Abscisic acid receptors are targeted for degradation by a family of E3 ubiquitin ligases at different subcellular locations, which modulates hormone signaling in plasma membrane, cytosol, and nucleus
Abstract
The turnover of abscisic acid (ABA) signaling core components modulates the plant’s response to ABA and is regulated by ubiquitination. We show that Arabidopsis (Arabidopsis thaliana) RING Finger ABA-Related1 (RFA1) and RFA4 E3 ubiquitin ligases, members of the RING between RING fingers (RBR)-type RSL1/RFA family, are key regulators of ABA receptor stability in root and leaf tissues, targeting ABA receptors for degradation in different subcellular locations. RFA1 is localized both in the nucleus and cytosol, whereas RFA4 shows specific nuclear localization and promotes nuclear degradation of ABA receptors. Therefore, members of the RSL1/RFA family interact with ABA receptors at plasma membrane, cytosol, and nucleus, targeting them for degradation via the endosomal/vacuolar RSL1-dependent pathway or 26S proteasome. Additionally, we provide insight into the physiological function of the relatively unexplored plant RBR-type E3 ligases, and through mutagenesis and biochemical assays we identified cysteine-361 in RFA4 as the putative active site cysteine, which is a distinctive feature of RBR-type E3 ligases. Endogenous levels of PYR1 and PYL4 ABA receptors were higher in the rfa1 rfa4 double mutant than in wild-type plants. UBC26 was identified as the cognate nuclear E2 enzyme that interacts with the RFA4 E3 ligase and forms UBC26-RFA4-receptor complexes in nuclear speckles. Loss-of-function ubc26 alleles and the rfa1 rfa4 double mutant showed enhanced sensitivity to ABA and accumulation of ABA receptors compared with the wild type. Together, our results reveal a sophisticated mechanism by which ABA receptors are targeted by ubiquitin at different subcellular locations, in which the complexity of the ABA receptor family is mirrored in the partner RBR-type E3 ligases.
The plant hormone abscisic acid (ABA) regulates many key processes in plants, including seed germination and development and various biotic and abiotic stress responses (Cutler et al., 2010; Finkelstein, 2013; Rodriguez et al., 2019). The ABA signaling pathway is initiated by ABA perception through the 14-member PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) family of proteins (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009b; Nishimura et al., 2010). Specific functions for some members of the PYR/PYL/RCAR family have emerged in recent years (Antoni et al., 2013; Zhao et al., 2016; Belda-Palazon et al., 2018; Dittrich et al., 2019). ABA perception by ABA receptors leads to interaction with and inactivation of clade A protein phosphatase type 2Cs (PP2Cs), such as ABA INSENSITIVE1 (ABI1) and ABI2, HYPERSENSITIVE TO ABA1 (HAB1) and HAB2, and PP2CA/ABA-HYPERSENSITIVE GERMINATION3 (PP2CA/AHG3), which are key negative regulators of ABA signaling (Ma et al., 2009; Park et al., 2009; Rubio et al., 2009; Santiago et al., 2009b; Umezawa et al., 2009; Vlad et al., 2009).
Structural and biochemical studies have revealed that PP2Cs are necessary ABA coreceptors able to monitor the occupancy of the ABA-binding pocket to achieve nanomolar affinity for ABA binding (Ma et al., 2009; Melcher et al., 2009; Miyazono et al., 2009; Santiago et al., 2009a; Yin et al., 2009; Dupeux et al., 2011; Moreno-Alvero et al., 2017). Comparison of the structure of ligand-bound crop ABA receptors in the absence and presence of clade A PP2Cs reveals their proactive role to form productive ternary complexes in which the phosphatase activity is efficiently inhibited (Moreno-Alvero et al., 2017). Hence, clade A PP2C-mediated inhibition on three ABA-activated SNF1-related protein kinases (SnRK2s; i.e. SnRK2.2/D, SnRK2.3/I, and SnRK2.6/E/OST1) is relieved (Umezawa et al., 2009; Vlad et al., 2009). These SnRK2s subsequently activate downstream signaling by phosphorylation of numerous players, including ABA-responsive transcription factors (Fujii et al., 2009; Fujii and Zhu, 2009; Nakashima et al., 2009), the chromatin-remodeling ATPase BRAHMA (Peirats-Llobet et al., 2016), ion and water channels (Geiger et al., 2009; Lee et al., 2009; Grondin et al., 2015), and other mediators/effectors involved in ABA signaling and action (Umezawa et al., 2013; Wang et al., 2013). Whereas transcription of some PYR/PYL/RCARs is repressed in response to ABA, that of PP2Cs is stimulated (Santiago et al., 2009b; Szostkiewicz et al., 2010), indicating that a negative feedback transcriptional mechanism is present to modulate ABA signaling by controlling transcript levels of core elements. Recently, it has been discovered that degradation of PP2Cs is a complementary mechanism to PYR/PYL/RCAR-mediated inhibition of PP2C activity (Kong et al., 2015; Wu et al., 2016; Belda-Palazon et al., 2019; Julian et al., 2019).
The ubiquitin (Ub)-26S proteasome system (UPS) plays a crucial role in plant hormone signaling (Santner and Estelle, 2009; Vierstra, 2009). Approximately 6% of the Arabidopsis (Arabidopsis thaliana) genome encodes components that can be connected to the UPS, representing one of the most elaborate and prevalent regulatory mechanisms in plants (Smalle and Vierstra, 2004; Vierstra, 2009). In addition to committing proteins to degradation by the 26S proteasome, ubiquitination (ubiquitylation) also serves proteasome-independent roles, including the endocytosis of membrane proteins (MacGurn et al., 2012; Kim et al., 2013; Gao et al., 2017; Yu and Xie, 2017). The turnover of ABA receptors follows a double mechanism involving both the UPS and the cellular pathway that comprises endocytosis, trafficking through the endosomal sorting complex required for transport (ESCRT), and vacuolar degradation (Bueso et al., 2014; Irigoyen et al., 2014; Belda-Palazon et al., 2016; Yu et al., 2016; García-León et al., 2019). The relative contribution of each pathway to regulate endogenous levels of ABA receptors remains to be investigated, particularly regarding degradation of ABA receptors through the UPS.
PYR/PYL/RCAR ABA receptors are subjected to ubiquitination, either at the nucleus through the CULLIN4-RING E3 ubiquitin ligase (CRL4) complex formed by COP10-DET1-DDB1 and the substrate adapter DDB1-ASSOCIATED1 (DDA1; Irigoyen et al., 2014) or at the plasma membrane through the monomeric E3 Ub ligase RING FINGER OF SEED LONGEVITY (RSL1; Bueso et al., 2014; Belda-Palazon et al., 2016). RSL1 bears a C-terminal transmembrane (TM) domain that targets the E3 ligase to the plasma membrane, where the interaction between RSL1 and ABA receptors occurs (Bueso et al., 2014; Belda-Palazon et al., 2016). The ubiquitination of PYL4 and PYR1 by RSL1 at the plasma membrane targets the receptors to the vacuolar degradation pathway (Belda-Palazon et al., 2016; Yu et al., 2016). We found that FYVE1/FREE1, which is a key component of the ESCRT-I machinery, interacts with RSL1-receptor complexes and recruits PYL4 to endosomal compartments (Belda-Palazon et al., 2016). Recently, a moonlight function in the nucleus of FYVE1/FREE1 to attenuate ABA signaling has also been reported (Li et al., 2019). Additionally, the ESCRT-I component VPS23A, which is a Ub-conjugating enzyme variant lacking the catalytic Cys, also recognizes ABA receptors for endosomal degradation (Yu et al., 2016). Finally, both FYVE1 and VPS23A interact with the ESCRT-III protein ALIX, which in turn interacts with ABA receptors and contributes to regulation of receptor turnover and stomatal aperture (García-León et al., 2019). Thus, although the ESCRT pathway had been assumed to be reserved for integral membrane proteins, it also mediates the degradation of ABA receptors, which can be found associated with membranes through CAR proteins or RSL1 but which are not integral membrane proteins (Rodriguez et al., 2014; Diaz et al., 2016). ESCRT-mediated sorting of proteins ubiquitinated at the plasma membrane leads to degradation in the vacuole after endosomal and multivesicular body trafficking (MacGurn et al., 2012). By contrast, proteins polyubiquitinated in cytosol or nucleus are degraded via the 26S proteasome (Smalle and Vierstra, 2004).
RSL1 belongs to a gene family composed of at least nine additional members, which we have named RFA1 (for RING finger ABA-related1) to RFA9 (Bueso et al., 2014). Initially, RSL1 was annotated as a RING-type E3 ligase (Bueso et al., 2014); however, further inspection of the gene family revealed that RSL1/RFAs are structurally characterized by the presence of three putative RING domains in tandem, named as RING1-IN BETWEEN RING (IBR)-RING2, and accordingly they belong to the RBR-type E3 ligase family (Marín, 2010; Callis, 2014). Molecular insights into the function of RBR-type E3 Ub ligases have been obtained mostly in humans from the study of Parkin, associated with autosomal recessive Parkinsonism, and Human Homolog of Ariadne (HHARI), involved in the regulation of translation (Dove et al., 2016). Thus, studies in mammals have shown that RBR E3s combine properties of both RING- and HECT-type E3s (Wenzel et al., 2011; Wenzel and Klevit, 2012). Noncovalent interaction with E2-Ub occurs at the RING1 domain as in RING/U-box proteins, but then the activated Ub is transferred to a conserved Cys residue in RING2. Finally, this cysteinyl residue of RING2 transfers Ub to target proteins in a HECT-type mechanism (Callis, 2014). Structural analyses of HHARI and Parkin have revealed that RING1 is the only domain with a cross-brace zinc-coordination topology, whereas the zinc-liganding residues in IBR and RING2 domains are arranged in a sequential fashion (Duda et al., 2013; Riley et al., 2013). Therefore, the RING nomenclature for IBR and RING2 does not reflect the canonical RING cross-brace structure that is only present in RING1.
Five members of the RSL1/RFA family (i.e. RSL1 and RFA6–RFA9) contain a TM domain at the C terminus of the protein, which suggests that they are membrane localized like RSL1 (Bueso et al., 2014; Belda-Palazon et al., 2016). In contrast, RFA1 to RFA5 lack the C-terminal TM domain, and their functional characterization, as well as their subcellular localization, have not yet been investigated. In this work, we investigated RFA1 and RFA4 function in ABA signaling. We identified UBIQUITIN CONJUGATING ENZYME26 (UBC26) as the cognate E2 enzyme for RFA4 and observed the formation of UBC26-ABA receptor-RFA4 complexes in nuclear speckles. This represents an additional pathway to the DDA1-CRL4 complex to promote the degradation of ABA receptors in the nucleus (Irigoyen et al., 2014). Altogether, our results reveal a sophisticated targeting of ABA receptors at different subcellular locations, where the complexity of the ABA receptor family is mirrored in the partner RBR-type E3 ligases.
RESULTS
Subcellular Localization of RFA1 and RFA4
In this work, we focused on RFA1 and RFA4 to investigate the branch of the RFA E3 family whose members lack the C-terminal TM domain, namely RFA1 to RFA5. Overall, the mRNA levels of RFA1 and RFA4 are higher than RFA2/3 genes in different tissues and developmental stages, but they are relatively similar to RFA5 (Supplemental Fig. S1A; http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Waese et al., 2017). The heat map viewer also shows a similar profile for the overall expression of RFA1/4/5 and PYR/PYL genes (Supplemental Fig. S1A). Additionally, we used data mining to analyze the expression of RFA1 and RFA4 in seedlings subjected to different abiotic stresses (Goda et al., 2008). As a result, we found that RFA4 expression was up-regulated in aerial tissue in response to cold, osmotic, salt, drought, and heat stress, whereas RFA1 expression was less consistently affected (Supplemental Fig. S1B). In contrast, analysis of microarray data in guard cells under different treatments (ABA, high CO2, darkness, and low humidity) did not reveal significant expression changes of RFA1 and RFA4 (Gene Expression Omnibus database accession nos. GSE41054 and GSE118520; Dittrich et al., 2019). Expression of RFA4 peaked at earlier stages of silique development and embryo globular/heart/torpedo stage, whereas expression of RFA1 increased at later stages, from embryo walking-stick to cotyledon stage (Supplemental Fig. S1C). A similar double peak in the expression of some ABA receptors (e.g. PYR1, PYL4, and PYL5) could be observed during seed and silique development, which suggests some overlapping between the expression of RFA1/4 and PYL genes (Supplemental Fig. S1C).
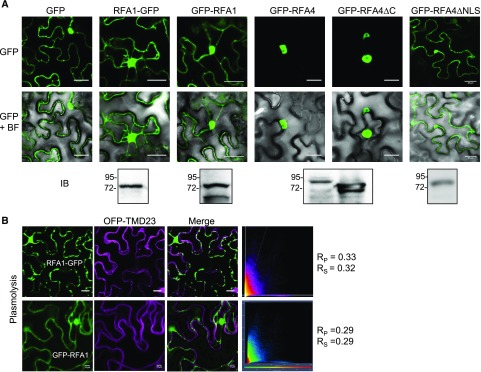
Alignment of RFA1 and RFA4 reveals the RING1-IBR-RING2 domains in tandem (Bueso et al., 2014; Supplemental Fig. S2). In the case of RFA4, after RING2, there is a very acidic C-terminal domain that contains more than 40 Asp residues (Supplemental Fig. S2). In contrast, RFA1 lacks this domain or the C-terminal TM domain of RSL1 (Supplemental Fig. S2). We generated 35S:GFP-RFA1, 35S:RFA1-GFP, 35S:GFP-RFA4, and 35S:GFP-RFA4 C-terminal deletion (RFA4ΔC, lacking amino acid residues 385–468) constructs and delivered them into leaf cells of Nicotiana benthamiana by agroinfiltration (Fig. 1A). Expression of the corresponding fusion proteins was verified by immunoblot analysis using anti-GFP antibodies (Fig. 1A). Expression of GFP-RFA4 was consistently lower than that of other GFP fusion proteins, likely because of the large acidic C-terminal domain. We found that RFA1 is apparently localized in cytosol and nucleus. The cytosolic localization of RFA1 was confirmed by the lack of colocalization with the plasma membrane marker OFP-TM23, both for RFA1-GFP and GFP-RFA1 proteins (Fig. 1B). This marker was previously used to show the plasma membrane localization of GFP-RSL1 (Bueso et al., 2014). In order to investigate whether the C-terminal acidic domain of RFA4 affects its subcellular location, we compared 35S:GFP-RFA4ΔC and 35S:GFP-RFA4 localization (Fig. 1A). Both RFA4ΔC and RFA4 were localized in the nucleus of N. benthamiana leaf cells, which suggests that the acidic domain of RFA is dispensable for nuclear localization of the protein. Indeed, a nuclear localization signal (NLS; type SV40 T antigen) composed of four basic residues (KKRR) was identified at the N terminus of RFA4 (Supplemental Fig. S2). Deletion of the NLS led to a change in RFA4 subcellular localization, and GFP-RFA4ΔNLS was apparently localized in cytosol and nucleus, similar to RFA1 (Fig. 1A).
Figure 1.
Subcellular localization of RFA1 and RFA4. A, RFA1 localizes to nucleus and cytosol, whereas RFA4 localizes to nucleus, upon transient expression in N. benthamiana leaf cells. Confocal images show transiently transformed epidermal cells expressing GFP, RFA1-GFP, GFP-RFA1, GFP-RFA4, GFP-RFA4ΔC, or GFP-RFA4ΔNLS. The merge of the GFP channel and bright-field microscopy imaging is abbreviated as GFP+BF. Immunoblotting analysis (IB) using anti-GFP was used to verify the expression of the corresponding fusion proteins. B, RFA1 does not localize to the plasma membrane. Confocal images show transiently transformed N. benthamiana leaf cells coexpressing RFA1-GFP or GFP-RFA1 and the plasma membrane marker OFP-TM23. The degree of colocalization between the two fluorescent signals was analyzed using merged images and Zeiss software (ZEN Lite 2012). N. benthamiana leaf cells were plasmolyzed using a 500 mm NaCl treatment for 30 min. Lack of colocalization with OFP-TM23 is shown in relative intensity (x and y axes) scatterplots and values of Pearson (RP) and Spearman (RS) coefficients. Bars = 30 µm.
The Interaction of RFA1, RFA4, and RSL1 with ABA Receptors Covers Different Subcellular Localizations in Plant Cells
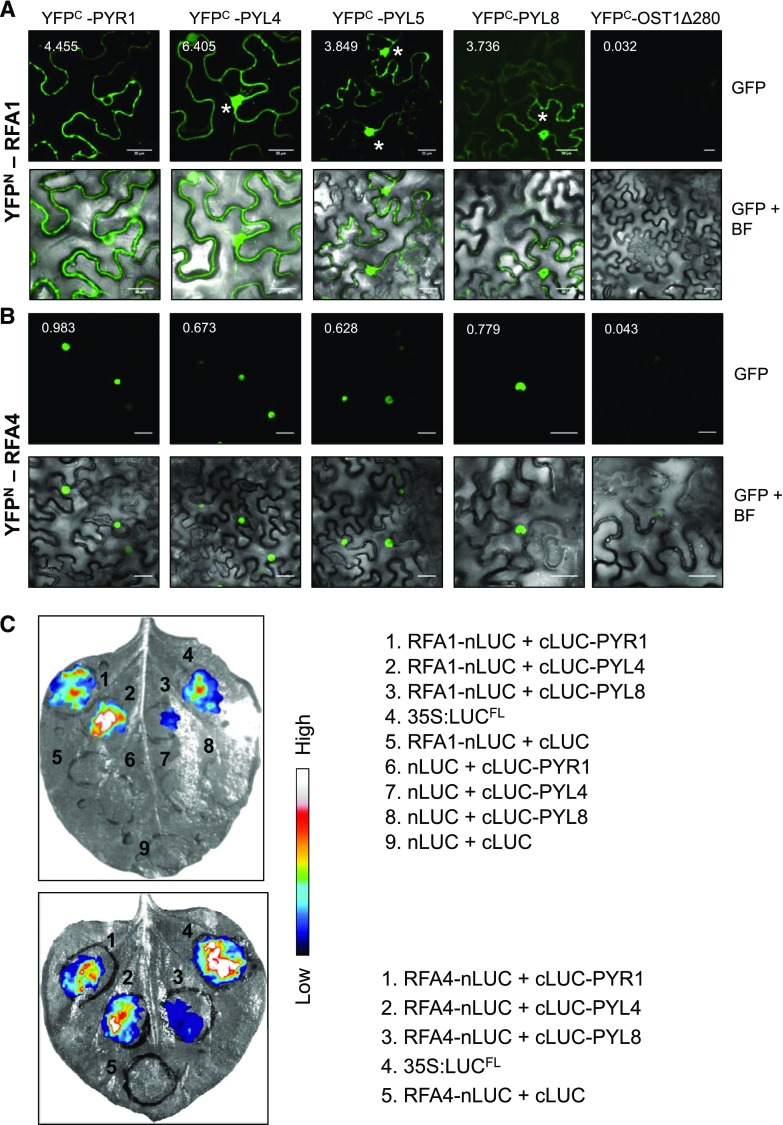
Comparison of RFA1, RFA4, and RSL1, excluding the C-terminal TM domain of RSL1 and the acidic domain of RFA4, reveals high sequence similarity (Supplemental Fig. S2), which suggests that they might all interact with ABA receptors. Only the interaction of RSL1 with ABA receptors has been analyzed previously (Bueso et al., 2014); therefore, we tested whether RFA1 and RFA4 interact with ABA receptors using bimolecular fluorescence complementation (BiFC) assays (Fig. 2). We coexpressed YFPN-RFA1 or YFPN-RFA4 with YFPC-PYR/PYLs using agroinfiltration and found that RFA1 interacts with different ABA receptors both at cytosol and nucleus of N. benthamiana leaf cells (Fig. 2A). On the other hand, the interaction of RFA4 and PYR/PYLs was localized exclusively in the nucleus, in agreement with the nuclear localization of GFP-RFA4 (Fig. 2B). Coexpression of YFPC-OST1Δ280, which is expressed in nucleus and cytosol (Vlad et al., 2009), with either YFPN-RFA1 or YFPN-RFA4 did not produce fluorescence reconstitution. We also investigated whether the very acidic C-terminal domain of RFA4 affects the interaction with ABA receptors. To this end, we tested the interaction of YFPN-RFA4ΔC with PYR/PYLs. The interaction of RFA4ΔC with PYR/PYLs did not differ from that of wild-type RFA4, which suggests that recognition of the target by RFA4 is not dependent on the C-terminal acidic domain (Supplemental Fig. S3A). We also verified that YFPN-RFA1, RFA4, and RFA4ΔC, as well as YFPC-PYR/PYLs or YFPC-OST1Δ280 proteins, were correctly expressed (Supplemental Fig. S3B). To validate the interactions observed using the BiFC assays, we performed a split-luciferase complementation assay in N. benthamiana leaves (Fig. 2C). In contrast to BiFC assays, the restoration of luciferase activity upon protein-protein interaction of the candidate proteins is reversible (Gehl et al., 2011). The coexpression of RFA1-nLUC and cLUC-PYR1, cLUC-PYL4, or cLUC-PYL8 reconstituted luciferase activity, although in the case of cLUC-PYL8 a weak signal was obtained (Fig. 2C, top). Luciferase activity was also reconstituted when RFA4-nLUC and cLUC-PYR1, cLUC-PYL4, or cLUC-PYL8 were coexpressed (Fig. 2C, bottom). As a positive control, we used the pGB0164 construct expressing full-length luciferase (Vazquez-Vilar et al., 2017), whereas expression of RFA1-nLUC or RFA4-nLUC with cLUC served as negative controls.
Figure 2.
RFA1 and RFA4 interact with ABA receptors. A, RFA1 and PYL proteins interact in nucleus (asterisks) and cytosol of N. benthamiana leaf cells. Confocal images show transiently transformed epidermal cells coexpressing YFPN-RFA1 and YFPC-PYR/PYL proteins. Interaction with PYR1 is observed in the nuclear envelope but excluded from the nucleus. The reconstituted YFP signal was quantified from 10 randomly chosen regions of interest of infiltrated leaves by ImageJ software, and the arbitrary units of fluorescence are indicated in numbers. The GFP channel shows the subcellular localization of the interaction, whereas bright-field (BF) microscopy imaging served to identify the nuclei. B, Nuclear interaction of RFA4 and ABA receptors. Confocal images show transiently transformed N. benthamiana leaf cells coexpressing YFPN-RFA4 and YFPC-PYR/PYL proteins. The reconstituted YFP signal was quantified from 10 randomly chosen regions of interest of infiltrated leaves by ImageJ software, and the arbitrary units of fluorescence are indicated in numbers. Bars = 30 µm. C, Split-luciferase complementation assay reveals interaction of RFA1 and RFA4 with PYR1, PYL4, and PYL8. The indicated construct pairs were coexpressed in N. benthamiana leaves (shown in gray) by A. tumefaciens-mediated infiltration, and 50 µm MG132 was applied into the infiltrated region 6 h before measurement of luciferase activity, which was performed 72 h after infiltration. The 35S:LUCFL construct served as a positive control. Luciferase activity was measured by applying 1 mm d-luciferin and imaging with a CCD imaging system. Luciferase signal was converted to false colors with ImageJ. The color scale represents luciferase activity. Three independent experiments were performed, and images correspond to representative leaves (n = 5 per experiment).
Since RSL1 interacts with ABA receptors at the plasma membrane (Bueso et al., 2014), the above-mentioned results indicate that other members of the RSL1/RFA family target PYR/PYLs at different subcellular locations. This was further investigated using multicolor BiFC by cloning PYR1 or PYL4 into p(MAS)-SCYCE(R), RSL1 into pDEST-SCYNE(R), and RFA4 into pDEST-VYNE(R) vectors, as described previously (Gehl et al., 2009). We coexpressed SCFPC-PYR1, SCFPN-RSL1, and VENUSN-RFA4 or SCFPC-PYL4, SCFPN-RSL1, and VENUSN-RFA4 in N. benthamiana cells. We observed the simultaneous formation of receptor-RSL1 and receptor-RFA4 complexes in plasma membrane and nucleus, respectively, of N. benthamiana cells (Supplemental Fig. S4). This suggests that the turnover of ABA receptors can be regulated differently by RSL1 and RFA4, targeting ABA receptors for degradation via the vacuolar or 26S proteasome pathway, respectively.
RFA1 and RFA4 Promote Ubiquitination of PYR1 in Vitro
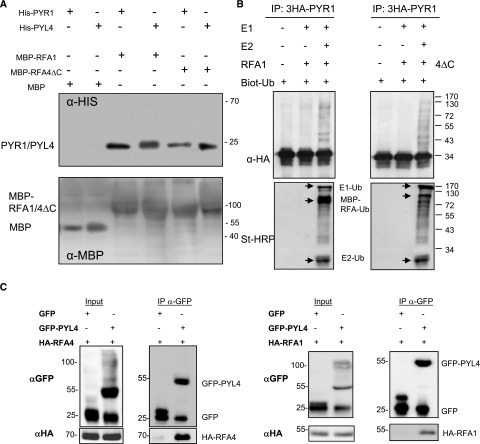
We generated maltose-binding protein (MBP)-RFA1 and MBP-RFA4ΔC fusion proteins to carry out pull-down and ubiquitination assays of ABA receptors. The C-terminal acidic domain of full-length RFA4 precluded efficient bacterial expression and purification of recombinant protein. Since RFA4ΔC could interact with ABA receptors in BiFC assays (Supplemental Fig. S3A), we used the MBP-RFA4ΔC fusion protein. Using a pull-down assay, we found that both MBP-RFA1 and MBP-RFA4ΔC could interact with PYR1 and PYL4 receptors, whereas MBP did not (Fig. 3A). In order to test whether RFA1 and RFA4ΔC catalyze the transfer of Ub to PYR1, we performed an enzymatic reaction in column using HA-PYR1 immunoprecipitated from Arabidopsis transgenic lines. The enzymatic reaction with immunoprecipitated HA-PYR1, biotinylated-Ub, ATP, human activating enzyme E1, Arabidopsis E2 UBC8, and either MBP-RFA1 or MBP-RFA4ΔC was performed as described previously (Bueso et al., 2014). We used the generic E2 UBC8 because in vitro activity was detected using this E2 together with the RBR-type enzymes RSL1 or ARI8, and UBC8 is the most commonly used E2, displaying high and promiscuous activity with different E3s (Kraft et al., 2005; Bueso et al., 2014; Kowarschik et al., 2018). The incorporation of biotinylated Ub in the reaction was monitored using streptavidin-horseradish peroxidase (St-HRP), and the incorporation of Ub into the substrate was confirmed using anti-HA antibody. RFA1 and RFA4 were able to catalyze the transfer of Ub to the PYR1 ABA receptor, as revealed using anti-HA immunoblotting (Fig. 3B). Using St-HRP, we could detect other proteins that were ubiquitinated during the reaction, which according to the expected molecular mass likely represent ubiquitinated components of the ubiquitination cascade (i.e. E1, E2, MBP-RFA1, or MBP-RFA4ΔC; Fig. 3B). In addition to in vitro biochemical assays, we performed coimmunoprecipitation experiments in plant extracts to further validate the observed interactions. To this end, we mixed Arabidopsis MG132-treated protein extracts obtained from lines expressing either HA-RFA1 or HA-RFA4 with N. benthamiana protein extracts that contain either GFP or GFP-PYL4 proteins (Fig. 3C). We immunoprecipitated GFP or GFP-PYL4 using paramagnetic beads coupled to anti-GFP antibody. Following anti-HA immunoblotting analysis, we could detect coimmunoprecipitation of HA-RFA1 or HA-RFA4 with GFP-PYL4 but not with the GFP control (Fig. 3C).
Figure 3.
RFA1 and RFA4 pull down ABA receptors and show E3 Ub ligase activity with PYR1. A, Interaction among PYR1/PYL4 with RFA1 or RFA4ΔC in a pull-down assay. Purified MBP, MBP-RFA1, or MBP-RFA4ΔC (5 μg each) and 5 μg of 6His-PYR1 or PYL4 protein were incubated for 1 h at 4°C with constant rocking in 0.5 mL of binding buffer (20 mm Tris-HCl, pH 7.5, 200 mm NaCl, 1 mm EDTA, and 0.5% [v/v] Tween 20). Next, MBP, MBP-RFA1, or MBP-RFA4DC protein was purified using amylose affinity chromatography, eluted, and analyzed by immunoblotting using anti-His and Ponceau staining. B, MBP-RFA1 or MBP-RFA4DC (2 µg) was assayed for E3 ligase activity in the presence of 100 ng of human E1, 250 ng of purified 6His-AtUBC8 (E2), 3HA-PYR1 immunoprecipitated from an Arabidopsis transgenic line, and 1 µg of biotinylated Ub. After incubation at 30°C for 2 h, the mixture was subjected to SDS-PAGE/blotting followed by detection using either St-HRP or anti-HA-HRP to detect either ubiquitylated or 3HA-tagged proteins, respectively. The components of the ubiquitination cascade (i.e. E1-Ub, E2-Ub, ubiquitinated MBP-RFA1, or MBP-RFA4DC) are indicated by arrows. C, PYL4 coimmunoprecipitates with RFA1 and RFA4. GFP or GFP-PYL4 proteins were expressed in N. benthamiana, and each protein extract was combined with MG132-treated Arabidopsis protein extracts obtained from lines expressing either HA-RFA1 or HA-RFA4. Proteins were immunoprecipitated using anti-GFP and next immunoblotted with anti-HA to detect coimmunoprecipitation of either HA-RFA1 or HA-RFA4.
RFA1 and RFA4 Are Ubiquitinated in Vivo
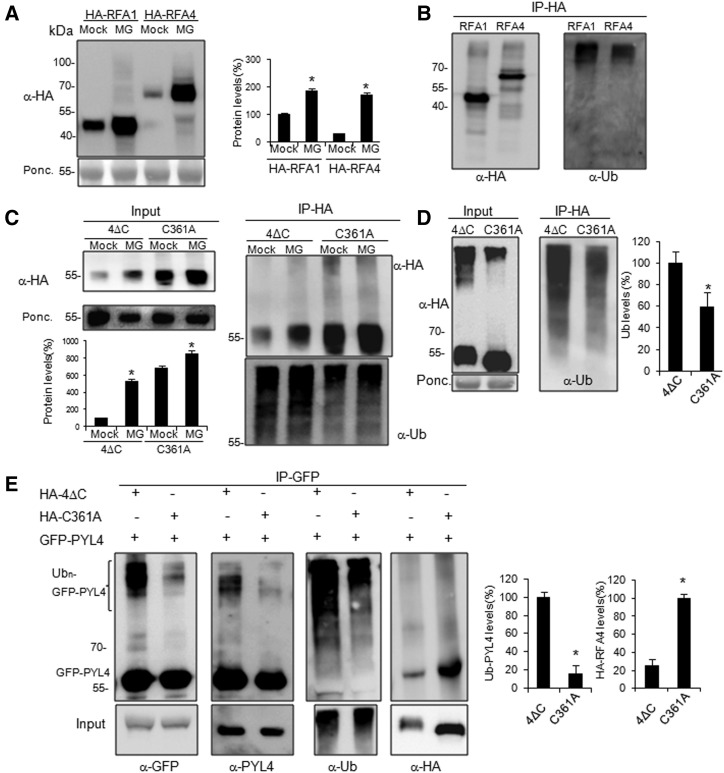
Studies done in mammals on the mechanism of action of RBR-type E3s have shown that their cognate E2 enzymes transfer the activated Ub to a conserved Cys residue of the E3 in RING2, which in turn transfers the Ub from this cysteinyl residue (active site Cys) to target Lys residues (Dove et al., 2016). Therefore, RBR-type E3s are ubiquitinated by the E2 during this process and subsequently RBRs ubiquitinate their targets through a mechanism similar to HECT-type E3 ligases (Callis, 2004). In order to investigate whether RFA1 and RFA4 are ubiquitinated in vivo, we constructed Arabidopsis lines expressing HA-tagged RFA1 and RFA4 from a 35S promoter (Fig. 4A; Supplemental Fig. S5). Interestingly, these lines showed reduced sensitivity to ABA-mediated inhibition of root growth, which is in common with the phenotype of RSL1-overexpressing lines that enhance degradation of ABA receptors (Bueso et al., 2014; Supplemental Fig. S5A). Protein extracts from Arabidopsis RFA1- and RFA4-overexpressing lines were prepared and immunoblotting analysis revealed that both RFA1 and RFA4 proteins reached higher levels upon MG132 treatment, which suggests that they are themselves subjected to ubiquitination and 26S proteasome degradation (Fig. 4A; Supplemental Fig. S5B). High-molecular-mass forms of RFA1 and RFA4 were detected in samples treated with MG132, which might correspond to ubiquitinated forms of the E3 ligases (Fig. 4B). To verify this, we immunoprecipitated HA-RFA1 and HA-RFA4 from MG132-treated samples, and immunoblot analysis using anti-Ub antibodies confirmed that both RFA1 and RFA4 are ubiquitinated in vivo (Fig. 4B).
Figure 4.
Analysis of RFA1 and RFA4 ubiquitination and the role of the Cys-361 residue of RFA4 in the ubiquitination and activity of the E3 ligase. A, Both RFA1 and RFA4 are themselves subjected to degradation via the 26S proteasome. Arabidopsis transgenic lines expressing HA-tagged RFA1 or RFA4 accumulate higher levels of these proteins after 50 μm MG132 treatment for 24 h. Values are averages ± sd obtained from three independent experiments (n = 5 plants in each experiment). *, P < 0.05 (Student’s t test) with respect to the corresponding mock-treated sample. B, Both RFA1 and RFA4 are ubiquitinated in vivo. The HA-RFA1 and HA-RFA4 proteins were immunoprecipitated from MG132-treated samples using anti-HA antibodies and subsequently analyzed by immunoblotting using anti-HA or anti-Ub (P4D1) antibody. C, The Cys-361Ala mutation markedly reduces the ubiquitination of RFA4 and stabilizes the protein compared with the wild type. Arabidopsis transgenic lines expressing HA-tagged RFA4ΔC or RFA4Cys-361AlaΔC accumulate higher levels of these proteins after 50 μm MG132 treatment for 24 h. Protein extracts were immunoprecipitated using anti-HA antibodies and subsequently analyzed by immunoblotting using anti-HA or anti-Ub (P4D1) antibodies. Values are averages ± sd obtained from three independent experiments (n = 5 plants in each experiment), and RFA4ΔC protein levels in mock-treated plants were taken as 100%. *, P < 0.05 (Student’s t test) compared with the corresponding mock-treated sample. D, Transient expression in N. benthamiana of HA-tagged RFA4ΔC or RFA4Cys-361AlaΔC reveals reduced ubiquitination of RFA4Cys-361AlaDC compared with the wild type. *, P < 0.05 (Student’s t test) compared with the wild type. E, In vivo ubiquitination of PYL4 by RFA4ΔC in N. benthamiana leaf cells. Agrobacteria encoding GFP-PYL4 were coinfiltrated in N. benthamiana cells with agrobacteria encoding either HA-RFA4ΔC or RFA4Cys-361AlaΔC. Protein extracts were immunoprecipitated using anti-GFP and subsequently immunoblotted with anti-GFP, anti-PYL4, anti-Ub, and anti-HA antibodies. *, P < 0.05 (Student’s t test) compared with the wild type. For D and E, values are averages ± sd obtained from three independent experiments (n = 5 leaves in each experiment).
The Cys-361 Residue of RFA4 Affects Its Own Ubiquitination and Capability to Ubiquitinate PYL4
In order to identify the putative catalytic cysteinyl residue of RFA4 involved in the transfer of Ub to target Lys residues, we performed a multiple sequence alignment of RFA4 RING2, HHARI RING2, and Parkin RING2 (Supplemental Fig. S6A). As a result, we identified Cys-361 in RFA4 as the putative active site Cys, which corresponds to the well-known active site Cys-431 in Parkin and Cys-357 in HHARI (Wenzel and Klevit, 2012; Duda et al., 2013; Riley et al., 2013), and we introduced a Cys-361Ala mutation in RFA4 that did not affect nuclear localization of the protein (Supplemental Fig. S6B). Additionally, we generated HA-tagged RFA4ΔC and RFA4Cys-361AlaΔC constructs, which were used to obtain stable lines in Arabidopsis (Fig. 4C) and that were delivered into leaf cells of N. benthamiana by agroinfiltration (Fig. 4D). Expression of HA-RFA4ΔC was lower than that of HA-RFA4Cys-361AlaΔC both in mock- and MG-treated samples obtained in Arabidopsis (Fig. 4C; Supplemental Fig. S5B). Upon immunoprecipitation with anti-HA antibodies and subsequent analysis with anti-Ub antibodies, we could detect more ubiquitination of HA-RFA4ΔC compared with HA-RFA4Cys-361AlaΔC (Fig. 4C). Similar results were obtained after transient expression in N. benthamiana cells, where expression of HA-RFA4ΔC was lower than that of HA-RFA4Cys-361AlaΔC (Fig. 4D). However, high-molecular-mass forms of HA-RFA4ΔC were more abundant than those of HA-RFA4Cys-361AlaΔC (Fig. 4D). In order to analyze the ubiquitination of HA-RFA4ΔC and HA-RFA4Cys-361AlaΔC, protein extracts were subjected to immunoprecipitation using anti-HA antibodies, and the immunoprecipitates were analyzed using anti-Ub antibodies. This revealed an ∼50% reduction of the ubiquitination in the Cys-361Ala mutant compared with the wild type (Fig. 4D). Taken together, the results obtained in Arabidopsis and N. benthamiana suggest that Cys-361 is involved in the ubiquitination of RFA4, although it is possible that other Lys or Cys residues of RFA4 could also be ubiquitinated.
Next, we examined whether the Cys-361Ala mutation affects the capability of RFA4 to ubiquitinate the PYL4 target. We set up an in vivo ubiquitination assay by coexpressing GFP-PYL4 in N. benthamiana together with HA-RFA4ΔC or HA-RFA4Cys-361AlaΔC (Fig. 4E). Two days after agroinfiltration, protein extracts from disc samples were prepared, immunoprecipitated using anti-GFP, immunoblotted, and analyzed with four different antibodies (Fig. 4E). In samples where GFP-PYL4 was coexpressed with HA-RFA4ΔC, the use of anti-GFP, anti-PYL4, and anti-Ub antibodies revealed more ubiquitinated PYL4 compared with samples where GFP-PYL4 was coexpressed with HA-RFA4Cys-361AlaΔC (Fig. 4E). These results indicate that Cys-361 is an important residue for the activity of RFA4. Interestingly, coimmunoprecipitation of HA-RFA4Cys-361AlaΔC with GFP-PYL4 was probably enhanced by the higher protein level of the Cys-361Ala mutant (Fig. 4E, α-HA).
RFA1 and RFA4 Promote Degradation of ABA Receptors in Vivo
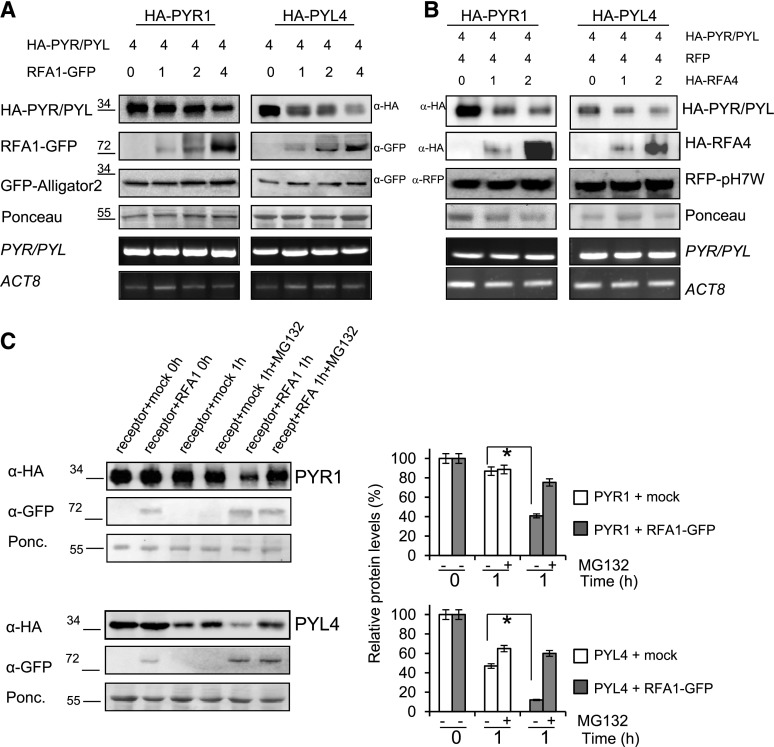
Once we confirmed the capability of RFA1 and RFA4 to interact with and ubiquitinate PYR1 and PYL4 ABA receptors, coinfiltration experiments in N. benthamiana were performed to test whether RFA1/RFA4 promote degradation of their targets in vivo (Zhao et al., 2013). Increasing amounts of the agrobacterium (Agrobacterium tumefaciens) that drives expression of the E3 ligase were coinfiltrated with different agrobacteria encoding constructs to express ABA receptors. Samples were collected for the detection of both protein and RNA levels (Fig. 5, A and B). Increasing amounts of RFA1 led to decreased levels of PYR1 and PYL4, whereas the internal GFP control was not significantly affected by increasing the amount of RFA1 (Fig. 5A). Similarly, increasing amounts of RFA4 led to degradation of PYR1 and PYL4, whereas the internal RFP control remained stable (Fig. 5B).
Figure 5.
Analysis of RFA1- and RFA4-promoted degradation of PYR1 and PYL4 by in vivo and semi-in vivo assays. A and B, In vivo degradation of either PYR1 or PYL4 was observed in coinfiltration experiments with increasing amounts of RFA1 or RFA4. The ratio of the relative concentration of agrobacteria used in the different coinfiltrations is indicated by numbers (top). In A, cell extracts were analyzed using anti-HA to detect HA-tagged PYR1/PYL4 and anti-GFP to detect GFP-RFA1, whereas in B, anti-HA was used to detect both HA-tagged PYR1/PYL4 and HA-RFA4. Anti-GFP or anti-RFP antibodies were used to detect the internal control of GFP or RFP, respectively. The mRNA expression levels of the target PYR1 and PYL4 genes and ACTIN8 (ACT8) were analyzed by reverse transcription-semiquantitative PCR. Molecular masses of marker proteins are indicated in kilodaltons. C, Cell-free experiments show HA-tagged PYR1/PYL4 degradation promoted by RFA1. Degradation of either HA-PYL4 or HA-PYR1 was performed by mixing N. benthamiana cell extracts from separated agroinfiltrations. HA-PYL4 or HA-PYR1 extract was mixed with RFA1-GFP extract or mock control extract and incubated at 4°C during 1 h either in the absence or presence of MG132. Proteins were detected by immunoblot as described above. Ponceau staining of the Rubisco protein is shown as a loading control. Graphs at right show the quantification of the experiment. Values are averages ± sd obtained from three independent experiments (n = 3 leaf extracts in each experiment). *, P < 0.05 (Student’s t test) compared with the corresponding mock-treated sample.
We also analyzed whether RFA1 promoted PYR1 and PYL4 degradation using a semi-in vivo degradation assay (Liu et al., 2010). RFA1 and each PYR/PYL were expressed separately via different agroinfiltrations. Then, we mixed together the N. benthamiana extracts containing RFA1 and each PYR/PYL and monitored protein levels after 1 h of incubation at 4°C (Fig. 5C). The addition of RFA1 enhanced the degradation of PYR/PYLs compared with the sample with nonagroinfiltrated extract (mock). When the MG-132 proteasome inhibitor was included, the level of PYR/PYL increased 2- to 3-fold with respect to the sample lacking the inhibitor (Fig. 5C), which indicates that RFA1 promotes the degradation of PYR1 and PYL4 through the 26S proteasome.
Identification of UBC26 as the Cognate Nuclear E2 Enzyme That Interacts with the RFA4 E3 Ligase
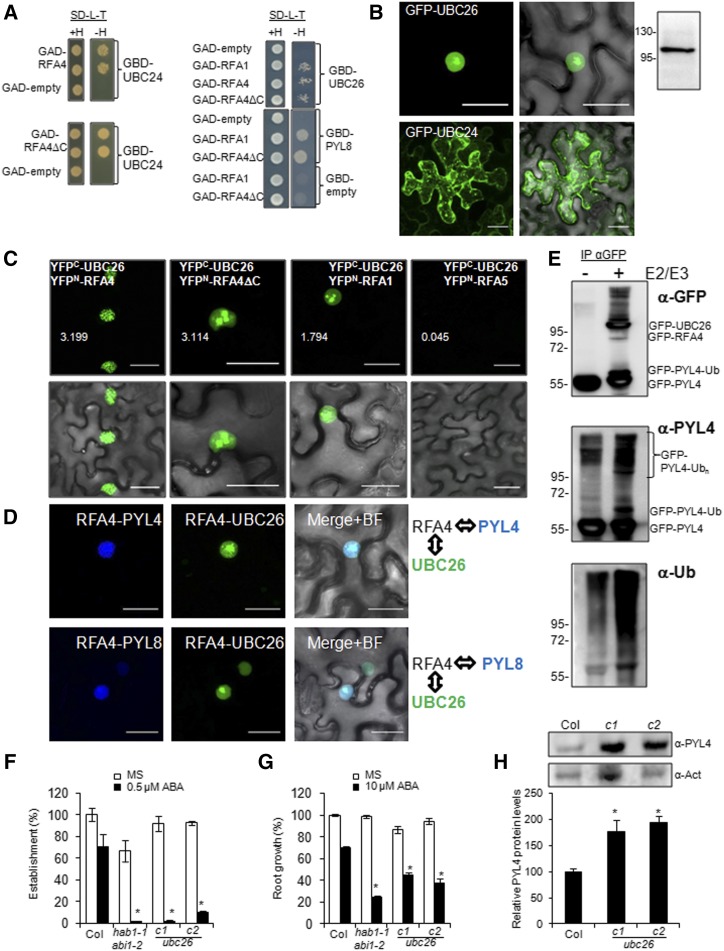
Attachment of Ub to substrate proteins is catalyzed by the concerted action of E1, E2, and E3 enzymes. E3 ligases mediate substrate specificity of the ubiquitination machinery; however, E2s also play a key role in mediating Ub chain assembly on the target (Callis, 2014; Kowarschik et al., 2018). In the case of U-box/RING-type E3 ligases, concerted action of the E2/E3 pair regulates substrate specificity and the type of Ub modification on the substrate proteins (Zhao et al., 2013). In the case of mammalian HECT- and RBR-type E3s, residues in the active site Cys determine the type of ubiquitination, although little is known for plant enzymes (Kim and Huibregtse, 2009; Dove et al., 2016). In plants, the cohort of E2s that function together with RBR-type ligases is unknown, as are the features and consequences of this interaction for the ubiquitination of the target. The Arabidopsis genome encodes 48 UBC domain-containing proteins; however, only 37 carry the active site Cys required for E2 Ub conjugase activity (Callis, 2014). We were particularly interested in the identification of nuclear E2s that could interact with RFA4 to form a nuclear E2-E3 complex; therefore, we conducted a yeast two-hybrid (Y2H) screen to identify cognate E2 enzymes for RFA4. To this end, either GAD-RFA4 or GAD-RFA4ΔC was used as a bait to find preys in a yeast pGBD library containing 30 E2s. As a result, we found that both UBC24 and UBC26, which belong to family XI of Arabidopsis E2 proteins, interacted with both RFA4 and RFA4ΔC (Fig. 6A; Supplemental Fig. S7). In contrast, the other E2s analyzed did not interact with RFA4 (Supplemental Fig. S7). We pursued further work with UBC26 because it contains an NLS (KKKTRKR) at the C terminus (Supplemental Fig. S8). Indeed, GFP-UBC26 is localized in the nucleus (Fig. 6B), whereas UBC24 has been reported to be mostly colocalized with endoplasmic reticulum and Golgi markers (Liu et al., 2012; Fig. 6B). In addition to RFA4, UBC26 also interacts with RFA1 in the Y2H assay; however, since RFA1 also shows cytosolic localization, it is likely that other unidentified UBCs interact with RFA1 (Fig. 6A). Additionally, using the Y2H assay, we also confirmed that RFA1 and RFA4ΔC could interact with ABA receptors (Fig. 6B; Supplemental Fig. S9A), which indicates that these E3s can interact in Y2H assays with both the E2 and the substrate.
Figure 6.
UBC26 and the UBC26-RFA4-ABA receptor complex are localized in the nucleus. A, Y2H interactions of RFA1 and RFA4. Transformed yeast cultures were grown overnight in liquid synthetic defined (SD) medium lacking Leu and Trp, and a dilution (10−1) of these cultures was dropped on either control medium lacking Leu and Trp (SD–Leu–Trp+His) or selective medium additionally lacking His (SD–Leu–Trp–His). Empty vectors were used as negative controls, and yeast was allowed to grow for 3 d at 30°C before interaction was scored. B, GFP-UBC26 localizes in the nucleus upon transient expression in N. benthamiana, whereas GFP-UBC24 seems to be membrane associated. Immunoblotting analysis using anti-GFP was used to verify the expression of GFP-UBC26. C, BiFC interaction of RFA4 and UBC26 in the nucleus of N. benthamiana epidermal cells. Confocal images show transiently transformed leaf cells coexpressing YFPC-UBC26 and YFPN-RFA4, RFA4ΔC, RFA1, or RFA5. Note that the E2-E3 interaction decorates nuclear speckles, whereas the PYL4-E3 interaction is absent in the nucleolus (Fig. 2A). Deletion of the acidic domain of RFA4 changes the nuclear pattern of the E2-E3 interaction compared with RFA4FL. YFPC-UBC26 does not interact with YFPN-RFA5, which is another member of the RFA family. Quantification of YFP signal was analyzed by ImageJ software, and the arbitrary units of fluorescence are indicated in numbers. D, Multicolor BiFC reveals the formation of E2-E3-PYL4/8 nuclear complexes. Confocal images show transiently transformed N. benthamiana epidermal cells coexpressing SCFPC-RFA4, SCFPN-PYL4/8, and VENUSN-UBC26. The RFA4-PYL4/8 interaction was visualized through reconstitution of the SCFP, whereas the RFA4-UBC26 interaction gave rise to SCFPC-VENUSN fluorescent protein. SCFPC-RFA4 coexpressed with SCFPN or VENUSN did not reconstitute fluorescent protein (Supplemental Fig. S3B). Constructs were delivered into N. benthamiana epidermal leaves through A. tumefaciens-mediated transfection. Leaves were examined using confocal laser scanning microscopy 48 to 72 h after infiltration. Bars = 30 μm in B to D. E, In vivo ubiquitination of PYL4 by UBC26-RFA4 in N. benthamiana leaf cells. Agrobacteria encoding GFP-PYL4 were coinfiltrated in N. benthamiana cells with agrobacteria lacking or encoding GFP-UBC26/GFP-RFA4 proteins (−/+ E2E3). Protein extracts were immunoprecipitated using anti-GFP and next immunoblotted with anti-GFP, anti-PYL4, and anti-Ub to detect ubiquitination of GFP-PYL4 in the absence or presence of GFP-UBC26/GFP-RFA4 proteins. Extraction of GFP-RFA4 for SDS-PAGE analysis seems to be inefficient because of the large C-terminal acidic domain. F, Enhanced sensitivity to ABA-mediated inhibition of seedling establishment. Approximately 100 seeds of each genotype (three independent experiments) were sown on Murashige and Skoog (MS) plates lacking or supplemented with 0.5 µm ABA and scored for the presence of both green cotyledons and the first pair of true leaves after 7d. Values are averages ± sd. *, P < 0.01 (Student’s t test) with respect to wild-type Columbia-0 (Col-0) assayed in the same conditions. G, Enhanced sensitivity to ABA-mediated inhibition of root growth. Quantification is shown for ABA-mediated inhibition of root growth in the indicated genotypes compared with the wild type. *, P < 0.01 (Student’s t test) with respect to wild-type Col-0 assayed in the same conditions. H, Accumulation of endogenous PYL4 protein in ubc26-c1 and ubc26-c2 alleles compared with the wild type. Seedlings of the indicated genotypes were grown for 7 d in MS medium, and total proteins were extracted and subjected to immunoblot analysis using the indicated antibodies. Actin was analyzed as a protein-loading control, and relative PYL4 protein levels were quantified. Values are averages ± sd obtained from three independent experiments (n > 20 seedlings in each experiment). *, P < 0.05 (Student’s t test) compared with wild-type Col-0.
To investigate the interaction in planta of RFA1 and RFA4 with UBC26, we performed BiFC assays (Fig. 6C). We coexpressed YFPN-RFA4, YFPN-RFA4ΔC, or YFPN-RFA1 with YFPC-UBC26 and found that they interact in the nucleus of N. benthamiana cells. In contrast, the closely related RFA5 protein did not interact with UBC26, even though it was expressed to similar levels to other RFAs (Fig. 6C; Supplemental Fig. S3B). The nuclear speckles formed by RFA4-UBC26 were different from those formed by RFA4ΔC-UBC26, which are more similar to RFA1-UBC26 speckles. These results suggest that although the C-terminal acidic domain of RFA4 is dispensable for interaction with the substrate and the E2, it determines the location of the interaction in a different nuclear territory. Finally, we performed a multicolor BiFC assay (Gehl et al., 2009) to analyze the possible formation of nuclear receptor-E3-E2 complexes. RFA4 interacted with both the E2 and ABA receptors when coexpressed in N. benthamiana cells (Fig. 6D).
UBC26 has been reported to be recalcitrant to expression in Escherichia coli (Zhao et al., 2013). Indeed, some E2s are not induced or are insoluble when expressed in bacteria, and in these cases, E2 activity can be recovered after expression in plants (Zhao et al., 2013). Since we had successfully expressed GFP-UBC26 by agroinfiltration in N. benthamiana cells and nuclear PYL4-RFA4-UBC26 complexes could be detected (Fig. 6D), we set up an in vivo ubiquitination assay involving GFP-PYL4, GFP-UBC26, and GFP-RFA4 (Fig. 6E). The GFP fusion proteins were immunoprecipitated and the ubiquitination of PYL4 was analyzed using anti-PYL4 and anti-Ub antibodies (Fig. 6E). High-molecular-mass forms of PYL4, recognized both by anti-PYL4 and anti-Ub antibodies, were notably enhanced when the E2/E3 proteins were coexpressed with PYL4, which suggests that ubiquitination of PYL4 was markedly enhanced by the action of UBC26 and RFA4 (Fig. 6E). Some ubiquitination of PYL4 in N. benthamiana cells could be detected in the absence of E2/E3 coexpression, which had been also previously observed in Arabidopsis lines expressing HA-PYL4 (Bueso et al., 2014) and might be explained by the endogenous endowment of E2/E3s. Finally, the expression of GFP-PYL4 plus only the E2 or E3 did not significantly enhance the basal ubiquitination of PYL4 (Supplemental Fig. S9B).
To provide genetic support to the possible role of UBC26 in ABA signaling, we generated CRISPR/Cas9 lines specifically impaired in UBC26 function (Supplemental Fig. S10). The ubc26-c1 and ubc26-c2 alleles contain one-base insertions at position 1,215 (counting from the ATG), which generates a frame-shift mutation and creates a stop codon at position 1,246. As a result, the Ub-conjugating E2 domain of UBC26 is disrupted (Supplemental Fig. S10, A and B). We analyzed ABA sensitivity of the ubc26-c1 and ubc26-c2 alleles in germination and root growth assays. We found that both alleles showed enhanced sensitivity to ABA-mediated inhibition of seedling establishment and root growth (Fig. 6, F and G; Supplemental Fig. S10C). We examined protein levels of PYL4, using the specific antibodies described by Yu et al. (2016), and found higher PYL4 levels in both ubc26-c1 and ubc26-c2 alleles compared with the wild type (Fig. 6H). This result suggests that UBC26 mediates the nuclear degradation of PYL4, likely through its interaction with RFA4.
The rfa1 rfa4 Double Mutant Accumulates More PYR1 and PYL4 Receptors
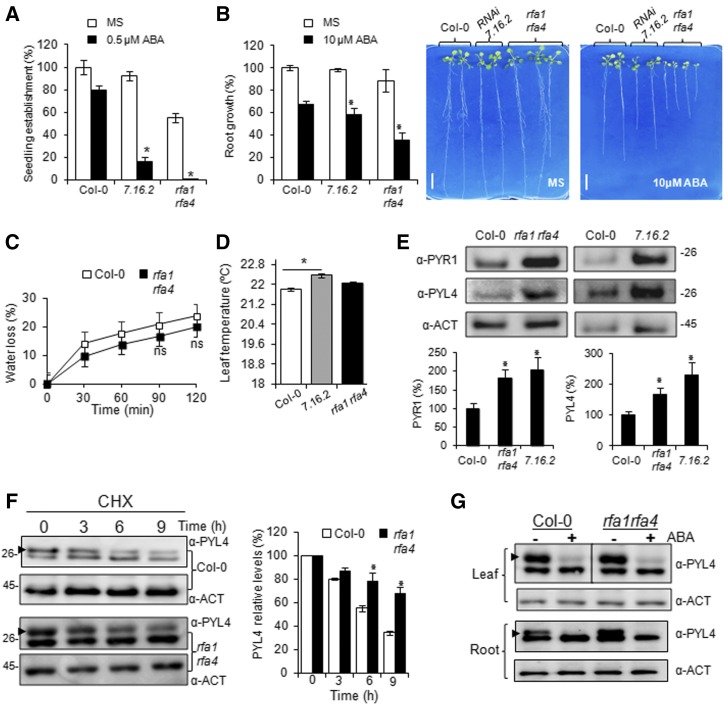
We screened the Arabidopsis Biological Resource Center (ABRC)/Nottingham Arabidopsis Stock Centre collection to identify T-DNA insertion rfa1 and rfa4 mutants (Supplemental Fig. S11A). The T-DNA insertion strongly impairs the expression of RFA1 and RFA4; however, single rfa1 and rfa4 mutants show wild-type sensitivity to ABA, which likely reflects a certain functional redundancy observed in multigene families (Supplemental Fig. S11, A–C). Therefore, we generated an rfa1 rfa4 double mutant and analyzed its sensitivity to ABA (Fig. 7A). As a control for this experiment, we used the ABA-hypersensitive RNA interference (RNAi) 7.16.2 line, where expression of at least three members of the RSL1/RFA1 family was impaired (i.e. RSL1, RFA1, and RFA3; Bueso et al., 2014). The rfa1 rfa4 double mutant was impaired in germination and showed enhanced sensitivity to ABA-mediated inhibition of seedling establishment and root growth compared with the wild type (Fig. 7, A and B; Supplemental Fig. S11, D and E). We also performed water-loss experiments, and detached leaves of rfa1 rfa4 showed no significant difference in water-loss assays compared with the wild type (Fig. 7C), which is in agreement with the low expression of RFA1 and RFA4 in guard cells (accession nos. GSE41054 and GSE118520, Gene Expression Omnibus database). Using infrared thermography of leaves to monitor transpiration, we did not observe significant differences in rfa1 rfa4 compared with the wild type; however, the RNAi 7.16.2 line showed a significant increase of leaf temperature (Fig. 7D). This suggests that ubiquitination and endocytosis of ABA receptors triggered by RSL1 might regulate stomatal aperture, as observed for the components of the ESCRT pathway FYVE1/FREE1 and ALIX (Belda-Palazon et al., 2016; García-León et al., 2019). Finally, we examined protein levels of two receptors, PYR1 and PYL4, that play a major role in ABA signaling and for which there are specific antibodies available (Fig. 7E; Supplemental Fig. S12). We found that rfa1 rfa4 accumulates higher levels of both PYR1 and PYL4 compared with the wild type, which was also observed in the RNAi 7.16.2 line. This result confirms that the RSL1/RFA family of E3 ligases regulates endogenous protein levels of ABA receptors. Additionally, we performed a time-course study of the PYL4 protein in the presence of cycloheximide (CHX) and observed that degradation of PYL4 was slower in rfa1 rfa4 compared with the wild type (Fig. 7F). Finally, we analyzed the effect of ABA treatment on PYL4 both in leaves and roots and found that ABA promotes the degradation of PYL4 both in the wild type and rfa1 rfa4. This result suggests that ABA-induced down-regulation of PYL4 can occur in the absence of RFA1 and RFA4, likely through transcriptional down-regulation of PYL4 expression in response to ABA (Goda et al., 2008; Santiago et al., 2009b) and protein degradation through the ESCRT/vacuolar pathway (Belda-Palazon et al., 2016; García-León et al., 2019).
Figure 7.
The rfa1 rfa4 double mutant shows enhanced sensitivity to ABA and accumulation of the PYR1 and PYL4 ABA receptors as compared with the wild type. A, Enhanced sensitivity to ABA-mediated inhibition of seedling establishment. Approximately 100 seeds of each genotype were sown on MS plates lacking or supplemented with 0.5 μm ABA and scored for the presence of both green cotyledons and the first pair of true leaves after 7 d. For A and B, values are averages ± sd obtained from three independent experiments (n > 100 seeds in A; n > 20 seedlings in B). *, P < 0.01 (Student’s t test) with respect to the wild type assayed in the same conditions. B, Enhanced sensitivity to ABA-mediated inhibition of root growth. Quantification of ABA-mediated inhibition of root growth is shown in the indicated genotypes compared with the wild type. The photographs show representative seedlings 10 d after the transfer of 4-d-old seedlings to MS plates lacking or supplemented with 10 µm ABA. Bars = 1 cm. C, Water loss in detached leaves of the indicated genotypes. Loss of fresh weight was measured in detached leaves from 15-d-old plants submitted to the drying atmosphere of a laminar flow hood. Values are averages ± sd obtained from three independent experiments (n = 5 plants in each experiment), and ns indicates nonsignificant difference when comparing (using Student’s t test) data of the wild type and rfa1 rfa4. D, Leaf temperature of the indicated genotypes was quantified by infrared thermography. Data are means ± sd (n = 5, approximately 1,000 measurements of square pixels from several leaves of each plant). *, P < 0.05 (Student’s t test) with respect to wild-type Col-0. E, Accumulation of endogenous PYR1 and PYL4 proteins in rfa1 rfa4 and RNAi 7.16.2 line compared with the wild type. Seedlings of the indicated genotypes were grown for 7 d in MS medium, and total proteins were extracted and subjected to immunoblot analysis using the indicated antibodies. Actin was analyzed as a protein-loading control, and PYR1 or PYL4 protein levels were quantified relative to Col-0 (taken as 100%). Values are averages ± sd obtained from three independent experiments (n = 10 seedlings per genotype in each experiment). *, P < 0.05 (Student’s t test) with respect to wild-type Col-0. F, Degradation of PYL4 is delayed in the rfa1 rfa4 mutant compared with wild-type Col-0. Seedlings of Col-0 or rfa1 rfa4 were grown in liquid MS medium for 10 d and then 50 mm CHX treated for the indicated time periods. Actin was analyzed as a loading control. Histograms show the quantification of PYL4 protein levels in CHX-treated samples relative to time 0, whose value was taken as 100% for Col-0 or rfa1 rfa4 samples. Values are averages ± sd obtained from three independent experiments (n = 10 seedlings per genotype in each experiment). *, P < 0.05 (Student’s t test) with respect to Col-0 at the same time point. G, ABA treatment leads to down-regulation of the PYL4 protein. Seedlings of Col-0 or rfa1 rfa4 were grown in liquid MS medium for 10 d and then were mock or 50 μm ABA treated for 6 h. Leaf and root tissues were separated, and protein extracts were analyzed by immunoblotting using anti-PYL4 and anti-ACT antibodies. Arrowheads indicate the band corresponding to PYL4. A new batch of the anti-PYL4 antibody recognized a second band below PYL4.
DISCUSSION
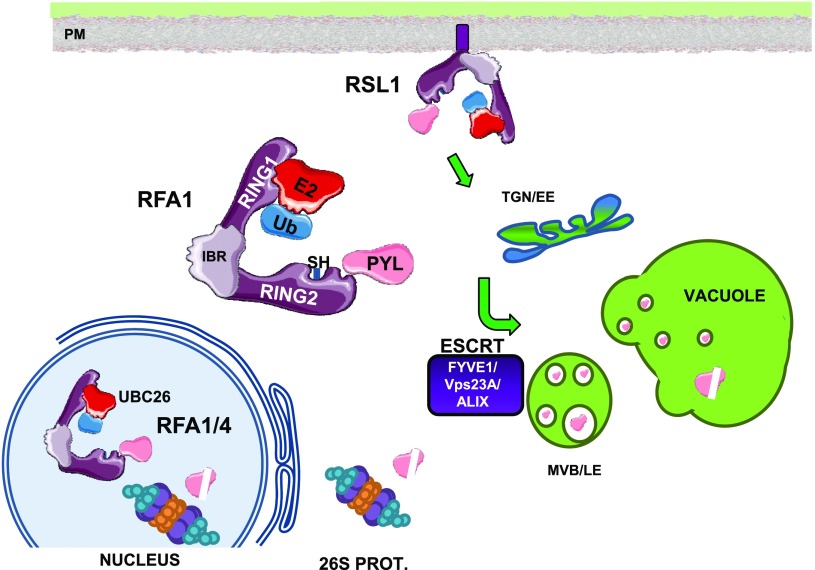
In this work, we gained insight into the biological functions of some RBR-type E3 ligases, which are poorly characterized enzymes in plants (Callis, 2014). There are 42 genes encoding RBR-type proteins in Arabidopsis, which are classified into four subfamilies: Plant II (22 members), Plant I/helicase (three members), ARA54 (one member), and ARIADNE (16 members; Marín, 2010). Compared with other families of E3 ligases, very little is known regarding the biological function of RBRs. The ARI12 and ARI14 members of the ARIADNE family have been implicated in UV-B signaling and fertilization, respectively (Ron et al., 2010; Lang-Mladek et al., 2012), but the rest of the family is hardly characterized. RSL1/RFAs belong to clade A of the Plant II subfamily, and it was demonstrated that RSL1 targets ABA receptors in plasma membrane (Bueso et al., 2014; Belda-Palazon et al., 2016). Since 14 ABA receptors exist in Arabidopsis, we hypothesized that their interacting E3s might also belong to multigene families (Bueso et al., 2014). We demonstrated that ABA receptors can be also targeted by RFA1 and RFA4, which are members of the RSL1/RFA family not associated with plasma membranes. We suggest that specific features of RSL1/RFA members, such as their particular subcellular localization or differential expression pattern, could notably increase the capability to recognize and regulate the stability of the whole family of ABA receptors. Interestingly, RFA4 shows specific nuclear localization, whereas RFA1 is localized both in nucleus and cytosol. Moreover, we demonstrated using multicolor BiFC that it is possible to detect the simultaneous formation of receptor-RSL1 and receptor-RFA4 complexes in different subcellular locations. Thus, the concerted action of several members of the RSL1/RFA family enables fine regulation of ABA receptor half-life in different subcellular compartments. Additionally, whereas RSL1-mediated ubiquitination promotes endosome-mediated vacuolar degradation of ABA receptors, RFA1 and RFA4 act likely through the 26S proteasome degradation pathway (Fig. 8). Therefore, our results further confirm that both 26S proteasome and non-26S Ub-dependent degradation pathways regulate the stability of key components of ABA signaling (Irigoyen et al., 2014; Belda-Palazon et al., 2016; Yu et al., 2016; Yu and Xie, 2017). Thus, the regulation of the turnover of ABA receptors can be achieved both in plasma membrane and nucleus, which are two key places of the cell to regulate ABA signaling either affecting ion/water transport or transcriptional response to ABA, respectively (Cutler et al., 2010).
Figure 8.
A working model depicting the targeting of ABA receptors by RSL1, RFA1, or RFA4 at different subcellular locations. Ubiquitination of ABA receptors at the plasma membrane by RSL1 triggers clathrin-mediated endocytosis, transit through the trans-Golgi network/early endosomes (TGN/EE), sorting of ubiquitinated cargo through the ESCRT pathway (involving FYVE1, VPS23A, and ALIX components), delivery to multivesicular bodies/late endosomes (MVB/LE), and finally to the vacuole for degradation. Ubiquitination of receptors at cytosol or nucleus leads to degradation by the 26S proteasome. The UBC26-RFA4-receptor complex regulates the half-life of ABA receptors in the nucleus. The cartoon represents the RING1-IBR-RING2 modular structure of RBR-type E3 ligases and the localization of the active site Cys-SH residue in RING2.
The interaction of RFA1/RFA4 with PYR1 and PYL4 ABA receptors was confirmed both in vitro and in vivo. Using agroinfiltration experiments, we demonstrated that increasing amounts of RFA1 or RFA4 correlated with enhanced degradation of PYR1 and PYL4. Moreover, using specific antibodies for PYR1 and PYL4, we could demonstrate that endogenous levels of these receptors were increased in the rfa1 rfa4 double mutant (Fig. 7). This finding strongly supports that endogenous ABA receptors are bona fide targets of RFA1 and RFA4. Additionally, we provided evidence that a previously described ABA-hypersensitive RNAi line that targets RSL1 and at least RFA1 and RFA3 also shows higher PYR1 and PYL4 levels than wild-type plants (Fig. 7). Endogenous PYL4 levels were also higher in the ubc26 mutant (Fig. 6H), which correlated with enhanced sensitivity to ABA in establishment and root growth assays (Fig. 6, F and G). This result, together with interaction and ubiquitination assays, suggests that UBC26 is the cognate E2 that forms ternary complexes with RFA4 and ABA receptors in the nucleus of plant cells. Interestingly, only family XI members of Arabidopsis E2 proteins were found to interact with RFA4 in yeast cells, and for UBC26, we confirmed that this interaction occurs in the nucleus of plant cells. UBC26, as well as other family XI E2 members, in addition to the C-terminal catalytic core domain present in all E2 enzymes, contains a large N-terminal domain that might be involved in the interaction with E3 (Supplemental Fig. S8). Future biochemical studies should further unravel the contribution of UBC26 to the ubiquitination properties of RFA4.
In addition to regulating the half-lives of their targets, some E3s have been reported to be ubiquitinated in vivo to regulate their own stability (Liu and Stone, 2010). For instance, the KEEP ON GOING (KEG) E3 ligase is autoubiquitinated in response to ABA, leading to ABA-induced degradation of KEG and accumulation of ABI5 (Liu and Stone, 2010). Specific antibodies are not available yet for RFAs; however, in epitope-tagged RFA1 and RFA4 lines, we observed that RFA1 and RFA4 are ubiquitinated and subjected to proteolytic degradation by the 26S proteasome (Fig. 4, A and B). Autocatalytic ubiquitination of RSL1, RFA1, and RFA4 has been observed in vitro (Bueso et al., 2014; Fig. 3B), which agrees with the mechanism of action of RBR-type E3s. In these enzymes, Ub is passed from the active site Cys of the E2 to that of the E3, which in turn catalytically transfers the activated Ub from its RING2 domain to Lys target residues. A prediction of this mechanism is that ubiquitinated forms of plant RBR-type E3s should be detected in vivo, which was confirmed both for RFA1 and RFA4 (Fig. 4, A–C). Additionally, the Cys-361Ala mutation impaired the capability of RFA4 to ubiquitinate the PYL4 target, which is in agreement with its predicted role as active site Cys (Fig. 4E).
Since degradation of ABA receptors can be achieved via the 26S proteasome and vacuolar pathways, it is expected that different types of Ub-linked chains and E2/E3 ligases are involved in these processes. For instance, K48-linked polyubiquitination promotes 26S proteasome degradation, monoubiquitination promotes internalization of plasma membrane proteins, and K63-linked polyubiquitination can promote subsequent endosome trafficking (Yu and Xie, 2017; Romero-Barrios and Vert, 2018). Indeed, both K48- and K63-linked polyubiquitination has been detected for PYL4 (Yu et al., 2016), which indicates that degradation of PYL4 occurs both through the UPS and vacuolar pathways. Degradation of ABA receptors can be differentially regulated by different members of the multigene RSL1/RFA family (Bueso et al., 2014; this work) or by different E3 ligases that have been reported to promote the degradation of ABA receptors (Irigoyen et al., 2014; Li et al., 2016, 2018; Zhao et al., 2017). However, in these latter studies, the protein levels of endogenous ABA receptors were not analyzed using specific antibodies. Now, the availability of specific antibodies against ABA receptors makes it possible to further analyze the contribution of the reported single- and multiple-subunit E3 ligases in regulating endogenous ABA receptor levels.
Recently, cooperation by E3-E3 pairs has been described in humans to regulate substrate ubiquitination (Scott et al., 2016). The CULLIN4-RING E3 ubiquitin ligase (CRL4) complex formed by COP10-DET1-DDB1 and the substrate adapter DDA1 are involved in nuclear degradation of ABA receptors; however, CRL4DDA1-mediated degradation of PYL8 was counteracted by ABA, which suggests that the CRL4DDA1 complex only works at basal ABA levels (Irigoyen et al., 2014). Contrary to PYL8, which is stabilized by ligand binding (Belda-Palazon et al., 2018), ABA prompts destabilization of PYL4 through the ESCRT/vacuolar pathway, since vp23a and alix1 mutants accumulate more PYL4 than Col-0 after ABA treatment (Yu et al., 2016; García-León et al., 2019). Recent studies in humans reveal that CRL- and RBR-type E3 ligases can work in unison to regulate substrate ubiquitination (Kelsall et al., 2013; Scott et al., 2016). For instance, it has been shown that the association of neddylated-CRLs with the RBR-type E3 ARIH1 leads to monoubiquitination of CRL client substrates by ARIH1 and a reciprocal role of RBRs as regulators of distinct CRLs (Kelsall et al., 2013; Scott et al., 2016). As a result, during the study of human E3 ligases, the concept of E3-E3 team tagging has emerged, which implies that different types of E3s can act successively on a common target (Scott et al., 2016). Thus, exquisite regulation of substrate ubiquitination can be achieved via team tagging. Future studies should address the possible cooperation of E3 ligases that target ABA receptors in the same cell compartment to jointly regulate substrate ubiquitination.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown as described by Pizzio et al. (2013). The rfa4 and rfa1 T-DNA insertion lines (SALK_208771C and SALK_005363, respectively) were obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk). To confirm and identify homozygous T-DNA individuals, seedlings of each insertion line were grown individually and DNA from each plant was extracted and submitted to PCR-mediated genotyping using the primers described in Supplemental Table S1. The rfa1 rfa4 double mutant was generated by crossing and genotyping of F2 individuals. The pAlligator2-35S:HA-RFA1 and pAlligator2-35S:HA-RFA4 constructs were transferred to Agrobacterium tumefaciens C58C1 (pGV2260; Deblaere et al., 1985) by electroporation and used to transform Col-0 wild-type plants by the floral dip method (Clough and Bent, 1998). T1 transgenic seeds were selected based on seed GFP fluorescence and sown in soil to obtain the T2 generation. Homozygous T3 progeny were used for further studies, and the expression of HA-tagged protein was verified by immunoblot analysis using anti-HA-HRP.
Generation of CRISPR/Cas9 Mutants
The single guide RNA (sgRNA) targeting UBC26 (At1g53025) was designed using the online tool CRISPR-PLANT (http://www.genome.arizona.edu/crispr/CRISPRsearch.html). A 19-bp sequence followed by the GGG PAM sequence was selected. A modified pDONR207 vector (GenBank accession no. MG917725.1) was used to clone the sgRNA, which contains the AtU6-26 promoter followed by the sgRNA scaffold and the RNA Polymerase III terminator. We introduced the sgRNA sequence by site-directed, ligase-independent mutagenesis-PCR as described previously by Chiu et al. (2004). After verifying the cloning by sequencing, a fragment including the AtU6-26 promoter, the sgRNA, the scaffold RNA, and the terminator was amplified by PCR, with primers containing HindIII and SpeI restriction sites (P1b-HindIII and P4b-SpeI). This fragment was cloned into the pHEE2E-TRI vector, which expresses Cas9 driven by the egg cell-specific EC1.2 promoter (Wang et al., 2015). The resulting vector pHEE2E-TRI, containing Cas9 and the sgRNA, was transferred to A. tumefaciens C58C1 (pGV2260; Deblaere et al., 1985) by electroporation and used to transform Col-0 wild-type plants by the floral dip method (Clough and Bent, 1998). Transformants were selected in medium supplemented with 20 μg mL−1 hygromycin. To analyze the mutations caused by CRISPR/Cas9, genomic DNA was extracted from each transgenic plant using the cetyl-trimethyl-ammonium bromide method. A 590-bp fragment containing the CRISPR target site was amplified using specific primers, cleaned with ExoSAP-IT (Affymetrix), and sequenced. T2 homozygous mutant plants were obtained, and their offspring were used in the indicated experiments.
Constructs
The OFP-TMD construct was obtained from Dr. Joerg Kudla (University of Münster), and the pENTR221-RFA4 clone was obtained from the ABRC. The open reading frame (ORF) of RFA1 was amplified by reverse transcription-PCR and cloned into pCR8/GW/TOPO. The pENTR221-RFA4 or pCR8-RFA1 constructs were recombined by LR reaction into pAlligator2 to generate 35S:HA-RFA4 or 35S:HA-RFA1 constructs, into pMDC43 to generate 35S:GFP-RFA4 or 35S:GFP-RFA1 constructs, or into pGADT7-GW to generate pGAD-RFA4 or pGAD-RFA1 constructs for Y2H assays. Additionally, we amplified the ORF of RFA1 lacking the stop codon and cloned it into pCR8/GW/TOPO. This construct was recombined by LR reaction into pMDC83 to generate the 35S:RFA1-GFP construct. The pENTR221-RFA4 and pCR8-RFA1 constructs were recombined by LR reaction into pYFPN43 for BIFC assays. The ORFs of RFA4ΔC, lacking residues 385 to 468, and RFA4ΔNLS, lacking residues 1 to 14, were amplified using the primers indicated in Supplemental Table S1 and cloned into pCR8/GW/TOPO. We introduced the Cys-361Ala mutation into the sequence of RFA4ΔC by site-directed, ligase-independent mutagenesis-PCR as described previously by Chiu et al. (2004).
Transient Protein Expression in Nicotiana benthamiana
A. tumefaciens infiltration of N. benthamiana leaves was performed basically as described by Saez et al. (2008). To investigate the interaction of RFA1 and RFA with ABA receptors in planta, we used the pYFPN43 and pYFPC43 vectors for BiFC assays (Belda-Palazón et al., 2012). To perform multicolor BiFC, we cloned PYR1/PYL4 into p(MAS)-SCYCE(R), RSL1 into pDEST-SCYNE(R), and RFA4 into pDEST-VYNE(R) vectors, as described by Gehl et al. (2009). In the case of RFA4 interactions with UBC26 and ABA receptors, we cloned RFA4 into p(MAS)-SCYCE(R), UBC26 into pDEST-SCYNE(R), and ABA receptors into pDEST-VYNE(R) vectors. The different binary vectors were introduced into A. tumefaciens C58C1 (pGV2260) by electroporation, and transformed cells were selected on Luria-Bertani plates supplemented with kanamycin (50 μg mL−1). Then they were grown in liquid Luria-Bertani medium to late exponential phase, and cells were harvested by centrifugation and resuspended in 10 mm MES-KOH, pH 5.6, containing 10 mm MgCl2 and 150 mm acetosyringone to an OD600 of 1. These cells were mixed with an equal volume of A. tumefaciens C58C1 (pCH32 35S:p19) expressing the silencing suppressor p19 of Tomato bushy stunt virus so that the final density of A. tumefaciens solution was about 1 (final concentration OD600 = 0.5, 0.33, or 0.25 each for bi, tri, or tetra infiltrations). Bacteria were incubated for 3 h at room temperature and then injected into young fully expanded leaves of 4-week-old N. benthamiana plants. Leaves were examined 48 to 72 h after infiltration using confocal laser scanning microscopy.
Split-Luciferase Complementation Assay
The coding sequences of RFA1 and RFA4 lacking the stop codon were amplified by PCR and cloned into pCR8/GW. Then they were recombined by LR reaction into pDEST-GWnLUC. For ABA receptors, coding sequences of PYR1, PYL4, and PYL8 were recombined by LR reaction into pDEST-cLUCGW. Split-luciferase complementation assay was performed by transient expression in leaves of N. benthamiana by A. tumefaciens-mediated infiltration as described above. MG132 (50 µm) was applied into the infiltrated region 6 h before inspection, which was performed 72 h after infiltration. To this end, leaves coexpressing different constructs were examined for luciferase activity by applying 1 mm d-luciferin and placed in the dark for 5 min before imaging. Luciferase complementation was observed with a CCD imaging system (LAS3000; Fujifilm) using 10-min exposures.
In Vivo and Semi-in Vivo Protein Degradation Assays
Protein degradation assays were performed as described by Liu et al. (2010) and Zhao et al. (2013) with small modifications. For in vivo protein degradation experiments, A. tumefaciens cultures containing constructs that express the indicated E3 ligase, 3HA-PYR1 or 3HA-PYL4, the indicated GFP or RFP internal control, and the silencing suppressor p19 were coinfiltrated at different ratios in N. benthamiana leaves. Three days after infiltration, samples were collected, ground in liquid nitrogen, and immediately placed in lysis buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% [v/v] Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail) on ice for protein extraction. Homogenates were cleared by centrifugation at 12,000g at 4°C for 15 min, and supernatants were used for protein immunoblot analysis. Samples were also collected for Actin and PYR1/PYL4 mRNA analyses to ensure that equal amounts of PYR1/PYL4 transcripts were expressed in different coinfiltrations.
For semi-in vivo protein degradation experiments, we expressed RFA1-GFP, 3HA-PYR1, and 3HA-PYL4 separately via different agroinfiltrations of N. benthamiana leaves. Samples were separately harvested, and proteins were extracted in native extraction buffer (50 mm Tris-HCl, pH 8, 0.5 m Suc, 1 mm MgCl2, 10 mm EDTA, 5 mm DTT, and protease inhibitor cocktail). Extracts were clarified by centrifugation as mentioned above, and a final concentration of 25 µm ATP was added to preserve the function of the 26S proteasome. Then equal amounts of HA-PYR1 or 3HA-PYL4 extracts were mixed with N. benthamiana protein extracts lacking (mock) or containing RFA1-GFP, and they were incubated at 4°C with rotation for 1 h in the presence or absence of 50 µm MG132. Samples were mixed with 5× Laemmli buffer and subjected to immunoblot analysis.
Protein Extraction, Analysis, Immunodetection, and Coimmunoprecipitation
Antibodies used in this work are listed in Supplemental Table S1. Protein extracts for immunodetection experiments were prepared from Arabidopsis transgenic lines expressing HA-tagged RFA1 or RFA4. Material (∼100 mg) for direct western-blot analysis was extracted in 2× Laemmli buffer (125 mm Tris-HCl, pH 6.8, 4% [w/v] SDS, 20% [v/v] glycerol, 2% [v/v] mercaptoethanol, and 0.001% [w/v] bromophenol blue), and proteins were run on a 10% SDS-PAGE gel and analyzed by immunoblotting. Proteins were transferred onto Immobilon-P membranes (Millipore) and probed with anti-HA-peroxidase (Roche). Immunodetection of ubiquitylated proteins was performed using anti-Ub antibody (Ub P4D1:sc-8017; Santa Cruz Biotechnology). Antibodies were used at a 1:1,000 dilution. Detection was performed using the ECL Advance Western Blotting Chemiluminiscent Detection Kit (GE Healthcare). Image capture was done using the image analyzer LAS3000, and quantification of the protein signal was done using Image Guache V4.0 software.
Coimmunoprecipitation experiments were performed by mixing protein extracts in lysis buffer from agroinfiltrated N. benthamiana leaves expressing GFP or GFP-PYL4 proteins with protein extracts from Arabidopsis transgenic lines expressing HA-tagged RFA1 or RFA4. GFP or GFP-PYL4 proteins were immunoprecipitated using superparamagnetic micro-MACS beads coupled to monoclonal anti-GFP antibody according to the manufacturer’s instructions (Miltenyi Biotec). Purified immunocomplexes were eluted in Laemmli buffer, boiled, and run on a 10% SDS-PAGE gel. Proteins immunoprecipitated with anti-GFP antibody were transferred onto Immobilon-P membranes (Millipore) and probed with anti-HA-peroxidase to detect coimmunoprecipitation of HA-tagged RFA1 or RFA4.
Pull-Down Assays
The construction, expression in bacteria, and purification of MBP fused to RFA1 or RFA4 lacking the C-terminal acidic domain was performed as described by Bueso et al. (2014). Purification of 6His-PYR1 was described by Santiago et al. (2009a). For pull-down assays, 5 μg of MBP-RFA1, MBP-RFA4ΔC, or MBP and 5 μg of either 6His-PYR1 or 6His-PYL4 were incubated 1 h at 4°C with constant rocking in 0.5 mL of binding buffer (20 mm Tris-HCl, pH 7.5, 200 mm NaCl, 1 mm EDTA, and 0.5% [v/v] Tween 20). Afterward, MBPs were purified using amylose affinity chromatography, eluted, and analyzed by SDS-PAGE, followed by western blotting and immunodetection using anti-His tag monoclonal antibodies (Roche).
In Vitro Ub Assay
Enzymatic reactions were performed on the column by using immunoprecipitated 3HA-PYR1 as a substrate, which was bound to superparamagnetic micro-MACS beads coupled to monoclonal anti-HA antibody. Immunoprecipitation of 3HA-PYR1 was performed according to the manufacturer’s instructions (Miltenyi Biotec) using Arabidopsis extracts of HA-tagged PYR1 lines containing 600 μg of total protein in 50 mm Tris-HCl, pH 8, 0.5 m Suc, 1 mm MgCl2, 10 mm EDTA, 5 mm DTT, 10 μm MG132, and protease inhibitor cocktail. The immobilized substrate was incubated at 30°C for 2 h in Ubi buffer (50 mm Tris, pH 7.5, 10 mm MgCl2, 0.2 mm DTT, 50 μm ZnCl2, and 5 mm ATP) containing 100 ng of human E1 (BostonBiochem), 250 ng of 6His-AtUBC8 (E2), 2 μg of either MBP-RFA1 or MBP-RFA4ΔC, and 1 µg of biotinylated Ub (Enzo Life Sciences). Reaction was stopped by washing the column with Ubi buffer followed by elution with Laemmli buffer. The eluted substrate was then subjected to SDS-PAGE/blotting followed by detection using either St-HRP or anti-HA-HRP to detect either ubiquitinated or 3HA-tagged proteins, respectively.
In Vivo Ubiquitination Assay in N. benthamiana
A. tumefaciens cultures containing constructs that express GFP-PYL4, GFP-RFA4 (diluted 1:4 with respect to the other cultures), GFP-UBC26, and the silencing suppressor p19 were coinfiltrated in N. benthamiana leaves. Control samples lacked GFP-RFA4 and GFP-UBC26. Three days after infiltration, samples were collected, ground in liquid nitrogen, and immediately placed in lysis buffer supplemented with 50 μm MG132 and inhibitors of deubiquitinases: 10 nm Ub aldehyde and 10 mm N-ethylmaleimide. GFP-tagged proteins were immunoprecipitated using superparamagnetic micro-MACS beads coupled to monoclonal anti-GFP antibody according to the manufacturer’s instructions (Miltenyi Biotec). Purified immunocomplexes were eluted in Laemmli buffer, boiled, and run on a 10% SDS-PAGE gel. Proteins immunoprecipitated with anti-GFP antibody were transferred onto Immobilon-P membranes (Millipore) and probed with anti-GFP, anti-PYL4, and anti-Ub antibodies (Supplemental Table S1).
Y2H Assays
Protocols were similar to those described previously (Saez et al., 2008). The screening to identify cognate E2s of RFA4 was performed as described previously (Cuéllar et al., 2013). In brief, the Saccharomyces cerevisiae PJ69-4A strain was cotransformed with pGADT7 and pGBKT7 vectors containing the bait and prey, respectively. Three individually transformed cultures were grown overnight in liquid SD medium lacking Leu and Trp, and a 10-fold dilution of these cultures was dropped on either control medium lacking Leu and Trp or selective medium additionally lacking His. Empty vectors were used as negative controls, and yeast was allowed to grow for 3 d at 30°C before interaction was scored.
Confocal Laser Scanning Microscopy
Confocal imaging was performed using a Zeiss LSM 780 AxioObserver.Z1 laser scanning microscope with a C-Apochromat 40×/1.20 W corrective water-immersion objective. The following fluorophores, which were excited and fluorescence emission detected by frame switching in the single or multitracking mode at the indicated wavelengths, were used in N. benthamiana leaf infiltration experiments: SCFP (405 nm/450–485 nm), GFP (488 nm/500–530 nm), YFP or SCFPC+VENUSN (488 nm/529–550 nm), OFP (561 nm/575–600 nm), and chlorophyll (561 nm/685–760 nm). For BiFC experiments involving the detection of two fluorescent proteins, sequential imaging of the fluorescent proteins was performed using the sequential channel acquisition mode. Pinholes were adjusted to 1 air unit for each wavelength. Postacquisition image processing was performed using ZEN (Zeiss Efficient Navigation) Lite 2012 imaging software and ImageJ (http://rsb.info.gov/ij/).
Seed Germination and Seedling Establishment Assays
After surface sterilization of the seeds, stratification was conducted in the dark at 4°C for 3 d. Approximately 100 seeds of each genotype were sown on MS plates supplemented with different ABA concentrations per experiment. To score seed germination, radicle emergence was analyzed at 72 h after sowing. Seedling establishment was scored as the percentage of seeds that developed green expanded cotyledons and the first pair of true leaves at 5 or 7 d.
Root Growth Assay
Seedlings were grown on vertically oriented MS plates for 4 to 5 d. Afterward, 20 plants were transferred to new MS plates lacking or supplemented with 10 µm ABA. The plates were scanned on a flatbed scanner after 10 d to produce image files suitable for quantitative analysis of root growth using the NIH Image software ImageJ.
Water-Loss Experiments
For water-loss analysis, plants were grown under greenhouse conditions (40% to 50% room humidity) and standard watering for 21 d. At least 10 similar leaves per individual genotype (n = 5) were excised and submitted to the drying atmosphere of a laminar flow hood. Gravimetric analysis of water loss was performed and represented as the percentage of fresh weight loss at each scored time point.
Infrared Thermography
Plants were grown in a controlled-environment growth chamber at 22°C under a 12-h-light/12-h-dark photoperiod at 100 μE m−2 s−1 and 40% to 50% room humidity. Philips bulbs were used (TL-D Super 8036W, white light 840, 4000K light code). Infrared thermography images of rosette leaves were acquired from 6-week-old plants with a thermal camera FLIR E95 equipped with a 42° lens. Images were processed and quantified with the FLIR tools software. For quantification, the average temperature of 15 different sections corresponding to four leaves per plant was calculated. Five plants per genotype were analyzed in each experiment. The mean temperature ± sd of all the plants for each genotype was reported. Statistical comparisons among genotypes were performed by pairwise Student’s t tests.
Statistics and Colocalization Analyses
Student’s t test was performed for single comparisons. Values are averages obtained from three independent experiments ± sd. Fluorescence colocalization analysis was performed using the PSC colocalization plug-in of ImageJ (French et al., 2008). Pearson’s and Spearman’s correlation coefficients in the range 0.4 to 1 indicate colocalization, whereas lower values indicate lack of colocalization.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers are as follows: RSL1, At2G26130; RFA1, At3G43750; RFA2, At3G45570; RFA3, At3G45580; RFA4, At2G21420; RFA5, At5G37560; RFA6, At3G45480; RFA7, At3G45460; RFA8, At3G45540; RFA9, At2g26135; UBC26, At1g53020; and UBC24, At2G33770.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression data of RFA genes compared with PYR/PYL genes.
Supplemental Figure S2. Amino acid sequence alignment of RSL1 (At2G26130), RFA1 (At3G43750), and RFA4 (At2G21420).
Supplemental Figure S3. Nuclear interaction of RFA4ΔC and ABA receptors.
Supplemental Figure S4. Simultaneous formation of receptor-RSL1 and receptor-RFA4 complexes in different subcellular locations revealed by multicolor BiFC assays.
Supplemental Figure S5. Analysis of Arabidopsis lines overexpressing different RSL1/RFA genes.
Supplemental Figure S6. Identification of Cys-361 as the putative active site of RFA4.
Supplemental Figure S7. Y2H assay between RFA4 or RFA4ΔC and a panel of 30 Arabidopsis E2 Ub-conjugating enzymes.
Supplemental Figure S8. Amino acid sequence alignment of UBC23, UBC24, UBC25, and UBC26 group XI UBCs.
Supplemental Figure S9. Y2H assay between RFA1 or RFA4ΔC and several ABA receptors.
Supplemental Figure S10. Generation and phenotype of CRISPR/CAS9-mediated knockout alleles of UBC26.
Supplemental Figure S11. The rfa1 and rfa4 single mutants show wild-type sensitivity to ABA.
Supplemental Figure S12. Specificity of α-PYR1 and α-PYL4 antibodies.
Supplemental Table S1. List of primers and antibodies used in this work.
Acknowledgments
We thank the ABRC and Nottingham Arabidopsis Stock Centre for providing DNA constructs and seeds. pHEE2E-TRI was a gift from Qi-Jun Chen (https://www.addgene.org/, plasmid #71288). We thank Marc Nishimura for sharing the unpublished vector GenBank accession number MG917725.
Footnotes
This work was supported by the Ministerio de Ciencia, Innovacion y Universidades(MICIU), Fondo Europeo de Desarrollo Regional, and Consejo Superior de Investigaciones Cientificas (grants BIO2014-52537-R and BIO2017-82503-R to P.L.R.), by a Juan de la Cierva contract from MICIU and by the Marie Skłodowska-Curie Action H2020-MSCA-IF-2015-707477 to J.L-J., by Programa VALi+d GVA APOSTD (2017/039 to B.B.-P.), by an FPI contract from MICIU (to J.J.) and by an FPU contract from MECD (to M.A.F.), and by the Belgian Science Policy Organization with postdoctoral fellowships to S.I.
Articles can be viewed without a subscription.
References
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Peirats-Llobet M, Pizzio GA, Fernandez MA, De Winne N, De Jaeger G, Dietrich D, Bennett MJ, et al. (2013) PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol 161: 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazon B, Gonzalez-Garcia MP, Lozano-Juste J, Coego A, Antoni R, Julian J, Peirats-Llobet M, Rodriguez L, Berbel A, Dietrich D, et al. (2018) PYL8 mediates ABA perception in the root through non-cell-autonomous and ligand-stabilization-based mechanisms. Proc Natl Acad Sci USA 115: E11857–E11863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazon B, Julian J, Coego A, Wu Q, Zhang X, Batistic O, Alquraishi SA, Kudla J, An C, Rodriguez PL (2019) ABA inhibits myristoylation and induces shuttling of the RGLG1 E3 ligase to promote nuclear degradation of PP2CA. Plant J 98: 813–825 [DOI] [PubMed] [Google Scholar]
- Belda-Palazon B, Rodriguez L, Fernandez MA, Castillo MC, Anderson EM, Gao C, Gonzalez-Guzman M, Peirats-Llobet M, Zhao Q, De Winne N, et al. (2016) FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell 28: 2291–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazón B, Ruiz L, Martí E, Tárraga S, Tiburcio AF, Culiáñez F, Farràs R, Carrasco P, Ferrando A (2012) Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS ONE 7: e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso E, Rodriguez L, Lorenzo-Orts L, Gonzalez-Guzman M, Sayas E, Muñoz-Bertomeu J, Ibañez C, Serrano R, Rodriguez PL (2014) The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J 80: 1057–1071 [DOI] [PubMed] [Google Scholar]
- Callis J. (2014) The ubiquitination machinery of the ubiquitin system. The Arabidopsis Book 12: e0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J, March PE, Lee R, Tillett D (2004) Site-directed, Ligase-Independent Mutagenesis (SLIM): A single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res 32: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cuéllar AP, Pauwels L, De Clercq R, Goossens A (2013) Yeast two-hybrid analysis of jasmonate signaling proteins. Methods Mol Biol 1011: 173–185 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J (1985) Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res 13: 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Sanchez-Barrena MJ, Gonzalez-Rubio JM, Rodriguez L, Fernandez D, Antoni R, Yunta C, Belda-Palazon B, Gonzalez-Guzman M, Peirats-Llobet M, et al. (2016) Calcium-dependent oligomerization of CAR proteins at cell membrane modulates ABA signaling. Proc Natl Acad Sci USA 113: E396–E405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich M, Mueller HM, Bauer H, Peirats-Llobet M, Rodriguez PL, Geilfus CM, Carpentier SC, Al Rasheid KAS, Kollist H, Merilo E, et al. (2019) The role of Arabidopsis ABA receptors from the PYR/PYL/RCAR family in stomatal acclimation and closure signal integration. Nat Plants 5: 1002–1011 [DOI] [PubMed] [Google Scholar]
- Dove KK, Stieglitz B, Duncan ED, Rittinger K, Klevit RE (2016) Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep 17: 1221–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Olszewski JL, Schuermann JP, Kurinov I, Miller DJ, Nourse A, Alpi AF, Schulman BA (2013) Structure of HHARI, a RING-IBR-RING ubiquitin ligase: Autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure 21: 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F, Santiago J, Betz K, Twycross J, Park SY, Rodriguez L, Gonzalez-Guzman M, Jensen MR, Krasnogor N, Blackledge M, et al. (2011) A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J 30: 4171–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. (2013) Abscisic acid synthesis and response. The Arabidopsis Book 11: e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP (2008) Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3: 619–628 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Zhuang X, Shen J, Jiang L (2017) Plant ESCRT complexes: Moving beyond endosomal sorting. Trends Plant Sci 22: 986–998 [DOI] [PubMed] [Google Scholar]
- García-León M, Cuyas L, El-Moneim DA, Rodriguez L, Belda-Palazón B, Sanchez-Quant E, Fernández Y, Roux B, Zamarreño AM, García-Mina JM, et al. (2019) Arabidopsis ALIX regulates stomatal aperture and turnover of abscisic acid receptors. Plant Cell 31: 2411–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C, Kaufholdt D, Hamisch D, Bikker R, Kudla J, Mendel RR, Hänsch R (2011) Quantitative analysis of dynamic protein-protein interactions in planta by a floated-leaf luciferase complementation imaging (FLuCI) assay using binary Gateway vectors. Plant J 67: 542–553 [DOI] [PubMed] [Google Scholar]
- Gehl C, Waadt R, Kudla J, Mendel RR, Hänsch R (2009) New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2: 1051–1058 [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Grondin A, Rodrigues O, Verdoucq L, Merlot S, Leonhardt N, Maurel C (2015) Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 27: 1945–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen ML, Iniesto E, Rodriguez L, Puga MI, Yanagawa Y, Pick E, Strickland E, Paz-Ares J, Wei N, De Jaeger G, et al. (2014) Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 26: 712–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian J, Coego A, Lozano-Juste J, Lechner E, Wu Q, Zhang X, Merilo E, Belda-Palazon B, Park SY, Cutler SR, et al. (2019) The MATH-BTB BPM3 and BPM5 subunits of Cullin3-RING E3 ubiquitin ligases target PP2CA and other clade A PP2Cs for degradation. Proc Natl Acad Sci USA 116: 15725–15734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsall IR, Duda DM, Olszewski JL, Hofmann K, Knebel A, Langevin F, Wood N, Wightman M, Schulman BA, Alpi AF (2013) TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes. EMBO J 32: 2848–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Scalf M, Smith LM, Vierstra RD (2013) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HC, Huibregtse JM (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol 29: 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Cheng J, Zhu Y, Ding Y, Meng J, Chen Z, Xie Q, Guo Y, Li J, Yang S, et al. (2015) Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat Commun 6: 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowarschik K, Hoehenwarter W, Marillonnet S, Trujillo M (2018) UbiGate: A synthetic biology toolbox to analyse ubiquitination. New Phytol 217: 1749–1763 [DOI] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang-Mladek C, Xie L, Nigam N, Chumak N, Binkert M, Neubert S, Hauser MT (2012) UV-B signaling pathways and fluence rate dependent transcriptional regulation of ARIADNE12. Physiol Plant 145: 527–539 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhang L, Li X, Kong X, Wang X, Li Y, Liu Z, Wang J, Li X, Yang Y (2018) AtRAE1 is involved in degradation of ABA receptor RCAR1 and negatively regulates ABA signalling in Arabidopsis. Plant Cell Environ 41: 231–244 [DOI] [PubMed] [Google Scholar]
- Li H, Li Y, Zhao Q, Li T, Wei J, Li B, Shen W, Yang C, Zeng Y, Rodriguez PL, et al. (2019) The plant ESCRT component FREE1 shuttles to the nucleus to attenuate abscisic acid signalling. Nat Plants 5: 512–524 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang L, Li D, Liu Z, Wang J, Li X, Yang Y (2016) The Arabidopsis F-box E3 ligase RIFP1 plays a negative role in abscisic acid signalling by facilitating ABA receptor RCAR3 degradation. Plant Cell Environ 39: 571–582 [DOI] [PubMed] [Google Scholar]
- Liu H, Stone SL (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Emr SD (2012) Ubiquitin and membrane protein turnover: From cradle to grave. Annu Rev Biochem 81: 231–259 [DOI] [PubMed] [Google Scholar]
- Marín I. (2010) Diversification and specialization of plant RBR ubiquitin ligases. PLoS ONE 5: e11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. (2009) Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Moreno-Alvero M, Yunta C, Gonzalez-Guzman M, Lozano-Juste J, Benavente JL, Arbona V, Menéndez M, Martinez-Ripoll M, Infantes L, Gomez-Cadenas A, et al. (2017) Structure of ligand-bound intermediates of crop ABA receptors highlights PP2C as necessary ABA co-receptor. Mol Plant 10: 1250–1253 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirats-Llobet M, Han SK, Gonzalez-Guzman M, Jeong CW, Rodriguez L, Belda-Palazon B, Wagner D, Rodriguez PL (2016) A direct link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol Plant 9: 136–147 [DOI] [PubMed] [Google Scholar]
- Pizzio GA, Rodriguez L, Antoni R, Gonzalez-Guzman M, Yunta C, Merilo E, Kollist H, Albert A, Rodriguez PL (2013) The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol 163: 441–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K, et al. (2013) Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun 4: 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez L, Gonzalez-Guzman M, Diaz M, Rodrigues A, Izquierdo-Garcia AC, Peirats-Llobet M, Fernandez MA, Antoni R, Fernandez D, Marquez JA, et al. (2014) C2-domain abscisic acid-related proteins mediate the interaction of PYR/PYL/RCAR abscisic acid receptors with the plasma membrane and regulate abscisic acid sensitivity in Arabidopsis. Plant Cell 26: 4802–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PL, Lozano-Juste J, Albert A (2019) PYR/PYL/RCAR ABA receptors In Seo M, and Marion-Poll A, eds, Advances in Botanical Research, Vol 92 Academic Press, London, p 51–82 [Google Scholar]
- Romero-Barrios N, Vert G (2018) Proteasome-independent functions of lysine-63 polyubiquitination in plants. New Phytol 217: 995–1011 [DOI] [PubMed] [Google Scholar]
- Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S (2010) Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev 24: 1010–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Rodrigues A, Santiago J, Rubio S, Rodriguez PL (2008) HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20: 2972–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA (2009a) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL (2009b) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Scott DC, Rhee DY, Duda DM, Kelsall IR, Olszewski JL, Paulo JA, de Jong A, Ovaa H, Alpi AF, Harper JW, et al. (2016) Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell 166: 1198–1214.e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E (2010) Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J 61: 25–35 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal 6: rs8. [DOI] [PubMed] [Google Scholar]
- Vazquez-Vilar M, Quijano-Rubio A, Fernandez-Del-Carmen A, Sarrion-Perdigones A, Ochoa-Fernandez R, Ziarsolo P, Blanca J, Granell A, Orzaez D (2017) GB3.0: A platform for plant bio-design that connects functional DNA elements with associated biological data. Nucleic Acids Res 45: 2196–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waese J, Fan J, Pasha A, Yu H, Fucile G, Shi R, Cumming M, Kelley LA, Sternberg MJ, Krishnakumar V, et al. (2017) ePlant: Visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 29: 1806–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, Zhu JK (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA 110: 11205–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Klevit RE (2012) Following Ariadne’s thread: A new perspective on RBR ubiquitin ligases. BMC Biol 10: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE (2011) UBCH7 reactivity profile reveals Parkin and HHARI to be RING/HECT hybrids. Nature 474: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang X, Peirats-Llobet M, Belda-Palazon B, Wang X, Cui S, Yu X, Rodriguez PL, An C (2016) Ubiquitin ligases RGLG1 and RGLG5 regulate abscisic acid signaling by controlling the turnover of phosphatase PP2CA. Plant Cell 28: 2178–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N (2009) Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol 16: 1230–1236 [DOI] [PubMed] [Google Scholar]
- Yu F, Lou L, Tian M, Li Q, Ding Y, Cao X, Wu Y, Belda-Palazon B, Rodriguez PL, Yang S, et al. (2016) ESCRT-I component VPS23A affects ABA signaling by recognizing ABA receptors for endosomal degradation. Mol Plant 9: 1570–1582 [DOI] [PubMed] [Google Scholar]
- Yu F, Xie Q (2017) Non-26S proteasome endomembrane trafficking pathways in ABA signaling. Trends Plant Sci 22: 976–985 [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhao L, Zhang M, Zafar SA, Fang J, Li M, Zhang W, Li X (2017) Arabidopsis E3 ubiquitin ligases PUB22 and PUB23 negatively regulate drought tolerance by targeting ABA receptor PYL9 for degradation. Int J Mol Sci 18: E1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Tian M, Li Q, Cui F, Liu L, Yin B, Xie Q (2013) A plant-specific in vitro ubiquitination analysis system. Plant J 74: 524–533 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y, et al. (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]