Transcriptome analysis of young ovules points to a receptor-like kinase that is preferentially expressed in archesporial and megaspore mother cells and functions in female germline cell specification in rice.
Abstract
Megasporogenesis is a key step during ovule development in angiosperms, but the small number and inaccessibility of these cells have hampered molecular and genome-wide studies. Thus, many questions remain regarding the molecular basis of cell specification, differentiation, and development in the female gametophyte. Here, taking advantage of the correlation between spikelet length and ovule development in rice (Oryza sativa), we studied the transcriptome dynamics of young ovules at three stages, the archesporial cell, the megaspore mother cell before meiosis, and the functional megaspore after meiosis, using expression profiling based on RNA sequencing. Our analysis showed that 5,274 genes were preferentially expressed in ovules during megasporogenesis as compared to ovules at the mature female gametophyte stage. Out of these, 958 (18.16%) genes were archesporial cell- and/or megaspore mother cell-preferential genes, and represent a significant enrichment of genes involved in hormone signal transduction and plant pathogen interaction pathways, as well as genes encoding transcription factors. The expression patterns of nine genes that were preferentially expressed in ovules of different developmental stages, including the OsERECTA2 (OsER2) receptor-like kinase gene, were confirmed by in situ hybridization. We further characterized the OsER2 loss-of-function mutant, which had an excessive number of female germline cells and an abnormal female gametophyte, suggesting that OsER2 regulates germline cell specification during megasporogenesis in rice. These results expand our understanding of the molecular control of megasporogenesis in rice and contribute to the functional studies of genes involved in megasporogenesis.
The male and female gametophytes are the key reproductive units of angiosperms and are critical for sexual reproduction (Yadegari and Drews, 2004; Yang et al., 2010; Shi and Yang 2011). Much of the reported research on plant gametophyte development has focused mainly on the male gametophyte, due in part to the large number of male gametophytes and how convenient they are to harvest. Numerous genes that control male gametophyte development in plants have been identified and functionally analyzed in detail (Wilson and Zhang, 2009; Gómez et al., 2015; Hafidh et al., 2016). In contrast, the number of female gametophytes is small, and the female gametophyte is deeply embedded within the ovule tissue, making it difficult to harvest and study. As a result, the understanding of the molecular mechanism underlying plant female gametophyte development lags far behind that of male gametophyte development.
In most flowering plants, including Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and maize (Zea mays), female germline development consists of two main phases, megasporogenesis and megagametogenesis. Megasporogenesis is initiated from a single subepidermal cell in the nucellus in the young ovule primordium, and this cell is specified and differentiates into the archesporial cell (AC), which subsequently elongates and develops into the megaspore mother cell (MMC). The MMC undergoes meiosis to yield a typically linear tetrad of megaspores. The three megaspores at the micropyle end degenerate, while the megaspore at the chalazal end differentiates and develops into a functional megaspore (FM), which marks the completion of megasporogenesis. After three rounds of mitosis, nuclear migration, cellularization, and differentiation, the FM develops into the mature female gametophyte, which contains an egg cell, two synergids, a binucleated central cell, and a variable number of antipodal cells, which completes megagametogenesis (Yadegari and Drews, 2004; Yang et al., 2010).
In Arabidopsis, female gametophyte development has been investigated extensively, and a number of essential genes regulating megasporogenesis and megagametogenesis have been identified, providing a basic understanding of these developmental processes (Yang et al., 2010; Zhao et al., 2014; Cao et al., 2018; Lora et al., 2019). It was demonstrated early on that the SPOROCYTELESS/NOZZLE (SPL/NZZ) gene acts as a transcriptional regulator of sporocyte development and germline cell fate in Arabidopsis. A mutation in SPL can prevent ACs from differentiating into sporocytes in both anther and ovule primordia, which results in male and female sterility (Schiefthaler et al., 1999; Yang et al., 1999). These results indicate that MMC formation requires SPL. ARGONAUTE 9 (AGO9) and RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) proteins were reported to control megasporocyte specification and female gamete formation through AGO9-dependent small RNA silencing in a non-cell-autonomous manner in Arabidopsis. Mutations in AGO9 and RDR6 lead to the differentiation of multiple MMCs capable of initiating gametogenesis (Olmedo-Monfil et al., 2010). A recent study showed that TRANSCRIPTION EXPORT1 (TEX1) protein prevents somatic cells from undergoing MMC formation non-cell-autonomously through Trans-Acting Small interfering RNA3 (TAS3)-mediated restriction of AUXIN RESPONSE FACTOR3 (ARF3) expression, and a mutation in TEX1 leads to the formation of multiple MMCs (Su et al., 2017). Zhao et al. (2017) found that RETINOBLASTOMA RELATED1 (RBR1) directly represses WUSCHEL (WUS) to mediate germline entry into meiosis by preventing overproliferation of megasporocytes in Arabidopsis, and the number of MMCs and subsequent embryo sacs in the ovule is regulated by a subset of redundantly acting KIP-RELATED PROTEIN (KRP/ICK) cyclin-dependent kinase (CDK) inhibitors. ICKs in Arabidopsis were also determined to play an important role in the degeneration of nonfunctional megaspores (Cao et al., 2018). The cytochrome P450 gene KLU (KLUH/CYP78A5), expressed in the inner integument, was found to non-cell-autonomously suppress megasporocyte cell fate in somatic cells surrounding the megasporocyte through activation of WRKY28 expression mediated by the chromatin remodeling complex SWi/snf2-Related1 (SWR1) in Arabidopsis (Zhao et al., 2018).
However, in cereals, female gametophyte development has not been investigated intensively and only a limited number of genes involved in female gametophyte development have been identified. MULTIPLE SPOROCYTE1 (MSP1) is known to control early sporogenic development in rice, and the msp1 mutant likely gives rise to an excessive number of ACs (Nonomura et al., 2003). MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1), a germline-specific member of the AGO gene family in rice, regulates premeiotic germ cell development and the progression of meiosis. In mel1 mutants, ovule development is arrested at various stages from premeiosis to tetrad formation, resulting in the failure of female gametophyte formation (Nonomura et al., 2007; Komiya et al., 2014). In maize, a mutation in AGO104, a gene expressed specifically in somatic cells surrounding the female meiocyte, results in the failure of MMC meiosis (Singh et al., 2011).
Although several genes involved in plant female gametophyte development have been identified, few key regulatory genes in rice have been cloned, and the molecular mechanisms underlying megasporogenesis in rice largely remain unclear. In recent years, several studies have employed microarrays or RNA sequencing (RNA-Seq) transcriptomic analysis to identify gene expression profiles in ovules during female gametophyte development in rice (Kubo et al., 2013; Wu et al., 2015). Kubo et al. (2013) first determined the gene expression profiles of rice ovules containing female germlines at six developmental stages, from the MMC at premeiotic and early meiosis stages to the mature female gametophyte stage, using microarrays. Wu et al. (2015) used RNA-Seq technology to analyze the transcriptome of rice ovules at four developmental stages including MMC during meiosis, megaspore mitosis, mature female gametophyte and postfertilization. These studies have identified a number of genes that showed dynamic or enriched expression patterns in developing rice ovules and highlighted many potential regulators that may play a role in female meiosis and megagametogenesis. However, detailed genetic information regarding MMC differentiation and specification, an important step during early megasporogenesis, have not been provided by previous studies due to the lack of gene expression profiling of young rice ovules at the AC stage. To improve our understanding of the dynamic gene expression profiling of ovules at early developmental stages, we performed transcriptome analysis of rice ovules during megasporogenesis including AC, premeiotic MMC, and postmeiotic FM stages and identified the genes that were specifically or preferentially expressed during early megaspore development, including the AC-stage preferentially expressed genes that have not been identified in previous studies. The expression patterns of nine genes preferentially expressed in ovules undergoing megasporogenesis were verified by in situ hybridization. We further conducted gene functional analysis using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated gene editing system and revealed that OsER2, which is preferentially expressed in AC- and MMC-stage ovules, plays a key role in MMC specification during megasporogenesis in rice.
RESULTS
Identification of Female Gametophyte Developmental Stages in Rice ‘ZH11’
Female gametophyte development in plants takes place in the ovule and comprises a series of developmental stages. Previous studies revealed the association between female gametophyte development and other rice reproductive organs (Lopez-Dee et al., 1999; Ito et al., 2004; Zeng et al., 2009). However, this correlation is not entirely consistent in different rice varieties. To ensure the accurate collection of different ovule stages, we systematically examined the morphology of the developing flower and characterized the female gametophyte structure by confocal microscopy. From this analysis, we generated a reproductive calendar for the rice variety ZH11 (Table 1) that allows the prediction of female gametophyte development on the basis of different floral organ parameters.
Table 1. Definition of the stages of female gametophyte development in ZH11.
Mono, Two, Four, and Eight indicate the different nuclear stages of the embryo sac.
| Stages | Cytological Characteristics | Length of Flower Organs (mm) | ||
|---|---|---|---|---|
| Spikelet | Gynoecium | Anther | ||
| AC | The AC enlarges, and integument primordium forms. | 1.5–2.0 | 0.4–0.45 | 0.4–0.5 |
| MMC | The AC differentiates into a MMC. The single integument primordium divides into inner and outer integuments. | 2.0–2.6 | 0.45–0.50 | 0.5–0.55 |
| Meiosis I | The MMC undergoes meiosis to form a dyad and the integuments elongate. | 2.6–3.2 | 0.50–0.60 | 0.55–0.65 |
| Meiosis II | The four linearly arranged megaspores are formed. | 3.2–3.8 | 0.60–0.70 | 0.65–0.75 |
| FM | Only the chalazal megaspore develops into a FM. The micropyle is formed. | 3.8–4.2 | 0.70–0.80 | 0.75–0.80 |
| Mono | The FM elongates and expands. The vacuole enlarges. | 4.2–4.6 | 0.8–0.9 | 0.8–1.0 |
| Two | First mitotic nuclear division. | 4.6–5.2 | 0.9–1.1 | 1.0–1.2 |
| Four | Second mitotic nuclear division. | 5.2–6.0 | 1.1–1.5 | 1.2–1.6 |
| Eight | Third mitotic nuclear division. | 6.0–7.0 | 1.5–1.9 | 1.6–1.9 |
| MO | Migration of nuclei and cellularization. | 7.0–8.0 | 1.9–2.1 | 1.9–2.2 |
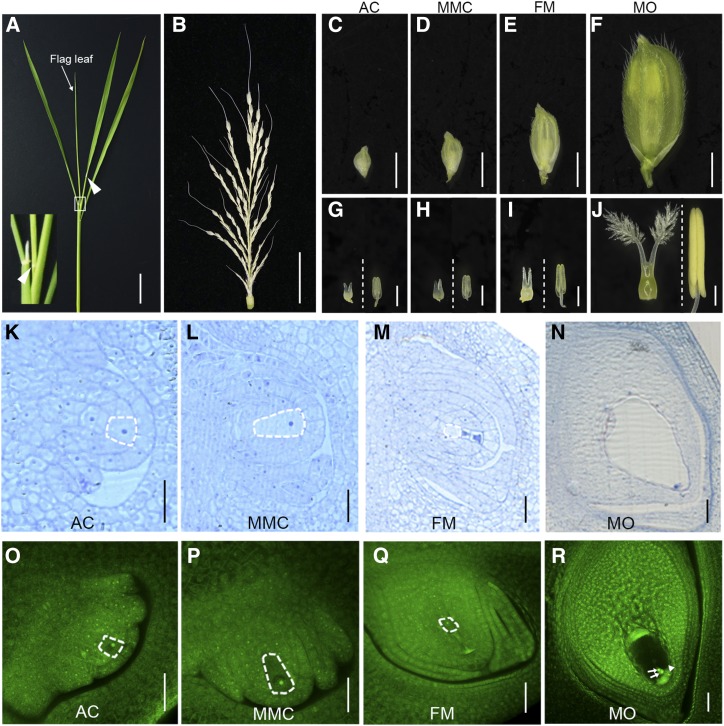
The female gametophyte developmental processes were divided into 10 stages: AC; MMC; meiosis I; meiosis II; FM; mono-, two-, four-, and eight-nucleate stages of the female gametophyte; and mature ovule (MO). When the flag leaf has not been completely drawn, and the distance between the collars of the flag leaf and the second upper leaf in a tiller is ∼3 cm (Fig. 1A), the length of the young panicle is 7–8 cm (Fig. 1B). The spikelets on this panicle are in the early stages of female gametophyte development, including the AC and MMC stages, ranging from the bottom to the top (Fig. 1, C and D). When the spikelet is 1.5–2.0 mm, the anther length is 0.4–0.5 mm and the gynoecium length is 0.4–0.45 mm (Fig. 1, C and G), with the AC differentiating from a single subepidermal cell and containing a large nucleus. Figure 1, K and O, shows the AC-stage ovule, in which the integument primordium is newly initiated. When the spikelet is 2.0–2.6 mm, the anther length is 0.50–0.55 mm and the gynoecium length is 0.45–0.50 mm (Fig. 1, D and H), with the AC elongating and differentiating into a MMC with dense protoplasm and a larger nucleus. Figure 1, L and P, shows the MMC-stage ovule. At this stage, the single integument primordium divides into inner and outer integuments. When the spikelet is 3.8–4.0 mm, the anther length is 0.75–0.80 mm and the gynoecium length is 0.70–0.80 mm (Fig. 1, E and I). At this stage, the MMC has completed meiosis, the three megaspores at the micropyle end degenerate, and the surviving FM is differentiated at the chalazal end. Figure 1, M and Q, shows the FM-stage ovule, in which the inner and outer integuments have fully enclosed and completed the formation of the micropyle. When the spikelet is 7.0–8.0 mm, the anther length is 1.9–2.2 mm and the gynoecium length is 1.9–2.1 mm (Fig. 1, F and J). At this stage, the female gametophyte has completed three rounds of mitosis, nuclear migration, and cellularization and develops into a mature embryo sac containing two polar nuclei, two synergid cells, and one egg cell. Figure 1, N and R, shows the MO-stage ovule.
Figure 1.
Morphological analysis of the developing spikelets and ovules in rice variety ZH11. A, The tiller at booting stage. The white box corresponds to the inset in the lower left corner. Arrowheads indicate the collars. B, The young panicle from the tiller in A. C to F, Spikelets at the AC (C), MMC (D), FM (E), and MO (F) stages. G to J, The pistil and anther (left and right, respectively, of the dotted line) at the AC (G), MMC (H), FM (I), and MO (J) stages. K to N, The structure of ovules observed by semithin sectioning at the AC (K), MMC (L), FM (M), and MO (N) stages. O to R, The structure of ovules observed by confocal microscopy at the AC (O), MMC (P), FM (Q), and MO (R) stages. The female germline cells in ZH11 ovules are outlined by the white dashed lines (K–M and O–Q). Arrows indicate the polar nuclei in the central cell, and the arrowhead indicates an egg cell (R). Scale bars = 2 cm (A and B), 2mm (C–F), 0.5 mm (G–J), 20 μm (K–N), and 30 μm (O–R).
Global Gene Expression Profiling During Megasporogenesis in a Rice Ovule
Gene expression profiling was performed to identify genes potentially involved in megasporogenesis. ZH11 rice ovules at the AC, MMC, and FM stages were isolated from the ovary for RNA extraction under a dissection microscope according to the spikelet and gynoecium lengths described above. Three biological replicates were conducted for these three ovule stages. In addition, FM-stage ovary wall (OW), FM-stage stigma/style (SS), and MO-stage ovules were also collected as control samples. Three biological replicates were also conducted for these three control samples. In total, 18 samples were harvested for RNA-Seq analysis in this study. The number of clean reads for the 18 samples ranged from 29.04 to 73.06 million (Supplemental Table S1). For these samples, 91.55% to 95.20% of the clean reads were mapped to the rice genome (O. sativa ssp. japonica; http://rice.plantbiology.msu.edu) using TopHat (Trapnell et al., 2012), with no more than two base pair mismatches in the alignment; 90.31% to 92.89% of the reads were uniquely mapped (Supplemental Table S1). These data indicate that the quantity and quality of the reads were sufficient for quantitative gene expression analysis.
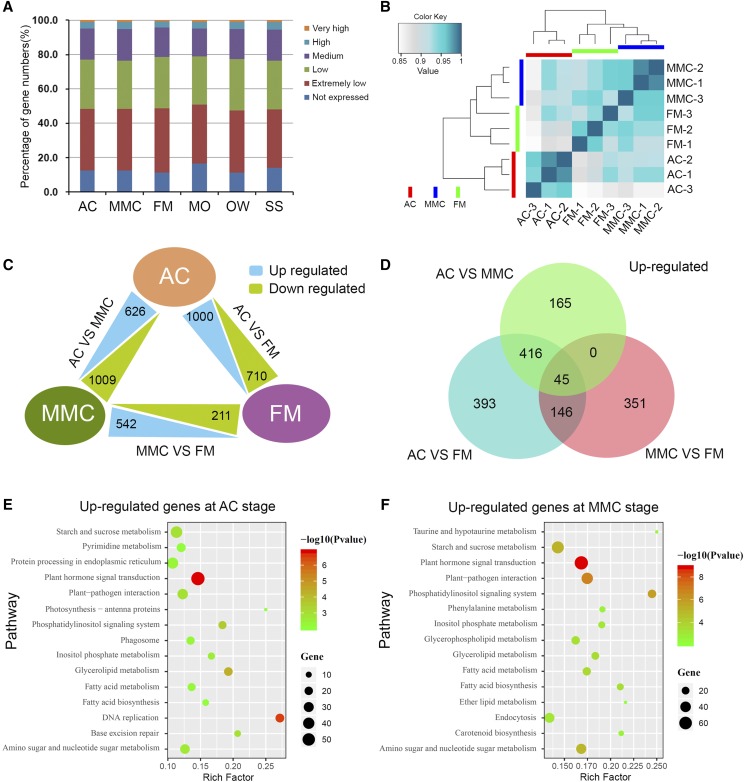
For global gene expression profiling of the different samples, the fragments per kilobase of transcript per million mapped reads (FPKM) method was used to assess the gene expression levels by normalizing the read counts. Genes with FPKM values >1.0 were considered expressed genes, and a total of 23,019 genes were expressed in the AC, MMC, FM, MO, OW, and SS tissues. We classified the expressed genes into five different groups according to their expression levels: extremely low-expression genes, with FPKM values < 10; low-expression genes, with 10–30 FPKM values; medium-expression genes, with 30–100 FPKM values; high-expression genes, with 100–300 FPKM values; and very high-expression genes, with FPKM values > 300. The number of expressed genes and the expression distribution pattern were highly similar among the six tissues (Fig. 2A; Supplemental Table S2). Hierarchical clustering of the transcriptomes from AC, MMC, and FM tissues showed that the three biological replicates of each tissue were grouped together with high correlation (Fig. 2B). AC was located on an independent branch, whereas MMC and FM were located on another branch, which was divided into two smaller branches. Similarly, the three biological replicates of each tissue from MO, OW, and SS were also classed together with a high correlation (Supplemental Fig. S1). These results indicated good reproducibility among the three biological replicates for each tissue in this study.
Figure 2.
Global expression levels, DEGs, and KEGG pathway analysis. A, Percentage of gene numbers in different categories according to their expression levels in each tissue, based on FPKM values. B, Hierarchical clustering of the nine samples during rice megasporogenesis using Pearson correlation coefficients. C, Numbers of differentially expressed genes among AC, MMC, and FM tissues. D, Numbers of up-regulated genes in AC versus MMC, AC versus FM, and MMC versus FM. E and F, KEGG pathway analysis for up-regulated genes in AC versus FM (E) and MMC versus FM (F).
Analysis of Differentially Expressed Genes in AC and MMC Ovules
To identify genes that are preferentially or specifically expressed during megasporogenesis, differential expression analysis was performed using Cufflinks and Cuffdiff (Trapnell et al., 2012). Genes with >2-fold changes and q < 0.05 in FPKM in different tissues were defined as differentially expressed genes (DEGs). In our initial analysis, when all three tissues (MO, OW, and SS) were used as controls, we identified 1,595 genes that were up-regulated DEGs in at least one tissue of AC, MMC, and FM (Supplemental Table S3). However, we found that several known genes that play a key role in rice megasporogenesis, such as OsMEL1 and OsIG1 (INDETERMINATE GAMETOPHYTE 1; Nonomura et al., 2007; Zhang et al., 2015), are not included in the identified up-regulated genes, suggesting that some key genes for rice megasporogenesis may be excluded in differential expression analysis using all three tissues— MO, OW, and SS—as controls. Then, we adjusted the strategy of analysis and only used MO as the control to screen the genes with up-regulated expression in the ovules at the AC, MMC, and/or FM stages.
Among the 23,019 genes, 5,274 were identified as having a significantly higher expression level in at least one of the megasporogenesis samples (AC, MMC, or FM) compared to MOs (Supplemental Table S4). These genes were designated as megasporogenesis up-regulated expression genes as compared to MOs. Of the 5,274 genes, 1,000 and 710 genes were significantly up-regulated and down-regulated in AC versus FM, respectively, and 542 and 211 genes were significantly up-regulated and down-regulated in MMC versus FM, respectively (Fig. 2C). The results in the Venn diagram showed that 45 genes were significantly up-regulated at the same time in the three comparison groups, and 416 genes were significantly up-regulated both in AC versus MMC and AC versus FM (Fig. 2D), and the total of 461 genes with significantly higher expression levels in the ovules at the AC stage compared to MMC, FM, and MO stages, were designated as AC-preferential genes. The 146 genes that were significantly up-regulated in both AC versus FM and MMC versus FM were designated as AC and MMC-preferential genes. There were 351 genes with up-regulated expression in MMC versus FM, designated as MMC-preferential genes, which had a significantly higher expression level in the ovules at the MMC stage compared to the AC, FM, and MO stages (Fig. 2D; Supplemental Table S5). These 958 genes were preferentially expressed in the developing ovules prior to meiosis in rice megasporogenesis, indicating that they may be involved in regulating early ovule development in rice.
To identify the key regulatory pathways in the early stage of rice ovule development, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for genes up-regulated at the AC and/or MMC stages was performed. The data showed that the enriched pathways for up-regulated genes at the two stages were involved in plant hormone signal transduction as well as plant-pathogen interaction (Fig. 2, E and F). Enrichment of the plant hormone signal transduction pathways at the AC and MMC stages was represented by 59 and 68 genes, respectively (Supplemental Table S6). And the enrichment of the plant-pathogen interaction pathways was represented by 33 and 47 genes, respectively (Supplemental Table S6). These results suggest that genes involved in plant hormone signal transduction and plant-pathogen interaction pathways might play an important role in young ovule development during rice megasporogenesis.
Auxin Signaling Pathway Genes Are Enriched in AC- and MMC-Preferential Genes
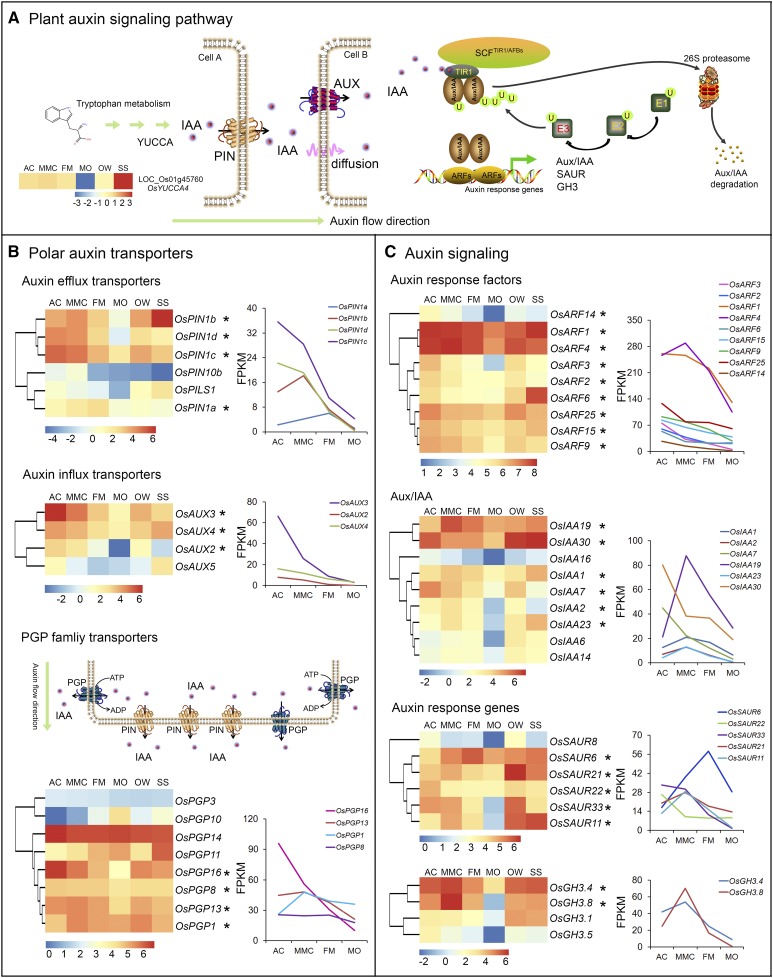
As indicated above, lots of genes related to plant hormone signaling were mostly enriched at the AC and MMC stages of rice megasporogenesis. Of these genes, >40 were in the auxin signaling pathway (Supplemental Table S7). It is well established that plant hormones, especially auxin, play key roles in many aspects of plant growth and development, including ovule development and sporophyte-gametophyte cross talk (Zhao, 2010; Lituiev et al., 2013; Chettoor and Evans, 2015; Su et al., 2017). Therefore, we focused on auxin signaling-related genes, including those involved in the biosynthesis, polar transport, and response of this hormone (Fig. 3A; Péret et al., 2012; Zhao et al., 2012; Panoli et al., 2015). The natural auxin in plants, indole-3-acetic acid (IAA), is mainly synthesized through the Trp-dependent auxin biosynthesis pathway, which is catalyzed by the YUCCA (YUC) family of flavin monooxygenases (Zhao, 2012). Our transcriptome data showed that only one rice YUC family gene, OsYUC4 (Os01g45670), was preferentially expressed at the MMC stage during megasporogenesis, and its highest expression was detected in the SS sample (Fig. 3A).
Figure 3.
Expression patterns of significant DEGs in the auxin signaling pathway in ovules. A, Diagram of the plant auxin signaling pathway. YUCCA4, YUCCA-LIKE GENE4; AUX, AUXIN INFLUX CARRIER; TIR1, TRANSPORT INHIBITOR RESPONSE1; SCF, SKP1-CULLIN-F-BOX COMPLEX; AFBs, AUXIN SIGNALING F-Boxes; ARFs, AUXIN RESPONSE FACTORs; Aux/IAA, Aux/IAA Family Protein; SAUR, SMALL AUXIN UPREGULATED RNA; GH3, GRETCHEN HAGEN3. B and C, Expression patterns of genes encoding polar auxin transporters (B) and genes in the auxin response pathway (C). Relative expression levels of the genes are shown in a heatmap diagram on the left and in a graph on the right. Asterisks indicate genes predominantly expressed in the early ovules at the AC and/or MMC stages.
Polar auxin transport is mediated by influx and efflux transporters located in the plasma membrane (Zazímalová et al., 2010). AUXIN1/LIKE-AUX1 (AUX1/LAX) and PIN-FORMED (PIN) family proteins have been reported to be auxin influx carriers (Péret et al., 2012) and efflux carriers (Petrásek et al., 2006), respectively. Six rice PIN genes and four rice AUX genes were included among the AC- and/or MMC-preferential genes (Fig. 3B). The two efflux carrier genes OsPIN1c and OsPIN1d were preferentially expressed at the AC and MMC stages during megasporogenesis and were significantly down-regulated in the other four samples examined. OsPIN1b was preferentially expressed at the MMC stage with the exception of high expression in the SS sample. OsPIN1a levels in the MMC and FM were higher than those in other samples. The influx carrier gene OsAUX3 was found to be preferentially expressed at the AC stage, similar to OsAUX2, OsAUX4, and OsAUX5. P-glycoproteins (PGPs), a B subfamily of the ATP-binding cassette transporters, also participate in intercellular auxin uptake and/or efflux as transporters (Kaneda et al., 2011). OsPGP14 was highly expressed in all six samples, and its expression level was highest at the AC stage. Similarly, OsPGP16 was preferentially expressed in the ovules at the AC stage and was significantly down-regulated in subsequent developmental stages during megasporogenesis. OsPGP13 expression levels in the AC, MMC, and FM were obviously higher than that in the MO. In contrast, OsPGP10 and OsPGP11 levels gradually increased from the AC stage to the MO stage during ovule development (Fig. 3B).
Several auxin signaling-related genes were also abundantly expressed at the AC and/or MMC stages during megasporogenesis (Fig. 3C). Nine genes in the rice ARF family were enriched in early ovules at the AC and MMC stages. The relative transcript levels of five ARF genes, OsARF2, OsARF3, OsARF9, OsARF14, and OsARF15, were significantly higher at the AC stage compared to the other stages. Similarly, OsIAA7 and OsIAA30 were preferentially expressed in early ovules at the AC stage. OsIAA19 was preferentially expressed at the MMC stage. These data suggest the potential involvement of these auxin signaling-related genes in rice megasporogenesis.
Expression Profiles of EPF-ER-MAPK Pathway Genes in Rice
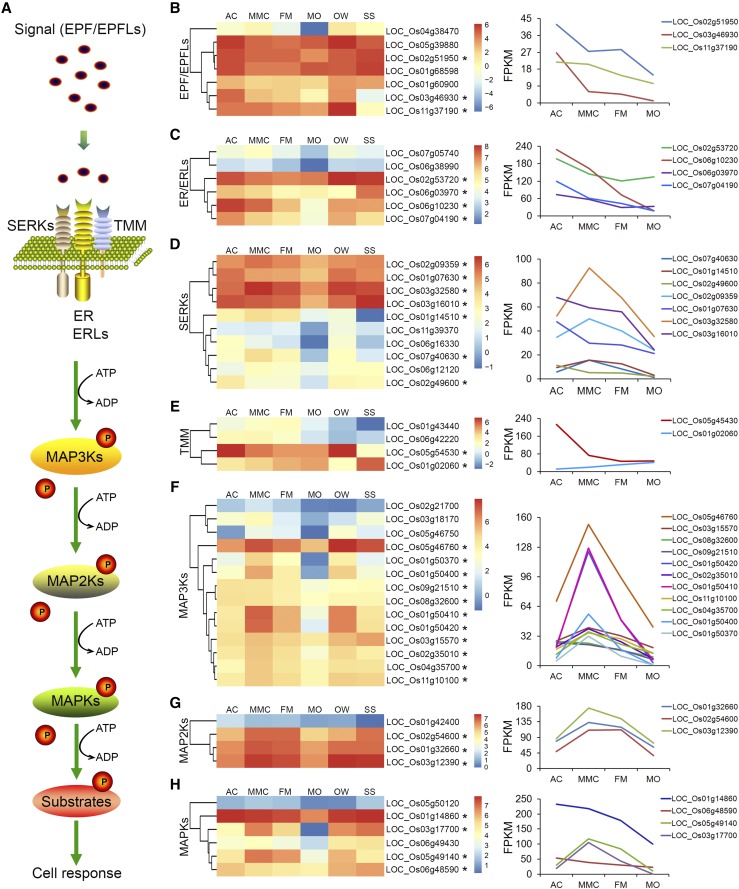
Our KEGG analysis of AC- and MMC-preferential genes, i.e. genes preferentially expressed in premeiotic stage ovules, showed an enrichment of genes involved in plant-pathogen interaction pathways (Fig. 2, E and F). Among these genes, we identified many genes associated with the EPIDERMAL PATTERNING FACTOR (EPF)-ERECTA (ER)-mitogen-activated protein kinase (MAPK) signaling pathway (Supplemental Table S8). Numerous models have been proposed to explain the actions of EPF, ER, and MAPKs in stomatal patterning and cellular responses to a wide range of stress stimuli (Lee et al., 2012; Tameshige et al., 2017), but their functions in rice ovule development have not been investigated.
EPF-ER-MAPK signaling pathway components (Fig. 4A) include EPF/EPF-LIKE (EPFL) small peptides that act as extramembranous ligands, ER/ER-LIKE (ERL) receptor kinases involved in transmembrane signal transduction, and MAPK enzymes important for cascade activation of downstream related proteins (Lee et al., 2012; Cai et al., 2017; Tameshige et al., 2017). In this study, expression profiling of genes related to the EPF-ER-MAPK signaling pathway showed that several OsEPF/EPFLs, OsER/ERLs, and OsMAPKs were preferentially expressed during rice megasporogenesis (Fig. 4, B–H). Three EPFL genes (Os02g51950, Os03g46930, and Os11g37190) as well as Os04g38470, Os05g39880, Os01g68598, and Os01g60900 were highly and preferentially expressed in early ovules at the AC and/or MMC stages (Fig. 4B). The transcript levels of four ER/ERL genes (Os02g53720, Os06g03970, Os06g10230, and Os07g04190) were significantly higher in ovules at the AC and/or MMC stages compared to other stages of megasporogenesis (Fig. 4C). Several rice SOMATIC EMBRYOGENESIS RECEPTOR KINASES (OsSERKs) and TOO MANY MOUTHS (OsTMMs), which encode putative coreceptors for many LEU-RICH REPEAT (LRR)-type receptor kinases such as ER in the plasma membrane, were found to be preferentially expressed in AC- and/or MMC-stage ovules, including Os02g09359, Os01g07630, Os03g32580, Os03g16010 (Fig. 4D), and Os05g54530 (Fig. 4E).
Figure 4.
Expression patterns of megasporogenesis-enriched genes in the EPF-ER-MAPK signaling pathway. A, Diagram of the plant EPF-ER-MAPK signaling pathway. B to H, Expression patterns of genes encoding EPF/EPFLs (B), ER/ERLs (C), SERKs (D), TMMs (E), MAP3Ks (F), MAP2Ks (G), and MAPKs (H). Heatmap diagrams of the relative expression levels of the genes are shown on the left, and the corresponding graphs are on the right. Asterisks (*) indicate genes predominantly expressed in the early ovules at the AC and/or MMC stages.
Transcript levels of MAPK cascade genes involved in the downstream signaling of ER were also obviously increased during megasporogenesis (Fig. 4, F–H). For example, the expression level of two OsMAPKKK (MEK KINASE) genes (Os09g21510 and Os08g32600) was higher in AC-stage ovules than in the other tissues examined. The OsMAPKKK genes Os01g50410 and Os01g50420 were preferentially expressed in MMC- and FM-stage ovules during megasporogenesis. Six OsMAPKKK genes (Os05g46760, Os01g50370, Os01g50400, Os02g35010, Os04g35700, and Os11g10100) were preferentially expressed in MMC-stage ovules (Fig. 4F). Similarly, three OsMAPKK genes (Os02g54600, Os01g32660, and Os03g12390) were preferentially expressed in MMC- and FM-stage ovules, and the OsMAPKK gene Os01g42400 was relatively highly expressed in AC-stage ovules (Fig. 4G). Furthermore, several OsMAPK genes were preferentially expressed in early developing ovules, including Os06g48590 at the AC stage; Os03g17700 and Os05g49140 at the MMC and FM stages; and Os01g14860 at the AC, MMC, and FM stages of megasporogenesis (Fig. 4H). Collectively, these data suggest that multiple genes associated with the EPF-ER-MAPK pathway may regulate megasporogenesis in rice.
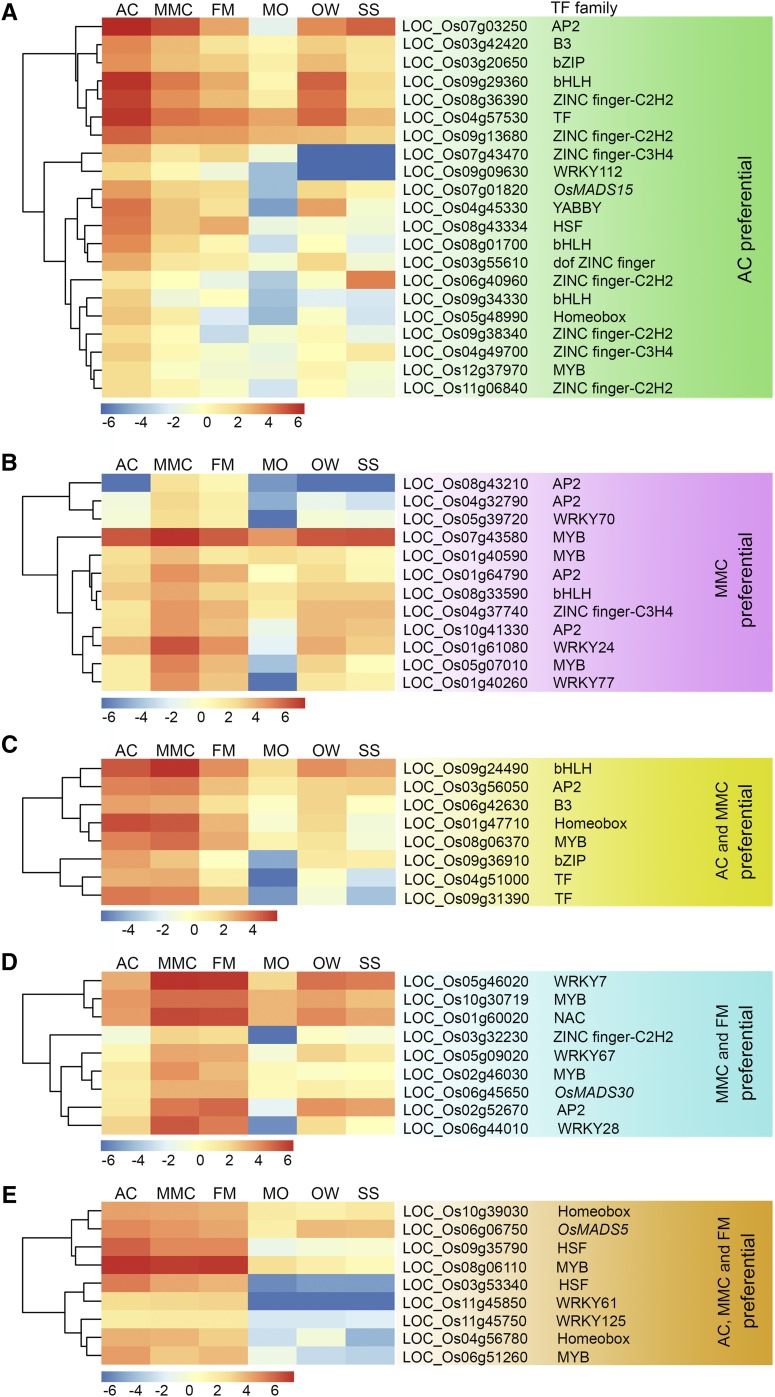
TF Genes Preferentially Expressed in Early Developing Ovules
The ∼1,430 putative transcription factor (TF) genes in rice ovules were identified from the Plant Transcription Factor Database (Pérez-Rodríguez et al., 2010) and the rice TF database (Kubo et al., 2013; Wu et al., 2015; http://drft.cbi.pku.edu.cn/). Our expression profiling analysis of early ovule development in rice identified 59 putative TF genes among the megasporogenesis preferentially expressed genes. The 59 genes represented diverse TF families, including MYB DOMAIN PROTEIN (MYB), BASIC LEU-ZIPPER (bZIP), BASIC HELIX-LOOP-HELIX (bHLH), WRKY DNA-BINDING PROTEIN (WRKY), and MADS-BOX TF FAMILY PROTEIN (MADS; Supplemental Table S9), and a subset of these TF genes that function in meristem identity specification or floral organ development were cloned and functionally analyzed in previous studies (Cui et al., 2010; Shao et al., 2019). However, whether these genes regulate female gametophyte development in rice is presently unknown.
The 59 TF genes were classified into five groups according to their expression patterns at the different stages of rice megasporogenesis. Of these genes, 21 had significantly higher expression in early ovules at the AC stage compared to the other five analyzed samples, and these were designated as AC-preferential TF genes (Fig. 5A; Supplemental Table S9). Twelve genes with significantly higher expression at the MMC stage compared to the other five stages were designated as MMC-preferential TF genes (Fig. 5B). Eight genes had significantly higher expression at the AC and MMC stages compared to the other analyzed stages, and these were designated as AC and MMC-preferential genes (Fig. 5C). The expression levels of nine genes at the MMC and FM stages were significantly higher than those in the other five tissues, and these genes were designated as MMC and FM-preferential TF genes (Fig. 5D). An additional nine genes were relatively highly expressed at the AC, MMC, and FM stages, and these were designated as AC, MMC, and FM-preferential TF genes (Fig. 5E).
Figure 5.
Hierarchical cluster analysis of the relative expression levels of megasporogenesis-enriched TF genes. A and B, TF genes expressed preferentially at the AC (A) and MMC (B) stages. C, TF genes expressed preferentially at both the AC and MMC stages. D, TF genes expressed preferentially at both the MMC and FM stages. E, TF genes expressed preferentially at AC, MMC, and FM stages during megasporogenesis in rice. AP2, AP2 domain containing protein; B3, B3 domain containing protein; bZIP, bZIP family transcription factor; HSF, heat shock factor; Homeobox, Homeobox domain containing protein.
MYB TFs are known to have important roles in plant reproductive development. Among the 59 megasporogenesis-enriched TF genes identified in this analysis, nine MYB genes were identified, including one AC-preferential gene (Fig. 5A), three MMC-preferential genes (Fig. 5B), one AC and MMC- preferential gene (Fig. 5C), two MMC and FM-preferential genes (Fig. 5D), and two AC, MMC, and FM-preferential genes (Fig. 5E). Another nine of the 59 TF genes were found to encode ZINC finger TFs, and most of these genes were AC-preferential genes or AC- and MMC-preferential genes. For example, Os08g36390 and Os09g13680 were relatively preferentially expressed in early ovules at the AC stage, and Os04g37740 was relatively highly expressed at the MMC stage (Fig. 5, A and B). These results indicate the potential involvement of MYB and ZINC finger TF genes in regulating female germline cell fate identity and MMC differentiation during megasporogenesis in rice.
In total, 41 of the 59 TF genes (i.e. 21 AC-preferential genes, 12 MMC-preferential genes, and 8 AC- and MMC-preferential genes) were preferentially expressed in early developing ovules before meiosis. Nine TF genes were preferentially expressed at the MMC and FM stages, while the remaining nine TF genes were relatively highly expressed at all three stages during megasporogenesis (Fig. 5). These data indicate that many AC- and MMC-preferentially expressed TF genes may be required to establish the regulatory network of gene expression in AC cell specification and MMC formation during megasporogenesis in rice, and this regulatory network remains largely uncharacterized.
Validation of Gene Expression Patterns by In Situ Hybridization
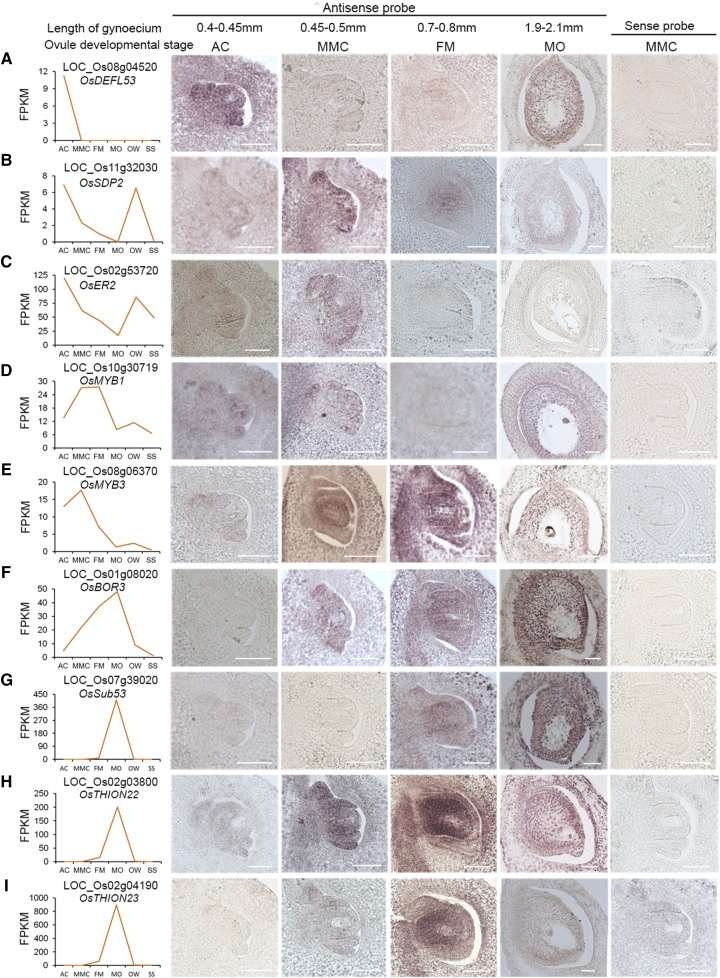
To validate the RNA-Seq results, we selected nine genes that were preferentially expressed at different stages of rice ovule development and performed in situ hybridization (Fig. 6). The nine selected genes encode the following: three small peptides (Fig. 6, A, H, and I), a short-chain dehydrogenases/reductase family member (Fig. 6B), a LRR receptor-like protein kinase (RLK; Fig. 6C), two MYB TFs (Fig. 6, D and E), a boron transporter protein (Fig. 6F), and a putative subtilisin homolog (Fig. 6G).
Figure 6.
Validation of the expression patterns of nine genes in ovules at different developmental stages by in situ hybridization. A to I, Expression patterns of OsDEFL53 (A), OsSDP2 (B), OsER2 (C), OsMYB1 (D), OsMYB3 (E), OsBOR3 (F), OsSub53 (G), OsTHION22 (H), OsTHION23(I). Images on the left show expression patterns based on the FPKM values from RNA-Seq analysis. Representative images of sense RNA probe hybridization for the nine genes are shown in the rightmost images. Scale bars = 50 μm.
A positive signal was detected with all nine antisense probes in the ovules at different developmental stages, while no hybridization signal was detected with the control sense probes (Fig. 6). The expression patterns of these nine genes during ovule development were similar to the RNA-Seq data. OsDEFL53 (DEFENSIN-LIKE 53, Os08g04520) encodes a defensin-like peptide and was detected specifically in early ovules at the AC stage (Fig. 6A). Two probes corresponding to genes encoding a short-chain dehydrogenases/reductase family member, rice SEX DETERMINATION PROTEIN2 (OsSDP2 [Os11g32030]) and a LRR RLK (OsER2 [Os02g53720]) showed strong signals in ovules at both the AC and MMC stages and lower signals in the FM and MO tissues, consistent with the RNA-Seq data (Fig. 6, B and C). The R2R3-type MYB TF (OsMYB1 [Os10g30719]) was detected preferentially in ovules at the AC, MMC, and MO stages (Fig. 6D). Another MYB gene (OsMYB3 [Os08g06370]) was detected at abundant levels at the MMC and FM stages (Fig. 6E). The signal of rice BORON TRANSPORTER3 (OsBOR3 [Os01g08020]) was increased from the AC to the MO stage, highly consistent with the RNA-Seq data (Fig. 6F). The last three genes were strongly detected at the FM and MO stages (Fig. 6, G–I–I). The different expression patterns observed for these selected genes suggest their diverse functions during rice ovule development. Moreover, the in situ hybridization results for these genes indicated that our gene profiling dataset has high spatiotemporal resolution, which will facilitate future functional validation of these genes and investigation of the molecular mechanisms underlying female gametophyte development in plants.
ER2 Regulates Female Germline Specification during Rice Megasporogenesis
The results described above focused on genes that were preferentially expressed during megasporogenesis compared to ovules harboring mature female gametophytes in rice and might therefore regulate associated developmental processes. We subsequently performed a functional analysis of the AC- and MMC-preferential gene OsER2 (Os02g53720) using the CRISPR/Cas9 gene-editing system (Ma et al., 2015). Through Agrobacterium tumefaciens-mediated transformation, the CRISPR/Cas9 constructs were introduced into the ZH11 cultivar, resulting in a set of transgenic plants (T1 generation).
PCR and sequencing were used to detect whether mutations were introduced in the targeted region in the T1 plants. All mutant plants in the T1 generation were heterozygotes, which were self-crossed to produce the T2 generation. Three homozygous mutant lines containing different mutations in the first exon of OsER2 were obtained in the T2 generation, and named oser2-1, oser2-2, and oser2-3 (Supplemental Fig. S2). oser2-1 contains a frameshift mutation resulting from a 1-bp insertion at position C32 bp of the coding sequence. oser2-2 contains a 2-bp deletion at positions C33 and C34 bp. oser2-3 has a 19-bp deletion at positions C28–C46 bp (Supplemental Fig. S2). These three mutations in the first exon of OsER2 resulted in altered protein sequences and a premature stop codon.
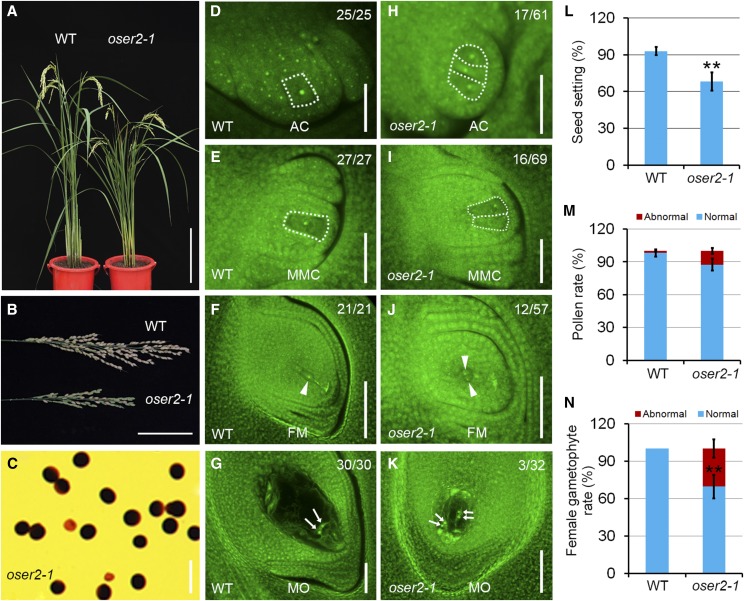
To assess the effects of disrupted OsER2 function on vegetative and reproductive growth, plant height, tiller number, panicle morphology, and seed set were investigated in the three oser2 lines at the mature stage. Compared to the wild type, all three mutants had obviously reduced plant height and panicle size as well as a significantly reduced seed setting rate (Fig. 7, A, B, and L, Supplemental Fig. S3). To determine the underlying cause of the decreased seed setting, we examined male and female gametophyte morphology in the oser2 lines and wild-type plants. Staining of pollen with iodine potassium iodide (I2-KI) solution, which indicates pollen viability, showed that ∼87.5% of the pollen could be stained in the oser2-1 mutant (n = 563; Fig. 7C), slightly lower than that in the wild type (98.1%, n = 517; Fig. 7M). While a significantly enhanced abnormality in the female gametophyte structure was detected in the mature oser2-1 ovules (28.1%, n = 32) as compared to the wild type (0%, n = 30; Fig. 7N). A mature ovule in the wild type harbors just one female gametophyte, which contains two synergid cells, an egg cell, one central cell with two polar nuclei, and a group of antipodal cells (Fig. 7G). In contrast, two types of abnormal female gametophytes were examined in mature ovules of the oser2-1 mutant. The abnormal female gametophytes in the oser2-1 mutant either composed two sets of female gametophytes within one ovule (Fig. 7K) or no female gametophyte (Supplemental Fig. S3S). Similar female gametophyte defects were also examined in oser2-2 and oser2-3 ovules (Supplemental Fig. S3, N, R, T, and U). These data indicate that abnormal female gametophyte development might be the main cause of reduced seed setting in the oser2 mutants.
Figure 7.
Phenotypic characterization and ovule development in wild-type (WT) and oser2-1 plants. A, Wild-type (left) and oser2-1 mutant (right) plants at maturity. B, Panicles from the wild type and the oser2-1 mutant. C, Viability of mature pollen grains in the oser2-1 mutant as assessed by I2-KI staining. D to G, Developing ovules in the wild type at the AC (D), MMC (E), FM (F), and MO (G) stages. H to K, Developing ovules in the oser2-1 mutant at the AC (H), MMC (I), FM (J), and MO (K) stages. Scale bars = 30 cm (A), 8 cm (B), 50 μm (C), and 20 μm (D–G and H–K). L, Seed setting rates in the wild type (n = 30) and oser2-1 mutant (n = 30). M, Viability of mature pollen grains in the wild type (n = 517) and oser2-1 mutant (n = 563). N, Percentage of abnormal female gametophytes at the mature stage in the wild type (n = 30) and oser2-1 mutant (n = 32). The female germline cells in wild-type and oser2-1 ovules are outlined by the white dashed lines. Arrowheads in F and J indicate the FM or FM-like cells. Arrows in G and K indicate the polar nuclei in the central cell. Numbers in the panels denote frequencies of the phenotypes shown. Data in L–N represent means ± sd. Double asterisks represent significant differences between the wild type and the mutant as determined by Student’s t test (**P < 0.01).
To determine the stage at which female gametophyte development in the oser2 ovule departed from that of the wild type, the ovules at early developmental stages were examined. In the wild-type ovule primordium, only a single subepidermal cell in the nucellus differentiates into the AC, which subsequently develops into a MMC and a FM (Fig. 7, D–F). In the oser2-1 mutant, more than one AC-like or MMC-like cell was observed at the AC or MMC stages with frequencies of 27.9% (n = 61) and 23.2% (n = 69), respectively (Fig. 7, H and I), indicating that female germline specification was defective in oser2-1 ovules during megasporogenesis. Similarly, 21.1% (n = 57) of oser2-1 ovules at the FM stage contained two FM-like cells within one ovule (Fig. 7J). The remaining 78.9% of oser2-1 ovules at the FM stage exhibited a wild-type-like phenotype containing one enlarged FM, suggesting that the lack of female gametophyte phenotype in mature oser2-1 ovules manifested after FM formation, likely during megagametogenesis. oser2-2 and oser2-3 mutant lines also exhibited more than one germline phenotype in ovules during megasporogenesis and double-female-gametophyte or no-gametophyte phenotypes in mature ovules (Supplemental Fig. S3). These data indicate that OsER2 plays an important role in MMC specification and subsequent female gametophyte development.
DISCUSSION
Identification of Many Preferential/Specific Genes in Rice Megasporogenesis by Transcriptome Analysis
The female gametophyte in the ovule of a flowering plant is the site of double fertilization, and it eventually develops into the embryo and endosperm of the seed, which is crucial for plant reproduction and human food production. Although female gametophyte development has been studied extensively, and many important genes controlling megasporogenesis and megagametogenesis have been identified in Arabidopsis (Ravi et al., 2008; Rabiger and Drews 2013; Qin et al., 2014; Cao et al., 2018; Zhao et al., 2018), the number of studies of female gametophyte development in rice is limited, and only a few key regulatory genes are known. Microarrays and RNA sequencing technology have been used previously to obtain transcriptome data for developing rice ovules from the MMC at the premeiotic to mature embryo sac stage (Ohnishi et al., 2011; Kubo et al., 2013; Wu et al., 2015), but limited information is available in terms of dynamic gene expression data for AC specification and MMC formation during megasporogenesis in rice.
In this study, in addition to gene expression profiles of ovules at the MMC, FM, and MO stages, a gene expression profile of ovules at the AC stage was obtained. During the analysis of up-regulated DEGs in the ovule during megasporogenesis, we found that when the MO, OW, and SS tissues were used as controls, several known genes that function in rice megasporogenesis, such as OsMEL1 and OsIG1 (Nonomura et al., 2007; Zhang et al., 2015), are not included in the identified up-regulated genes. In consideration of the fact that some genes have multiple roles in different developmental tissues and might not show tissue-specific expression patterns (for example, OsIG1 regulates female gametophyte development and floral organ number and is expressed not only in ovule primordia but also in floret organs such as in the palea and lemma [Zhang et al., 2015]), we adjusted the strategy of analysis and only used the MO as the control to screen the genes with up-regulated expression in the ovules during megasporogenesis. The known key genes, such as OsMEL1 and OsIG1 for rice megasporogenesis, were indeed included in the up-regulated DEGs when only the MO tissue was used as a control (Supplemental Table S4). Therefore, these genes were designated as megasporogenesis genes with up-regulated expression compared to MOs and were used for further analysis. This MO-only comparison tends to identify megasporogenesis stage preferential or specific genes. Whether they show a different expression pattern in the surrounding ovule tissue, such as integuments and nucellus, is not known, and their functions in megasporogenesis also remain to be further validated.
Our RNA-Seq analysis identified 5,274 genes that were up-regulated in rice early ovules during megasporogenesis as compared to MOs; 958 of these genes were preferentially expressed in young ovules at the AC and/or MMC stages before meiosis, as compared to ovules at the FM and mature stages (Fig. 2D). KEGG pathway analysis of the AC-preferential and MMC-preferential genes, i.e. genes preferentially expressed in young ovules, indicated an enrichment of genes associated with plant hormone signal transduction and plant-pathogen interactions. There were also a large number of genes related to starch and Suc biosynthesis (Fig. 2, E and F). These results were consistent with a previous gene expression profiling study of developing rice ovules by Wu et al. (2015). Among the megasporogenesis preferentially expressed genes identified in this study, 41 encoded auxin signaling-related genes, including six auxin efflux carrier genes, four influx carrier genes, four PGP genes, nine ARF genes, nine Aux/IAA genes, and 10 auxin response genes (Fig. 3; Supplemental Table S6). Among the genes associated with plant-pathogen interaction pathways, >45 were related to EPF-ER-MAPK signaling, including seven EPF/EPFL-encoding genes, 20 genes encoding RLKs, and 24 genes encoding MAPK cascade factors (Fig. 4; Supplemental Table S8).
Among the 59 megasporogenesis-enriched TF genes, 41 were AC- and/or MMC-preferential genes, and these included MYB, bZIP, bHLH, WRKY, and MADS TF family members (Fig. 5, A–C). In particular, three MYB genes (Os12g37970, Os01g40590, and Os05g07010) and four bHLH genes (Os09g29360, Os08g01700, Os09g34330, and Os08g33590) were preferentially expressed in the early ovules at the AC or MMC stage (Fig. 5, A and B). In Arabidopsis, MYB64 and MYB119 are expressed in the nucleus of the female gametophyte during the FG5 transition, and most gametophytes fail to initiate the FG5 transition in the myb64 myb119 double mutant, resulting in enlarged coenocytic gametophytes with supernumerary nuclei (Rabiger and Drews, 2013). Our rice megasporogenesis transcriptome analysis provides gene expression data for developing rice ovules; it may therefore help identify key megasporogenesis regulators and shed light on our understanding of the mechanisms underlying ovule development in plants.
Expression Patterns of Auxin Signaling-Related Genes in Early Ovules Highlight Their Potentially Important Roles in Rice Megasporogenesis
The phytohormone auxin plays a fundamental role during plant growth and development and in the response to environmental conditions. The effect of auxin reflects the interaction of auxin biosynthesis, polar transportation, and signal transduction, and a defect in any of these three processes disrupts plant vegetative and reproductive growth (Péret et al., 2012; Panoli et al., 2015). Studies in Arabidopsis have established that these three processes are also required for ovule initiation, including the emergence of the ovule primordia from the placenta, AC cell specification, and MMC formation (Robert et al., 2015). Nevertheless, the regulatory role of auxin during ovule development in rice remains unknown, and genes related to auxin signaling have not been reported to control megasporogenesis or female gametophyte development in rice.
In Arabidopsis, it was proposed that repression of the YUCCA auxin biosynthesis genes by the MADS-box TF SPL/NZZ may compromise MMC formation during early ovule development (Schiefthaler et al., 1999; Yang et al., 1999; Li et al., 2008). Down-regulation or mutation of the auxin efflux carrier PIN1 severely affects ovule polarity and blocks progression of female gametophyte development (Ceccato et al., 2013). The auxin influx carriers AUX1, LAX1, and LAX2 reportedly function redundantly to ensure normal female gametophyte development, and the aux1 lax1 lax2 triple mutant exhibits abnormal ovule development (Panoli et al., 2015). In our rice ovule transcriptome data, several auxin influx and efflux carrier genes were found to be preferentially expressed in early ovules at the AC and/or MMC stages, including OsPIN1b, OsPIN1c, OsPIN1d, OsAUX2, OsAUX3, and OsAUX4 (Fig. 3B). Considering the sequence similarity of homologous genes in rice and Arabidopsis (Wang et al., 2009; Zhao et al., 2012), it is reasonable to hypothesize that these AC and/or MMC-preferential auxin carrier genes might regulate megasporogenesis in rice ovules.
Auxin signal transduction is an important part of the physiological function of auxin, and its components include ARFs and Aux/IAA repressors. Down-regulation of SlARF6 and SlARF8 by microRNA167 leads to abnormal floral development and female sterility in tomato (Solanum lycopersicum; Liu et al., 2014). Our recent study demonstrated that AtARF3 is specifically expressed in the endodermal cells of the chalaza in early ovules; ectopic expression of AtARF3 in lateral epidermal cells induces supernumerary MMC formation, indicating that AtARF3 regulates MMC specification in Arabidopsis (Su et al., 2017). The RNA-Seq analysis described here showed that several rice auxin response factors, including OsARF2, OsARF3, OsARF14, and OsARF15, were specifically expressed in early ovules at the AC stage in rice (Fig. 3C). Phylogenetically, these four OsARFs and AtARF3 are present on the same evolutionary branch with high sequence similarity (Wang et al., 2007a). These results suggest that these OsARFs may regulate meiocyte formation and early ovule development, in a manner similar to that observed for AtARF3.
EPF-ER-MAPK Pathway Genes May Regulate Rice Megasporogenesis
Several studies have shown that the ligand-receptor signaling system is critical for the specification of germline cell fate and number in plants (Sheridan et al., 1999; Nonomura et al., 2003; Yang et al., 2010). The ligand-receptor signaling system comprises three components, extracellular ligands, membrane receptors, and the intracellular kinase cascade, and is relatively conserved among plant species. It was previously reported that the extracellular ligands EPFL4, EPFL5, and EPFL6 may be sensed by ER family (ERf) receptors in developing ovules, and the epfl4 epfl5 epfl6 triple mutant has reduced fertility (Abrash et al., 2011; Shpak 2013; Uchida and Tasaka, 2013; Tameshige et al., 2017). Our KEGG pathway analysis highlighted a set of EPF/EPFL genes with relatively high expression in early ovules at the AC and/or MMC stages (Fig. 4B), suggesting their potential involvement as signals in the regulation of megasporogenesis and ovule development in rice.
The three ERf genes in Arabidopsis, ER, ERL1, and ERL2, encode LRR-RLKs. Beyond their functions in stomatal development and stress response, ERf genes also regulate reproductive organ development, including integument development during early ovule morphogenesis. The ovules in the haploinsufficient er erl1 erl2/+ mutant develop abnormally, with decreased cell proliferation in the integuments and an abortive gametophyte (Pillitteri et al., 2007; Shpak 2013). The ovule defects of the er mutant are substantially enhanced by a mutation affecting the chromatin remodeling complex SWR1 in Arabidopsis (Cai et al., 2017). Recently, three ERfs have been identified in the rice genome, including OsER1 (Os06g10230), OsER2 (Os02g53720), and OsERL(Os06g03970); the mutation in OsER2 resulted in a significantly reduced seed setting rate, and the evolutionary analysis indicated that the ER family may be highly conserved across different species (Zhang et al., 2018). Our expression profiling showed that the three ERf genes, OsER1, OsER2, and OsERL, were preferentially expressed in early-stage ovules before meiosis (Fig. 4C). Moreover, the expression pattern of OsER2 was confirmed by in situ hybridization (Fig. 6C). Loss-of-function oser2 mutants had more than one AC-like cell at the AC stage, as well as supernumerary MMCs and FMs (Fig. 7), suggesting that the rice ER gene family might participate in restricting germline cell fate transition.
Rice MULTIPLE SPOROCYTE 1 (MSP1) encodes a membrane-bound LRR-RLK with crucial roles in determining AC fate by preventing the surrounding cells from developing into germline cells. Specifically, OsMSP1 transcripts are detected in somatic cells surrounding the germline cells but not in the developing germline cells, and ovules in the msp1 mutant have more than one AC and an excessive number of embryo sacs (Nonomura et al., 2003). Yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays showed that rice TAPETUM DETERMINANT LIKE1A (OsTDL1A) could bind the extracellular domain of OsMSP1 as a putative ligand, and the ovules of OsTDL1A RNA interference plants exhibited phenotypic defects similar to those of the msp1 mutant (Zhao et al., 2008). The present transcriptome analysis identified a number of RLK-encoding genes that were specifically or preferentially expressed in ovules during megasporogenesis as compared to ovules harboring mature female gametophytes, indicating their potential involvement in rice megasporogenesis.
In the EPF-ER-MAPK pathway, ER/ERL receptors activate the downstream MAPK signaling cascade after sensing the extramembrane signal (Shpak 2013). Previous studies indicate that the MAPK signaling cascade consisting of YODA (YDA), MEK 4 (MKK4), MKK5, MAPK 3 (MPK3), and MPK6 regulates integument development in young ovules (Wang et al., 2007b; Meng et al., 2012). AtMPK3 and AtMPK6, which encode two closely related MAPKs in Arabidopsis, facilitate cell division in the integument specifically during early ovule development. Similar to the er erl1 erl2/+ mutant, the mpk3/+ mpk6 mutant exhibits abnormal integument development with arrested cell division, resulting in physical restriction of embryo sacs and sterility of the female gametophyte (Wang et al., 2008). Many genes encoding MAPK cascade proteins were found in this study to be predominantly expressed during rice megasporogenesis (Fig. 4, F–H). Combined with information from previous studies, the present findings suggest that these genes might also have a prominent role in early ovule differentiation and development in rice. Further analyses are required to validate the functions of these genes in rice megasporogenesis.
MATERIALS AND METHODS
Plant Materials and Rice Ovule Observation
Rice (Oryza sativa) ssp. japonica ‘Zhong Hua 11’ (ZH11) and the mutant oser2 were grown in the greenhouse of Fujian Agriculture and Forestry University at 22°C to 32°C and 80% to 90% humidity with a 14-h light/10-h dark photoperiod. To assess the correlation between the morphological features of rice spikelets and female gametophyte development, spikelets at various stages were carefully collected from young panicles of 70-d-old seedlings before the collar of the flag leaf emerged from the sheath of the second upper leaf. For the analysis, spikelet, anther, and gynoecium length were measured. At the same time, the spikelets were fixed and observed to certify the ovule developmental stages using semithin sections and whole-mount Eosin B-staining confocal laser scanning microscopy (CLSM), as described by Zeng et al. (2007, 2009). After fixation, Eosin B staining, and clearing, the ovaries were placed at the bottom of a glass cell culture dish (Nest) and scanned using a Leica SP8 CLSM (Leica Microsystems). The excitation wavelength was 514 nm, and the emission wavelengths were 550–630 nm. Ovary images were recorded using the software accompanying the microscope.
RNA Extraction and Transcriptome Sequencing
Ovules from 1.5- to 2-mm spikelets (anther length 0.4–0.5 mm), 2- to 2.6-mm spikelets (anther length 0.5–0.55 mm), and 3.8- to 4.2-mm spikelets (anther length 0.75–0.8 mm) were collected using microdissection needles, immediately frozen in liquid nitrogen, and stored at −80°C for total RNA extraction. Moreover, the ovary wall and the ovary style and stigma from 3.8- to 4.5-mm spikelets (anther length 0.75–1.0 mm) and the ovules from mature spikelets before flowering (anther length 1.9–2.2 mm) were collected as controls. Three biological replicates of each tissue were conducted individually. RNA from the different samples was extracted using the Plant RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen). One microgram of RNA was used for cDNA synthesis, and the final product was diluted five times in a total volume of 100 μL. The cDNA libraries for sequencing were constructed using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturer’s suggested procedures. Transcriptome sequencing was performed on the HiSeq 2500 platform using the 100-bp paired-end protocol. The RNA-seq raw data entitled “Transcriptome analysis of rice ovules during megasporogenesis” were deposited in the BioProject repository with accession number PRJNA591969 (https://www.ncbi.nlm.nih.gov/bioproject/591969).
RNA-Seq Data Processing and DEG Identification
A standard protocol was used for the RNA-Seq data preprocessing. The fastq format raw sequencing reads from all samples were collected and initially processed using fastp (Chen et al., 2018), and the adapter and low-quality reads were removed. The clean reads were mapped to the rice reference genome (MSU7.0; http://rice.plantbiology.msu.edu/index.shtml) using TopHat (Trapnell et al., 2012), then the transcripts were assembled and quantified using Cufflinks. DEGs were analyzed using Cuffdiff (Trapnell et al., 2012). Genes with ≥2-fold up-regulated or down-regulated expression and q ≤ 0.05 (counted using Cuffdiff) were regarded as up-regulated or down-regulated genes. KEGG analysis of the DEGs was performed to predict the enrichment pathway by using the proteome data Kobas 3.0 (kobas.cbi.pku.edu.cn). After being downloaded from the MSU7.0, the rice proteome file was submitted into the Kobas 3.0 and annotated functionally. Then, the protein sequences of up-regulated genes at the AC or MMC stages were searched against the annotated rice proteome file, and the statistical significance of the enrichment of KEGG pathways was examined using Fisher’s exact test, followed by the false discovery rate correction set at P < 0.05 using the method of Benjamini and Hochberg (1995). The enriched pathways were identified and drawn by the bubble chart using the R program (http://www.R-project.org/).
RNA In Situ Hybridization
The spikelets and ovaries at various developmental stages in wild-type cv ZH11 were fixed in RNase-free 4% (w/v) paraformaldehyde for 16 h at 4°C, dehydrated through an ethanol series, then embedded in Paraplast Plus (Sigma). All samples were sectioned at 8 μm thickness using an RM2245 rotary microtome (Leica). The target cDNA was amplified and subcloned into the pGEMR-T Easy vector (Promega), which was used as a template to generate both the antisense and sense RNA probes. The primer sequences of the nine genes selected are listed in Supplemental Table S10. The segment was transcribed in vitro under control of the T3 or T7 promoter with RNA polymerase using the DIG RNA Labeling Kit (Promega), and the mixture was prepared for the DIG-labeled RNA antisense or sense probe. The concentration of the probe for each gene was 1.5 ng/mL. RNA hybridization and immunological detection of the hybridized probes were performed as described previously (Brewer et al., 2006; Yu et al., 2017). The different hybridizations were treated in the same way: hybridization at 58°C for 36 h, and color reaction at room temperature for 2–3 h. Images were obtained using an Olympus IX71 microscope.
CRISPR/Cas9-Mediated Mutation and Phenotype Association Assay
To investigate OsER2 function, we generated a guide RNA (gRNA) construct with the gRNA (5′ CCACACTGGGCACAAGTTCCCAA 3′) and plant-optimized Cas9 driven by the rice OsU6a and maize pUBI-H promoters, respectively (Ma et al., 2015). The plasmid was introduced into wild-type ZH11, and DNA isolated from the leaves of transgenic plants was subjected to PCR and sequencing analysis with the primer set OsER2-gRNA-seq (forward primer, 5′GCCGTTGGGAACTTGTGCCCAGTG3′; reverse primer, 5′AAACCACTGGGCACAAGTTCCCAA3′). To evaluate whether a mutation was present and whether the candidate mutation site in OsER2 was associated with a female sterile phenotype, we observed the target site sequence of all the CRISPR/Cas9 transgenic plants via sequencing of the PCR products. To further observe the sterility phenotype in the mutants, we examined female gametophytes in ovules at different stages using whole-mount stain-clearing CLSM (Zeng et al., 2009).
Accession Numbers
Sequence data from this article for the cDNA and genomic DNA of OsER2 can be found in the Rice Genome Annotation Project data libraries under accession number LOC_Os02g53720.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Hierarchical clustering of the nine control samples using Pearson correlation coefficients.
Supplemental Figure S2. Identification of three CRISPR/Cas9-mediated mutations in OsER2.
Supplemental Figure S3. Phenotypic characterization and ovule development in wild-type, oser2-2, and oser2-3 plants.
Supplemental Table S1. Number of reads from the 18 samples in this study.
Supplemental Table S2. Classification of gene expression levels from the six tissues in this study.
Supplemental Table S3. Preferentially expressed genes in at least one tissue of AC, MMC, and FM, as compared to MO, OW, and SS, during megasporogenesis.
Supplemental Table S4. Up-regulated genes in at least one tissue during megasporogenesis compared to MO.
Supplemental Table S5. Genes preferentially expressed in early developing ovules before meiosis.
Supplemental Table S6. Enriched genes related to plant hormone signaling and plant-pathogen interaction pathways.
Supplemental Table S7. Megasporogenesis-enriched genes related to the auxin signaling pathway.
Supplemental Table S8. Megasporogenesis-enriched genes related to the EPF-ER-MAPK signaling pathway.
Supplemental Table S9. Megasporogenesis-enriched TF genes.
Supplemental Table S10. Primers of the nine genes used for in situ hybridization in this work.
Footnotes
This work was supported by the National Natural Science Foundation of China (NSFC; U1605212, 31761130074, 31970333, and 31571264), a Guangxi Distinguished Experts Fellowship, a Newton Advanced Fellowship (NA160391), and the Fujian Natural Science Foundation (2019J01425).
References
- Abrash EB, Davies KA, Bergmann DC(2011) Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23: 2864–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y(1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Brewer PB, Heisler MG, Hejátko J, Friml J, Benková E(2006) In situ hybridization for mRNA detection in Arabidopsis tissue sections. Nat Protoc 1: 1462–1467 [DOI] [PubMed] [Google Scholar]
- Cai H, Zhao L, Wang L, Zhang M, Su Z, Cheng Y, Zhao H, Qin Y(2017) ERECTA signaling controls Arabidopsis inflorescence architecture through chromatin-mediated activation of PRE1 expression. New Phytol 214: 1579–1596 [DOI] [PubMed] [Google Scholar]
- Cao L, Wang S, Venglat P, Zhao L, Cheng Y, Ye S, Qin Y, Datla R, Zhou Y, Wang H(2018) Arabidopsis ICK/KRP cyclin-dependent kinase inhibitors function to ensure the formation of one megaspore mother cell and one functional megaspore per ovule. PLoS Genet 14: e1007230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccato L, Masiero S, Sinha Roy D, Bencivenga S, Roig-Villanova I, Ditengou FA, Palme K, Simon R, Colombo L(2013) Maternal control of PIN1 is required for female gametophyte development in Arabidopsis. PLoS One 8: e66148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J(2018) fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettoor AM, Evans MM(2015) Correlation between a loss of auxin signaling and a loss of proliferation in maize antipodal cells. Front Plant Sci 6: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z(2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61: 767–781 [DOI] [PubMed] [Google Scholar]
- Gómez JF, Talle B, Wilson ZA(2015) Anther and pollen development: A conserved developmental pathway. J Integr Plant Biol 57: 876–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafidh S, Fíla J, Honys D(2016) Male gametophyte development and function in angiosperms: A general concept. Plant Reprod 29: 31–51 [DOI] [PubMed] [Google Scholar]
- Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM(2004) The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Kaneda M, Schuetz M, Lin BSP, Chanis C, Hamberger B, Western TL, Ehlting J, Samuels AL(2011) ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J Exp Bot 62: 2063–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R, Ohyanagi H, Niihama M, Watanabe T, Nakano M, Kurata N, Nonomura K(2014) Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J 78: 385–397 [DOI] [PubMed] [Google Scholar]
- Kubo T, Fujita M, Takahashi H, Nakazono M, Tsutsumi N, Kurata N(2013) Transcriptome analysis of developing ovules in rice isolated by laser microdissection. Plant Cell Physiol 54: 750–765 [DOI] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU(2012) Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev 26: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Qin GJ, Tsuge T, Hou XH, Ding MY, Aoyama T, Oka A, Chen Z, Gu H, Zhao Y, et al. (2008) SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytol 179: 751–764 [DOI] [PubMed] [Google Scholar]
- Lituiev DS, Krohn NG, Müller B, Jackson D, Hellriegel B, Dresselhaus T, Grossniklaus U(2013) Theoretical and experimental evidence indicates that there is no detectable auxin gradient in the angiosperm female gametophyte. Development 140: 4544–4553 [DOI] [PubMed] [Google Scholar]
- Liu N, Wu S, Van Houten J, Wang Y, Ding B, Fei Z, Clarke TH, Reed JW, van der Knaap E(2014) Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J Exp Bot 65: 2507–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Dee ZP, Wittich P, Enrico Pè M, Rigola D, Del Buono I, Gorla MS, Kater MM, Colombo L(1999) OsMADS13, a novel rice MADS-box gene expressed during ovule development. Dev Genet 25: 237–244 [DOI] [PubMed] [Google Scholar]
- Lora J, Yang X, Tucker MR(2019) Establishing a framework for female germline initiation in the plant ovule. J Exp Bot 70: 2937–2949 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, et al. (2015) A Robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Meng X, Wang H, He Y, Liu Y, Walker JC, Torii KU, Zhang S(2012) A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24: 4948–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N(2003) The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15: 1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N(2007) A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Takanashi H, Mogi M, Takahashi H, Kikuchi S, Yano K, Okamoto T, Fujita M, Kurata N, Tsutsumi N(2011) Distinct gene expression profiles in egg and synergid cells of rice as revealed by cell type-specific microarrays. Plant Physiol 155: 881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP(2010) Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panoli A, Martin MV, Alandete-Saez M, Simon M, Neff C, Swarup R, Bellido A, Yuan L, Pagnussat GC, Sundaresan V(2015) Auxin import and local auxin biosynthesis are required for mitotic divisions, cell expansion and cell specification during female gametophytedevelopment in Arabidopsis thaliana. PLoS One 10: e0126164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Swarup K, Ferguson A, Seth M, Yang Y, Dhondt S, James N, Casimiro I, Perry P, Syed A, et al. (2012) AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B(2010) PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res 38: D822–D827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubes M, Covanová M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Bemis SM, Shpak ED, Torii KU(2007) Haploinsufficiency after successive loss of signaling reveals a role for ERECTA-family genes in Arabidopsis ovule development. Development 134: 3099–3109 [DOI] [PubMed] [Google Scholar]
- Qin Y, Zhao L, Skaggs MI, Andreuzza S, Tsukamoto T, Panoli A, Wallace KN, Smith S, Siddiqi I, Yang Z, et al. (2014) ACTIN-RELATED PROTEIN6 regulates female miosis by modulating meiotic gene expression in Arabidopsis. Plant Cell 26: 1612–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiger DS, Drews GN(2013) MYB64 and MYB119 are required for cellularization and differentiation during female gametogenesis in Arabidopsis thaliana. PLoS Genet 9: e1003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M, Marimuthu MPA, Siddiqi I(2008) Gamete formation without meiosis in Arabidopsis. Nature 451: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Robert HS, Crhak Khaitova L, Mroue S, Benková E(2015) The importance of localized auxin production for morphogenesis of reproductive organs and embryos in Arabidopsis. J Exp Bot 66: 5029–5042 [DOI] [PubMed] [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K(1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G, Lu Z, Xiong J, Wang B, Jing Y, Meng X, Liu G, Ma H, Liang Y, Chen F, et al. (2019) Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol Plant 12: 1090–1102 [DOI] [PubMed] [Google Scholar]
- Sheridan WF, Golubeva EA, Abrhamova LI, Golubovskaya IN(1999) The mac1 mutation alters the developmental fate of the hypodermal cells and their cellular progeny in the maize anther. Genetics 153: 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi DQ, Yang WC(2011) Ovule development in Arabidopsis: Progress and challenge. Curr Opin Plant Biol 14: 74–80 [DOI] [PubMed] [Google Scholar]
- Shpak ED.(2013) Diverse roles of ERECTA family genes in plant development. J Integr Plant Biol 55: 1238–1250 [DOI] [PubMed] [Google Scholar]
- Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D(2011) Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23: 443–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Zhao L, Zhao Y, Li S, Won S, Cai H, Wang L, Li Z, Chen P, Qin Y, et al. (2017) The THO complex non-cell-autonomously represses female germline specification through the TAS3-ARF3 module. Curr Biol 27: 1597–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameshige T, Ikematsu S, Torii KU, Uchida N(2017) Stem development through vascular tissues: EPFL-ERECTA family signaling that bounces in and out of phloem. J Exp Bot 68: 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L(2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Tasaka M(2013) Regulation of plant vascular stem cells by endodermis-derived EPFL-family peptide hormones and phloem-expressed ERECTA-family receptor kinases. J Exp Bot 64: 5335–5343 [DOI] [PubMed] [Google Scholar]
- Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y(2007a) Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394: 13–24 [DOI] [PubMed] [Google Scholar]
- Wang H, Liu Y, Bruffett K, Lee J, Hause G, Walker JC, Zhang S(2008) Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20: 602–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S(2007b) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JR, Hu H, Wang GH, Li J, Chen JY, Wu P(2009) Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Mol Plant 2: 823–831 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Zhang DB(2009) From Arabidopsis to rice: Pathways in pollen development. J Exp Bot 60: 1479–1492 [DOI] [PubMed] [Google Scholar]
- Wu Y, Yang L, Cao A, Wang J(2015) Gene expression profiles in rice developing ovules provided evidence for the role of sporophytic tissue in female gametophyte development. PLoS One 10: e0141613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R, Drews GN(2004) Female gametophyte development. Plant Cell 16(Suppl): S133–S141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Shi DQ, Chen YH(2010) Female gametophyte development in flowering plants. Annu Rev Plant Biol 61: 89–108 [DOI] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V(1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Ma T, Zhang Y, Hu Y, Yu K, Chen Y, Ma H, Zhao J(2017) Identification and analysis of the stigma and embryo sac-preferential/specific genes in rice pistils. BMC Plant Biol 17: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazímalová E, Murphy AS, Yang H, Hoyerová K, Hosek P(2010) Auxin transporters—Why so many? Cold Spring Harb Perspect Biol 2: a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YX, Hu CY, Lu YG, Li JQ, Liu XD(2007) Diversity of abnormal embryo sacs in indica/japonica hybrids in rice demonstrated by confocal microscopy of whole ovary. Plant Breed 126: 574–580 [Google Scholar]
- Zeng YX, Hu CY, Lu YG, Li JQ, Liu XD(2009) Abnormalities occurring during female gametophyte development result in the diversity of abnormal embryo sacs and leads to abnormal fertilization in indica/japonica hybrids in rice. J Integr Plant Biol 51: 3–12 [DOI] [PubMed] [Google Scholar]
- Zhang J, Tang W, Huang Y, Niu X, Zhao Y, Han Y, Liu Y(2015) Down-regulation of a LBD-like gene, OsIG1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice. J Exp Bot 66: 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li S, Xue S, Yang S, Huang J, Wang L(2018) Phylogenetic and CRISPR/Cas9 studies in deciphering the evolutionary trajectory and phenotypic impacts of rice ERECTA genes. Front Plant Sci 9: 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ma H, Yu L, Wang X, Zhao J(2012) Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PLoS One 7: e49210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Cai H, Su Z, Wang L, Huang X, Zhang M, Chen P, Dai X, Zhao H, Palanivelu R, et al. (2018) KLU suppresses megasporocyte cell fate through SWR1-mediated activation of WRKY28 expression in Arabidopsis. Proc Natl Acad Sci USA 115: E526–E535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, He J, Cai H, Lin H, Li Y, Liu R, Yang Z, Qin Y(2014) Comparative expression profiling reveals gene functions in female meiosis and gametophyte development in Arabidopsis. Plant J 80: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Bramsiepe J, Van Durme M, Komaki S, Prusicki MA, Maruyama D, Forner J, Medzihradszky A, Wijnker E, Harashima H, et al. (2017) RETINOBLASTOMA RELATED1 mediates germline entry in Arabidopsis. Science 356: eaaf6532. [DOI] [PubMed] [Google Scholar]
- Zhao X, de Palma J, Oane R, Gamuyao R, Luo M, Chaudhury A, Hervé P, Xue Q, Bennett J(2008) OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J 54: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.(2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.(2012) Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]