Abstract
RALF isoforms play many biological roles, and their specific functions are defined by combinatorial interactions with dynamic receptor complexes that vary more than initially thought.
A family of secreted peptides known as the RALFs (rapid alkalinization factors) induce rapid alkalinization of the extracellular compartment of plant cells. RALFs are perceived by a family of Catharanthus roseus Receptor Like Kinase 1 Like proteins (CrRLK1Ls), a structurally related family of plasma membrane-enriched receptors. RALF perception coincides with the downregulation of plasma membrane H+-ATPase function, which reduces energy for solute uptake and results in growth inhibition. In this review, we discuss the initial discovery of this ubiquitous family of peptide growth regulators and their cognate receptors. We describe recent detailed structural studies of RALF-CrRLK1Ls interactions and the identification of a subset of downstream signaling components. We then summarize the current state of RALFs-CrRLK1L research with emphasis on the Arabidopsis (Arabidopsis thaliana) RALFs (AtRALFs) and their roles in cellular regulation of plant growth and development and in biotic and abiotic stress responses.
RALF DISCOVERY, STRUCTURE, AND DIVERSITY
The origins of the RALF field lie in the work of Clarence “Bud” Ryan’s group at Washington State University, with the discovery of the first RALF peptide using an activity assay originally designed to detect systemins, which are small, ∼18 amino acid peptide hormones involved in the wound response in tobacco (Nicotiana tabacum) plants. This assay targeted the alkalinization caused by systemins and was used to test HPLC-fractionated tobacco leaf extracts. Surprisingly, an HPLC fraction eluting later than systemins elicited an unexpectedly fast and prominent increase in the pH of the culture medium. Edman degradation later confirmed that this activity was a larger peptide of approximately 5 kDa (Pearce et al., 2001). This new peptide failed to induce proteinase inhibitors when applied to cut stems of small tomato (Solanum lycopersicum) plants, suggesting it was not involved in the wound response. However, germinating seeds of tomato and Arabidopsis experienced immediate root growth arrest when transferred to media spiked with the peptide versus unspiked media, and this effect was mainly perceived in the elongation zone of the root.
A search of sequence databases using the original 17 N-terminal amino acids obtained from peptide sequencing revealed that RALFs are ubiquitous in the plant kingdom and identified conserved motifs. For example, a pair of Arg residues (RR), a feature typically found in animal prohormones processed by convertases, is located just upstream from the N terminus of the active peptide in almost all RALFs (Pearce et al., 2001). Furthermore, the dibasic site RR not only functions as the cleavage site for prohormone processing enzymes but is also central to the involvement of AtRALFs in coordinating the immune response (Matos et al., 2008; Srivastava et al., 2009; Stegmann et al., 2017).
It was established early on that the N-terminal YISY motif of the mature peptide is required for productive binding of the peptide to its target receptor; this was experimentally demonstrated in truncation and mutagenesis experiments on synthetic RALF (Pearce et al., 2010). Importantly, the flanking tyrosines and internal I/L of this motif show the strongest conservation, whereas the internal Ser shows moderate conservation, with occasional substitution by Gly or Thr. GASYY and the C-terminal RCRR(S) motifs also occur in many mature AtRALF sequences, but their role remains to be experimentally determined. However, truncation experiments that demonstrated the requirement of the C-terminal portion of the AtRALF1 peptide for activity suggest that these motifs may play a role in peptide structure or binding (Pearce et al., 2010).
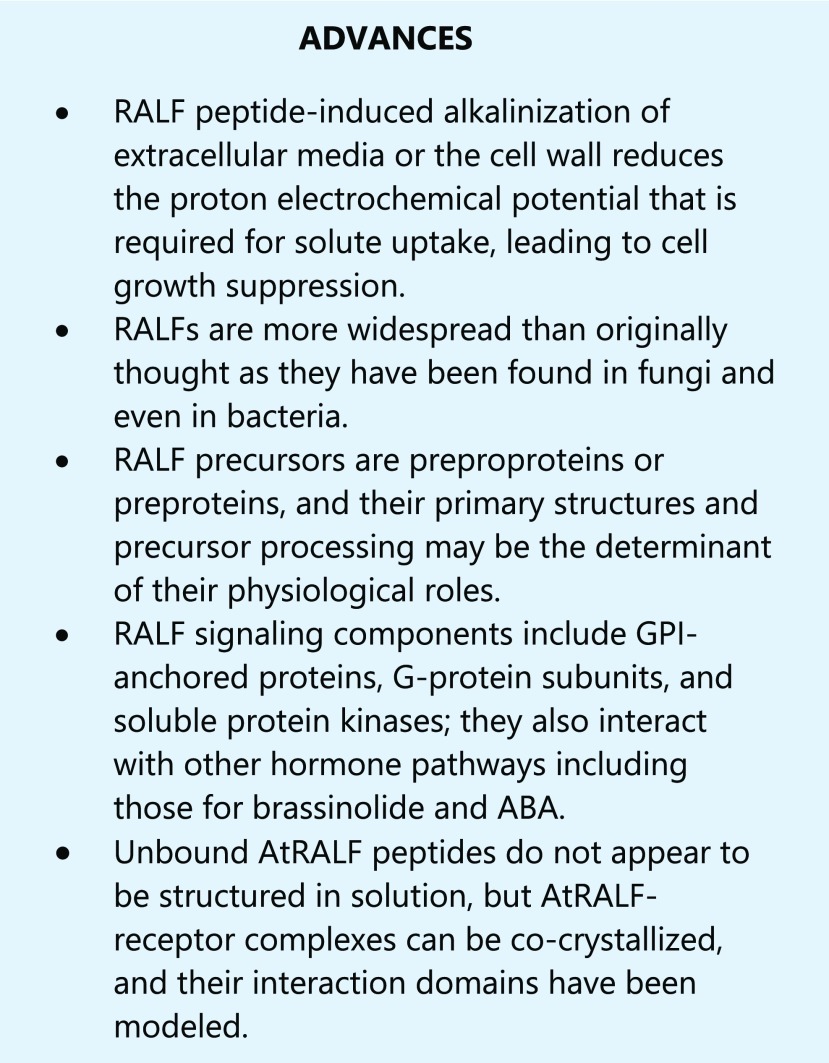
Notably, many AtRALF sequences contain four Cys residues that are positionally conserved and absolutely required for biological activity (Pearce et al., 2001; Haruta et al., 2008). For these peptides, oxidative folding of the cleaved peptides also occurs to generate the mature AtRALF peptide, and mutation or blocking (alkylation) of these cysteines is known to disrupt physiological activity. Despite the presence of two functionally important disulfide bridge sites, the oxidatively folded mature peptide appears to be largely unstructured due to the strikingly minimal length of the peptide loop between each pair of bridged cysteines (Pearce et al., 2001). This configuration was independently validated with the recently solved solution structure of AtRALF8, which indicates an entirely unstructured peptide except where movement of the peptide backbone is constrained in the immediate vicinity of each disulfide bridge (Frederick et al., 2019; Fig. 1A, inset). It is possible that all Cys-rich AtRALFs adopt this configuration because the relative amino acid positions of those Cys residues are conserved among AtRALF peptide sequences.
Figure 1.
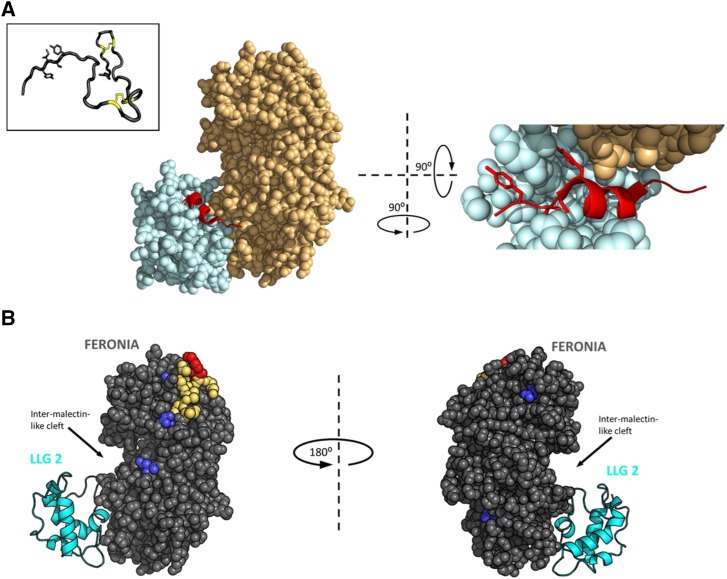
RALF peptides adopt an induced fold when complexed with a CrRLK1L protein. A, the left shows the crystal structure of the heterotypic complex among LLG2 (pale blue), RALF23 (red), and the ectodomain of FERONIA (pale orange). Density for the full-length RALF23 peptide was obtained for residues from RYISY to the first conserved Cys of the mature peptide. The right shows 900 pivot along the y axis by an ∼900 tilt out of the page and zoomed in, with ∼40 residues of the N-terminal domain of ectoFER removed to clear the view of RALF23. The mature peptide hormone intercalates tightly between the juxtamembrane domain of FERONIA (light orange) and the flexible loops of LLG2 (light blue), adopting an induced alpha helix. YISY sidechains are shown as sticks for emphasis. PDB ID: 6A5E (Xiao et al., 2019). The inset shows the solution NMR structure of RALF8 confirms a mostly unstructured mature peptide with a minimal loop-spanning disulfide configuration formed from the four invariant cysteines. Bridged Cys sidechains are represented as yellow sticks. YITY sidechains are shown as sticks for emphasis. PDB ID: 6NU4 (Frederick et al., 2019). B, Chemical cross linking and footprinting data from RALF1 complexed with the extracellular domain of FERONIA (gray spheres) in vitro, in the absence of an LLG protein, mapped to the crystal structure 6A5E. LLG2 (cyan cartoon) is included in the image for spatial reference but was not part of the experiment. Dark blue, residues that gain protection from the bulk solvent in the presence of full-length RALF1 (EDC-GEE labeling); red, Lys 60 (K60) on FERONIA, which cross linked to the C-terminal Lys of RALF1 (BS3 cross linking); yellow, the peptide containing K60, identified by mass spectrometry (Liu et al., 2018).
Given these conserved motifs, it has been possible to demonstrate that RALF-related sequences are much more universal than originally thought, as peptide sequences with RALF motifs have been isolated from phytopathogenic fungi and even from bacteria (Masachis et al., 2016; Thynne et al., 2017). On the other hand, the diversity of RALF sequences within and among species suggests that the RALF signaling paradigm is complex. In Arabidopsis alone, the AtRALF genes comprise a multigene family of more than 30 members. The exact number varies according to the criteria used to establish AtRALF identity, and current reported numbers range from 33 to 37 (Cao and Shi, 2012; Campbell and Turner, 2017).
Dozens of other AtRALF homologs are present in other species of terrestrial plants. Phylogenetic analysis of nearly 800 identified RALF peptides across the plant kingdom found that much of the sequence variability of the AtRALF peptides occurs in the sequence of mature RALF peptides, and this variability further divides the mature peptides into four distinct clades (Campbell and Turner, 2017).
Since their discovery, an enormous amount of research has extended our understanding of the RALF family, its interactions with other proteins, and its biological functions across the plant kingdom. Despite the ubiquity of the RALF peptides, Arabidopsis emerged as the natural model to study their functions and contains nine AtRALF sequences closely related to the originally isolated tobacco RALF. For this reason, the following sections will largely discuss the RALF family in the context of Arabidopsis biology.
THE DISCOVERY OF RALF RECEPTORS AND ANALYSIS OF RALF-RECEPTOR INTERACTIONS
The first study to investigate AtRALF-receptor interactions used a synthetic, radiolabeled tomato RALF (LeRALF) peptide modified with a photolabile azido handle to probe for LeRALF receptors. When cross linking was induced in Lycopersicon peruvianum cell suspension cultures spiked with this synthetic LeRALF, two membrane proteins at approximately 25 and 120 kDa were identified (Scheer et al., 2005). The RALF receptor was first identified in 2014 in the laboratory of Michael R. Sussman at the University of Wisconsin-Madison in a phosphoproteomic study that screened for AtRALF1-dependent changes using stable-isotope labeling combined with mass spectrometry of proteins that could be coimmunoprecipitated with AtRALF1 (Haruta et al., 2014).
This phosphoproteomic screen detected an increased abundance of phosphopeptides derived from a plasma membrane H+-ATPase 2 (AHA2) and calcium-dependent protein kinase 9 (CPK9). This coincided with increases in the phosphorylation of FERONIA (FER), a member of CrRLK1L family of receptor-like kinases (Haruta et al., 2014). AtRALF1 was also shown to elevate cytoplasmic calcium and suppress root growth in Arabidopsis seedlings in a FER-dependent manner, because fer mutants were insensitive to AtRALF1 in root growth suppression and cytoplasmic calcium mobilization, further supporting the hypothesis that the AtRALF1 peptide is sensed by the FER receptor kinase. These results suggested the FER signaling axis as a molecular mechanism for AtRALF-induced alkalinization of the extracellular media and elevation of cytoplasmic calcium concentrations.
FER is localized at the plasma membrane, where it functions as an AtRALF receptor in root cell growth regulation and is involved in many diverse pathways in plant physiology, including female gamete control of pollen tube rupture upon arrival of a female gametophyte to release sperm for fertilization (Huck et al., 2003; Rotman et al., 2003), rosette morphology, and guard cell movement (Yu and Assmann, 2018), immune responses (Stegmann et al., 2017), and abiotic stress responses (Chen et al., 2016; Xu et al., 2019). FER interacts with a RAC/ROP GTPase in the cellular signaling pathway regulating root hair development (Duan et al., 2010). FER also interacts with the GPI-anchored protein (GPI-AP) LORELEI (LRE) and LRE-LIKE GPI-AP1 (LLG1), proteins that act as chaperones that correctly target FER to the plasma membrane (Li et al., 2015). Besides their chaperone role, LLG proteins also bind AtRALF23 and function as a coreceptor for FER (Li et al., 2015; Xiao et al., 2019). In retrospect, after more than 15 years of ongoing research in the AtRALF field, it is possible that the original proteins identified by Scheer et al. (2005) are tomato homologs of Arabidopsis LLG proteins and CrRLK1Ls.
The presence of a protein complex containing AtRALF1, FER, and LLG1 was shown in vivo, and LLG1 was proposed to function as a coreceptor for FER in AtRALF1 perception (Li et al., 2015). Recent detailed structural studies report a submicromolar affinity (KD) for AtRALF binding in AtRALF-receptor complexes, including AtRALF1 or AtRALF23-FER interactions, indicating that this diverse family of peptides interacts with target receptors with high specificity (Liu et al., 2018; Xiao et al., 2019). An analysis of AtRALF23 cocrystallized with FER and LLG2 demonstrated that AtRALF23 adopts a helical fold when complexed with FER and LLG2 (Xiao et al., 2019; Fig. 1A). Unstructured, fully reduced AtRALF peptide (with four free thiols) was as active as correctly oxidized AtRALF peptide, suggesting the unusual possibility that the disulfide configuration itself can be induced during or right before receptor binding (Pearce et al., 2010).
In the study of Xiao et al. (2019), AtRALF23 was found to directly interact with the coreceptor LLG2 through tight interactions between its N terminus, which forms an alpha helix in crystallo, and disordered loops in LLG2 (Fig. 1A). Notably, this complex packs against the juxtamembrane domain of FER, and the YISY motif of AtRALF23 intercalates within the loops of LLG2 through a combination of polar and hydrophobic interactions. This suggests a general mechanism for how YISY, within the N-terminal α-helical domain, is minimally required for AtRALF peptides to bind their target receptors and coreceptors and supports the notion that AtRALF peptides become structured upon binding to their receptors (Xiao et al., 2019).
AtRALF peptide binds its receptor independently of a GPI-anchored coreceptor like LLG2. Recent in vitro chemical cross linking studies of AtRALF1 complexed with the extracellular domain of FER have suggested that the AtRALF1 peptide occupies a region spanning a large hydrophobic cleft on FER and that the C terminus of AtRALF1 is involved in binding to FER (Liu et al., 2018). Specifically, chemical cross linking with the amine-reactive cross linker bis-sulfosuccinimidyl suberate (BS3) identified an N-terminal Lys on AtRALF1 that cross links with a Lys on the ectodomain of FER near the hydrophobic cleft, and protein footprinting demonstrated that changes in solvent exposure occurred on residues in the vicinity of the cleft and this Lys on FER in response to AtRALF1 binding (Fig. 1B). Similar AtRALF1-FER interprotein cross linking was also observed in vitro when other amine reactive chemical cross linkers, disuccinimidyl sulfoxide, or dithiobis (succinimidylpropionate) were tested. Fluorescence polarization experiments examining the interaction of Alexa 488 labeled AtRALF1 and various truncated forms of FER ectodomain (ectoFER) showed increases in fluorescence polarization values with wild-type ectoFER-bound form of Alexa 488 AtRALF1, but not in those with truncated forms. Thus, the experiments demonstrated that both malectin-like domains of FER are required for AtRALF1 binding, suggesting that the ligand binding site of AtRALF1 spans the cleft and both malectin-like domains (Liu et al., 2018).
It is worth noting that other CrRLK1L ectodomain structures have also been published for ANXUR1 (ANX1) and ANX2, a pair of CrRLKL1 receptor kinases most closely related to FER (Du et al., 2018; Moussu et al., 2018) and that these models indicate that the cleft between the malectin-like domains of these CrRLK1Ls have a different residue composition than the intermalectin-like cleft of FER, suggesting that the AtRALF1-FER interaction is not necessarily mirrored by ANX1/2 and AtRALF4/19 interactions. Since three-dimensional structures of ANX1 and ANX2 were obtained as unbound forms, it is yet to be examined how closely AtRALF23-FER and AtRALF4/19-ANX1/2 complex structures are related.
These recent studies provide evidence about how an AtRALF peptide—specifically, the behavior of the N- and C-terminal portions of the peptide—binds its receptor and is an important first step to understanding the biochemical and structural basis of RALF perception. However, additional high-resolution structural and biochemical studies of these complex interactions remain to be conducted to resolve apparent inconsistencies between the findings of Liu et al. (2018) and Xiao et al. (2019).
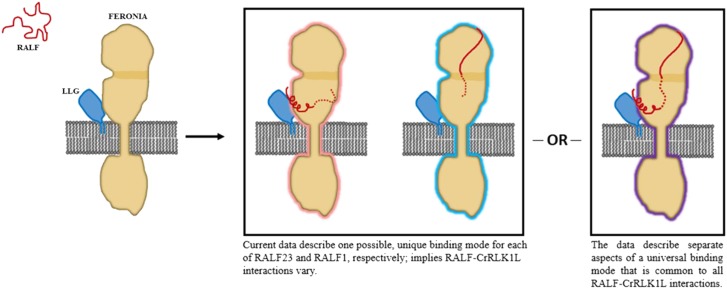
As with any in vitro experiment, the observed interactions in these structural studies do not necessarily reflect the physiological environment and could be prone to mixing or crystallization artifacts. Furthermore, the role of correct disulfide configurations was not explicitly addressed in either study, and it remains to be determined how the conserved Cys of each AtRALF contributes to ligand-receptor interactions. In addition, N-Hydroxysuccinimide-based cross linkers, such as BS3 or disuccinimidyl sulfoxide, rely on the availability of Lys residues located at or near the protein interaction interface, and thus AtRALF1-FER interaction sites modeled by the cross linking technique may not necessarily reflect the domains or amino acid residues that could be located at the closest contact of the two proteins. Ultimately, a unifying mechanism governing the specificity of RALF-receptor interactions remains to be determined and is a major open question in the RALF field (Fig. 2).
Figure 2.
Two proposed models considering available data on the induced fold of AtRALF and AtRALF-CrRLK1L protein binding interactions. To date, the solution NMR structure of AtRALF8 (Frederick et al., 2019) remains the only structure of an unbound AtRALF peptide and suggests that most Cys-rich peptides are similarly disordered. Note that LLG is shown here associated with FER before AtRALF binding for simplicity, but direct experimental evidence of AtRALF-independent LLG-FER interactions has not yet been demonstrated. Model 1 shows ehe crystal structure of the ectodomain of FERONIA complexed with LLG2 and AtRALF23 is currently the only structure of an AtRALF-receptor complex and suggests that the N terminus of AtRALF23 assumes an induced fold during binding and that its YISY motif is highly involved in AtRALF binding (Xiao et al., 2019). Meanwhile, the chemical cross linking and protein footprinting study of the AtRALF1/FER complex in the absence of an LLG protein suggests that the C terminus of AtRALF1 spans the intermalectin-like cleft. In the case of AtRALF23, binding occurs in an LLG-dependent manner near the juxtamembrane domain while AtRALF1 binding occurs independently of LLG at the malectin-like domains, suggesting two distinct, AtRALF-specific binding mechanisms. Model 2 shows, alternatively, these two binding modes reflect an interaction that is generalizable to many mature AtRALF peptides. Until more comprehensive data are available on AtRALF-receptor interactions, this remains a possibility.
How do individual AtRALF peptides coordinate so many complex and specific responses, especially when considering the CrRLK1L family of receptors that bind the structurally similar AtRALF peptides? This question remains the subject of intense research efforts, and there is particular interest in solving the structure of a mature AtRALF peptide in a complex with its receptor. To date, RALF ligand interactions with CrRLK1Ls ectodomains are demonstrated for AtRALF1 and FER, AtRALF23 and FER, BUDDHA’S PAPER SEAL1/2 (BUPS1/2) and AtRALF4/19, and ANX1/2 and AtRALF4/19 (Haruta et al., 2014; Ge et al., 2017; Xiao et al., 2019). AtRALF ligand perception is mediated by the CrRLK1L family and LLG coreceptor family, but their signaling output is likely a complex interplay of different AtRALFs playing roles as agonists and antagonists (Ge et al., 2017; Stegmann et al., 2017).
THE RALF SIGNALING AXIS EXTENDS TO OTHER DIVERSE PROTEINS AND FUNCTIONS
Both RALFs and CrRLK1Ls interact with numerous proteins that are implicated in RALF signaling. A pollen-specific RALF from tomato (SlPRALF) was first identified as a potential interactor of the LRR domain of a tomato pollen LEUCINE-RICH REPEAT EXTENSIN (LRX) protein in a Y2H screen (Covey et al., 2010). Pollen-expressed AtRALF4 and 19 interact with pollen-expressed proteins of the LRX family, which regulates pollen tube integrity and growth (Mecchia et al., 2017; Ge et al., 2019a). In roots, the amino terminus of LRX4 interacts with both AtRALF1 and the amino terminus of FER to regulate cell elongation by controlling vacuolar expansion (Dünser et al., 2019). Similarly, the LRR domains of LRX3 and LRX4 can interact with both AtRALF22/23 and FER to regulate salt stress response (Zhao et al., 2018). Notably, it was recently found that AtRALF1 also interacts in a Ca2+ and pH-dependent manner with calmodulin-like 38 (CML38), another secreted protein that is now understood to be an important component of the AtRALF1 root growth signaling pathway, further complicating the current understanding of AtRALF1 specificity (Campos et al., 2018).
Molecular genetic studies have shown that FER is involved in plant cell growth and reproduction, mechanosensing, pathogen defense, salt, and nutrient stress responses (Table 1; Li et al., 2016; Stegmann et al., 2017; Feng et al., 2018; Xu et al., 2019). Since fer knockout mutants display a wide range of phenotypes, it has been proposed that FER may be acting as a scaffold in an array of protein complexes. It is possible that CrRLK1L receptors act as heterodimers in such complexes. This idea is supported by the detection of BUPS1/2-ANX1/2 or BUPS1-BUPS2 complex formation in both an in vitro protein interaction assay and a yeast two-hybrid assay; specifically, these results suggested that BUPS and ANXs form heterodimers to regulate pollen tube elongation (Ge et al., 2017; Zhu et al., 2018). A more recent study further suggested that LLG2/LLG3 are also acting as coreceptors for RALF4 and RALF19 perception by BUPS1, BUPS2, ANX1, and ANX2 receptors in maintaining pollen tube integrity (Ge et al., 2019c).
Table 1. Physiological overlap of AtRALF signaling and other biological pathways.
| Physiological Conditions | Interaction with RALF Signaling | Cellular Mechanisms | References |
|---|---|---|---|
| Brassinolide (BL) (DWF4) | Antagonistic effect between BR and AtRALF1 (or AtRALF23) | EXPANSIN5 (AtEXPA5), DWARF gene expression BR downregulates RALF1 or 23 expression | (Srivastava et al., 2009; Bergonci et al., 2014a, 2014b) |
| Abscisic acid (ABA) | Cross talk between AtRALF1 and ABA signaling and stress responses | ABI2 protein phosphatase dependence | (Chen et al., 2016) |
| FER interaction with G protein β-subunit (AGB1) | (Yu and Assmann, 2018) | ||
| Pseudomonas syringae infection (elicitors flg22 or elf18) | Negative regulation of innate immunity by AtRALF23 | Reduced protein interaction of FLS2 or EFR with BAK1 | (Stegmann et al., 2017) |
| Carbon/nitrogen nutrient ratio | AtRALF1 and FER suppress stress response caused by high carbon/nitrogen | Phosphorylation of ATL6 E3 ubiquitin ligase and reduced stability of 14-3-3 protein | (Xu et al., 2019) |
REGULATION OF CELL PHYSIOLOGY, GROWTH, AND DEVELOPMENTAL PROCESSES BY RALF PEPTIDES
In the immediate aftermath of the discovery of the RALF peptides in tomato, tobacco, alfalfa (Medicago truncatula), and hybrid poplar (Populus trichocarpa × Populus deltoides), only two cellular activities—alkalinization and MAP kinase activation—were known in addition to the inhibition of root growth as physiological roles (Pearce et al., 2001; Haruta and Constabel, 2003). All of the original RALF peptides, whether biochemically isolated, recombinantly produced, or chemically synthesized, exhibit alkalization activity in cell suspension cultures (Pearce et al., 2001, 2010; Haruta and Constabel, 2003; Haruta et al., 2008; Mingossi et al., 2010; Morato do Canto et al., 2014). Additionally, AtRALFs were shown to transiently increase the concentration of the intracellular second messenger Ca2+ in Arabidopsis seedlings expressing the intracellular calcium reporter, aequorin; this suggested that AtRALFs exhibit growth factor-like coupling (Haruta et al., 2008; Morato do Canto et al., 2014).
Strikingly, alkalinization activity does not appear to be universally essential for the AtRALFs’ physiological roles, as some of these peptides elicit activities that are independent or partially independent of alkalinization (Dressano et al., 2017; Stegmann et al., 2017; Campos et al., 2018). Intriguingly, when NaRALF of Nicotiana attenuata was silenced by an inverted repeat RNA construct, the irRALF plants unexpectedly retained the ability to acidify the external media (Wu et al., 2007). In Arabidopsis cell cultures, AtRALFs tested so far all show robust extracellular pH alkalinization activities, except the pollen-expressed AtRALF4 (Morato do Canto et al., 2014). In contrast, AtRALF19 is also expressed in pollen but is a strong alkalinization factor, showing a 4.6 nm half-maximal activity in the assay (Morato do Canto et al., 2014). Those observations may suggest that seven amino acid substitutions between AtRALF4 and AtRALF19 contribute to their differential alkalinization activity as well as that endogenous AtRALF19 or other AtRALFs’ expression levels influence AtRALF4 effect on cells. AtRALF alkalinization of the extracellular media or apoplast may be a prerequisite for other AtRALFs to associate with their receptors, as has been suggested by the pH dependence of AtRALF34 for binding to the ectodomain of the THESEUS1 (THE1) receptor (Gonneau et al., 2018). The pH dependence of AtRALF binding could also be related to the affinity of AtRALFs binding to the receptors, but this possibility has yet to be investigated.
The initial discovery of RALFs identified the inhibition of root elongation as an important function (Pearce et al., 2001). A clearer role for AtRALFs in the inhibition of cell expansion emerged from the work on AtRALF1, which remains one of the most studied RALF peptides. Overexpression of AtRALF1 causes bushy, semidwarf plants with small leaves, short roots, and a reduction in the number of both lateral roots and small cells in roots (Matos et al., 2008; Bergonci et al., 2014a). On the other hand, knockdown or knockout mutants of the AtRALF1 gene showed the opposite phenotype: plants with long roots and long hypocotyls, increased lateral root number, and large root cells (Bergonci et al., 2014a). Biochemically isolated and Escherichia coli-produced AtRALF1 was active in the typical cytoplasmic Ca2+assay and inhibited cell expansion and root growth (Haruta et al., 2008; Mingossi et al., 2010; Morato do Canto et al., 2014).
Other RALF peptides are also involved in regulating growth (overexpression of two other AtRALF sequences, AtRALF8 and AtRALF23, also resulted in semidwarf plants; Srivastava et al., 2009; Atkinson et al., 2013). The AtRALF23 gene was found to be downregulated by brassinolide (BL) treatment, and its overexpression also resulted in semidwarf plants with reduced root growth and a compromised BL response (Nemhauser et al., 2004; Srivastava et al., 2009). Antagonism between AtRALFs and BL was also demonstrated when AtRALF-induced genes involved in cell wall rearrangement were identified (Table 1; Bergonci et al., 2014a, 2014b). Brassinosteroids (BRs) are important regulators of growth and development and play a critical role in cell expansion. BRs are mainly perceived by the cell membrane receptor kinase BRASSINOSTEROID INSENSITIVE1 (BRI1) that, upon binding of BR, associates with its coreceptor BRI1-ASSOCIATED KINASE1 (BAK1) to up-regulate BR cellular responses (Belkhadir and Jaillais, 2015). AtRALF1 also binds BAK1, and BAK1 is critical for the inhibition of root growth and cell expansion. The antagonistic relationship between AtRALF and BRs could well be regulated by switching both signaling pathways on and off through BAK1 (Dressano et al., 2017).
The specific developmental stages or environmental conditions in which AtRALF1-FER signaling contributes to root growth regulation remains unclear. One plausible hypothesis states that the AtRALF1-FER pair controls growth suppression of roots under dim light, because fer seedlings display a faster growth phenotype than wild type in these conditions. FER and other members of CrRLK1Ls may also be involved in cell growth regulation during photomorphogenesis, because those genes were shown to be transcriptionally induced by BL, a growth regulator that overlaps with light signaling (Li et al., 1996; Guo et al., 2009a, 2009b; Clouse, 2011; Zhu et al., 2013). Additionally, fer-4 root elongation was not completely suppressed when tested at higher AtRALF1 concentrations (Haruta et al., 2014), suggesting that another member of the CrRLK1L family is also involved in sensing AtRALF1 during root growth and development.
FER was originally discovered as the female determinant of sperm cell release from the pollen tube as it penetrates the synergid cell of the ovule (Escobar-Restrepo et al., 2007). The discovery of FER as the AtRALF1 receptor and the abundance of RALF peptides in the reproductive tissues led to a concerted effort in the research community to unravel the roles of these peptides in plant reproduction. The interest in reproduction-associated RALFs began when a family of AtRALF-like sequences was identified in the expression sequence tag library derived from the ovule of Solanum chacoense (Germain et al., 2005), which led to the critical finding that silencing ScRALF3 expression by RNA interference caused developmental arrest in pollen mitosis and abnormal nuclear distribution in the embryo sac (Chevalier et al., 2013; Loubert-Hudon et al., 2020). ScRALF3 mRNA expression and promoter activity assay results suggested that ScRALF3 functions as a secreted signal regulating cell-to-cell communication between the embryo sac and the sporophytic tissue surrounding the ovule, demonstrating the first example of RALF physiology occurring outside of the roots. Additional research efforts in Arabidopsis led to the discovery of another pair of RALF-CrRLK1L interactors: pollen-expressed AtRALF4 and AtRALF19 are peptide ligands of the pollen-expressed CrRLK1L BUPS1/2 and ANX1/2 receptors. These receptor-bound complexes are responsible for maintaining cell integrity during pollen tube germination and growth (Ge et al., 2017). As pollen tubes approach ovules, ovule-expressed AtRALF34 peptide competes with pollen-expressed AtRALF4 and AtRALF19 for binding to ANX1/2 or BUPS1/2, allowing sperm cell discharge (Ge et al., 2017).
Recently, two CrRLK1L receptor-like kinases, HERCULES1 (HERK1) and ANJEA (ANJ), were identified as female determinants of pollen tube reception (Galindo-Trigo et al., 2020). Both HERK1 and ANJ interact with FER and, although not yet proved, an analogous mechanism was proposed where pollen-derived RALFs would act upon receptor heterocomplexes made of FER, HERK1, and ANJ assembled on female tissues to control pollen tube reception (Galindo-Trigo et al., 2020). Fertilization is the result of an interplay between signal molecules, their receptors, second messengers, and cell wall components. Establishing RALFs, CrRLK1Ls, and LLG coreceptor pairs involved in each part of this process and their relation to each of the intracellular events is presently a matter of intense research interest (Ge et al., 2019a). It is also essential to understand whether a RALF-CrRLK1L-LLG ternary complex multimerizes into higher-order complexes that may be composed of different sets of RALFs, CrRLK1Ls, and LLGs (Ge et al., 2019b).
REGULATION OF BIOTIC AND ABIOTIC STRESS RESPONSES BY RALF PEPTIDES
Recently, several physiological studies have implicated AtRALFs-FER signaling in biological pathways including innate immune responses, stress responses mediated by abscisic acid (ABA), and a nutrient stress condition (Table 1).
It has been suggested that FER may act as a scaffold protein for the assembly of an immune receptor complex (Chakravorty et al., 2018; Duan and Cheung, 2018; Haruta et al., 2018a). An AtRALF23-FER interaction potentially coordinates the assembly of an immune complex involving EF-TU RECEPTOR (EFR), FLAGELLIN SENSING 2 (FLS2), and BAK1 (Stegmann et al., 2017). Interestingly, a search for mutants that could recover the immune function lost in bak1-5 mutants resulted in the isolation of the subtilase SITE-1 PROTEASE (S1P)/SBT6.1, the prohormone convertase that processes AtRALF23 (Srivastava et al., 2009; Stegmann et al., 2017). This finding is striking because it shows that proteolytic processing of AtRALF23 from its precursor inhibits plant immune responses that are activated by cellular perception of flg22 or elf18 elicitor peptides by FLS2 or EFR. RALF was known to affect the level of reactive oxygen species (ROS) production in N. attenuata, and decreased levels of ROS accumulation were found in irRALF plants, although that observation was related to root hair development (Wu et al., 2007). AtRALF23 and AtRALF33 are suppressors of ROS production when elicited by elf18, a peptide derived from the bacterial elongation factor Tu (Table 1); however AtRALF17, a peptide whose precursor lacks a canonical convertase processing site, induces ROS production instead of suppressing it, further suggesting the importance of S1P in RALF-mediated immune responses (Stegmann et al., 2017). AtRALF32 neither induces nor suppresses ROS production elicited by elf18 and also lacks a conserved processing site yet has a propeptide region (Stegmann et al., 2017). This further suggests that different AtRALFs have different biological effects, which might be contingent on the biological context (such as tissue type) and how the mature peptide is processed.
MYC2 transcription factor, which was identified from a transcriptome analysis of the fer mutant and promoter motif analysis, was also shown to be a FER kinase substrate and act as a negative regulator of immunity to Pseudomonas syringae pv. tomato DC3000 (pst DC3000; Guo et al., 2018). Arabidopsis plants treated with AtRALF23 suppress the induction of immune responses, while plants overexpressing the AtRALF23 gene are more susceptible to pathogenic infection. Thus, AtRALF23 ligand itself is a negative regulator of immune response. Meanwhile, Arabidopsis fer mutants showed hypersensitivity to this pathogen, indicating that the normal function of FER is to up-regulate the immune response by acting as a scaffold for proteins involved in pathogen-associated molecular pattern-triggered immunity and by suppressing jasmonate-responsive signaling.
As a Ser/Thr receptor kinase, FER has been proposed to activate itself by autophosphorylation (Escobar-Restrepo et al., 2007), and it exhibits kinase activity in vitro on both itself and on synthetic substrates (Minkoff et al., 2017; Haruta et al., 2018a). To date, there is evidence of additional substrates of FER, including ARABIDOPSIS TOXICOS EN LEVADURA6 (ATL6) and ERBB3-BINDING PROTEIN1 (EBP1; Li et al., 2018; Xu et al., 2019).
AtRALF-CrRLK1L signaling also plays a role in stomatal opening and stress responses. AtRALF8 was originally identified as one of the genes that were transcriptionally upregulated in Arabidopsis plants exposed to a combination of water deficit and nematode stresses (Atkinson et al., 2013). FER was identified in a screen for protein interactors of AGB1, a G protein β-subunit that is involved in ABA-mediated regulation of stomatal movements. AtRALF1 was then shown to regulate stomatal aperture, which is important in both immune and drought responses, through FER (Table 1; Yu and Assmann, 2018). This work was the first to demonstrate that AtRALF1 inhibits stomatal opening and induces its closure through FER, which functions like a G protein-coupled receptor. Because FER and AGB1 are both known to be involved in salt stress responses mediated by ABA, the mutant phenotypes of fer and agb1 were compared in two different cell types, guard cells and roots, to examine epistatic relationship of the two genes (Yu and Assmann, 2018). Both fer and agb1 mutants perform poorly in high-salt conditions and the double mutant demonstrated an even more severe phenotype of hypersensitivity to the salt stress (Yu and Assmann, 2018). When salt was combined with AtRALF1 treatment, these plants also demonstrated a hypersensitivity phenotype. This phenotype was observed again when AtRALF22 was overexpressed (Zhao et al., 2018). Interestingly, in a different study, atralf1 mutant lines did not show any phenotypic difference from control plants when exposed to high-salt conditions, and AtRALF1 overexpressors showed increased tolerance to salt (Feng et al., 2018). However, contrasting results for AtRALF-treated plants or for plants in which endogenous levels of expression are genetically manipulated have been reported (Wieghaus et al., 2019).
CONCLUSION
The initial biochemical screening for extracellular alkalinization activity led to the discovery of the first RALF peptide nearly 20 years ago. As the proton gradient at the cell surface is key to cell growth, this peptide family has long been thought to regulate cell expansion. Consistent with this prediction, both biochemical and genetic experiments showed that a plasma membrane H+-ATPase acts downstream of AtRALF1 and FER action in cellular signaling suppressing root growth (Haruta et al., 2018b). Moreover, an increasing body of literature supports cell growth regulation by RALF ligands and their cognate receptor complexes in other tissues. As demonstrated by molecular, genetic, and biochemical studies of the RALF peptide and CrRLK1L receptor families, many physiological responses interact with cell elongation signaling in the plant. The wealth of knowledge that has been produced on RALF biology over the last two decades has also opened many new questions about this complex area of plant physiology (see Outstanding Questions). To fully understand how this complex signaling mechanism is regulated, experimental approaches with high spatial and/or temporal resolution will be needed. We believe that many additional fruitful RALF-CrRLK1L studies will follow to understand plant cell growth regulation mechanisms initiated at the interface of plasma membrane and cell wall.
Acknowledgments
The authors thank the reviewers for critical reading of the manuscript and suggesting substantial improvements. The authors apologize for not citing other important studies in this review due to page limitations.
Footnotes
This work was supported by the National Science Foundation (MCB MSN203469 to M.H. and PGRP MSN187204 to M.R.B) and the São Paulo Research Foundation (2014/14487-1 to D.S.M).
Articles can be viewed without a subscription.
References
- Atkinson NJ, Lilley CJ, Urwin PE(2013) Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol 162: 2028–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y(2015) The molecular circuitry of brassinosteroid signaling. New Phytol 206: 522–540 [DOI] [PubMed] [Google Scholar]
- Bergonci T, Ribeiro B, Ceciliato PHO, Guerrero-Abad JC, Silva-Filho MC, Moura DS(2014a) Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. J Exp Bot 65: 2219–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonci T, Silva-Filho MC, Moura DS(2014b) Antagonistic relationship between AtRALF1 and brassinosteroid regulates cell expansion-related genes. Plant Signal Behav 9: e976146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L, Turner SR(2017) A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front Plant Sci 8: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos WF, Dressano K, Ceciliato PHO, Guerrero-Abad JC, Silva AL, Fiori CS, Morato do Canto A, Bergonci T, Claus LAN, Silva-Filho MC, et al. (2018) Arabidopsis thaliana rapid alkalinization factor 1-mediated root growth inhibition is dependent on calmodulin-like protein 38. J Biol Chem 293: 2159–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Shi F(2012) Evolution of the AtRALF gene family in plants: Gene duplication and selection patterns. Evol Bioinform Online 8: 271–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty D, Yu Y, Assmann SM(2018) A kinase-dead version of FERONIA receptor-like kinase has dose-dependent impacts on rosette morphology and RALF1-mediated stomatal movements. FEBS Lett 592: 3429–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yu F, Liu Y, Du C, Li X, Zhu S, Wang X, Lan W, Rodriguez PL, Liu X, et al. (2016) FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc Natl Acad Sci USA 113: E5519–E5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier E, Loubert-Hudon A, Matton DP(2013) ScRALF3, a secreted RALF-like peptide involved in cell-cell communication between the sporophyte and the female gametophyte in a solanaceous species. Plant J 73: 1019–1033 [DOI] [PubMed] [Google Scholar]
- Clouse SD.(2011) Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey PA, Subbaiah CC, Parsons RL, Pearce G, Lay FT, Anderson MA, Ryan CA, Bedinger PA(2010) A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol 153: 703–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressano K, Ceciliato PHO, Silva AL, Guerrero-Abad JC, Bergonci T, Ortiz-Morea FA, Bürger M, Silva-Filho MC, Moura DS(2017) BAK1 is involved in AtRALF1-induced inhibition of root cell expansion. PLoS Genet 13: e1007053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S, Qu LJ, Xiao J(2018) Crystal structures of the extracellular domains of the CrRLK1L receptor-like kinases ANXUR1 and ANXUR2. Protein Sci 27: 886–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Cheung AY(2018) Context-specific dependence on FERONIA kinase activity. FEBS Lett 592: 2392–2394 [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM(2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dünser K, Gupta S, Herger A, Feraru MI, Ringli C, Kleine-Vehn J(2019) Extracellular matrix sensing by FERONIA and Leucine-Rich Repeat Extensins controls vacuolar expansion during cellular elongation in Arabidopsis thaliana. EMBO J 38: e100353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U(2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660 [DOI] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu M-C, Maman J, Steinhorst L, Schmitz-Thom I, et al. (2018) The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol 28: 666–675.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick RO, Haruta M, Tonelli M, Lee W, Cornilescu G, Cornilescu CC, Sussman MR, Markley JL(2019) Function and solution structure of the Arabidopsis thaliana RALF8 peptide. Protein Sci 28: 1115–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo-Trigo S, Blanco-Touriñán N, DeFalco TA, Wells ES, Gray JE, Zipfel C, Smith LM(2020) CrRLK1L receptor-like kinases HERK1 and ANJEA are female determinants of pollen tube reception. EMBO Rep 21: e48466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Bergonci T, Zhao Y, Zou Y, Du S, Liu MC, Luo X, Ruan H, García-Valencia LE, Zhong S, et al. (2017) Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358: 1596–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Cheung AY, Qu LJ(2019a) Pollen tube integrity regulation in flowering plants: insights from molecular assemblies on the pollen tube surface. New Phytol 222: 687–693 [DOI] [PubMed] [Google Scholar]
- Ge Z, Dresselhaus T, Qu LJ(2019b) How CrRLK1L receptor complexes perceive RALF signals. Trends Plant Sci 24: 978–981 [DOI] [PubMed] [Google Scholar]
- Ge Z, Zhao Y, Liu MC, Zhou LZ, Wang L, Zhong S, Hou S, Jiang J, Liu T, Huang Q, et al. (2019c) LLG2/3 are coreceptors in BUPS/ANX-RALF signaling to regulate Arabidopsis pollen tube integrity. Curr Biol 29: 3256–3265.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain H, Chevalier E, Caron S, Matton DP(2005) Characterization of five RALF-like genes from Solanum chacoense provides support for a developmental role in plants. Planta 220: 447–454 [DOI] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Martin M, Doblas VG, Bacete L, Miart F, Sormani R, Hématy K, Renou J, Landrein B, et al. (2018) Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis. Curr Biol 28: 2452–2458.e4 [DOI] [PubMed] [Google Scholar]
- Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y(2009a) Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci USA 106: 7648–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Nolan TM, Song G, Liu S, Xie Z, Chen J, Schnable PS, Walley JW, Yin Y(2018) FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Curr Biol 28: 3316–3324.e6 [DOI] [PubMed] [Google Scholar]
- Guo H, Ye H, Li L, Yin Y(2009b) A family of receptor-like kinases are regulated by BES1 and involved in plant growth in Arabidopsis thaliana. Plant Signal Behav 4: 784–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Constabel CP(2003) Rapid alkalinization factors in poplar cell cultures. Peptide isolation, cDNA cloning, and differential expression in leaves and methyl jasmonate-treated cells. Plant Physiol 131: 814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Gaddameedi V, Burch H, Fernandez D, Sussman MR(2018a) Comparison of the effects of a kinase-dead mutation of FERONIA on ovule fertilization and root growth of Arabidopsis. FEBS Lett 592: 2395–2402 [DOI] [PubMed] [Google Scholar]
- Haruta M, Monshausen G, Gilroy S, Sussman MR(2008) A cytoplasmic Ca2+ functional assay for identifying and purifying endogenous cell signaling peptides in Arabidopsis seedlings: Identification of AtRALF1 peptide. Biochemistry 47: 6311–6321 [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR(2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Tan LX, Bushey DB, Swanson SJ, Sussman MR(2018b) Environmental and genetic factors regulating localization of the plant plasma membrane H+-ATPase. Plant Physiol 176: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck N, Moore JM, Federer M, Grossniklaus U(2003) The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159 [DOI] [PubMed] [Google Scholar]
- Li C, Liu X, Qiang X, Li X, Li X, Zhu S, Wang L, Wang Y, Liao H, Luan S, et al. (2018) EBP1 nuclear accumulation negatively feeds back on FERONIA-mediated RALF1 signaling. PLoS Biol 16: e2006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang L, Cui Y, He L, Qi Y, Zhang J, Lin J, Liao H, Lin Q, Yang T, et al. (2016) Two FERONIA-like receptor (FLR) genes are required to maintain architecture, fertility, and seed yield in rice. Mol Breed 36: 151 [Google Scholar]
- Li C, Yeh FL, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L, et al. (2015) Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4: e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J(1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Liu P, Haruta M, Minkoff BB, Sussman MR(2018) Probing a plant plasma membrane receptor kinase’s three-dimensional structure using mass spectrometry-based protein footprinting. Biochemistry 57: 5159–5168 [DOI] [PubMed] [Google Scholar]
- Loubert-Hudon A, Mazin BD, Chevalier É, Matton DP(2020) The ScRALF3 secreted peptide is involved in sporophyte to gametophyte signalling and affects pollen mitosis I. Plant Biol (Stuttg) 22: 13–20 [DOI] [PubMed] [Google Scholar]
- Masachis S, Segorbe D, Turrà D, Leon-Ruiz M, Fürst U, El Ghalid M, Leonard G, López-Berges MS, Richards TA, Felix G, et al. (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat Microbiol 1: 16043. [DOI] [PubMed] [Google Scholar]
- Matos JL, Fiori CS, Silva-Filho MC, Moura DS(2008) A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Lett 582: 3343–3347 [DOI] [PubMed] [Google Scholar]
- Mecchia MA, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V, Martínez-Bernardini A, Fabrice TN, Ringli C, Muschietti JP, et al. (2017) RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358: 1600–1603 [DOI] [PubMed] [Google Scholar]
- Mingossi FB, Matos JL, Rizzato AP, Medeiros AH, Falco MC, Silva-Filho MC, Moura DS(2010) SacRALF1, a peptide signal from the grass sugarcane (Saccharum spp.), is potentially involved in the regulation of tissue expansion. Plant Mol Biol 73: 271–281 [DOI] [PubMed] [Google Scholar]
- Minkoff BB, Makino SI, Haruta M, Beebe ET, Wrobel RL, Fox BG, Sussman MR(2017) A cell-free method for expressing and reconstituting membrane proteins enables functional characterization of the plant receptor-like protein kinase FERONIA. J Biol Chem 292: 5932–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morato do Canto A, Ceciliato PHO, Ribeiro B, Ortiz Morea FA, Franco Garcia AA, Silva-Filho MC, Moura DS(2014) Biological activity of nine recombinant AtRALF peptides: implications for their perception and function in Arabidopsis. Plant Physiol Biochem 75: 45–54 [DOI] [PubMed] [Google Scholar]
- Moussu S, Augustin S, Roman AO, Broyart C, Santiago J(2018) Crystal structures of two tandem malectin-like receptor kinases involved in plant reproduction. Acta Crystallogr D Struct Biol 74: 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J(2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA Jr.(2001) RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci USA 98: 12843–12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Yamaguchi Y, Munske G, Ryan CA(2010) Structure-activity studies of RALF, Rapid Alkalinization Factor, reveal an essential—YISY—motif. Peptides 31: 1973–1977 [DOI] [PubMed] [Google Scholar]
- Rotman N, Rozier F, Boavida L, Dumas C, Berger F, Faure JE(2003) Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr Biol 13: 432–436 [DOI] [PubMed] [Google Scholar]
- Scheer JM, Pearce G, Ryan CA(2005) LeRALF, a plant peptide that regulates root growth and development, specifically binds to 25 and 120 kDa cell surface membrane proteins of Lycopersicon peruvianum. Planta 221: 667–674 [DOI] [PubMed] [Google Scholar]
- Srivastava R, Liu JX, Guo H, Yin Y, Howell SH(2009) Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J 59: 930–939 [DOI] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C(2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- Thynne E, Saur IML, Simbaqueba J, Ogilvie HA, Gonzalez-Cendales Y, Mead O, Taranto A, Catanzariti AM, McDonald MC, Schwessinger B, et al. (2017) Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Mol Plant Pathol 18: 811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieghaus A, Prüfer D, Schulze Gronover C(2019) Loss of function mutation of the Rapid Alkalinization Factor (RALF1)-like peptide in the dandelion Taraxacum koksaghyz entails a high-biomass taproot phenotype. PLoS One 14: e0217454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Kurten EL, Monshausen G, Hummel GM, Gilroy S, Baldwin IT(2007) NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. Plant J 52: 877–890 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Stegmann M, Han Z, DeFalco TA, Parys K, Xu L, Belkhadir Y, Zipfel C, Chai J(2019) Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572: 270–274 [DOI] [PubMed] [Google Scholar]
- Xu G, Chen W, Song L, Chen Q, Zhang H, Liao H, Zhao G, Lin F, Zhou H, Yu F(2019) FERONIA phosphorylates E3 ubiquitin ligase ATL6 to modulate the stability of 14-3-3 proteins in response to the carbon/nitrogen ratio. J Exp Bot 70: 6375–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Assmann SM(2018) Inter-relationships between the heterotrimeric Gβ subunit AGB1, the receptor-like kinase FERONIA, and RALF1 in salinity response. Plant Cell Environ 41: 2475–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zayed O, Yu Z, Jiang W, Zhu P, Hsu CC, Zhang L, Tao WA, Lozano-Durán R, Zhu JK(2018) Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc Natl Acad Sci USA 115: 13123–13128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Sae-Seaw J, Wang ZY(2013) Brassinosteroid signalling. Development 140: 1615–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Chu LC, Liang Y, Zhang XQ, Chen LQ, Ye D(2018) The Arabidopsis CrRLK1L protein kinases BUPS1 and BUPS2 are required for normal growth of pollen tubes in the pistil. Plant J 95: 474–486 [DOI] [PubMed] [Google Scholar]