High acetylation in the promoter of a transcription factor gene is associated with sucrose accumulation in pear fruit by activating the expression of a sucrose transport gene.
Abstract
Sugar content is an important trait of fleshy fruit, and elevating Suc levels is a major goal in horticultural crop breeding. Here, we examined the sugar content in two varieties of the Ussurian pear (Pyrus ussuriensis), ‘Nanguo’ (NG) and its bud sport (BNG), and we found that Suc content was higher in BNG fruit than in NG fruit. We compared the transcriptomes of the two varieties using RNA sequencing and identified a SWEET (Sugars Will Eventually be Exported Transporter) gene, PuSWEET15, expressed at higher levels in BNG fruit. Heterologous expression of PuSWEET15 in a SUSY7/ura yeast (Saccharomyces cerevisiae) strain showed that PuSWEET15 is an active Suc transporter. Overexpression of PuSWEET15 in NG pear fruit increased Suc content, while silencing of PuSWEET15 in BNG fruit decreased Suc content. The WRKY transcription factor PuWRKY31 was also expressed more highly in BNG fruit than in NG fruit, and we found that PuWRKY31 bound to the PuSWEET15 promoter and induced its transcription. The histone acetylation level of the PuWRKY31 promoter was higher in BNG fruit, suggesting a mechanism by which Suc levels can be elevated.
In plants, the three major soluble sugars are Suc, Glc, and Fru. Of these, Suc is the main carbohydrate transported from the photosynthetic source tissues to heterotrophic sink tissues, and so is central to the resource allocation system (Rennie and Turgeon, 2009; Eom et al., 2012; Braun et al., 2014). Suc represents a metabolic resource for carbon skeleton construction and energy, allowing growth and development, and is also an important contributor to the sweetness and flavor of many fleshy fruits (Braun et al., 2014). Sweetness is one of the main factors of fruit quality and it has been recognized as an important driver of consumer preference (Jaeger et al., 1998). Thus, an understanding of the mechanisms involved in Suc transport and the enhancement of sugar accumulation in fruit is of both fundamental and applied importance.
Suc accumulation in fruits depends on its transportation and metabolism. The enzymes involved in the metabolism of Suc are Suc phosphate synthase (SPS), Suc synthase (SS), and invertase (INV; Stitt et al., 1988; Moriguchi et al., 1992; Sturm et al., 1999). Suc movement between cells can be passive, through plasmodesmata along a concentration gradient, or active, involving transporters such as membrane-localized Suc transporters (SUTs) that translocate Suc from mesophyll cells into the phloem in leaves (Riesmeier et al., 1992; Lemoine, 2000). It has also been shown that intracellular Suc is transported from mesophyll cells to the apoplast by the SWEET (Sugars Will Eventually be Exported Transporter) proteins (Chen et al., 2012). The role of SWEET genes in Suc transport was first identified in Arabidopsis (Arabidopsis thaliana), in which a double mutation of AtSWEET11 and AtSWEET12 caused severe growth retardation and reduced Suc content in the vascular bundles but increased Suc levels in the leaves (Chen et al., 2012). These results demonstrated that the SWEET genes play important roles in Suc phloem loading, and led to subsequent identification and characterization of SWEET family members in other plant species, including rice (Oryza sativa), soybean (Glycine max), grape (Vitis vinifera), apple (Malus domestica), sorghum (Sorghum bicolor), and pear (Pyrus bretschneideri; Yuan and Wang, 2013; Chong et al., 2014; Wei et al., 2014; Patil et al., 2015; Mizuno et al., 2016; Li et al., 2017a). SWEET transporters are predicted to have seven transmembrane segments (TMSs) with two distinct repeated units of three TMSs and a connecting fourth TMS (Xuan et al., 2013). There are 21 SWEET genes in the rice genome, among which OsSWEET11 (also called Os8N3/Xa13) and OsSWEET14 (Os11N3) encode proteins that are localized to the plasma membrane and so likely affect sugar levels in the apoplast. Knocking out OsSWEET11 causes smaller seeds, reduced pollen viability, defective stamens, and decreased Suc content (Chu et al., 2006; Yang et al., 2006; Ma et al., 2017; Gao et al., 2018). In Arabidopsis, AtSWEET15 localizes to the plasma membrane, and its transcript levels are significantly higher during water stress, suggesting a role in Suc apoplastic unloading (Durand et al., 2016). AtSWEET17 is a Fru transporter (Guo et al., 2014). In soybean, GmSWEET15 mediates Suc export from endosperm to early embryo, and in the gmsweet15 mutant, the Suc and Glc contents are significantly decreased in all seed parts compared with the wild type (Wang et al., 2019). Moreover, a Medicago truncatula MtSWEET1b transporter supplies Glc for Arbuscular mycorrhizal (An et al., 2019a). These findings suggest a broad role for SWEET genes in sugar transportation.
Pear is a very important horticultural crop in the world. With the published pear genome, genes related to many quality traits such as stone cells, sugar, acid, volatiles, color, and ripening have been identified (Wu et al., 2013; Chagné et al., 2014; Dong et al., 2019). This provides plenty of resources to study the formation of quality traits in pear. However, information regarding sugar accumulation in pear fruit is still lacking. In this study, we characterized the basis of sweetness in the ‘Nanguo’ (NG) clonal variety of Ussurian pear (Pyrus ussuriensis). NG is highly valued by growers and consumers because of its cold resistance, taste, and aroma (Huang et al., 2014). In perennial fruits, a new variety that derives from shoot cells of the parent, presumably through genetic or epigenetic alterations, is called a bud sport variety (Furiya et al., 2009). A bud sport variety of NG (BNG) was identified on a NG tree in the 1980s on a farm in the Anshan region in Liaoning province. The skin color of BNG fruit is similar to that of NG fruit in the early stage (before 40 d after full bloom [DAFB]) and thereafter turns to brown (Fig. 1A), and this phenotype is stable after being propagated clonally (Supplemental Fig. S1, A and B). More interestingly, BNG is sweeter tasting than NG, but the underlying mechanism is unknown. We compared the sugar content of NG and BNG fruits and found that the Suc content was higher in BNG fruit. A Suc transporter, PuSWEET15, was more highly expressed in BNG fruit than in NG fruit. We also determined that a WRKY transcription factor, PuWRKY31, which was also expressed at higher levels in BNG fruit, bound to the PuSWEET15 promoter and upregulated its expression.
Figure 1.
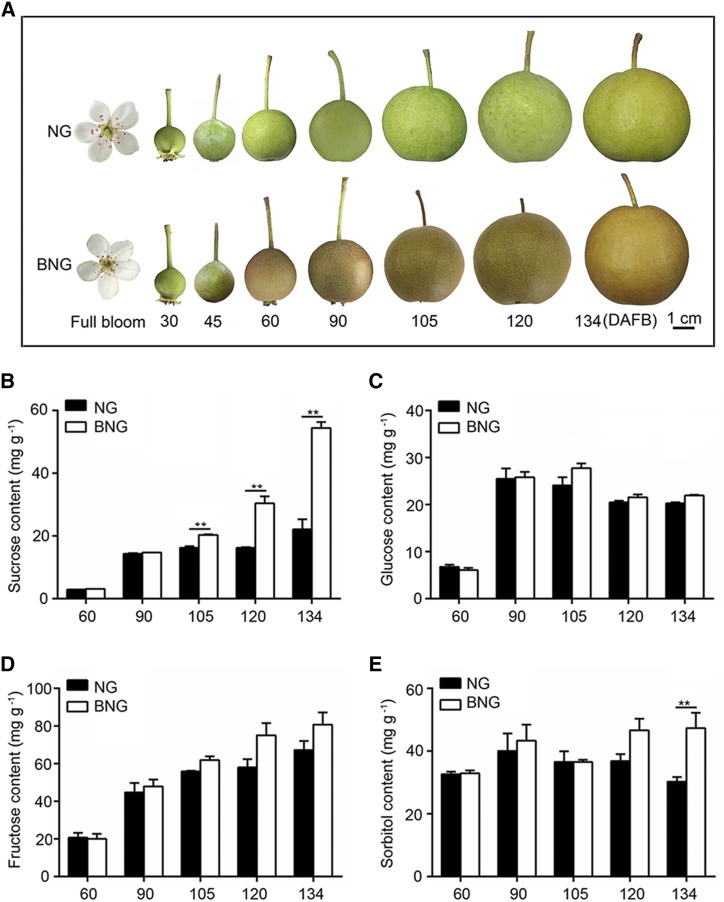
Phenotype and sugar content of NG and BNG fruits during development. A, Flowers and fruits of NG and BNG. Pictures were taken at different DAFB in 2014. B to E, Sugar content of NG and BNG fruits during development. HPLC was used to measure the content of Suc (B), Glc (C), Fru (D), and sorbitol (E) in fruit collected at the indicated DAFB in 2014. Commercial harvest day was 134 DAFB (September 4, 2014). Numbers under the x axes indicate the DAFB. Three biological replicates were analyzed, and the error bars represent the se. Asterisks indicate significant difference as determined by Student’s t test (**P < 0.01).
Plant WRKY proteins participate in developmental processes and respond to various biotic and abiotic stresses (Zhou et al., 2008; Ren et al., 2010; Sun et al., 2019). The expression of WRKY genes is also strongly induced during senescence; for example, overexpression of AtWRKY45 significantly accelerates the expression of SENESCENCE ASSOCIATED GENEs (Chen et al., 2017). AtWRKY57 interacts with repressors of the jasmonate and auxin signaling pathways, affecting jasmonate-induced leaf senescence in Arabidopsis (Jiang et al., 2014). In addition, AtWRKY75 interacts with DELLA proteins and may function as a component of the gibberellin (GA)-mediated signaling pathway to positively regulate Arabidopsis flowering (Zhang et al., 2018). WRKY proteins are also reported to participate in regulation of quality traits in proanthocyanidin and anthocyanin biosynthesis (Lloyd et al., 2017). For example, a WRKY transcription factor (TRANSPARENT TESTA GLABRA2 [TTG2]) interacts with the MYB-bHLH-WD40 (MBW) complex to regulate proanthocyanidin biosynthesis in Arabidopsis seed (Gonzalez et al., 2016). Overexpressing MdWRKY11 significantly promotes anthocyanin accumulation and increases the expression of MYB transcription factors and structural genes of anthocyanin in apple (Liu et al., 2019). MdWRKY40 interacts with MdMYB1 physically, thus enhancing the binding of MdMYB1 to its target genes to induce wounding-induced anthocyanin biosynthesis in apple fruit (An et al., 2019b). WRKY transcription factors respond to sugar treatment by activating the expression of sugar-responsive genes in Arabidopsis (Chen et al., 2019). However, to date, involvement of WRKYs in sugar transport has not been reported. We show here that increased histone acetylation in the PuWRKY31 promoter is associated with its higher expression in BNG fruit.
RESULTS
Suc Levels Are Significantly Higher in BNG Fruit than in NG Fruit
To investigate the basis of the sweeter taste of BNG fruit, we first compared the content of total soluble solids in BNG and NG fruits. Based on measurements from two years (2014 and 2018), we found that the total soluble solid content was higher in BNG fruit than in cv NG fruit (Supplemental Fig. S2, A and B). To determine which sugars were present at higher levels in BNG, Suc, Glc, Fru, and sorbitol levels were measured in fruit at different developmental stages using HPLC. We observed a significantly higher Suc content in BNG fruit than in NG fruit from 105 to 134 DAFB (Fig. 1B), while no significant differences were observed for Glc and Fru (Fig. 1, C and D). These results were consistent with data from a 2018 study (Supplemental Fig. S2, C–E). Sorbitol content was significantly higher in BNG than in NG fruit only at the time of commercial harvest (134 DAFB; Fig. 1E), while no difference was found in the 2018 samples (Supplemental Fig. S2F). These findings suggested that the higher BNG sugar content and sweeter taste were due to a higher accumulation of Suc.
The Sugar Transporter PuSWEET15 Is Highly Expressed in BNG Fruit
To identify genes that might contribute to higher Suc accumulation in BNG fruit, we compared the transcriptomes of NG and BNG fruits harvested at 134 DAFB (commercial harvest) using RNA sequencing (RNA-seq). Genes known to be involved in Suc transport and metabolism, such as SUT, SPS, SS, and INV, did not show differential expression between NG and BNG fruits. However, the RNA-seq analysis revealed that a SWEET gene showed ∼11 fold higher expression in BNG fruit than in NG fruit (Supplemental Fig. S3; Supplemental Dataset S1). We cloned this gene from both varieties and in both cases the coding region of PuSWEET was completely identical over 918 bp. The predicted amino acid sequence was most similar to the AtSWEET15 protein, with 50% identity (Supplemental Fig. S4), and so it was named PuSWEET15.
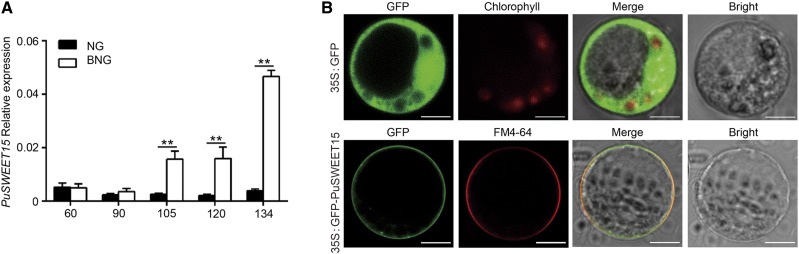
The expression profile of PuSWEET15 was investigated in NG and BNG fruits during development, and we found that it was expressed at significantly higher levels in BNG fruit from 105 to 134 DAFB (Fig. 2A; Supplemental Fig. S5), consistent with the change in Suc content (Fig. 1B). To determine the intracellular localization of PuSWEET15, its coding sequence (CDS) was fused downstream of a GFP tag driven by the CaMV35S promoter (35S:GFP-PuSWEET15) in the pRI101 vector. The recombinant plasmid (35S:GFP-PuSWEET15), or a plasmid encoding GFP alone, was transiently expressed in protoplasts of maize (Zea mays) leaves. GFP alone was detected in both the membrane and nucleus, while GFP-PuSWEET15 only localized to the plasma membrane (Fig. 2B).
Figure 2.
Expression of PuSWEET15 in NG and BNG fruits and its subcellular localization. A, Relative expression of PuSWEET15 during NG and BNG fruit development as determined by RT-qPCR. Fruit samples were collected in 2014. Numbers under the x axes indicate the DAFB. Three biological replicates were analyzed, and the error bars represent the se. Asterisks indicate significant difference as determined by Student’s t test (**P < 0.01). B, Subcellular localization of PuSWEET15. 35S:GFP-PuSWEET15 was transiently expressed in protoplasts of maize leaves. Transient expression of GFP alone (35S:GFP) was used as a control. FM4-64 was used as a plasma membrane marker. Scale bars = 5 μm.
Functional Characterization of PuSWEET15 by Heterologous Expression in Yeast Cells
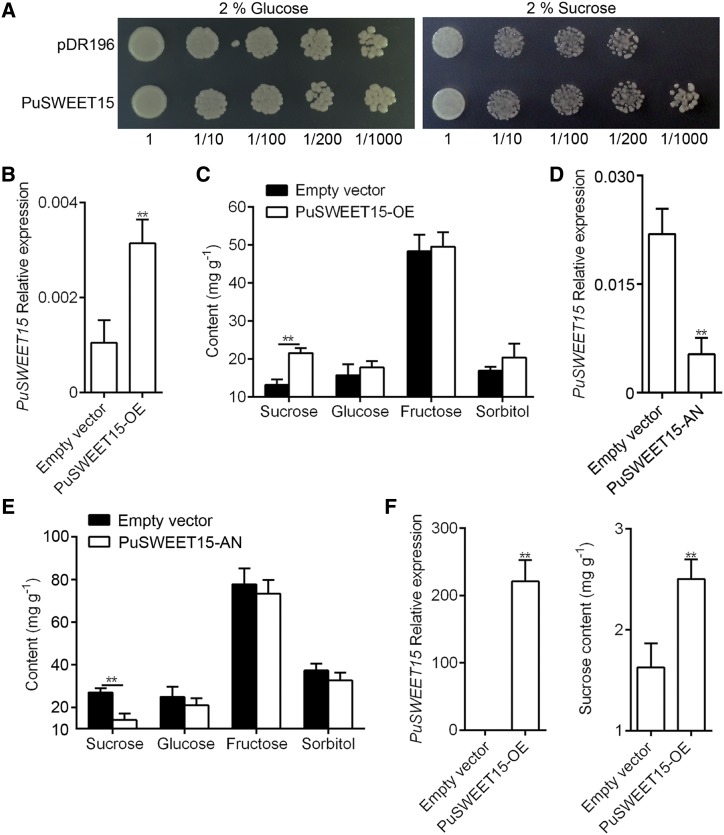
To investigate whether PuSWEET15 encodes a functional Suc transporter, we ligated its CDS into the pDR196 vector and expressed it in a yeast (Saccharomyces cerevisiae) mutant strain, SUSY7/ura, that is deficient in the wild-type Suc uptake mechanism in yeast (invertase-mediated hydrolysis of Suc with uptake of the resulting monosaccharides) and has a plant-derived Suc synthase activity to metabolize any Suc taken up by foreign Suc transporters. The mutant strain carrying an empty pDR196 vector was used as a control. All transformants were grown on synthetic-deficient (SD) solid medium containing Glc or Suc as the sole carbon source without uracil. Compared with control cells, the yeast cells containing PuSWEET15 survived well on SD/−uracil solid medium containing Suc as the sole carbon source (Fig. 3A), suggesting that PuSWEET15 is a typical Suc transporter.
Figure 3.
Functional analysis of PuSWEET15. A, Heterologous expression of PuSWEET15 in yeast strain SUSY7/ura. Yeast cells with pDR196-PuSWEET15 or pDR196 vector (as a negative control) were grown on SD/−uracil solid medium containing 2% (w/v) Glc or Suc as the sole carbon source. Numbers under the images indicate the dilution fold. B and C, PuSWEET15 was overexpressed in NG pear fruit using Agrobacterium tumefaciens-mediated infiltration. The expression of PuSWEET15 was detected by RT-qPCR (B) and the sugar content was measured by HPLC (C). D and E, PuSWEET15 was silenced in BNG pear fruit using A. tumefaciens-mediated infiltration. The expression of PuSWEET15 was detected by RT-qPCR (D) and the sugar content was measured by HPLC (E). F, PuSWEET15 was overexpressed in N. benthamiana leaves using A. tumefaciens-mediated infiltration. The expression of PuSWEET15 (left) was detected by RT-qPCR, and the sugar content (right) was measured by HPLC. Three biological replicates were analyzed, and the error bars represent the se. Asterisks indicate significant difference as determined by Student’s t test (**P < 0.01).
PuSWEET15 Is Essential for Suc Accumulation in Pear Fruit
To identify the function of PuSWEET15 in pear fruit, we overexpressed PuSWEET15 under the control of the CaMV35S promoter in NG fruit using Agrobacterium tumefaciens-mediated infiltration. The empty pRI101 vector was used as a control. Higher expression of PuSWEET15 was detected in PuSWEET15-overexpressing fruit (PuSWEET15-OE; Fig. 3B), and the Suc content was significantly higher than in control fruit, while no significant difference was observed for the other three sugars investigated (Fig. 3C). Then we silenced PuSWEET15 expression in BNG pear fruit using A. tumefaciens-mediated infiltration. Lower expression of PuSWEET15 was detected in PuSWEET15-silenced fruit (PuSWEET15-AN; Fig. 3D), and the Suc content was significantly lower than in control fruit, while no significant difference was observed for the other three sugars investigated (Fig. 3E), suggesting that PuSWEET15 is essential for Suc accumulation in pear fruit.
To provide further evidence for PuSWEET15 functioning as a Suc transporter, we examined the putative role of PuSWEET15 in Suc transport using A. tumefaciens-mediated infiltration of Nicotiana benthamiana leaves. Following treatment with 1% Suc for 6 d, PuSWEET15 was highly expressed (Fig. 3F), and significantly higher Suc levels were detected in PuSWEET15-OE leaves than in those of the wild type (Fig. 3F). These results were all consistent with PuSWEET15 contributing to Suc transport.
The Transcription Factor PuWRKY31 Is Highly Expressed in BNG Fruit
To elucidate the PuSWEET15 expression profiles in NG and BNG fruits, we compared their PuSWEET15 CDSs; however, no difference was found. Moreover, no differences were observed in the PuSWEET15 promoter regions (1,177 bp from the translation initiation site) from NG and BNG, and the methylation levels (+1 to −1,107) and histone acetylation levels (−60 to −409 and −895 to −1,167) of the promoters were also almost identical (Supplemental Fig. S6).
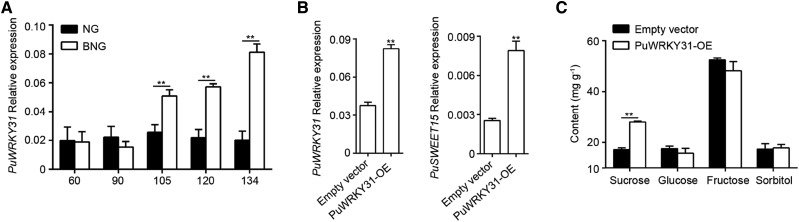
We then analyzed the cis-elements of the PuSWEET15 promoter (1,177 bp) and identified binding sites of transcription factors such as WRKY, DNA-binding one finger (DOF), and MYB. In combination with the RNA-seq results, a WRKY transcription factor, PuWRKY31, was more highly expressed in BNG fruit than in NG fruit (Supplemental Fig. S7). This was confirmed by reverse transcription quantitative PCR (RT-qPCR; Fig. 4A; Supplemental Fig. S8). We then focused on the characterization of PuWRKY31.
Figure 4.
Functional analysis of PuWRKY31. A, Expression of PuWRKY31 during NG and BNG fruit development. Fruit samples were the same as in Figure 1. Numbers under the x axis indicate the DAFB. B and C, PuWRKY31 was overexpressed in NG pear fruit using Agrobacterium tumefaciens-mediated infiltration. The relative expression of PuWRKY31 and PuSWEET15 was detected by RT-qPCR (B) and the sugar content in PuWRKY31-OE and control fruit was measured by HPLC (C). Three biological replicates were analyzed, and the error bars represent the se. Asterisks indicate significant difference as determined by Student’s t test (**P < 0.01).
To investigate the function of PuWRKY31, we cloned the corresponding CDS into the pRI101 vector to allow its expression under the control of the CaMV35S promoter and as a fusion with a MYC peptide tag. This construct was overexpressed in NG fruit (PuWRKY31-OE), and the higher expression of PuWRKY31 in PuWRKY31-OE fruit was verified by RT-qPCR (Fig. 4B). We detected that the Suc content in PuWRKY31-OE fruit was significantly higher than that in control fruit (Fig. 4C). Notably, the expression level of PuSWEET15 was also higher in PuWRKY31-OE fruit (Fig. 4B), suggesting that PuWRKY31 might play a role in Suc transport by regulating the expression of PuSWEET15.
PuWRKY31 Binds to the Promoter of PuSWEET15 and Upregulates its Transcription
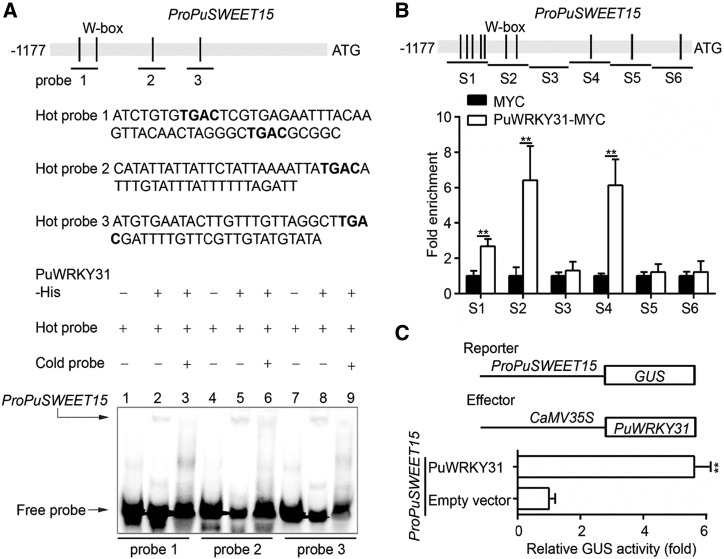
To investigate whether PuSWEET15 is a direct target of PuWRKY31, we performed an electrophoretic mobility shift assay (EMSA) with three biotin-labeled fragments of the PuSWEET15 promoter containing four W-box motifs (TGAC, the binding site of WRKY) as the labeled probe. His-tagged PuWRKY31 (PuWRKY31-His) was purified and used for DNA-binding assays. As shown in Figure 5A, PuWRKY31 bound to the PuSWEET15 promoter (Fig. 5A, lanes 2, 5, and 8). When an unlabeled probe was added as a competitor, the binding of PuWRKY31 to the PuSWEET15 promoter was reduced (Fig. 5A, lanes 3, 6, and 9), confirming that PuWRKY31 bound to the PuSWEET15 promoter in vitro.
Figure 5.
PuWRKY31 promotes PuSWEET15 transcription. A, EMSA analysis of PuWRKY31 binding to the PuSWEET15 promoter. The hot probe was a biotin-labeled PuSWEET15 promoter, while the cold probe was a nonlabeled competitive probe (with a 100-fold higher concentration than the hot probe). PuWRKY31-His was purified and used for DNA-binding assays. The sequence of the biotin labeled probe is shown and the W-box motif is highlighted in bold. B, ChIP-PCR showing the in vivo binding of PuWRKY31 to the PuSWEET15 promoter. Cross-linked chromatin samples were extracted from PuWRKY31-MYC-overexpressing NG pear fruit and precipitated with an anti-MYC antibody. Eluted DNA was used to amplify the sequences neighboring the W-box by qPCR. Six regions (S1–S6) were analyzed. Fruit overexpressing GFP were used as negative controls. The ChIP assay was repeated three times and the enriched DNA fragments in each ChIP were used as one biological replicate for qPCR. C, Schematic representation of the GUS reporter vector containing the PuSWEET15 promoter and the effector vector containing PuWRKY31. The effector reporter vectors were infiltrated into Nicotiana benthamiana leaves to analyze the regulation of GUS activity. Three independent infiltrations were performed, and the error bars represent the se. Asterisks indicate significant difference as determined by Student’s t test (**P < 0.01).
Next, we used chromatin immunoprecipitation (ChIP) PCR to investigate the in vivo binding of PuWRKY31 to the PuSWEET15 promoter. Cross-linked chromatin samples were extracted from the PuWRKY31-OE fruit (Fig. 4B) and precipitated with an anti-MYC antibody. Eluted DNA was used to amplify the sequences neighboring the W-box by qPCR. Fruits overexpressing the GFP sequence were used as negative controls. Figure 5B shows that the presence of PuWRKY31 substantially enhanced the PCR-based detection of the PuSWEET15 promoter, indicating in vivo binding of PuWRKY31 to the PuSWEET15 promoter.
We then investigated regulation of the PuSWEET15 promoter by PuWRKY31 using a GUS activation assay in N. benthamiana leaves, following coinfiltration with Pro-35S:PuWRKY31 and ProPuSWEET15:GUS. Pro-35S:GUS was used as a control. When Pro-35S:PuWRKY31 was coinfiltrated with ProPuSWEET15:GUS, PuSWEET15 promoter activity increased significantly compared with the control (Fig. 5C), suggesting that PuWRKY31 is a transcriptional activator of PuSWEET15. Collectively, these results suggested that PuWRKY31 binds to the PuSWEET15 promoter and promotes its transcription.
The Expression Profile of PuWRKY31 Correlates with Histone Acetylation Levels
To investigate the PuWRKY31 expression profiles in NG and BNG fruits, we compared the CDSs, promoter sequences (1,550 bp from the translation initiation site), and methylation levels of its promoter regions (Supplemental Fig. S9). However, no significant differences were observed.
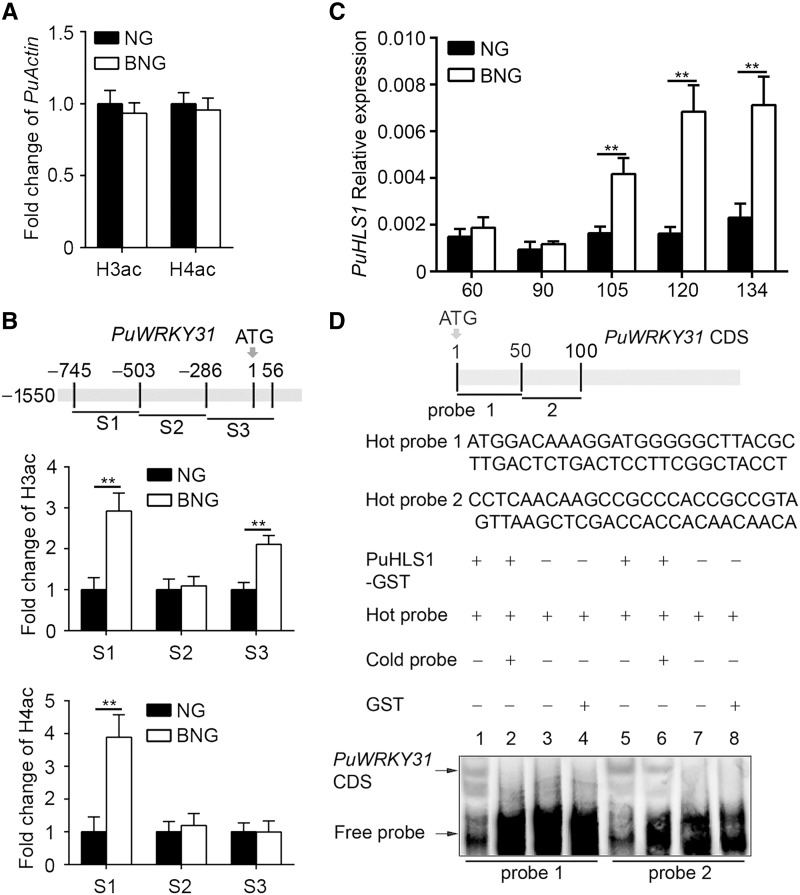
We hypothesized that the PuWRKY31 expression pattern might correlate with a change in histone modification, and so examined the PuWRKY31 histone acetylation levels in NG and BNG fruits by ChIP-PCR, using antiacetyl-histone H3 (H3ac) and H4ac antibodies. As a control, the change in histone acetylation (H3ac and H4ac) of the PuActin housekeeping gene was also analyzed. No significant changes in H3ac and H4ac were found in NG or BNG for the PuActin gene (Fig. 6A), and it was used to normalize the subsequent ChIP-PCR results. Three regions (S1–S3) of pear genomic DNA including the PuWRKY31 promoter and CDS were examined, and these regions were predicted to be easily acetylated (Zhou et al., 2013; Han et al., 2016). The acetylation levels of regions S1 and S3 detected by histone H3ac, and region S1 detected by H4ac, were significantly higher in BNG than in NG fruit (Fig. 6B).
Figure 6.
PuWRKY31 histone acetylation and PuHLS1 expression between NG and BNG fruits. A, The histone acetylation level of PuActin chromatin by H3ac or H4ac between NG and BNG fruits. The results were normalized to the amount of input DNA. B, The histone acetylation level at different regions of the PuWRKY31 chromatin by H3ac or H4ac between NG and BNG fruits as determined by ChIP-PCR. Fruit harvested at commercial harvest day in 2014 were used. The results were normalized relative to the amount of PuActin. Each experiment was repeated three times. The ChIP assay was repeated three times and the enriched DNA fragments in each ChIP were used as one biological replicate for qPCR. Error bars represent the se, and asterisks indicate significant difference as determined by Student’s t test (**P < 0.01). C, Expression of PuHLS1 during NG and BNG fruit development as determined by RT-qPCR. Fruit samples were the same as in Figure 1. Numbers under the x axis indicate DAFB. Three biological replicates were analyzed, and the error bars represent the se. Asterisks indicate significant difference as determined by Student’s t test (**P < 0.01). D, EMSA analysis of PuHLS1 binding to the CDS of PuWRKY31. The hot probe was biotin-labeled PuWRKY31 CDS, while the cold probe was a nonlabeled competitive probe (with a 100-fold higher concentration than the hot probe). PuHLS1-GST was purified and used for DNA-binding assays. The sequence of the biotin labeled probe is shown.
To investigate what causes the higher acetylation level of PuWRKY31 in BNG fruit, we identified a histone acetyltransferase (HAT) gene, HOOKLESS1 (PuHLS1; Liao et al., 2016), from the RNA-seq results (Supplemental Dataset S1). PuHLS1 expression was higher in BNG fruit than in NG fruit (Fig. 6C; Supplemental Fig. S10). A previous report has shown that HLS1 binds to the transcription start site and the 3′-CDS of AtWRKY33 in Arabidopsis (Liao et al., 2016). We investigated whether PuHLS1 directly interacts with the CDS of PuWRKY31 using EMSA analysis with two biotin-labeled fragments of the CDS of PuWRKY31 (probe 1, 1–50; probe 2, 50–100) as the hot probe. GST-tagged PuHLS1 (PuHLS1-GST) was purified and used for DNA-binding assays. As shown in Figure 6D, the GST alone did not bind to the PuWRKY31 CDS (Fig. 6D, lanes 4 and 8), but PuHLS1 did (Fig. 6D, lanes 1 and 5). When an unlabeled probe was added as a competitor, the binding of PuHLS1 to the PuWRKY31 CDS was reduced (Fig. 6D, lanes 2 and 6), confirming that PuHLS1 bound to the PuWRKY31 CDS in vitro. To elucidate the PuHLS1 expression profiles in NG and BNG fruits, we compared the CDS, promoter sequences (2,032 bp from the translation initiation site), and methylation levels of its promoter regions, which were almost identical in NG and BNG (Supplemental Fig. S11).
DISCUSSION
Suc is the main photosynthesis product transported in most plants (Ayre, 2011). By comparing the contents of different sugars in NG and BNG fruits, we found that only the Suc content was significantly higher in BNG fruit than in NG fruit (Fig. 1). Although sorbitol is important for transport of photosynthesis products in tree fruit crops of the Rosaceae family (Priestley, 1983; Zhang et al., 2014), our data showed that the sorbitol content was higher in BNG fruit than in NG fruit only in samples collected at 134 DAFB in 2014 (Fig. 1E), and no difference was observed in samples collected in 2018 (Supplemental Fig. S2F). The difference in sorbitol content between years might be caused by climatic conditions such as rainfall, light, or temperature. However, importantly, the difference in Suc content between BNG and NG fruits did not vary between years. These results suggest that BNG is a bud sport variety of NG pear with higher Suc accumulation.
SWEET proteins have been widely identified as sugar transporters in plants, especially for Suc transport (Chen et al., 2012). Here, PuSWEET15 was observed to transport Suc in pear fruit tissue and N. benthamiana leaves when expressed heterologously (Fig. 3). This is consistent with the function of AtSWEET11, AtSWEET12, and AtSWEET15 from Arabidopsis, in which a double mutation of AtSWEET11 and AtSWEET12 causes defects in phloem Suc loading (Chen et al., 2012). AtSWEET15 was shown to transport Suc by expressing SWEET15 in Xenopus laevis oocytes and measuring [14C]Suc uptake (Chen et al., 2015).
In plants, the WRKY family is one of the largest transcription factor families (Zhang and Wang, 2005; Rushton et al., 2010), but functional characterization has mostly focused on their roles in various biotic and abiotic stresses and developmental processes (Rushton et al., 2010). For example, WsWRKY1 regulates nitrogen stress tolerance through modulation of phytosterol and defense pathways in Withania somnifera, and soybean GmWRKY16 enhances drought and salt tolerance in Arabidopsis through an abscisic acid-mediated pathway (Pal et al., 2017; Singh et al., 2017; Ma et al., 2019). Another recent report showed that VaWRKY33 is involved in cold tolerance in Amur grape (Vitis amurensis; Sun et al., 2019). A more recent study reported that AtWRKY18 and AtWRKY53 directly bind to the promoter of sugar response genes and activate their expression in response to Glc treatment in Arabidopsis (Chen et al., 2019). In our study, we showed that PuWRKY31 was expressed at significantly higher levels in BNG fruit than in NG fruit (Fig. 4A). Moreover, PuWRKY31 positively regulated the expression of PuSWEET15 by binding to its promoter (Fig. 5). Importantly, overexpression of PuWRKY31 in pear fruit led to increased Suc content (Fig. 4C), suggesting the involvement of PuWRKY31 in Suc transport in pear fruit.
Bud sport varieties occasionally occur in tree fruit crops and are usually caused by a small number of presumably genetic or epigenetic alterations (Whitham and Slobodchikoff, 1981; Furiya et al., 2009). BNG was found by our colleague in the 1980s on a NG tree. BNG showed phenotypes similar to those of NG in leaf, flower, and fruit shape (Fig. 1A; Supplemental Fig. S1). Unfortunately, we do not have the original picture showing both NG and BNG fruits on different branches of the same tree. However, BNG maintained stable phenotypes when grafted onto a NG tree (Supplemental Fig. S1A) or when propagated clonally and cultivated in different regions (Supplemental Fig. S1, A and B). Moreover, we analyzed the genomic DNA of both NG and BNG using 17 pairs of simple sequence repeat (SSR) primers, but failed to detect any polymorphic bands between two varieties (Supplemental Fig. S1C). These findings indicated that BNG and NG share high similarity in genetic background. In addition to sweetness and Suc content, BNG differs from NG in other characteristics, such as fruit skin color (Fig. 1A; Supplemental Fig. S1). It will be quite interesting to explore whether the skin color is related to the high Suc content in BNG fruit.
We set out to determine why PuWRKY31 was expressed differentially in NG and BNG fruits. We investigated the CDS and promoter sequence, as well as the promoter methylation levels of two varieties, but found no differences. So we compared the PuWRKY31 histone acetylation level, and found that the higher PuWRKY31 expression level in BNG fruits is associated with a higher level of histone acetylation in its promoter and CDS regions (Fig. 6B). A high histone acetylation level can be regulated by various coactivators, which recruit HATs to enhance the acetylation of Lys residues, which in turn can neutralize the positive charge of histone proteins. This causes an unwinding of the chromatin structure and exposure of binding sites in the promoter, thereby increasing the accessibility for transcription factors (Shahbazian and Grunstein, 2007). In Arabidopsis, a HAT HLS1 mediates histone acetylation on AtWRKY33 chromatin, and the histone H3 acetylation level at AtWRKY33 chromatin is significantly lower in the hls1 mutant than in the wild type (Liao et al., 2016). Moreover, in Arabidopsis, hls1 mutants accumulate less total soluble sugar than the wild type (Ohto et al., 2006). In our study, a HAT PuHLS1 showed significantly higher expression in BNG fruit than in NG fruit, and PuHLS1 could bind to the CDS of PuWRKY31 (Fig. 6, C and D). Therefore, we propose that the higher expression level of PuHLS1 might cause the higher histone acetylation level of PuWRKY31, resulting in higher Suc accumulation in BNG fruit.
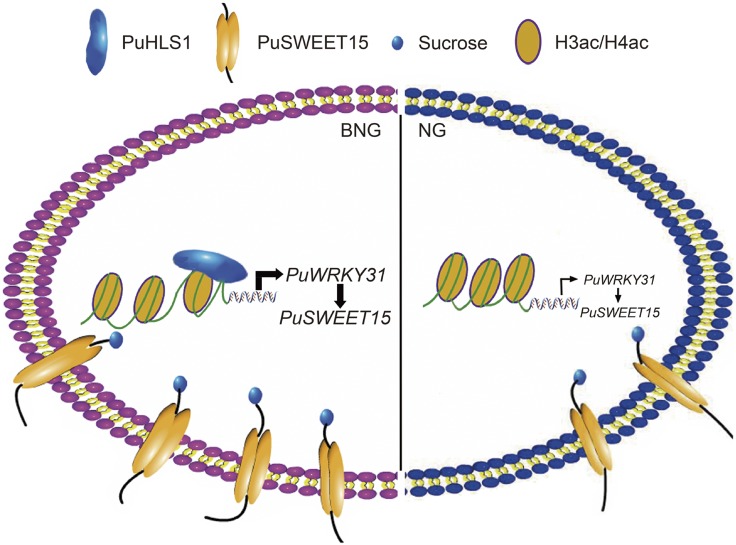
In conclusion, PuSWEET15 showed higher expression in BNG fruit than in NG fruit and PuWRKY31 bound to the PuSWEET15 promoter to induce its expression. Moreover, the high acetylation level of the PuWRKY31 promoter was associated with its high expression level in BNG fruit (Fig. 7).
Figure 7.
Model showing the molecular mechanism of differential Suc accumulation in NG and BNG fruits. In the fruit of BNG, a bud sport of NG with high accumulation of Suc, the high acetylation level of the PuWRKY31 promoter is associated with its high expression, and PuWRKY31 binds to the promoter of PuSWEET15, an active Suc transporter, to induce its expression, resulting in high levels of Suc.
MATERIALS AND METHODS
Plant Material and Treatments
Fruits of Ussurian pear (Pyrus ussuriensis) ‘Nanguo’ (NG) and its bud sport variety (BNG) were sampled from mature trees growing on the experimental farm of the Liaoning Pomology Institute. Fruits were harvested at 60, 90, 105, 120, and 134 DAFB (commercial harvest day) in 2014, and 60, 90, 120, and 137 DAFB (commercial harvest day) in 2018, and immediately transported to the laboratory. At each sampling point, three fruits of each variety were selected for measuring sugar content. The flesh of those fruits was cut into pieces, frozen in liquid nitrogen, and stored at −80°C for further use.
Nicotiana benthamiana plants used in this study were grown with potting medium in a growth chamber (25°C, 16 h light, 8 h dark).
Measurements of Soluble Solids and Sugar Content
At each sampling point, the fruit flesh was homogenized with a homogenizer and filtered through a cell strainer (Cat. no. CSS010040, Jet Biofil; https://www.jetbiofil.com), and the soluble solids content of the filtrate was measured with a sugar meter (PAL-1, Atago). The soluble sugar content was measured by HPLC (1260 Series, Agilent Technologies), as described in Jia et al. (2011). Briefly, samples were ground to a fine powder in liquid nitrogen. Three grams of the powder was mixed with 10 mL of 80% (v/v) ethanol, incubated in a water bath for 30 min at 80°C, and then centrifuged at 10,000g for 5 min in a 50-mL centrifuge tube and the supernatant collected. The above step was repeated twice to reextract the pellets, the supernatants were combined, and the samples were evaporated in boiling water. After drying in a 50-mL centrifuge tube, the samples were dissolved in 1 mL of ultrapure water and passed through a 0.45 μm membrane, and the soluble sugar content of the filtrate was measured. HPLC (Agilent 1260) was then performed with the following components and parameters: a 7.8 × 300 mm Carbomix Ca-NP column (Sepax); ultrapure water as the mobile phase, at a flow rate of 1 mL min–1; a column temperature of 80°C; a refractive index detector temperature of 35°C; and an injection volume of 10 μL. At each sampling point, at least nine fruits were randomly selected and divided into three groups as three biological replicates. The flesh in each group was pooled for measuring soluble solids and sugar content.
RNA-Seq
Total RNA was extracted from NG and BNG fruits harvested at the commercial harvest day (134 DAFB in 2014). RNA-seq analysis, including library construction, sequencing, and bioinformatics analysis was performed as in Huang et al. (2014) by Biomarker (www.biomarker.com.cn). Sequencing was performed on an Illumina HiSeq 2000 system. The total RNA was extracted from the three groups of fruit, as mentioned in the previous section, as three biological replicates for RNA-seq. All the raw data were deposited into the NCBI Sequence Read Archive (SRA) under accession number PRJNA545020. The heat maps for differentially expressed genes between NG and BNG fruits were constructed according to the log2 (FC) value from the RNA-Seq data using online software (https://console.biocloud.net/static/index.html#/drawtools/intoDrawTools/heatmap/input).
Gene Cloning and Expression Analysis
Total RNA extraction was conducted as in Li et al. (2015), and first-strand complementary DNA (cDNA) was synthesized from 700 ng of total RNA using the M-MLV RTase cDNA Synthesis Kit (cat. no. D6130, TaKaRa). The cDNA was then used as a template for RT-qPCR and standard RT-PCR assays, using sequence information for each gene derived from the RNA-seq data. Standard RT-PCR was performed according to Li et al. (2015), with 4 μL of each PCR product separated on a 1% (w/v) agarose gel and imaged on a GelDoc XR System (Bio-Rad). RT-qPCR was performed using the SYBR Premix ExTaq II Kit (cat. no. RR820, TaKaRa) on an Applied Biosystems 7500 Real-Time PCR System, as previously described (Li et al., 2015). The pear Actin gene was used as an internal control and total RNA was extracted from the three groups of fruit as three biological replicates, as described in the previous two sections. All primers were designed using the Primer3 software (http://frodo.wi.mit.edu/) and are listed in Supplemental Dataset S2.
SSR Analysis of NG and BNG Pear Fruit
Genomic DNA was isolated from the fruit samples harvested in 2014 as described in Wang et al. (2013). SSR primers with polymorphism were selected from previous reports (Yamamoto et al., 2002; Jiang et al., 2009). Standard PCR was conducted and the PCR products were analyzed on 6% (w/v) denaturing polyacrylamide gel with silver staining according to Bao et al. (2007).
Subcellular Localization of PuSWEET15
The protoplasts of maize (Zea mays) leaves were prepared as described previously (Yoo et al., 2007). The PuSWEET15 coding region was cloned into the BamHI and SacI sites downstream of GFP in the pRI101 vector (TaKaRa) to form the Pro-35S:GFP:PuSWEET15 construct. Pro-35S:GFP was used as a control. The constructs were transformed into the protoplasts of maize leaves according to a previous report (Yoo et al., 2007). The fluorescence was observed using a fluorescence microscope 16 h after transformation under a confocal microscope (TCS SP8, Leica). N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide (FM4-64; cat. no. T3166, Thermo Fisher Scientific) was used as a cell membrane dye. All transient expression assays were repeated at least three times. The primers used are listed in Supplemental Dataset S2.
Heterologous Expression of PuSWEET15 in Yeast Cells
For the complementation assay in yeast (Saccharomyces cerevisiae) cells, the CDS of PuSWEET15 was cloned into the yeast expression vector pDR196 (cat. no. VT8007; YouBio, http://www.youbio.cn/) using Sma1 and Sal1 restriction enzyme sites to form the pDR196-PuSWEET15 construct. The empty pDR196 vector was used as a negative control. The constructs were transformed into yeast mutant strain SUSY7/ura (Li et al., 2017c; Riesmeier et al., 1992) using the lithium acetate method (Soni et al., 1993). The transformants were cultured in liquid SD medium (cat. no. PM2271, Coolaber; http://www.coolaber.com/) containing 2% (w/v) Glc (Sigma) as the sole carbon source without uracil by shaking at 180 rpm under 30°C to OD600 0.5. The culture was then diluted by different fold values (×10, ×100, ×200, and ×1000), and 6 μL of dilution was dropped on SD/−uracil solid medium containing 2% (w/v) Glc or 2% (w/v) Suc (Sigma) as the sole carbon source at pH 4.0. Yeast cells on medium with Glc were grown at 30°C for 2 d, and those on medium with Suc were grown at 30°C for 4 d.
EMSA
The PuWRKY31 CDS was cloned and inserted into the pEASY-E1 vector (Transgen Biotech, http://www.transgen.com.cn/), resulting in its fusion to a His-tag, and the CDS of PuHLS1 was cloned and inserted downstream of GST in the pGEX4T-1 vector (GE Healthcare; http://www3.gehealthcare.com/) before being transformed into Escherichia coli BL21-competent cells (DE3, Transgen Biotech). Purification of the His-tagged and GST-tagged fusion proteins was performed as previously described (Li et al., 2016). For EMSA, the 3′ biotin end-labeled double-stranded DNA probes were prepared by annealing complementary oligonucleotides. The oligonucleotides were heated at 95°C for 5 min, then at 72°C for 20 min, and left to cool to room temperature before use. The biotin-labeled PuSWEET15 promoter and PuWRKY31 CDS sequences are shown in Figures 5A and 6D. EMSA was performed as previously described (Li et al., 2016) using the LightShift Chemiluminescent EMSA Kit (cat. no. 20148, Thermo Scientific).
GUS Analysis
The PuSWEET15 promoter sequence (1,177 bp upstream of the translation start site) was cloned into the SalI and SmaI sites upstream of the GUS reporter gene in the pBI101 vector to generate a reporter construct. The PuWRKY31 CDS was introduced into the pRI101 vector through restriction enzyme sites (SalI and KpnI) to form the effector construct. The infiltration of the reporter and effector constructs into N. benthamiana leaves and the measurement of GUS activity were performed as previously described (Li et al., 2016). The infiltration was repeated independently at least three times. The primers used are listed in Supplemental Dataset S2.
Methylation Analysis
Genomic DNA was isolated from the fruit samples harvested in 2014 as described in Wang et al. (2013). McrBC-PCR was used to analyze the methylation of relative sequences. One μg of DNA isolated from fruit was digested with McrBC (cat. no. M0272, New England Biolabs) according to the manufacturer’s instructions. Three biological replicates were analyzed. For the control, water was added instead of GTP. After digestion, DNA was used as a template for standard PCR analysis. The thermal cycling conditions were 3 min at 95°C; 27 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C; and 5 min at 72°C as a final extension. The PCR product was separated in 0.5% (w/v) agarose gel and photographed with the GelDoc XR System (BioRad). Four regions of the PuSWEET15 or PuWRKY31 promoter and five regions of the PuHLS1 promoter were examined (Supplemental Figs. S6, S9, and S11). The amount of PCR product was used to estimate the degree of methylation of the promoter region. The PCR bands were quantified by ImageJ software.
Agrobacterium-Mediated Infiltration
To overexpress PuSWEET15 in N. benthamiana leaves, its CDS was cloned into the pRI101 plant transformation vector using BamHI and SacI restriction enzyme sites to form the Pro-35S:PuSWEET15 construct. The recombinant plasmid was transformed into Agrobacterium tumefaciens strain EHA105 for infiltration of N. benthamiana leaves, as previously described (Li et al., 2017b). Briefly, the suspension for infiltration was injected into mature leaves of N. benthamiana grown in potting medium when the plants were 5 weeks old. After infiltration, the potting medium was irrigated with 1% (w/v) Suc every 2 d. The plants were collected 6 d after infiltration for further use.
To overexpress PuSWEET15 or PuWRKY31 in NG pear fruit, the CDS regions were separately cloned into the SalI and KpnI sites upstream of the MYC tag in the pRI101 vector to form Pro-35S:PuSWEET15-MYC and Pro-35S:PuWRKY31-MYC, respectively. To silence PuSWEET15 expression in BNG pear fruit, a partial PuSWEET15 CDS (686–898 bp) was ligated into the pRI101 vector in the reverse direction to generate the antisense PuSWEET15 construct (PuSWEET15-AN). These plasmids were transformed into A. tumefaciens strain EHA105, and the preparation of infiltration buffer and fruit infiltration were performed as previously described (Li et al., 2016). Briefly, 100 μL of the infiltration buffer was taken with a 1-mL sterile syringe and injected into on-tree fruit at a depth of 0.3 cm at 120 DAFB. For each fruit, one side was used for infiltrating target constructs, and the other side for infiltrating empty pRI101 as control. Three injections were performed on each side of each fruit. The infiltrated fruits were harvested 6 d after infiltration, and the fruit flesh around the infiltrated area was sampled for further use. One fruit was used as a biological replicate, and at least three biological replications were performed. The overexpression of PuSWEET15 and PuWRKY31 was performed on NG fruit, and silencing of PuSWEET15 on BNG fruit.
ChIP-PCR
The recombinant Pro-35S:PuWRKY31-MYC construct was transformed into NG pear fruit as described above and ChIP assays were performed using the EpiQuik Plant ChIP Kit (cat. no. P-2014, Epigentek; https://www.epigentek.com/) according to the manufacturer’s instructions. An anti-MYC antibody (Transgen Biotech) was used to verify the binding of PuWRKY31 to the PuSWEET15 promoter in vivo as previously described (Li et al., 2017b). The amount of immunoprecipitated chromatin was determined by qPCR as previously described (Li et al., 2017b), with 0.5 μL of immunoprecipitated chromatin as template. Each ChIP assay was repeated three times and the enriched DNA fragments in each ChIP sample were used as one biological replicate for qPCR. Three regions of the PuSWEET15 promoter were analyzed to assess enrichment. Primers used are listed in Supplemental Dataset S2.
Analysis of Histone Acetylation Levels
NG and BNG fruits harvested at commercial harvest day in 2014 were used for analyzing the histone acetylation levels. The chromatin was prepared as above and immunoprecipitated with specific antibodies including antiacetyl-histone H3 and H4 (Millipore). ChIP-PCR analysis to measure the histone acetylation level of the PuWRKY31 or PuSWEET15 promoter was performed as described by Li et al. (2017b). PuActin was used as an internal control to normalize the ChIP enrichment signal. Three regions of the PuWRKY31 or PuSWEET15 promoter were analyzed to assess enrichment. Primers used are listed in Supplemental Dataset S2.
Accession Numbers
Sequence data from this article can be found in GenBank libraries under accession numbers MK940530 (PuSWEET15), MK940531 (PuWRKY31), MN201566 (PuHLS1), and AF386514 (PuActin).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotype of Nanguo pear fruit (NG) and its bud sport (BNG).
Supplemental Figure S2. Sugar contents of NG and BNG fruits during development.
Supplemental Figure S3. Heat map of sugar transporter genes with differential expression between NG and BNG fruits from the RNA-seq data.
Supplemental Figure S4. Sequence alignment of PuSWEET15 and AtSWEET15 amino acid sequences.
Supplemental Figure S5. Expression of PuSWEET15 in NG and BNG fruits sampled in 2018 as determined by RT-qPCR.
Supplemental Figure S6. Methylation and histone acetylation levels of PuSWEET15 promoter regions between NG and BNG fruits.
Supplemental Figure S7. Heat map of transcription factors with differential expression between NG and BNG fruits from the RNA-seq data.
Supplemental Figure S8. Expression of PuWRKY31 in NG and BNG fruits sampled in 2018 as determined by RT-qPCR.
Supplemental Figure S9. Methylation level of PuWRKY31 promoter regions in NG and BNG fruits determined using McrBC-PCR.
Supplemental Figure S10. Expression of PuHLS1 in NG and BNG fruits sampled in 2018 as determined by RT-qPCR.
Supplemental Figure S11. Methylation level of PuHLS1 promoter regions in NG and BNG fruits determined using McrBC-PCR.
Supplemental Dataset S1. Differentially expressed genes between NG and BNG fruits from RNA-seq data.
Supplemental Dataset S2. List of primers used in this study.
Acknowledgments
We thank the editors and anonymous reviewers for their efforts on our article and PlantScribe (http://www.plantscribe.com) for editing the article.
Footnotes
This work was supported by the National Natural Science Foundation of China (31722047), the National Key Research and Development Program of China (2018YFD1000105), and the LiaoNing Revitalization Talents Program (XLYC1802019).
Articles can be viewed without a subscription.
References
- An J, Zeng T, Ji C, de Graaf S, Zheng Z, Xiao TT, Deng X, Xiao S, Bisseling T, Limpens E, et al. (2019a) A Medicago truncatula SWEET transporter implicated in arbuscule maintenance during arbuscular mycorrhizal symbiosis. New Phytol 224: 396–408 [DOI] [PubMed] [Google Scholar]
- An JP, Zhang XW, You CX, Bi SQ, Wang XF, Hao YJ(2019b) MdWRKY40 promotes wounding‐induced anthocyanin biosynthesis in association with MdMYB1 and undergoes MdBT2‐mediated degradation. New Phytol 244: 380–395 [DOI] [PubMed] [Google Scholar]
- Ayre BG.(2011) Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol Plant 4: 377–394 [DOI] [PubMed] [Google Scholar]
- Bao L, Chen K, Zhang D, Cao Y, Yamamoto T, Teng Y(2007) Genetic diversity and similarity of pear (Pyrus L.) cultivars native to East Asia revealed by SSR (simple sequence repeat) markers. Genet Resour Crop Evol 54: 959–971 [Google Scholar]
- Braun DM, Wang L, Ruan YL(2014) Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J Exp Bot 65: 1713–1735 [DOI] [PubMed] [Google Scholar]
- Chagné D, Crowhurst RN, Pindo M, Thrimawithana A, Deng C, Ireland H, Fiers M, Dzierzon H, Cestaro A, Fontana P, et al. (2014) The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’). PLoS One 9: e92644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB(2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211 [DOI] [PubMed] [Google Scholar]
- Chen L, Xiang S, Chen Y, Li D, Yu D(2017) Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol Plant 10: 1174–1189 [DOI] [PubMed] [Google Scholar]
- Chen LQ, Lin IW, Qu XQ, Sosso D, McFarlane HE, Londoño A, Samuels AL, Frommer WB(2015) A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Xu X, Xu D, Zhang H, Zhang C, Li G(2019) WRKY18 and WRKY53 coordinate with HISTONE ACETYLTRANSFERASE1 to regulate rapid responses to sugar. Plant Physiol 180: 2212–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Piron MC, Meyer S, Merdinoglu D, Bertsch C, Mestre P(2014) The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J Exp Bot 65: 6589–6601 [DOI] [PubMed] [Google Scholar]
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, et al. (2006) Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev 20: 1250–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Wang Z, Tian L, Zhang Y, Qi D, Huo H, Xu J, Li Z, Liao R, Shi M, et al. (2019) De novo assembly of a wild pear (Pyrus betuleafolia) genome. Plant Biotechnol J 18: 581–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand M, Porcheron B, Hennion N, Maurousset L, Lemoine R, Pourtau N(2016) Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiol 170: 1460–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom JS, Choi SB, Ward JM, Jeon JS(2012) The mechanism of phloem loading in rice (Oryza sativa). Mol Cells 33: 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furiya T, Suzuki S, Sueta T, Takayanagi T(2009) Molecular characterization of a bud sport of Pinot gris bearing white berries. Am J Enol Vitic 60: 66–73 [Google Scholar]
- Gao Y, Zhang C, Han X, Wang ZY, Ma L, Yuan P, Wu JN, Zhu XF, Liu JM, Li DP, et al. (2018) Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease. Mol Plant Pathol 19: 2149–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Brown M, Hatlestad G, Akhavan N, Smith T, Hembd A, Moore J, Montes D, Mosley T, Resendez J, et al. (2016) TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev Biol 419: 54–63 [DOI] [PubMed] [Google Scholar]
- Guo WJ, Nagy R, Chen HY, Pfrunder S, Yu YC, Santelia D, Frommer WB, Martinoia E(2014) SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiol 164: 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YC, Kuang JF, Chen JY, Liu XC, Xiao YY, Fu CC, Wang JN, Wu KQ, Lu WJ(2016) Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol 171: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Li T, Li X, Tan D, Jiang Z, Wei Y, Li J, Wang A(2014) Comparative transcriptome analysis of climacteric fruit of Chinese pear (Pyrus ussuriensis) reveals new insights into fruit ripening. PLoS One 9: e107562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger SR, Andani Z, Wakeling IN, MacFie HJ(1998) Consumer preferences for fresh and aged apples: A cross-cultural comparison. Food Qual Prefer 9: 355–366 [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY(2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yang S, Yu D(2014) Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26: 230–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Tang F, Huang H, Hu H, Chen Q(2009) Assessment of genetic diversity of Chinese sand pear landraces (Pyrus pyrifolia Nakai) using simple sequence repeat markers. HortScience 44: 619–626 [Google Scholar]
- Lemoine R.(2000) Sucrose transporters in plants: Update on function and structure. Biochim Biophys Acta 1465: 246–262 [DOI] [PubMed] [Google Scholar]
- Li J, Qin M, Qiao X, Cheng Y, Li X, Zhang H, Wu J(2017a) A new insight into the evolution and functional divergence of SWEET transporters in Chinese white pear (Pyrus bretschneideri). Plant Cell Physiol 58: 839–850 [DOI] [PubMed] [Google Scholar]
- Li T, Jiang Z, Zhang L, Tan D, Wei Y, Yuan H, Li T, Wang A(2016) Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J 88: 735–748 [DOI] [PubMed] [Google Scholar]
- Li T, Tan D, Liu Z, Jiang Z, Wei Y, Zhang L, Li X, Yuan H, Wang A(2015) Apple MdACS6 regulates ethylene biosynthesis during fruit development involving ethylene-responsive factor. Plant Cell Physiol 56: 1909–1917 [DOI] [PubMed] [Google Scholar]
- Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, Wang A(2017b) The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29: 1316–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Feng S, Ma S, Sui X, Zhang Z(2017c) Spatiotemporal expression and substrate specificity analysis of the cucumber SWEET gene family. Front Plant Sci 8: 1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CJ, Lai Z, Lee S, Yun DJ, Mengiste T(2016) Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. Plant Cell 28: 1662–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wang Y, Yu L, Jiang H, Guo Z, Xu H, Jiang S, Fang H, Zhang J, Su M, et al. (2019) MdWRKY11 participates in anthocyanin accumulation in red-fleshed apples by affecting MYB transcription factors and the photoresponse factor MdHY5. J Agric Food Chem 67: 8783–8793 [DOI] [PubMed] [Google Scholar]
- Lloyd A, Brockman A, Aguirre L, Campbell A, Bean A, Cantero A, Gonzalez A(2017) Advances in the MYB-bHLH-WD Repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol 58: 1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhang D, Miao Q, Yang J, Xuan Y, Hu Y(2017) Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol 58: 863–873 [DOI] [PubMed] [Google Scholar]
- Ma Q, Xia Z, Cai Z, Li L, Cheng Y, Liu J, Nian H(2019) GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in Arabidopsis thaliana. Front Plant Sci 9: 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Kasuga S, Kawahigashi H(2016) The sorghum SWEET gene family: Stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol Biofuels 9: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi T, Abe K, Sanada T, Yamaki S(1992) Levels and role of sucrose synthase, sucrose-phosphate synthase, and acid invertase in sucrose accumulation in fruit of Asian pear. J Am Soc Hortic Sci 117: 274–278 [Google Scholar]
- Ohto MA, Hayashi S, Sawa S, Hashimoto-Ohta A, Nakamura K(2006) Involvement of HLS1 in sugar and auxin signaling in Arabidopsis leaves. Plant Cell Physiol 47: 1603–1611 [DOI] [PubMed] [Google Scholar]
- Pal S, Yadav AK, Singh AK, Rastogi S, Gupta MM, Verma RK, Nagegowda DA, Pal A, Shasany AK(2017) Nitrogen treatment enhances sterols and withaferin A through transcriptional activation of jasmonate pathway, WRKY transcription factors, and biosynthesis genes in Withania somnifera (L.) Dunal. Protoplasma 254: 389–399 [DOI] [PubMed] [Google Scholar]
- Patil G, Valliyodan B, Deshmukh R, Prince S, Nicander B, Zhao M, Sonah H, Song L, Lin L, Chaudhary J, et al. (2015) Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics 16: 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestley C.(1983) Interconversions of 14C-labelled sugars in Apple Tree Tissues12. J Exp Bot 34: 1740–1747 [Google Scholar]
- Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, Hong X, Zhu JK, Gong Z(2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J 63: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie EA, Turgeon R(2009) A comprehensive picture of phloem loading strategies. Proc Natl Acad Sci USA 106: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB(1992) Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J 11: 4705–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ(2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M(2007) Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 76: 75–100 [DOI] [PubMed] [Google Scholar]
- Singh AK, Kumar SR, Dwivedi V, Rai A, Pal S, Shasany AK, Nagegowda DA(2017) A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol 215: 1115–1131 [DOI] [PubMed] [Google Scholar]
- Soni R, Carmichael JP, Murray JA(1993) Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simplified procedure. Curr Genet 24: 455–459 [DOI] [PubMed] [Google Scholar]
- Stitt M, Wilke I, Feil R, Heldt HW(1988) Coarse control of sucrose-phosphate synthase in leaves: Alterations of the kinetic properties in response to the rate of photosynthesis and the accumulation of sucrose. Planta 174: 217–230 [DOI] [PubMed] [Google Scholar]
- Sturm A, Hess D, Lee HS, Lienhard S(1999) Neutral invertase is a novel type of sucrose‐cleaving enzyme. Physiol Plant 107: 159–165 [Google Scholar]
- Sun X, Zhang L, Wong DCJ, Wang Y, Zhu Z, Xu G, Wang Q, Li S, Liang Z, Xin H(2019) The ethylene response factor VaERF092 from Amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J 99: 988–1002 [DOI] [PubMed] [Google Scholar]
- Wang S, Yokosho K, Guo R, Whelan J, Ruan YL, Ma JF, Shou H(2019) The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo. Plant Physiol 180: 2133–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Meng D, Wang A, Li T, Jiang S, Cong P, Li T(2013) The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol 162: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Liu F, Chen C, Ma F, Li M(2014) The Malus domestica sugar transporter gene family: Identifications based on genome and expression profiling related to the accumulation of fruit sugars. Front Plant Sci 5: 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham TG, Slobodchikoff CN(1981) Evolution by individuals, plant-herbivore interactions, and mosaics of genetic variability: The adaptive significance of somatic mutations in plants. Oecologia 49: 287–292 [DOI] [PubMed] [Google Scholar]
- Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, et al. (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23: 396–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan YH, Hu YB, Chen LQ, Sosso D, Ducat DC, Hou BH, Frommer WB(2013) Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc Natl Acad Sci USA 110: E3685–E3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF(2006) Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci USA 103: 10503–10508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Kimura T, Sawamura Y, Manabe T, Kotobuki K, Hayashi T, Ban Y, Matsuta N(2002) Simple sequence repeats for genetic analysis in pear. Euphytica 124: 129 [Google Scholar]
- Yoo SD, Cho YH, Sheen J(2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yuan M, Wang S(2013) Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant 6: 665–674 [DOI] [PubMed] [Google Scholar]
- Zhang HP, Wu JY, Tao ST, Wu T, Qi KJ, Zhang SJ, Wang JZ, Huang WJ, Wu J, Zhang SL(2014) Evidence for apoplasmic phloem unloading in pear fruit. Plant Mol Biol Report 32: 931–939 [Google Scholar]
- Zhang L, Chen L, Yu D(2018) Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol 176: 790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L(2005) The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol Biol 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY(2008) Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 6: 486–503 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Tan B, Luo M, Li Y, Liu C, Chen C, Yu CW, Yang S, Dong S, Ruan J, et al. (2013) HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25: 134–148 [DOI] [PMC free article] [PubMed] [Google Scholar]