Abstract
MYO6 is known as a genetic cause of autosomal dominant and autosomal recessive inherited hearing loss. In this study, to clarify the frequency and clinical characteristics of hearing loss caused by MYO6 gene mutations, a large-scale genetic analysis of Japanese patients with hearing loss was performed. By means of massively parallel DNA sequencing (MPS) using next-generation sequencing for 8074 Japanese families, we found 27 MYO6 variants in 33 families, 22 of which are novel. In total, 2.40% of autosomal dominant sensorineural hearing loss (ADSNHL) in families in this study (32 out of 1336) was found to be caused by MYO6 mutations. The present study clarified that most cases showed juvenile-onset progressive hearing loss and their hearing deteriorated markedly after 40 years of age. The estimated hearing deterioration was found to be 0.57 dB per year; when restricted to change after 40 years of age, the deterioration speed was accelerated to 1.07 dB per year. To obtain supportive evidence for pathogenicity, variants identified in the patients were introduced to MYO6 cDNA by site-directed mutagenesis and overexpressed in epithelial cells. They were then assessed for their effects on espin1-induced microvilli formation. Cells with wildtype myosin 6 and espin1 co-expressed created long microvilli, while co-expression with mutant constructs resulted in severely shortened microvilli. In conclusion, the present data clearly showed that MYO6 is one of the genes to keep in mind with regard to ADSNHL, and the molecular characteristics of the identified gene variants suggest that a possible pathology seems to result from malformed stereocilia of the cochlear hair cells.
Keywords: MYO6, myosin, non-syndromic hearing loss, DFNA22, DFNB37, autosomal dominant, prevalence, genotype–phenotype correlation, hearing progression
1. Introduction
Genetic etiology is the most common cause of sensorineural hearing loss (SNHL), with over 50% of congenital hearing loss attributed to genetic causes in the US [1], a figure that is likely similar in all industrialized nations. More than 100 genes have been reported to be associated with non-syndromic hearing loss [2] (hereditary hearing loss home page https://hereditaryhearingloss.org/). Among them, several myosin genes are known to cause both syndromic and non-syndromic hearing loss (MYO7A, DFNA11 [3]; DFNB2 [4]; Usher syndrome type 1B [5]; MYH9, DFNA17 [6]; MYH9-related disorder [7]; MYH14, DFNA4 [8]; peripheral neuropathy, myopathy, hoarseness, and hearing loss [9]; MYO6, DFNA22 [10], DFNB37 [11]; MYO3A, DFNB30 [12]; MYO15A, DFNB3 [13]).
The human MYO6 gene is located on chromosome 6q13 and its gene mutations can cause either autosomal dominant (AD) inherited non-syndromic hearing loss (DFNA22) or autosomal recessive (AR) inherited non-syndromic hearing loss (DFNB37), possibly reflecting differences in pathophysiology among the mutations. The MYO6 gene encodes myosin 6, which is known to play a crucial role as a motor protein for cargo transportation [14]. In the mouse cochlea, myosin 6 proteins are expressed in outer and inner hair cells, particularly in the basal regions of the stereocilia [15,16]. Interestingly, it is also known that only type 6 myosin among the myosin family moves towards the basal end of stereocilia rootlets [17,18]. Seki et al. showed that the stereocilia of MYO6 mutant mice degenerated gradually and formed fused or branched stereocilia [19]. Therefore, myosin 6 is likely to be involved in the maintenance of stereocilia.
Despite its importance in the inner ear, a limited number of MYO6-associated hearing loss cases have been published. We have recently reported seven families with MYO6 mutations and showed their clinical features [20].
In this study, to obtain a more complete picture of the mutational spectrum, frequency of mutations, and characteristic clinical features of the patients, we undertook an analysis using a larger (n = 8074) cohort. In addition, we also performed in vitro functional analysis of novel missense MYO6 mutations identified in this study to clarify the cellular effects of the mutations.
2. Materials and Methods
2.1. Subjects
A total of 8074 Japanese hearing loss patients (AD, 1336; AR or sporadic, 5564; unknown, 1174) from 67 otolaryngology departments nationwide participated in this study. Our previous reported subjects (n = 1120) were included in the 8074 subjects [20]. Written informed consent was obtained from all subjects (or from their next of kin, caretaker, or guardian in the case of minors/children) prior to enrollment in the project. This study was approved by the Shinshu University Ethical Committee as well as the respective ethical committees of the other participating institutions listed hereinafter: Niigata University Ethical Committee, Toranomon Hospital Ethical Committee, Shiga University Ethical Committee, Kyoto Prefectural University Ethical Committee, Kyushu University Ethical Committee, Saitama Medical University Ethical Committee, Kobe University Ethical Committee, Osaka Medical Center and Research Institute for Maternal and Children Health Ethical Committee, and Hokkaido University Ethical Committee. All methods were performed in accordance with the Guidelines for Genetic Tests and Diagnoses in Medical Practice of the Japanese Association of Medical Sciences and the Declaration of Helsinki as required by Shinshu University, and the protocol was approved by the Ethics Committee of Shinshu University School of Medicine (No. 387—4 September 2012 and No. 576—2 May 2017).
2.2. Clinical Evaluation
Clinical data, including the onset age, progressiveness of hearing loss, pedigree, and episodes of vertigo, were collected based on their anamnestic records. Pure-tone audiometry was performed on patients over the age of 5. For very young children under the aged 4 or under, the auditory steady state response (ASSR), conditioned orientation response audiometry (COR) or play audiometry, was performed. The pure-tone average (PTA) was calculated from the audiometric thresholds at four frequencies (0.5, 1, 2, and 4 kHz). Severity of hearing loss was categorized into four groups: mild (PTA 20–40 dB hearing level (HL)), moderate (41–70 dBHL), severe (71–90 dBHL), and profound (>91 dBHL) [21].
2.3. Amplicon Library Preparation
Amplicon libraries were prepared using an Ion AmpliSeq Custom Panel (Thermofisher Scientific, Waltham, MA, USA), in accordance with the manufacturer’s instructions, for 63 genes reported to cause non-syndromic hearing loss [22]. After preparation, emulsion PCR and sequencing were performed according to the manufacturer’s instructions. The detailed protocol has been described elsewhere [22]. In brief, massively parallel DNA sequencing (MPS) was performed with an Ion Torrent Personal Genome Machine (PGM) system using an Ion PGM 200 Sequencing Kit and an Ion 318 Chip (Thermofisher Scientific, Waltham, MA, USA) or an Ion Proton system with an Ion PI Chip and Ion Hi-Q Chef Kit (Thermofisher Scientific, Waltham, MA, USA).
The sequence data were mapped against the human genome sequence (build GRCh37/hg19) with a Torrent Mapping Alignment Program. After sequence mapping, the DNA variant regions were piled up with Torrent Variant Caller plug-in software. After variant detection, their effects were analyzed using the ANNOVAR software. The missense, nonsense, insertion/deletion, and splicing variants were selected from among the identified variants. Variants were further selected as less than 1% of (1) the 1000 genome database, (2) the 6500 exome variants, (3) the Human Genetic Variation Database (dataset for 1208 Japanese exome variants), and (4) the 333 in-house Japanese normal hearing controls by using our database software. Direct sequencing was utilized to confirm the selected variants.
The pathogenicity of the candidate variants was interpreted based on the standards and guidelines of the ACMG (American College of Medical Genetics) [23]. Co-segregation analysis was performed for each proband and their family members by using direct sequencing.
2.4. Mutagenesis
Wild-type MYO6 cDNA, fused with the flexi Halotag, was purchased from Kazusa Genome Technologies (pFN21A-MYO6, clone id hj00061, KGT, Chiba, Japan). The identified mutations were introduced by Infusion HD cloning (Takara-Bio, Shiga, Japan) according to the manufacturer’s protocol. Briefly, site-specific primers with the mutant variations were constructed (Supplemental Table S1) (Sigma-Aldrich, St. Louis, MO, USA) and PCR was performed for 9 mutations; c.374C > A, c.604A > G, c.614G > A, c.647A > T, c.1376G > A, c.1455T > A, c.2111G > A, c.2438G > C, and c.3746T > C, under the conditions of 35 cycles of 98 °C for 10 s, 58 °C for 30 s, and 72 °C for 8 min. Template DNA was then digested by DPN1 and the PCR product was purified with a PCR Purification kit (Qiagen, Hilden, Germany). PCR products were then re-circularized with the Infusion HD enzyme and transformed into TOP10 competent cells (Thermofisher Scientific, Waltham, MA, USA). Plasmid DNA was amplified and confirmed to possess the targeted mutations by Sanger sequencing (Supplemental Table S1). Plasmid DNA was also confirmed to not contain off-target mutations by Sanger sequencing.
2.5. Transfection
LLC-PK1-CL4 porcine kidney epithelial cells (CL4 cells) were acquired from the James Bartles lab [24]. The day before transfection, CL4 cells were seeded at 50% confluency in DMEM with 10% FBS (Thermofisher Scientific, Waltham, MA, USA) and grown overnight at 37 °C in a 5% CO2 incubator in a 12-well plate on 15-mm glass coverslips (Matsunami Glass, Osaka, Japan) precoated with 2% gelatin. Wild-type and mutant MYO6 plasmid constructs were co-transfected with GFP-espin1 (obtained from the James Bartles lab) [24] with Lipofectamine 3000 (Thermofisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Briefly, 1 μg of DNA for each construct was prediluted in OptiMEMmedium (Thermofisher Scientific, Waltham, MA, USA) and combined with a pre-diluted Lipofectamine reagent, incubated at RT for 5 min, and then added to the cells. After 36 h, cells were checked for GFP expression by epifluorescent microscopy and fixed for immunocytochemistry.
2.6. Immunocytochemistry
Cells on coverslips were fixed in 4% paraformaldehyde in PBS for 10 min and then washed three times briefly in PBS, permeabilized for 20 min at room temperature with 0.1% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA) in PBS, washed in PBS 3 times, and then blocked with 3% goat serum (Thermofisher Scientific, Waltham, MA, USA) in PBS for 20 min. Cells were then incubated for 1 h at room temperature with a 1:200 dilution of the rabbit anti-Halotag antibody (Promega, Madison, WI, USA). Cells were washed three times with PBS, and then incubated for 1 h at room temperature with a 1:200 dilution of Alexa Fluor 546 goat anti-rabbit antibody and a 1/1000 dilution of DAPI. Cells were again washed in PBS 3 times, then the coverslips were mounted onto a slide glass with a Pro-long Gold Antifade mounting medium (Thermofisher Scientific, Waltham, MA, USA). Images were taken with an Olympus Fluoview FV-10i confocal microscope (Olympus, Okaya, Japan). The lengths of the microvilli with incorporated GFP-espin1 were measured with ImageJ software (NIH, Bethesda, MD, USA) and statistical significance assessed by Student’s t-test.
3. Results
3.1. Identified Mutations and Pathogenicity Interpretation
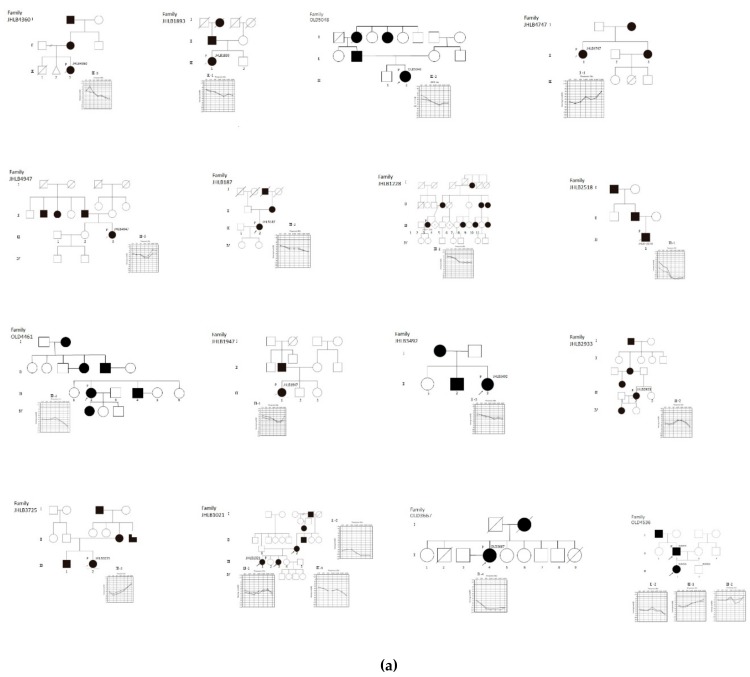
A total of 27 possibly disease-causing MYO6 candidate variants (six nonsense variants, five frameshift variants, seven splicing variants, one non-frameshift deletion, and eight missense variants) were identified from 32 AD inherited families and a single proband known to be affected with unknown inheritance in a heterozygous states (Table 1). Among these 33 MYO6-associated hearing loss families, seven families were reported in our previous paper. Thus, the prevalence of MYO6-associated hearing loss was 2.40% (32/1336) of Japanese autosomal dominant sensorineural hearing loss (ADSNHL) patients and 0.41% (33/8074) of all inheritance modes of Japanese hearing loss patients. All identified variants were confirmed by Sanger sequencing and the pathogenicity of these variants was interpreted in accordance with the ACMG guidelines [23]. As a result, three variants were classified as “pathogenic”, 16 as “likely pathogenic”, and 8 as “uncertain significance”. In cases where family member DNA samples were also obtained, segregation analyses were performed (Figure 1).
Table 1.
Possibly disease-causing MYO6 candidate variants.
| Family | Subject | Exon | Base Change | AA Change | Onset (y.o) |
Age (y.o) |
Progression | Vertigo | Audiogram (dB) | SIFT | PP2HV | LRT | Mut Taser | Mut Assesor | CADD Phred | ACMG Criteria |
Heredity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| JHLB4360 | Ⅲ-3 | 3 | c.187_187del | p.C63fs | under 10 | 9 | NA | NA | NA | likely pathogenic | AD | |||||||
| JHLB1893 | Ⅲ-1 | 4 | c.201delT | p.Y67fs | 20 | 31 | + | + | 40 | likely pathogenic | AD | |||||||
| OLD5048 | Ⅲ-2 | 4 | c.238C > T | p.R80X | 18 | 37 | + | – | 55 | D | A | 39 | pathogenic | AD | ||||
| JHLB4747 | Ⅱ-1 | 5 | c.374C > A | p.P125H | 30 | 60 | + | – | 47.5 | D | D | D | D | H | 25.9 | uncertain significance | AD | |
| JHLB4947 | Ⅲ-3 | 6 | c.429_431del | p.143_144del | 20 | 40 | + | – | 41.25 | likely pathogenic | AD | |||||||
| JHLB187 | Ⅲ-2 | 7 | c.553 + 1G > T | splicing aberrant | 20 | 41 | + | – | 40 | D | 24.9 | likely pathogenic | AD | |||||
| JHLB1228 | Ⅲ-2 | 8 | c.577delG | p.G193fs | NA | 45 | – | – | 35 | likely pathogenic | AD | |||||||
| JHLB2518 | Ⅲ-1 | 8 | c.604A > G | p.N202D | 5 | 60 | + | – | 101.25 | D | D | D | D | H | 24.2 | uncertain significance | AD | |
| OLD4461 | Ⅲ-2 | 8 | c.614G > A | p.R205Q | 50 | 65 | + | – | 55 | uncertain significance | AD | * | ||||||
| JHLB1947 | Ⅲ-1 | 10 | c.863_866del | p.D288fs | 8 | 20 | – | – | 37.5 | pathogenic | AD | |||||||
| JHLB3492 | Ⅱ-3 | 10 | c.863_866del | p.D288fs | NA | NA | NA | NA | NA | pathogenic | NA | |||||||
| JHLB2933 | Ⅲ-2 | 12 | c.1079 – 2A > G | splicing aberrant | 27 | 39 | + | + | 40 | D | 24.3 | likely pathogenic | AD | |||||
| JHLB3725 | Ⅲ-2 | 13 | c.1376G > A | p.G459D | 12 | 23 | – | – | 65 | D | D | D | D | H | 25.4 | uncertain significance | AD | |
| JHLB1021 | Ⅲ-1 | 14 | c.1455T > A | p.N485K | 27 | 50 | + | – | 53.75 | D | D | N | D | H | 23.2 | uncertain significance | AD | |
| JHLB1021 | Ⅱ-2 | 14 | c.1455T > A | p.N485K | 20s | 79 | + | – | NA | D | D | N | D | H | 23.2 | uncertain significance | AD | |
| JHLB1021 | Ⅲ-3 | 14 | c.1455T > A | p.N485K | elementary school | 46 | NA | – | NA | D | D | N | D | H | 23.2 | uncertain significance | AD | |
| OLD3667 | Ⅱ-4 | 19 | c.1975C > T | p.R659X | 50 | 76 | + | + | 103.8 | D | A | 48 | likely pathogenic | AD | * | |||
| OLD4536 | Ⅱ-2 | 19 | c.1975C > T | p.R659X | 28 | 43 | + | – | 72.5 | 48 | likely pathogenic | AD | * | |||||
| OLD4536 | Ⅲ-1 | 19 | c.1975C > T | p.R659X | 9 | 9 | + | – | 37.5 | 48 | likely pathogenic | AD | * | |||||
| OLD4536 | Ⅲ-2 | 19 | c.1975C > T | p.R659X | pre-critical | 11 | NA | – | 17.5 | 48 | likely pathogenic | AD | * | |||||
| JHLB4574 | Ⅲ-2 | 20 | c.2077 + 3A > G | splicing aberrant | 70 | 73 | + | – | 55 | likely pathogenic | AD | |||||||
| OLD4674 | Ⅲ-1 | 21 | c.2111G > A | p.G704D | 6 | 14 | – | – | 33.75 | D | D | D | D | H | 24.4 | uncertain significance | AD | |
| JHLB940 | Ⅳ-3 | 22 | c.2209 – 2A > G | splicing aberrant | 20 | 21 | – | + | 37.5 | D | 25 | likely pathogenic | AD | |||||
| JHLB433 | Ⅲ-1 | 23 | c.2287 – 2A > G | splicing aberrant | 6 | 10 | NA | – | 45 | D | 24.8 | likely pathogenic | AD | |||||
| JHLB3986 | Ⅲ-1 | 23 | c.2416 + 5G > A | splicing aberrant | 20 | 57 | + | + | 80 | likely pathogenic | AD | |||||||
| JHLB1589 | Ⅳ-1 | 24 | c.2438G > C | p.R813P | 0 | 7 | – | – | 68.75 | D | D | D | D | M | 27.4 | uncertain significance | AD | |
| OLD4362 | Ⅲ-3 | 25 | c.2563_2564insT | p.E855fs | 6 | 26 | + | – | 70 | likely pathogenic | AD | |||||||
| OLD3510 | Ⅲ-3 | 26 | c.2839C > T | p.R947X | 24 | 35 | + | – | 53.75 | D | A | 38 | likely pathogenic | AD | ||||
| OLD2267 | Ⅲ-2 | 28 | c.2998C > T | p.Q1000X | 30 | 43 | NA | NA | 56.25 | D | A | 42 | likely pathogenic | AD | ||||
| OLD2267 | Ⅱ-2 | 28 | c.2998C > T | p.Q1000X | NA | 60s | NA | NA | NA | D | A | 42 | likely pathogenic | AD | ||||
| JHLB1235 | Ⅱ-1 | 32 | c.3361A > T | p.K1121X | 26 | 41 | + | – | 41.25 | D | A | 50 | likely pathogenic | AD | * | |||
| JHLB530 | Ⅲ-4 | 34 | c.3496C > T | p.R1166X | 29 | 40 | NA | – | 43.75 | D | A | 47 | pathogenic | AD | * | |||
| JHLB315 | Ⅳ-2 | 34 | c.3496C > T | p.R1166X | 35 | 37 | + | – | 32.5 | D | A | 47 | pathogenic | AD | * | |||
| JHLB193 | Ⅲ-1 | 34 | c.3496C > T | p.R1166X | 31 | 32 | NA | NA | 32.5 | D | A | 47 | pathogenic | AD | * | |||
| JHLB3296 | Ⅱ-2 | 34 | c.3496C > T | p.R1166X | NA | 36 | + | – | 65 | D | A | 47 | pathogenic | sporadic | ||||
| JHLB3050 | Ⅱ-2 | 34 | c.3496C > T | p.R1166X | 30 | 48 | + | – | 93.75 | D | A | 47 | pathogenic | AD | ||||
| OLD2149 | Ⅲ-2 | 35 | c.3659 – 2A > G | splicing aberrant | 39 | 43 | + | – | 42.5 | D | 23.4 | likely pathogenic | AD | |||||
| JHLB3236 | Ⅳ-3 | 35 | c.3746T > C | p.F1249S | 0 | 15 | – | – | 46.25 | D | D | D | D | M | 25.6 | uncertain significance | AD |
AAChange, amino acid change; PP2HV, PolyPhen2 HumVar; MutTaster, MutationTaster; MutAssessor, MutationAssesor, ACMG, American College of Medical Genetics; D, deleterious or probably damaging; A, disease-causing-automatic; M, medium; H, high; N, neutral; AD, autosomal dominant; NA, data was not available. *: previously reported case [20].
Figure 1.
Pedigrees and audiograms of patients with MYO6-associated hearing loss. Arrowheads indicate family members receiving genetic testing, and arrowheads accompanying a “P” indicate the probands. (a) Filled symbols indicate affected individuals, and open symbols indicate unaffected individuals. (b) Pedigrees and audiograms of patients with MYO6-associated hearing loss.
Among the “pathogenic” variants, p.R1166X was previously reported as a cause of autosomal recessive sensorineural hearing loss (ARSNHL) and ADSNHL [11,20]. In this study, this p.R1166X variant was identified from four cases with AD inheritance and a single proband known to be affected with unknown inheritance.
3.2. Clinical Characteristics of MYO6-Associated Hearing Loss
The clinical features of 38 cases from 33 families with MYO6 variants are summarized in Table 1.
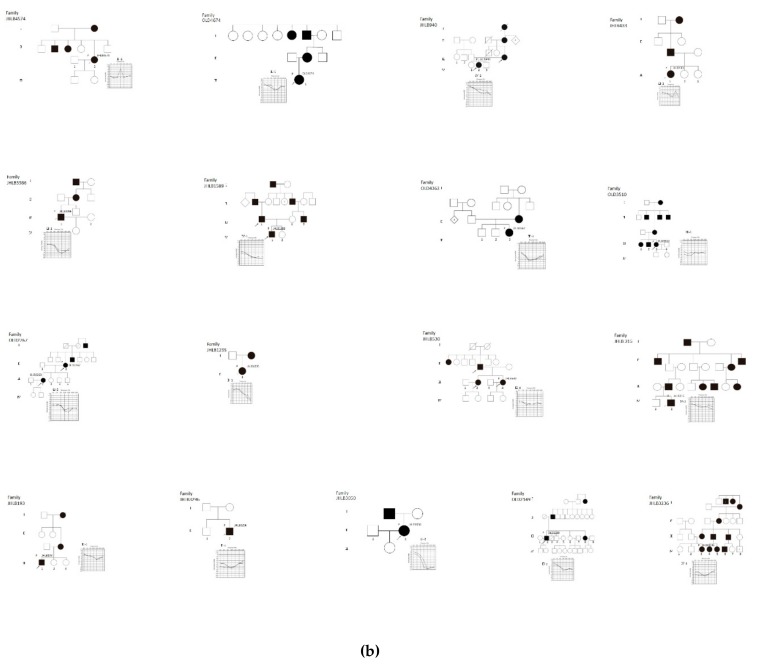
The average onset age is 24.0 years (range, 0 to 70 years). Among the 24 cases in which the age of onset was clarified, 22 cases occurred at the age of 40 or younger (Figure 2). Most of the cases suffered from mild-to-moderate hearing loss. However, three cases suffered from severe hearing loss and three cases suffered from profound hearing loss. Of 29 cases, an anamnestic evaluation indicated that 22 cases (75%) showed a progression of hearing loss.
Figure 2.
Distribution of the onset age of MYO6-associated hearing loss cases. Histogram of 33 cases segregated into 10-year intervals or congenital.
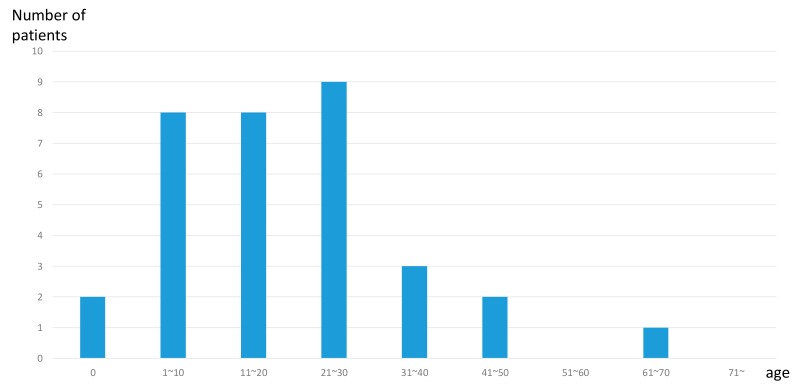
The pure-tone averages of the patients with MYO6-associated hearing loss and their ages when their hearing was tested are plotted in Figure 3A. The estimated hearing deterioration speed was 0.56 dB overall per year. However, when the analysis is restricted to over 40-year-old patients, the deterioration speed was increased to 1.07 dB per year. The averaged hearing thresholds for each frequency for every 20 years is indicated in Figure 3B. Patients who were 0−40 years old showed mild-to-moderate mid-frequency hearing loss; however, in the 41- to 60 age group, hearing deteriorated mainly in the higher frequencies and showed moderate high frequency-associated hearing loss. Furthermore, regarding the 61−80 age group, hearing deteriorated in all frequencies and showed a severe flat-type hearing loss. Most patients did not experience vertigo, with only five cases reporting episodes.
Figure 3.
Hearing loss deterioration in patients with MYO6 mutations. (A) Correlation between age and hearing. Pure-tone averages of each patient were plotted by dB hearing level (HL) and age at the time of the hearing test. (B) Average hearing threshold for each 20-year age group for patients with MYO6 mutations. A total of 8 cases were indicated in the 0−20, 11 in the 21−40, 12 in the 41−60, and 4 in the 60 or older.
As shown in Supplementary Figure S1, there were 11 cases with multiple hearing test results, showing the actual progression of hearing loss.
3.3. In Vitro Analysis of the Identified MYO6 Variants
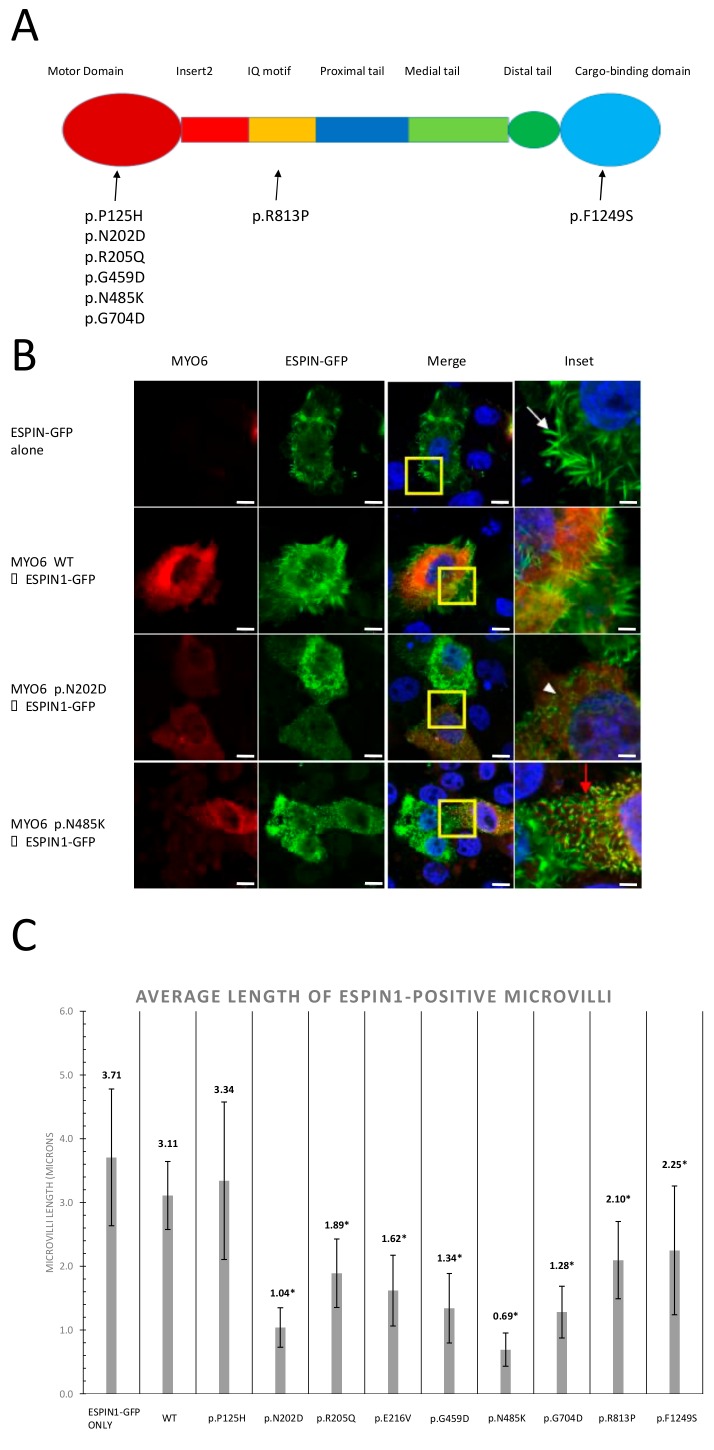
To clarify the functional effects of the MYO6 candidate missense variants identified in this study and the previous studies [20], we constructed a wild-type MYO6 plasmid and incorporated the following identified variants by site-directed mutagenesis: p.P125H, p.N202D, p.R205Q, p.E216V, p.G459D, p.N485K, p.G704D, p.R813P, and p.F1249S (Figure 4).
Figure 4.
Mutations in MYO6 resulted in shorter microvilli. (A) Locations of targeted mutations in the myosin 6 domain structure as indicated. Most mutations were located in the motor domain. (B) Confocal images of immunocytochemistry of CL4 epithelial cells transfected with GFP-espin1 alone or co-transfected with Halo-tagged MYO6 constructs as indicated. The yellow boxes are magnified as the insets. MYO6 overexpression was detected using an anti-Halotag antibody. GFP-espin1 induced long microvilli (white arrow), but these were shorter in cells with mutant myosin 6 (white arrowhead). MYO6 expression can be detected at the base of each microvilli (red arrow). Scale bars = 10 µm. (C) Graph indicating the average length of GFP-espin1-positive microvilli. Microvilli length of cells with each mutation were measured in ImageJ (µm). For each sample, 50−70 microvilli were measured for length. Asterisks indicates statistically significant differences compared to the WT (p < 0.001).
We then transfected them into CL4 cells with a GFP-espin1 plasmid [25]. The microvilli of the CL4 cells were elongated by espin overexpression and provided a useful structure with which to investigate stereocilia-related proteins.
As a result, when we transfected only the GFP-espin1 plasmid or GFP-espin1 with the wild-type MYO6 plasmid, cells exhibited the formation of long microvilli, indicating that the overexpression of MYO6 itself does not affect microvilli formation (Figure 4B). However, when we incorporated GFP-espin1 with most candidate variants of MYO6, only shorter microvilli were formed (Figure 4B, Supplementary Figure S2). In addition, all mutated myosin 6 proteins were accumulated at the base of the microvilli. The lengths of the microvilli formed in transfected cells were measured and statistical analysis was performed (Figure 4C). All mutations, except p.P125H, showed significantly shorter microvilli compared to wild-type MYO6.
4. Discussion
This study expanded our previous study [20] and identified 33 families with candidate MYO6 gene variants (Table 1) and, using the largest number (n = 8074) of HL patients with MYO6-associated hearing loss to date, clarified the precise data regarding frequency, mutation spectrum, and clinical characteristics of associated hearing loss.
The frequency of MYO6-associated hearing loss was 2.40% in the Japanese AD hearing loss patients. This frequency is compatible with our previous study (2.6%; 7/266) [20] and is lower than the most frequent ADSNHL gene (6.6% for KCNQ4 [26]) but comparable with other ADSNHL genes, such as 3.2% for TECTA [27], 2.5% for POU4F3 [28], and 2.5% for WFS1 [29]. Therefore, MYO6 is considered an important causative gene for Japanese patients with ADSNHL. However, this is not a population-specific phenomenon as Seco et al. recently also reported that the MYO6 gene is a frequent cause of AD hearing loss in the population in the Netherlands [30].
It is noted that, as shown in Figure 2, MYO6-associated hearing loss is mainly found in late-onset mild hearing loss cases, who are easily overlooked. MPS is useful in identifying the causative gene mutations in such juvenile-onset mild-to-moderate hearing loss cases. Indeed, herein, we report that MYO6-associated hearing loss is characterized by juvenile-onset mild to moderate hearing loss up to age 40. Thirty out of thirty-three cases in this study also showed juvenile-onset before the age of 40, which is consistent with our previous reports (Figure 2). We previously reported that the onset of hearing loss in MYO6-associated cases could be detected between the ages of 5 and 50, with the hearing deteriorating to profound hearing loss by the 6th decade [20]. We have shown that, in half of all individuals, hearing loss first appeared in the lower and mid frequencies and progressed to flat or sloping severe-to-profound hearing loss [20].
Most MYO6-associated hearing loss reported in previous papers showed progressive hearing loss [20,31,32]. Our results were consistent with such previous reports and most patients in this study also showed progressive hearing loss. It is noteworthy that the hearing thresholds were maintained until the age of 40, but rapidly deteriorated after that, worsening to severe hearing loss after the age of 60 (Figure 3B). In this study, multiple audiograms were obtained in 11 cases showing progressive hearing loss, providing supporting evidence of the progressive nature of the hearing loss due to MYO6 gene variants (Supplementary Figure S1). When the analysis was restricted to patients over 40 years of age, the deterioration speed increased to 1.07 dB per year. The reason for such an accelerated rate of deterioration for patients over 40 years old patients is unknown, but it may be related to the type of variants found in this study. Most of the variants found are non-functional variants (nonsense, frame shift, or splicing variants), and, therefore, it is plausible that any dysfunctions may be due to haploinsufficiency. If non-functional or unstable proteins were produced by these mutant alleles, such conditions may become more critical with age.
MYO6 is known to cause ARSNHL (DFNB37) [11] and ADSNHL (DFNA22) [10]. The p.R1166X variant is an interesting one as it was first reported as a cause of AR hearing loss (DFNB 37) [11], but was later reported in three independent ADSNHL (DFNA22) families [20]. An in vitro functional study of the p.R1166X variant using RPE cells showed the pathogenicity of this mutation, indicating that this variant may cause cellular dysfunction [33].
In the present large cohort, the p.R1166X variant was found in four ADSNHL families and a single proband known to be affected with unknown inheritance, but unfortunately, we could not obtain audiograms from the parents in the sporadic family.
A possible explanation of how the p.R1166X variant causes both recessive and dominant inherited hearing loss involves autosomal semi-dominant inheritance. Indeed, an autosomal semi-dominant inheritance mode has been previously reported in MYO6-related hearing loss [34]. In the reported family, the family member with homozygotes for a splice mutation, c.897G>T, showed a more severe phenotype with early-onset, compared with the family members with heterozygotes with late-onset progressive hearing loss [34].
It should be noted that the phenotype of the DFNB 37 family [11] is based on anamnestic evaluation. Based on our clinical data, the phenotype caused by the p.R1166X variant is mild, and there is a risk that hearing loss could be overlooked unless audiometric evaluation is performed, or the parents are too young to develop hearing loss.
Since the p.R1166X variant has been reported in other populations with different origins [11,20,35], this mutation may be present due to a hot spot rather than a common ancestor phenomenon.
In vitro analysis showed that the espin1-induced microvilli of CL4 cells become shorter when patient-identified missense MYO6 variants were overexpressed. This observation could provide supportive evidence of the pathogenicity of the identified MYO6 mutations. A mutant mouse model with a homozygous MYO6 gene splicing mutation showed fused and/or branched stereocilia [10]. It is possible to cause such morphological abnormalities as a result of the inhibition of the stereocilia extension. Structural protein interactions at the base of stereocilia requires proper myosin 6 function. RDX, CLIC5, TPRN, and PTPRQ are the components which interact with MYO6 to create protein complexes regulating stereocilia formation, maintenance, and mechano-transduction of the sound [36,37]. Mutations in the MYO6 motor domain have been shown to inhibit MYO6 localization in hair cells [36]. Interestingly, we found that mutant myosin 6 proteins were also localized in the basolateral region of the espin1-induced microvilli. These results indicate that the ability of the mutant myosin 6 to move to the basolateral end of microvilli was not disrupted. It has also been shown that myosin 6 is endogenously expressed in CL4 cells [38] and is estimated to have the ability to form the basal complex with myosin 6 proteins. When CLIC5 was overexpressed in CL4 cells, it directly interacted with TPRN and endogenous ezrin protein [36], indicating the proper functioning of this basal complex. From these observations and our results, mutations in MYO6 may still allow myosin 6 proteins to maintain their motor function, as they are located in the basal region of microvilli, but disrupt complex formation or binding to other component proteins. In this paper, most of the candidate mutations were found to result in shorter microvilli; however, we could not identify any correlation between the length of the microvilli and the severity of hearing loss. Thus, further functional studies are required to be elucidated this phenotype–genotype correlation and molecular function.
5. Conclusions
In summary, using a large cohort, we could obtain a more complete pictures of the mutational spectrum, frequency of mutations, and characteristic clinical features of patients suffering from hearing loss caused by MYO6 variants. The better prediction of the hearing loss progression speed allows us to provide more appropriate intervention for the patients, such as hearing aids or cochlear implants, with optimal timing. Careful attention should be paid to the patients with late-onset mild-to-moderate progressive hearing loss as there is the potential for marked hearing deterioration to occurs after the age of 40.
Acknowledgments
We thank the participants of the Deafness Gene Study Consortium for providing samples and clinical information. We also thank Sachiko Matsuda and Fumiko Tomioka for their technical assistance with this research.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/3/273/s1, Table S1: List of primers used for site-directed mutagenesis and sequencing. At www.mdpi.com/xxx/fs1, Figure S1: Audiogram for cases where serial testing was performed. Figure legends show age at testing as indicated. At www.mdpi.com/xxx/fs2, Figure S2: Confocal images of immunocytochemistry of CL4 epithelial cells co-transfected with GFP-espin1 and the mutant Halo-tagged MYO6 constructs as indicated. Myosin 6 overexpression was detected using an anti-Halotag antibody (red). Yellow boxes are magnified as the insets.
Author Contributions
Conceptualization, S.-y.N. and S.-i.U.; methodology, S.-y.N.; software, S.-y.N.; validation, S.-i.O., T.F.D., S.-i.K., H.M., and M.M.; formal analysis, S.-i.O. and S.-i.K.; investigation, S.-i.U.; resources, S.M., S.I., T.I., S.A., J.N., M.H., N.O., N.U., and C.O.; data curation, S.-i.O., T.F.D., S.-i.K., H.M., and S.-y.N.; writing—original draft preparation, S.-i.O. and S.-i.K.; writing—review and editing, S.-i.O., T.F.D., S.-i.K., and S.-y.N.; visualization, S.-i.O. and T.D.; supervision, S.-i.U.; project administration, S.-i.U.; funding acquisition, S.-i.U. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a Health and Labor Sciences Research Grant for Research on Rare and Intractable Diseases and Comprehensive Research on Disability Health and Welfare from the Ministry of Health, Labor and Welfare of Japan (S.-i.U. H29-Nanchitou(Nan)-Ippan-031), a Grant-in-Aid from Japan Agency for Medical Research and Development (AMED) (S.-i.U. 16kk0205010h001, 18ek0109363h0001), and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (S.-i.U. 15H02565; T.D. 19K18802).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Morton C., Nance W. Newborn hearing screening—A silent revolution. N. Engl. J. Med. 2006;18:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Hereditary Hearing Loss Homepage. [(accessed on 23 January 2020)]; Available online: http://hereditaryhearingloss.org/
- 3.Luijendijk M.W., Van Wijk E., Bischoff A.M., Krieger E., Huygen P.L., Pennings R.J., Brunner H.G., Cremers C.W., Cremers F.P., Kremer H. Identification and molecular modelling of a mutation in the motor head domain of myosin VIIA in a family with autosomal dominant hearing impairment (DFNA11) Hum. Genet. 2004;115:149–156. doi: 10.1007/s00439-004-1137-3. [DOI] [PubMed] [Google Scholar]
- 4.Riazuddin S., Nazli S., Ahmed Z.M., Yang Y., Zulfiqar F., Shaikh R.S., Zafar A.U., Khan S.N., Sabar F., Javid F.T., et al. Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum. Mutat. 2008;29:502–511. doi: 10.1002/humu.20677. [DOI] [PubMed] [Google Scholar]
- 5.Adato A., Weil D., Kalinski H., Pel-Or Y., Ayadi H., Petit C., Korostishevsky M., Bonne-Tamir B. Mutation profile of all 49 exons of the human myosin VIIA gene, and haplotype analysis, in Usher 1B families from diverse origins. Am. J. Hum. Genet. 1997;61:813–821. doi: 10.1086/514899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lalwani A.K., Goldstein J.A., Kelley M.J., Luxford W., Castelein C.M., Mhatre A.N. Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9. Am. J. Hum. Genet. 2000;67:1121–1128. doi: 10.1086/321212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pecci A., Ma X., Savoia A., Adelstein R.S. MYH9: Structure, functions and role of non-muscle myosin IIA in human disease. Gene. 2018;664:152–167. doi: 10.1016/j.gene.2018.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donaudy F., Snoeckx R., Pfister M., Zenner H.P., Blin N., Di Stazio M., Ferrara A., Lanzara C., Ficarella R., Declau F., et al. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4) Am. J. Hum. Genet. 2004;74:770–776. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi B.O., Kang S.H., Hyun Y.S., Kanwal S., Park S.W., Koo H., Kim S.B., Choi Y.C., Yoo J.H., Kim J.W., et al. A complex phenotype of peripheral neuropathy, myopathy, hoarseness and hearing loss is linked to an autosomal dominant mutation in MYH14. Hum. Mutat. 2011;32:669–677. doi: 10.1002/humu.21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melchionda S., Ahituv N., Bisceglia L., Sobe T., Glaser F., Rabionet R., Arbones M.L., Notarangelo A., Di Iorio E., Carella M., et al. MYO6, the human homologue of the gene responsible for deafness in Snell’s waltzer mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am. J. Hum. Genet. 2001;69:635–640. doi: 10.1086/323156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmed Z.M., Morell R.J., Riazuddin S., Gropman A., Shaukat S., Ahmad M.M., Mohiddin S.A., Fananapazir L., Caruso R.C., Husnain T., et al. Mutations of MYO6 are associated with recessive deafness, DFNB37. Am. J. Hum. Genet. 2003;72:1315–1322. doi: 10.1086/375122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh T., Walsh V., Vreugde S., Hertzano R., Shahin H., Haika S., Lee M.K., Kanaan M., King M.C., Avraham K.B. From flies’ eyes to our ears: Mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc. Natl. Acad. Sci. USA. 2002;99:7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang A., Liang Y., Fridell R.A., Probst F.J., Wilcox E.R., Touchman J.W., Morton C.C., Morell R.J., Noben-Trauth K., Camper S.A., et al. Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science. 1998;280:1447–1451. doi: 10.1126/science.280.5368.1447. [DOI] [PubMed] [Google Scholar]
- 14.Hasson T. Myosin VI: Two distinct roles in endocytosis. J. Cell Sci. 2003;116:3453–3461. doi: 10.1242/jcs.00669. [DOI] [PubMed] [Google Scholar]
- 15.Hasson T., Gillespie P.G., Garcia J.A., MacDonald R.B., Zhao Y., Yee A.G., Mooseker M.S., Corey D.P. Unconventional myosins in inner-ear sensory epithelia. J. Cell Biol. 1997;137:1287–1307. doi: 10.1083/jcb.137.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Self T., Sobe T., Copeland N.G., Jenkins N.A., Avraham K.B., Steel K.P. Role of myosin VI in the differentiation of cochlear hair cells. Dev. Biol. 1999;214:331–341. doi: 10.1006/dbio.1999.9424. [DOI] [PubMed] [Google Scholar]
- 17.Wells A.L., Lin A.W., Chen L.Q., Safer D., Cain S.M., Hasson T., Carragher B.O., Milligan R.A., Sweeney H.L. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 18.Rock R.S., Rice S.E., Wells A.L., Purcell T.J., Spudich J.A., Sweeney H.L. Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seki Y., Miyasaka Y., Suzuki S., Wada K., Yasuda S.P., Matsuoka K., Ohshiba Y., Endo K., Ishii R., Shitara H., et al. A novel splice site mutation of myosin VI in mice leads to stereociliary fusion caused by disruption of actin networks in the apical region of inner ear hair cells. PLoS ONE. 2017;12:e0183477. doi: 10.1371/journal.pone.0183477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyagawa M., Nishio S.Y., Kumakawa K., Usami S.I. Massively parallel DNA sequencing successfully identified seven families with deafness-associated MYO6 mutations: The mutational spectrum and clinical characteristics. Ann. Otol. Rhinol. Laryngol. 2015;124(Suppl. 1):148S–157S. doi: 10.1177/0003489415575055. [DOI] [PubMed] [Google Scholar]
- 21.Mazzoli M., Van Camp G., Newton V., Giarbini N., Declau F., Parving A. Recommendations for the description of genetic and audiological data for families with nonsyndromic hereditary hearing impairment. Audiol. Med. 2009;1:148–150. [Google Scholar]
- 22.Nishio S.Y., Usami S.I. Deafness gene variations in a 1120 nonsyndromic hearing loss cohort: Molecular epidemiology and deafness mutation spectrum of patients in Japan. Ann. Otol. Rhinol. Laryngol. 2015;124(Suppl. 1):49S–60S. doi: 10.1177/0003489415575059. [DOI] [PubMed] [Google Scholar]
- 23.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loomis P.A., Zheng L., Sekerková G., Changyaleket B., Mugnaini E., Bartles J.R. Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. J. Cell Biol. 2003;163:1045–1055. doi: 10.1083/jcb.200309093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng L., Beeler D.M., Bartles J.R., Zheng L., Beeler D.M., Bartles J.R. Characterization and regulation of an additional actin-filament-binding site in large isoforms of the stereocilia actin-bundling protein espin. J. Cell Sci. 2014;127:1306–1317. doi: 10.1242/jcs.143255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naito T., Nishio S.Y., Iwasa Y., Yano T., Kumakawa K., Abe S., Ishikawa K., Kojima H., Namba A., Oshikawa C., et al. Comprehensive genetic screening of KCNQ4 in a large autosomal dominant nonsyndromic hearing loss cohort: Genotype-phenotype correlations and a founder mutation. PLoS ONE. 2013;8:e63231. doi: 10.1371/journal.pone.0063231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasukawa R., Moteki H., Nishio S.Y., Ishikawa K., Abe S., Honkura Y., Hyogo M., Mihashi R., Ikezono T., Shintani T., et al. The prevalence and clinical characteristics of TECTA-associated autosomal dominant hearing loss. Genes (Basel) 2019;10:E744. doi: 10.3390/genes10100744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitano T., Miyagawa M., Nishio S.Y., Moteki H., Oda K., Ohyama K., Miyazaki H., Hidaka H., Nakamura K.I., Murata T., et al. POU4F3 mutation screening in Japanese hearing loss patients: Massively parallel DNA sequencing-based analysis identified novel variants associated with autosomal dominant hearing loss. PLoS ONE. 2017;12:e0177636. doi: 10.1371/journal.pone.0177636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi M., Miyagawa M., Nishio S.Y., Moteki H., Fujikawa T., Ohyama K., Sakaguchi H., Miyanohara I., Sugaya A., Naito Y., et al. WFS1 mutation screening in a large series of Japanese hearing loss patients: Massively parallel DNA sequencing-based analysis. PLoS ONE. 2018;13:e0193359. doi: 10.1371/journal.pone.0193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zazo Seco C., Wesdorp M., Feenstra I., Pfundt R., Hehir-Kwa J.Y., Lelieveld S.H., Castelein S., Gilissen C., de Wijs I.J., Admiraal R.J., et al. The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in the Netherlands. Eur. J. Hum. Genet. 2017;25:308–314. doi: 10.1038/ejhg.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian T., Lu Y., Yao J., Cao X., Wei Q., Li Q. Identification of a novel MYO6 mutation associated with autosomal dominant non-syndromic hearing loss in a Chinese family by whole-exome sequencing. Genes Genet. Syst. 2018;93:171–179. doi: 10.1266/ggs.18-00006. [DOI] [PubMed] [Google Scholar]
- 32.Cheng J., Zhou X., Lu Y., Chen J., Han B., Zhu Y., Liu L., Choy K.W., Han D., Sham P.C., et al. Exome sequencing identifies a novel frameshift mutation of MYO6 as the cause of autosomal dominant nonsyndromic hearing loss in a Chinese family. Ann. Hum. Genet. 2014;78:410–423. doi: 10.1111/ahg.12084. [DOI] [PubMed] [Google Scholar]
- 33.Arden S.D., Tumbarello D.A., Butt T., Kendrick-Jones J., Buss F. Loss of cargo binding in the human myosin VI deafness mutant (R1166X) leads to increased actin filament binding. Biochem. J. 2016;473:3307–3319. doi: 10.1042/BCJ20160571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brownstein Z., Abu-Rayyan A., Karfunkel-Doron D., Sirigu S., Davidov B., Shohat M., Frydman M., Houdusse A., Kanaan M., Avraham K.B. Novel myosin mutations for hereditary hearing loss revealed by targeted genomic capture and massively parallel sequencing. Eur. J. Hum. Genet. 2014;22:768–775. doi: 10.1038/ejhg.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanggaard K.M., Kjaer K.W., Eiberg H., Nürnberg G., Nürnberg P., Hoffman K., Jensen H., Sørum C., Rendtorff N.D., Tranebjaerg L. A novel nonsense mutation in MYO6 is associated with progressive nonsyndromic hearing loss in a Danish DFNA22 family. Am. J. Med. Genet. A. 2008;146A:1017–1025. doi: 10.1002/ajmg.a.32174. [DOI] [PubMed] [Google Scholar]
- 36.Hertzano R., Shalit E., Rzadzinska A.K., Dror A.A., Song L., Ron U., Tan J.T., Shitrit A.S., Fuchs H., Hasson T., et al. A Myo6 mutation destroys coordination between the myosin heads, revealing new functions of myosin VI in the stereocilia of mammalian inner ear hair cells. PLoS Genet. 2008;4:e1000207. doi: 10.1371/journal.pgen.1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salles F.T., Andrade L.R., Tanda S., Grati M., Plona K.L., Gagnon L.H., Johnson K.R., Kachar B., Berryman M.A. CLIC5 stabilizes membrane-actin filament linkages at the base of hair cell stereocilia in a molecular complex with radixin, taperin, and myosin VI. Cytoskeleton. 2015;71:61–78. doi: 10.1002/cm.21159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasson T., Mooseker M.S. Porcine myosin-VI: Characterization of a new mammalian unconventional myosin. J. Cell Biol. 1994;127:425–440. doi: 10.1083/jcb.127.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.