Abstract
Background
Auxin may have a positive effect on plants under drought stress. White clover is widely cultivated and often prone to water shortages. In the present study, we investigated the effects of exogenous indole − 3-acetic acid (IAA) on growth and physiological changes of white clover under drought stress condition. The contents of endogenous IAA and other hormones including ABA, CTK, JA, GA, IAA, and SA were assayed. Moreover, expressions of auxin-responsive genes, drought-responsive genes and leaf senescence-associated genes were detected in response to exogenous IAA.
Results
Compared to control, drought stress alone significantly diminished stem dry weigh, relative water content (RWC) and total chlorophyll content (Chl). Exogenous IAA treatment significantly increased RWC and Chl, whereas L-AOPP treatment drastically decreased stem dry weight, RWC and Chl under drought stress condition. Additionally, exogenous IAA treatment significantly increased ABA content and JA content, up-regulated expression of auxin responsive genes (GH3.1, GH3.9, IAA8), drought stress responsive genes (bZIP11, DREB2, MYB14, MYB48, WRKY2, WRKY56, WRKY108715 and RD22), and down-regulated expressions of auxin-responding genes (GH3.3, GH3.6, IAA27) and leaf senescence genes (SAG101 and SAG102) in the presence of PEG. Contrarily, L-AOPP treatment significantly reduced contents of ABA, GA3 and JA, down-regulated expressions of GH3.1, GH3.9, IAA8, bZIP11, DREB2, MYB14, MYB48, WRKY2, WRKY56, WRKY108715, ERD and RD22, and up-regulated SAG101 and SAG102.
Conclusions
Exogenous IAA improved drought tolerance of white clover possibly due to endogenous plant hormone concentration changes and modulation of genes involving in drought stress response and leaf senescence. These results provided useful information to understand mechanisms of IAA improved drought tolerance in white clover.
Keywords: Drought stress, IAA, Phytohormone, Auxin, Transcription factor, Drought responsive gene, Senescence-associated gene
Background
Water is a vital component for plants as it allows plants to survive [1]. Drought stress adversely impacts plant growth and has attracted researchers’ attention due to its effects on a variety of physiological and metabolic mechanisms in plants [2–4]. It has been estimated that up to 30% of the plants in the world are subjected to varying degrees of drought (https://www.un.org/en/events/desertification_decade/whynow.shtml). To maintain growth and development, plants adjust their morphological and physiological characteristics in response to drought, which improve plants’ ability to withstand water deprivation in arid environments [5–8]. For example, plants accumulate low-molecular-weight osmolytes to recruit potassium and other nutrient ions, and increase root elongation to enhance water uptake. Additionally, phytohormones, such as auxins (IAA), abscisic acid (ABA), cytokinin (CTK), salicylic acid (SA), gibberellin (GA) and jasmonic acid (JA), could modulate the plant tolerance to drought stress. Studies approved the alleviation of drought stress in plants after application of hormones [9–11], however, the changes of endogenous hormones were not illustrated. Among the phytohormones mentioned above, IAA regulates many processes during plant growth and development [7, 12]. Recently, accumulating evidences indicate the possible link between IAA and other hormones [13–15], indicating the cross-talks among phytohormones might play key roles during plant stress response.
To date, some drought-responsive genes have been identified, such as Early Responsive to Dehydration (ERD) that are rapidly activated by drought stress [16, 17] and Senescence-Associated Genes (SAG) that regulate chlorophyll degradation and cytoplasmic destruction [18]. In Arabidopsis, ERD1 encodes a protein, which is induced by dehydration and does not respond to ABA [19]. ERD10 and ERD14 are strongly induced in Arabidopsis by dehydration and ABA, but not by 2, 4-D and GA [16, 20]. However, the effect of IAA on the expression of ERD is unknown. The expression of SAG13 gene in Arabidopsis increases during senescence [21]. Therefore, the expression of SAG gene could be an indicator of plant senescence. Plant hormones are involved in the regulation of plant senescence. Therefore, the application of IAA possibly modulates the expression of SAG gene and needs to be studied.
In addition, the expression of plant transcription factor (TFs) genes, including basic region/leucine zipper (bZIP) motif, dehydration response element-binding (DREB), myeloblastosis (MYB), and WRKYs were also modulated after drought stress treatment [22–25]. Overexpression of OsbZIP72 and OsbZIP46 significantly increased the drought resistance of rice by elevating the expression of ABA response gene [26, 27]. DREB2 has been reported to play a crucial role in enhancing the abiotic stress tolerance of plants by interacting with a cis-element present in the promoter region of various abiotic stress-responsive genes [24]. It has been reported that PbrMYB21 could promote drought tolerance of tobacco due to the modulation of polyamine synthesis by regulating the arginine decarboxylase expression [28]. The up-regulated OsMYB48–1 promoted drought tolerance of rice by means of regulating the expression of OsNCED4 and OsNCED5 (ABA biosynthesis genes), OSRK1 and OsPP2C68 (early signaling genes) and some late responsive genes such as OsLEA3 and RAB21 [29]. Overexpression of ZmWRKY58, which interacted with ZmCaM2, enhanced the drought tolerance in transgenic rice [30]. Both TaWRKY1 and TaWRKY33 activated several stress-related genes and promoted root growth in Arabidopsis under various stresses, and increased the drought resistance in Arabidopsis [25]. In previous studies, auxin also regulated gene expression of auxin responding genes [31–33] and Auxin-Response transcription Factor (ARF) family mediated the roles of IAA during plant growth [34]. Also, there is very little evidence available to support that auxin regulates the expresion of ARFas well as expressions of TFs genes.
White clover (Trifolium repens) is one of the essential forages. It is widely cultivated and grazed to many animals in the world due to its high yield and quality. However, it is susceptible to drought stress and shows significant changes in dehydration during drought [35]. In addition, the effects of IAA on white clover under drought stress are not fully understood. The objects of the research are to reveal: (1) whether IAA has crosstalk with other plant hormones in white clover under drought stress condition; (2) how could IAA activate auxin signal transduction pathway and modulate expressions of drought responsive genes.
Results
Effects of exogenous IAA on drought tolerance of white clover
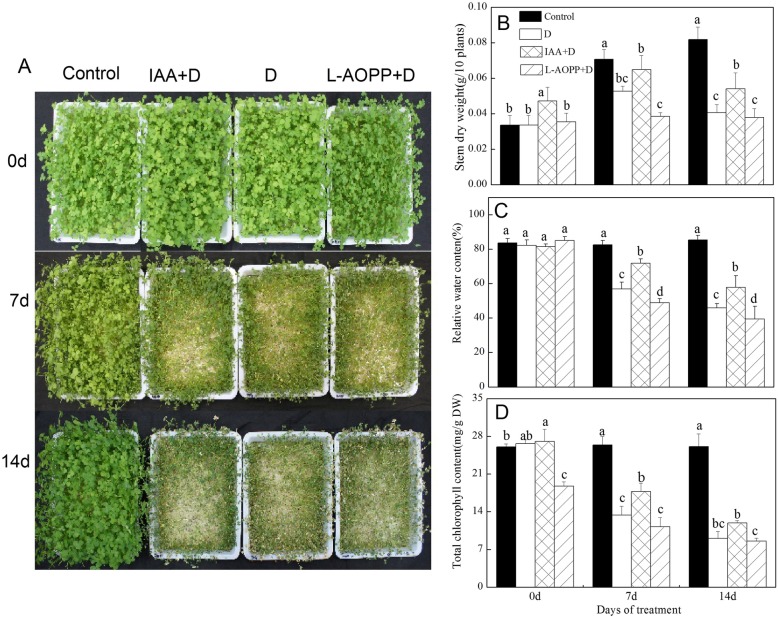
The morphological appearance of white clover was shown in Fig. 1a. On 0 d (no PEG stress), exogenous IAA significantly increased stem dry weight and Chl (Fig. 1b-d) and had no effect on RWC. L-AOPP significantly reduced Chl (Fig. 1d). On 7 d and 14 d, in all PEG sets, IAA increased stem dry weight, RWC and Chl, while L-AOPP decreased these indicators (Fig. 1b-d). Meanwhile, our results showed that RWC and Chl of white clover in control remained stable.
Fig. 1.
Morphological appearance (a), Stem dry weight (b), relative water content (c) and total chlorophyll content (d) of white clover in different sets. Vertical columns represent Mean + STD (n = 4). The same letter indicates no significant difference and the different letters indicate a significant difference (Fisher’s LSD, P < 0.05) in a pairwise comparison between sets at the same time
Content of ABA, CTK (iPAs and ZRs), GA3, JA and SA
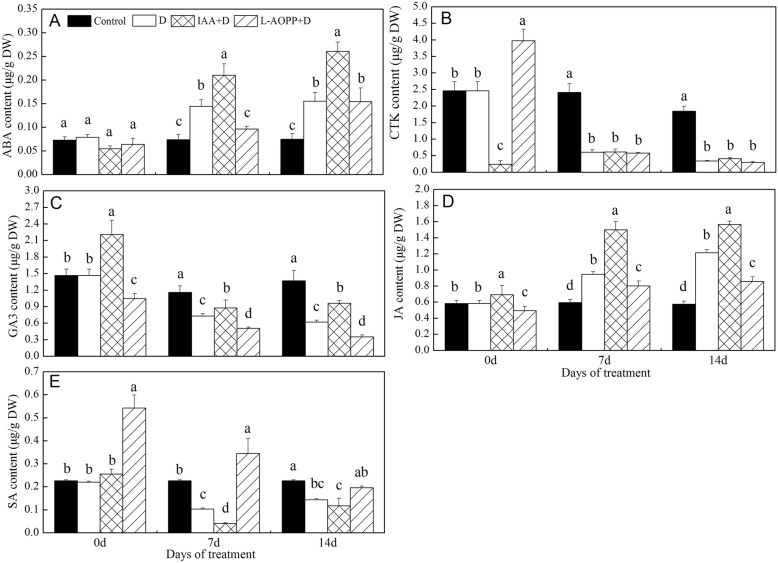
On day 0, the IAA reduced CTK by 90.2% and increased GA3 and JA by 45.2 and 18.4%, respectively (Fig. 2b-d). Nevertheless, it has no effect on ABA and SA content (Fig. 2a, e). However, L-AOPP increased CTK by 61.4% and SA by 130% (Fig. 2b, e), and reduced GA3 and JA contents by 28.8 and 13.8% (Fig. 2c, d). Meanwhile, it does not affect ABA content (Fig. 2a).
Fig. 2.
Content of other major phytohormones of white clover leaves in different sets. ABA content (a), CTK content (b), GA content (c), JA content (d) and SA content (e). Vertical columns represent Mean+/−std (n = 4). The same letter indicates no significant a difference and the different letters indicate significant difference (Fisher’s LSD, P < 0.05) in a pairwise comparison between sets at the same time
On days 7 and 14 (PEG stress), in all PEG treatments, all ABA, GA3, and JA in IAA pre-treatment were significantly higher than those in other treatment groups (Fig. 2a, c, d), while the SA content was significantly lower than that in other groups (Fig. 2e). In contrast, L-AOPP pre-treatment reduced the content of ABA, GA3 and JA (Fig. 2a, c, d) and increased the content of SA (Fig. 2e). Interestingly, CTK is not affected by IAA and L-AOPP.
Endogenous IAA content and relative the expression of auxin-responsive genes
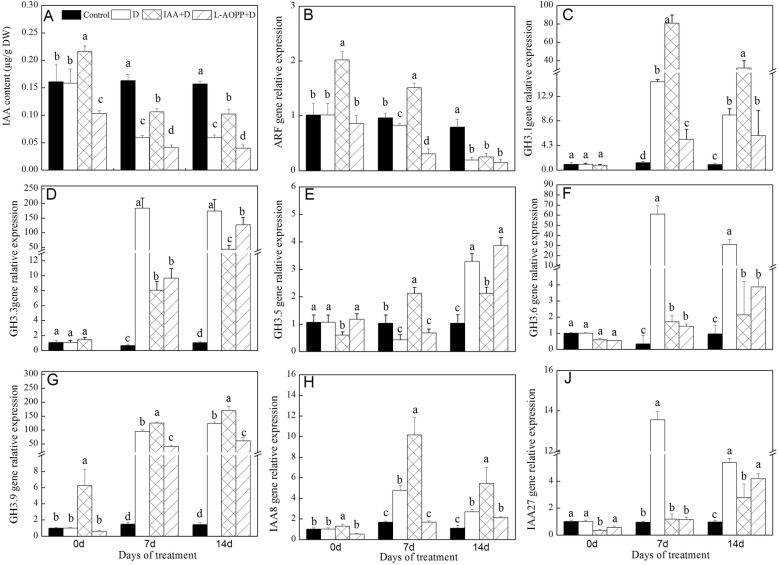
Clearly, the endogenous IAA content in control remained stable at all sampling times (Fig. 3a). On day 0, exogenous IAA increased the endogenous IAA content of white clover, while L-AOPP decreased it (Fig. 3a). On days 7 and 14, the content of endogenous IAA decreased significantly in groups treated with PEG (Fig. 3a), the content of endogenous IAA increased in the IAA pre-treatment group, while decreased in the L-AOPP group (Fig. 3a).
Fig. 3.
Endogenous IAA content (a) and relative expression of auxin response genes of white clover in different sets. ARF (b), GH3.1 (c), GH3.3 (d), GH3.5 (e), GH3.6 (f), GH3.9 (g), IAA8 (h), IAA27 (j) and Vertical columns represent Mean + STD (n = 4). The same letter indicates no significant difference and the different letters indicate a significant difference (Fisher’s LSD, P < 0.05) in a pairwise comparison between sets at the same time
On day 0, exogenous IAA improved expression of ARF, GH3.9 and IAA8 (Fig. 3b, g, h), and decreased the expression of GH3.5 and IAA27 (Fig. 3e, j), but did not affect the expression of GH3.1, GH3.3, and GH3.6 (Fig. 3c, d, f). L-AOPP inhibited the expression of GH3.1 and GH3.3 (Fig. 3c, d), but did not affect the expression of other genes.
On days 7 and 14, in all PEG treatments, IAA significantly increased the expression of GH3.1, GH3.9 and IAA8 (Fig. 3c, g, h), and decreased the expression of GH3.3, GH3.6, and IAA27 (Fig. 3d, f, j). However, L-AOPP significantly down-regulated the expression of all genes except for GH3.5 (Fig. 3b-j).
The expression of transcription factor (TF) genes responding to drought
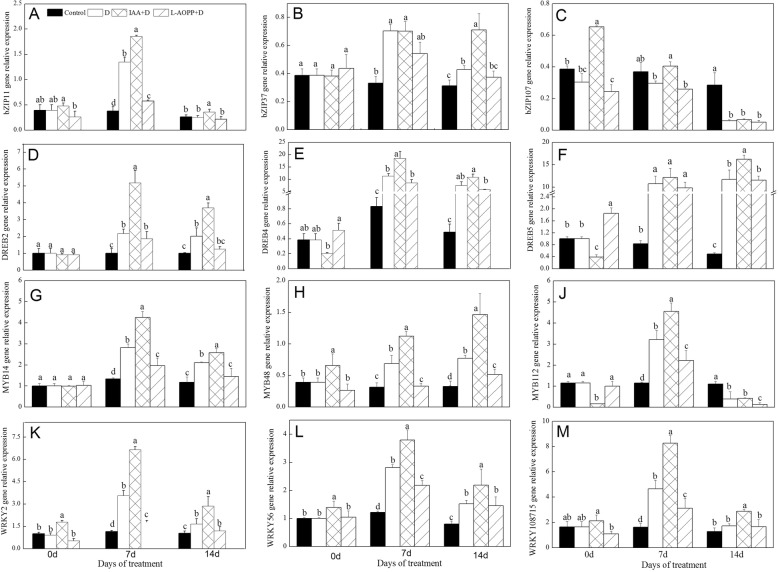
In this experiment, we selected 3 genes from the bZIP, DREB, MYB, and WRKY transcription factor families, respectively. On day 0, IAA significantly up-regulated the expression of bZIP107, MYB48, WRKY2 and WRKY56 (Fig. 4c, h, k, l), and down-regulated the expression of DREB5 and MYB112 (Fig. 4f, j), but did not affect the expression of bZIP11, bZIP37, DREB2, DREB4, MYB14 and WRKY108715. L-AOPP reduced the expression of bZIP107 (Fig. 4c) and increased the expression of DREB5 (Fig. 4f).
Fig. 4.
Expression of drought-induced transcriptional factors of white clover leaves in different treatments. bZIP11 (a), bZIP 37 (b), bZIP 107 (c), DREB2 (d), DREB4 (e), DREB5 (f), MYB14 (g), MYB48 (H), MYB112 (j), WRKY2 (k), WRKY56 (l) and WRKY108715 (m). Vertical columns represent Mean + STD (n = 4). The same letter indicates no significant difference and the different letters indicate a significant difference (Fisher’s LSD, P < 0.05) in a pairwise comparison between sets at the same time
In all PEG treatments, on days 7 and 14, IAA up-regulated the expression of all transcription factor genes in most cases, while L-AOPP reduced the expression of these genes except bZIP37 and bZIP107 (Fig. 4b, c).
Expression of drought-response genes and senescence-associated genes
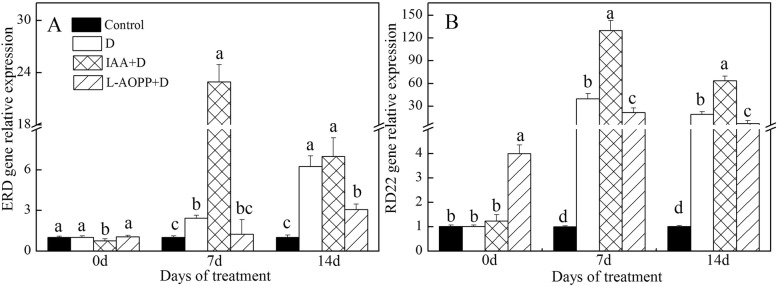
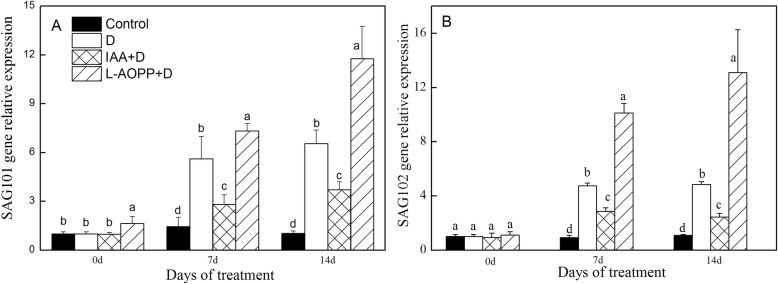
On day 0, exogenous IAA decreased the ERD expression (Fig. 5a), and L-AOPP improved the expression of RD22 and SAG101 (Figs. 5b, 6a). Both IAA and L-AOPP had no effects on the expression of SAG102.
Fig. 5.
Expression of drought-induced genes of white clover leaves in different treatments. ERD (a) and RD22 (b). Vertical columns represent Mean + STD (n = 4). The same letter indicates no significant difference and the different letters indicate a significant difference (Fisher’s LSD, P < 0.05) in a pairwise comparison between sets at the same time
Fig. 6.
Expression of senescence-associated genes of white clover leaves in different treatments. SAG101 (a) and SAG102 (b). Vertical columns represent Mean + STD (n = 4). The same letter indicates no significant difference and the different letters indicate a significant difference (Fisher’s LSD, P < 0.05) in a pairwise comparison between sets at the same time
On days 7 and 14, in all PEG treatments, IAA significantly enhanced the expression of ERD and RD22 (Fig. 5a, b), and significantly decreased the expression of SAG101 and SAG102 (Fig. 6a, b). L-AOPP, however, significantly decreased the expression of ERD and RD22 (Fig. 5a, b), and significantly increased the expression of SAG101 and SAG102.
Discussion
Effects of IAA on white clover under no PEG stress
In the absence of PEG stress, IAA significantly increased stem dry weight of white clover, indicating that IAA pre-treatment caused white clover to accumulate more organic matter during the same growth time. At this time, the chlorophyll content of white clover leaves also increased significantly. Subsequently, increased content of chlorophyll contributed to an increase in photosynthetic rate.
As far as IAA and GA were concerned, some researchers proved that an average level of bioactive GA1 required normal level of IAA in elongating pea stems [36], and IAA promoted GA1 synthesis [37]. Here, our results showed that exogenous IAA increased GA3 (Fig. 2c). This probably because of IAA’s activation in enzymes related to GA3 syntheses, like IAA’s promotion on GA1 synthesis [37].
In zinnia, the transcript level of IAA8 was induced by auxin [38]. Here, we found that exogenous IAA up-regulated the expression of IAA8 in white clover. Therefore, it may be true that IAA is essential for the expression of IAA8 gene. IAA8 was found to play a crucial role in floral organ development and abnormal formation of floral in IAA8-mutation Arabidopsis could be changed through JA application, meaning that a strong link between IAA8 and JA [39]. Fortunately, exogenous IAA significantly increased the expression of IAA8 and JA levels in our findings. For the ARF gene, IAA significantly increased the expression of it in the present study. One study also found that auxin treatment could affect the transcript abundance of several OsARF genes, and these ARF genes might play crucial roles in varied metabolic pathways and some cellular processes in rice [40].
Several studies also showed that the high expression level of ERD and RD22 subserved plant resistance to drought [41, 42]. ERD genes are a group of genes that are rapidly induced (in 1 h) under stress [43]. Our results showed that IAA significantly reduced the expression of ERD gene. It may be inferred that the ERD gene was not necessary for plants suffering no PEG stress, and IAA also inhibited the expression of it.
Improved growth and physiologies in white clover under PEG stress
Under PEG stress, the results showed that exogenous IAA mitigated plant dehydration, and L-AOPP worsened it (Fig. 1a). IAA improved stem dry weight, relative water content, and total chlorophyll content in leaves, however, L-AOPP decreased all of them (Fig. 1b-d). Studies have shown that IAA is related to drought tolerance in plants, and wild Arabidopsis plants pre-treated with IAA exhibited enhanced drought resistance [44]. The application of IAA could ease the adverse effects brought by PEG stress and enhanced barley growth [45]. IAA conferred white clover with the better morphological and physiological state in the IAA + D group than that in the D group (Fig. 1), suggesting that IAA had a positive effect in improving drought tolerance of white clover.
Changes in Phytohormones and subsequent effects in white clover under PEG stress
Transcriptome data revealed that an increase of ABA content activated expression of many drought-resistant genes [46]. ABA regulated downstream response of RD29B (dehydration stress gene) by regulating the bZIP gene [47]. In our studies, we also found that there was a consistent correlation between ABA and expression of RD22 under PEG stress (Figs. 2a, 5b), suggesting that ABA also probably regulated expression of RD22 gene in white clover and increased drought resistance.
It has been found that the interaction between IAA and ABA promoted the development of lateral roots in plants, and this pattern of root growth regulation was necessary for plants to respond to severe drought stress [48]. Besides, exogenous ABA enhanced the recovery of photosynthetic rate in upland rice under PEG stress [49]. Based on these experimental results and Figs. 1 and 2 A, we could speculate that an increase in ABA content might enhance drought resistance through multiple ways, such as improved RD22 expression, higher total chlorophyll, and more stem dry weight. Moreover, the opposite effects L-AOPP on these indicators further confirmed that these changes arose from IAA.
GA3 application reduced cell permeability and electrolyte leakage under drought stress [50]. Therefore, increased content of GA3 could enhance drought resistance in plants. In the present study, the results showed that IAA significantly increased the content of GA3 in white clover under PEG stress, and improved its resistance to PEG stress.
And the strong interaction between JA and ABA was observed [51]. Some researchers have shown that JA was upstream of ABA biosynthesis, and the accumulation of JA at an early stage led to an accumulation of jasmonic acid isoleucine, which was one necessary condition for ABA synthesis under drought stress [52]. Here, our results showed that exogenous IAA increased the content of both JA (Fig. 2d) and ABA (Fig. 2a). And we could conclude that IAA might regulate ABA synthesis via JA in white clover.
In summary, we believed that plant hormones had reached a new homeostasis after applying exogenous IAA under PEG stress. Changes in these plant hormones may promote plant drought resistance through specific signal transduction and gene regulatory pathways.
Expressions of genes responding to IAA and TF genes under PEG stress
Transcriptome data showed that rice AUX/IAA genes were induced by exogenous IAA and drought [53]. AUX/IAA1 in Sorghum was also up-regulated by drought [54]. In this experiment, PEG stress significantly increased the expression of IAA8 and IAA27. Interestingly, IAA significantly increased the expression of IAA8 rather than IAA27. It was found that IAA8 was involved in lateral root formation in Arabidopsis [55]. White clover under PEG stress would instinctively improve the expression of IAA8 to form more lateral root to get more water, and IAA may enhance its expression. Some researchers revealed that the Sl-IAA27 gene was down-regulated by auxin [56]. Our result of IAA27 was in line with this finding. Tomato transgenic plants with under-expression of the Sl-IAA27 gene showed multiple phenotypes interrelated to vegetative growth. Here, the down-regulation of IAA27 may have multiple effects on growth and root development in white clover.
GH3 family genes were also involved in plants responding to biotic and abiotic stress. Our studies showed that expression of GH3.1, GH3.3, GH3.6, and GH3.9 were induced by drought stress (Fig. 3c, d, f, and g), denoting that these GH family genes could respond to drought stress. Besides, exogenous IAA also prompted expressions of GH3.1 and GH3.9 genes (Fig. 3c and g), showing that these two genes may have a relation to endogenous IAA content. Arabidopsis thaliana seedlings pre-treated with IAA showed an improved drought tolerance, and a variety of expressions of GH genes related to stress were regulated by exogenous IAA [57]. It was found that decreased endogenous IAA content in rice mutants accompanied by a deficiency in carotenoid and transgenic plants over-expressing OsGH3.2 showed the sensitivity to drought [22]. Activation of OsGH3.13 enhanced drought resistance in Rice [58]. Exogenous IAA activated responsive gene GH3.9 and resulted in the strong drought resistance in plants [59]. These results showed that exogenous IAA could enhance drought resistance in white clover, and GH3.1 and GH3.9 gene was involved in drought tolerance.
For bZIPs, only a few members were identified to play roles in plant growth and development, abiotic stress, and hormone signal transduction, but their potential molecular mechanisms were still unknown and need further exploration [60]. An earlier study has shown that OsbZIP23 in maize is involved in ABA signaling and regulates drought stress [61]. Other researchers found that bZIP11 in Arabidopsis interacted with one adapted proteins via an amino-terminal activation domain to recruit the histone acetylation system to specific auxin-responsive genes [62]. bZIP37 expressed in the salt-stressed plant activating downstream of ABA-induced gene expression [63]. We also found that expression of bZIP11 was also induced by exogenous IAA (Fig. 4a), and that of bZIP37 was induced by PEG-6000 (Fig. 4b).
Also, DREBs (dehydration-responsive element-binding proteins) play essential roles in plant response to drought stress and were found to be activated in ways dependent on ABA [64]. It was shown that exogenous IAA enhanced expression of DREB2 and DREB4, and L-AOPP negatively regulated expression of DREB2 and DREB4 (Fig. 4d, e) in our studies. Another study has shown that DREBs regulating the expression of many downstream genes of drought resistance and over-expression of the DREB gene can enhance drought resistance in plants [65]. Our results showed that improved drought resistance of white clover by exogenous IAA could be associated with the expression of the DREB2 and DREB4.
MYBs are also essential in regulating plant growth, development, metabolism, and stress response, and almost all eukaryotes have MYB transcription factors. The response mechanisms of MYBs in the stress environment are not very clear. Our studies found that both exogenous IAA and drought stress-regulated expression of MYB14 and MYB48 and L-AOPP decreased their expressions (Fig. 4g, h). Xiong found that the over-expression of MYB48–1 promoted biosynthesis of ABA and improved drought resistance of transgenic rice [29]. AtMYB60 regulated stomatal movement and promoted Arabidopsis thaliana to respond to drought stress [66]. Different MYBs showed varied functions in the progress of responding to drought and improved drought resistance.
At present, research of WRKY transcription factors in abiotic stress has been progressed. It was found that WRKYs were involved in plant stress regulatory networks, and WRKY proteins were induced by drought stress [67]. WRKYs also played essential roles in plant drought stress and regulated plant response to abiotic stress through interaction with hormones and protein kinases [68]; however, the molecular mechanisms of their regulations were still limited. WRKY transcription factor ABO3 induced expression of drought resistance genes, such as RD29A and COR47, and improved drought resistance [69]. In terms of WRKY2, WRKY56, and WRKY108715 gene, we found that drought induced their expressions, and exogenous IAA also up-regulated the expression of WRKY family genes in white clover.
Expression of stress gene and senescence-associated gene under PEG stress
ERD and RD22 subserved plant resistance to drought [41, 42], and our results suggested that both PEG stress and exogenous IAA up-regulated the expression of them in white clover, but L-AOPP decreased them. Besides, the expression of ERD11 and ERD13 gene was induced by dehydration, but not influenced by GA, ABA, 6-BA, and 2,4-D [70]. Other studies also found that the RD22 gene was doubled by both ABA and MYB proteins [71].
As far as SAG101 and SAG102 gene was concerned, our study found that PEG stress and L-AOPP increased their expressions, but IAA decreased them. SAG101 in Arabidopsis encoded an Acyl hydrolase involved in leaf senescence [72]. It was found that exogenous IAA inhibited the transcription level of SAG12 [73] and retarded the senescence of leaves. The plant with over-expression of the YUCCA6 gene improved the content of endogenous IAA and hindered senescence of plant by down-regulating expression of SAG12 [74]. Similarly, the decreased expression of SAG101 and SAG102 by IAA could play a role in delaying senescence resulted from PEG stress in white clover.
Conclusion
Above all, this study highlighted the protective role of exogenous IAA during drought tolerance in white clover. Drought responsive plant hormones, such as ABA, JA and GA, transcription factors (bZIP11, DREB2, MYB14, MYB48 and WRKYs,) and related genes (ERD, RD22, SAG101 and SAG102) could play a positive role in alleviating drought stress damage .
Methods
Plant material and growth condition
Seeds of ‘Pixie’ (Trifoliumrepens cv.) were purchased from Beijing Mammoth Seed Industry Company, Beijing, China. The seeds were sterilized with 1% (w/v) sodium hypochlorite solution for 3 min and then rinsed 3 times with sterile water. The seeds were planted in pots (9 cm depth, 18 cm width, 24 cm length) filled with sterilized quartz sand. The pots were placed in a growth chamber (23 °C/16 h during the day and 19 °C/8 h at night, with an irradiance of approximately 300 μmol quanta·m− 2·s− 1, and relative humidity of 75%) and 50 mL deionized water was added daily in each pot. After germination, seedlings were transplanted in Hoagland’s nutrient solution. Positions of the pots were rearranged daily in order to reduce the environmental impact.
Experimental design
The seedlings at the two-leaf stage were divided into four groups: (1) control, Hoagland’s nutrient solution; (2) PEG-6000 treatment (labeled as D), Hoagland’s nutrient solution containing 15% PEG-6000 (W/V,-0.3Mpa); (3) IAA pre-treatment + PEG-6000 treatment (labeled as IAA + D), first pre-treated with Hoagland’s nutrient solution containing 1 μM IAA for 7 days and then added 15% PEG-6000 in Hoagland’s nutrient solution; (4) L-AOPP pre-treatment+PEG-6000 treatment (labeled as L-AOPP+D), first pre-treated with Hoagland nutrient solution containing 100 μML-AOPP for 7 days and then added 15% PEG-6000 in Hoagland’s nutrient solution. Each treatment had four biological replications. Samples were taken at 0 (labeled with 0 d), 7 (labeled as 7 d) and 14 (labeled as 14 d) days after treating with PEG-6000.
Measurement of physiological indicators
Relative water content (RWC), stem dry weight and Total chlorophyll content (Chl)
For RWC, the 0.2 g fresh leaves were taken as fresh weight (FW). Then leaves were placed in distilled water at 4 °C for 24 h and weighed to get saturated weight (SW). Subsequently, samples were dried at 105 °C for 30 min, followed by drying at 75 °C for 48 h and weighed to record dry weight (DW). RWC was calculated by the formula: RWC(%) = (FW-DW)/ (SW-DW) × 100% [75].
The total chlorophyll contents were extracted by incubating ~ 0.1 g fresh leaves with a 10 mL solution of 80% acetone: 95% methanol (1:1, V/V) in the dark until the leaves became colorless. The light absorption values of Chl a and Chl b were measured at 645 nm and 663 nm, respectively. The total chlorophyll contents were calculated according to the formula:
Chl (a + b)(mg/g) = (20.2 × OD645 + 8.02 × OD663)/(DW × 1000) [75].
Quantification of IAA, ABA, CTK (iPAs and ZRs), GA3, JA and SA
HPLC–ESI–MS method (minor modification) [76] was used to quantify the phytohormones. 0.2 g leaves were transferred to a 5 ml screw-cap tube, freeze in liquid nitrogen for 10 min and grounded to powder form using Plant Tissue Breaker. Firstly, 200 μl working solution of internal standards was added to each tube, then 2 mL extraction solvent was added and shook at the rate of 100 rpm for 30 min at 4 °C; subsequently, 2 mL dichloromethane was added to each tube and shook as before. Then samples were centrifuged at the rate of 12,000 rpm for 5 min at 4 °C and transferred ~ 1.8 ml of the solvent from the lower phase into a screw-cap vial followed by concentrating through nitrogen evaporator. For analysis, samples were redissolved with 0.2 mL methanol by injecting 20 μl of the methanol solution into the C18 (reverse-phase) HPLC column. The HPLC–ESI–MS (HPLC system, LC-10 AD series, Shimadzu, Japan; ESI-MS system, ABI 4000 QTRAP) conditions and settings were employed as reported previously [76]. Applied Biosystems Analyst software version 1.5.1 was used to control the MS system and to perform data analysis and data management.
Expression analysis of genes by realtime qRT-PCR
Real-time quantitative PCR (qRT-PCR) was used to test the expression of GH3.1, GH3.3, GH3.5, GH3.6, GH3.9, IAA8, IAA27, and ARF in leaf. Total RNAs of the leaf was extracted through Plant RNA Kits (TIANGEN Biochemical Technology Co., Ltd), and cDNAs used in qRT-PCR were synthesized using iScriptTM cDNA Synthesis Kit (Bio-Rad Laboratories, Inc). 10 μL qRT-PCR reaction system consisted of 1 μL cDNA, 0.5 μL each of upstream and downstream primers, 5 μL SYBR Green SuperMix and 3 μL ddH2O. qRT-PCR program was: pre-denaturation 30 s at 95 °C, denaturation 10 s at 95 °C, annealing 15 s at 59.3 °C (GH3.1), 61 °C (GH3.5), 60 °C (GH3.3, GH3.6, GH3.9, IAA8, IAA27) and 58.9 °C (ARF), extension 5 s at 72 °C, 40 cycles, then last extension 10 min at 72 °C. GAPDH was used to calculate the relative expression level of each concerned gene through the formula of 2-ΔΔCt [77]. Primer sequences of the genes related to IAA and their corresponding GeneBank accession numbers were listed in Additional file 1: Table 1.
10 μL qRT-PCR reaction system was the same as mentioned above. The reaction program was: pre-denaturation 30 s at 95 °C, denaturation 10 s at 95 °C, annealing 15 s at 57.2 °C (bZIP37、bZIP107、MYB48、MYB112), 58 °C (DREB2、DREB4、DREB5 、MYB14), 58.4 °C (WRKY108715), 61 °C (WRKY2、WRKY56、bZIP11), 56.4 °C (RD22) and 59.5 °C (ERD), 55.5 °C (SAG101/SAG102), extension 5 s at 72 °C, 40 cycles, then last extension 10 min at 72 °C. The reference gene also was GAPDH, and calculation was the formula of 2-ΔΔCt [77]. Primer sequences of drought-induced transcription factors and drought-induced genes and their corresponding Gene Bank accession numbers were listed in Additional file 1: Table 2.
Statistic
In this paper, Origin 8.5.1 was employed to generate the histograms, SPSS 19.0 to the analysis of variance (ANOVA) at the 0.05 probability level. Data were transformed to meet normality and homogeneity of variance. Fisher’s LSD was used to determine differences between groups.
Supplementary information
Additional file 1: Table 1 Primer sequences of the genes related to IAA and their corresponding GeneBank accession numbers. Table 2 Primer sequences of drought-induced transcription factors and drought-induced genes and their corresponding GeneBank accession numbers.
Acknowledgments
Not applicable.
Abbreviations
- IAA
Indole-3-acetic acid
- L-AOPP
L-2-aminooxy-3-phenyl propionic acid
- ABA
Abscisic acid
- JA
Jasmonic acid
- CTK
Cytokinin
- GA(3)
Gibberellin(3)
- SA
Salicylic acid
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- FW
Fresh weight
- SW
Saturated weight
- DW
Dry weight
- ERD
Early-response to dehydration
- RD
Response to dehydration
- SAG
Senescence-associated genes
- TF(s)
Transcriptional factor(s)
- bZIP
Basic region/leucine-zipper
- DREB
Dehydration responsive element binding
- MYB
Myeloblastosis
- ARF
Auxin response transcription factor
- PEG-6000
Polyethylene glycol-6000
- RWC
Relative water content
- Chl
Total chlorophyll content
Authors’ contributions
YP and ZLconceived the research and designed the experiments; YL performed the experiments; YZ analyzed the data; YZ wrote the manuscript; YZ, YL, ZL, and YP discussed the results, MJH reviewed the manuscript. All authors have approved the final version of the manuscript.
Funding
This research was supported by Grant No. 2018HH0067 from the International Cooperation Project of Sichuan Province, Provincial Industry Independent Innovation Ability Project in Jilin Province: Construction of Independent Innovation Ability of Applied Eco-engineering Laboratory in Jilin Province by Grant No.2018c002, and Sichuan Forage Innovation Team Program by Grant No.SCCXTD-2020-16. The funding agency had no role in the design of the study, collection, analysis, and interpretation of data, or in writing the manuscript.
Availability of data and materials
The data used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Youzhi Zhang and Yaping Li contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-02354-y.
References
- 1.Blatt MR, Chaumont F, Farquhar G. Focus on water. Plant Physiol. 2014;164:1553–1555. doi: 10.1104/pp.114.900484. [DOI] [Google Scholar]
- 2.Harb A, Krishnan A, Ambavaram MMR, Pereira A. Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 2010;154:1254–1271. doi: 10.1104/pp.110.161752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi H, Ye T, Chan Z. Comparative proteomic responses of two bermudagrass (Cynodon dactylon (L). Pers.) varieties contrasting in drought stress resistance. Plant Physiol Bioch. 2014;82:218–228. doi: 10.1016/j.plaphy.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Zhang Y, Zhang X, Merewitz E, Peng Y, Ma X, et al. Metabolic pathways regulated by chitosan contributing to drought resistance in white clover. J Proteome Res. 2017;16:3039–3052. doi: 10.1021/acs.jproteome.7b00334. [DOI] [PubMed] [Google Scholar]
- 5.Cutler SR, Rodriguez PL, Finkelstein RR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 6.Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- 7.Qin F, Shinozaki K, Yamaguchi-Shinozaki K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011;52:1569–1582. doi: 10.1093/pcp/pcr106. [DOI] [PubMed] [Google Scholar]
- 8.Krasensky J, Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot. 2012;63:1593–1608. doi: 10.1093/jxb/err460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Jia W, Yang J. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 2006;97:111–119. doi: 10.1016/j.fcr.2005.08.018. [DOI] [Google Scholar]
- 10.Li Z, Lu GY, Zhang XK. Improving drought tolerance of germinating seeds by exogenous application of gibberellic acid (GA3) in rapeseed (Brassica napus L.) Seed Sci Technol. 2010;38:432–440. doi: 10.15258/sst.2010.38.2.16. [DOI] [Google Scholar]
- 11.Akter N, Islam MR, Karim MA. Alleviation of drought stress in maize by exogenous application of gibberellic acid and cytokinin. J Crop Bio. 2014;17:41–48. doi: 10.1007/s12892-013-0117-3. [DOI] [Google Scholar]
- 12.Du H, Wu N, Fu J. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J Exp Bot. 2012;63:6467–6480. doi: 10.1093/jxb/ers300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Junichi U, Kensuke M, Miyako A. Jasmonic acid inhibits the IAA-induced elongation of oat coleoptile segments: a possible mechanism involving the metabolism of cell wall polysaccharides. Plant Cell Physiol. 1994;35:1065–1070. [Google Scholar]
- 14.Brian J, Sara AG, Sara VP. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell. 2010;22:2956–2969. doi: 10.1105/tpc.110.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li MS, Yan XF. Jasmonic acid signaling in plants and its biological functions in relation to environment. Acta Ecol Sin. 2014;34:6779–6788. [Google Scholar]
- 16.Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Mol Bio. 1994;25:791–798. doi: 10.1007/BF00028874. [DOI] [PubMed] [Google Scholar]
- 17.Rahma JJ, Mohamed ME. Physiological responses of transgenic tobacco plants expressing the dehydration responsive RD22 gene of Vitis vinifera to salt stress. Turk J Bot. 2014;38:268–280. doi: 10.3906/bot-1301-53. [DOI] [Google Scholar]
- 18.Gan S, Amasino RM. Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence) Plant Physio. 1997;113:313. doi: 10.1104/pp.113.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of a cDNA for a dehydration-inducible gene that encodes a Clp a, B-like protein in Arabidopsis thaliana L. Biochem Biophys Res Commun. 1993;196:1214–1220. doi: 10.1006/bbrc.1993.2381. [DOI] [PubMed] [Google Scholar]
- 20.Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in Arabidopsis thaliana. Plant Cell Physiol. 1994;35:225–231. [PubMed] [Google Scholar]
- 21.Gan S, Amasino RM. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270:1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- 22.Yang S, Xu K, Chen S. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol. 2019;19:260. doi: 10.1186/s12870-019-1872-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charu L, Manoj P. Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot. 2011;62:4731–4748. doi: 10.1093/jxb/err210. [DOI] [PubMed] [Google Scholar]
- 24.He GH, Xu JY, Wang YX. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016;16:116. doi: 10.1186/s12870-016-0806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elena B, Annamaria G, Eleonora C. Plant MYB Transcription factors: their role in drought response mechanisms. Int J Mol Sci. 2015;16:15811–15851. doi: 10.3390/ijms160715811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu G, Gao C, Zheng X, Han B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta. 2009;229:605–615. doi: 10.1007/s00425-008-0857-3. [DOI] [PubMed] [Google Scholar]
- 27.Tang N, Zhang H, Li X, Xiao J, Xiong L. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 2012;158:1755–1768. doi: 10.1104/pp.111.190389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li K, Xing C, Yao Z, Huang X. PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol J. 2017;15:1186. doi: 10.1111/pbi.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong H, Li J, Liu P, Duan J, Zhao Y, Guo X, et al. Overexpression of OsMYB48–1 , a novel MYB-related transcription factor, enhances drought and salinity tolerance in Rice. PLoS One. 2014;9:e92913. doi: 10.1371/journal.pone.0092913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai RH, Zhao Y, Wang YF. Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice. Plant Cell Tissue Organ Cult. 2014;119:565–577. doi: 10.1007/s11240-014-0556-7. [DOI] [Google Scholar]
- 31.Ulmasov T, Murfett J, Hagen G. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 33.Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136(6):1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of arabidopsis auxin repsonse factor17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrs HD, Weatherley PE. A re-examination of the relative turgidity techniques for estimating water deficits in leaves. Australian J Biol Sci. 1962;15:413–428. doi: 10.1071/BI9620413. [DOI] [Google Scholar]
- 36.Zhou Q. Experimental guidance of plant physiology [M] Beijing: China Agriculture Press; 2000. [Google Scholar]
- 37.Pan XQ, Welti R, Wang XM. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat Protocl. 2010;5:985–991. doi: 10.1038/nprot.2010.37. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Ross J, O’Neill D. New interactions between classical plant hormones. Trends Plant Sci. 2001;6(1):1–3. doi: 10.1016/S1360-1385(00)01795-7. [DOI] [PubMed] [Google Scholar]
- 40.Saibo NJ, Vriezen WH, Beemster GT, van Der Straeten D. Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J. 2003;33:989–1000. doi: 10.1046/j.1365-313X.2003.01684.x. [DOI] [PubMed] [Google Scholar]
- 41.Groover AT, Pattishall A, Jones AM. IAA8 expression during vascular cell differentiation. Plant Mol Biol. 2003;51:427–435. doi: 10.1023/A:1022039815537. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Yan DW, Yuan TT. A gain-of-function mutation in IAA8 alters Arabidopsis floral organ development by change of jasmonic acid level. Plant Mol Biol. 2013;82:771–783. doi: 10.1007/s11103-013-0039-y. [DOI] [PubMed] [Google Scholar]
- 43.Wang DK, Pei KM, Fu YP. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa) Gene. 2007;394:13–24. doi: 10.1016/j.gene.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Almazroue HA. Identification, cloning, and expression of tobacco responsive to dehydration like protein (RD22), SBIP-355 and its role in SABP2 mediated SA pathway in plant defense [D] 2014. [Google Scholar]
- 45.Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to drought stress [M] Amsterdam: Elsevier Science BV; 1998. [Google Scholar]
- 46.Devi K, Prathima PT, Gomathi R. Gene expression profiling in sugarcane genotypes during drought stress and rehydration. Sugar Tech volume. 2019;21:717–733. doi: 10.1007/s12355-018-0687-y. [DOI] [Google Scholar]
- 47.Shi HT, Chen L, Ye TT, Liu XD. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol Bioch. 2014;82:209–217. doi: 10.1016/j.plaphy.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Ashraf MY, Azhar N, Hussain M. Indole acetic acid (IAA) induced changes in growth, relative water contents and gas exchange attributes of barley (Hordeum vulgare L.) grown under water stress conditions. Plant Growth Regul. 2006;50:85–90. doi: 10.1007/s10725-006-9130-6. [DOI] [Google Scholar]
- 49.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and toletance to dehydrationand cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 50.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. P Natl Acad Sci USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saini S, Sharma I, Kaur N. Auxin: a master regulator in plant root development. Plant Cell Rep. 2013;32:741–757. doi: 10.1007/s00299-013-1430-5. [DOI] [PubMed] [Google Scholar]
- 52.Teng K, Li J, Liu L. Exogenous ABA induces drought tolerance in upland rice: the role of chloroplast and ABA biosynthesis-related gene expression on photosystem II during PEG stress. Acta Physiol Plant. 2014;36:2219–2227. doi: 10.1007/s11738-014-1599-4. [DOI] [Google Scholar]
- 53.Yang CJ, Zhang XK, Zou CS. Effects of drought simulated by PEG-6000 on germination and seedling growth of rapeseed (Brassica napus L.) Oil Crop Science. 2007;29:425–430. [Google Scholar]
- 54.Fernández-Arbaizar A, Regalado JJ, Lorenzo O. Isolation and characterization of novel mutant loci suppressing the ABA hypersensitivity of the Arabidopsis coronatine insensitive 1-16 (coi1-16) mutant during germination and seedling growth. Plant Cell Physiol. 2012;53:53–63. doi: 10.1093/pcp/pcr174. [DOI] [PubMed] [Google Scholar]
- 55.Deollas C, Arbona V, GóMez-Cadenas A. Jasmonoyl isoleucine accumulation is needed for abscisic acid build-up in roots of Arabidopsis under water stress conditions. Plant Cell Environ. 2015;38:2157–2170. doi: 10.1111/pce.12536. [DOI] [PubMed] [Google Scholar]
- 56.Cheol Park H, Cha JY, Yun DJ. Roles of YUCCAs in auxin biosynthesis and drought stress responses in plant. Plant Signal Behav. 2013;8:337–349. doi: 10.4161/psb.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang SK, Bai YH, Shen CJ. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomic. 2010;10:533–546. doi: 10.1007/s10142-010-0174-3. [DOI] [PubMed] [Google Scholar]
- 58.Arase F, Nishitani H, Egusa M, Nishimoto N, Sakurai S, Sakamoto N, et al. IAA8 involved in lateral root formation interacts with the TIR1 auxin receptor and ARF transcription factors in Arabidopsis. PLoS One. 2012;7:e43414. doi: 10.1371/journal.pone.0043414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bassa C, Etemadi M, Combier JP. Sl-IAA27 gene expression is induced during arbuscular mycorrhizal symbiosis in tomato and in Medicago truncatula. Plant Signal Behav. 2013;8:e25637. doi: 10.4161/psb.25637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foyer CH, Descourvieres P, Kunert KJ. Protection against oxygen radicals: an important defence mechanism studied in transgenic plants. Plant Cell Environ. 1994;17:507–523. doi: 10.1111/j.1365-3040.1994.tb00146.x. [DOI] [Google Scholar]
- 61.Zhang SW, Li CH, Cao J. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 2009;151:1889–1901. doi: 10.1104/pp.109.146803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khan S, Stone JM. Arabidopsis thaliana GH3.9 influences primary root growth. Planta. 2007;226:21–34. doi: 10.1007/s00425-006-0462-2. [DOI] [PubMed] [Google Scholar]
- 63.Golldack D, Lüking I, Yang O. Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011;30:1383–1391. doi: 10.1007/s00299-011-1068-0. [DOI] [PubMed] [Google Scholar]
- 64.Xiang Y, Tang N, Du H. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008;148:1938–1952. doi: 10.1104/pp.108.128199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiste C, Dröge-Laser W. The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat Commun. 2014;5:1–12. doi: 10.1038/ncomms4883. [DOI] [PubMed] [Google Scholar]
- 66.Fan GQ, Wang LM, Deng MJ. Changes in transcript related to osmosis and intracellular ion homeostasis in paulownia tomentosa under salt stress. Front Plant Sci. 2016;7:384. doi: 10.3389/fpls.2016.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Todaka D, Shinozaki K, Yamaguchi-Shinozaki K. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front Plant Sci. 2015;6:84. doi: 10.3389/fpls.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Q, Kasuga M, Sakuma Y. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eleonora C, Galbiati M, Vavasseur A. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 70.Tripathi P, Rabara RC, Rushton PJ. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta. 2014;239:255–266. doi: 10.1007/s00425-013-1985-y. [DOI] [PubMed] [Google Scholar]
- 71.Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 72.Ren X, Chen Z, Liu Y. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010;63:417–429. doi: 10.1111/j.1365-313X.2010.04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomohiro K. KazukoYS, Kazuo S. characterization of two cDNAs (ERD11 and ERD13) for dehydration-inducible genes that encode putative glutathione S-transferases in Arabidopsis thaliana L. FEBS Lett. 1993;335:189–192. doi: 10.1016/0014-5793(93)80727-C. [DOI] [PubMed] [Google Scholar]
- 74.Abe H, Yamaguchi-Shinozaki K, Urao T. Role of arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He YH, Gan SH. A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell. 2002;14:805–815. doi: 10.1105/tpc.010422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Noh YS, Amasino RM. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol Biol. 1999;41:181–194. doi: 10.1023/A:1006342412688. [DOI] [PubMed] [Google Scholar]
- 77.Kim JI, Murphy AS, Baek D, Lee SW, Yun DJ, Bressan RA, et al. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J Exp Bot. 2011;62:3981–3992. doi: 10.1093/jxb/err094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table 1 Primer sequences of the genes related to IAA and their corresponding GeneBank accession numbers. Table 2 Primer sequences of drought-induced transcription factors and drought-induced genes and their corresponding GeneBank accession numbers.
Data Availability Statement
The data used and/or analyzed during the current study are available from the corresponding author on reasonable request.