Abstract
Apoptotic cell clearance (efferocytosis) elicits an anti-inflammatory response by phagocytes, but the mechanisms underlying this response are still being defined. Here, we uncover a chloride-sensing signaling pathway that controls both the phagocyte appetite and its anti-inflammatory response. Efferocytosis transcriptionally altered the genes coding for solute carrier (SLC) proteins SLC12A2 and SLC12A4. Interfering with SLC12A2 expression or function led to significantly enhanced corpse uptake per phagocyte, while loss of SLC12A4 inhibited corpse uptake. In SLC12A2-deficient phagocytes the canonical anti-inflammatory program was replaced by pro-inflammatory and oxidative stress-associated gene programs. This ‘switch’ to pro-inflammatory sensing of apoptotic cells was due to disruption of the chloride-sensing pathway (and not corpse overload or poor degradation,) and the chloride-sensing kinases WNK1-OSR1-SPAK that function upstream of SLC12A2 similarly affected efferocytosis. Collectively, the WNK1-OSR1-SPAK-SLC12A2/SLC12A4 chloride-sensing pathway and chloride flux in phagocytes act as key modifiers of how a phagocyte interprets the engulfed apoptotic corpse.
Every day, we turnover >200 billion cells in the body via apoptosis as part of normal homeostasis1-4. These apoptotic cells are removed by the process of ‘efferocytosis’, which involves specific recognition and uptake by professional phagocytes (such as macrophages and dendritic cells) and non-professional phagocytes (such as epithelial cells)5-9. Research from a number of laboratories has identified a series of steps in efferocytosis, including: the release of soluble factors from apoptotic cells (‘find-me’ signals or the ‘smell’ phase) that help the phagocyte sense the corpses, the specific ligands on the apoptotic cell surface (‘eat-me’ signals) that are recognized by specific receptors on the phagocytes (the ‘taste’ phase), and corpse internalization involving extensive cytoskeletal reorganization and subsequent digestion of corpse contents (the ‘ingestion’ phase)10-15.
It is now well-established that defects or failures in proper removal of apoptotic cells can result in inflammation with a predilection toward autoimmune disease, including systemic lupus erythematosus and chronic inflammation-associated diseases, such as atherosclerosis and ulcerative colitis1, 3, 16, 17. Thus, understanding the key molecules and mechanisms that regulate the distinct steps of efferocytosis can help target these players and pathways, and potentially modulate efferocytosis and inflammation. Ingesting a corpse with potentially ‘dangerous’ contents and managing this cargo while maintaining homeostasis is a unique challenge for the phagocyte. An added complexity is that most tissues have fewer phagocytes than the number of cells undergoing apoptosis, requiring individual phagocytes to successively ingest multiple apoptotic targets1, 2, 4. While significant progress has been made in deciphering key membrane receptors on phagocytes and ligands on apoptotic cells, our knowledge of molecular players regulating the phagocyte response after a corpse is ingested is just emerging. Efferocytic phagocytes also perform two concurrent actions: dampen pro-inflammatory mediators and activate production of anti-inflammatory mediators (i.e. ’immunosuppressive’)1. Thus, defining molecular players that regulate phagocyte responses after corpse uptake and from becoming pro-inflammatory are important, and a focus of this work.
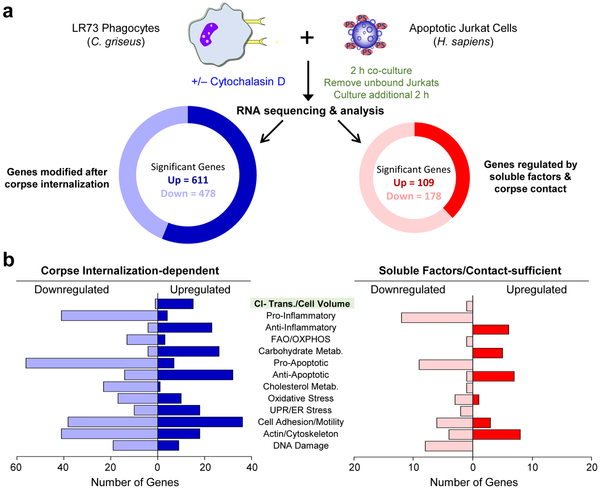
While some genes in phagocytes are induced by specific find-me signals18, others can be initiated only by phagocyte:apoptotic cell contact (without corpse internalization)19-22. However, our knowledge of specific transcriptional programs initiated after a corpse is internalized is limited. To address this, we performed RNAseq analysis, comparing RNA from phagocytes either eating apoptotic cells normally, or phagocytes capable of binding but not internalizing corpses (Fig. 1a). Also, to better distinguish the phagocyte-derived RNA (from that of the corpse), we used hamster LR73 fibroblasts as phagocytes and apoptotic human Jurkat T cells as targets. We utilized LR73 fibroblasts to overcome the issue of corpse-derived RNA contamination in the phagocyte RNAseq datasets 23; comparison of hamster versus human coding genomes allowed us to specifically focus on phagocyte-specific gene modulation (which was not feasible with mouse versus human comparison). In subsequent analyses, we addressed gene programs induced after corpse internalization from those induced by corpse contact/soluble factors (Fig. 1a). About 5x more genes were modulated in phagocytes by corpse internalization compared to apoptotic cell supernatant/contact (Fig. 1a). Among the corpse internalization-specific genes, phagocytes upregulated gene programs linked to chloride transport/cell volume, carbohydrate metabolism, anti-apoptotic responses, cell adhesion/motility, actin/cytoskeleton rearrangement, and anti-inflammatory responses (Fig. 1b), while downregulating genes associated with fatty acid oxidation, oxidative phosphorylation, pro-apoptotic responses, cholesterol metabolism, oxidative stress, and pro-inflammatory responses (Fig. 1b). Thus, upon engulfing an apoptotic corpse, phagocytes induce distinct gene programs.
Fig. 1:
Identification of corpse internalization-dependent signals in engulfing phagocytes
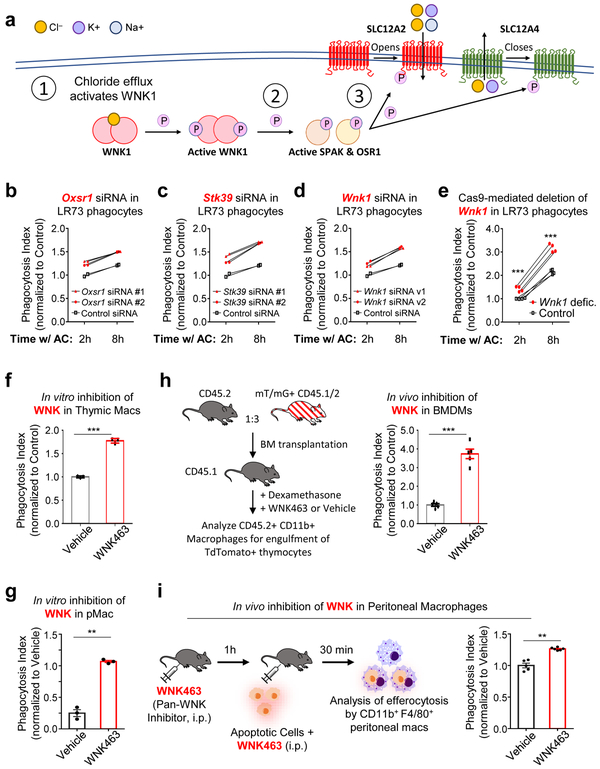
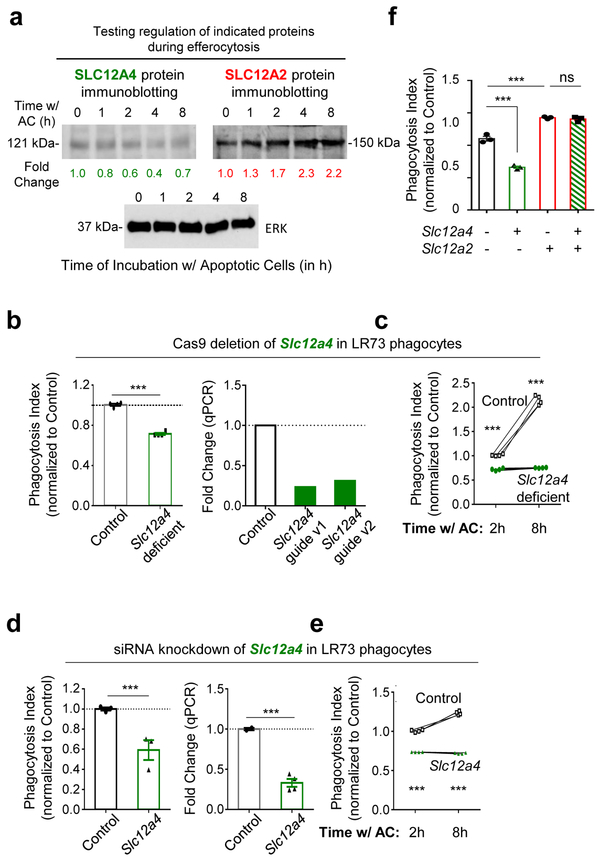
For further in-depth analysis, we picked SLC12 (solute carrier family 12) pathway genes that were part of the cell volume/cell shape gene program for several reasons. First, modulation of a number of SLC family members during efferocytosis was recently noted15, although the function of SLC12 family is not defined. Second, within the SLC12 family, multiple genes were coordinately regulated (Supplementary Table 1), including the transporter Slc12a2 (encoding SLC12A2, also called NKCC1), Slc12a4 (encoding SLC12A4, also called KCC1), and the upstream kinases that regulate SLC12A2 and SLC12A4, namely Oxsr1 (encoding OSR1) and Stk39 (encoding SPAK). Third, SLC12A2 and SLC12A4 are linked to chloride flux into and out of cells, in particular in the kidney24, and little is known about the potential role of chloride flux during efferocytosis. After confirming the transcriptional changes noted in the RNAseq within the SLC12 pathway genes in phagocytes by qPCR (Extended Data Fig. 1a), we addressed the functional relevance of this pathway and made several striking observations.
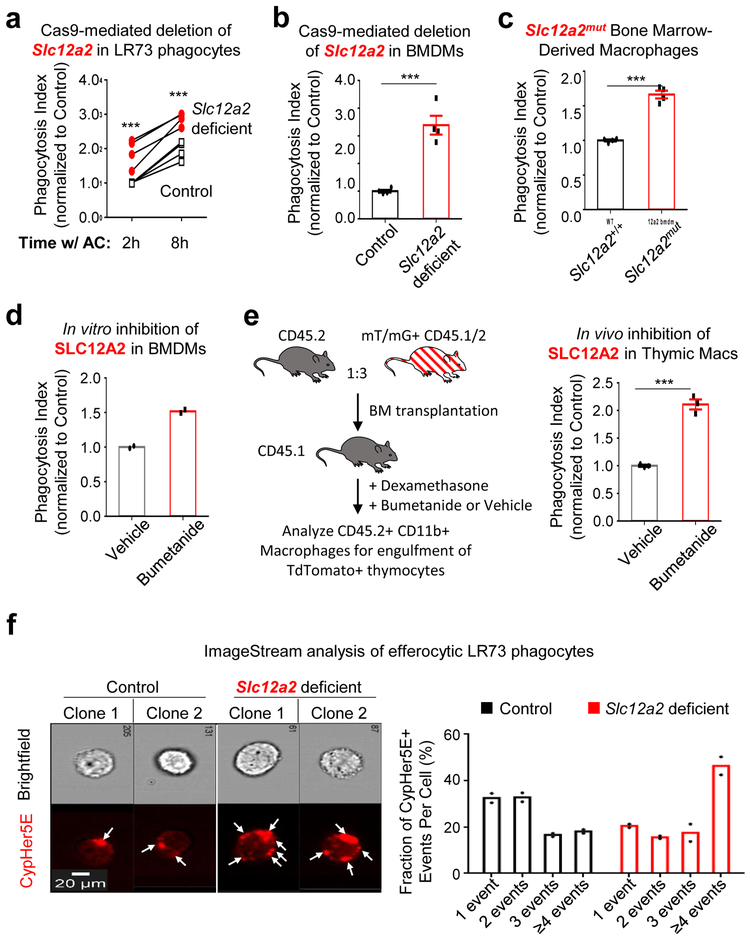
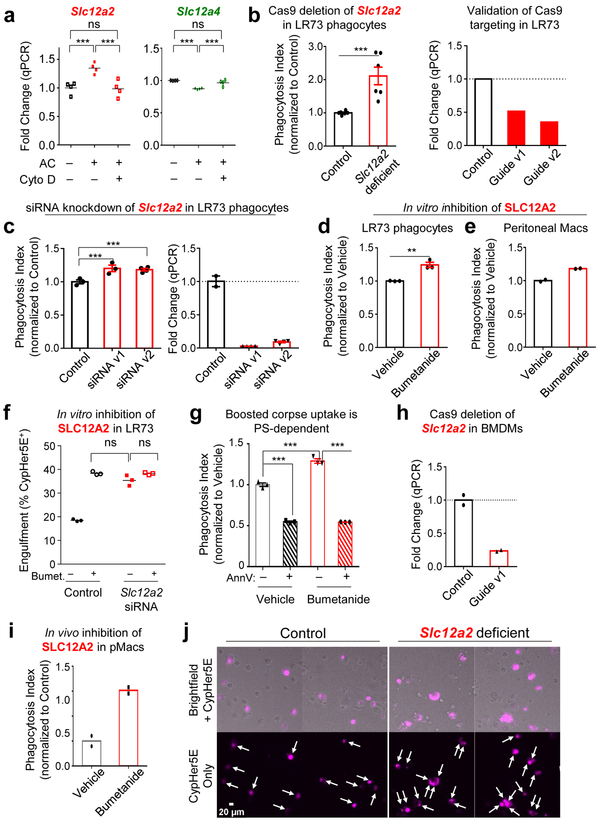
Surprisingly, targeted deletion of Slc12a2 via CRISPR/Cas9 or siRNA-mediated knockdown in LR73 phagocytes resulted in significantly enhanced uptake of apoptotic corpses (Fig. 2a and Extended Data Fig. 1b,c). Further, treatment of phagocytes with bumetanide, an inhibitor of SLC12A2 transporter activity, also led to increased corpse uptake (Extended Data Fig. 1d,e); bumetanide did not further increase efferocytosis in phagocytes in Slc12a2-deficient phagocytes (Extended Data Fig. 1f). This greater uptake was dependent on recognition of PtdSer exposed on apoptotic cells (Extended Data Fig. 1g) and was seen in Slc12a2-deficient LR73 phagocytes over an extended period of co-culture with apoptotic cells (Fig. 2a).
Fig. 2:
SLC12A2 functions as a ‘brake’ on apoptotic cell uptake
We next tested SLC12A2 function in professional phagocytes such as macrophages via multiple approaches. First, we performed genetic deletion of Slc12a2 ex vivo via CRISPR/Cas9 in ER-Hoxb825 immortalized primary bone marrow-derived macrophages, and the loss of SLC12A2 significantly increased efferocytosis (Fig. 2b and Extended Data Fig. 1h). Second, loss-of-function mutations in SLC12 family protein members (including SLC12A2) result in an array of human diseases24, 26, 27, and incorporating a 11-bp deletion in exon 22 observed in a human patient into the mouse Slc12a2 gene locus (Slc12a2mut) results in multiple phenotypes28-30 and early lethality in homozygous mice. Using bone marrow from this mouse strain29, we derived macrophages (BMDMs) that displayed similarly increased efferocytosis (Fig. 2c). Third, treatment of wild type BMDMs (Fig. 2d) or peritoneal macrophages (pMacs; Extended Data Fig. 1e) in vitro with the SLC12A2 inhibitor bumetanide also enhanced efferocytosis. Fourth, using two different in vivo models, we assessed whether disruption of SLC12A2 function would lead to increased efferocytosis. Thymic macrophage engulfment of apoptotic thymocytes induced by dexamethasone after bone marrow transplantation (see experimental model; Fig. 2e), and apoptotic cell uptake by peritoneal macrophages (Extended Data Fig. 1i) were both enhanced after inhibition of SLC12A2. Imaging flow cytometry (ImageStreamX) of Slc12a2-deficient LR73 phagocytes showed more corpse uptake on a per cell basis (Fig. 2f); whereas control phagocytes mostly engulfed 1 to 2 corpses under these assay conditions, ~45% of efferocytic Slc12a2-deficient phagocytes contained 4 or more corpses (Fig. 2f). Similarly, fluorescent microscopy of Slc12a2-deficient macrophages revealed significantly increased efferocytosis on a per-cell basis (Extended Data Fig. 1j). Collectively, these data suggest that SLC12A2 acts as a natural brake on apoptotic cell uptake, and that interfering with SLC12A2 function leads to increased efferocytosis by both professional and non-professional phagocytes.
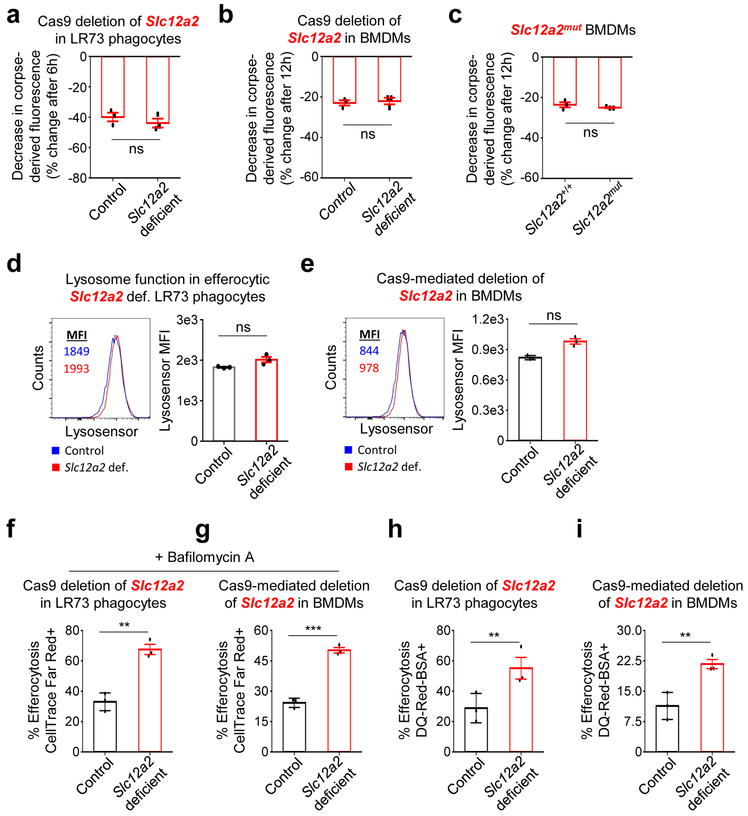
The enhanced corpse-derived fluorescence seen with Slc12a2-deficient phagocytes was not due to defective corpse degradation based on several criteria. First, we fed apoptotic cells labeled with TAMRA, a dye that directly labels peptides and proteins in apoptotic cells, and monitored degradation of TAMRA signal via flow cytometry in both LR73 phagocytes and macrophages (Fig. 3a-c). Although rates of degradation varied between LR73 phagocytes and macrophages (i.e. non-professional vs. professional phagocytes), when paired control and Slc12a2-deficient phagocytes are compared (Fig. 3a-c), the loss of SLC12A2 did not affect the rates of corpse degradation. Second, by time lapse microscopy, we observed similar kinetics of corpse degradation between control and Slc12a2-deficient phagocytes (Supplementary Videos 1-6). There was no significant difference in lysosomal pH in efferocytic phagocytes between control and Slc12a2-deficient LR73 phagocytes or macrophages (Fig. 3d, e), and inhibition of lysosomal acidification did not alter the Slc12a2-deficient LR73 phagocyte or macrophage phenotype of enhanced apoptotic cell uptake (Fig. 3f, g). Finally, labeling apoptotic cells with the reagent DQ-Red-BSA, whose cleavage by cathepsin protease activity within the acidic phagolysosome induces the DQ-Red fluorescence, revealed comparable phagolysosomal activity in the control and SLC12A2-deficient LR73 cells and macrophages; we also noted increased total signal in Slc12a2-deficient phagocytes relative to control phagocytes (Fig. 3h, i). Thus, our data suggest that SLC12A2 dampens phagocyte uptake on a per cell basis, without an apparent effect on lysosomal acidification or corpse degradation.
Fig. 3:
Boosted engulfment in SLC12A2 deficient phagocytes is not due to failure to degrade apoptotic cells
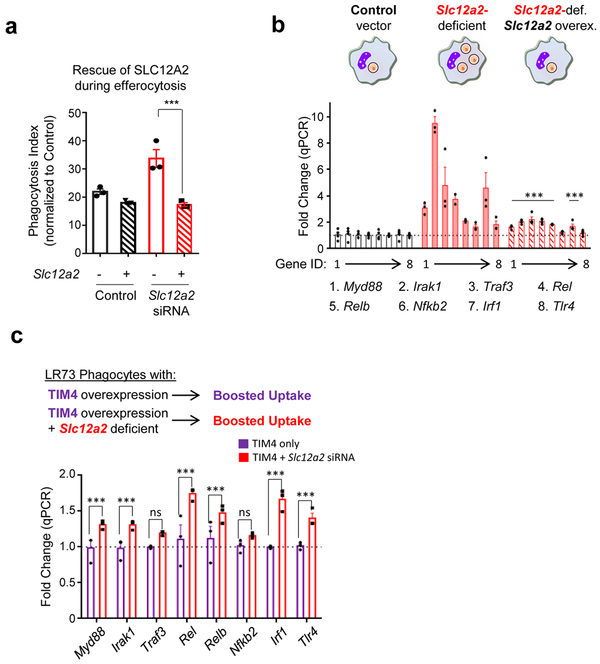
Interestingly, another SLC12 family member, SLC12A4, acts in opposition to SLC12A2 during regulatory volume change 31. Although Slc12a4 was only modestly downmodulated transcriptionally and by total protein after corpse internalization (Extended Data Fig. 1a and 2a), targeting of Slc12a4 by CRISPR/Cas9-mediated deletion or siRNA-mediated silencing in LR73 phagocytes led to a reduction in apoptotic cell uptake (Extended Data Fig. 2b-e). This reduced uptake was not a delay in eating, as this was seen even after 8-hour co-culture with apoptotic cells (Extended Data Fig. 2c, e). Thus, SLC12A2 and SLC12A4 may functionally counterbalance each other during efferocytosis. Interestingly, when we performed double targeting of Slc12a2 and Slc12a4 by siRNA-mediated knockdown of Slc12a4 in the context of Slc12a2-deficient LR73 cells, double-deficient phagocytes still showed greater efferocytosis (Extended Data Fig. 2f), suggesting that SLC12A2 deficiency is a dominant phenotype over the loss of SLC12A4.
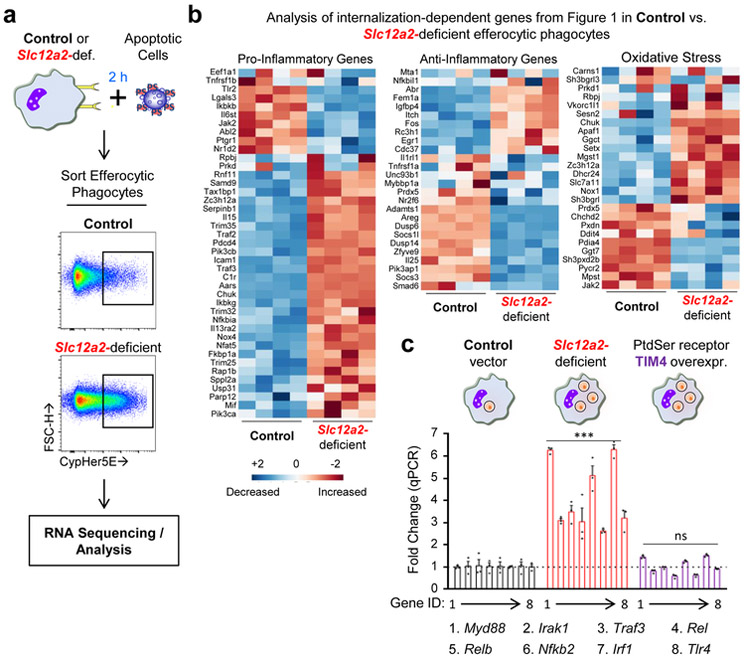
To address the consequence of Slc12a2 deficiency in efferocytic phagocytes, we performed RNAseq analysis of efferocytic control and Slc12a2-deficient phagocytes (Fig. 4a,b). Normally, efferocytosis elicits multiple distinct transcriptional programs in engulfing phagocytes (Fig. 1b). Strikingly, Slc12a2-deficient phagocytes showed unexpected changes in several of these signatures, including reversals of the anti-inflammatory and oxidative stress signatures, as well as a strong increase in a pro-inflammatory gene program (Fig. 4b), suggesting that loss of SLC12A2 affects how a phagocyte normally interprets apoptotic cell uptake as anti-inflammatory. Interestingly, many of the anti-inflammatory and pro-inflammatory genes found to be dysregulated in engulfing Slc12a2-deficient phagocytes have genetic /functional linkages with chronic inflammatory, auto-inflammatory, and autoimmune diseases (Supplementary Table 2).
Fig. 4.
SLC12A2 enforces a homeostatic efferocytosis signature
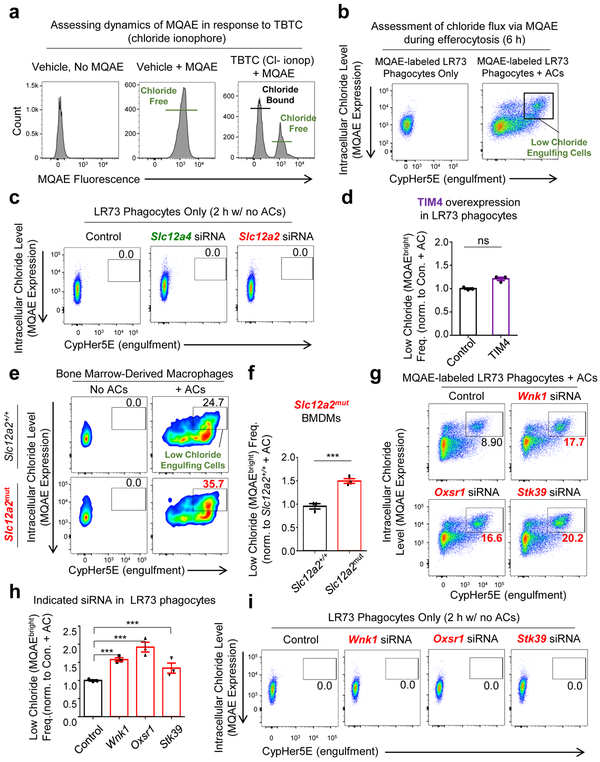
When we re-introduced Slc12a2 cDNA into Slc12a2-deficient efferocytic phagocytes, this reversed the greater corpse uptake and pro-inflammatory gene induction, confirming that the observed pro-inflammatory signature was in fact due to the absence of SLC12A2 (Extended Data Fig. 3a,b). The pro-inflammatory signature noted above could be a general property of phagocytes over-eating more corpses. To address this, we used LR73 phagocytes overexpressing the PtdSer receptor TIM-4, which increases uptake on a per cell basis 32-35. We then sorted engulfing (i.e. CypHer5E+) phagocytes from three conditions: control phagocytes with normal uptake, TIM4 overexpressing phagocytes with increased uptake, and phagocytes lacking SLC12A2 that also have enhanced uptake. Despite eating more corpses, the TIM4-overexpressing phagocytes did not show upregulation of pro-inflammatory signature genes, contrasting with Slc12a2-deficient phagocytes (Fig. 4c). Further, siRNA-mediated knockdown of Slc12a2 in TIM4 overexpressing cells induced upregulation of many of the pro-inflammatory signature genes (Extended Data Fig. 3c). This latter phenotype was less pronounced than Slc12a2-deficient phagocytes, likely because TIM4 is stably overexpressed and siRNA knockdown efficiency can vary between cells in a population. Thus, the pro-inflammatory gene signature of Slc12a2-deficient efferocytic phagocytes is not simply a corpse-overload effect, and that loss of Slc12a2 alters the anti-inflammatory response during efferocytosis.
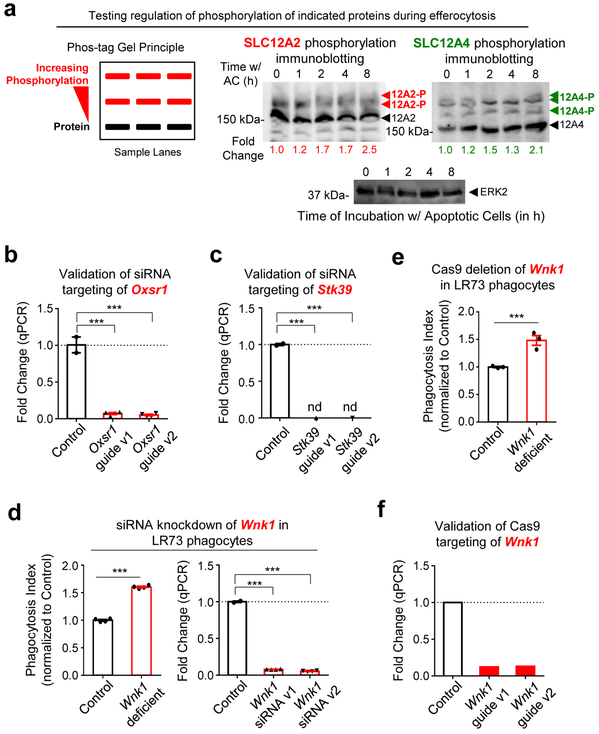
Both SLC12A2 and SLC12A4 are regulated by upstream kinases, as part of a pathway for chloride sensing and chloride flux in cells. When there is chloride efflux from a cell, it is sensed by one of the four members of the ‘With-No-lysine Kinase’ (WNK) family of Ser/Thr kinases (Fig. 5a)24. WNK1, a broadly and ubiquitously expressed member, becomes activated by chloride efflux and subsequently phosphorylates the downstream serine/threonine kinases OSR1 and SPAK (encoded by the genes Oxsr1 and Stk39, respectively; Fig. 5a)36-41. In our RNAseq, we noted that both Oxsr1 and Stk39 are transcriptionally upregulated in phagocytes after corpse internalization (Fig. 1c). OSR1 and SPAK, in turn, phosphorylate both SLC12A4 and SLC12A2 leading to decreased SLC12A4-mediated chloride efflux and increased SLC12A2-mediated chloride influx (Fig. 5a)31, 39. Although phosphorylation on multi-pass transmembrane proteins are difficult to detect, we could observe increased phosphorylation on both SLC12A2 and SLC12A4 proteins in a time course of efferocytosis (Extended Data Fig. 4a). Therefore, we next addressed the functional relevance of OSR1, SPAK, and WNK1. Knockdown of either OSR1 or SPAK resulted in greater apoptotic cell uptake both at early and late time points (Fig. 5b,c and Extended Data Fig. 4b,c). Furthermore, deletion/reduction of Wnk1 expression either via CRISPR/Cas9 deletion or siRNA-mediated knockdown in LR73 phagocytes resulted in significantly greater corpse uptake, essentially phenocopying Slc12a2 deficiency (Fig. 5d,e and Extended Data Fig. 4d-f).
Fig. 5.
SLC12 family-associated kinases also regulate rate of corpse uptake
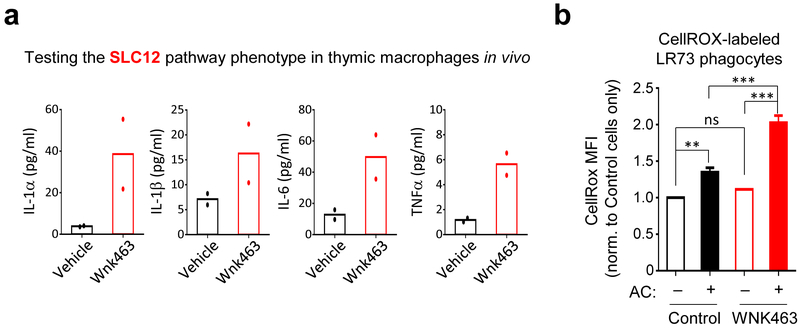
We then assessed whether WNK proteins regulate rate of apoptotic cell engulfment in vivo. After confirming that BMDMs or peritoneal macrophages treated with a pan-WNK inhibitor WNK463 42 in vitro enhanced efferocytosis (Fig. 5f,g), we injected WNK463 into the peritoneum of mice prior to dexamethasone injection (thymic engulfment model; Fig. 5h) or injection of apoptotic Jurkat cells (peritoneal engulfment model; Fig. 5i); in both cases we saw increased apoptotic cell uptake by resident macrophages in vivo. Thus, the WNK1–SLC12A2 chloride sensing pathway acts as a brake in vitro and in vivo to control the ‘appetite’ of phagocytes for apoptotic cells.
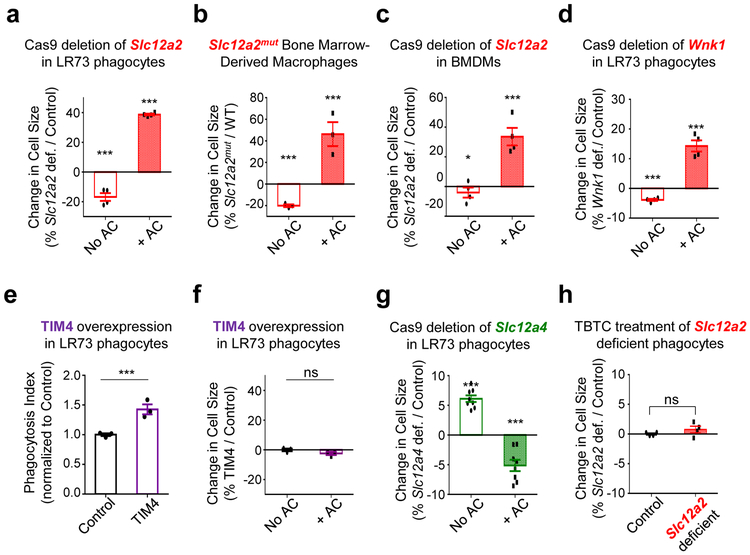
As chloride is important for many biological functions 43-45, we hypothesized that chloride flux may also affect different aspects of efferocytosis. As chloride flux is well-linked to cell volume regulation, we initially tested the possible impact of chloride on phagocyte cell size during efferocytosis. We analyzed the forward scatter area (FSC-A) of engulfing phagocytes by flow cytometry, a commonly used indicator of cell size change, which allowed us to gate specifically on engulfing phagocytes and assess FSC-A changes in thousands of phagocytes. Interestingly, control phagocytes eating corpses routinely had a modest but detectable decrease in cell size, suggesting that despite engulfing another cell, the phagocyte volume is actively controlled. While Slc12a2-deficient LR73 cells were slightly smaller in size basally, engulfing Slc12a2-deficient phagocytes became 40% larger compared to controls (Extended Data Fig. 5a). This was not unique to LR73 phagocytes, as BMDMs from Slc12a2mut mice and Slc12a2-deficient BMDMs were also slightly smaller in size basally, but became larger during efferocytosis (Extended Data Fig. 5b,c). Similarly, engulfing Wnk1-deficient phagocytes were also larger than control Wnk1-sufficient phagocytes (Extended Data Fig. 5d). Importantly, the larger size of Slc12a2- or Wnk1-deficient phagocytes does not appear to be due to ‘over-eating’ per se, as LR73 phagocytes overexpressing the PtdSer receptor TIM4 do ingest more corpses, yet do not show larger cell size (Extended Data Fig. 5e,f). When we compared Slc12a2 and Slc12a4 deficient phagocytes, Slc12a4-deficient cells were modestly larger in size prior to addition of apoptotic corpses, and although efferocytosis was attenuated by loss of Slc12a4, these Slc12a4-deficient phagocytes that did engulf apoptotic cells were smaller than control phagocytes (Extended Data Fig. 5g). Thus, the SLC12 members and upstream regulatory kinases can influence phagocyte cell size during efferocytosis.
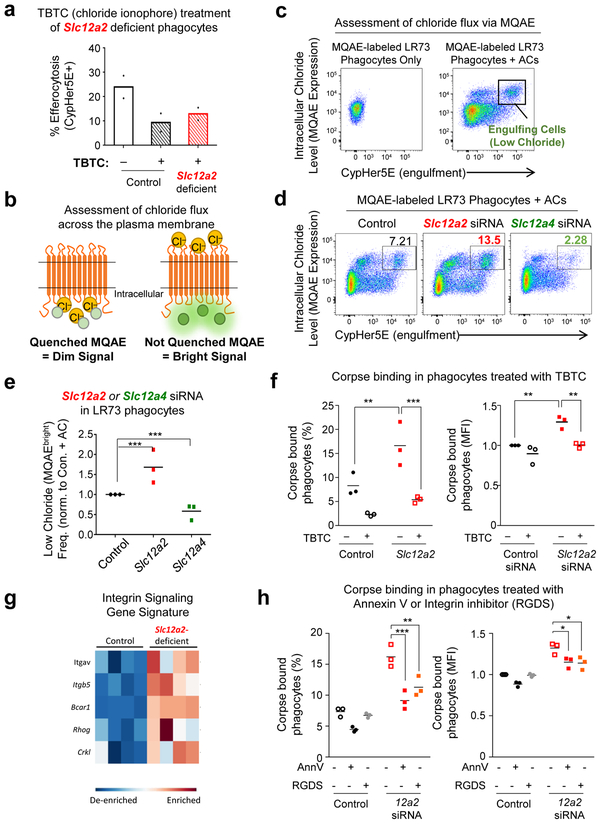
We next tested if enhanced engulfment in Slc12a2-deficient phagocytes required chloride flux across the plasma membrane. We treated phagocytes with a chloride ionophore tributyltin chloride (TBTC), which forces extracellular chloride into the phagocyte cytosol. Treatment with TBTC reversed the increased uptake and larger FSC-A/ cell size phenotype after corpse uptake observed in Slc12a2-deficient phagocytes (Fig. 6a and Extended Data Fig. 5h). To directly assess chloride flux during efferocytosis, we used the cell permeable chloride-sensitive fluorescent dye MQAE 46, 47. MQAE reversibly binds chloride in the cytosol, with the fluorescence signal for MQAE being quenched (i.e. decreased), at higher cytosolic chloride concentrations, while its fluorescence increases at lower chloride concentrations (Fig. 6b)46, 47. Further, MQAE fluorescence is not affected by pH or bicarbonate47, allowing its use during phagocytosis assays. We first confirmed that phagocytes can be labeled with MQAE and the baseline MQAE fluorescence was quenched with forced entry of chloride through TBTC (Extended Data Fig. 6a). During efferocytosis, phagocytes maximally engulfing CypHer5E-labeled apoptotic targets (CypHer5E ‘bright’) showed less intracellular chloride that is revealed as MQAEhigh fluorescence in the flow cytometry readout (Fig. 6b,c and Extended Data Fig. 6b). These data suggest that the efflux of cytosolic chloride from the engulfing phagocytes is a component of normal efferocytosis.
Fig. 6.
Chloride efflux and influx act as “accelerator” and “brake” during efferocytosis
Next, we hypothesized that in engulfing phagocytes, the loss of SLC12A2 would result in less cytosolic chloride, while loss of SLC12A4 would result in more cytosolic chloride. siRNA mediated knockdown of Slc12a2 resulted in significantly more engulfing phagocytes with significantly lower cytosolic chloride level (Fig. 6d, quantification in Fig. 6e and Extended Data Fig. 6c). In contrast, TIM4 overexpressing phagocytes that also take up more corpses had similar numbers of efferocytic phagocytes with lower MQAE levels (Extended Data Fig. 6d). A similar reduced chloride level (i.e. more MQAEhigh cells) was also seen in Slc12a2mut bone marrow-derived macrophages actively eating apoptotic cells (Extended Data Fig. 6ef). In contrast, targeting Slc12a4 via siRNA resulted in fewer engulfing phagocytes with MQAEhigh phenotype (Fig. 6d,e and Extended Data Fig. 6c). We then assessed chloride flux in efferocytic phagocytes where the upstream kinases (WNK1/OSR1/SPAK) in the chloride-sensing pathway were knocked down. siRNA targeting of Wnk1, Oxsr1, or Stk39 phenocopied the Slc12a2-deficient phenotype, displaying many more engulfing phagocytes with a MQAEhigh / low cytosolic chloride profile (Extended Data Fig. 6g, quantification in Fig. S6g, and Extended Data Fig. 6i). Disruption of any one of the upstream kinases manifested similar results to the loss of SLC12A2-mediated chloride influx, further correlating with the observation seen in Extended Data Fig. 4a. These data suggest that components of the chloride-sensing pathway including SLC12A2, SLC12A4, and the kinases WNK1, OSR1, and SPAK contribute to chloride flux into and out of phagocytes during efferocytosis, with chloride influx acting as a ‘brake’ on corpse uptake.
In addressing a possible mechanism for increased uptake, we found that Slc12a2-deficient phagocytes bound more corpses compared to control (Fig. 6f). Increased binding of corpses by Slc12a2-deficient LR73 phagocytes was indicated by both percentage of phagocytes that bound corpses and the number of targets bound to phagocytes via MFI. Interestingly, addition of the chloride ionophore TBTC, followed by the binding assay, showed reversal of the increased corpse binding to Slc12a2-deficient phagocytes (Fig. 6f). While performing these binding experiments, we noted that a 6 h TBTC treatment was more effective in seeing significant reversal of binding in Slc12a2-deficient phagocytes. This prompted us to go back to the RNAseq data and re-analyze expression of genes coding for engulfment receptors in the Slc12a2-deficient phagocytes. In Slc12a2-deficient LR73 phagocytes, there was an increase in gene expression for integrin αv and integrin β5 that have been linked to efferocytosis, as well as molecules linked to integrin dependent signaling such as p130Cas (BCAR1), and the small GTPase RhoG and CrkI linked genetically and biochemically to efferocytosis via αvβ5 integrin (Fig. 6g) 48-53. When we tested whether integrin-dependent interaction might contribute to the increased efferocytosis observed with Slc12a2-deficient phagocytes, an RGDS peptide added prior to addition of apoptotic targets partially inhibited the increased corpse binding to Slc12a2-deficient phagocytes (almost similar to the positive control for blocking with annexin V) (Fig. 6h). Collectively, these data support a mechanism that the altered chloride efflux in the Slc12a2-deficient phagocytes leads to enhanced apoptotic cell uptake, in part, via increased expression of integrins and greater binding of apoptotic targets. While this increased integrin expression is clearly one mechanism, there could also be other modalities that remain to be explored.
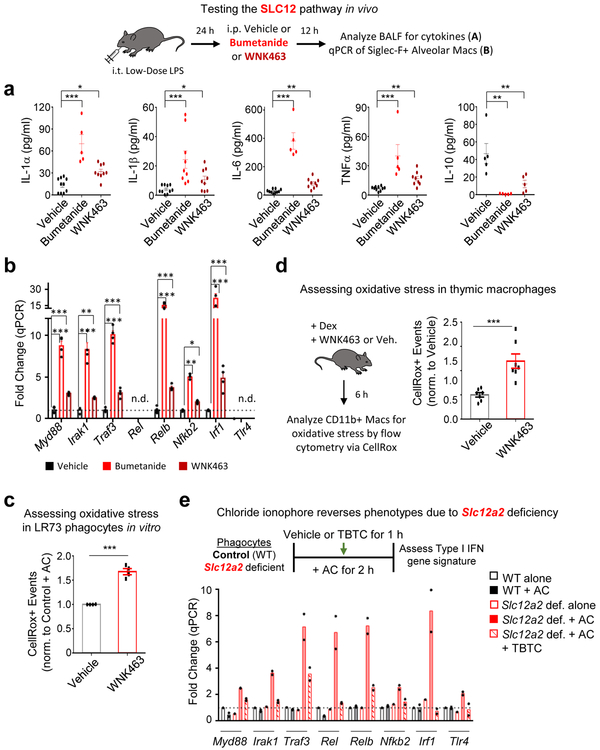
To test the in vivo linkage between this chloride-sensing pathway and the interpretation of apoptotic cells as anti- vs. pro-inflammatory, we used a well-established apoptotic cell clearance model of low-dose LPS-induced acute lung inflammation54. In this model (schematic in Fig. 7), low-dose LPS is administered intranasally, eliciting neutrophil and monocyte infiltration, with induction of pro-inflammatory cytokines including TNFα, IL-6, and IL-1. After 24 h, neutrophils undergo apoptosis and are engulfed by lung macrophages, leading to resolution of the inflammatory response54. At 24 h post LPS inoculation, after the initial onset of the LPS-induced response, we intraperitoneally administered either the SLC12A2 inhibitor bumetanide or the pan-WNK kinase inhibitor WNK463. After an additional 12 h, bronchoalveolar fluid (BALF) was assessed for the resolution of the inflammatory response. Strikingly, mice treated with the SLC12A2 inhibitor bumetanide had significant levels of the pro-inflammatory cytokines IL-1α, IL-1β, IL-6, and TNFα, which were absent in vehicle control-treated mice (Fig. 7a). Additionally, bumetanide treated mice failed to upregulate the pro-resolution cytokine IL-10 (Fig. 7a). The pan-WNK inhibitor WNK463 had a similar effect, albeit less pronounced, on inflammation resolution (Fig. 7a). We also isolated Siglec F+ alveolar macrophages from the same mice, and detected upregulation of the pro-inflammatory gene signature in macrophages treated with either bumetanide or WNK463 (Fig. 7b). The increased inflammatory cytokine production in the BALF correlated with efferocytic thymic macrophages treated with WNK463 ex vivo producing significant amounts of these pro-inflammatory cytokines (Extended Data Fig. 7a).
Fig. 7.
The SLC12 pathway and chloride flux are required to prevent inflammatory apoptotic cell clearance
RNAseq of efferocytic Slc12a2-deficient phagocytes showed a robust change in gene programs associated with oxidative stress (Fig. 4b). Oxidative stress, induced by accumulation of reactive oxygen species (ROS), is known to induce pro-inflammatory cytokine production, including IL-1β55. Consistent with our RNAseq data, inhibition of the chloride-sensing pathway resulted in increased accumulation of ROS in efferocytic phagocytes in vitro (Fig. 7c and Extended Data Fig. 7b). Thymic macrophages treated with WNK463 during dexamethasone-induced apoptotic thymocyte clearance also showed significantly higher accumulation of ROS than vehicle treated controls (Figure 7d). Thus, the chloride-sensing pathway in phagocytes involving WNK and SLC12A2 is an important part of ROS regulation and normal suppression of the inflammatory response during efferocytosis in vivo.
Lastly, we asked whether forcing entry of extracellular chloride can reverse the pro-inflammatory responses. We allowed control or Slc12a2-deficient phagocytes to engulf apoptotic cells normally for 1 hour (by which time significant number of phagocytes have already engulfed but not digested corpses) and added the chloride ionophore TBTC. Treatment with TBTC significantly suppressed many of the pro-inflammatory genes in Slc12a2-deficient phagocytes, with the others (Myd88, Nfkb2, and Tlr4) being modestly decreased (Fig. 7e). Thus, forced chloride influx was sufficient to suppress the pro-inflammatory gene signature induced by Slc12a2 deficiency during efferocytosis.
The data presented in this manuscript have several implications for efferocytosis in general and specifically for understanding how phagocytes interpret ingested apoptotic corpses as anti-inflammatory. First, this work identifies a previously unknown regulatory step that is induced within phagocytes after a corpse is internalized that involves SLC12A2 and kinases in the chloride-sensing pathway. Second, this chloride-sensing pathway controls cytosolic chloride flux during efferocytosis. Third, chloride flux during efferocytosis impacts the appetite of a given phagocyte, acting as a natural brake in efferocytosis. Fourth, when a phagocyte has disruption in the function of SLC12A2 or the upstream kinases, phagocytes fail to properly induce an anti-inflammatory gene program, a hallmark of efferocytosis. Further, Slc12a2-deficient phagocytes actively switch from a homeostatic efferocytosis signature to a pro-inflammatory program; this is not simply due to corpse overload as phagocytes engulfing multiple corpses in response to PtdSer receptor overexpression do not show a similar pro-inflammatory signature. A concept that has emerged from these studies is that the SLC12A2 pathway and chloride flux represent a ‘reversible switch’ controlling the anti- vs. pro-inflammatory response of a phagocyte after apoptotic cell uptake and could potentially be targeted for both dampening and promoting an immune response. Interestingly, monogenic human diseases, such as Bartter’s Syndrome or Gitelman’s Syndrome, are caused by mutations in the SLC12 family of proteins as well as their regulators. The primary symptoms manifested in this constellation of syndromes are typically treatable; however, several case reports have suggested inflammatory disease sequelae that arise at later stages56-59. Whether routine efferocytosis is differentially ‘interpreted’ due to mutations in SLC12 pathway members is a possibility worthy of further exploration. In a recent study, treatment of RAW 264.7 macrophages with LPS resulted in increased uptake of fluorescent E. coli bioparticles, as well as increased proinflammatory cytokine release and this was attenuated via bumetanide pre-treatment60. This earlier work focused on SLC12A2 function in bacterial clearance/LPS responses (not efferocytosis); nevertheless, future studies are required to determine how the SLC12 pathway functions in different forms of phagocytosis and the associated immune responses, including bacterial, antibody-mediated, and complement-mediated clearance.
Materials and methods
Mice
C57BL/6J mice were obtained from Jackson Laboratories and used for in vivo peritoneal apoptotic cell clearance assays. The identification of the de novo mutation and the generation of Slc12a2mut mice was reported previously 28, 30. For bone marrow-derived macrophage derivation, peritoneal macrophage isolation and all in vivo experiments, mice between the ages of 8 and 12 weeks were used. All animal procedures were performed according to the protocols provided by the Institutional Animal Care and Use Committee (IACUC) of the University of Virginia. This study is compliant with all relevant ethical regulations regarding animal research.
RNA sequencing
In order to distinguish between apoptotic cell cargo-derived and phagocyte-derived RNA, we utilized a mixed species approach using C. griseus (hamster) LR73 fibroblasts and H. sapien Jurkat lymphoma cells. LR73 cells were co-cultured with apoptotic Jurkat cells for 2 h in the presence or absence of Cytochalasin D. After 2 h, unbound Jurkat cells were washed away, and LR73 cells were rested in culture medium (with or without Cytochalasin D) for an additional 2h. For experiments related to Slc12a2-deficient cells, GFP+ Slc12a2-sufficient or -deficient LR73 cells were co-cultured with CypHer5E+-labeled apoptotic Jurkat cells for 2 h. After 2 h, phagocytes were removed with Trypsin + EDTA, and the efferocytic phagocytes (CypHer5E+, GFP+ LR73 cells) were sorted via multi-color flow cytometry into fresh tubes containing lysis buffer and β-mercaptoethanol and preserved for RNA isolation. Total RNA was isolated using the NucleoSpin RNA isolation kit with on-column rDNase digestion (Machery-Nagel) and mRNA libraries were prepared via the Illumina TruSeq technology. Libraries were then sequenced by an Illumina NextSeq sequencer at 75 bp, paired-end reads with approximately 20 million reads per sample. Four independent experiments were sequenced for each condition.
Macrophage generation and isolation
Bone-marrow derived macrophages (BMDM) were derived from bone marrow obtained from femurs and tibias that were removed from 8-week-old C57BL/6J mice and flushed with 10mL alpha-MEM containing 10% fetal bovine serum and 1% penicillin/streptomycin/glutamine (PSQ). After red blood cell lysis, the remaining bone marrow cells were cultured in 10 cm dishes in alpha-MEM containing 10% fetal bovine serum, 1% PSQ, and 10% L929 cell-conditioned media. Media was replaced at day 3 post-culture once bone marrow cells have attached, then harvested for experiments day 6–8 post-culture. Mouse peritoneal macrophages were obtained by flushing the peritoneal cavity of 8–12-week-old C57BL/6J mice with 10 mL of PBS containing 10% FBS. Collected cells were resuspended in X-VIVO 10 and plated on non-tissue culture-treated 24-well plates at 5×105 total peritoneal cells per well and used for engulfment assays the following day.
In vitro efferocytosis assay
LR73 fibroblasts, peritoneal macrophages or bone marrow-derived macrophages were seeded in a 24-well plate at a density of 1×105 cells per well 18–20 h prior to starting the engulfment assay. In some experiments, Jurkat cells were used as apoptotic cell targets, and were induced to undergo apoptosis using 150 mJ cm−2 ultraviolet C irradiation followed by incubation for 4 h at 37 °C and 5% CO2. In chloride transport experiments, thymocytes were used as apoptotic cell targets. Thymocytes were isolated from 4 to 6-week-old mice and treated with 50 μM dexamethasone for 4 h to induce apoptosis. Apoptotic Jurkat cells or thymocytes were then stained with 1 μM CypHer5E (GE Healthcare) in serum-free HBSS for 45 min at 37 °C, washed, then incubated in serum-complete assay media for an additional 25min. Apoptotic cells were co-cultured with phagocytes at a 1:10 phagocyte to target ratio. For all pharmacological studies, phagocytes were pre-incubated with the compounds listed below for one hour prior to addition of apoptotic targets: Cytochalasin D (Sigma-Aldrich, 1 μM), DCPIB (Tocris; 12.5–100 μM), Bumetanide (Tocris, 10–50 μM), WNK463 (Selleckchem, 10 μM), Bafilomycin A1 (Cayman chemicals; 100 nM), Tritbutyltin Chloride (Sigma; 0.1–10 μM). Phagocytes were examined to ensure no gross morphological changes or cell death occurred due to drug treatment. Apoptotic cells were removed via three cold PBS washes and phagocytes were harvested via trypsin/EDTA and assessed for efferocytosis by flow cytometry. Cell size was determined using the Forward Scatter Area measure, and the FSC-A of CypHer5E+ (i.e. efferocytic cells) was compared relative to non-efferocytic (e.g. CypHer5E negative) phagocytes, or phagocytes co-cultured in media alone. The lysosomal pH indicator LysoSensor (ThermoFisher) was used according to manufacturer protocol. To assess late phagolysosomal function (e.g. cathepsin protease activity; ThermoFisher), apoptotic cells were first stained using the reagent DQ-Red-BSA, washed twice, then co-cultured with CFSE labeled phagocytes. In some experiments, samples were imaged and analyzed on an ImageStreamX™ imaging flow cytometer. When indicated, cellular ROS was determined using the reagent CellROX (ThermoFisher) according to manufacturer protocol.
Assay for scoring corpse binding to phagocytes
GFP expressing LR73 fibroblasts were seeded in a 24-well plate at a density of 0.2×105 cells per well. Next day, cells were treated with Lipofectamine 2000 (Thermo Fisher) with specific siRNAs according to the manufacturer’s protocol, two days prior to efferocytosis assays. Thymocytes were isolated from 4 to 6-week-old mice and treated with 25 μM dexamethasone for 4 h to induce apoptosis. Apoptotic thymocytes were then stained with 10 μM TAMRA in serum-free HBSS for 45 min at 37 °C, washed, and incubated in serum-containing assay media for an additional 25min. Phagocytes were pre-incubated with Cytochalasin D (Sigma-Aldrich, 1 μM) for 1 h and, in some experiments, Tritbutyltin chloride (Sigma; 10 μM for 6 h), or with RGDS peptide for 30 min (Tocris 100 μM); in some cases apoptotic cells were incubated with Annexin V (Abcam, 10 μg/ml for 30 min). Apoptotic cells were subsequently co-cultured with phagocytes, in the presence of vehicle or indicated inhibitor, at a 1:10 phagocyte to target ratio for 30 minutes. After the incubation, apoptotic cells were removed via three cold PBS washes and phagocytes were harvested via trypsin/EDTA and assessed for the percentage and MFI of GFP+ TAMRA+ LR73 cells by flow cytometry.
In vivo efferocytosis assays
For in vivo peritoneal efferocytosis assays, Jurkat cells were induced to undergo apoptosis by treatment with 150 mJ cm−2 ultraviolet C irradiation (Stratalinker), and then incubated for 4 h at 37 °C and 5% CO2. Apoptotic cells were then collected, stained with CypHer5E, resuspended in X-VIVO 10 media, and injected into the peritoneal cavity of mice at 6×106 cells per mouse. Thirty minutes post-injection, total peritoneal cells were collected, stained with CD11b (eBioscience, Clone: M1/70) and F4/80 (eBioscience, Clone: BM8), and analyzed by flow cytometry. For in vivo thymocyte engulfment assays, bone marrow (BM) was obtained by flushing donor humerus, tibia, and femur. BM was then RBC lysed and T cell-depleted by labeling cells with PE anti-CD4 and anti-CD8 and anti-PE microbeads, followed by magnetic cell separation using an AutoMACS (Miltenyi Biotech). 1.5 ×106 CD45.2 and 4.5×106 mT/mG CD45.1/2 cells were mixed and injected into lethally irradiated CD45.1 C57BL/6J recipient mice. Mice were maintained on antibiotic water one day prior and two weeks after transplantation. Mice were allowed to recover for 8 weeks, then treated with 50 μM dexamethasone. After 6 h, thymi were harvested, and CD11b+ thymic macrophages were isolated and analyzed by flow cytometry. Over 99% chimerism was confirmed by analyzing for CD45.1 and CD45.2. In some experiments, the SLC12A2 inhibitor Bumetanide (0.5 mg/kg; Cayman chemicals) or the pan-WNK inhibitor WNK463 (1 mg/kg; MRC PPU) were injected intraperitoneally 1 h before and concurrently with injection of apoptotic cells (peritoneal engulfment) or administration of dexamethasone (thymocyte engulfment). When indicated, cellular ROS was determined using the reagent CellROX according to manufacturer protocol. Flow cytometry data was collected on a FACS Canto I (Becton Dickinson) or Attune NxT (ThermoFisher) and analyzed with FlowJo version 10 (Treestar, Inc).
CRISPR/Cas9 Deletion, siRNA knockdown, and overexpression
Stable, individual clones of Cas9/GFP-expressing LR73 cells or Hoxb8-ER BMDMs were generated by lentiCas9-EGFP plasmid via lentiviral transduction and a protocol adapted from Zhang and colleagues 55 followed by single cell cloning of GFP+ cells and verification of Cas9 expression. Slc12a4, Slc12a2 and Wnk1 were deleted from LR73 cells using two independent Cas9/GFP LR73 cell clones and using the Zhang lab lentiGuide-Puro sgRNA plasmid with two unique guides for each gene and two unique small guides. LentiCas9-EGFP was a gift from Phil Sharp & Feng Zhang (Addgene plasmid #63592) and lentiGuide-Puro was a gift from Feng Zhang (Addgene plasmid #52963). Guide RNAs targeting hamster Slc12a4 were generated using the following oligo pairs:
Guide 1:
5’---CACCGATAAGCGTGGTGCTCCAGTA-−-3’
3’---CTATTCGCACCACGAGGTCATCAAA-−-5’
Guide 2:
5’---CACCGCACCGCGTGAGGTGTCCCGG-−-3’
3’---CGTGGCGCACTCCACAGGGCCCAAA-−-5’
Guide RNAs targeting hamster Slc12a2 were generated using the following oligo pairs:
Guide 1:
5’---CACCGTTAATATCACAATGGCGAAC-−-3’
3’---CAATTATAGTGTTACCGCTTGCAAA-−-5’
Guide 2:
5’---CACCGAACTTTGGACCAGATTTCCG-−-3’
3’---CTTGAAACCTGGTCTAAAGGCCAAA-−-5’
Guide RNAs targeting mouse Slc12a2 were generated using the following oligo pairs:
Guide 1:
5’---CACCGCGTGAGCTTCCAGAACGGCG-−-3’
3’---CGCACTCGAAGGTCTTGCCGCCAAA-−-5’
Guide 2:
5’---CACCGCGACCACAGCATCTCTGGT-−-3’
3’---CGCTGGTGTCGTAGAGACCACAAA-−-5’
Guide RNAs targeting Wnk1 were generated using the following oligo pairs:
Guide 1:
5’---CACCGCAATCTTGACTGAGCCCGTA-−-3’
3’---CGTTAGAACTGACTCGGGCATCAAA-−-5’
Guide 2:
5’---CACCGCCGGAGTTACCACTAGCAC-−-3’
3’---CGGCCTCAATGGTGATCGTGCAAA-−-5’
For siRNA and plasmid transduction experiments, LR73 cells were treated with Lipofectamine 2000 (Thermo Fisher) with specific siRNAs according to the manufacturer’s protocol 2 days prior to efferocytosis assays. Murine Timd4 was cloned into the pEBB-GFP vector and used for overexpression experiments. siRNAs targeting mRNAs were customized by GE Healthcare Dharmacon.
siRNAs targeting Slc12a4
5’--- AGACAGUUAUGGUGGACAAUU -−-3’
5’--- CAGCAAGGUUCGAGAGGGAUU -−-3’
siRNAs targeting Slc12a2
5’--- UGCUAAAGGUUAUGGGAAAUU -−-3’
5’--- CCAAGGAUGUGGUGGUAAAUU -−-3’
siRNAs targeting Wnk1
5’--- GCAAAGGACACAUGAAUUAUU -−-3’
5’--- GGAAGAAGGCGGAGACCUAUU -−-3’
siRNAs targeting Oxsr1
5’--- GGAUAAACCUUGAGAAAUGUU -−-3’
5’--- UGGAAGGGAUUUAGUAAUAUU-−-3’
siRNAs targeting Stk39
5’--- GAGAAGUCACGAAGAGUAAUU -−-3’
5’--- AAACUGGAGUAGAGGAUAAUU -−-3’
Generation of Hoxb8-ER primary BMDMs
Immortalized Hoxb8-ER primary BMDM precursors were generated as previously reported 25 from mice bearing constitutive expression of Cas9 and GFP56 on the C57BL/6 background. Precursor cells are maintained in alpha-MEM containing 100 ng/ml GM-CSF and 0.5 μM β-estradiol (Sigma) and differentiated into BMDMs using L929-conditioned BMDM media (see above).
Quantitative RT-PCR
Total RNA was extracted from cells using the RNeasy Mini Kit (Qiagen) or NucleoSpin RNA isolation kit (Machery-Nagel) and cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen) according to manufacturers’ instructions. Quantitative gene expression for hamster and mouse genes was performed using Taqman probes (Applied Biosystems) run on a StepOnePlus Real Time PCR System (Applied Biosystems). Probes are listed in Supplementary Table 3.
Immunoblotting
LR73 phagocytes or BMDMs were seeded in 10 cm plates at a concentration of 2×106 cells per plate. In some experiments, apoptotic Jurkat cells were added to LR73 cells at a 10:1 ratio and incubated for the indicated times. After washing unbound Jurkat cells, the remaining cells were lysed in RIPA buffer and used in total protein Western blots or phosphorylation blots (Phos-Tag; Wako). The blots were probed using SLC12A2 (Cell Signaling Technology), SLC12A4 (Abcam), and total ERK2 (Santa Cruz Biotechnology, #sc-154-G) antibodies in Can Get Signal solution (TOYOBO) followed by chemiluminescence detection.
Luminex assessment of cytokine production
For in vitro experiments, phagocytes were cultured with apoptotic cells for 2 h. Apoptotic cells were then removed, 500 μl of fresh media was added, and phagocytes were cultured an additional 2 h. Supernatant was then collected in 1.5 ml Eppendorf tube, spun to collect cells and debris, then transferred to a fresh tube and frozen for downstream Luminex analysis. For in vivo LPS experiments, 1 ml of PBS was lavaged into the lung via the trachea, and the resultant bronchoalveolar lavage fluid was collected in a 1.5 ml Eppendorf tube, spun to collect cells, then transferred to a new Eppendorf tube and frozen for downstream Luminex analysis.
Intracellular chloride detection
In indicated efferocytosis assays, LR73 cells, peritoneal macrophages, or bone marrow-derived macrophages were labeled with N-[ethoxycarbonylmethyl]-6-methoxy-quinolinium bromide (MQAE) for 2 h in cell-type specific complete media. Cells were then washed and allowed to recover for 1 h in complete media prior to the start of the efferocytosis assay. MQAE signal emits optimally at 460nm and is easily detected by flow cytometry. Additionally, MQAE is not sensitive to pH, bicarbonate, borate, nitrate, or sulfate anions, making it suitable for detecting chloride changes regardless of other physiological cell changes 47.
Code Availability
The codes used for analyses are available upon request.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism 7, SPSS v22, and R v3.3.2. Significance was determined via unpaired Student’s two-tailed t-test, Fisher’s exact test. non-parametric Mann-Whitney U tests, one-way ANOVA or two-way ANOVA, according to data requirements. Rv3.3.2 was used for graphical and statistical analyses and the R package DESeq2 was used for differential gene expression analysis of RNAseq data. All genes were curated using a combination of literature mining and function determination (known or predicted) via Uniprot. Pathway analyses were performed using the Molecular Signatures Database (MSigDB) compendium of previously defined pathways. Fisher’s exact tests were performed to determine significant pathway enrichment. All biologically independent samples were included and used for downstream relative analyses (e.g. phagocytosis index) and statistical analyses. No statistical method was used to predetermine sample size and no data was excluded from this manuscript even in circumstances where magnitude of difference was low. The statistical source data are included with this manuscript.
All experiments were performed independently at least three times except for Fig. 2d that was performed twice as the experiment simply provided additional support for experiments in Fig. 2a-c, Fig. 2f that was performed twice, as this was an imaging experiment were each individual replicate contained over 200 unique cells analyzed, Fig. 5b-d that was performed twice, as these experiments consisted of two independent siRNAs, and Fig. 7e that was performed twice, as this experiment consisted of 8 unique genes over 5 different conditions.
Data Availability.
All RNA-seq data for this experiment have been submitted to the Gene Expression Omnibus under accession number GSE131860. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Extended Data
Extended Data Fig. 1. SLC12A2 acts as a ‘brake’ on apoptotic cell clearance.
(a) qPCR of Slc12a2 and Slc12a4 during efferocytosis. Data shown as mean ± SEM ***p < .001, n=4 independent experiments.
(b) Slc12a2 or control LR73s were fed CypHer5E-labeled apoptotic Jurkat cells for 2h. Data are from n=6 independent experiments. Data shown as mean ± SEM. ***p < .001. Validation of Slc12a2 deletion by qPCR for two independent small guides.
(c) (Left panel) Slc12a2 siRNA-treated LR73 phagocytes were incubated with CypHer5E-labeled apoptotic Jurkat cells for 2h. Data represent n=3 independent experiments. Data shown as mean ± SEM. *p < .05, ns = not significant. (Right panel) siRNA targeting efficiency of Slc12a2 in LR73 phagocytes via qPCR. ***p < .001.
(d, e) (d) LR73 phagocytes or (e) peritoneal macrophages were pre-treated for 1h with bumetanide then incubated with CypHer5E-labeled apoptotic Jurkat cells for 2h (e), or 1h (f). Data represent n=3 (d, ***p < .001, mean ± SEM) or n=2 independent experiments (e).
(f) Control or Slc12a2-deficient LR73 phagocytes were pre-treated for 1h with bumetanide then incubated with CypHer5E-labeled apoptotic Jurkat cells for 2h. Data shown as mean ± SD, n=3 independent experiments. ns = not significant.
(g) Control or Slc12a2-deficient LR73 phagocytes were cultured with apoptotic cells, with or without Annexin V for 2h. Data are n=3 independent experiments. Data shown as mean ± SEM. ***p < .001.
(h) qPCR determination of the knockdown efficiency of Slc12a2 in ER-Hoxb8 immortalized bone marrow-derived macrophages used in Fig. 2b. n=2 independent experiments.
(i) Mice were injected i.p. with the SLC12A2 inhibitor bumetanide. After 1h, CypHer5E-labeled apoptotic Jurkat cells were injected. Peritoneal cells were collected and engulfment by CD11b+ F4/80+ macrophages assessed by flow cytometry. Data represent n=2 independent experiments.
(j) SLC12A2-deficient macrophages were cultured for 3h with apoptotic cells prior to imaging. Efferocytosis was assessed by fluorescent microscopy. Images represent four independent experiments. scale bar= 20 microns. Statistics source data are provided in Source Data Extended Data Figure 1.
Extended Data Fig. 2. SLC12A4 acts as an ‘accelerator’ during apoptotic cell clearance.
(a) LR73 phagocytes were incubated with apoptotic Jurkat cells for the indicated times. Apoptotic cells were thoroughly washed away with cold PBS, and total LR73 cell lysate was collected on ice and analyzed by immunoblotting using the indicated antibodies and the band intensities were quantitated. After normalization to ERK (control protein for each sample), the fold changes in band intensity relative to the 0 min time point are indicated.
(b) (Left) LR73 cells with CRISPR/Cas9-mediated deletion of Slc12a4 (or control cells with Cas9-GFP but scramble guide RNAs) were co-cultured with CypHer5E-labeled apoptotic Jurkat cells for 2 h, and efferocytosis assessed. Data from n=2 independent experiments with 2 unique clones per condition (n=4 independent experiments). Data shown as mean ± SEM. ***p < .001. (Right) Also shown are the knockdown efficiencies of two small guide RNA used to target SLC12A4.
(c) LR73 cells were co-cultured with apoptotic cells for 2 h or 8 h and assessed for efferocytosis by flow cytometry. Data are from n=4 independent experiments. Data shown as mean ± SEM. ***p < .001.
(d) (Left) LR73 phagocytes were transfected with siRNAs targeting Slc12a4 and incubated with CypHer5E-labeled apoptotic Jurkat cells for 2 h. ***p < .001. Data represent n=3 independent experiments and are shown as mean ± SEM. (Right) qPCR of siRNA-mediated Slc12a4 knockdown efficiency in LR73 phagocytes. ***p < .001.
(e) Similar to (d), but for 2 h and 8 h. Data are representative of n=4 independent experiments and are shown as mean ± SEM. ***p < .001.
(f) Control or Slc12a2-deficient phagocytes were treated with siRNA targeting Slc12a4 or control siRNA for 48h and incubated with apoptotic cells for 2 h. Data represent n=3 independent experiments. ns = not significant, ***p < .001. Statistics source data are provided in Source Data Extended Data Figure 2. Unprocessed blots are provided in Unprocessed Blots Extended Data Figure 3.
Extended Data Fig. 3. SLC12A2 deficiency overrides homeostatic efferocytosis signature.
(a and b) Overexpression of SLC12A2 restores appetite suppression and reverses the pro-inflammatory gene signature in engulfing Slc12a2-deficient phagocytes. A control vector or Slc12a2 cDNA was overexpressed in control or Slc12a2-deficient LR73 phagocytes. Phagocytes were then mixed with CypHer5E-labeled apoptotic Jurkat cells for 2 h, cDNA+ CypHer5E+ phagocytes were sorted (a) and assessed for the presence of a pro-inflammatory gene signature by qPCR (b). Data represent n=3 independent experiments. Data shown as mean ± SEM. ns = not significant, ***p < .001.
(c) Slc12a2 knockdown induces pro-inflammatory genes in normally anti-inflammatory efferocytosis after TIM-4 overexpression. TIM4 stably expressing LR73 phagocytes were transfected with control siRNA or Slc12a2 siRNA, and then co-cultured with apoptotic cells for 2 h. RNA was subsequently isolated from the phagocytes, and the pro-inflammatory gene signature was assessed via qPCR. Data represent n=3 independent experiments. Data shown as mean ± SEM. ns = not significant, ***p < .001. Statistics source data are provided in Source Data Extended Data Figure 3.
Extended Data Fig. 4. WNK1, OSR1, & SPAK deficiency phenocopies SLC12A2 during apoptotic cell clearance.
(a) Phosphorylation of SLC12A2 and SLC12A4 during efferocytosis. LR73 phagocytes were mixed with apoptotic Jurkat cells for the indicated times, and apoptotic cells thoroughly washed. Total lysates of LR73 cells were analyzed using phosphate affinity electrophoresis (see diagram). 12A2-P and 12A4-P denote the phospho-mobility shift, while 12A2 and 12A4 denote the non-phosphorylated forms. Quantification was based on the intensity and size of the bands followed by subtracting background value. Fold change relative to time point 0 is shown after normalization to ERK2 loading control. Molecular size marker does not show their aciculate sizes due to the modified gels.
(b, c) Validation of Oxsr1 or Stk39 knockdown in LR73 phagocytes. Knockdown efficiency of siRNAs targeting Oxsr1 (OSR1) or (C) Stk39 (SPAK) were determined by qPCR. ***p < .001, n=3 (Stk39) or 4 (Osr1) independent experiments, mean ± SEM.
(d) siRNA targeting of Wnk1 results in increased efferocytosis. (Left) LR73 phagocytes were transfected with siRNAs targeting Wnk1 and incubated with CypHer5E-labeled apoptotic Jurkat cells for 2 h. Data are presented as phagocytosis index and represent n=4 independent experiments. Data shown as mean ± SEM. (Right) Wnk1 knockdown efficiency of two independent siRNAs, determined by qPCR. ***p < .001.
(e) Wnk1 deficiency phenocopies Slc12a2 deficiency in phagocytes during efferocytosis. GFP+ Wnk1-deficient LR73 cells were co-cultured with CypHer5E-labeled apoptotic Jurkat cells for 2 h. ***p < .001. Data are from n=3 independent experiments. Data shown as mean ± SEM.
(f) qPCR validation of CRISPR/Cas9-mediated targeting of Wnk1 in LR73 phagocytes. *p < .001. The drop in mRNA expression due to individual small guide used to target Wnk1 are shown. Statistics source data are provided in Source Data Extended Data Figure 4. Unprocessed blots are provided in Unprocessed Blots Extended Data Figure 3.
Extended Data Fig. 5. SLC12 pathway regulates cell size of efferocytic phagocytes.
(a) From experiments in Fig. S1b, average Forward Scatter – Area (FSC-A) of 10,000 phagocytes was determined and presented as change in size of Slc12a2-deficient cells relative to control LR73 cells, with or without apoptotic cells. Data represent analysis of at least 2,000 engulfing (CypHer5E+) events from n=4 independent experiments. Data shown as mean ± SEM. ***p < .001.
(b) Similar to (a), cell size analysis via FSC-A from experiments in Fig. 2c. Data represent n=3 independent experiments. Data shown as mean ± SEM. ***p < .001.
(c) Similar to (a), cell size analysis via FSC-A from experiments in Fig. 2b). Data represent n=4 independent experiments. Data shown as mean ± SEM. * p < .05, ***p < .001.
(d) Similar to (a), cell size analysis via FSC-A from experiments in Supplementary Fig. S4e. Data represent n=4 independent experiments with 2 biological replicates per condition. Data shown as mean ± SEM. ***p < .001.
(e, f) LR73 phagocytes overexpressing the PtdSer receptor TIM4 maintain their cell size while engulfing more apoptotic cells. LR73 phagocytes transfected with control or Timd4 (TIM4) plasmid were incubated with CypHer5E-labeled apoptotic Jurkat cells for 2h and their efferocytosis (f) and cell size by FSC-A (g) assessed. Data represent n=3 independent experiments. Data shown as mean ± SEM. ***p < .001. ns = not significant.
(g) Similar to (a), cell size analysis via FSC-A from experiments in Supplementary Fig. S2b. Data from n=4 independent experiments.
(h) Chloride ionophore TBTC reverses cell size increase in Slc12a2-deficient phagocytes. Average cell size was quantified by FSC-A from the experiments performed in Fig. 6a. ns = not significant. Data shown as mean ± SEM, n=4 independent experiments. Statistics source data are provided in Source Data Extended Data Figure 5.
Extended Data Fig. 6. Chloride flux during efferocytosis.
(a) LR73 phagocytes were labeled with MQAE for 2h, washed, treated with tributyltin chloride (TBTC), a chloride ionophore. Treatment with TBTC results in a significant quenching/decrease of MQAE fluorescence due to greater chloride levels. Data represent two independent experiments.
(b) Chloride flux during efferocytosis monitored by MQAE. Performed as in Fig. 6c. Shown are representative plots of MQAE+ phagocytes cultured without or with apoptotic cells. Gates indicates phagocytes actively engulfing apoptotic cells (CypHer5Ehigh), displaying relatively low chloride levels (MQAEbright).
(c) Slc12a2- and Slc12a4-deficient phagocytes have comparable MQAE signal prior to efferocytosis. From experiments in Fig. 6d and 6e. LR73 phagocytes treated with control, Slc12a4, or Slc12a2 siRNA were labeled with MQAE and the basal MQAE plots without incubation with apoptotic cells are shown.
(d) TIM4 overexpression leaves chloride flux unaffected. LR73 phagocytes transfected with control or Timd4 (TIM4) plasmid were labeled with MQAE and incubated with CypHer5E-labeled apoptotic thymocytes for 2h and assessed for chloride flux. Data represent n=3 independent experiments. ns = not significant.
(e and f) Macrophages from Slc12a2mut mice show decreased chloride influx during apoptotic cell clearance. Bone marrow-derived macrophages from Slc12a2mut mice or control littermates were labeled with MQAE and incubated with apoptotic thymocytes for 1h. Data represent n=3 independent experiments. ***p < .001.
(g and h) WNK1, OXSR1, and SPAK contribute to influx of extracellular chloride during efferocytosis. Wnk1, Oxsr1 (OSR1), and Stk39 (SPAK) were targeted by siRNA and analyzed as in Fig. 6e above (g). A summary plot of CypHer5E+ MQAE 'bright' phagocytes is shown (h). Data represent n=3 independent experiments, shown as mean ± SEM. ***p < .001.
(i) WNK1, OSR1, and SPAK have comparable basal MQAE signal prior to efferocytosis. Controls for Fig. S6g and S6h, showing basal MQAE signal from LR73 cells incubated without apoptotic cells. Data represent three independent experiments. Statistics source data are provided in Source Data Extended Data Figure 6.
Extended Data Fig. 7. SLC12 pathway regulates ROS levels and pro-inflammatory cytokine production during efferocytosis.
(a) Inhibition of SLC12 pathway during efferocytosis results in increased pro-inflammatory cytokine production by thymic macrophages ex vivo. Performed as in Fig. 7d, except thymic macrophages were isolated and cultured for 6 h, then supernatants were collected and analyzed for pro-inflammatory cytokine production. Data are representative of n=2 independent experiments.
(b) Inhibition of SLC12 pathway induces accumulation of ROS in efferocytic phagocytes. Data are from Fig. 7c, but instead normalized to vehicle + no AC control to demonstrate parity between control and WNK463-treated phagocytes not exposed to ACs, n=4. Data shown as mean ± SEM. **p < .01, ***p < .001, ns = not significant. Statistics source data are provided in Source Data Extended Data Figure 7.
Supplementary Material
Supplementary Video 1 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Video 2 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Video 3 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Video 4 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Video 5 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Video 6 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Table 1 - Cell Volume Associated Genes Listed are members of the SLC12 (electroneutral chloride transporter) pathway genes with altered expression (based on adjusted p value and log2 fold change as determined via DESeq2) after corpse internalization, but not due to soluble factors/corpse-contact.
Supplementary Table 2 - Anti- and Pro-Inflammatory Genes List of genes associated with autoimmunity/chronic inflammatory disease that arose from Slc12a2-deficient efferocytic phagocytes (see Fig. 4).
Supplementary Table 3 – qPCR TaqMan Probes List of all hamster and mouse TaqMan probes used.
Acknowledgments
The authors thank members of the Ravichandran laboratory for discussions and critical reading of this manuscript; J.S.A.P. was supported by Cancer Research Institute – Mark Foundation Fellowship, NCI 1K99CA237728–01, Burroughs Wellcome PDEP award, and NCI Cancer Research Training Award 5T32CA009109–39. S.M. is supported by grants from the Mishima-Kaiun Memorial Foundation and The Kanae Foundation for the Promotion of Medical Science. This work was supported by grants to K.S.R. from the NIGMS (GM064709), NIMH (MH096484), NHLBI (P01HL120840), and the Center for Cell Clearance at the University of Virginia, Pilot funding from the UVa Brain Institute, Odysseus I award from the FWO, and an EOS grant from the FWO. E.D. is funded by NIH grants R21GM118944 and R01DK093501. C.M. is supported by the Pannexin Program Award through NHLBI (P01HL120840). M.H.R. was supported by the UVA Neuroscience Training Program 4T32GM008328–25. J.I.E. is supported by NIAID Training Award 5T32AI007496. C.D.L. is supported by an award from The Wellcome Trust (206566/Z/17/Z).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Arandjelovic S & Ravichandran KS Phagocytosis of apoptotic cells in homeostasis. Nat Immunol 16, 907–917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henson PM Cell Removal: Efferocytosis. Annual Review of Cell and Developmental Biology 33, 127–144 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Gordon S Phagocytosis: An Immunobiologic Process. Immunity 44, 463–475 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Blander JM The many ways tissue phagocytes respond to dying cells. Immunological Reviews 277, 158–173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Penberthy KK & Ravichandran KS Apoptotic cell recognition receptors and scavenger receptors. Immunological Reviews 269, 44–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott MR, Koster KM & Murphy PS Efferocytosis Signaling in the Regulation of Macrophage Inflammatory Responses. The Journal of Immunology 198, 1387–1394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green DR, Oguin TH & Martinez J The clearance of dying cells: table for two. Cell Death And Differentiation 23, 915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies LC, Jenkins SJ, Allen JE & Taylor PR Tissue-resident macrophages. Nature Immunology 14, 986 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canton J, Neculai D & Grinstein S Scavenger receptors in homeostasis and immunity. Nature Reviews Immunology 13, 621 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Elliott Michael R. & Ravichandran Kodi S. The Dynamics of Apoptotic Cell Clearance. Developmental Cell 38, 147–160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochreiter-Hufford A & Ravichandran KS Clearing the Dead: Apoptotic Cell Sensing, Recognition, Engulfment, and Digestion. Cold Spring Harbor Perspectives in Biology 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segawa K & Nagata S An Apoptotic ‘Eat Me’ Signal: Phosphatidylserine Exposure. Trends in Cell Biology 25, 639–650 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Levin R, Grinstein S & Canton J The life cycle of phagosomes: formation, maturation, and resolution. Immunological Reviews 273, 156–179 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Medina CB & Ravichandran KS Do not let death do us part: ‘find-me’ signals in communication between dying cells and the phagocytes. Cell Death And Differentiation 23, 979 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morioka S et al. Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosurgi L et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. 356, 1072–1076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morioka S, Maueröder C & Ravichandran KS Living on the Edge: Efferocytosis at the Interface of Homeostasis and Pathology. Immunity 50, 1149–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo B et al. Erythropoeitin Signaling in Macrophages Promotes Dying Cell Clearance and Immune Tolerance. Immunity 44, 287–302 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Kim S, Elkon KB & Ma X Transcriptional Suppression of Interleukin-12 Gene Expression following Phagocytosis of Apoptotic Cells. Immunity 21, 643–653 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Chung EY et al. Interleukin-10 Expression in Macrophages during Phagocytosis of Apoptotic Cells Is Mediated by Homeodomain Proteins Pbx1 and Prep-1. Immunity 27, 952–964 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucas M et al. Requirements for Apoptotic Cell Contact in Regulation of Macrophage Responses. The Journal of Immunology 177, 4047–4054 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Fond AM, Lee CS, Schulman IG, Kiss RS & Ravichandran KS Apoptotic cells trigger a membrane-initiated pathway to increase ABCA1. The Journal of Clinical Investigation 125, 2748–2758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvey CM et al. SIRPA-Inhibited, Marrow-Derived Macrophages Engorge, Accumulate, and Differentiate in Antibody-Targeted Regression of Solid Tumors. Current Biology 27, 2065–2077.e2066 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shekarabi M et al. WNK Kinase Signaling in Ion Homeostasis and Human Disease. Cell Metabolism 25, 285–299 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Wang GG et al. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nature Methods 3, 287 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Orlov SN, Koltsova SV, Kapilevich LV, Gusakova SV & Dulin NO NKCC1 and NKCC2: The pathogenetic role of cation-chloride cotransporters in hypertension. Genes & Diseases 2, 186–196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delpire E & Gagnon Kenneth B.E. SPAK and OSR1: STE20 kinases involved in the regulation of ion homoeostasis and volume control in mammalian cells. Biochemical Journal 409, 321–331 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Flores B, Schornak C, Wolfe L, Adams D & Delpire E Functional Characterization of the First Known Mutation of the Human SLC12A2 (NKCC1) Gene. The FASEB Journal 30, 12241222 (2016). [Google Scholar]
- 29.Delpire E et al. A patient with multisystem dysfunction carries a truncation mutation in human SLC12A2, the gene encoding the Na-K-2Cl cotransporter, NKCC1. Molecular Case Studies 2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koumangoye R, Omer S & Delpire E Mistargeting of a truncated Na-K-2Cl cotransporter in epithelial cells. American Journal of Physiology-Cell Physiology 0, null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kahle KT et al. WNK Protein Kinases Modulate Cellular Cl− Flux by Altering the Phosphorylation State of the Na-K-Cl and K-Cl Cotransporters. Physiology 21, 326–335 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Miyanishi M et al. Identification of Tim4 as a phosphatidylserine receptor. Nature 450, 435 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Park D, Hochreiter-Hufford A & Ravichandran KS The Phosphatidylserine Receptor TIM-4 Does Not Mediate Direct Signaling. Current Biology 19, 346–351 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Lee J et al. A scaffold for signaling of Tim-4-mediated efferocytosis is formed by fibronectin. Cell Death & Differentiation (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanagihashi Y, Segawa K, Maeda R, Nabeshima Y. i. & Nagata S Mouse macrophages show different requirements for phosphatidylserine receptor Tim4 in efferocytosis. 114, 8800–8805 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anselmo AN et al. WNK1 and OSR1 regulate the Na+, K+, 2Cl− cotransporter in HeLa cells. Proceedings of the National Academy of Sciences 103, 10883–10888 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vitari Alberto C., Deak M, Morrice Nick A. & Alessi Dario R. The WNK1 and WNK4 protein kinases that are mutated in Gordon’s hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochemical Journal 391, 17–24 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moriguchi T et al. WNK1 Regulates Phosphorylation of Cation-Chloride-coupled Cotransporters via the STE20-related Kinases, SPAK and OSR1. Journal of Biological Chemistry 280, 42685–42693 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Vitari, Alberto C. et al. Functional interactions of the SPAK/OSR1 kinases with their upstream activator WNK1 and downstream substrate NKCC1. Biochemical Journal 397, 223–231 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinehart J et al. Sites of Regulated Phosphorylation that Control K-Cl Cotransporter Activity. Cell 138, 525–536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zagórska A et al. Regulation of activity and localization of the WNK1 protein kinase by hyperosmotic stress. The Journal of Cell Biology 176, 89–100 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada K et al. Small-molecule WNK inhibition regulates cardiovascular and renal function. Nature Chemical Biology 12, 896 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Arroyo JP, Kahle KT & Gamba G The SLC12 family of electroneutral cation-coupled chloride cotransporters. Molecular Aspects of Medicine 34, 288–298 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Busetto S, Trevisan E, Decleva E, Dri P & Menegazzi R Chloride Movements in Human Neutrophils during Phagocytosis: Characterization and Relationship to Granule Release. The Journal of Immunology 179, 4110–4124 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Wang G Chloride flux in phagocytes. Immunological Reviews 273, 219–231 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Koncz C & Daugirdas JT Use of MQAE for measurement of intracellular [Cl-] in cultured aortic smooth muscle cells. American Journal of Physiology-Heart and Circulatory Physiology 267, H2114–H2123 (1994). [DOI] [PubMed] [Google Scholar]
- 47.Verkman AS, Sellers MC, Chao AC, Leung T & Ketcham R Synthesis and characterization of improved chloride-sensitive fluorescent indicators for biological applications. Analytical Biochemistry 178, 355–361 (1989). [DOI] [PubMed] [Google Scholar]
- 48.Albert ML, Kim J-I & Birge RB αvβ5 integrin recruits the CrkII–Dock180–Rac1 complex for phagocytosis of apoptotic cells. Nature Cell Biology 2, 899–905 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Reddien PW & Horvitz HR CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nature Cell Biology 2, 131 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Leverrier Y & Ridley AJ Requirement for Rho GTPases and PI 3-kinases during apoptotic cell phagocytosis by macrophages. Current Biology 11, 195–199 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Tosello-Trampont A-C, Brugnera E & Ravichandran KS Evidence for a Conserved Role for CrkII and Rac in Engulfment of Apoptotic Cells. 276, 13797–13802 (2001). [DOI] [PubMed] [Google Scholar]
- 52.deBakker CD et al. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr Biol 14, 2208–2216 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Nakaya M, Tanaka M, Okabe Y, Hanayama R & Nagata S Opposite Effects of Rho Family GTPases on Engulfment of Apoptotic Cells by Macrophages. 281, 8836–8842 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Lucas CD et al. Downregulation of Mcl-1 has anti-inflammatory pro-resolution effects and enhances bacterial clearance from the lung. Mucosal Immunology 7, 857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cruz CM et al. ATP Activates a Reactive Oxygen Species-dependent Oxidative Stress Response and Secretion of Proinflammatory Cytokines in Macrophages. 282, 2871–2879 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mishima E et al. Inherited, not acquired, Gitelman syndrome in a patient with Sjögren’s syndrome: importance of genetic testing to distinguish the two forms. CEN Case Rep 6, 180–184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu X, Su Z, Chen M, Xu Y & Wang Y Acquired Gitelman syndrome in a primary Sjögren syndrome patient with a SLC12A3 heterozygous mutation: A case report and literature review. 22, 652–655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou H et al. Complicated Gitelman syndrome and autoimmune thyroid disease: a case report with a new homozygous mutation in the SLC12A3 gene and literature review. 18, 82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kusuda T et al. Acquired Gitelman Syndrome in an Anti-SSA Antibody-positive Patient with a SLC12A3 Heterozygous Mutation. Internal Medicine 55, 3201–3204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hung C-M, Peng C-K, Wu C-P & Huang K-L Bumetanide attenuates acute lung injury by suppressing macrophage activation. Biochemical Pharmacology 156, 60–67 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Video 2 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Video 3 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Video 4 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Video 5 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Video 6 Control (Videos 1–3) or Slc12a2-deficient (Videos 4–6) ER-Hoxb8 BMDMs were cultured for 3 h with apoptotic cells labeled with CypHer5E prior to imaging. Apoptotic cells were then washed away and actively engulfing phagocytes, as determined by microscopy, were imaged for the loss of CypHer5E signal over time. All videos are over a 5 h time course with frame intervals of 10 min. Videos are representative of two independent experiments with two replicates per condition.
Supplementary Table 1 - Cell Volume Associated Genes Listed are members of the SLC12 (electroneutral chloride transporter) pathway genes with altered expression (based on adjusted p value and log2 fold change as determined via DESeq2) after corpse internalization, but not due to soluble factors/corpse-contact.
Supplementary Table 2 - Anti- and Pro-Inflammatory Genes List of genes associated with autoimmunity/chronic inflammatory disease that arose from Slc12a2-deficient efferocytic phagocytes (see Fig. 4).
Supplementary Table 3 – qPCR TaqMan Probes List of all hamster and mouse TaqMan probes used.
Data Availability Statement
All RNA-seq data for this experiment have been submitted to the Gene Expression Omnibus under accession number GSE131860. All other data supporting the findings of this study are available from the corresponding author on reasonable request.