Xylem cell expansion and fiber intrusive tip growth require the activity of enzymes that rearrange xyloglucan in cell walls, and their deficiency not only affects cell expansion but alters subsequent secondary wall formation via cell wall integrity-sensing mechanisms.
Abstract
Xyloglucan is the major hemicellulose of dicotyledon primary cell walls, affecting the load-bearing framework with the participation of xyloglucan endo-transglycosylase/hydrolases (XTHs). We used loss- and gain-of function approaches to study functions of XTH4 and XTH9 abundantly expressed in cambial regions during secondary growth of Arabidopsis (Arabidopsis thaliana). In secondarily thickened hypocotyls, these enzymes had positive effects on vessel element expansion and fiber intrusive growth. They also stimulated secondary wall thickening but reduced secondary xylem production. Cell wall analyses of inflorescence stems revealed changes in lignin, cellulose, and matrix sugar composition indicating an overall increase in secondary versus primary walls in mutants, indicative of higher xylem production compared with the wild type (since secondary walls were thinner). Intriguingly, the number of secondary cell wall layers compared with the wild type was increased in xth9 and reduced in xth4, whereas the double mutant xth4x9 displayed an intermediate number of layers. These changes correlated with specific Raman signals from the walls, indicating changes in lignin and cellulose. Secondary walls were affected also in the interfascicular fibers, where neither XTH4 nor XTH9 was expressed, indicating that these effects were indirect. Transcripts involved in secondary wall biosynthesis and cell wall integrity sensing, including THESEUS1 and WALL ASSOCIATED KINASE2, were highly induced in the mutants, indicating that deficiency in XTH4 and XTH9 triggers cell wall integrity signaling, which, we propose, stimulates xylem cell production and modulates secondary wall thickening. Prominent effects of XTH4 and XTH9 on secondary xylem support the hypothesis that altered xyloglucan affects wood properties both directly and via cell wall integrity sensing.
The plant cell wall is composed of cellulose microfibrils embedded in a matrix of hemicelluloses and pectins, structural glycoproteins, and, in some cell types, lignin. Xyloglucan (XG) is an abundant hemicellulose present in all lineages of plant species studied to date as well as in green algae (Popper et al., 2011). In dicotyledons, including Arabidopsis (Arabidopsis thaliana), XG constitutes ∼20% of dry weight in primary walls, but the mature secondary walls have not been reported to contain XG (Scheller and Ulvskov, 2010).
The XG backbone is made up of β-(1,4)-linked Glc substituted with α-(1,6)-linked Xyl, which can be further decorated with β-(1,2)-Gal residues with or without substitution by α-(1,2)-Fuc (reviewed by Pauly and Keegstra [2016]). Acetyl groups are usually present on Gal. XG substitutions are plant species and organ-/tissue-/cell type specific and can vary during cell development. In the majority of dicots, XXXG-type XGs are present where three out of four backbone residues are xylosylated and the two last Xyl residues are β-1,2-galactosylated, with the last Gal frequently α-1,2-fucosylated. In some tissues, like root hairs, GalUA can be attached to Xyl at position 2 (Peña et al., 2012).
XG coats hydrophobic microfibril surfaces (Park and Cosgrove, 2012, 2015). Earlier models suggested that XG cross-links adjacent microfibrils (Hayashi et al., 1994). Recently, however, it was proposed to form biomechanical hotspots instead, which are the links between adjacent microfibrils with XG embedded in between (Park and Cosgrove, 2012, 2015). XG also covalently links with pectins (Thompson and Fry, 2000; Popper and Fry, 2005, 2008) and with ARABINOXYLAN PECTIN ARABINOGALACTAN PROTEIN1 (Tan et al., 2013).
XG biosynthesis starts in the Golgi apparatus as a team effort of a glucan synthase, CELLULOSE SYNTHASE-LIKE C4, three XG xylosyltransferases (XXT1, XXT2, and XXT5), two galactosyltransferases (MURUS3 and XYLOGLUCAN L-SIDE CHAIN GALACTOSYLTRANSFERASE POSITION 2), and a fucosyltransferase (FUT1; Pauly and Keegstra, 2016). Nascent XG chains are packaged into the secretory vesicles and secreted into the apoplast, where they are incorporated into the existing XG-cellulose network nonenzymatically and/or by the xyloglucan endo-transglycosylase (XET) activity. XET enzymes were discovered in 1992 by three independent groups (Farkas et al., 1992; Fry et al., 1992; Nishitani and Tominaga, 1992; Okazawa et al., 1993). They are grouped in carbohydrate-active enzyme family GH16, along with xyloglucan endo-hydrolases (XEHs), and thus the family has been named as XTH for xyloglucan endo-transglycosylase/hydrolase (Rose et al., 2002). The XTH family includes three major clades, I/II, IIIA, and IIIB, and XEH activity has been reported only in clade IIIA (Baumann et al., 2007; Eklöf and Brumer, 2010; Kaewthai et al., 2013). XETs cleave the XG backbone (donor) endolytically, the enzyme-bound XG fragment is then transferred to the nonreducing end of another XG chain (acceptor), and the glycosidic linkage is reformed (Eklöf and Brumer, 2010). Few XTH members are able to use substrates other than XG or water acceptors, including mixed-link glucan (Hrmova et al., 2007; Simmons et al., 2015; Simmons and Fry, 2017), or to use amorphous cellulose as acceptor and donor (Shinohara et al., 2017).
XETs are thought to regulate cell wall plasticity along with expansins and pectin-digesting enzymes, but the molecular action of their activity in cell walls is not fully understood (Van Sandt et al., 2007; Park and Cosgrove, 2015). XET activity has been demonstrated to incorporate the fluorescently labeled XG oligosaccharides to a fraction aligned with cellulose microfibrils, possibly by transglycosylation of the XG loose ends or cross-links, thus probably resulting in wall loosening, as well as to a cell wall fraction inaccessible to the XG-degrading enzymes (Vissenberg et al., 2005a). The latter could possibly constitute the biomechanical hotspots, and XET activity would then contribute to wall strengthening. Analysis of several xth mutants revealed reduced cell sizes (Osato et al., 2006; Liu et al., 2007; Sasidharan et al., 2010; Ohba et al., 2011), whereas the overexpression or exogenous application of XET proteins either stimulated (Shin et al., 2006; Ohba et al., 2011; Miedes et al., 2013) or decreased (Maris et al., 2009) cell expansion. Other studies have shown that XETs could be involved in either wall loosening or strengthening, depending on the acceptor size (Takeda et al., 2002).
XETs are known to be highly expressed, both in primary (Xu et al., 1995; Antosiewicz et al., 1997; Oh et al., 1998; Dimmer et al., 2004; Romo et al., 2005; Vissenberg et al., 2005b; Jiménez et al., 2006; Hara et al., 2014) and in secondary (Bourquin et al., 2002; Nishikubo et al., 2007; Goulao et al., 2011) vascular tissues, but their roles in these tissues are not fully understood. Only one gene, AtXTH27, has been functionally characterized in vascular tissues, shown to affect tracheary element development in minor veins of rosette leaves, and proposed to mediate XG degradation in these cells (Matsui et al., 2005). In hybrid aspen (Populus tremula × Populus tremuloides), PtxtXTH34 protein and XET activity were highly expressed in developing xylem fibers, coinciding with CCRC-M1 antibody (recognizing the terminal Fuc of XG) signals (Puhlmann et al., 1994), and suggesting a deposition of XG to primary wall layers (compound middle lamella) through the developing secondary cell wall layers (Bourquin et al., 2002; Nishikubo et al., 2011). In support of this hypothesis, several genes similar to XG xylosyltransferases were found to be expressed during secondary wall formation (Sundell et al., 2017), raising the possibility of continuous XG deposition and XTH function in this process. Indeed, the overexpression of PtxtXTH34 resulted in more CCRC-M1 signals in the compound middle lamella and more cell wall-tightly bound XG at early stages of secondary xylem cell differentiation. But the later stages of xylogenesis did not show increased XG anymore, and the role of such XET-induced XG deposition in xylem cells remained elusive.
To address the role of XTH genes during secondary xylem development, we analyzed patterns of XTH gene family expression in developing wood using the AspWood database for aspen (Populus tremula; Sundell et al., 2017). For functional analyses, we selected two Arabidopsis genes, AtXTH4 and AtXTH9, similar to abundant aspen XTH transcripts exhibiting the most frequently observed expression pattern, and tested if these two genes are involved in xylem cell expansion or in other aspects of xylem cell differentiation. Mutant analysis revealed that AtXTH4 and AtXTH9 not only regulate xylem cell expansion but also influence several characteristics of secondary growth, including secondary xylem production and secondary wall deposition. The deficiency in these two genes was additive for some traits, suggesting their partially redundant or additive roles for those traits, while it was unique or even opposite for other traits. The up-regulation of several cell wall integrity-related genes in these xth mutants and their non-cell-autonomous effects suggest that some of them are induced by the cell wall integrity signaling. These analyses indicate new and diverse roles for XTH genes in secondary xylem cell differentiation.
RESULTS
AtXTH4 and AtXTH9 Are Homologs of Major Secondary Vascular Tissue XET-Encoding Genes, PtXTH34 and PtXTH35
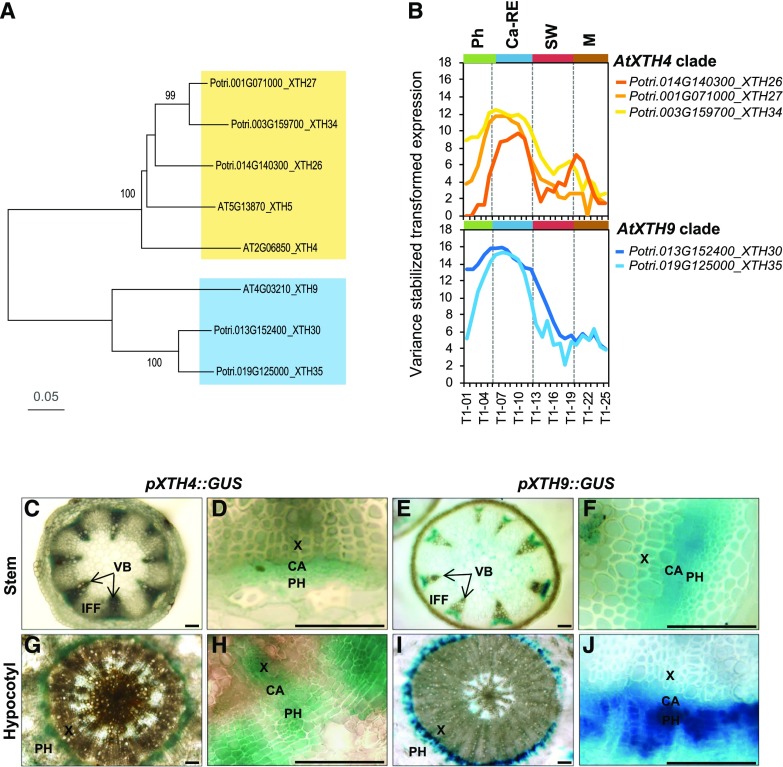
To illustrate the importance of XTH genes in secondary growth, we analyzed the expression patterns of the XTH family members across the wood developmental zones available in the AspWood database (http://aspwood.popgenie.org; Sundell et al., 2017). Out of the recently updated census of 43 Populus XTH genes (Kumar et al., 2019), 26 were found in AspWood and the majority belonged to cluster e (Supplemental Fig. S1), which groups genes with peak expression in the cambium and radial expansion zone, coinciding with the peak of XET activity (Bourquin et al., 2002). The subclade of PtXTH34 (also known as PtXET16A) and the subclade of PtXTH35 include the most highly expressed genes of this cluster, with documented (Kallas et al., 2005) and predicted (Baumann et al., 2007) XET activity, respectively. Arabidopsis AtXTH4 and AtXTH9 genes known to be highly expressed in stems and seedlings (Yokoyama and Nishitani, 2001), similar to PtXTH34 and PtXTH35, respectively, were selected (Fig. 1A) for functional studies during secondary growth. Promoters of AtXTH4 and AtXTH9 were active in developing secondary vascular tissues in secondarily thickened hypocotyls and basal stems, where secondary growth occurs. The signals were observed in the vascular cambium, and in adjacent developing secondary xylem and phloem, but not in the interfascicular fibers (Fig. 1, C–J). This pattern matches the expression of their homologous clades in aspen (Fig. 1B), supporting their conserved functions in secondary growth in the two species.
Figure 1.
Clades AtXTH4 and AtXTH9 in Arabidopsis and Populus trichocarpa. A, Phylogenic tree constructed using the neighbor-joining method of MEGA7 in default mode using MUSCLE-aligned protein sequences (http://phylogeny.lirmm.fr/phylo_cgi/index.cgi), with bootstrap test of 1,000 replicates shown in percentages beside branches. B, The expression patterns of P. trichocarpa members of AtXTH4 and AtXTH9 clades in different wood developmental zones (http://aspwood.popgenie.org). Ca-RE, Cambium-radial expansion zone; M, maturation zone; Ph, phloem; SW, secondary wall formation zone. C to J, AtXTH4 and AtXTH9 promoter activity in Arabidopsis mature inflorescence stems and hypocotyls as visualized by GUS histochemistry. In the inflorescence stems (C–F), the expression of both genes was detected in vascular bundles, whereas interfascicular fibers did not show any expression (C and E); the closeup vascular bundles (D and F) show signals in the vascular cambium, developing xylem, and developing and differentiated phloem. In hypocotyls (G–J), both genes were expressed in the region of secondary vascular tissue formation (G and I), encompassing the vascular cambium, developing secondary xylem and phloem, and recently differentiated phloem cells (H and J). CA, Vascular cambium; IFF, interfascicular fibers; PH, phloem; VB, vascular bundle; X, xylem. Bars = 50 µm.
Loss- and Gain-of-Function XET Lines Show Altered Growth and XG Signals
Lines with T-DNA insertions in AtXTH4 and AtXTH9 genes (Fig. 2A) obtained from the Nottingham Arabidopsis Stock Centre were purified by repeated back-crossing until single inserts with segregation ratio 3:1 were obtained. Reverse transcription PCR analysis in basal inflorescence stems and secondarily thickened hypocotyls of flowering plants detected no transcripts of AtXTH4 in xth4 or those of AtXTH9 in xth9. The mutants were crossed to generate the double mutant xth4x9. As expected, there was no expression of AtXTH4 in the double mutant; however, a low level of residual expression of AtXTH9 was detected in this genetic background in the hypocotyl, indicating that the xth9 mutant was conditionally slightly leaky. There was also a compensatory up-regulation of AtXTH9 and AtXTH4 in single xth4 and xth9 mutants, respectively. For gain-of-function studies, the highly expressing line 18 (OE18) with the coding sequence of hybrid aspen PtxtXTH34 (AF515607) under the control of the 35S promoter (Miedes et al., 2013) was used.
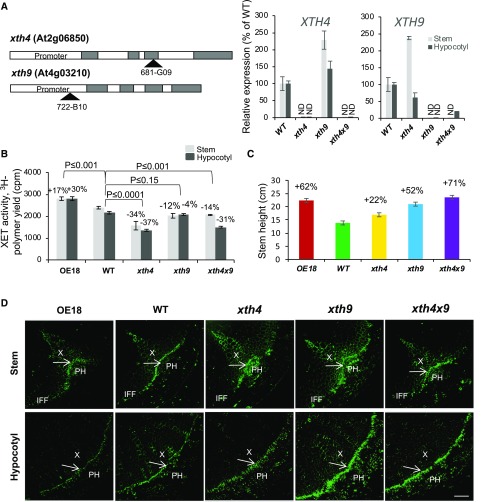
Figure 2.
Mutations in AtXTH4 and AtXTH9 affect XET activity in stem and hypocotyl, stem height, and XG signals in secondary vascular tissues. A, Positions of T-DNA inserts in AtXTH4 and AtXTH9 genes, and expression of target genes in mutant single insert lines and in the double mutant analyzed in inflorescence stem base and in secondarily thickened hypocotyls by reverse transcription quantitative PCR (RT-qPCR). ND, Not detected; WT wild type. Error bars indicate se. B, XET activity in protein extracts from the basal part of inflorescence stems and in hypocotyls of 6-week-old xth4 and xth9 mutants, double mutant xth4x9, and line OE18 with strong heterologous expression of PtxtXTH34 determined by the incorporation of 3H-labeled XG oligosaccharide acceptor to insoluble residue and normalized to tissue fresh weight. C, Stem height in 6-week-old plants grown in long-day conditions. In B, P values for the post-ANOVA Student’s t test are shown, and in C, significant differences from the wild type are shown by percentage change (post-ANOVA Student’s t test, P ≤ 5%). Values are means ± se; n = 5 and 30, respectively. D, Immunolocalization of XG with CCRC-M1 antibody in vascular tissues of basal stem and hypocotyl of 6-week-old plants. Arrows point to cambium. IFF, Interfascicular fibers; PH, phloem; X, xylem. Bar = 50 µm.
XET activity in protein extracts from stems and hypocotyls was reduced in xth4 and xth4x9, not significantly affected in xth9, and increased in the OE18 line as compared with the wild type (Fig. 2B). Small impact of the single mutations and overexpression on activity in the protein extract was not surprising, considering that many other XTH family genes could be contributing to the measured activity and that the xth9 mutation was conditionally leaky, and AtXTH4 and AtXTH9 exhibited compensatory reciprocal transcript activation in the single mutant background (Fig. 2A). Despite the modest changes in the XET activity of extracts, the lines exhibited clearly detectable phenotypic changes as compared with the wild type. Stem height was stimulated in both single and double mutants as well as in OE18 plants (Fig. 2C). Second, there were clear changes in cell wall XG signals from the monoclonal antibody CCRC-M1 in the cambium region tissues, as detected by immunofluorescence. Compared with the wild type, signals were decreased in the OE18 line and increased in the xth9 mutant in stem and hypocotyl sections and, to a small degree, increased in the double mutant xth4x9 in the hypocotyl and in the xth4 mutant in the stem (Fig. 2D), suggesting that XET activity either affects the amount of primary walled tissues or the content of XG epitopes in these tissues, with defects in AtXTH4 and AtXTH9 being nonadditive.
AtXTH4 and AtXTH9 Mediate Xylem Cell Expansion and Inhibit Secondary Xylem Production
We have previously reported that overexpression of PtxtXTH34 in hybrid aspen increased vessel diameter (Nishikubo et al., 2011). To study the role of AtXTH4 and AtXTH9 in xylem cell expansion, the sizes of secondary xylem cells were measured in hypocotyls of 6-week-old xth4, xth9, and xth4x9 mutants and OE18 Arabidopsis plants. Vessel element diameter was reduced in the mutants and increased in OE18 (Fig. 3A). Vessel element length was also reduced in all three mutants, and the reduction was primarily caused by the shortened tails. This is interesting, since the length of tails partially depends on the placement of the perforation plate and partially on the tail intrusive growth. Therefore, we analyzed the length of fibers that uniquely elongate by intrusive growth and found reductions in all three mutants but no marked effect was observed in the OE line. This suggests that both genes, AtXTH4 and AtXTH9, positively regulate intrusive xylem cell elongation and vessel expansion. Interestingly, XET has not previously been implicated in intrusive growth. Fiber diameters, which enlarge by symplastic expansion, were not consistently affected by XET activity, being enlarged in both the OE18 line and in the double mutant (Fig. 3A).
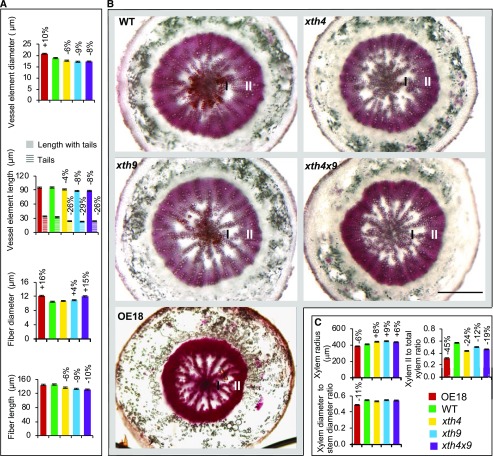
Figure 3.
Effects of altered XET expression on secondary xylem anatomy in hypocotyls of 6-week-old Arabidopsis plants. A, Sizes of secondary xylem cells measured in wood macerates from 10 plants. B, Light microscopy images of hypocotyl cross sections stained with phloroglucinol. I, Xylem I; II, xylem II; WT, wild type. Bar = 500 µm. C, Morphometric analysis based on sections of 10 plants. Secondary xylem radius, xylem diameter-to-stem diameter ratio, and the ratio of xylem II radial width to total xylem radius are shown. In A and C, statistically significant differences from the wild type are shown by percentage change compared with the wild type (post-ANOVA Student’s t test, P ≤ 5%). Values are means ± se; n = 100 for fibers, 300 for vessel elements, and 10 for xylem and stem.
Anatomy of secondarily thickened hypocotyls revealed changes in xylem radius; it was stimulated in the mutants and inhibited in the OE18 line (Fig. 3, B and C). The radius reflects the number of cells in radial files and their diameters. Since in Arabidopsis hypocotyls, vessels tend to be arranged in radial files, some of which extend from the cambium to the pith (Fig. 3B), and the diameters of vessel elements were decreased in the mutants and increased in the OE18 line, it can be concluded that the number of xylem cells per radial file must have been increased in the mutants and decreased in the OE plants. This implicates XET in the negative control of periclinal cell division in xylem mother cells. This conclusion is further supported by the reduced ratio of secondary xylem diameter to total stem diameter in OE18 plants (Fig. 3C).
Moreover, the secondary xylem development was affected in the analyzed genotypes. Secondary xylem development is known to have two distinct growth phases: the early phase, when only vessel elements and primary walled parenchyma cells are produced (xylem I); and the later stage, when the differentiation of secondary walled fibers occurs (xylem II). The ratio of xylem II to total xylem was reduced in both mutants and OE plants compared with the wild type, with the strongest reduction (by 45%) observed in the OE18 line (Fig. 3, B and C). This result implicates XET in the control of the transition between xylem I and xylem II phases, although these responses did not follow XET activity consistently.
XET Affects Cell Wall Thickness and Ultrastructure in Mature Xylem Cells
To test if XET has any influence on xylem cell wall ultrastructure, we studied secondarily thickened hypocotyls and stem bases by transmission electron microscopy (TEM). In woody plants, the secondary wall typically includes three layers, S1, S2, and S3, which can be observed by TEM (Mellerowicz et al., 2001). Similar ultrastructure was reported for Arabidopsis interfascicular fibers (Zhong and Ye, 2015). We have also observed mostly three partite secondary walls in wild-type Arabidopsis plants, in the fibers and vessel elements of stems and hypocotyls as well as in the interfascicular fibers (Fig. 4, A and B; Supplemental Figs. S2–S4). Unexpectedly, the XET-modified plants exhibited severe alterations in the number of secondary cell wall layers in these cell types. In all types of fibers (hypocotyl secondary xylem fibers, stem fascicular fibers, and stem interfascicular fibers), typically only two secondary wall layers were observed in xth4 mutant, whereas many more layers than three (up to 11) were present in xth9 mutant (Fig. 4, A and B; Supplemental Fig. S2). In the double mutant xth4x9, the number of secondary wall layers was intermediate between those observed in xth4 and xth9. The OE18 line had unchanged or reduced numbers of layers in the hypocotyl and increased numbers of layers in the stem, compared with the wild type (Fig. 4B). Increased cell wall layer numbers were observed in the vessel elements of the xth9 mutant as well, in both hypocotyls and stems, but the change was much less pronounced than in the fibers (Supplemental Figs. S3 and S4).
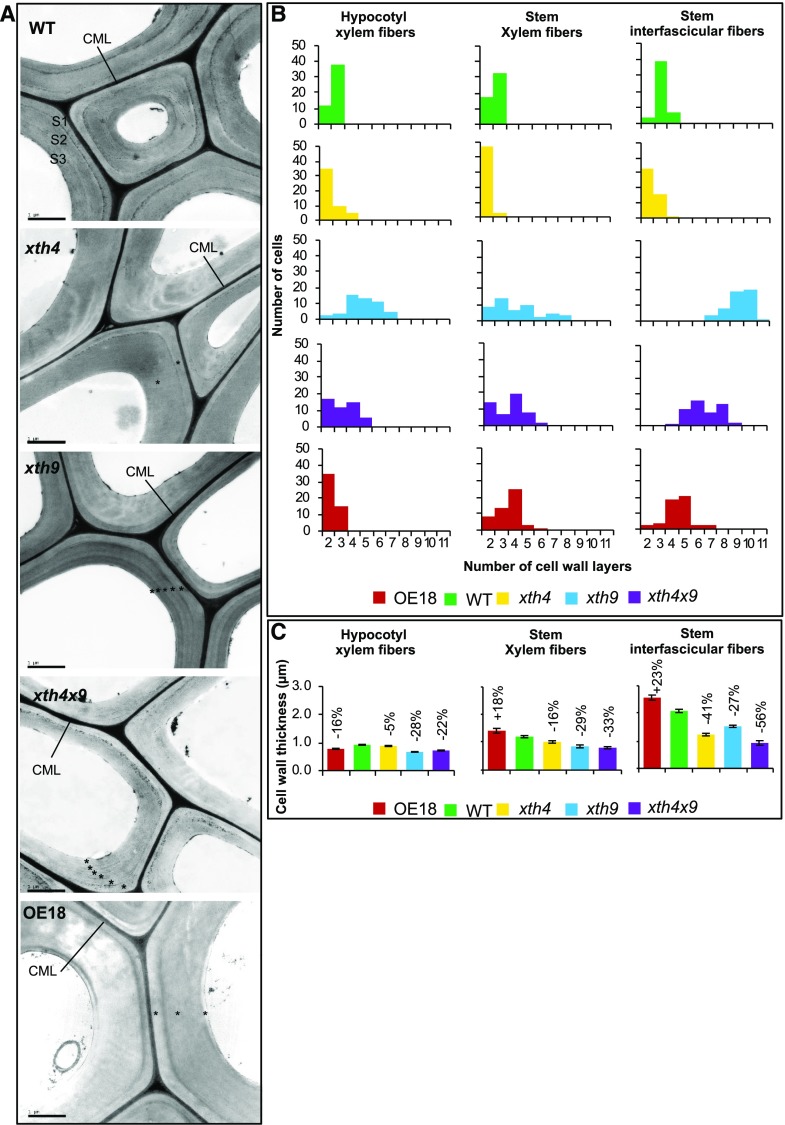
Figure 4.
Effects of altered XET levels on secondary cell wall morphology of fibers as visualized by TEM in 8-week-old Arabidopsis plants. A, TEM micrographs of hypocotyl secondary xylem fibers showing cell wall ultrastructure. CML, Compound middle lamella; S1, S2, and S3, secondary cell wall layers; WT, wild type. Note the abnormal secondary wall layering in the mutants and OE plants shown by asterisks. Bars = 1 μm. B, Frequency distributions of cells with different numbers of cell wall layers in hypocotyl and stem xylem fibers and interfascicular stem fibers. Twenty-five cells were scored in each of two plants. C, Secondary cell wall thickness in the different types of fibers. Statistically significant differences from the wild type are shown by percentage change above the bars (post-ANOVA Student’s t test, P ≤ 5%). Values are means ± se; n = 100 for hypocotyl and 15 for stem fibers.
Cell wall thickness in the hypocotyl and stem fibers and vessel elements was reduced in all the mutants, whereas in the line with strong heterologous expression of PtxtXTH34, it was either reduced (in hypocotyl fibers and stem vessel elements) or increased (in stem fibers; Fig. 4C; Supplemental Figs. S3 and S4).
XET Affects Cell Wall Composition in Basal Inflorescence Stems
To test if there was any change in the chemical composition of cell walls correlating with the morphological changes observed in xylem cells, alcohol-insoluble residue (AIR1) was prepared from the basal segments of inflorescence stems and analyzed by several techniques.
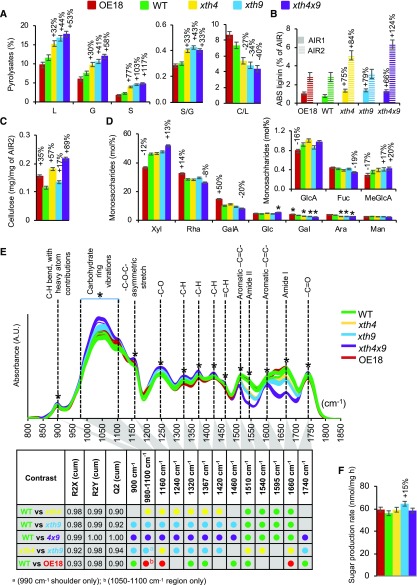
Pyrolysis-mass spectrometry (MS) analysis detected substantial increases in lignin (total, by up to 53%; G, by up to 58%; and S, by up to 117%) in all three mutants, with most prominent effects in the double mutant xth4x9, whereas no difference compared with the wild type was found in OE18 plants (Fig. 5A). Consequently, there was a significant increase in S/G ratio in all mutants, the most prominent in xth9 (by 43%). Concomitantly, significant decreases in carbohydrate-to-lignin ratios were evident in all mutants, by up to −40% in xth4x9.
Figure 5.
XET affects cell wall chemistry in basal inflorescence stem. A, Pyrolysis gas chromatography-MS of AIR1. C, Carbohydrates; G, guaiacyl lignin; L, total lignin; S, syringyl lignin. B, ABS lignin in AIR1 before (AIR1) and after (AIR2) amylase treatment. C, Updegraff cellulose content in AIR2. D, Monosaccharide composition of AIR2 determined by methanolysis and trimethylsilyl derivatization. E, Diffuse reflectance FTIR spectra of AIR1. Asterisks denote bands that are significantly different (50% or greater correlation) in the different contrasts shown in the table, according to OPLS-DA (orthogonal projections to latent structures-discriminant analysis) models using one + two (predictive + orthogonal) components. Colored dots indicate genotypes with significantly higher signals. Score plots and loadings for the models are shown in Supplemental Figure S5. AU, Arbitrary units. F, Sugar yields of dried stems in saccharification analysis after alkaline pretreatment. Data are means ± se; n = 3 for A to D and n = 5 for F. Percentage changes or asterisks are given for means significantly different from the wild type (WT; post-ANOVA Student’s t test, P ≤ 0.05).
Acetyl bromide-soluble (ABS) lignin content was determined in AIR1 directly as well as after the amylase treatment (AIR2). ABS lignin was substantially increased in AIR1 in all the mutants (by up to 79% in xth9) while it was unaffected in the OE line, in agreement with pyrolysis-MS results (Fig. 5B). However, lignin amounts in the destarched samples increased only in xth4 and xth4x9 mutants, but not in xth9, which was most affected in the AIR1 fraction. This indicates that the xth9 mutant either contained less starch in AIR1 than other lines (contributing to artifactually high ABS lignin content when expressed per AIR1 weight) or that the lignin induced by the xth9 mutation is vulnerable to amylase treatment (e.g. by containing relatively more water-soluble components).
Crystalline cellulose content in the amylase-treated AIR2 was increased in the double mutant (by 89%) and to a lesser degree in the single mutants (by 57% in xth4 and 17% in xth9; Fig. 5C). Interestingly, the OE18 line also had higher cellulose content (by 35%) than the wild type. Matrix polysaccharide composition analysis of AIR2 revealed increased Xyl and 4-O-methyl GlcA in the double mutant xth4x9, indicative of increased glucuronoxylan content, and decreased Rha, GalUA, Gal, and Ara, indicative of reduced pectin content (Fig. 5D). Opposite changes were observed in the OE18 line, indicative of decreased glucuronoxylan and increased pectin content as compared with the wild type. Taken together, these changes suggested that there could be a change in relative contributions from primary and secondary walls in the basal stems of the double mutant and the OE18 line, the former having more secondary walls and the latter more primary walls than the wild type.
To reveal other changes in cell walls, diffuse reflectance Fourier-transform infrared (FTIR) spectra of AIR1 were compared among the genotypes, revealing clear differences (Fig. 5E; Supplemental Fig. S5). The most affected peak was 1,660 cm−1, where signals were substantially reduced in the xth mutants, with the largest decrease in xth4x9, a larger decrease in xth9 than in xth4, and a significant increase in OE18. The 1,660 cm−1 band corresponds to the amide I vibration of proteins and as such can be connected to XET proteins covalently attached to XG in the cell wall. The protein origin of this band was confirmed by similar trends observed for the 1,540 cm−1 band (amide II). Bands associated to aromatic -C=C- skeletal vibrations (approximately 1,510 and 1,595 cm−1), which are typically interpreted as lignin signals, were also affected, but these bands overlap with the amide bands. Thus, they are highly susceptible to strong changes in the amide bands observed in the mutants and OE plants. This interpretation is consistent with the wet chemistry data obtained from the same material (Fig. 5, A and B). The mutants also showed significant increases in bands traditionally associated with polysaccharidic compounds (e.g. carbohydrate ring vibrations between 980 and 1,100 cm−1, a -C-O-C- asymmetrical stretch at 1,160 cm−1, and diverse -C-H-related vibrations at 1,320 and 1,420 cm−1), indicating a higher proportion of carbohydrates, potentially cellulose (1,160 cm−1; Dokken et al., 2005) and xylan (1,320 and 1,420 cm−1; Kačuráková et al., 1999), as compared with the wild type. The cellulose-associated -C-O-C- band (1,160 cm−1) also increased in the OE18 line compared with the wild type and in xth4 compared with xth9 (Fig. 5E; Supplemental Fig. S5). These carbohydrate-related signals were consistent with the wet chemistry data (Fig. 5, C and D).
Considering substantial alterations in cell wall chemistry in the basal stems and in the ultrastructure of secondary walls in the studied xth mutants and OE18 plants, we examined their saccharification characteristics. However, the sugar production rate from the dried stems after alkaline pretreatment (Gomez et al., 2010) only improved in the xth9 mutant (Fig. 5F).
In Situ FTIR Microspectroscopy Data Reveal Chemical Differences in Secondary Walls of xth Mutants and OE18 Plants
The changes in cell wall composition in the xth mutants and OE18 plants revealed by the bulk analyses of basal inflorescence stem tissues suggested relative changes in the amounts of primary and secondary walls. To reveal any potential chemical changes specifically in secondary walls, we carried out in situ FTIR microspectroscopy analysis of fascicular and interfascicular fibers, which are the two cell types highly contributing secondary cell wall material in the basal inflorescence stem. In addition, we analyzed xylem fibers in hypocotyls to verify if similar changes are induced in this organ.
In situ FTIR spectra of either stem or hypocotyl fiber walls did not reveal differences in xth4 compared with the wild type (Supplemental Fig. S6, A–D). In contrast, xth9 mutant showed differences in signal intensities of several bands; notably, there was an increase in the 1,510 cm−1 signal and a decrease in the 1,320 cm−1 signal in both analyzed organs. The increased intensity of the 1,510 cm−1 band, which was also seen in hypocotyls in the double mutant and which was observed in xth9 compared with xth4, could be related to a more cross-linked form of lignin (Faix, 1991). Several bands were altered in the OE18 plants, most of them being consistent between stems and hypocotyls (Supplemental Fig. S6, A–D). Besides the 1,510 cm−1 band (indicative of aromatics, although the influence of the neighboring amide II band cannot excluded) and the 1,460 cm−1 band (observed for both lignin and hemicellulose, potentially originating from =C-H functionalities) that had increased proportional intensities, we observed increased intensities of the 1,240 cm−1 (-C-O) and 1,420 cm−1 (-C-H) bands and a shift of the 1,740 cm−1 band (-C=O) to higher wave numbers (indicating higher energy vibrations of this functional group). Together, the increases of -C-H and -C-O band intensities and the shifted -C=O band may indicate an increased level of esterification. In conclusion, in situ FTIR microspectroscopy revealed clear chemical changes in the chemical composition of mature fiber walls.
In Situ Raman Microspectroscopy Data Reveal Differences in Lignin in Secondary Wall Cells of xth Mutants and OE18 Plants
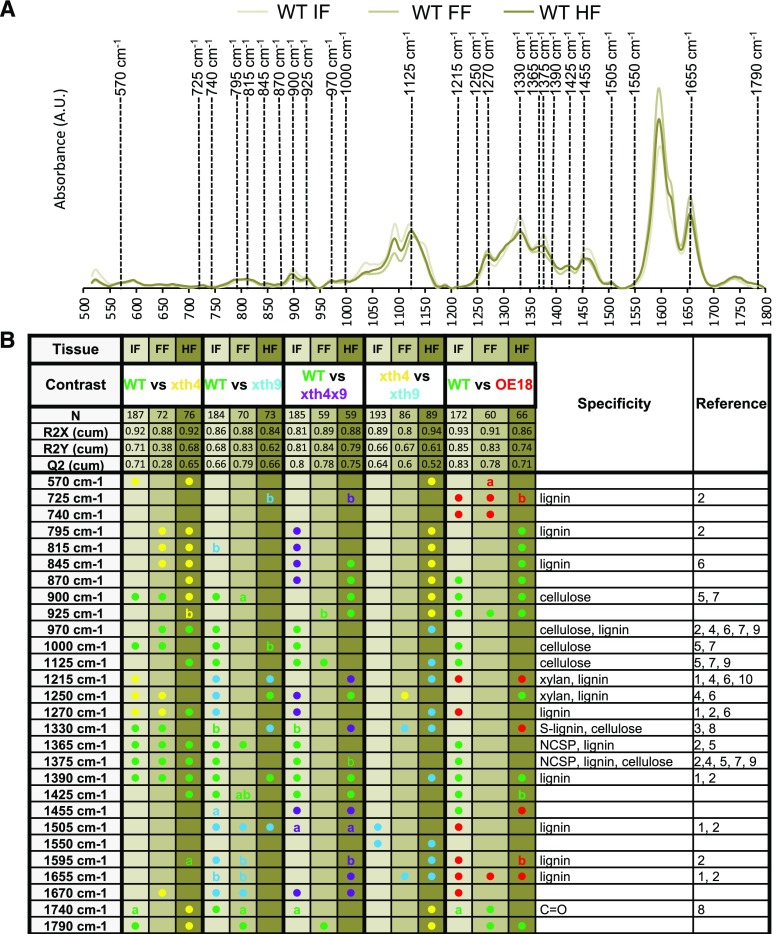
To obtain more support for the chemical changes in cell walls of mature fibers, we carried out in situ Raman microspectroscopy, which provides a higher spatial resolution and spectra with narrower bands and significantly reduced contributions from hemicelluloses and pectins compared with FTIR spectroscopy. The spectra of interfascicular fibers, where neither AtXTH4 nor AtXTH9 was expressed, and fascicular fibers, where both these genes were expressed (Fig. 1, C–J), were taken separately to address the question of cell-autonomous versus non-cell-autonomous effects. Pairwise comparisons by cell type, between each genotype and the wild type, revealed significant differences among genotypes (Supplemental Fig. S7, A–D). Considering only the differences between genotypes that were consistent between at least two cell types, we noted higher signals in xth4 compared with the wild type from several bands corresponding to lignin, such as 595, 845, and 1,270 cm−1, or corresponding to xylan and lignin, such as – 1,250 cm−1 (Fig. 6). In contrast, bands corresponding to cellulose were reduced in fiber cell walls in this genotype (900, 970, and 1,000 cm−1) together with bands corresponding to noncellulosic polysaccharides, cellulose, or lignin (1,330, 1,365, and 1,390 cm−1). Several of these bands also appeared similarly affected in other mutants, but only in one tissue type. Higher signals were observed in the xth9 mutant compared with the wild type for signals around 1,215 cm−1, corresponding to xylan and lignin, and at 1,505 cm−1, corresponding to aromatics (lignin). These bands showed increased intensities in xth9 than in xth4, but only in one of the studied tissues. Other lignin-related bands at 725 and 1,655 cm−1 were increased in OE18. This result supports the conclusions of an altered lignin structure/composition in the fibers in the xth mutants and OE18 plants, compared with the wild type, and indicates that distinct lignin alterations have occurred in xth4, xth9, and OE18. Intriguingly, the changes were most frequently observed in interfascicular fibers, suggesting their non-cell-autonomous induction.
Figure 6.
In situ Raman microspectroscopy of cell walls in fibers in three different tissues in xth mutants and OE18 plants. A, Representation of average wild-type (WT) spectra of interfascicular fibers (IF), fascicular fibers (FF), and hypocotyl fibers in secondary xylem (HF), with the bands that were significantly contributing to the separations in at least two of the studied tissues in any of the constructs shown in B. AU, Arbitrary units. B, Summary of OPLS-DA models of pairwise comparisons using one + two (predictive + orthogonal) components. Colored dots indicate genotypes with higher levels of given signals. Details of the models are given in Supplemental Figure S7. References are as follows: 1, Agarwal (1999); 2, Agarwal et al. (2011); 3, Agarwal and Atalla (2010); 4, Agarwal and Ralf (1997); 5, Kačuráková et al. (1999); 6, Larsen and Barsberg (2010); 7, Schenzel and Fischer (2001); 8, Schulz and Baranska (2007); 9, Wiley and Atalla (1987); 10, Zeng et al. (2016). a, Low end (i.e. red shift); b, high end (i.e. blue shift); NCSP, noncellulosic, structural polysaccharides.
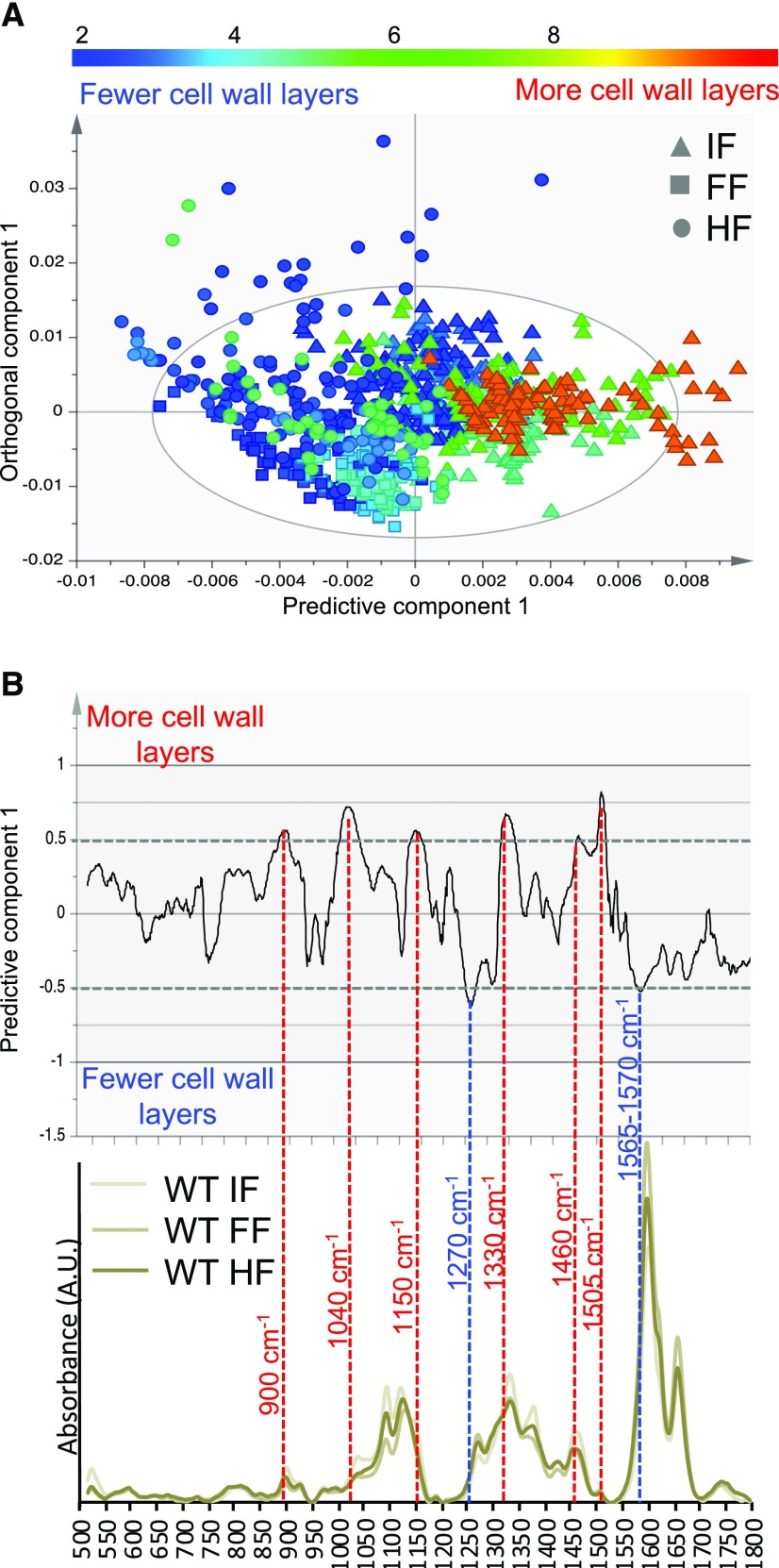
To assess if any chemical change could be related to the layering phenotype of secondary walls, we used the cell wall layers as Y (response) variables in an OPLS model of Raman spectra. The average number of cell wall layers was obtained from TEM analyses for interfascicular, fascicular, and hypocotyl fibers in each genotype. The resulting scatterplot (Fig. 7A) shows that the predictive component could identify spectral changes correlated to the number of cell wall layers, while the first orthogonal component was mainly related to tissue type. Thus, there were specific spectral features that were correlated to the numbers of secondary wall layers, irrespective of tissue type. The corresponding loadings (Fig. 7B) identified these features as bands at 900, 1,040, 1,150, 1,330, 1,460, and 1,505 cm−1, correlated with the high layer number, and bands at 1,270 and 1,565 to 1,570 cm−1, correlated to low numbers of layers. The band at 900 cm−1 (correlated to high numbers of cell wall layers) can be found in the spectra of all major polysaccharidic polymers, with cellulose and hemicellulose among the most common associations (Gierlinger, 2017). The band at 1,040 cm−1 is located firmly in the region that is dominated by carbohydrate ring vibrations often used to assess their proportions (Gierlinger, 2017). The band at 1,150 cm−1 is usually attributed to -C-O-C- vibrations but it is rather unspecific, being present in both polysaccharidic but also lignin polymers. The bands at 1,330 and 1,505 cm−1, on the other hand, while also correlated to high numbers of layers, have been assigned to different lignins (Gierlinger, 2017). The band at 1,460 cm−1 (correlated to high numbers of cell wall layers) is attributed to -C-H, and the one at 1,270 cm−1 (correlated to low numbers of cell wall layers) is attributed to -C-O vibrations, but they are seldom specific. The shoulder between 1,565 and 1,570 cm−1 (correlated to low numbers of layers) is in the spectral region of C=X vibrations, where X is either another carbon (often aromatic, making it a lignin-related band) or an oxygen. Both exclude the possibility of this band originating from cellulose. Thus, bands that can be assigned to cellulose are found correlated to a higher number of cell wall layers, while lignin-associated bands are found correlated to both higher and lower numbers of cell wall layers, indicating an altered lignin composition. Overall, these data indicate specific changes in lignin and cellulose in secondary walls as a function of the number of secondary wall layers, irrespective of cell type.
Figure 7.
OPLS model correlating average number of cell wall layers to Raman microspectroscopic features in three tissues: interfascicular fibers (IF), fascicular fibers (FF), and hypocotyl secondary xylem fibers (HF). A, Scatterplot showing separation according to the number of cell wall layers (predictive component) and tissue type (orthogonal component). B, The corresponding loadings plot and representative wild-type (WT) spectra for the three tissues. The spectral bands with more than 50% correlation levels (red dashed lines) are marked by band numbers. Bands at 900, 1,040, 1,150, 1,330, 1,460, and 1,505 cm−1 are proportionally more intensive when there are more cell wall layers, whereas bands at 1,270 cm−1 and a shoulder at 1,565 to 1,570 cm−1 are proportionally more intensive when there are fewer cell wall layers. The model details are as follows: 820 spectra, one + three (predictive + orthogonal) components; R2X(cum) = 0.928; R2Y(cum) = 0.383; Q2(cum) = 0.377. AU, Arbitrary units.
Effects of Altered AtXTH4 and AtXTH9 on Gene Expression in Lignin and Cellulose Biosynthesis and Cell Wall Integrity-Sensing Pathways
To test if the reported changes in secondary walls in xth mutants and OE18 plants were induced indirectly by the activation of biosynthetic and signaling pathways, we analyzed transcripts of selected sets of genes related to secondary wall biosynthesis and to cell wall integrity-sensing pathways in the basal part of inflorescence stems.
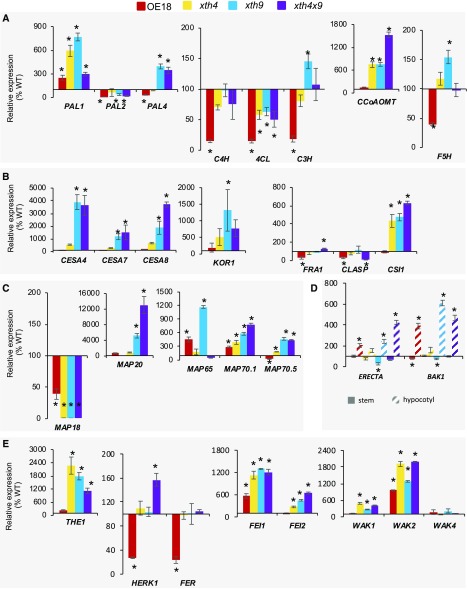
Among the genes representing six enzymatic activities essential for monolignol biosynthesis, we detected strong up-regulation in the xth mutants in genes of the early pathway, PHENYLALANINE AMMONIA-LYASE1 (PAL1) and PAL4, down-regulation in all mutants in 4-COUMARATE:COENZYME A LIGASE (4CL), and up-regulation of genes representing the late pathway, p-COUMARATE 3-HYDROXYLASE (C3H), CAFFEOYL-COENZYME A O-METHYLTRANSFERASE1 (CCoAOMT1), and FERULATE 5-HYDROXYLASE (F5H; Fig. 8A). PAL1, PAL4, C3H, and F5H were especially highly up-regulated in the xth9 mutant. The up-regulation of these monolignol biosynthetic genes could explain the increase in lignin content in the mutants and a distinct lignin composition in xth9 detected by pyrolysis and wet chemistry analyses (Fig. 5A). The OE18 plants, on the other hand, exhibited many lignin biosynthesis genes that were down-regulated, including PAL2, PAL4, CINNAMATE 4-HYDROLASE (C4H), 4CL, C3H, and F5H.
Figure 8.
Expression levels of genes affected in the xth mutants and OE18 plants. Genes related to lignin biosynthesis (A), secondary wall cellulose biosynthesis (B), microtubule dynamics (C), secondary xylem regulation in hypocotyl (D), and cell wall integrity sensing (E) were determined by RT-qPCR. Expression levels were normalized to the wild type (WT; 100%). Data are means ± se; n = 3 biological replicates. Asterisks indicate means significantly different from the wild type (post-ANOVA Student’s t test, P ≤ 0.05).
The genes encoding cellulose synthase complex proteins, such as secondary wall CESAs (CESA4, CESA7, and CESA8), KORRIGAN1 (KOR1), and CELLULOSE SYNTHASE INTERACTING1 (CSI1), were greatly up-regulated in xth9 and xth4x9 mutants, and in the latter case, also in the xth4 mutant (Fig. 8B). FRAGILE FIBER1 (FRA1), encoding a kinesin-like protein regulating cellulose microfibril angle (Zhong et al., 2002), and CLIP170-ASSOCIATED PROTEIN (CLASP) that promotes microtubule stability (Ambrose et al., 2007), showed some variability in xth4x9 and mild down-regulation in the OE18 line. Similarly, four genes encoding microtubule-binding proteins with known function in secondary wall cellulose biosynthesis (Korolev et al., 2007; Rajangam et al., 2008; Pesquet et al., 2010) were highly up-regulated in the mutants, especially MAP65 in xth9, MAP20 in xth9 and xth4x9, and MAP70.1 and MAP70.5 in all mutants (Fig. 8C). Some variability was also observed in the OE18 line in MAP65, MAP70.1, and MAP70.5. In contrast, MAP18, encoding a microtubule-destabilizing protein regulating cell growth in expanding cells (Wang et al., 2007), was down-regulated in all mutants and OE18 plants.
One way these changes in biosynthetic secondary wall-related genes could be induced is by the triggering of the cell wall integrity pathway. We found several key players in cell wall integrity sensing affected in the mutants and OE18 plants. Notably, the genes encoding plasma membrane-localized receptor-like kinases, such as THESEUS1 (THE1), HERKULES1 (HERK1), FERONIA (FER), WALL-ASSOCIATED KINASES (WAKs; Li et al., 2016), and the ERECTA-related receptor-like kinases FEI1 and FEI2 (Xu et al., 2008), were affected. THE1 was significantly up-regulated in all mutants, HERK1 was significantly up-regulated in xth4x9, and HERK1 and FER were down-regulated in OE18 (Fig. 8E). Among tested WAKs, WAK1 and WAK2 were up-regulated in all mutants and WAK2 was also up-regulated in OE18. FEI1 and FEI2 were up-regulated in all mutants, and FEI1 was up-regulated in OE18 plants. These data indicate the stimulation of cell wall integrity pathways in the basal part of inflorescence stems by altered XET expression.
Moreover, since the cell wall integrity sensors eventually would need to affect global master regulatory switches to affect vascular development seen in the mutants and OE18 plants (Fig. 3), we tested one such switch, ERECTA, which is known as a negative regulator of xylem II differentiation in hypocotyls (Ikematsu et al., 2017), and its putative coreceptor BAK1 (Jordá et al., 2016). We found both ERECTA and BAK1 up-regulated in hypocotyls of OE18 plants and in xth9 and xth4x9 mutants (Fig. 8D). This can provide a direct explanation of the suppressed xylem II differentiation in these plants.
DISCUSSION
XETs have been implicated in plant growth and development as well as stress reactions to mechanical stimuli, wounding, and cold (reviewed by Rose et al. [2002]). In spite of the growing knowledge about their enzymatic activities, their mode of action in plant tissues is still being debated. XETs were observed to be expressed in growing and expanded vascular tissues and were hypothesized to mediate both cell expansion and cell wall development in these tissues for over two decades (Xu et al., 1995; Antosiewicz et al., 1997; Oh et al., 1998; Bourquin et al., 2002; Dimmer et al., 2004; Jiménez et al., 2006). However, data from overexpression or from knockout/suppression studies demonstrating the aspects of vascular development affected by XET are scarce (Matsui et al., 2005; Nishikubo et al., 2011). Here, we have analyzed secondary vascular development in mutants of two vascular-expressed XETs, AtXTH4 and AtXTH9, in their double mutant xth4x9, and in plants ectopically expressing hybrid aspen PtxtXTH34, which is closely related to AtXTH4. We demonstrate the influence of these mutations and heterologous expression on different aspects of secondary growth, including secondary wall formation. The action of AtXTH4 and AtXTH9 genes was found to be overlapping in some cases and distinct or even opposite in others. Moreover, we identified non-cell-autonomous effects of altered XET expression. Here, we discuss the most prominent effects observed.
AtXTH4 and AtXTH9 Mediate Secondary Xylem Cell Diffuse Expansion and Intrusive Tip Growth
A previous study on transgenic hybrid aspen overexpressing PtxtXTH34 indicated that XET positively regulates vessel radial expansion (Nishikubo et al., 2011). Our results for Arabidopsis confirmed and extended these findings, providing evidence for the positive effect of AtXTH4 and AtXTH9 not only on vessel element radial expansion but also on tail elongation and fiber intrusive tip growth (Fig. 3A). Fiber diameter growth, similar to vessel element radial expansion, was stimulated by XET overexpression, but in contrast to vessel elements, it was also increased in the fibers of xth9 and xth4x9 mutants. Since vessel elements develop precociously relative to fibers (Mellerowicz et al., 2001), it is likely that narrower vessels in the xth9 and xth4x9 mutants create more room for fiber radial expansion.
Whereas the role of XET in cell wall expansion by diffuse growth has been supported by many observations (Vissenberg et al., 2000; Osato et al., 2006; Shin et al., 2006; Liu et al., 2007; Ohba et al., 2011; Miedes et al., 2013), the role of these enzymes in the tip growth and intrusive growth has not yet been clarified. Whereas in some species, including Arabidopsis, XET activity and expression of some XTH genes have been observed in tip-growing root hairs (Vissenberg et al., 2003, 2005b), defects in root hair elongation have not been reported so far for any XTH mutants, possibly due to redundancy. Our results for xth4 and xth9 mutants provide evidence for their involvement in tip growth of fibers and vessel element tails. It is not clear whether the effects of XETs are direct, mediating the mechanical properties of cell walls at elongating tips, or indirect. Nevertheless, XETs may be considered as targets for fiber length improvement programs in forest tree species. An example of an association between PtoXTH34 (formerly PtoXET16A) alleles and fiber length has been already reported for Populus tomentosa (Wang and Zhang, 2014).
Deficiencies in AtXTH4 and AtXTH9 Stimulate the Production of Secondary Wall Tissues But Reduce Secondary Wall Thickness Non Cell Autonomously
Chemical analyses of the stem bases clearly showed increased lignin, crystalline cellulose, and xylan-related monosaccharide contents in the xth mutants, indicative of increased overall secondary wall content in the stem basal tissues (Fig. 5, A–C). This was supported by an increase in the expression of genes involved in secondary wall biosynthesis (Fig. 8, A–C). In the OE18 plants, on the other hand, changes in matrix polysaccharides suggested decreased xylan and increased pectins in this genotype (Fig. 5D), pointing to the overall decreased contribution of secondary walls. Consistently, several lignin biosynthetic genes appeared to be down-regulated (Fig. 8A), suggesting an overall decreased lignification in the basal stem tissues. Curiously, the effects on secondary cell wall thickness were opposite: the fiber wall thickness was reduced in the xth mutants and increased in the OE18 plants (Fig. 4C). Thus, the stimulation of secondary wall biosynthesis in xth mutants, evident from bulk chemical analyses and gene expression, must have occurred at the whole tissue level by the up-regulation of the number of cells with secondary cell walls rather than by the stimulation of secondary wall thickening. This interpretation of the data concerning the stem bases is supported by direct observations in hypocotyls, where the production of secondary xylem was induced in the mutants (Fig. 3B) with simultaneous suppression of the secondary wall thickness (Fig. 4C; Supplemental Fig. S4B). Thus, the effects of XET on secondary xylem production and thickening of secondary walls are independent. This conclusion is in agreement with the current models of independent regulation of meristematic activity leading to xylem production and secondary wall thickening activity of differentiating xylem cells (Ramachandran et al., 2017). AtXTH4 and AtXTH9 deficiency can inhibit secondary wall thickening non cell autonomously (i.e. outside of their expression range), and the effects are apparently additive for the two genes (Fig. 4C).
xth4 and xth9 Mutants Display Opposite Changes in Secondary Wall Layers
Secondary wall layering defects seen in xth mutants and OE18 plants are the most striking phenotypes reported in this study (Fig. 4; Supplemental Figs. S2–S4). These defects were evident in all cells with secondary walls but were most apparent in the interfascicular fibers, where neither AtXTH4 nor AtXTH9 was expressed. This indicates that the secondary wall layering is induced non cell autonomously by AtXTH4 and AtXTH9 deficiency. Curiously, deficiency in AtXTH4 resulted in the reduction of secondary cell wall layers and the opposite phenotype was induced by deficiency in AtXTH9. The intermediate effects on cell wall layering seen in the double mutant support opposite effects of the two mutants. XET overexpression also affected secondary wall layering with different outcomes, depending on cell and organ type.
The opposite effects of AtXTH4 and AtXTH9 deficiency on secondary cell wall layering are puzzling, since their mutants had additive phenotypes in other measured parameters, like secondary wall thickening. They could be related to subtle differences in AtXTH4 and AtXTH9 enzymatic properties, which are currently unknown, or to differences in expression pattern in developing xylem cells, as seen for their aspen orthologs (Fig. 1B). Differential expression is supported by separate coexpression networks of AtXTH4 and AtXTH9 in Arabidopsis, as revealed by GeneMANIA analysis (http://www.genemania.org; Warde-Farley et al., 2010), and for PtXTH34 and PtXTH35 clades in the AspWood database (http://aspwood.popgenie.org; Sundell et al., 2017).
Secondary wall layering is normally related to changes in cellulose microfibril angle, which are matched and affected by the changes in cortical microtubules (Mellerowicz et al., 2001). Currently, it is unknown whether XET deficiency or surplus affects cortical microtubules in the studied genotypes. Although we have seen changes in the expression of several genes encoding microtubule-associated proteins that play essential roles in microtubule dynamics, these changes may be due to overall secondary wall induction in the stems of mutants, as they paralleled the expression of secondary wall CESAs (Fig. 8, B and C). There are other reports suggesting a possible coupling between XTHs or XG, cortical microtubules, and cellulose biosynthesis (Takeda et al., 2002; Sasidharan et al., 2014; Xiao et al., 2016; Armezzani et al., 2018), but it is unclear how this coupling mechanistically occurs.
The secondary wall-layering defects were observed in different plant species when cinnamoyl-CoA reductase was down-regulated (Ruel et al., 2002; Leplé et al., 2007). This was accompanied by reductions in noncondensed lignin and by the presence of abnormal lignin units. Our in situ FTIR and Raman analyses demonstrated increased lignin content and changes in signals related to aromatic components in the studied genotypes (Fig. 6; Supplemental Fig. S6), whereas the wet chemistry analyses (Fig. 5, A and B) and gene expression data (Fig. 8A) revealed distinct changes in xth9 mutants in lignin quality and biosynthesis compared with xth4. This could be the basis of opposite wall-layering phenotypes in these mutants. The more direct proof of distinct chemistries in secondary walls in these mutants comes from the models correlating cell wall numbers and chemistry based on in situ Raman signals (Fig. 7), which indicated changes both in polysaccharidic compounds (e.g. increasing cellulose proportions with higher numbers of cell wall layers) and lignin (mixed correlations to cell wall layer numbers, indicating altered lignin composition/structure). While the exact natures of these changes are hard to elucidate from our data, they clearly point to an overall change in the cell wall matrix.
XET Role in the Perception of Mechanical Stimuli?
XETs have been implicated in various mechanoperception responses such as thigmomorphogenesis (Xu et al., 1995; Lee et al., 2005; Martin et al., 2014), gravitropism (Cui et al., 2005), and tension wood formation (Nishikubo et al., 2007; Baba et al., 2009; Kaku et al., 2009; Gerttula et al., 2015). XET activity conceivably could be involved in these responses by generating signaling XG oligosaccharides, although their mode of action is poorly understood (York et al., 1984; McDougall and Fry, 1988, 1989; Kaida et al., 2010; González-Pérez et al., 2012, 2014). XET could also be involved in these processes by preventing a buildup of tension among polymers in newly deposited cell wall layers, as has been proposed for the xylan endo-transglycosylase PtxtXYN10A (Derba-Maceluch et al., 2015). In such cases, XET deficiency would lead to increased tension in the cell wall that could be perceived by plant cells as mechanical stress, thus probably acting via cell wall integrity-sensing mechanisms (Li et al., 2016; Wolf, 2017). Interestingly, the observed symptoms of AtXTH4 and AtXTH9 deficiency, stimulation of xylem mother cell division, suppression of secondary wall thickening in xylem cells, and increased height growth, mimic those observed in hybrid aspen when secondary wall xylan synthase members PtxtGT43B and PtxtGT43C were suppressed (Ratke et al., 2018). Moreover, the altered expression of several receptor-like kinases, including THE1 and WAK2, known to mediate cell wall integrity sensing, observed in xth9, xth4, and xth4x9 mutants as well as in plants with strong heterologous XET expression, strongly supports the modulation of the signaling via a cell wall integrity-sensing pathway in these genotypes (Fig. 8E). Up-regulation of some of these receptors, such as WAK2, in both mutants and OE18 plants, could explain how XET deficiency and excess could lead to similar phenotypes such as stem height stimulation (Fig. 2C), hypocotyl xylem II-to-total xylem ratio (Fig. 3, B and C), and others observed in this study. Moreover, the up-regulation of ERECTA and BAK1 in hypocotyls (Fig. 8D) could provide a direct explanation for the observed reduction in xylem II (Fig. 3, B and C). Although the signaling cascades involving these different receptor-like kinases are currently not known, clearly the cell wall integrity signaling could be responsible for many XET effects that are indirect and non-cell autonomous as well as those involving microtubule network modulation.
XET as a Target for Biomass Improvement
Considering the extensive effects of XET on secondary growth revealed by this study, XETs may become targets for woody biomass improvement. In support, our analysis demonstrated 15% gains compared with the wild type in sugar production in xth9 mutants (Fig. 5F), which also exhibited over 50% growth increase (Fig. 2C). Our data with Arabidopsis XET-modified genotypes indicate that several wood traits are affected by XET. Indeed, Wang and Zhang (2014) identified single-nucleotide polymorphisms in PtoXTH34 in P. tomentosa associated with lignin content, stem volume, and microfibril angle, which explained up to 11% of the phenotypic variance. Our data in Arabidopsis suggest causality behind these associations.
The results with XET-affected plants indicate that other manipulation of XG metabolism could lead to strong effects on wood properties. This conclusion is supported by studies with the constitutive heterologous expression of Aspergillus aculeatus XYLOGLUCANASE2 in trees, which ectopically decreases XG content (Park et al., 2004). Such modified trees exhibited several developmental alterations, including stem height and diameter increase (Park et al., 2004; Hartati et al., 2011), increased cellulose content, density, and elastic modulus (Park et al., 2004), increased cellulose microfibril width (Yamamoto et al., 2011), and remarkable improvement in saccharification for both Glc yields (50% gains) and cellulose conversion (60% gains; Kaida et al., 2009). Such gains are among the highest reported so far for hemicellulose-modified tree species (Donev et al., 2018). Thus, the manipulation of XG metabolism is a vital route for tree improvement despite its low content in wood cell walls.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Sterile seeds of Arabidopsis (Arabidopsis thaliana) Columbia-0 (wild type), transgenic plants, and the mutants were vernalized in 0.1% (w/v) agar at 4°C for 3 d and planted in soil with vermiculite (3:1). Plants were grown at 22°C and light intensity of 120 to 150 µE m−2 s−1 under long-day conditions (16 h of light/8 h of light) for 6 to 8 weeks. For phenotyping secondarily thickened hypocotyls, plants were first subjected to short days (8 h of light/16 h of dark) for 4 weeks and then to long days for an additional 2 to 4 weeks.
Production and Selection of T-DNA Insertion Lines and PtxtXTH34 OE Lines
T-DNA mutant insertion lines (SAIL 681-G09 and SAIL 722-B10; Fig. 2A) obtained from the Nottingham Arabidopsis Stock Centre were backcrossed several times until single-locus (as determined by the segregation ratio) homozygotic plants were obtained. OE lines carrying PtxtXTH34 were obtained as described previously (Miedes et al., 2013).
Histochemical GUS Activity Tests for Promotor Activity Studies
pXTH4::GUS plants were obtained by transforming Columbia-0 with the vector containing 5′ upstream sequence (1 kb) from the start codon of XTH4 (At2g06850). The primers used were 5′-caccTATTTTGATAGTAGAGAGTTGGAATACCAT-3′ and 5′-AACAGTCATGGTTGTGTTTTAGAAAGAGAT-3′. The fragment was inserted into pBGGUS (Kubo et al., 2005). At least three independent lines were analyzed before selecting the representative line for final analyses. The pXTH9::GUS line was described by Becnel et al. (2006). Cross sections of the mature hypocotyls and basal parts of the stems of pXTH4::GUS and pXTH9::GUS lines were incubated for 4 h in a mixture of 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid cyclohexylammonium salt, 1 mm K33Fe(CN)6, 1 mm K44Fe(CN)6, 50 mm sodium phosphate buffer, pH 7.2, and 0.1% (v/v) Triton X-100 at 37°C, fixed in 5% (v/v) formaldehyde, 5% (v/v) acetic acid, and 50% (v/v) ethanol for 10 min, rinsed several times in 50% and then in 100% ethanol until the chlorophyll was completely removed, and observed with an Axioplan 2 microscope connected with an AxioVision camera (Carl Zeiss).
XET Activity by Radiometric Tritium Assay
Stems and hypocotyls of three 6-week-old plants were ground in dry ice and then added to 0.5 m NaCl (0.5 mg tissue 1 µL−1 salt). The slurry was centrifuged at 5,000g for 5 min at 4°C, and supernatants were analyzed for XET activity using radiometric assay (Fry et al., 1992). Ten microliters of each extract was incubated for 2 h at room temperature in a volume of 40 µL of reaction mixture containing 50 mm MES-Na, pH 6, 1.4 kBq of tritium-labeled XLLGol, and 2 mg mL−1 XG. The reaction was stopped by adding 100 µL of 20% (v/v) formic acid. The resulting 140-µL mixture was spotted onto a 5-cm × 5-cm piece of 3M paper and allowed to air dry overnight. The 3M paper was washed with running water at an ∼45° angle for 2 h and air dried. The scintillation was carried out with 2 mL of scintillant in a Tri-Carb 2100 liquid scintillation analyzer. Five biological replicates (pools of three plants) were used for each line.
Immunodetection of XG
The basal parts of the inflorescence stems and hypocotyls of 6-week-old plants were cross sectioned at 60 µm thickness using a vibratome (VT 1000S; Leica) and fixed in 4% (w/v) paraformaldehyde dissolved in 0.5 m PIPES buffer, pH 7, for 30 min. The sections were then washed five times for 3 min in phosphate-buffered saline with 0.1% (v/v) Tween 20 (PBS-T), blocked in 5% (w/v) skim milk in phosphate-buffered saline buffer for 1 h at room temperature, incubated in primary antibody CCRC-M1 (a gift from Dr. Michael Hahn) for 2 h at room temperature, washed five times for 3 min in PBS-T, and incubated in secondary antibody Alexa Fluor 488 anti-mouse gold (Nanoprobes) for 2 h in the dark. After removal of the secondary antibody by washing five times for 3 min in PBS-T, the sections were observed by a laser scanning confocal microscope (Carl Zeiss; LSM 510) with a 488-nm argon-krypton laser.
Histological Analysis
The hand cross sections of mature hypocotyls were stained with 1% (w/v) phloroglucinol in 18% (v/v) HCl and viewed with an Axioplan 2 microscope equipped with an AxioVision camera (Carl Zeiss). The proportions between tissues were measured for 10 hypocotyls from each line using image analysis (AxioVision). Four measurements per section were made along four equidistant radii around the hypocotyl circumference.
Wood Maceration
Ten hypocotyls of each line were put in a tube and macerated in a mixture of 3% (v/v) hydrogen peroxide and 50% (v/v) glacial acetate acid at 95°C for 8 h, washed in water, and neutralized. The individual cells were released by vigorous shaking of tubes and viewed with differential interference contrast in an Axioplan microscope (Carl Zeiss). For each analyzed line, the length and width of 100 fibers and 300 vessel elements were measured using AxioVision image-analysis software (Carl Zeiss).
TEM
Approximately 3-mm-long segments from the basal part of the inflorescence stems and from hypocotyls of two 8-week-old plants for each line (wild type, xth4, xth9, xth4x9, and OE18) were fixed in 4% (w/v) paraformaldehyde and 0.05% (v/v) glutaraldehyde (Analytical Standards) in 25 mm phosphate buffer, pH 7.2. They were dehydrated in a graded ethanol series, embedded in LR-White resin (TAAB), and polymerized at 60°C for 16 to 20 h. Cross sections were cut using an ultramicrotome and observed with a JEM-1230 transmission electron microscope (JEOL). Measurements of the cell wall thickness (without the compound middle lamella) were performed in 100 cells (50 cells per plant) for hypocotyl fibers and 15 cells (seven to eight cells per plant) for the other cell types, at all contacts with neighboring cells, and counts of secondary cell wall layers were performed for 50 cells of each type (25 cells per plant), including interfascicular fibers, fascicular fibers, fascicular vessel elements in the stems, and secondary xylem fibers and vessel elements in hypocotyls.
Cell Wall Analyses
Approximately 30-d-old plants having 20-cm-long stems were used for cell wall analyses. Basal 10-cm-long segments were collected and pooled to make one biological replicate. They were immediately frozen in liquid N2, freeze-dried for 36 h, and ball milled to fine powder in stainless steel jars with one 12-mm grinding ball at 30 Hz for 2 min, using an MM400 bead mill (Retsch). The AIR1 was prepared from wood powder as described earlier and analyzed by pyrolysis gas chromatography-MS and FTIR spectroscopy (Gandla et al., 2015). AIR1 material was destarched to obtain AIR2 and used for the determination of monosugar composition after methanolysis and trimethylsilyl derivatization and cellulose content (Gandla et al., 2015). AIR1 and AIR2 fractions were used to analyze lignin content (Foster et al., 2010).
Vibrational Microspectroscopy
Segments (0.5 cm) from hypocotyls and from the basal part of side branches from 6-week-old plants were immediately frozen in liquid N2 and stored in −20°C. Frozen material was mounted in O.C.T. compound (361603E; VWR Chemicals) and sectioned using an HM505E (Microm) cryostat. Ten-micrometer-thick cross sections were washed in water and transferred to a Raman and infrared transparent CaF2 slides. The sections were dried in a desiccator for at least 48 h. The same section was analyzed by in situ FTIR and Raman microspectroscopy.
FTIR Microspectroscopy
FTIR microspectroscopic measurements and data analyses were performed as described by Gorzsás et al. (2011). Spectra were recorded using 32 scans at a spectral resolution of 4 cm−1 on a Bruker Tensor 27 spectrometer equipped with a Hyperion 3000 microscopy accessory, including a liquid nitrogen-cooled 64 × 64 mercury cadmium telluride focal plane array detector (Bruker Optik). Five plants were used for each line, and four to seven positions were selected in each section for hyperspectral imaging, representing entire cross sections. Suboptimal quality images (such as saturated spectra due to folding upon drying, etc.) were excluded from further analyses. Ten spectra per image and cell type were extracted, compiled, and exported as .mat files using OPUS (version 7.0.129; Bruker Optik). The spectral region of 900 to 1,800 cm−1 was used in subsequent analyses.
Raman Microspectroscopy
Raman microspectroscopy (mapping) was performed as described by Gorzsás (2017) on a Renishaw InVia Raman spectrometer equipped with a CCD detector, using a 514-nm Ar+ ion laser with 2-s irradiation time and 100% laser power set by the software (WiRE, versions 3.0–3.4), in static mode, centered at 1,190 cm−1 (resulting in a spectral region of approximately 510–1,803 cm−1 with approximately 1 cm−1 spectral resolution using a 2,400-line grating). A 50× magnification lens was used, with 1.5-μm step sizes (xy direction), in normal confocal mode, recording images consisting of a minimum of 100 spectra per image. Raman shifts were calibrated using the built-in Si standard of the instrument. Cosmic rays were removed, and the hyperspectral data noise was filtered using the chemometrics package of WiRE (version 3.4). Five plants were analyzed for each line, recording two independent hyperspectral images per plant, and using five (fascicular fibers and hypocotyl secondary xylem fibers) or 10 (interfascicular fibers) spectra per image for multivariate analyses. Suboptimal quality images were excluded.
Processing and Data Evaluation of Vibrational Spectra
Spectra were preprocessed using the open-source MCR-ALS script (run in Matlab environments, versions 14a–18a; Mathworks), as provided by the Vibrational Spectroscopy Core Facility (http://www.kbc.umu.se/english/visp/download-visp/). Baseline correction was performed using asymmetrical least squares (Eilers, 2004), with λ varying between 100,000 (Raman spectra) and 10,000,000 (diffuse reflectance FTIR spectra), while P was always kept at a minimum (0.001). Spectra were smoothed by Savitzky-Golay filtering (Savitzky and Golay, 1964) using a first-order polynomial and a frame rate of 5. Spectra were total area normalized in the spectral region used for the analyses and analyzed by SIMCA-P (versions 13.0–14.0; Umetrics). Spectral data were centered, while y variables (average cell wall layer numbers) were unit variance scaled. Initial principal component analysis models were created using the Autofit function for overview purposes and identifying potential outliers before OPLS-DA models. OPLS-DA models used a fixed number of components, as listed for each model, kept to a minimum to avoid overfitting.
Gene Expression Analyses
Total RNA was extracted from frozen ground basal stem and hypocotyl tissues using the RNeasy Plant Mini Kit (Qiagen), DNase treated using the DNA‐free Kit (Invitrogen), and cDNA was synthesized using the iScript cDNA synthesis kit (Bio‐Rad) following the manufacturer’s protocol. RT-qPCR was performed using Light Cycler 480 SYBR Green I master mix (Roche) in a Light Cycler 480 II instrument (Roche) as per the manufacturer’s instructions. Primers used for RT-qPCR are listed in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of secondary growth-related XTH genes in Populus trichocarpa and Arabidopsis.
Supplemental Figure S2. Effects of altered XET level on cell wall ultrastructure in inflorescence stem fascicular and interfascicular fibers as visualized by TEM.
Supplemental Figure S3. Effects of altered XET level on cell wall ultrastructure in inflorescence stem xylem vessel elements as visualized by TEM.
Supplemental Figure S4. Effects of altered XET level on cell wall morphology in hypocotyl xylem vessel elements as visualized by TEM.
Supplemental Figure S5. OPLS-DA models (pairwise comparisons) based on diffuse reflectance FTIR spectra from basal inflorescence stem AIR of different genotypes.
Supplemental Figure S6. In situ FTIR microspectroscopy of cell walls in fibers of inflorescence stems and hypocotyls in xth mutants and plants with strong heterologous expression of PtxtXTH34.
Supplemental Figure S7. OPLS-DA models (pairwise comparisons) for interfascicular fibers based on Raman microspectroscopic data.
Supplemental Table S1. List of primers used in RT-qPCR.
Acknowledgments
We thank Dr. Takahisa Hayashi for helpful comments on the article, Dr. Michael Hahn for CCRC-M1 antibody, Dr. Hans Stenlund for advice on analyses of microspectroscopic data, the Nottingham Arabidopsis Seed Centre for the SAIL lines, and the Umeå Core Facility for Electron Microscopy for help and advice.
Footnotes
This work was supported by the Chemical Biological Centre and the Department of Chemistry of Umeå University for the Vibrational Spectroscopy Core Facility, Bio4Energy, TC4F for the Plant Analytical Biopolymer platform, and Vinnova (the Swedish Governmental Agency for Innovation Systems) and the Knut and Alice Wallenberg Foundation for the plant growth facility. This work was also supported by the Swedish Foundation for Strategic Research (ValueTree project RBP14-0011), the Swedish Research Council Formas, and the Swedish Research Council (grants to E.J.M.).
Articles can be viewed without a subscription.
References
- Agarwal UP, Atalla RH (2010) Vibrational spectroscopy In Heitner C, Dimmel DR, and Schmidt JA, eds, Lignin and Lignans: Advances in Chemistry. CRC Press, Boca Raton, FL, pp 103–136 [Google Scholar]
- Agarwal UP, McSweeny JD, Ralf SA (2011) FT-Raman investigation of milled-wood lignins: Softwood, hardwood, and chemically modified black spruce lignins. J Wood Chem Technol 31: 324–344 [Google Scholar]
- Agarwal UP, Ralf SA (1997) FT-Raman spectroscopy of wood: Identifying contributions of lignin and carbohydrate polymers in the spectrum of black spruce (Picea mariana). Appl Spectrosc 51: 1648–1655 [Google Scholar]
- Agarwal UP. (1999) An overview of Raman spectroscopy as applied to lignocellulosic materials In Argyropoulos D, ed, Advances in Lignocellulosics Characterization. Tappi Press, Atlanta, GA, pp 201–205 [Google Scholar]
- Ambrose JC, Shoji T, Kotzer AM, Pighin JA, Wasteneys GO (2007) The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19: 2763–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antosiewicz DM, Purugganan MM, Polisensky DH, Braam J (1997) Cellular localization of Arabidopsis xyloglucan endotransglycosylase-related proteins during development and after wind stimulation. Plant Physiol 115: 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armezzani A, Abad U, Ali O, Andres Robin A, Vachez L, Larrieu A, Mellerowicz EJ, Taconnat L, Battu V, Stanislas T, et al. (2018) Transcriptional induction of cell wall remodelling genes is coupled to microtubule-driven growth isotropy at the shoot apex in Arabidopsis. Development 145: dev162255. [DOI] [PubMed] [Google Scholar]
- Baba K, Park YW, Kaku T, Kaida R, Takeuchi M, Yoshida M, Hosoo Y, Ojio Y, Okuyama T, Taniguchi T, et al. (2009) Xyloglucan for generating tensile stress to bend tree stem. Mol Plant 2: 893–903 [DOI] [PubMed] [Google Scholar]
- Baumann MJ, Eklöf JM, Michel G, Kallas Å, Teeri TT, Czjzek M, Brumer H III (2007) Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: Biological implications for cell wall metabolism. Plant Cell 19: 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel J, Natarajan M, Kipp A, Braam J (2006) Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol 61: 451–467 [DOI] [PubMed] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14: 3073–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui D, Neill SJ, Tang Z, Cai W (2005) Gibberellin-regulated XET is differentially induced by auxin in rice leaf sheath bases during gravitropic bending. J Exp Bot 56: 1327–1334 [DOI] [PubMed] [Google Scholar]
- Derba-Maceluch M, Awano T, Takahashi J, Lucenius J, Ratke C, Kontro I, Busse-Wicher M, Kosik O, Tanaka R, Winzéll A, et al. (2015) Suppression of xylan endotransglycosylase PtxtXyn10A affects cellulose microfibril angle in secondary wall in aspen wood. New Phytol 205: 666–681 [DOI] [PubMed] [Google Scholar]
- Dimmer E, Roden L, Cai D, Kingsnorth C, Mutasa-Göttgens E (2004) Transgenic analysis of sugar beet xyloglucan endo-transglucosylase/hydrolase Bv-XTH1 and Bv-XTH2 promoters reveals overlapping tissue-specific and wound-inducible expression profiles. Plant Biotechnol J 2: 127–139 [DOI] [PubMed] [Google Scholar]
- Dokken KM, Davis LC, Marinkovic NS (2005) Use of infrared microspectroscopy in plant growth and development. Appl Spectrosc Rev 40: 301–326 [Google Scholar]
- Donev E, Gandla ML, Jönsson LJ, Mellerowicz EJ (2018) Engineering non-cellulosic polysaccharides of wood for the biorefinery. Front Plant Sci 9: 1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers PH. (2004) Parametric time warping. Anal Chem 76: 404–411 [DOI] [PubMed] [Google Scholar]
- Eklöf JM, Brumer H (2010) The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol 153: 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix O. (1991) Classifications of lignins from different botanical origins by FT-IR spectroscopy. Holzforschung 45(Suppl): 21–27 [Google Scholar]
- Farkas V, Sulova Z, Stratilova E, Hanna R, Maclachlan G (1992) Cleavage of xyloglucan by nasturtium seed xyloglucanase and transglycosylation to xyloglucan subunit oligosaccharides. Arch Biochem Biophys 298: 365–370 [DOI] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part I: Lignin. J Vis Exp 37: e1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ (1992) Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J 282: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandla ML, Derba-Maceluch M, Liu X, Gerber L, Master ER, Mellerowicz EJ, Jönsson LJ (2015) Expression of a fungal glucuronoyl esterase in Populus: Effects on wood properties and saccharification efficiency. Phytochemistry 112: 210–220 [DOI] [PubMed] [Google Scholar]
- Gerttula S, Zinkgraf M, Muday GK, Lewis DR, Ibatullin FM, Brumer H, Hart F, Mansfield SD, Filkov V, Groover A (2015) Transcriptional and hormonal regulation of gravitropism of woody stems in Populus. Plant Cell 27: 2800–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierlinger N. (2017) New insights into plant cell walls by vibrational microspectroscopy. Appl Spectrosc Rev 53: 517–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez LD, Whitehead C, Barakate A, Halpin C, McQueen-Mason SJ (2010) Automated saccharification assay for determination of digestibility in plant materials. Biotechnol Biofuels 3: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Pérez L, Perrotta L, Acosta A, Orellana E, Spadafora N, Bruno L, Bitonti BM, Albani D, Cabrera JC, Francis D, et al. (2014) In tobacco BY-2 cells xyloglucan oligosaccharides alter the expression of genes involved in cell wall metabolism, signalling, stress responses, cell division and transcriptional control. Mol Biol Rep 41: 6803–6816 [DOI] [PubMed] [Google Scholar]
- González-Pérez L, Vázquez Glaría A, Perrotta L, Acosta Maspons A, Scriven SA, Herbert R, Cabrera JC, Francis D, Rogers HJ (2012) Oligosaccharins and Pectimorf stimulate root elongation and shorten the cell cycle in higher plants. Plant Growth Regul 68: 211–221 [Google Scholar]
- Gorzsás A. (2017) Chemical imaging of xylem by Raman microspectroscopy. Methods Mol Biol 1544: 133–178 [DOI] [PubMed] [Google Scholar]
- Gorzsás A, Stenlund H, Persson P, Trygg J, Sundberg B (2011) Cell-specific chemotyping and multivariate imaging by combined FT-IR microspectroscopy and orthogonal projections to latent structures (OPLS) analysis reveals the chemical landscape of secondary xylem. Plant J 66: 903–914 [DOI] [PubMed] [Google Scholar]
- Goulao LF, Vieira-Silva S, Jackson PA (2011) Association of hemicellulose- and pectin-modifying gene expression with Eucalyptus globulus secondary growth. Plant Physiol Biochem 49: 873–881 [DOI] [PubMed] [Google Scholar]
- Hara Y, Yokoyama R, Osakabe K, Toki S, Nishitani K (2014) Function of xyloglucan endotransglucosylase/hydrolases in rice. Ann Bot 114: 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartati S, Sudarmonowati E, Kaku T, Ikegaya H, Kaida R, Baba K, Hayashi T (2011) Overexpression of xyloglucanase (AaXEG2) accelerates heteroblastic development in mangium leaves. J Wood Sci 57: 463–469 [Google Scholar]
- Hayashi T, Takeda T, Ogawa K, Mitsuishi Y (1994) Effects of the degree of polymerization on the binding of xyloglucans to cellulose. Plant Cell Physiol 35: 893–899 [PubMed] [Google Scholar]
- Hrmova M, Farkas V, Lahnstein J, Fincher GB (2007) A barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1,3;1,4)-β-D-glucans. J Biol Chem 282: 12951–12962 [DOI] [PubMed] [Google Scholar]
- Ikematsu S, Tasaka M, Torii KU, Uchida N (2017) ERECTA-family receptor kinase genes redundantly prevent premature progression of secondary growth in the Arabidopsis hypocotyl. New Phytol 213: 1697–1709 [DOI] [PubMed] [Google Scholar]
- Jiménez T, Martín I, Labrador E, Dopico B (2006) The immunolocation of a xyloglucan endotransglucosylase/hydrolase specific to elongating tissues in Cicer arietinum suggests a role in the elongation of vascular cells. J Exp Bot 57: 3979–3988 [DOI] [PubMed] [Google Scholar]
- Jordá L, Sopeña-Torres S, Escudero V, Nuñez-Corcuera B, Delgado-Cerezo M, Torii KU, Molina A (2016) ERECTA and BAK1 receptor like kinases interact to regulate immune responses in Arabidopsis. Front Plant Sci 7: 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kačuráková M, Wellner N, Ebringerová A, Hromádková Z, Wilson RH, Belton PS (1999) Characterisation of xylan-type polysaccharides and associated cell wall components by FT-IR and FT-Raman spectroscopies. Food Hydrocoll 13: 35–41 [Google Scholar]
- Kaewthai N, Gendre D, Eklöf JM, Ibatullin FM, Ezcurra I, Bhalerao RP, Brumer H (2013) Group III-A XTH genes of Arabidopsis encode predominant xyloglucan endohydrolases that are dispensable for normal growth. Plant Physiol 161: 440–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida R, Kaku T, Baba K, Oyadomari M, Watanabe T, Nishida K, Kanaya T, Shani Z, Shoseyov O, Hayashi T (2009) Loosening xyloglucan accelerates the enzymatic degradation of cellulose in wood. Mol Plant 2: 904–909 [DOI] [PubMed] [Google Scholar]
- Kaida R, Sugawara S, Negoro K, Maki H, Hayashi T, Kaneko TS (2010) Acceleration of cell growth by xyloglucan oligosaccharides in suspension-cultured tobacco cells. Mol Plant 3: 549–554 [DOI] [PubMed] [Google Scholar]
- Kaku T, Serada S, Baba K, Tanaka F, Hayashi T (2009) Proteomic analysis of the G-layer in poplar tension wood. J Wood Sci 55: 250–257 [Google Scholar]
- Kallas Å, Piens K, Denman SE, Henriksson H, Fäldt J, Johansson P, Brumer H, Teeri TT (2005) Enzymatic properties of native and deglycosylated hybrid aspen (Populus tremula × tremuloides) xyloglucan endotransglycosylase 16A expressed in Pichia pastoris. Biochem J 390: 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev AV, Buschmann H, Doonan JH, Lloyd CW (2007) AtMAP70-5, a divergent member of the MAP70 family of microtubule-associated proteins, is required for anisotropic cell growth in Arabidopsis. J Cell Sci 120: 2241–2247 [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Hainaut M, Delhomme N, Mannapperuma C, Immerzeel P, Street NR, Henrissat B, Mellerowicz EJ (2019) Poplar carbohydrate-active enzymes: Whole-genome annotation and functional analyses based on RNA expression data. Plant J 99: 589–60931111606 [Google Scholar]
- Larsen KL, Barsberg S (2010) Theoretical and Raman spectroscopic studies of phenolic lignin model monomers. J Phys Chem B 114: 8009–8021 [DOI] [PubMed] [Google Scholar]
- Lee D, Polisensky DH, Braam J (2005) Genome-wide identification of touch- and darkness-regulated Arabidopsis genes: A focus on calmodulin-like and XTH genes. New Phytol 165: 429–444 [DOI] [PubMed] [Google Scholar]
- Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, Naumann A, Kang KY, Kim H, Ruel K, et al. (2007) Downregulation of cinnamoyl-coenzyme A reductase in poplar: Multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wu HM, Cheung AY (2016) FERONIA and her pals: Functions and mechanisms. Plant Physiol 171: 2379–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YB, Lu SM, Zhang JF, Liu S, Lu YT (2007) A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in Arabidopsis. Planta 226: 1547–1560 [DOI] [PubMed] [Google Scholar]
- Maris A, Suslov D, Fry SC, Verbelen JP, Vissenberg K (2009) Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J Exp Bot 60: 3959–3972 [DOI] [PubMed] [Google Scholar]
- Martin L, Decourteix M, Badel E, Huguet S, Moulia B, Julien JL, Leblanc-Fournier N (2014) The zinc finger protein PtaZFP2 negatively controls stem growth and gene expression responsiveness to external mechanical loads in poplar. New Phytol 203: 168–181 [DOI] [PubMed] [Google Scholar]
- Matsui A, Yokoyama R, Seki M, Ito T, Shinozaki K, Takahashi T, Komeda Y, Nishitani K (2005) AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J 42: 525–534 [DOI] [PubMed] [Google Scholar]
- McDougall GJ, Fry SC (1988) Inhibition of auxin-stimulated growth of pea stem segments by a specific nonasaccharide of xyloglucan. Planta 175: 412–416 [DOI] [PubMed] [Google Scholar]
- McDougall GJ, Fry SC (1989) Anti-auxin activity of xyloglucan oligosaccharides: The role of groups other than the terminal a-L-fucose residue. J Exp Bot 40: 233–238 [Google Scholar]
- Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W (2001) Unravelling cell wall formation in the woody dicot stem. Plant Mol Biol 47: 239–274 [PubMed] [Google Scholar]
- Miedes E, Suslov D, Vandenbussche F, Kenobi K, Ivakov A, Van Der Straeten D, Lorences EP, Mellerowicz EJ, Verbelen JP, Vissenberg K (2013) Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. J Exp Bot 64: 2481–2497 [DOI] [PubMed] [Google Scholar]
- Nishikubo N, Awano T, Banasiak A, Bourquin V, Ibatullin F, Funada R, Brumer H, Teeri TT, Hayashi T, Sundberg B, et al. (2007) Xyloglucan endo-transglycosylase (XET) functions in gelatinous layers of tension wood fibers in poplar: A glimpse into the mechanism of the balancing act of trees. Plant Cell Physiol 48: 843–855 [DOI] [PubMed] [Google Scholar]
- Nishikubo N, Takahashi J, Roos AA, Derba-Maceluch M, Piens K, Brumer H, Teeri TT, Stålbrand H, Mellerowicz EJ (2011) Xyloglucan endo-transglycosylase-mediated xyloglucan rearrangements in developing wood of hybrid aspen. Plant Physiol 155: 399–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K, Tominaga R (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267: 21058–21064 [PubMed] [Google Scholar]
- Oh MH, Romanow WG, Smith RC, Zamski E, Sassa J, Clouse SD (1998) Soybean BRU1 encodes a functional xyloglucan endotransglycosylase that is highly expressed in inner epicotyl tissues during brassinosteroid promoted elongation. Plant Cell Physiol 39: 124–130 [Google Scholar]
- Ohba T, Takahashi S, Asada K (2011) Alteration of fruit characteristics in transgenic tomatoes with modified expression of a xyloglucan endotransglucosylase/hydrolase gene. Plant Biotechnol 28: 25–32 [Google Scholar]
- Okazawa K, Sato Y, Nakagawa T, Asada K, Kato I, Tomita E, Nishitani K (1993) Molecular cloning and cDNA sequencing of endoxyloglucan transferase, a novel class of glycosyltransferase that mediates molecular grafting between matrix polysaccharides in plant cell walls. J Biol Chem 268: 25364–25368 [PubMed] [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K (2006) A principal role for AtXTH18 in Arabidopsis thaliana root growth: A functional analysis using RNAi plants. J Plant Res 119: 153–162 [DOI] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2012) A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol 158: 1933–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2015) Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol 56: 180–194 [DOI] [PubMed] [Google Scholar]
- Park YW, Baba K, Furuta Y, Iida I, Sameshima K, Arai M, Hayashi T (2004) Enhancement of growth and cellulose accumulation by overexpression of xyloglucanase in poplar. FEBS Lett 564: 183–187 [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K (2016) Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu Rev Plant Biol 67: 235–259 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Kong Y, York WS, O’Neill MA (2012) A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell 24: 4511–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E, Korolev AV, Calder G, Lloyd CW (2010) The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Curr Biol 20: 744–749 [DOI] [PubMed] [Google Scholar]
- Popper ZA, Fry SC (2005) Widespread occurrence of a covalent linkage between xyloglucan and acidic polysaccharides in suspension-cultured angiosperm cells. Ann Bot 96: 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Fry SC (2008) Xyloglucan-pectin linkages are formed intra-protoplasmically, contribute to wall-assembly, and remain stable in the cell wall. Planta 227: 781–794 [DOI] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Hervé C, Domozych DS, Willats WG, Tuohy MG, Kloareg B, Stengel DB (2011) Evolution and diversity of plant cell walls: From algae to flowering plants. Annu Rev Plant Biol 62: 567–590 [DOI] [PubMed] [Google Scholar]
- Puhlmann J, Bucheli E, Swain MJ, Dunning N, Albersheim P, Darvill AG, Hahn MG (1994) Generation of monoclonal antibodies against plant cell-wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal alpha-(1→2)-linked fucosyl-containing epitope. Plant Physiol 104: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajangam AS, Kumar M, Aspeborg H, Guerriero G, Arvestad L, Pansri P, Brown CJ, Hober S, Blomqvist K, Divne C, et al. (2008) MAP20, a microtubule-associated protein in the secondary cell walls of hybrid aspen, is a target of the cellulose synthesis inhibitor 2,6-dichlorobenzonitrile. Plant Physiol 148: 1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Carlsbecker A, Etchells JP (2017) Class III HD-ZIPs govern vascular cell fate: An HD view on patterning and differentiation. J Exp Bot 68: 55–69 [DOI] [PubMed] [Google Scholar]
- Ratke C, Terebieniec BK, Winestrand S, Derba-Maceluch M, Grahn T, Schiffthaler B, Ulvcrona T, Özparpucu M, Rüggeberg M, Lundqvist SO, et al. (2018) Downregulating aspen xylan biosynthetic GT43 genes in developing wood stimulates growth via reprograming of the transcriptome. New Phytol 219: 230–245 [DOI] [PubMed] [Google Scholar]
- Romo S, Jiménez T, Labrador E, Dopico B (2005) The gene for a xyloglucan endotransglucosylase/hydrolase from Cicer arietinum is strongly expressed in elongating tissues. Plant Physiol Biochem 43: 169–176 [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Ruel K, Montiel MD, Goujon T, Jouanin L, Burlat V, Joseleau JP (2002) Interrelation between lignin deposition and polysaccharide matrices during the assembly of plant cell walls. Plant Biol 4: 2–8 [Google Scholar]
- Sasidharan R, Chinnappa CC, Staal M, Elzenga JT, Yokoyama R, Nishitani K, Voesenek LA, Pierik R (2010) Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol 154: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R, Keuskamp DH, Kooke R, Voesenek LACJ, Pierik R (2014) Interactions between auxin, microtubules and XTHs mediate green shade-induced petiole elongation in Arabidopsis. PLoS ONE 9: e90587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitzky A, Golay MJE (1964) Smoothing and differentiation of data by simplified least-squares procedures. Anal Chem 36: 1627–1639 [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Schenzel K, Fischer S (2001) NIR FT Raman spectroscopy: A rapid analytical tool for detecting the transformation of cellulose polymorphs. Cellulose 8: 49–57 [Google Scholar]
- Schulz H, Baranska M (2007) Identification and quantification of valuable plant substances by IR and Raman spectroscopy. Vib Spectrosc 43: 13–25 [Google Scholar]
- Shin YK, Yum H, Kim ES, Cho H, Gothandam KM, Hyun J, Chung YY (2006) BcXTH1, a Brassica campestris homologue of Arabidopsis XTH9, is associated with cell expansion. Planta 224: 32–41 [DOI] [PubMed] [Google Scholar]
- Shinohara N, Sunagawa N, Tamura S, Yokoyama R, Ueda M, Igarashi K, Nishitani K (2017) The plant cell-wall enzyme AtXTH3 catalyses covalent cross-linking between cellulose and cello-oligosaccharide. Sci Rep 7: 46099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons TJ, Fry SC (2017) Bonds broken and formed during the mixed-linkage glucan: Xyloglucan endotransglucosylase reaction catalysed by Equisetum hetero-trans-β-glucanase. Biochem J 474: 1055–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons TJ, Mohler KE, Holland C, Goubet F, Franková L, Houston DR, Hudson AD, Meulewaeter F, Fry SC (2015) Hetero-trans-β-glucanase, an enzyme unique to Equisetum plants, functionalizes cellulose. Plant J 83: 753–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundell D, Street NR, Kumar M, Mellerowicz EJ, Kucukoglu M, Johnsson C, Kumar V, Mannapperuma C, Delhomme N, Nilsson O, et al. (2017) AspWood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29: 1585–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Furuta Y, Awano T, Mizuno K, Mitsuishi Y, Hayashi T (2002) Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc Natl Acad Sci USA 99: 9055–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, et al. (2013) An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25: 270–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Fry SC (2000) Evidence for covalent linkage between xyloglucan and acidic pectins in suspension-cultured rose cells. Planta 211: 275–286 [DOI] [PubMed] [Google Scholar]
- Van Sandt VS, Suslov D, Verbelen JP, Vissenberg K (2007) Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann Bot 100: 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Fry SC, Pauly M, Höfte H, Verbelen JP (2005a) XTH acts at the microfibril-matrix interface during cell elongation. J Exp Bot 56: 673–683 [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Martinez-Vilchez IM, Verbelen JP, Miller JG, Fry SC (2000) In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell 12: 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Oyama M, Osato Y, Yokoyama R, Verbelen JP, Nishitani K (2005b) Differential expression of AtXTH17, AtXTH18, AtXTH19 and AtXTH20 genes in Arabidopsis roots: Physiological roles in specification in cell wall construction. Plant Cell Physiol 46: 192–200 [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Van Sandt V, Fry SC, Verbelen JP (2003) Xyloglucan endotransglucosylase action is high in the root elongation zone and in the trichoblasts of all vascular plants from Selaginella to Zea mays. J Exp Bot 54: 335–344 [DOI] [PubMed] [Google Scholar]
- Wang B, Zhang D (2014) Association of allelic variation in PtoXET16A with growth and wood properties in Populus tomentosa. Int J Mol Sci 15: 16949–16974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu L, Liu B, Wang C, Jin L, Zhao Q, Yuan M (2007) Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. (2010) The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38: W214–W220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley JH, Atalla RH (1987) Band assignments in the Raman spectra of celluloses. Carbohydr Res 160: 113–129 [Google Scholar]
- Wolf S. (2017) Plant cell wall signalling and receptor-like kinases. Biochem J 474: 471–492 [DOI] [PubMed] [Google Scholar]
- Xiao C, Zhang T, Zheng Y, Cosgrove DJ, Anderson CT (2016) Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiol 170: 234–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, Kieber JJ (2008) Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20: 3065–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Saito T, Isogai A, Kurita M, Kondo T, Taniguchi T, Kaida R, Baba K, Hayashi T (2011) Enlargement of individual cellulose microfibrils in transgenic poplars overexpressing xyloglucanase. J Wood Sci 57: 71–75 [Google Scholar]
- Yokoyama R, Nishitani K (2001) A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol 42: 1025–1033 [DOI] [PubMed] [Google Scholar]
- York WS, Darvill AG, Albersheim P (1984) Inhibition of 2,4-dichlorophenoxyacetic acid-stimulated elongation of pea stem segments by a xyloglucan oligosaccharide. Plant Physiol 75: 295–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yarbrough JM, Mittal A, Tucker MP, Vinzant TB, Decker SR, Himmel ME (2016) In situ label-free imaging of hemicellulose in plant cell walls using stimulated Raman scattering microscopy. Biotechnol Biofuels 9: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Morrison WH III, Ye ZH (2002) A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14: 3101–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2015) Secondary cell walls: Biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 56: 195–214 [DOI] [PubMed] [Google Scholar]