Sequence differences among ALCOHOL ACYLTRANSFERASES1s likely account for differences in the levels of the aroma compound γ-decalactone between high- and low-aroma peach cultivars.
Abstract
Aroma contributes to the unique flavors of fruits and is important for fruit quality evaluation. Among the many volatiles in peach (Prunus persica) fruits, γ-decalactone has the greatest contribution to the characteristic peach aroma. Some peach cultivars have γ-decalactone contents that are too low to detect. Comparison of the transcriptomes and metabolomes of a high-aroma cultivar, Fenghuayulu, and a low-aroma cultivar, Achutao, suggested that amino acid substitutions in ALCOHOL ACYLTRANSFERASE (PpAAT1) are responsible for the undetectable levels of γ-decalactone in cv Achutao fruit. Modeling and molecular docking analysis of PpAAT1 indicated that the substituted residues might determine substrate recognition or act as control channels to the active site. In vitro enzyme assays on PpAAT1 heterologously expressed and purified from Escherichia coli and in vivo assays using transient PpAAT1 expression in Nicotiana benthamiana or the oleaginous yeast Yarrowia lipolytica indicated that PpAAT1 from high-aroma cultivars was more efficient than PpAAT1 from low-aroma cultivars in catalyzing the conversion of 4-hydroxydecanoyl-coenzyme A into γ-decalactone. Examination of loss-of-function mutations of PpAAT1 generated by CRISPR/Cas9 in cv Fenghuayulu showed that fruits with PpAAT1 mutations had significantly lower γ-decalactone contents. Expression of the version of PpAAT1 from cv Fenghuayulu in cv Achutao restored γ-decalactone levels to those measured in ‘Fenghuayulu’, confirming the specific contribution of PpAAT1 to the formation of this key aroma compound. These results show how the biosynthesis of the peach aroma compound γ-decalactone is compromised in some low-aroma cultivars and illustrate the physiological role of PpAAT1 in plant lactone biosynthesis.
Fruit aromas attract seed-dispersal agents (Lomáscolo et al., 2010; Hodgkison et al., 2013; Rodríguez et al., 2013; Nevo et al., 2016) and are critical for promoting consumer consumption of fruits (Gilbert et al., 1996; Ball et al., 1998; Cipollini, 2000; Goff and Klee, 2006; Steingass et al., 2015). Unlike other fruit characteristics, fruit aroma is not amenable to high-throughput assays, and breeding for aroma is beyond the capabilities of most breeding programs. Therefore, breeding programs have historically focused on yield, disease resistance, and fruit firmness (Cao et al., 2014); improvement of aroma quality has not been a top priority (Gutterson, 1993; Lauxmann et al., 2014). In the process of constructing a peach (Prunus persica) core germplasm collection, we analyzed the characteristics of peach cultivars in the National Fruit Germplasm Repository (Nanjing, China) and found that, since the 1980s, some varieties have lost their peach-like aroma (Yu et al., 2010; Shen et al., 2013).
The special aromas of some fruits depend on a mixture of several compounds, as in tomato (Solanum lycopersicum; Buttery et al., 1990). By contrast, in other fruits, single compounds are almost entirely responsible for a characteristic aroma, as with the key compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone, which is responsible for the aroma of strawberries (Fragaria spp.; Larsen and Poll, 1992; Song et al., 2016). γ-Decalactone was identified in peach fruit in 1964, and its aroma was found to be like that of peach jam (Jennings and Sevenants, 1964). Subsequent studies showed that γ-decalactone is the main lactone component of the volatiles from peach fruits (Sevenants and Jennings, 1966; Do et al., 1969). Gas chromatography (GC) and sensory analysis of volatiles from cling peaches showed that γ-decalactone is the major contributor to the peachy aroma (Spencer et al., 1978). Subsequent analyses of peach fruit volatiles identified more than 100 compounds and confirmed γ-decalactone as the most important component contributing to peach aroma (Horvat et al., 1990; Jia and Okamoto, 2001; Eduardo et al., 2010; Sánchez et al., 2012; Li et al., 2015; Zhang et al., 2017b). Given that γ-decalactone itself produces the typical peach-like aroma, this compound is currently the most important lactone in the food and fragrance industry, with a market volume of several hundred tons per year (Braga and Belo, 2016).
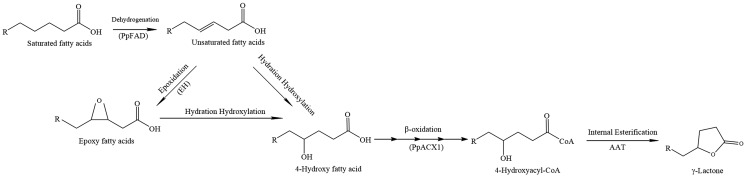
The lactone biosynthetic pathway in plants has not been fully elucidated. The current understanding is that all lactones are fatty acid-derived flavor alkanolides originating from saturated fatty acids, which go through steps of dehydrogenation, epoxidation, hydration hydroxylation, shortening by β-oxidation, and then internal esterification with hydroxyacetyl-coenzyme A (hydroxyacyl-CoA; Fig. 1; Schöttler and Boland, 1996; Schwab et al., 2008; Aragüez and Valpuesta, 2013; Sánchez et al., 2013). Some enzymes specifically involved in the formation of γ-decalactone in peach fruits have been reported, including fatty acid desaturase, epoxide hydrolases, and acyl-CoA oxidase (Vecchietti et al., 2009; Sánchez et al., 2013; Zhang et al., 2017b). Recent studies have shown that the alcohol acyltransferases (AATs), which catalyze the transfer of an acyl group from a CoA donor to an alcohol acceptor, are critical for the biosynthesis of fruit aroma in apple (Malus × domestica; Souleyre et al., 2014), banana (Musa sapientum; Beekwilder et al., 2004), tomato (Goulet et al., 2015), papaya (Carica papaya; Balbontín et al., 2010), and kiwifruit (Actinidia chinensis; Souleyre et al., 2011). Gene expression studies of peach fruits during postharvest ripening have suggested that AAT1 is associated with aroma formation (Zhang et al., 2010), and enhanced expression of AAT1 prevents aroma loss due to chilling injury (Xi et al., 2012). However, to the best of our knowledge, functional verification of the AAT in peach has not been reported.
Figure 1.
The putative pathway of γ-lactone biosynthesis. EH, Epoxide hydrolase; PpACX1, acyl-CoA oxidase1; PpFAD, fatty acid desaturase.
In the putative γ-lactone pathway, internal esterification, a reaction that can be catalyzed by AAT, is the last step of γ-lactone biosynthesis. AATs are capable of combining various alcohols and acyl-CoAs to produce a wide range of flavor esters (D’Auria, 2006; Schwab et al., 2008; Aragüez and Valpuesta, 2013). To date, the characterization of AAT enzymes has focused mainly on ester synthesis (Aharoni et al., 2000; Souleyre et al., 2005, 2014; Goulet et al., 2015). Whether AATs in peach are capable of catalyzing the synthesis of lactones using hydroxyacyl-CoA as a substrate remains to be demonstrated.
To examine peach aroma, we compared a variety with strong aroma and a variety with little aroma, taking advantage of the diversity of peach cultivars in core collections. To date, more than 600 peach cultivars have been conserved and used to establish the core collection at the National Fruit Germplasm Repository in China; moreover, 60 morphological indexes have been recorded from these cultivars (Shen et al., 2013). In the standard description for peach, strong, intermediate, and slight are used to describe aroma intensity (Wang and Zhu, 2005). We found a high-aroma-producing cultivar, Fenghuayulu, and a low-aroma-producing cultivar, Achutao; the aromas of these cultivars were consistently described as strong and slight, respectively, over 10 years of cultivation in the germplasm repository. In this study, we assessed these two cultivars through combined metabolome and transcriptome analysis to understand how γ-decalactone biosynthesis is compromised in low-aroma cultivars. We also functionally characterized PpAAT1 variants found in high- and low-aroma-producing peach varieties in vitro and in vivo. This study reports the physiological role of PpAAT1 in plant lactone biosynthesis and provides a solid foundation for the improvement of peach cultivars by increasing levels of the peach aroma compound γ-decalactone.
RESULTS
PpAAT1 Is Associated with γ-Decalactone Production in High- and Low-Aroma Peach Varieties
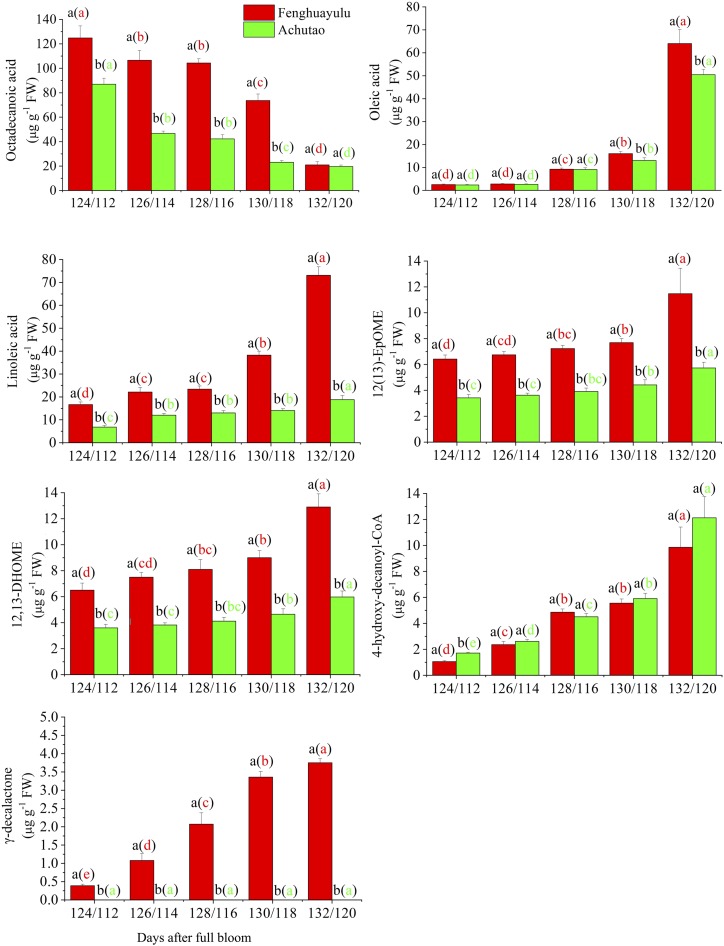
To gain insight into how the biosynthesis of the peach aroma compound γ-decalactone is compromised in some low-aroma cultivars, we undertook metabolic and transcriptomic profiling in the high-aroma cv Fenghuayulu and the low-aroma cv Achutao. To identify a suitable sampling time, we first measured the γ-decalactone contents in ‘Fenghuayulu’ peach fruits at 10 sampling points during development. The GC results showed no γ-decalactone in any of the three biological replicates at each sampling point before 124 d after full bloom (DAFB). At 124 DAFB, γ-decalactone accumulated to levels detectable by GC in all three biological replicates, and its abundance progressively increased until 132 DAFB (Fig. 2). Therefore, we selected five developmental stages at which γ-decalactone was present to use in sampling for metabolome and transcriptome analysis. For each time point, samples from the two cultivars were matched to ensure similar ripeness, based on the index of absorbance difference (IAD; Zhang et al., 2017a); therefore, the cv Fenghuayulu samples were collected at 124 to 132 DAFB and the cv Achutao samples were collected at 112 to 120 DAFB (for additional details, see “Materials and Methods”).
Figure 2.
Accumulation profiles of intermediates in the putative γ-decalactone pathway in cvs Fenghuayulu and Achutao. Data are presented as means ± se (n = 3). Letters indicate statistical differences at P ≤ 0.05 as determined using ANOVA followed by Fisher’s lsd test. Letters outside parentheses indicate differences between the two cultivars at each developmental stage, and those inside parentheses indicate differences in one cultivar across developmental stages. FW, Fresh weight.
Ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) combined with GC-MS detected 151 metabolites in cv Fenghuayulu, but UPLC-MS only detected 89 metabolites in cv Achutao and GC-MS only detected 20 metabolites (Supplemental Tables S1 and S2). The metabolome results showed no detectable γ-decalactone in any samples from cv Achutao (Fig. 2).
To further assess which steps in γ-decalactone biosynthesis are compromised in cv Achutao, we identified putative intermediates and genes in the lactone biosynthesis process in cv Fenghuayulu. We then calculated Pearson’s correlation coefficients among the metabolites from cv Fenghuayulu. With a cutoff of r > 0.6, 20 compounds showed positive correlations and six showed negative correlations with γ-decalactone (Supplemental Table S3).
To more accurately assess the putative intermediates in the γ-decalactone biosynthesis pathway, we used Paintomics 3.0 to assign the 26 compounds to nine biosynthesis pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG; Supplemental Table S4), but four compounds (2-methylbutanoic acid, 4-hydroxydecanoic acid, 4-hydroxydecanoyl-CoA, and 1-octen-3-one) could not be integrated into the KEGG pathway. The lactone components are mainly fatty acid-derived flavor alkanolides (Schwab et al., 2008; Aragüez and Valpuesta, 2013). Therefore, we selected 11 compounds in the fatty acid metabolic and linoleic acid metabolic pathways for further analysis. We also selected 4-hydroxydecanoyl-CoA because it had a high positive correlation with γ-decalactone (Supplemental Table S3). Thus, we chose 12 compounds in total for the subsequent analysis.
We performed RNA sequencing (RNA-seq) analysis of the two cultivars using samples from the same five time points as in the metabolome analysis, with three independent replicates for each. The data have been deposited in the National Center for Biotechnology Information Short Read Archive database (SRP156184). A total of 6.71 Gb of sequencing reads were generated on average for each sample. After read filtering, we obtained an average of 44,627,256 clean reads, of which 76.24% were uniquely mapped to the peach reference genome. We next calculated the gene expression levels for each sample using RSEM (RNA-Seq expression estimation by Expectation-Maximization) and used these results to identify differentially expressed genes (DEGs) between samples (adjusted P < 0.05). Ten randomly selected DEGs with different expression levels were subjected to reverse transcription quantitative PCR (RT-qPCR) validation, and the results from the RT-qPCR were highly correlated with the reads per kilobase of exon model per million mapped reads values from the RNA-seq analysis, which confirmed the robustness of the RNA-seq data (Supplemental Table S5).
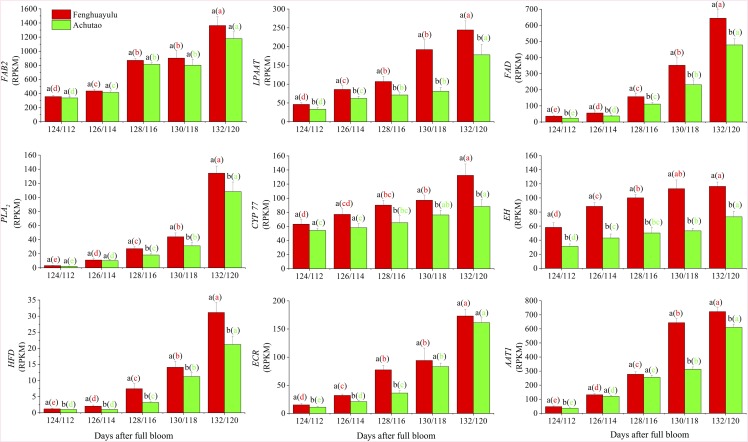
To generate candidate genes likely to be involved in γ-decalactone biosynthesis, we considered for subsequent analysis those DEGs whose expression level was higher than 1 reads per kilobase of exon model per million mapped reads and that were specified in the KEGG pathway for γ-decalactone formation in cv Fenghuayulu. We then subjected the 38 DEGs identified by Paintomics 3.0 to further correlation analysis with the above compounds (Supplemental Table S6). With cutoff values of r > 0.6 (Supplemental Table S7), this process selected six compounds and nine unigenes as components of the putative γ-decalactone pathway for subsequent analysis. The compounds were as follows: a free saturated fatty acid (octadecanoic acid), two free unsaturated fatty acids (oleic acid and linoleic acid), a free epoxy acid [12(13)-EpOME], a free hydroxy acid (12,13-DHOME), and hydroxyacyl-CoA. The nine unigenes encode fatty acid biosynthesis2 (FAB2), lysophosphatidic acid acyltransferase (LPAAT), fatty acid desaturase (FAD), phospholipase A2 (PLA2), a cytochrome P450 family 77 member (CYP77), epoxide hydrolase (EH), hydroxy fatty dehydratase (HFD), enoyl-CoA reductase (ECR), and alcohol acyltransferase1 (AAT1).
The putative intermediates and genes in cv Fenghuayulu were also found in cv Achutao and showed similar trends during development: in both cultivars, the levels of putative intermediates (except octadecanoic acid) tended to increase, and the transcript levels in all of the putative genes in both cultivars also tended to increase (Figs. 2 and 3). For subsequent analysis, we focused on the gene AAT (referred to as PpAAT1), encoding AAT, which we believed might catalyze the last step in lactone formation.
Figure 3.
Gene expression in the putative γ-decalactone pathway in cvs Fenghuayulu and Achutao. Data are presented as means ± se (n = 3), and letters represent significant differences at P ≤ 0.05 as determined using ANOVA followed by Fisher’s lsd test. Letters outside parentheses indicate differences between the two cultivars at each developmental stage, and those inside parentheses indicate differences in one cultivar across developmental stages. RPKM, Reads per kilobase of exon model per million mapped reads.
PpAAT1 Sequences in High- and Low-Aroma Cultivars Showed Differences in Amino Acids Potentially Involved in Substrate Binding
To identify whether the loss of γ-decalactone in cv Achutao was due to sequence variation in PpAAT1, we cloned and sequenced the PpAAT1 coding sequences (CDSs) from the two cultivars. PpAAT1 encodes a predicted polypeptide of 450 amino acids. The PpAAT1 proteins from cvs Fenghuayulu and Achutao (referred to as fhPpAAT1 and acPpAAT1, respectively) both contained an HXXXD-type acyltransferase-like motif (HAMCD), similar to other known AATs (D’Auria, 2006). However, our sequence analysis identified mutations encoding three amino acid substitutions in acPpAAT1 compared with fhPpAAT1 (Lys-30→Glu, Ile-49→Val, and Ala-343→Val). Phylogenetic analysis of the amino acid sequences of these and other AATs indicated that fhPpAAT1 and acPpAAT1 clustered together and had a close relationship with the AATs from Fragaria × ananassa (Supplemental Fig. S1).
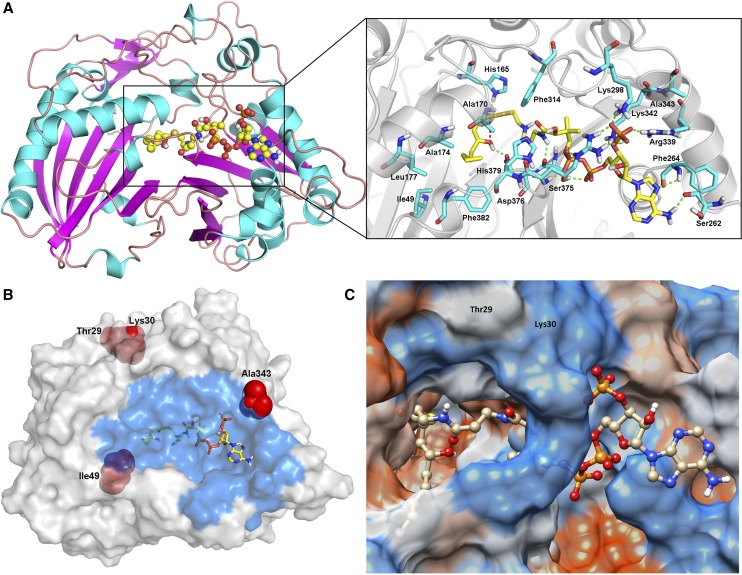
We performed computer modeling of PpAAT1 and molecular docking studies to predict the putative binding mechanism of 4-hydroxydecanoyl-CoA with PpAAT1 (Fig. 4). To specifically analyze which amino acids participate in substrate-enzyme recognition, we used the combination mode of the molecular docking software, which selected the most reasonable binding conformations based on energy minimization. The results showed that during substrate binding to the active site of PpAAT1, the amino acid residues primarily involved in recognition are Ile-49, His-165, Ala-170, Ala-174, Leu-177, Ser-262, Phe-264, Lys-298, Phe-314, Arg-339, Lys-342, Ala-343, Ser-375, Asp-376, His-379, and Phe-382 (Fig. 4A). These include two (Ile-49 and Ala-343) of the three residues that differ between fhPpAAT1 and acPpAAT1 (Fig. 4B). The third residue, Lys-30, is located at the entrance to the active site of PpAAT1 (Fig. 4C).
Figure 4.
Orientation of important residues during the internal esterification reaction. A, Docking analysis with substrate (the ball-and-stick model represents 4-hydroxydecanoyl-CoA). B, Amino acid substitutions in the PpAAT1 variants of low-aroma cultivars (the ball-and-stick model represents 4-hydroxydecanoyl-CoA, and the blue region represents the residues involved in the recognition). C, Thr-29 and Lys-30 are located at the entrance of PpAAT1.
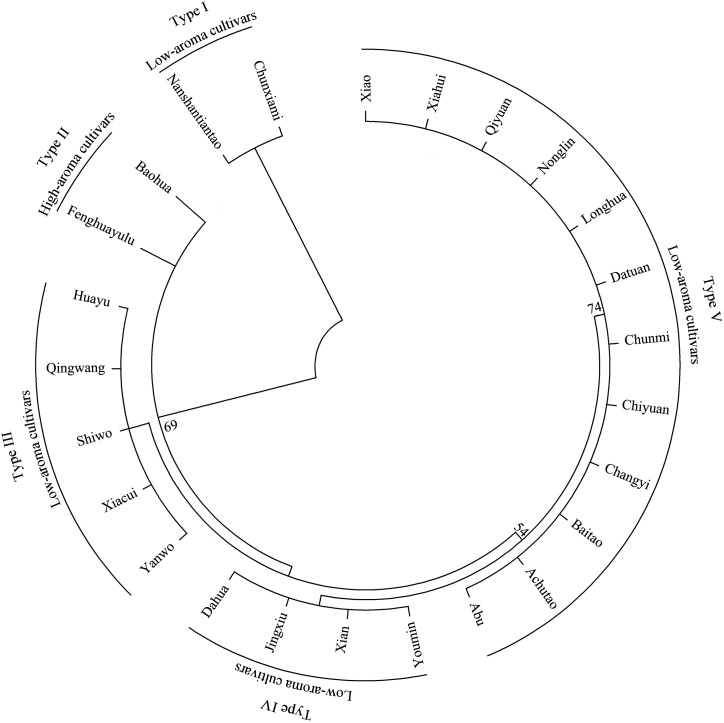
Next, we were interested in determining whether sequence differences in PpAAT1 correlated with the absence of peach aroma in other low-aroma cultivars. Therefore, we selected 22 low-aroma peach cultivars, as well as another high-aroma cultivar, from the National Fruit Germplasm Repository for sequence analysis. The results showed that all of the low-aroma cultivars we tested had amino acid substitutions in PpAAT1 compared with the sequence of fhPpAAT1, but another high-aroma cultivar had no substitutions. Phylogenetic analysis showed that the cultivars could be divided into five clusters and the two high-aroma cultivars clustered together (clade II; Fig. 5). There were four substitution types (clade I, III, IV, and V) for PpAAT1 in low-aroma cultivars. In type I, the substitution occurred at amino acid residue 29 (Thr-29→Asn). Types III and IV each showed one of the substitutions seen in cv Achutao (Ile-49→Val and Ala-343→Val, respectively), and type V showed all three of the same substitutions as cv Achutao.
Figure 5.
Phylogenetic analysis of the amino acid sequences of PpAAT1 from different peach cultivars. Phylogenetic analysis was performed using the maximum likelihood method with 1,000 bootstrap replicates using MEGA 6.06. The numbers on the nodes are support values. High-aroma cultivars are as follows: Fenghuayulu and Baohua. Low-aroma cultivars are as follows: Nanshantiantao, Chunxiami, Xiao, Xiahui, Qiyuan, Nonglin, Longhua, Datuan, Chunmi, Chiyuan, Changyi, Baitao, Achutao, Abu, Youmin, Xian, Jingxiu, Dahua, Yanwo, Xiacui, Shiwo, Qingwang, and Huayu.
In Vitro Enzymatic Characterization of the Five PpAAT1 Types Showed Higher Activity in Types from High-Aroma Cultivars
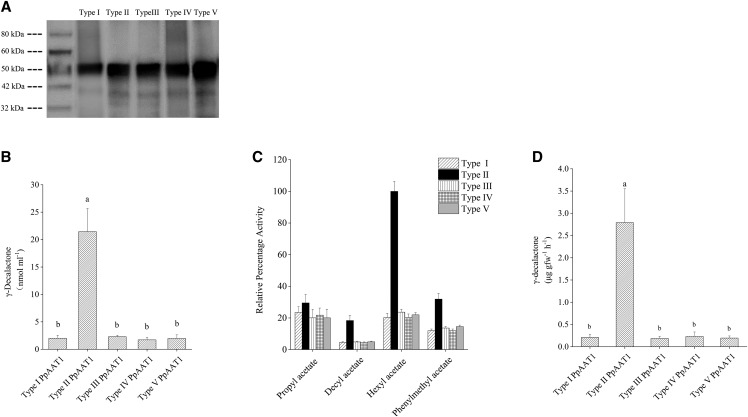
To test whether the variations in sequence affected the internal esterification capabilities of the enzymes, we isolated the CDSs of five PpAAT1 genes and functionally evaluated them in vitro. The predicted molecular masses of the recombinant His-tagged PpAAT1 proteins were 50.99, 50.96, 50.96, 50.96, and 50.97 kD, and the western-blot bands detected using an anti-His antibody were within the expected size ranges (Fig. 6A). In addition, liquid chromatography-tandem mass spectrometry (LC-MS/MS) data showed 94.3%, 93.91%, 91.66%, 93.7%, and 90.16% sequence coverage rates for the PpAAT1 His-tag protein hydrolyzed peptide segments, respectively, confirming the identity of the recombinant enzymes.
Figure 6.
Enzymatic activity of the five PpAAT1 variants. A, Western analysis of the five PpAAT1 variants. B, The content of γ-decalactone produced by the five kinds of PpAAT1 in vitro. C, Comparison of esterification activity of the five PpAAT1 variants ± se (n = 6). The differences in activity between the type II and other PpAAT1s are significant for all the esters (P ≤ 0.05) except propyl acetate. The 100% relative activity corresponds to 0.22 μmol min–1 mg–1 hexyl acetate. D, γ-Decalactone contents detected in transgenic N. benthamiana leaves expressing the five PpAAT1 variants. Data are presented as means ± se (n = 6), and letters represent significant differences at P ≤ 0.05 as determined using ANOVA followed by Fisher’s lsd test.
To evaluate the catalytic activity of the recombinant enzymes, we assessed their production of γ-decalactone when provided with 4-hydroxydecanoyl-CoA as a substrate. After the reaction, GC-MS analysis showed the formation of a single major peak, corresponding to γ-decalactone, that was not detected in a control reaction with nonfunctional (boiled) protein. The results indicated that all of the PpAAT1s could catalyze the internal esterification of 4-hydroxydecanoyl-CoA to γ-decalactone, but the type II PpAAT1 produced significantly more γ-decalactone (Fig. 6B). Kinetic measurements were performed to better understand the enzymatic characteristics of PpAAT1 (Table 1). The type II enzyme exhibited a lower Km value and a much higher Kcat/Km ratio for the 4-hydroxydecanoyl-CoA substrate test than the other four PpAAT1s. Thus, type II PpAAT1s are more catalytically active with the 4-hydroxydecanoyl-CoA substrate than the other four types of PpAAT1.
Table 1. Enzyme kinetics of the five recombinant PpAAT1s using 4-hydroxydecanoyl-CoA as substrate.
Data are presented as means ± se. Letters indicate significant differences at P ≤ 0.05 (n = 6) as determined using ANOVA followed by Fisher’s lsd test.
| Enzymes | Km | Kcat | Kcat/Km |
|---|---|---|---|
| mm | s–1 | ||
| Type I | 6.5211 ± 0.4321 b | 0.1552 ± 0.0112 b | 0.0232 ± 0.0011 b |
| Type II | 2.4144 ± 0.1516 a | 2.2411 ± 0.0991 a | 0.9507 ± 0.0661 a |
| Type III | 6.3314 ± 0.5611 b | 0.1446 ± 0.0191 b | 0.0224 ± 0.0031 b |
| Type IV | 7.0852 ± 0.6911 b | 0.1720 ± 0.0160 b | 0.0220 ± 0.0013 b |
| Type V | 6.2014 ± 0.4952 b | 0.1730 ± 0.0120 b | 0.0271 ± 0.0014 b |
In some fruits, AAT is also able to synthesize esters, which also affect fruit aroma (Aharoni et al., 2000; Souleyre et al., 2005, 2014; Goulet et al., 2015). Therefore, we assessed the esterification activity of the PpAAT1 variants toward different substrates. We used different alcohols and acetyl-CoA that could potentially produce the esters found in cv Fenghuayulu (Supplemental Table S2) in this study. The results showed that a similar amount of propyl acetate, but greater amounts of hexyl acetate, decyl acetate, and phenylmethyl acetate, were produced in the presence of type II PpAAT1 as compared with the other four types (Fig. 6C).
Transient Expression of PpAAT1 Types in Nicotiana benthamiana Leaves Confirmed the PpAAT1 Activity in Planta
To examine the activity of the PpAAT1 enzymes in planta, we transiently expressed them in N. benthamiana leaves. When provided with 4-hydroxydecanoyl-CoA substrate, N. benthamiana plants expressing type II PpAAT1 produced significantly more γ-decalactone (Fig. 6D) in their leaves than the other four transgenic lines. Meanwhile, there was no detectable γ-decalactone in the empty vector-transformed or negative control N. benthamiana leaves. Thus, the products derived from PpAAT1s in plant tissues confirmed the differing in vitro activities of the five PpAAT1s, in particular their different γ-decalactone synthase activities.
Functional Tests of PpAAT1 Types in Oleaginous Yeast Confirm Their Differing Abilities to Produce γ-Decalactone
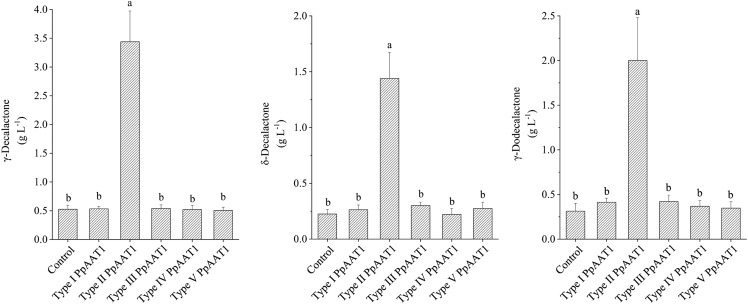
To examine the catalytic activities of the AATs encoded by the five types of PpAAT1 genes, we isolated the CDSs of the PpAAT1 genes and tested them by heterologous expression in oleaginous yeast (Yarrowia lipolytica). We detected three lactones in the control cells transformed with the empty vector (Fig. 7). The five strains of recombinant oleaginous yeast containing the five PpAAT1 genes showed different γ-decalactone production abilities. The transformants containing the type II PpAAT1 gene, but not PpAAT1 genes of the four other types, had significantly higher lactone contents (Fig. 7).
Figure 7.
GC-MS analysis of lactone production from oleaginous yeast strain Po1g containing different types of PpAAT1. Data are presented as means ± se (n = 8), and letters represent significant differences at P ≤ 0.05 as determined using ANOVA followed by Fisher’s lsd test.
CRISPR/Cas9 Mutagenesis of PpAAT1 in cv Fenghuayulu Reduces Fruit γ-Decalactone Contents
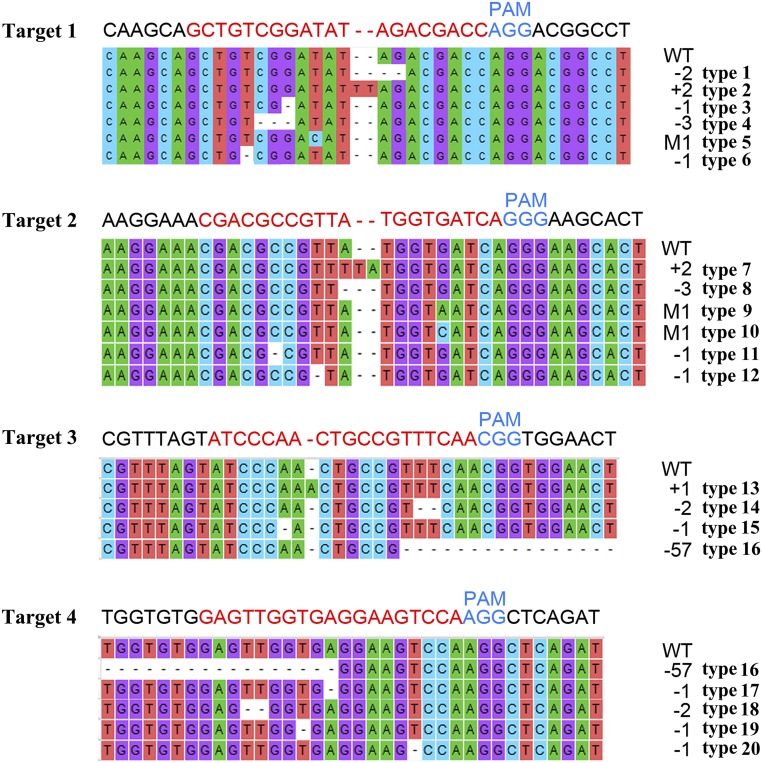
To determine whether these substitutions were responsible for the lack of γ-decalactone in cv Achutao fruit, we used CRISPR/Cas9 to generate PpAAT1 loss-of-function mutations in cv Fenghuayulu fruit. We chose four targets in the region where substitutions occurred in the low-aroma cultivars (Supplemental Fig. S2) and selected four sequences (three 20-bp targets and one 19-bp target) in the PpAAT1 locus with tandem guanosine nucleotides (protospacer-adjacent motifs) as single guide RNA (sgRNA) complementary sites (Fig. 8). Then, we constructed a CRISPR/Cas9 vector harboring these four sgRNA cassettes. To deliver the CRISPR/Cas reagents, Agrobacterium tumefaciens containing either this construct or the empty vector was injected into developing fruits of ‘Fenghuayulu’ peach trees.
Figure 8.
Specific mutations of fhPpAAT1 introduced into peach fruits using CRISPR/Cas9. Numbers in black at right represent the numbers of mutant bases. PAM, Protospacer-adjacent motif; WT, wild type.
When the transgenic, empty vector-injected, and negative control (uninjected cv Fenghuayulu) fruits reached the best harvesting time (determined based on the IAD [IAD = 0.1]), we conducted GC-MS analysis. As compared with the control fruits, 70 out of 200 fruits injected with A. tumefaciens containing the CRISPR/Cas9 vector harboring the four sgRNA cassettes showed significantly lower levels of γ-decalactone and the other four lactones (γ-hexalactone, δ-decalactone, γ-dodecalactone, and δ-dodecalactone; Table 2). However, there were no obvious differences in lactone content in the empty vector-injected fruits compared with the uninjected fruits.
Table 2. Lactones, esters, alcohols, acids, aldehydes, and 4-hydroxydecanoyl-CoA in the cv Fenghuayulu CRISPR/Cas9 transgenic fruits and controls.
Data are presented as means ± se (n = 6). Letters indicate significant differences at P ≤ 0.05 as determined using ANOVA followed by Fisher’s lsd test.
| Metabolic Compounds | High-Mutation-Rate Fruits | Low-Mutation-Rate Fruits | Empty Vector Control | Wild Type (Uninjected Fruits) |
|---|---|---|---|---|
| Lactone | ||||
| γ-Decalactone | 0.903 ± 0.411 b | 2.790 ± 0.842 a | 3.599 ± 0.411 a | 3.726 ± 0.390 a |
| γ-Hexalactone | 0.194 ± 0.072 b | 0.477 ± 0.191 a | 0.532 ± 0.092 a | 0.546 ± 0.089 a |
| δ-Decalactone | 0.329 ± 0.107 b | 0.913 ± 0.296 a | 1.103 ± 0.198 a | 1.017 ± 0.1490 a |
| γ-Dodecalactone | 0.413 ± 0.116 b | 1.129 ± 0.388 a | 1.473 ± 0.191 a | 1.489 ± 0.151 a |
| δ-Dodecalactone | 0.561 ± 0.169 b | 1.336 ± 0.429 a | 1.614 ± 0.177 a | 1.706 ± 0.113 a |
| Total lactones | 2.4 | 6.645 | 8.321 | 8.484 |
| Ester | ||||
| Phenylmethyl acetate | 0.093 ± 0.015 b | 0.311 ± 0.083 a | 0.379 ± 0.045 a | 0.331 ± 0.071 a |
| Decyl acetate | 0.061 ± 0.011 b | 0.196 ± 0.036 a | 0.221 ± 0.031 a | 0.234 ± 0.021 a |
| Hexyl acetate | 0.422 ± 0.129 b | 1.245 ± 0.327 a | 1.405 ± 0.119 a | 1.397 ± 0.108 a |
| Propyl acetate | 0.068 ± 0.041 b | 0.221 ± 0.037 a | 0.249 ± 0.019 b | 0.233 ± 0.027 b |
| Total esters | 0.644 | 1.973 | 2.254 | 2.195 |
| Alcohol | ||||
| Hexanol | 2.271 ± 0.659 a | 0.608 ± 0.091 b | 0.466 ± 0.059 b | 0.429 ± 0.061 b |
| Benzyl alcohol | 23.482 ± 5.021 a | 11.334 ± 4.108 b | 9.521 ± 1.108 b | 9.022 ± 0.808 b |
| Decanol | 2.854 ± 0.763 a | 1.238 ± 0.341 b | 0.929 ± 0.104 b | 0.950 ± 0.058 b |
| Propanol | 1.401 ± 0.422 a | 0.582 ± 0.105 b | 0.453 ± 0.061 b | 0.401 ± 0.022 b |
| 3-Hexen-1-ol | 1.142 ± 0.233 | 1.122 ± 0.276 | 1.100 ± 0.154 | 1.111 ± 0.135 |
| Total alcohols | 31.15 | 14.984 | 12.469 | 11.913 |
| Acid | ||||
| Heptanoic acid | 3.315 ± 0.411 | 3.395 ± 0.341 | 3.195 ± 0.241 | 3.257 ± 0.308 |
| Octanoic acid | 1.149 ± 0.189 | 1.051 ± 0.182 | 1.027 ± 0.116 | 1.039 ± 0.132 |
| Hexanoic acid | 1.314 ± 0.245 | 1.312 ± 0.400 | 1.612 ± 0.100 | 1.416 ± 0.182 |
| Butanoic acid, 2-methyl- | 0.834 ± 0.097 | 0.921 ± 0.115 | 0.823 ± 0.095 | 0.901 ± 0.104 |
| Acetic acid | 0.115 ± 0.022 | 0.135 ± 0.031 | 0.155 ± 0.021 | 0.144 ± 0.015 |
| 2-Hexenoic acid | 1.511 ± 0.206 | 1.461 ± 0.132 | 1.501 ± 0.132 | 1.612 ± 0.182 |
| Total acids | 8.238 | 8.275 | 8.313 | 8.369 |
| Aldehyde | ||||
| Hexanal | 0.867 ± 0.291 a | 0.231 ± 0.084 b | 0.161 ± 0.024 b | 0.152 ± 0.020 b |
| Benzaldehyde | 49.877 ± 9.179 a | 30.528 ± 7.195 b | 22.109 ± 3.246 b | 23.126 ± 3.278 b |
| 3-Hexenal | 0.547 ± 0.251 | 0.509 ± 0.117 | 0.496 ± 0.073 | 0.477 ± 0.061 |
| Octanal | 7.730 ± 1.189 | 6.992 ± 1.024 | 6.230 ± 0.889 | 6.324 ± 0.979 |
| Total aldehydes | 59.021 | 38.260 | 28.996 | 30.079 |
| 4-Hydroxydecanoyl-CoA | 41.903 ± 15.651 a | 13.633 ± 4.718 b | 9.533 ± 1.106 b | 9.732 ± 0.956 b |
To examine the PpAAT1 alleles produced, 20 clones from each of the 200 transgenic fruits and empty vector-injected fruits were randomly selected for sequencing. The sequencing results showed that 151 of the transgenic fruits carried mutations, whereas 49 had no detectable mutations in PpAAT1. A total of 20 mutant fhPpAAT1 gene variants were detected (Fig. 8) and all of the mutant fruits were found to be mosaic, containing one to 12 mutations. No mutations were detected in the fruits containing the empty vector. Most of the fhPpAAT1 mutations obtained were small insertions or deletions at the target sites, caused by nonhomologous end-joining repairs following sgRNA-directed Cas9 cleavage. Type 5, type 9, and type 10 were synonymous mutations.
To better understand the efficiency of the loss of function caused by CRISPR/Cas9, we calculated the mutation rates in the 151 transgenic fruits. For example, if six mutant fhPpAAT1 genes were found in the 20 clones from one fruit, the mutation rate was defined as 30%. Synonymous mutants were treated as 0. The samples showed varying mutation rates, from 0% to 60%. The higher mutation percentages (25% to 60%) in the fruits from those 70 mutated varieties resulted in significant reductions in lactone content, whereas the fruits with low mutation rates (0% to 20%) showed no significant changes in lactone content (Table 2).
We also analyzed the transgenic fruits for the presence of the principal esters, alcohols, acids, aldehydes, and 4-hydroxydecanoyl-CoA (Table 2). The GC-MS results showed that only those transgenic fruits with high mutation rates showed a clear reduction in total esters. In contrast, the levels of total alcohols and aldehydes were higher in those transgenic fruits than in the controls, while the levels of acids were similar. Interestingly, the precursor of γ-decalactone, 4-hydroxydecanoyl-CoA, was also increased in those high-mutation-rate transgenic lines.
Expression of fhPpAAT1 in Non-γ-Decalactone-Producing Fruits Rescues γ-Decalactone Production
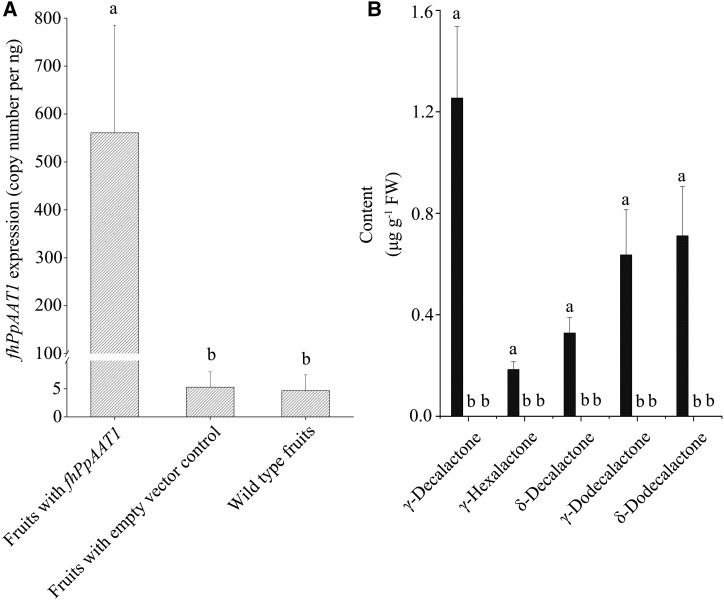
We carried out a functional assay to test the ability of type II PpAAT1 from the high-aroma cv Fenghuayulu (fhPpAAT1) to produce high levels of γ-decalactone in the low-aroma cv Achutao, which normally does not accumulate γ-decalactone to detectable levels. We assessed fhPpAAT1 expression through RT-qPCR based on the TaqMan assay, using a TaqMan MGB probe that was specific for the fhPpAAT1 sequence. The expression of fhPpAAT1 was significantly higher in all 100 transgenic fruits relative to empty vector-transformed and uninjected controls (which showed little or no FAM signal; Fig. 9A). The very weak signals detected in control fruits might be due to false priming or primer-dimer formation (Billard et al., 2012). Lactone accumulation also increased in the transgenic fruits and was undetectable in the control fruits (Fig. 9B), and the levels of total esters were higher in the transgenic fruits than in the control fruits (Table 3). In contrast, the total alcohol and aldehyde contents were significantly lower in the transgenic fruits than in the controls, as was the content of the precursor of γ-decalactone, 4-hydroxydecanoyl-CoA.
Figure 9.
Overexpression of fhPpAAT1 in cv Achutao increased the lactone content. A, Transcript levels of fhPpAAT1 in fruits relative to the wild type and empty vector controls. B, Lactone accumulation also increased in the transgenic fruits and was undetectable in the wild type and empty vector controls. Data are presented as means ± se (n = 10), and letters represent significant differences at P ≤ 0.05 as determined using ANOVA followed by Fisher’s lsd test. FW, Fresh weight.
Table 3. Esters, alcohols, acids, aldehydes, and 4-hydroxydecanoyl-CoA in the cv Achutao fhPpAAT1-overexpressing fruits and controls.
Data are presented as means ± se (n = 6). N.D., Below the limit of quantification, which is defined as signal/noise > 5. Letters indicate significant differences at P ≤ 0.05 as determined using ANOVA followed by Fisher’s lsd test.
| Metabolic Compounds | fhPpAAT1 Overexpressing | Empty Vector Transformed | Wild Type (Uninjected Fruits) |
|---|---|---|---|
| Ester | |||
| Phenylmethyl acetate | 0.101 ± 0.041 | N.D. | N.D. |
| Hexyl acetate | 0.269 ± 0.079 | N.D. | N.D. |
| Propyl acetate | 0.591 ± 0.106 a | 0.253 ± 0.033 b | 0.262 ± 0.041 b |
| Total esters | 0.961 | 0.253 | 0.262 |
| Alcohol | |||
| Hexanol | 0.317 ± 0.106 b | 1.068 ± 0.232 a | 1.014 ± 0.115 a |
| Benzyl alcohol | 0.932 ± 0.249 b | 1.875 ± 0.139 a | 1.901 ± 0.116 a |
| Propanol | 0.146 ± 0.083 | 0.429 ± 0.052 | 0.488 ± 0.033 |
| Total alcohols | 1.395 | 2.925 | 2.915 |
| Acid | |||
| Heptanoic acid | 4.104 ± 0.503 | 4.116 ± 0.425 | 4.279 ± 0.517 |
| Octanoic acid | 0.806 ± 0.121 | 0.816 ± 0.047 | 0.791 ± 0.019 |
| Hexanoic acid | 1.915 ± 0.203 | 1.899 ± 0.252 | 1.985 ± 0.142 |
| Butanoic acid, 2-methyl- | 0.817 ± 0.022 | 0.919 ± 0.071 | 0.818 ± 0.091 |
| Acetic acid | 0.192 ± 0.024 | 0.196 ± 0.021 | 0.189 ± 0.012 |
| 2-Hexenoic acid | 1.942 ± 0.303 | 1.941 ± 0.117 | 1.886 ± 0.193 |
| Total acids | 9.776 | 9.887 | 9.948 |
| Aldehyde | |||
| Hexanal | 0.079 ± 0.011 b | 0.281 ± 0.027 a | 0.281 ± 0.027 a |
| Benzaldehyde | 6.132 ± 1.031 b | 9.859 ± 0.731 a | 9.974 ± 0.912 a |
| Octanal | 3.851 ± 0.632 | 4.077 ± 0.359 | 4.317 ± 0.426 |
| Total aldehydes | 10.062 | 14.217 | 14.572 |
| 4-Hydroxydecanoyl-CoA | 2.426 ± 0.581 b | 12.121 ± 1.708 a | 12.331 ± 1.621 a |
DISCUSSION
In this study, we compared the transcriptomes and metabolomes of high- and low-aroma peach varieties and found that differences in the enzymatic activity of the AATs of high- and low-aroma peach varieties affect γ-decalactone production. We also established that three amino acid substitutions in PpAAT1 in low-aroma peach varieties such as ‘Achutao’ result in an undetectable level of γ-decalactone. Our sequencing of other low-aroma cultivars indicated that substitutions might cause the absence of γ-decalactone. Studies in other species such as papaya, apple, and tomato have observed that substitutions in AATs are responsible for decreases in aroma compounds (Morales-Quintana et al., 2013; Souleyre et al., 2014; Goulet et al., 2015). A 3D model analysis showed that these substitutions in low-aroma cultivars may affect the enzyme’s ability to interact with substrates and might affect the entrance to the active site. Substitutions affecting the entrance were also observed with the AATs of Cucumis melo and C. papaya (Morales-Quintana et al., 2012; Galaz et al., 2013). Molecular docking results identified 16 key amino acids that participate in γ-decalactone biosynthesis; further work will be required to establish their individual contributions to the catalytic efficiency of the enzyme.
To date, the model plants chosen for wide study, such as tomato, rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana), lack lactones (Sánchez et al., 2013; Zhang et al., 2017b). The functions of the genes and enzymes involved in the lactone biosynthesis pathway are still poorly understood. In this study, we elucidated the biological significance of PpAAT1 through recombinant enzyme activity analysis in vitro, transient expression in N. benthamiana, heterologous expression in Y. lipolytica, and loss- and gain-of-function testing in peach fruits.
We found that PpAAT1s are capable of catalyzing internal esterification at the hydroxyl (-OH) and -CoA groups of 4-hydroxydecanoyl-CoA in vitro and that the type II PpAAT1 from high-aroma cultivars was more catalytically active than the type I, III, IV, and V PpAAT1s from low-aroma cultivars. Furthermore, transient expression in N. benthamiana and heterologous expression in oleaginous yeast confirmed that the presence of substitutions in PpAAT1 genes corresponds to the absence of γ-decalactone in low-aroma peach cultivars. γ-Decalactone, the primary component of peach aroma, is difficult to detect in low-aroma peach fruits, possibly because the mutant PpAAT1s produce so little of it. Because other esters also affect peach fruit aroma (Eduardo et al., 2010), we investigated other esterification activities of the PpAAT1s and found that type II PpAAT1 produced significantly greater amounts of other esters as well. This indicates that PpAAT1 is capable of catalyzing the synthesis of a broad range of esters, like the AATs found in other fruits (Aharoni et al., 2000; Souleyre et al., 2005; Schwab et al., 2008; Morales-Quintana et al., 2012; Galaz et al., 2013). This may explain why the content of aroma volatiles in general was far greater in the high-aroma peach cultivars. For an in vivo loss-of-function test, we used the CRISPR/Cas9 system to knock out fhPpAAT1 in the high-aroma cv Fenghuayulu, confirming that substitutions in PpAAT1 have led to loss of γ-decalactone in some low-aroma cultivars. Overexpression of fhPpAAT1 in the low-aroma cv Achutao fruits validated the role of PpAAT1 in γ-decalactone production, as the levels of γ-decalactone were higher in transgenic fruits compared with nontransgenic controls. These in vivo observations are in agreement with the catalytic capabilities of PpAAT1 determined in vitro.
In this work, we found that the last step in the biosynthesis of the peach aroma compound γ-decalactone is compromised in some low-aroma cultivars and identified and functionally characterized PpAAT1 as catalyzing the internal esterification in the putative γ-decalactone biosynthesis pathway. To fully elucidate the γ-decalactone biosynthesis pathway, we will continue to investigate other genes in the pathway.
CONCLUSION
In peach fruits, γ-decalactone is the most important chemical contributor to aroma. We conducted transcriptomic and metabolomic comparisons between a high-aroma and a low-aroma peach cultivar. Sequence substitutions in PpAAT1 in low-aroma cultivars were found to be responsible for their undetectable levels of γ-decalactone. The results of our functional tests of PpAAT1 and loss- and gain-of-functions studies both in vitro and in vivo provide a foundation for improving peach cultivars through alterations in the content of the peach-like aroma compound γ-decalactone.
MATERIALS AND METHODS
Plant Material
Peach fruits (Prunus persica, ‘Fenghuayulu’ and ‘Achutao’) were harvested from the National Peach Germplasm Repository (Nanjing, China). Other peach cultivars used in this study were also collected from National Peach Germplasm Repository.
The development of peach fruits can be divided into four distinct maturity stages (S1–S4), with γ-decalactone usually becoming detectable during S4 (Lombardo et al., 2011). Peach fruit maturity was determined based on the IAD, which measures the loss of chlorophyll as fruits ripen by comparing absorbances at 670 and 720 nm (Zhang et al., 2017a). Cultivator Fenghuyulu fruits were sampled at time points corresponding to the final two maturity stages (S3 and S4) at 114 (IAD = 1.3), 116 (IAD = 1.2), 118 (IAD = 1.1), 120 (IAD = 0.9), and 122 (IAD = 0.7) DAFB (S3) and at 124 (IAD = 0.5), 126 (IAD = 0.4), 128 (IAD = 0.3), 130 (IAD = 0.2), and 132 (IAD = 0.1) DAFB (S4). The best harvest time for cv Fenghuyulu was found to be 132 DAFB. The sampling time points for cv Achutao were selected to correspond to the same fruit maturity point as used for cv Fenghuayulu based on IAD. Therefore, cv Achutao was sampled at 102, 104, 106, 108, and 110 DAFB (S3) and at 112, 114, 116, 118, and 120 DAFB (S4). At each sampling point, three biological replicates, with six fruits in each, were sampled. Following this, the samples were mechanically peeled, cored, sliced, frozen in liquid nitrogen, and stored at −80°C for use in further experiments.
Analysis of γ-Decalactone Content
The γ-decalactone content in the peach fruits of the two cultivars harvested at each sampling point was measured as previously described (Eduardo et al., 2010; Xi et al., 2012; Zhang et al., 2017b). The analysis was performed on a 7890 A Agilent GC device equipped with a DB-WAX column (0.32 mm, 30 m, 0.25 μm; J&W Scientific). Frozen mesocarp tissues were ground to a powder in liquid nitrogen. A 600-mg sample of frozen tissue powder from each replicate was sealed in a 15-mL vial. Then, the volatile compounds were extracted by adsorption to a fiber coated with 65 μm of polydimethylsiloxane and divinylbenzene (Supelco). Conditions for the GC analysis were as follows: injector 230°C, initial oven temperature 40°C held for 2 min, increased by 5°C min−1 to 240°C, and then held for 2 min. Nitrogen was used as a carrier gas at 1 mL min−1. A commercial γ-decalactone standard (Sigma-Aldrich) was used to confirm the compound annotation. A standard curve for γ-decalactone was generated by analyzing a standard concentration series ranging from 2 μg mL−1 to 5 mg mL−1.
Metabolome Analysis
In line with the foregoing results, samples from the two cultivars were taken at the stages at which γ-decalactone was present and then subjected to metabolome analysis. In order to obtain as many intermediates in the γ-decalactone biosynthesis pathway as possible, GC-MS (Agilent 7890 A gas chromatograph coupled with a 5975 C mass spectrometer detector; Agilent Technologies) and UPLC-MS (2777C UPLC system coupled with a Xevo G2-XS QTOF mass spectrometer detector; Waters) were used to perform untargeted metabolomic analyses.
The GC-MS analysis was performed essentially as described by Sánchez et al. (2012) with some modifications. For each sample, 20 μL of 2-octanol (20 mg mL−1; Sigma-Aldrich) was added as an internal standard for GC-MS analysis, and GC was conducted using the same capillary column and chromatographic conditions as for the previous GC analysis. The mass spectra conditions were as follows: ionizing electron energy 70 eV, transfer line temperature 280°C, electron-impact ion source temperature 230°C, and scan range mass-to-charge ratio 50 to 550. The GC-MS data were acquired using MSD ChemStation software (Agilent Technologies) in the NIST 11 mass spectral library. The quantification analysis was conducted using the internal standard curves method described by Eduardo et al. (2010) combined with the quality control–based robust LOESS (locally estimated scatterplot smoothing) signal correction (QC-RLSC protocol; Dunn et al., 2011).
The UPLC-MS analyses were conducted according to Arbona et al. (2009) with some modifications. A 180-mg sample of frozen powdered fruit tissue, 1,000 μL of cold methanol:water (1:1, v/v), and 100 μm ricinolic acid (internal standard) were mixed per replicate. Then, the samples were centrifuged at 12,000g for 30 min at 4°C and the supernatants were transferred to new tubes. The injection volume for each sample was 10 μL. Each sample was analyzed three times so that a higher level of precision could be achieved. An ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm, 1.7 μm; Waters) was used for the reverse phase separation. The mobile phase consisted of 0.1% (v/v) formic acid in water (solvent A) and 0.1% (v/v) formic acid in acetonitrile (solvent B). A constant flow rate of 0.4 mL min−1 was used with the following gradient elution conditions: initially 100% A, held for 0 to 2 min; then 2 to 9 min 0% B to 100% B; and then 100% B, held for 9 to 11 min. A high-resolution tandem mass spectrometer (Xevo G2-XS QTOF; Waters) was used to detect the metabolites eluted from the column. The QTOF device was operated in both positive and negative ion modes. For the MS/MS detection, all precursors were fragmented using 20 to 40 eV, and the scan time was 0.2 s. The MS data were analyzed using the Progenesis QI program (version 2.2; Waters). The KEGG database was used to align the molecular mass data (mass-to-charge ratio) to annotate metabolites. The quantitative analyses used the internal standard method (Sadre et al., 2016) combined with the QC-RLSC protocol (Dunn et al., 2011). The qualitative and quantitative determination of 4-hydroxydecanoyl-CoA (Accela ChemBio) was carried out using the standard sample and standard curve in LC analysis by Shanghai Accela ChemBio.
Pearson correlation coefficients between metabolites were calculated using the programming language R, and the metabolites that met the criterion (r > 0.6) for γ-decalactone were located in the KEGG pathway with Paintomics 3.0 (García-Alcalde et al., 2011).
Transcriptome Analysis
We performed RNA-seq analysis of the two cultivars using samples from the same time points as in the metabolome analysis. Total RNA was extracted from frozen tissue powder using a TaKaRa MiniBEST Plant RNA Extraction Kit and treated with RNase-free DNase I (Takara). RNA quantity and quality were evaluated using a NanoPhotometer spectrophotometer (Implen) and an RNA Nano 6000 Assay Kit, which is part of the Bioanalyser 2100 system (Agilent Technologies). RNA-seq was performed at the Beijing Genomics Institute on an Illumina HiSeq 4000 platform, and 150-bp single-end reads were generated. The clean reads were obtained after filtering out the reads with low quality, containing adaptors, or having a high content of unknown bases (N). HISAT (version v0.1.6-beta) was used to map the clean reads to the reference genome, which was obtained from the Genome Database for the Rosaceae (http://www.rosaceae.org/peach/genome). Gene expression levels were calculated by RSEM (version v1.2.12). Then, DEseq2 and PossionDis were used to detect DEGs from the different samples with the default parameters. The DEGs were located in the KEGG pathways using Paintomics 3.0 (García-Alcalde et al., 2011).
Combined Transcriptome and Metabolome Analysis
To identify the possible intermediates and candidate genes involved in the putative γ-decalactone biosynthesis pathway, Pearson correlation coefficients between candidate compounds and genes, specified in the KEGG pathway for γ-decalactone formation, were calculated using R.
RT-qPCR
RT-qPCR was performed on an ABI-7500 system (Applied Biosystems) using SYBR Green qPCR Master Mix (Takara). The first-strand cDNA was synthesized using a PrimeScript II First Strand cDNA Synthesis Kit (Takara) according to the manufacturer’s instructions. All cDNA samples were stored at –20°C and diluted 10-fold with RNase-free water before being used as a template for the RT-qPCR and further analyses. The primers used for the RT-qPCR are listed in Supplemental Table S5. The thermal cycling conditions were according to Zhang et al. (2012).
Sequence Analysis and Gene Isolation of PpAAT1
Gene-specific primers for the full-length PpAAT1 were designed using its homologous gene sequence in the Genome Database for the Rosaceae (Supplemental Table S8). The PpAAT1 genes were amplified using Mastercycler pro (Eppendorf), and the cDNAs were used as the templates. The PCR products were cloned into PMD 18-T (Takara) and sequenced by Tsingke. The amino acid sequences produced were translated using MEGA 6.06 (Tamura et al., 2013). Then, the amino acid sequences for PpAAT1, along with other known AATs, were phylogenetically analyzed using the maximum likelihood method with 1,000 bootstrap replicates using MEGA 6.06. The 3D models of PpAAT1 proteins were built by homology modeling. The crystal structures were retrieved from the Protein Data Bank with the National Center for Biotechnology Information’s Protein BLAST tool. The crystal structure of an Arabidopsis (Arabidopsis thaliana) acyltransferase protein (Protein Data Bank no. 5KJT) was selected as the template for PpAAT1 based on the high-resolution crystal structures of homologous proteins. Homology modeling of the PpAAT1 proteins was carried out using modeler v9.19 (http://salilab.org/modeller/). Further molecular mechanics optimization was carried out in order to obtain the reasonable 3D structural model resulting from modeler. The optimized protein model was evaluated using the PROCHECK program (Laskowski et al., 1993). Docking studies were performed using AutoDock 4.2.6 (Morris et al., 2009) and optimized using the MOPAC program (Stewart, 1990) to predict the putative binding of 4-hydroxydecanoyl-CoA.
Recombinant Protein Preparation
The full-length cDNAs of the different PpAAT1 variants were cloned into the pET6xHN Expression Vector (Takara) according to the supplier’s recommendations. The expression and purification of recombinant proteins from Escherichia coli were performed using the pET Express & Purify Kits (Takara) according to the manufacturer’s instructions. Confirmation of the identity of the recombinant proteins was carried out using LC-MS/MS by Shanghai Applied Protein Technology. Concentrations of PpAAT1 proteins were determined by densitometry using BSA as a standard.
PpAAT1 Enzyme Assay
The specific activities of PpAAT1s for γ-decalactone formation were assayed using 4-hydroxydecanoyl-CoA (Accela ChemBio) as a substrate. Preliminary assays were conducted to determine the optimum pH and temperature for the enzyme activity. In vitro PpAAT1 enzyme assays were conducted in a total reaction volume of 1 mL with 50 mm Tris-HCl (pH 8.2), 5.1 μg of protein, and 10 mm hydroxydecanoyl-CoA. Reactions were conducted at 31°C for 40 min under agitation, after which the product was collected and analyzed by GC-MS as described by Zhang et al. (2017b). All the assays were performed in sextuplicate. Control assays using boiled (nonfunctional) protein were also performed. To characterize the esterification activity of the PpAAT1s, propanol, hexanol, benzyl alcohol, decanol, and acetyl-CoA (Sigma-Aldrich) were used as substrates to produce esters found in fruits. Reactions were conducted as described by Goulet et al. (2015). Origin 2019 (www.originlab.com) software was used to calculate the Km and Kcat values.
Transient Expression in Nicotiana benthamiana
The PpAAT1 cDNA clone was transferred into the pMDC32-HPB (Addgene no. 32078) plasmid vector using the Gateway LR Clonase II Enzyme mix (Thermo Fisher Scientific) and transformed into Agrobacterium tumefaciens LBA4404 Electro-Cells (Takara) according to the manufacturer’s instructions. The tomato bushy stunt virus P19 suppressor gene (Qu and Morris, 2002) was prepared for coinfiltration to prevent posttranscriptional gene silencing by small interfering RNAs. A. tumefaciens strain LBA4404 infiltration (agroinfiltration) was performed as described by Souleyre et al. (2005) with minor modifications. Briefly, leaves from 4-week-old N. benthamiana plants were agroinfiltrated with the A. tumefaciens strain harboring the PpAAT1 vector in combination with the strain harboring the P19 vector at a ratio of 2:1, respectively. For negative controls, strains for the expression of PpAAT1 were replaced with a GFP-expressing strain. After 7 d, leaves were infiltrated again with both vectors together with 4-hydroxydecanoyl-CoA (10 mm). After 10 h, the infiltrated leaves were harvested for GC-MS analysis to quantify γ-decalactone produced, as described earlier. Leaves expressing GFP were also viewed under UV light to confirm successful agroinfiltration and gene expression.
Heterologous Expression in Oleaginous Yeast
The oleaginous yeast (Yarrowia lipolytica) mutant Po1g strain (Leu–, ΔAEP, ΔAXP, Suc+ pBR platform) and the pINA1296 expression vector, which were kindly provided by Catherine Madzak, were used for heterologous expression of PpAAT1. The construction steps were performed as follows. First, five variants of PpAAT1 were isolated and were each flanked with the SfiI/KpnI (Supplemental Table S8) restriction sites. Then, the SfiI and KpnI restriction enzymes (New England Biolabs) were used to digest both the PCR fragments and the pINA1296 plasmid. Next, the modified PCR fragments and the linear plasmid were mixed with T4 DNA ligase (New England Biolabs). Then, the constructs were amplified in E. coli DH5α Electro-Cells (Takara) and extracted with a TIANprep Mini Plasmid Kit (Tiangen). The cloning plasmids were digested with NotI restriction enzyme (New England Biolabs) according to the procedure of Blazeck et al. (2014).
Transformants were selected in Leu-deficient medium containing yeast nitrogen base. Then, Po1g strain transformants were grown in yeast peptone dextrose medium at 28°C overnight. In order to prevent the oleaginous yeast from consuming the γ-decalactone produced, we use the method to produce γ-decalactone as described by Braga and Belo (2013). The γ-decalactone was extracted and detected as described by Braga and Belo (2013), and each construct was tested using eight Po1g transformants.
Mutagenesis of PpAAT1 in Peach Fruit
The fhPpAAT1 gene was selected as the target for the CRISPR/Cas9 system. The potential Cas9 target sites in fhPpAAT1 were identified using CRISPRdirect (http://crispr.dbcls.jp/). Four of the output target sites for the sgRNA sequences (Supplemental Table S9), including one in region I, one in region II, and two in region III, were selected based on their locations in the gene and off-target likelihoods. The four gRNA cassettes, driven by LacZ-AtU3b, AtU3d, AtU6-1, and AtU6-29, respectively, were combined with a Gibson Assembly Cloning Kit (New England Biolabs) to form a multiplex genome-targeting vector system. Then, this multiplex targeting vector was inserted into the pYLCRISPR/Cas9Pubi-B plasmid. Finally, a CRISPR/Cas9 construct harboring the four sgRNA cassettes was generated (Supplemental Fig. S2). All of the sgRNA and binary CRISPR/Cas9 vectors were kindly provided by Yao-Guang Liu.
CRISPR/Cas9 constructs harboring either the four sgRNA cassettes or the empty vector were transferred into A. tumefaciens LBA4404 Electro-Cells (Takara). The transformation of A. tumefaciens was performed according to the manufacturer’s instructions. A. tumefaciens cells containing the construct were grown overnight to an OD600 of 0.6 and then resuspended at a final OD600 of 0.8 in liquid MS medium containing 2% (w/v) Suc. Two hundred healthy ‘Fenghuayulu’ peach fruits were randomly selected and injected with the A. tumefaciens construct, and another 200 healthy fruits were injected with A. tumefaciens containing the empty vector during their early development stages before γ-decalactone was present. The A. tumefaciens culture suspended in infiltration buffer was infiltrated into the whole fruit using a syringe. The fruits were then left on the trees to grow. All peach fruits were harvested at the best harvesting time and were matched to ensure similar ripeness based on the IAD (Zhang et al., 2017a). When the transgenic fruits had reached the best harvesting time, the γ-decalactone content was measured as described above (three different parts of each fruit were selected for analysis). The genomic DNA was extracted from the transgenic fruits with a TaKaRa MiniBEST Plant Genomic DNA kit (Takara) so that the fhPpAAT1 mutant gene in the transgenic peach fruits could be analyzed. The target DNA fragments were amplified by PCR, and the PCR products were sequenced by Tsingke.
Overexpression Assays in Non-γ-Decalactone Fruits
The full-length CDS of fhPpAAT1 (a type II PpAAT1) was amplified using cDNA from cv Fenghuayulu fruit as described earlier. After sequencing confirmed the correct sequence, this CDS was ligated into the pMDC32-HPB vector as described earlier.
The overexpression vector and empty vector were transferred into A. tumefaciens LBA4404 Electro-Cells. The growing conditions for A. tumefaciens containing the overexpression vector were the same as above, and an A. tumefaciens suspension with an OD600 of 0.8 was injected into ‘Achutao’ fruits at 116 DAFB. The A. tumefaciens culture with an OD600 of 0.8 suspended in infiltration buffer (1:1) was infiltrated into the whole fruit using a syringe. When the transgenic fruits reached the best harvesting time (evaluated by IAD), the γ-decalactone content was measured. TaqMan MGB assays were also conducted to quantify fhPpAAT1 expression in ‘Achutao’ fruits. Primers (forward, 5′-GGCGAAGCCAACGCCTAT-3′; reverse, 5′-GGCCGTCCTGGTCGTCTAT-3′) and a probe that was specific for fhPpAAT1 (5′-FAM-AAACCAAGCAGCTGTC-MGB-3′; the specific site for fhPpAAT1 is underlined) were designed using Primer Express 3.0 software (Applied Biosystems). The TaqMan MGB RT-qPCR was performed using a One Step PrimeScript RT-PCR Kit (Takara), and the protocol followed the manufacturer’s instructions with minor exceptions, which were that each reaction (20 μL) contained 100 ng of RNA, 0.2 μm primers, and 0.3 μm probes. The thermal cycling conditions followed the manufacturer’s instructions.
Statistical Analyses
Pearson correlation coefficients between metabolites were calculated using the programming language R. Significant differences were statistically analyzed by IBM SPSS Statistics 23. The comparison analysis was estimated by Fisher’s lsd test (P ≤ 0.05). Origin 2019 (www.originlab.com) software was used to calculate the Km and Kcat values. Histograms were prepared with Origin 2019.
Accession Numbers
RNA-seq raw sequence data generated from this study can be found in the National Center for Biotechnology Information Short Read Archive database under the accession number SRP156184. The sequences of fhPpAAT1 and acPpAAT1 can be found in the National Center for Biotechnology Information database under the following numbers (MH700174 and MH700175).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of PpAAT1 and other plant AATs.
Supplemental Figure S2. Schematic of target site selection in the fhPpAAT1 gene and the construction of the Cas9/sgRNA expression vector.
Supplemental Table S1. Metabolic compounds detected by UPLC-MS.
Supplemental Table S2. Metabolic compounds detected by GC-MS.
Supplemental Table S3. Pearson correlation coefficient among metabolites.
Supplemental Table S4. KEGG pathway analysis of the selected compounds.
Supplemental Table S5. DEGs used for RT-qPCR and the validation results with RNA-seq data.
Supplemental Table S6. KEGG pathway analysis of the selected unigenes.
Supplemental Table S7. Pearson correlation analysis between candidate genes and intermediates.
Supplemental Table S8. Primer pairs for CDS amplification and heterologous expression in oleaginous yeast.
Supplemental Table S9. Primers used for CRISPR/Cas9.
Acknowledgments
We thank Catherine Madzak (Centre de Biotechnologies Agro-Industrielles, Institut National Agronomique Paris) for providing the oleaginous yeast expression system: mutant Po1g strain (Leu–, ΔAEP, ΔAXP, Suc+ pBR platform) and the pINA1296 expression vector, and for technical assistance. We thank Yao-Guang Liu (South China Agriculture University) for providing the sgRNA expression cassettes, the binary CRISPR/Cas9 vector containing LacZ-AtU3b, AtU3d, AtU6-1, and AtU6-29, the pYLCRISPR/Cas9Pubi-B plasmid, and for technical assistance. We thank the Paintomics Team for providing the bioinformatics resources for peach in the Paintomics 3.0 data sets and for technical assistance. We thank Xingxu Huang and Dr. Jiankui Zhou (School of Life Science, Technology, Shanghai Tech University) for support with CRISPR.
Footnotes
This work was supported by the China Agriculture Research System (CARS-30) and by the Jiangsu Agriculture Science and Technology Innovation Fund (grant no. CX-15-1020).
References
- Aragüez I, Valpuesta V (2013) Metabolic engineering of aroma components in fruits. Biotechnol J 8: 1144–1158 [DOI] [PubMed] [Google Scholar]
- Arbona V, Iglesias DJ, Talón M, Gómez-Cadenas A (2009) Plant phenotype demarcation using nontargeted LC-MS and GC-MS metabolite profiling. J Agric Food Chem 57: 7338–7347 [DOI] [PubMed] [Google Scholar]
- Aharoni A, Keizer LCP, Bouwmeester HJ, Sun Z, Alvarez-Huerta M, Verhoeven HA, Blaas J, van Houwelingen AMML, De Vos RCH, van der Voet H, et al. (2000) Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell 12: 647–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbontín C, Gaete-Eastman C, Fuentes L, Figueroa CR, Herrera R, Manriquez D, Latché A, Pech JC, Moya-León MA (2010) VpAAT1, a gene encoding an alcohol acyltransferase, is involved in ester biosynthesis during ripening of mountain papaya fruit. J Agric Food Chem 58: 5114–5121 [DOI] [PubMed] [Google Scholar]
- Ball RD, Murray SH, Young H, Gilbert JM (1998) Statistical analysis relating analytical and consumer panel assessments of kiwifruit flavour compounds in a model juice base. Food Qual Prefer 9: 255–266 [Google Scholar]
- Beekwilder J, Alvarez-Huerta M, Neef E, Verstappen FW, Bouwmeester HJ, Aharoni A (2004) Functional characterization of enzymes forming volatile esters from strawberry and banana. Plant Physiol 135: 1865–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billard A, Laval V, Fillinger S, Leroux P, Lachaise H, Beffa R, Debieu D (2012) The allele-specific probe and primer amplification assay, a new real-time PCR method for fine quantification of single-nucleotide polymorphisms in pooled DNA. Appl Environ Microbiol 78: 1063–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazeck J, Hill A, Liu L, Knight R, Miller J, Pan A, Otoupal P, Alper HS (2014) Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat Commun 5: 3131. [DOI] [PubMed] [Google Scholar]
- Braga A, Belo I (2013) Immobilization of Yarrowia lipolytica for aroma production from castor oil. Appl Biochem Biotechnol 169: 2202–2211 [DOI] [PubMed] [Google Scholar]
- Braga A, Belo I (2016) Biotechnological production of γ-decalactone, a peach like aroma, by Yarrowia lipolytica. World J Microbiol Biotechnol 32: 169. [DOI] [PubMed] [Google Scholar]
- Buttery RG, Teranishi R, Ling LC, Turnbaugh JG (1990) Quantitative and sensory studies on tomato paste volatiles. J Agric Food Chem 38: 336–340 [Google Scholar]
- Cao K, Zheng Z, Wang L, Liu X, Zhu G, Fang W, Cheng S, Zeng P, Chen C, Wang X, et al. (2014) Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol 15: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollini ML. (2000) Secondary metabolites of vertebrate-dispersed fruits: Evidence for adaptive functions. Rev Chil Hist Nat 73: 421–440 [Google Scholar]
- D’Auria JC. (2006) Acyltransferases in plants: A good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Do JY, Salunkhe DK, Olson LE (1969) Isolation, identification and comparison of the volatiles of peach fruit as related to harvest maturity and artificial ripening. J Food Sci 34: 618–621 [Google Scholar]
- Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, et al. (2011) Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc 6: 1060–1083 [DOI] [PubMed] [Google Scholar]
- Eduardo I, Chietera G, Bassi D, Rossini L, Vecchietti A (2010) Identification of key odor volatile compounds in the essential oil of nine peach accessions. J Sci Food Agric 90: 1146–1154 [DOI] [PubMed] [Google Scholar]
- Galaz S, Morales-Quintana L, Moya-León MA, Herrera R (2013) Structural analysis of the alcohol acyltransferase protein family from Cucumis melo shows that enzyme activity depends on an essential solvent channel. FEBS J 280: 1344–1357 [DOI] [PubMed] [Google Scholar]
- García-Alcalde F, García-López F, Dopazo J, Conesa A (2011) Paintomics: A web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics 27: 137–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JM, Young H, Ball RD, Murray SH (1996) Volatile flavor compounds affecting consumer acceptability of kiwifruit. J Sens Stud 11: 247–259 [Google Scholar]
- Goff SA, Klee HJ (2006) Plant volatile compounds: Sensory cues for health and nutritional value? Science 311: 815–819 [DOI] [PubMed] [Google Scholar]
- Goulet C, Kamiyoshihara Y, Lam NB, Richard T, Taylor MG, Tieman DM, Klee HJ (2015) Divergence in the enzymatic activities of a tomato and Solanum pennellii alcohol acyltransferase impacts fruit volatile ester composition. Mol Plant 8: 153–162 [DOI] [PubMed] [Google Scholar]
- Gutterson NC. (1993) Molecular breeding for color, flavor and fragrance. Sci Hortic (Amsterdam) 55: 141–160 [Google Scholar]
- Hodgkison R, Ayasse M, Häberlein C, Schulz S, Zubaid A, Mustapha WAW, Kunz TH, Kalko EKV, Hare D (2013) Fruit bats and bat fruits: The evolution of fruit scent in relation to the foraging behaviour of bats in the New and Old World tropics. Funct Ecol 27: 1075–1084 [Google Scholar]
- Horvat RJ, Chapman GW, Robertson JA, Meredith FI, Scorza R, Callahan AM, Morgens P (1990) Comparison of the volatile compounds from several commercial peach cultivars. J Agric Food Chem 38: 234–237 [Google Scholar]
- Jennings WG, Sevenants MR (1964) Volatile components of peach. J Food Sci 29: 796–801 [Google Scholar]
- Jia H, Okamoto G (2001) Distribution of volatile compounds in peach fruit. J Jpn Soc Hortic Sci 70: 223–225 [Google Scholar]
- Larsen M, Poll L (1992) Odour thresholds of some important aroma compounds in strawberries. Z Lebensm Unters Forsch 195: 120–123 [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst 26: 283–291 [Google Scholar]
- Lauxmann MA, Borsani J, Osorio S, Lombardo VA, Budde CO, Bustamante CA, Monti LL, Andreo CS, Fernie AR, Drincovich MF, et al. (2014) Deciphering the metabolic pathways influencing heat and cold responses during post-harvest physiology of peach fruit. Plant Cell Environ 37: 601–616 [DOI] [PubMed] [Google Scholar]
- Li XW, Jiang J, Zhang LP, Yu Y, Ye ZW, Wang XM, Zhou JJ, Chai MI, Zhang HQ, Arús P, et al. (2015) Identification of volatile and softening-related genes using digital gene expression profiles in melting peach. Tree Genet Genomes 11: 71 [Google Scholar]
- Lomáscolo SB, Levey DJ, Kimball RT, Bolker BM, Alborn HT (2010) Dispersers shape fruit diversity in Ficus (Moraceae). Proc Natl Acad Sci USA 107: 14668–14672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo VA, Osorio S, Borsani J, Lauxmann MA, Bustamante CA, Budde CO, Andreo CS, Lara MV, Fernie AR, Drincovich MF (2011) Metabolic profiling during peach fruit development and ripening reveals the metabolic networks that underpin each developmental stage. Plant Physiol 157: 1696–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Quintana L, Moya-León MA, Herrera R (2012) Molecular docking simulation analysis of alcohol acyltransferases from two related fruit species explains their different substrate selectivities. Mol Simul 38: 912–921 [Google Scholar]
- Morales-Quintana L, Nuñez-Tobar MX, Moya-León MA, Herrera R (2013) Molecular dynamics simulation and site-directed mutagenesis of alcohol acyltransferase: A proposed mechanism of catalysis. J Chem Inf Model 53: 2689–2700 [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo O, Heymann EW, Schulz S, Ayasse M (2016) Fruit odor as a ripeness signal for seed-dispersing primates? A case study on four neotropical plant species. J Chem Ecol 42: 323–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Morris TJ (2002) Efficient infection of Nicotiana benthamiana by tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol Plant Microbe In 15: 193–202 [DOI] [PubMed] [Google Scholar]
- Rodríguez A, Alquézar B, Peña L (2013) Fruit aromas in mature fleshy fruits as signals of readiness for predation and seed dispersal. New Phytol 197: 36–48 [DOI] [PubMed] [Google Scholar]
- Sadre R, Magallanes-Lundback M, Pradhan S, Salim V, Mesberg A, Jones AD, DellaPenna D (2016) Metabolite diversity in alkaloid biosynthesis: A multilane (diastereomer) highway for camptothecin synthesis in Camptotheca acuminata. Plant Cell 28: 1926–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez G, Besada C, Badenes ML, Monforte AJ, Granell A (2012) A non-targeted approach unravels the volatile network in peach fruit. PLoS ONE 7: e38992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez G, Venegas-Calerón M, Salas JJ, Monforte A, Badenes ML, Granell A (2013) An integrative “omics” approach identifies new candidate genes to impact aroma volatiles in peach fruit. BMC Genomics 14: 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttler M, Boland W (1996) Biosynthesis of dodecano-4-lactone in ripening fruits: Crucial role of an epoxide-hydrolase in enantioselective generation of aroma components of the nectarine (Prunus persica var. nucipersica) and the strawberry (Fragaria ananassa). Helv Chim Acta 79: 1488–1496 [Google Scholar]
- Schwab W, Davidovich-Rikanati R, Lewinsohn E (2008) Biosynthesis of plant-derived flavor compounds. Plant J 54: 712–732 [DOI] [PubMed] [Google Scholar]
- Sevenants MR, Jennings WG (1966) Volatile components of peach. II. J Food Sci 31: 81–86 [Google Scholar]
- Shen ZJ, Ma RJ, Yu ML, Cai ZX, Xu JL (2013) Establishment of peach primary core collection based on accessions conserved in National Fruit Germplasm Repository of Nanjing. Yuan Yi Xue Bao 40: 125–134 (in Chinese) [Google Scholar]
- Song C, Hong X, Zhao S, Liu J, Schulenburg K, Huang FC, Franz-Oberdorf K, Schwab W (2016) Glucosylation of 4-hydroxy-2,5-dimethyl-3(2H)-furanone, the key strawberry flavor compound in strawberry fruit. Plant Physiol 171: 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souleyre EJ, Chagné D, Chen X, Tomes S, Turner RM, Wang MY, Maddumage R, Hunt MB, Winz RA, Wiedow C, et al. (2014) The AAT1 locus is critical for the biosynthesis of esters contributing to ‘ripe apple’ flavour in ‘Royal Gala’ and ‘Granny Smith’ apples. Plant J 78: 903–915 [DOI] [PubMed] [Google Scholar]
- Souleyre EJ, Greenwood DR, Friel EN, Karunairetnam S, Newcomb RD (2005) An alcohol acyl transferase from apple (cv. Royal Gala), MpAAT1, produces esters involved in apple fruit flavor. FEBS J 272: 3132–3144 [DOI] [PubMed] [Google Scholar]
- Souleyre EJ, Günther CS, Wang MY, Newcomb RD, Marsh KB (2011) Ester biosynthesis in kiwifruit: From genes to enzymes to pathways. Acta Hortic 913: 205–211 [Google Scholar]
- Spencer MD, Pangborn RM, Jennings WG (1978) Gas chromatographic and sensory analysis of volatiles from cling peaches. J Agric Food Chem 26: 725–732 [Google Scholar]
- Steingass CB, Dell C, Lieb V, Mayer-Ullmann B, Czerny M, Carle R (2015) Assignment of distinctive volatiles, descriptive sensory analysis and consumer preference of differently ripened and post-harvest handled pineapple (Ananas comosus [L.] Merr.) fruits. Eur Food Res Technol 242: 33–43 [Google Scholar]
- Stewart JJP. (1990) MOPAC: A semiempirical molecular orbital program. J Comput Aided Mol Des 4: 1–105 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchietti A, Lazzari B, Ortugno C, Bianchi F, Malinverni R, Caprera A, Mignani I, Pozzi C (2009) Comparative analysis of expressed sequence tags from tissues in ripening stages of peach (Prunus persica L. Batsch). Tree Genet Genomes 5: 377–391 [Google Scholar]
- Wang LR, Zhu GR (2005) Descriptors and Data Standard for Peach (Prunus persica L.). China Agriculture Press, Beijing (in Chinese) [Google Scholar]
- Xi WP, Zhang B, Shen JY, Sun CD, Xu CJ, Chen KS (2012) Intermittent warming alleviated the loss of peach fruit aroma-related esters by regulation of AAT during cold storage. Postharvest Biol Technol 74: 42–48 [Google Scholar]
- Yu ML, Ma RJ, Shen ZJ, Cai ZX (2010) Research advances in peach germplasm in China. Jiangsu J Agr Sci 26: 1418–1423 (in Chinese) [Google Scholar]
- Zhang B, Peng B, Zhang C, Song Z, Ma R (2017a) Determination of fruit maturity and its prediction model based on the pericarp index of absorbance difference (IAD) for peaches. PLoS ONE 12: e0177511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Shen JY, Wei WW, Xi WP, Xu CJ, Ferguson I, Chen K (2010) Expression of genes associated with aroma formation derived from the fatty acid pathway during peach fruit ripening. J Agric Food Chem 58: 6157–6165 [DOI] [PubMed] [Google Scholar]
- Zhang C, Shen Z, Zhang Y, Han J, Ma R, Korir NK, Yu M (2012) Cloning and expression of genes related to the sucrose-metabolizing enzymes and carbohydrate changes in peach. Acta Physiol Plant 35: 589–602 [Google Scholar]
- Zhang L, Li H, Gao L, Qi Y, Fu W, Li X, Zhou X, Gao Q, Gao Z, Jia H (2017b) Acyl-CoA oxidase 1 is involved in γ-decalactone release from peach (Prunus persica) fruit. Plant Cell Rep 36: 829–842 [DOI] [PubMed] [Google Scholar]