Abstract
Newly assembled herpesvirus nucleocapsids traverse the intact nuclear envelope by a vesicle-mediated nucleo-cytoplasmic transport for final virion maturation in the cytoplasm. For this, they bud at the inner nuclear membrane resulting in primary enveloped particles in the perinuclear space (PNS) followed by fusion of the primary envelope with the outer nuclear membrane (ONM). While the conserved viral nuclear egress complex orchestrates the first steps, effectors of fusion of the primary virion envelope with the ONM are still mostly enigmatic but might include cellular proteins like SUN2 or ESCRT-III components. Here, we analyzed the influence of the only known AAA+ ATPases located in the endoplasmic reticulum and the PNS, the Torsins (Tor), on nuclear egress of the alphaherpesvirus pseudorabies virus. For this overexpression of wild type and mutant proteins as well as CRISPR/Cas9 genome editing was applied. Neither single overexpression nor gene knockout (KO) of TorA or TorB had a significant impact. However, TorA/B double KO cells showed decreased viral titers at early time points of infection and an accumulation of primary virions in the PNS pointing to a delay in capsid release during nuclear egress.
Keywords: herpesvirus, pseudorabies virus, nuclear egress, AAA+ ATPase, Torsin, CRISPR/Cas9
1. Introduction
Herpesviruses are double-stranded DNA viruses, which use the host cell nucleus and the cytoplasm for replication and morphogenesis. While transcription, DNA replication, assembly of viral capsids as well as viral genome encapsidation take place in the nucleus, the nucleocapsid has to be transferred to the cytoplasm for final virion maturation. With a diameter of approximately 125 nm, its size far exceeds the 40 nm threshold for passage through intact nuclear pores [1]. However, no evidence for a significant impairment or alteration of barrier and gating functions of nuclear pores was found even at late time points after infection [2] demonstrating continued integrity of the nuclear envelope.
In eukaryotic cells, the nuclear envelope (NE) separates the nuclear contents from the cytoplasm. It consists of two concentric lipid bilayers designated as the inner (INM) and outer nuclear membrane (ONM) which are separated by the perinuclear space (PNS). The PNS is contiguous with the lumen of the endoplasmic reticulum (ER) as is the ONM with the ER membrane. In contrast, the INM harbors a unique set of membrane proteins distinct from that of the ONM and ER. INM and ONM are fused at sites where nuclear pore complexes (NPCs) are inserted, which also allow the import of the herpesviral genome at early stages of infection. Traffic into and out of the nucleus is thought to occur exclusively through NPCs (reviewed in Adam [3], Knockenhauer and Schwartz [4]). Interestingly, herpesvirus nucleocapsids are translocated through the nuclear envelope (NE) by a vesicle-mediated process designated as nuclear egress (reviewed in [5,6,7]).
Budding of herpesvirus nucleocapsids at the INM is driven by the nuclear egress complex (NEC) composed of two conserved herpesviral proteins designated as pUL31 and pUL34 in the alphaherpesviruses pseudorabies virus (PrV) and herpes simplex viruses (HSV-1, -2) [5,6,7]. The NEC is not only required for efficient nuclear egress, thereby generating primary enveloped virions in the PNS, but also sufficient for vesicle formation and scission from artificial lipid membranes and the INM, [8,9,10,11]. In a subsequent step, this primary envelope fuses with the ONM to release the nucleocapsids into the cytoplasm (reviewed in [12]).
Budding of nucleocapsids at the INM is quite well understood at the molecular level, while the fusion process of the primary envelope with the ONM remains mostly enigmatic. In contrast to reports for HSV-1 [13], the viral fusion machinery which is active during entry of herpesviruses is not involved in nuclear egress of PrV [14]. In addition, a variety of different PrV gene deletion mutants studied so far showed no detectable effect on nuclear egress arguing against a virus-encoded fusion machinery active at the NE. Only mutants lacking the alphaherpesvirus specific protein kinase pUS3 showed an impairment of nuclear translocation. In the absence of pUS3 [15,16,17] or by impairment of its kinase function [18,19,20] primary enveloped virions accumulate in herniations of the INM. However, pUS3 is not essential for viral replication and viral titers are only approx. 10-fold reduced. Based on these data, we speculated that herpesviruses might use a cellular machinery either already present in or recruited to the NE.
Although vesicle-mediated transport processes between cytoplasmic organelles and the plasma membrane are well studied, knowledge on vesicular transport and fusion events at the NE is poor. An example of such a cellular mechanism would be the fusion of the INM with the ONM that occurs during NPC insertion in a growing interphase nucleus (reviewed in Otsuka and Ellenberg [21]). This process is thought to involve an inside-out extrusion of the INM into and across the PNS followed by its subsequent fusion with the ONM [22,23]. Although the molecular mechanism underlying INM-ONM fusion remains incompletely understood, several cellular proteins have been implicated in this process including the multi-subunit endosomal sorting complex required for transport III (ESCRT-III) and the ATPase-associated with various cellular activities (AAA+) protein Vps4 (reviewed in Otsuka and Ellenberg [21]). Recent work suggests that ESCRT-III and Vps4 are also important for herpesvirus nuclear egress [24,25,26], but conflicting findings have been reported [27,28].
Two other potential cellular mediators of INM-ONM fusion during interphase NPC insertion are Torsin A (TorA) and TorB. As similar to Vps4, Torsins belong to the AAA+ ATPase superfamily. They function as molecular chaperones, which use energy derived from ATP-hydrolysis to remodel their target molecules and are involved in numerous processes including budding and fission of vesicles, and assembly as well as disassembly of protein complexes [29,30,31,32,33,34,35]. Torsins are composed of a N-terminal signal peptide, followed by a hydrophobic stretch (TorA and TorB) upstream of the Walker A and B motifs, which mediate ATP binding and hydrolysis [32]. TorA (TOR1A) and TorB (TOR1B) are atypical AAA+ proteins for the following three reasons. First, they contain a non-canonical Walker A motif [36]. Second, they and the related Tor2 (TOR2A), Tor3 (TOR3A), and Tor4 (TOR4A) proteins are the only AAA+ proteins known to reside within the contiguous ER lumen and PNS of the nuclear envelope [32,37,38,39]. Third, they lack the conserved ATP-hydrolysis-promoting arginine finger [35,40]. Consistent with the lack of an arginine finger, purified Tor proteins are unable to hydrolyze ATP in vitro [41]. Instead, they need to be activated by the direct interaction with the luminal domain of one of two known regulatory protein cofactors: the INM lamina-associated polypeptide 1 (LAP1) or the ER/ONM protein luminal domain-like LAP1 (LULL1). Mutations in TorA lead to an autosomal dominant disease in humans, called early-onset torsion dystonia 1 (DYT1/TOR1A dystonia) [42]. The most frequent disease linked form TorA∆E302/303 lacks a single glutamic acid residue at position Glu302 or Glu303 in the C-terminus of the protein [43,44,45]. None of the other Torsins are implicated in human disease. Nonetheless, EQ mutations in the Walker B domain of TorA or TorB lead to expression of ATP hydrolysis-deficient Torsin molecules [33,45] which exert dominant-negative effects [38,46,47]. While the substrates of TorA and TorB remain unknown, they and the related proteins Tor2 and Tor3 appear to function in a partially redundant manner [48,49,50]. Furthermore, in previous studies, TorA has been shown to play a role in NE maintenance. Specifically, severe defects in NE architecture with “blebbing” of the INM in neuronal tissue are observed in knockout (KO) mice or mice lacking proper Torsin function, by expression of the dystonia-related allele TorAΔE302/303 [37,45,50,51,52,53]. This phenotype is reminiscent of the INM herniations, which were observed in cells overexpressing the NEC components [8] or in cells infected with US3-deletion mutants [15,16,17].
Consistent with a potential role during herpesvirus nuclear egress, overexpression of TorA or TorB resulted in slightly reduced HSV-1 titers in neuron-like and epithelial cells, as well as in the appearance of primary enveloped virions in cytoplasmic vesicles [54]. Moreover, HSV-1 replication was reduced in HeLa cells lacking both TorA and TorB [55]. To date, the molecular mechanism underlying the contribution of TorA and TorB to herpesvirus nuclear egress as well as interphase NPC biogenesis remains poorly defined. Nevertheless, a growing body of evidence supports the hypothesis that TorA is required for the assembly of functional linker of nucleoskeleton and cytoskeleton (LINC) complexes [39,48,56,57,58,59]. This conserved NE-spanning molecular bridge is present in all nucleated cells [60,61] and mechanically integrates the nucleus with the cytoskeleton mediating several fundamental cellular processes including cell division, DNA damage repair, meiotic chromosome pairing, mechano-regulation of gene expression, and nuclear positioning (reviewed in Meinke and Schirmer [62]).
LINC complexes are composed of ONM Klarsicht/ANC-1/SYNE homology (KASH)-domain and INM Sad1/UNC-84 homology (SUN)-domain containing proteins [63,64]. Although the LINC complex is involved in many essential cellular processes, it is still unknown how assembly and disassembly is achieved. TorA is reported to have affinity for the KASH domains of nesprin-1, -2, and -3 [56]. In addition, TorA was shown to interact with SUN1 and SUN2 in a heterologous system [48], while its localization to the NE was found to be SUN1-dependent [65]. Furthermore, a knockdown of TorA disrupted the localization of KASH proteins [66]. Interestingly, recent evidence proposed a role for Torsins in the translocation of large ribonucleoprotein (RNP) particles from the nucleus into the cytoplasm in neuromuscular junctions in Drosophila [67] through a pathway which mechanistically resembles nuclear egress of herpesvirus [68].
For PrV, we recently demonstrated that expression of the luminal SUN2 domain, which was described to disturb normal function in a dominant-negative (dn) manner [64], resulted in lower virus titers, a severe dilation of the PNS and the ER, and an escape of primary enveloped virions from the PNS into the ER [69]. Since this was similar to the effect reported for TorA overexpression on HSV-1 [54], we were interested to study the function of TorA and B in PrV infection. Here, we overexpressed GFP-tagged wild type or mutant proteins and used the CRISPR/Cas9 genome editing system for generation of cell lines lacking TorA, TorB and TorA/B to examine how modulation of their expression affects PrV replication with special focus on nuclear egress.
2. Material and Methods
2.1. Cells and Virus
Rabbit kidney cells (RK13, CCLV-Rie 109) were cultivated in Dulbecco’s modified Eagle’s minimum essential medium supplemented with 10% fetal calf serum, provided by the Friedrich-Loeffler-Institute bio bank (Greifswald, Insel Riems, Germany). PrV strain Kaplan (PrV-Ka) [70] was propagated on RK13 cells. RK13 cells were used throughout this study since (I) they propagate PrV to high titers; (II) are easy to transfect; (III) tolerate a wide panel of foreign protein expression; and (IV) are intensively studied in our laboratory for many years.
2.2. DNA Constructs
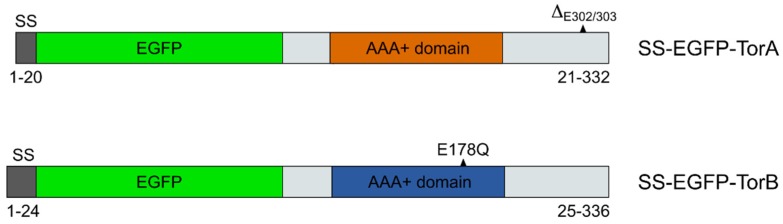
SS-EGFP-TorAWT, SS-EGFP-TorAΔE302/303, SS-EGFP-TorBWT, and SS-EGFP-TorBE178Q constructs used in this work had been described [37,44,52,57]. Plasmid pDsRed2-ER was purchased from Takara Bio Europe, Inc. Constructs used to perform CRISPR/Cas9-mediated genome editing were generated as follows. Guide RNAs (gRNAs) were designed by targeting the first exon of TorA (TOR1A) or TorB (TOR1B) as predicted in the rabbit genome OryCun2.0 (Oryctolagus cuniculus, ensemble.org [71]) with the help of the online tool (http://crispr.mit.edu/). Four gRNAs with the highest score and the lowest probability for off-target effects were selected for each gene (Table 1). gRNAs were ordered as unmodified DNA oligonucleotides (MWG Eurofins, Ebersberg, Germany) with BbsI restriction overhang, hybridized and inserted into the BbsI-digested vector pX330-NeoR (kindly provided by Dr. W. Fuchs), which is a modified version of pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene, Watertown, MA, USA, #42230) carrying an additional expression cassette for a G418 resistance for selection (previously described in Hübner, et al. [72]). The correct cloning of gRNAs was verified by Sanger sequencing at the Friedrich-Loeffler-Institut with HU6-F primer (5′-ATAATTTCTTGGGTAGTTTGCAG-3′).
Table 1.
Oligonucleotide sequences. Compatible 5′ overhangs for restriction enzyme BbsI used for cloning are underlined.
| Name | Sequence (5′–3′) |
|---|---|
| TorA_gRNA#1_Fwd | CACCCTGGCGGTAGCGCCGGTCGG |
| TorA_gRNA#1_Rev | AAACCCGACCGGCGCTACCGCCAG |
| TorA_gRNA#2_Fwd | CACCTGTCTGGCGGTAGCGCCGGT |
| TorA_gRNA#2_Rev | AAACACCGGCGCTACCGCCAGACA |
| TorA_gRNA#3_Fwd | CACCCGCCGGTCGGTGGTCAGCGC |
| TorA_gRNA#3_Rev | AAACCGCTGACCACCGACCGGCGG |
| TorA_gRNA#4_Fwd | CACCGTTCCTGCGCTGACCACCGA |
| TorA_gRNA#4_Rev | AAACGTCGGTGGTCAGCGCAGGAA |
| TorB_gRNA #1_Fwd | CACCGTGATTCTGAAGGCGCTGAC |
| TorB_gRNA #1_Rev | AAACGTCAGCGCCTTCAGAATCAC |
| TorB_gRNA #2_Fwd | CACCCGCCTTCAGAATCACTTCCG |
| TorB_gRNA #2_Rev | AAACCGGAAGTGATTCTGAAGGCG |
| TorB_gRNA #3_Fwd | CACCTTTTTGGTTTTTGGTAACGA |
| TorB_gRNA #3_Rev | AAACTCGTTACCAAAAACCAAAAA |
| TorB_gRNA #4_Fwd | CACCGAAGCTGTTCGGACAGCATC |
| TorB_gRNA #4_Rev | AAACGATGCTGTCCGAACAGCTTC |
2.3. Transfection of Cells for Co-Localization Studies
RK13 cells were seeded on coverslips in a 24-well dish and transiently co-transfected by calcium phosphate-precipitation [73] with an ER marker protein plasmid (pDsRed2-ER, Takara Bio Europe Saint-Germain-en-Laye, France) and plasmids expressing the GFP-tagged constructs. We used the calcium phosphate-coprecipitation method, although it is not very efficient, because it is milder to the cells and therefore allows to capture qualitative images later on.
2.4. Immunoblotting
Cells were transfected with 1 µg of plasmid DNA using polyethylenimine (PEI) [74], and harvested 24 h post transfection by scraping into the medium, pelleted, washed twice with phosphate-buffered saline (PBS) and lysed in SDS-containing sample buffer (0.13 M Tris-HCl, pH 6.8; 4% SDS; 20% glycerin; 0.01% bromophenol blue; 10% 2-mercaptoethanol). Here we used PEI transfection, instead of calcium phosphate-precipitation method, because PEI transfection is more efficient. Proteins were separated in SDS 10% polyacrylamide gels and after transfer to nitrocellulose membranes, blots were probed with a rabbit anti-GFP serum (kindly provided by Dr. G. M. Keil, FLI, Insel Riems, Germany) and a monoclonal antibody specific for alpha-tubulin (Sigma-Aldrich, Munich Germany, T5168) as loading control. After incubation with secondary peroxidase-labelled antibodies and substrate (Clarity ECL western Blot substrate, Bio-Rad, Feldkirchen, Germany), chemiluminescence was recorded in a Bio-Rad Versa Doc imager.
2.5. Generation of Stably Expressing RK13 Cell Lines
For generation of cells stably overexpressing wild type or mutant forms of Torsins A and B, cells in a 6-well dish were transfected by calcium phosphate-coprecipitation [73] using 1.5 µg of plasmid DNA expressing protein constructs schematically depicted in Figure 1. Two days after transfection, cells were transferred to 10 cm plates (Corning, Kaiserslautern, Germany) and selected in medium containing 500 µg/mL G418 (Invitrogen, Schwerte, Germany). Ten to 14 days after transfection GFP-positive cell colonies were picked by aspiration and further analyzed. Cell clones were seeded on cover slips in a 24-well plate for analysis of protein localization.
Figure 1.
Schematic depiction of expression constructs used in this study. All constructs carried EGFP at the N-terminus. Mutations resulting in loss-of-function in TorA and B are indicated. Numbers given below represent the corresponding amino acid residues of the Torsins in the constructs used. SS: signal sequence.
2.6. Generation of Stable RK13 Knockout Cell Lines
Stable KO cell lines were generated by co-transfection of all four gRNA-containing pX330-NeoR constructs (1.5 µg per plasmid) using calcium phosphate-co-precipitation [73]. For DKO, all eight plasmids were co-transfected simultaneously. Two days after transfection in 6 well dishes, cells were transferred to 10 cm plates (Corning) and selected in medium containing 500 µg/mL G418 (Invitrogen). Ten to 14 days after transfection cell colonies were picked by aspiration and tested for KO by sequencing of the targeted gene sequence.
2.7. Test for Bi-Allelic Gene Knockout
DNA of the potential KO cell clones was isolated using Bradley Lysis Buffer (10 mM Tris, 10 mM EDTA, 0.5% SDS, 10 mM NaCl) with Pronase (1 mg/mL) and following ethanol precipitation. The targeted gene region was amplified with Phusion® High-Fidelity DNA Polymerase (NEB, Frankfurt am Main, Germany) and primers given in Table 2. The gel purified phosphorylated PCR products were then blunt-end cloned into EcoRV-digested and dephosphorylated pBluescript II SK (+) (Stratagene, Darmstadt, Germany). Ten white colonies each were randomly picked [75], plasmid DNA was isolated and sequenced using the vector specific T7 primer by Sanger sequencing. In cases where all ten sequenced plasmids carried identical inserts, plasmids of five additional bacterial clones were isolated and sequenced. Mutations induced by Cas9 nuclease were identified by nucleotide sequence alignments with the rabbit genome (OryCun2.0) using Geneious 11.1.5 (https://www.geneious.com).
Table 2.
Primers used for amplification of targeted gene.
| Name | Sequence (5′–3′) |
|---|---|
| TorA_seq_Fwd | CACCGGAGACAGCTATAGCC |
| TorA_seq_Rev | GACCTTCTTGGCCAGATGCT |
| TorB_seq_Fwd | CCGCGCGAATGTGAAGTGCGCCCCCGTGGAAC |
| TorB_seq_Rev | GTCTTGTGCTCATGCGGGAAGTGCAGTGTG |
2.8. PrestoBlue Assay
Cell viability of modified and knockout cells was determined using Presto Blue™ Reagent (Thermo Scientific, Dreieich, Germany), a resazurin-based metabolic assay. RK13 wild type cells were used as control. 1 × 104 cells in 90 µL volume were seeded in a black 96-well plate with a flat and clear bottom (Corning). At 24, 48 and 72 h after seeding 10 µL Presto Blue Reagent was added to the cells and resuspended. The samples were incubated for 30 min at 37 °C. For each time point, cells were measured in triplicates and eight medium containing wells were included for background estimation. Before bottom-read measuring of fluorescence in a Tecan Reader at Ex560/Em590, the plate was shaken for 5 sec. Multiple reads per well (3 × 3) were performed using the i-control™ microtiter reader software. Blank-reduced raw data (fluorescence intensities) are given in the corresponding figures. For standardization, we used the following formula: (measured value – mean)/standard deviation. Statistics was applied on the standardized values.
2.9. In Vitro Replication Studies
To test the efficiency of PrV propagation in the generated cell lines, cells were infected with PrV-Ka at a multiplicity of infection (MOI) of 5. Cells and supernatants were harvested at different time points after infection (0, 4, 8, 12, 24 and 30 h p.i.). To determine the infectious virus titer, samples were thawed, cell debris was removed by centrifugation (2 min, 15,000 rpm), the supernatant was serially diluted (10−1 to 10−6) and used to infect RK13 cells in 24 well culture plates. After incubation for 1 h, the inoculum was replaced by a semi sold medium allowing only direct cell-to cell spread of the virus. Cells were fixed after 2 days with formaldehyde and stained with crystal violet. Virus plaques, which were detectable as holes in the blue-stained cell monolayer, were counted in at least two different wells and mean values were calculated as plaque forming units per milliliter (pfu/mL).
Shown are mean values of three (EGFP-TorA, -B overexpressing cells) or six (knockout cells) independent experiments with corresponding standard deviations. To exclude clonal and putative second site effects at least three different cell clones were tested initially for each mutated cell line.
2.10. Statistics
For each assay at least three independent experiments were performed. The statistical significance of the data presented in Figures 5, 6, 7 and 8 was determined by a two-way ANOVA followed by Dunnett’s multiple comparison test. All statistical tests were performed using GraphPad Prism version 8.1.0 (GraphPad Software, La Jolla, CA, USA). We compared the mean of each time point with the mean of the corresponding parental RK13 cells. A p-value ≤ 0.05 was considered significant and is presented in Figures 6 and 8 by the presence of asterisks (*, p ≤ 0.05, **, p ≤ 0.01, ****, p ≤ 0.0001).
2.11. Laser Scanning Confocal Microscopy
For confocal microscopy, we used stably expressing RK13 cells and RK13 cells transiently co-expressing the GFP-tagged plasmids and an ER-marker plasmid [73]. In addition, RK13 and Torsin knockout cells were infected with 250 pfu of PrV-Ka. Cells in 24 well dishes were fixed with 4% paraformaldehyde for 15 min one day after seeding for the stable expressing cells or two days after transient transfection. Infected cells were analyzed 18 h p.i. Fixed cells were washed three times and then incubated for 30 min with 50 mM NH4Cl in 1X PBS to quench the free aldehyde groups after PFA fixation. The GFP-tagged proteins and the DsRed-ER marker proteins were directly visualized via their autofluorescence. After permeabilization with 0.1% Triton X-100 in 1x PBS and subsequent blocking for 20 min with 0.25% skimmed milk the viral antigen was stained with a polyclonal rabbit serum specific for pUL34 (1:500, [76]). Alexa-Fluor 568-conjugated goat anti-rabbit IgG (dilution 1:1000, Invitrogen) was used to detect bound antibody. The nuclei were counterstained with 300 mM DAPI for 5 min and cells were mounted in a drop of Kaiser’s glycerol gelatin (Merck, Darmstadt, Germany). Samples were analyzed using with a confocal laser scanning microscope (Leica DMI 6000 TCS SP5, 63× oil-immersion objective, NA = 1.4; Leica, Wetzlar, Germany). Representative images were processed using the Fiji software [77,78]. Scale bars indicate 10 µm.
2.12. Ultrastructural Analyses
RK13 and KO cell lines were infected with PrV-Ka at an MOI of 1 for 14 h and processed for transmission electron microscopy as described previously [76]. Numbers of primary virions present in the PNS in infected RK13 and RK13-TorA/BDKO were counted in 10 different sections each.
3. Results
3.1. Influence of Torsin Overexpression on PrV Replication
To test whether overexpression of either the GFP-tagged wild type or mutated forms of (human) Torsins A and B has an effect on PrV replication, the different expression constructs (Figure 1) were transfected into RK13 cells for transient expression and generation of stably expressing cell lines. Torsins are well conserved in metazoans [34] and functional expression of the same constructs in murine cells was reported [66].
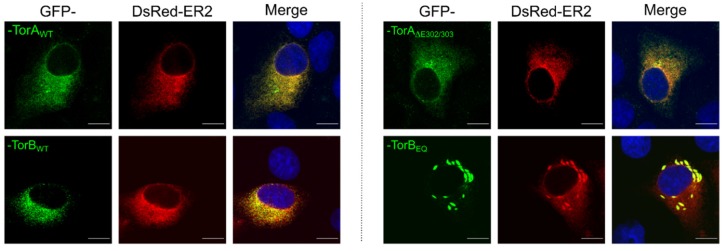
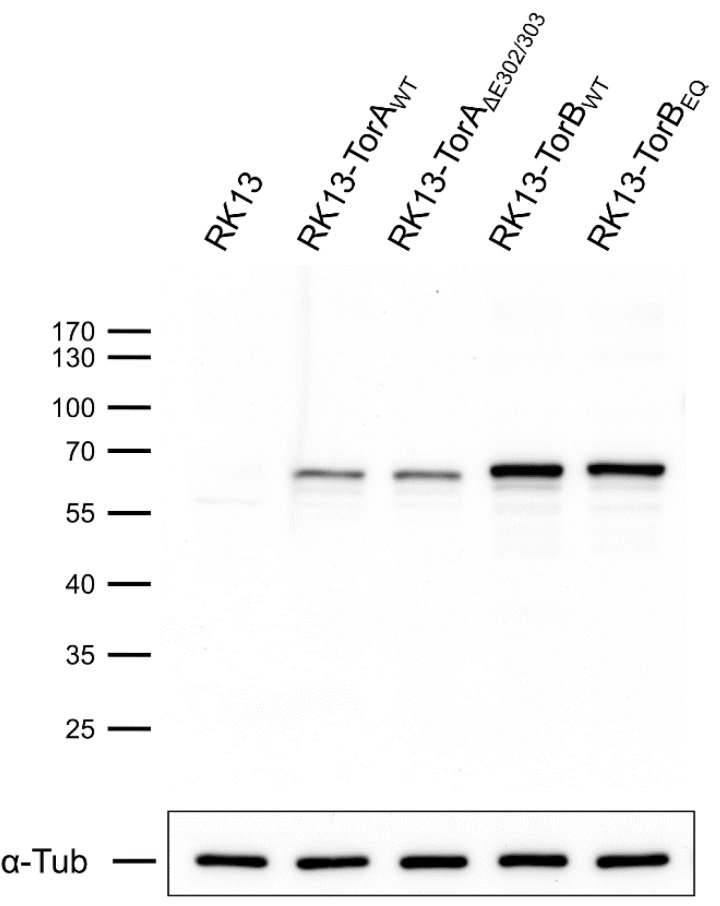
As expected, each of these constructs was targeted to the ER/NE when transiently expressed in RK13 cells (Figure 2) showing a clear colocalization with the DsRed2-tagged ER marker (DsRed2-ER) [38,46,47]. Consistent with previous reports, the expression of SS-EGFP-TorBE178Q resulted in the appearance of dense protein accumulations within the ER [46]. Furthermore, each of the proteins was expressed in RK13 cells at the predicted molecular mass evaluated in western blot analysis (Figure 3). The SS-EGFP-tagged TorA and TorB constructs were ~65–70 kDa, with expression levels of TorB slightly higher than for TorA. Taken together, these results demonstrate that the GFP-Torsin constructs are expressed properly in RK13 cells.
Figure 2.
Localization of GFP-tagged constructs in RK13 cells. Representative images of RK13 cells transiently expressing the GFP-tagged constructs. Cells were co-transfected with plasmids expressing the DsRed2-ER marker and the GFP-tagged Torsins. Nuclei were counterstained with DAPI and autofluorescence was detected with a confocal laser scanning microscope. Scale bars indicate 10 µm.
Figure 3.
Expression of GFP-tagged Torsins in RK13 cells. Lysates of transfected cells were harvested, and proteins were separated in SDS 10%-polyacrylamide gels. Blots were probed with a GFP-specific rabbit antiserum and a monoclonal antibody against α-tubulin as loading control. Molecular masses of marker proteins (in kDa) are indicated on the left.
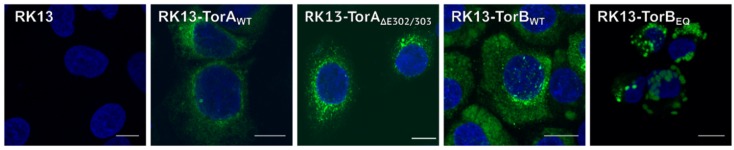
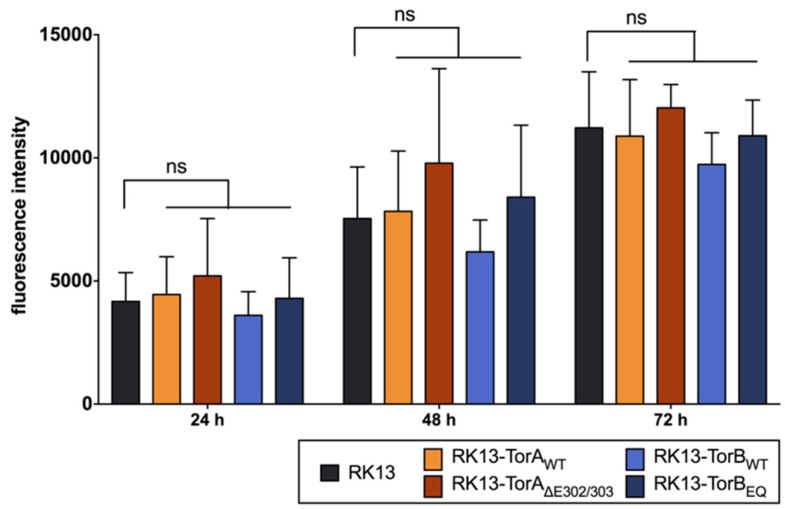
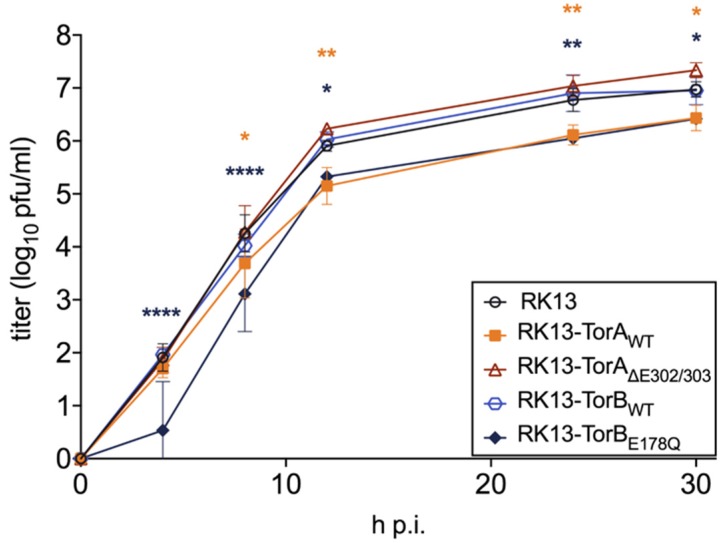
Stably expressing cell lines were selected for homogeneous GFP expression. The subcellular localization of each protein in these cell lines was indistinguishable from what was observed in transient expression (Figure 4). Ultrastructurally, cells overexpressing TorB showed a significant expansion of the rough ER with either diffuse matter (TorBWT) or filled with protein filaments (TorBE178Q) (data not shown) but no sinusoidal ER structures as reported for HeLa cells [46]. No deleterious effects on cellular metabolic activity were observed from overexpression of the GFP-tagged cellular genes (Figure 5). To test whether overexpression of these constructs influences virus replication, stably expressing cells as well as parental RK13 cells were infected with PrV strain Kaplan (PrV-Ka) [70] at an MOI of 5 and harvested at different time points after infection. As shown in Figure 6 small but significant 3- to 5-fold titer reduction was found after infection of RK13-TorAWT at all time points later than 8 h after infection, while cells expressing the mutant form TorA∆E302/303 supported PrV replication to similar titers as non-transgenic RK13 cells. For RK13-TorBWT cells there were no significant changes in viral titers compared to parental RK13 cells, while infection of TorBE178Q expressing cells resulted in 4 to 7-fold titer decrease at all time points after infection.
Figure 4.
Localization of stably expressed RK13-GFP-TorA and -TorB. Representative merged images show the intracellular localization of RK13 cells stably expressing the GFP-tagged mutant and wild type proteins. Nuclei were stained with DAPI, and GFP autofluorescence was detected with a confocal laser scanning microscope. Scale bars indicate 10 µm.
Figure 5.
Cell viability of the modified cell lines. RK13 control and stably expressing RK13-GFP-TorA and -TorB cells were seeded with 1 × 104 cells per well and at 24, 48, and 72 h post seeding the mitochondrial activity was measured (in fluorescence intensities) via the Presto Blue Assay. Shown is the mean of three independent experiments, ns = statistically not significant.
Figure 6.
Effects of overexpression of Torsins on PrV replication. Stably expressing cell lines and parental RK13 cells were infected with PrV-Ka (MOI of 5) and harvested at different time points. Progeny virus titers were determined on RK13 cells. Given are mean values of three independent experiments with corresponding standard deviations. Statistically significant differences compared to the parental RK13 were determined by GraphPad Prism software and are indicated by asterisks in the same color as the corresponding graphs (*, p ≤ 0.05, **, p ≤ 0.01, ****, p ≤ 0.0001).
3.2. Torsin A and Torsin B Are Required for Efficient PrV Replication in RK13 Cells
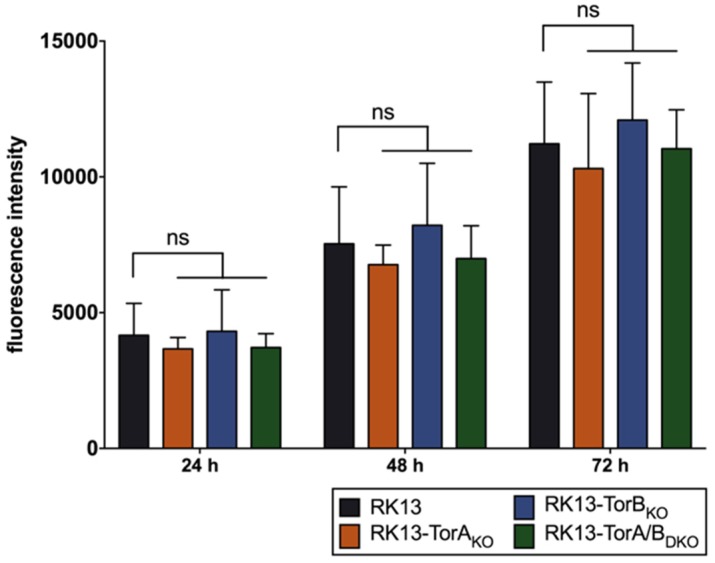
Torsin A and B are suggested to be functionally redundant [52]. Surprisingly, overexpression of the wild type TorA and the mutant TorBE178Q slightly impaired PrV replication while the other two forms had no significant impact pointing to different mechanisms. We were interested to analyze whether this effect might be more pronounced when both proteins are targeted simultaneously. Since equivalent simultaneous expression of both proteins in cell lines is difficult to achieve and maintain, we decided to use the CRISPR/Cas9 genome editing system to generate single and double KO cell lines for TorA and TorB. Four guide RNAs per gene were designed (Table 1), cloned into vector pX330-NeoR [72], and transfected simultaneously into RK13 cells. Genomic DNA of several cell clones was isolated, and the target region was amplified by PCR using primers given in Table 2. The PCR products were cloned into pBluescript SK+ and plasmid DNA from at least ten bacterial colonies each was isolated and sequenced. Wild type sequences in comparison to the mutations found in the different plasmids are summarized in Table 3. All cloned PCR products derived from RK13-TorBKO and RK13-TorA/BDKO exhibited only a single type of mutation indicating that both alleles carry the same deletion. Two different allelic variants were present in RK13-TorAKO cells. In-frame deletions were found in the TorB allele of RK13-TorA/BDKO cells (∆30 bp, both alleles). Unfortunately, the tested antisera, which are specific for human Torsins, did not detect the corresponding homologs in RK13 cells (data not shown). As similar to the stably expressing cells described above, no deleterious effects on metabolic activity were observed in the KO cell lines (Figure 7).
Table 3.
Summary of mutations detected in the KO cell lines.
| Mutant | Genotype | Sequence | Mutations | |
|---|---|---|---|---|
| TorAKO | wild type | GAACCGGAAGAGCGTGTCTGGCGGTAGCGCCG..GTCGGTGGTCAGCGCAGGAAGGCGCGGGGAGGCG | ||
| knockout |
GAACCGGAAGAGCGTGTCTGGCGGTAGCGCGGTGGT----GGTCAGCGCAGGAAGGCGCGGGGAGGCG (3)
GAACCGGAAGAGCGTGTCTGGCGGTAGCGCCG..GT----GGTCAGCGCAGGAAGGCGCGGGGAGGCG (7) |
1 nt Ex, 2 bp In, 4 bp Del4 bp Del | ||
| TorBKO | wild type | CTTGGAGAAGCTGTTCGGACAGCATCTGGCCACGGAAGTGATTCTGAAGGCGCTGACCGGCTTCAAGA | ||
| knockout | CTTGGAGAAGCTGTTCGGACAGC-----------------------------CTGACCGGCTTCAAGA (15) | 29 bp Del | ||
| TorA/BDKO | TorA | wild type | GAACCGGAAGAGCGTGTCTGGCGGTAGCGCCGGTCGGTGGTCAGCGCAGGAAGGCGCGGGGAGGCGCG | |
| knockout | GAACCGGAAGAGCGTGTCTGGCGGTAGCGCCCG-------TCAGCGCAGGAAGGCGCGGGGAGGCGCG (15) | 1 nt Ex, 7 bp Del | ||
| TorB | wild type | CTTGGAGAAGCTGTTCGGACAGCATCTGGCCACGGAAGTGATTCTGAAGGCGCTGACCGGCTTCAAGA | ||
| knockout | CTTGGAGAAGCTGTTCGGACA------------------------------GCTGACCGGCTTCAAGA (15) | 30 bp Del | ||
The sequence for each targeted gene region was compared to the sequence of the parental RK13 sequence. Numbers in brackets indicate the frequency of InDel (Insertion or Deletion) mutations found in the clones sequenced. Deletion (Del) of a base pairs (bp) is shown as hyphen, insertion (In) of a bp is marked by a dot in the parental sequence and nucleotide (nt) exchanges (Ex) are shown in bold characters.
Figure 7.
Cell viability of generated modified cell lines. RK13 control together with TorsinKO cell lines were seeded with 1 × 104 cells per well and at 24, 48, and 72 h post seeding the mitochondrial activity was measured (in fluorescence intensities) with the Presto Blue Assay. Shown is the mean of three independent experiments, ns = statistically not significant.
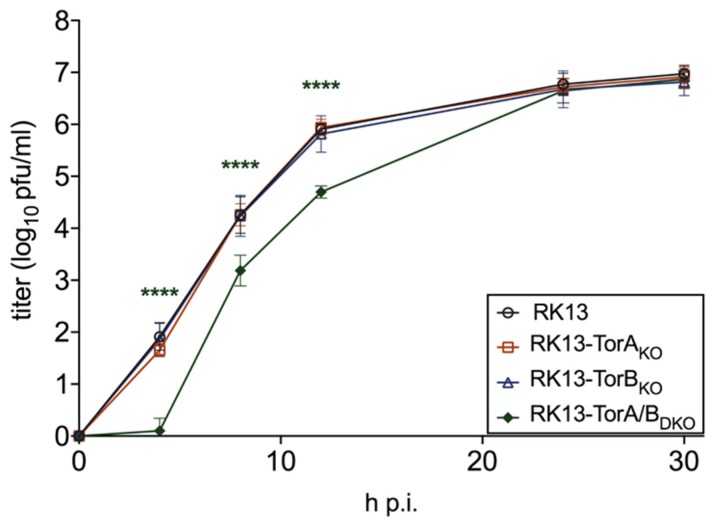
The generated KO and DKO cell lines were tested for virus propagation by infection with PrV-Ka at an MOI of 5 and harvested at different times after infection. As shown in Figure 8, no significant differences in viral progeny titers were found for RK13-TorAKO and RK13-TorBKO cells. In TorA/BDKO cells a significant drop in virus titer compared to the parental RK13 cells was observed at 4, 8 and 12 h after infection which disappeared at later time points.
Figure 8.
Effect of gene knockout on PrV replication. RK13 and TorsinKO cell lines were infected with PrV-Ka (MOI of 5) and harvested at different time points after infection. Progeny virus titers were determined on RK13 cells. Shown are mean values of six independent experiments. Statistics were done with GraphPad Prism software and asterisks indicate statistically significant differences compared to the parental RK13 in the same color as the corresponding graph (****, p ≤ 0.0001).
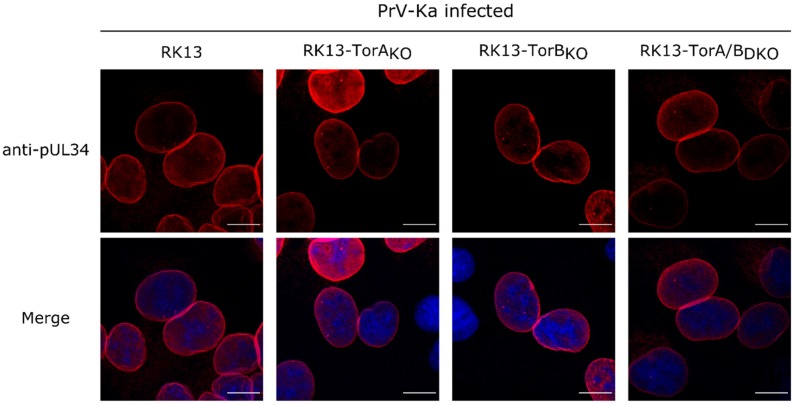
To test whether the absence of Torsin A and/or B influences nuclear envelope localization of the viral NEC component pUL34, parental RK13 and Torsin knockout cells were infected with PrV-Ka under plaque assay conditions. Cells were fixed after 18 h and stained with the monospecific anti-pUL34 rabbit serum [76]. As shown in Figure 9, no difference in nuclear rim staining for pUL34 was obvious independent of presence or absence of Torsins A, B or both proteins.
Figure 9.
Localization of the NEC component pUL34 in PrV-Ka infected RK13 and TorsinKO cells. Representative images showing undisturbed localization of the NEC component pUL34 (red) in infected RK13-TorsinKO cells. Nuclei were counterstained with DAPI (blue) and the fluorescence was imaged with a confocal laser scanning microscope. Scale bars indicate 10 µm.
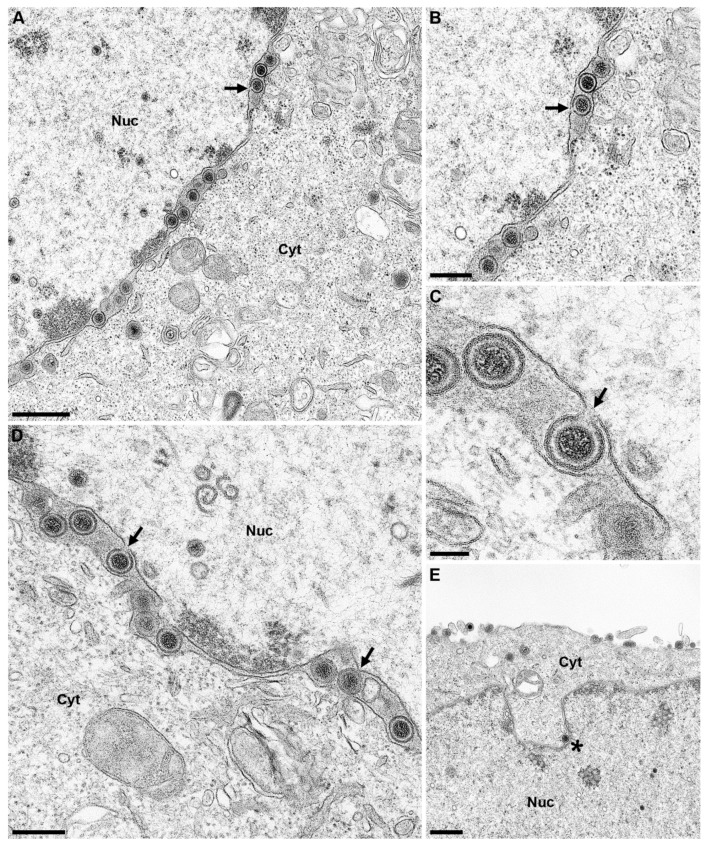
Although no drastic effect on viral titers was observed, we analyzed nuclear egress at the ultrastructural level. All KO cell lines were infected with PrV-Ka at an MOI of 1 and processed for electron microscopy. We did not observe any impairment in nuclear egress or virion morphogenesis in infected RK13-TorAKO and RK13-TorBKO (data not shown). In contrast to previous studies [45,51,52,53], nuclear envelope blebbing in the Torsin KO cells was not obvious. However, in PrV infected RK13-TorA/BDKO cells primary enveloped virions accumulated in the PNS which is in contrast to PrV infected parental RK13 cells, exhibiting only rare single virions in the PNS (Figure 10E; marked by an asterisk). We counted the number of primary enveloped virions in 10 sections each of PrV-Ka infected RK13 and RK13-TorA/BDKO cells. While in RK13 cells 5 primary enveloped virions could be detected in 30 nuclei, infected RK13-TorA/BDKO cells contained 593 primary virions in 52 nuclei. Compared to the accumulations observed in mutants lacking the pUS3 protein kinase [15], primary virions did not preferentially accumulate in herniations of the INM but were mainly found lined up in the PNS (Figure 10). Fission from the INM seemed to be less efficient in the absence of TorA/B since primary virions were frequently found still attached to the INM by a small neck (Figure 10, arrows).
Figure 10.
TorA/B DKO results in accumulation of primary enveloped virions in the PNS. RK13, RK13-TorAKO, RK13-TorBKO and RK13-TorA/BDKO cells were infected with PrV-Ka (MOI of 1) and processed for electron microscopic imaging 14 h p.i. No obvious effect was found for the single KO cells (data not shown), while the TorA/BDKO showed accumulations of primary virions within the PNS, many of them still connected to the INM by a short neck (indicated by arrows) (A–D). Panel (B) shows a higher magnification of the infected cell in panel (A), while panel (C) shows a higher magnification of panel (D). Panel (E) shows a rare case of a primary virion in the PNS in parental RK13 cells (marked by asterisk). Scale bars indicate 600 nm in panel (A,E), 300 nm in panel (B,C) and 100 nm in panel (D). Nuc: nucleus, Cyt: cytoplasm.
4. Discussion
Herpesvirus nucleocapsids rely on a vesicular pathway engaging the NE for nuclear egress. While the viral NEC orchestrates budding at and scission from the INM, no viral protein essential for de-envelopment at the ONM could be identified yet, while the pUS3 protein kinase exhibits a regulatory role (reviewed in Mettenleiter, Klupp and Granzow [5], Mettenleiter, Muller, Granzow and Klupp [7], and Johnson and Baines [6]).
Here, we analyzed a possible converging role for Torsin in herpesvirus nuclear egress by overexpressing them singly, and by generation of single and double KO cells using CRISPR/Cas9 based mutagenesis in the rabbit kidney cell line RK13. Torsins A and B seem to be ubiquitously expressed and are at least partially functionally redundant, complicating interpretation of experimental data generated by targeting only one form. Overexpression of TorAWT or TorBE178Q, which carries a mutation rendering the protein unable to hydrolyze ATP [46], resulted in a small but significant drop in virus titers compared to the non-transgenic RK13 control. A similar drop in virus titer was also observed for HSV-1 after infection of a neuronal cell line expressing TorAWT [54]. Primary enveloped virions escaped into the lumen of the ER as it was shown after overexpression of a dominant-negative SUN2 [69]. These data support the notion that Torsins regulate the SUN/nesprin interaction and that an intact LINC complex/Torsin relationship is necessary to restrict primary virions in the PNS [54]. However, neither TorAWT nor TorBE178Q expression resulted in an obvious increase of the number of primary virions in cytoplasmic structures, alteration of the nuclear envelope or the spacing between the INM and ONM (data not shown). Due to the lack of antibodies detecting SUN proteins in RK13 cells, an influence of Tor expression on the LINC could not be tested.
We also generated single and double knockout cells by CRISPR/Cas9 mutagenesis, which was verified by sequencing of the corresponding gene regions. No obvious differences in growth or metabolic activity were observed between the modified cell lines stably expressing the respective Torsin constructs or in Torsin KO cell lines, compared to parental RK13 cells. In addition, we did not detect any morphological defects, indicating that the targeted proteins are non-essential for cellular proliferation under our cell culture conditions. Since we could not exclude second site effects, we always included different cell clones for each KO in the preliminary screens. In none of the KO cell lines wild-type sequences could be identified. Unfortunately, the available antisera against the corresponding proteins of human origin did not react with the rabbit homologs in the RK13 cell lysates. The used gRNAs were designed to target exons that are shared by all transcript variants of the gene of interest to minimize the chance that shorter but still functional protein isoforms might be expressed. Although the RK13-TorA/BDKO cell line carries an in-frame deletion (aa 76–85) upstream of the Walker A motif (aa 105–112) in both alleles of the TorB gene and we cannot exclude expression of a truncated protein, the observed effects indicate a significant loss of function.
In contrast to the overexpression experiments, in which a slight but significant drop in progeny virus titers was found after infection of RK13-TorAWT and RK13-TorBEQ cells, none of the single KO cells showed a significant effect on infectious virus production or on localization of the NEC component pUL34. In ultrastructural analyses, we could not detect perturbations of the NE in cells lacking TorA or B, and primary virions were only rarely detected in the PNS or in cytoplasmic vesicles arguing against an impairment of nuclear egress. However, after infection of RK13-TorA/BDKO cells a drop in virus titer occurred at early time points after infection. High-resolution imaging revealed striking accumulations of primary enveloped virions lined-up in the PNS, while accumulations in herniations of the ONM were only rarely detected. Many of the primary envelopes seemed to be connected to the INM indicating that in the absence of Torsin A and B scission might be impaired (Figure 10). In line with this, in a HeLa cell line where all four known Torsins had been eliminated simultaneously, NE blebs still connected to the INM had been described [50]. Unfortunately, these cell lines were not tested for effects on herpesvirus nuclear egress. It is tempting to speculate that not only Torsin A and B, but also other Torsins might be involved in this process. Incomplete scission of primary HSV-1 virions from the INM was also reported after depletion of ESCRT-III proteins [24]. It might be speculated that recruitment of an AAA+ ATPase (Torsin/Vps4) might alleviate the scission process mediated by the NEC proteins.
It should be noted that we did not observe clear blebs of the INM into the PNS of our TorA/BDKO cells, which contrasts previous reports from fibroblasts [52], HeLa cells [50], or neurons [53].
Torsins are supposed to function as regulators of the LINC complex [56] and an intact LINC complex may be required for efficient nuclear egress by restricting primary virions to the PNS in close proximity to the NE for efficient fusion of the primary envelope with the ONM [69]. In our TorA/BDKO cells no impact on spacing between INM and ONM was apparent indicating no impairment of LINC.
In summary, we demonstrate that Torsins A and B, which might be involved in proper functioning of the LINC complex, play a role during nuclear egress of herpesvirus capsids. These results together with our previous findings on SUN2 impairment highlight the importance of this complex for vesicle-mediated herpesvirus capsid transport through the nuclear envelope.
Acknowledgments
We thank Karla Günther, Cindy Krüper, and Petra Meyer for technical support, Mandy Jörn for help with electron microscopic images and Susanne Amler for help with statistics. The pX330-NeoR vector was kindly provided by Walter Fuchs, the anti-EGFP serum by Günther M. Keil, the pDsRed2-ER marker plasmid by Birke A. Tews and the PrestoBlue Reagent by Anja Bauer. Further, we thank the Friedrich-Loeffler-Institut Collection of Cell Lines for their assistance in supplying cell lines and media.
Author Contributions
Conceptualization, T.C.M., B.G.K. and J.E.H.; methodology, J.E.H. and K.F.; software, J.E.H.; validation, T.C.M., B.G.K. and J.E.H.; formal analysis, J.E.H.; investigation, J.E.H.; resources, G.W.G.L.; data curation, T.C.M., B.G.K. and J.E.H.; writing—original draft preparation, J.E.H. and B.G.K.; writing—review and editing, T.C.M., B.G.K. and G.W.G.L.; visualization, J.E.H. and K.F.; supervision, T.C.M. and B.G.K.; project administration, T.C.M. and B.G.K.; funding acquisition, T.C.M. and B.G.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Deutsche Forschungsgemeinschaft (DFG ME 854/12-2 to TCM) and the National Institutes of Health (GM129374 to GWGL).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Pante N., Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hofemeister H., O’Hare P. Nuclear pore composition and gating in herpes simplex virus-infected cells. J. Virol. 2008;82:8392–8399. doi: 10.1128/JVI.00951-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adam S.A. The nuclear pore complex. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-reviews0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knockenhauer K.E., Schwartz T.U. The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell. 2016;164:1162–1171. doi: 10.1016/j.cell.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mettenleiter T.C., Klupp B.G., Granzow H. Herpesvirus assembly: An update. Virus Res. 2009;143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Johnson D.C., Baines J.D. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 2011;9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 7.Mettenleiter T.C., Muller F., Granzow H., Klupp B.G. The way out: What we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 2013;15:170–178. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- 8.Klupp B.G., Granzow H., Fuchs W., Keil G.M., Finke S., Mettenleiter T.C. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. USA. 2007;104:7241–7246. doi: 10.1073/pnas.0701757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bigalke J.M., Heuser T., Nicastro D., Heldwein E.E. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat. Commun. 2014;5:4131. doi: 10.1038/ncomms5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai P.J., Pryce E.N., Henson B.W., Luitweiler E.M., Cothran J. Reconstitution of the Kaposi’s sarcoma-associated herpesvirus nuclear egress complex and formation of nuclear membrane vesicles by coexpression of ORF67 and ORF69 gene products. J. Virol. 2012;86:594–598. doi: 10.1128/JVI.05988-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorenz M., Vollmer B., Unsay J.D., Klupp B.G., Garcia-Saez A.J., Mettenleiter T.C., Antonin W. A single herpesvirus protein can mediate vesicle formation in the nuclear envelope. J. Biol. Chem. 2015;290:6962–6974. doi: 10.1074/jbc.M114.627521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mettenleiter T.C. Herpesvirus assembly and egress. J. Virol. 2002;76:1537–1547. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farnsworth A., Wisner T.W., Webb M., Roller R., Cohen G., Eisenberg R., Johnson D.C. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. USA. 2007;104:10187–10192. doi: 10.1073/pnas.0703790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klupp B., Altenschmidt J., Granzow H., Fuchs W., Mettenleiter T.C. Glycoproteins required for entry are not necessary for egress of pseudorabies virus. J. Virol. 2008;82:6299–6309. doi: 10.1128/JVI.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klupp B.G., Granzow H., Mettenleiter T.C. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 2001;82:2363–2371. doi: 10.1099/0022-1317-82-10-2363. [DOI] [PubMed] [Google Scholar]
- 16.Wagenaar F., Pol J.M., Peeters B., Gielkens A.L., de Wind N., Kimman T.G. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. Pt 7J. Gen. Virol. 1995;76:1851–1859. doi: 10.1099/0022-1317-76-7-1851. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds A.E., Wills E.G., Roller R.J., Ryckman B.J., Baines J.D. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 2002;76:8939–8952. doi: 10.1128/JVI.76.17.8939-8952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher D., McKinney C., Kaufer B.B., Osterrieder N. Enzymatically inactive U(S)3 protein kinase of Marek’s disease virus (MDV) is capable of depolymerizing F-actin but results in accumulation of virions in perinuclear invaginations and reduced virus growth. Virology. 2008;375:37–47. doi: 10.1016/j.virol.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato A., Liu Z., Minowa A., Imai T., Tanaka M., Sugimoto K., Nishiyama Y., Arii J., Kawaguchi Y. Herpes simplex virus 1 protein kinase Us3 and major tegument protein UL47 reciprocally regulate their subcellular localization in infected cells. J. Virol. 2011;85:9599–9613. doi: 10.1128/JVI.00845-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sehl J., Portner S., Klupp B.G., Granzow H., Franzke K., Teifke J.P., Mettenleiter T.C. Roles of the different isoforms of the pseudorabies virus protein kinase pUS3 in nuclear egress. J. Virol. 2020 doi: 10.1128/JVI.02029-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otsuka S., Ellenberg J. Mechanisms of nuclear pore complex assembly—Two different ways of building one molecular machine. FEBS Lett. 2018;592:475–488. doi: 10.1002/1873-3468.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D’Angelo M.A., Anderson D.J., Richard E., Hetzer M.W. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- 23.Dultz E., Ellenberg J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J. Cell Biol. 2010;191:15–22. doi: 10.1083/jcb.201007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arii J., Watanabe M., Maeda F., Tokai-Nishizumi N., Chihara T., Miura M., Maruzuru Y., Koyanagi N., Kato A., Kawaguchi Y. ESCRT-III mediates budding across the inner nuclear membrane and regulates its integrity. Nat. Commun. 2018;9:3379. doi: 10.1038/s41467-018-05889-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C.P., Liu G.T., Kung H.N., Liu P.T., Liao Y.T., Chow L.P., Chang L.S., Chang Y.H., Chang C.W., Shu W.C., et al. The Ubiquitin Ligase Itch and Ubiquitination Regulate BFRF1-Mediated Nuclear Envelope Modification for Epstein-Barr Virus Maturation. J. Virol. 2016;90:8994–9007. doi: 10.1128/JVI.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee C.P., Liu P.T., Kung H.N., Su M.T., Chua H.H., Chang Y.H., Chang C.W., Tsai C.H., Liu F.T., Chen M.R. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr Virus. PLoS Pathog. 2012;8:e1002904. doi: 10.1371/journal.ppat.1002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crump C.M., Yates C., Minson T. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 2007;81:7380–7387. doi: 10.1128/JVI.00222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kharkwal H., Smith C.G., Wilson D.W. Blocking ESCRT-mediated envelopment inhibits microtubule-dependent trafficking of alphaherpesviruses in vitro. J. Virol. 2014;88:14467–14478. doi: 10.1128/JVI.02777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 30.Erzberger J.P., Berger J.M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 31.Laudermilch E., Schlieker C. Torsin ATPases: Structural insights and functional perspectives. Curr. Opin. Cell Biol. 2016;40:1–7. doi: 10.1016/j.ceb.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breakefield X.O., Kamm C., Hanson P.I. TorsinA: Movement at many levels. Neuron. 2001;31:9–12. doi: 10.1016/S0896-6273(01)00350-6. [DOI] [PubMed] [Google Scholar]
- 33.Hanson P.I., Whiteheart S.W. AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 34.Rose A.E., Brown R.S., Schlieker C. Torsins: Not your typical AAA+ ATPases. Crit. Rev. Biochem. Mol. Biol. 2015;50:532–549. doi: 10.3109/10409238.2015.1091804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sosa B.A., Demircioglu F.E., Chen J.Z., Ingram J., Ploegh H.L., Schwartz T.U. How lamina-associated polypeptide 1 (LAP1) activates Torsin. eLife. 2014;3:e03239. doi: 10.7554/eLife.03239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy M., Wu H.C., Liu Z., Kedzierska-Mieszkowska S., Zolkiewski M. Walker-A threonine couples nucleotide occupancy with the chaperone activity of the AAA+ ATPase ClpB. Protein Sci. 2009;18:287–293. doi: 10.1002/pro.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jungwirth M., Dear M.L., Brown P., Holbrook K., Goodchild R. Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum. Mol. Genet. 2010;19:888–900. doi: 10.1093/hmg/ddp557. [DOI] [PubMed] [Google Scholar]
- 38.Kustedjo K., Bracey M.H., Cravatt B.F. Torsin A and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J. Biol. Chem. 2000;275:27933–27939. doi: 10.1074/jbc.M910025199. [DOI] [PubMed] [Google Scholar]
- 39.Vander Heyden A.B., Naismith T.V., Snapp E.L., Hodzic D., Hanson P.I. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol. Biol. Cell. 2009;20:2661–2672. doi: 10.1091/mbc.e09-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown R.S., Zhao C., Chase A.R., Wang J., Schlieker C. The mechanism of Torsin ATPase activation. Proc. Natl. Acad. Sci. USA. 2014;111:E4822–E4831. doi: 10.1073/pnas.1415271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao C., Brown R.S., Chase A.R., Eisele M.R., Schlieker C. Regulation of Torsin ATPases by LAP1 and LULL1. Proc. Natl. Acad. Sci. USA. 2013;110:E1545–E1554. doi: 10.1073/pnas.1300676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozelius L.J., Hewett J.W., Page C.E., Bressman S.B., Kramer P.L., Shalish C., de Leon D., Brin M.F., Raymond D., Corey D.P., et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- 43.Gonzalez-Alegre P. Advances in molecular and cell biology of dystonia: Focus on torsinA. Neurobiol. Dis. 2019;127:233–241. doi: 10.1016/j.nbd.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Goodchild R.E., Dauer W.T. Mislocalization to the nuclear envelope: An effect of the dystonia-causing torsinA mutation. Proc. Natl. Acad. Sci. USA. 2004;101:847–852. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naismith T.V., Heuser J.E., Breakefield X.O., Hanson P.I. TorsinA in the nuclear envelope. Proc. Natl. Acad. Sci. USA. 2004;101:7612–7617. doi: 10.1073/pnas.0308760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose A.E., Zhao C., Turner E.M., Steyer A.M., Schlieker C. Arresting a Torsin ATPase reshapes the endoplasmic reticulum. J. Biol. Chem. 2014;289:552–564. doi: 10.1074/jbc.M113.515791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hewett J., Gonzalez-Agosti C., Slater D., Ziefer P., Li S., Bergeron D., Jacoby D.J., Ozelius L.J., Ramesh V., Breakefield X.O. Mutant torsinA, responsible for early-onset torsion dystonia, forms membrane inclusions in cultured neural cells. Hum. Mol. Genet. 2000;9:1403–1413. doi: 10.1093/hmg/9.9.1403. [DOI] [PubMed] [Google Scholar]
- 48.Chalfant M., Barber K.W., Borah S., Thaller D., Lusk C.P. Expression of TorsinA in a heterologous yeast system reveals interactions with lumenal domains of LINC and nuclear pore complex components. Mol. Biol. Cell. 2019;30:530–541. doi: 10.1091/mbc.E18-09-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pappas S.S., Liang C.C., Kim S., Rivera C.O., Dauer W.T. TorsinA dysfunction causes persistent neuronal nuclear pore defects. Hum. Mol. Genet. 2018;27:407–420. doi: 10.1093/hmg/ddx405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laudermilch E., Tsai P.L., Graham M., Turner E., Zhao C., Schlieker C. Dissecting Torsin/cofactor function at the nuclear envelope: A genetic study. Mol. Biol. Cell. 2016;27:3964–3971. doi: 10.1091/mbc.E16-07-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodchild R.E., Kim C.E., Dauer W.T. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 52.Kim C.E., Perez A., Perkins G., Ellisman M.H., Dauer W.T. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc. Natl. Acad. Sci. USA. 2010;107:9861–9866. doi: 10.1073/pnas.0912877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanabe L.M., Liang C.C., Dauer W.T. Neuronal Nuclear Membrane Budding Occurs during a Developmental Window Modulated by Torsin Paralogs. Cell Rep. 2016;16:3322–3333. doi: 10.1016/j.celrep.2016.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maric M., Shao J., Ryan R.J., Wong C.S., Gonzalez-Alegre P., Roller R.J. A functional role for TorsinA in herpes simplex virus 1 nuclear egress. J. Virol. 2011;85:9667–9679. doi: 10.1128/JVI.05314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner E.M., Brown R.S., Laudermilch E., Tsai P.L., Schlieker C. The Torsin Activator LULL1 Is Required for Efficient Growth of Herpes Simplex Virus 1. J. Virol. 2015;89:8444–8452. doi: 10.1128/JVI.01143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nery F.C., Zeng J., Niland B.P., Hewett J., Farley J., Irimia D., Li Y., Wiche G., Sonnenberg A., Breakefield X.O. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J. Cell Sci. 2008;121:3476–3486. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saunders C.A., Harris N.J., Willey P.T., Woolums B.M., Wang Y., McQuown A.J., Schoenhofen A., Worman H.J., Dauer W.T., Gundersen G.G., et al. TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement. J. Cell Biol. 2017;216:657–674. doi: 10.1083/jcb.201507113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gill N.K., Ly C., Kim P.H., Saunders C.A., Fong L.G., Young S.G., Luxton G.W.G., Rowat A.C. DYT1 Dystonia Patient-Derived Fibroblasts Have Increased Deformability and Susceptibility to Damage by Mechanical Forces. Front. Cell. Dev. Biol. 2019;7:103. doi: 10.3389/fcell.2019.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dominguez Gonzalez B., Billion K., Rous S., Pavie B., Lange C., Goodchild R. Excess LINC complexes impair brain morphogenesis in a mouse model of recessive TOR1A disease. Hum. Mol. Genet. 2018;27:2154–2170. doi: 10.1093/hmg/ddy125. [DOI] [PubMed] [Google Scholar]
- 60.Zhou X., Graumann K., Evans D.E., Meier I. Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J. Cell Biol. 2012;196:203–211. doi: 10.1083/jcb.201108098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Razafsky D., Hodzic D. Bringing KASH under the SUN: The many faces of nucleo-cytoskeletal connections. J. Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meinke P., Schirmer E.C. LINC’ing form and function at the nuclear envelope. FEBS Lett. 2015;589:2514–2521. doi: 10.1016/j.febslet.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 63.Starr D.A., Fridolfsson H.N. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., Hodzic D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jungwirth M.T., Kumar D., Jeong D.Y., Goodchild R.E. The nuclear envelope localization of DYT1 dystonia torsinA-DeltaE requires the SUN1 LINC complex component. BMC Cell Biol. 2011;12:24. doi: 10.1186/1471-2121-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saunders C.A., Luxton G.W. LINCing defective nuclear-cytoskeletal coupling and DYT1 dystonia. Cell. Mol. Bioeng. 2016;9:207–216. doi: 10.1007/s12195-016-0432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jokhi V., Ashley J., Nunnari J., Noma A., Ito N., Wakabayashi-Ito N., Moore M.J., Budnik V. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 2013;3:988–995. doi: 10.1016/j.celrep.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Speese S.D., Ashley J., Jokhi V., Nunnari J., Barria R., Li Y., Ataman B., Koon A., Chang Y.T., Li Q., et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149:832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klupp B.G., Hellberg T., Granzow H., Franzke K., Dominguez Gonzalez B., Goodchild R.E., Mettenleiter T.C. Integrity of the Linker of Nucleoskeleton and Cytoskeleton Is Required for Efficient Herpesvirus Nuclear Egress. J. Virol. 2017;91 doi: 10.1128/JVI.00330-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaplan A.S., Vatter A.E. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959;7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 71.Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Giron C.G., et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hübner A., Petersen B., Keil G.M., Niemann H., Mettenleiter T.C., Fuchs W. Efficient inhibition of African swine fever virus replication by CRISPR/Cas9 targeting of the viral p30 gene (CP204L) Sci. Rep. 2018;8:1449. doi: 10.1038/s41598-018-19626-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Graham F.L., van der Eb A.J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 74.Boussif O., Lezoualc’h F., Zanta M.A., Mergny M.D., Scherman D., Demeneix B., Behr J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ni W., Qiao J., Hu S., Zhao X., Regouski M., Yang M., Polejaeva I.A., Chen C. Efficient gene knockout in goats using CRISPR/Cas9 system. PLoS ONE. 2014;9:e106718. doi: 10.1371/journal.pone.0106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klupp B.G., Granzow H., Mettenleiter T.C. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 2000;74:10063–10073. doi: 10.1128/JVI.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]