ER quality control mediated by ERdj3B is important for anther development at high temperature in Arabidopsis.
Abstract
Pollen development is highly sensitive to heat stress, which impairs cellular proteostasis by causing misfolded proteins to accumulate. Therefore, each cellular compartment possesses a dedicated protein quality control system. An elaborate quality control system involving molecular chaperones, including immunoglobulin-binding protein (BiP), heat shock protein70, and regulatory J domain-containing cochaperones (J proteins), allows the endoplasmic reticulum (ER) to withstand a large influx of proteins. Here, we found that Arabidopsis (Arabidopsis thaliana) mutants of ER-localized DnaJ family 3B (ERdj3B), one of three ER-resident J proteins involved in ER quality control, produced few seeds at high temperatures (29°C) due to defects in anther development. This temperature-sensitive fertility defect is specific to the defective interactions of BiP with ERdj3B but not with the other two J proteins, indicating functional differences between ERdj3B and the other J proteins. RNA sequencing analysis revealed that heat stress affects pollen development in both wild-type and mutant buds, but the erdj3b mutant is more susceptible, possibly due to defects in ER quality control. Our results highlight the importance of a specific ER quality control factor, ERdj3B, for plant reproduction, particularly anther development, at high temperatures.
Plants are exposed to various environmental stresses, including high temperature, drought, and salinity. Plant reproduction, particularly the development and functioning of the male gametophyte, pollen, is the most temperature-sensitive process during a plant’s life cycle. Exposure to both short-term high temperatures and long-term mildly elevated temperatures leads to defects in pollen development (Rieu et al., 2017). High-temperature stress affects protein structures and can lead to the accumulation of misfolded proteins. These misfolded proteins, which have a serious impact on cellular proteostasis, must be handled by the protein quality control systems that operate in each cellular compartment.
The endoplasmic reticulum (ER) is the entry site of the secretory pathway for approximately 30% of cellular proteins and is thus exposed to large protein influxes (Vembar and Brodsky, 2008). The ER is therefore equipped with an elaborate quality control system to constantly monitor the folding states of newly synthesized and imported proteins and to prevent their potential misfolding in the ER (Ellgaard and Helenius, 2003). ER quality control is one of the most important mechanisms for thermotolerance during pollen development (Fragkostefanakis et al., 2016; Rieu et al., 2017). The accumulation of unfolded or misfolded proteins in the ER lumen leads to increased expression of the components of the ER quality control system via a mechanism known as the unfolded protein response (UPR; Iwata and Koizumi, 2012), involving the ER-localized sensor protein, inositol-requiring enzyme1 (IRE1). The UPR is affected by heat stress in plants, and the Arabidopsis (Arabidopsis thaliana) ire1 mutant shows defects in male reproduction at high temperatures (Deng et al., 2016).
Molecular chaperones in the ER, including immunoglobulin-binding protein (BiP), calnexin, and calreticulin, play important roles in ER quality control. BiP, a major heat shock protein70 (Hsp70) molecular chaperone in the ER, plays key roles in ER quality control (Nishikawa et al., 2005). Hsp70 chaperones bind to and dissociate from their client proteins via an ATP-regulated cycle. This chaperone cycle is regulated by cofactors of Hsp70 (Bukau et al., 2006). J domain-containing cochaperones (J proteins) are a major class of cofactors of Hsp70, which interact with Hsp70 through the well-conserved J domain (Kampinga and Craig, 2010). Arabidopsis has three luminal ER-resident J proteins, ERdj3A, ERdj3B, and P58IPK, which are thought to function in ER quality control as cofactors of BiP (Yamamoto et al., 2008).
In this study, we analyze Arabidopsis T-DNA mutants of the ERDJ3A, ERDJ3B, and P58IPK genes. The erdj3b mutant showed defects in anther development, which resulted in reduced seed production at an elevated temperature of 29°C. This seed production defect was observed in the erdj3b mutant but not in erdj3a or p58ipk, suggesting that ER quality control mediated by a specific cofactor of BiP is involved in thermotolerance during pollen development. RNA sequencing (RNA-seq) analysis showed that a high temperature caused reduced expression of pollen development-related genes in both wild-type and erdj3b anthers, but the erdj3b anthers were highly susceptible to heat stress. Therefore, the heat-induced low seed production phenotype of the erdj3b mutant is most likely caused by a combination of heat injury during pollen development and the increased vulnerability of erdj3b anthers to heat due to defects in ER quality control.
RESULTS
The erdj3b Mutants Exhibit Reduced Seed Production at a High Temperature
Pollen development and functioning are among the most heat-sensitive processes in the plant life cycle (Rieu et al., 2017). High-temperature stress affects protein structures; therefore, we analyzed the reproductive growth phenotypes of Arabidopsis mutants deficient in J proteins in the ER lumen at high temperature. We found that the erdj3b mutants produced small siliques with very reduced seed sets when they were exposed to the high temperature of 29°C during their reproductive growth phase (Fig. 1A). While wild-type plants grown at 29°C produced 28.1 ± 6.1 seeds per silique (n = 8), the erdj3b-1 and erdj3b-2 mutants produced only 3.6 ± 1.5 and 6.3 ± 2.3 seeds per silique (n = 8), respectively (Supplemental Fig. S1). The mutant plants did not show any obvious defects in flower development or fertility at 22°C (Supplemental Figs. S1 and S2, A and B; Yamamoto et al., 2008; Maruyama et al., 2014). The seed production defect of the erdj3b-1 mutant at 29°C was alleviated by introducing a construct expressing the ERDJ3B gene from the ERDJ3B promoter (3Bpro:3B; Supplemental Figs. S1 and S3A), indicating that the erdj3b mutation caused the low seed yield phenotype at 29°C.
Figure 1.
Phenotypes of wild-type, erdj3b-1, and erdj3b-2 plants grown at 29°C. A, Inflorescences of wild-type (WT), erdj3b-1 (3b-1), and erdj3b-2 (3b-2) plants grown at 29°C. Seed numbers per silique are shown in Supplemental Figure S1. Bar = 10 mm. B, Flowers observed by stereomicroscopy. Bar = 0.5 mm. C, Mature anthers observed by stereomicroscopy. Note that erdj3b mutant anthers dehisce normally and pollen grains are observed on the anthers. Bar = 100 μm. D, Self-pollinated stigmas stained by DiOC2 and observed using differential interference contrast microscopy. Pollen grains were visualized by DiOC2 staining, which stains pollen exine. Arrows indicate pollen grains on the mutant stigmas (Regan and Moffatt, 1990). Bar = 100 μm. E, Number of pollen grains on stigmas of wild-type (n = 14), erdj3b-1 (n = 16), and erdj3b-2 (n = 15) plants grown at 29°C. Each dot shows the number of pollen grains on each stigma; the circle indicates an outlier. Statistical differences were calculated using the Tukey-Kramer method. Different letters indicate statistically significant differences between results at P < 0.05.
Production of seeds in the erdj3a and p58ipk mutants, which were deficient in other ER-resident luminal J proteins, was similar to that in wild-type plants at 29°C (Supplemental Fig. S3B). The erdj3b mutant lines harboring constructs that expressed ERdj3A or P58IPK under the control of the ERDJ3B promoter had short siliques and exhibited low seed production at 29°C (Supplemental Figs. S1 and S3C). Immunoblotting showed expression of ERdj3A and P58IPK from the ERDJ3B promoter in flower buds of the transgenic plants at both 22°C and 29°C (Supplemental Fig. S3D). These results indicate that expression of ERdj3A or P58IPK did not suppress the reduced fertility phenotype of the erdj3b mutant at 29°C; ERdj3B is functionally distinct from other J proteins in the ER lumen and is required to maintain fertility at 29°C.
erdj3b Mutant Plants Grown at 29°C Are Defective in Pollen Release and Accumulate Aberrant Materials on the Pollen Grain Surface
When grown at 29°C, the erdj3b mutants exhibited normal floral development (Fig. 1, B and C). Although the mutant flowers contained dehisced anthers with pollen grains (Fig. 1C), only a few pollen grains were found on the stigmas of self-pollinated mutant flowers grown at 29°C (Fig. 1D); while 44.1 ± 14.4 pollen grains were found on the stigmas of wild-type flowers (n = 14), only 4.4 ± 7.1 and 2.6 ± 3.2 pollen grains were found on the stigmas of erdj3b-1 (n = 16) and erdj3b-2 (n = 15) flowers, respectively (Fig. 1E). By contrast, when plants were grown at 22°C, the numbers of pollen grains found on the stigmas of self-pollinated erdj3b-1 and erdj3b-2 mutants were similar to those found on wild-type plants (Supplemental Fig. S2C). These results strongly suggest that the low seed yield in the erdj3b mutant at 29°C is due to pollination defects. Similar numbers of pollen grains were found on the stigmas of heterozygous erdj3b/+ and wild-type plants grown at 29°C (Supplemental Fig. S4), indicating that lesions in sporophytic tissue(s) caused the pollination defect in erdj3b.
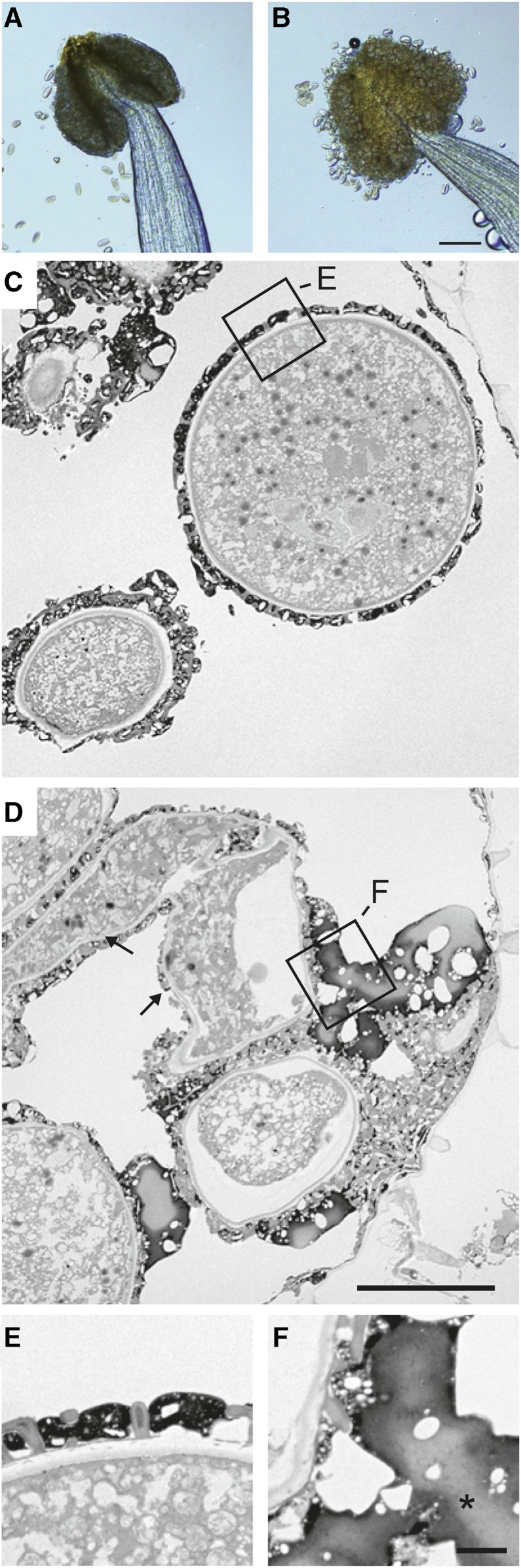
Whereas pollen grains were released from wild-type anthers (Fig. 2A), pollen grains in mutant anthers tightly adhered to anther tissues (Fig. 2B), indicating that the pollination defect seen at 29°C in the erdj3b mutant was mainly caused by defective pollen release from the anthers. We used transmission electron microscopy to analyze dehisced anthers (anther stage 13) from wild-type and erdj3b mutant plants grown at 29°C. We observed collapsed pollen grains in anthers from the erdj3b mutant (Fig. 2D; Supplemental Fig. S5, B and C, arrows) but not in wild-type anthers (Fig. 2C; Supplemental Fig. S5A). We sometimes observed pollen grains and collapsed pollen grains in contact with the anther wall in erdj3b mutants, suggesting pollen adhesion to anther tissue (Fig. 2D; Supplemental Fig. S5, B and C). Transmission electron microscopy also revealed abnormalities in the pollen coat of the erdj3b mutant grown at 29°C (Fig. 2, D and F); large clumps of electron-dense materials were deposited on the surfaces of pollen grains, which were noticeably absent from wild-type pollen grains (Fig. 2, C and E). These electron-dense materials were continuous with the pollen coat deposited within the cavities of the exine layer, suggesting that they were related to the pollen coat. However, these materials were markedly different from those of the wild-type pollen coat (Fig. 2E). Electron-lucent regions were occasionally observed inside the electron-dense clumps (Fig. 2F, asterisk). The large clumps appeared to join pollen grains together and to join pollen grains to anther walls (Fig. 2, D and F). The surface of pollen grains in erdj3b anthers lacked parts of the pollen coat. We analyzed three independent sections of erdj3b anthers and observed such abnormal pollen grains and pollen coats in all cases (Fig. 2D; Supplemental Fig. S5, B and C).
Figure 2.
erdj3b anthers show defects in pollen release. Mature anthers of wild-type (A, C, and E) and erdj3b-1 (B, D, and F) plants grown at 29°C are shown. A and B, Pollen grains released from an anther observed by light microscopy. Bar = 100 μm. C to F, Thin sections of mature anthers observed by transmission electron microscopy. The regions boxed in C and D are magnified in E and F, respectively. In D, arrows indicate collapsed pollen grains. In F, the asterisk indicates aberrant deposition. Bars = 10 μm (C and D) and 1 μm (E and F).
We next analyzed germination of the pollen grains found on the stigmas of erdj3b-1 and erdj3b-2 plants grown at 29°C. Pollen tubes were stained with Aniline Blue, which binds specifically to β-1,3-glucan (callose; Kho and Baër, 1968). Pistils of self-pollinated flowers were collected from wild-type, erdj3b-1, and erdj3b-2 plants grown at 22°C or 29°C at 1 d postflowering and fixed and stained with Aniline Blue. When plants were grown at 22°C, approximately 100 pollen tubes were observed on each stigma of wild-type, erdj3b-1, and erdj3b-2 plants (Supplemental Fig. S6, A and B). No significant differences were observed between wild-type, erdj3b-1, and erdj3b-2 pistils in the number of pollen tubes per stigma (Supplemental Fig. S6B). The number of pollen tubes observed on a pistil from a wild-type plant grown at 29°C was 56.1 ± 18.1 (Supplemental Fig. 6, A and B), which was similar to the number of pollen grains found on the stigma (Fig. 1E). We also counted the number of pollen tubes developing on the pistils of mutant plants grown at 29°C (Supplemental Fig. S6A) and found that the mean numbers of pollen tubes on the pistils of erdj3b-1 and erdj3b-2 plants were 3.1 ± 6.2 and 8 ± 8.7, respectively (Supplemental Fig. S6B). Notably, these values were similar to the numbers of pollen grains found on the stigmas of mutant plants grown at 29°C (Fig. 1E). These results indicate that pollen grains deposited on the stigmas of erdj3b-1 and erdj3b-2 plants retained their viability and could germinate.
ERdj3B Is Important for Anther Development at 29°C
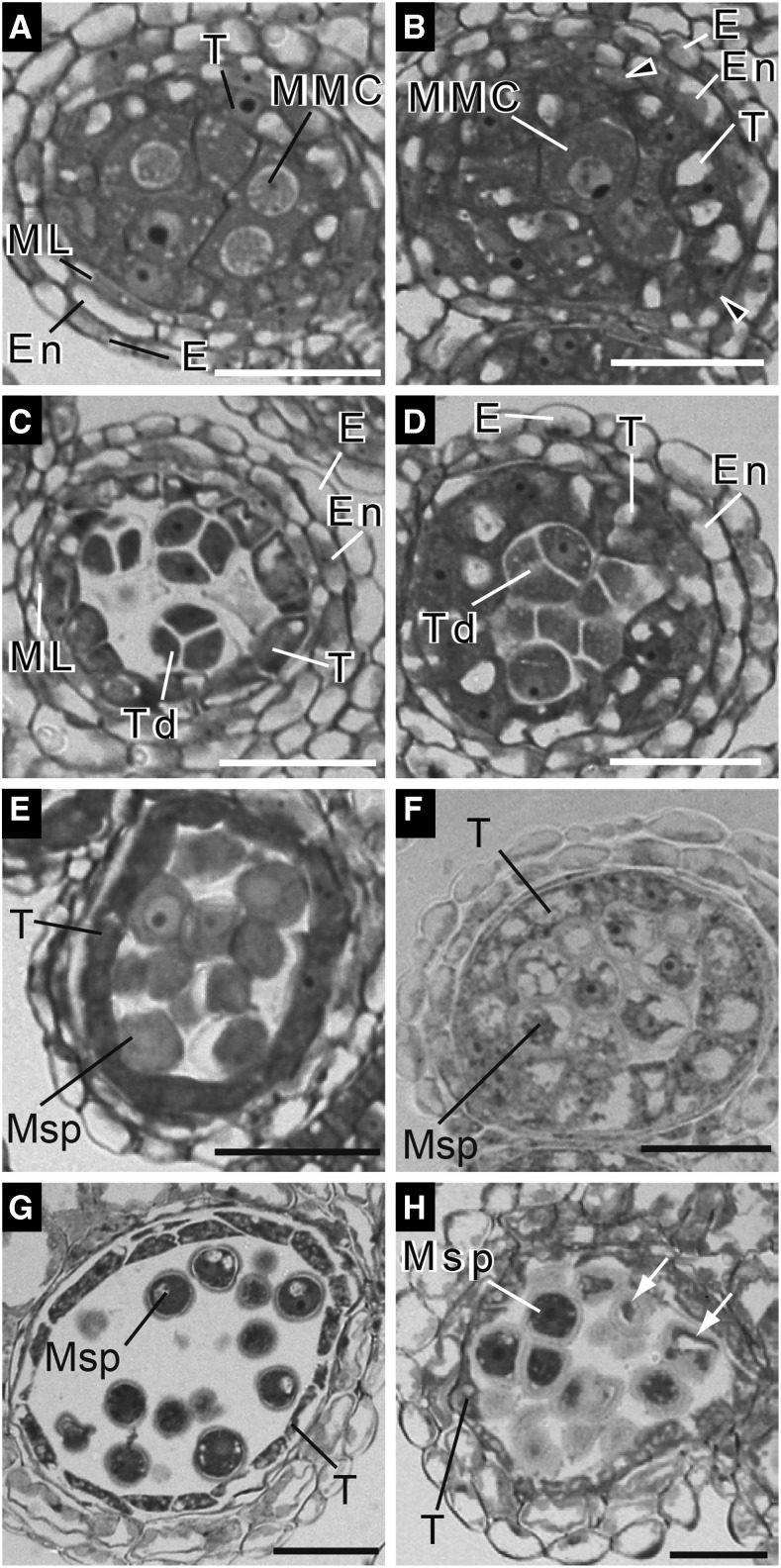
We analyzed anther development in wild-type and erdj3b plants grown at 29°C by examining semithin cross sections to detect the pollination defect of the erdj3b mutant (Fig. 3). Anther development in wild-type Arabidopsis can be divided into 14 stages (Sanders et al., 1999). Stage 5 anthers contain microspore mother cells surrounded by four layers of different cell types, including (from inside to outside) the tapetum, middle layer, endothecium, and epidermis (Fig. 3A; Supplemental Fig. S7A). Meiosis occurs between stages 5 and 7, generating tetrads consisting of four haploid microspores surrounded by callose walls (Fig. 3C; Supplemental Fig. S7C). At stage 8 of anther development, the callose walls degenerate and microspores are released into the anther locule (Fig. 3E). In anthers of wild-type plants grown at both 22°C and 29°C, tapetum cells were vacuolated from stage 6 onward and started to collapse at stage 9 of anther development (Fig. 3, C, E, and G; Supplemental Fig. S7, A, C, and E; Sanders et al., 1999). Anther development in the erdj3b mutant grown at 22°C was indistinguishable from that in wild-type plants (Supplemental Fig. S7, B, D, and F).
Figure 3.
Developing anthers of the erdj3b mutant grown at 29°C show tapetal hypertrophy. Semithin sections of anthers prepared from wild-type (A, C, E, and G) and erdj3b-1 (B, D, F, and H) plants grown at 29°C, stained with Toluidine Blue, and observed by differential interference contrast microscopy are shown. Sections of anthers at stage 5 (A and B), stage 7 (C and D), stage 8 (E and F), and stage 9 (G and H) are shown. E, Epidermis; En, endothecium; ML, middle layer; MMC, microspore mother cells; Msp, microspores; T, tapetum cell; Td, tetrads. Arrowheads in B indicate aberrant middle layer-like cells. In H, arrows indicate collapsed pollen grains. Bars = 30 μm.
In stage 5 to 8 anthers of the erdj3b mutant grown at 29°C, tapetum cells became abnormally enlarged and vacuolated (Fig. 3, B, D, and F). The mutant tapetum layer in stage 9 anthers was even thicker than that of wild-type cells at the same stage (Fig. 3, G and H). The mutant anther walls at stages 5 and 7 consisted of a tapetum, endothecium, and epidermis but lacked the middle layer (Fig. 3, B and D). We occasionally observed cells morphologically similar to those in the middle layer between the tapetum and endothecium layers (Fig. 3B, arrowheads). However, the middle layer-like cells did not form a layer. Microspore mother cells underwent meiosis in mutant anthers, producing microspore tetrads at stage 7 (Fig. 3D). Callose degeneration also took place at stage 8, releasing microspores into the anther locule (Fig. 3F). In contrast to wild-type anthers, collapsed or shrunken microspores were visible in mutant anthers at stage 9 (Fig. 3H, white arrows).
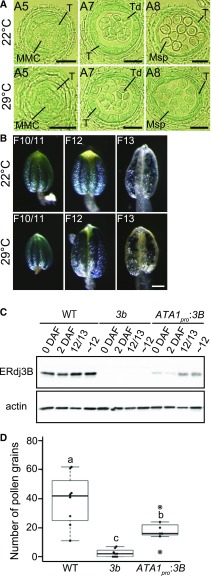
Since abnormal anther development was observed in the erdj3b mutant at 29°C, we compared the expression patterns of ERDJ3B in the developing anthers of plants grown at 22°C and 29°C using transgenic plants expressing GUS driven by the endogenous ERDJ3B promoter (Fig. 4). A cross section of GUS-stained anthers from a plant grown at 22°C showed GUS signals in all anther tissues, including tapetum cells, microspores, and anther locules, at anther stage 5 (Fig. 4A). Strong GUS signals in tapetum cells were observed at anther stages 7 and 8 (Fig. 4A). GUS signals were also observed in microspores at anther stages 7 and 8 (Fig. 4A). Although abnormal phenotypes were observed in anthers at stages 5, 7, and 8 from erdj3b mutants grown at 29°C (Fig. 3, B, D, and F), the GUS staining patterns in anthers of transgenic plants grown at 29°C were indistinguishable from those in anthers of transgenic plants grown at 22°C (Fig. 4A). We also analyzed the pattern of GUS expression in anthers at later stages of development. Strong GUS signals were observed in anthers at flower stage 10/11, which corresponds to anther stages 9 and 10, in plants grown at either 22°C or 29°C (Fig. 4B, left images; Sanders et al., 1999). GUS signals were observed mainly in pollen grains at flower stage 13 (Fig. 4B, middle), consistent with tapetum degeneration during flower stage 12 (Sanders et al., 1999). The patterns of GUS staining were similar in anthers of plants grown at 22°C and 29°C (Fig. 4B), which suggested that plant growth at an elevated temperature did not affect ERDJ3B expression.
Figure 4.
Expression of ERDJ3B in tapetum cells. A, Semithin sections of GUS-stained anthers at various stages of development from transformants grown at 22°C (top row) or 29°C (bottom row), observed using differential interference contrast microscopy. Anthers at anther stage 5 (A5), anther stage 7 (A7), and anther stage 8 (A8) are shown. MMC, Microspore mother cells; Msp, microspores; T, tapetum cell; Td, tetrads. Bars = 20 μm. B, The expression patterns of anthers at flower stage 10/11 (F10/11), flower stage 12 (F12), and flower stage 13 (F13) in transformants expressing the GUS gene driven by the ERDJ3B promoter grown at 22°C (top row) or 29°C (bottom row). Bar = 100 μm. C, Immunoblots of total proteins extracted from samples collected 0 d after flowering (0 DAF) and 2 DAF, from flower buds at flower stages 12 and 13 (12/13), and from flower buds prior to flower stage 12 (∼12) from the wild type (WT), erdj3b-1 [3b], and erdj3b-1 harboring the ATA1pro:3B transgene [ATA1pro:3B] grown at 22°C. Samples were analyzed by SDS-PAGE and immunoblotting with anti-ERdj3B and anti-actin antibodies. D, Number of pollen grains on stigmas of the wild type, erdj3b-1, and erdj3b-1 harboring the ATA1pro:3B transgene; plants were grown at 29°C (n = 8, 8, and 7, respectively). Dots represent the number of pollen grains on each stigma; circles indicate outliers. Statistical differences were calculated using the Tukey-Kramer method. P < 0.05 is indicated by different letters.
ERDJ3B was strongly expressed in tapetum cells at anther stages 8 and 9; therefore, we investigated whether the functions of ERdj3B in tapetum cells are important for fertility at 29°C. We constructed a fusion gene, ATA1pro:3B, which allowed ERDJ3B to be expressed under the control of the ATA1 promoter, which is active in the tapetum at anther stages 7 to 10 (Lebel-Hardenack et al., 1997). Immunoblotting revealed ERdj3B expression in flower buds at flower stage 12/13 and before flower stage 12 (Fig. 4C). We analyzed the numbers of pollen grains on the stigmas of self-pollinated plants grown at 29°C. A mean of 18.1 pollen grains were attached to the self-pollinated stigmas of erdj3b plants harboring the ATA1pro:3B transgene (n = 7), whereas 2.5 pollen grains were found on stigmas of erdj3b (n = 8; Fig. 4D). This result indicates that ERDJ3B expression in tapetum cells partially suppresses the pollination defects of erdj3b at 29°C [Fig. 4D, 3b(ATA1pro:3B)]. However, ERDJ3B expression from the ATA1 promoter was not sufficient to suppress the seed production defect of the erdj3b mutant grown at 29°C (Supplemental Fig. S1).
Abrogation of ERdj3B-BiP Interactions Causes a Fertility Defect at High Temperature
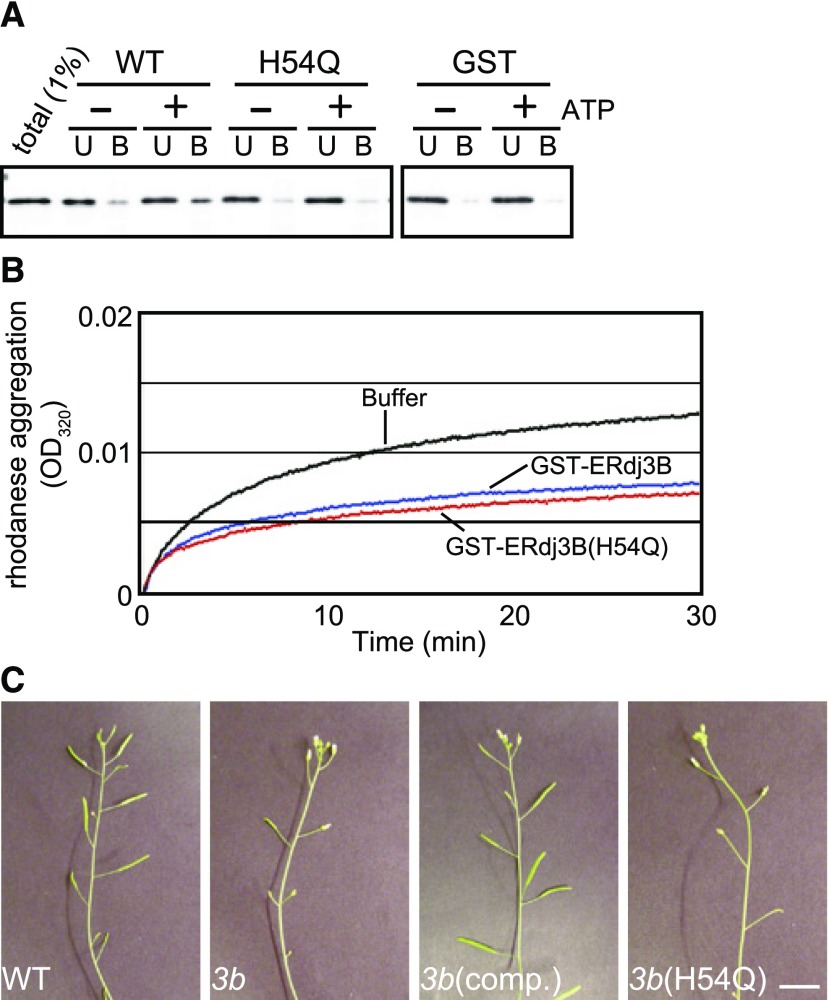
We next asked whether the role of ERdj3B as a partner for BiP is required for pollination at 29°C. J proteins generally interact with Hsp70 molecular chaperones through the well-conserved J domain and bind to client proteins of Hsp70 as well. To determine whether ERdj3B would exhibit such Hsp70-assisting activities, we expressed a fusion protein comprising glutathione S-transferase (GST) and ERdj3B lacking its signal sequence (residues 23–346) in Escherichia coli cells and purified it by affinity chromatography (Fig. 5). Pull-down assays showed that BiP was recovered from detergent extracts of Arabidopsis seedlings in the presence of ATP with GST-ERdj3B but not with GST (Fig. 5A). A highly conserved His-Pro-Gln sequence in the J domain plays an essential role in the interaction between J proteins and Hsp70 (Walsh et al., 2004), and a His-to-Gln substitution of this sequence abolishes the interactions between J proteins and Hsp70, including yeast (Saccharomyces cerevisiae) Ydj1/Ssa1 and Jem1/BiP (Tsai and Douglas, 1996: Makio et al., 2008). The H54Q substitution in the His-Pro-Asp sequence of GST-ERdj3B substantially reduced the amount of coprecipitated BiP (Fig. 5A). In addition to binding to BiP, we investigated whether the recombinant GST-ERdj3B is functional as a cochaperone by performing a chaperone assay with rhodanese. Chemically unfolded rhodanese produces aggregates after being diluted in buffer lacking a denaturant, and this aggregation is suppressed by the E. coli DnaJ protein (Langer et al., 1992). Purified GST-ERdj3B also suppressed the aggregation of chemically denatured rhodanese (Fig. 5B). GST-ERdj3B containing the H54Q mutation also suppressed the aggregation of rhodanese. Therefore, the H54Q mutation impairs the interaction between ERdj3B and BiP but does not affect its binding to client proteins. We next examined whether these observations are also relevant in planta. We found that ERDJ3B containing the H54Q mutation failed to complement the impaired fertility of erdj3b plants at 29°C (Fig. 5C; Supplemental Fig. S1). Therefore, the interaction of ERdj3B with BiP is essential for maintaining fertility at 29°C.
Figure 5.
Interactions between ERdj3B and BiP are responsible for fertility at 29°C. A, GST-ERdj3B protein (WT), GST-ERdj3B(H54Q) mutant protein (H54Q), or GST alone was immobilized on glutathione Sepharose beads and incubated with total lysate prepared from Arabidopsis seedlings with (+) or without (−) 2 mm ATP. The beads were washed as described in “Materials and Methods,” and the eluate was analyzed by SDS-PAGE and immunoblotting using anti-BiP antibodies. U indicates proteins recovered in the unbound (1%) fraction; B indicates proteins recovered in the glutathione Sepharose beads (100%) fraction. B, Rhodanese was denatured at 46 μm in 6 m guanidinium-HCl for 60 min at room temperature. Unfolded rhodanese was diluted 100-fold (0.46 μm final concentration) in 50 mm Tris-HCl, pH 7.4, 1 mm CaCl2, and 150 mm NaCl at 23°C in the absence (black line) or presence of 2.3 μm GST-ERdj3B (blue line) or GST-ERdj3B(H54Q) (red line). Aggregation (turbidity) was measured at 320 nm. C, Inflorescences of wild-type, erdj3b-1 [3b], and erdj3b-1 plants transformed with a construct expressing ERdj3B [3b(comp.)] and a construct expressing ERdj3B(H54Q) [3b(H54Q)] grown at 29°C. Note that the expression of ERdj3B(H54Q) failed to restore the temperature-sensitive low seed yield phenotype of the erdj3b-1 mutant. Seed numbers per silique are shown in Supplemental Figure S1. Bar = 10 mm.
Loss of ERdj3B Function Affects the Expression of Pollen Coat Protein Genes
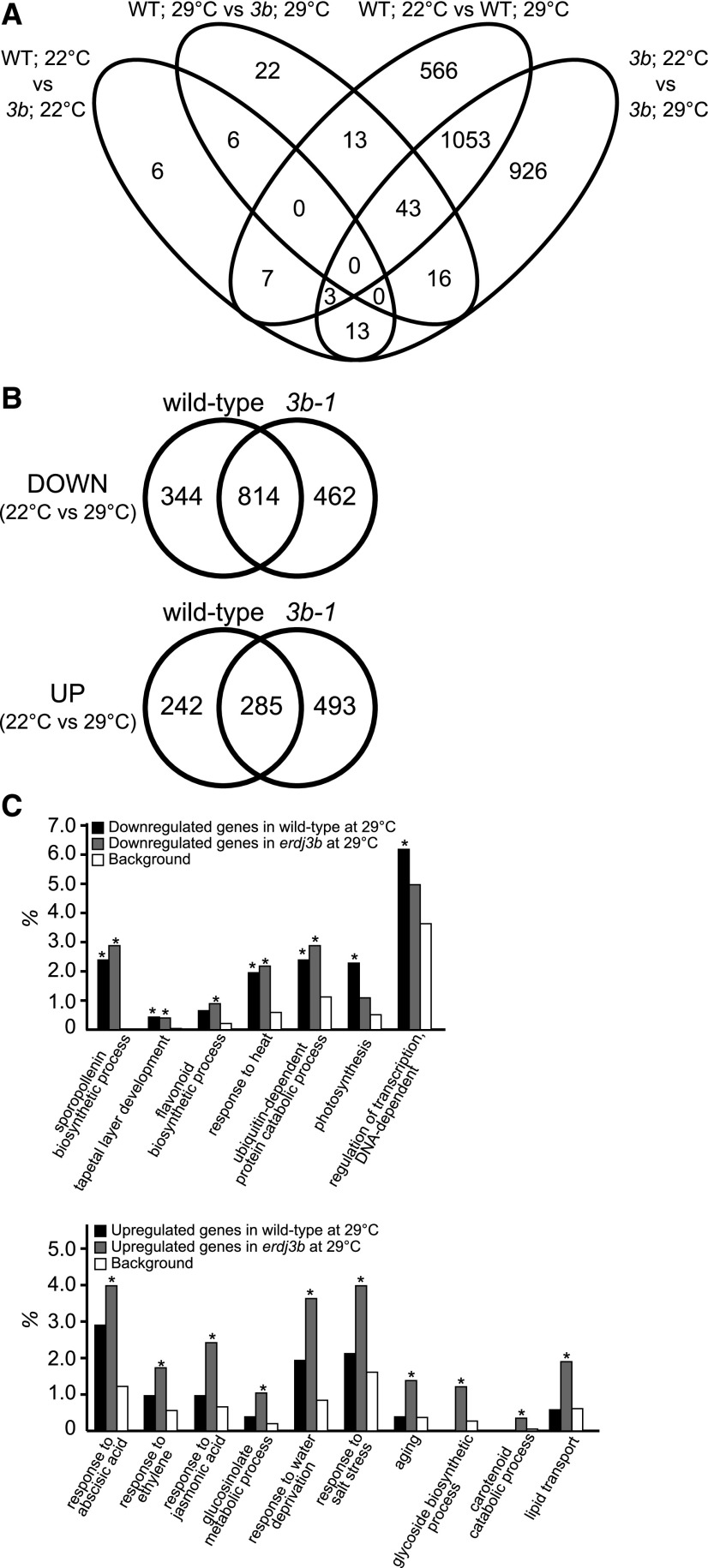
We used a transcriptomic approach to identify mechanisms underlying the anther developmental defect of the erdj3b mutant under elevated temperature. We analyzed the effects of elevated temperature on gene expression in wild-type and erdj3b mutant anthers during pollen development (Fig. 6). Total RNA fractions were prepared from anthers from stage 8 to 13 flowers of wild-type and erdj3b mutant plants grown at 22°C and 29°C and subjected to RNA-seq. We included three biological replicates for each condition, and the results were highly reproducible (Supplemental Fig. S8). When plants were grown at 22°C, only 35 genes were identified as differentially expressed between wild-type and erdj3b plants, with a false discovery rate (FDR) < 0.01; when plants were grown at 29°C, this number increased to 100 (Fig. 6A). By contrast, we identified 1,685 genes as differentially expressed between wild-type plants grown at 22°C and 29°C and 2,054 genes as differentially expressed between erdj3b mutants grown at 22°C and 29°C, with an FDR < 0.01 (Fig. 6A; Supplemental Data Sets S1–S6). Since growth temperature had a greater effect on gene expression than genotype, we conducted a further analysis of differential gene expression in wild-type and erdj3b plants grown at 22°C and 29°C. In wild-type plants, growth at 29°C resulted in 1,158 and 527 genes being down- and up-regulated, respectively, relative to their expression in plants grown at 22°C. In erdj3b mutants, growth at 29°C resulted in 1,276 and 778 genes being down- and up-regulated, respectively, relative to expression in plants grown at 22°C (Fig. 6B). Among these, 344 and 242 genes were down- and up-regulated, respectively, specifically in wild-type plants, while 462 and 493 genes were down- and up-regulated, respectively, specifically in erdj3b plants.
Figure 6.
Functional biological processes affected in the erdj3b mutant at 29°C, as revealed by RNA-seq analysis. A, Total RNA fractions prepared from anthers from stage 8 to 13 flowers of wild-type and erdj3b-1 plants grown at 22°C or 29°C and subjected to RNA-seq analysis. The Venn diagram indicates the numbers of genes differentially expressed between wild-type and erdj3b plants grown at 22°C (WT; 22°C vs 3b; 22°C), wild-type and erdj3b plants grown at 29°C (WT; 29°C vs 3b; 29°C), wild-type plants grown at 22°C and 29°C (WT; 22°C vs WT; 29°C), and erdj3b plants grown at 22°C and 29°C (3b; 22°C vs 3b; 29°C). B, Venn diagrams showing the numbers of genes down- and up-regulated in wild-type and erdj3b (3b-1) plants grown at 29°C compared with the corresponding plants grown at 22°C. C, Overrepresented GO terms of down-regulated (top graph) and up-regulated (bottom graph) genes in wild-type and erdj3b plants grown at 29°C compared with the corresponding plants grown at 22°C. Asterisks indicate corrected P values adjusted by Benjamini and Hochberg’s method (P < 0.05). Background means all genes in the Arabidopsis genome.
Gene Ontology (GO) enrichment analysis of the down-regulated genes showed significant enrichment of GO terms associated with pollen formation and anther development, such as sporopollenin biosynthetic process, tapetal layer development, and biosynthetic process of flavonoid, in both wild-type and erdj3b plants (Fig. 6C; Supplemental Data Sets S7 and S8). Genes annotated with the GO term tapetal layer development include TDF1, AMS, MS188/MYB103/MYB80, and MS1, which encode transcription factors required for this process (Li et al., 2017). These GO terms were not enriched when we performed GO enrichment analysis of down-regulated genes specifically in wild-type plants (Supplemental Data Set S9). GO enrichment analysis of genes that were down-regulated specifically in erdj3b plants showed no significant enrichment of GO terms. These results suggest that growth under temperature stress at 29°C generally affected pollen/anther development in Arabidopsis. Genes annotated as biosynthetic process of flavonoid, an important pollen coat component, were enriched among down-regulated genes in erdj3b but not in the wild type, which is consistent with the pollen coat abnormality of the mutant (Fig. 2, D and F). Interestingly, genes annotated as response to heat, including several Hsp genes, were down-regulated in both wild-type and erdj3b plants at 29°C (Supplemental Table S1). We selected four genes, ROF1, At1g54050, cpHSC70-2, and HSP17.8, which were annotated as response to heat, and analyzed their expression by real-time quantitative PCR. Expression of these four genes was confirmed to be lower in anthers of both wild-type and erdj3b plants grown at 29°C than of plants grown at 22°C (Supplemental Table S2). We also analyzed the expression of other Hsp genes, seven Hsp100 family genes, six Hsp90 family genes, 11 Hsp70 family genes, 36 Hsp20 family genes, and 18 Hsf family genes, all of which had not been identified as differentially expressed genes, with an FDR < 0.01. RNA-seq showed that these genes were not significantly up-regulated in either wild-type or erdj3b plants grown at 29°C (Supplemental Table S3). It is likely that the chronic exposure of male reproductive organs to heat stress does not result in a heat shock response, instead repressing the expression of heat shock genes. Genes annotated as photosynthesis and regulation of transcription DNA-dependent were enriched among down-regulated genes in wild-type plants (Fig. 6C).
GO enrichment analysis of up-regulated genes in wild-type plants showed no significant enrichment of GO terms. By contrast, GO terms related to phytohormone responses, including response to abscisic acid, response to ethylene, and response to jasmonic acid, were enriched among up-regulated genes in the erdj3b mutant at 29°C (Fig. 6C; Supplemental Data Set S10). The GO terms glucosinolate metabolic process, response to water deprivation, and response to salt stress were also significantly enriched among the up-regulated genes in erdj3b. Abscisic acid, ethylene, and glucosinolate contribute to high-temperature stress tolerance (Larkindale and Knight, 2002), suggesting that strong heat and drought stress responses occurred in erdj3b anthers at 29°C. The GO terms glycoside biosynthetic process, carotenoid catabolic process, and lipid transport were also significantly enriched among up-regulated genes in erdj3b plants at 29°C (Fig. 6C). Most of these GO terms were also enriched among genes up-regulated specifically in the erdj3b mutant at 29°C (Supplemental Data Set S11).
Accumulation of misfolded proteins in the ER induces the expression of genes targeted by the UPR, including BiP, ERDJ3A, ERDJ3B, and P58IPK. The Arabidopsis ire1 mutant, which is defective in the UPR, is sterile at elevated temperatures (Deng et al., 2016). To examine whether the UPR was activated in the erdj3b mutant, we investigated whether the expression of UPR-targeted genes was altered in erdj3b anthers. RNA-seq showed that genes encoding ER chaperones, including BiP, ERdj3A, and P58IPK, were not up-regulated in the erdj3b mutant at 22°C or 29°C (Supplemental Table S4). No significant differences in levels of IRE1A and IRE1B expression were observed between wild-type and erdj3b plants at either 22°C or 29°C (Supplemental Table S4). When we compared our RNA-seq results with data from the ire1a ire1b double mutant reported by Deng et al. (2016), in which RNA samples were prepared from flowers at stages 10 and 11, we found no substantial overlaps in down- or up-regulated genes between the erdj3b mutant and ire1a ire1b (Supplemental Fig. S9). Expression levels of ERDJ3B in ire1a ire1b or bzip60 flowers resembled those in wild-type flowers (Deng et al., 2016), and it is therefore unlikely that the developmental defects of the anther seen in erdj3b are related to the UPR. Our results suggest that a different signaling pathway from the UPR operates in developing anthers to mitigate heat stress.
DISCUSSION
ER-resident J proteins function in various processes as functional partners for BiP, the major Hsp70 in the ER. Here, we found that ERdj3B, an ER-luminal resident J protein in Arabidopsis involved in ER quality control, is required for maintaining fertility, especially pollination, at a high temperature (29°C). The number of pollen grains on self-pollinated stigmas of the erdj3b mutant was dramatically reduced compared with the wild type. Pollination and anther developmental defects were also observed in male gametogenesis impaired anthers, a loss-of-function mutant of a P5-ATPase in the ER and secretory vesicles (Jakobsen et al., 2005). Cod1/Spf1, a yeast homolog of MALE GAMETOGENESIS IMPAIRED ANTHERS, functions in ER quality control (Cronin et al., 2000, 2002; Vashist et al., 2002). Therefore, ER quality control plays an important role in anther development.
Among mutants of the three ER-luminal resident J proteins, ERdj3A, ERdj3B, and P58IPK, the temperature-sensitive seed production defect was observed only in erdj3b plants grown at 29°C. ERDJ3A or P58IPK expression driven by the ERDJ3B promoter did not suppress the reduced seed yield phenotype of erdj3b plants grown at 29°C, indicating that the functions of ERdj3B differ from those of ERdj3A and P58IPK. Mammalian cells contain four ER-luminal resident J proteins: ERdj3, ERdj4, ERdj5, and P58IPK (Gidalevitz et al., 2013). Although these J proteins function in protein folding and ER quality control, they exhibit differences in substrate recognition. For example, ERdj4 and ERdj5, but not ERdj3, bind to misfolded surfactant protein C (Dong et al., 2008). Differences in substrate recognition similar to those in mammalian cells might explain the difference between the requirement for ERdj3B versus ERdj3A and P58IPK in Arabidopsis grown at 29°C.
Yang et al. (2009) reported that tms1-1, a mutant allele of ERDJ3A, shows male sterility at 30°C. However, we did not observe such a fertility defect in erdj3a-1 or erdj3a-2 grown at 29°C. The erdj3a-1 mutant has a T-DNA insertion in the fourth exon of ERDJ3A (Yamamoto et al., 2008). Immunoblotting showed that no truncated ERdj3A protein is expressed in erdj3a-1, indicating that this mutant is a null allele (Maruyama et al., 2014). Thus, it is unlikely that erdj3a-1 has retained partial ERdj3A activity and is fertile at 29°C. A possible explanation for this discrepancy is the difference in the Arabidopsis ecotype backgrounds of the mutants; tms1-1 is in the Landsberg erecta background and erdj3a-1 and erdj3a-2 are in the Columbia background, and recent genome-wide association analysis revealed that the heat sensitivity of male fertility in Arabidopsis varies among ecotypes (Bac-Molenaar et al., 2015).
Analysis of the temperature-sensitive fertility defects of erdj3b revealed aberrant anther development in the mutant plants at 29°C. Both transmission electron microscopy and light microscopy revealed collapsed pollen grains in erdj3b mutants grown at 29°C, suggesting that pollen viability was affected. On the other hand, we observed pollen germination on stigmas of erdj3b mutants grown at 29°C, indicating that some fraction of the pollen grains retained their viability at this temperature; consistent with this observation, the erdj3b mutant produced seeds at 29°C.
The observed pollen coat-related abnormality in erdj3b at 29°C was likely to have been caused by tapetum defects. Aberrant tapetum morphology, including hypertrophy and vacuolation, was only observed in erdj3b anthers when plants were grown at 29°C. ERDJ3B is expressed in the tapetum cells in developing anthers. Both the pollination defects and the reduced seed yield phenotype seen in erdj3b plants grown at 29°C were partially suppressed by tapetum-specific expression of ERdj3B. This points to an important role of ERdj3B in the tapetum. Growth at 29°C reduced seed production to some extent, even in wild-type plants; by contrast, no significant differences in anther developmental phenotypes were observed between plants grown at 29°C and 22°C. We also observed reductions in the numbers of pollen grains and pollen tubes in the stigmas of wild-type plants grown at 29°C compared with those grown at 22°C. RNA-seq analysis revealed that expression of genes related to pollen formation and anther development was reduced in wild-type plants grown at 29°C, indicating that chronic mild heat stress affected anther development, resulting in reduced pollination.
By contrast, RNA-seq analysis indicated that genes encoding major heat shock proteins were not up-regulated in anthers of either wild-type or erdj3b plants grown at 29°C relative to their expression in plants grown at 22°C, indicating that the major heat shock response was not induced by chronic heat stress at 29°C. The expression patterns of ERDJ3B in developing anthers were similar in plants grown at 22°C and 29°C. RNA-seq analysis also showed that the UPR did not occur in wild-type and erdj3b plants grown at 29°C. Heat stability varies from protein to protein, and it is possible that ERdj3B functions to stabilize a fraction of heat-labile proteins under a mild heat stress of 29°C. The severe pollination defect of erdj3b plants grown at 29°C most likely resulted from the combined effects of heat injury on anther development and a protein homeostasis defect in the ER that is specific to this mutant.
Reduced crop yields at high temperature is a crucial problem in several crops (Hatfield et al., 2011). Heat stress-induced male sterility is associated with disturbed tapetal development in several other plants (Dolferus et al., 2011). The erdj3b mutant also showed tapetum abnormalities at 29°C. Thus, it would be interesting to analyze whether ER quality control in the tapetum is related to thermotolerance in crop plants in the future.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used as the wild type. The T-DNA insertional mutants used in this study included erdj3a-1 (SALK_103280), erdj3a-2 (SALK_027193), erdj3b-1 (SALK_113364), erdj3b-2 (SALK_055599), p58ipk-1 (SALK_140273), and p58ipk-2 (SALK_080901; Yamamoto et al., 2008). Seeds were either sown in soil or surface sterilized with chlorine gas and sown on Murashige and Skoog medium (Wako) containing 0.7% (w/v) agar and 1% (w/v) Suc. The Arabidopsis plants were grown at 22°C or 29°C under continuous light (50 μmol m−2 s−1).
Construction of Transgenic Plants
The oligonucleotide primers used in plasmid construction are listed in Supplemental Table S5. The plasmids used for the expression of ERDJ3B from the ERDJ3B or ATA1 promoters were constructed as follows. A 3.3-kb genomic DNA fragment containing the ERDJ3B coding region and 1 kb of the region downstream of the stop codon of ERDJ3B was amplified using primers 62600F2 and 62600R2, digested with SmaI and HindIII, and inserted into the SmaI and HindIII sites of pCAMBIA1300 (Cambia) to produce pMYP022. Gateway reading frame cassette A (Life Technologies Japan) was inserted into the SmaI site of pMYP022 to produce pMYP023. DNA fragments corresponding to 2 kb of the region upstream of the start codon of ERDJ3B or 1.59 kb of the region upstream of the start codon of ATA1 were amplified from Arabidopsis genomic DNA by PCR using primer sets 62600F3/62600R3 and ATA1F1/ATA1R1, respectively, and cloned into pENTER/D-TOPO (Life Technologies Japan). The promoter region was inserted upstream of ERDJ3B in pMYP023 using LR Clonase II (Life Technologies Japan). The ERDJ3B promoter:ERDJ3A fusion gene was constructed by nested PCR as follows. DNA fragments corresponding to the promoter of ERDJ3B and the coding region of ERDJ3A together with 1 kb of the region downstream of the stop codon of ERDJ3A were amplified by PCR using primer sets 62600F3/62600R4 and 08970F1/08970R1, respectively. The amplified DNA fragments were mixed and used as templates for PCR using primers 62600F3 and 08970R2. Similarly, the ERDJ3B promoter:P58IPK fusion gene was constructed by nested PCR using primer sets 62600F2/62600R5 and 03160F1/03160R1 for the first-round PCR and 62600F3/03160R2 for the second-round PCR. The amplified DNA fragments were cloned into pENTER/D-TOPO. The fusion genes were introduced into pGWB1 (Nakagawa et al., 2007) using LR Clonase II. The plasmid used for the complementation test of ERDJ3B was described by Maruyama et al. (2014). The H54Q mutation was introduced into ERDJ3B by PCR using primers 62600(HQ)F1 and 62600(HQ)R1. Plasmid construction for ERDJ3B promoter:GUS fusion genes was described by Maruyama et al. (2014). The resulting plasmids were transformed into Arabidopsis plants by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101.
Histochemical Analyses
The number of pollen grains that adhered to pistils was counted as described by Zinkl et al. (1999), except that self-pollinated pistils were washed with 200 μL of 50 mm potassium phosphate, pH 7.4, before staining. To observe semithin sections of anthers, bud clusters were fixed with 2% (w/v) glutaraldehyde in 20 mm sodium cacodylate, pH 7.4, overnight at 4°C. The samples were dehydrated through an ethanol series and embedded in Technovit 7100 resin (Kulzer) according to the manufacturer’s instructions. Sections were cut to a thickness of 2 μm using a Leica RM2255 microtome (Leica Biosystems), stained with 0.5% (w/v) Toluidine Blue O in 0.1% (w/v) Na2CO3, and observed with a BX51 microscope (Olympus) with bright-field illumination. Each sample was analyzed at least in two biological and two technical repeats with similar results.
Histochemical GUS staining was performed as described by Preuss et al. (1994). To prepare sections, GUS-stained samples were embedded in Technovit 7100 resin (Kulzer). Each sample was analyzed at least in two biological and two technical repeats with similar results.
Electron Microscopy
For transmission electron microscopy, anthers were fixed in 2% (w/v) glutaraldehyde, 2% (w/v) paraformaldehyde, and 50 mm sodium cacodylate, pH 7.4, for 3 d at 4°C. The tissue was washed with 50 mm sodium cacodylate, pH 7.4, and postfixed for 8 h in 2% (w/v) aqueous osmium tetroxide at 4°C. The tissue was then dehydrated in a graded acetone series, transferred into propylene oxide, infiltrated, and embedded in Quetol 651. Thin sections (70 nm) were stained with 2% (v/v) aqueous uranyl acetate and lead citrate and examined with a JEOL JEM 1200 EX electron microscope at 80 kV.
GST-ERdj3B Fusion Proteins
Plasmids used for the expression of wild-type or H54Q mutant GST-ERdj3B in Escherichia coli were constructed as follows. A DNA fragment corresponding to residues 23 to 346 of ERdj3B was amplified by PCR using primers 62600F4 and 62600R6 from the ERDJ3B complementary DNA clone (Yamamoto et al., 2008) or the ERDJ3B complementary DNA clone containing the H54Q mutation and cloned into pGEX4T-2 (GE Healthcare) to produce pMYA010 and pMYA011, respectively. E. coli strain BL21(DE3) harboring pMYA010, pMYA011, or pGEX4T-2 was grown in Luria-Bertani medium containing 50 μg mL−1 ampicillin at 37°C to OD600 of 0.5 to 0.6. Isopropyl β-d-thiogalactopyranoside was added to the medium at a final concentration of 1 mm, and the culture was further incubated overnight at 18°C. Cells were harvested by centrifugation at 3,400g for 5 min at 4°C, suspended in 540 mm NaCl, 2.7 mm KCl, 8.1 mm Na2HPO4, 1.5 mm KH2PO4, 1 mm MgCl2, 1 mm EDTA, 1 mm DTT, pH 7.4, and protease inhibitor cocktail [1 mm phenylmethylsulfonyl fluoride, 2 μg mL−1 chymostatin, 10 μg mL−1 N-tosyl-l-phenylmethyl-chloromethyl ketone, 200 μg mL−1 p-aminobenzamidine hydrochloride, 5 μg mL−1 pepstatin A, 5 μg mL−1 E-64, 1 mm 6-aminohexanoic acid, 2 μg mL−1 aprotinin, 2 μg mL−1 antipain, 2 μg mL−1 leupeptin, and 20 μg mL−1 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride], and disrupted by sonication for 1 min (2 s on/1 s off pulsed periods, 40% duty cycle) using an Astorason XL2020 sonicator with a microtip (Heat Systems-Ultrasonic; Misonix). The cell lysate was clarified by two sequential centrifugations at 13,000g for 10 min at 4°C and 48,000g for 1 h at 4°C. The resulting supernatant was applied onto a glutathione Sepharose 4B column (GE Healthcare; 2-mL bed volume). The column was washed with 30 mL of 540 mm NaCl, 2.7 mm KCl, 8.1 mm Na2HPO4, 1.5 mm KH2PO4, 1 mm MgCl2, 1 mm EDTA, and 0.1% (v/v) Tween 20, pH 7.4, and with 30 mL of 540 mm NaCl, 2.7 mm KCl, 8.1 mm Na2HPO4, and 1.5 mm KH2PO4, pH 7.4. Proteins were eluted from the column with 50 mm Tris-HCl, pH 8, and 10 mm glutathione and dialyzed against 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 1 mm MgCl2. Concentrations of the prepared proteins were determined by Coomassie Brilliant Blue staining after SDS-PAGE (Laemmli, 1970) using BSA as a standard.
Chaperone Activity Analysis
Rhodanese from bovine liver (Sigma-Aldrich) was dissolved at 46 μm in 6 m guanidinium-HCl, 50 mm Tris-HCl, pH 7.4, and 5 mm DTT and denatured by 60 min of incubation at room temperature. Denatured rhodanese was diluted 100-fold in 50 mm Tris-HCl, pH 7.4, 1 mm CaCl2, and 150 mm NaCl containing 2.3 μm GST-ERdj3B, GST-ERdj3B(H54Q), or GST. Protein aggregation was measured by monitoring the A320 at 23°C. The experiment was repeated four times with similar results.
Total protein extracts were prepared from erdj3b-1 mutant seedlings by grinding the tissue in 100 mm HEPES-KOH, pH 7.4, 5 mm EGTA, 5 mm MgCl2, 300 mm Suc, and protease inhibitors. The homogenate was centrifuged at 13,000g for 20 min at 4°C to remove cell debris. The homogenate was diluted to 0.1 μg protein mL−1 in 20 mm HEPES-KOH, pH 7.4, 400 mm KCl, 1 mm EDTA, 1.5 mm MgCl2, 2 mm DTT, 15% (w/v) glycerol, and 0.65% (w/v) CHAPS, and insoluble materials were removed by centrifugation at 100,000g for 1 h at 4°C. The supernatant was mixed with an equal volume of 20 mm HEPES-KOH, pH 7.4, 1 mm EDTA, 1.5 mm MgCl2, 2 mm DTT, 15% (w/v) glycerol, and 0.65% (w/v) CHAPS. Glutathione Sepharose 4B beads (10 μL of slurry; GE Healthcare) prebound to 3 μg of GST-ERdj3B, GST-ERdj3B(H54Q), or GST proteins were incubated with 1 mL of protein extract (50 μg) in the presence (2 mm) or absence of ATP for 1 h at 4°C. The reaction mixtures were centrifuged at 21,000g for 20 s at 4°C to remove unbound proteins. The glutathione Sepharose 4B beads were washed four times with 140 mm NaCl, 2.7 mm KCl, 8.1 mm Na2HPO4, 1.5 mm KH2PO4, 1 mm MgCl2, pH 7.4, with or without 2 mm ATP, and proteins retained on the beads were eluted in SDS sampling buffer (Laemmli, 1970) at 95°C. Interaction between BiP and GST-ERdj3B was reproducibly observed in two independent experiments.
Western-Blot Analysis
Proteins were extracted from bud clusters or flowers from Arabidopsis inflorescences. Samples were frozen in liquid nitrogen, ground to a fine powder, suspended in SDS-PAGE sampling buffer (Laemmli, 1970), heated at 94°C for 5 min, and centrifuged to remove insoluble materials. The extracted proteins were subjected to SDS-PAGE (Laemmli, 1970), followed by transfer to Immobilon-P membranes (Millipore) as described by Towbin et al. (1979). Primary antibodies were used at the following dilutions for immunoblotting: anti-ERdj3A, 1:1,000; anti-ERdj3B, 1:1,000; anti-P58IPK, 1:1,000; anti-BiP, 1:5,000 (Yamamoto et al., 2008); and anti-actin, 1:2,000 (Affinity BioReagents). Horseradish peroxidase-labeled anti-rabbit IgG or anti-mouse IgG (GE Healthcare) was used as a secondary antibody at 1:5,000 dilution. Signals were detected using an ECL Prime western blotting detection kit (GE Healthcare) and an LAS4000 mini image analyzer (Fujifilm).
RNA-Seq Analysis
Total RNA was prepared from anthers of stage 8 to 13 flowers (∼180 anthers) using an RNeasy Plant Mini kit (Qiagen). All samples were treated with DNase I using an RNase-Free DNase Set (Qiagen) and quantified. RNA samples prepared from three independent plants were used for subsequent analysis. For library preparation, 0.5 μg of total RNA from samples with an RNA Integrity Number ≥ 8 determined by the Agilent 2100 Bioanalyzer was used. All libraries were prepared using a TruSeq Stranded mRNA LT Sample kit (Illumina) according to the manufacturer’s protocol. Multiplex sequencing was performed on a NextSeq 500 (Illumina), and 75-bp-long single-end reads were obtained. The reads were mapped to the reference Arabidopsis genome (The Arabidopsis Information Resource 10) using TopHat2 (Kim et al., 2013) and counted using the htseq-counts script in the HTSeq library (Anders et al., 2015). Count data were subjected to trimmed mean of M values normalization, and differentially expressed genes were defined using edgeR (Robinson et al., 2010; McCarthy et al., 2012). Genes with FDR < 0.01 were classified as differentially expressed genes. Principal component analysis was performed on the differentially expressed genes data set using the built-in R function (R Core Team, 2013). GO enrichment analysis was performed using the Biological Networks Gene Ontology (BiNGO) tool (Maere et al., 2005). Overrepresented GO terms of corrected P < 0.05 calculated from the hypergeometric test and adjusted by the Benjamini and Hochberg method were selected.
Real-Time Reverse Transcription Quantitative PCR Analysis
First-strand DNA was synthesized from the RNA sample used for RNA-seq analysis using a ReverTra Ace qPCR RT kit (Toyobo) according to the manufacturer’s protocol. The gene-specific primers used for reverse transcription quantitative PCR are listed in Supplemental Table S6. Reverse transcription quantitative PCR was performed using an Applied Biosystems 7300 Real-Time PCR System (Life Technologies Japan) and THUNDERBIRD SYBR qPCR mix (Toyobo) with 40 cycles of denaturation at 95°C for 15 s and extension at 60°C for 1 min. ABI sequence detection software (version 1.4; Life Technologies Japan) was used for quantification. UBQ10 was used as an internal control to normalize the RNA quantity. Each sample was analyzed in three biological and three technical repeats.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_187509 (ERdj3A/At3g08970), NP_191819 (ERdj3B/At3g62600), and NP_195936 (P58IPK/At5g03160). The Arabidopsis Genome Initiative codes of the genes analyzed by real-time PCR are provided in Supplemental Table S2. The RNA-seq data reported here are available in the DNA Data Bank of Japan Sequenced Read Archive under accession number DRA006470.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Seed number per silique in Arabidopsis wild-type, erdj3b, and transgenic plants grown at 22°C and 29°C.
Supplemental Figure S2. erdj3b mutant plants show no obvious defect in flower development at 22°C.
Supplemental Figure S3. The role of ERdj3B in the fertility of Arabidopsis plants grown at 29°C cannot be rescued by either ERdj3A or P58IPK.
Supplemental Figure S4. The erdj3b heterozygous mutant does not show pollination defects at 29°C.
Supplemental Figure S5. Transmission electron micrographs showing anther locules of plants grown at 29°C.
Supplemental Figure S6. Pollen tubes of stigmas of wild-type and erdj3b plants.
Supplemental Figure S7. Sections of anthers from plants grown at 22°C.
Supplemental Figure S8. Principal component analysis for all of the RNA-seq libraries.
Supplemental Figure S9. The UPR does not occur in erdj3b at 29°C.
Supplemental Table S1. Genes annotated as response to heat, which were down-regulated in wild-type and erdj3b plants grown at 29°C.
Supplemental Table S2. Real-time reverse transcription quantitative PCR analysis of down-regulated genes annotated as response to heat in wild-type and erdj3b anthers.
Supplemental Table S3. Expression of HSP and HSF in anthers of wild-type and erdj3b plants.
Supplemental Table S4. Genes targeted by the UPR and IRE1 are not up-regulated in erdj3b anthers.
Supplemental Table S5. PCR primers used for plasmid construction.
Supplemental Table S6. PCR primers used for real-time reverse transcription quantitative PCR.
Supplemental Data Set S1. Down-regulated genes in both wild-type and erdj3b anthers under high-temperature conditions.
Supplemental Data Set S2. Down-regulated genes in wild-type anthers under high-temperature conditions.
Supplemental Data Set S3. Down-regulated genes in erdj3b anthers under high-temperature conditions.
Supplemental Data Set S4. Up-regulated genes in both wild-type and erdj3b anthers under high-temperature conditions.
Supplemental Data Set S5. Up-regulated genes in wild-type anthers under high-temperature conditions.
Supplemental Data Set S6. Up-regulated genes in erdj3b anthers under high-temperature conditions.
Supplemental Data Set S7. Selected GO terms (biological process category) overrepresented among down-regulated genes in erdj3b anthers, as identified using the BiNGO tool.
Supplemental Data Set S8. Selected GO terms (biological process category) overrepresented among down-regulated genes in wild-type anthers, as identified using the BiNGO tool.
Supplemental Data Set S9. Selected GO terms (biological process category) overrepresented among down-regulated genes in wild-type but not in erdj3b anthers, as identified using the BiNGO tool.
Supplemental Data Set S10. Selected GO terms (biological process category) overrepresented among up-regulated genes in erdj3b anthers, as identified using the BiNGO tool.
Supplemental Data Set S11. Selected GO terms (biological process category) overrepresented among up-regulated genes in erdj3b but not in wild-type anthers, as identified using the BiNGO tool.
Acknowledgments
We thank Tsuyoshi Nakagawa for the pGWB vectors, Shiori Kato and Akiko Yoshii for their assistance with sample preparation, Ravi Palanivelu and Rob Swanson for critically reading the article, and members of the Endo laboratory for useful discussions.
Footnotes
This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas from the Japan Society for the Promotion of Science (grant nos. 23120512, 25120711, 17H05837, and 19H04857 to S.N. and grant nos. 16H01472, 18H04787, and 18H04844 to S.K.), Grants-in Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant nos. 16K07394 and 19K06704 to S.N. and grant no. 16K07408 to S.K.), the NOVARTIS Foundation (Japan) for Promotion of Science (grant no. 12-109 to S.N.), the Sasaki Environment Technology Foundation (to S.N.), and the MEXT Supported Program for the Strategic Research Foundation at Private Universities from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant no. S1511023 to S.K.). M.Y. was a Research Fellow at the Japan Society for the Promotion of Science.
Articles can be viewed without a subscription.
References
- Anders S, Pyl PT, Huber W (2015) HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bac-Molenaar JA, Fradin EF, Becker FF, Rienstra JA, van der Schoot J, Vreugdenhil D, Keurentjes JJ (2015) Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis thaliana. Plant Cell 27: 1857–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A (2006) Molecular chaperones and protein quality control. Cell 125: 443–451 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cronin SR, Khoury A, Ferry DK, Hampton RY (2000) Regulation of HMG-CoA reductase degradation requires the P-type ATPase Cod1p/Spf1p. J Cell Biol 148: 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin SR, Rao R, Hampton RY (2002) Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J Cell Biol 157: 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Quilichini TD, Dong H, Bao Y, Horner HT, Howell SH (2016) IRE1, a component of the unfolded protein response signaling pathway, protects pollen development in Arabidopsis from heat stress. Plant J 88: 193–204 [DOI] [PubMed] [Google Scholar]
- Dolferus R, Ji X, Richards RA (2011) Abiotic stress and control of grain number in cereals. Plant Sci 181: 331–341 [DOI] [PubMed] [Google Scholar]
- Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE (2008) ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol Biol Cell 19: 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A (2003) Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4: 181–191 [DOI] [PubMed] [Google Scholar]
- Fragkostefanakis S, Mesihovic A, Hu Y, Schleiff E (2016) Unfolded protein response in pollen development and heat stress tolerance. Plant Reprod 29: 81–91 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Stevens F, Argon Y (2013) Orchestration of secretory protein folding by ER chaperones. Biochim Biophys Acta 1833: 2410–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield JL, Boote KJ, Kimball BA, Ziska LH, Izaurralde RC, Ort D, Thomson AM, Wolfe D (2011) Climate impacts on agriculture: Implications for crop production. Agron J 103: 351–370 [Google Scholar]
- Iwata Y, Koizumi N (2012) Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci 17: 720–727 [DOI] [PubMed] [Google Scholar]
- Jakobsen MK, Poulsen LR, Schulz A, Fleurat-Lessard P, Møller A, Husted S, Schiøtt M, Amtmann A, Palmgren MG (2005) Pollen development and fertilization in Arabidopsis is dependent on the MALE GAMETOGENESIS IMPAIRED ANTHERS gene encoding a type V P-type ATPase. Genes Dev 19: 2757–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho YO, Baër J (1968) Observing pollen tubes by means of fluorescence. Euphytica 17: 298–302 [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU (1992) Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356: 683–689 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Knight MR (2002) Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol 128: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel-Hardenack S, Ye D, Koutnikova H, Saedler H, Grant SR (1997) Conserved expression of a TASSELSEED2 homolog in the tapetum of the dioecious Silene latifolia and Arabidopsis thaliana. Plant J 12: 515–526 [DOI] [PubMed] [Google Scholar]
- Li DD, Xue JS, Zhu J, Yang ZN (2017) Gene regulatory network for tapetum development in Arabidopsis thaliana. Front Plant Sci 8: 1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Makio T, Nishikawa S, Nakayama T, Nagai H, Endo T (2008) Identification and characterization of a Jem1p ortholog of Candida albicans: Dissection of Jem1p functions in karyogamy and protein quality control in Saccharomyces cerevisiae. Genes Cells 13: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Maruyama D, Yamamoto M, Endo T, Nishikawa S (2014) Different sets of ER-resident J-proteins regulate distinct polar nuclear-membrane fusion events in Arabidopsis thaliana. Plant Cell Physiol 55: 1937–1944 [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Brodsky JL, Nakatsukasa K (2005) Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD). J Biochem 137: 551–555 [DOI] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW (1994) Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- R Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
- Regan SM, Moffatt BA (1990) Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I, Twell D, Firon N (2017) Pollen development at high temperature: From acclimation to collapse. Plant Physiol 173: 1967–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Douglas MG (1996) A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem 271: 9347–9354 [DOI] [PubMed] [Google Scholar]
- Vashist S, Frank CG, Jakob CA, Ng DT (2002) Two distinctly localized P-type ATPases collaborate to maintain organelle homeostasis required for glycoprotein processing and quality control. Mol Biol Cell 13: 3955–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL (2008) One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9: 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P, Bursać D, Law YC, Cyr D, Lithgow T (2004) The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep 5: 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Maruyama D, Endo T, Nishikawa S (2008) Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant Cell Physiol 49: 1547–1562 [DOI] [PubMed] [Google Scholar]
- Yang KZ, Xia C, Liu XL, Dou XY, Wang W, Chen LQ, Zhang XQ, Xie LF, He L, Ma X, et al. (2009) A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis. Plant J 57: 870–882 [DOI] [PubMed] [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D (1999) Pollen-stigma adhesion in Arabidopsis: A species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126: 5431–5440 [DOI] [PubMed] [Google Scholar]