three-component BSMV system was used for virus-induced gene silencing (VIGS) of TaZFP1B in wheat [126]. This system uses three different plasmids (pCaBS-α, pCaBS-β, pCaBS-γ) that respectively carry DNA sequences encoding the three genomic RNAs (RNAα, RNAβ, RNAγ) of the BSMV strain ND18. The siRNA against TaZFP1B was designed using GenScript siRNA Target Finder (https://www.genscript.com/tools/sirna-target-finder). The selected sense-loop-antisense DNA sequence (AAGAGTATTGCGGATCTGAAGttgatatccgCTTCAGATCCGCAATACTCtttttttt) was used to generate the siRNA chimeric construct. To ensure that this siRNA is specific to TaZFP1B, BLAST analysis was run against the NCBI and EnsemblPlant databases (release 36) for the wheat genome. The only detectable targets were close members of the same transcription factor family. The siRNA fragment was amplified (see Additional file 5: Table S2 for primers) and subcloned into pCaBS-γ by ligation-independent cloning (LIC). The resulting pCaBS-γ:1B-siRNA vector was used with pCaBS-α and pCaBS-β for the silencing of TaZFP1B in wheat (Fig. 1b).
Agroinfiltration of Nicotiana benthamiana and viral inoculation in wheat
Viral extracts to be used either for overexpression or silencing of TaZFP1B in wheat were produced in N. benthamiana. The plasmids (pCaBS-α, pCaBS-β, pCaBS-γ:TaZFP1B-siRNA for silencing; pCaBS-α, pCaBS-β, pCaBS-γ1:TaZFP1B and pCaBS-γ2:TaZFP1B for overexpression), were transformed individually into Agrobacterium tumefaciens strain EHA105 by electroporation. Single colonies were grown overnight at 28 °C in LB containing rifampicin (25 μg/ml) and kanamycin (50 μg/ml). The cultures were diluted 1:100 with LB containing the same antibiotics, 10 mM 2-(N-morpholino) ethanesulfonic acid (MES) pH 5.2 and 20 μM acetosyringone, and grown at 28 °C for 12 h. Bacterial cells were pelleted at 2200 g and equal amounts of the three or four Agrobacterium cultures (OD600 = 0.700) were mixed and incubated for 3–5 h at 28 °C in infiltration buffer (10 mM MES, pH 5.2, 10 mM MgCl2 and 0.1 mM acetosyringone) as described previously [126]. Agroinfiltration of N. benthamiana leaves and preparation of the homogenates containing the viral extracts were done as described previously [63, 127]. These extracts were aliquoted in small volumes and stored at − 20 °C for later use. As control for BSMV infection, empty vectors (pCaBS-γ1:00 and pCaBS-γ2:00) were used with the pCaBS-α, pCaBS-β vectors. Wheat seeds were inoculated with viral extracts for 3 days during seed imbibition, as previously described [63, 127]. Infected seeds were sown in potting mixture at a density of 10 per pot.
Four different types of plants were used in this study: wild-type, plants transformed with the BSMV vectors not containing the TaZFP1B cDNA (empty-vector), plants overexpressing TaZFP1B (1B-OEX) and plants under expressing TaZFP1B (1B-siRNA).
Determination of growth parameters
Plants were harvested at different times as described for each experiment. Entire plants (roots and shoots) separately were weighed before and after drying at 70 °C for 3 days, and the following growth parameters were assessed: dry weight, plant height, shoot-root ratio calculated from dry weight, second leaf width, and relative water content (RWC). RWC was measured as described previously [128]. Survival rates were calculated from wheat plants that were able to regrow after water withholding and rewatering. The effect of drought stress on grain yield (number and weight) was assessed in the spring wheat cultivar Dakosta by applying drought stress during the booting stage as described above.
Imaging of chlorophyll luminescence
Chlorophyll luminescence was imaged using an in vivo imaging system that includes a light-tight cabinet and a charged-coupled-device (CCD) camera (NightOWL II LB983, Berthold Technologies) with an exposure time of 0.6 s. Image analysis was performed with the IndiGO software (Berthold).
RNA isolation, RNA-Seq library preparation, sequencing and analyses
Total RNA samples of wild-type and BSMV-infected plants (cv. Atlas 66) were harvested from the second leaf of three 21 day-old well-watered wheat plants (control) and three 14 day-old plants that were drought-stressed for 7 days (total of 21 days of growth). The tissue was flash-frozen in liquid nitrogen and total RNA was extracted with the RNeasy Plant Mini kit (QIAGEN) according to the manufacturer’s instructions. RNA samples were treated with on-column RNase-free DNase (QIAGEN) to remove any contaminating genomic DNA. The quality of RNA samples with OD260/OD280 values greater than 2.0 was assessed by electrophoresis. Determination of RNA Integrity Number (RIN) value, preparation and sequencing of RNA libraries were performed at Novogene (California, USA). Paired-end (2 × 150 b) sequencing was performed on RNA-Seq libraries on the Illumina NovaSeq platform, and we analysed the raw data provided by Novogene. Each read of the paired-end sequences was analysed separately and the transcript per million values were then compared to exclude inconsistent results (more than five-fold difference). The quality of the raw data was verified with FastQC [129]. Adapters (TruSeq3 pair ended) were trimmed using Trimmomatic (version 0.36.3) [130]. Reads were globally filtered to retain reads with base quality exceeding Q30. The RNA-Seq data were cleaned to remove adapters, low read quality and artifact sequences. Transcriptome analysis was performed using the Galaxy pipeline https://usegalaxy.org [131]. Sequence artifacts were removed using a FASTX-toolkit available on the Galaxy platform. The similarity between RNA-Seq datasets was evaluated using plotCorrelation [132]. A heat map plotting type based on the Spearman correlation method was generated to visualize correlations between the libraries and reads from both ends of the paired-end sequencing. To quantify the expression of transcripts, Salmon (version 0.8.2) was used in quasi-mapping-mode [133]. Kmer size was set at 31. Wheat transcriptomic data (Database version 90.3) was retrieved from Ensembl Plants (ftp://ftp.ensemblgenomes.org/pub/plants/release-37/fasta/triticum_aestivum/cdna/) and selected as reference transcriptome for transcript quantification [134]. The transcript per million values of the transcripts generated by Salmon on the RNA-Seq data was used to calculate the splicing ratio. Genes with zero or low counts were excluded by applying a feature selection (Additional file 3: Table S1 and Additional file 6: Table S3). For analysis, we selected genes that are overexpressed 5-fold or more in 1B-OEX plants compared to wild-type plants either under well-watered conditions or under drought stress. Genes that showed similar expression in the empty vector and 1B-OEX RNA-Seq data were excluded. We also selected a few genes of particular interest such as ROS detoxifying enzymes that were overexpressed less than 5-fold in the RNA-Seq data. In this case, their expression level was quantified by qRT-PCR. The heatmapper program (http://www.heatmapper.ca/expression/) was used to draw the heatmap of the expression data. Annotation of transcripts was done based on the wheat annotated genomic transcripts (version release 37) available on Ensembl Plants (ftp://ftp.ensemblgenomes.org/pub/plants/release-37/fasta/triticum_aestivum/) or by BLAST search against the NCBI nucleotide collection (nr/nt). De novo transcriptome assemblies were performed using Trinity [135] to determine the real sequences of genes in wheat cv. Atlas 66. The RNA-Seq data were deposited at NCBI Gene Expression Omnibus (GEO) under the accession number GSE136683.
Quantitative RT-PCR analysis
Total RNA used for quantitative real-time polymerase chain reaction (qRT-PCR) analysis was extracted from the second leaf of wheat (cv. Atlas 66) plants as described above. Total RNA (0.5 μg) was reverse-transcribed in a total volume of 20 μl using the SensiFAST™ cDNA Synthesis Kit (Bioline). Real-time quantitative RT-PCR was performed on a CFX96 Touch™ Thermal Cycler (Bio-Rad) using the Luna® Universal qPCR Master Mix (New England Biolabs). To quantify specific transcripts from homoeologous genes, primers in Additional file 5: Table S2 were designed based on sequences retrieved from RNA-Seq data of cv. Atlas 66. For each gene, the reference gene sequence was retrieved from Ensembl Plants database and aligned to the assembled transcriptome generated by Trinity. The sequence of interest was identified based on the lowest E-value. Amplification was performed as follows: 5 min at 95 °C followed by 35 cycles of 95 °C for 15 s, 58 °C for 30 s. The target gene transcript level was normalized against the 18S rRNA level. Fold changes of RNA transcripts were calculated using the 2-ΔΔCt method [136].
ROS detection
Each leaf sample (0.1 g) was ground in liquid nitrogen and the powder was mixed in a microtube with 1 mL of 10 mM Tris-HCl, pH 7.2, then centrifuged at 12,000 g for 20 min at 4 °C. The supernatant was transferred to a fresh microtube and used for total ROS and H2O2 detection. Detection of total ROS was performed using the fluorogenic dye 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA), which is deacetylated by cellular esterases to a compound that can be oxidized by ROS to 2′,7′-dichlorofluorescein (DCF) [137]. DCF fluorescence was measured using a microplate reader with excitation and emission wavelengths of 495 nm and 529 nm, respectively. The non-specific background was subtracted from experimental values by performing other measurements after treatment with catalase (300 units/mL) at room temperature for 10 min. The corrected fluorescence values were expressed as relative fluorescence units/mg of protein extract. Detection of hydrogen peroxide (H2O2) was carried out using the Hydrogen Peroxide Fluorescent Detection Kit (Arbor Assays, USA), using excitation and emission wavelengths of 570 nm and 590 nm, respectively. H2O2 concentration was calculated from a standard curve and expressed as μmol/g of fresh weight. The DCF and H2O2 data were normalized to corresponding well-watered wild-type plants.
Lipid peroxidation detection
Oxidative damage to lipids in leaf tissue was estimated using the thiobarbituric acid reactive substances (TBARS) test which determines the content of malondialdehyde (MDA). Each leaf sample (0.5 g) was ground in a mortar with 0.5% (w/v) trichloroacetic acid (TCA) and the homogenate was centrifuged at 12,000 g for 20 min. The supernatant was transferred to a glass tube and mixed with a solution containing 40% TCA and 1% (w/v) thiobarbituric acid (TBA). The mixture was incubated in boiling water for 30 min and the reaction was stopped by moving the tube to an ice bath. The mixture was then mixed with 1-butanol and vortexed vigorously for 10 s, then phases were separated by centrifugation (4000 g, 5 min at room temperature). The upper phase was carefully transferred to a new tube and absorbance values were read at 532 nm and 600 nm. The latter value represents non-specific absorption and was subtracted. The content of MDA-TBA complex in the samples was calculated as MDA equivalent from an MDA standard curve (Cayman Chemical, USA). The MDA data was normalized to corresponding well-watered wild-type plants.
Determination of antioxidant components: enzymes and metabolites
Total activity of superoxide dismutase (SOD; EC 1.15.1.1) was assayed by the inhibition of photochemical reduction of nitroblue tetrazolium (NBT) as described previously [138]. Each leaf sample (0.5 g) was ground with liquid nitrogen and suspended in 1.5 mL of homogenization buffer: 50 mM sodium phosphate (pH 7.8), 1 mM ethylenediaminetetraacetic acid (EDTA) and 2% (w/v) polyvinylpolypyrrolidone (PVPP). The homogenate was centrifuged at 12,000 g for 30 min at 4 °C and the supernatant was immediately used for the SOD assay. The 3.0 mL assay medium contained 50 mM sodium phosphate (pH 7.8), 0.66 mM EDTA, 10 mM L-methionine, 33 μM nitroblue tetrazolium (NBT) and 3.3 μM riboflavin. The reaction was started by placing the tubes under light (600 μmol m− 2 s− 1) for 10 min at 25 °C. Reduction of NBT was measured at 560 nm. One enzyme unit of SOD is defined as the amount of protein causing a 50% inhibition of NBT photoreduction. The results were expressed as units/mg of soluble proteins.
Total ascorbate peroxidase activity (APX; EC 1.11.1.11) was determined by following the decrease in A290 for 2 min at 25 °C [139]. Each leaf sample (0.5 g) was ground with liquid nitrogen and suspended in 1.5 mL of homogenization buffer containing 50 mM sodium phosphate (pH 7.0), 2% (w/v) PVPP, 0.1 mM EDTA and 2 mM ascorbate. The homogenate was centrifuged at 12,000 g for 30 min at 4 °C. The reaction mixture (1 mL) contained 50 mM sodium phosphate, 0.1 mM EDTA, 0.5 mM ascorbate and 0.5 mM H2O2 and the reaction rate (ascorbate oxidized/min) was measured at 22 °C. The molar extinction coefficient of ascorbate (ε) used for calculation was 2.8 mM− 1 cm− 1. The results were expressed as units of APX (μmol ascorbate) per mg of soluble proteins.
Catalase activity (CAT; EC 1.11.1.6) was determined by following the consumption of H2O2 at 240 nm for 2 min at 25 °C [140]. Each leaf sample (0.5 g) was ground with liquid nitrogen and suspended in 1.5 mL of homogenization buffer containing 50 mM Tris-HCl (pH 7.8), 0.1 mM EDTA, 0.2% (v/v) Triton X-100, 1 mM phenylmethylsulphonyl fluoride (PMSF) and 2 mM dithiothreitol (DTT). The homogenate was centrifuged at 12,000 g for 30 min at 4 °C and the supernatant was used for the CAT assay. The reaction mixture contained 50 mM sodium phosphate (pH 7.0) and 0.1% H2O2 and the reaction rate (μmol H2O2 consumed/min) was measured at 22 °C. The molar extinction coefficient of H2O2 (ε) used for calculation was 39.4 mM− 1 cm− 1. The results were calculated as units of CAT (μmol H2O2) per mg soluble proteins.
For glutathione determination, each leaf tissue (0.1 g) was ground in liquid nitrogen and the powder was placed in a microtube with 1 mL of 10 mM Tris-HCl, pH 7.2 and centrifuged at 12,000 g for 20 min at 4 °C. The supernatant was transferred to a fresh microtube. Reduced glutathione (GSH) and oxidized glutathione (GSSG) levels were determined using the Glutathione (GSH) Fluorescent Detection Kit (Arbor Assays, USA) according to the manufacturer’s protocol. A standard curve of GSH was established to determine the concentrations of free GSH and total GSH in samples. The results were expressed as μmol per mg soluble proteins.
Protein quantification
Proteins were quantified using the Bio-Rad Protein Assay Dye Reagent (Bio-Rad, Canada). Absorbance was read at 595 nm with a microplate reader, and protein concentration was determined using a standard curve prepared with bovine serum albumin (BSA).
Statistical analyses and validation of RNA-Seq data
All experiments with the exception of RNA-Seq were repeated at least four times independently and the value presented are means ± standard deviations (SD). Comparisons of means were conducted using one-way analysis of variance (ANOVA). Differences among means were analyzed using Tukey’s post hoc test at p values < 0.05. Statistical analyses were performed using InStat 3.0. Pairwise correlation was performed to analyze the relationship between libraries, and a heat map was generated to verify the correlation between both ends of paired end sequencing of each treatment (Additional file 7: Fig. S4). The heatmap reveals a close relationship between the wild-type and the empty vector treatment, indicating that there is no major change in gene expression due to the BSMV infection itself. The analysis shows that the well-watered and drought libraries have a low correlation, indicating profound changes in wheat transcriptome profiles when drought stress is applied. Additionally, the correlation between the 1B-OEX and 1B-siRNA libraries is the lowest whether comparing well-watered or drought-treated plants, indicating significant differences in gene expression profiles. This suggests that several genes that are regulated by the expression of TaZFP1B are likely regulated in an opposite manner in the 1B-siRNA plants. For the qRT-PCR validation of RNA-Seq results, 16 genes up-regulated in drought-stressed 1B-OEX were randomly selected from the lists of differentially regulated transcripts in the RNA-Seq data and expression levels were analysed using specific primers between drought-stressed 1B-OEX and wild-type plants (Additional file 5: Table S2). Scatterplots were generated by comparing the log2 fold change (OEX drought/wild-type drought). The results show that the correlation between the expression patterns obtained by qPCR and the RNA-Seq data (R2 = 0.6374, p > 0.01) (Additional file 8: Fig. S5) is similar to what is observed between qPCR and microarray data.
Supplementary information
Additional file 1: Figure S1. Silencing of TaZFP1B affects relative expression of the closest TaZFP1B relatives. The different types of wheat plants (see Fig. 1) were grown for 14 days then were either well-watered for an additional 7 days or drought-stressed by withholding water for 7 days, and expression levels were determined by qRT-PCR. Data are the mean expression ± SD of four biological replicates. Different letters indicate statistically significant differences between samples (P < 0.05 by Tukey’s test).
Additional file 2: Figure S2. Chlorophyll autofluorescence from wheat leaves. The different types of wheat plants (see Fig. 1) were grown for 14 days then were either well-watered for an additional 10 or 14 days (top leaf in the panels) or drought-stressed by withholding water for 10 or 14 days (bottom leaf in the panels). Fluorescence was captured using a NightOWL II imaging cabinet.
Additional file 3: Table S1. List of genes up-regulated by TaZFP1B overexpression.
Additional file 4: Figure S3. Validation of RNA-Seq data by qRT-PCR. The different types of wheat plants (see Fig. 1) were grown for 14 days then were either well-watered for an additional 7 days or drought-stressed by withholding water for 7 days. Expression levels are relative to the well-watered wild-type group. Numbers refer to the corresponding genes in Tables 2, 3, 4 and 5. Data are mean expression ± SD of four biological replicates. Different letters indicate statistically significant differences between samples (P < 0.05 by Tukey’s test).
Additional file 5: Table S2. List of genes down-regulated by TaZFP1B overexpression.
Additional file 6: Table S3. Primers used in this study.
Additional file 7: Figure S4 Heatmap of correlations between RNA-seq libraries. The different types of wheat plants (see Fig. 1) were grown for 14 days then were either well-watered for an additional 7 days or drought-stressed by withholding water for 7 days. RNA-Seq libraries were prepared and paired-end sequencing was performed. Each read generated from paired-end sequencing was analyzed individually. The hierarchical clustering was generated using Spearman correlation coefficient. The color scale indicates the degree of correlation.
Additional file 8: Figure S5. Pearson correlation between the RNA-seq and qRT-PCR data. The qRT-PCR log2 value of the expression ratio (drought-treated wild-type/drought-treated 1B-OEX) (y-axis) was plotted from the RNA-seq log2 value of expression ratio (drought-treated wild-type/drought-treated 1B-OEX) (x-axis). Genes used to calculate the correlation are listed in supplementary Table 1. All qRT-PCR data were collected from three biological replicates. --- represents the 95% confidence interval. The calculated correlation value (R2) is shown along with the regression line.
Acknowledgments
The authors thank Dawei Li (State Key Laboratory of Agro-biotechnology at China Agriculture University) for providing the pCaBS-α, pCaBS-β and pCaBS-γ vectors.
Abbreviations
- ABA
Abscisic acid
- APX
Ascorbate peroxidase
- ATP
Adenosine triphosphate
- bHLH
basic helix-loop-helix protein
- BSA
Bovine serum albumin
- BSMV
Barley stripe mosaic virus
- bZIP
basic leucine zipper
- CaM1
Calmodulin 1
- CAT
Catalase
- CBF
C-repeat binding factors
- DCF
2′,7′-dichlorofluorescein
- DREB
Drought response element binding protein
- DTT
Dithiothreitol
- EDTA
Ethylenediaminetetraacetic acid
- ERF
Ethylene responsive factor
- GRP
Glycine-rich proteins
- GSH
Reduced glutathione
- GSSG
Oxidized glutathione
- GST
Glutathione S-transferase
- H2O2
Hydrogen peroxide
- HD-ZIP
Homeodomain-leucine zipper transcription factor
- JA
Jasmonic acid
- Laz 1
Lazarus 1
- LEA
Late embryogenesis abundant
- LOL1
Lsd one like 1
- LRR-RLK
Leucine-rich repeat receptor-like kinases
- MDA
Malondialdehyde
- MYB
Myeloblastosis oncogene
- NAC
NAM, ATAF1/2 and CUC
- NADPH
Reduced nicotinamide adenine dinucleotide phosphate
- NBT
Nitroblue tetrazolium
- NF-Y
Nuclear factor Y
- PCD
Programmed cell death
- PMSF
Phenylmethylsulphonyl fluoride
- PVPP
Polyvinylpolypyrrolidone
- ROS
Reactive oxygen species
- RPK 1
Receptor-like kinase 1
- RUBISCO
Ribulose-1,5-bisphosphate carboxylase/oxygenase
- RWC
Relative water content
- SLAC
Slow anion channel
- SOD
Superoxide dismutase
- SWEET
Sugars will eventually be exported transporters
- TBA
Thiobarbituric acid
- TBARS
Thiobarbituric acid reactive substances
- TCA
Trichloroacetic acid
- VIGS
Virus-induced gene silencing
- VOX
Virus-mediated overexpression
- WRKY
Proteins containing the highly conserved amino acid sequence WRKYGQK
- XTH
Xyloglucan endotransglucosylase/hydrolase
- ZFP
Zinc finger proteins
Authors’ contributions
AC performed the experiments; AC and MH designed the experiments, interpreted the data and wrote the manuscript; FO interpreted the data and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by a Natural Sciences and Engineering Research Council of Canada grant (RGPIN-2017-05256) to M.H.
Availability of data and materials
The RNA-Seq datasets generated and/or analysed during the current study are available in the GEO repository under the accession number GSE136683, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136683. The TaZFP1B sequence is available at GenBank under accession number MN577972 (https://www.ncbi.nlm.nih.gov/nuccore/MN577972).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-02355-x.
References
- 1.FAO . Global agriculture towards 2050. Rome: FAO; 2009. [Google Scholar]
- 2.Valin H, Sands RD, van der Mensbrugghe D, Nelson GC, Ahammad H, Blanc E, Bodirsky B, Fujimori S, Hasegawa T, Havlik P, et al. The future of food demand: understanding differences in global economic models. Agric Econ. 2014;45(1):51–67. doi: 10.1111/agec.12089. [DOI] [Google Scholar]
- 3.Farooq M, Hussain M, Wahid A, Siddique KHM. Plant Responses to Drought Stress. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012. Drought stress in plants: An overview; pp. 1–33. [Google Scholar]
- 4.Vicente-Serrano SM, Beguería S, Camarero JJ. Drought severity in a changing climate. In: Handbook of drought and water scarcity: CRC Press; Boca Raton, USA, 2017. p. 279–303.
- 5.Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167(2):313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeandroz S, Lamotte O. Editorial: plant responses to biotic and abiotic stresses: lessons from cell signaling. Front Plant Sci. 2017;8:1772. doi: 10.3389/fpls.2017.01772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Distelfeld A, Pearce SP, Avni R, Scherer B, Uauy C, Piston F, Slade A, Zhao R, Dubcovsky J. Divergent functions of orthologous NAC transcription factors in wheat and rice. Plant Mol Biol. 2012;78(4–5):515–524. doi: 10.1007/s11103-012-9881-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldman M, Levy AA. Genome evolution due to allopolyploidization in wheat. Genetics. 2012;192(3):763–774. doi: 10.1534/genetics.112.146316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316(5833):1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61(6):1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- 11.Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9(10):490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi S, Seki M, Ishida J, Satou M, Sakurai T, Narusaka M, Kamiya A, Nakajima M, Enju A, Akiyama K. Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Mol Biol. 2004;56(1):29–55. doi: 10.1007/s11103-004-2200-0. [DOI] [PubMed] [Google Scholar]
- 13.Batista R, Fonseca C, Planchon S, Negrao S, Renaut J, Oliveira MM. Environmental stress is the major cause of transcriptomic and proteomic changes in GM and non-GM plants. Sci Rep. 2017;7(1):10624. doi: 10.1038/s41598-017-09646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruz de Carvalho MH. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal Behav. 2008;3(3):156–165. doi: 10.4161/psb.3.3.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan JM. Osmoregulation and water stress in higher plants. Annu Rev Plant Physiol. 1984;35(1):299–319. doi: 10.1146/annurev.pp.35.060184.001503. [DOI] [Google Scholar]
- 16.Lamaoui M, Jemo M, Datla R, Bekkaoui F. Heat and drought stresses in crops and approaches for their mitigation. Front Chem. 2018;6:26. doi: 10.3389/fchem.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Gall H, Philippe F, Domon JM, Gillet F, Pelloux J, Rayon C. Cell wall metabolism in response to abiotic stress. Plants (Basel) 2015;4(1):112–166. doi: 10.3390/plants4010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhargava S, Sawant K. Drought stress adaptation: metabolic adjustment and regulation of gene expression. Plant Breed. 2013;132(1):21–32. doi: 10.1111/pbr.12004. [DOI] [Google Scholar]
- 19.Ijaz R, Ejaz J, Gao S, Liu T, Imtiaz M, Ye Z, Wang T. Overexpression of annexin gene AnnSp2, enhances drought and salt tolerance through modulation of ABA synthesis and scavenging ROS in tomato. Sci Rep. 2017;7(1):12087. doi: 10.1038/s41598-017-11168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdeen A, Schnell J, Miki B. Transcriptome analysis reveals absence of unintended effects in drought-tolerant transgenic plants overexpressing the transcription factor ABF3. BMC Genomics. 2010;11:69. doi: 10.1186/1471-2164-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai C, Lee Y, Lee IC, Nam HG, Kwak JM. Calmodulin 1 regulates senescence and ABA response in Arabidopsis. Front Plant Sci. 2018;9:803. doi: 10.3389/fpls.2018.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson S, Kudoyarova GR, Veselov DS, Arkhipova TN, Davies WJ. Plant hormone interactions: innovative targets for crop breeding and management. J Exp Bot. 2012;63(9):3499–3509. doi: 10.1093/jxb/ers148. [DOI] [PubMed] [Google Scholar]
- 23.Kurahashi Y, Terashima A, Takumi S. Variation in dehydration tolerance, ABA sensitivity and related gene expression patterns in D-genome progenitor and synthetic hexaploid wheat lines. Int J Mol Sci. 2009;10(6):2733–2751. doi: 10.3390/ijms10062733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao X, Xiong W, Ye T, Wu Y. Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis. J Exp Bot. 2012;63(7):2579–2593. doi: 10.1093/jxb/err433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003;22(11):2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arias CL, Pavlovic T, Torcolese G, Badia MB, Gismondi M, Maurino VG, Andreo CS, Drincovich MF, Gerrard Wheeler MC, Saigo M. NADP-dependent malic enzyme 1 participates in the abscisic acid response in Arabidopsis thaliana. Front Plant Sci. 2018;9:1637. doi: 10.3389/fpls.2018.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Wu L, Chang W, Li Z, Miao M, Li Y, Yang J, Liu Z, Tan J. Overexpression of the maize E3 ubiquitin ligase gene ZmAIRP4 enhances drought stress tolerance in Arabidopsis. Plant Physiol Biochem. 2018;123:34–42. doi: 10.1016/j.plaphy.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 28.Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell. 2005;17(4):1105–1119. doi: 10.1105/tpc.104.027474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulik A, Wawer I, Krzywinska E, Bucholc M, Dobrowolska G. SnRK2 protein kinases--key regulators of plant response to abiotic stresses. OMICS. 2011;15(12):859–872. doi: 10.1089/omi.2011.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lou D, Wang H, Liang G, Yu D. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front Plant Sci. 2017;8:993. doi: 10.3389/fpls.2017.00993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dey A, Samanta MK, Gayen S, Maiti MK. The sucrose non-fermenting 1-related kinase 2 gene SAPK9 improves drought tolerance and grain yield in rice by modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression. BMC Plant Biol. 2016;16(1):158. doi: 10.1186/s12870-016-0845-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149(1):88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciftci-Yilmaz S, Mittler R. The zinc finger network of plants. Cell Mol Life Sci. 2008;65(7–8):1150–1160. doi: 10.1007/s00018-007-7473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller G, Shulaev V, Mittler R. Reactive oxygen signaling and abiotic stress. Physiol Plant. 2008;133(3):481–489. doi: 10.1111/j.1399-3054.2008.01090.x. [DOI] [PubMed] [Google Scholar]
- 35.Kiełbowicz-Matuk A. Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Sci. 2012;185:78–85. doi: 10.1016/j.plantsci.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 36.Yang S, Vanderbeld B, Wan J, Huang Y. Narrowing down the targets: towards successful genetic engineering of drought-tolerant crops. Mol Plant. 2010;3(3):469–490. doi: 10.1093/mp/ssq016. [DOI] [PubMed] [Google Scholar]
- 37.Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants. 2013;19(3):307–321. doi: 10.1007/s12298-013-0179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joshi R, Wani SH, Singh B, Bohra A, Dar ZA, Lone AA, Pareek A, Singla-Pareek SL. Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci. 2016;7:1029. doi: 10.3389/fpls.2016.01029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6(5):410–417. doi: 10.1016/S1369-5266(03)00092-X. [DOI] [PubMed] [Google Scholar]
- 40.Roy S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal Behav. 2016;11(1):e1117723. doi: 10.1080/15592324.2015.1117723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, Wang T, Bartholomew E, Black K, Dong M, Zhang Y, Yang S, Cai Y, Xue S, Weng Y, et al. Comprehensive analysis of NAC transcription factors and their expression during fruit spine development in cucumber (Cucumis sativus L.) Hortic Res. 2018;5(1):31. doi: 10.1038/s41438-018-0036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kidokoro S, Watanabe K, Ohori T, Moriwaki T, Maruyama K, Mizoi J. Myint Phyu sin Htwe N, Fujita Y, Sekita S, Shinozaki K: soybean DREB 1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 2015;81(3):505–518. doi: 10.1111/tpj.12746. [DOI] [PubMed] [Google Scholar]
- 43.Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J. WRKY transcription factors in plant responses to stresses. J Integr Plant Biol. 2017;59(2):86–101. doi: 10.1111/jipb.12513. [DOI] [PubMed] [Google Scholar]
- 44.Wang F, Chen HW, Li QT, Wei W, Li W, Zhang WK, Ma B, Bi YD, Lai YC, Liu XL, et al. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015;83(2):224–236. doi: 10.1111/tpj.12879. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol. 2007;65(6):799–815. doi: 10.1007/s11103-007-9244-x. [DOI] [PubMed] [Google Scholar]
- 46.Yang A, Dai X, Zhang WH. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot. 2012;63(7):2541–2556. doi: 10.1093/jxb/err431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SB, Kim H, Kim RJ, Suh MC. Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Rep. 2014;33(9):1535–1546. doi: 10.1007/s00299-014-1636-1. [DOI] [PubMed] [Google Scholar]
- 48.Sun SJ, Guo SQ, Yang X, Bao YM, Tang HJ, Sun H, Huang J, Zhang HS. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot. 2010;61(10):2807–2818. doi: 10.1093/jxb/erq120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt R, Mieulet D, Hubberten HM, Obata T, Hoefgen R, Fernie AR, Fisahn J, San Segundo B, Guiderdoni E, Schippers JH, et al. Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell. 2013;25(6):2115–2131. doi: 10.1105/tpc.113.113068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004;136(1):2734–2746. doi: 10.1104/pp.104.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davletova S, Schlauch K, Coutu J, Mittler R. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 2005;139(2):847–856. doi: 10.1104/pp.105.068254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006;580(28–29):6537–6542. doi: 10.1016/j.febslet.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, Warren D, Cushman J, Connolly EL, Mittler R. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem. 2007;282(12):9260–9268. doi: 10.1074/jbc.M611093200. [DOI] [PubMed] [Google Scholar]
- 54.Shi H, Wang X, Ye T, Chen F, Deng J, Yang P, Zhang Y, Chan Z. The cysteine2/histidine2-type transcription factor zinc finger of Arabidopsis thaliana 6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and c-repeat-binding factor genes in arabidopsis. Plant Physiol. 2014;165(3):1367–1379. doi: 10.1104/pp.114.242404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu DQ, Huang J, Guo SQ, Yang X, Bao YM, Tang HJ, Zhang HS. Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.) FEBS Lett. 2008;582(7):1037–1043. doi: 10.1016/j.febslet.2008.02.052. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, Ni L, Liu Y, Wang Y, Zhang A, Tan M, Jiang M. The C2H2-type zinc finger protein ZFP182 is involved in abscisic acid-induced antioxidant defense in rice. J Integr Plant Biol. 2012;54(7):500–510. doi: 10.1111/j.1744-7909.2012.01135.x. [DOI] [PubMed] [Google Scholar]
- 57.Zhang H, Liu Y, Wen F, Yao D, Wang L, Guo J, Ni L, Zhang A, Tan M, Jiang M. A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J Exp Bot. 2014;65(20):5795–5809. doi: 10.1093/jxb/eru313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell. 2005;17(1):268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rizhsky L, Davletova S, Liang H, Mittler R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem. 2004;279(12):11736–11743. doi: 10.1074/jbc.M313350200. [DOI] [PubMed] [Google Scholar]
- 60.Cheuk A, Houde M. Genome wide identification of C1-2i zinc finger proteins and their response to abiotic stress in hexaploid wheat. Mol Gen Genomics. 2016;291(2):873–890. doi: 10.1007/s00438-015-1152-1. [DOI] [PubMed] [Google Scholar]
- 61.Ali-Benali MA, Badawi M, Houde Y, Houde M. Identification of oxidative stress-responsive C2H2 zinc fingers associated with Al tolerance in near-isogenic wheat lines. Plant Soil. 2012;366(1–2):199–212. [Google Scholar]
- 62.Sun B, Zhao Y, Shi S, Yang M, Xiao K. TaZFP1, a C2H2 type-ZFP gene of T. aestivum, mediates salt stress tolerance of plants by modulating diverse stress-defensive physiological processes. Plant Physiol Biochem. 2019;136:127–142. doi: 10.1016/j.plaphy.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 63.Cheuk A, Houde M. A new barley stripe mosaic virus allows large protein overexpression for rapid function analysis. Plant Physiol. 2018;176(3):1919–1931. doi: 10.1104/pp.17.01412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burgess P, Huang B, Hossain MA, Wani SH, Bhattacharjee S, Burritt DJ. Mechanisms of hormone regulation for drought tolerance in plants. In: Tran L-SP, editor. Drought Stress Tolerance in Plants, Vol 1. Switzerland: Springer International Publishing; 2016. pp. 45–75. [Google Scholar]
- 65.Noctor G, Veljovic-Jovanovic S, Driscoll S, Novitskaya L, Foyer CH: Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration? Ann Bot. 2002;89 Spec No:841–850. [DOI] [PMC free article] [PubMed]
- 66.Foyer CH, Shigeoka S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011;155(1):93–100. doi: 10.1104/pp.110.166181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ayala A, Munoz MF, Arguelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxidative Med Cell Longev. 2014;2014:360438. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trchounian A, Petrosyan M, Sahakyan N. Plant cell redox homeostasis and reactive oxygen species. In: Gupta DK, Palma JM, Corpas FJ, editors. Redox state as a central regulator of plant-cell stress responses. Cham: Springer International Publishing; 2016. pp. 25–50. [Google Scholar]
- 69.Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol. 2004;54(5):767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- 70.Kurepin LV, Dahal KP, Savitch LV, Singh J, Bode R, Ivanov AG, Hurry V, Huner NP. Role of CBFs as integrators of chloroplast redox, phytochrome and plant hormone signaling during cold acclimation. Int J Mol Sci. 2013;14(6):12729–12763. doi: 10.3390/ijms140612729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsubayashi Y, Ogawa M, Kihara H, Niwa M, Sakagami Y. Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 2006;142(1):45–53. doi: 10.1104/pp.106.081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gabaldón T, Koonin EV. Functional and evolutionary implications of gene orthology. Nat Rev Genet. 2013;14(5):360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marshall A, Aalen RB, Audenaert D, Beeckman T, Broadley MR, Butenko MA, Cano-Delgado AI, de Vries S, Dresselhaus T, Felix G, et al. Tackling drought stress: receptor-like kinases present new approaches. Plant Cell. 2012;24(6):2262–2278. doi: 10.1105/tpc.112.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dievart A, Perin C, Hirsch J, Bettembourg M, Lanau N, Artus F, Bureau C, Noel N, Droc G, Peyramard M, et al. The phenome analysis of mutant alleles in Leucine-rich repeat receptor-like kinase genes in rice reveals new potential targets for stress tolerant cereals. Plant Sci. 2016;242:240–249. doi: 10.1016/j.plantsci.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 75.Causier B, Ashworth M, Guo W, Davies B. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 2012;158(1):423–438. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baxter A, Mittler R, Suzuki N. ROS as key players in plant stress signalling. J Exp Bot. 2014;65(5):1229–1240. doi: 10.1093/jxb/ert375. [DOI] [PubMed] [Google Scholar]
- 77.Qi J, Song CP, Wang B, Zhou J, Kangasjärvi J, Zhu JK, Gong Z. ROS signaling and stomatal movement in plant responses to drought stress and pathogen attack. J Integr Plant Biol. 2018;60(9):805–826. doi: 10.1111/jipb.12654. [DOI] [PubMed] [Google Scholar]
- 78.Liu Y, He C. A review of redox signaling and the control of MAP kinase pathway in plants. Redox Biol. 2017;11:192–204. doi: 10.1016/j.redox.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sinha AK, Jaggi M, Raghuram B, Tuteja N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal Behav. 2011;6(2):196–203. doi: 10.4161/psb.6.2.14701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen XC, Kim SH, Lee K, Kim KE, Liu XM, Han HJ, Hoang MH, Lee SW, Hong JC, Moon YH, et al. Identification of a C2H2-type zinc finger transcription factor (ZAT10) from Arabidopsis as a substrate of MAP kinase. Plant Cell Rep. 2012;31(4):737–745. doi: 10.1007/s00299-011-1192-x. [DOI] [PubMed] [Google Scholar]
- 81.Liu XM, Nguyen XC, Kim KE, Han HJ, Yoo J, Lee K, Kim MC, Yun DJ, Chung WS. Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem Biophys Res Commun. 2013;430(3):1054–1059. doi: 10.1016/j.bbrc.2012.12.039. [DOI] [PubMed] [Google Scholar]
- 82.Shen G, Kuppu S, Venkataramani S, Wang J, Yan J, Qiu X, Zhang H. Ankyrin repeat-containing protein 2A is an essential molecular chaperone for peroxisomal membrane-bound ascorbate peroxidase 3 in Arabidopsis. Plant Cell. 2010;22(3):811–831. doi: 10.1105/tpc.109.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu XM, Lin H, Maple J, Bjorkblom B, Alves G, Larsen JP, Moller SG. The Arabidopsis DJ-1a protein confers stress protection through cytosolic SOD activation. J Cell Sci. 2010;123(Pt 10):1644–1651. doi: 10.1242/jcs.063222. [DOI] [PubMed] [Google Scholar]
- 84.Li Z, Huang J, Wang Z, Meng F, Zhang S, Wu X, Zhang Z, Gao Z. Overexpression of arabidopsis nucleotide-binding and leucine-rich repeat genes RPS2 and RPM1(D505V) confers broad-spectrum disease resistance in rice. Front Plant Sci. 2019;10:417. doi: 10.3389/fpls.2019.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hell R, Wirtz M. Molecular biology, biochemistry and cellular physiology of cysteine metabolism in Arabidopsis thaliana. Arabidopsis Book. 2011;9:e0154. doi: 10.1199/tab.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shu K, Yang W. E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 2017;58(9):1461–1476. doi: 10.1093/pcp/pcx071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Park JJ, Yi J, Yoon J, Cho LH, Ping J, Jeong HJ, Cho SK, Kim WT, An G. OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. Plant J. 2011;65(2):194–205. doi: 10.1111/j.1365-313X.2010.04416.x. [DOI] [PubMed] [Google Scholar]
- 88.Liu YC, Wu YR, Huang XH, Sun J, Xie Q. AtPUB19, a U-box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol Plant. 2011;4(6):938–946. doi: 10.1093/mp/ssr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bemer M, van Dijk ADJ, Immink RGH, Angenent GC. Cross-family transcription factor interactions: An additional layer of gene regulation. Trends Plant Sci. 2017;22(1):66–80. doi: 10.1016/j.tplants.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 90.Agarwal P, Jha B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol Plant. 2010;54(2):201–212. doi: 10.1007/s10535-010-0038-7. [DOI] [Google Scholar]
- 91.Lata C, Yadav A, Prasad M. Abiotic stress response in plants-physiological, biochemical and genetic perspectives. Rijeka: InTech; 2011. Role of plant transcription factors in abiotic stress tolerance; pp. 269–296. [Google Scholar]
- 92.Häusler RE, Hirsch HJ, Kreuzaler F, Peterhänsel C. Overexpression of C4-cycle enzymes in transgenic C3 plants: a biotechnological approach to improve C3-photosynthesis. J Exp Bot. 2002;53(369):591–607. doi: 10.1093/jexbot/53.369.591. [DOI] [PubMed] [Google Scholar]
- 93.Doubnerova Hyskova V, Miedzinska L, Dobra J, Vankova R, Ryslava H. Phosphoenolpyruvate carboxylase, NADP-malic enzyme, and pyruvate, phosphate dikinase are involved in the acclimation of Nicotiana tabacum L. to drought stress. J Plant Physiol. 2014;171(5):19–25. doi: 10.1016/j.jplph.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 94.Laporte MM, Shen B, Tarczynski MC. Engineering for drought avoidance: expression of maize NADP-malic enzyme in tobacco results in altered stomatal function. J Exp Bot. 2002;53(369):699–705. doi: 10.1093/jexbot/53.369.699. [DOI] [PubMed] [Google Scholar]
- 95.Cummins I, Dixon DP, Freitag-Pohl S, Skipsey M, Edwards R. Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metab Rev. 2011;43(2):266–280. doi: 10.3109/03602532.2011.552910. [DOI] [PubMed] [Google Scholar]
- 96.Kumar S, Trivedi PK. Glutathione S-transferases: role in combating abiotic stresses including arsenic detoxification in plants. Front Plant Sci. 2018;9(751):751. doi: 10.3389/fpls.2018.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dixon DP, Lapthorn A, Edwards R. Plant glutathione transferases. Genome Biol. 2002;3(3):REVIEWS3004. doi: 10.1186/gb-2002-3-3-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci U S A. 2007;104(49):19631–19636. doi: 10.1073/pnas.0709453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Farrant JM. Plant desiccation tolerance. 2007. Mechanisms of desiccation tolerance in angiosperm resurrection plants; pp. 51–90. [Google Scholar]
- 100.Zhang X, Liu S, Takano T. Overexpression of a mitochondrial ATP synthase small subunit gene (AtMtATP6) confers tolerance to several abiotic stresses in Saccharomyces cerevisiae and Arabidopsis thaliana. Biotechnol Lett. 2008;30(7):1289–1294. doi: 10.1007/s10529-008-9685-6. [DOI] [PubMed] [Google Scholar]
- 101.Jarzyniak KM, Jasinski M. Membrane transporters and drought resistance - a complex issue. Front Plant Sci. 2014;5:687. doi: 10.3389/fpls.2014.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pandey JK, Dash SK, Biswal B. Loss in photosynthesis during senescence is accompanied by an increase in the activity of beta-galactosidase in leaves of Arabidopsis thaliana: modulation of the enzyme activity by water stress. Protoplasma. 2017;254(4):1651–1659. doi: 10.1007/s00709-016-1061-0. [DOI] [PubMed] [Google Scholar]
- 103.Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25(12):1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- 104.Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006;18(5):1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124(4):1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lata C, Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot. 2011;62(14):4731–4748. doi: 10.1093/jxb/err210. [DOI] [PubMed] [Google Scholar]
- 107.Checker VG, Khurana P. Molecular and functional characterization of mulberry EST encoding remorin (MiREM) involved in abiotic stress. Plant Cell Rep. 2013;32(11):1729–1741. doi: 10.1007/s00299-013-1483-5. [DOI] [PubMed] [Google Scholar]
- 108.Zhao MR, Li F, Fang Y, Gao Q, Wang W. Expansin-regulated cell elongation is involved in the drought tolerance in wheat. Protoplasma. 2011;248(2):313–323. doi: 10.1007/s00709-010-0172-2. [DOI] [PubMed] [Google Scholar]
- 109.Gechev TS, Dinakar C, Benina M, Toneva V, Bartels D. Molecular mechanisms of desiccation tolerance in resurrection plants. Cell Mol Life Sci. 2012;69(19):3175–3186. doi: 10.1007/s00018-012-1088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen X, Goodwin SM, Boroff VL, Liu X, Jenks MA. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and WAX production. Plant Cell. 2003;15(5):1170–1185. doi: 10.1105/tpc.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jager K, Fabian A, Eitel G, Szabo L, Deak C, Barnabas B, Papp I. A morpho-physiological approach differentiates bread wheat cultivars of contrasting tolerance under cyclic water stress. J Plant Physiol. 2014;171(14):1256–1266. doi: 10.1016/j.jplph.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 112.Pan Y, Li J, Jiao L, Li C, Zhu D, Yu J. A non-specific Setaria italica lipid transfer protein gene plays a critical role under abiotic stress. Front Plant Sci. 2016;7(1752):1752. doi: 10.3389/fpls.2016.01752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen YS, Qiu XB. Ubiquitin at the crossroad of cell death and survival. Chin J Cancer. 2013;32(12):640–647. doi: 10.5732/cjc.012.10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Love AJ, Milner JJ, Sadanandom A. Timing is everything: regulatory overlap in plant cell death. Trends Plant Sci. 2008;13(11):589–595. doi: 10.1016/j.tplants.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 115.Rogers HJ. Cell death and organ development in plants. Curr Top Dev Biol. 2005;71:225–261. doi: 10.1016/S0070-2153(05)71007-3. [DOI] [PubMed] [Google Scholar]
- 116.Yoda H, Hiroi Y, Sano H. Polyamine oxidase is one of the key elements for oxidative burst to induce programmed cell death in tobacco cultured cells. Plant Physiol. 2006;142(1):193–206. doi: 10.1104/pp.106.080515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Malinovsky FG, Brodersen P, Fiil BK, McKinney LV, Thorgrimsen S, Beck M, Nielsen HB, Pietra S, Zipfel C, Robatzek S, et al. Lazarus1, a DUF300 protein, contributes to programmed cell death associated with Arabidopsis acd11 and the hypersensitive response. PLoS One. 2010;5(9):e12586. doi: 10.1371/journal.pone.0012586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Epple P, Mack AA, Morris VR, Dangl JL. Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc Natl Acad Sci U S A. 2003;100(11):6831–6836. doi: 10.1073/pnas.1130421100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL. A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell. 1997;88(5):685–694. doi: 10.1016/S0092-8674(00)81911-X. [DOI] [PubMed] [Google Scholar]
- 120.Wituszynska W, Slesak I, Vanderauwera S, Szechynska-Hebda M, Kornas A, Van Der Kelen K, Muhlenbock P, Karpinska B, Mackowski S, Van Breusegem F, et al. Lesion simulating disease1, enhanced disease susceptibility1, and phytoalexin deficient4 conditionally regulate cellular signaling homeostasis, photosynthesis, water use efficiency, and seed yield in Arabidopsis. Plant Physiol. 2013;161(4):1795–1805. doi: 10.1104/pp.112.208116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Szechynska-Hebda M, Czarnocka W, Hebda M, Bernacki MJ, Karpinski S. PAD4, LSD1 and EDS1 regulate drought tolerance, plant biomass production, and cell wall properties. Plant Cell Rep. 2016;35(3):527–539. doi: 10.1007/s00299-015-1901-y. [DOI] [PubMed] [Google Scholar]
- 122.Czolpinska M, Rurek M. Plant glycine-rich proteins in stress response: An emerging, still prospective story. Front Plant Sci. 2018;9:302. doi: 10.3389/fpls.2018.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang DH, Kwak KJ, Kim MK, Park SJ, Yang KY, Kang H. Expression of Arabidopsis glycine-rich RNA-binding protein AtGRP2 or AtGRP7 improves grain yield of rice (Oryza sativa) under drought stress conditions. Plant Sci. 2014;214:106–112. doi: 10.1016/j.plantsci.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 124.Mangeon A, Magioli C, Menezes-Salgueiro AD, Cardeal V, de Oliveira C, Galvao VC, Margis R, Engler G, Sachetto-Martins G. AtGRP5, a vacuole-located glycine-rich protein involved in cell elongation. Planta. 2009;230(2):253–265. doi: 10.1007/s00425-009-0940-4. [DOI] [PubMed] [Google Scholar]
- 125.Carver B, Whitmore W, Smith E, Bona L. Registration of four aluminum-tolerant winter wheat germplasms and two susceptible near-isolines. Crop Sci. 1993;33(5):1113–1114. doi: 10.2135/cropsci1993.0011183X003300050060x. [DOI] [Google Scholar]
- 126.Yuan C, Li C, Yan L, Jackson AO, Liu Z, Han C, Yu J, Li D. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One. 2011;6(10):e26468. doi: 10.1371/journal.pone.0026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cheuk A, Houde M. A rapid and efficient method for uniform gene expression using the barley stripe mosaic virus. Plant Methods. 2017;13(1):24. doi: 10.1186/s13007-017-0175-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xiao FH, Xue GP. Analysis of the promoter activity of late embryogenesis abundant protein genes in barley seedlings under conditions of water deficit. Plant Cell Rep. 2001;20(7):667–673. doi: 10.1007/s002990100384. [DOI] [Google Scholar]
- 129.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. [Google Scholar]
- 130.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goecks J, Nekrutenko A, Taylor J, Galaxy T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F. Manke T: deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Clavijo BJ, Venturini L, Schudoma C, Accinelli GG, Kaithakottil G, Wright J, Borrill P, Kettleborough G, Heavens D, Chapman H, et al. An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 2017;27(5):885–896. doi: 10.1101/gr.217117.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 137.Jambunathan N. Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Methods Mol Biol. 2010;639:292–298. doi: 10.1007/978-1-60761-702-0_18. [DOI] [PubMed] [Google Scholar]
- 138.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44(1):276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 139.Nakano Y, Asada K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981;22(5):867–880. [Google Scholar]
- 140.Aebi H. Catalase. In: Bergmeyer HU, editor. Methods of enzymatic analysis. 2nd ed: Academic Press, Cambridge, USA; 1974. p. 673–84.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Silencing of TaZFP1B affects relative expression of the closest TaZFP1B relatives. The different types of wheat plants (see Fig. 1) were grown for 14 days then were either well-watered for an additional 7 days or drought-stressed by withholding water for 7 days, and expression levels were determined by qRT-PCR. Data are the mean expression ± SD of four biological replicates. Different letters indicate statistically significant differences between samples (P < 0.05 by Tukey’s test).
Additional file 2: Figure S2. Chlorophyll autofluorescence from wheat leaves. The different types of wheat plants (see Fig. 1) were grown for 14 days then were either well-watered for an additional 10 or 14 days (top leaf in the panels) or drought-stressed by withholding water for 10 or 14 days (bottom leaf in the panels). Fluorescence was captured using a NightOWL II imaging cabinet.
Additional file 3: Table S1. List of genes up-regulated by TaZFP1B overexpression.
Additional file 4: Figure S3. Validation of RNA-Seq data by qRT-PCR. The different types of wheat plants (see Fig. 1) were grown for 14 days then were either well-watered for an additional 7 days or drought-stressed by withholding water for 7 days. Expression levels are relative to the well-watered wild-type group. Numbers refer to the corresponding genes in Tables 2, 3, 4 and 5. Data are mean expression ± SD of four biological replicates. Different letters indicate statistically significant differences between samples (P < 0.05 by Tukey’s test).
Additional file 5: Table S2. List of genes down-regulated by TaZFP1B overexpression.
Additional file 6: Table S3. Primers used in this study.
Additional file 7: Figure S4 Heatmap of correlations between RNA-seq libraries. The different types of wheat plants (see Fig. 1) were grown for 14 days then were either well-watered for an additional 7 days or drought-stressed by withholding water for 7 days. RNA-Seq libraries were prepared and paired-end sequencing was performed. Each read generated from paired-end sequencing was analyzed individually. The hierarchical clustering was generated using Spearman correlation coefficient. The color scale indicates the degree of correlation.
Additional file 8: Figure S5. Pearson correlation between the RNA-seq and qRT-PCR data. The qRT-PCR log2 value of the expression ratio (drought-treated wild-type/drought-treated 1B-OEX) (y-axis) was plotted from the RNA-seq log2 value of expression ratio (drought-treated wild-type/drought-treated 1B-OEX) (x-axis). Genes used to calculate the correlation are listed in supplementary Table 1. All qRT-PCR data were collected from three biological replicates. --- represents the 95% confidence interval. The calculated correlation value (R2) is shown along with the regression line.
Data Availability Statement
The RNA-Seq datasets generated and/or analysed during the current study are available in the GEO repository under the accession number GSE136683, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136683. The TaZFP1B sequence is available at GenBank under accession number MN577972 (https://www.ncbi.nlm.nih.gov/nuccore/MN577972).