Abstract
Lateral root development progresses through different steps with, the peptides and receptors involved in each of these steps triggering downstream mechanisms upon peptide perception.
Plant root systems enable nutrient and water uptake as well as anchorage to the substrate. Nutrient and water availability are known to affect root system architecture, and certain root traits can be linked to nutrient and water use efficiency (Comas et al., 2013; Uga et al., 2013; Li et al., 2016; Duque and Villordon, 2019; Sandhu et al., 2019). A better understanding of how root system architecture is determined could therefore be extremely valuable for crop yield enhancement. Regular spacing of lateral roots (LRs), as well as initiation and development of LR primordia, is tightly regulated in Arabidopsis (Arabidopsis thaliana). However, LR development is readily influenced by external cues, ensuring the root system architecture is highly adaptable to different environmental conditions (Tian et al., 2014; Satbhai et al., 2015). To achieve such strict regulation while maintaining a high degree of flexibility, LR development relies on strong intercellular communication networks, mediated by the exchange of molecular messengers over both short and long distances. Several types of molecular messengers are known to regulate plant development, including phytohormones, reactive oxygen species, mobile transcription factors, and small noncoding RNAs (van Norman et al., 2011). However, it is becoming increasingly clear that a multitude of secreted signaling peptides and their transmembrane receptors are involved in the control of numerous physiological and developmental processes, including LR development. Over the past few years, involvement of multiple peptide signaling pathways has been uncovered in several aspects of LR development. Here, we provide an overview of the peptide-receptor modules currently known to play a role in different steps of the LR developmental process in Arabidopsis.
SIGNALING PEPTIDES CONTROLLING LR INITIATION
TOLS2 Signaling Controls the Lateral Inhibition of LR Founder Cell Specification
The regions of the primary root that experience a peak of DR5:LUCIFERASE (DR5:LUC) expression in the elongation zone (Box 1) are relatively broad and typically encompass at least 10 pericycle cells from which only one pair will become LR founder cells (LRFCs; de Smet et al., 2007; Moreno-Risueno et al., 2010). This has led to the assumption that from early on, inhibitory mechanisms are at play that would avoid overproliferation of the pericycle and prevent initiation of surplus LR primordia. It has recently been shown that TARGET OF LBD SIXTEEN2 (TOLS2) and a close homolog called PLASMA MEMBRANE INTRINSIC PROTEIN2 (PIP2), together with their transmembrane receptor RECEPTOR-LIKE KINASE7 (RLK7), are involved in the selection of LRFCs within this region (Toyokura et al., 2019). TOLS2 is expressed in LRFCs and developing LR primordia and its transcription is auxin-inducible via the activity of LATERAL ORGAN BOUNDARIES-DOMAIN16 (LBD16), a transcriptional activator with a demonstrated role in LR formation (Okushima et al., 2007; Lee et al., 2009; Goh et al., 2012). TOLS2 overexpression and treatment with synthetic TOLS2 or PIP2 peptides reduces the density of prebranch sites, resulting in a reduction in total LR primordium density. RLK7 is expressed in the pericycle, endodermis, and cortex from the differentiation zone onwards. Its expression is strongest in the pericycle but is absent in LRFCs and LR primordia, where TOLS2 is expressed. Loss-of-function rlk7 mutants exhibit an increased prebranch site density and prebranch sites are often found in close proximity to each other, a phenotype also observed in tols2pip2 double mutants. These findings indicate that TOLS2, PIP2, and RLK7 are negative regulators of LRFC specification, which is in agreement with the presumed role of LBD16 during prebranch site formation (Goh et al., 2012). Prebranch site analysis using the DR5:LUC marker indicated that the regions of the primary root that were primed in the elongation zone often contain two pairs of LRFCs in close proximity to each other, even in wild-type roots, but that one of them is typically transient and disappears so that LR initiation is inhibited at this site. This inhibition is dependent on the activity of TOLS2 and RLK7. Transcriptome analysis has shown that PUCHI, a transcription factor known to control cell division patterning during LR primordium development (Hirota et al., 2007; Kang et al., 2013; Trinh et al., 2019), is induced by TOLS2 in an RLK7-dependent manner. In agreement with this, puchi mutants display increased prebranch site densities and increased frequencies of paired prebranch sites. Furthermore, puchi mutants show a reduced sensitivity to TOLS2 peptide treatments. Consistent with the expression patterns of RLK7, PUCHI is not expressed in LRFCs but is preferentially expressed in the adjacent cells in which RLK7 is thought to be activated by TOLS2 to induce its transcription. Taken together, the TOLS2 signaling peptide thus seems to be involved in a lateral inhibition mechanism that prevents LR primordia from developing in close proximity to each other. This is achieved via the transcriptional activation of PUCHI in the regions flanking a LRFC pair through its receptor RLK7. In the case where a new prebranch site is established in close proximity to a pre-existing one, the TOLS2 signaling pathway represses LRFC identity in one of them, thereby ensuring proper LR spacing (Fig. 1). Interestingly, the increased prebranch site density in rlk7 mutants does not result in an increase in LR density as it does in puchi mutants, suggesting that an additional lateral inhibition mechanism is at play after prebranch site formation that might also work via PUCHI.
Figure 1.
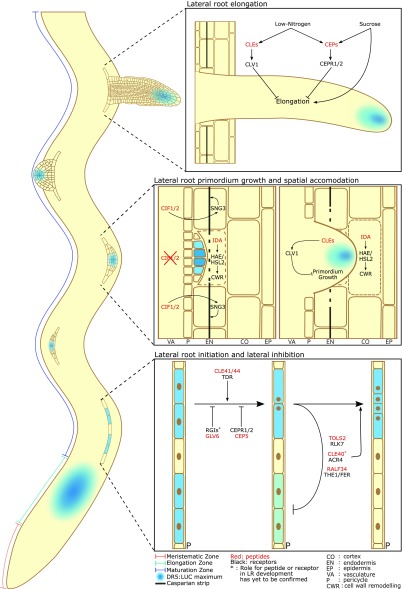
Peptide-receptor signaling throughout LR development. Overview of the different steps of LR development and the peptide-receptor pairs currently known to be involved in each of these steps. During a DR5:LUC maximum in the elongation zone, XPP cells are primed for LR formation. In the maturation zone, LR initiation takes place, a process that is influenced by the stimulatory and inhibitory effects of multiple peptide signaling pathways. Additionally, several signaling peptides repress LR development in the vicinity of pre-existing primordia in a process called “lateral inhibition.” During later developmental stages, several peptide-receptor pathways affect LR primordium growth and its spatial accommodation in the surrounding tissues, allowing for emergence from the primary root. Finally, signaling peptides also control the elongation of LRs in response to environmental stimuli.
Apart from TOLS2, other peptides from the same family were also found to show an inhibitory effect on LR density (Ghorbani et al., 2015), but whether they affect the same pathway or trigger an independent one has yet to be discovered.
CLE Peptides Play Diverse Roles during LR Initiation
The CLAVATA3/ESR-RELATED1 (CLE) family is a large family of peptides, the most well studied of which is CLAVATA3 (CLV3), which plays an important role in the control of stem cell differentiation in the shoot apical meristem (Clark et al., 1997; Fletcher et al., 1999; Brand et al., 2000; Schoof et al., 2000; Ogawa et al., 2008; Nimchuk et al., 2011). Several CLE genes show distinct but often overlapping expression patterns throughout LR primordium development and some were found to be expressed at the base of the elongation zone, which may indicate a role in the priming of xylem pole pericycle (XPP) cells (Jun et al., 2010; Czyzewicz et al., 2015). Synthetic CLE1, CLE4, CLE7, CLE26, and CLE27 peptide treatments were found to increase emerged LR densities, suggesting a stimulatory role on LR initiation (Czyzewicz et al., 2015). However, these results should be interpreted with caution because this increase coincided with a strong decrease in primary root length, rendering LR density measurements rather unreliable. Furthermore, as will be discussed below, overexpression of CLE1, CLE4, and CLE7 has been reported to result in a strong reduction in emerged LR density without affecting primary root length (Araya et al., 2014). Nonetheless, CLE26 peptide treatments can induce the formation of a limited number of LRs in arf7arf19 double mutants, in which the canonical auxin signaling required for LR initiation is almost completely inhibited (Okushima et al., 2005), suggesting a stimulatory role of CLE26 on LR initiation downstream of AUXIN RESPONSE FACTOR7 (ARF7) and ARF19.
Two other CLE peptides, CLE41 and CLE44, both encoding the same mature peptide known as TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF), have also been shown to positively regulate LR initiation (Cho et al., 2014). TDIF and its receptor TDIF RECEPTOR (TDR) are known to suppress vascular stem cell differentiation into xylem (Hirakawa et al., 2008) and have been shown to stimulate LR initiation by regulating the transcriptional activity of ARF7 and ARF19 (Cho et al., 2014). TDIF peptide treatments increase emerged LR density in a TDR-dependent manner while tdr loss-of-function mutants show reduced emerged LR densities. The kinase domain of TDR was found to interact with BRASSINOSTEROID-INSENSITIVE2 (BIN2), which has been implicated in the regulation of auxin and brassinosteroid signaling (Pérez-Pérez et al., 2002; Vert et al., 2008). BIN2 gain-of-function mutants show an increased LR density while loss-of-function mutants exhibit a reduction, indicating that BIN2 positively regulates LR development. Furthermore, BIN2 gain-of-function mutants were found to be hypersensitive to auxin (Pérez-Pérez et al., 2002; Cho et al., 2014). BIN2 can directly interact with ARF7 and is able to phosphorylate both ARF7 and ARF19. This BIN2-mediated phosphorylation attenuates the interaction of these ARFs with AUXIN/INDOLE-3-ACETIC ACID INDUCIBLE proteins (Aux/IAA), thereby enlarging the pool of free ARF7 and ARF19 transcriptional activators that are able to bind their target gene promoters, including LBD16 and LBD29. BIN2 is expressed in XPP, epidermis, and cortex cells, but is restricted to the vasculature in the elongation zone. During LR development, BIN2 expression was observed in LR initiation sites and the basal part of dome-shaped LR primordia. Similarly, TDR is expressed in the vasculature, the pericycle and during LR initiation. TDIF peptides, on the other hand, are mainly expressed in the phloem and their expression can be induced by auxin (Goda et al., 2008), suggesting that auxin-induced TDIF in the phloem travels to the pericycle where it stimulates TDR-induced activation of BIN2, which in turn attenuates the inhibitory activity of Aux/IAAs on ARF7 and ARF19, thereby reinforcing auxin signaling for the initiation of LR development.
The receptor-like kinase ARABIDOPSIS CRINKLY4 (ACR4) was identified as an auxin-inducible regulator of the first asymmetric anticlinal cell division upon LR initiation and is expressed specifically in the small central daughter cells resulting from this initial asymmetric cell division (Malamy and Benfey, 1997; de Smet et al., 2008). A significant increase in LR primordium density can be observed in acr4 mutants as a result of an increased density in LR initiation events. Furthermore, LR primordia are often initiated close to one another, sometimes even opposing each other, and double-layered stretches of pericycle cells or fused primordia have also been observed. Despite the increase in total LR primordium density, emerged LR densities are significantly reduced in arc4 mutants. Conversely, ectopic expression of ACR4 specifically in the XPP results in an increase in emerged LR density. Similar to the lateral inhibition mechanism that has been described for the TOLS2-RLK7 signaling module, ACR4 is believed to repress divisions in pericycle cells surrounding LRFCs, while simultaneously promoting the correct organization of the initial LRFC divisions, thereby ensuring proper LR spacing and initiation. A peptide ligand for ACR4 during LR initiation has not yet been identified. However, CLE40 has been proposed as a ligand for ACR4 in the root apical meristem where the CLE40-ACR4/CLV1 signaling module regulates the fate of root stem cells (Stahl et al., 2009, 2013; Berckmans et al., 2020).
RALF34 Is Involved in LR Initiation and Lateral Inhibition
RAPID ALKALINIZATION FACTOR (RALF) peptides are known to be involved in the regulation of various processes, primarily as inhibitors of cell elongation (Bergonci et al., 2014; Haruta et al., 2014). T-DNA knock-down lines of RALF34 display an increase in total LR primordium density, primarily due to an increased density of stage-I primordia (Murphy et al., 2016). Additionally, primordia were often found in close proximity to one another and aberrant pericycle cell divisions flanking LR primordia were regularly observed. RALF34 starts to be expressed in XPP cells before there is any visible sign of LR initiation and continues to be expressed in LR primordia during the whole developmental process. Additionally, RALF34 is expressed in the pericycle cells flanking LR primordia. This flanking expression, in combination with the increased density of stage-I primordia and the aberrant spacing of LR primordia in ralf34 T-DNA knock-down mutants, suggests that RALF34 serves as a negative regulator of LR initiation and probably acts to prevent LR initiation in close proximity to existing primordia. Once again, this points to the involvement of a peptide signaling pathway in a lateral inhibition mechanism that mediates the spatial distribution of LRs along the primary root. The malectin receptor kinase THESEUS1 (THE1) has been identified as a receptor for RALF34 (Gonneau et al., 2018). THE1 is expressed throughout the stele, including the pericycle, and in developing LR primordia, and the1 loss-of-function mutants display the same defects as ralf34 knock-down mutants. In addition to THE1, RALF34 signaling also seems to require FERONIA (FER), a receptor kinase that is known to perceive other RALF peptides as well (Haruta et al., 2014; Gonneau et al., 2018). In agreement with this, it has recently been shown that fer mutants have increased LR densities (Dong et al., 2019). Although parallel THE1 and FER pathways can currently not be ruled out, it is likely that the THE1 and FER pathways downstream of RALF34 somehow converge, possibly already at the level of these receptors themselves. The the1-4 mutant, which lacks the cytosolic domain, is still sensitive to RALF34 peptide treatment, indicating that its intracellular domain is not required for RALF34 signaling. THE1 might thus rather act as a coreceptor for FER, allowing it to interact with the RALF34 peptide. The downstream mechanism via which the RALF34-THE1/FER module regulates LR initiation has not yet been uncovered. However, it has been shown that RALF1-FER signaling results in the phosphorylation of the proton exporter PLASMA MEMBRANE H+-ATPase2 (AHA2), thereby reducing its activity and preventing acidification of the apoplast, which is required for cell wall loosening and cell expansion during primary root growth (Haruta et al., 2014). Because swelling of LRFCs is important for LR initiation (Ramakrishna et al., 2019; Vilches Barro et al., 2019), a similar mechanism could potentially be at play downstream of RALF34 during LR development. Alternatively, RALF34 induced changes in apoplastic pH might interfere with auxin flows, thereby impacting LRFC divisions. It has been shown that aha2 mutants show significantly reduced frequencies of LR initiation (Młodzińska et al., 2015), further suggesting that it could be part of the RALF34-THE1/FER pathway. In agreement with the inhibitory effect of RALF34 on LR initiation, RALF1 and RALF8 have also been found to reduce LR densities (Atkinson et al., 2013; Bergonci et al., 2014), suggesting that other family members share the same function.
GLV6 Disrupts the Essential Asymmetry of the First LRFC Division
GOLVEN (GLV) peptides, also known as ROOT GROWTH FACTORS or CLE-LIKE peptides, are mainly known for their role in primary root growth and root apical meristem maintenance, but their expression patterns and overexpression phenotypes indicate that at least some GLV peptides also play an role during LR initiation (Meng et al., 2012; Fernandez et al., 2013, 2015). Out of the 11 GLV genes that are encoded in the Arabidopsis genome, eight are expressed during LR primordium development. These GLV genes are sequentially induced at different stages (Malamy and Benfey, 1997) of primordium development in a fixed order, with GLV6 and GLV10 as the earliest ones, already being expressed upon LR initiation (Fernandez et al., 2015). Overexpression of several GLV genes triggers aberrant anticlinal cell divisions throughout the whole pericycle resulting in a strong reduction in LR densities (Meng et al., 2012; Fernandez et al., 2013). This effect can be mimicked with GLV peptide treatments and is strongest for GLV peptides that are expressed early on during LR development. Due to the early onset of its expression upon LR initiation, the role of GLV6 during LR development has been analyzed more thoroughly (Fernandez et al., 2015). GLV6 starts to be expressed in LRFCs before the onset of nuclear migration (see Box 1). As is the case for several other GLVs, GLV6 overexpression and peptide treatments trigger supernumerary anticlinal divisions throughout the XPP. Ectopic expression of GLV6 in different root tissue layers indicated that this effect can almost be completely replicated upon expression in the XPP alone but is a lot weaker when expressed in the overlying tissues. This suggests that GLV6 peptides function as autocrine signals that are produced as well as perceived in the XPP. Careful analysis of the effect of GLV6 during LR initiation indicated that the nuclear migration in LRFCs is disrupted upon treatment with GLV6 peptide. As a result, the first anticlinal cell division of LRFCs loses its essential asymmetry, preventing further LR primordium development. GLV6 peptide signaling thus seems to be required for proper cell patterning upon LR initiation. The downstream mechanisms via which GLV6 exerts its effect on LR initiation have yet to be discovered. However, five Leucine-rich repeat receptor-like kinases (LRR-RLKs), called “ROOT GROWTH FACTOR INSENSITIVE” (RGI1, RGI2, RGI3, RGI4, RGI5), have been identified as receptors for GLV peptides, and GLV-RGI signaling was found to stimulate the expression of PLETHORA1 (PLT1) and PLT2 in the root apical meristem, both at the transcriptional and posttranscriptional level (Ou et al., 2016; Shinohara et al., 2016; Song et al., 2016). GLV peptide signaling is thus required for the installation of the typical PLT protein gradient in the root apical meristem, which is essential for the maintenance of the root stem cell niche and transit amplifying cell proliferation (Galinha et al., 2007). Because several PLT transcription factors also play a role during LR development (Hofhuis et al., 2013; Du and Scheres, 2017), it is possible that their expression is modulated via GLV peptide signaling during this process. In addition, GLV peptides were shown to affect root gravitropism by stimulating PIN-FORMED2 trafficking to the plasma membrane, thereby disrupting the asymmetric auxin distribution in the root tip required for a gravitropic response (Whitford et al., 2012). Because correct distribution of auxin is crucial for LR development, the GLV overexpression phenotypes could potentially result from the disruption of normal auxin fluxes upon LR initiation. As discussed above, several other signaling peptides that inhibit LR initiation upon overexpression or peptide treatment seem to be involved in the lateral inhibition of LR development. Thorough analysis of the LR phenotypes of glv and rgi mutants should determine whether this is also the case for the GLV6-RGI pathway.
CEPs Serve as Negative Regulators of LR Initiation
Several members of the C-TERMINALLY ENCODED PEPTIDE (CEP) family are expressed during LR development (Roberts et al., 2013). As will be discussed below, they have mainly been studied for their role in nitrogen (N) starvation responses and the associated inhibition of LR elongation, but some of them have also been shown to affect LR initiation. Under N-limiting conditions, cep3 loss-of-function mutants show increased total LR primordium numbers, while CEP3 peptide treatments result in a decrease, pointing to an inhibitory effect of CEP3 on LR development (Delay et al., 2013). No significant changes in the proportions of the different developmental stages were observed, suggesting that CEP3 inhibits LR initiation but does not affect further development once initiation has taken place. However, the authors report LR primordium numbers instead of densities, and because cep3 knock-out and CEP3 peptide treatment increase and decrease primary root length respectively, the reported changes in LR numbers might be an effect of changes in primary root length rather than LR initiation.
The role of CEP5 during LR initiation in Arabidopsis has been studied more extensively (Roberts et al., 2016). CEP5 is downregulated by auxin and its expression can be detected in the root tip at the start of the elongation zone and in association with LR primordia. In both cases, CEP5 is predominantly expressed in phloem pole pericycle cells and to a lesser extent in the adjacent phloem. Knocking down CEP5 expression causes an increased density of stage-I primordia and a faster progression through the LR developmental stages upon gravistimulation-induced LR initiation. CEP5 overexpression and synthetic peptide treatments, on the other hand, result in a decrease in total LR density, indicating a reduction in LR initiation events. Furthermore, aberrant pericycle divisions, as well as fused and closely spaced LR primordia were often observed upon overexpression or peptide treatment. As discussed above for several other peptides, the occurrence of these fused and clustered primordia might suggest that CEP5 is also involved in a lateral inhibition mechanism that prevents the development of LR primordia in close proximity to each other. However, in contrast to these other cases (TOLS2, ACR4, and RALF34), the occurrence of closely spaced LR primordia does not coincide with an increase in total LR density. Conversely, the increased occurrence of closely spaced primordia in CEP5-overexpressing seedlings is accompanied by a decrease in total LR density, suggesting another mechanism to be at play that requires further investigation. The LRR-RLKs CEP RECEPTOR1 (CEPR1)/XYLEM INTERMIXED WITH PHLOEM1 and CEPR2 serve as receptors for CEP1, CEP3, and CEP5 and potentially also other members of the CEP family (Tabata et al., 2014). In the root, CEPR1 starts to be expressed in the phloem pole pericycle and the adjacent phloem from the start of the elongation zone onwards and can only be detected in LRs after emergence. Loss-of-function cepr1 mutants were found to show a reduced sensitivity to CEP5 peptide treatments, supporting the role of CEPR1 as a CEP5 receptor during LR development. However, these mutants display a reduction in total LR density resulting from a reduction in LR initiation events (Roberts et al., 2016), reminiscent of the CEP5 overexpression phenotype. It was therefore suggested that CEP5 functions as a negative regulator of CEPR1 activity in the context of LR initiation (Roberts et al., 2016). According to this hypothesis, CEPR1 serves to promote LR initiation while its activity is suppressed upon binding with CEP5 peptide ligands. Conversely, another study reported that cepr1cepr2 double mutants showed an increase in emerged as well as nonemerged LR primordium densities (Dimitrov and Tax, 2018), suggesting that CEPRs might act as negative regulators of LR initiation. Thorough analysis of the LR phenotypes of different CEPR mutants in combination with CEP5 peptide treatments is required to resolve these contradictory results. Furthermore, because both CEP5 and CEPR1 are expressed in phloem pole pericycle cells, it is currently unclear how they affect LR initiation in XPP cells. A downstream mobile signal that travels from the phloem pole pericycle to the XPP might be involved but has yet to be identified.
SIGNALING PEPTIDES REGULATING THE SPATIAL ACCOMMODATION OF LR DEVELOPMENT
IDA-Induced Cell Separation Enables Growth of LR Primordia through the Overlying Tissues
The INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) signaling peptide and its receptors HAESA (HAE) and HAESA-LIKE2 (HSL2) are known to stimulate the cell separation that is required for the abscission of floral organs after pollination (Butenko et al., 2003; Stenvik et al., 2008). Because LRs originate from a tissue layer deep within the primary root, developing LR primordia have to grow through the overlying tissues, a process that was also found to rely on IDA-HAE/HSL2-induced cell separation (Kumpf et al., 2013; Zhu et al., 2019). Significant reductions in emerged LR densities can be observed in ida, hae, and hsl2 single mutants as well as haehsl2 double mutants. Gravistimulation experiments indicated that early stages of LR development are not affected in these mutants, but that a smaller percentage of LRs is able to emerge. Microscopic analysis indicated that LR primordia in these mutants have difficulty growing through the overlying endodermal, cortical, and epidermal tissues. Furthermore, primordia are often flattened and get stuck in tissue layers that have usually already been penetrated by the corresponding developmental stage in wild-type conditions. In agreement with these loss-of-function phenotypes, IDA, HAE, and HLS2 are expressed in the tissue layers covering developing LR primordia. Expression of IDA can already be detected in endodermal cells on top of stage-I primordia. Induction of IDA expression can subsequently be observed in the cortical and epidermal cells overlying a primordium right before it enters these tissue layers. Similarly, HAE and HSL2 start to be expressed in the cells overlying LR primordia from stages II to III onwards. Cell separation in the overlying tissues is induced by auxin coming from the developing primordium and accumulating in the cells above. Auxin entering the endodermis triggers the degradation of IAA3 while LIKE-AUX1 3 (LAX3)-mediated auxin influx in the cortex and epidermis triggers the degradation of IAA14 (Swarup et al., 2008). The breakdown of these Aux/IAAs releases the ARFs in these tissues, leading to the induction of cell wall remodeling genes, resulting in cell separation and spatial accommodation of the underlying LR primordium (Swarup et al., 2008; Vermeer et al., 2014). In agreement with this, expression of IDA is auxin-inducible, and the onset of IDA expression is delayed in lax3 mutants. Furthermore, the degradation of pectin in the middle lamella between endodermis and cortex cells is hampered in ida, hae, and haehsl2 mutant roots, and expression levels of several cell wall remodeling enzymes were found to be reduced. IDA signaling in floral organ abscission requires the activity of a MITOGEN-ACTIVATED PROTEIN KINASE (MPK) phosphorylation cascade, composed of MPKK4/5 and MPK6/3 (Cho et al., 2008; Meng et al., 2016) and it was recently discovered that this MPK module is also required for IDA-induced cell separation during the spatial accommodation of LR development (Zhu et al., 2019). IDA peptide treatment is able to induce MPK6 and MPK3 phosphorylation, but this effect is suppressed in mpkk4mpkk5 and haehsl2 double mutants. Furthermore, pectin degradation in the cell wall between endodermis and cortex cells covering LR primordia does not take place in these mutants and transcript levels of several cell wall remodeling genes are significantly reduced. The IDA-HAE/HSL2 signaling pathway thus seems to function as an auxin-inducible mechanism that triggers cell wall remodeling genes through a downstream MPK cascade in the cells that surround LR primordia, thereby enabling growing primordia to penetrate the overlying tissues.
Local Inactivation of CIF Signaling Might Be Required for Primordium Growth through the Endodermis
While the IDA-HAE/HSL2 peptide signaling pathway stimulates LR formation by accommodating its development in the overlying tissues, CASPARIAN STRIP INTEGRITY FACTOR (CIF) peptides seem to have the opposite effect (Ghorbani et al., 2015). Treatment with CIF2, and to a lesser extent also with CIF1 peptides, results in a marked decrease in emerged LR density, while the overall LR primordium density is not affected. Moreover, the density of stage-V primordia is unusually high in CIF2-treated roots indicating that LR development is often halted or slowed down at this developmental stage. The transition from a stage-IV to a stage-V primordium usually entails the transition from a flat-topped to a more dome-shaped primordium. In contrast, the majority of primordia in CIF2-treated roots appear flattened and have difficulty penetrating the overlying tissues, reminiscent of ida and haehls2 mutant phenotypes. CIF2 peptide signaling thus seems to inhibit LR primordium growth by preventing its spatial accommodation in the overlying tissues. The downstream mechanisms via which CIF peptides affect LR development have yet to be elucidated. However, it is known that CIF1 and CIF2 control the formation of an extracellular hydrophobic barrier on endodermal cells referred to as the “Casparian strip” (Doblas et al., 2017; Nakayama et al., 2017). CIF1 and CIF2 are expressed in the stele, including the pericycle, and are perceived by the LRR-RLKs SCHENGEN3/GASSHO1 (SGN3/GSO1) and GSO2, both expressed in the endodermis. Together with the receptor-like cytoplasmic kinase SGN1, SGN3, and GSO2 stimulate localized lignification and suberization upon perception of CIF peptides, thereby ensuring the formation of continuous Casparian strips. To allow growing LR primordia to penetrate the endodermis, this strong and tight seal around the endodermal cells has to be degraded. In agreement with this, CIF2 and SGN3 expression is absent at sites of LR initiation (Racolta et al., 2014; Ghorbani et al., 2015; Nakayama et al., 2017). Downregulation of CIFs and SGN3 upon LR initiation, possibly due to local auxin accumulation, might thus be required for the local breakdown of Casparian strips, thereby allowing LR primordia to grow through the endodermis.
SIGNALING PEPTIDES CONTROLLING LR PRIMORDIUM GROWTH AND LR ELONGATION
CEP Peptides Regulate LR Elongation in Response to Environmental Stimuli
As discussed above, several CEP peptides have an inhibitory effect on LR initiation. However, some CEPs were also found to regulate the growth of LR primordia after initiation as well as the elongation of mature LRs. Furthermore, they seem to provide a mechanism to integrate information about the prevailing environmental conditions and adapt the root system accordingly. Several CEP genes are expressed during LR development and it has been shown that CEP1 overexpression as well as treatments with synthetic CEP5 peptides result in the inhibition of LR elongation due to a reduction in cell numbers and cell size in the meristematic zone (Ohyama et al., 2008; Delay et al., 2013; Roberts et al., 2013; Tabata et al., 2014; Chapman et al., 2019). In agreement with this, LR elongation is enhanced in cepr1cepr2 double receptor mutants. The molecular mechanisms via which CEPs and their receptors regulate LR elongation are not completely clear, but the data that has been reported so far suggests there is a strong link with the role of these peptides as regulators of a systemic N-starvation response mechanism (Delay et al., 2013; Tabata et al., 2014; Ohkubo et al., 2017). CEP gene expression is mainly associated with the vasculature and several CEPs are induced in the root upon NO3−-starvation. These low N-induced CEP peptides are believed to travel from the root to the shoot via the xylem, resulting in their accumulation in the vasculature of cotyledons and mature leaves, where they bind the CEPR1 and CEPR2 receptors in the phloem. Perception of CEPs by their receptors triggers the local expression of CEP DOWNSTREAM1 (CEPD1) and CEPD2, two nonsecreted polypeptides that travel via the phloem from the shoot to the root, where they trigger the expression of NITRATE TRANSPORTER2.1 (NRT2.1) specifically in the parts of the root system located in a N-rich environment. It is possible that this pathway is also responsible for the effects of CEP peptides on LR development. In agreement with this, cepd1cepd2 double mutants display increased LR lengths, as observed in cepr1cepr2 receptor mutants, suggesting that the CEP-induced inhibition of LR elongation is established through the activity of the CEPRs as well as their downstream targets CEPD1 and CEPD2. Interestingly, it has been shown that in low-N conditions, NRT1.1 reduces auxin accumulation in LRs, thereby preventing their growth (Krouk et al., 2010; Mounier et al., 2014). The CEP-induced inhibition of LR elongation could thus potentially be an effect of the downstream upregulation of NRTs. However, CEPs might also regulate LR elongation in an NRT-independent manner.
CEP peptides are also involved in the regulation of LR elongation in response to Suc (Dimitrov and Tax, 2018; Chapman et al., 2019). Addition of Suc to the growth medium typically enhances LR elongation, but this effect is even stronger in the cepr1 single mutants and cepr1cepr2 double mutants, which seem hypersensitive to Suc addition. Moreover, cepr1 mutants show increased LR lengths in high light conditions, suggesting that CEPR1 normally represses LR elongation in response to photosynthesis-derived sugars. Accordingly, expression of several CEP genes is induced upon Suc addition and CEP5 peptide treatments were shown to reduce LR length in a CEPR1-dependent manner. CEP-CEPR signaling thus seems to work as a control mechanism that attenuates LR elongation in response to Suc to balance growth with resource availability.
CLE Peptides Inhibit LR Primordium Growth and LR Elongation upon N-Deficiency
Apart from CEPs, CLE peptides have also been found to regulate LR primordium development and LR elongation depending on N-availability (Araya et al., 2014). The expression of several CLE peptides (CLE1, CLE3, CLE4, and CLE7) is induced upon N-deficiency and overexpression of these CLEs results in reduced LR lengths without affecting primary root growth, an effect that is strongest in moderate to high-N conditions. Additionally, emerged LR density is strongly decreased in these overexpression lines due to an increase in the frequency of nonemerged LR primordia (stages I–IV) while total LR primordium density was not affected. This indicates that these CLE peptides do not affect LR initiation but inhibit LR primordium growth and LR elongation. Mutants in CLV1, the LRR-RLK that is responsible for the perception of CLV3 in the shoot apical meristem (Fletcher et al., 1999; Ogawa et al., 2008), have elongated LRs and increased emerged LR densities under low-N conditions. Furthermore, CLE3 overexpression under its endogenous promoter only causes slight reductions in LR length in the clv1 mutant background while a strong inhibition of LR elongation is triggered in wild-type seedlings. These findings indicate that CLV1 serves as the receptor for these N-responsive CLE peptides in the context of LR primordium growth and LR elongation. Additionally, clv1 mutants were found to overaccumulate CLE2, CLE3, CLE4, and CLE7 transcripts, suggesting that expression of these CLE peptides is kept in check via a negative feedback loop. CLE1, CLE2, CLE3, CLE4, and CLE7 are mainly expressed in the pericycle, while CLV1 is expressed in the phloem companion cells. CLE peptides are thus expected to diffuse into the stele where they activate CLV1 in the plasma membrane of phloem companion cells, which triggers a yet-unknown downstream mechanism that regulates LR primordium growth and mature LR elongation. The localization of CLV1 in phloem companion cells suggests that downstream signals are transferred through the phloem for the systemic regulation of LR architecture. Similar to the role of CEP peptides, the CLE-CLV1 signaling module thus seems to support an important mechanism for the prevention of the expansion of the root system in N-deficient patches in the soil.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
The development of LRs is a tightly coordinated process during which auxin serves as a well-known master regulator. However, it is now clear that from the priming of LRFCs onwards, peptide-receptor signaling plays an important role in the modulation of all subsequent steps of LR development.
One of the most striking common themes that emerged from recent studies is the role of several peptide signaling pathways in lateral inhibition mechanisms that prevent the development of LR primordia in close proximity to each other, indicating that this is an important and tightly regulated process. Interestingly, the involvement of signaling peptides in lateral inhibition seemingly coincides with a role during LR initiation. This suggests that lateral inhibition is one of the first events in the LR developmental process and occurs rather early in the young part of the differentiation zone, probably around the same time at which LR initiation takes place.
Some signaling peptides are not merely required for normal LR development but appear to actively modulate the process depending on the environmental conditions. Several peptides from both the CLE and the CEP family prevent the expansion of the root system in N-deficient patches in the soil by inhibiting LR primordium development and/or LR elongation in these conditions. Additionally, some CEPs attenuate the elongation of LRs in response to photosynthesis-derived sugars. Furthermore, expression levels of several CEP peptides were also found to respond to osmotic- and salt stress as well as high CO2 levels (Delay et al., 2013), suggesting that environmental cues are integrated at the level of these peptides to modulate LR development accordingly. These peptide-receptor modules thus seem to serve as molecular mechanisms that underlie the developmental plasticity of the root system, allowing plants to adapt their root system architecture to the environment and to balance growth with their energetic and nutritional status.
Signaling peptide families are usually rather big and functional redundancy within peptide families has often been reported. Because single knock-out mutants rarely show striking phenotypes, most of our knowledge about the activity of these peptides is based on gain-of-function studies. Further elucidation of the roles of these redundant molecules during LR development will thus require the analysis of higher-order mutants. On the other hand, it should be noted that peptides from the same family do not necessarily perform the same function, as illustrated by the differing effects of different CLE peptides on primary and LR development. Future research should thus determine which peptides within a family act redundantly, which perform a different function, and what the underlying molecular mechanism is for these differences. Additionally, it should be noted that multiple signaling peptides and receptors from different families are also often involved in the regulation of the same LR developmental process. However, because peptide-receptor pathways have until now only been studied in isolation, it is not clear whether peptides and receptors from different families can also act redundantly with one another. Future research should thus determine whether functional overlap and crosstalk between different peptide-receptor pathways exists when they regulate the same process. Therefore, it will be important to unravel the mechanisms that are activated downstream of each peptide-receptor pair during LR development, information that is currently lacking for most peptide signaling pathways.
Several peptides and receptors were found not to be expressed in XPP cells or LR primordia, leaving it unclear how they affect LR development. Furthermore, several peptides affect the cells in which they are produced, raising the question as to why a secreted signal is employed in the first place. For a proper understanding of peptide-receptor pathways as mechanisms for intercellular communication, more detailed knowledge of when and where peptides are secreted and perceived is required.
Finally, it will be interesting to investigate the involvement of other peptide-receptor pathways in LR development. The Arabidopsis genome is estimated to encode more than 1,000 signaling peptides and over 600 RLKs (Shiu and Bleecker, 2001; Lease and Walker, 2006), of which only a fraction has been studied so far. Furthermore, it will be paramount to determine whether the peptide-receptor modules that govern LR development in Arabidopsis are conserved in other plant species. It is known that the CLE and CEP peptide families are also represented in Medicago truncatula and that some of these peptides have similar effects on LR development as described in Arabidopsis (Imin et al., 2013; Huault et al., 2014; Patel et al., 2018). However, the role of peptide-receptor signaling during LR development in crop plants remains unexplored.
Acknowledgments
We thank Maria F. Njo for her help with the artwork.
Footnotes
This work was supported by the Research Foundation Flanders (FWO) Fellowship accredited to J.J. (1168218N).
Articles can be viewed without a subscription.
References
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wirén N, Takahashi H (2014) CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc Natl Acad Sci USA 111: 2029–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson NJ, Lilley CJ, Urwin PE (2013) Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol 162: 2028–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B, Kirschner G, Gerlitz N, Stadler R, Simon R (2020) CLE40 signalling regulates the fate of root stem cells in Arabidopsis. Plant Physiol 182: 1776–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonci T, Ribeiro B, Ceciliato PHO, Guerrero-Abad JC, Silva-Filho MC, Moura DS (2014) Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. J Exp Bot 65: 2219–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik G-E, Amundsen SS, Mandal A, Aalen RB (2003) Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K, Taleski M, Ogilvie HA, Imin N, Djordjevic MA (2019) CEP-CEPR1 signalling inhibits the sucrose-dependent enhancement of lateral root growth. J Exp Bot 70: 3955–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Ryu H, Rho S, Hill K, Smith S, Audenaert D, Park J, Han S, Beeckman T, Bennett MJ, et al. (2014) A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat Cell Biol 16: 66–76 [DOI] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC (2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Comas LH, Becker SR, Cruz VM, Byrne PF, Dierig DA (2013) Root traits contributing to plant productivity under drought. Front Plant Sci 4: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyzewicz N, Shi CL, Vu LD, van de Cotte B, Hodgman C, Butenko MA, de Smet I (2015) Modulation of Arabidopsis and monocot root architecture by CLAVATA3/EMBRYO SURROUNDING REGION 26 peptide. J Exp Bot 66: 5229–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay C, Imin N, Djordjevic MA (2013) CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. J Exp Bot 64: 5383–5394 [DOI] [PubMed] [Google Scholar]
- de Smet I, Tetsumura T, de Rybel B, Frei dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- de Smet I, Vassileva V, de Rybel B, Levesque MP, Grunewald W, van Damme D, van Noorden G, Naudts M, van Isterdael G, et al. (2008) Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597 [DOI] [PubMed] [Google Scholar]
- Dimitrov I, Tax FE (2018) Lateral root growth in Arabidopsis is controlled by short and long distance signaling through the LRR RLKs XIP1/CEPR1 and CEPR2. Plant Signal Behav 13: e1489667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblas VG, Smakowska-Luzan E, Fujita S, Alassimone J, Barberon M, Madalinski M, Belkhadir Y, Geldner N (2017) Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355: 280–284 [DOI] [PubMed] [Google Scholar]
- Dong Q, Zhang Z, Liu Y, Tao LZ, Liu H (2019) FERONIA regulates auxin-mediated lateral root development and primary root gravitropism. FEBS Lett 593: 97–106 [DOI] [PubMed] [Google Scholar]
- Du Y, Scheres B (2017) PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root outgrowth. Proc Natl Acad Sci USA 114: 11709–11714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque LO, Villordon A (2019) Root branching and nutrient efficiency: Status and way forward in root and tuber crops. Front Plant Sci 10: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Nguyen A, Beeckman T, Madder A, Hilson P (2013) Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiol 161: 954–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Vassileva V, Madder A, Beeckman T, Hilson P (2015) The GLV6/RGF8/CLEL2 peptide regulates early pericycle divisions during lateral root initiation. J Exp Bot 66: 5245–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Ghorbani S, Lin YC, Parizot B, Fernandez A, Njo MF, van de Peer Y, Beeckman T, Hilson P (2015) Expanding the repertoire of secretory peptides controlling root development with comparative genome analysis and functional assays. J Exp Bot 66: 5257–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, Fukaki H (2012) The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139: 883–893 [DOI] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Martin M, Doblas VG, Bacete L, Miart F, Sormani R, Hématy K, Renou J, Landrein B, et al. (2018) Receptor kinase THESEUS1 is a Rapid Alkalinization Factor 34 receptor in Arabidopsis. Curr Biol 28: 2452–2458.e4 [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H (2008) Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota A, Kato T, Fukaki H, Aida M, Tasaka M (2007) The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell 19: 2156–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofhuis H, Laskowski M, Du Y, Prasad K, Grigg S, Pinon V, Scheres B (2013) Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr Biol 23: 956–962 [DOI] [PubMed] [Google Scholar]
- Huault E, Laffont C, Wen J, Mysore KS, Ratet P, Duc G, Frugier F (2014) Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PLoS Genet 10: e1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA (2013) The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J Exp Bot 64: 5395–5409 [DOI] [PubMed] [Google Scholar]
- Jun J, Fiume E, Roeder AHK, Meng L, Sharma VK, Osmont KS, Baker C, Ha CM, Meyerowitz EM, Feldman LJ, et al. (2010) Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol 154: 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang NY, Lee HW, Kim J (2013) The AP2/EREBP gene PUCHI Co-Acts with LBD16/ASL18 and LBD18/ASL20 downstream of ARF7 and ARF19 to regulate lateral root development in Arabidopsis. Plant Cell Physiol 54: 1326–1334 [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Kumpf RP, Shi C-L, Larrieu A, Stø IM, Butenko MA, Péret B, Riiser ES, Bennett MJ, Aalen RB (2013) Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc Natl Acad Sci USA 110: 5235–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease KA, Walker JC (2006) The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol 142: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim NY, Lee DJ, Kim J (2009) LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol 151: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zeng R, Liao H (2016) Improving crop nutrient efficiency through root architecture modifications. J Integr Plant Biol 58: 193–202 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Meng L, Buchanan BB, Feldman LJ, Luan S (2012) CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc Natl Acad Sci USA 109: 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhou J, Tang J, Li B, de Oliveira MVV, Chai J, He P, Shan L (2016) Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Reports 14: 1330–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Młodzińska E, Kłobus G, Christensen MD, Fuglsang AT (2015) The plasma membrane H+-ATPase AHA2 contributes to the root architecture in response to different nitrogen supply. Physiol Plant 154: 270–282 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier E, Pervent M, Ljung K, Gojon A, Nacry P (2014) Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ 37: 162–174 [DOI] [PubMed] [Google Scholar]
- Murphy E, Vu LD, van den Broeck L, Lin Z, Ramakrishna P, van de Cotte B, Gaudinier A, Goh T, Slane D, Beeckman T, et al. (2016) RALFL34 regulates formative cell divisions in Arabidopsis pericycle during lateral root initiation. J Exp Bot 67: 4863–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Shinohara H, Tanaka M, Baba K, Ogawa-Ohnishi M, Matsubayashi Y (2017) A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355: 284–286 [DOI] [PubMed] [Google Scholar]
- Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM (2011) Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol 21: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Ohkubo Y, Tanaka M, Tabata R, Ogawa-Ohnishi M, Matsubayashi Y (2017) Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat Plants 3: 17029. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Ogawa M, Matsubayashi Y (2008) Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J 55: 152–160 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Lu X, Zi Q, Xun Q, Zhang J, Wu Y, Shi H, Wei Z, Zhao B, Zhang X, et al. (2016) RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Res 26: 686–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N, Mohd-Radzman NA, Corcilius L, Crossett B, Connolly A, Cordwell SJ, Ivanovici A, Taylor K, Williams J, Binos S, et al. (2018) Diverse peptide hormones affecting root growth identified in the Medicago truncatula secreted peptidome. Mol Cell Proteomics 17: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Ponce MR, Micol JL (2002) The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 242: 161–173 [DOI] [PubMed] [Google Scholar]
- Racolta A, Bryan AC, Tax FE (2014) The receptor-like kinases GSO1 and GSO2 together regulate root growth in Arabidopsis through control of cell division and cell fate specification. Dev Dyn 243: 257–278 [DOI] [PubMed] [Google Scholar]
- Ramakrishna P, Ruiz Duarte P, Rance GA, Schubert M, Vordermaier V, Vu LD, Murphy E, Vilches Barro A, Swarup K, Moirangthem K, et al. (2019) EXPANSIN A1-mediated radial swelling of pericycle cells positions anticlinal cell divisions during lateral root initiation. Proc Natl Acad Sci USA 116: 8597–8602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts I, Smith S, de Rybel B, van den Broeke J, Smet W, de Cokere S, Mispelaere M, de Smet I, Beeckman T (2013) The CEP family in land plants: Evolutionary analyses, expression studies, and role in Arabidopsis shoot development. J Exp Bot 64: 5371–5381 [DOI] [PubMed] [Google Scholar]
- Roberts I, Smith S, Stes E, de Rybel B, Staes A, van de Cotte B, Njo MF, Dedeyne L, Demol H, Lavenus J, et al. (2016) CEP5 and XIP1/CEPR1 regulate lateral root initiation in Arabidopsis. J Exp Bot 67: 4889–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu N, Subedi SR, Singh VK, Sinha P, Kumar S, Singh SP, Ghimire SK, Pandey M, Yadaw RB, Varshney RK, et al. (2019) Deciphering the genetic basis of root morphology, nutrient uptake, yield, and yield-related traits in rice under dry direct-seeded cultivation systems. Sci Rep 9: 9334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satbhai SB, Ristova D, Busch W (2015) Underground tuning: quantitative regulation of root growth. J Exp Bot 66: 1099–1112 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Shinohara H, Mori A, Yasue N, Sumida K, Matsubayashi Y (2016) Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proc Natl Acad Sci USA 113: 3897–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Liu L, Wang J, Wu Z, Zhang H, Tang J, Lin G, Wang Y, Wen X, Li W, et al. (2016) Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res 26: 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, Kühnemuth R, Weidtkamp-Peters S, Pinto KG, Kirschner GK, Schmid JB, Wink RH, et al. (2013) Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol 23: 362–371 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R (2009) A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA (2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, de Smet I, Vanneste S, et al. (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y (2014) Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346: 343–346 [DOI] [PubMed] [Google Scholar]
- Tian H, De Smet I, Ding Z (2014) Shaping a root system: Regulating lateral versus primary root growth. Trends Plant Sci 19: 426–431 [DOI] [PubMed] [Google Scholar]
- Toyokura K, Goh T, Shinohara H, Shinoda A, Kondo Y, Okamoto Y, Uehara T, Fujimoto K, Okushima Y, Ikeyama Y, et al. (2019) Lateral inhibition by a peptide hormone-receptor cascade during Arabidopsis lateral root founder cell formation. Dev Cell 48: 64–75.e5 [DOI] [PubMed] [Google Scholar]
- Trinh D-C, Lavenus J, Goh T, Boutté Y, Drogue Q, Vaissayre V, Tellier F, Lucas M, Voß U, Gantet P, et al. (2019) PUCHI regulates very long chain fatty acid biosynthesis during lateral root and callus formation. Proc Natl Acad Sci USA 116: 14325–14330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45: 1097–1102 [DOI] [PubMed] [Google Scholar]
- van Norman JM, Breakfield NW, Benfey PN (2011) Intercellular communication during plant development. Plant Cell 23: 855–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JEM, von Wangenheim D, Barberon M, Lee Y, Stelzer EHK, Maizel A, Geldner N (2014) A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343: 178–183 [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL (2008) Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA 105: 9829–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilches Barro A, Stöckle D, Thellmann M, Ruiz-Duarte P, Bald L, Louveaux M, von Born P, Denninger P, Goh T, Fukaki H, et al. (2019) Cytoskeleton dynamics are necessary for early events of lateral root initiation in Arabidopsis. Curr Biol 29: 2443–2454.e5 [DOI] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, Tejos R, Pérez AC, Kleine-Vehn J, Vanneste S, Drozdzecki A, Leitner J, Abas L, Aerts M, et al. (2012) GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev Cell 22: 678–685 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Shao Y, Ge S, Zhang M, Zhang T, Hu X, Liu Y, Walker J, Zhang S, Xu J (2019) A MAPK cascade downstream of IDA-HAE/HSL2 ligand-receptor pair in lateral root emergence. Nat Plants 5: 414–423 [DOI] [PubMed] [Google Scholar]