Columella stem cell fate is controlled by factors acting independently of WUSCHEL RELATED HOMEOBOX 5 and the main role of the CLAVATA3/ESR-RELATED 40 peptide is the regulation of quiescent center activity and position.
Abstract
The quiescent center (QC) of the Arabidopsis (Arabidopsis thaliana) root meristem acts as an organizer that promotes stem cell fate in adjacent cells and patterns the surrounding stem cell niche. The stem cells distal from the QC, the columella stem cells (CSCs), are maintained in an undifferentiated state by the QC-expressed transcription factor WUSCHEL RELATED HOMEOBOX5 (WOX5) and give rise to the columella cells. Differentiated columella cells provide a feedback signal via secretion of the peptide CLAVATA3/ESR-RELATED40 (CLE40), which acts through the receptor kinases ARABIDOPSIS CRINKLY4 (ACR4) and CLAVATA1 (CLV1) to control WOX5 expression. Previously, it was proposed that WOX5 protein movement from the QC into CSCs is required for CSC maintenance, and that the CLE40/CLV1/ACR4 signaling module restricts WOX5 mobility or function. Here, these assumptions were tested by exploring the function of CLE40/CLV1/ACR4 in CSC maintenance. However, no role for CLE40/CLV1/ACR4 in constricting the mobility of WOX5 or other fluorescent test proteins was identified. Furthermore, in contrast to previous observations, WOX5 mobility was not required to inhibit CSC differentiation. We propose that WOX5 acts mainly in the QC, where other short-range signals are generated that not only inhibit differentiation but also promote stem cell division in adjacent cells. Therefore, the main function of columella-derived CLE40 signal is to position the QC at a defined distance from the root tip by repressing QC-specific gene expression via the ACR4/CLV1 receptors in the distal domain and promoting WOX5 expression via the CLV2 receptor in the proximal meristem.
A specific feature of plants is their extensive postembryonic development. This continuous growth style is only possible if a pool of stem cells is maintained that replenishes and restores the meristem cell pool. Stem cells are maintained in a niche. The two major stem cell niches within a plant, the apical meristems, must remain active for extended periods of time, up to many years (Stahl and Simon, 2005). In Arabidopsis (Arabidopsis thaliana) meristems, stem cells receive maintenance signals from adjacent cell groups, namely the organizing center (OC) in the shoot meristem and the quiescent center (QC) in the root meristem (Aichinger et al., 2012). Stem cell divisions give rise to rapidly dividing daughter cells. The daughter cells are displaced from the meristematic regions and ultimately differentiate into specialized cell types or tissues (Greb and Lohmann, 2016). Several layers of feedback act between the niche and the stem cells and their differentiating descendants. These feedback mechanisms operate via signaling molecules such as mobile transcription factors and secreted peptides (Pierre-Jerome et al., 2018).
The shoot apical meristem utilizes a short-range communication mechanism between the OC and the overlying stem cells that creates a finely tuned balance of stem cell maintenance and cell differentiation. Specifically, the homeodomain transcription factor WUSCHEL (WUS) is expressed in the OC and moves toward the stem cells, where it promotes stem cell fate and the expression of the 13-amino acid CLAVATA3 (CLV3) peptide, which is then secreted from stem cells (Fletcher et al., 1999; Brand et al., 2000; Ohyama et al., 2009; Daum et al., 2014). WUS moves through plasmodesmata (PDs), the cytoplasmic bridges between neighboring cells (Daum et al., 2014). CLV3 is recognized by several plasma membrane-localized receptor kinases, among them the Leu-rich repeat (LRR) receptor kinase CLV1, the LRR receptor protein CLV2, and the membrane-localized pseudokinase CORYNE (CRN), resulting in repression of WUS in the OC. The interaction between WUS and CLV3 creates negative feedback regulation that serves to balance stem cell maintenance and cell differentiation in the shoot meristem (Fletcher et al., 1999; Brand et al., 2000; Ogawa et al., 2008; Ohyama et al., 2009; Yadav et al., 2011).
In the root meristem, stem cells (or initials) surround the QC. Division of the stem cells generates daughter cells that are arranged in ordered cell files, creating the radial symmetry of the root. The vascular initials are located proximal to the QC, while lateral stem cells give rise to the endodermis, cortex, epidermis, and lateral root cap. Distal to the QC lie the columella cell (CC) initials, also called columella stem cells (CSCs), which generate the CCs. Although the shoot and root meristems are structurally different, there is strong evidence that the molecular components of the stem cell-controlling pathways are closely related (Stahl and Simon, 2010). The WUS homolog WUSCHEL RELATED HOMEOBOX 5 (WOX5) is expressed in the QC (Sarkar et al., 2007). Starch granules, a hallmark for cellular differentiation in CCs, accumulate in the CSCs of wox5-1 mutants, indicating that WOX5, like WUS, promotes stem cell maintenance in a non-cell-autonomous manner (Sarkar et al., 2007). The peptide CLAVATA3/ESR-RELATED 40 (CLE40), which is synthesized by and secreted from CCs, promotes differentiation of CSCs, an activity that requires the receptor-like kinase ARABIDOPSIS CRINKLY4 (ACR4; Stahl et al., 2009). Both CLE40 and ACR4 are expressed in the distal domain of root meristems, and cle40 and acr4 mutant plants fail to confine CSC fate to only one cell layer. Furthermore, acr4 mutants are resistant to exogenous addition of CLE40 peptide (CLE40p), indicating that ACR4 must perceive the CLE40 signal to restrict CSC maintenance (Stahl et al., 2009). CLE40 therefore serves as a feedback signal to confine stem cell activities when sufficient numbers of stem cell daughters are present. Based on parallels drawn to the regulation of the stem cells in the shoot, WOX5 expression in the QC was thought to be a downstream target of a CLE40/ACR4 signaling module, since application of CLE40p to root meristems not only promoted CSC differentiation, but also simultaneously triggered a rearrangement of the WOX5 expression domain from the QC toward the vascular initials (Stahl et al., 2009). In addition to ACR4, CLV1 was found to contribute to CSC control, because clv1 mutants (like acr4 or cle40 mutants) also carry an increased number of CSC layers compared to wild-type (ecotype Columbia of Arabidopsis [Col-0]) plants. Interestingly, CLV1 and ACR4 can form different homo- and heteromeric complexes at the plasma membrane that concentrate at PDs (Stahl et al., 2013), indicating that they could act jointly in a signaling cascade that restricts stem cell fate.
Together, these results led to an exciting, but also highly speculative, model for the non-cell-autonomous control of stem cell behavior. In this model, the receptor kinases ACR4 and CLV1, residing at PDs, establish a “gating” mechanism that restricts the intercellular movement of stemness factors between the QC and adjacent stem cells (Fig. 1A; Stahl and Simon, 2013). Activation of the receptor kinase complex and the gating depends on the secreted peptide CLE40p. Analogous to WUS moving from the OC into the apical shoot stem cell domain, WOX5 could be the prime candidate for a QC-derived stem cell factor whose mobility is controlled by the CLV1/ACR4 gating mechanism. Consistent with this notion, WOX5 protein was previously shown to move from the QC into the CSCs (Pi et al., 2015). Furthermore, using fusions to fluorescent proteins in order to either track the WOX5 protein or to interfere with its mobility, movement of WOX5 from the QC to the CSCs was found to be crucial for CSC maintenance (Pi et al., 2015). In the CSCs, WOX5 appears to act at least in part by directly repressing expression of the transcription factor CDF4, which normally promotes cell differentiation (Pi et al., 2015). Normal repression of CDF4 by WOX5 causes two distinct cell expression patterns: high expression of CDF4 in the differentiated CCs, which lack WOX5, and low expression in the CSCs and the QC, where WOX5 is present. These complementary patterns could further balance stem cell maintenance with cell differentiation in the distal root meristem (Pi et al., 2015). However, the role of WOX5 in controlling distal stem cell fate could be less predominant than assumed from these experiments, because CSCs can be maintained even in the absence of WOX5 activity if CLE40p signaling is abolished, and mutations in other differentiation-promoting factors (such as SOMBRERO and ARF10/ARF16) showed similar effects (Bennett et al., 2014; Richards et al., 2015; Rahni et al., 2016).
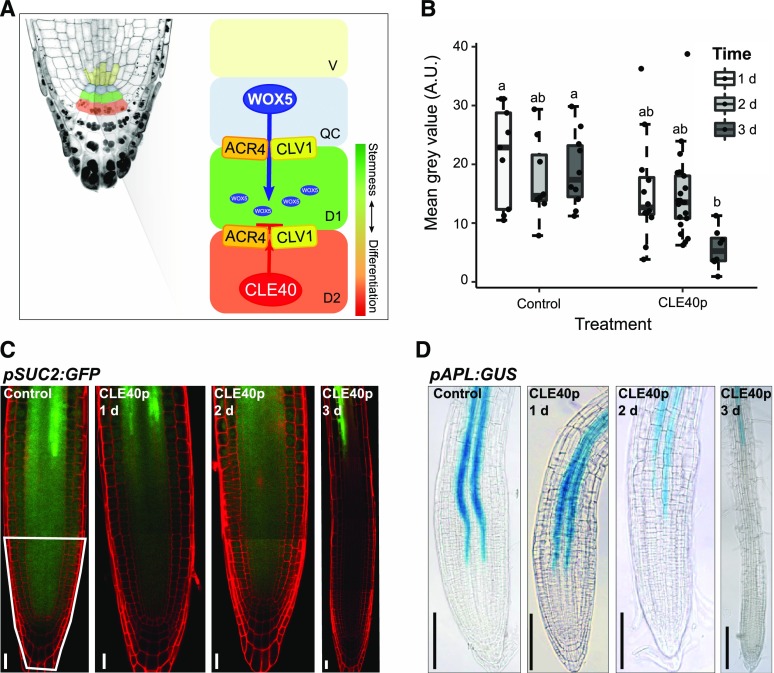
Figure 1.
Regulatory mechanism involved in root stem cell maintenance and how CLE40p delays protophloem development. A, Schematic overview of the current hypothesis of the regulation mechanism involved in root stem cell maintenance. WOX5 is synthesized in the QC and moves from there through PDs to the underlying CSCs, maintaining their fate. ACR4 and CLV1 are believed to be involved in the regulation of this movement due to their localization at the sites of PDs. CLE40p is synthesized from the differentiated CCs. Binding to the ACR4-CLV1 complex could lead to a conformational change, making WOX5 unable to move to the next cell layer, which will lead to a differentiation of the CSCs. B, Fluorescence intensity (depicted as the mean gray value in the root tip, indicated by the white outline in C), of plants expressing pSUC2:GFP under the control condition and after 1, 2, or 3 d of CLE40p treatment. Significant differences (P < 0.05), as determined by two-way ANOVA with Tukey's post hoc analysis, are indicated by lowercase letters above the bars (n ≥ 25). A.U., Arbitrary units. C, pSUC2:GFP in the root meristem under the control condition and after 1, 2, or 3 d of CLE40p (1 μm) treatment. Scale bars = 20 µm. The region outlined in white was used for fluorescence intensity measurements in B. D, pAPL:GUS marking the phloem in the root meristem under the control condition and after 1, 2, or 3 d of CLE40p (1 μm) treatment. Scale bars = 100 µm.
Here we investigated WOX5 function in maintaining CSC fate and the factors controlling WOX5 mobility. The “gating model” whereby CLE40p signaling through the receptor kinases CLV1 and ACR4 controls WOX5 mobility was tested by analyzing, with a high spatial and temporal resolution, whether the CLE40p/CLV1/ACR4 signaling module affects transport and cytoplasmic connectivity through PDs and whether the mobility of WOX5 is controlled by the CLE40p/CLV1/ACR4 module. We found that neither increased nor reduced signaling by the CLE40p/CLV1/ACR4 module affects intercellular connectivity in the stem cell niche. Furthermore, we could show that although WOX5 can move from the the QC into the CSCs, this movement is neither affected by CLE40p/CLV1/ACR4 nor required to maintain distal stem cells. Finally, we found that as an immediate response to increased CLE40p signaling via the CLV2 receptor, WOX5 expression is up-regulated in a new vascular domain, and not repressed as suggested previously. These combined results do not support the previous elegant model for the control of CSC fate in Arabidopsis by mobile WOX5 and the antagonistic activities of CLE40 and CDF4 (Sarkar et al., 2007; Stahl et al., 2009). We instead propose that factors acting independently of WOX5 in the QC control CSC fate and that the main roles of the CLE40p/CLV1/ACR4 module are in the regulation of QC activity and position.
RESULTS
Does CLE40p Signaling Restrict Intercellular Protein Movement through PDs?
To analyze if CLE40p signaling can affect intercellular mobility of proteins, we used Arabidopsis plants expressing GFP from the phloem companion cell-specific SUC-H+ SYMPORTER 2 gene promoter (pSUC2:GFP). As observed previously by others (Stadler et al., 2005a; Vatén et al., 2011; Ross-Elliott et al., 2017), the expressed GFP diffuses from the companion cells into the phloem and is then symplastically unloaded via PDs from phloem cells into sink tissues. Therefore, the observed GFP fluorescence intensities in the different sink tissues, such as the root meristem, can be used as a proxy for cellular connectivity (Imlau et al., 1999). Incubation of seedlings on solid growth medium supplemented with 1 μm CLE40p resulted in a continuous reduction of GFP unloading into the basal root meristem from day 1 to day 3 relative to untreated plants (Fig. 1, B and C). External application of several different root-active CLE peptides was previously shown to perturb protophloem development and to lead to a proximal shift of the developing protophloem sieve elements (Hazak et al., 2017). Using expression of GUS under control of the ALTERED PHLOEM DEVELOPMENT (APL) promoter as a protophloem marker, we found that CLE40p application delays protophloem differentiation, leading to a proximal shift of pAPL:GUS expression (Fig. 1D). Since AtSUC2 promoter activity is restricted to metaphloem companion cells, the CLE-dependent delay in phloem differentiation will affect expression of the GFP reporter (Stadler et al., 2005a; Ross-Elliott et al., 2017). Therefore, reduced GFP intensity cannot be unequivocally assigned to a reduction in PD-dependent transport capacity, since cellular differentiation in the meristem is simultaneously affected (Pallakies and Simon, 2014). To analyze intercellular connectivity and transport specifically in the stem cell niche and independent of the vascular transport system, we employed characteristics of the DRONPA-s fluorophore. DRONPA-s is a fluorophore that can reversibly photoswitch between a bright and a dark state (Ando et al., 2004; Habuchi et al., 2005; Lummer et al., 2011). In vivo de novo synthesized DRONPA is, by default, in its “on” state, emitting bright green fluorescence when excited with 488-nm laser light. High 488-nm laser power leads to a photoswitch of DRONPA-s to its nonfluorescent “off” state. Irradiation at 405 nm can then restore the fluorescent on state (Supplemental Fig. S1, A–H). We used a two-photon laser for highly cell-type-specific switching of the DRONPA-s fluorophore in deep tissue layers (Gerlitz et al., 2018). Fluorescent DRONPA-s expressed from the 35S promoter was first switched to the off state in the entire root stem cell niche using 488 nm excitation (Supplemental Fig. S1, B and F). Using 405 nm excitation in individual cells of the QC or CSC domain, DRONPA-s was activated into the fluorescence on state (Supplemental Fig. S1, C and G), while changes in fluorescence intensity were monitored in the excited cell and the immediate surrounding cells 5 min after excitation (Supplemental Fig. S1, D and H). The diffusion of fluorescent DRONPA-s was analyzed by first determining the fluorescence signal in the excited region and in the adjacent cells, and then calculating the ratio between these values at time 0 (excitation of DRONPA-s) and time 5 (after 5 min). When exciting either the QC or CSC, an ∼50% to 60% decrease in this ratio was observed within 5 min (Supplemental Fig. S1, I), caused by rapid outward diffusion of DRONPA-s from the excited cells. To assay whether CLE40 signaling can alter intercellular connectivity, we pretreated seedlings by incubating them for 1 d on growth medium containing 1 μm CLE40 peptide. After excitation of DRONPA-s fluorescence in either the QC or CSC, we found a similar decrease in the fluorescence intensity in QC and CSC cells (Supplemental Fig. S1, I). We concluded that CLE40p does not generally affect the intercellular connectivity of the QC or CSC with adjacent cell groups at a detectable level. Nevertheless, proteins other than DRONPA-s and those that act as mobile stemness factors might behave differently and be directly or indirectly affected in their mobility by CLE40.
Inhibition of WOX5 Movement Does Not Compromise Stem Cell Maintenance
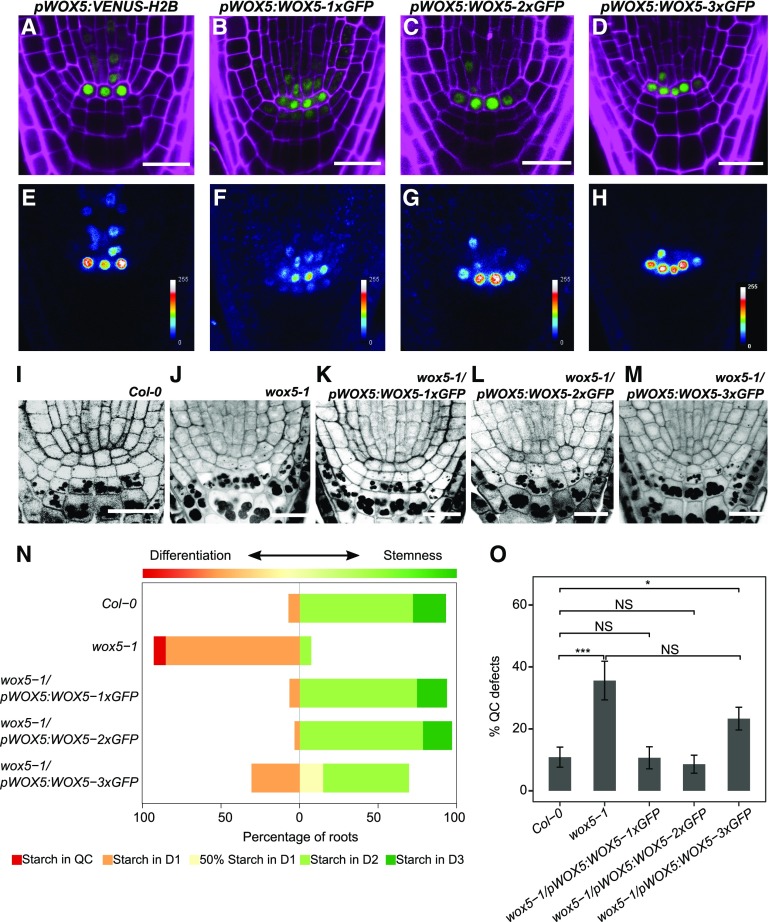
Because no general effect of CLE40 signaling on intercellular protein movement could be detected, we next asked whether CLE40p can affect the previously reported mobility of WOX5 as a potential stemness factor and target of a CLE40-dependent pathway. WOX5 RNA is expressed in the QC and, at a lower level, in some vascular initials. wox5-1 mutant plants show premature differentiation of cell layers located distal to the QC (Stahl et al., 2009), which we labeled according to their position as Distal 1 (D1), D2, D3, etc. In the wild type, D1 corresponds to the CSC layer, and the D2–D5 positions are occupied by differentiated CCs (Fig. 1A). To assess the mobility of WOX5, we first observed the expression pattern of WOX5 using the fluorescence reporter Venus fused to Histone 2B (H2B) for nuclear targeting (pWOX5:VENUS-H2B) and compared this with the translational reporter pWOX5:WOX5-1xGFP (or pWOX5:WO5-GFP). We found the WOX5 promoter to be active mainly in the QC and to a much lesser extent in the vascular initials, in accordance with results obtained via RNA in situ hybridization and consistent with previously published expression data (Fig. 2, A and E; Sarkar et al., 2007; Pi et al., 2015). In comparison, WOX5-GFP was detected in the QC as well as in the immediately surrounding cells, including the D1 layer (Fig. 2, B and F), indicating that the WOX5-GFP fusion protein can move from the QC into adjacent cells, as suggested previously. We then generated a fusion between the WOX5 protein and two copies of the GFP protein under the control of the WOX5 promoter, pWOX5:WOX5-2xGFP, which should be too large to move into adjacent cells. WOX5-2xGFP was only detected in the QC and the vasculature, similar to the pWOX5:VENUS-H2B expression pattern (Fig. 2, C and G), indicating that a larger WOX5 fusion protein becomes immobile and cannot leave the cells where it is initially expressed. For further analysis, after subtracting background fluorescence, the fluorescence intensities were quantified as the mean gray value in the D1 and D2 regions relative to values in the QC region within the same root. Calculating the D1/QC or D2/QC ratio allowed us to compare the relative fluorescence intensities in D1 and D2 between the different constructs and transgenic lines. For pWOX5:WOX5-1xGFP, the D1/QC ratio was 35.6% ± 11.7% (mean ± sd, n = 31). For pWOX5:WOX5-2xGFP, the D1/QC was 7.2% ± 3.7% (mean ± sd, n = 35), which was not significantly different from the D1/QC ratio in pWOX5:VENUS-H2B at 1.9% ± 0.9% (mean ± sd, n = 29) or from the D2/QC ratios across the three different constructs (0.34% ± 0.15%, 2.1% ± 7.5%, and 3.8% ± 3.3% for pWOX5:VENUS-H2B, pWOX5:WOX5-GFP, and pWOX5:WOX5-2xGFP, respectively; Supplemental Fig. S2A). Since WOX5-2xGFP was not detectable at significant levels in the D1 layer, it could be used to study whether WOX5 is necessary within the QC and/or within the CSCs to maintain CSC fate. To address this question, both pWOX5:WOX5-1x-GFP and pWOX5:WOX5-2xGFP complementation experiments were performed by introducing each construct into a wox5-1 mutant background. Loss of WOX5 function leads to differentiation of the D1 layer and more frequent division of the cells at the QC position (Sarkar et al., 2007; Bennett et al., 2014). To distinguish between the cell-autonomous function of WOX5 in the QC and its potential function in the CSCs, both the QC and CSC phenotypes were separately analyzed in the rescued lines. Complementation of the CSC phenotype was assessed by performing modified pseudo-Schiff-propidium iodide (mPS-PI) staining, which, after fixation of the sample, stains both the cell wall and starch granules. Differentiated CCs are characterized by the accumulation of starch granules, which are lacking in the undifferentiated CSC. The frequency of roots with QC divisions was also counted. Surprisingly, both pWOX5:WOX5-1xGFP and pWOX5:WOX5-2xGFP were able to rescue the wox5-1 QC and CSC mutant phenotypes (Fig. 2, I–L, N, and O). These results indicate that a nonmobile, QC-localized WOX5 protein is sufficient to restore CSC fate.
Figure 2.
Inhibition of WOX5 movement does not compromise CSC maintenance. A to H, Images of roots from 5-d-old pWOX5:VENUS-H2B (A and E), pWOX5:WOX5-1xGFP (B and F), pWOX5:WOX5-2xGFP (C and G), and pWOX5:WOX5-3xGFP (D and H) seedlings. The top row (A–D) shows GFP (green) and PI (magenta) fluorescence. False-color images are shown in E–H. Scale bars = 20 µm. I to M, Images of mPS-PI staining of 5-d-old root tips of Col-0 (I), wox5-1 (J), wox5-1/pWOX5:WOX5-1xGFP (K), wox5-1/pWOX5:WOX5-2xGFP (L), and wox5-1/pWOX5:WOX5-3xGFP (M). Scale bars = 20 µm. N, Frequency of roots showing starch granule accumulation in the indicated cell layer (n ≥ 25). O, Frequency of roots of the indicated lines showing a defect in QC organization (e.g. QC divisions or unorganized arrangement). Statistical significance was determined by two-proportion Z-test (*P < 0.05, ***P < 0.001). Data represent the mean ± se (n ≥ 25).
Previous research documented that a WOX5-3xGFP fusion protein was unable to move from the QC and could not complement the wox5-1 CSC mutant phenotype (Pi et al., 2015). Our data here clearly contrast these observations. Due to this apparent discrepancy, we decided to perform further studies to determine whether WOX5 presence in the CSC is necessary for its maintenance. We created a WOX5-3xGFP fusion protein and expressed it under its own promoter in both Col-0 and wox5-1 backgrounds. Similar to WOX5-2xGFP, the WOX5-3xGFP fusion protein localized only to the QC and vascular initials, overlapping the WOX5 expression domain (Fig. 2, D and H). In the wox5-1 background, the pWOX5:WOX5-3xGFP transgene resulted in a partial complementation of the CSC phenotype (Fig. 2, M and N), with 30.43% of the analyzed roots revealing no CSCs and 54.34% carrying at least one CSC distal to the QC layer (Fig. 2N), compared to 85% and 7.31%, respectively, in wox5-1 mutants. Importantly, the QC defects in the wox5-1 background were only partially rescued by the transgene, and cells expressing pWOX5:WOX5-3xGFP often formed an irregular QC compared to the Col-0 background (Fig. 2O; Supplemental Fig. S2, B and C), with 23.30% of 3xGFP roots showing abnormal QC divisions compared to 10.08% in Col-0. Complementation of wox5-1 with pWOX5:WOX5-1xGFP or pWOX5:WOX5-2xGFP restored the frequency of QC divisions to wild-type levels. We thus found that the WOX5-3xGFP fusion protein is not fully functional, even within the QC. We hypothesize that since the Mr of WOX5 is around the same as that of GFP (∼25 kD), fusing a tag three times its own size could impair WOX5 protein function. It is worthy of note that WOX5 functions, such as QC maintenance, inhibition of QC divisions, and maintenance of CSCs, requires direct protein-protein interaction, for example with members of the HAM-protein family (Zhou et al., 2015).
To further test whether the 3xGFP tag interferes with WOX5 functionality, we designed an experiment similar to that in Sarkar et al. (2007), which showed that overexpression of a dexamethasone (DEX)-inducible WOX5 led to accumulation of undifferentiated CSCs. Here both WOX5-GFP and WOX5-3xGFP proteins tagged with the hormone-binding domain of the rat glucocorticoid receptor (GR) were generated under the control of the CaMV35S promoter. For both constructs, 1 d of DEX induction in a Col-0 background led to a similar accumulation of CSC layers (Supplemental Fig. S3, A, B, and E). However, in the wox5-1 background, a clear difference could be seen after 1 d of DEX induction, where 35S:WOX5-1xGFP-GR led to an increased amount of CSC layers (15.02 ± 2.40; mean ± sd, n = 21) compared to Col-0 (12.61 ± 2.31; mean ± sd, n = 40,). 35S:WOX5-3xGFP-GR had an average of 7.9 ± 1.43 (mean ± sd, n = 40) extra CSC layers in the wox5-1 background, but in the Col-0 background it was similar to the 1xGFP construct, at 11.5 ± 2.28 (mean ± sd, n = 47) CSC layers (Supplemental Fig. S3, C–E). We have to mention that these constructs were leaky and already showed overexpression under control conditions (Supplemental Fig. S3, F and G). However, we are convinced that this does not influence the conclusion that the effect of ectopic expression of WOX5-1xGFP and WOX5-3xGFP depends on the background. It is likely that endogenous WOX5 somehow controls the ectopic divisions, whereas in a wox5-1 mutant background this control is absent, leading to more irregular divisions and a lateral expansion of the root meristem (Supplemental Fig. S3C). In addition, our observations show that in the absence of endogenous WOX5, the WOX5-3xGFP-GR fusion protein is unable to create extra CSC layers.
The inability of the WOX5-3xGFP fusion to fully rescue the wox5-1 mutant could be caused by the large triple GFP tag interfering with the functionality of the protein and not only with its mobility, as reported earlier. In addition, the partial rescue of CSC fate by a large and nonmobile WOX5-3xGFP protein reveals that although movement of WOX5 from the QC to CSCs is possible, such a movement is not essential to regulate CSC fate. We therefore conclude that additional QC-derived signals, whose activity or expression might depend on WOX5 function in the QC, act non-cell-autonomously to control CSC fate.
WOX5 Interdomains Control Intracellular Localization But Not Mobility
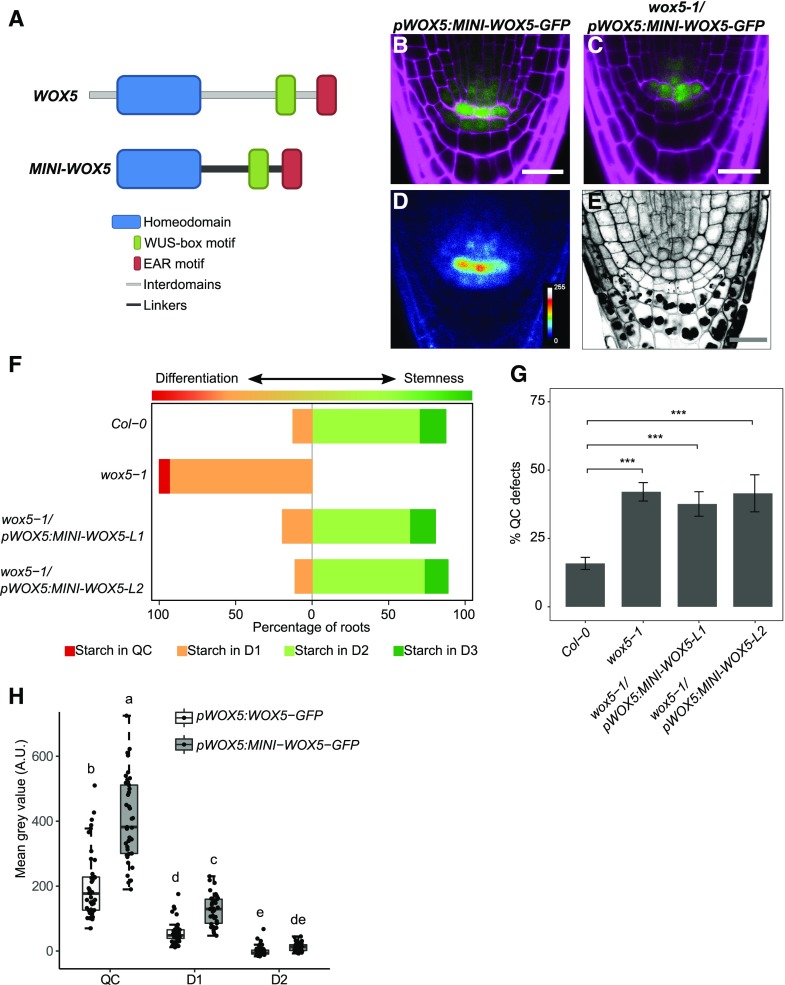
The mobility of WUS is restricted by a nonconserved amino acid stretch located between the conserved WUS homeodomain and the WUS-box (Daum et al., 2014). A minimal WUS version, in which the conserved homeodomain, WUS box, and EAR-like domain were connected by structurally unrelated linkers, has an increased mobility range in the shoot apical meristem, leading to stem cell overproliferation and shoot apical meristem expansion (Daum et al., 2014). This indicated that restricting protein mobility may be important for local stem cell fate and homeostasis. Although our analysis has so far shown that WOX5 movement is not required for CSC maintenance, we asked whether restriction of movement contributes to patterning in the root stem cell niche. For example, retention of WOX5 protein in the QC could serve to maintain a higher concentration of the protein in these cells or to hinder adjacent cells from accumulating too much WOX5. To identify a possible role of WOX5 mobility in stem cell maintenance, we investigated whether a minimal version of WOX5 (MINI-WOX5) would affect root stem cell homeostasis or its ability to move. A more mobile WOX5 could reach more distal CC layers and inhibit their differentiation or be retained in the QC only at lower levels. The MINI-WOX5 was created following the strategy for the minimal WUS version (Daum et al., 2014), maintaining the conserved homeodomain, WUS-box, and EAR-like domain of WOX5 and exchanging the nonconserved region for linker sequences (Fig. 3A). This MINI-WOX5 was fused to GFP and introduced under the WOX5 promoter into Col-0 or wox5-1 mutant backgrounds. The MINI-WOX5-GFP protein showed less nuclear localization and a redistribution to both cytoplasm and nucleus compared to WOX5-GFP (Figs. 2, B and F, and 3, B and D). We expected that a relative increase in its cytoplasmic localization would also increase diffusion toward the neighboring cells; however, MINI-WOX5-GFP signal was still restricted to the QC, the CSC, and the first vascular cell layer in a localization pattern similar to that of WOX5-GFP (Fig. 3, B and D). This indicates that, in contrast to WUS, the interdomain regions of WOX5 are not involved in restricting its mobility but do determine its nuclear localization. So far, no classical nuclear localization signal (NLS) has been identified for WOX-family proteins (van der Graaff et al., 2009), indicating that a yet uncharacterized NLS signal must reside in the interdomain regions or that this region is required for interaction with another protein that confers nuclear localization. In a wox5-1 mutant background, the introduction of the pWOX5:MINI-WOX5-GFP construct was able to revert the wox5-1 CSC phenotype (Fig. 3, C, E, and F) but did not rescue the QC defects (Fig. 3G), indicating that the MINI-WOX5-GFP protein did not fully retain its function. Since the interdomains of WUS are important for homodimerization and/or protein-protein interaction (Daum et al., 2014), we propose that they play a similar role for the WOX5 protein. The D1/QC and D2/QC fluorescence intensity ratios of pWOX5:MINI-WOX5-GFP were not different from those of pWOX5:WOX5-GFP (Fig. 3H). However, the measured GFP intensities in the QC and D1 cells of the pWOX5:MINI-WOX5-GFP line were higher compared to those of pWOX5:WOX5-GFP plants (Fig. 3H), which could indicate that portions of the interdomains could be involved in the turnover of the WOX5 protein. In agreement with this, several phosphorylated peptides of WOX5 were identified previously, among which some fall within the interdomains of WOX5 (Meyer et al., 2015).
Figure 3.
MINI-WOX5-GFP is not restricted to the nucleus and can complement the wox5-1 CSC phenotype. A, Schematic representation of the MINI-WOX5 design. B and D, Images of 5-d-old roots of pWOX5:MINI-WOX5-GFP in the Col-0 background, showing GFP (green) and PI (magenta) fluorescence (B) and the false-color image (D). C and E, Images of 5-d-old roots of pWOX5:MINI-WOX5-GFP in the wox5-1 background, showing GFP (green) and PI (magenta) fluorescence (C) and mPS-PI staining (E). Scale bars = 20 µm. F, Starch granule accumulation frequency in the indicated cell layers of roots (n ≥ 25). G, Frequency of roots of the indicated lines showing QC divisions or unorganized arrangement. Statistical significance was determined by two-proportion Z-test (***P < 0.05). Data represent the mean ± se (n ≥ 25). In F and G, L1 and L2 indicate two independent lines. H, Fluorescence intensity depicted as the mean gray value in the indicated lines in the QC, D1, and D2 cells. Significant differences (P < 0.05) as determined by two-way ANOVA with Tukey's post hoc analysis are indicated by lowercase letters above box plots (n ≥ 25). A.U., Arbitrary units.
In contrast with a minimal WUS version, removal of the interdomains of WOX5 does not increase its mobility range. The MINI-WOX5, however, does show increased cytoplasmic localization compared to the full-length version. Furthermore, our experiments indicated that the interdomains could play a role in the turnover of WOX5.
Is WOX5 Expression or Localization Controlled by the CLE40/CLV1/ACR4 Signaling Module?
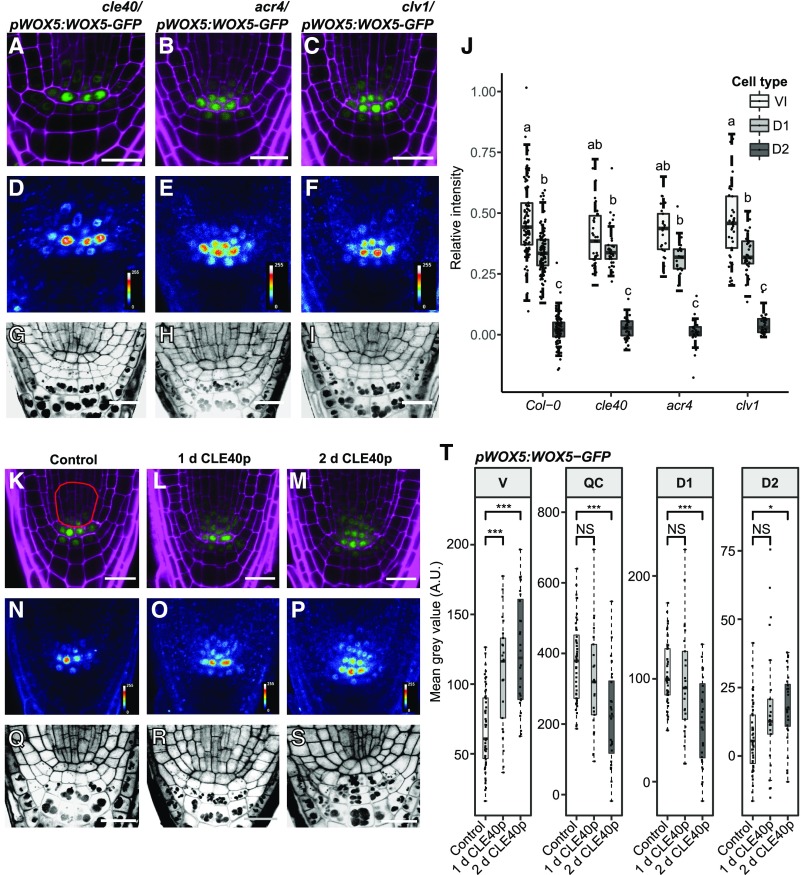
We previously proposed that the CLE40/CLV1/ACR4 module confines WOX5 to the QC and D1 layers and that mutations in the module allow higher expression of WOX5 or movement of the protein to the D2 layer (Stahl et al., 2009, 2013). To test this hypothesis, we compared the localization of WOX5-GFP in Col-0 roots with cle40, acr4, and clv1 mutant backgrounds (Fig. 4, A–F). We compared the pWOX5:WOX5-GFP fluorescence intensity ratios in the D1 and D2 layers to those in the QC in the mutants. The cle40, acr4, and clv1 mutants were characterized by a higher frequency of CSCs at the D2 position (Fig. 4, G–I–I; Supplemental Fig. S2E). However, this CSC identity did not correlate with a significant increase in WOX5-GFP signal intensity compared to the D2 layer of Col-0, which normally differentiates into CCs (Fig. 4J). Furthermore, WOX5-GFP signals in the vascular initials were similar in the mutants and Col-0 (Fig. 4J). We conclude that in the cle40, acr4, and clv1 mutants, extra CSC layers are not caused by the presence of more WOX5 protein in these cells. We also conclude that it is not WOX5 itself, but WOX5-dependent or WOX5-independent factors, that promote CSC identity in the distal root meristem.
Figure 4.
CLE40p up-regulates WOX5 expression in the vascular initials, but WOX5 mobility is not dependent on CLE40p, ACR4, or CLV1. A to I, Images of 5-d-old roots of cle40/pWOX5:WOX5-GFP, acr4/pWOX5:WOX5-GFP, and clv1/pWOX5:WOX5-GFP, showing GFP (green) and PI (magenta) fluorescence (A–C), false color images (D–F), and mPS-PI staining (G–I) of the indicated lines. Scale bars = 20 µm. J, Fluorescence ratio between the indicated regions—vascular initials (VI), D1, D2, and the QC—of the indicated lines. Significant differences (P < 0.05) as determined by two-way ANOVA with Tukey's post hoc analysis are indicated by lowercase letters above the bars (n ≥ 25). K to S, Images of 5-d-old roots of pWOX5:WOX5-GFP under control conditions or treated with CLE40p for 1 or 2 d. Shown are GFP (green) and PI (magenta) fluorescence (K–M), false-color images (N–P), and mPS-PI staining (Q–S) of the indicated lines. The red outline in K indicates the vascular region used for intensity measurements in T. Scale bars = 10 µm. T, Fluorescence intensity depicted as the mean gray value of WOX5-GFP under control conditions and after 1 or 2 d of CLE40p treatment in the indicated cell types. Significant differences as determined by two-sided t test (NS, not significant, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; n ≥ 25). A.U., Arbitrary units; V, vascular region.
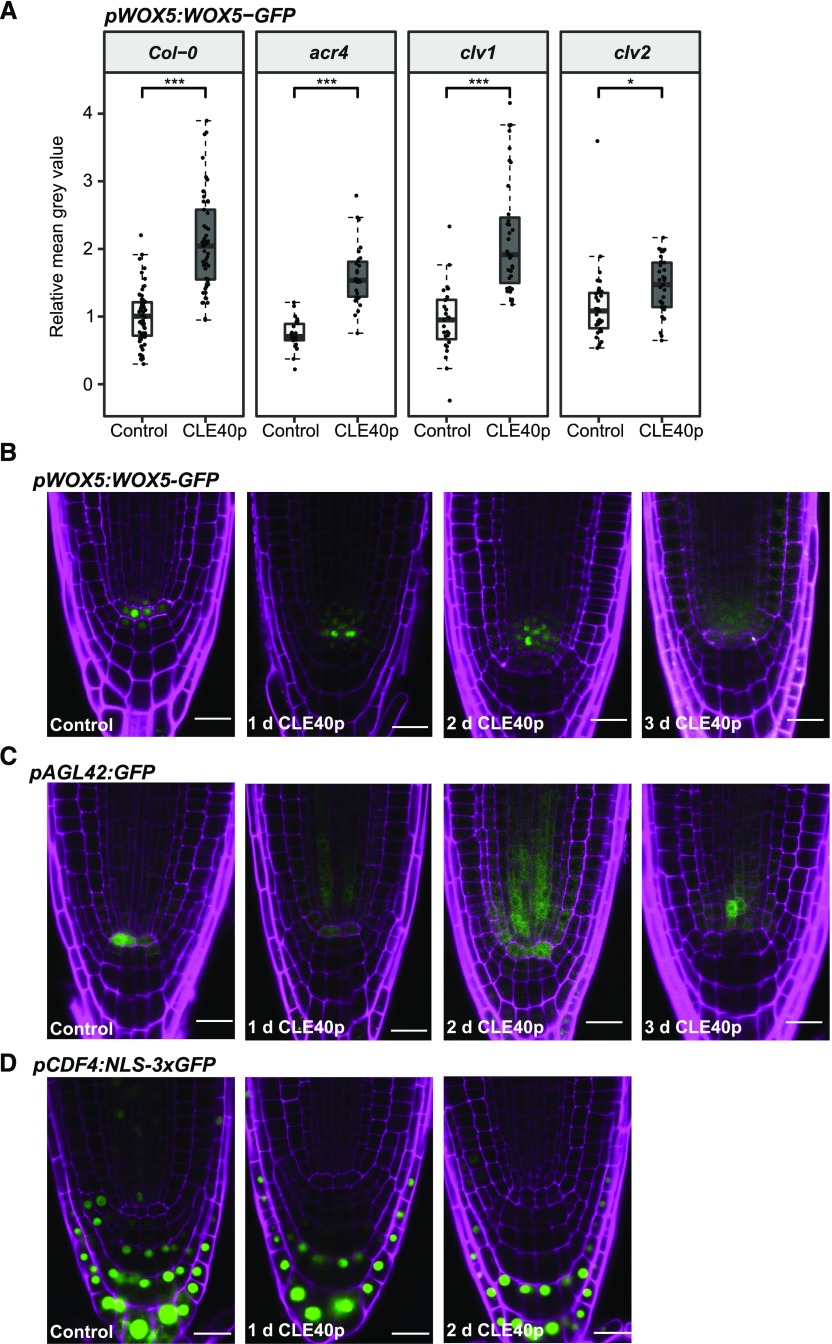
We next tested whether increased signaling through the CLE40/CLV1/ACR4 module can affect WOX5 gene expression or protein localization. Increased CLE40 signaling was previously shown to cause a reduction of WOX5 RNA levels in the QC, a proximal shift of the WOX5 gene expression domain toward the vascular initials, and differentiation of the CSC (Stahl et al., 2013). In this experiment, the effect of increased CLE40 signaling on WOX5-GFP protein levels and localization was analyzed at higher spatial and temporal resolutions, and the fluorescence intensities in the vasculature (Fig. 4T, V), QC, D1, and D2 layers were quantified. The region of interest (ROI) in the vasculature was extended (Fig. 4K, red outline) as treatment with CLE40p visually led to an expansion of the WOX5-GFP domain (Fig. 4, K–Q). The mean gray values were corrected against background levels obtained in the same cell types in CLE40p-treated Col-0 plants. After 1 d of CLE40p treatment, there was a significant increase in WOX5-GFP fluorescence intensity in vascular cells (Fig. 4, L, O, and T), while expression in the QC was not affected, nor was CSC differentiation yet observable (Fig. 4R; Supplemental Fig. S4A). After 2 d of CLE40p treatment, WOX5-GFP levels further increased in the vascular cells (Fig. 4, M, P, and T), but were reduced in both the QC and the D1 layer (Fig. 4T), coinciding with differentiation of the CSC (Fig. 4S; Supplemental Fig. S4A). The increase in WOX5-GFP is at least in part due to an increase in WOX5 expression upon CLE40p treatment, as an increase in expression of the transcriptional reporter line pWOX5:VENUS-H2B was observed as well (Supplemental Fig. S4B). However, we cannot exclude that CLE40p could have an additional effect on WOX5-GFP protein stability.
Our analyses showed that WOX5 mobility is not dependent on CLE40, ACR4, or CLV1, since no change in the distribution of WOX5 could be seen in any of the mutants. Treatment with CLE40p led to a shift of the WOX5 expression domain, most prominently toward the proximal vascular region, as early as 1 d after CLE40p treatment. However, a decrease in GFP signal in QC and D1 only became visible after 2 d of CLE40p treatment, a time point at which ∼50% more treated plants showed starch granule accumulation in the D1 compared to Col-0 plants. This indicates that the disappearance of WOX5 from D1 and QC cells upon increased CLE40 signaling is not the cause of increased columella differentiation, but rather a consequence thereof. Importantly, CLE40 first triggers an increase in WOX5 expression in the vasculature, which precedes its down-regulation in the QC.
CLE40p Leads to a Loss of QC Identity
The first response triggered by CLE40p treatment was increased expression of WOX5 in the vascular cells. Surprisingly, CLE40p treatment also caused a rapid increase of pWOX5:WOX5-GFP expression in the vascular cells of acr4 or clv1 mutants, resembling the response noted in the wild-type background (Fig. 5A). This is consistent with the observation that neither ACR4 nor CLV1 is expressed in the vascular initials, and that other receptors are involved in CLE40p perception in these cells (Stahl et al., 2009). Interestingly, the response to CLE40p was strongly reduced in a clv2 background (Fig. 5A), indicating that CLV2 is involved in the CLE40p-dependent up-regulation of WOX5 expression in the vascular region. The results so far indicated that WOX5 expression was not directly regulated by the CLE40 pathway. To address whether CLE40 interferes with QC identity or position, we compared the responses of pWOX5:WOX5-GFP and pAGL42:GFP, a known QC marker, upon prolonged CLE40p treatment. Similar to the effect on WOX5, CLE40p treatment caused down-regulation of pAGL42:GFP in the QC and up-regulation of pAGL42:GFP in the vascular region within 1 to 2 d (Fig. 5, B and C; Supplemental Fig. S5, A and B). Similar results were obtained with the QC-specific marker QC25:GFP (Supplemental Fig. S6).
Figure 5.
CLV2 is involved in CLE40p-dependent WOX5 up-regulation in the vascular region, and CLE40p leads to a loss in QC identity. A, Fluorescence intensity in the vascular region (outlined in red in Fig. 4K) depicted as the mean gray value of WOX5-GFP under control conditions and after 1 d of CLE40p treatment in the indicated backgrounds transformed with pWOX5:WOX5-GFP. Significant differences were determined by two-sided t test (***P ≤ 0.001, ***P ≤ 0.001, n ≥ 25). B – to D, Images of roots of 5-d-old pWOX5:WOX5-GFP (B), pAGL42:GFP (C), and pCDF4:NLS-3xGFP (D) seedlings under control conditions and after 1, 2, or 3 d of 1 μm CLE40p treatment, showing GFP (green) and PI (magenta) fluorescence. Scale bars = 20 µm.
This suggests that CLE40p first inhibits QC identity and then triggers the formation of a new QC in the vascular region.
CLE40p-Induced Reduction of WOX5 in QC and D1 Does Not Lead to Increased CDF4 Expression
WOX5 was previously shown to repress expression of the differentiation factor CDF4 in CSCs (Pi et al., 2015). CDF4 is expressed in a gradient with the highest expression in the differentiated CCs, the lowest expression in the CSC, and no expression in the QC. In wox5-1 mutants, CDF4 is also expressed in the CSCs and the QC. This leads to a model in which two opposing gradients—of WOX5 and CDF4—pattern the distal root stem cell domain (Pi et al., 2015). However, our experiments so far had already indicated that WOX5 movement is not a key requirement for CSC maintenance. The model would suggest that CLE40p-triggered differentiation of CSCs is mediated by ectopic expression of CDF4 in CSCs, which was tested by measuring the fluorescence intensity of pCDF4:NLS-3XGFP within the QC and the D1–D4 layers under control conditions and upon CLE40p treatment. However, CLE40p-mediated reduction of WOX5 within 2 d in the CSC and the QC did not lead to an increase of CDF4 within these cell types (Fig. 5D; Supplemental Fig. S5C). In contrast, we observed that CLE40p reduced CDF4 expression, most prominently in D4 after 2 d of CLE40p treatment. These results indicated that a derepression of CDF4 via reduction of WOX5 expression is not necessary for differentiation of CSCs. Furthermore, the data suggest that the balance between WOX5 and CDF4 is not a key factor for stem cell maintenance or differentiation.
WOX5-Dependent Dedifferentiation of CCs Requires Signals from the QC
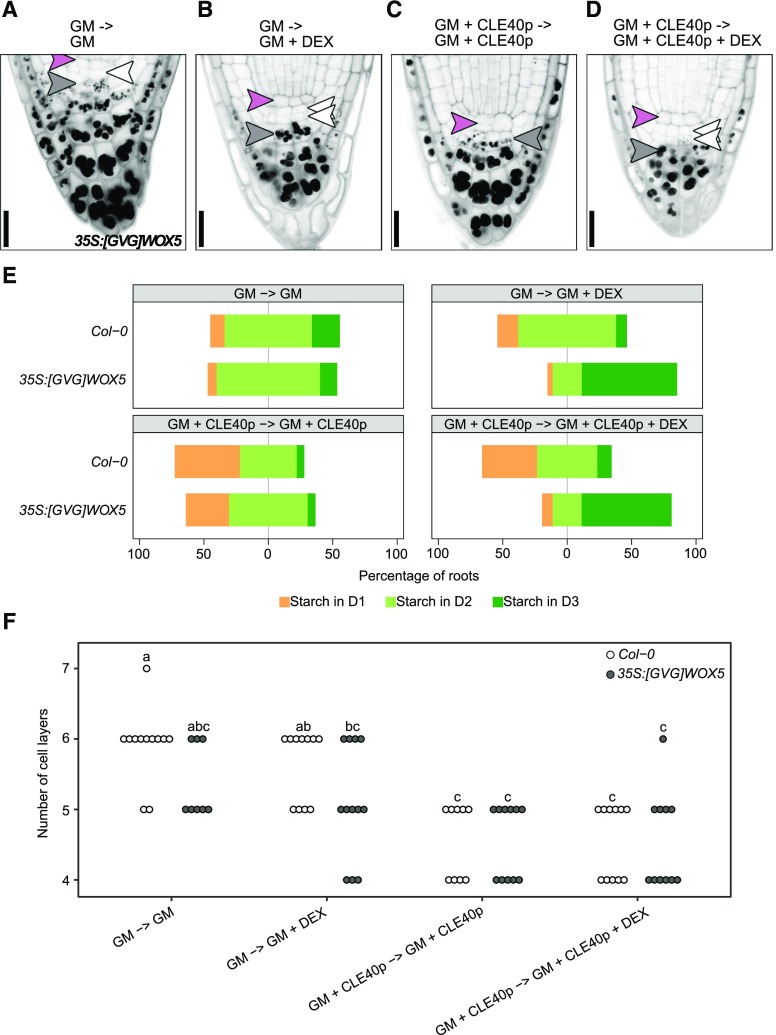
Our results demonstrated that WOX5 is not necessary within the CSCs to maintain stem cell fate. Earlier research by others indicated that high-level expression of WOX5 is sufficient to induce CSC fate (Sarkar et al., 2007). Since treating roots with CLE40p leads to the opposite phenotype (Stahl et al., 2009), we asked how the roles of WOX5 and CLE40 relate to each other. To test this, Col-0 and 35S:[GVG]WOX5 (Sarkar et al., 2007) seedlings were grown for 4 d on medium with or without 1 μm CLE40p. Seedlings were then transferred for 24 h to plates containing the same medium with or without 2 μm DEX to induce WOX5 overexpression. Col-0 seedlings showed an increased degree of CSC differentiation when grown on CLE40p, and were not affected by DEX treatment (Fig. 6E; Stahl et al., 2009). Similar to Col-0, 35S:[GVG]WOX5 seedlings reacted to CLE40p with a rapid differentiation of the CSCs (Fig. 6, A, C, and E). In contrast, when 35S:[GVG]WOX5 seedlings were grown on CLE40p medium with DEX, WOX5 overexpression counteracted any CSC differentiation caused by CLE40p (Fig. 6, B, D, and E) and induced the formation of additional CSCs compared to Col-0. We then asked whether these additional CSCs are derived from additional cell divisions distal to the QC induced by WOX5 overexpression, or from dedifferentiated CCs that were already established before the overexpression commenced. To distinguish between these possibilities, the cell layers distal to the QC were counted. More cell layers over time would be expected if the second CSC layer is derived from additional cell division, whereas no change in total cell layer numbers would be expected if the second CSC layer arises from dedifferentiating CCs. In the 35S:[GVG]WOX5 line, the number of roots carrying two CSC layers increased within 24 h (Fig. 6E), but the total number of cell layers distal to the QC did not (Fig. 6F). Thus, the additional CSC layer is first derived by dedifferentiation of CCs. After only 2 d of growth on the induction (DEX) medium, the overexpression of WOX5 also led to an increase in the number of cells distal to the QC (Supplemental Fig. S7).
Figure 6.
WOX5-dependent dedifferentiation of CCs requires signals from the QC. A to D, Representative images after mPS-PI staining of 35S:[GVG]WOX5 seedlings grown for 4 d on GM without (A and B) or with (C and D) 1 μm CLE40p and then shifted to GM ± CLE40p plates further supplemented with 0 (A and C) or 2 μm (B and D) DEX for 24 h. Pink, white, and gray arrowheads represent QC position, CSC position (D1), and CC position (D2), respectively. Double white arrowheads indicate CSC fate in D2, whereas absence of a white arrowhead indicates CC fate in the D1 position. Scale bars = 20 µm. E, Frequency of roots showing starch granule accumulation in the indicated cell layer after the indicated treatment (n ≥ 30). F, Number of cell layers distal to the QC of the indicated roots. Significant differences (P < 0.05) as determined by two-way ANOVA with Tukey's post hoc analysis are indicated by lowercase letters above bars (n ≥ 25).
We conclude that increased WOX5 expression initially triggers dedifferentiation and that later additional cell divisions generate additional CSC layers. Importantly, only CCs next to the CSCs can dedifferentiate (Supplemental Fig. S7), indicating that ectopic WOX5 expression alone is not sufficient for CSC fate and that dedifferentiation is only triggered in those cells that receive additional signals from the QC.
DISCUSSION
Non-cell-autonomous activities of transcription factors play important roles in diverse plant signaling processes (Gallagher et al., 2014) and are important for controlled formative cell divisions and the regulation of stem cell homeostasis (Stahl et al., 2013; Daum et al., 2014; Pi et al., 2015; Kitagawa and Jackson, 2017). Movement of transcription factors from cell to cell is mediated by PDs, but how their mobility and that of other informational molecules such as small RNAs is controlled is not understood (Skopelitis et al., 2018). The discovery of receptor-like kinases (RLKs) that are localized at PDs and regulate cell fate decisions stimulated the development of explanatory models that integrated apoplastic signals with selective symplastic trafficking. For example, the spread of an immune response based on small interfering RNAs can be interrupted by viral proteins such as the C4 protein of the tomato yellow leaf curl virus, which was shown to bind the intracellular domains of the PD-localized RLKs BAM1 and BAM2, which are homologs of the stem cell regulatory RLK CLV1 (Rosas-Diaz et al., 2018). Bacterial pathogens trigger the modification of PD functions through the pathogen-associated molecular pattern-dependent activation of the RLKs FLS2 and LYM2 (Stahl and Faulkner, 2016). The STRUBBELIG RLK, together with the C2-domain protein QUIRKY, interact at PDs where they mediate communication between adjacent meristem cell layers, possibly via selective control of PD conductivity (Vaddepalli et al., 2014, 2017). In the root meristem, CSC fate in the cell layer abutting the QC was shown to depend on the CLE40/CLV1/ACR4 signaling pathway, involving two RLKs that accumulate and interact at PDs, and the antagonistic activities and complementary expression patterns of the transcription factors CDF4 and WOX5 (Pi et al., 2015). We now investigated whether CLE40 signaling affects the connectivity within the root stem cell domain, and whether mobility of the hub transcription factor WOX5 is regulated by the CLE40/CLV1/ACR4 signaling module.
CLE40 Signaling Does Not Affect General Conductivity of PDs
Our experiments using pSUC2:GFP and 35S:DRONPA-s reporter lines did not provide clear evidence that CLE40p influences cell-to-cell connectivity within the root meristem. Unloading of GFP expressed in phloem companion cells is often used as an assay system for symplastic connectivity. However, both increased and decreased CLE40 signaling drastically affected general root morphology by interfering with phloem development through a CLV2/CRN-dependent pathway (Hazak et al., 2017), which confounded any potential role in regulation of PD selectivity or function. DRONPA-s can be toggled between its fluorescent on and off states in a cell type-specific manner, permitting the analysis of symplastic connectivity with high spatial and temporal resolution (Gerlitz et al., 2018). We incubated root tissue for only 1 d with CLE40p, since longer treatments lead to a disruption of stem cell niche organization, which again would make it difficult to assign any observed effect to a function of CLE40p in the regulation of cell connectivity. For DRONPA-s, no statistically significant differences between CLE40p-treated and control roots was observed, indicating that CLE40 signaling cannot control connectivity in general, but must have more specific (if any) functions at the PDs. Since CLE40 had been implicated in the regulation of CSC fate, we next investigated whether CLE40p alters the mobility of WOX5 in its movement from the QC toward the CSC.
WOX5 Movement Is Not Controlled by the CLE40/ACR4/CLV1 Signaling Module
WOX5 was previously proposed to move from its site of synthesis in the QC to the distal cell layer and induce CSC fate at this position (Pi et al., 2015). Using a WOX5-GFP expressed from the WOX5 promoter, we quantified WOX5 protein amounts using the GFP signal intensity as a proxy, and found that the majority of the WOX5-GFP localized to the QC, and a minor amount to the CSC layer of wild-type plants. In cle40, acr4, or clv1 mutant lines carrying an extra layer of CSCs, WOX5-GFP levels were not increased, and WOX5-GFP was absent in the more distal CSCs, indicating that WOX5-GFP is not required in CSCs, and that the increased stemness of cle40, acr4, and clv1 mutants is caused by other factors. This is in agreement with the observation that in a cle40 wox5-1 double mutant, the wox5-1 mutant phenotype is partially rescued, indicating that even in the absence of WOX5, CSC layers can still be formed (Bennett et al., 2014; Richards et al., 2015). Application of CLE40p led to decreased WOX5-GFP expression in CSCs, but this was notable only after 2 d of CLE40p treatment, by which time most of the CSCs were differentiated (Stahl et al., 2009). Thus, reduced WOX5 expression is rather a consequence and not causal for differentiation of CSCs to CCs. Furthermore, after CLE40p treatment we observed a proximal shift of WOX5-GFP protein localization toward the vascular initials, as described previously (Stahl et al., 2009). Due to the high connectivity of vascular cells, increased amounts of WOX5 protein would cause the protein to spread out into a larger domain. Importantly, we found that this increased expression in the vasculature of WOX5 upon CLE40p treatment did not depend on the ACR4 receptor, but on CLV2.
These results allow the following conclusions. First, the concept that WOX5 mobility from the QC toward the distal stem cells is required for CSC identity cannot explain our findings that extra CSC layers are present in cle40, acr4, or clv1 mutants, without increased levels of WOX5 protein in these cells. Second, WOX5 expression in the QC is not directly repressed by CLE40 signaling; instead, WOX5 down-regulation is a consequence of CLE40 affecting WOX5 regulators. Third, CLE40 can positively regulate WOX5 expression and the establishment of a new QC from vascular initials via a CLV2-dependent signaling pathway.
Movement of WOX5 from the QC Is Not Necessary to Maintain Distal Stemness
Our experiments showed that the presence of WOX5 protein only in the QC is sufficient for CSC maintenance. This is in strong contradiction to previously reported experiments, which suggested that the movement of WOX5 to the CSC is necessary for its stem cell fate. We found that fusing WOX5 to a 2xGFP tag was sufficient to prevent detectable amounts of WOX5 movement toward the CSC; nevertheless, this construct was able to rescue the wox5-1 CSC and QC mutant phenotypes. Pi et al. (2015) had used a 3xYFP fusion protein to study the necessity of WOX5 mobility, and stated that WOX5-3xYFP is immobile. When we repeated this experimental approach, we observed that the WOX5-3xGFP fusion protein is only partially able to complement the typical QC defects of wox5-1 mutants, indicating that this huge tag not only might restrict mobility, but could also interfere with some of the WOX5 functions, for example, its capacity to interact with itself or with other transcription factors, such as the HAM proteins or TPL (Long et al., 2002; Busch et al., 2010; Zhou et al., 2015). The Mr of GFP or YFP is similar to that of WOX5, and adding a large tag three times the size of the transcription factor not only may render the protein immobile, but could also inhibit protein-protein interactions, and therefore function. We cannot entirely exclude that small (undetectable) amounts of WOX5 are released from the fusion proteins, and are thereby able to move to the CSC and rescue the CSC phenotype in wox5-1 mutants. However, in this case we would expect that WOX5 would be cleaved off the 3xGFP tag as well, and hence would also be able to fully rescue the wox5-1 CSC phenotype. Therefore, based on the partial rescue of the wox5-1 CSC phenotype with WOX5-3xGFP, in contrast to a full rescue for the WOX5-2xGFP fusion protein, and the fact that both WOX5-2xGFP and WOX5-3xGFP are restricted to the same cells, we believe that the difference in rescuing the wox5-1 CSC phenotype is caused by interference with WOX5 function due to the massive size of the fluorescent tag. We also observed that ectopic high-level expression (so that mobility of the protein is not required) of a WOX5 protein in a wox5-1 mutant background caused overproliferation of the distal root meristem when WOX5 was fused to a 1xGFP tag, but not in fusions with 3xGFP, indicating that size matters and that very large fusion proteins cannot exert normal functions.
In order to further explore possible roles for WOX5 mobility, we created a minimal version of the WOX5 protein (MINI-WOX5), similar to the approach taken previously for WUS (Daum et al., 2014). The conserved homeodomain, WUS box, and EAR domain were retained in MINI-WOX5, while interdomains where exchanged by linkers. Expression of MinimeWUS from the WUS promoter had caused overproliferation of the shoot apical meristem, which was assigned to a more extended movement of MinimeWUS compared to the endogenous WUS (Daum et al., 2014). Although MINI-WOX5-GFP was able to rescue the wox5-1 CSC phenotype, the QC was often disorganized, indicating that MINI-WOX5 was not fully functional. MINI-WOX5-GFP, however, showed a more equal intracellular distribution between the nucleus and cytoplasm, which was not reported for MinimeWUS, and MINI-WOX5-GFP localized similar to WOX5-GFP. Furthermore, no overproliferation of root cells was observed. We conclude that the mobility of MINI-WOX5-GFP was not increased by the amino acid replacements. The missing interdomains might be involved in protein-protein interactions or posttranslational modifications, which are necessary for full WOX5 function. We also observed that the amount of MINI-WOX5-GFP (judged by analyses of fluorescence intensities) was higher than that of WOX5-GFP, which could indicate that MINI-WOX5-GFP is less prone to rapid turnover.
Our results strongly indicate that WOX5 presence in the QC is sufficient for CSC maintenance, although WOX5 can move through PDs into adjacent cells, which was also observed for other transcription factors (Lozano-Elena et al., 2018). While WOX5 acts within the QC to repress CYCD3;3 and CYCD1;1 expression, and thereby cell divisions, it contributes to CSC maintenance by keeping CSCs in a division-active state (Forzani et al., 2014).
Stem Cell Fate Is Regulated by Multiple Intertwined Signaling Pathways
CLE40 and WOX5 act antagonistically upon CSC differentiation; however, our misexpression experiments uncovered that these functions are context-dependent. Increased CLE40 signaling caused starch granule accumulation also in the CSCs, converting them into CCs, but WOX5 expression triggered dedifferentiation only in CCs abutting the QC, indicating that other QC-derived factors are required. More CSC layers were then generated by proliferation of the existing CSCs. Again, these results suggest a prominent role for WOX5 to set the divisional status of root stem cells.
The displacement of WOX5 expression toward the vascular initials upon CLE40p treatment coincides with a proximal shift of other QC specific markers, such as AGL42. Similarly, destruction of the QC by laser ablation caused the underlying CSCs to differentiate, and formation of a new QC in the proximal vascular cells (van den Berg et al., 1997; Sabatini et al., 1999). It is plausible that prolonged CLE40p treatment leads to a loss of QC cell fate, which is compensated by the root through formation of a new QC. The change in WOX5 expression and localization is then a secondary effect, whereby the loss of QC identity might be the triggering factor for CSC differentiation.
The differentiation factor CDF4 was identified as a direct target of repression by WOX5. A gradient model was proposed where high levels of WOX5 in the QC and CSC maintain stem cell fate, while CDF4 accumulates in CCs and promotes their differentiation. Consistent with this model, CFD4 is ectopically expressed in the QC in the wox5-1 mutant background. The opposing gradient between CDF4 and WOX5 is, however, not maintained upon CLE40p treatment. The CLE40p-induced proximal shift of WOX5 expression and CSC differentiation did not coincide with a shift in the CDF4 gradient toward CSCs and/or the QC, indicating that CDF4 is still regulated by other factors as well and that derepression of CDF4 is not necessary for differentiation of CSCs.
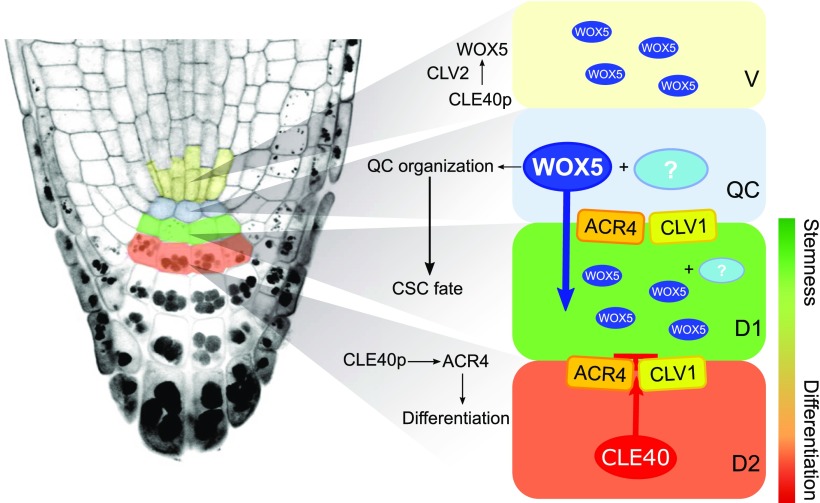
Our results and conclusions require a reevaluation of current models for root stem cell homeostasis (Fig. 7). We observed that WOX5, like many other transcription factors, is mobile and can therefore move between cells that are connected via PDs, but we found no conclusive evidence that WOX5 mobility is necessary for CSC maintenance. Moreover, WOX5 mobility is not influenced by CLE40p/CLV1/ACR4. We conclude that WOX5 acts within the QC to maintain its identity by repressing cell divisions, as previously reported, and regulates CSC maintenance in combination with other factors, for example by controlling the production of another, non-cell-autonomous signal. Overall, the CLE40 pathway antagonizes the activity of the QC, so that increased signaling of CLE40 triggers the loss of QC identity and the establishment of a new QC at the site of the vascular initials. Regional expression of different receptor kinases (ACR4 and CLV1 in the distal stem cell niche, CLV2 and CRN in the proximal domain) results in specific interpretations of the CLE40 signal, which acts as a feedback signal from differentiating cells to fine-tune the activity and position of the QC. Finally, the proposed gradient model where the opposing levels of WOX5 and CDF4 determine stem cell homeostasis does not hold up when CLE40 signaling is increased, since absence of WOX5 does not cause up-regulation of CDF4, indicating that CDF4 does not participate in cell differentiation in this scenario.
Figure 7.
Model summarizing the findings presented in this work. WOX5 acts within the QC, maybe together with an unknown factor(s), to maintain its organization. WOX5 presence within the QC is sufficient to regulate CSC maintenance, probably in combination with other factors, whose activity or expression might depend on WOX5 and could act as a non-cell-autonomous signal. WOX5 can move through PDs into adjacent cells, where its exact function is not clear. The CLE40p/CLV1/ACR4 signaling pathway overall antagonizes the activity of the QC, promoting CSC differentiation. Increased signaling of CLE40 leads to an up-regulation of WOX5 expression in a new vascular domain, which is dependent on the CLV2 receptor. This allows establishment of a new QC at the sites of the vascular initials upon loss of QC identity after prolonged CLE40 signaling. V, vascular region.
Our work presented here supports the view that stem cell regulation is not locally controlled by single entities, but rather is achieved by a complex interacting network of multiple pathways (Rahni et al., 2016). A main feature of this emerging model is the antagonistic activity of two opposing axes that control each other’s differentiation, and a distributed regulation of localized stem cells. The search for a single stemness-controlling factor might therefore bear no fruits.
Finally, how the CLE40p/CLV1/ACR4 signaling pathway contributes to stem cell control, and the role of receptor kinases localized at PDs is still not understood. Single cell profiling approaches of the root stem cell niche seem to be a promising approach to obtain some fresh answers to these questions.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) and the T-DNA insertion lines clv1-20 (clv1; SALK008670), acr4-2 (acr4; SAIL_240_B04), wox5-1 (SALK_038262), and cle40-2 (cle40), clv2-gabi (clv2; GK-686A09), which were described earlier and obtained from the Nottingham Arabidopsis Stock Centre (NASC; Gifford et al., 2003; Sarkar et al., 2007; Stahl et al., 2009; Kleinboelting et al., 2012). The pCDF4:NLS-3xGFP construct was described previously and provided by Thomas Laux (Pi et al., 2015). pAGL42:GFP was provided by Tom Beeckman (Nawy et al., 2005). pSUC2:GFP and 35S:DRONPA were provided by Ruth Stadler (Stadler et al., 2005b; Gerlitz et al., 2018).
Arabidopsis seeds were surface-fume sterilized in a sealed container with 100 mL bleach (chlorine gas) supplemented by 3 mL of 37% (v/v) HCl for 3 h, then suspended in 0.1% (w/v) agarose and plated on a growth medium (GM) consisting of half-strength Murashige and Skoog (MS) salts (Duchefa), 1% (w/v) Suc, 0.8% (w/v) plant agar, and MES (2-(N-Morpholino)ethanesulfonic acid; pH 5.8). The seeds were stratified for 2 d at 4°C in a dark room, and then placed vertically in a growth chamber under constant light conditions at 22°C. For CLE40p (RQVPTGSDPLHHK; the underlined P is Hyp) treatments, seeds were germinated on GM for 3 d and transferred to medium supplemented with 1 μm CLE40p or to MS plates for control conditions. For DEX treatment, seedlings were transferred to media containing 2 μm DEX or to control medium after 3 d of germination. For mPS-PI experiments with the 35S:[GVG] WOX5 line, seedlings were grown on MS medium for 4 d, supplemented with either no peptide or 1 μm CLE40p. Seedlings were then transferred to plates containing MS supplied with either no peptide and 2 μm DEX or 2 μm DEX and 1 μm CLE40p for 24 h. For the Lugol staining experiment, the 35S:[GVG]WOX5 seedlings were grown on GM for 2 d, after which they were transferred to plates containing GM supplied with 2 μm DEX or 2 μm DEX + 1 μm CLE40p.
Cloning and Generation of Transgenic Lines
Standard molecular biology protocols and Gateway (Invitrogen) technology were followed to obtain expression clones. Open reading frames were amplified from a genomic DNA template with Pfu DNA Polymerase (Promega). All primers used for open reading frame and promoter isolation are listed in Supplemental Table S1.
For creating the pWOX5:WOX5-GFP construct, 4,603 bp upstream of the start codon was cloned into the pENTR-5′-TOPO vector according to the pENTR 5′-TOPO TA cloning kit (Invitrogen) to obtain pENTR-5′-TOPO-pWOX5. The WOX5 genomic region was cloned into pENTR/D-TOPO according to the pENTR/D-TOPO Cloning Kit (Invitrogen) to generate pENTR-TOPO-WOX5. P2R-P3-GFP was obtained from PSB-UGent. The different entry modules were recombined by multisite Gateway LR cloning (Invitrogen) with pKm34GW,0 (PSB-UGent). The structure and sequence of P2R-P3:GFP and pKm34GW,0 were as described (Karimi et al., 2002, 2007) and are accessible online (http://www.psb.ugent.be/gateway/).
The pWOX5:VENUS-H2B and pWOX5:WOX5-2xGFP constructs were built by PCR amplification of a 4,603-bp fragment upstream of the start codon of WOX5 and cloning by restriction and ligation via an AscI site into either pMDC32 (Curtis and Grossniklaus, 2003) or a modified MDC32 with C-terminal 2xeGFP in frame after the attR2 site, respectively. The WOX5 coding region without stop codon and VENUS-H2B were inserted downstream of the promoter by Gateway cloning from the pENTR-TOPO-WOX5 and a pENTR-TOPO-VENUS-H2B entry clone, respectively.
MINI-WOX5-GFP was synthesized and cloned into pDONR221 by GeneArt (Invitrogen), according to the design of MINIMe, as described before (Daum et al., 2014). All nonconserved sequences were replaced by structurally unrelated linkers, with the region N-terminal of the homeodomain (amino acids 1–21) exchanged for the first 9 aa of GFP to ensure efficient protein translation and stability and with the stretches between the homeodomain and WUS-Box and EAR-like domain replaced by Ser-Gly linkers. pDONR221-MINI-WOX5-GFP was cloned by Gateway cloning into the modified pMDC32 vector with N-terminal pWOX5 inserted into the AscI cloning site.
The pWOX5:WOX5-3xGFP, p35S:WOX5-GFP-GR, and p35S:WOX5-3xGFP-GR constructs were designed through Greengate cloning as described before (Lampropoulos et al., 2013). For this, the promoter of WOX5 was cloned into pGGA000, from which the BsaI restriction sites were removed by PCR-based mutagenesis. Briefly, the 5′TOPO-ENTRY clone containing the pWOX5 promoter was amplified with primers bearing the mutated BsaI sites. After degradation of the methylated (parental) DNA with DpnI (1 h at 37°C), the mutated plasmid was transformed in Escherichia coli, and the presence of the mutation was confirmed by sequencing. The genomic coding region of WOX5 was cloned into both pGGB00 and pGGC00, where a BsaI restriction site was also removed by PCR-based mutagenesis. pGGD-GR was designed by amplifying GR from the pGEM-GAL4-VP16-GR vector available in the lab. pGGAD04 (35S), pGGC025 (3xGFP), pGGC014 (GFP), pGGE009 (UBQ10 terminator), pGGF005 (pUBQ10:HygrR), pGGD002 (d-dummy), and the destination vector pGGZ001 were obtained from the Lohmann lab (Lampropoulos et al., 2013).
Subsequent transformation of Arabidopsis Col-0 plants was carried out with the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on MS medium containing hygromycin (15 mg/mL) or kanamycin (50 mg/mL). All experiments were performed with at least two independent transgenic lines.
For crossing Arabidopsis, the heterozygous F1 seeds were grown on soil, allowed to self, and harvested. The F2 plants were analyzed for mutant alleles and fluorescent markers. All experiments were performed on plants homozygous for the mutant allele and fluorescent marker.
Tissue Staining and Microscopy
For GUS staining, seedlings were incubated in 80% (v/v) acetone for 30 min, washed in phosphate buffer, and incubated in the GUS staining solution (1 mg/mL of 5-bromo-4-chromo-3-indolyl β-d-glucuronide, 2 mm ferricyanide, and 0.5 mm ferrocyanide in 100 mm phosphate buffer [pH 7.4]) at 37°C in the dark for 2 h. The material was cleared in 70% (v/v) ethanol and a chloral hydrate-10% (v/v) glycerol solution and examined under a light microscope.
mPS-PI stainings were done according to the mPS-PI method (Truernit et al., 2008) and imaged with a 40× water objective with a NA of 1.20 using a Zeiss LSM 780 laser scanning microscope. PI was excited with a 561 nm Argon laser with emission detection at 566–718 nm. mPS-PI images were used to score starch accumulation and QC defects.
The different reporter lines with GFP or VENUS fluorophores were examined with a 40× water objective with a NA of 1.20 using the Zeiss LSM 780. Yellow fluorescence was excited using a 514 nm Argon laser and emission was detected between 519 and 620 nm. Green fluorescence was excited with a 488 nm Argon laser with emission detection at 490–544 nm. Counterstaining with PI (10 μm) was performed before mounting the samples on the slides. PI was excited with a 561 nm Argon laser with emission detection at 566–718 nm.
For DRONPA experiments, DRONPA-s was first deactivated by irradiation with 488‐nm laser light for 30 s at 70% total laser intensity (argon laser, ∼20 mW, Leica Microsystems). DRONPA activation occurred by setting the ROI in the center of the target cell without contacting the cell wall, to avoid fluorescence activation in adjacent cells. The ROIs were exposed to 800 nm light for 20 to 40 s using two‐photon equipment (Mai Tai HP two‐photon laser, Newport Spectra‐Physics). DRONPA-s detection occurred immediately after activation and after 5 min of single-cell activation. DRONPA-s was excited with 488‐nm laser light (argon laser) and emission detection occurred at 500–541 nm. The pinhole was kept at Airy 1. Saturation was avoided during all experiments. Successful single‐cell activation was controlled optically during all experiments. Counterstaining with PI (10 μm) was performed before mounting the samples on the slides. PI was excited with a 561 nm Argon laser with emission detection at 566–718 nm.
Data Analysis
Fluorescence intensity was determined using Fiji software (https://fiji.sc/) on images acquired with a single in-focus plane using equal laser power, detector gain, and pinhole. A ROI covering the cell type was drawn to measure the mean gray value. For background subtractions, the average mean gray value (for 10 roots) was determined in roots without fluorescent signal in the same region.
All plots were created in R (R Core Team, 2018). Diverged stacked bar charts for starch granule accumulation were created with the likert function of the R package “HH” (Heiberger and Robbins, 2014). Box plots and bar charts were created with the ggplot2 package (Wickham, 2016). The box plots indicate the 25th, 50th, and 75th percentiles, whereby the whiskers are extended to the most extreme data point that is no more than 1.5× the interquartile range from the edge of the box (Tukey style).
For statistical significance tests, the two-tailed Student's t test with the indicated P-value was done using the geom_signif() function within the ggsignif package (Ahlmann-Eltze, 2017). For QC defects, the sd was calculated as sd = sqrt(p * (1 − p)/n). Significance values were calculated using the Z-test, which was done by using the prop.test function, with a confidence interval of 95%, within the “stats base” package of R. To perform two-way ANOVA tests using the aov() function, the stats base package of R was used, after which groups were determined with a Tukey’s test as post hoc analysis (P < 0.05) using the HSD.test function within the agricolae package (De Mendiburu, 2017).
For all experiments, n denotes the minimum amount of samples/measurements that were used to perform the statistical analysis. All experiments were done with at least two independent lines and repeated at least twice, yielding comparable results. Results are shown for one or two independent lines, as including more lines would greatly distort the readability of the graphs.
Accession Numbers
Sequence data for the genes used in this article can be found in The Arabidopsis Information Resource (TAIR) database under the following accession numbers: AT3G11260 (WOX5), AT5G12990 (CLE40), AT3G59420 (ACR4), AT1G75820 (CLV1), AT1G65380 (CLV2), AT2G34140 (CDF4), and AT1G22710 (SUC2).
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Overview and results of the DRONPA-s fluorophore experiments.
Supplemental Figure S2. Fluorescence intensity values of the different WOX5 constructs, confocal images of wox5-1 complemented with pWOX5:WOX5-3xGFP, and starch granule accumulation in cle40, acr4, and clv1 lines.
Supplemental Figure S3. Confocal images and number of cell layers of DEX and dimethyl sulfoxide-treated Col-0 and wox5-1 lines containing the WOX5-1xGFP-GR or WOX5-3xGFP-GR construct.
Supplemental Figure S4. Starch granule accumulation upon CLE40p treatment and up-regulation of pWOX5:VENUS upon CLE40p treatment.
Supplemental Figure S5. Fluorescence intensity values in pAGL42:GFP, pWOX5:WOX5-GFP, and pCDF4:NLS-3xGFP lines after different days of CLE40p treatment.
Supplemental Figure S6. Fluorescence intensity values in QC25:GFP after different durations of CLE40p treatment.
Supplemental Figure S7. Confocal images of 35S:[GVG]WOX5 lines after DEX and DMSO treatment.
Supplemental Table S1. List of oligonucleotides.
Acknowledgments
We are grateful to Karine Pinto, Cornelia Gieseler, Silke Winters, and Carin Theres for technical support. We thank Ruth Stadler for sharing the pSUC2:GFP and 35S:DRONPA lines. We thank Thomas Laux for sharing the pCDF4:NLS-3xGFP and 35S:[GVG]WOX5 lines. We furthermore acknowledge the Center of Advanced Imaging (CAi) at the Heinrich Heine University Düsseldorf for help with microscopy.
Footnotes
This work was supported by the Alexander von Humboldt Foundation (B.B.) and the Deutsche Forschungsgemeinschaft (Si947/10-1).
References
- Ahlmann-Eltze (2017) Package “ggsignif” Type Package Title Significance Brackets for “ggplot2”. R package version 0.6.0. https://CRAN.R-project.org/package=ggsignif
- Aichinger E, Kornet N, Friedrich T, Laux T (2012) Plant stem cell niches. Annu Rev Plant Biol 63: 615–636 [DOI] [PubMed] [Google Scholar]
- Ando R, Mizuno H, Miyawaki A (2004) Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science 306: 1370–1373 [DOI] [PubMed] [Google Scholar]
- Bennett T, van den Toorn A, Willemsen V, Scheres B (2014) Precise control of plant stem cell activity through parallel regulatory inputs. Development 141: 4055–4064 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A, et al. (2010) Transcriptional control of a plant stem cell niche. Dev Cell 18: 849–861 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Medzihradszky A, Suzaki T, Lohmann JU (2014) A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc Natl Acad Sci USA 111: 14619–14624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mendiburu F. (2017) Agricolae: Statistical Procedures for Agricultural Research. R Packag version 12-8. https//CRANR-project.org/package=agricolae 153
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Forzani C, Aichinger E, Sornay E, Willemsen V, Laux T, Dewitte W, Murray JAH (2014) WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr Biol 24: 1939–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KL, Sozzani R, Lee C-M (2014) Intercellular protein movement: Deciphering the language of development. Annu Rev Cell Dev Biol 30: 207–233 [DOI] [PubMed] [Google Scholar]
- Gerlitz N, Gerum R, Sauer N, Stadler R (2018) Photoinducible DRONPA-s: A new tool for investigating cell-cell connectivity. Plant J 94: 751–766 [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean S, Ingram GC (2003) The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development 130: 4249–4258 [DOI] [PubMed] [Google Scholar]
- Greb T, Lohmann JU (2016) Plant stem cells. Curr Biol 26: R816–R821 [DOI] [PubMed] [Google Scholar]
- Habuchi S, Ando R, Dedecker P, Verheijen W, Mizuno H, Miyawaki A, Hofkens J (2005) Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc Natl Acad Sci USA 102: 9511–9516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazak O, Brandt B, Cattaneo P, Santiago J, Rodriguez-Villalon A, Hothorn M, Hardtke CS (2017) Perception of root-active CLE peptides requires CORYNE function in the phloem vasculature. EMBO Rep 18: 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiberger RM, Robbins NB (2014) Design of diverging stacked bar charts for Likert scales and other applications. J Stat Softw 57: 1–3225400517 [Google Scholar]
- Imlau A, Truernit E, Sauer N (1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P, Hilson P (2007) Recombinational cloning with plant gateway vectors. Plant Physiol 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Jackson D (2017) Plasmodesmata-mediated cell-to-cell communication in the shoot apical meristem: How stem cells talk. Plants (Basel) 1: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J (2013) GreenGate—A novel, versatile, and efficient cloning system for plant transgenesis. PLoS One 8: e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Woody S, Poethig S, Meyerowitz EM, Barton MK (2002) Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development 129: 2297–2306 [DOI] [PubMed] [Google Scholar]
- Lozano-Elena F, Planas-Riverola A, Vilarrasa-Blasi J, Schwab R, Caño-Delgado AI (2018) Paracrine brassinosteroid signaling at the stem cell niche controls cellular regeneration. J Cell Sci 131: jcs204065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lummer M, Humpert F, Steuwe C, Caesar K, Schüttpelz M, Sauer M, Staiger D (2011) Reversible photoswitchable DRONPA-s monitors nucleocytoplasmic transport of an RNA-binding protein in transgenic plants. Traffic 12: 693–702 [DOI] [PubMed] [Google Scholar]
- Meyer MR, Shah S, Zhang J, Rohrs H, Rao AG (2015) Evidence for intermolecular interactions between the intracellular domains of the Arabidopsis receptor-like kinase ACR4, its homologs and the Wox5 transcription factor. PLoS One 10: e0118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee J-Y, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Pallakies H, Simon R (2014) The CLE40 and CRN/CLV2 signaling pathways antagonistically control root meristem growth in Arabidopsis. Mol Plant 7: 1619–1636 [DOI] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T (2015) Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell 33: 576–588 [DOI] [PubMed] [Google Scholar]
- Pierre-Jerome E, Drapek C, Benfey PN (2018) Regulation of division and differentiation of plant stem cells. Annu Rev Cell Dev Biol 34: 289–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ [Google Scholar]
- Rahni R, Efroni I, Birnbaum KD (2016) A case for distributed control of local stem cell behavior in plants. Dev Cell 38: 635–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Wink RH, Simon R (2015) Mathematical modelling of WOX5- and CLE40-mediated columella stem cell homeostasis in Arabidopsis. J Exp Bot 66: 5375–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Diaz T, Zhang D, Fan P, Wang L, Ding X, Jiang Y, Jimenez-Gongora T, Medina-Puche L, Zhao X, Feng Z, et al. (2018) A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc Natl Acad Sci USA 115: 1388–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Elliott TJ, Jensen KH, Haaning KS, Wager BM, Knoblauch J, Howell AH, Mullendore DL, Monteith AG, Paultre D, Yan D, et al. (2017) Phloem unloading in Arabidopsis roots is convective and regulated by the phloem-pole pericycle. eLife 6: e24125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Skopelitis DS, Hill K, Klesen S, Marco CF, von Born P, Chitwood DH, Timmermans MCP (2018) Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat Commun 9: 3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Lauterbach C, Sauer N (2005a) Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol 139: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Wright KM, Lauterbach C, Amon G, Gahrtz M, Feuerstein A, Oparka KJ, Sauer N (2005b) Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. Plant J 41: 319–331 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Faulkner C (2016) Receptor complex mediated regulation of symplastic traffic. Trends Plant Sci 21: 450–459 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, Kühnemuth R, Weidtkamp-Peters S, Pinto KG, Kirschner GK, Schmid JB, Wink RH, Hülsewede A, et al. (2013) Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol 23: 362–371 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Simon R (2013) Gated communities: Apoplastic and symplastic signals converge at plasmodesmata to control cell fates. J Exp Bot 64: 5237–5241 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Simon R (2010) Plant primary meristems: Shared functions and regulatory mechanisms. Curr Opin Plant Biol 13: 53–58 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Simon R (2005) Plant stem cell niches. Int J Dev Biol 49: 479–489 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R (2009) A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui J-C (2008) High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaddepalli P, Fulton L, Wieland J, Wassmer K, Schaeffer M, Ranf S, Schneitz K (2017) The cell wall-localized atypical β-1,3 glucanase ZERZAUST controls tissue morphogenesis in Arabidopsis thaliana. Development 144: 2259–2269 [DOI] [PubMed] [Google Scholar]
- Vaddepalli P, Herrmann A, Fulton L, Oelschner M, Hillmer S, Stratil TF, Fastner A, Hammes UZ, Ott T, Robinson DG, et al. (2014) The C2-domain protein QUIRKY and the receptor-like kinase STRUBBELIG localize to plasmodesmata and mediate tissue morphogenesis in Arabidopsis thaliana. Development 141: 4139–4148 [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B (1997) Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Laux T, Rensing SA (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof Y-D, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21: 1144–1155 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2016) Ggplot2 : Elegant Graphics for Data Analysis. Springer-Verlag, New York [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM (2015) Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517: 377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]