Abstract
Germline variants in DNA repair genes are associated with aggressive prostate cancer (PrCa). The aim of this study was to characterize germline variants in DNA repair genes associated with lethal PrCa in Finnish and Swedish populations. Whole-exome sequencing was performed for 122 lethal and 60 unselected PrCa cases. Among the lethal cases, a total of 16 potentially damaging protein-truncating variants in DNA repair genes were identified in 15 men (12.3%). Mutations were found in six genes with CHEK2 (4.1%) and ATM (3.3%) being most frequently mutated. Overall, the carrier rate of truncating variants in DNA repair genes among men with lethal PrCa significantly exceeded the carrier rate of 0% in 60 unselected PrCa cases (p = 0.030), and the prevalence of 1.6% (p < 0.001) and 5.4% (p = 0.040) in Swedish and Finnish population controls from the Exome Aggregation Consortium. No significant difference in carrier rate of potentially damaging nonsynonymous single nucleotide variants between lethal and unselected PrCa cases was observed (p = 0.123). We confirm that DNA repair genes are strongly associated with lethal PrCa in Sweden and Finland and highlight the importance of population-specific assessment of variants contributing to PrCa aggressiveness.
Keywords: prostate cancer, DNA repair genes, lethal cancer
1. Introduction
Prostate cancer (PrCa), the most common male cancer worldwide, has a wide spectrum of clinical behavior that ranges from decades of indolence to rapid metastatic progression and lethality [1]. PrCa is also among the most heritable human cancers, with 57% of the interindividual variation in risk attributed to genetic factors [2]. Genome-wide association studies (GWAS) have thus far confirmed ~170 susceptibility loci that account for over 30% of the familial relative risk [3]. However, the risk variants identified using case-control designs show little or no ability to discriminate between indolent and fatal forms of this disease [4]. Therefore, studies contrasting patients with more and less aggressive disease and those exploring associations with disease progression and prognosis should be more effective at detecting genetic risk factors for aggressive PrCa with prognostic potential.
Inherited and acquired defects in DNA repair genes are a common hallmark of cancer and, to date, numerous inherited DNA repair gene mutations that increase cancer risk has been identified [5]. In particular, mutations in BRCA1 and BRCA2 genes, both associated with several DNA repair pathways, confer a strikingly increased risk of breast and ovarian cancer [6]. In addition, it is now recognized that the downregulation of DNA repair response is necessary for tumor progression into a more aggressive phenotype [5]. Accumulating evidence suggests that pathogenic germline variants in known cancer-predisposing genes such as BRCA2 can increase the risk of developing PrCa, especially the more aggressive form of the disease [7]. Likewise, several other genes that were initially implicated as high-risk genes in cancers other than PrCa, such as CHEK2 and BRIP1, have subsequently been shown to increase the risk of PrCa as well [8,9,10]. Recent studies have reported a high carrier rate of inherited DNA repair gene mutations among men with metastatic PrCa (11.8%), significantly exceeding the prevalence (4.6%) among men with localized PrCa [11].
In this study, we evaluated germline variants of DNA repair genes in men who died of PrCa. The aim of our study was to identify and investigate the frequency of pathogenic germline variants in men with the lethal form of the disease.
2. Materials and Methods
2.1. Study Subjects
Genomic DNA from a total of 122 lethal PrCa patients was collected from an ongoing collection of Finnish PrCa patients (TAMPERE, n = 47) and the Swedish Cancer of Prostate in Sweden (CAPS, n = 75) study. To create an extremely aggressive phenotype, the inclusion criterion for lethal PrCa cases was that the patient should have died due to PrCa before the age of 65. All of the Finnish patients were recruited in the Pirkanmaa Hospital District as part of a hereditary PrCa family collection or through collection of sporadic cases treated at the regional hospital [12]. The Swedish CAPS study is a population-based case-control study that enrolled participants between 2001 and 2003 [13]. An additional 70 PrCa patients from the TAMPERE population, not selected for disease aggressiveness or young age at death (hereby denoted unselected cases), with whole-exome sequencing data available were also included to contrast against the lethal cases. Clinical information, such as clinical stage, pathologic grade, nodal or distant metastases, and diagnostic serum levels of PSA and vital status, including cause of death, was obtained through medical records and national cancer registries. All samples were collected with written and signed informed consent. The project was approved by the research ethics committee at Pirkanmaa Hospital District (R03203), the Finnish National Supervisory Authority of Welfare and Health (5569/32/300/05) and by the ethics committees at the Karolinska Institutet (04-449/4 and 06-381/32).
2.2. Sample Preparation, Sequencing and Genotyping
Genomic DNA was extracted from whole blood by standard methods. For the 122 lethal cases, exome capture was performed using Agilent SureSelect Human All Exon 50 M kit (Agilent Technologies, Inc., Santa Clara, CA, USA) according to standard protocol and sequenced at the Science for Life laboratory (Stockholm, Sweden). Of the 70 unselected cases 25 samples were sequenced by BGI Tech Solutions (Hong Kong, China) with exome capture performed by the SureSelect Human All Exon 50 M kit while the remaining 45 unselected cases were sequenced at Mayo Clinic, Rochester, MN, USA with exome capture performed using Agilent SureSelect Human All Exon 50Mb or V4+UTR kits. At each site samples were sequenced using the Illumina Hiseq (Illumina, Inc, San Diego, CA, USA).
2.3. Sample Quality Control and Variant Calling
The reads were aligned against the hg19 genome build retrieved from UCSC using BWA [14]. BEDtools [15] was used to calculate the genome-wide coverage for each sample where samples with less than 30% of bases covered by at least 20 reads were excluded. The PCR duplicates were marked using PICARD [16], and the base score recalibration was performed using GATK [17]. Subsequently, GATK was used to call the variants and genotypes following the GATK best practices protocol for germline exome-sequencing data [18,19]. The candidate false-positive variants were initially filtered using the variant quality score recalibration procedure using the tranche threshold 99.0. Furthermore, variants having an allele fraction of less than 0.3 or a coverage of less than 12 were filtered out. Finally, variants with a readPosRankSum less than or equal to −1.7 were discarded. The variants were annotated using ANNOVAR [20].
2.4. Variant Prioritization
Variants found in 175 DNA repair genes [21,22,23] were selected for further analysis. To prioritize variants for validation, we utilized a similar approach to that introduced by Mijuskovic and coworkers [7]. The intergenic and common (minor allele frequency > 0.01) variants were filtered out. The remaining rare variants were classified into two categories: potentially damaging and neutral. The potentially damaging variants were further classified into two categories (Tier 1 and Tier 2) based on their impact. The classification was performed utilizing a database of reported associations of variants to clinical phenotypes (ClinVar) provided by ANNOVAR and two tools for pathogenicity prediction, CADD [24] and REVEL [25], of which the latter is specifically designed for discovery of rare deleterious variants. Moreover, the known protein domains from the UniProt [26] database were utilized to assess the pathogenicity of protein truncating variants.
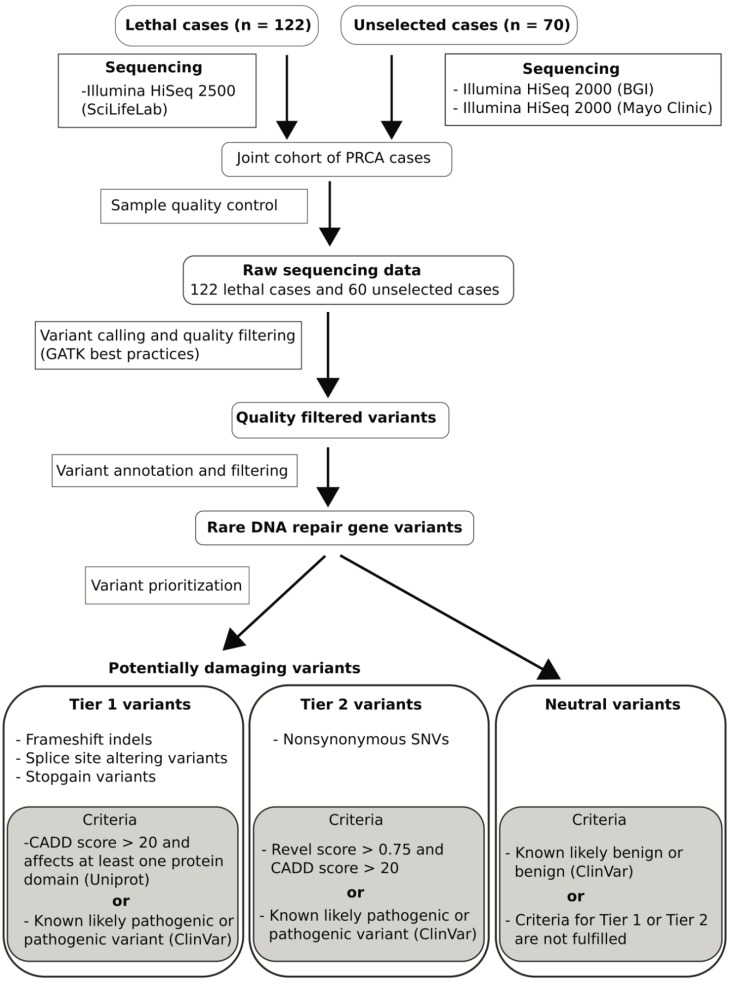
Those variants that are reported as likely benign or benign in ClinVar were classified as neutral. Protein truncating variants (stopgain, frameshift indels or splicing site altering variants) were classified as Tier 1 variants if they had a CADD phred score ≥ 20. Furthermore, the variants were required to be reported to be pathogenic or likely pathogenic by the ClinVar database or alternatively known to affect a protein domain reported in Uniprot (e.g., occurring before or within a protein domain). All nonsynonymous single nucleotide variants (missense variants) reported to be pathogenic or likely pathogenic by ClinVar or had a CADD phred score ≥ 20 and REVEL score ≥ 0.75 were classified as Tier 2 variants. The same prioritization criteria were applied to both case cohorts. The full workflow including details of the sequencing data analysis is illustrated in Figure 1.
Figure 1.
Flow chart describing processing of whole exome sequencing, quality control, variant calling and annotation, and variant prioritizing. PRCA: prostate cancer; ClinVar: database of reported associations of variants to clinical phenotypes; CADD: combined annotation dependent depletion; Revel: rare exome variant ensemble learner.
2.5. Population Frequencies
To explore the expected population allele frequencies of pathogenic variants in the discovered DNA repair genes, we extracted data from two subsets of the Exome Aggregation Consortium (ExAC) browser [27], one set comprising 6192 Swedish population controls and one set comprising 3307 Finnish individuals unselected for cancer history. Full details of the data processing, variant calling and resources have been described previously [27]. Variant prioritization among these population controls was performed by the same filtering algorithm as described above for the PrCa cases.
2.6. Statistical Analysis
Baseline characteristics were described using the median (interquartile range [IQR]) for continuous variables and absolute and relative frequencies for categorical variables. The frequency of potentially damaging DNA repair gene mutation carriers among the lethal PrCa patients was compared to the frequency in unselected PrCa patients and the two control populations with the use of a two-sided Fisher’s exact test. For the control populations, the frequency of mutation carriers in a specific gene was calculated on the basis of the total number of persons for whom sequence coverage was adequate for the given allele, under the assumption that each individual carried at most one deleterious mutation in the explored gene. This assumption may have introduced a slight overestimation in the carrier frequency in the control populations. In all analyses, Tier 1 and Tier 2 mutations were assessed separately. No adjustment was made for multiple testing, and p values less than 0.05 were considered to indicate statistical significance.
3. Results
We performed a comprehensive genetic assessment of DNA repair genes in 122 PrCa cases selected for very aggressive disease and 70 PrCa cases unselected for disease aggressiveness. After exclusion of 10 samples due to insufficient sequencing coverage, 122 lethal cases and 60 unselected cases remained for analysis (Figure 1)—see Table 1 for the clinical characteristics of case cohorts.
Table 1.
Clinical characteristics of patients.
| Lethal PrCa (n = 122) |
Unselected PrCa (n = 60) |
|
|---|---|---|
| Age at diagnosis, median (IQR) | 57.0 (55.1–58.2) | 66.5 (57.8–73.8) |
| Diagnostic PSA level (ng/mL), median (IQR) | 56.2 (17.9–247.2) | 10.8 (7.0–18.8) |
| Clinical T-stage, n (%) | ||
| TX | 2 (1.8) | 0 (0.0) |
| T1 | 8 (7.3) | 20 (38.5) |
| T2 | 18 (16.4) | 15 (28.8) |
| T3 | 61 (55.5) | 15 (28.8) |
| T4 | 21 (19.1) | 2 (3.8) |
| NA | 12 | 8 |
| Clinical N-stage, n (%) | ||
| NX | 86 (78.2) | 52 (100.0) |
| N0 | 9 (8.2) | 0 (0.0) |
| N1 | 15 (13.6) | 0 (0.0) |
| NA | 12 | 8 |
| Clinical M-stage, n (%) | ||
| MX | 11 (10.0) | 14 (26.9) |
| M0 | 45 (40.9) | 32 (61.5) |
| M1 | 54 (49.1) | 6 (11.5) |
| NA | 12 | 8 |
| Gleason score, n (%) | ||
| 2–6 | 11 (10.5) | 16 (47.1) |
| 7 | 36 (34.3) | 7 (20.6) |
| 8–10 | 58 (55.2) | 11 (32.4) |
| NA | 17 | 26 |
| Death due to PrCa, n (%) | 122 (100.0) | 15 (25.0) |
| Age at death, median (IQR) | 60.0 (57.9–62.9) | 79.5 (69.5–84.5) |
PrCa: prostate cancer; PSA: prostate-specific antigen; NA: not available.
In total, 22,850,167 variants were discovered and variant prioritization yielded 31 potentially damaging variants distributed across 17 DNA repair genes among the cases (Table 2).
Table 2.
Potentially damaging mutations identified in men with lethal prostate cancer.
| Gene | RSID | Type | Ref | Alt | Protein Change | ClinVar | CADD/REVEL | MAF | Tier |
|---|---|---|---|---|---|---|---|---|---|
| ATM | rs758081262 | stopgain | C | T | Q852X | 5 | 35/- | 2.5 × 10−5 | 1 |
| ATM | rs761486324 | frameshift ins | - | TG | H1082fs | - | -/- | - | 1 |
| ATM | rs767099464 | frameshift del | C | - | H1083fs | - | -/- | - | 1 |
| ATM | rs769142993 | missense | G | C | A2524P | 4 | 31/0.89 | 2.5 × 10−5 | 2 |
| ATM | - | frameshift del | AGTAG | - | S2611fs | - | -/- | - | 1 |
| ATM | rs753961188 | frameshift ins | - | T | L2885fs | 5,4 | -/- | 4.2 × 10−5 | 1 |
| ATM | rs376676328 | missense | A | G | R2912G | 3 | 29/0.88 | 3.0 × 10−4 | 2 |
| BRCA1 | rs41293459 | missense | C | T | R1699Q | 5,4,3 | 35/0.79 | 2.5 × 10−5 | 2 |
| CHEK2 | rs555607708 | frameshift del | G | - | T367fs | 5 | -/- | 1.8 × 10−3 | 1 |
| CHEK2 | rs137853007 | missense | G | A | R145W | 5,4 | 33/0.81 | 3.3 × 10−5 | 2 |
| CHEK2 | rs730881700 | frameshift ins | - | T | E457fs | 5,4 | -/- | 5.0 × 10−5 | 1 |
| CHEK2 | rs28909982 | missense | T | C | R117G | 5,4 | 27/0.93 | 1.0 × 10−4 | 2 |
| ERCC3 | rs753182861 | frameshift del | T | - | Q586fs | - | -/- | 2.0 × 10−4 | 1 |
| ERCC3 | rs145267069 | missense | A | G | F297S | - | 30/0.82 | 2.5 × 10−5 | 2 |
| FAN1 | rs778927800 | missense | G | A | R749Q | - | 34/0.89 | 8.3 × 10−6 | 2 |
| FANCM | rs147021911 | stopgain | C | T | Q1701X | 4 | 35/0.12 | 1.3 × 10−3 | 1 |
| HLTF | rs184046773 | missense | C | T | G1886A | - | 33/0.81 | 2.0 × 10−4 | 2 |
| MRE11A | rs372000848 | missense | G | A | R305W | 4,3 | 33/0.85 | 5.0 × 10−5 | 2 |
| MUTYH | rs34126013 | missense | G | A | R238W | 5,4 | 33/0.79 | 9.2 × 10−5 | 2 |
| NEIL1 | rs5745906 | missense | G | A | G169D | - | 27/0.86 | 1.3 × 10−3 | 2 |
| NTHL1 | rs150766139 | stopgain | G | A | Q90X | 5,3 | 35/- | 1.5 × 10−3 | 1 |
| POLG | rs761584617 | missense | G | A | A1115V | - | 23/0.80 | 2.5 × 10−5 | 2 |
| POLG | rs113994097 | missense | C | G | W748S | 5,3 | 33/0.91 | 8.0 × 10−4 | 2 |
| POLG | rs113994096 | missense | G | A | P587L | 5,3 | 28/0.80 | 1.7 × 10−3 | 2 |
| POLG | rs121918052 | missense | C | G | Q497H | 5,3 | 26/0.71 | 2.0 × 10−4 | 2 |
| POLL | rs139871590 | missense | C | T | G356S | - | 34/0.83 | 1.0 × 10−3 | 2 |
| RAD18 | rs138830303 | stopgain | T | A | K197X | - | 36/- | 1.0 × 10−4 | 1 |
| RECQL | rs149937760 | missense | C | T | C414Y | - | 33/0.84 | 2.0 × 10−4 | 2 |
| RECQL5 | rs768705080 | missense | T | G | Y362S | - | 32/0.76 | 8.2 × 10−6 | 2 |
| TP53 | rs876660754 | missense | C | T | V173M | 5,4 | 28/0.89 | - | 2 |
| TP53 | rs779000871 | missense | G | A | T170M | 3 | 24/0.87 | 8.2 × 10−5 | 2 |
Note: ClinVar clinical significance score defines as: 5 = pathogenic, 4 = likely pathogenic, 3 = uncertain significance. Minor allele frequency of variants derived from the Exome Aggregation Consortium. Ref: reference allele; Alt: alternative allele; ClinVar: database of reported associations of variants to clinical phenotypes; CADD: combined annotation dependent depletion; REVEL: rare exome variant ensemble learner; MAF: minor allele frequency; ins: insertion; del: deletion.
Screening of those 17 genes among the population controls revealed 157 potentially damaging variants (Supplementary Table S1) of which 137 were only discovered in the control populations, giving a total of 168 potentially damaging variants. In total, 79 of these variants were known to be pathogenic or likely pathogenic according to ClinVar, while the remaining variants were considered potentially damaging due to their truncating effects on protein domains or by having a REVEL score ≥ 0.75 and a CADD score ≥ 20. Of the 168 potentially damaging variants, 47 were classified as Tier 1 variants and 121 as Tier 2 variants. In total, 21 of the 47 Tier 1 variants were stopgain, 16 were frameshift indels, and 10 were splicing site altering variants.
In exploring the final 168 variants among the 122 lethal cases, 15 men (12.3%) carried at least one potentially damaging Tier 1 germline mutation in a DNA repair gene (one man carried two different Tier 1 mutations in the ATM gene), which was significantly higher than that observed in unselected cases (0%, p = 0.003, Table 3).
Table 3.
Carrier rates of potentially damaging mutations, stratified by Tier 1 and Tier 2 classification, in men with lethal prostate cancer, unselected prostate cancer, and population controls.
| Lethal PrCa (n = 122) | Unselected PrCa (n = 60) | p Value | Finnish Controls (n = 3307) | p Value | Swedish Controls (n = 6192) |
p Value | |
|---|---|---|---|---|---|---|---|
| Tier 1 | |||||||
| ERCC3, n (%) | 1 (0.82) | 0 | 1.000 | 0 | 0.036 | 3 (0.05) | 0.075 |
| RAD18, n (%) | 1 (0.82) | 0 | 1.000 | 0 | 0.036 | 0 | 0.019 |
| ATM, n (%) | 4 (3.28) | 0 | 0.304 | 4 (0.12) | <0.001 | 10 (0.16) | <0.001 |
| FANCM, n (%) | 2 (1.64) | 0 | 1.000 | 89 (2.69) | 0.772 | 44 (0.71) | 0.223 |
| NTHL1, n (%) | 2 (1.64) | 0 | 1.000 | 24 (0.73) | 0.236 | 39 (0.63) | 0.187 |
| CHEK2, n (%) | 5 (4.10) | 0 | 0.173 | 60 (1.81) | 0.080 | 5 (0.08) | <0.001 |
| All, n (%) | 15 (12.30) | 0 | 0.003 | 177 (5.35) | 0.004 | 101 (1.63) | <0.001 |
| Tier 2 | |||||||
| MUTYH, n (%) | 0 | 1 (1.67) | 0.330 | 34 (1.03) | 0.633 | 75 (1.21) | 0.406 |
| ERCC3, n (%) | 1 (0.82) | 1 (1.67) | 0.552 | 5 (0.15) | 0.195 | 4 (0.06) | 0.093 |
| HLTF, n (%) | 1 (0.82) | 0 | 1.000 | 20 (0.60) | 0.534 | 9 (0.15) | 0.177 |
| POLL, n (%) | 1 (0.82) | 0 | 1.000 | 15 (0.45) | 0.441 | 28 (0.45) | 0.433 |
| MRE11A, n (%) | 1 (0.82) | 0 | 1.000 | 0 | 0.036 | 0 | 0.019 |
| ATM, n (%) | 2 (1.64) | 0 | 1.000 | 13 (0.39) | 0.098 | 28 (0.45) | 0.114 |
| RECQL, n (%) | 1 (0.82) | 0 | 1.000 | 0 | 0.036 | 13 (0.21) | 0.239 |
| FAN1, n (%) | 1 (0.82) | 0 | 1.000 | 2 (0.06) | 0.103 | 16 (0.26) | 0.283 |
| NEIL1, n (%) | 1 (0.82) | 0 | 1.000 | 3 (0.09) | 0.135 | 16 (0.26) | 0.283 |
| POLG, n (%) | 5 (4.10) | 0 | 0.173 | 197 (5.96) | 0.555 | 190 (3.07) | 0.429 |
| TP53, n (%) | 2 (1.64) | 0 | 1.000 | 3 (0.09) | 0.012 | 7 (0.11) | 0.012 |
| BRCA1, n (%) | 1 (0.82) | 0 | 1.000 | 2 (0.06) | 0.103 | 5 (0.08) | 0.111 |
| RECQL5, n (%) | 1 (0.82) | 0 | 1.000 | 3 (0.09) | 0.135 | 1 (0.02) | 0.038 |
| CHEK2, n (%) | 1 (0.82) | 1 (1.67) | 0.552 | 2 (0.06) | 0.103 | 28 (0.45) | 0.433 |
| All, n (%) | 16 (13.11) | 3 (5.00) | 0.123 | 299 (9.04) | 0.148 | 420 (6.78) | 0.011 |
PrCa: prostate cancer. P value: the frequency of potentially damaging DNA repair gene mutation carriers among the lethal PrCa patients was compared to the frequency in unselected PrCa patients and the two control populations with the use of a two-sided Fisher’s exact test.
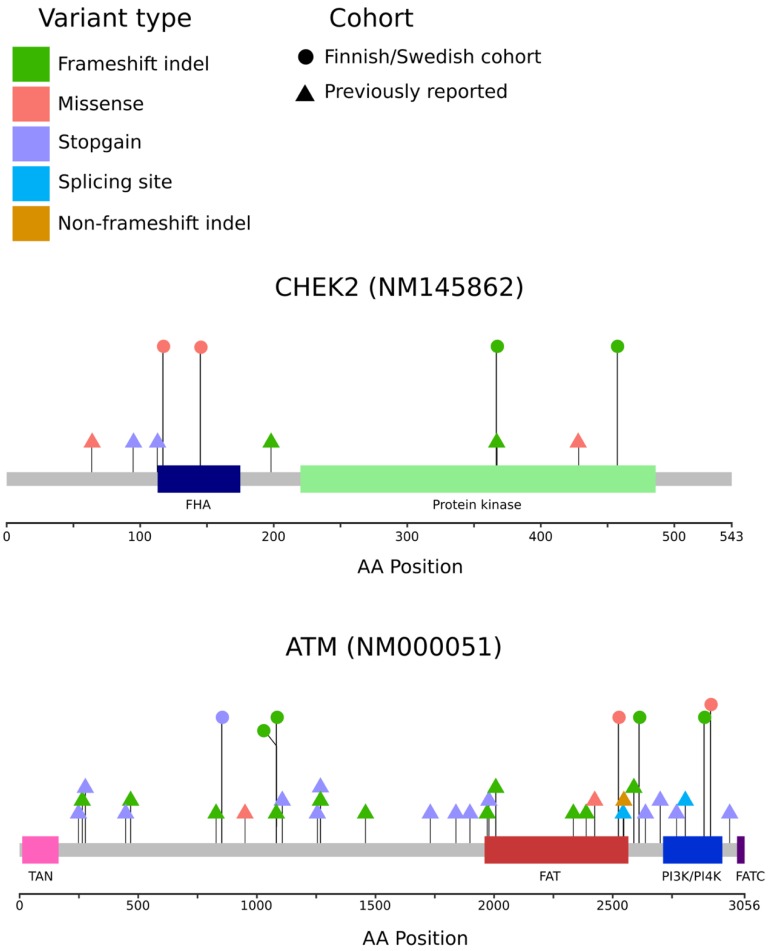
No significant difference in the Tier 1 mutation carrier rate was observed between Swedish (13.3%) and Finnish (10.6%, p = 0.781) lethal cases. The two most frequently mutated genes were CHEK2 (4.1%) and ATM (3.3%, Table 3, Figure 2). The observed carrier rate of Tier 1 mutations was significantly higher in the lethal cases compared to the prevalence in the Swedish (1.6%, p < 0.001) and the Finnish (5.4%, p = 0.040) population controls.
Figure 2.
Potentially damaging variants found in the CHEK2 and ATM genes. Locations of variants are shown as lollipop structures. The variants found in the Finnish/Swedish lethal or unselected cases are indicated by circles, and variants found in selected previous studies [7,11,18,28] are indicated by triangles. The variant type is indicated by the color.
The observed carrier rate of potentially damaging Tier 2 germline mutations was higher in the lethal cases (13.1%) compared to that of the unselected cases (5.0%); however, the difference was not statistically significant (p = 0.123, Table 3). Compared to Swedish controls (6.8%, p = 0.011), a higher mutation rate was observed among the lethal cases; however, there was no statistically significant difference in the carrier rate of Tier 2 mutations between the lethal cases and the Finnish population controls (9.0%, p = 0.148). No significant difference in the Tier 2 mutation carrier rate was observed between Swedish and Finnish lethal cases (p = 0.102).
No potentially damaging variants, neither Tier 1 nor Tier 2, were observed in the BRCA2 gene in any of the PrCa cases. In the population controls, we observed a carrier rate of Tier 1 BRCA2 mutations of 0.68% and 0.64% in Sweden and Finland, respectively.
4. Discussion
In this study, we characterized the germline variants occurring in the DNA repair pathway from 122 lethal and 60 unselected PrCa patients. In total, 16 potentially damaging protein truncating variants (Tier 1) were identified in 15 men (12.3%) among the lethal cases significantly exceeding the carrier rate of 0% in the unselected cases as well as the population prevalence of 1.6% and 5.4% in Swedish and Finnish population controls. In contrast, the frequency of potentially damaging nonsynonymous single nucleotide variants (Tier 2) showed similar frequencies among lethal cases, unselected cases and population controls.
Previous studies focusing on aggressive and metastatic PrCa cases have found higher frequencies of deleterious germline variants in BRCA2 than in any other DNA repair gene and thus considered it to be the major contributor among DNA repair genes to the aggressive phenotype [7,11,29]. However, we observed a frequency of zero pathogenic BRCA2 variants in our lethal cases, suggesting that BRCA2 does not play a major role in aggressive and lethal PrCa in the Swedish and Finnish populations. This agrees with earlier studies in which BRCA1 and BRCA2 were not found to have a significant contribution to PrCa susceptibility or aggressiveness in Finland or Sweden [30,31]. In a recent study by Mayrhofer and coworkers, sequencing of 217 metastatic PrCa cases from Sweden revealed only two pathogenic BRCA2 mutation carriers (0.93% carrier rate, [31]). Assuming the same carrier rate among our lethal cases, we would expect to find, on average, 1.1 carriers of BRCA2 mutations in our study, and our null finding is therefore not surprising. In general, the frequencies of established prostate cancer susceptibility variants deviate from population to population. One such case is the known cancer susceptibility variant G84E in HOXB13, which has been shown to have a mutation frequency approximately three-fold higher in Sweden and Finland compared to the mutation frequency in North America [32,33,34].
ATM and its role in pancreatic cancer was recently reviewed [35] and germline mutations in ATM have been associated with predisposition for several cancer forms [36] including PrCa [3]. Several studies have particularly reported potentially damaging variants in ATM in aggressive PrCa cases [7,9,29,31]. We also found high frequencies of potentially damaging variants in our lethal cohort (3.28% and 1.64% for Tier 1 and 2 variants, respectively), while in the unselected cases, the frequencies of these variants were found to be very low, similar to those of the population controls. These data support the evidence that deleterious variants in ATM are associated with the lethal phenotype of the disease. ATM is known to have a predominant role in the DNA damage response, but it also plays a role in maintaining the overall functionality of the cell [37]. ATM mutations that cause its inactivation or deficiency have shown a variety of pathological manifestations, including oxidative stress, metabolic syndrome, mitochondrial dysfunction and neurodegeneration. Recently ATM deficiency was shown to promote the progression of castration-resistant PrCa by enhancing the Warburg effect, suggesting that ATM mutation contributes through a metabolic—in addition to DNA repair—mechanism [38].
CHEK2 variants have been associated with PrCa predisposition in several studies [9,10], and we found that this gene was the most frequently mutated Tier 1 gene in our study (4.1%). In a recent study of 217 metastatic PrCa patients from Sweden [31], CHEK2 was also the most frequently mutated DNA repair gene (3.8%), highlighting the importance of CHEK2 mutations for aggressive PrCa in the Nordic population. Of note, in both the present study and the study by Mayrhofer and coworkers [31], c.1100delC was the most commonly observed mutation in CHEK2 (3.2% and 1.9%, respectively). Wu and coworkers also assessed the frequencies of potentially damaging CHEK2 variants in lethal cases and in cases with localized low-risk PrCa from the US [39]. Overall, no association was found between CHEK2 mutation status and lethal disease, but one variant, c.1100delC, was found to have a significantly higher frequency in the lethal cases (1.3%) compared to that of the low-risk PrCa patients (0.2%, p = 0.004), supporting the importance of this mutation for lethal PrCa. The c.1100delC has been shown to trigger nonsense-mediated mRNA decay, and subsequent protein analyses suggested that the truncated protein is likely highly unstable [40]. No mechanistic data are available for PrCa, but patients with CHEK2 mutations are among those showing a high response rate to treatment with the poly-ADP ribose polymerase inhibitor Olaparib when cancers were no longer responding to standard treatments [41].
Of note, only heterozygous carriers of protein-truncating variants were observed in our study conforming to the classical two-hit model for tumor suppressor genes [42,43]. No novel candidate genes within the DNA repair pathway were found in our study. The lack of novel findings is not surprising considering the limited sample size of the study. Moreover, we applied a relatively strict approach for prioritizing variants, which may have led us to underestimate the role of some genes or even to completely miss potential candidate genes.
We pooled Finnish and Swedish lethal cases to improve the statistical power of the association analysis. No adjustment for possible confounding, for example by population stratification, PSA screening history or family history of PrCa, was performed. Population stratification is always of importance in genetic association studies. However, genotypes from genome-wide single nucleotide polymorphisms were not available for all cases and we were therefore not able to adjust for possible population stratification through principal components in the current study. PSA screening is known to decrease PrCa-specific mortality [44,45] and it is possible that screening history may have confounded our analysis. However, for this to be the case PSA screening history must be associated with carrying pathogenic mutations in DNA repair genes which we find unlikely. Finally, Pritchard and coworkers [11] reported that deleterious mutation frequencies of DNA repair genes did not differ according to whether a family history of PrCa was present among 692 men with metastatic PrCa. Therefore, we argue that confounding by family history is of limited concern in our study.
5. Conclusions
In conclusion, germline variants in DNA repair genes have been shown to be associated with the aggressive form of PrCa—a finding that is supported by our study. Unlike previous studies, we did not observe high numbers of potentially damaging germline variants in BRCA2. Instead, mutations in ATM and CHEK2 were found to be most frequent among the lethal cases, highlighting the importance of the population-specific assessment of the variants contributing to the aggressiveness of PrCa.
Acknowledgments
Kirsi Rouhento is thanked for her assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/3/314/s1, Table S1: Potentially damaging variants discovered in control populations.
Author Contributions
Conceptualization, T.W., J.S. and F.W.; Data curation, T.R. and J.L.; Formal analysis, T.R. and F.W.; Funding acquisition, F.W.; Investigation, J.S. and F.W.; Methodology, T.R., J.L. and M.N.; Project administration, A.K., C.H. and J.L.; Resources, T.W., A.K., C.H., J.L., T.L.J.T., J.S. and F.W.; Software, T.R. and F.W.; Supervision, J.S. and F.W.; Validation, J.S. and F.W.; Visualization, J.S. and F.W.; Writing—original draft, T.R.; Writing—review and editing, T.R., J.S. and F.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Sigrid Juselius Foundation (to J.S.), the Academy of Finland (#251074 and #310115 to J.S.), the Cancer Foundation Finland (to J.S. and T.R.), the National Cancer Institute Grant U01 CA 89600 (support for the ICPCG), the Tampere Graduate Program in Biomedicine and Biotechnology (to T.R.), the Swedish Cancer Society (CAN 2016/818), and the Nordic Cancer Union.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Mucci L.A., Hjelmborg J.B., Harris J.R., Czene K., Havelick D.J., Scheike T., Graff R.E., Holst K., Moller S., Unger R.H., et al. Familial risk and heritability of cancer among twins in nordic countries. JAMA. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schumacher F.R., Al Olama A.A., Berndt S.I., Benlloch S., Ahmed M., Saunders E.J., Dadaev T., Leongamornlert D., Anokian E., Cieza-Borrella C., et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szulkin R., Karlsson R., Whitington T., Aly M., Gronberg H., Eeles R.A., Easton D.F., Kote-Jarai Z., Al Olama A.A., Benlloch S., et al. Genome-wide association study of prostate cancer-specific survival. Cancer Epidemiol. Biomarkers Prev. 2015;24:1796–1800. doi: 10.1158/1055-9965.EPI-15-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeggo P.A., Pearl L.H., Carr A.M. DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer. 2016;16:35–42. doi: 10.1038/nrc.2015.4. [DOI] [PubMed] [Google Scholar]
- 6.Friedenson B. The BRCA1/2 pathway prevents hematologic cancers in addition to breast and ovarian cancers. BMC Cancer. 2007;7:152. doi: 10.1186/1471-2407-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mijuskovic M., Saunders E.J., Leongamornlert D.A., Wakerell S., Whitmore I., Dadaev T., Cieza-Borrella C., Govindasami K., Brook M.N., Haiman C.A., et al. Rare germline variants in DNA repair genes and the angiogenesis pathway predispose prostate cancer patients to develop metastatic disease. Br. J. Cancer. 2018;119:96–104. doi: 10.1038/s41416-018-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kote-Jarai Z., Jugurnauth S., Mulholland S., Leongamornlert D.A., Guy M., Edwards S., Tymrakiewitcz M., O’Brien L., Hall A., Wilkinson R., et al. A recurrent truncating germline mutation in the BRIP1/FANCJ gene and susceptibility to prostate cancer. Br. J. Cancer. 2009;100:426–430. doi: 10.1038/sj.bjc.6604847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulo P., Maia S., Pinto C., Pinto P., Monteiro A., Peixoto A., Teixeira M.R. Targeted next generation sequencing identifies functionally deleterious germline mutations in novel genes in early-onset/familial prostate cancer. PLoS Genet. 2018;14:e1007355. doi: 10.1371/journal.pgen.1007355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seppala E.H., Ikonen T., Mononen N., Autio V., Rokman A., Matikainen M.P., Tammela T.L., Schleutker J. CHEK2 variants associate with hereditary prostate cancer. Br. J. Cancer. 2003;89:1966–1970. doi: 10.1038/sj.bjc.6601425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pritchard C.C., Mateo J., Walsh M.F., De Sarkar N., Abida W., Beltran H., Garofalo A., Gulati R., Carreira S., Eeles R., et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N. Engl. J. Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schleutker J., Matikainen M., Smith J., Koivisto P., Baffoe-Bonnie A., Kainu T., Gillanders E., Sankila R., Pukkala E., Carpten J., et al. A genetic epidemiological study of hereditary prostate cancer (HPC) in Finland: Frequent HPCX linkage in families with late-onset disease. Clin. Cancer Res. 2000;6:4810–4815. [PubMed] [Google Scholar]
- 13.Lindmark F., Zheng S.L., Wiklund F., Bensen J., Balter K.A., Chang B., Hedelin M., Clark J., Stattin P., Meyers D.A., et al. H6D polymorphism in macrophage-inhibitory cytokine-1 gene associated with prostate cancer. J. Natl. Cancer Inst. 2004;96:1248–1254. doi: 10.1093/jnci/djh227. [DOI] [PubMed] [Google Scholar]
- 14.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinlan A.R. BEDTools: The swiss-army tool for genome feature analysis. Curr. Protoc. Bioinform. 2014;47:11–12. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.PICARD. [(accessed on 6 February 2019)]; Available online: http://broadinstitute.github.io/picard/
- 17.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange S.S., Takata K., Wood R.D. DNA polymerases and cancer. Nat. Rev. Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood R.D., Mitchell M., Lindahl T. Human DNA repair genes, 2005. Mutat. Res. 2005;577:275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Wood R.D., Mitchell M., Sgouros J., Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 24.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.UniProt C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hart S.N., Ellingson M.S., Schahl K., Vedell P.T., Carlson R.E., Sinnwell J.P., Barman P., Sicotte H., Eckel-Passow J.E., Wang L., et al. Determining the frequency of pathogenic germline variants from exome sequencing in patients with castrate-resistant prostate cancer. BMJ Open. 2016;6:e010332. doi: 10.1136/bmjopen-2015-010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Na R., Zheng S.L., Han M., Yu H., Jiang D., Shah S., Ewing C.M., Zhang L., Novakovic K., Petkewicz J., et al. Germline Mutations in ATM and BRCA1/2 Distinguish Risk for Lethal and Indolent Prostate Cancer and are Associated with Early Age at Death. Eur. Urol. 2017;71:740–747. doi: 10.1016/j.eururo.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikonen T., Matikainen M.P., Syrjakoski K., Mononen N., Koivisto P.A., Rokman A., Seppala E.H., Kallioniemi O.P., Tammela T.L., Schleutker J. BRCA1 and BRCA2 mutations have no major role in predisposition to prostate cancer in Finland. J. Med. Genet. 2003;40:e98. doi: 10.1136/jmg.40.8.e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayrhofer M., De Laere B., Whitington T., Van Oyen P., Ghysel C., Ampe J., Ost P., Demey W., Hoekx L., Schrijvers D., et al. Cell-free DNA profiling of metastatic prostate cancer reveals microsatellite instability, structural rearrangements and clonal hematopoiesis. Genome Med. 2018;10:85. doi: 10.1186/s13073-018-0595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ewing C.M., Ray A.M., Lange E.M., Zuhlke K.A., Robbins C.M., Tembe W.D., Wiley K.E., Isaacs S.D., Johng D., Wang Y., et al. Germline mutations in HOXB13 and prostate-cancer risk. N. Engl. J. Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laitinen V.H., Wahlfors T., Saaristo L., Rantapero T., Pelttari L.M., Kilpivaara O., Laasanen S.L., Kallioniemi A., Nevanlinna H., Aaltonen L., et al. HOXB13 G84E mutation in Finland: Population-based analysis of prostate, breast, and colorectal cancer risk. Cancer Epidemiol. Biomarkers Prev. 2013;22:452–460. doi: 10.1158/1055-9965.EPI-12-1000-T. [DOI] [PubMed] [Google Scholar]
- 34.Xu J., Lange E.M., Lu L., Zheng S.L., Wang Z., Thibodeau S.N., Cannon-Albright L.A., Teerlink C.C., Camp N.J., Johnson A.M., et al. HOXB13 is a susceptibility gene for prostate cancer: Results from the International Consortium for Prostate Cancer Genetics (ICPCG) Hum. Genet. 2013;132:5–14. doi: 10.1007/s00439-012-1229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nanda N., Roberts N.J. ATM serine/threonine kinase and its role in pancreatic risk. Genes. 2020;11:108. doi: 10.3390/genes11010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi M., Kipps T., Kurzrock R. ATM mutations in cancer: Therapeutic implications. Mol. Cancer Ther. 2016;15:1781–1791. doi: 10.1158/1535-7163.MCT-15-0945. [DOI] [PubMed] [Google Scholar]
- 37.Guleria A., Chandna S. ATM kinase: Much more than a DNA damage responsive protein. DNA Repair (Amst) 2016;39:1–20. doi: 10.1016/j.dnarep.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Xu L., Ma E., Zeng T., Zhao R., Tao Y., Chen X., Groth J., Liang C., Hu H., Huang J. ATM deficiency promotes progression of CRPC by enhancing Warburg effect. Endocr. Relat. Cancer. 2019;26:59–71. doi: 10.1530/ERC-18-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y., Yu H., Zheng S.L., Na R., Mamawala M., Landis T., Wiley K., Petkewicz J., Shah S., Shi Z., et al. A comprehensive evaluation of CHEK2 germline mutations in men with prostate cancer. Prostate. 2018;78:607–615. doi: 10.1002/pros.23505. [DOI] [PubMed] [Google Scholar]
- 40.Anczukow O., Ware M.D., Buisson M., Zetoune A.B., Stoppa-Lyonnet D., Sinilnikova O.M., Mazoyer S. Does the nonsense-mediated mRNA decay mechanism prevent the synthesis of truncated BRCA1, CHK2, and p53 proteins? Hum. Mutat. 2008;29:65–73. doi: 10.1002/humu.20590. [DOI] [PubMed] [Google Scholar]
- 41.Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., Nava Rodrigues D., Robinson D., Omlin A., Tunariu N., et al. DNA-Repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knudson A.G., Jr. Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L.H., Wu C.F., Rajasekaran N., Shin Y.K. Loss of tumor suppressor gene function in human cancer: An overview. Cell Physiol. Biochem. 2018;51:2647–2693. doi: 10.1159/000495956. [DOI] [PubMed] [Google Scholar]
- 44.Schroder F.H., Hugosson J., Roobol M.J., Tammela T.L., Ciatto S., Nelen V., Kwiatkowski M., Lujan M., Lilja H., Zappa M., et al. Screening and prostate-cancer mortality in a randomized European study. N. Engl. J. Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 45.Schroder F.H., Hugosson J., Roobol M.J., Tammela T.L., Zappa M., Nelen V., Kwiatkowski M., Lujan M., Maattanen L., Lilja H., et al. Screening and prostate cancer mortality: Results of the European randomised study of screening for prostate cancer (ERSPC) at 13 years of follow-up. Lancet. 2014;384:2027–2035. doi: 10.1016/S0140-6736(14)60525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.