Repression of miR159 in tobacco triggers a constitutive defense response that confers immunity to Phytophthora parasitica.
Abstract
MicroR159 (miR159) regulation of GAMYB expression is highly conserved in terrestrial plants; however, its functional role remains poorly understood. In Arabidopsis (Arabidopsis thaliana), although GAMYB-like genes are constitutively transcribed during vegetative growth, their effects are suppressed by strong and constitutive silencing by miR159. GAMYB expression occurs only if miR159 function is inhibited, which results in detrimental pleiotropic defects, questioning the purpose of the miR159-GAMYB pathway. Here, miR159 function was inhibited in tobacco (Nicotiana tabacum) and rice (Oryza sativa) using miRNA MIM159 technology. Similar to observations in Arabidopsis, inhibition of miR159 in tobacco and rice resulted in pleiotropic defects including stunted growth, implying functional conservation of the miR159-GAMYB pathway among angiosperms. In MIM159 tobacco, transcriptome profiling revealed that genes associated with defense and programmed cell death were strongly activated, including a suite of 22 PATHOGENESIS-RELATED PROTEIN (PR) genes that were 100- to 1,000-fold upregulated. Constitutive expression of a miR159-resistant GAMYB transgene in tobacco resulted in phenotypes similar to that of MIM159 tobacco and activated PR gene expression, verifying the dependence of the above-mentioned changes on GAMYB expression. Consistent with the broad defense response, MIM159 tobacco appeared immune to Phytophthora infection. These findings suggest that the tobacco miR159-GAMYB pathway functions in the biotic defense response, which becomes activated upon miR159 inhibition. However, PR gene expression was not upregulated in Arabidopsis or rice when miR159 was inhibited, suggesting that miR159-GAMYB pathway functional differences exist between species, or factors in addition to miR159 inhibition are required in Arabidopsis and rice to activate this broad defense response.
MicroRNAs (miRNAs) are a class of small RNAs that mediate the silencing of target genes via base pairing to highly complementary binding sites. In plants, many miRNA-target relationships are ancient and have been strongly conserved throughout the plant kingdom (Axtell and Bartel, 2005). These miRNAs control regulatory processes that appear fundamental to development of land plants (Jones-Rhoades, 2012). For example, miR156 regulation of SPL controls the vegetative phase change (Wu and Poethig, 2006); miR165/miR166 regulation of the PHV/PHB/REV family of genes controls leaf polarity, enabling the formation of a laminar leaf (Emery et al., 2003); and miR319 regulation of the TCP family controls leaf morphogenesis (Palatnik et al., 2003). miRNA regulation is fundamental for the highly complex temporal and spatial expression patterns of each of these regulatory genes (Li et al., 2014a; D’Ario et al., 2017).
Similarly, the miR159-GAMYB pathway is also ancient, as indicated by its presence in basal vascular plants to angiosperms (Axtell and Bartel, 2005). All evidence points to the GAMYB or GAMYB-like genes being the main conserved targets of miR159 (Millar et al., 2019). Although degradome data has found other targets for miR159, miR159 regulation of these genes does not appear conserved in diverse species (Addo-Quaye et al., 2008; Li et al., 2010). GAMYB or GAMYB-like genes encode conserved R2R3 MYB domain transcription factors, which are named for their role in positive transduction of the gibberellin (GA) signal in the seed aleurone (Gubler et al., 1995). Similarly, GAMYB also transduces the GA signal in the tapetum of the anther (Aya et al., 2009), and in both aleurone and tapetum tissues, GAMYB activity promotes programmed cell death (PCD; Millar and Gubler, 2005, Guo and Ho, 2008, Aya et al., 2009; Alonso-Peral et al., 2010). By contrast, strong GAMYB expression only appears to occur in the vegetative parts of the plant when miR159 function is inhibited. For instance, in the Arabidopsis (Arabidopsis thaliana) mir159ab double mutant (Allen et al., 2007), or rice (Oryza sativa) plants expressing a Short Tandem Target MIMIC against miR159 (Zhang et al., 2017: Zhao et al., 2017), inhibition of miR159 function results in strong deregulated GAMYB expression. In both instances, this results in pleiotropic deleterious defects throughout the plant, including stunted growth and phenotypes counterintuitive to a role in GA signaling. Moreover, the isolation of gamyb mutants and their analysis have clearly shown that GAMYB does not play a functional role in GA-regulated growth and development in vegetative tissues (Kaneko et al., 2004; Millar and Gubler, 2005; Alonso-Peral et al., 2010).
To date, the miR159-GAMYB pathway has been best characterized in Arabidopsis. Its major target genes are the GAMYB-like genes MYB33 and MYB65, indicated by the suppression of mir159ab mutant growth and developmental defects in a mir159ab.myb33.myb65 quadruple mutant (Allen et al., 2007). Both miR159 and MYB33/MYB65 appear constitutively transcribed throughout vegetative tissues (Palatnik et al., 2007; Li et al., 2016). However, based on the lack of expression of a MYB33:GUS translational fusion in vegetative tissues, indistinguishable phenotypes of wild-type and myb33.myb65 double-mutant plants, and indistinguishable transcriptomes of wild-type and myb33.myb65 plants, the MYB33/MYB65 genes appear fully silenced (Alonso-Peral et al., 2010). Underpinning this strong silencing are conserved RNA secondary structures associated with the miR159-binding sites of MYB33 and MYB65, which have been shown by a structure/function analysis to be required for this strong silencing in vegetative tissues of Arabidopsis (Zheng et al., 2017). Interestingly, these structures are not present in GAMYB-like homologs that are predominantly transcribed in seeds and anthers (Allen et al., 2007, 2010; Leydon et al., 2013), suggesting that these structures are under strong selection pressure in GAMYB-like homologs transcribed in vegetative tissue in order to ensure strong silencing. Currently, the only evidence that these GAMYB-like genes are expressed in the rosette is a slight change to the vegetative phase change of the myb33 mutant (Guo et al., 2017). Indeed, an Arabidopsis mir159ab.myb33.myb65 quadruple mutant appears phenotypically indistinguishable from the wild type, even when grown under many different abiotic stress conditions (Li et al., 2016). This raises the question of what the biological function of the miR159-GAMYB pathway is, especially given that failed miR159 repression of GAMYB has such dramatic deleterious consequences for the plant (Millar et al., 2019).
In this study, we investigated the role of the miR159-GAMYB pathway by expressing a miR159 decoy in different species to generate miR159 loss-of-function phenotypes. To this end, we chose to use MIM159 (Todesco et al., 2010), as it has been previously shown to be the most effective decoy against miR159 (Reichel et al., 2015), and expressed this in tobacco (Nicotiana tabacum) and rice, which are representative model species for dicot and monocot plants. Our data show that, as in Arabidopsis, inhibition of miR159 results in pleiotropic developmental defects due to widespread deregulation of GAMYB expression. However, at the molecular level, tobacco has a very different response, whereby major upregulated genes are those associated with disease resistance and PCD. Subsequently, we show that MIM159 tobacco plants are highly resistant to Phytophthora infection. These results suggest that the fundamental role for the miR159-GAMYB pathway is in response to pathogens, which provides the rationale for the ubiquitous nature of the miR159-GAMYB pathway in plants.
RESULTS
Characterization of the miR159-GAMYB Pathway in Tobacco
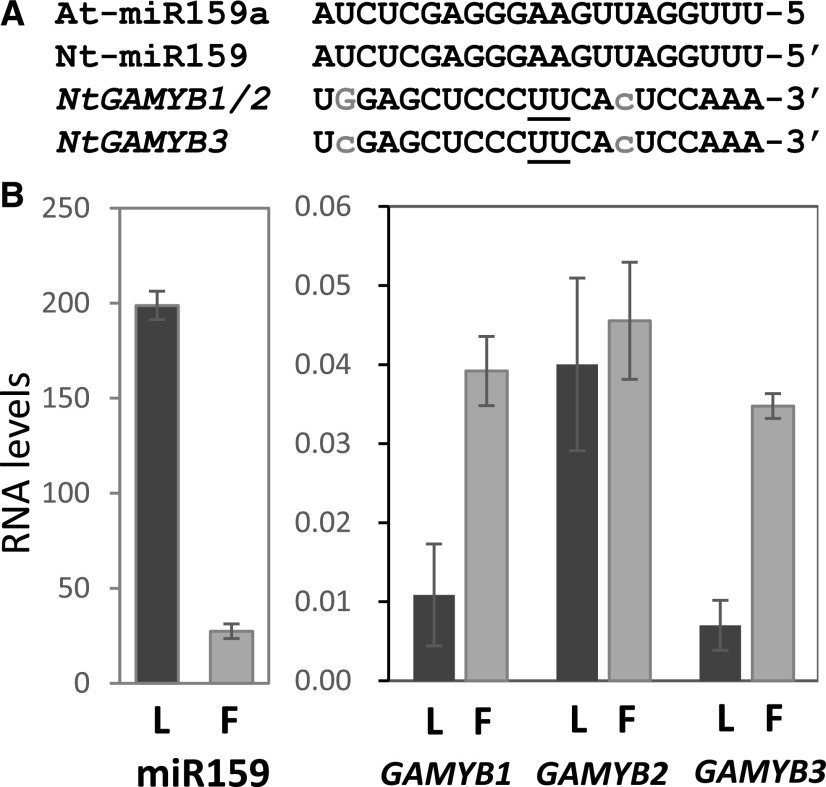
To study the functional diversity of miR159 in plants, we characterized the miR159-GAMYB pathway in tobacco. To begin with, we determined whether miR159 and GAMYB mRNA are present in both vegetative and floral tissues. According to miRBase (Kozomara and Griffiths-Jones, 2014), tobacco has a single miR159 isoform, whose 21-nucleotide sequence is identical to that of Arabidopsis miR159a (Fig. 1A). Using Taqman miRNA assays, miR159 was found to be highly abundant in leaves, and to a lesser extent in flowers (Fig. 1B). For the GAMYB family members, there are three homologs in tobacco, which are designated here NtGAMYB1–NtGAMYB3 (NCBI Reference accessions XM_016589328, XM_016599515, and XM_016629135, respectively). Based on nucleotide and amino acid sequence alignments, NtGAMYB1 and NtGAMYB2 share a greater sequence similarity, with NtGAMYB3 exhibiting more sequence differences (Supplemental Fig. S1). All three homologs contain putative miR159 binding sites (Fig. 1A). To determine the transcript abundances of NtGAMYB1–NtGAMYB3, reverse transcription quantitative PCR (RT-qPCR) was performed with primers corresponding to their 3′-untranslated regions (UTRs) to ensure gene-specific amplicons. As the amplicon does not span the miR159 cleavage site, these primers will measure both cleaved and uncleaved GAMYB transcripts. NtGAMYB1 and NtGAMYB3 transcripts were more abundant in flowers than in leaves, whereas NtGAMYB2 had similar transcript levels in leaves and flowers (Fig. 1B). Next, to determine whether the three NtGAMYB homologs undergo miR159-mediated cleavage, miRNA-cleavage assays were performed for all three GAMYB homologs on RNA prepared from tobacco leaves and flowers. Analysis revealed that all three NtGAMYB homologs are cleaved by miR159 at the expected canonical miR159 cleavage site (Supplemental Fig. S2). Although these cleavage assays are nonquantitative, they experimentally validate that the miR159-GAMYB pathway is active in both vegetative and reproductive tissues of tobacco.
Figure 1.
MiR159 and GAMYB homologs in tobacco. A, Sequence alignments of tobacco mature miR159 and the miR159 binding sites in NtGAMYB transcripts. Underlined nucleotides represent the cleavage site, lowercase gray letters represent mismatches to Nt-miR159, and G:U pairing is shown in uppercase gray letters. B, RT-qPCR measurement of miR159 and GAMYB transcript levels in tobacco. RNA was extracted from pools of tissue from two plants, each composed of two young leaves (L) or five mature open flowers (F). The mRNA levels of the NtGAMYB genes were normalized to PROTEIN PHOSPHATASE2A and the miR159 RNA levels were normalized to snoR101. Measurements are the average of three biological replicates, with error bars representing the se.
35S-MIM159 Transgenic Tobacco Displays Pleiotropic Developmental Defects
To investigate the functional role of miR159 in tobacco, a 35S-MIM159 transgene (Todesco et al., 2010) was stably transformed into tobacco. Of 15 independent T0 MIM159 transgenic tobacco plants, seven lines displayed similar developmental defects, which were inherited in T1 progeny plants. As compared to the wild type, defects of MIM159 tobacco included stunted growth with a reduced apical dominance (Fig. 2, A and B) and leaves that were smaller and crinkled, with sectorized chlorosis in the older leaves (Fig. 2, C and D). Additionally, MIM159 transgenic tobacco had smaller flowers with pale petals and shorter anther filaments (Fig. 2E). MIM159 tobacco also exhibited a delayed flowering time compared to the wild type (Fig. 2F) and produced seeds that had reduced weight compared to wild-type seeds (Fig. 2G). Based on these observations, inhibition of miR159 function in tobacco results in phenotypes similar to those of Arabidopsis with inhibited miR159 function (Allen et al., 2007; Todesco et al., 2010).
Figure 2.
Expression of MIM159 in transgenic tobacco results in strong phenotypic defects. A, Multiple T1 independent MIM159 transgenic lines (L2, L4, and L8) displayed smaller growth stature compared to the wild type (WT). B, Eight-week-old wild-type and MIM159 transgenic tobacco grown under identical conditions. C, Close-up view of the MIM159 transgenic plant depicted in B. Arrows indicate sectorized chlorosis on leaves. D, Young leaf of 8-week-old wild-type and MIM159 transgenic plants. E, MIM159 plants produce smaller flowers, paler petals, and shorter anther filaments compared to wild-type plants. In D and E, the various organs were digitally extracted for comparison. F, Flowering time of wild-type and MIM159 transgenic plants. All plants were grown without antibiotic selection. The bar charts represent the mean of flowering time of eight wild-type plants and 13 MIM159 plants from three independent lines. Error bars represent the se, and the asterisk indicates statistically significant difference determined by Student’s t test. G, Seed weight of wild-type and MIM159 transgenic plants. The bar charts represent the average seed weight of >100 seeds from five wild-type and five MIM159 plants from three independent lines. Error bars represent the se, and the asterisk indicates statistically significant difference determined by Student’s t test.
NtGAMYB Expression Is Deregulated in MIM159 Tobacco
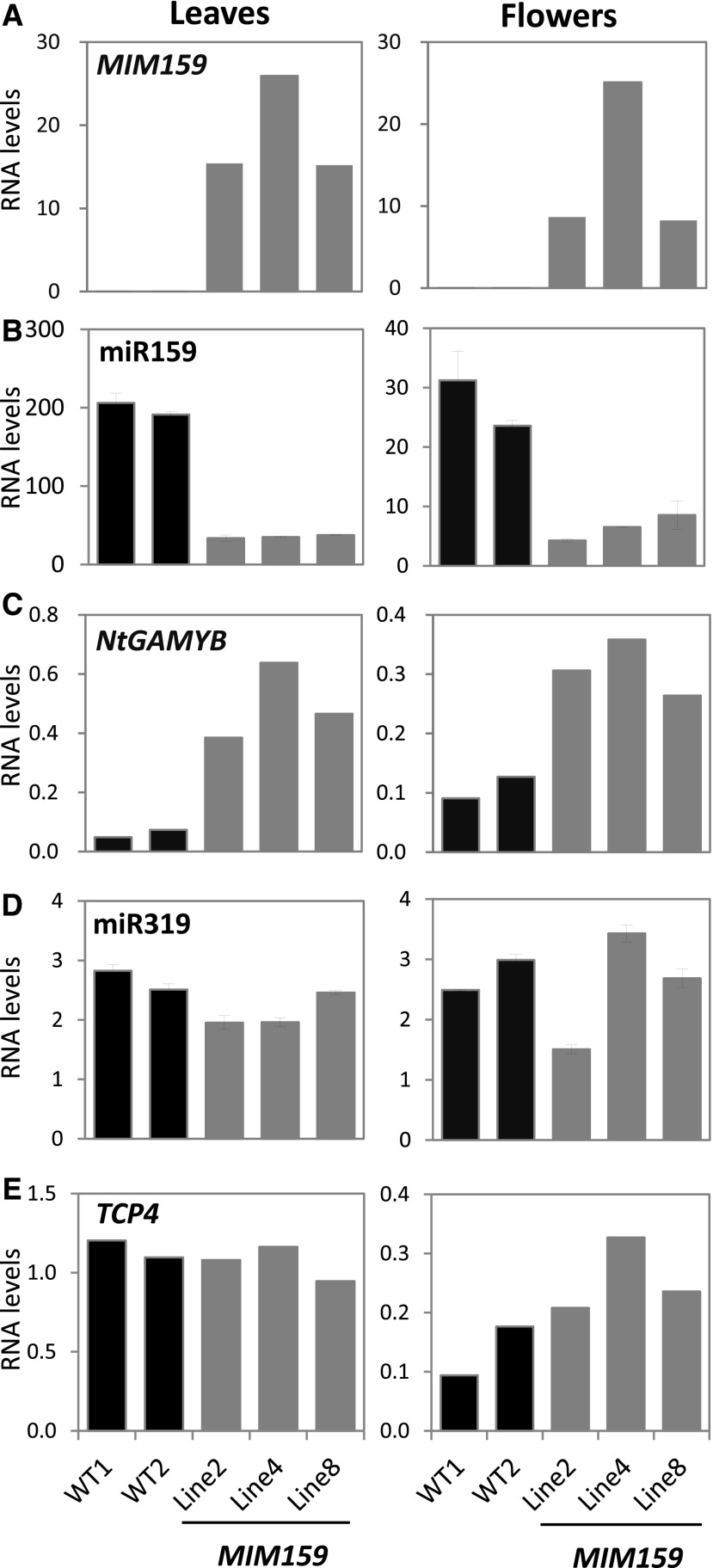
To confirm that the MIM159 transgene was being expressed, RT-qPCR analysis was performed, revealing high levels of MIM159 RNA in both leaves and flowers of MIM159 plants (Fig. 3A; Supplemental Fig. S3A). Mature miR159 levels were strongly reduced in these MIM159 transgenic lines (Fig. 3B), and the transcript levels of all three NtGAMYB homologs were higher in MIM159 transgenic lines than in the wild type (Fig. 3C; Supplemental Fig. S3B). This expression analysis was performed with a set of primers corresponding to the GAMYB coding region that spanned the miRNA cleavage site and could amplify all three GAMYB homologs simultaneously. To determine which of the three NtGAMYB homologs are deregulated, 3′-UTR gene-specific primers were used. The mRNA levels of NtGAMYB1 and NtGAMYB2 were higher in leaves and flowers of MIM159 transgenic lines than in those of the wild type (Supplemental Fig. S4), with the exception of NtGAMYB1 mRNA levels in MIM159 Line 8, which were lower than those in wild-type leaves and flowers (Supplemental Fig. S4A). The mRNA levels of NtGAMYB3 in leaves of MIM159 lines was higher than in the wild type, but was similar in flowers (Supplemental Fig. S4C). Overall, this analysis found that the MIM159 transgene successfully inhibits miR159 in MIM159 tobacco, resulting in deregulation of all three NtGAMYB homologs.
Figure 3.
RNA profiling in MIM159 tobacco. RT-qPCR measurements in leaves (left) and flowers (right) of MIM159 (A), miR159 (B), NtGAMYB1-3 (total NtGAMYB; C), miR319 (D), and TCP4 (E) of wild-type (WT1 and WT2) and three independent T1 MIM159 lines (Lines 2, 4, and 8). RNA was extracted from either two young leaves of 8-week-old plants (Leaves) or five mature open flowers of 12-week-old plants (Flowers). The transcript levels of total NtGAMYBs include all three NtGAMYB homologs, as NtGAMYB amplicons spanned the conserved sequences flanking the miR159 binding site. The MIM159, NtGAMYBs, and TCP4 RNA levels were normalized to PROTEIN PHOSPHATASE2A, whereas miR159 and miR319 levels were normalized to snoR101. Measurements are the averages of three technical replicates.
It has been shown that MIM159 can result in off-target inhibition of miR319 in Arabidopsis (Reichel and Millar, 2015), as miR319 and miR159 are highly similar (17 of 21 nucleotides are identical; Palatnik et al., 2007). However, analysis of MIM159 tobacco lines did not present any strong evidence of a decrease of miR319 levels or deregulation of a miR319 target, such as TCP4 (Fig. 3, D and E). This suggests that the phenotypic defects in MIM159 tobacco are mainly derived from miR159 inhibition.
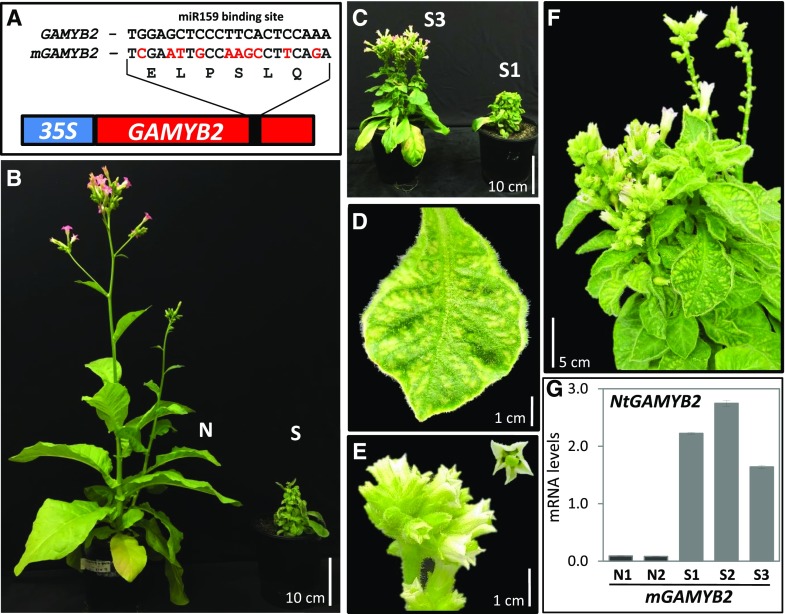
mGAMYB2 Expression in Tobacco Exhibits Phenotypic Defects Similar to MIM159
To test the hypothesis that the MIM159 phenotype is mainly due to deregulated GAMYB expression, tobacco was transformed with either a 35S-GAMYB2 transgene or a miR159-resistant version. The latter, namely 35S-mGAMYB2, contained synonymous mutations within its miR159 binding site (Fig. 4A). Eight GAMYB2 transgenic lines were generated; however, none displayed any obvious developmental defects. By contrast, of 10 independent T0 mGAMYB2 tobacco lines, four displayed strong phenotypic defects (S lines) compared to six lines that had no obvious phenotypic defects and resembled the wild type (N lines). The mGAMYB2-S lines displayed similar phenotypic defects to MIM159 plants (Fig. 4, B and C), including stunted growth, reduced apical dominance, upward leaf curl, leaf chlorosis, and smaller flowers with developmental defects (Fig. 4, D–F). Of the four mGAMYB2-S lines, only one line (mGAMYB2-S3) produced seeds. Therefore, the phenotypic defects of these mGAMYB2-S plants were more severe than those of MIM159 tobacco, which may reflect higher mGAMYB2 expression due to the strength of the 35S promoter.
Figure 4.
Expression of mNtGAMYB2 in tobacco results in severe phenotypic defects. A, Schematic representation of the 35S-GAMYB2 and 35S-mGAMYB2 transgenes. B, mGAMYB2 tobacco plants displayed either no (N) or strong (S) phenotypic defects. C, Two independent mGAMYB2-S lines (S1 and S3), with only the S3 line exhibiting flowers and seed setting. D, Detail of a leaf exhibiting chlorosis. E, Detail of mGAMYB2-S flowers that are white and display strong developmental defects. For D and E, the various organs were digitally extracted for comparison. F, Defects displayed by a 12-month old mGAMYB2-S plant. G, RT-qPCR measurement of GAMYB2 transcript levels in two independent mGAMYB2-N lines (N1 and N2) and three independent mGAMYB2-S lines (S1–S3). RNA was extracted from two young leaves of 12-week-old plants and GAMYB2 mRNA levels were normalized to PROTEIN PHOSPHATASE2A. Measurements are the average of three technical replicates.
To determine whether mGAMYB2 transcript levels correlated with phenotypic severity, RT-qPCR analysis was performed on leaves from five mGAMYB2 lines. GAMYB2 transcript levels were considerably higher in the strong lines S1–S3 compared to N1 and N2 lines, which displayed no obvious developmental defects (Fig. 4G). The transcript level of mGAMYB2 was 5- to 10-fold higher than that of any of the GAMYB genes in the MIM159 plants, likely explaining the more severe phenotypes of the mGAMYB2 plants. Together, the data suggest that the observed phenotypes in MIM159 tobacco are due to the deregulation of GAMYB expression.
Transcriptional Networks Activated in MIM159 Tobacco
As GAMYB genes encode MYB R2R3 transcription factors, RNA sequencing was performed on leaves of wild-type and MIM159 tobacco to identify differentially expressed genes and downstream pathways possibly under NtGAMYB control. RNA was isolated from young leaves of 8-week-old wild-type and MIM159-transgenic plants. For each sample, three biological replicates were prepared: three different wild-type plants and plants from three independent transgenic MIM159 lines. The six isolated RNA samples were used to construct complementary DNA (cDNA) libraries on which Illumina deep sequencing (Novogene) was performed. Sequencing reads were aligned to the reference tobacco TN90 genome (Sierro et al., 2014). The alignments were analyzed by Cuffdiff software (cufflinks version 2.2.1) and sequence data were statistically analyzed to detect genes that were differentially expressed between MIM159 and wild-type tobacco. Using a 2-fold change cut-off, 12,418 genes were upregulated and 9,431 genes were downregulated in MIM159 compared to the wild type (Supplemental Dataset S1). This suggests that the inhibition of miR159 function in tobacco facilitates global changes to the transcriptome.
NtGAMYB1–NtGAMYB3 (gene_47587, gene_56181, and gene_42641, respectively) were each found to be upregulated in MIM159 tobacco leaves by levels that are consistent with the RT-qPCR data (∼3-, 9-, and 6-fold, respectively). To gain insight into which biological pathways were altered in the MIM159 plants, Gene Ontology (GO) analysis of the differentially expressed genes was performed. As the tobacco genome has not been completely annotated, the GO enrichment analysis was determined by using the closest homologs from Arabidopsis. Given that so many differentially expressed genes were detected at a 2-fold level, the GO enrichment analysis was restricted to include only those genes differentially expressed at a >5-fold level, reducing the analysis to 1,478 upregulated genes and 2,345 downregulated genes in MIM159.
Of the top 20 enriched pathways ranked by P-value significance represented by upregulated genes, three general themes emerged. First, “defense response” was the highest-enriched category alongside another four defense-related categories that were also among the top 20 enriched pathways, including “defense response to fungus” and “bacterium” (Table 1). The second highest category was “programmed cell death” (PCD), which was also accompanied by a further four related categories in the top 20 enriched pathways (Table 1). This high representation is consistent with previous reports demonstrating that GAMYB promotes PCD in the seed aleurone and the anther tapetum (Aya et al., 2009; Alonso-Peral et al., 2010). Finally, six of the top 20 enriched pathways were related to responses to hormone and environmental stimulus (Table 1).
Table 1. Top 20 enriched pathways of upregulated genes in MIM159 tobacco.
Analysis was based on the 1,478 genes that were upregulated in MIM159 tobacco leaves >5-fold compared to the wild type. These genes were referred to by the Arabidopsis counterpart sharing the highest sequence similarity, and the GO enrichment analysis was performed by agriGO (Du et al., 2010). The highest 20 enriched pathways were ranked by P-value. A pathway with a P-value ≤0.05 is considered significantly enriched.
| Pathway | GO Term | P-Value |
|---|---|---|
| Defense response | GO:0006952 | 1.20E−15 |
| PCD | GO:0012501 | 1.30E−10 |
| Phosphate metabolic process | GO:0006796 | 3.80E−10 |
| Response to carbohydrate stimulus | GO:0009743 | 7.60E−09 |
| Response to hormone stimulus | GO:0009725 | 2.10E−08 |
| Response to oxidative stress | GO:0006979 | 6.50E−08 |
| Apoptosis | GO:0006915 | 6.70E−08 |
| Response to chitin | GO:0010200 | 1.50E−07 |
| Defense response to fungus | GO:0050832 | 2.80E−07 |
| Immune system process | GO:0002376 | 1.40E−06 |
| Defense response, incompatible interaction | GO:0009814 | 6.80E−06 |
| Response to abscisic acid stimulus | GO:0009737 | 1.80E−05 |
| Aging | GO:0007568 | 3.90E−05 |
| Response to temperature stimulus | GO:0009266 | 5.80E−05 |
| Ion transport | GO:0006811 | 1.10E−04 |
| Cellular nitrogen compound metabolic process | GO:0034641 | 1.10E−04 |
| Senescence | GO:0010149 | 1.20E−04 |
| Host programmed cell death induced by symbiont | GO:0034050 | 1.30E−04 |
| Cellular amino acid derivative biosynthetic process | GO:0042398 | 1.30E−04 |
| Response to bacterium | GO:0009617 | 2.40E−04 |
For the 2,345 genes that were downregulated in MIM159 tobacco leaves, the top 20 enriched pathways identified from GO analysis included several that were associated with cell cycle, cell size, and morphogenesis (Table 2). The top-ranking pathway, namely “microtubule-based processes”, is also potentially related to cell division and development (Hamada, 2014). Downregulation of such pathways may underlie the stunted growth and leaf phenotypes of MIM159 tobacco plants.
Table 2. Top 20 enriched pathways of downregulated genes in MIM159 tobacco.
Analysis was based on the 2345 genes that were downregulated in MIM159 tobacco leaves more than 5-fold compared to wild type. These genes were referred to by the Arabidopsis counterpart sharing the highest level of sequence similarity, and the GO enrichment analysis was performed by agriGO (Du et al., 2010). The highest 20 enriched pathways were ranked by P-value. A pathway with a P-value ≤ 0.05 is considered as significantly enriched.
| Pathway | GO Term | P-Value | |
|---|---|---|---|
| Microtubule-based process | GO:0007017 | 6.60E−13 | |
| Cell cycle | GO:0007049 | 7.70E−12 | |
| Lipid localization | GO:0010876 | 4.90E−11 | |
| Postembryonic development | GO:0009791 | 1.50E−10 | |
| Transmembrane receptor protein Tyr kinase signaling pathway | GO:0007169 | 2.00E−10 | |
| Cell surface receptor linked signaling pathway | GO:0007166 | 2.80E−09 | |
| Cellular developmental process | GO:0048869 | 7.30E−09 | |
| Postembryonic morphogenesis | GO:0009886 | 1.80E−08 | |
| Cellular component morphogenesis | GO:0032989 | 9.50E−08 | |
| Tissue development | GO:0009888 | 2.10E−07 | |
| Cell morphogenesis | GO:0000902 | 2.40E−07 | |
| Response to radiation | GO:0009314 | 4.00E−07 | |
| Regulation of cell size | GO:0008361 | 7.40E−07 | |
| Regulation of cell cycle | GO:0051726 | 7.90E−07 | |
| Cell wall polysaccharide metabolic process | GO:0010383 | 8.30E−07 | |
| Regulation of cellular component size | GO:0032535 | 9.10E−07 | |
| Cellular component organization | GO:0016043 | 9.50E−07 | |
| Lipid metabolic process | GO:0006629 | 1.60E−06 | |
| Unidimensional cell growth | GO:0009826 | 1.70E−06 | |
| Developmental growth involved in morphogenesis | GO:0060560 | 1.70E−06 | |
Many Classes of Defense Genes Are Strongly Upregulated in MIM159 Tobacco
Next, the 50 most upregulated genes were identified using a BLAST search against the tobacco transcriptome (Supplemental Table S1). Of these 50 genes, 22 encode PATHOGENESIS-RELATED PROTEINS (PR; Table 3), which are involved in plant defense against pathogens. The mRNA levels of these PR genes were strongly upregulated in MIM159 tobacco leaves, specifically 360- to 2020-fold higher than in the wild type (Table 3). These genes included homologs from seven PR subfamilies, such as the PR-2 (seven genes) and the PR-Q'-like (two genes) subfamilies, which encode β-1,3-glucanases, and the PR-Q (two genes) and PR-R (two genes) major form and endochitinase subfamilies, which all encode chitinases. Also upregulated were PR5 subfamily members (two genes) that encode osmotin, a homolog of thaumatin that has an antifungal function by breaking fungal membranes, leading to osmotic rupture (Hakim et al., 2018).
Table 3. PR genes display strongly upregulated expression in MIM159 tobacco leaves.
A list of the PR genes identified in the top 50 upregulated genes in MIM159 tobacco is shown. The tobacco gene identifier (ID) is provided alongside fold-level expression upregulation in MIM159 relative to the wild type. The PR genes are grouped in their seven PR subfamilies. WT, wild type.
| Gene ID | Fold Change (MIM159/WT) | Description (Protein Family) | Function |
|---|---|---|---|
| Gene_56557 | 2,020 | PR-1a | Antifungal |
| Gene_12559 | 1,451 | PR-1a | |
| Gene_61952 | 500 | PR-1b | |
| Gene_40024 | 2,143 | PR-1c | |
| Gene_14265 | 1,640 | PR-1c | |
| Gene_5026 | 1,937 | PR-2, β-1,3-glucanase | Cleaves β-1,3-glucans |
| Gene_39617 | 1,262 | PR-2, β-1,3-glucanase | |
| Gene_59805 | 1,069 | PR-2, β-1,3-glucanase | |
| Gene_66775 | 811 | PR-2, β-1,3-glucanase | |
| Gene_59815 | 610 | PR-2, β-1,3-glucanase | |
| Gene_15771 | 570 | PR-2, β-1,3-glucanase | |
| Gene_59819 | 392 | PR-2, β-1,3-glucanase | |
| Gene_26030 | 546 | PR-Q'-like, β-1,3-glucanase | Cleaves β-1,3-glucans |
| Gene_1355 | 516 | PR-Q'-like, β-1,3-glucanase | |
| Gene_8988 | 578 | PR-Q, acidic chitinase | Endochitinase |
| Gene_46441 | 400 | PR-Q, acidic chitinase | |
| Gene_32773 | 369 | Acidic endochitinase | Endochitinase |
| Gene_4867 | 360 | Acidic endochitinase | |
| Gene_16878 | 731 | PR-R major form, chitinase | Antifungal and chitinase |
| Gene_27076 | 403 | PR-R major form, chitinase | |
| Gene_59471 | 1,056 | PR5, osmotin | Respond to osmotic stress, antifungal |
| Gene_58140 | 493 | PR5, osmotin |
Other genes associated with defense response were also strongly upregulated in MIM159 tobacco. PHOTOASSIMILATE-RESPONSIVE PROTEIN 1c (PAR-1c) homologs (gene_46853 and gene_13483) were ∼500-fold upregulated in MIM159 tobacco leaves (Supplemental Table S1). PAR transcription is induced by viruses and its expression is coordinated with PR genes (Herbers et al., 1995). Likewise, homologs of SAR8.2 (gene_60737 and gene_69711), which are induced by salicylic acid in tobacco and associated with systemic acquired resistance (SAR), were ∼500-fold upregulated in MIM159 tobacco (Supplemental Table S1). SAR8.2 genes are involved in defense response against fungal, bacterial, and viral diseases (Alexander et al., 1992; Song and Goodman, 2002).
Finally, there were 27 predicted NUCLEOTIDE-BINDING SITE AND LEU-REPEAT (NBS-LRR) genes that were upregulated 5- to 350-fold in MIM159 tobacco leaves (Supplemental Table S2). NBS-LRR genes are upstream elicitors of the hypersensitive response (HR) of plant immunity (Coll et al., 2011; Spoel and Dong, 2012; Weiberg et al., 2014). Many of these NBS-LRR genes are predicted to encode resistance proteins belonging to two major classes: Tobacco Mosaic Virus (TMV) resistance N-like proteins and the late blight resistance protein homolog R1, the latter of which confers resistance to late blight disease resulting from Phytophthora infestans infection (Ballvora et al., 2002; Fry, 2008).
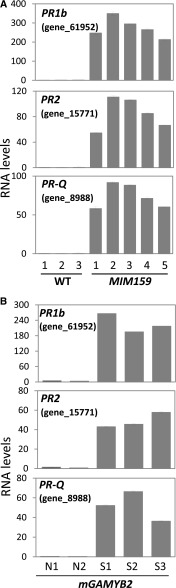
To validate the upregulation of the PR genes in MIM159 tobacco, RT-qPCR analysis was performed on leaves of multiple MIM159 lines and wild-type plants. Compared to the wild type, PR-1b, PR-2, and PR-Q transcript levels were strongly increased in all MIM159 transgenic lines tested (Fig. 5A). Likewise, PR-1b, PR-2, and PR-Q transcript levels were strongly increased in mGAMYB2-S lines (Fig. 5B). Therefore, upregulation of GAMYB expression, through either inhibition of miR159 or expression of a miR159-resistant GAMYB2 transgene, leads to strong upregulation of PR expression in tobacco leaves. Whether these PR genes are directly downstream of GAMYB or their overexpression is an indirect result of reduced fitness of these transgenic lines remains unknown. However, GAMYB expression by itself is insufficient to induce high PR mRNA levels, since although wild-type tobacco anthers have relatively high GAMYB mRNA levels, the mRNA levels of the PR genes were observed to be orders of magnitude lower than in leaves of MIM159 plants (Supplemental Fig. S5). This suggests that other specific factors in combination with GAMYB expression are required for PR induction in MIM159 leaves.
Figure 5.
PR transcript levels are upregulated in MIM159 and mNtGAMYB2 tobacco. A, RT-qPCR measurement of PR gene mRNA levels in the wild type (WT; three lines) and MIM159 (five lines), using leaves of 8-week-old plants. B, The same levels measured by RT-qPCR in mGAMYB2-N (N1 and N2) and mGAMYB2-S (S1–S3), using leaves of 12-week-old plants. RNA levels were normalized to PROTEIN PHOSPHATASE2A, and measurements are the average of three technical replicates.
MIM159 Tobacco Has Increased Resistance to Phytophthora parasitica Infection
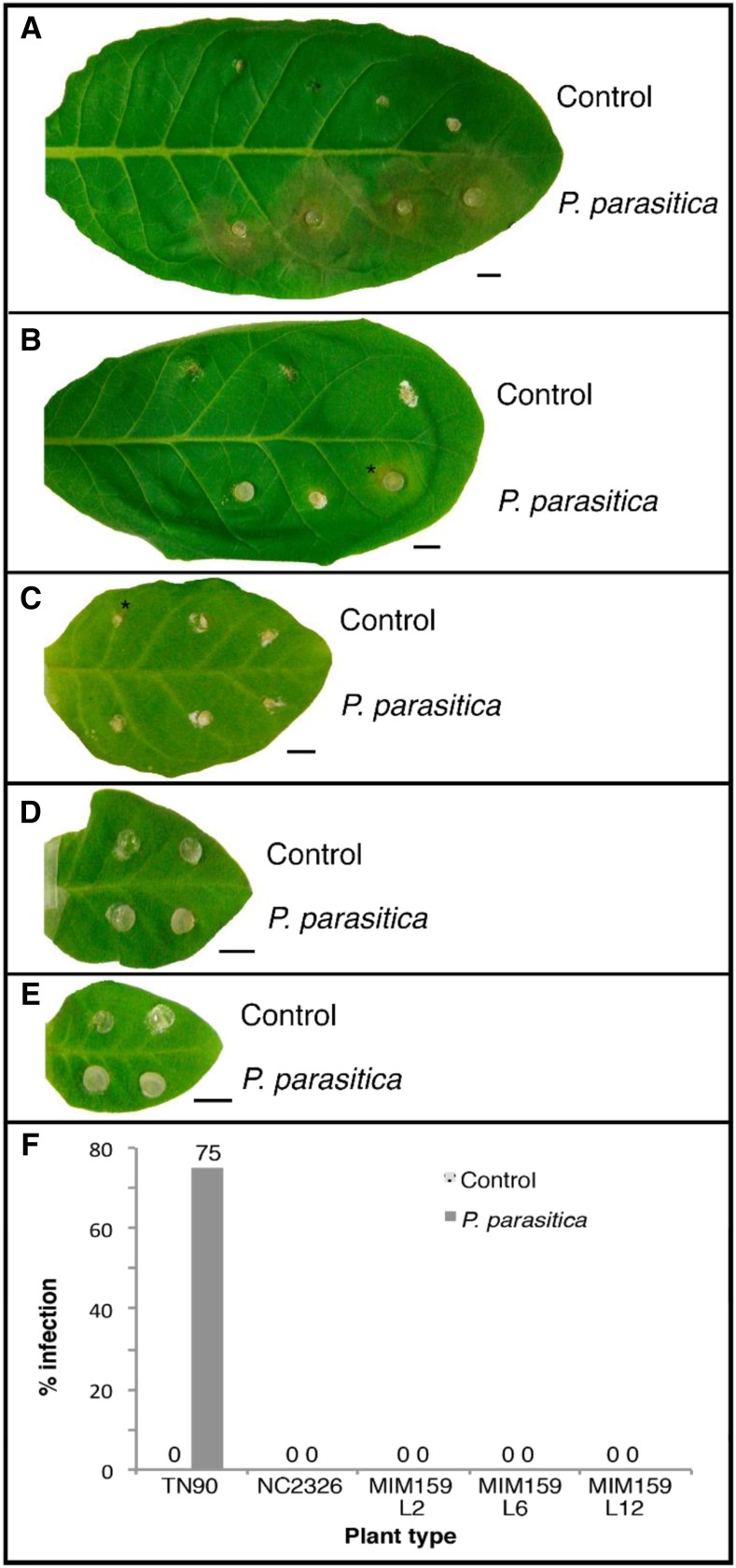
To determine the impact of expression upregulation of those genes associated with disease resistance, the susceptibility of MIM159-transgenic plants to the oomycete pathogen P. parasitica was tested in three independent lines (Lines 2, 6, and 12). Infection assays on multiple detached tobacco leaves showed that wild-type tobacco plants were highly susceptible to P. parasitica, with the infection visible at 3–4 d postinoculation by clear necrosis occurring around the infection site (Fig. 6A). Of note, in a few cases, P. parasitica infection did not occur at some wild-type inoculation sites. Leaves from the resistant NC2326 tobacco cultivar generally showed no sign of infection (Fig. 6B). In all experiments, including preliminary assays (data not shown), the three tobacco MIM159 lines did not display signs of P. parasitica infection (Fig. 6, C–E). On some leaves, a small amount of cell death around the agar plug was visible, but as there was no difference observed between the mock and P. parasitica inoculated plugs, this was presumed to be a response to wounding (Fig. 6, B and C). Thus, the MIM159 lines were strongly resistant to P. parasitica infection compared to the wild type (Fig. 6F).
Figure 6.
P. parasitica infection of susceptible and resistant tobacco cultivars alongside transformed MIM159 lines. A to E, Leaves 3 d postinoculation with agar (Control) on one side and P. parasitica hyphae on the other are shown for susceptible tobacco NT90 (A), resistant tobacco NC2326 (B), MIM159 transgenic line L2 (C), MIM159 transgenic line L6 (D), and MIM159 transgenic line L12 (E). Scale bars = 10 mm. Asterisks in A–C indicate cell death associated with wounding or HR. F, Percentage of infection in leaves inoculated with P. parasitica or control V8 agar (n = 12–15).
Investigating the miR159-GAMYB Pathway in Rice
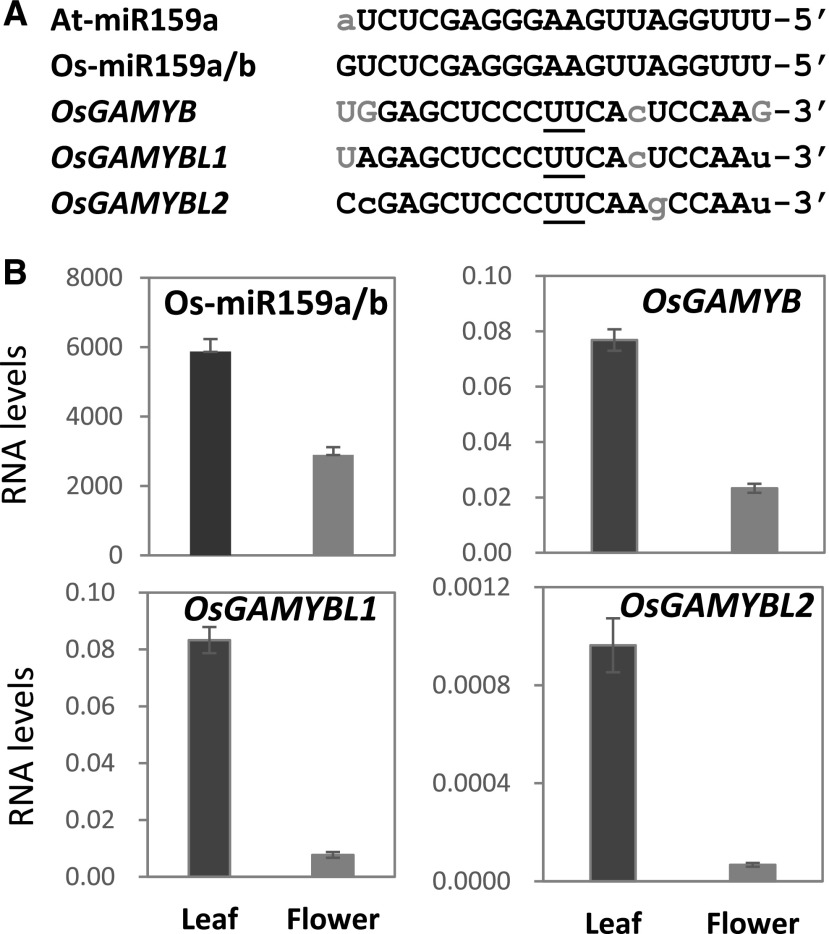
We extended the study of miR159 function to a more divergent plant species, namely monocot rice. Based on miRBase (Kozomara and Griffiths-Jones, 2014), rice Os-miR159a and Os-miR159b have identical mature sequences which correspond to the most abundant isoform in rice, and differ from Arabidopsis At-miR159a at only one nucleotide (Fig. 7A). In rice, one GAMYB and two GAMYB-like genes have been identified, all of which have a conserved miR159 binding site that is highly complementary to Os-miR159a/b (Fig. 7A; Tsuji et al., 2006).
Figure 7.
The miR159-GAMYB pathway in rice. A, Sequence alignments of Arabidopsis and rice miR159 and the miR159-binding site in OsGAMYB homologs. Underlined nucleotides represent the cleavage site, lowercase gray letters represent mismatches to Os-miR159a/b, and G:U pairings are shown in uppercase gray letters. B, RT-qPCR measurement of miR159 and GAMYB transcript levels in rice. RNA was extracted from pools of O. sativa plants, with tissue analyzed from three flag leaves (Leaf) or 30 florets (Flower). The mRNA levels of the OsGAMYB genes were normalized to ACTIN and the miR159 RNA levels were normalized to snoR14. Measurements are the average of three biological replicates, with error bars representing the se.
RT-qPCR analysis revealed that Os-miR159 is expressed in both leaves and flowers of rice (Fig. 7B), consistent with a previous report (Tsuji et al., 2006). For the OsGAMYB homologs, primers were designed to amplify their 3′-UTR regions to ensure gene-specific amplicons. These primers do not span the miR159 cleavage site so were able to quantify both cleaved and uncleaved OsGAMYB transcripts. The transcript levels of all three OsGAMYB homologs were substantially higher in leaves than in flowers (Fig. 7B). This result differs from a previous report that, using OsGAMYB promoter-GUS reporter genes, found that OsGAMYB and OsGAMYBL1 were not transcribed in vegetative tissues (Tsuji et al., 2006). To verify that the OsGAMYB homologs are regulated by miR159, 5′-RACE miRNA cleavage assays were performed on RNA from rice leaves and flowers. These assays found miR159-guided cleavage products for OsGAMYB and OsGAMYBL1, but not for OsGAMYBL2 (Supplemental Fig. S6), indicating that OsGAMYB and OsGAMYBL1 are cleaved by miR159 in both rice leaves and flowers. In conclusion, similar to Arabidopsis and tobacco, the miR159-GAMYB pathway appears active in both vegetative and floral tissues in rice.
The Functional Role of miR159 in Rice
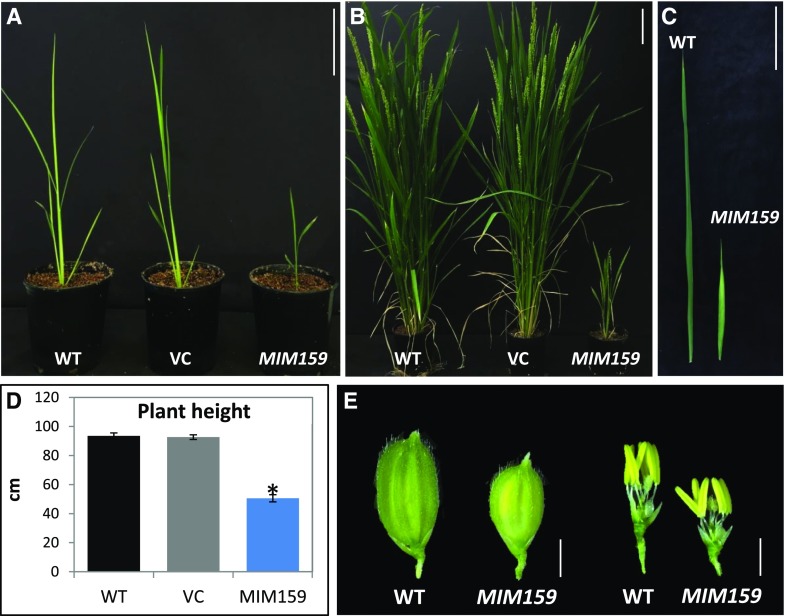
To investigate the functional role of miR159 in rice, a MIM159 construct was placed under control of the maize ubiquitin promoter and transformed into Oryza sativa Japonica. A corresponding empty vector control (VC) was also transformed into rice. Of 24 independent T0 MIM159 transgenic rice plants, more than half displayed a smaller growth stature compared to the wild type and VC plants. To determine whether this stunted growth phenotype was heritable, T1 progeny from multiple independent lines were analyzed. Compared to the wild type, the MIM159 lines displayed pleiotropic phenotypic defects throughout development. These included a smaller growth stature (Fig. 8, A–D), with shorter flag leaves (Fig. 8C) and smaller florets (Fig. 8E). These stunted growth phenotypes of MIM159 rice are analogous to the growth defects of MIM159 Arabidopsis and tobacco plants.
Figure 8.
Phenotyping of MIM159 rice. A, Three-week-old wild-type (WT), VC, and MIM159-transgenic rice. Scale bar = 10 cm. B, Eleven-week-old MIM159 transgenic plants displayed smaller growth stature compared to wild-type and VC plants. Scale bar = 10 cm. C, Flag leaves of wild-type and MIM159 plants. Scale bar = 10 cm. D, Average plant heights of wild-type plants, VC T1 plants, and MIM159 T1 plants (n = 10, 10, and 20, respectively). The VC and MIM159 T1 plants analyzed were derived from multiple independent transgenic lines. Error bars represent the se and the asterisk indicates a statistically significant difference compared to the wild type as determined by Student’s t test. E, Floret, anther, and pistil phenotypes of wild type and MIM159 transgenic plants. Scale bars = 2 mm. The different plant parts were digitally extracted for comparison.
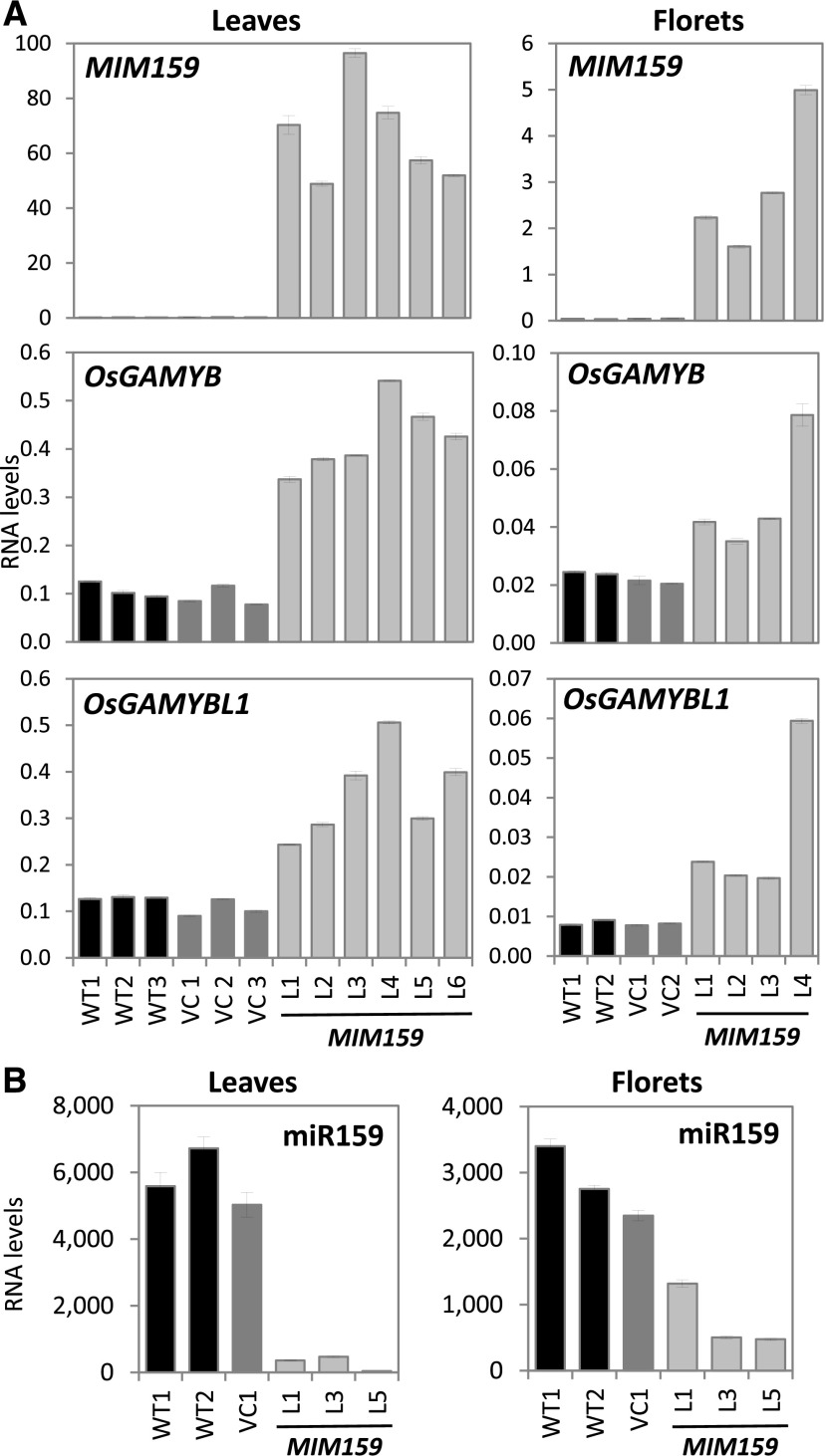
Transcript profiling of the MIM159 transgene, miR159, and the GAMYB homologs was performed on wild-type, VC, and MIM159 plants. Expression of MIM159 resulted in a repression of miR159 in both leaves and florets of MIM159 lines (Fig. 9). Correspondingly, OsGAMYB and OsGAMYBL1 transcript levels were higher in both leaves and florets of MIM159 lines (Fig. 9A), implying that GAMYB and GAMYBL1 are deregulated in MIM159 lines. Therefore, similar to Arabidopsis and tobacco, inhibition of miR159 in rice vegetative tissues results in deregulated GAMYB expression, a phenomenon that appears conserved across the monocot and dicot divide.
Figure 9.
Transcript profiling of MIM159 transgenic rice. A, RT-qPCR measurement of MIM159, OsGAMYB, and OsGAMYBL1 expression. RNA was extracted from wild-type (WT, 2–3 lines), VC (2–3 lines) and MIM159 (4–6 transgenic lines) lines, with analyzed samples comprising three flag leaves (Leaves) or 30 florets (Flowers) and RNA levels normalized to ACTIN. B, MiR159 levels in a subset of the lines in A. RNA levels were normalized to snoR14. Measurements are the average of three technical replicates.
PR Transcript Levels Were Not Upregulated in miR159 Loss-of-Function Arabidopsis and Rice
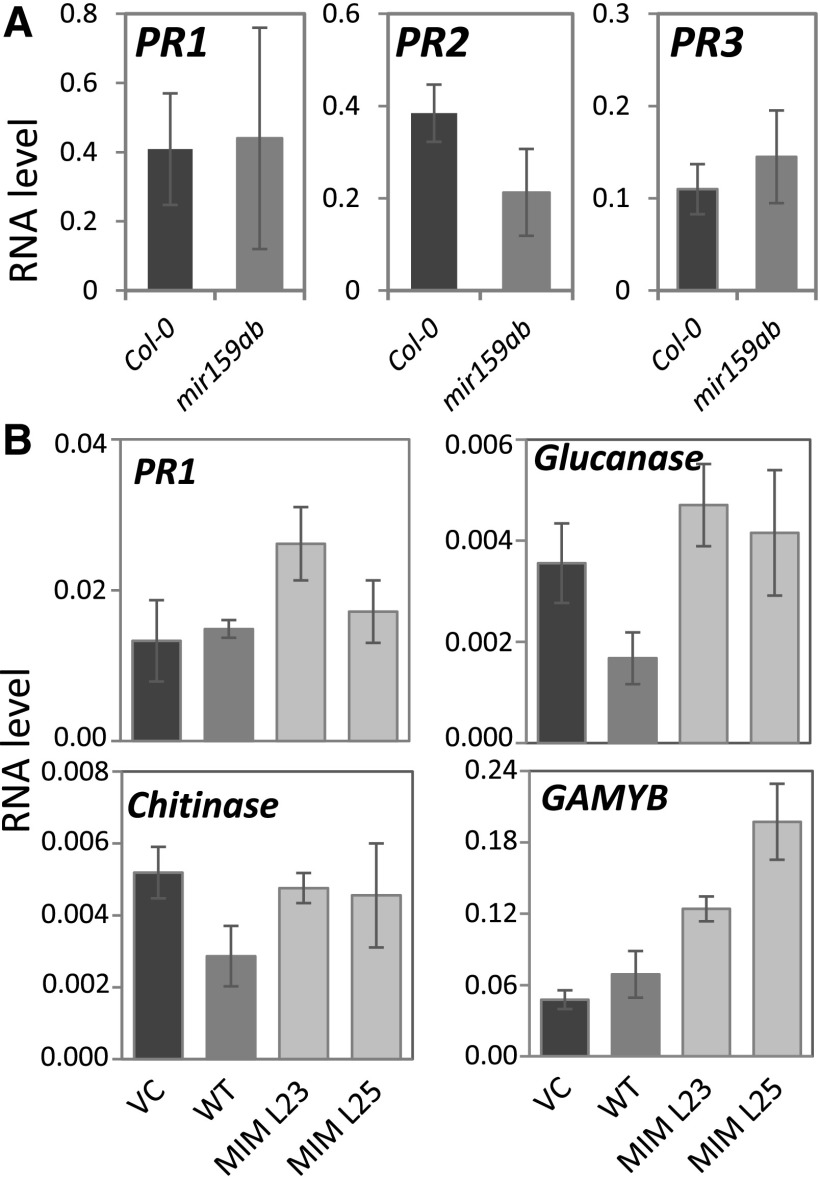
As PR mRNA levels were dramatically upregulated in MIM159 tobacco leaves, it was investigated whether PR mRNA levels are also upregulated in the Arabidopsis mir159ab mutant and in MIM159 rice. However, RT-qPCR analysis found that the corresponding PR homologs in Arabidopsis and rice were not upregulated in these miR159 loss-of-function plants (Fig. 10). This suggests that upregulation of the PR genes due to inhibition of miR159 is not conserved.
Figure 10.
Measurement of PR mRNA levels in Arabidopsis and rice. A, RT-qPCR measurement of PR1 (At2g14610), PR2 (At3g57260), and PR3 (At3g12500) in wild-type Arabidopsis (Col-0) and the mir159ab mutant. RNA levels were normalized to CYCLOPHILIN. B, RT-qPCR measurement of PR homologs (PR1a [EF061246], endo-1,3-β-glucosidase [AK318559], and chitinase [AB016497]) and GAMYB in VC, wild-type (WT), and two independent MIM159 rice lines (MIM L23 and MIM L25). Measurements were normalized to ACTIN. Measurements are the average of three biological replicates, with error bars representing the se.
GAMYB Proteins from Diverse Plant Species Activate Similar Pathways When Expressed in Arabidopsis
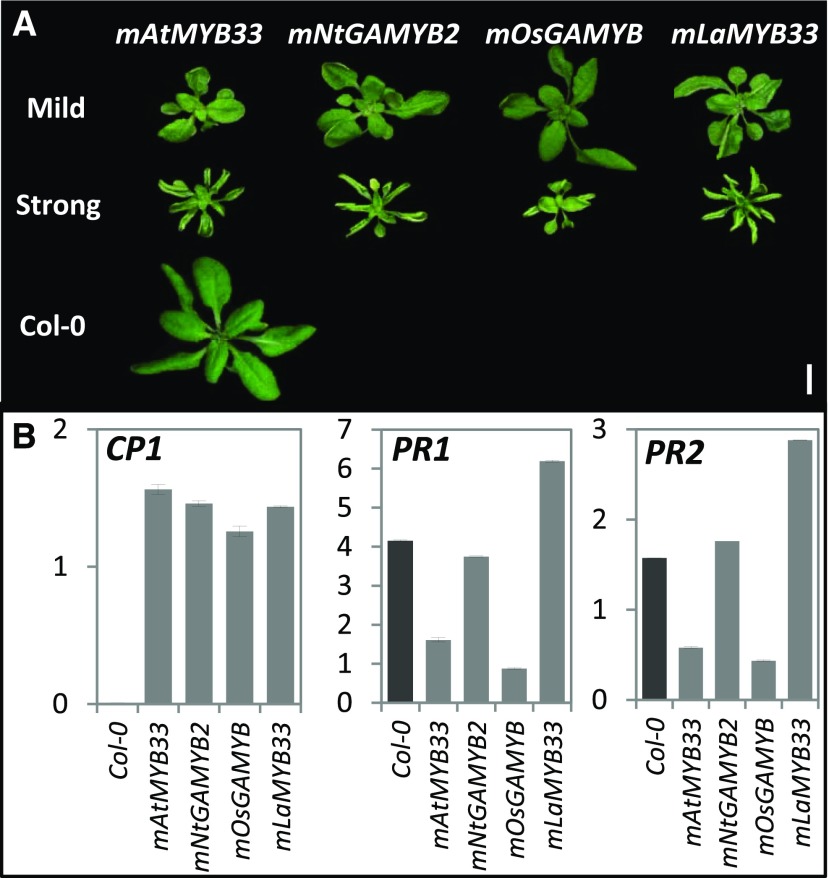
The differential regulation of PR genes between tobacco and Arabidopsis/rice with respect to GAMYB expression raises the question of whether the tobacco GAMYB protein has a divergent function compared to the Arabidopsis and rice GAMYB proteins. To investigate this, homologs of GAMYB (or GAMYB-like) proteins from representative plant species were aligned to analyze amino acid sequence identity. The proteins were found to be strongly conserved within the R2R3 region, but highly divergent in all other regions, especially within the C-terminal portion (Supplemental Fig. S7). This raises the possibility that the divergent protein sequences in GAMYB homologs could activate different downstream pathways. To investigate this, the coding regions of GAMYB-like genes from tobacco, rice, and Japanese larch (Larix kaempferi; Li et al., 2013a), namely mNtGAMYB2, mOsGAMYB, and mLaMYB33, respectively, were constitutively expressed in Arabidopsis via the 35S promoter. As these three GAMYB-like genes all contained a highly conserved miR159 binding site (Figs. 1A and 7A), synonymous mutations were introduced to generate miR159-resistant mGAMYB transgenes. These 35S-mGAMYB transgenes were individually transformed into the Arabidopsis ecotype Columbia (Col-0) wild-type plants and multiple primary transformants were generated and analyzed for each construct.
Expression of mNtGAMYB2, mOsGAMYB, and mLaMYB33 in Arabidopsis led to similar developmental rosette defects of stunted growth with mild to strong upward leaf curl, similar to mAtMYB33 transformants (Fig. 11A). The mRNA level of a downstream marker gene of Arabidopsis GAMYB protein activity, CYS PROTEINASE1 (CP1; Alonso-Peral et al., 2010), was found to be upregulated in mNtGAMYB2, mOsGAMYB, and mLaMYB33 transformants, at levels similar to those observed for the mAtMYB33 transformants (Fig. 11B). This suggests that GAMYB proteins from diverse plant species can activate similar downstream genes when expressed in Arabidopsis. However, PR mRNA levels in all of the mGAMYB lines, even the mNtGAMYB2 Arabidopsis transformants, remained low, (Fig. 11B). Therefore, it appears that NtGAMYB only upregulates PR expression in tobacco.
Figure 11.
GAMYB proteins from diverse plant species activate similar pathways when expressed in Arabidopsis. A, Three-week-old Arabidopsis transformants constitutively expressing mAtMYB33 (Arabidopsis), mNtGAMYB2 (Nicotiana tabacum), mOsGAMYB (Oryza sativa), and mLaMYB33 (Larix kaempferi) all display similar phenotypes. Scale bar = 1 cm. The rosettes were digitally extracted for comparison. B, Transcript levels of the CYSTEINIE PROTEINASE1 (CP1), PR1, and PR2 in 35S-mGAMYB primary transformants measured by RT-qPCR. RNA was extracted from rosette tissues of 15 randomly selected, 3-week-old primary transformants. Col-0 was used as the wild-type control and mRNA levels were normalized to CYCLOPHILIN. Measurements are the averages of three technical replicates.
DISCUSSION
The Deleterious Impact of GAMYB Expression in Vegetative Tissues Is Conserved
Although the miR159-GAMYB regulatory relationship is ancient, the functional role for this pathway during vegetative growth and development has remained elusive. Its role has now been investigated in distantly related flowering plant species, including Arabidopsis (Allen et al., 2007; Alonso-Peral et al., 2010), rice (Figs. 7–10; Zhao et al., 2017), and tobacco (Figs. 1–5). In all species, miR159 is required to strongly silence GAMYB expression in vegetative tissues; inhibition of miR159 in tobacco, rice, or Arabidopsis results in the universal outcome of deregulated GAMYB expression and strong deleterious impacts on growth and development, leading to dwarf stature. Consistent with this, in MIM159 tobacco, genes related to cell cycle, cell size, and cell morphogenesis are significantly downregulated (Table 2). STTM159 rice has decreased cell layers in parenchymatous tissue in stems and reduced numbers of small veins in the flag leaf (Zhao et al., 2017). Likewise, rosette leaves of mir159ab Arabidopsis have decreased cell number, altered cell morphology, and simpler venation (Alonso-Peral et al., 2010). As these phenotypes also occur via expression of miR159-resistant GAMYB transgenes in tobacco and Arabidopsis (Figs. 4 and 11; Millar and Gubler, 2005; Zheng et al., 2017), these phenotypes can be attributed to deregulated GAMYB activity. Currently, the only reported exception to this is in Gloxinia, where inhibition of miR159 function does not result in deleterious impacts, but rather in accelerated flowering (Li et al., 2013b). Given the conserved impact of GAMYB activity found here and described widely, it is difficult to reconcile these differences.
A Conserved miR159-GAMYB Regulatory Futile Cycle in Vegetative Tissues?
A recurrent theme in the plants examined is that the miR159-GAMYB pathway is present in vegetative tissues, but widespread GAMYB transcription is strongly silenced by miR159, resulting in no or little net phenotypic impact on the plant.
First, miR159 appears abundant in vegetative tissues, with expression levels higher in leaves than in flowers in both tobacco and rice. Similarly, all three isoforms of GAMYB mRNA are readily detectable in vegetative tissues in both species (Figs. 1 and 7). Indeed, in rice, the mRNA levels of the GAMYB homologs are much higher in vegetative tissues than in florets. However, for at least OsGAMYB, this mRNA is likely to be strongly silenced, as an osgamyb (gamyb-1) mutant does not display any obvious phenotypic differences from wild-type plants at the vegetative stage (Kaneko et al., 2004). It appears that only when miR159 is inhibited, as is the case in our MIM159 rice and in the previously reported STTM159 rice, can these GAMYB homologs be expressed, resulting in strong pleiotropic developmental defects (Fig. 8; Zhang et al., 2017; Zhao et al., 2017).
Similarly, constitutive transcription of GAMYB in tobacco via a 35S-GAMYB2 transgene resulted in no obvious phenotypic defects, which was in stark contrast to expression of a 35S-mGAMYB2 transgene that resulted in severe growth and developmental phenotypes (Fig. 4). This contrast supports that the 35S-GAMYB2 transgene is being strongly and constitutively silenced by miR159. Consistent with this notion, the NtGAMYB and OsGAMYB homologs contain the conserved nucleotides that are predicted to form the RNA secondary structure adjacent to the miR159 binding site that has been shown to promote silencing of the GAMYB-like homolog, MYB33, in vegetative tissues of Arabidopsis (Zheng et al., 2017). Based on all these data, it appears that miR159 strongly silences GAMYB mRNA in vegetative tissues to phenotypically insignificant levels.
Therefore, similar to Arabidopsis, the miR159-GAMYB pathway appears active in vegetative tissues of rice and tobacco, but only when miR159 is inhibited can the transcribed GAMYB mRNA be expressed, resulting in a phenotypic impact. This raises the question of what is the selective pressure to conserve GAMYB transcription in vegetative tissues of these plant species, since it is usually strongly silenced by miR159. Without a clear rationale, the miR159-GAMYB pathway appears to be a regulatory futile cycle, where production of both miR159 and GAMYB mRNA results in no clear phenotypic outcome. This conundrum is extenuated by the fact that expression of GAMYB protein in the vegetative parts of the plant is deleterious.
Hypothesis: A Fundamental Role for miR159-GAMYB in Defense Response
Our work on the tobacco miR159-GAMYB pathway may have given an insight into the potential evolutionary role for this pathway in vegetative tissues. In MIM159 tobacco, a broad suite of defense genes, including PR, PAR-1, Sar-8.2, NBS-LRR, and METACASPASE classes, were strongly upregulated. This molecular response can confer a strong resistance to plant pathogens, as MIM159 plants appeared immune to the pathogen P. parasitica. Such resistance is consistent with the upregulation of genes such as PR1a and Sar-8.2, which have previously been shown to increase resistance to P. parasitica (Alexander et al., 1993; Song and Goodman, 2002). The fact that such a broad defense response is triggered upon miR159 inhibition argues that it may play a fundamental role in plant defense. Although PR gene expression was elevated and leaf chlorosis was observed in both MIM159 and mNtGAMYB2 tobacco plants, it is unknown whether GAMYB is directly inducing these phenotypes. We cannot rule out the possibility that the reduced growth of MIM159 and mNtGAMYB tobacco induces leaf chlorosis, which in turn results in the upregulation of the PR genes. Further experiments will be needed to address this issue.
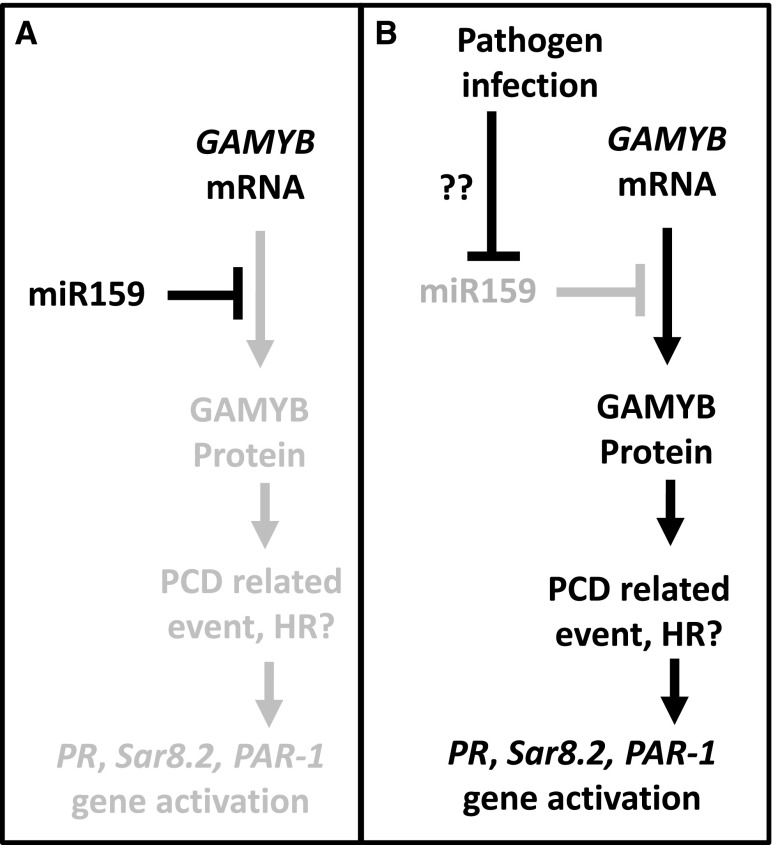
We hypothesize that GAMYB mRNA is being consistently transcribed throughout vegetative tissues, but is under constant repression by miR159 (Fig. 12). Upon pathogen infection, inhibition of miR159 occurs that enables GAMYB activity, triggering the defense response and PCD to limit pathogen infection. Such a scenario would explain the selective pressure for GAMYB transcription throughout vegetative tissues, despite its potential deleterious impacts. This scenario may explain why conserved RNA secondary structures associated with the miR159 binding sites of GAMYB genes have arisen, namely to ensure strong GAMYB silencing. Moreover, this scenario is consistent with the highly conserved nature of the miR159-GAMYB pathway, which appears ubiquitously present in terrestrial plants.
Figure 12.
Hypothesized role of the miR159-GAMYB pathway in tobacco. A, GAMYB mRNA in vegetative tissues. The presence of miR159 strongly repressed GAMYB expression, resulting in no obvious phenotypic impact. B, Upon pathogen infection, miR159 is repressed (via an unknown mechanism, potentially silencing suppressors), enabling the GAMYB mRNA to be expressed. The transcription factor activates defense response pathways, including the HR, causing cell death to limit pathogen infection.
However, some of the results reported here do not support the above hypothesis. Whereas the miR159-GAMYB systems in rice and Arabidopsis share many features with that of tobacco, such as the strong and characteristic phenotypic defects of MIM159 rice and mir159ab Arabidopsis, defense gene expression does not appear to be triggered in these species by GAMYB activity. This was shown here for several of the PR genes, and more broadly by transcriptome profiling experiments for STTM159 rice (Zhao et al., 2017) and mir159ab Arabidopsis (Alonso-Peral et al., 2010). Additionally, PR mRNA levels were not upregulated when tobacco GAMYB was expressed in Arabidopsis (Fig. 11). Therefore, GAMYB appears to upregulate PR mRNA levels only when expressed in tobacco. It may be that tobacco has additional factors that enable GAMYB activity to trigger this response, or that additional signals are required for the defense molecular response to be triggered in rice and Arabidopsis. Thus, despite the conserved nature of the miR159-GAMYB pathway, there are clear significant species-specific differences, for which considerable work will be required to understand their underlying complexity. However, it is worth investigating whether the miR159-inhibited or GAMYB-upregulated Arabidopsis and rice lines have increased induction of PR genes upon pathogen infection, as it is possible that the miR159-GAMYB pathway may function specifically during pathogen infections in these plant species.
Presently, the best characterized function for GAMYB is in the seed (aleurone) and anther (tapetum), where the common role is to promote PCD. Here, we provide evidence that GAMYB activity may also influence PCD-related events in leaves. MIM159 tobacco displays leaf chlorosis, and the MIM159-upregulated genes NBS-LRR and METACASPASE-1 are known to positively regulate HR, which is a form of PCD at the site of pathogen invasion that includes chloroplast disruption (Coll et al., 2011; Mur et al., 2008; Teh and Hofius, 2014). Given this, it is tempting to speculate that the MIM159 leaf chlorosis may be a consequence of constitutive HR. Such a phenotype, in addition to stunted growth and high levels of PR expression without pathogen triggers, is reminiscent of phenotypes of autoimmune mutants (Gou and Hua, 2012; Spoel and Dong, 2012). However, the fact that MIM159 rice and mir159ab Arabidopsis also display stunted growth without the molecular defense response argues against the involvement of the autoimmune response in the stunted phenotype.
Currently, there is no clear evidence linking miR159 or GAMYB expression changes to biotic stress in any plant species. However, inhibition of GAMYB expression by miR159 involves a translational repression component (Li et al., 2014b). Therefore, via regulation of miR159 function, biotic stress may induce changes in GAMYB protein levels in the absence of strong changes in mRNA abundance. For instance, many pathogens, including Phytophthora (Qiao et al., 2015), encode silencing suppressors that may inhibit miR159 function but may not dramatically affect the level of miR159 in infected tissues. Alternatively, GAMYB derepression could occur via disruption of the conserved secondary RNA structures in GAMYB that are required for strong miR159-mediated silencing (Zheng et al., 2017). This could potentially explain the function of these highly conserved RNA elements. However, in Arabidopsis, neither transgenic expression of silencing suppressors nor infection with a Turnip Mosaic Virus that contains the silencing suppressor HC-Pro were able to strongly perturb miR159 silencing of MYB33/MYB65 (Li et al., 2016).
To verify and understand the potential role of the tobacco miR159-GAMYB module as a pathway in biotic defense, future studies should identify host and pathogen factors that inhibit miR159 function leading to deregulated GAMYB expression and induced plant defense response. Tomato infected by P. infestans showed decreased miR159 levels and increased MYB levels (Luan et al., 2015), suggesting that the miR159-GAMYB module might be involved in Phytophthora resistance in tomato. However, as infection of tomato with leaf curl New Delhi virus increases miR159 levels and decreases GAMYB mRNA, this is unlikely to be a general response to all pathogens (Naqvi et al., 2010). Another obvious question to ask is whether GAMYB-mediated plant defense response is specific to Solanaceae species or is conserved across other plant families. These and many other questions remain to be addressed to gain a greater understanding of the highly conserved miR159-GAMYB pathway in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Nicotiana tabacum TN90 seeds were sterilized by exposure to chlorine gas for 2–3 h in a desiccator jar and sown on agar plates containing Murashige and Skoog (MS) Basal medium and 3% (w/v) Suc, followed by stratification for 48 h at 4°C in the dark. After ∼3 weeks, tobacco plants were transferred to soil (Debco Plugger soil mixed with Osmocote Extra Mini fertilizer at 7 g/L). Tobacco plants were grown under long-day conditions (16 h light/8 h dark, at 500 μmol m−2 s−2 at 25°C).
Japonica rice (Oryza sativa) ‘Kitaake’ was used in experiments and transformation. After glumes were removed, rice seeds were sterilized by washing once with 70% (v/v) ethanol and twice with 33% to 50% (v/v) commercial bleach, followed by three washes with sterilized water. Sterilized and dry seeds were placed in Gellan Gum culture containing MS medium and 3% Suc. Two-week-old rice plants were transplanted on Rice Mix soil (80% [v/v] peat, 10% [v/v] perlite, 10% [v/v] vermiculite, dolomite lime, and macro and micro nutrients; mixed with Osmocote Extra Mini fertilizer at 7 g/L). Rice plants were grown under 12 h light/12 h dark, at 400 μmol m−2 s−2 at 28°C.
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) seed was sterilized with chlorine gas for 4–8 h in a desiccator jar and sown on soil (Debco Plugger soil mixed with Osmocote Extra Mini fertilizer at 3.5 g/L) or on MS agar plates. Seeds were stratified for 24 h at 4°C in the dark. Plants were grown under long-day conditions (16 h light/8 h dark, at 150 μmol m−2 s−2 at 22°C).
Generation of the MIM159 Decoy Constructs
The artificial target mimic MIM159 (Todesco et al., 2010) was obtained from the Nottingham Arabidopsis Stock Centre (NASC) and subcloned into pDONR/Zeo via Gateway BP reaction. Sequencing-confirmed entry clones of MIM159 were then integrated into the Gateway destination vector pGWB602Ω for tobacco transformation (Nakamura et al., 2010) via Gateway LR reactions (Invitrogen). For the rice MIM159 construct, first, a 661-bp rice IPS1 gene (AY568759) was synthesized by Integrated DNA Technologies and subsequently subcloned into pDONR/Zeo via Gateway BP reaction. Next, site-directed mutagenesis was performed on the pENTRY vector to generate the rice miR159 binding site with a 3-nucleotide bulge at the cleavage site (Supplemental Table S3) and this vector was then recombined into the modified destination vector pMDC32-Ubi (in which a ubiquitin promoter had replaced a 35S promoter) via Gateway LR reactions.
Generation of 35S-GAMYB or 35S-mGAMYB Constructs
Tobacco GAMYB2 (NtGAMYB2) and rice GAMYB (OsGAMYB) gene sequences were amplified with gene-specific primers, which contained attB1 and attB2 recombination sequences, on the tobacco and rice cDNA templates, respectively (Supplemental Tables S3 and S4). PCR amplification was performed using high-fidelity KOD Hot Start DNA Polymerase (Merck), with the following cycling conditions: one cycle of 95°C for 2 min, 35 cycles of 95°C for 20 s, 55°C for 10 s, and 70°C for 20 s/kb extension time according to amplicon size, and one cycle of 70°C for 10 min. PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega). Gateway BP reactions were performed with BP ClonaseTM II enzyme mix (Invitrogen) to integrate the purified PCR products into the pDONOR/ZEO vector (Invitrogen). The LaMYB33 construct was obtained from the Liwang Qi Lab (Laboratory of Cell Biology, Research Institute of Forestry, Chinese Academy of Forestry) and subcloned into the pDONR/Zeo vector. The miR159-resistant versions mGAMYB versions were generated by performing site-directed mutagenesis of the miR159 binding site on the verified pENTRY-GAMYB vectors (Supplemental Tables S3 and S4) and using KOD Hot Start DNA Polymerase. All entry clones were integrated into the destination vector pGWB602Ω via Gateway LR reactions (Invitrogen) to generate the binary vectors expressing 35S-GAMYB or 35S-mGAMYB transgenes.
Plant Transformation
For tobacco transformation, the MIM159, NtGAMYB2, and mNtGAMYB2 expression vectors were individually transformed into the Agrobacterium tumefaciens strain GV3101 by electroporation (Hellens et al., 2000) and selected on Luria-Bertani (LB) plates (50 μg/mL rifamycin, 25 μg/mL gentamicin, and 50 μg/mL spectinomycin). Transformed Agrobacterium was used to transform tobacco following the protocol of Horsch et al. (1985).
For rice transformation, the empty vector and the MIM159 binary vector were transformed into the Agrobacterium strain AGL1 by electroporation (Hellens et al., 2000) and selected on LB plates (25 g/L rifamycin and 50 g/L kanamycin). Transformed Agrobacterium was used to transform rice following the protocol of Hiei and Komari (2006), using 35 g/L hygromycin as selection. Transformants were verified via PCR genotyping of the hygromycin-resistant gene.
For Arabidopsis transformation, Gateway expression vectors were transformed into the Agrobacterium tumefaciens strain GV3101 by electroporation (Hellens et al., 2000), and then grown on LB plates (50 μg/mL rifamycin and 25 μg/mL gentamicin) containing the appropriate antibiotic for plasmid selection at 28°C for 48 h. Arabidopsis was transformed using the standard floral dipping procedure (Clough and Bent, 1998).
RNA Analysis
TRIzol (Invitrogen) was used to extract total RNA from plant tissues as previously described (Li et al., 2014b). Of the total RNA, 30 μg was then treated with RQ1 RNase-Free DNase (Promega) and RNaseOut Recombinant RNase Inhibitor (Invitrogen) in a 100-μL reaction volume following the manufacturer’s protocol. Treated RNA was then purified using RNeasy Plant Mini Kit (Qiagen).
TaqMan small RNA (sRNA) assays (Applied Biosystems) were then used to measure mature miRNA levels according to the manufacturer’s instructions, and with modifications as previously described (Allen et al., 2010). For each RNA sample, the retrotranscription was multiplexed with looped-RT primers for both the miRNA and the control tobacco sRNA snoR101 and rice sRNA snoR14, respectively. Each cDNA was assayed in three technical replicates using a Rotor-Gene Q real-time PCR machine (Qiagen). Expression levels of all miRNAs were normalized to corresponding sRNAs using the comparative quantitation program provided by the Rotor-Gene Q software (Qiagen).
For RT-qPCR of mRNA, cDNA synthesis was carried out using SuperScript III Reverse Transcriptase (Invitrogen) and an oligo dT primer according to the manufacturer’s protocol. For each sample, 2 μg of total RNA was used. The 10 μL reaction was then diluted 50 times in nuclease-free distilled water and used for subsequent qPCR, where 9.6 μL of each diluted cDNA sample was added to 10 μL of SensiFAST SYBR (Bioline) mix with 0.4 μL of qPCR primers at 10 μmol. The qPCR reactions were carried out on a Rotor-Gene Q real time PCR machine (Qiagen) in triplicate, under the following cycling conditions: one cycle of 95°C for 5 min and 45 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 20 s, followed by a 55°C to 99°C melting cycle. CYCLOPHILIN (AT2G29960) was used to normalize Arabidopsis mRNA levels; PROTEIN PHOSPHATASE 2A was used to normalize tobacco mRNA levels (Liu et al., 2012); and ACTIN (Os03g50890) was used to normalize rice mRNA levels (Caldana et al., 2007), using the comparative quantitation program in the Rotor-Gene Q software package. The average values of three technical replicates were used for analysis.
RNA Sequencing
RNA was isolated from three biological replicates of young leaves of 8-week-old wild-type and MIM159-transgenic plants. Each sample consisted of tissues from three different plants; the MIM159 sample tissues came from multiple independent lines. RNA samples were treated with DNase (Promega) and purified using RNeasy Plant Mini Kit (Qiagen) as described above, then used to construct cDNA libraries on which Illumina deep sequencing was performed by Novogene using an Illumina HiSeq platform with a paired-end 150-bp sequencing strategy. Novogene performed sample quality control, library construction, library quality control, mRNA sequencing, and data quality control.
Clean RNA sequencing reads were mapped to the reference tobacco TN90 genome (Sierro et al., 2014) by Tophat V2.1.1 software (Trapnell et al., 2009). The alignment results were analyzed by Cuffdiff software (cufflinks V2.2.1; Trapnell et al., 2012) to determine differentially expressed genes between wild-type and MIM159 tobacco. The ≥2-fold change and the false discovery rate (FDR) of ≤0.01 were used to define the significantly differentially expressed genes.
Genes differentially expressed at the level of >5-fold or 10-fold in MIM159 tobacco leaves compared to the wild type were subjected to GO enrichment analysis using agriGO (http://bioinfo.cau.edu.cn/agriGO/). As the tobacco genome is not fully annotated, but provides annotations from the closest Arabidopsis homologs, the homologs were used to determine the GO enrichment of the biological pathways. Pathways with a P-value of ≤0.05 were considered significantly enriched and were ranked by P-value.
Tobacco Infection Assay
Phytophthora parasitica isolate H1111 (ATCC MYA-141, also known as Phytophthora nicotianae), initially isolated by Dr. David Guest (University of Sydney), was grown on V8 agar (10% [v/v] V8 juice, 1.7% [w/v] agar, 0.01% [w/v] CaCO3, and 0.002% [v/v] β-sitosterol) at 25°C for 4–6 d in the dark. Tobacco infection assays were performed on detached leaves with a number of variations to limit dehydration and wound-induced stress. Leaves of tobacco TN90, the tobacco cultivar NC2326, which is resistant to P. parasitica (Blackman and Hardham, 2008), and three independent transformed MIM159 lines (L2, L6, and L12) were excised from ∼5-week-old plants and kept hydrated until use. The second leaf from plant apexes gave the most consistent results in preliminary experiments and these were used thereafter. Leaves were inoculated by placing explants of agar (6-mm diameter) from the leading edge of the P. parasitica colony and explants of control V8 agar on sites on the upper surface of leaves where the cuticle had been removed by gentle abrasion with glass powder. Each leaf had one to four control agar explants on one side of the midvein and a similar number of hyphae/agar explants on the other side, the number depending on the size of the leaf. Leaves were placed in chambers lined with wet filter paper to limit dehydration and infection was monitored for 6 d under 24 h light at 23°C. The infection assay used 3–10 leaves from three different plants, the number depending on the size of the leaves.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers MI0021329 (Nt-miR159), XM_016589328 (NtGAMYB1), XM_016599515 (NtGAMYB2), XM_016629135 (NtGAMYB3), MI0001092 (Os-miR159a), MI0001093 (Os-miR159b), CAA67000 (OsGAMYB), AB212075 (OsGAMYBL1), and AAT76349 (OsGAMYBL2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence similarity of tobacco GAMYB homologs.
Supplemental Figure S2. MiR159-mediated cleavage of NtGAMYB genes.
Supplemental Figure S3. Transcript profiling in MIM159 tobacco.
Supplemental Figure S4. Transcript profiling of individual GAMYB homologs in MIM159 tobacco.
Supplemental Figure S5. RNA profiling in tobacco anthers.
Supplemental Figure S6. MiR159-mediated cleavage of OsGAMYB.
Supplemental Figure S7. Sequence analysis of GAMYB proteins.
Supplemental Table S1. Top 50 most upregulated genes in MIM159 tobacco leaves.
Supplemental Table S2. NBS-LRR genes upregulated in MIM159 tobacco leaves.
Supplemental Table S3. Primers used for rice-related work.
Supplemental Table S4. Primers used for tobacco-related work.
Supplemental Dataset S1. Differential gene expression between wild-type and MIM159 tobacco.
Acknowledgments
We thank the Sumie Ishiguro Laboratory, Department of Biological Mechanisms and Functions, Graduate School of Bioagricultural Sciences, Nagoya University for the Gateway Destination pGWB602Ω vector, and the Liwang Qi Laboratory, Laboratory of Cell Biology, Research Institute of Forestry, Chinese Academy of Forestry for the pinetree LaMYB33 DNA sample. Also we thank Riya Kuruvilla, Deyun Qiu, and Rosemary Birch for help and training with rice and tobacco transformation.
Footnotes
This work was supported by the Research School of Biology (RSB), Australian National University, and a RSB International student PhD scholarship (to Z.Z.).
Articles can be viewed without a subscription.
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D, Goodman RM, Gut-Rella M, Glascock C, Weymann K, Friedrich L, Maddox D, Ahl-Goy P, Luntz T, Ward E, et al. (1993) Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc Natl Acad Sci USA 90: 7327–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D, Stinson J, Pear J, Glascock C, Ward E, Goodman RM, Ryals J (1992) A new multigene family inducible by tobacco mosaic virus or salicylic acid in tobacco. Mol Plant Microbe Interact 5: 513–515 [DOI] [PubMed] [Google Scholar]
- Allen RS, Li J, Alonso-Peral MM, White RG, Gubler F, Millar AA (2010) MicroR159 regulation of most conserved targets in Arabidopsis has negligible phenotypic effects. Silence 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RS, Li J, Stahle MI, Dubroué A, Gubler F, Millar AA (2007) Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 104: 16371–16376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Peral MM, Li J, Li Y, Allen RS, Schnippenkoetter W, Ohms S, White RG, Millar AA (2010) The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol 154: 757–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Bartel DP (2005) Antiquity of microRNAs and their targets in land plants. Plant Cell 17: 1658–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballvora A, Ercolano MR, Weiss J, Meksem K, Bormann CA, Oberhagemann P, Salamini F, Gebhardt C (2002) The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J 30: 361–371 [DOI] [PubMed] [Google Scholar]
- Blackman LM, Hardham AR (2008) Regulation of catalase activity and gene expression during Phytophthora nicotianae development and infection of tobacco. Mol Plant Pathol 9: 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S (2007) A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ario M, Griffiths-Jones S, Kim M (2017) Small RNAs: Big impact on plant development. Trends Plant Sci 22: 1056–1068 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Fry W. (2008) Phytophthora infestans: The plant (and R gene) destroyer. Mol Plant Pathol 9: 385–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M, Hua J (2012) Complex regulation of an R gene SNC1 revealed by auto-immune mutants. Plant Signal Behav 7: 213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV (1995) Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7: 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo WJ, Ho THD (2008) An abscisic acid-induced protein, HVA22, inhibits gibberellin-mediated programmed cell death in cereal aleurone cells. Plant Physiol 147: 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Xu Y, Shi M, Lai Y, Wu X, Wang H, Zhu Z, Poethig RS, Wu G (2017) Repression of miR156 by miR159 regulates the timing of the juvenile-to-adult transition in Arabidopsis. Plant Cell 29: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim, Ullah A, Hussain A, Shaban M, Khan AH, Alariqi M, Gul S, Jun Z, Lin S, Li J, et al. (2018) Osmotin: A plant defense tool against biotic and abiotic stresses. Plant Physiol Biochem 123: 149–159 [DOI] [PubMed] [Google Scholar]
- Hamada T. (2014) Microtubule organization and microtubule-associated proteins in plant cells. Int Rev Cell Mol Biol 312: 1–52 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Komari T (2006) Improved protocols for transformation of indica rice mediated by Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult 85: 271–283 [Google Scholar]
- Hellens R, Mullineaux P, Klee H (2000) Technical Focus: A guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5: 446–451 [DOI] [PubMed] [Google Scholar]
- Herbers K, Mönke G, Badur R, Sonnewald U (1995) A simplified procedure for the subtractive cDNA cloning of photoassimilate-responding genes: Isolation of cDNAs encoding a new class of pathogenesis-related proteins. Plant Mol Biol 29: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hofmann NL, Eichholtz D, Rogers SGR, Fraley T (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW. (2012) Conservation and divergence in plant microRNAs. Plant Mol Biol 80: 3–16 [DOI] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, Hattori T, Miyao A, Hirochika H, Ashikari M, et al. (2004) Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S (2014) miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leydon AR, Beale KM, Woroniecka K, Castner E, Chen J, Horgan C, Palanivelu R, Johnson MA (2013) Three MYB transcription factors control pollen tube differentiation required for sperm release. Curr Biol 23: 1209–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Reichel M, Li Y, Millar AA (2014a) The functional scope of plant microRNA-mediated silencing. Trends Plant Sci 19: 750–756 [DOI] [PubMed] [Google Scholar]
- Li J, Reichel M, Millar AA (2014b) Determinants beyond both complementarity and cleavage govern microR159 efficacy in Arabidopsis. PLoS Genet 10: e1004232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WF, Zhang SG, Han SY, Wu T, Zhang JH, Qi LW (2013a) Regulation of LaMYB33 by miR159 during maintenance of embryogenic potential and somatic embryo maturation in Larix kaempferi (Lamb.) Carr. Plant Cell Tissue Organ Cult 113: 131–136 [Google Scholar]
- Li X, Bian H, Song D, Ma S, Han N, Wang J, Zhu M (2013b) Flowering time control in ornamental gloxinia (Sinningia speciosa) by manipulation of miR159 expression. Ann Bot 111: 791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Alonso-Peral M, Wong G, Wang M-B, Millar AA (2016) Ubiquitous miR159 repression of MYB33/65 in Arabidopsis rosettes is robust and is not perturbed by a wide range of stresses. BMC Plant Biol 16: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R (2010) Transcriptome-wide identification of microRNA targets in rice. Plant J 62: 742–759 [DOI] [PubMed] [Google Scholar]
- Liu D, Shi L, Han C, Yu J, Li D, Zhang Y (2012) Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One 7: e46451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y, Cui J, Zhai J, Li J, Han L, Meng J (2015) High-throughput sequencing reveals differential expression of miRNAs in tomato inoculated with Phytophthora infestans. Planta 241: 1405–1416 [DOI] [PubMed] [Google Scholar]
- Millar AA, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Lohe A, Wong G (2019) Biology and function of miR159 in plants. Plants (Basel) 8: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E (2008) The hypersensitive response; the centenary is upon us but how much do we know? J Exp Bot 59: 501–520 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Mano S, Tanaka Y, Ohnishi M, Nakamori C, Araki M, Niwa T, Nishimura M, Kaminaka H, Nakagawa T, et al. (2010) Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci Biotechnol Biochem 74: 1315–1319 [DOI] [PubMed] [Google Scholar]
- Naqvi AR, Haq QM, Mukherjee SK (2010) MicroRNA profiling of tomato leaf curl New Delhi virus (tolcndv) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol J 7: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E, Dezulian T, Huson D, et al. (2007) Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell 13: 115–125 [DOI] [PubMed] [Google Scholar]
- Qiao Y, Shi J, Zhai Y, Hou Y, Ma W (2015) Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc Natl Acad Sci USA 112: 5850–5855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel M, Li Y, Li J, Millar AA (2015) Inhibiting plant microRNA activity: Molecular SPONGEs, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol J 13: 915–926 [DOI] [PubMed] [Google Scholar]
- Reichel M, Millar AA (2015) Specificity of plant microRNA target MIMICs: Cross-targeting of miR159 and miR319. J Plant Physiol 180: 45–48 [DOI] [PubMed] [Google Scholar]
- Sierro N, Battey JN, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV (2014) The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5: 3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Goodman RM (2002) Cloning and identification of the promoter of the tobacco Sar8.2b gene, a gene involved in systemic acquired resistance. Gene 290: 115–124 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Teh OK, Hofius D (2014) Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. J Exp Bot 65: 1297–1312 [DOI] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D (2010) A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Aya K, Ueguchi-Tanaka M, Shimada Y, Nakazono M, Watanabe R, Nishizawa NK, Gomi K, Shimada A, Kitano H, et al. (2006) GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J 47: 427–444 [DOI] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Bellinger M, Jin H (2014) Small RNAs: A new paradigm in plant-microbe interactions. Annu Rev Phytopathol 52: 495–516 [DOI] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Yan J, Gou F, Mao Y, Tang G, Botella JR, Zhu JK (2017) Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc Natl Acad Sci USA 114: 5277–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wen H, Teotia S, Du Y, Zhang J, Li J, Sun H, Tang G, Peng T, Zhao Q (2017) Suppression of microRNA159 impacts multiple agronomic traits in rice (Oryza sativa L.). BMC Plant Biol 17: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Reichel M, Deveson I, Wong G, Li J, Millar AA (2017) Target RNA secondary structure is a major determinant of miR159 efficacy. Plant Physiol 174: 1764–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]