Abstract
Centromeres are central to chromosome segregation and genome stability, and thus their molecular foundations are important for understanding their function and the ways in which they go awry. Human centromeres typically form at large megabase-sized arrays of alpha satellite DNA for which there is little genomic understanding due to its repetitive nature. Consequently, it has been difficult to achieve genome assemblies at centromeres using traditional next generation sequencing approaches, so that centromeres represent gaps in the current human genome assembly. The role of alpha satellite DNA has been debated since centromeres can form, albeit rarely, on non-alpha satellite DNA. Conversely, the simple presence of alpha satellite DNA is not sufficient for centromere function since chromosomes with multiple alpha satellite arrays only exhibit a single location of centromere assembly. Here, we discuss the organization of human centromeres as well as genomic and functional variation in human centromere location, and current understanding of the genomic and epigenetic mechanisms that underlie centromere flexibility in humans.
Keywords: satellite DNA, repetitive DNA, epiallele, haplotype, dicentric chromosome, kinetochore
The centromere is the chromosomal locus that ensures chromosome inheritance through cell division. The kinetochore, the multi-protein structure that attaches chromosomes to spindle microtubules, is assembled at the centromere, locally coordinating chromosome movement during cell division. Each chromosome must have a centromere; without one, the chromosome will be lost, leading to an aneuploid karyotype. A locus with such an essential role in genome stability in all organisms would be expect to exhibit similar genomic characteristics among organisms. Surprisingly, centromeric DNAs differ among organisms and even between different chromosomes of the same organism (see Talbert and Henikoff in this issue 2019). Centromeres range in size from small point centromeres (~125bp) in budding yeasts to large regional centromeres (100kb – 5Mb) in humans and plants. Despite these sequence disparities, the proteins of eukaryotic centromeres are related, emphasizing the functional importance of the locus. Centromeres are defined by specialized nucleosomes containing the histone H3 variant CENP-A. Clusters of CENP-A nucleosomes are interspersed with groups of canonical H3 nucleosomes to create a unique type of centromeric (CEN) chromatin, also sometimes referred to as centrochromatin, that differentiates the centromere from the rest of the chromosome. CEN chromatin serves as the foundation of the kinetochore, interacting with CENP-C and other members of the constitutive centromere associated network (CCAN) to assemble the protein network between the DNA and the microtubules. CEN chromatin assembly occurs on DNA sequences that differ among organisms and even within the same organism, suggesting a general lack of sequence specificity for CENP-A and other centromere proteins and pointing to CENP-A and/or CEN chromatin as an important epigenetic determinant of centromere identity and maintenance. The lack of sequence similarities at eukaryotic centromeres led to the current view of centromere identity as an epigenetic process, with little contribution from the underlying DNA.
The genomics of human centromeres
Native human centromeres are formed at regions of alpha satellite, a repetitive DNA that is defined by a 171bp monomeric sequence unit. Individual monomers that are 50–70% identical are arranged tandemly (Fig. 1a). A defined number of monomers create a higher order repeat (HOR) unit that is repeated hundreds to thousands of times to produce a large homogenous array in which the HOR units are 97–100% identical. The number and order of monomers within a HOR unit confers chromosome specificity, so that even though the same monomers are present at every centromere in the genome, their placement within a HOR unit contributes to individual alpha satellite arrays that can be molecularly distinguished [1]. For instance, the human X chromosome (HSAX) centromere is defined by a HOR of 12 monomers (DXZ1, 12-mer), whereas the human Y (HSAY) chromosome centromere is defined by 34-mer HOR (DYZ3) [2,3]. Since the order and sequence of the monomers vary between chromosome-specific HORs, alpha satellite arrays on each chromosome can be distinguished molecularly by PCR, Southern blotting, and fluorescence in situ hybridization (FISH) under stringent conditions [4–6]. Despite basic structural and organizational features to differentiate alpha satellite arrays, the human genome assembly lacks contiguous alpha satellite sequences at centromeres on every chromosome. Although monomers are present within short-read sequence datasets, they are present in hundreds to thousands of copies within the genome and at individual centromeres, so it is difficult to accurately and confidently assign a monomeric read to a HOR unit or multi-megabase array (reviewed in [7]). We do not have a complete understanding of alpha satellite organization and structure on individual chromosomes within the population, nor of genotype-function correlations, owing to the lack of contiguous centromeric reference genome assemblies.
Figure 1. Genomic organization and variation of alpha satellite DNA at human centromeres.
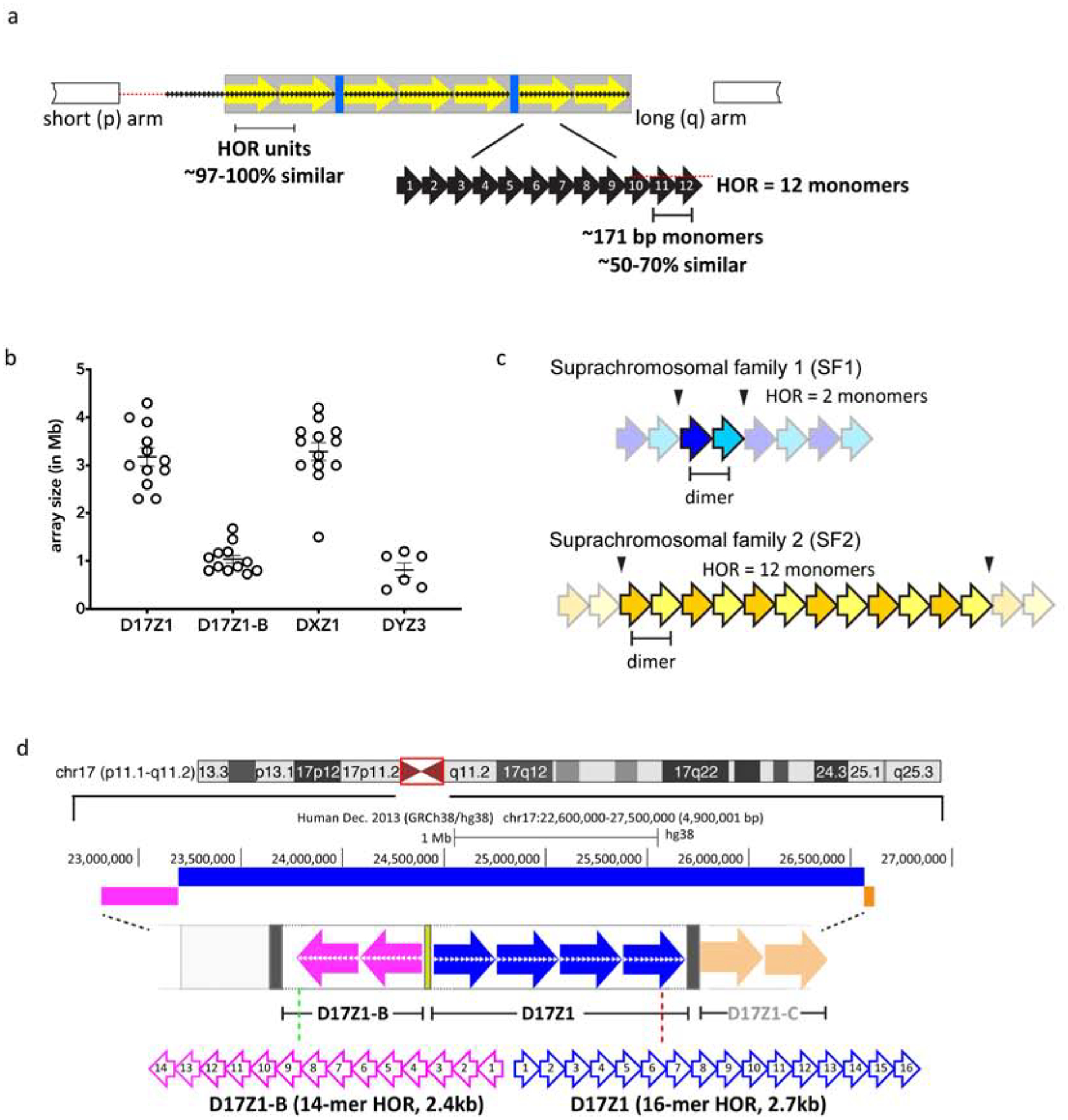
a. Schematic of the general organization of an alpha satellite DNA array at human centromere regions. Alpha satellite is based on ~171bp monomeric repeat units (black arrows with white numbers) that are 50–70% identical in sequence and arranged tandemly to form a HOR unit; shown here as a 12 monomer HOR (yellow array). Monomers are numbered by their position within the HOR and not based on their homology between two distinct HORs. The HORs are repeated hundreds to thousands of times to create homogenous arrays in which HOR within a given array are 97–100% identical. The HOR array is flanked by degenerate alpha satellite DNA monomers (small black arrows) that lack hierarchical structure and separate the HOR array from the chromosome arms. HOR arrays are interrupted by other repetitive elements, such as transposable elements (TEs, blue), but the extent of TE distribution across specific alpha satellite arrays is currently unclear due the lack of linear, contiguous alpha satellite assemblies.
b. The number of times a chromosome-specific HOR unit is repeated to create an extensive homogenous array varies between homologs and individuals. The distribution of total array sizes from a subset of the population is shown for our distinct alpha satellite arrays, including D17Z1 and D17Z1-B, two arrays from Homo sapiens chromosome 17 (HSA17), as well as DXZ1 and DYZ3, the alpha satellite arrays from HSAX and HSAY. Each open circle represents an independent chromosome/individual.
c. Individual alpha satellite monomers are distributed into five suprachromosomal groups or families, based on the concentration of specific monomer types (based on sequence homology) on distinct chromosomes. Suprachromosomal families 1 and 2 (SF1, SF2) represent the most abundant HOR configuration within human centromere regions and are defined by distinct dimeric configurations (shown as blue or yellow. HOR units on some chromosomes like HSA1 are defined by the two dimers themselves while larger HOR units on chromosomes like HSA18 are comprised of multiple alternating copies of the same two monomers. The HOR units are then repeated hundreds to thousands of times to produce a large homogenous satellite array. d. Over half of native human chromosomes contain more than one distinctive alpha satellite array. For example, HSA17 has three arrays that are ~92% identical and defined by different HOR units. D17Z1, the largest array (blue) is defined by a canonical 16 monomer (16-mer) HOR that is repeated thousands of times to produce total array sizes that range from 2.3–4.2Mb in the population. D17Z1-B (pink) and D17Z1-C (light orange) flank D17Z1 and are each defined by different 14-mer HOR units. Centromere assembly can occur at either D17Z1 or D17Z1-B.
Genomic variation within human centromeres
Sequence variation is a source of functional diversity that is typically studied in the context of gene expression and non-coding regulatory regions. It has been difficult to define the amounts and types of variation that are present within alpha satellite without genome assemblies. However, early molecular studies of HOR unit organization of some alpha satellite arrays have several types of variation at human centromeres: 1) total size of the array, 2) structure of the HOR units within a given array, 3) sequence variation within the same HOR unit of a distinct array, and 4) number of independent arrays on a single chromosome [8–10].
Total size variation of chromosome-specific alpha satellite arrays
Alpha satellite is frequently thought to be identical at all human centromeres, but although each array is built from the same pool of monomers, it has its own distinctive organization within the human genome. Additionally, degrees of variation exist within an array. On a given chromosome, the number of times a HOR unit is repeated differs within the population, giving rise to a range of total array lengths between homologs in the same individual and among homologs within the different individuals. For example, on individual HSAXs in the population, the 2.0 kb DXZ1 HOR is repeated 750–2100 times, yielding total array size lengths that range from 1.5Mb to 4.2 Mb (mean of 3.0Mb) (Fig. 1b) [3,11,12]. Alpha satellite arrays on autosomes also exhibit similar inter-homolog and inter-individual array size polymorphisms, such that population array lengths vary 10- to 20-fold. However, within a given family, alpha satellite array sizes are heritable and stable through meiosis, to the extent that specific chromosome homologs can be tracked molecularly through families based solely on alpha satellite array sizes [13–15]. Alpha satellite array sizes of the same chromosome have not been routinely compared among more than two tissues, but appear to be largely stable in phenotypically normal individuals [16]. Whether this remains true for all cells, including undifferentiated or aging cells, remains to be explored.
Structure and sequence variation of HOR units within specific arrays
Alpha satellite monomers differ in sequence by 10–40%. Two adjacent monomers may differ in sequence, but similarities in monomer sequence and their order in the HOR unit, are shared among different chromosomes. Individual monomers are derived from twelve consensus alpha satellite monomers that are distributed among five suprachromosomal groups or families, based on the combination and concentration of these monomers on specific chromosomes. Suprachromosomal families 1 through 3 (SF1–3) represent the majority of “functional” alpha satellite HORs and are typically enriched for centromere proteins [17,18]. SF1 and SF2 are defined by distinct types of dimeric (two monomer) configurations (Fig. 1c) and are present on all chromosomes with the exception of HSA1, HSA11, HSA17, and HSAX that are defined by the pentameric (five monomer) HOR configuration of SF3 [18–20]. While both monomers of dimeric HOR unit arrays are enriched for centromere proteins, not all monomers within pentameric HOR arrays are equally associated with centromere proteins [21].
The binding of Centromere Protein B (CENP-B) to alternate monomers in dimeric HOR subfamilies SF1 and SF2 confers enhanced binding and integrity of the constitutive centromere associated-network complex (CCAN). CENP-B is a DNA binding protein that recognizes a 17-bp sequence motif called the CENP-B box that is present in a subset of alpha satellite monomers [22,23]. The density of CENP-B boxes within HOR alpha satellite arrays has been correlated with stronger CENP-A enrichment, and led to the model that dimeric arrays exhibit the highest CENP-B box density [18]. Within pentameric HOR units of SF3, CENP-B boxes are irregularly spaced. Furthermore, the density of CENP-B boxes is also influenced total array size, such that a 3Mb dimeric HOR array will have the same number of CENP-B boxes as a 3Mb pentameric HOR array that has irregularly spaced CENP-B boxes. A lower density of CENP-B boxes within an alpha satellite array may not entirely disqualify it for centromere assembly. Indeed, centromere assembly occurs on minor arrays of HOR alpha satellite even when a nearby major HOR array on the same chromosome has twice or thrice the number of CENP-B boxes [15,17,24].
Genomic variation within HORs of specific alpha satellite arrays
The suprachromosomal family classifications illustrate that variation within the alpha satellite DNA is common and complex, due to monomeric differences and chromosome-specific differences in HOR unit size and monomer order and organization. Although HOR units are largely thought to be highly identical within a given chromosome-specific array [25], some HOR units within the array contain structural variants such as monomer insertions or deletions, monomer or entire HOR unit inversions, and insertion of non-satellite elements such as transposons [26–28] Thus, alpha satellite arrays actually exist as complex arrays of variant and canonical HORs punctuated with other repetitive elements [1,29].
HSA17 is one chromosomal example of HOR unit polymorphisms within an alpha satellite array. The predominant HOR unit on D17Z1 is a 16-monomer (16-mer) (Fig. 2a), and makes up most of the HOR units in D17Z1 within the population. However, single or multiple monomeric deletions within the canonical 16-mer HOR unit have produced variant D17Z1 HOR units including 15-mers, 14-mers, 13-mers, 12-mers, and very rare 11-mers. These monomer deletions presumably arose by unequal recombination or gene conversion during meiosis [30,31]. Within humans, 15-, 14- and 12-mer HOR units represent a small fraction of the total HOR units within a given D17Z1 array. However, the 13-mer HOR unit is the most abundant variant HOR unit within D17Z1 arrays. As such, D17Z1 arrays in the population are defined by two haplotypes: 1) Haplotype I: arrays consisting of 16-mers, with occasional 15- and 14-mers, that are found on 65% of HSA17s, and 2) Haplotype II, the variant haplotype that is comprised of arrays that contain 13-mer HOR units and are present on 35% of HSA17s (Fig. 2a). The proportion of variant 13-mer HOR units within D17Z1 arrays ranges from as little as 1% to over 75% of the array, depending on the chromosome and the individual [15].
Figure 2. Functional centromeric epialleles on Homo sapiens chromosome 17 (HSA17) are influenced by genomic variation within alpha satellite DNA.
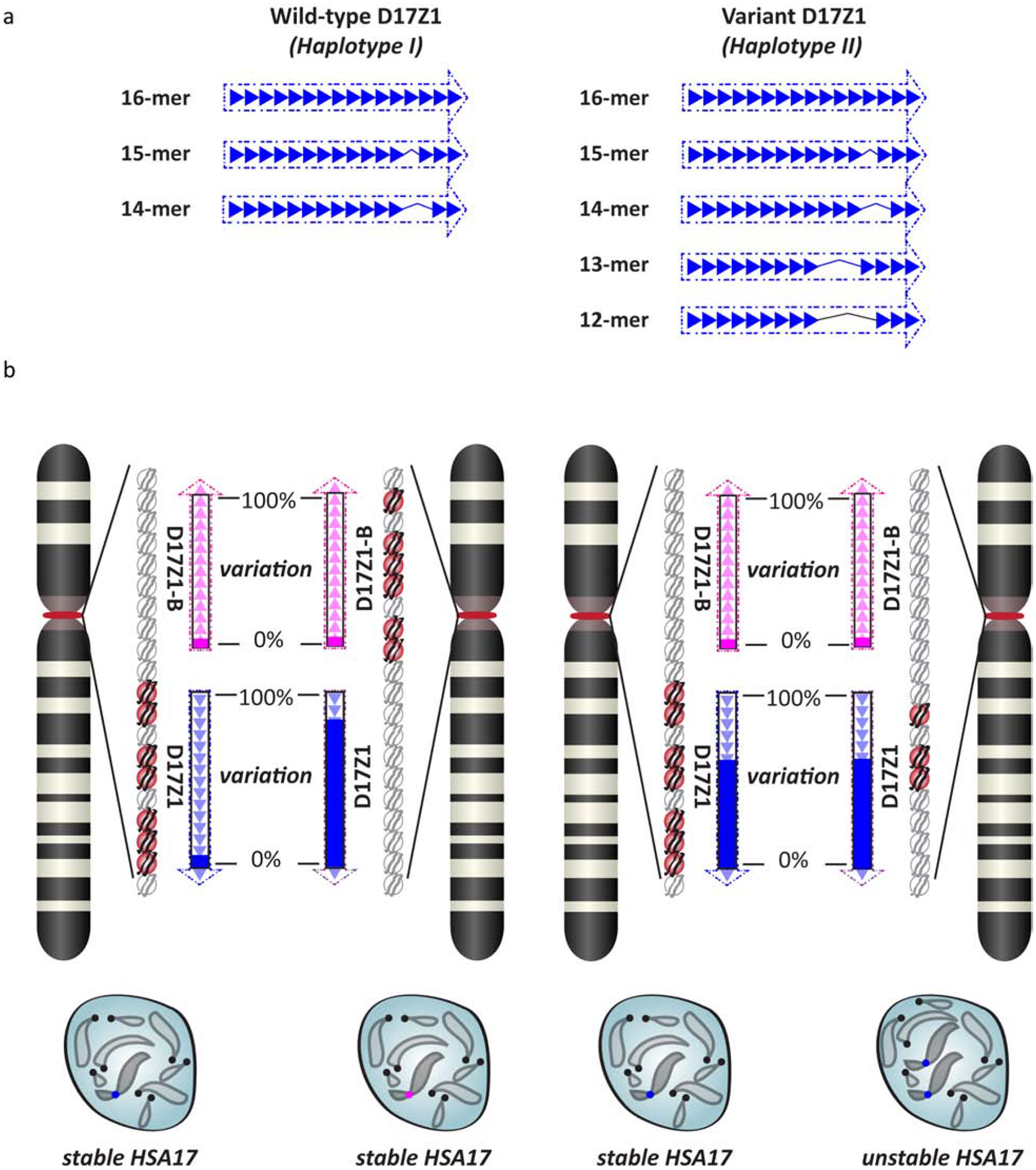
a. The large array D17Z1 on HSA17 is defined by a canonical (wild-type) 16-mer HOR unit. However, D17Z1 is highly polymorphic such that single and multiple monomer deletions produce HOR variants that differ in length by an integral number of monomers. In the general population, HOR variants range from 15-mers to 12-mers. Two major haplotypes (Haplotype I - wildtype; Haplotype II – variant) exist in the population, distinguished by the presence or absence of the 13-mer HOR unit.
b. HSA17 exhibits centromeric epialleles (i.e. multiple sites of centromere assembly) based on the amount of variation within D17Z1. Arrays containing predominantly wild-type 16-mer HORs are preferred locations for centromere assembly, denoted by CENP-A (red circles) and other centromere proteins, and are associated with stable HSA17s. When D17Z1 arrays are composed of more than 70% variant 13-mer HORs, centromere assembly occurs on neighboring D17Z1-B (pink arrows) and chromosome stability is comparable to HSA17s with wild-type arrays. However, centromere assembly occurs on D17Z1 arrays exhibiting intermediate levels of variation (40–60%) but leads to two distinct chromosome phenotypes: stable and unstable. Variant arrays exhibiting the unstable HSA17 phenotype are associated with reduced numbers of centromere proteins.
In addition, to HOR unit variation, single nucleotide changes have also been mapped to specific monomers within some of the HOR units of alpha satellite arrays [3,4,25,32]. The functional significance of size and sequence variants in alpha satellite is unclear. Centromere-specific/centromere-proximal haplotypes (cenhaps) are present within human populations [33], and might be associated with different long-term chromosome stability outcomes. Ultra-long (UL) read sequence assemblies of DXZ1 (HSAX) and DYZ3 (HSAY) have revealed multiple types of structural variation within alpha satellite, including single and multiple monomeric insertions and deletions that changed HOR size and in the case of DXZ1, a single LINE-1 (L1H) insertion [3,26].
Multi- array chromosomes exhibit variation in the number of independent alpha satellite arrays within the human karyotype
Although it was noted in the 1990s that the acrocentric chromosomes (HSA13, 14, 15, 21, and 22) had more than one distinct alpha satellite, it has been more recently appreciated that over half of human chromosomes, such as HSA1, HSA7, HSA17, HSA18, HSA19, and HSA20, contain two or even three HOR arrays [17,20]. HSA17 for example contains three distinct arrays, D17Z1, D17Z1-B, and D17Z1-C (Fig. 1d). The presence of multiple arrays on the same chromosome has led to two models: 1) one array is “active/live” and the other is “inactive/dead”; and 2) native chromosomes are arranged as structurally dicentric or tricentric chromosomes with arrays that retain functional potential. Studies using human artificial chromosomes (HACs) to test the functional potential of “live” or “dead” arrays argue against the first model for chromosomes such as HSA7 and HSA17 [17,34] (see section on “Functional variation of centromeres”). Arrays lacking HOR unit structure are incapable of supporting de novo centromere function, so there appears to be a clear distinction between the functional potential of HOR arrays and monomeric arrays [17,35].
Epigenomic variation of human centromeres
Human centromeres have an epigenomic RNA component
Centromeres and heterochromatin were long thought to be transcriptionally inert because they can effectively silence transgenes or genes that are inserted in or nearby the centromere [36]. However, studies in the fission yeast S. pombe first revealed that small RNAs produced from centromere regions are required to establish and reinforce heterochromatin at the centromere and pericentromere [37–40]. Mammalian pericentromeres also have an RNA component that appears to be linked to the cell cycle [38,39,41].
Alpha satellite DNA is incorporated into both CEN chromatin and pericentric heterochromatin [5,42]. CEN chromatin covers 30–45% of a single array of alpha satellite, at native chromosomes or centromeres of human artificial chromosomes (HACs) [5,43–45]. Within CEN chromatin, the H3 nucleosomes within CEN chromatin are marked by dimethylation on lysine 4 (K4me2) and lysine 36 (K36me2), post-translational histone modifications that are typically associated with euchromatin. The remaining portion of the alpha satellite array is assembled into heterochromatin marked by histone modifications such as H3K9me2, H3K9me3, and H3K27me3 [5,43–46]. Each chromosome is associated with sequence-specific, array-specific non-coding transcripts. The alpha satellite non-coding transcripts are produced in cis and are complexed with centromere proteins CENP-A and CENP-C, as well as the alpha satellite DNA binding protein CENP-B [24,47]. Although CENP-C is a known RNA-binding protein, the specific binding sites for alpha satellite RNAs on CENP-C and other centromere proteins have not yet been identified. However, non-coding alpha satellite RNAs also recruit the histone lysine methyltransferases SUV39H1/2 that add methyl groups to H3 at lysine 9 to create a binding site for heterochromatin protein 1 (HP1) [40,48]. How a single alpha satellite array is apportioned into multiple chromatin domains is not clear, although the cell cycle timing, structure, or long-term stability of alpha satellite transcripts may be a factor in their ultimate function. Alpha satellite transcripts have been proposed to exist as RNA:DNA hybrids or as single-stranded RNAs [40,49], so it is possible that distinct sets of transcripts or their local concentrations at various regions of the alpha satellite array feed into a CEN chromatin versus heterochromatin assembly pathway.
DNA methylation within centromere regions
Centromere regions are also enriched for DNA methylation that can be linked to histone methylation. For instance, methylation of mouse minor (centromere) and major (pericentromere) satellite DNAs are important for chromosome and satellite array stability [50–52]. Alpha satellite DNA is hyper-methylated, presumably through the action of DNMT3B that interacts with CENP-C [53,54]. Variation in enrichment of DNA methylation between different alpha satellite arrays in the same or different individuals is not well understood, but technological advancements may help resolve this gap in knowledge. Nanopore and PacBio sequencing technologies can detect modifications to DNA bases, such as cytosine methylation, through either alternation in normal electrolytic current signals or polymerase synthesis rates, respectively, when a methylated cytosine is encountered. The use of nanopore UL-reads to fully assemble the human X chromosome has provided information on DNA methylation within the DXZ1 alpha satellite array [26]. Although DXZ1 was generally methylated throughout the array, a 60kb region of hypomethylation was detected. A similarly sized (75kb) region of hypomethylation was also detected within the manually assembled D8Z2 array on HSA8. Similar hypomethylation of the satellite repeats in Arabidopsis thaliana and Zea mays has been reported previously [55]. The functional relevance of hypermethylated and hypomethylated regions within centromeres remains to be fully determined, although it possible hypomethylation demarcates distinct chromatin domains within centromere regions or ensures the faithful binding of centromere proteins [56].
Functional variation of human centromeres
Human centromere function varies in a few notable ways. The most drastic example is the occurrence of neocentromeres, atypical centromeres that arise spontaneously at non-canonical (i.e. non alpha satellite) sequences [57–59], either on broken chromosomes or when the native centromere loses function or is functionally mutated. The basis of such functional variation in centromere location remains a topic of intense interest, and approaches to engineer neocentromeres are aimed at exploring (epi)genomic features that promote neocentromere formation. Neocentromeres are discussed in detail by Murillo-Pineda and Jansen in this volume (ref to specific chapter from Special Issue). Here, we focus on variability in native centromere function.
Stability of native chromosomes as a measure of centromere fitness
One alpha satellite array on each chromosome has the highest local CENP-A concentration and is where new CENP-A is deposited each cell cycle [44,60]. Thus, once established by CENP-A incorporation, a centromere is maintained at the same location on a portion of alpha satellite DNA [44]. Approximately 35% of any given array is assembled into CEN chromatin (CENP-A plus H3K4me2 nucleosomes combined). Considering the range of alpha satellite array lengths in the population, CEN chromatin domain lengths on a chromosome-specific array can range from 500kb to 1.5Mb [6,43]. Although differences in the amount of total centromere proteins between large and small chromosomes and large and small alpha satellite arrays have been reported [61,62], quantitative analyses of CENP-A nucleosomes at native centromeres have concluded that the number of CENP-A molecules does not vary largely among all centromeres [44,60]. These results suggest that perhaps the sizes and/or number of CENP-A-containing and H3-containing subdomains differ on individual chromosome-specific arrays. Although patterns of subdomain organization remain to be validated experimentally at native human centromeres, ChIP studies of a neocentromere formed on HSA10 (mardel10) showed a range of CENP-A domain sizes across the CEN chromatin domain [63]. Thus, not only could subdomain sizes differ between individual arrays, they might also differ on the same array.
Variation in CENP-A and H3 subdomain sizes may reflect inherent natural flexibility in centromere organization and normal homeostasis of CENP-A enrichment on a range of alpha satellite array lengths. The establishment and maintenance of CENP-A domain sizes may be dictated by sequence and organization of the HOR units within an array. Several recent studies suggest that under perturbing conditions that challenge chromosome stability, the genomic composition of centromeres influences the sensitivity of the centromere [64,65]. A subset of alpha satellite monomers contain a 17bp motif called the CENP-B box, a binding site for centromere protein B (CENP-B) [66]. The location of CENP-B boxes varies based on chromosome-specific HOR and is linked to the HOR structure [22]. Some HORs have multiple CENP-B boxes, while others like the 16-mer HOR D7Z2 on HSA7, have only a single CENP-B box (reviewed in [20,67]). The DYZ3 array on HSAY completely lacks CENP-B boxes [22,68]. Overall, the range of total array lengths in the population also reflect differences in the total number of functionally significant CENP-B boxes. A recent study reported that alpha satellite arrays with more CENP-B boxes and thus greater CENP-B binding were less likely to mis-segregate [65]. The increased number of CENP-B boxes correlated with increased binding of CENP-C, indicating that centromeres that are less sensitized to destabilizing conditions contain more functional CENP-B boxes that recruit CENP-B and CENP-C [69]. Longer alpha satellite arrays might be predicted to build stronger centromeres because they theoretically have more CENP-B boxes, but the correlation between centromere length and centromere strength was less significant. Not all CENP-B boxes in alpha satellite arrays are incorporated into kinetochore chromatin, since a portion of the array is also assembled into heterochromatin [5,70]. Regulation of the binding of CENP-B and CENP-C across the alpha satellite array, perhaps through methylation of CENP-B boxes that inhibits CENP-B binding and/or controls satellite transcription [54,56], may influence or control centromere strength.
Centromeric epialleles: variation in alpha satellite array location of centromere assembly
As mentioned above, HSA17 is unique in that it has three alpha satellite arrays D17Z1, D17Z1-B and D17Z1-C. HAC technology showed that D17Z1 and D17Z1-B arrays can independently support de novo centromere formation [17,34], suggesting that in vivo, centromere location might vary on native chromosomes. Indeed, on HSA17, either D17Z1 or D17Z1-B can be the site of centromere assembly [15,17,34]. These centromeric epialleles are mitotically and meiotically stable [15]. They also are not exclusive to HSA17, since recent studies have identified centromeric epialleles on multiple chromosomes [20,24,71,72], indicating that flexibility in centromere location is an inherent, and previously unappreciated, property of native human chromosomes.
An intriguing question is how centromere location is determined on multi-array chromosomes. On HSA17, D17Z1 is a large array that ranges in length from 2–4Mb and contains many CENP-B boxes, compared to D17Z1-B that is a smaller array (0.5–1.5Mb) and contains fewer (but perhaps a proportionately equal number of) CENP-B boxes. Although D17Z1 is predominantly the site of centromere assembly [15,71], even when it is not the site of centromere assembly, its array length exceeds that of D17Z1-B. Like recent studies, this suggests that centromere length does not completely predict centromere location. Instead, the variation within D17Z1 arrays (described above in the section Structure and sequence variation of HOR units) appears to influence centromere location on HSA17. On HSA17s in which D17Z1 contains little to no variation, the centromere forms at D17Z1; conversely when HSA17 has a D17Z1 array with greater than 70% variation (i.e. more 13-mer HORs versus wild-type 16-mers), centromere assembly occurs on D17Z1-B (Fig. 2b). Centromeres formed on wild-type D17Z1 arrays or on D17Z1-B arrays when D17Z1 is highly variant appear to be highly efficient and are rarely associated with HSA17 instability or aneuploidy.
Nevertheless, the presence of variation within D17Z1 does not absolutely correlate with centromeric epialleles and formation of the centromere at D17Z1-B. In fact, it has been shown variant D17Z1 arrays can be the site of centromere assembly, but many of these HSA17s exhibit chromosome instability and increased aneuploidy long-term [15]. Variant, active D17Z1 arrays suggest that the amount of variation within an alpha satellite array is functionally important. Centromere assembly occurs on D17Z1 arrays with moderate variation (40–60% of the array contains variant HOR units), but there is a functional cost (Fig. 2b). Unstable HSA17s that build the centromere on variant D17Z1 arrays are associated with reduced amounts of CENP-A and CENP-C, suggesting that variant alpha satellite DNA assemble a kinetochore that is defective in architecture and/or composition [15]. However, not every variant D17Z1 array is associated with a defective kinetochore and/or chromosome mis-segregation, raising the possibility that differences in long-range organization (i.e. where specific HOR units are located with respect to one another within an array) and perhaps even transcription of distinct variant arrays are linked to a range of centromere strength and quality outcomes.
The molecular basis for reduced association of CENPs and inefficient centromere function on variant D17Z1 alpha satellite arrays remains to be specifically determined. However, studies of HSA17 and other human centromeres point toward DNA-dependent alpha satellite factors, including alpha satellite sequence variation, that strongly affect centromere assembly and chromosome stability. The opportunity now exists to combine the newest ultra-long read sequencing technologies and optical mapping approaches with molecular and functional assays to comprehensively capture alpha satellite organization and variation within the population and test how specific (epi)genomic configurations of alpha satellite impact centromere assembly and chromosome stability.
Acknowledgements
This work was supported by NIH grants R01 GM124041, R01 GM129263, and R21 CA238758 to B.A.S. We are especially grateful to Rachel J. O’Neill (University of Connecticut, Storrs) for conceptualization and design ideas for Figure 2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Willard HF, Waye JS, Hierarchical order in chromosome-specific human alpha satellite DNA, Trends Genet. (1987). 10.1016/0168-9525(87)90232-0. [DOI] [Google Scholar]
- [2].Waye JS, Willard HF, Chromosome-specific alpha satellite DNA: Nucleotide sequence analysis of the 2.0 kilobasepair repeat from the human X chromosome, Nucleic Acids Res. 13 (1985) 2731–43. 10.1093/nar/13.8.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ, Centromere reference models for human chromosomes X and Y satellite arrays, Genome Res. 24 (2014) 697–707. 10.1101/gr.159624.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Warburton PE, Greig GM, Haaf T, Willard HF, PCR amplification of chromosome-specific alpha satellite DNA: definition of centromeric STS markers and polymorphic analysis, Genomics. 11 (1991) 324–333. [DOI] [PubMed] [Google Scholar]
- [5].Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA, Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA, Proc. Natl. Acad. Sci. U S A 103 (2006) 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA, Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells, Chromosome Res. 19 (2011) 457–470. 10.1007/s10577-011-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Miga KH, Completing the human genome: the progress and challenge of satellite DNA assembly, Chromosome Res. 23 (2015) 421–6. 10.1007/s10577-015-9488-2. [DOI] [PubMed] [Google Scholar]
- [8].Willard HF, Waye JS, Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat, J Mol Evol. 25 (1987) 207–214. [DOI] [PubMed] [Google Scholar]
- [9].Rudd MK, Willard HF, Analysis of the centromeric regions of the human genome assembly, Trends Genet. 20 (2004) 529–533. [DOI] [PubMed] [Google Scholar]
- [10].Willard HF, Centromeres of mammalian chromosomes, Trends Genet. 6 (1990) 410–416. [DOI] [PubMed] [Google Scholar]
- [11].Mahtani MM, Willard HF, Pulsed-field gel analysis of alpha-satellite DNA at the human X chromosome centromere: high-frequency polymorphisms and array size estimate, Genomics. 7 (1990) 607–613. [DOI] [PubMed] [Google Scholar]
- [12].Miga KH, Chromosome-specific centromere sequences provide an estimate of the ancestral chromosome 2 fusion event in hominin genomes, in: J. Hered, 2017. 10.1093/jhered/esw039. [DOI] [PubMed] [Google Scholar]
- [13].Wevrick R, Willard HF, Long-range organization of tandem arrays of alpha satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability, Proc. Natl. Acad. Sci. U S A 86 (1989) 9394–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Willard HF, Greig GM, Powers VE, Waye JS, Molecular organization and haplotype analysis of centromeric DNA from human chromosome 17: implications for linkage in neurofibromatosis, Genomics. 1 (1987) 368–373. [DOI] [PubMed] [Google Scholar]
- [15].Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA, Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles, Genome Res. 26 (2016) 1301–1311. 10.1101/gr.206706.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wevrick R, Earnshaw WC, Howard-Peebles PN, Willard HF, Partial deletion of alpha satellite DNA associated with reduced amounts of the centromere protein CENP-B in a mitotically stable human chromosome rearrangement, Mol Cell Biol. 10 (1990) 6374–6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hayden KE, Strome ED, Merrett SL, Lee HR, Rudd MK, Willard HF, Sequences associated with centromere competency in the human genome, Mol Cell Biol. 33 (2013) 763–772. 10.1128/MCB.01198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Henikoff JG, Thakur J, Kasinathan S, Henikoff S, A unique chromatin complex occupies young a-satellite arrays of human centromeres, Sci. Adv 1 (2015) e1400234 10.1126/sciadv.1400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mashkova TD, Akopian TA, Romanova LY, Mitkevich SP, Yurov YB, Kisselev LL, Alexandrov IA, Genomic organization, sequence and polymorphism of the human chromosome 4-specific alpha-satellite DNA, Gene. 140 (1994) 211–217. [DOI] [PubMed] [Google Scholar]
- [20].McNulty SM, Sullivan BA, Alpha satellite DNA biology: finding function in the recesses of the genome, Chromosome Res. 26 (2018) 115–138. 10.1007/s10577-018-9582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE, The octamer is the major form of CENP-A nucleosomes at human centromeres, Nat Struct Mol Biol. 20 (2013) 687–695. 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T, Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box, J Cell Biol. 116 (1992) 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T, A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite, J. Cell Biol 109 (1989) 1963–73. 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].McNulty SM, Sullivan LL, Sullivan BA, Human Centromeres Produce Chromosome-Specific and Array-Specific Alpha Satellite Transcripts that Are Complexed with CENP-A and CENP-C, Dev Cell. 42 (2017) 226–240. 10.1016/j.devcel.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Durfy SJ, Willard HF, Patterns of intra- and interarray sequence variation in alpha satellite from the human X chromosome: Evidence for short-range homogenization of tandemly repeated DNA sequences, Genomics. 5 (1989) 810–21. 10.1016/0888-7543(89)90123-7. [DOI] [PubMed] [Google Scholar]
- [26].Miga KH, Koren S, Rhie A, Vollger MR, Gershman A, Bzikadze A, Brooks S, Howe E, Porubsky D, Logsdon GA, Schneider VA, Potapova T, Wood J, Chow W, Armstrong J, Fredrickson J, Pak E, Tigyi K, Kremitzki M, Markovic C, Maduro V, Dutra A, Bouffard GG, Chang AM, Hansen NF, Thibaud-Nissen F, Schmitt AD, Belton J-M, Selvaraj S, Dennis MY, Soto DC, Sahasrabudhe R, Kaya G, Quick J, Loman NJ, Holmes N, Loose M, Surti U, ana Risques R, Lindsay TAG, Fulton R, Hall I, Paten B, Howe K, Timp W, Young A, Mullikin JC, Pevzner PA, Gerton JL, Sullivan BA, Eichler EE, Phillippy AM, Telomere-to-telomere assembly of a complete human X chromosome, BioRxiv. (2019). 10.1101/735928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Miga KH, Centromeric satellite DNAs: Hidden sequence variation in the human population, Genes (Basel). 10 (2019) E352 10.3390/genes10050352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zahn J, Kaplan MH, Fischer S, Dai M, Meng F, Saha AK, Cervantes P, Chan SM, Dube D, Omenn GS, Markovitz DM, Contreras-Galindo R, Expansion of a novel endogenous retrovirus throughout the pericentromeres of modern humans, Genome Biol. 16 (2015) 74 10.1186/s13059-015-0641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Waye JS, Willard HF, Molecular analysis of a deletion polymorphism in alpha satellite of human chromosome 17: evidence for homologous unequal crossing-over and subsequent fixation, Nucleic Acids Res. 14 (1986) 6915–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Warburton PE, Waye JS, Willard HF, Nonrandom localization of recombination events in human alpha satellite repeat unit variants: implications for higher-order structural characteristics within centromeric heterochromatin., Mol. Cell. Biol 13 (1993) 6520–9. 10.1128/mcb.13.10.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Warburton PE, Willard HF, Interhomologue sequence variation of alpha satellite DNA from human chromosome 17: Evidence for concerted evolution along haplotypic lineages, J. Mol. Evol 41 (1995) 1006–15. 10.1007/BF00173182. [DOI] [PubMed] [Google Scholar]
- [32].Durfy SJ, Willard HF, Molecular analysis of a polymorphic domain of alpha satellite from the human X chromosome, Am. J. Hum. Genet 41 (1987) 391–401. [PMC free article] [PubMed] [Google Scholar]
- [33].Langley SA, Miga KH, Karpen GH, Langley CH, Haplotypes spanning centromeric regions reveal persistence of large blocks of archaic DNA, Elife. (2019). 10.7554/eLife.42989.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Maloney KA, Sullivan LL, Matheny JE, Strome ED, Merrett SL, Ferris A, Sullivan BA, Functional epialleles at an endogenous human centromere, Proc. Natl. Acad. Sci. U S A 109 (2012) 13704–13709. 10.1073/pnas.1203126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H, Construction of YAC-based mammalian artificial chromosomes, Nat Biotechnol. 16 (1998) 431–439. [DOI] [PubMed] [Google Scholar]
- [36].Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC, Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres, Cell. 91 (1997) 1021–1032. [DOI] [PubMed] [Google Scholar]
- [37].Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M, Dicer is essential for formation of the heterochromatin structure in vertebrate cells, Nat Cell Biol. 6 (2004) 784–791. [DOI] [PubMed] [Google Scholar]
- [38].Guenatri M, Bailly D, Maison C, Almouzni G, Mouse centric and pericentric satellite repeats form distinct functional heterochromatin, J Cell Biol. 166 (2004) 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lu J, Gilbert DM, Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin, J Cell Biol. 179 (2007) 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Johnson WL, Yewdell WT, Bell JC, McNulty SM, Duda Z, O’Neill RJ, Sullivan BA, Straight AF, RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin, Elife. 6 (2017). 10.7554/eLife.25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Maison C, Bailly D, Peters AHFM, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G, Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component, Nat. Genet 30 (2002) 329–34. 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- [42].Sullivan BA, Karpen GH, Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin, Nat Struct Mol Biol. 11 (2004) 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mravinac B, Sullivan LL, Reeves JW, Yan CM, Kopf KS, Farr CJ, Schueler MG, Sullivan BA, Histone modifications within the human X centromere region, PLoS One. 4 (2009) e6602 10.1371/journal.pone.0006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ross JE, Woodlief KS, Sullivan BA, Inheritance of the CENP-A chromatin domain is spatially and temporally constrained at human centromeres, Epigenetics Chromatin. 9 (2016) 20 10.1186/s13072-016-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Spence JM, Critcher R, Ebersole TA, Valdivia MM, Earnshaw WC, Fukagawa T, Farr CJ, Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array, EMBO J. 21 (2002) 5269–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bergmann JH, Martins NM, Larionov V, Masumoto H, Earnshaw WC, HACking the centromere chromatin code: insights from human artificial chromosomes, Chromosome Res. 20 (2012) 505–519. 10.1007/s10577-012-9293-0. [DOI] [PubMed] [Google Scholar]
- [47].Quenet D, Dalal Y, A long non-coding RNA is required for targeting centromeric protein A to the human centromere, Elife. 3 (2014) e03254 10.7554/eLife.03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T, Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins, Nature. 410 (2001) 116–20. 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- [49].Kabeche L, Nguyen HD, Buisson R, Zou L, A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation, Science. 359 (2018) 108–114. 10.1126/science.aan6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH, Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin, Curr Biol. 13 (2003) 1192–1200. [DOI] [PubMed] [Google Scholar]
- [51].Martens JHA, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T, The profile of repeat-associated histone lysine methylation states in the mouse epigenome, EMBO J. 24 (2005) 800–12. 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jaco I, Canela A, Vera E, Blasco MA, Centromere mitotic recombination in mammalian cells, J. Cell Biol 181 (2008) 885–92. 10.1083/jcb.200803042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Scelfo, Fachinetti, Keeping the Centromere under Control: A Promising Role for DNA Methylation, Cells. 8 (2019) E912 10.3390/cells8080912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD, DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions, Hum Mol Genet. 18 (2009) 3178–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhang W, Lee HR, Koo DH, Jiang J, Epigenetic modification of centromeric chromatin: Hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize, Plant Cell. 20 (2008) 25–34. 10.1105/tpc.107.057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tanaka Y, Kurumizaka H, Yokoyama S, CpG methylation of the CENP-B box reduces human CENP-B binding, FEBS J. 272 (2005) 282–289. 10.1111/j.1432-1033.2004.04406.x. [DOI] [PubMed] [Google Scholar]
- [57].du Sart D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KH, A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA, Nat Genet. 16 (1997) 144–153. [DOI] [PubMed] [Google Scholar]
- [58].Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, Van Dyke DL, Willard HF, Zinn AB, Schwartz S, Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA, Hum Mol Genet. 6 (1997) 1195–1204. [DOI] [PubMed] [Google Scholar]
- [59].Voullaire LE, Slater HR, Petrovic V, Choo KH, A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere?, Am J Hum Genet. 52 (1993) 1153–1163. [PMC free article] [PubMed] [Google Scholar]
- [60].Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, V Shah J, Jansen LE, The quantitative architecture of centromeric chromatin, Elife. 3 (2014) e02137 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].V Irvine D, Amor DJ, Perry J, Sirvent N, Pedeutour F, Choo KH, Saffery R, Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres, Chromosome Res. 12 (2004) 805–815. [DOI] [PubMed] [Google Scholar]
- [62].Lo AW, Liao GC, Rocchi M, Choo KH, Extreme reduction of chromosome-specific alpha-satellite array is unusually common in human chromosome 21, Genome Res. 9 (1999) 895–908. [DOI] [PubMed] [Google Scholar]
- [63].Lo AW, Magliano DJ, Sibson MC, Kalitsis P, Craig JM, Choo KH, A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA, Genome Res. 11 (2001) 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Worrall JT, Tamura N, Mazzagatti A, Shaikh N, van Lingen T, Bakker B, Spierings DCJ, Vladimirou E, Foijer F, McClelland SE, Non-random Mis-segregation of Human Chromosomes, Cell Rep. 23 (2018) 3366–3380. 10.1016/j.celrep.2018.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Dumont M, Gamba R, Gestraud P, Klaasen S, Worrall JT, De Vries SG, Boudreau V, Salinas-Luypaert C, Maddox PS, Lens SM, Kops GJ, McClelland SE, Miga KH, Fachinetti D, Human chromosome-specific aneuploidy is influenced by DNA-dependent centromeric features., EMBO J. 39 (2019) e102924 10.15252/embj.2019102924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T, A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite, J Cell Biol. 109 (1989) 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Dumont M, Fachinetti D, DNA Sequences in Centromere Formation and Function, Prog. Mol. Subcell. Biol 56 (2017) 305–336. 10.1007/978-3-319-58592-5_13. [DOI] [PubMed] [Google Scholar]
- [68].Haaf T, Mater AG, Wienberg J, Ward DC, Presence and abundance of CENP-B box sequences in great ape subsets of primate-specific alpha-satellite DNA, J Mol Evol. 41 (1995) 487–491. [DOI] [PubMed] [Google Scholar]
- [69].Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW, DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function, Dev Cell. 33 (2015) 314–327. 10.1016/j.devcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H, CENP-B Controls Centromere Formation Depending on the Chromatin Context, Cell. 131 (2007) 1287–1300. [DOI] [PubMed] [Google Scholar]
- [71].Contreras-Galindo R, Fischer S, Saha AK, Lundy JD, Cervantes PW, Mourad M, Wang C, Qian B, Dai M, Meng F, Chinnaiyan A, Omenn GS, Kaplan MH, Markovitz DM, Rapid molecular assays to study human centromere genomics, Genome Res. 27 (2017) 2040–2049. 10.1101/gr.219709.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Pironon N, Puechberty J, Roizès G, Molecular and evolutionary characteristics of the fraction of human alpha satellite DNA associated with CENP-A at the centromeres of chromosomes 1, 5, 19, and 21, BMC Genomics. 11 (2010) 195 10.1186/1471-2164-11-195. [DOI] [PMC free article] [PubMed] [Google Scholar]


