A unique cytosol-localized geranyl diphosphate synthase supporting a large production of shikonin has evolved from farnesyl diphosphate synthase in Lithospermum erythrorhizon.
Abstract
Geranyl diphosphate (GPP) is the direct precursor of all monoterpenoids and is the prenyl source of many meroterpenoids, such as geranylated coumarins. GPP synthase (GPPS) localized in plastids is responsible for providing the substrate for monoterpene synthases and prenyltransferases for synthesis of aromatic substances that are also present in plastids, but GPPS activity in Lithospermum erythrorhizon localizes to the cytosol, in which GPP is utilized for the biosynthesis of naphthoquinone pigments, which are shikonin derivatives. This study describes the identification of the cytosol-localized GPPS gene, LeGPPS, through EST- and homology-based approaches followed by functional analyses. The deduced amino acid sequence of the unique LeGPPS showed greater similarity to that of farnesyl diphosphate synthase (FPPS), which generally localizes to the cytosol, than to plastid-localized conventional GPPS. Biochemical characterization revealed that recombinant LeGPPS predominantly produces GPP along with a trace amount of FPP. LeGPPS expression was mainly detected in root bark, in which shikonin derivatives are produced, and in shikonin-producing cultured cells. The GFP fusion protein in onion (Allium cepa) cells localized to the cytosol. Site-directed mutagenesis of LeGPPS and another FPPS homolog identified in this study, LeFPPS1, showed that the His residue at position 100 of LeGPPS, adjacent to the first Asp-rich motif, contributes to substrate preference and product specificity, leading to GPP formation. These results suggest that LeGPPS, which is involved in shikonin biosynthesis, is recruited from cytosolic FPPS and that point mutation(s) result in the acquisition of GPPS activity.
Terpenoids represent the largest class of plant specialized (secondary) metabolites, with over 40,000 compounds described to date (Zwenger and Basu, 2008; Tholl, 2015). This class of metabolites has a wide range of physiological functions, acting, for example, as pest repellents (Kessler and Heil, 2011) and pollinator attractants (Byers et al., 2014), as well as in intra-/interplant signaling (Arimura et al., 2011).
Terpenoids are grouped according to the number of carbon atoms, as hemi- (C5), mono- (C10), sesqui- (C15), di- (C20), sester- (C25), tri- (C30), tetra- (C40), and poly- (>C40) terpenoids. In addition, composite type natural products, called meroterpenoids, consist of terpenoid residues on phenolic skeletons. All terpenoid moieties are built from the condensation of the universal C5 units, isopentenyl diphosphate (IPP) and its allylic isomer dimethylallyl diphosphate (DMAPP), with the sequential condensation of these units yielding a series of prenyl diphosphates of different chain lengths. Plant cells possess two independent terpenoid pathways, the mevalonate (MVA) pathway in the cytosol and the methylerythritol phosphate (MEP) pathway in plastids (Liao et al., 2016).
Monoterpenoids provide the major class of plant-derived volatile compounds, along with several sesquiterpenoids and a limited number of phenolic substances, such as phenylpropenes (Abbas et al., 2017). Geranyl diphosphate (GPP) is the direct and common precursor of all monoterpenoids. In general, GPP synthase (GPPS), the enzyme responsible for the synthesis of GPP, is localized in plastids. This enzyme utilizes equimolar amounts of IPP and DMAPP to form GPP, which is utilized by monoterpenoid synthases, such as limonene synthase, which are also localized to plastids (Yuba et al., 1996). GPPS in mint (Mentha piperita) cells is heterotetrameric (Chang et al., 2010), consisting of a small subunit (SSU) lacking the first/second Asp-rich motif (FARM/SARM) widely conserved in trans-prenyl diphosphate synthases (Burke and Croteau, 2002a), and a large subunit (LSU) showing similarity to geranylgeranyl diphosphate synthase (GGPPS). Similar types of GPPSs functioning as a heterodimer have also been reported in snapdragon (Antirrhinum majus) and Clarkia breweri (Tholl et al., 2004), whereas homodimeric GPPSs have also been identified from Abies grandis (Burke and Croteau, 2002b), Picea abies, and Quercus rober (Schmidt and Gershenzon, 2008). All of these GPPSs localize to plastids.
By contrast, a unique GPPS localized to the cytosol was reported in a medicinal plant Lithospermum erythrorhizon (Sommer et al., 1995). This plant produces shikonin derivatives, consisting of red naphthoquinone meroterpenoids as the ester forms of various fatty acids, in the roots (Supplemental Fig. S1). In the 1980s, cultured L. erythrorhizon cells were first used in the industrial production of shikonin, making this the first use of dedifferentiated plant cells for the production of secondary metabolites. Shikonin derivatives accumulated in the root epidermis of intact L. erythrorhizon plants and had antibacterial activity against soil-borne microorganisms, suggesting that these pigments play defensive roles in L. erythrorhizon (Brigham et al., 1999). Because L. erythrorhizon roots have antibacterial (Tanaka and Odani, 1972), anti-inflammatory (Lu et al., 2011), and antitumor (Sankawa et al., 1977) properties, these roots have been used as a crude drug in traditional Asian medicine.
Shikonin is produced using a two-stage culture system of L. erythrorhizon cells. These cells are cultured in cell growth medium, followed by culture in shikonin production medium (M9), resulting in the production of ∼10% shikonin per dry weight of L. erythrorhizon cells (Fujita et al., 1981b). This two-stage cell culture system has led to the identification of many physical and chemical regulators of shikonin production (Yazaki, 2017). For example, indole acetic acid and Cu2+ enhance shikonin production (Tabata et al., 1974; Fujita et al., 1981a), whereas light and NH4+ strongly suppresses shikonin biosynthesis (Tabata et al., 1974; Mizukami et al., 1977). One key regulatory step in shikonin biosynthesis is the coupling of a geranyl moiety with p-hydroxybenzoic acid (PHB), a reaction catalyzed by PHB:geranyltransferase (Yazaki et al., 2002). This geranylated intermediate is subsequently cyclized to form a naphthoquinone skeleton, leading to shikonin formation (Heide et al., 1989; Yazaki, 2017).
GPPS has been partially purified from cultured L. erythrorhizon cells, with its activity detected in the cytosolic fraction (Heide and Berger, 1989; Sommer et al., 1995). In addition, tracer experiments showed that the geranyl moiety of shikonin originates from the MVA pathway (Li et al., 1998), further indicating that GPPS localizes to the cytosol, not to plastids, in these plants.
Four hypotheses have been proposed to explain the unique localization of GPPS in the cytosol of L. erythrorhizon. The first is that plastidial homomeric GPPS lost its transit peptide remaining in the cytosol, the second is that a heteromeric GPPS along with a SSU, which are lacking the transit peptide to alter the localization from plastid to cytosol as reported in Arabidopsis (Arabidopsis thaliana; Wang and Dixon, 2009; Ruiz-Sola et al., 2016), the third is that a regulatory subunit that alters the enzymatic activity of farnesyl diphosphate synthase (FPPS) may be present in the cytosol, and the fourth is that a cytosol-localized FPPS acquired mutations leading to different specificities, both for the substrate and the product, to provide GPP. In this study, a cytosolic GPPS was identified in L. erythrorhizon (LeGPPS) by screening EST libraries of cultured L. erythrorhizon cells, followed by biochemical characterization using recombinant proteins. Moreover, site-directed mutagenesis was performed to gain insights into the molecular evolution of this enzyme family.
RESULTS
Search for LeGPPS Candidates from EST Data of L. erythrorhizon
We first performed a BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for a homolog of trans-prenyl diphosphate synthases in 13,242 independent clones of an EST library constructed from cultured L. erythrorhizon cells. However, we could not identify any sequences showing significant homology with known GPPSs. This indicated that our first hypothesis, that a plastidial GPPS had lost its transit peptide, remaining in the cytosol, was rather unlikely. The enzymatic activity of GPPS in cultured cells was consistently strong, with GPPS expression being high enough to support the production of large amounts of shikonin (Heide and Berger, 1989). Despite the expression level of FPPS usually being low in various cultured plant cells in our experiences, being present in 1 to 2 per 10,000 ESTs, seven clones annotated as FPPS were identified in our EST library. This unusually high representation of FPPS-like genes suggested that a FPPS homolog might be involved in shikonin biosynthesis, suggesting that a unigene represented by these seven clones was a candidate for the putative GPPS gene in L. erythrorhizon (LeGPPS). The full coding sequence of the FPPS homolog was isolated by 5′ RACE, and its complementary DNA (cDNA) was recloned from the cDNA pool of cultured L. erythrorhizon cells. The coding sequence consisted of 1053 bp, encoding a protein of 350 amino acids (Supplemental Fig. S2).
Functional Expression of the Recombinant FPPS Homolog
The coding sequence of the FPPS homolog was subcloned into the pET22b (+) vector, which was introduced into Escherichia coli Origami B strain. The expression of the recombinant protein was induced by the addition of isopropyl-β-d-thiogalactopyranoside. Utilizing a C-terminal His-tag, the recombinant protein was purified from the cell lysate by nickel-nitrilotriacetic acid affinity chromatography. SDS-PAGE of the eluate fraction showed a single band at approximately 40 kD (Supplemental Fig. S3), a molecular mass similar to that predicted from its amino acid sequence (∼40 kD). Prior to functional analysis, the elution buffer was replaced with the assay buffer for determination of enzyme activity (see “Materials and Methods”). The enzymatic activity of the purified protein was evaluated by assessing prenyltransferase-catalyzed chain elongation through the detection of dephosphorylated reaction products by gas chromatography-mass spectrometry (GC-MS).
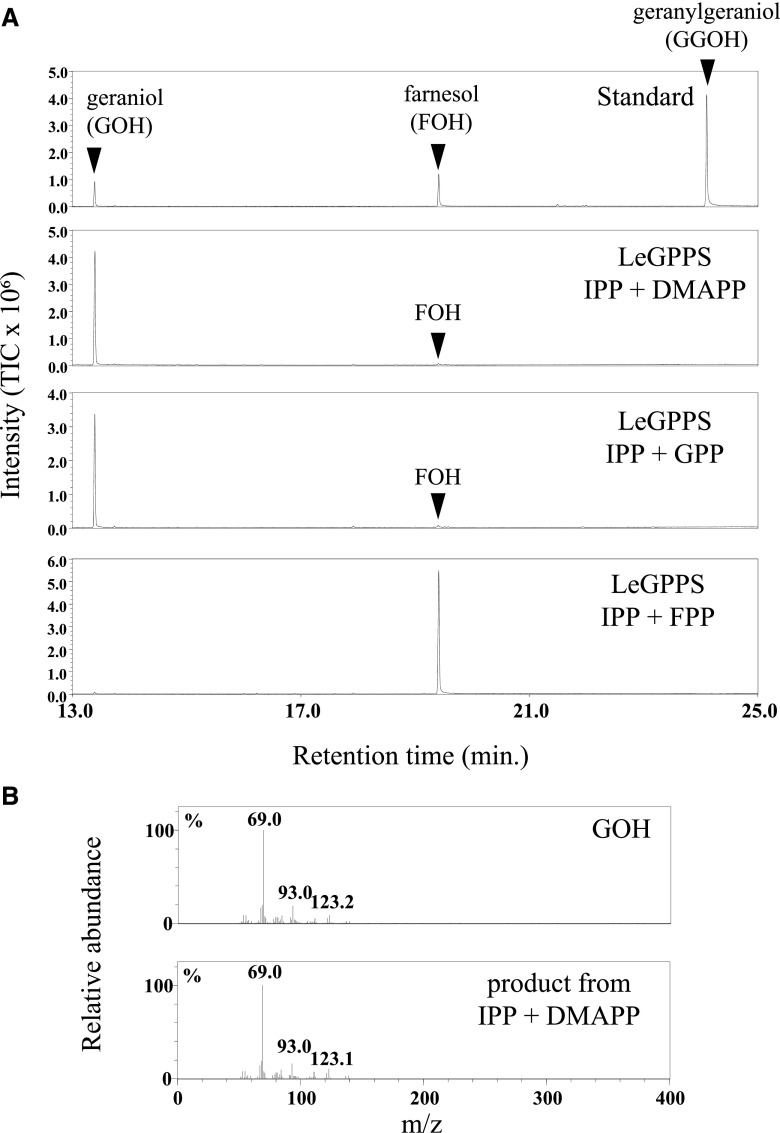
Surprisingly, GC-MS analysis clearly showed that the recombinant FPPS homolog catalyzed the formation of GPP (C10) in the presence of IPP and DMAPP, but was unable to further elongate the prenyl chain to FPP (C15) from these substrates (Fig. 1). Even when GPP was the prenyl acceptor substrate, its FPPS activity was barely detectable. Similarly, no GGPPS activity was detected when IPP and FPP were substrates (Fig. 1A, bottom). The FPPS homolog was designated LeGPPS (accession no., LC427363).
Figure 1.
Enzyme activity of recombinant FPPS homolog (LeGPPS) from L. erythrorhizon. The enzyme reaction products were analyzed by GC-MS after dephosphorylation by acid/alkaline phosphatases. A, Total ion chromatograms of reaction products of recombinant LeGPPS with DMAPP, GPP, or FPP as acceptor substrates in the presence of IPP, the donor substrate. The prenyl acceptor substrates GPP and FPP for FPPS and GGPPS, respectively (bottom two chromatograms), were detected in their respective assay mixtures. B, Mass fragmentation patterns of a standard specimen of geraniol and the enzyme reaction product of recombinant LeGPPS with IPP and DMAPP as substrates.
Expression of LeGPPS in Intact Plants and Cultured L. erythrorhizon Cells
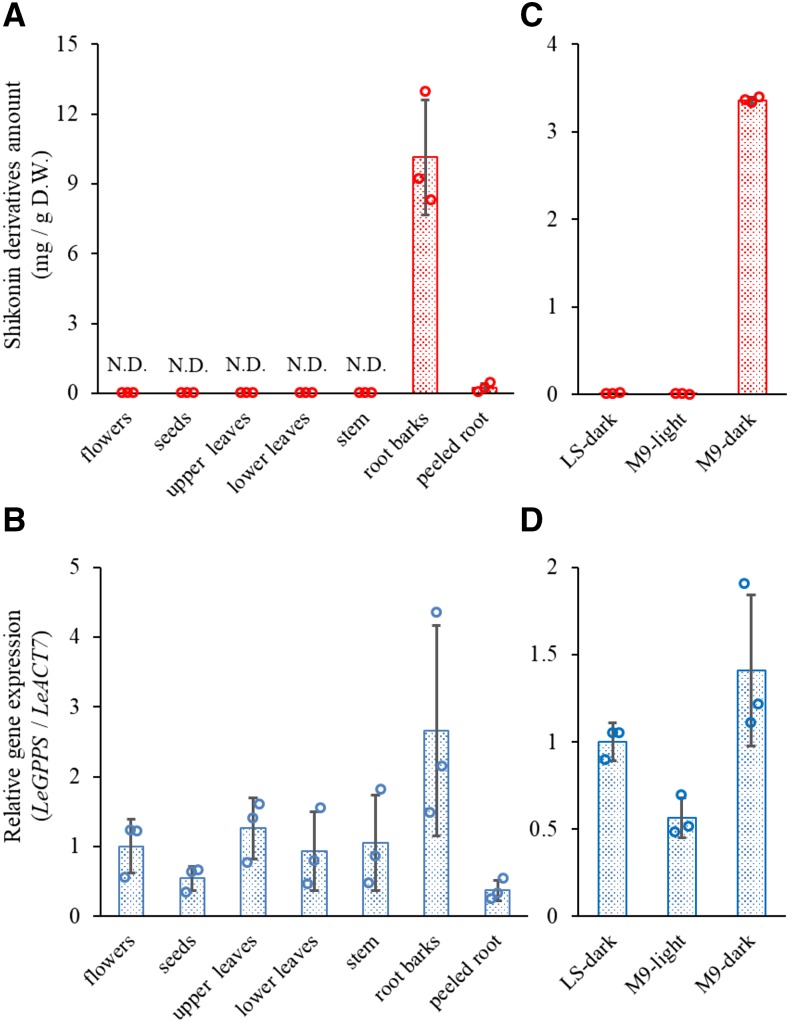
Intact L. erythrorhizon plants produce large quantities of shikonin derivatives, which accumulate almost exclusively in the root bark, with trace amounts in peeled roots (Fig. 2A). To analyze the expression profile of LeGPPS, total RNA was extracted from seven organs of intact plants: flowers, seeds, upper leaves, lower leaves, stems, root barks, and peeled roots. After synthesis of each cDNA by reverse transcription, reverse transcription quantitative PCR was performed. The highest level of LeGPPS expression was observed in root bark, the major site of accumulation of shikonin derivatives, although detectable levels of LeGPPS transcript were observed in all organs tested (Fig. 2B).
Figure 2.
Quantification of shikonin derivatives and relative expression of the LeGPPS gene in L. erythrorhizon. A, Organ-specific accumulation of shikonin derivatives in intact L. erythrorhizon plants. N.D., Not determined; D.W., dry weight. B, Relative transcript level of LeGPPS in intact L. erythrorhizon plants. C, Production of shikonin derivatives in L. erythrorhizon cells cultured in LS or M9 medium. A negative control is the cell cultures in M9 medium under illumination. D, Relative transcript level of LeGPPS in cultured L. erythrorhizon cells. Bars indicate mean ± sd of three biological replicates. Open circles indicate the individual values in each analysis.
The amount of shikonin derivatives and transcript levels of LeGPPS were also measured in cultured L. erythrorhizon cells. Cultured cells grown in Linsmaier and Skoog (LS) medium (Linsmaier and Skoog, 1965) did not produce detectable amounts of shikonin derivatives (Fig. 2C). Shikonin was produced by culturing these cells in M9 medium in the dark, but not when these cells were exposed to light, even when cultured in M9 medium (Fig. 2C). The LeGPPS transcript level was higher in shikonin-producing than shikonin-nonproducing cells cultured in M9 medium (Fig. 2D). The level of LeGPPS expression detected in shikonin nonproducing cells was in agreement with a report showing high GPPS activity in LS-cultured white cells of L. erythrorhizon, from which the enzyme was partially purified (Heide and Berger, 1989). Moreover, our results provide further evidence that GPPS activity is not a rate-limiting step for shikonin biosynthesis. Interestingly, L. erythrorhizon cells in LS medium produce an appreciable amount of the benzoquinone derivative dihydroechinofuran, another GPP-derived compound (Supplemental Fig. S4; Fukui et al., 1992). LeGPPS expression in LS medium is therefore necessary to provide the GPP used for dihydroechinofuran biosynthesis.
Subcellular Localization of GFP Fusion Protein in Onion Epidermal Cells
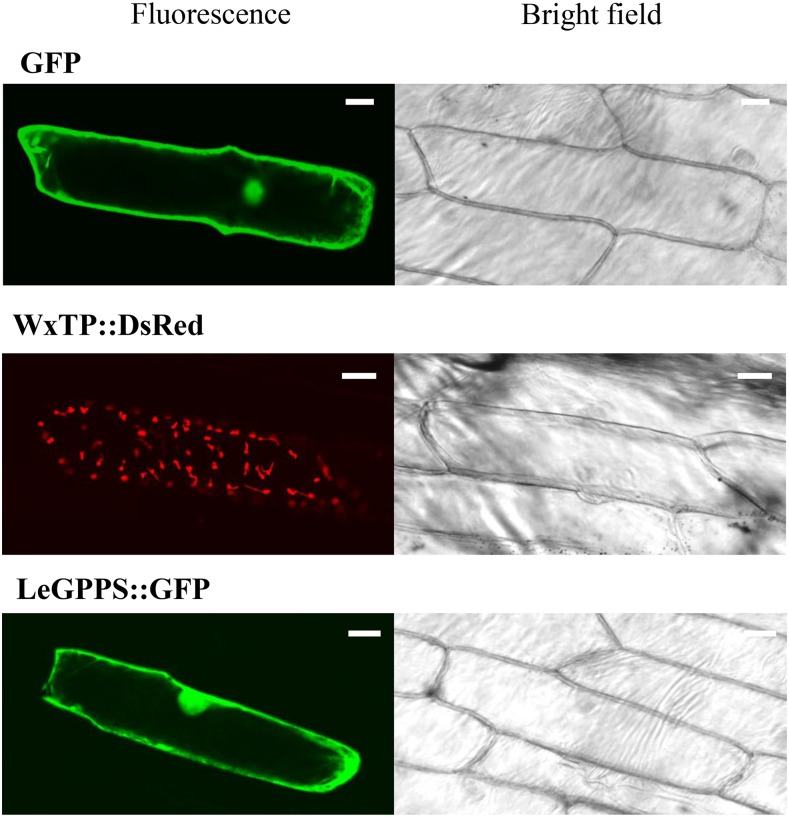
Alignment of LeGPPS with known trans-prenyl diphosphate synthases clearly showed that LeGPPS and known FPPSs lack N-terminal extension, while both monomeric and heteromeric GPPSs (LSU and SSU) have putative transit peptide sequences (Supplemental Fig. S5). To examine the subcellular localization, a vector for transient expression of LeGPPS fused to GFP at its C terminus was prepared and introduced into onion (Allium cepa) epidermal cells by particle bombardment. Free GFP was used as a cytosol marker, and WxTP::DsRed as a plastid marker (Kitajima et al., 2009). The fluorescence pattern of LeGPPS::GFP was localized to the cytosol, similar to free GFP (Fig. 3). The red fluorescence of WxTP::DsRed showed a typical plastid pattern that did not match that of LeGPPS::GFP. These results suggest that LeGPPS localizes to the cytosol, consistent with a report showing that GPPS activity was present in the cytosol of fractionated L. erythrorhizon cells (Sommer et al., 1995). Taken together, the enzyme activity, expression profiles, and subcellular localization of LeGPPS indicated that the LeGPPS gene is responsible for the synthesis of cytosolic GPP that contributes to the biosynthesis of shikonin, as well as to dihydroechinofuran in LS medium.
Figure 3.
Subcellular localization of the GFP fusion protein of LeGPPS. Confocal microscopic images and bright-field pictures of onion cells expressing free GFP (cytosol control), WxTP::DsRed (plastid control), and LeGPPS::GFP. Bars = 30 µm.
Identification of Genuine FPPS from L. erythrorhizon
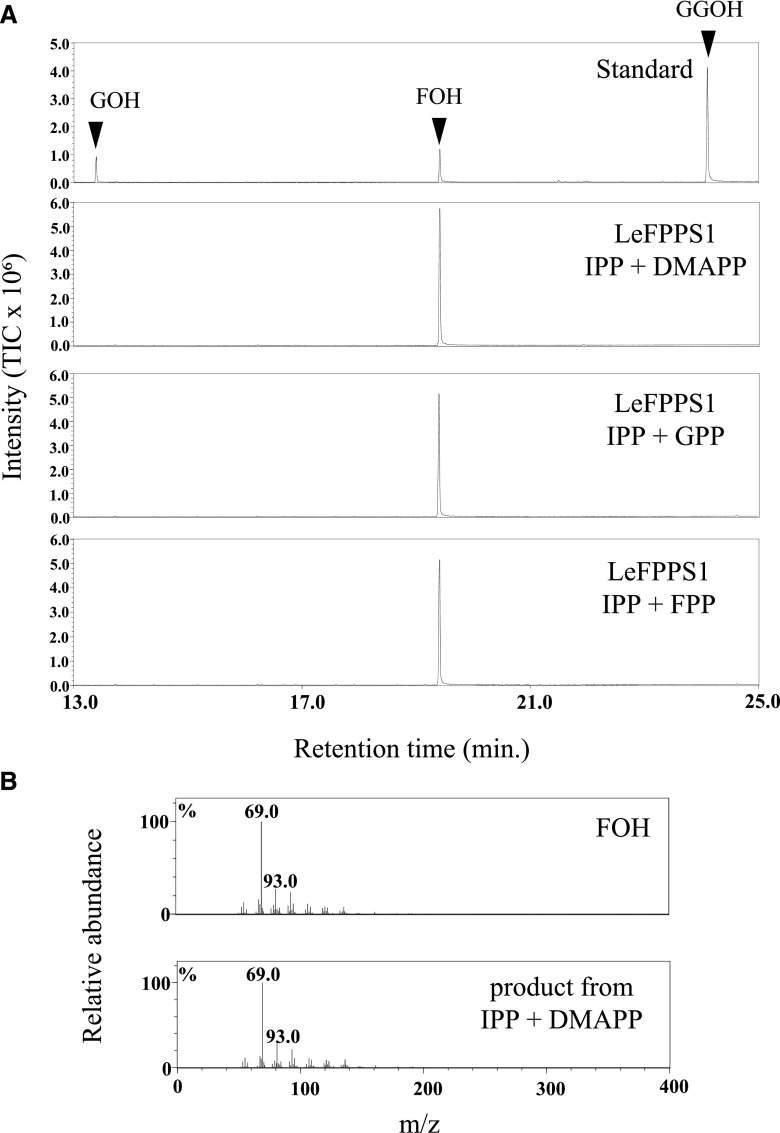
The strong sequence similarity between LeGPPS and FPPS suggested that this GPP-specific enzyme evolved from FPPS. FPP, however, is also required by these cells to produce important primary metabolites, such as sterols. Because LeGPPS produced trace amounts of FPP (Fig. 1), a genuine FPPS is needed to provide the FPP necessary to maintain plant life. Although we did not find any other FPPS homologs in the EST library, a search of multiple omics data of L. erythrorhizon led to the detection of 11 open reading frames for the trans-prenyl diphosphate synthase gene (Takanashi et al., 2019), among which there were two FPPS homologs (comp87014 and comp89799), with one having a higher fragments per kilobase of exon per million mapped reads (FPKM) value than the other (Supplemental Table S1). The FPPS homolog with the higher FPKM value (comp89799) was cloned, and the enzyme activity of the recombinant protein was measured in a manner similar to that of LeGPPS. This FPPS homolog produced FPP from DMAPP or GPP in the presence of a prenyl donor substrate, IPP, suggesting that this gene product is a genuine FPPS required by L. erythrorhizon for primary metabolism (Fig. 4). This enzyme has been designated LeFPPS1 (accession no., LC427365).
Figure 4.
Enzyme activity of recombinant FPPS homolog (LeFPPS1) from L. erythrorhizon. The reaction products were analyzed by GC-MS after dephosphorylation by acid/alkaline phosphatases. A, Total ion chromatograms of reaction products of recombinant LeFPPS1 using IPP as donor substrate and DMAPP, GPP, and FPP as acceptor substrates. B, Mass fragmentation patterns of a standard specimen of farnesol and the enzyme reaction product of recombinant LeFPPS1 with IPP and DMAPP as substrates.
To study the kinetic parameters, we used ultra-performance liquid chromatography-mass spectrometry/mass spectrometry (UPLC-MS/MS) to detect the diphosphate products directly (Supplemental Fig. S6). The kinetic parameters for LeGPPS and LeFPPS1 were examined as summarized in Tables 1 and 2. The Km value of LeGPPS for DMAPP was approximately 4-fold higher than that of LeFPPS1, showing that the substrate affinity of LeFPPS1 for DMAPP is greater than that of LeGPPS. For IPP, almost no difference in the Km values was observed between LeGPPS and LeFPPS1. The kcat value (turnover number) of LeFPPS1 for DMAPP was slightly higher than that of LeGPPS, and also a similar tendency was seen with IPP for both enzymes. On the other hand, the Km value of LeFPPS1 for GPP was extraordinarily low, showing that LeFPPS1 has a clear preference to use GPP rather than DMAPP for the FPP production.
Table 1. Kinetic Parameters for LeGPPS.
The enzyme reaction products were quantitated by UPLC-MS analysis. Data are shown as means ± SD of three technical replicates.
| Substrate | LeGPPS | ||
|---|---|---|---|
| Km (µm) | kcat (min−1) | kcat/Km (μm−1⋅min−1) | |
| DMAPP (at 100 µm IPP) | 8.17 ± 1.35 | 2.24 ± 0.15 | 0.27 ± 0.03 |
| GPP (at 100 µm IPP) | – | – | – |
| IPP (at 60 µm DMAPP) | 2.69 ± 0.60 | 2.83 ± 0.46 | 1.06 ± 0.08 |
Table 2. Kinetic Parameters for LeFPPS1.
The enzyme reaction products were quantitated by UPLC-MS analysis. Data are shown as means ± SD of three technical replicates. Asterisk indicates the value calculated from near detection limit of UPLC-MS analysis because of the high affinity of LeFPPS1 to GPP.
| Substrate | LeFPPS1 | ||
|---|---|---|---|
| Km (µm) | kcat (min−1) | kcat/Km (μm−1⋅min−1) | |
| DMAPP (at 100 µm IPP) | 2.20 ± 0.48 | 3.48 ± 0.21 | 1.58 ± 0.25 |
| GPP (at 100 µm IPP) | 0.10 ± 0.03* | 2.49 ± 0.04 | 26.14 ± 9.65 |
| IPP (at 60 µm DMAPP) | 3.03 ± 0.91 | 3.90 ± 0.67 | 1.29 ± 0.16 |
Site-Directed Mutagenesis of LeGPPS
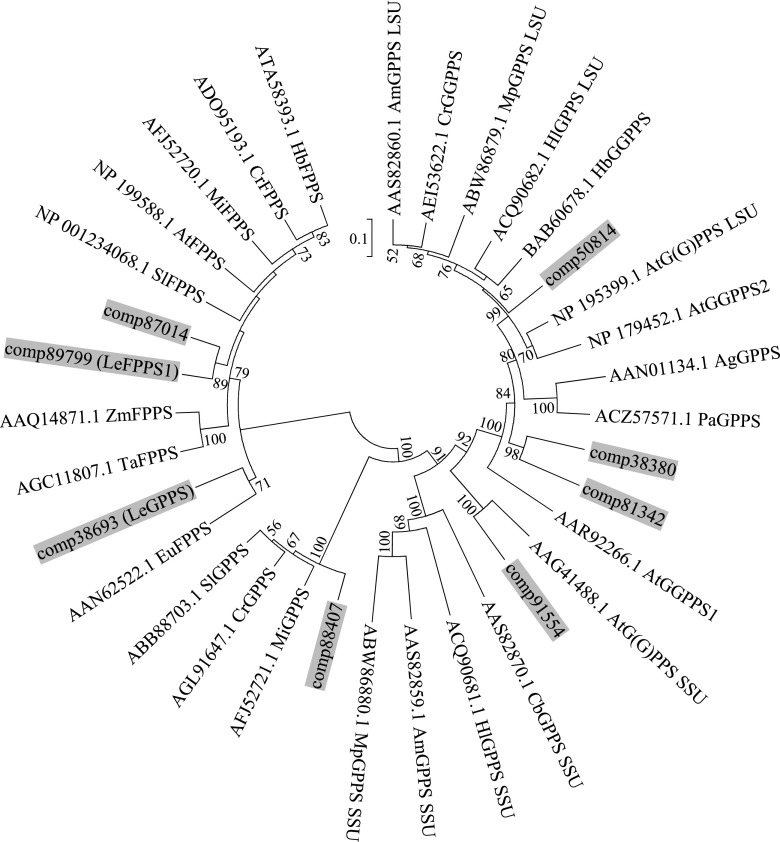
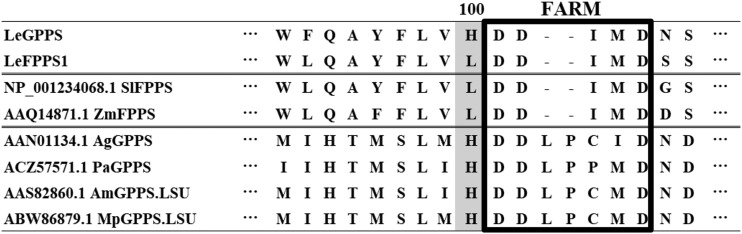
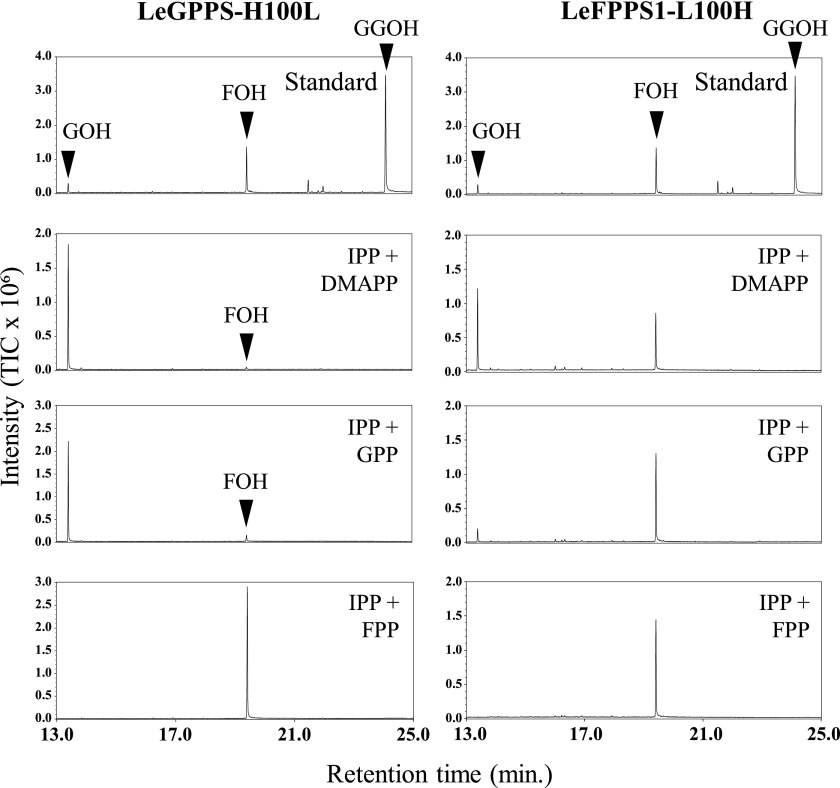
A phylogenetic tree showed that LeGPPS is grouped into the clade of plant FPPSs (Fig. 5). To identify the amino acid residues responsible for the GPPS activity of LeGPPS, the amino acid sequences of LeGPPS and LeFPPS1 were aligned with those of known GPPSs and FPPSs (Fig. 6). Careful comparisons showed the importance of the residue at position 100, adjacent to the N terminus of the FARM sequence conserved among all trans-prenyl diphosphate synthases (Chen et al., 1994). LeGPPS and other GPPSs have a His residue at this position (H100), whereas all FPPSs, including LeFPPS1, have a Lys or Ala residue. Substitution of another amino acid residue at this position was found to influence substrate preference in avian (Gallus gallus) and yeast (Saccharomyces cerevisiae) FPPSs (Stanley Fernandez et al., 2000; Rubat et al., 2017). To test the contribution of H100 to the product specificity of LeGPPS, LeGPPS and LeFPPS1 were subjected to site-directed mutagenesis, yielding two types of point mutants, LeGPPS-H100L and LeFPPS1-L100H, respectively. Although the enzymatic properties of LeGPPS-H100L were almost identical to those of wild-type LeGPPS, the properties of LeFPPS-L100H were altered, leading to the production of GPP (Fig. 7). These findings suggest that LeGPPS acquired its GPPS activity through point mutations in FPPS, with H100 playing a key role.
Figure 5.
Unrooted phylogenetic analysis of the deduced amino acid sequences of trans-prenyl diphosphate synthase homologs in L. erythrorhizon and other plant species. The prenyl diphosphate synthases of L. erythrorhizon are highlighted in gray. Three partial sequences of trans-prenyl diphosphate synthase homologs found in RNA-seq data of L. erythrorhizon were not included. The tree was inferred by the bootstrap neighbor-joining method using the program MEGA7 (http://evolgen.biol.se.tmu.ac.jp/MEGA/) with default settings. The percentage of bootstraps, calculated by 1000 repeats, is shown next to each branch, with bootstrap values less than 50% omitted. The GenBank accession numbers are shown for each enzyme. The first two letters indicate the scientific names of each species: At, Arabidopsis; Ag, Abies grandis; Am, Antirrhinum majus; Eu, Eucommia ulmoides; Cb, Clarkia breweri; Cr, Catharanthus roseus; Hb, Hevea brasiliensis; Hl, Humulus luplus; Mi, Mangifera indica; Mp, Mentha x piperita; Pa, Picea abies; Sl, Solanum lycopersicum; Ta, Triticum aestivum; Zm, Zea mays.
Figure 6.
Multiple alignment of deduced amino acid sequences of transprenyl diphosphate synthases from L. erythrorhizon and other plant species. The FARM, conserved among all prenyl diphosphate synthases, is boxed. The target site of point mutation is highlighted in gray. The GenBank accession numbers are shown for each enzyme. The first two letters indicate the scientific names of each species: Ag, Abies grandis; Am, Antirrhinum majus; Mp, Mentha × piperita; Pa, Picea abies; Sl, Solanum lycopersicum; Zm, Zea mays.
Figure 7.
Biochemical assays of recombinant LeGPPS-H100L and LeFPPS1-L100H. The enzyme reaction products were analyzed by GC-MS after dephosphorylation by acid/alkaline phosphatases. Total ion chromatograms of reaction products are shown for LeGPPS-H100L and LeFPPS1-L100H using IPP as donor substrate and DMAPP, GPP, and FPP as acceptor substrates.
DISCUSSION
This study describes the identification of a cytosol-localized GPPS in L. erythrorhizon. This enzyme, LeGPPS, mainly produces GPP from IPP and DMAPP and belongs to the FPPS family. Originally, we proposed four hypotheses to explain the development of cytosol-localized GPPS necessary for shikonin biosynthesis: (1) the conventional plastid-localized homomeric GPPS lost its transit peptide, remaining in the cytosol; (2) the heteromeric GPPS composed of LSUs and SSUs lost its transit peptides to localize in the cytosol; (3) the cytosol contains a regulatory subunit that interacts with FPPS, converting its enzyme activity to GPPS; and (4) cytosol-localized FPPS acquired mutations altering its enzymatic activity to GPPS. From L. erythrorhizon transcriptome data, we found 11 contigs for trans-prenyl diphosphate synthases, and eight of them appeared to have full coding sequences, with which a phylogenetic tree was made (Fig. 5). This shows that L. erythrorhizon has homologs of GGPPS, GPPS.LSU, homomeric GPPS, GPPS.SSU, and FPPS, while FPKM values for homomeric GPPS homolog (comp88407), GPPS.LSU (comp50814), and GPPS.SSU homolog (comp91554) were decisively lower than LeGPPS (Supplemental Table S1), suggesting that these are insufficient to support the large production of shikonin derivatives. In conclusion, while the third speculation is not completely denied, our experimental data strongly support the fourth hypothesis. To date, plants have been shown to contain homomeric and heteromeric plastidial GPPSs, consisting of catalytic LSUs and noncatalytic SSUs (Tholl, 2015). The LeGPPS identified in this study is none of type (1), (2), and (3); rather, it is the first FPPS-like GPPS identified in plants. Because homomeric GPPS and GPPS.SSU homologs are present in L. erythrorhizon transcriptome data, a possible existence of another functional GPPS that may participate in the biosynthesis of other metabolites like monoterpenes cannot be excluded.
Multiple omics data of L. erythrorhizon indicated that this plant has three types of FPPS homolog: LeGPPS, LeFPPS1, and LeFPPS2 (accession no., LC519333). The latter was not characterized in this study due to its low expression level. Each of these enzymes likely has differential physiological functions. The enzymatic activity and expression patterns of LeGPPS suggest its involvement in shikonin biosynthesis, whereas the product specificity of LeFPPS1 suggests its function in primary metabolism, such as the biosynthesis of sterols. TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/) predicts that LeFPPS2 (comp87014), with a lower FPKM value than LeFPPS1, localizes to the mitochondria (Supplemental Table S1). Mitochondrial FPPS genes have been detected in several organisms, although their physiological functions are not yet fully understood (Martín et al., 2007). For example, FPPS1 from Arabidopsis encodes two isoforms: FPPS1L, which localizes to the mitochondria, and FPPS1S, which, localizes to the cytosol (Cunillera et al., 1997). Tomato (Solanum lycopersicum) FPPS2 harboring a mitochondrial signal peptide was expressed predominantly in pollen and was involved in unilateral incompatibility (Qin et al., 2018). LeFPPS2 may be associated with the biosynthesis of ubiquinone and heme, which are localized to the mitochondria.
In contrast to L. erythrorhizon, some other plant species appear to have recruited FPPS family members for the biosynthesis of various terpenoids in plastids. For example, the big sagebrush Artemisia tridentata ssp. spiciformis contains three FPPS homologs, FDS-1, FDS-2, and FDS-5. FDS-1 and FDS-2 possess FPPS activity, whereas FDS-5, which contains a transit peptide, shows no FPPS activity but produces an irregular monoterpenoid, chrysanthemyl diphosphate, with GPP as a byproduct (Hemmerlin et al., 2003). Chrysanthemyl diphosphate is an important biosynthetic precursor of pyrethrins, which are insecticidal meroterpenoids produced by some Asteraceae plants (Matsuda et al., 2005). Rhododendron dauricum produces the FPP-derived meroterpenoid daurichromenic acid in plastids, a production enhanced by treatment of these plants with mevastatin, which inhibits the MVA pathway (Saeki et al., 2018). These findings suggested that the subcellular localization of a FPPS in R. dauricum was converted from the cytosol to plastids for the biosynthesis of this specialized metabolite (Saeki et al., 2018). Our findings with LeGPPS demonstrate that the evolution of the FPPS family plays important roles in the evolution of the terpenoid pathway. To highlight how His-100 is conserved among FPPs of other plant species, we made a phylogenetic tree solely of FPP homologs and found some Rosaceous FPPS homologs (strawberry [Fragaria × ananassa], apple [Malus domestica], and peach [Prunus persica]) formed a clade with LeGPPS (Supplemental Fig. S7). The multiple alignment reveals that His-100 is indeed conserved in these FPPS-like proteins (Supplemental Fig. S8), while these plant species have other FPPS homologs grouped with LeFPPS1 of L. erythrorhizon, the genuine FPPS (Supplemental Fig. S7). In fact, the formation of GPP-derived terpenoids in the cytosol has been reported in several Rosaceae plants (Francis and O’Connell, 1969; Hampel et al., 2006). The identification of LeGPPS as the first cytosolic GPPS will provide important insights into the biosynthetic mechanisms underlying the diversification of terpenoid compounds in plants.
Large-scale transcriptome data enabled us to find putative terpenoid synthases expressed in L. erythrorhizon, i.e. we found seven contigs coding for proteins sharing similarities with known terpene synthases. Among them, three contigs (comp63758, comp71249, and comp91292) showed similarities with limonene, geraniol, and alpha-terpineol synthases, respectively (Supplemental Table S2). Although their FPKM values are generally very low and the occurrence of monoterpene is not reported in L. erythrorhizon to our knowledge, we analyzed the extracts of each aerial organ with GC-MS to assess a possibility that LeGPPS expressed in the aerial organs might be involved in the monoterpene biosynthesis (Supplemental Fig. S9). However, no detectable level of monoterpenes was found in leaves, stems, and seeds. While some small peaks were found in flowers, their retention times were not identical with the above three monoterpenes. These data suggest that the involvement of LeGPPS in monoterpene biosynthesis in the aerial part of this plant is rather unlikely.
Localization of GPPS in the cytosol may allow better regulation of shikonin biosynthesis, resulting in its effective production. A key regulatory step in shikonin biosynthesis is the formation of m-geranyl-p-hydroxybenzoic acid, with GPP and PHB being the specific substrates involved in the biosynthesis of this intermediate (Heide et al., 1989). The enzyme responsible for this rate-limiting step is PHB:geranyltransferase, whose expression is strongly inhibited by all negative regulators, such as light and NH4+ (Yazaki et al., 2002). Light inhibition is the reason that shikonin is produced solely in root tissues of intact plants. Similarly, shikonin is produced by cultured cells only in the dark, at a yield of up to 10% of shikonin per cell dry weight (Fujita and Hara, 1985). As PHB formation is insensitive to light, PHB overaccumulates as its glucoside form under shikonin-nonproducing conditions (Yazaki et al., 1986), with excess amounts of GPP not observed in white cells. Upon induction of shikonin synthesis, the GPP supply is increased to match its use by geranyltransferase (Yazaki, 2017). Other monoterpenes are induced under light conditions, with the precursor GPP provided by the MEP pathway in plastids and the upstream precursors of GPP being photosynthates, pyruvate, and glyceraldehyde-3-phosphate (Vranová et al., 2013). Because shikonin is produced only in the dark, however, MEP pathway-derived GPP cannot support the high production of shikonin. Rather, the MVA pathway originating from acetyl-CoA may be a more appropriate source for the large amount of GPP produced in the dark. PHB:geranyltransferase is a membrane-bound enzyme located in the endoplasmic reticulum, making it advantageous for the efficient supply of cytosolic GPP as a substrate (Yazaki et al., 2002).
Many site-directed mutagenesis studies of trans-prenyl transferases have identified several key residues for chain-length determinations (CLDs; Tarshis et al., 1996; Lee et al., 2005). The artificial conversion of FPPS to GPPS activity was demonstrated in nonplant organisms, including Geobacillus stearothermophilus, G. gallus, and S. cerevisiae (Narita et al., 1999; Stanley Fernandez et al., 2000; Fischer et al., 2011). These studies found that the first and fourth positions downstream as well as the 23rd position upstream of FARM influenced product preference (Narita et al., 1999; Stanley Fernandez et al., 2000; Fischer et al., 2011). A unique FPPS homolog (IPPS) in the green peach aphid, Myzus persicae, showed both GPPS and FPPS activity (Vandermoten et al., 2008). Point mutations of aphid IPPS indicated that its catalytic activity was due primarily to the first and fourth positions upstream of FARM (Vandermoten et al., 2009). A SARM may also be involved, as the crystal structure of FPS-5 in A. tridentata indicated that the first position of the conserved motif SARM was also a key residue for CLD (Lee et al., 2017). Utilizing the information, we attempted to identify putative key residue(s) of CLD(s) in LeGPPS (Supplemental Fig. S10). However, the only residue for CLD identified was H100. Future analysis of the tertiary structure of crystallized LeGPPS may provide additional information about the structure-function relationship oftrans-prenyl transferases.
Interactions among serial biosynthetic enzymes, as in ubiquinone biosynthesis, are often called “metabolons.” Similar to shikonin biosynthesis, a membrane-bound prenyltransferase involved in the synthesis of coenzyme Q (COQ2) accepting PHB as a substrate serves as a membrane anchor, binding a series of enzymes responsible for the biosynthesis of ubiquinone and forming a multisubunit complex (Gin and Clarke, 2005). The prenyl chain-elongating enzyme COQ1, a trans-prenyltransferase, is a member of this biosynthetic complex. Because of the interactions of the involved biosynthetic enzymes, intermediates are not usually released, making metabolic turnover very rapid. To prove the existence of the metabolon in shikonin biosynthesis, all enzymes involved in the biosynthetic route should be identified in the future.
Membrane dynamics common to vesicle secretion processes have been reported as involved in the apoplastic accumulation of shikonin derivatives (Tatsumi et al., 2016), although the mechanisms responsible for secretion remain largely unknown. The geranyl moiety provides hydrophobicity to the intermediate PHB, suggesting that the succeeding biosynthetic reactions take place in membrane systems. It is therefore of great interest to determine whether other GPP-derived metabolites, such as monoterpenoids and geranylated coumarins, are secreted by the cells of other plant species and accumulate in the subcuticular cavity of glandular trichomes of Lamiaceae and oil glands of Rutaceae plants (Lange, 2015). L. erythrorhizon cell cultures may help uncover the biochemical and molecular mechanisms involved in secretion, due to the visibility, regulatory features, and high productivity of shikonin.
MATERIALS AND METHODS
Plant Materials
Cultured Lithospermum erythrorhizon cells (line T-TOM) were maintained in LS medium containing 10−6 m indole acetic acid and 10−5 m kinetin at 25°C, 80 rpm in the dark, and subcultured at 2-week intervals. For shikonin production, these cells were transferred to M9 medium supplemented with the same auxin and cytokinin combination as above and cultured in the dark under the same agitation conditions. Two-year-old intact plants, kindly provided by Amato Pharmaceutical Products, were used for both gene expression analysis of LeGPPS and quantification of shikonin derivatives.
Construction of an EST Library and Screening of Candidate Genes
Total RNA was extracted from cultured L. erythrorhizon cells 9 d after inoculation in M9 medium, using RNeasy plant mini kits (Qiagen), according to the manufacturer’s standard protocol. Poly(A)+ RNA was purified with Takara Oligotex-dT30 < Super > mRNA purification kits (TaKaRa). A yeast (Saccharomyces cerevisiae)-Escherichia coli shuttle vector pDR196 was used for library construction because of its advantage in cloning genes encoding proteins toxic to E. coli, as the promoter of this vector, plasma membrane ATPase, does not show leaky activity in E. coli (Rentsch et al., 1995). Using pDR196 as a final vector, EST libraries were constructed with cDNA Synthesis Kits (Stratagene, Agilent Technologies) according to the manufacturer’s instructions. The 13,242 independent clones were sequenced and annotated by BLAST-based similarity. Based on their annotation as trans-prenyl diphosphate synthases, seven clones were selected as candidates of LeGPPS.
Cloning of LeGPPS
The sequence upstream of LeGPPS was obtained by 5′ RACE with a GeneRacer kit (Invitrogen) according to the manufacturer’s instructions. The full-length cDNA was amplified using KOD plus polymerase (Toyobo) and the primers LeGPPS-Fw1 and LeGPPS-Rv1 (Supplemental Table S3). The amplification protocol consisted of an initial denaturation at 95°C for 4 min, 30 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, extension at 68°C for 1.3 min, and a final extension at 68°C for 4 min. After the addition of adenine to the cDNA ends using Go Taq DNA polymerase (Promega), the fragment was subcloned into pT7 vector using T4 DNA ligase (TaKaRa), and the insert was verified by sequencing.
Heterologous Expression of LeGPPS and LeFPPS1
The coding sequences of LeGPPS and LeFPPS1 without stop codons were amplified using the primer pairs LeGPPS-Fw2 and LeGPPS-Rv2 and LeFPPS1-Fw and LeFPPS1-Rv, respectively (Supplemental Table S3). These amplicons were inserted individually upstream of the 6× His-tag in the E. coli expression vector pET-22b (+) (Novagene), yielding the plasmids pET-22b::LeGPPS::His and pET-22b::LeFPPS1::His, respectively. The resulting plasmids were used to transform E. coli Origami B strain. Each E. coli transformant was precultured overnight at 37°C in lysogeny broth medium containing 80 mg/mL ampicillin and 50 mg/ml kanamycin. The preculture was inoculated into new lysogeny broth medium (50 mL) and grown at 37°C to an optical density at 600 nm of 0.6. Overexpression of each recombinant protein was induced by the addition of isopropyl-β-d-thiogalactopyranoside (final concentration, 1 mm) and cultured at 16°C for 18 h on a rotary shaker (200 rpm). The bacteria expressing recombinant protein were collected by centrifugation (4°C, 10,000 rpm, 5 min) and resuspended in wash buffer (150 mm NaCl, 10 mm Tris-HCl, pH 8.0). After subsequent centrifugation (4°C, 10,000 rpm, 5 min), the bacterial pellet was stored at −80°C until use.
Affinity Purification and Buffer Exchange
His-tagged protein was purified with QIAexpress@ Ni-NTA Fast Start Kits (Qiagen), according to the manufacturer’s instructions, with a slight modification, i.e. the bacterial pellet was additionally disrupted on ice using an ultrasonic homogenizer (Branson Sonifier, 5 × 10 s, duty cycle 40%, output constant). The purified protein was electrophoresed by SDS-PAGE and visualized by Coomassie Brilliant Blue staining to verify purification. Before enzyme assay, the protein buffer was exchanged on a PD-10 desalting column (GE Healthcare) to assay buffer (25 mm 3-morpholinopropanesulfonic acid, 10% [v/v] glycerol, 5 mm dithiothreitol [DTT], 10 mm MgCl2, 5 mm sodium ascorbate, pH 7.5) for the analyses of enzyme activities.
Enzyme Assay and Reaction Product Analysis by GC-MS
A 1-mL mixture containing 20 µg His-tagged protein, 6.25 mm 3-morpholinopropanesulfonic acid (MOPS), 2.5% (v/v) glycerol, 1.25 mm DTT, 2.5 mm MgCl2, 1.25 mm sodium ascorbate, 100 µm IPP, and 100 µm DMAPP/GPP/FPP was incubated at 30°C for 4 h, followed by incubation with acid phosphatase (Wako) and alkaline phosphatase (TaKaRa) at 37°C for 1 h. The released alcohols were extracted with 1 mL hexane, and the organic phase was concentrated to approximately 30 µL and analyzed by GC-MS on a GC-2010 (Shimadzu) coupled with GC-MS-QP 2010 Plus (Shimadzu) using a DB-5ms column (Agilent Technologies, 30 m length, 0.25 mm inner diameter, 0.25 µm film). Helium was used as a carrier gas and samples were injected at 240°C and a flow rate of 1.9 mL/min. Volatile compounds were separated by a temperature gradient, consisting of 50°C for 5 min, 10°C/min to 240°C, and a final 6 min hold at 240°C.
Enzyme Assay with UPLC-MS/MS
A 40-µL mixture containing 1 µg His-tagged protein, 6.25 mm MOPS, 2.5% (v/v) glycerol, 1.25 mm DTT, 2.5 mm MgCl2, 1.25 mm sodium ascorbate, 40 µm IPP, and 40 µm DMAPP, GPP, or FPP was incubated at 30°C for 30 min, followed by heat denaturation at 95°C for 3 min. The reaction products were analyzed by UPLC-MS/MS on an ACQUITY UPLC system (Waters) coupled with Xevo TQD (Waters) using an ACQUITY UPLC BEH C18 1.7 µm (2.1 × 50 mm; Waters) connected to an ACQUITY UPLC BEH C18 VanGuard Precolumn (2.1 × 5 mm; Waters). The mobile phase consisting of solution A (5 mm ammonium bicarbonate, pH 10) and solution B (acetonitrile) was used at the flow rate of 0.4 min/mL under the following gradient conditions: 0 to 1.0 min isocratic with 95% (v/v) solution A, 1.0 to 6.0 min linear gradient from 95% (v/v) to 40% (v/v) solution A, 6.0 to 7.5 min isocratic with 5% (v/v) solution A, 7.5 to 9.0 min wash with 100% (v/v) solution B, 9.0 to 12.5 min equilibration with 95% (v/v) solution A. The injection volume was 2 µL and column temperature was 40°C. For the detection of prenyl diphosphates, MS/MS was performed using negative ion mode and multiple reaction monitoring mode (m/z of precursor/product ion, GPP, 313.2/78.8; FPP, 381.3/78.8; GGPP, 449.4/78.8). To optimize the detectability, the MS parameters were adjusted as follows: capillary voltage 3.0 kV, cone voltage 35 V, collision energy 15 V, source temperature 200°C, desolvation temperature 200°C, cone gas flow 50 L/h, and desolvation gas flow 500 L/h.
Kinetic Analysis
Six concentrations, each of DMAPP (1-20 µm) or GPP (0.5-3 µm) in the presence of 100 µm IPP, or IPP (1-12 µm) in the presence of or 60 µm DMAPP, were used for the enzyme reactions, respectively. A reaction mixture containing 0.1 to 0.5 µg of purified His-tagged protein, 6.25 mm MOPS, 2.5% (v/v) glycerol, 1.25 mm DTT, 2.5 mm MgCl2, 1.25 mm sodium ascorbate, IPP, and DMAPP or GPP was incubated for 3 to 5 min at 30°C. The reaction was terminated with liquid nitrogen. The detection of reaction products was performed in the same manner as mentioned above. For the calculation of kinetic constants, analytical curves of GPP and FPP were made from peak area of authentic standards in a series of concentrations. The kinetic parameters of LeGPPS and LeFPPS1 were determined based on Lineweaver-Burk plot.
Extraction and Quantification of Shikonin Derivatives
Shikonin derivatives were extracted from roots of L. erythrorhizon with 4 mL hexane. Then 2 mL of 2.5% (w/v) KOH was added to the organic extract to collect shikonin derivatives into the aqueous phase. The concentration of shikonin was determined by measuring the A650 on a UV-1280 spectrophotometer (Shimadzu), with standard curves generated from the A650 of a standard shikonin dilution series. In cell suspension cultures, 3 mL of liquid paraffin and 3 mL hexane were added to 30 mL of culture medium to trap shikonin derivatives in the organic layer. The organic phase was collected and partitioned with 2 mL of 2.5% (w/v) KOH to solubilize shikonin derivatives in the aqueous phase. The concentration of shikonin derivatives in aqueous phase was analyzed by measuring the A650.
Qualitative Analysis of L. erythrorhizon Root Extract by UPLC-MS/MS
Shikonin derivatives were extracted from approximately 0.1 g of dried roots of L. erythrorhizon with 2 mL methanol. The dried roots were prepared by incubating the fresh roots in an oven of 50°C for 1 d. The diluted extract (1:10) was analyzed by UPLC-MS/MS with the same system mentioned above. The mobile phase consisting of solution A (0.1% [v/v] formic acid) and solution B (acetonitrile) was used at the flow rate of 0.3 min/mL under the isocratic condition (A:B = 3:7). The injection volume was 2 µL and column temperature was 40°C. Each shikonin derivative was detected by MS/MS using negative ion mode and multiple reaction monitoring mode (m/z of precursor/product ion:acetylshikonin, 329.1/269.1; deoxyshikonin, 272.2/203.1; β,β-dimethylacrylshikonin, 369.5/269.1; β-hydroxyisovalerylshikonin, 387.6/269.1; isobutyrylshikonin, 357.4/269.1; α-methyl-n-butyrylshikonin, 371.5/269.1; shikonin, 287.2/218.1). The MS parameters were adjusted as follows: capillary voltage 2.5 kV, cone voltage 40 V, collision energy 20 V, source temperature 100°C, desolvation temperature 450°C, cone gas flow 50 L/h, and desolvation gas flow 500 L/h.
Expression Analysis of LeGPPS
Two-year-old intact plants were dissected into seven organs: flowers, seeds, upper leaves (on lateral stems), lower leaves (on main stem), stems, root barks, and peeled roots. Cultured L. erythrorhizon cells were grown under three conditions (LS medium in the dark and M9 medium in the light or dark). Total RNA was extracted from the samples using RNeasy plant mini kits (QIAGEN), with RNAs extracted from root bark and peeled root samples further extracted with 2× cetyltrimethylammonium bromide buffer (55 mm cetyltrimethylammonium bromide, 0.1 m Tris-HCl, 20 mm EDTA, 1.4 m NaCl, and 2% [v/v] 2-mercaptoethanol, pH 8.0), chloroform, and 2.5 m LiCl to remove phenolic metabolites and polysaccharides. cDNA was synthesized from each 0.1 µg RNA sample with ReverTra Ace qPCR RT master mix and gDNA Remover (Toyobo) in a 10-µL reaction volume. The relative abundance of LeGPPS in diluted cDNA (1:50) was assessed by reverse transcription quantitative PCR, performed on a CFX96 Deep Well real-time system (Bio-Rad) with Thunderbird SYBR qPCR mix (Toyobo) and the primers LeGPPS-qPCR-Fw and LeGPPS-qPCR-Rv (Supplemental Table S3). As an internal standard, a partial sequence of LeACT7 was amplified using the primers LeACT7-Fw and LeACT7-Rv (Supplemental Table S3). The amplification protocol consisted of an initial denaturation at 95°C for 1 min and 40 cycles of denaturation at 95°C for 10 s, annealing at 55°C for 15 s, and extension at 72°C for 30 s A melting curve was generated through additional cycles at 65°C to 95°C for 5 s each to determine primer specificity. The relative expression level of LeGPPS was calculated according to the ΔΔCt method.
Transient Expression of GFP Fusion Protein in Onion (Allium cepa) Epidermal Cells
The full coding sequence of LeGPPS cDNA without a stop codon was amplified using the primers LeGPPS-Fw3 and LeGPPS-Rv3 (Supplemental Table S3) and subcloned into pT7 vector (Invitrogen). After confirmation of the sequence, LeGPPS was fused with soluble-modified red-shifted GFP (Davis and Vierstra, 1998) to yield Cauliflower mosaic virus 35S::LeGPPS::GFP. The cytosolic control GFP consisted of Cauliflower mosaic virus 35S-driven soluble-modified red-shifted GFP, whereas the plastidic control pWxTP::DsRed has been described (Kitajima et al., 2009). Particle bombardment and microscopic analysis were performed as described (Sasaki et al., 2008).
Phylogenetic Analysis and Multiple Sequence Alignment
The amino acid sequences of LeGPPS and LeFPPS1 were deduced by ExPASy translation tools (https://web.expasy.org/translate/), whereas the amino acid sequences of the other transprenyl diphosphate synthases were obtained from GenBank (https://www.ncbi.nlm.nih.gov/genbank) and Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). Multiple sequence alignment was performed by ClustalW through BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) with default settings, and a neighbor-joining tree and maximum likelihood trees were constructed using MEGA7 (http://evolgen.biol.se.tmu.ac.jp/MEGA) in default mode.
Site-Directed Mutagenesis of LeGPPS and LeFPPS1
His-100 in LeGPPS was mutated using a QuikChange site-directed mutagenesis kit (Stratageme, Agilent Technologies) according to the manufacturer’s instructions with the primers LeGPPS-H100L-Fw and LeGPPS-H100L-Rv (Supplemental Table S3). Leu-100 in LeFPPS1 was mutated by overlap extension PCR using the primers LeFPPS1-L100H-Fw and LeFPPS1-L100H-Rv (Supplemental Table S3) and KOD-Plus-Neo (TOYOBO). The obtained amplicons were subcloned into pET22b(+), and the identity of each insert was verified by sequencing.
GC-MS Analysis for Volatile Compounds
The extracts were prepared from flowers, seeds, upper leaves, lower leaves, and stems of intact L. erythrorhizon plants with 1 to 2 mL hexane that were analyzed by GC-MS on a GC-2010 (Shimadzu) coupled with GCMS-QP 2010 Plus (Shimadzu) using a DB-5ms column (Agilent Technologies, 30 m length, 0.25 mm inner diameter, 0.25 µm film). Helium was used as a carrier gas, and samples were injected at 240°C and a flow rate of 1.9 mL/min. Volatile compounds were separated by a temperature gradient, consisting of 50°C for 5 min, 10°C/min to 240°C, and a final 6 min hold at 240°C.
Accession Numbers
DDBJ/GenBank/EMBL accession numbers for the genes identified in this article are LC427363 for LeGPPS, and LC427365 and LC519333 for LeFPPS1 and LeFPPS2, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Shikonin derivatives produced in roots of L. erythrorhizon.
Supplemental Figure S2. N-terminal sequence of LeGPPS cDNA.
Supplemental Figure S3. SDS-PAGE and CBB staining of LeGPPS recombinant protein.
Supplemental Figure S4. Biosynthetic pathway of shikonin and dihydroechinofuran.
Supplemental Figure S5. N-terminal sequence alignment of transprenyl diphosphate synthases.
Supplemental Figure S6. Detection of transprenyl diphosphate after enzyme assay by UPLC-MS/MS.
Supplemental Figure S7. Unrooted phylogenetic analysis of deduced amino acid sequences of plant FPPS homologs.
Supplemental Figure S8. Multiple alignment of deduced amino acid sequences of LeGPPS and LeFPPS1 with FPPS homologs from Rosaceae plants.
Supplemental Figure S9. GC-MS analysis of plant extracts of L. erythrorhizon.
Supplemental Figure S10. Additional alignment of amino acid sequences of LeGPPS and LeFPPS1 with transprenyl diphosphate synthases.
Supplemental Table S1. List of putative transprenyl diphosphate synthase genes in L. erythrorhizon.
Supplemental Table S2. List of putative terpenoid synthase genes in L. erythrorhizon.
Supplemental Table S3. Primer sequences used in this study.
Acknowledgments
We thank Dr. Wolf B. Frommer of the Carnegie Institution for Science, Toshiaki Mitsui of Niigata University, and Amato Pharmaceutical Products for providing the pDR196 vector, pWxTP::DsRed vector, and intact L. erythrorhizon plants, respectively. We also thank the DASH system of the Research Institute for Sustainable Humanosphere (RISH), Kyoto University, for providing the GC-MS facility.
Footnotes
This work was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS; no. 19H05638 to K.Y.), a grant from the New Energy and Industrial Technology Development Organization (NEDO) project (no. 16100890 to K.Y.), and a grant from mission research of RISH, Kyoto University.
Articles can be viewed without a subscription.
References
- Abbas F, Ke Y, Yu R, Yue Y, Amanullah S, Jahangir MM, Fan Y (2017) Volatile terpenoids: Multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta 246: 803–816 [DOI] [PubMed] [Google Scholar]
- Arimura G, Ozawa R, Maffei ME (2011) Recent advances in plant early signaling in response to herbivory. Int J Mol Sci 12: 3723–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigham LA, Michaels PJ, Flores HE (1999) Cell-specific production and antimicrobial activity of naphthoquinones in roots of Lithospermum erythrorhizon. Plant Physiol 119: 417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke C, Croteau R (2002a) Interaction with the small subunit of geranyl diphosphate synthase modifies the chain length specificity of geranylgeranyl diphosphate synthase to produce geranyl diphosphate. J Biol Chem 277: 3141–3149 [DOI] [PubMed] [Google Scholar]
- Burke C, Croteau R (2002b) Geranyl diphosphate synthase from Abies grandis: cDNA isolation, functional expression, and characterization. Arch Biochem Biophys 405: 130–136 [DOI] [PubMed] [Google Scholar]
- Byers KJ, Bradshaw HD Jr., Riffell JA (2014) Three floral volatiles contribute to differential pollinator attraction in monkeyflowers (Mimulus). J Exp Biol 217: 614–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TH, Hsieh FL, Ko TP, Teng KH, Liang PH, Wang AHJ (2010) Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation. Plant Cell 22: 454–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Kroon PA, Poulter CD (1994) Isoprenyl diphosphate synthases: protein sequence comparisons, a phylogenetic tree, and predictions of secondary structure. Protein Sci 3: 600–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunillera N, Boronat A, Ferrer A (1997) The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J Biol Chem 272: 15381–15388 [DOI] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36: 521–528 [DOI] [PubMed] [Google Scholar]
- Fischer MJC, Meyer S, Claudel P, Bergdoll M, Karst F (2011) Identification of a lysine residue important for the catalytic activity of yeast farnesyl diphosphate synthase. Protein J 30: 334–339 [DOI] [PubMed] [Google Scholar]
- Francis MJO, O’Connell M (1969) The incorporation of mevalonic acid into rose petal monoterpenes. Phytochemistry 8: 1705–1708 [Google Scholar]
- Fujita Y, Hara Y (1985) The effective production of shikonin by cultures with an increased cell population. Agric Biol Chem 49: 2071–2075 [Google Scholar]
- Fujita Y, Hara Y, Ogino T, Suga C (1981a) Production of shikonin derivatives by cell suspension cultures of Lithospermum erythrorhizon: I. Effects of nitrogen sources on the production of shikonin derivatives. Plant Cell Rep 1: 59–60 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Hara Y, Suga C, Morimoto T (1981b) Production of shikonin derivatives by cell suspension cultures of Lithospermum erythrorhizon: II. A new Medium for the production of shikonin derivatives. Plant Cell Rep 1: 61–63 [DOI] [PubMed] [Google Scholar]
- Fukui H, Tani M, Tabata M (1992) An unusual metabolite, dihydroechinofuran, released from cultured cells of Lithospermum erythrorhizon. Phytochemistry 31: 519–521 [Google Scholar]
- Gin P, Clarke CF (2005) Genetic evidence for a multi-subunit complex in coenzyme Q biosynthesis in yeast and the role of the Coq1 hexaprenyl diphosphate synthase. J Biol Chem 280: 2676–2681 [DOI] [PubMed] [Google Scholar]
- Hampel D, Mosandl A, Wüst M (2006) Biosynthesis of mono- and sesquiterpenes in strawberry fruits and foliage: 2H labeling studies. J Agric Food Chem 54: 1473–1478 [DOI] [PubMed] [Google Scholar]
- Heide L, Berger U (1989) Partial purification and properties of geranyl pyrophosphate synthase from Lithospermum erythrorhizon cell cultures. Arch Biochem Biophys 273: 331–338 [DOI] [PubMed] [Google Scholar]
- Heide L, Nishioka N, Fukui H, Tabata M (1989) Enzymatic regulation of shikonin biosynthesis in Lithospermum erythrorhizon cell cultures. Phytochemistry 28: 1873–1877 [Google Scholar]
- Hemmerlin A, Rivera SB, Erickson HK, Poulter CD (2003) Enzymes encoded by the farnesyl diphosphate synthase gene family in the Big Sagebrush Artemisia tridentata ssp. spiciformis. J Biol Chem 278: 32132–32140 [DOI] [PubMed] [Google Scholar]
- Kessler A, Heil M (2011) The multiple faces of indirect defences and their agents of natural selection. Funct Ecol 25: 348–357 [Google Scholar]
- Kitajima A, Asatsuma S, Okada H, Hamada Y, Kaneko K, Nanjo Y, Kawagoe Y, Toyooka K, Matsuoka K, Takeuchi M, et al. (2009) The rice α-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 21: 2844–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM. (2015) The evolution of plant secretory structures and emergence of terpenoid chemical diversity. Annu Rev Plant Biol 66: 139–159 [DOI] [PubMed] [Google Scholar]
- Lee JS, Pan JJ, Ramamoorthy G, Poulter CD (2017) Structure-function studies of Artemisia tridentata farnesyl diphosphate synthase and chrysanthemyl diphosphate synthase by site-directed mutagenesis and morphogenesis. J Am Chem Soc 139: 14556–14567 [DOI] [PubMed] [Google Scholar]
- Lee PC, Petri R, Mijts BN, Watts KT, Schmidt-Dannert C (2005) Directed evolution of Escherichia coli farnesyl diphosphate synthase (IspA) reveals novel structural determinants of chain length specificity. Metab Eng 7: 18–26 [DOI] [PubMed] [Google Scholar]
- Li SM, Hennig S, Heide L (1998) Shikonin: A geranyl diphosphate-derived plant hemiterpenoid formed via the mevalonate pathway. Tetrahedron Lett 39: 2721–2724 [Google Scholar]
- Liao P, Hemmerlin A, Bach TJ, Chye ML (2016) The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol Adv 34: 697–713 [DOI] [PubMed] [Google Scholar]
- Linsmaier EM, Skoog F (1965) Organic growth factor requirements of tobacco tissue cultures. Physiol Plant 18: 100–127 [Google Scholar]
- Lu L, Qin A, Huang H, Zhou P, Zhang C, Liu N, Li S, Wen G, Zhang C, Dong W, et al. (2011) Shikonin extracted from medicinal Chinese herbs exerts anti-inflammatory effect via proteasome inhibition. Eur J Pharmacol 658: 242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín D, Piulachs MD, Cunillera N, Ferrer A, Bellés X (2007) Mitochondrial targeting of farnesyl diphosphate synthase is a widespread phenomenon in eukaryotes. Biochim Biophys Acta 1773: 419–426 [DOI] [PubMed] [Google Scholar]
- Matsuda K, Kikuta Y, Haba A, Nakayama K, Katsuda Y, Hatanaka A, Komai K (2005) Biosynthesis of pyrethrin I in seedlings of Chrysanthemum cinerariaefolium. Phytochemistry 66: 1529–1535 [DOI] [PubMed] [Google Scholar]
- Mizukami H, Konoshima M, Tabata M (1977) Effect of nutritional factors on shikonin derivative formation in Lithospermum callus cultures. Phytochemistry 16: 1183–1186 [Google Scholar]
- Narita K, Ohnuma S, Nishino T (1999) Protein design of geranyl diphosphate synthase. Structural features that define the product specificities of prenyltransferases. J Biochem 126: 566–571 [DOI] [PubMed] [Google Scholar]
- Qin X, Li W, Liu Y, Tan M, Ganal M, Chetelat RT (2018) A farnesyl pyrophosphate synthase gene expressed in pollen functions in S-RNase-independent unilateral incompatibility. Plant J 93: 417–430 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB (1995) NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370: 264–268 [DOI] [PubMed] [Google Scholar]
- Rubat S, Varas I, Sepúlveda R, Almonacid D, González-Nilo F, Agosin E (2017) Increasing the intracellular isoprenoid pool in Saccharomyces cerevisiae by structural fine-tuning of a bifunctional farnesyl diphosphate synthase. FEMS Yeast Res 17: fox032. [DOI] [PubMed] [Google Scholar]
- Ruiz-Sola MÁ, Barja MV, Manzano D, Llorente B, Schipper B, Beekwilder J, Rodriguez-Concepcion M (2016) A single arabidopsis gene encodes two differentially targeted geranylgeranyl diphosphate synthase isoforms. Plant Physiol 172: 1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H, Hara R, Takahashi H, Iijima M, Munakata R, Kenmoku H, Fuku K, Sekihara A, Yasuno Y, Shinada T, et al. (2018) An aromatic farnesyltransferase functions in biosynthesis of the anti-HIV meroterpenoid daurichromenic acid. Plant Physiol 178: 535–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankawa U, Ebizuka Y, Miyazaki T, Isomura Y, Otsuka H (1977) Antitumor activity of shikonin and its derivatives. Chem Pharm Bull (Tokyo) 25: 2392–2395 [DOI] [PubMed] [Google Scholar]
- Sasaki K, Mito K, Ohara K, Yamamoto H, Yazaki K (2008) Cloning and characterization of naringenin 8-prenyltransferase, a flavonoid-specific prenyltransferase of Sophora flavescens. Plant Physiol 146: 1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Gershenzon J (2008) Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies). Phytochemistry 69: 49–57 [DOI] [PubMed] [Google Scholar]
- Sommer S, Severin K, Camara B, Heide L (1995) Intracellular localization of geranylpyrophosphate synthase from cell cultures of Lithospermum erythrorhizon. Phytochemistry 38: 623–627 [Google Scholar]
- Stanley Fernandez SM, Kellogg BA, Poulter CD (2000) Farnesyl diphosphate synthase. Altering the catalytic site to select for geranyl diphosphate activity. Biochemistry 39: 15316–15321 [DOI] [PubMed] [Google Scholar]
- Tabata M, Mizukami H, Hiraoka N, Konoshima M (1974) Pigment formation in callus cultures of Lithospermum erythrorhizon. Phytochemistry 13: 927–932 [Google Scholar]
- Takanashi K, Nakagawa Y, Aburaya S, Kaminade K, Aoki W, Saida-Munakata Y, Sugiyama A, Ueda M, Yazaki K (2019) Comparative proteomic analysis of Lithospermum erythrorhizon reveals regulation of a variety of metabolic enzymes leading to comprehensive understanding of the shikonin biosynthetic pathway. Plant Cell Physiol 60: 19–28 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Odani T (1972) [Pharmacodynamic study on “Shiunko”. I. Antibacterial effect of “Shiunko”]. Yakugaku Zasshi 92: 525–530 [DOI] [PubMed] [Google Scholar]
- Tarshis LC, Proteau PJ, Kellogg BA, Sacchettini JC, Poulter CD (1996) Regulation of product chain length by isoprenyl diphosphate synthases. Proc Natl Acad Sci USA 93: 15018–15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi K, Yano M, Kaminade K, Sugiyama A, Sato M, Toyooka K, Aoyama T, Sato F, Yazaki K (2016) Characterization of Shikonin derivative secretion in Lithospermum erythrorhizon hairy roots as a model of lipid-soluble metabolite secretion from plants. Front Plant Sci 7: 1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D. (2015) Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol 148: 63–106 [DOI] [PubMed] [Google Scholar]
- Tholl D, Kish CM, Orlova I, Sherman D, Gershenzon J, Pichersky E, Dudareva N (2004) Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell 16: 977–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermoten S, Charloteaux B, Santini S, Sen SE, Béliveau C, Vandenbol M, Francis F, Brasseur R, Cusson M, Haubruge E (2008) Characterization of a novel aphid prenyltransferase displaying dual geranyl/farnesyl diphosphate synthase activity. FEBS Lett 582: 1928–1934 [DOI] [PubMed] [Google Scholar]
- Vandermoten S, Santini S, Haubruge E, Heuze F, Francis F, Brasseur R, Cusson M, Charloteaux B (2009) Structural features conferring dual geranyl/farnesyl diphosphate synthase activity to an aphid prenyltransferase. Insect Biochem Mol Biol 39: 707–716 [DOI] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W (2013) Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 64: 665–700 [DOI] [PubMed] [Google Scholar]
- Wang G, Dixon RA (2009) Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc Natl Acad Sci USA 106: 9914–9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K. (2017) Lithospermum erythrorhizon cell cultures: Present and future aspects. Plant Biotechnol (Tokyo) 34: 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K, Fukui H, Tabata M (1986) Accumulation of p-O-β-D-glucosylbenzoic acid and its relation to shikonin biosynthesis in Lithospermum cell cultures. Phytochemistry 25: 1629–1632 [Google Scholar]
- Yazaki K, Kunihisa M, Fujisaki T, Sato F (2002) Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon. Cloning and characterization of a ket enzyme in shikonin biosynthesis. J Biol Chem 277: 6240–6246 [DOI] [PubMed] [Google Scholar]
- Yuba A, Yazaki K, Tabata M, Honda G, Croteau R (1996) cDNA cloning, characterization, and functional expression of 4S-(-)-limonene synthase from Perilla frutescens. Arch Biochem Biophys 332: 280–287 [DOI] [PubMed] [Google Scholar]
- Zwenger S, Basu C (2008) Plant terpenoids : Applications and future potentials. Biotechnol Mol Biol Rev 3: 1–7 [Google Scholar]