Loss of a trans-Golgi network clathrin adaptor impairs immunity to Pseudomonas bacteria and defense signaling, correlating with lower receptor kinase and coreceptor levels in the plasma membrane.
Abstract
The plasma membrane (PM) provides a critical interface between plant cells and their environment to control cellular responses. To perceive the bacterial flagellin peptide flg22 for effective defense signaling, the immune receptor FLAGELLIN SENSING2 (FLS2) needs to be at its site of function, the PM, in the correct abundance. However, the intracellular machinery that controls PM accumulation of FLS2 remains largely undefined. The Arabidopsis (Arabidopsis thaliana) clathrin adaptor EPSIN1 (EPS1) is implicated in clathrin-coated vesicle formation at the trans-Golgi network (TGN), likely aiding the transport of cargo proteins from the TGN for proper location; but EPS1’s impact on physiological responses remains elusive. Here, we identify EPS1 as a positive regulator of flg22 signaling and pattern-triggered immunity against Pseudomonas syringae pv tomato DC3000. We provide evidence that EPS1 contributes to modulating the PM abundance of defense proteins for effective immune signaling because in eps1, impaired flg22 signaling correlated with reduced PM accumulation of FLS2 and its coreceptor BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 (BAK1). The eps1 mutant also exhibited reduced responses to the pathogen/damage-associated molecular patterns elf26 and AtPep1, which are perceived by the coreceptor BAK1 and cognate PM receptors. Furthermore, quantitative proteomics of enriched PM fractions revealed that EPS1 was required for proper PM abundance of a discrete subset of proteins with different cellular functions. In conclusion, our study expands the limited understanding of the physiological roles of EPSIN family members in plants and provides novel insight into the TGN-associated clathrin-coated vesicle trafficking machinery that impacts plant PM-derived defense processes.
The plasma membrane (PM) serves as a central contact point between a cell and its environment. Proteins at this location perform critical functions to facilitate signal perception as well as initiation, amplification, and attenuation of responses. Overall, eukaryotic cells share similar strategies for perceiving extracellular stimuli; however, plants have independently evolved plant-specific PM proteins (Cock et al., 2002; Nürnberger et al., 2004) to cope with biotic and abiotic stimuli that are distinct from those pertinent to animals. For many biotic stresses, including immunity against the pathogenic flagellated bacterium Pseudomonas syringae pv tomato (Pto) DC3000, the cellular mechanisms and molecular machinery regulating the accumulation of plant PM proteins required for effective responses remain largely elusive. One strategy that plant cells utilize to control the protein composition of the host PM is protein cargo trafficking to and from the PM by vesicular trafficking (for review, see Ben Khaled et al., 2015; Paez Valencia et al., 2016; Ekanayake et al., 2019).
Vesicular trafficking involves the movement of cargo proteins in small vesicles from a donor to a target membrane. The trans-Golgi network/early endosome (TGN/EE) has emerged as a central station for protein sorting to and from the plant cell surface (Gendre et al., 2015; Uemura, 2016). In contrast to animals, the plant TGN and EE functionally overlap and serve as a point of convergence for protein secretion, vacuolar trafficking, and endocytosis (Viotti et al., 2010; Gendre et al., 2015; Uemura, 2016). Protein sorting at the TGN/EE does not appear to occur via simple bulk-flow processes, because different cargo proteins seem to utilize distinct trafficking routes with unique vesicle components (Nomura et al., 2011; Gu and Innes, 2012; Sauer et al., 2013; Gendre et al., 2015). The importance of the TGN/EE in plant immunity is underscored by the fact that mutations in genes encoding TGN/EE-resident proteins result in altered susceptibility to plant pathogens (for review, see Uemura, 2016; Underwood, 2016; LaMontagne and Heese, 2017). Furthermore, pathogen effectors target TGN/EE-associated trafficking components to interfere with the plant’s ability to mount effective immunity (Mukhtar et al., 2011; Nomura et al., 2011; Gu and Innes, 2012; Weßling et al., 2014; LaMontagne and Heese, 2017). The plant TGN/EE also contributes to sorting of immune cargo proteins, such as pathogen-related proteins and proteins for callose deposition, to their correct subcellular locations for effective immunity (Gu and Innes, 2011, 2012; Nomura et al., 2011; Wu et al., 2015).
Another cargo protein that has roles in plant defense and utilizes the TGN/EE for trafficking to and from the PM is the receptor kinase FLAGELLIN SENSING2 (FLS2; Beck et al., 2012; Choi et al., 2013). FLS2 belongs to a class of pattern recognition receptors (PRRs) that serve as a first line of defense, in that PRRs act as sentinels to detect pathogen- or host-derived molecular patterns, termed microbial/pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), respectively (Xin and He, 2013; Couto and Zipfel, 2016; Yu et al., 2017). FLS2 forms a ligand-induced complex with BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 (BAK1; Chinchilla et al., 2007; Heese et al., 2007), which serves as a coreceptor for multiple, diverse PRRs and their cognate ligands, thereby linking the perception of structurally diverse D/PAMPs to the initiation of a conserved immune signaling network (Chinchilla et al., 2007; Roux et al., 2011; Tang et al., 2015; Yasuda et al., 2017).
FLS2 needs to be at the PM so that it can utilize its extracellular Leu-rich repeat domain (Sun et al., 2013) to detect the bacterial PAMP flagellin or the active peptide-derivative flg22 (Gómez-Gómez and Boller, 2000; Robatzek and Wirthmueller, 2012; Yu et al., 2017). The perception of flg22 initiates a plethora of immune responses including the production of reactive oxygen species (ROS) as well as the activation of calcium-dependent protein kinases (CDPKs) and mitogen-activated protein kinases (MAPKs), which either directly or indirectly alter the accumulation of thousands of transcripts as part of early, intermediate, and late signaling (Boutrot et al., 2010; Couto and Zipfel, 2016; Yu et al., 2017). These responses are not linear but rather fall into at least three different branches of the flg22 signaling network (Boudsocq et al., 2010; Korasick et al., 2010; Smith et al., 2014a, 2014b; Xu et al., 2014). Notably, absent or reduced levels of functional FLS2 compromise flg22 signaling (Zipfel et al., 2004; Boutrot et al., 2010; Mersmann et al., 2010). Increasing evidence shows that FLS2 requires vesicular trafficking to localize (1) correctly to its site of function (the PM) and (2) in the correct abundance to induce effective defense signaling (for review, see Ben Khaled et al., 2015; Ekanayake et al., 2019). However, the identity and role(s) of TGN/EE-associated vesicle components that function in FLS2 trafficking to modulate flg22 signaling remain ill-defined.
In this study, we utilized cross-disciplinary approaches including immune assays, live-cell imaging, biochemical fractionation, and quantitative proteomics to identify Arabidopsis (Arabidopsis thaliana) EPSIN1 (EPS1) as a modulator of plant immunity. More specifically, we provide evidence that EPS1 contributes to (1) pattern-triggered immunity (PTI) against bacterial Pto DC3000 strains and (2) modulating the PM abundance of a finite set of proteins, including FLS2 and BAK1, for effective D/PAMP responses. Arabidopsis EPS1, also called EpsinR1 (Lee et al., 2007), is a monomeric clathrin adaptor previously shown to localize to the TGN/EE and Golgi (Song et al., 2006). In animals and yeast, EPSINs and EPSIN-related proteins act as key components of vesicular trafficking by helping to recruit distinct clathrin coat, adaptor, and accessory proteins to either the PM or the TGN, ultimately to initiate the budding of cargo-containing vesicles to send them to their target membranes (Duncan and Payne, 2003; Legendre-Guillemin et al., 2004; De Craene et al., 2012). Thus, our results offer novel insights into the clathrin-associated trafficking machinery at the TGN/EE that affects plant PM-derived immune responses.
RESULTS
EPS1 Is Required for Robust PTI against P. syringae Strains
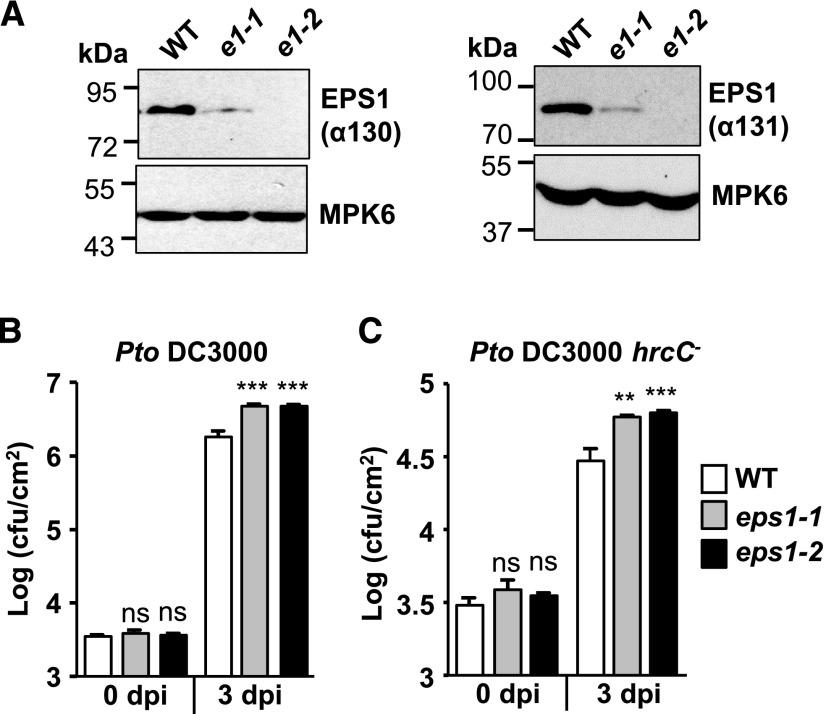
Previously, we performed phosphoproteomic screens (Peck et al., 2001; Nühse et al., 2003, 2007) to uncover vesicle components with novel roles in flg22 signaling and plant immunity (Kalde et al., 2007; Smith et al., 2014a). Expanding on this work, we focus here on another phosphocandidate, namely the Arabidopsis clathrin adaptor EPS1 (At5g11710). To test whether EPS1 has roles in plant immunity against flagellated bacterial strains and/or flg22 signaling, we utilized a previously published Arabidopsis knockdown allele with the T-DNA insertion in its promotor region (Song et al., 2006) that we termed eps1-1 (Supplemental Fig. S1A). In addition, we isolated a second, independent allele, eps1-2, with a T-DNA insertion between its fourth and fifth exons (Supplemental Fig. S1A). With two different affinity-purified polyclonal peptide antibodies made against different epitopes in EPS1 (α130 and α131; Supplemental Fig. S1B), we detected reduced or no detectable levels of EPS1 protein in total protein extract from eps1-1 (knockdown) or eps1-2 (knockout), respectively, compared with Columbia-0 (Col-0), the wild type (Fig. 1A; Supplemental Fig. S1, C and D). Reduced levels of EPS1 protein in eps1-1 had also been reported previously (Song et al., 2006). In accordance with Kalthoff et al. (2002) for animal epsin1 and Song et al. (2006) for Arabidopsis EPS1, EPS1 migrated more slowly with an apparent molecular mass of about 80 kD when probing total protein extracts from seedlings with either α130 or α131 antibody (Fig. 1A; Supplemental Fig. S1, C and D) than its expected molecular mass of about 60 kD. In agreement with the antibodies made against EPS1-specific peptides, no additional cross-reacting protein bands were detected in the wild type or the two independent eps1 mutant alleles on larger immunoblots (Supplemental Fig. S1, C and D). Except for slightly smaller leaves, eps1 plants did not exhibit any gross developmental defects (Supplemental Fig. S1E).
Figure 1.
eps1 mutants are hypersusceptible to Pto DC3000 and Pto DC3000 hrcC− infection. A, Using two different affinity-purified peptide antibodies (α130 and α131) made against different EPS1 epitopes (Supplemental Fig. S1B), immunoblot analyses of total protein seedling extract indicate that eps1-1 (e1-1) is a knockdown mutant and eps1-2 (e1-2) is a null mutant compared with the Col-0 wild type (WT). Molecular mass markers are indicated in kilodaltons (kD). B and C, Leaves of 5-week-old eps1 mutant or Col-0 wild-type plants were syringe infiltrated with Pto DC3000 (OD600 = 0.0005; B) or Pto DC3000 hrcC− (OD600 = 0.02; C). Bacterial growth was assessed by serial dilution plating as colony-forming units (cfu) at 0 and 3 dpi. n = 8 samples per genotype, with each n representing a biological sample taken from an individual leaf from four different plants for each genotype and time point. Values are means ± se. Asterisks indicate statistically significant differences compared with the wild type of the same dpi by two-tailed Student’s t test: **, P < 0.01; ***, P < 0.001; ns, no statistically significant difference. Experiments were performed at least three times with similar results.
With two independent eps1 alleles in hand, we first monitored the growth of virulent, flagellated bacterium Pto DC3000 after syringe infiltration into mature 5- to 6-week-old leaves. Bacterial growth was assessed at 0 and 3 d postinfection (dpi) by bacterial dilution plating as described in Korasick et al. (2010). At 0 dpi, no differences in bacterial levels were observed between eps1 mutants and Col-0 plants (Fig. 1B), confirming that equal amounts of bacteria were initially delivered into leaves of the different plant lines. After 3 dpi, both eps1 alleles exhibited increased growth of Pto DC3000 compared with Col-0 (Fig. 1B). Loss of EPS1 also resulted in increased bacterial growth after infiltration with Pto DC3000 hrcC− (Fig. 1C), a bacterial strain that elicits PAMP signaling but is hypovirulent because its defective type III secretion system fails to suppress PTI (Xin and He, 2013). In conclusion, eps1 mutant alleles showed increased susceptibility to both pathogenic (Pto DC3000) and nonpathogenic (Pto DC3000 hrcC−) flagellated bacteria, with the latter indicating that EPS1 functions as a positive regulator in PTI. Importantly, similar results were obtained with two independent eps1 mutant alleles, confirming that the increased susceptibility to these Pto strains was specific to mutations in EPS1.
EPS1 Function Contributes to D/PAMP-Induced Immune Responses
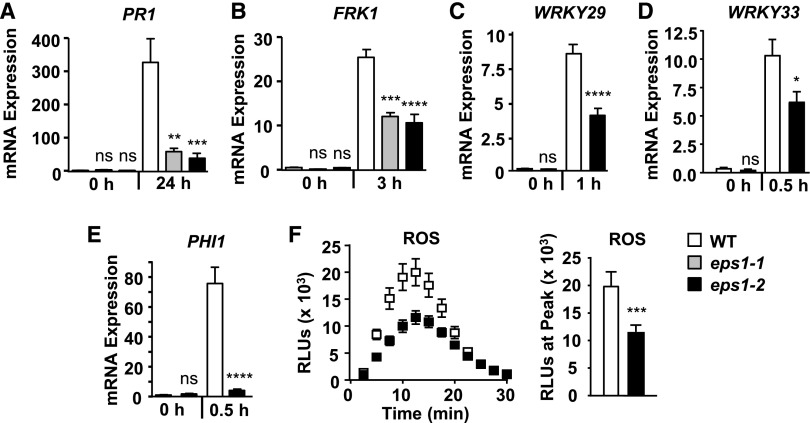
To investigate the basis of defective PTI in eps1 mutants (Fig. 1C), we examined whether the loss of EPS1 resulted in altered molecular PTI responses. First, we assessed mRNA accumulation of early, intermediate, and late defense marker genes (Yu et al., 2017) in response to flg22 using reverse transcription quantitative PCR (RT-qPCR) in eps1 mutant alleles compared with Col-0 (Fig. 2, A–E). Because mutations in genes encoding vesicular trafficking proteins can affect at least three distinct branches of the flg22 signaling network differently (Korasick et al., 2010; Smith et al., 2014a), we tested changes in the expression of marker genes that represent those different branches. First, we focused on PATHOGENESIS RELATED1 (PR1), a late defense marker downstream of the plant hormone salicylic acid, and the MAPK-dependent marker gene FLG22-INDUCED RECEPTOR-LIKE KINASE1 (FRK1). Both independent eps1-1 and eps1-2 mutant alleles showed reduced PR1 (Fig. 2A) and FRK1 (Fig. 2B) mRNA levels after flg22 elicitation, providing evidence that it was indeed mutations in the EPS1 gene that were responsible for impaired immune responses in these mutant plants (Fig. 1, B and C). Based on these results, we utilized the eps1-2 null mutant allele in all subsequent assays. Loss of EPS1 also resulted in decreased flg22-induced mRNA accumulation of the early MAPK-dependent marker genes WRKY29 (Fig. 2C) and WRKY33 (Fig. 2D) as well as the early CDPK-dependent marker gene PHI1 (Fig. 2E). Thus, eps1 mutant plants showed attenuated flg22-induced accumulation of all transcripts tested. Furthermore, loss of EPS1 function led to defects in another early Ca2+-dependent response, namely impaired ROS production, after flg22 elicitation (Fig. 2F).
Figure 2.
EPS1 is required for robust flg22 signaling. A, Leaves of 5- to 6-week-old eps1 mutant or Col-0 wild-type (WT) plants were syringe infiltrated with 1 µm flg22 for 0 or 24 h. Using RT-qPCR, relative mRNA levels of PR1 were measured and normalized to the reference gene At2g28390. n = 6 samples per genotype per treatment, with each biological sample (n) consisting of three leaf discs taken from four to six different plants for each genotype and treatment. B to E, Eight-day-old seedlings were treated with 10 nm flg22 for the indicated times in hours. Using RT-qPCR, relative mRNA levels of FRK1 (B), WRKY29 (C), WRKY33 (D), and PHI1 (E) were measured and normalized to the reference gene At2g28390. For each genotype and treatment, n = 3 biological samples, with each sample consisting of three to four seedlings. F, Time course (left) and peak (right) of flg22-triggered (10 nm) ROS production over 30 min from leaves of 5- to 6-week-old plants. n = 10 samples per genotype, with each sample (n) consisting of one leaf disc half taken from three to four individual plants. Peak flg22-triggered ROS production (right) is shown as an example for statistical analysis and represents data from the same time-course experiment (left). RLU, Relative light units. For all experiments, values are means ± se. Asterisks indicate statistically significant differences compared with the wild type by two-tailed Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, no statistically significant difference. All experiments were performed at least three times with similar results.
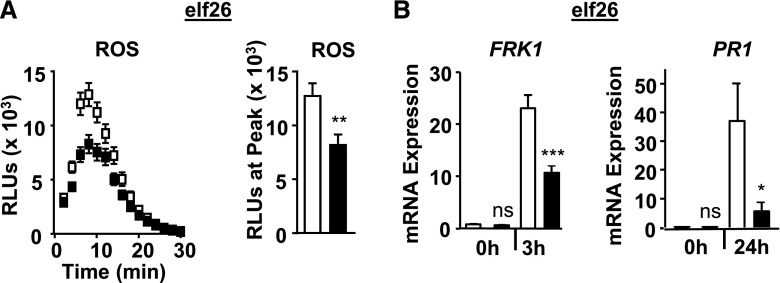
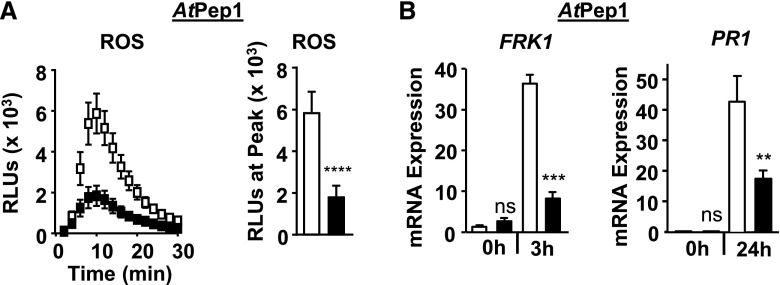
Because loss or reduced levels of FLS2 function alone does not have a substantial effect on Pto DC3000 growth when these pathogenic bacteria are syringe infiltrated for apoplastic infection (Supplemental Fig. S2A; Zipfel et al., 2004; Boutrot et al., 2010; Mersmann et al., 2010), we tested next whether EPS1 functions in immune responses to unrelated D/PAMPs that are recognized by Arabidopsis PRRs other than FLS2. For these experiments, we focused on responses that represented early/Ca2+-, intermediate/MAPK-, and late salicylic acid-dependent immune signaling branches using the eps1-2 null mutant allele. As was observed for responses to flg22, loss of EPS1 resulted in reduced ROS production (Fig. 3A) as well as PR1 and FRK1 mRNA accumulation (Fig. 3B) after elicitation with elf26, the bacterial PAMP recognized by the host EF-TU RECEPTOR (EFR; Zipfel et al., 2006; Krol et al., 2010; Yamaguchi et al., 2010). Similar results were obtained in response to the plant DAMP AtPep1 (Fig. 4), which is perceived by the PLANT ELICITOR PEPTIDE RECEPTOR1 (PEPR1) and PEPR2 (Zipfel et al., 2006; Krol et al., 2010; Yamaguchi et al., 2010; Ortiz-Morea et al., 2016). Taking these results together, EPS1 is required for PTI signaling in response to multiple D/PAMPs, consistent with EPS1’s positive role in PTI.
Figure 3.
EPS1 is required for robust elf26 signaling. A, Time course and peak for elf26-triggered (10 nm) apoplastic ROS production from leaves of 5- to 6-week-old plants for eps1-2 (black squares or bars) and the Col-0 wild type (white squares or bars). n = 15 samples per genotype per treatment, with each sample (n) consisting of one leaf disc half taken from three to four individual plants for each genotype and treatment. RLU, Relative light units. B, For FRK1 mRNA accumulation, 8-d-old seedlings were treated with 100 nm elf26 for the indicated times in hours. n = 3 to 4 samples per genotype per treatment, with each biological sample (n) containing three to four seedlings. For PR1 mRNA accumulation, leaves of 5- to 6-week old plants were infiltrated with 1 µm elf26. n = 6 samples per genotype per treatment. Each sample (n) represents three leaf discs taken from three to four different plants for each genotype and treatment. Relative mRNA levels of FRK1 and PR1 were measured and normalized to the reference gene At2g28390. Values represent means ± se. Asterisks indicate statistically significant differences compared with the wild type by two-tailed Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no statistically significant difference. All experiments were performed at least three times with similar results.
Figure 4.
EPS1 is required for robust AtPep1 signaling. A, Time course and peak for AtPep1-triggered (10 nm) apoplastic ROS production from 8-d-old seedlings. n = 20 samples per genotype per treatment, with each sample (n) consisting of one cotyledon half obtained from five or more individual seedlings for each genotype and treatment. The experiment was performed twice in seedlings and twice in leaf discs with similar results. RLU, Relative light units. B, For FRK1 mRNA accumulation, 8-d-old seedlings were treated with 100 nm AtPep1 for the indicated times in hours. n = 3 to 4 samples per genotype per treatment, with each biological sample (n) containing three to four seedlings. For PR1 mRNA accumulation, leaves of 5- to 6-week-old plants were infiltrated with 1 µm AtPep1 for the indicated times. n = 6 samples per genotype per treatment, with each sample (n) containing three leaf discs taken from three to four different plants for each genotype and treatment. Relative mRNA levels of FRK1 and PR1 were measured and normalized to the reference gene At2g28390. Values represent means ± se. For all experiments, eps1-2 is represented by black squares or bars and Col-0 is represented by white squares or bars. Asterisks indicate statistically significant differences compared with the wild type by two-tailed Student’s t test: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, no statistically significant difference. Unless specified otherwise, all experiments were performed at least three times with similar results.
EPS1 Is Required for Correct FLS2 Abundance at the PM for Effective flg22 Response
Signaling defects in early, intermediate, and late responses that represented the three branches of the immune signaling network (Figs. 2–4) indicated a potential impairment at a very early point of defense signaling, such as at the level of the cognate PRRs. Because reduced steady-state mRNA levels of FLS2 can cause reduced flg22 responses due to reduced steady-state protein accumulation of FLS2 (Boutrot et al., 2010; Mersmann et al., 2010), we compared basal FLS2 mRNA and FLS2 protein levels in the eps1-2 null mutant with wild-type seedlings, but eps1-2 did not show any significant differences in mRNA accumulation of FLS2 (Supplemental Fig. S2B) or total cellular FLS2 protein (Figs. 5A and 6B). Similarly, no defects in the mRNA levels of EFR, PEPR1, and their coreceptor BAK1 were detected (Supplemental Fig. S2B).
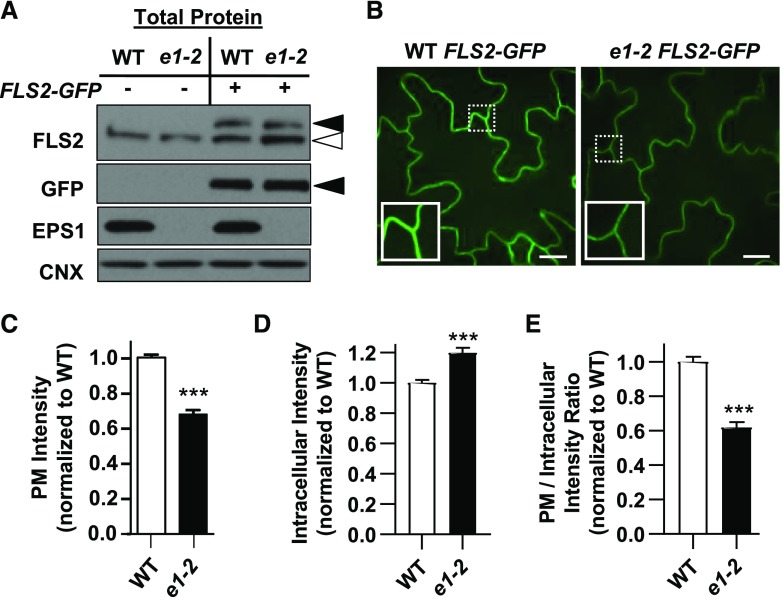
Figure 5.
Loss of EPS1 results in reduced PM and increased intracellular accumulation of FLS2-GFP. A, In total protein extracts, eps1-2 FLS2-GFP accumulated FLS2-GFP to similar levels as wild-type (WT) Col-0 FLS2-GFP. Total proteins were extracted from Col-0 (wild type) and eps1-2 (e1-2) 7-d-old seedlings that did (+) or did not (−) express pFLS2::FLS2-GFP and subjected to immunoblot analysis with the indicated antibodies. Col-0 FLS2-GFP and eps1-2 FLS2-GFP were from the same homozygous F4 seedlings used for SDCM and pixel intensity analyses in B to E. Antibody against FLS2 detected both endogenous FLS2 (white arrowhead) and FLS2-GFP (black arrowhead); antibody against GFP identified FLS2-GFP (black arrowhead); antibody against EPS1 confirmed the eps1-2 mutant; and antibody against calnexin (CNX; an endoplasmic reticulum membrane marker) served as a loading control. B, Representative single Z-plane images and zoomed insets (10 × 10 μm in size) for wild-type FLS2-GFP and eps1-2 FLS2-GFP cotyledons using SDCM. Bars = 10 μm. C to E, Quantification of FLS2-GFP pixel intensity from selected regions of the PM (C), in the intracellular region (D), and as a PM-to-intracellular ratio (E) for wild-type FLS2-GFP (white bars) and eps1-2 FLS2-GFP (e1-2; black bars). Data for eps1-2 are normalized to the wild type with n = 48 images from 24 seedlings per genotype. Values are means ± se. Asterisks indicate statistically significant differences compared with the wild type by two-tailed Student’s t test: ***, P < 0.0001. All experiments were performed three times with similar results.
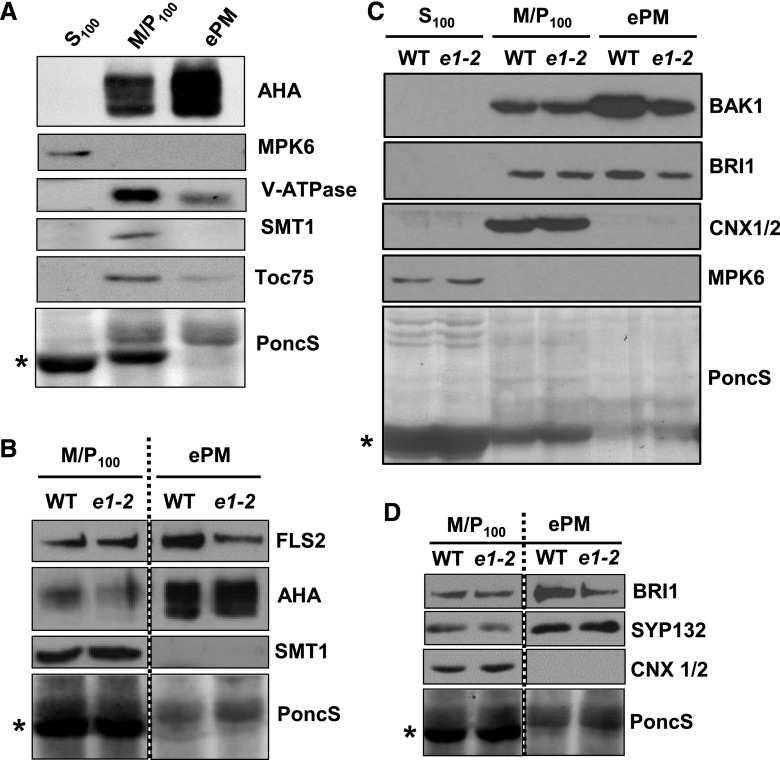
Figure 6.
Loss of EPS1 results in reduced protein levels of FLS2, BAK1, and BRI1 in enriched PM fractions but not in microsomal fractions. A, Efficacy of PM enrichment in Col-0 seedlings using immunoblot analysis. Soluble (S100), microsomal (M/P100), and ePM fractions were probed for the following subcellular marker proteins: AHA (PM), MPK6 (cytosol), Toc75 (chloroplast), V-ATPase (tonoplast), SMT1 (endoplasmic reticulum membrane), and PoncS (Ponceau S; a general protein dye). *, PoncS shows staining for stromal Rubisco (known to be partly released during cell lysis into soluble fraction S100). B, Immunoblot analysis of MP100 and ePM proteins from eps1-2 (e1-2) and the wild type (WT) using antibodies against FLS2. AHA (PM) and SMT1 (endoplasmic reticulum) serve as PM enrichment and endoplasmic reticulum depletion markers, respectively. C, Immunoblot analysis of S100, M/P100, and ePM proteins from eps1-2 and the wild type probed with antibodies against BAK1 and BRI1. Calnexin1/2 (CNX1/2) and MPK6 served as endoplasmic reticulum membrane and soluble marker proteins, respectively. D, Immunoblot analysis of SYP132 and BRI1 (both PM proteins) for eps1-2 and the wild type. CNX1/2 and PoncS served as loading controls. All experiments were performed in 7- to 8-d-old seedlings and were repeated at least three times with similar results. *, For all experiments, PoncS served as a loading control and showed depletion of Rubisco in ePM. Stippled lines indicate immunoblots derived from the same gel.
Because clathrin-coated vesicles are implicated in protein trafficking from the TGN/EE to the PM (Gendre et al., 2015), we examined whether the loss of EPS1 affected FLS2 protein abundance at the PM, the site of flg22 perception. First, we utilized live-cell imaging via spinning-disc confocal microscopy (SDCM; Smith et al., 2014a; Leslie and Heese, 2017) to compare PM levels of FLS2-GFP between eps1-2 null mutant and wild-type sibling seedlings expressing pFLS2::FLS2-myc-eGFP (FLS2-GFP; Beck et al., 2012; Smith et al., 2014a). First, we confirmed that FLS2-GFP levels in total protein extracts were similar in eps1-2/FLS2-GFP and wild type/FLS2-GFP (Fig. 5A). However, when comparing PM intensity measurements using SDCM (Smith et al., 2014a), steady-state levels of FLS2-GFP at the PM in eps1-2/FLS2-GFP were reduced compared with wild type/FLS2-GFP (Fig. 5, B and C). We did not detect any obvious accumulation of FLS2-GFP in defined intracellular puncta in eps1-2 compared with the wild type (Fig. 5B), but we observed an apparent diffuse intracellular FLS2-GFP labeling (Fig. 5B), in agreement with increased intracellular pixel intensity in eps1 over the wild type (Fig. 5D). Furthermore, the PM-to-intracellular ratio of FLS2-GFP signals in eps1-2/FLS2-GFP was lower compared with that in wild type/FLS2-GFP (Fig. 5E), indicating that loss of EPS1 function resulted in greater intracellular accumulation of FLS2-GFP. Note that seedlings used for the immunoblot shown in Figure 5A were the same homozygous eps1-2/FLS2-GFP and wild-type/FLS2-GFP seedlings imaged by SDCM and analyzed for FLS2-GFP pixel intensities in the PM and intracellular areas (Fig. 5, B–E). EPS1 did not have any apparent role in ligand-induced endocytosis of FLS2 because we did not observe any statistical difference in the number of FLS2-GFP endosomal puncta between eps1-2/FLS2-GFP and wild-type/FLS2-GFP seedlings (Supplemental Fig. S3) when using SDCM to quantify ligand-induced endocytosis of FLS2-GFP (Smith et al., 2014a; Leslie and Heese, 2017).
As an independent approach to assess the subcellular localization of FLS2, we used a simple and rapid biochemical method to analyze the PM-associated accumulation of FLS2 protein in eps1-2 null mutant and wild-type seedlings. This method uses differential centrifugation and the detergent Brij-58 to enrich for PM proteins by depleting contaminating organelles (Zhang and Peck, 2011; Zhang et al., 2013; Collins et al., 2017). Fractionation efficacy in whole seedling samples was verified by immunoblot analyses of soluble, microsomal, and enriched PM (ePM) subcellular fractions using compartment-specific antibodies (Fig. 6). ePM fractions from whole seedlings were enriched for PM marker protein AHA H+-ATPases while showing reduced levels of cytosolic (MPK6) and organellar marker proteins, including the endoplasmic reticulum membrane STEROL METHYLTRANSFERASE (SMT1), plastid 75 KDA CHLOROPLAST MEMBRANE TRANSLOCON (TOC75), plastid Rubisco, and tonoplastic VACUOLAR-ATPase (V-ATPase; Fig. 6A). Importantly, while no significant difference in FLS2 accumulation was observed between microsomal fractions, FLS2 levels were lower in ePM fractions of eps1-2 compared with the wild type (Fig. 6B). Thus, loss of EPS1 did not affect total FLS2 protein accumulation but resulted specifically in reduced levels of FLS2 at the PM.
Taking these results together, using two independent approaches, live-cell imaging (Fig. 5) and biochemical fractionation of ePM (Fig. 6B), we provided evidence that EPS1 contributes to correct FLS2 accumulation in the PM (its site of function), which in turn is necessary for effective flg22 signaling (Fig. 2).
EPS1 Modulates PM Abundance of a Distinct Subset of Proteins Including BAK1 and BRI1
To determine if other PM proteins are similarly affected by the absence of EPS1, we probed microsomal and ePM fractions for the FLS2 coreceptor, BAK1, using an αBAK1-specific affinity-purified peptide antibody (Supplemental Fig. S4). We observed that BAK1 protein also accumulated to reduced levels in the enriched PM for eps1-2 mutant seedlings (Fig. 6C, ePM) while accumulating to similar levels in the microsomal fraction (Fig. 6C, M/P100). Because BAK1 is the coreceptor for a number of plant PRRs (e.g. FLS2, EFR, and PEPR1), we concluded that in eps1, reduced BAK1 protein accumulation in ePM may contribute to impaired flg22, elf26, and AtPep1 responses in eps1 (Figs. 2, 3, and 4, respectively). Similar to FLS2 and BAK1, BRASSINOSTEROID INSENSITIVE1 (BRI1), the receptor kinase for brassinosteroid signaling and also a protein implicated in plant immunity (Albrecht et al., 2012; Belkhadir et al., 2012; Xiong et al., 2020), showed reduced protein accumulation in ePM but not in microsomal fractions of eps1 (Fig. 6, C and D). Notably, loss of EPS1 did not affect the accumulation of all PM proteins with immune function, because PM levels of AHA H+-ATPase (Elmore and Coaker, 2011) and SYNTAXIN OF PLANTS132 (SYP132; Kalde et al., 2007) were not altered in eps1-2 (Fig. 6, B and D).
To obtain a more comprehensive overview of the extent of PM proteins affected by loss of EPS1, we combined ePM fractionation with quantitative liquid chromatography-tandem mass spectrometry (LC-MS/MS) using previously established methodology (Zhang and Peck, 2011). The mass spectrometry data did not detect or provide a clear quantification for the PRRs or for BAK1 (Supplemental Data Set S1). However, LC-MS/MS of ePM fractions from eps1-2 and the wild type confirmed our immunoblot analyses of ePM (Fig. 6, C and D) that the PM abundance of BRI1 was reduced whereas SYP132 was not affected in eps1-2 (Table 1; Supplemental Data Set S1). Overall, the proteomic analysis revealed that only 1.7% of the detected ePM proteome was decreased in eps1-2 while the large majority of proteins remained unaffected (Table 1; Supplemental Data Set S1). Gene Ontology (GO) analysis of the ePM proteins reduced in eps1 indicated that they did not segregate into any obvious, distinct structural, molecular function or protein class categories (Supplemental Fig. S5; Supplemental Data Set S2). More specifically, Fisher’s exact test with false discovery rate (FDR) correction indicated that no terms were overrepresented relative to their representation in the whole Arabidopsis genome to a statistically significant degree.
Table 1. Representative list of proteins with decreased and similar PM abundance in eps1-2 compared with the Col-0 wild type measured by quantitative mass spectrometry.
| Protein Identifier | Gene Identifier | Expected Size (kD) | Average Log2 Fold Change | Predicted Localization |
|---|---|---|---|---|
| ABCB14: ABC Transporter Member14 | At1g28010 | 136 | −2.581 | PM |
| BRI1: Brassinosteroid Insensitive1 | At4g39400 | 131 | −2.497 | PM |
| FLA1: Fasciclin-Like Arabinogalactan | At5g55730 | 45 | −2.931 | PM |
| PDR9: Pleiotropic Drug Resistance9 | At3g53480 | 164 | −4.684 | PM |
| S-Lectin Protein Kinase | At1g11330 | 94 | −2.579 | PM |
| ACA11: Autoinhibited Ca2+-ATPase11 | At3g57330 | 112 | −0.179 | PM |
| LRR-RLK: Leucine-Rich Repeat Receptor-Like Kinase | At2g26730 | 72 | −0.008 | PM |
| PIP2A: PM Intrinsic Protein2A | At3g53420 | 30 | 0.319 | PM |
| SYP121: Syntaxin of Plants121/PEN1 | At3g11820 | 38 | −0.294 | PM |
| SYP132: Syntaxin of Plants132 | At5g08080 | 34 | −0.029 | PM |
In conclusion, our quantitative proteomics results implicate EPS1 in trafficking of a specific subset of proteins rather than globally regulating the transport of all PM proteins, consistent with eps1 plants exhibiting no gross developmental growth defects (Supplemental Fig. S1E).
DISCUSSION
The correct subcellular localization of proteins rather than their mere presence within a cell determines biological outcomes. Thus, delineating molecular components and cellular mechanisms that regulate the PM proteome is critical to our understanding of how eukaryotic cells interact with and respond to their environment. This work defines EPS1, the TGN/Golgi-associated clathrin adaptor previously implicated in vacuolar trafficking (Song et al., 2006), as a modulator in PM accumulation of FLS2 for effective flg22 responses. EPS1 is not the only known vesicle component with immune function reported to affect more than one trafficking pathway that originates from the TGN/EE. Notably, KEEP ON GOING, a TGN/EE-localized RING E3 ligase required for plant immunity against fungal powdery mildew (Gu and Innes, 2011, 2012), participates in regulating multiple endomembrane trafficking routes, including cargo transport to both the vacuole and PM (Gu and Innes, 2012; Wu et al., 2015), with TGN/EE-to-PM trafficking potentially being clathrin dependent (Wu et al., 2015).
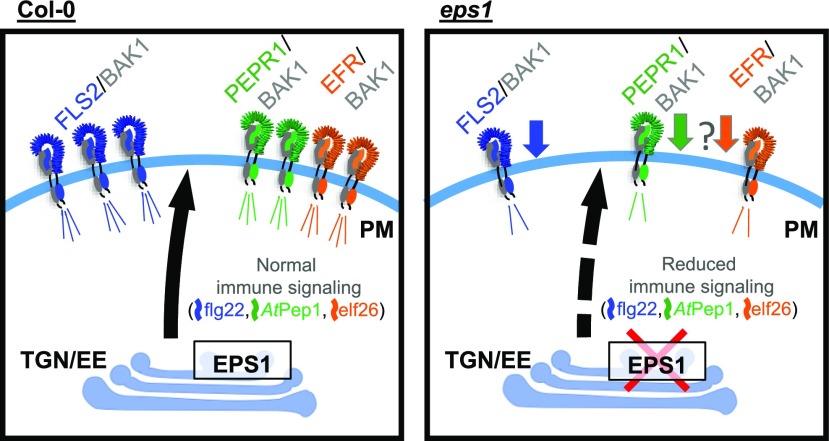
Based on two independent techniques, namely live-cell imaging using SDCM (Fig. 5) and biochemical enrichment of PM (Fig. 6), loss of EPS1 resulted in reduced accumulation of FLS2 in the PM. Consistent with having reduced but not abolished PM abundance of FLS2, eps1 null mutant plants displayed impairment rather than lack of flg22 signaling (Figs. 2 and 7). In eps1, the reduced PM accumulation of BAK1 (Fig. 6C) likely contributed to diminished flg22 signaling (Fig. 7). The fact that BAK1 also serves as the coreceptor for EFR (Chinchilla et al., 2007; Roux et al., 2011; Yasuda et al., 2017) and PEPR1/2 (Tang et al., 2015; Yasuda et al., 2017) may explain why elf26 and AtPep1 responses were compromised in eps1 mutants (Figs. 3, 4, and 7). Another possible explanation for the decreased elf26 and AtPEP1 signaling in eps1 is that in addition to FLS2, the PM accumulation of their cognate PRRs, namely EFR and PEPR1/2, may also be at least in part controlled by EPS1 (Fig. 7). In Arabidopsis, lack or reduced levels of FLS2 alone (Supplemental Fig. S2A), of individual other PRRs, or of their coreceptor BAK1 alone do not result in enhanced susceptibility to Pto DC3000 strains during apoplastic defenses (Zipfel et al., 2004, 2006; Nekrasov et al., 2009; Roux et al., 2011). Thus, the scenario in which EPS1 governs the PM abundance of multiple PRRs and/or other PM proteins with roles in plant immunity would be consistent with our findings that eps1 mutant plants supported increased growth of Pto DC3000 strains after syringe infiltration. Furthermore, we cannot exclude the possibility that EPS1 has roles in trafficking of yet unknown protein(s) to the PM and potentially the vacuole that may contribute to the plant’s ability to mount resistance against Pto DC3000 strains.
Figure 7.
Model depicting EPS1-dependent immune signaling defects due to reduced PM levels of PRRs and BAK1. Loss of the TGN/EE-associated clathrin adaptor EPS1 perturbs PM abundance of FLS2 (the PRR for flg22) and its coreceptor, BAK1. Consistent with reduced preexisting levels of FLS2 and BAK1 proteins, eps1 mutants display decreased immune responses upon elicitation with flg22. Based on impaired elf26 and AtPep1 responses, EPS1 may regulate the PM abundance of other PRRs including EFR and PEPR1, respectively. EPS1 also impacts a small subset of additional PM proteins with immune and other cellular functions (not depicted in this model).
The fact that FLS2 (Figs. 5A and 6B) accumulated to similar levels in total protein extract or microsomal fractions of eps1 null mutant and the wild type indicated that in eps1, the reduced PM accumulation of FLS2 was unlikely due to a general defect in protein expression or accumulation. Furthermore, we did not detect any obvious difference in steady-state FLS2 mRNA expression between eps1 mutant and the wild type, suggesting that gene expression did not appear to be the cause for the differences in PM protein accumulation between these genotypes. For eps1, the decrease in the PM accumulation of FLS2-GFP was accompanied by an increased intracellular accumulation (Fig. 5, D and E), which is consistent with the notion that EPS1 modulates FLS2 trafficking to the PM. Because EPS1 has been postulated to serve as a clathrin adaptor for trafficking cargo proteins out of the TGN/EE (Song et al., 2006), an expected consequence of the loss of EPS1 may have been the accumulation of a cargo protein, such as FLS2-GFP, in TGN/EE-associated puncta. However, despite careful inspection of the eps1 SDCM images, we did not detect any difference from the wild type in defined intracellular puncta accumulating FLS2-GFP. We observed, however, an apparent increase in diffuse intracellular labeling of FLS2-GFP. Because EPS1 has been localized to the TGN/EE as well as the Golgi apparatus (Song et al., 2006), cargo proteins that require EPS1 function for effective accumulation in the PM may be backed up and distribute diffusely throughout the endomembrane and/or secretory system in eps1 mutants. In the future, it will be interesting to determine where within the plant cell FLS2 may accumulate in eps1 mutants.
Arabidopsis EPS1 belongs to the ENTH/ANTH/VHS superfamily of proteins that span across kingdoms, and its members contain an ENTH (EPSIN N-terminal homology), an ANTH (AP180 N-terminal homology), or a VHS (Vps27, Hrs, and STAM) domain at their N terminus (Duncan and Payne, 2003; Legendre-Guillemin et al., 2004; De Craene et al., 2012). The Arabidopsis genome encodes 35 ENTH/ANTH/VHS domain family members, including six with an ENTH domain referred to as EPS1 to EPS6 (Holstein and Oliviusson, 2005; Zouhar and Sauer, 2014). These six EPSs form a subfamily that is distinct from the divergent ENTH domain-containing MODIFIED TRANSPORT TO THE VACUOLE, with roles in TGN/EE-to-vacuolar cargo trafficking (Sauer et al., 2013). The Arabidopsis ENTH subfamily also clusters away from the large ANTH protein family that includes EPSIN-LIKE CLATHRIN ADAPTOR4 (ECA4), PHOSPHATIDYLINOSITOL BINDING CLATHRIN ASSEMBLY PROTEIN5A (PICALM5; also called ECA2), PICALM5b, and AP180, for which physiological roles have been recently described using loss-of-function mutants (Sauer et al., 2013; Zouhar and Sauer, 2014; Muro et al., 2018; Nguyen et al., 2018; Kaneda et al., 2019). Thus, our identification of EPS1 modulating immune responses expands the short list of Arabidopsis ENTH/ANTH/VHS domain family members with known physiological functions.
In mammalian and yeast EPSINs, the ENTH domains participate in lipid binding and protein-protein interaction for recruiting EPSINs to the TGN or PM and inducing membrane curvature. Furthermore, sequence divergence in the ENTH domain and differences in peptide/domain architecture are indicative of diverse cellular functions, including the recruitment of different cargo, coat, adaptor, and accessory components to distinct subcellular compartments (for review, see Ford et al., 2002; Duncan and Payne, 2003; Mills et al., 2003; Legendre-Guillemin et al., 2004; Itoh and De Camilli, 2006; Lee et al., 2007). When expanding on previous studies (Holstein and Oliviusson, 2005; De Craene et al., 2012; Zouhar and Sauer, 2014), we found that Arabidopsis EPS1 shares relatively high amino acid sequence identity as well as similar domain and motif architecture with EPS1 orthologs from flowering plants (angiosperms; 50% to 88% identity; Supplemental Fig. S6), indicating similar cellular functions. In addition to a highly conserved ENTH domain, EPS1 and its orthologs shared the same peptide motif architecture that includes a confirmed (LIDL; Song et al., 2006) and a putative (LADV) clathrin-interacting motif as well as three putative α-adaptin-binding motifs (DPF), of which two appear to be relatively well conserved (Supplemental Fig. S6). Such similarities point toward potentially similar cellular roles of EPS1 orthologs across plant species. We also observed a C-terminal poly-Gln stretch that is only present in EPS1 orthologs of the Brassicaceae family but not in other plant EPS1 orthologs and paralogs (Supplemental Figs. S6–S8). Although the function of this domain is unknown for Brassicaceae EPS1s, poly-Gln domains have been implicated in protein aggregation and cell toxicity contributing to neurodegenerative disease in animals (Adegbuyiro et al., 2017; Silva et al., 2018). In contrast, EPS1 shares a less conserved ENTH domain and differs in peptide motifs and domain architecture from the five Arabidopsis paralogs (20% to 38%; Supplemental Fig. S7) as well as from the animal or yeast EPSINS (26% to 30%; Supplemental Fig. S8), with trafficking roles from the TGN (Legendre-Guillemin et al., 2004). Arabidopsis EPS2 (also referred to as EpsinR2; Lee et al., 2007) and EPS3 share a degenerate LADV motif with EPS1 but lack DPF or LIDL peptide motifs, and the other three Arabidopsis ENTH proteins are composed of little more than a divergent ENTH domain (Supplemental Fig. S7). We propose that the low sequence similarity and diverse peptide motif structure may indicate distinct cellular and/or physiological functions of the Arabidopsis EPSINs.
So far, it remains unknown how Arabidopsis EPS1 or related family members may recognize cargo proteins and whether such recognition occurs through direct or indirect interaction(s). In support of the latter, EPS1 interacts with γ-adaptin-related protein and VACUOLAR SORTING RECEPTOR1 (Song et al., 2006), both of which belong to protein families with cargo-binding properties (Ahmed et al., 2000; daSilva et al., 2005), but so far, the physiological role(s) of these protein-protein interactions have not been elucidated. In mammals and yeast, two different types of EPSINs localize to two distinct locations, the PM and the TGN. EPSINs that localize to the PM generally contain a C-terminal ubiquitin-interacting motif (UIM) that enables interaction with ubiquitylated cargoes to target these cargoes for endocytic degradation (Hawryluk et al., 2006; Dores et al., 2010; Chen et al., 2011). In contrast, TGN-localized mammalian EpsinR or yeast EPSIN Ent3p lack a C-terminal UIM (Duncan and Payne, 2003). By these distinctions and consistent with EPS1’s localization to the TGN/EE (Song et al., 2006), Arabidopsis EPS1 more closely resembles the TGN type in that it also lacks a UIM (Figs. 1 and 2B; Holstein and Oliviusson, 2005; Zouhar and Sauer, 2014 ). Thus, it is unlikely that EPS1 recognizes ubiquitylated cargo proteins directly. This scenario, however, does not eliminate the possibility that EPS1 may recruit yet unknown UIM-containing protein(s) for cargo recognition (Isono et al., 2010).
In conclusion, by identifying novel functions for Arabidopsis EPS1 in plant immunity against bacteria and modulating FLS2 abundance in the PM for effective flg22 responses, we advanced the limited information available for plant EPSINs and ENTH/ANTH/VHS superfamily members and their roles in physiological responses. Beyond FLS2 and the multitasking coreceptor BAK1, quantitative comparison between the ePM proteome of eps1 mutant and the wild type identified BRI1 and a small subset of structurally diverse PM proteins that require EPS1 for their correct PM accumulation, implicating EPS1 in contributing to potential cellular functions in addition to plant immunity. Because EPS1 serves as a monomeric clathrin adaptor involved in trafficking from the TGN/EE, analyses of EPS1 may aid in uncovering novel function(s) for clathrin at the TGN/EE that are distinct from those at the PM.
MATERIALS AND METHODS
Plant Material and Growth Conditions
T-DNA insertion lines of Arabidopsis (Arabidopsis thaliana) eps1-1 (SALK_049204; Song et al., 2006) and eps1-2 (SAIL_394_G02; this study) were obtained from the Nottingham Arabidopsis Stock Centre or the Arabidopsis Biological Resource Center at Ohio State University, respectively. bak1-4 (SALK_034523) and fls2Δ (SALK_093905) were previously described (Heese et al., 2007). All mutants were in the Arabidopsis ecotype Col-0 background. Plants were genotyped for homozygosity using allele-specific primers (Supplemental Table S1). eps1-2/FLS2-GFP plants were generated by crossing eps1-2 with FLS2pro:FLS2-3xMyc-EGFP (Beck et al., 2012; Smith et al., 2014a) and genotyped using primers (Supplemental Table S1). Surface-sterilized seeds were grown on 0.5× Murashige and Skoog medium + 1% (w/v) Suc solidified with 0.6% (w/v) agar at 22°C under continuous light as described (Korasick et al., 2010). After transplanting of seedlings, plants were grown in individual pots at 22°C with an 8-h-light/16-h-dark photoperiod at 82 μmol m−2 s−1. Except when noted, 7- to 8-d-old seedlings or fully expanded rosette leaves from 5- to 6-week-old plants were used for assays as described (Smith et al., 2014a, 2014b).
D/PAMP Peptides
The peptides flg22 (QRLSTGSRINSAKDDAAGLQIA), elf26 (SKEKFERTKPHVNVGTIGHVDHGKTT), and AtPep1 (ATKVKAKQRGKEKVSSGRPGQHN) were synthesized by Genscript and used for elicitation at the indicated concentrations and for the indicated times as described.
Gene Structure and Protein Domain Structure
Schematics for gene and protein domain structures were created using GSDS 2.0 (Hu et al., 2015) and IBS 1.01 (Liu et al., 2015), respectively.
Bacterial Pathogen Infections
Syringe infiltration with Pseudomonas syringae pv tomato DC3000 lux (OD600 = 0.0005) or Pto DC3000 hrcC− (OD600 = 0.02), collection of leaf discs, and quantification of bacterial growth using serial dilution plating were done as described previously (Korasick et al., 2010), except that leaf discs were ground in 100 µL of distilled water per leaf disc before serial dilution plating.
RNA Isolation and RT-qPCR
RNA isolation, cDNA synthesis, and RT-qPCR were performed as previously described (Libault et al., 2007; Korasick et al., 2010; Anderson et al., 2011). Unless stated otherwise, for each sample, three to five seedlings were elicited with the indicated concentration of D/PAMP and placed at 22°C. For PR1, leaves of 5- to 6-week-old plants were syringe infiltrated with the indicated concentration of D/PAMP, allowed to dry, and placed at 22°C. Tissue was flash frozen in liquid nitrogen at the indicated time points. Total RNA was isolated from tissue using Trizol reagent (Sigma-Aldrich) according to the manufacturer’s protocol. RT-qPCR was performed on cDNA using a 7500 Realtime PCR system (Applied Biosystems) using gene-specific primers (Supplemental Table S1) and normalized to the reference SAND gene (At2g28390; Libault et al., 2007).
Apoplastic ROS Production
Luminol-based ROS production was performed as previously described (Leslie and Heese, 2014) with the following modifications. Seedling cotyledons or leaf discs (1.5 cm2) from 5- to 6-week-old plants were cut in half and placed onto a 96-well plate for subsequent elicitation with D/PAMP peptides at the indicated concentrations. All ROS experiments shown in the same figure section were performed at the same time on a single 96-well plate to allow for direct comparison.
SDCM for Quantification of PM and Cytoplasmic Intensity, and Ligand-Induced Endocytosis of FLS2-GFP
SDCM was performed as described (Smith et al., 2014a; Leslie and Heese, 2017). Briefly, Col-0 FLS2-GFP and eps1-2 FLS2-GFP seedlings were directly mounted in distilled water for imaging. FLS2-GFP was imaged in the epidermal pavement cell layer of the adaxial (top) cotyledon surface. For each experiment, at least 12 seedlings were imaged per genotype, with two fields of view/cotyledon/genotype using a custom Olympus IX-71 inverted microscope equipped with a Yokogawa CSU-XI 5,000-rpm spinning-disc unit, Andor iXon Ultra 897 High Speed EMCCD camera, PZ-2000 XYZ series automated stage with Piezo Z-axis top plate (Applied Scientific Instrumentation), and a 60× silicon oil objective (Olympus UPlanSApo 60×/1.30 Sil). GFP was excited with a Spectra Physics 488-nm diode laser, and fluorescence was collected through a series of Semrock Brightline 488-nm single-edge dichroic beam splitter and 500- to 550-nm bandpass filters. Camera exposure time was set to 150 ms.
PM intensity measurements were performed as previously described (Smith et al., 2014a). In brief, single Z-plane images of epidermal pavement cells were captured independently using consistent experimental and imaging parameters across all samples. Brightness and contrast were adjusted consistently between all images using Fiji software. Bright sections of the PM were selected using the Oval Selection tool and analyzed for mean pixel intensity using ImageJ (https://imagej.net/Fiji/Downloads). FLS2-GFP PM intensity for each image was calculated as the average value of the pixel intensity measurements from four selected regions. For each genotype, FLS2-GFP PM intensities are reported relative to the FLS2-GFP PM intensity of Col-0 FLS2-GFP.
For quantifying intracellular compared with PM pixel intensity, single-plane confocal images from 7-d-old seedling cotyledons were analyzed for pixel intensity using ImageJ (https://imagej.net/Fiji/Downloads) as performed previously (Kitakura et al., 2011; Nguyen et al., 2018). Briefly, the Oval Selection tool was used to select regions of the cell cytoplasm adjacent to the PM and measure the mean pixel intensity. A quotient was calculated using these cytoplasmic values and the PM measurements before normalizing to the average wild-type control for each data set.
FLS2-GFP endocytosis experiments were performed as described previously (Smith et al., 2014a; Leslie and Heese, 2017). Z-stacks were collected from the adaxial surface of cotyledons in response to 1 μm flg22 in water for either 0 or 50 to 60 min as described above. Images were displayed as maximum intensity projections (MIPs) using ImageJ before brightness and contrast were adjusted uniformly. Stomata were removed from each image using the Freehand Selection Tool before detecting FLS2-GFP puncta using the Advanced Weka Segmentation plug-in from Fiji. Briefly, a single MIP with visible puncta was used to create a Vesicle Classifier that was applied to all other MIPs in an experiment. Puncta were analyzed in the resulting binary images using the Analyze Particles selection in Fiji with the following parameters: particle size = 0.25 to 2.5 μm2 and circularity = 0.25 to 1. Each MIP image was manually inspected for accuracy and, if necessary, adjustments were made to particle detection.
Protein Sample Preparation, Immunoblot Analysis, and Antibodies
Sample preparation and immunoblot analysis of total, soluble, and microsomal proteins were performed as previously described (Heese et al., 2007; Smith et al., 2014b; LaMontagne et al., 2016) with 30 µg loaded per well for seedling extracts. Affinity-purified, peptide-specific antibodies were produced in rabbit against EPS1 (#130, CSLSNQRYQSGGFKQ; #131, CFADSKPQHLQKKDPF; Eurogentec) or BAK1 (CRQDFNYPTHHPAVS; Sigma Genosys) according to the manufacturers’ specifications. The following antibody dilutions were used: 1:2,500 αEPS1#130 (this study), 1:2,500 αEPS1#131 (this study), 1:500 αBAK1 (this study), 1:500 αBRI1 (a kind gift of Marisa Otegui; Wu et al., 2011), 1:3,000 αMPK6 (Merkouropoulos et al., 2008), 1:500 αFLS2 (Heese et al., 2007), 1:3,000 αGFP (JL-8; Clontech Laboratories), 1:3,000 αSYP132 (Kalde et al., 2007), 1:10,000 αAHA (AS07 260; Agrisera), 1:1,000 αSMT1 (AS07 266; Agrisera), 1:3,000 αCNX1/2 (AS12 2365; Agrisera), 1:1,000 αV-ATPase (AS07 213; Agrisera), and 1:1,000 αToc75 (AS06 150; Agrisera).
PM Enrichment from Seedlings
Enrichment of PMs from whole seedlings was performed as described (Collins et al., 2017). Briefly, approximately 140 seedlings per genotype were homogenized in buffer H (250 mm Suc, 50 mm HEPES-KOH, pH 7.5, 5% [v/v] glycerol, 50 mm NaPP, 1 mm Na2MoO4, 25 mm NaF, 10 mm EDTA, and 0.5% [w/v] polyvinylpyrrolidone). Homogenate was filtered through two layers of Miracloth and centrifuged for 10 min at 8,000g. Supernatant was then ultracentrifuged for 30 min at 100,000g to obtain the microsomal pellet. Pellets were resuspended in buffer H, and microsomal protein was quantified. Microsomal protein was then incubated for 30 min on ice with 0.02% (w/v) Brij-58 at a protein-to-detergent ratio of 2 μL of 0.02% (w/v) Brij-58 solution per 1 μg of microsomal protein followed by centrifugation at 100,000g. This Brij-58 incubation and centrifugation step was repeated a second time. Pellets were then washed with buffer H (no detergent) followed by centrifugation at 100,000g. Final pellets were resuspended for immunoblot analysis.
Quantitative PM Proteomics
PM proteomics was performed as described (Zhang and Peck, 2011; Zhang et al., 2013). Briefly, equal amounts of PM-enriched proteins from each genotype were separated using 8% SDS-PAGE. Each gel lane was cut into 20 slices, and proteins in each slice were digested using trypsin, extracted, and lyophilized. Peptides were resuspended in 1% (v/v) formic acid before analysis using an LTQ-Orbitrap LC-MS/MS device (Thermoelectron) controlled by XCalibur v. 2.2.1. Mass spectrometer measurements were obtained (Zhang and Peck, 2011; Zhang et al., 2013). Mascot distiller 2.0 (Matrix Science) was used to deconvolute the mass spectra, and then Mascot (server v. 2.3; Matrix Science) and X! Tandem (v. 2007.01.01.1) were used to identify peptides. Scaffold software (v. 4.0; Proteome Software) was used to assign peptide identities at greater than 95% probability for a given peptide (Keller et al., 2002). Proteins were identified when at least two peptides (at greater than 99% probability) were found (Nesvizhskii et al., 2003). Normalized spectral counts for each genotype were determined using Scaffold software (v. 4.0; Proteome Software). For each genotype, the mean and sd were calculated for the spectral counts from each biological replicate. For each experiment, proteins for which the fold difference between genotypes was greater than one to two times the coefficient of variance (CV; CV = sd/mean) were considered changing. For replicate 1, CV = 1.2; for replicate 2, CV = 1.06; and for replicate 3, CV = 1.43.
Bioinformatic Analysis
The PANTHER classification system (version 13.1, released February 3, 2018) was used to categorize protein hits into Molecular Function and Protein Class using GO-Slim functional classification tools (Thomas et al., 2003). To determine if any categories were enriched relative to their representation in the whole Arabidopsis genome, a statistical overrepresentation test was performed (PANTHER, released December 5, 2017), specifically Fisher’s exact test with FDR multiple test correction. Categories with FDR < 0.05 were considered statistically significant. Structural classifications were as annotated in UniProt and TAIR.
Sequence Alignment
Orthologous or paralogous sequences were determined using amino acids 27 to 152 of Arabidopsis EPS1 (denoted as AthEPS1 in Supplemental Figs. S6–S8) for UniProt BLAST. Full-length amino acid sequences were aligned using the Clustal Omega Multiple Sequence Alignment tool. Sequence conservation compared with AthEPS1 was annotated in ExPASy Boxshade using AthEPS1 as the consensus sequence. The fraction of sequences that must agree for shading was set to 0.1.
Statistical Analysis
For each experiment, mutant samples were compared with Col-0 wild-type samples, and statistical analyses were performed using n values, with each n representing a biological sample as detailed in each figure legend. Statistical significance was based on unpaired two-tailed Student’s t test determined using GraphPad Prism 4.03 or GraphPad Prism 7.02 software. GraphPad QuickCalcs outlier calculator (https://www.graphpad.com/quickcalcs/Grubbs1.cfm) with α = 0.05 (standard) was used to identify outliers. Unless stated otherwise, each experiment represents a biological replicate and was performed at least three independent times with similar results.
Accession Numbers
Accession numbers are as follows: EPSIN1 (At5g11710), FLS2 (At5g46330), BAK1 (At4g33430), EFR (At5g20480), PEPR1 (At1g73080), and BRI1 (At4g39400).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Isolation and confirmation of eps1 mutant alleles.
Supplemental Figure S2. Increased susceptibility of eps1-2 null mutant to Pto DC3000 compared with the wild type and fls2Δ mutant and no difference in mRNA accumulation of PRRs and BAK1.
Supplemental Figure S3. Ligand-induced endocytosis of FLS2-GFP is not affected in eps1-2 null mutant.
Supplemental Figure S4. BAK1 antibody specificity.
Supplemental Figure S5. Proteins reduced in eps1-2 ePM fraction do not segregate into any distinct biological or structural categories.
Supplemental Figure S6. EPS1 is conserved across plant species.
Supplemental Figure S7. Protein sequence alignment of ENTH domain-containing proteins from Arabidopsis.
Supplemental Figure S8. Protein sequence alignment of Arabidopsis EPS1 with orthologous TGN-localized ENTH proteins from nonplant species.
Supplemental Table S1. Primer information.
Supplemental Data Set S1. Quantitative proteomics.
Supplemental Data Set S2. GO term and structure analysis.
Acknowledgments
We thank Dr. Marisa Otegui for αBRI1 antibody, Dr. Chris Pires and Makenzie Mabry for sequence analysis discussion, and former and present Heese lab members for technical help and discussions, in particular Dr. Michelle Leslie for training in and discussion on live-cell imaging.
Footnotes
This work was supported by the National Science Foundation (grant nos. IOS-1025837 to A.H., IOS-1456256 to S.C.P., and GRF-1443129 to E.D.L.), the Diane P. and Robert E. Sharp Fund Fellowship (to G.E.), the University of Missouri CAFNR Alexander Undergraduate Research Internships (to A.S.C. and L.A.B.), and the HHS|NIH|National Institute of General Medical Sciences (grant no. T32 GM008396 to D.J.S.).
Articles can be viewed without a subscription.
References
- Adegbuyiro A, Sedighi F, Pilkington AW IV, Groover S, Legleiter J(2017) Proteins containing expanded polyglutamine tracts and neurodegenerative disease. Biochemistry 56: 1199–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed SU, Rojo E, Kovaleva V, Venkataraman S, Dombrowski JE, Matsuoka K, Raikhel NV(2000) The plant vacuolar sorting receptor AtELP is involved in transport of NH2-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J Cell Biol 149: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C(2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA 109: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Bartels S, González Besteiro MA, Shahollari B, Ulm R, Peck SC(2011) Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J 67: 258–268 [DOI] [PubMed] [Google Scholar]
- Beck M, Zhou J, Faulkner C, MacLean D, Robatzek S(2012) Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24: 4205–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão-Pires E, Dangl JL, Chory J(2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci USA 109: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Khaled S, Postma J, Robatzek S(2015) A moving view: Subcellular trafficking processes in pattern recognition receptor-triggered plant immunity. Annu Rev Phytopathol 53: 379–402 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J(2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP(2010) Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci USA 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Dores MR, Grimsey N, Canto I, Barker BL, Trejo J(2011) Adaptor protein complex-2 (AP-2) and epsin-1 mediate protease-activated receptor-1 internalization via phosphorylation- and ubiquitination-dependent sorting signals. J Biol Chem 286: 40760–40770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T(2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Choi SW, Tamaki T, Ebine K, Uemura T, Ueda T, Nakano A(2013) RABA members act in distinct steps of subcellular trafficking of the FLAGELLIN SENSING2 receptor. Plant Cell 25: 1174–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, Vanoosthuyse V, Gaude T(2002) Receptor kinase signalling in plants and animals: Distinct molecular systems with mechanistic similarities. Curr Opin Cell Biol 14: 230–236 [DOI] [PubMed] [Google Scholar]
- Collins CA, Leslie ME, Peck SC, Heese A (2017) Simplified enrichment of plasma membrane proteins from Arabidopsis thaliana seedlings using differential centrifugation and Brij-58 treatment. In Russinova E, Caño-Delgado AI, eds, Brassinosteroids. Methods in Molecular Biology, Vol 1564 Humana Press, New York, pp 155–168 [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C(2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- daSilva LL, Taylor JP, Hadlington JL, Hanton SL, Snowden CJ, Fox SJ, Foresti O, Brandizzi F, Denecke J(2005) Receptor salvage from the prevacuolar compartment is essential for efficient vacuolar protein targeting. Plant Cell 17: 132–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene JO, Ripp R, Lecompte O, Thompson JD, Poch O, Friant S(2012) Evolutionary analysis of the ENTH/ANTH/VHS protein superfamily reveals a coevolution between membrane trafficking and metabolism. BMC Genomics 13: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores MR, Schnell JD, Maldonado-Baez L, Wendland B, Hicke L(2010) The function of yeast epsin and Ede1 ubiquitin-binding domains during receptor internalization. Traffic 11: 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MC, Payne GS(2003) ENTH/ANTH domains expand to the Golgi. Trends Cell Biol 13: 211–215 [DOI] [PubMed] [Google Scholar]
- Ekanayake G, LaMontagne ED, Heese A(2019) Never walk alone: Clathrin-coated vesicle (CCV) components in plant immunity. Annu Rev Phytopathol 57: 387–409 [DOI] [PubMed] [Google Scholar]
- Elmore JM, Coaker G(2011) The role of the plasma membrane H+-ATPase in plant-microbe interactions. Mol Plant 4: 416–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT(2002) Curvature of clathrin-coated pits driven by epsin. Nature 419: 361–366 [DOI] [PubMed] [Google Scholar]
- Gendre D, Jonsson K, Boutté Y, Bhalerao RP(2015) Journey to the cell surface: The central role of the trans-Golgi network in plants. Protoplasma 252: 385–398 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T(2000) FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gu Y, Innes RW(2011) The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol 155: 1827–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Innes RW(2012) The KEEP ON GOING protein of Arabidopsis regulates intracellular protein trafficking and is degraded during fungal infection. Plant Cell 24: 4717–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk MJ, Keyel PA, Mishra SK, Watkins SC, Heuser JE, Traub LM(2006) Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic 7: 262–281 [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP(2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein SE, Oliviusson P(2005) Sequence analysis of Arabidopsis thaliana E/ANTH-domain-containing proteins: Membrane tethers of the clathrin-dependent vesicle budding machinery. Protoplasma 226: 13–21 [DOI] [PubMed] [Google Scholar]
- Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G(2015) GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31: 1296–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E, Katsiarimpa A, Müller IK, Anzenberger F, Stierhof YD, Geldner N, Chory J, Schwechheimer C(2010) The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell 22: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, De Camilli P(2006) BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta 1761: 897–912 [DOI] [PubMed] [Google Scholar]
- Kalde M, Nühse TS, Findlay K, Peck SC(2007) The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc Natl Acad Sci USA 104: 11850–11855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalthoff C, Alves J, Urbanke C, Knorr R, Ungewickell EJ(2002) Unusual structural organization of the endocytic proteins AP180 and epsin 1. J Biol Chem 277: 8209–8216 [DOI] [PubMed] [Google Scholar]
- Kaneda M, van Oostende-Triplet C, Chebli Y, Testerink C, Bednarek SY, Geitmann A(2019) Plant AP180 N-terminal homolog proteins are involved in clathrin-dependent endocytosis during pollen tube growth in Arabidopsis thaliana. Plant Cell Physiol 60: 1316–1330 [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R(2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kitakura S, Vanneste S, Robert S, Löfke C, Teichmann T, Tanaka H, Friml J(2011) Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, McMichael C, Walker KA, Anderson JC, Bednarek SY, Heese A(2010) Novel functions of Stomatal Cytokinesis-Defective 1 (SCD1) in innate immune responses against bacteria. J Biol Chem 285: 23342–23350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KAS, et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem 285: 13471–13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMontagne ED, Collins CA, Peck SC, Heese A(2016) Isolation of microsomal membrane proteins from Arabidopsis thaliana. Curr Protoc Plant Biol 1: 217–234 [DOI] [PubMed] [Google Scholar]
- LaMontagne ED, Heese A(2017) Trans-Golgi network/early endosome: A central sorting station for cargo proteins in plant immunity. Curr Opin Plant Biol 40: 114–121 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Kim H, Kang H, Jang M, Lee DW, Lee S, Hwang I(2007) EpsinR2 interacts with clathrin, adaptor protein-3, AtVTI12, and phosphatidylinositol-3-phosphate: Implications for EpsinR2 function in protein trafficking in plant cells. Plant Physiol 143: 1561–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS(2004) ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci 117: 9–18 [DOI] [PubMed] [Google Scholar]
- Leslie ME, Heese A (2014) A re-elicitation assay to correlate flg22-signaling competency with ligand-induced endocytic degradation of the FLS2 receptor. In Otegui M, ed, Plant Endosomes. Methods in Molecular Biology, Vol 1209 Humana Press, New York, pp 149–162 [DOI] [PubMed] [Google Scholar]
- Leslie ME, Heese A (2017) Quantitative analysis of ligand-induced endocytosis of FLAGELLIN-SENSING 2 using automated image segmentation. In Shan L, He P, eds, Plant Pattern Recognition Receptors. Methods in Molecular Biology, Vol 1578 Humana Press, New York, pp 39–54 [DOI] [PubMed] [Google Scholar]
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G(2007) Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact 20: 900–911 [DOI] [PubMed] [Google Scholar]
- Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y, Zhao Y, et al. (2015) IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 31: 3359–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkouropoulos G, Andreasson E, Hess D, Boller T, Peck SC(2008) An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a substrate of MAP kinases 3 and 6. J Biol Chem 283: 10493–10499 [DOI] [PubMed] [Google Scholar]
- Mersmann S, Bourdais G, Rietz S, Robatzek S(2010) Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol 154: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills IG, Praefcke GJK, Vallis Y, Peter BJ, Olesen LE, Gallop JL, Butler PJG, Evans PR, McMahon HT(2003) EpsinR: An AP1/clathrin interacting protein involved in vesicle trafficking. J Cell Biol 160: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro K, Matsuura-Tokita K, Tsukamoto R, Kanaoka MM, Ebine K, Higashiyama T, Nakano A, Ueda T(2018) ANTH domain-containing proteins are required for the pollen tube plasma membrane integrity via recycling ANXUR kinases. Commun Biol 1: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, et al. (2009) Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R(2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Nguyen HH, Lee MH, Song K, Ahn G, Lee J, Hwang I(2018) The A/ENTH domain-containing protein AtECA4 is an adaptor protein involved in cargo recycling from the trans-Golgi network/early endosome to the plasma membrane. Mol Plant 11: 568–583 [DOI] [PubMed] [Google Scholar]
- Nomura K, Mecey C, Lee YN, Imboden LA, Chang JH, He SY(2011) Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc Natl Acad Sci USA 108: 10774–10779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse TS, Boller T, Peck SC(2003) A plasma membrane syntaxin is phosphorylated in response to the bacterial elicitor flagellin. J Biol Chem 278: 45248–45254 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Bottrill AR, Jones AM, Peck SC(2007) Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J 51: 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T, Brunner F, Kemmerling B, Piater L(2004) Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol Rev 198: 249–266 [DOI] [PubMed] [Google Scholar]
- Ortiz-Morea FA, Savatin DV, Dejonghe W, Kumar R, Luo Y, Adamowski M, Van den Begin J, Dressano K, Pereira de Oliveira G, Zhao X, et al. (2016) Danger-associated peptide signaling in Arabidopsis requires clathrin. Proc Natl Acad Sci USA 113: 11028–11033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez Valencia J, Goodman K, Otegui MS(2016) Endocytosis and endosomal trafficking in plants. Annu Rev Plant Biol 67: 309–335 [DOI] [PubMed] [Google Scholar]
- Peck SC, Nühse TS, Hess D, Iglesias A, Meins F, Boller T(2001) Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell 13: 1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Wirthmueller L(2012) Mapping FLS2 function to structure: LRRs, kinase and its working bits. Protoplasma 250: 671–681 [DOI] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C(2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Delgadillo MO, Zouhar J, Reynolds GD, Pennington JG, Jiang L, Liljegren SJ, Stierhof YD, De Jaeger G, Otegui MS, et al. (2013) MTV1 and MTV4 encode plant-specific ENTH and ARF GAP proteins that mediate clathrin-dependent trafficking of vacuolar cargo from the trans-Golgi network. Plant Cell 25: 2217–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A, de Almeida AV, Macedo-Ribeiro S(2018) Polyglutamine expansion diseases: More than simple repeats. J Struct Biol 201: 139–154 [DOI] [PubMed] [Google Scholar]
- Smith JM, Leslie ME, Robinson SJ, Korasick DA, Zhang T, Backues SK, Cornish PV, Koo AJ, Bednarek SY, Heese A(2014a) Loss of Arabidopsis thaliana Dynamin-Related Protein 2B reveals separation of innate immune signaling pathways. PLoS Pathog 10: e1004578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Salamango DJ, Leslie ME, Collins CA, Heese A(2014b) Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol 164: 440–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Lee MH, Lee GJ, Yoo CM, Hwang I(2006) Arabidopsis EPSIN1 plays an important role in vacuolar trafficking of soluble cargo proteins in plant cells via interactions with clathrin, AP-1, VTI11, and VSR1. Plant Cell 18: 2258–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J(2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628 [DOI] [PubMed] [Google Scholar]
- Tang J, Han Z, Sun Y, Zhang H, Gong X, Chai J(2015) Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res 25: 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A(2003) PANTHER: A library of protein families and subfamilies indexed by function. Genome Res 13: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T.(2016) Physiological roles of plant post-Golgi transport pathways in membrane trafficking. Plant Cell Physiol 57: 2013–2019 [DOI] [PubMed] [Google Scholar]
- Underwood W.(2016) Contributions of host cellular trafficking and organization to the outcomes of plant-pathogen interactions. Semin Cell Dev Biol 56: 163–173 [DOI] [PubMed] [Google Scholar]
- Viotti C, Bubeck J, Stierhof YD, Krebs M, Langhans M, van den Berg W, van Dongen W, Richter S, Geldner N, Takano J, et al. (2010) Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling R, Epple P, Altmann S, He Y, Yang L, Henz SR, McDonald N, Wiley K, Bader KC, Gläßer C, et al. (2014) Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Liu S, Zhao Y, Wang W, Kong Z, Tang D(2015) ENHANCED DISEASE RESISTANCE4 associates with CLATHRIN HEAVY CHAIN2 and modulates plant immunity by regulating relocation of EDR1 in Arabidopsis. Plant Cell 27: 857–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Wang X, Li X, Kamiya Y, Otegui MS, Chory J(2011) Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal 4: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, He SY(2013) Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51: 473–498 [DOI] [PubMed] [Google Scholar]
- Xiong J, He R, Yang F, Zou L, Yi K, Lin H, Zhang D(2020) Brassinosteroids are involved in ethylene-induced Pst DC3000 resistance in Nicotiana benthamiana. Plant Biol J 22: 309–316 [DOI] [PubMed] [Google Scholar]
- Xu J, Xie J, Yan C, Zou X, Ren D, Zhang S(2014) A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J 77: 222–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA(2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Okada K, Saijo Y(2017) A look at plant immunity through the window of the multitasking coreceptor BAK1. Curr Opin Plant Biol 38: 10–18 [DOI] [PubMed] [Google Scholar]
- Yu X, Feng B, He P, Shan L(2017) From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol 55: 109–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Voothuluru P, Yamaguchi M, Sharp RE, Peck SC(2013) Developmental distribution of the plasma membrane-enriched proteome in the maize primary root growth zone. Front Plant Sci 4: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZJ, Peck SC(2011) Simplified enrichment of plasma membrane proteins for proteomic analyses in Arabidopsis thaliana. Proteomics 11: 1780–1788 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G(2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T(2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
- Zouhar J, Sauer M(2014) Helping hands for budding prospects: ENTH/ANTH/VHS accessory proteins in endocytosis, vacuolar transport, and secretion. Plant Cell 26: 4232–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]