Abstract
The genetic basis underlying the phenotype of purple tomatoes guides an understanding of these varieties which were introduced over 10 years ago.
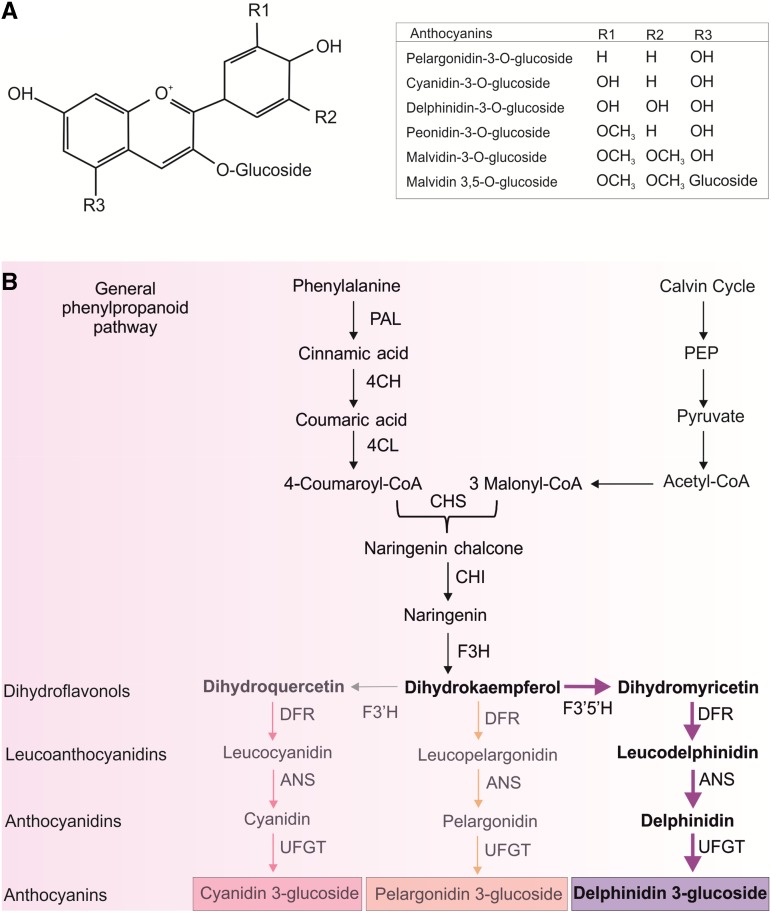
The term “anthocyanins” was first used in 1835 by L.C. Marquart to indicate the blue pigments of flowers (Shibata et al., 1919). In the early 20th century, these pigments, whose colors range from red to purple to blue, were first isolated and chemically characterized by Richard Willstatter (Robinson and Robinson, 1931). A wide range of different anthocyanins have subsequently been extracted from flowers, fruits, seeds, roots, and leaves, thus increasing the size of this subclass of polyphenolic compounds belonging to the larger class of flavonoids (Koes et al., 2005). Anthocyanins are glycosylated, polyhydroxy or polymethoxy molecules, formed by two aromatic rings connected by a C3 bridge to a benzene ring (Fig. 1A). These pigments are water-soluble and mainly accumulate in the vacuole of plant cells: they are odorless and flavorless, although their presence in edible fruits and vegetables gives a slightly astringent taste (Martín et al., 2017). A total of 702 different anthocyanins and 27 anthocyanidins (aglycone forms of anthocyanins) have been classified (Krga and Milenkovic, 2019). All these phenolic compounds vary from each other in the different groups of carbon substitutions on the structural rings (Fig. 1A; Liu et al., 2018).
Figure 1.
Anthocyanin biosynthesis in higher plants. A, Chemical structure of the general anthocyanin molecule and main anthocyanin pigments synthesized in plants, according to the different nature of the R lateral groups. B, Anthocyanin biosynthetic pathway. Enzyme abbreviations: PAL, Phenylalanine ammonia-lyase; C4H, cinnamate-4-hydroxylase; 4CL, 4-coumaroyl:CoA-ligase; CHS, chalcone synthase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′5′-hydroxylase; ANS, anthocyanin synthase; UFGT, anthocyanidin 3-o-glucosyltransferase. In the final part of the biosynthetic way, from dihydroflavonols to anthocyanins, the pathway which is predominant in tomato plants is highlighted.
Anthocyanins play a major role during plant life and development. Initially, they were seen as having just one main function—i.e. their colored pigments attracted pollinating insects and seed-spreading animals (Harborne and Williams, 2000; Hoballah et al., 2007). It then became clear that plants also produce anthocyanins to protect themselves from a wide variety of biotic and abiotic stimuli (Chalker-Scott, 1999). When plants are exposed to different types of stresses, such as high light, heavy metals, salty soils, drought, high/low temperatures, as well as pathogen or insect attacks, there is typically an increase in reactive oxygen species (ROS) production, which may then be counterbalanced through the generation of antioxidant molecules (Sharma et al., 2019). Anthocyanins, which primarily accumulate in the subepidermal cell layers of vegetative tissues, act as a photoprotective light screen, by absorbing potentially damaging UV-B radiation (Gould, 2004). They can also act as antioxidant compounds through different chemical mechanisms, such as releasing hydrogen atoms from the hydroxyl groups or as metal-chelating agents, thus efficiently scavenging free radicals and ROS (Gould, 2004). Through the same mechanisms, anthocyanins can delay senescence processes. In deciduous species, for example, they can protect senescing leaves by acting as ROS scavengers and as a light screen against photoinhibitory irradiances that can damage the photosynthetic machinery while it is losing its photoprotective capacity (Hoch et al., 2001).
Anthocyanins are beneficial metabolites not only for plants; they are widespread in edible fruits and vegetables and thanks to their antioxidant and anti-inflammatory actions they can provide pharmacological activity (Li et al., 2017; Liu et al., 2017). Integrating high phenolic content foods into the human diet is believed to help prevent and control diseases such as cardiovascular disorders (Mink et al., 2007; Mauray et al., 2010; Cassidy et al., 2013), diabetes and obesity (Muraki et al., 2013; Wang et al., 2013), and cancer (Li et al., 2017). Anthocyanins may also have a neuroprotective activity (Youdim et al., 2004), and promising results have been demonstrated in animal models (Wang et al., 2012a, 2012b). Plant research has thus moved toward understanding the regulatory mechanisms of anthocyanin synthesis to produce anthocyanin-enriched versions of horticultural crops so as to increase the daily uptake of these health-promoting compounds (Martin et al., 2011).
Because it is one of the most consumed vegetable products in the world, tomato (Solanum lycopersicum) has been extensively used to achieve this goal. Unlike other Solanaceae such as eggplant (Solanum melongena) or pepper (Capsicum spp.), tomato fruits do not normally accumulate anthocyanins due to the incomplete activation of the flavonoid pathway (Povero et al., 2011). To increase their content, genetic engineering methods (Butelli et al., 2008) or genotype selection arising from natural breeding (Mes et al., 2008; Gonzali et al., 2009) have thus been exploited with excellent results. This review summarizes and discusses recent research into breeding purple varieties of tomato.
REGULATION OF THE ANTHOCYANIN BIOSYNTHETIC PATHWAY
Anthocyanin biosynthesis is well characterized in higher plants and consists of a strongly conserved pathway (Holton and Cornish, 1995; Fig. 1B). It shares the first part of the required biosynthetic enzymes with other flavonoids, such as flavonols and flavanones, and represents a branch of the general phenylpropanoid pathway, whose main precursor is Phe, which itself comes from the shikimic acid pathway (Rasmussen and Dixon, 1999). From the condensation of one molecule of 4-coumaroyl CoA (from Phe) and three of malonyl CoA (from fatty acid metabolism), through the combined consecutive actions of three different enzymes—chalcone synthase, chalcone isomerase (CHI), and flavanone 3-hydroxylase the dihydroflavonol dihydrokaempferol is produced. Acting in the first part of the biosynthetic pathway, the genes that encode these enzymes are named early biosynthetic genes (EBGs). Dihydrokaempferol can then be hydroxylated, through the action of flavonoid 3′-hydroxylase or flavonoid 3′-5′-hydroxylase (F3′5′H), to produce the other two dihydroflavonols, dihydroquercetin and dihydromyricetin, respectively. Downstream, the dihydroflavonols are reduced by dihydroflavonol 4-reductase (DFR) into different leucoanthocyanidins, which are converted into the corresponding anthocyanidins (leucopelargonidin, leucocyanidin, and leucodelphynidin) by the action of anthocyanin synthase (also called LDOX; Holton and Cornish, 1995; Springob et al., 2003). The colorless anthocyanidins are then glycosylated at C3, giving rise to the corresponding colored anthocyanin compounds (pelargonidin, cyanidin, and delphinidin), which are chemically stable and water-soluble (Sasaki et al., 2014). There are several forms of additional glycosylation, as well as acylation, which produce many different molecules. In addition to the three compounds mentioned above, other common anthocyanins found in plants are peonidin and petunidin (from methylation at the 3′ hydroxyl group of cyanidin and delphinidin, respectively), and malvidin (from methylation at the 3′ and 5′ hydroxyl groups of delphinidin). The final step of the pathway is the transport into the vacuole, through the action of glutathione S-transferase (GST) or putative anthocyanin transporter (Gonzali et al., 2009). The genes encoding the enzymes acting downstream of the dihydroflavonols are called late biosynthetic genes (LBGs), and are those specifically regulated for anthocyanin production.
Many plants exhibit a limited range of anthocyanin colors due to the lack of an essential gene or to the substrate specificity of a biosynthetic enzyme. Although in most cases DFR enzymes can utilize all three dihydroflavonols as substrates, in some species they show substrate specificity due to minor changes in their molecular structure (Johnson et al., 2001). Similar to petunia (Petunia x hybrida) and other Solanaceae, in tomato the action of DFR is mainly carried out on dihydromyricetin. The delphinidin-type anthocyanins are thus the most abundant in this species (Fig. 1B; Bovy et al., 2007).
Complex regulatory mechanisms induced by developmental and environmental stimuli activate and/or repress the anthocyanin biosynthetic pathway (Albert et al., 2014). A central role is played by specific transcription factors (TFs), which, induced or repressed by different signals, can act individually or in multiprotein complexes to activate the transcription of EBGs and/or LBGs, thus modulating the synthesis of anthocyanins to the different needs of the plant. Among these TFs, R2R3 MYBs cooperate with basic Helix–Loop–Helix (bHLH) and WD-repeat (WDR) proteins to form ternary MYB–bHLH–WDR (MBW) complexes (Koes et al., 2005; Ramsay and Glover, 2005; Albert et al., 2014; Liu et al., 2015; Montefiori et al., 2015; Xu et al., 2015). Some of the TFs are constitutively expressed in plant cells, whereas others require transcriptional activation. The R2R3 MYB factors are induced by the triggering signals (developmental or environmental) and arrange themselves into the MBW complex, by binding constitutively expressed or transcriptionally induced bHLH and WDR proteins (Albert et al., 2014). The MBW complex is the key regulator of the pathway because it triggers the transcription of biosynthetic genes, LBGs in particular, and thus the final part of the biosynthetic pathway (Koes et al., 2005). It can also activate the expression of negative regulators, such as R3 MYB proteins, which inhibit anthocyanin production through a negative feedback mechanism (Koes et al., 2005; Albert et al., 2014; Colanero et al., 2018). Several R2R3 MYB proteins, bHLH, WDR, and R3 MYBs, have recently been characterized in plants (Koes et al., 2005; Hichri et al., 2011; Zhang et al., 2019), thus providing an increasingly comprehensive picture of how the regulation of the anthocyanin biosynthetic pathway works.
SWITCHING ON THE ANTHOCYANIN PATHWAY IN TOMATO FRUITS
Tomato was imported to Europe in the 16th century from central and southwestern America. It was initially used as an ornamental plant and then gradually became an economically important crop for human consumption (Peralta and Spooner, 2007). During domestication, much of the genetic variability was lost (Bai and Lindhout, 2007) and the selection toward new productivity traits had a negative impact on some other important features. The size and color of the fruit also changed over time (Tanksley, 2004).
Tomatoes provide humans with key nutrients in their daily diet such as lycopene, the most abundant carotenoid in the ripe fruit, which has a strong antioxidant activity that is beneficial in the prevention of a wide range of diseases (Martí et al., 2016; Mozos et al., 2018). Moreover, tomatoes contain a smaller amount of other carotenoids and a high amount of soluble sugars, organic acids, amino acids, vitamins, and volatile compounds, which all affect the characteristic flavor (Tieman et al., 2017; Wang and Seymour, 2017).
Anthocyanins are generally not found in tomatoes, whereas other flavonoids, such as flavonols (mainly quercetin, myricetin, and kaempferol) and flavanones (narigenin), are detectable (Scarano et al., 2018). There is thus growing interest in expanding the repertoire of health-promoting compounds in tomato fruit to further enhance its already high nutraceutical potential. For this reason, molecular biologists and plant breeders are producing tomato lines with increased anthocyanin content in their fruits (Butelli et al., 2008; Mes et al., 2008; Gonzali et al., 2009). In the next subsections we will discuss how this has been achieved.
Metabolic Engineering Studies Led To the Production of Purple Tomato Fruits
Initial attempts to increase the flavonoid content in tomato fruits started in the early 2000s with the overexpression of structural genes encoding enzymes of the pathway. Given the low level of expression of the gene encoding the CHI enzyme compared to the other EBGs, the petunia chalcone isomerase gene was overexpressed in tomato plants to enhance the basal level of flavonoids (Muir et al., 2001). The transgenic plants showed an increase of up to 78-fold in fruit peel flavonols, mainly due to the accumulation of quercetin glycoside and lower amounts of kaempferol glycoside (Muir et al., 2001). No anthocyanins were detected and the whole biosynthetic pathway appeared to be switched off in the fruit flesh (Muir et al., 2001). Using a similar approach, Schijlen et al. (2006) generated transgenic tomato plants overexpressing structural flavonoid genes from different plant sources. High levels of stilbenes, deoxychalcones, and flavones were obtained in these plants, in relation to the structural gene introduced (Schijlen et al., 2006). Although the overexpression of the EBGs did not affect the anthocyanin branch of the biosynthetic pathway, it strongly increased the total antioxidant activity measured in the fruit peel extracts (Schijlen et al., 2006). Such studies indicated that to trigger anthocyanin synthesis, increasing the expression of the EBGs is not enough. Specific regulatory TFs are necessary to trigger LBG expression.
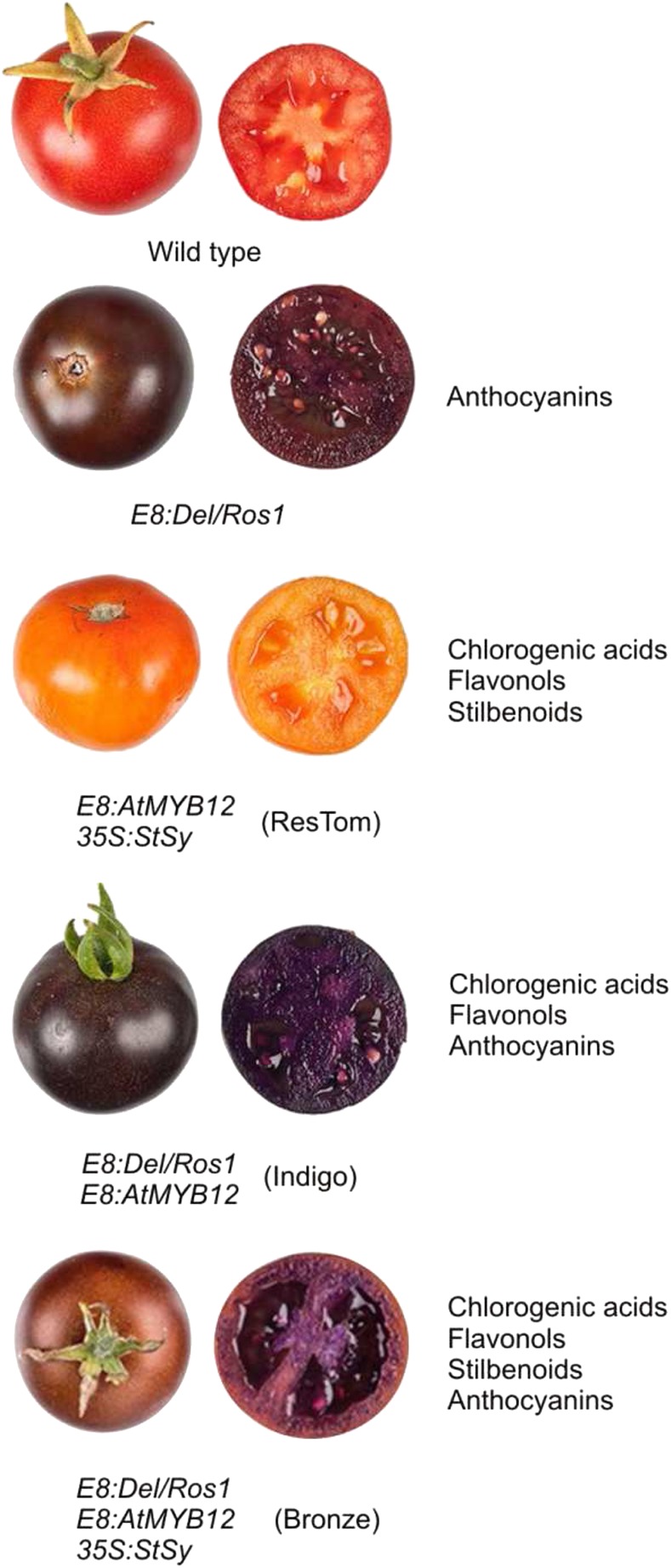
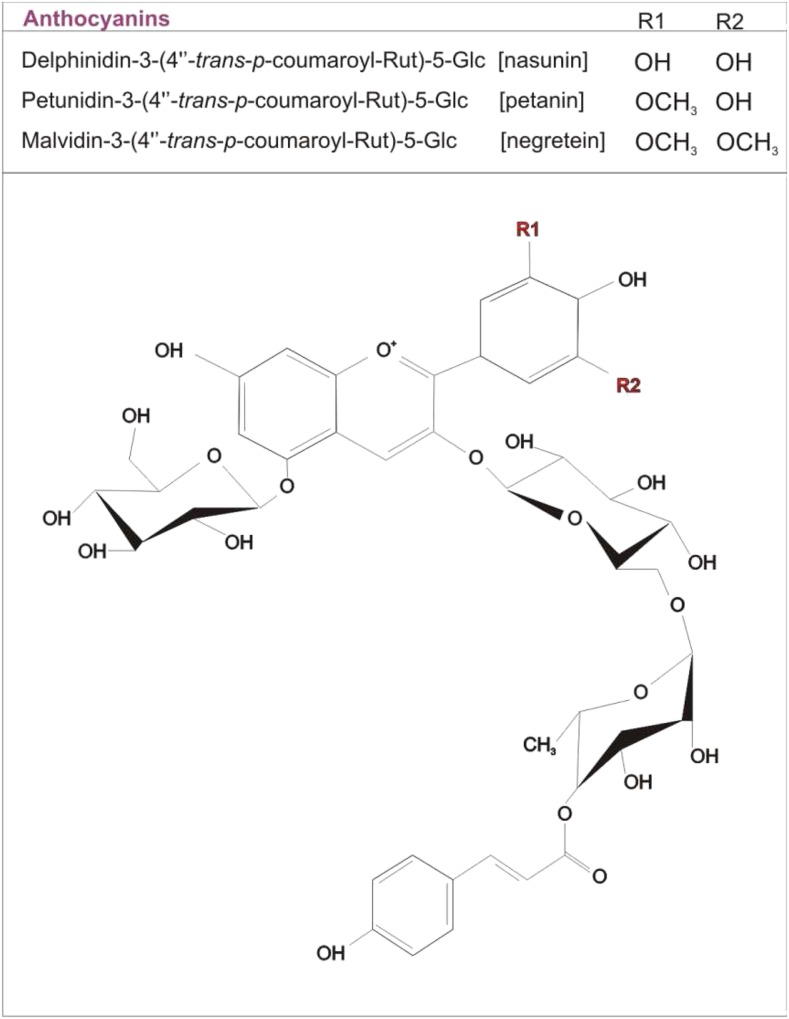
Bovy et al. (2002) tried to overexpress in tomato two anthocyanin biosynthetic regulatory genes of maize (Zea mays), the MYB gene C1 and the bHLH gene LC, which in other species ectopically activated anthocyanin synthesis. The transgenic tomatoes showed high level of flavonols in both the peel and flesh (Bovy et al., 2002). These two regulatory genes were thus sufficient to activate the flavonoid pathway in the flesh of the fruits. However, no change in the anthocyanin content was measured (Bovy et al., 2002). This was mainly due to the inability of the two maize TFs to activate the F3′5′H structural gene, which is necessary to produce dihydromyricetin, which, as explained above, represents the main dihydroflavonol of the anthocyanin biosynthetic pathway in tomato (Fig. 1B). The overexpression of other MYB-type TFs, both endogenous, such as SlANT1 (Mathews et al., 2003), and heterologous, such as AtPAP1 (Zuluaga et al., 2008), led to the first partial successes with tomatoes characterized by an anthocyanin-spotted fruit peel. Finally, anthocyanin-enriched tomatoes were produced when Butelli et al. (2008) overexpressed the regulatory genes Delila (Del; bHLH) and Rosea1 (Ros1; R2R3 MYB) from snapdragon (Antirrhinum majus) under the control of the tomato fruit-specific E8 promoter, resulting in completely purple fruits not only in the peel but also in the flesh (Fig. 2). Del and Ros1 were able to activate not only LBGs but also EBGs, such as Phe ammonia-lyase and CHI (Butelli et al., 2008), strongly promoting flavonoid biosynthesis. The anthocyanin content was comparable to the levels found in blackberries and blueberries (>2 mg g−1 fresh weight; Butelli et al., 2008). Seven different compounds were identified, with delphinidin-3–(4′′–trans–p–coumaroyl)-Rut-5-Glc and petunidin-3-(4′′–trans–p–coumaroyl)-Rut-5-Glc (petanin) as major molecules (Tohge et al., 2015; Fig. 3). The Del/Ros1 tomato extracts were used to feed cancer-susceptible Trp-53 −/− knockout mice, leading to an extended life span, suggesting that the anthocyanin levels contained in these transgenic tomatoes were sufficient to exert a protective role against cancer progression (Butelli et al., 2008). Remarkably, enrichment of anthocyanins significantly extended the shelf life of the purple tomato fruits, likely suppressing processes of overripening and reducing susceptibility to Botrytis cinerea, one of the most important postharvest pathogens (Zhang et al., 2013).
Figure 2.
Purple tomatoes produced by genetic engineering. The main classes of polyphenols synthesized in each tomato line are reported. Abbreviations: AtMYB12, A. thaliana MYB12; StSy, V. vinifera Stilbene Synthase. The photographs of wild-type, Del/Ros1, ResTom, and Indigo fruits are from Zhang et al. (2015; CC BY 4.0, http://creativecommons.org/licenses/by/4.0/). The photographs of Bronze tomatoes are from Scarano et al. (2018; CC BY 4.0, https://creativecommons.org/licenses/by/4.0/). No changes were made to the photographs.
Figure 3.
Main anthocyanins synthesized in purple tomato fruits. Petanin and delphinidin-3–(4′′–trans–p–coumaroyl)-Rut-5-Glc are the most abundant anthocyanins identified in Del/Ros1 tomato fruits. Petanin and malvidin-3-(4′′–trans–p–coumaroyl)-Rut-5-Glc are the most abundant molecules identified in Aft/Aft atv/atv tomato fruits. The chemical structure was redrawn from Blando et al. (2019).
Some years later, in a multilevel engineering approach, Del/Ros1 plants were crossed with a tomato line overexpressing the Arabidopsis (Arabidopsis thaliana) MYB12 TF, known to be able to strongly activate the flavonoid EBGs (Luo et al., 2008). The new tomato line was called “Indigo” (Zhang et al., 2015). AtMYB12 was able to bind in tomato the promoters of genes encoding enzymes of both primary and secondary metabolism strongly promoting flavonoid biosynthesis. The simultaneous presence of this TF with Del and Ros1 thus significantly increased the contents of the main phenylpropanoids (chlorogenic acids, flavonols, and anthocyanins) in Indigo tomatoes in comparison with the previous lines (Zhang et al., 2015; Fig. 2).
Finally, after the same multilevel approach, Scarano et al. (2018) developed a Bronze tomato characterized by three different classes of polyphenols (flavonols, anthocyanins, and stilbenoids), by introducing, in the Indigo line, the Stilbene Synthase gene from Vitis vinifera (Fig. 2). Using a mouse model for inflammatory bowel disease, Scarano et al. (2018) showed that supplementing the diet with Bronze tomato significantly reduced bowel inflammation more than the high anthocyanin or high stilbene tomatoes, demonstrating synergistic effects of different classes of polyphenols for reducing inflammation.
The above studies clearly indicate how the various metabolic engineering strategies for the enrichment of anthocyanins and other flavonoid compounds have been very successful in tomato and have increased the range of polyphenolic metabolites that can be produced. However, the purple tomato lines thus produced are not currently commercially available mainly due to the public’s worries about the safety of genetically modified food.
Genetic Mapping of Loci Associated with High-Anthocyanin Content in Tomato Fruits: from the Breeding of Purple Fruit-Lines to the Understanding of the Genetic Loci Involved
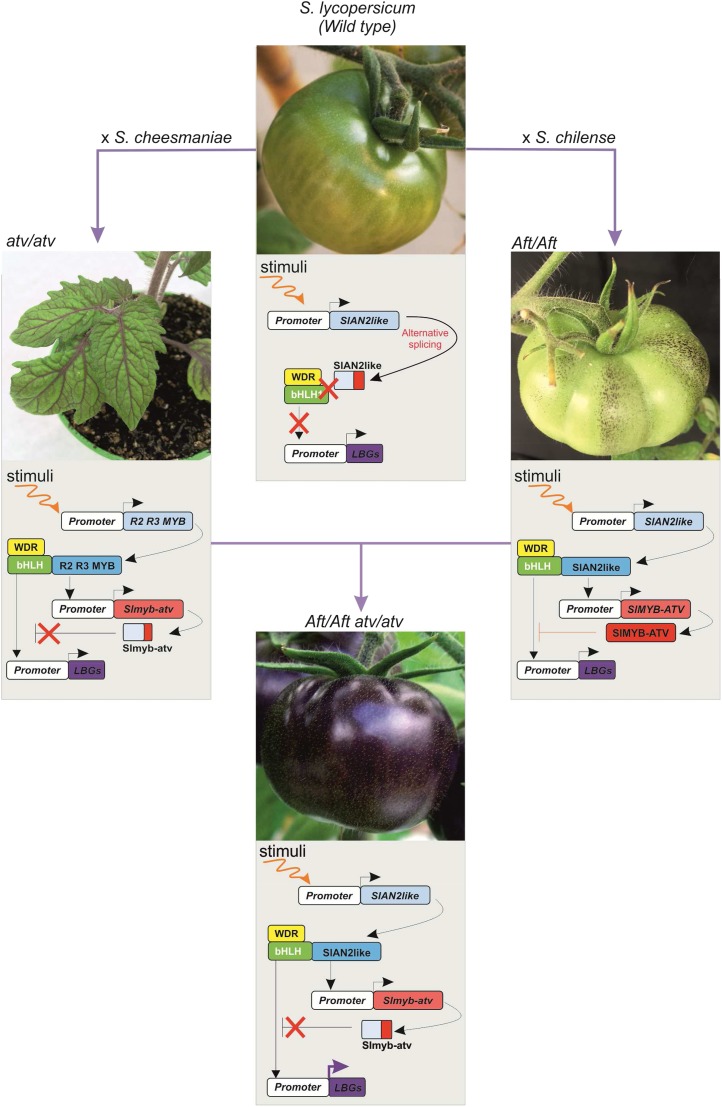
Although cultivated varieties do not synthesize anthocyanins in the fruits, some wild tomato species (e.g. Solanum chilense, Solanum peruvianum, Solanum lycopersicoides) produce green fruits which, upon suitable light conditions, can accumulate anthocyanins in the peel (Bedinger et al., 2011). Some of the related traits have been introgressed into the cultivated species through interspecific crosses, which in some cases occurred spontaneously (Mes et al., 2008; Gonzali et al., 2009; Myers, 2012). New tomato accessions were thus obtained showing anthocyanins in fruit peel, such as Anthocyanin fruit (Aft; Box 1) and Aubergine (Abg), or anthocyanin accumulation in vegetative tissues, such as atroviolacea (atv; Rick et al., 1968, 1994; Georgiev, 1972). These accessions have been used in subsequent breeding programs to further enrich the anthocyanin content of the fruits. The cross between Aft and atv or between Abg and atv produced Aft/Aft atv/atv and Abg/− atv/atv, respectively, which are two new lines with a strong purple anthocyanin pigmentation in the fruit peel (Mes et al., 2008; Gonzali et al., 2009). The more stable Aft/Aft atv/atv genetic combinations were also patented as nongenetically modified purple tomatoes and commercialized under different brands (Myers, 2012; Mazzucato et al., 2013). The strong purple phenotype that these lines can exhibit is due to the synergistic combination of the Aft and atv traits that allows a sustained activation of many EBGs and most of the LBGs (Povero et al., 2011). More than 75% of the anthocyanin profile in Aft/Aft atv/atv fruits is constituted by petanin and malvidin-3-(4′′–trans–p–coumaroyl)-Rut-5-Glc (Fig. 3; Box 2; Blando et al., 2019), both of them already identified in Del/Ros1 fruits. Some of the characteristics that anthocyanins conferred to Del/Ros1 purple tomatoes, such as extended shelf life and reduced susceptibility to postharvest pathogens, were also found in the Aft/Aft atv/atv fruits (Bassolino et al., 2013). This indicated that the peel-specific accumulation of anthocyanins, which is predominant in these latter tomatoes, is sufficient to provide them with these important properties.
The attempts to shed light on the regulation of the anthocyanin pathway in tomato started with the same genotypes as those used in breeding programs. For a long time, however, the genes responsible for the Aft and atv phenotypes were not identified, although, due to the specific inheritance of the two traits (Aft being dominant and atv recessive), the involvement of an activator and of a repressor of the pathway were hypothesized (Povero et al., 2011). Only in the last three years, also as a result of the greater understanding of the regulation of the anthocyanin biosynthetic pathway, have the natural allelic variations been uncovered that are responsible for the Aft and atv phenotypes. These studies also confirmed that the combination of specific alleles of these genes is indeed responsible for the complete and uniform purple fruit pigmentation of the Aft/Aft atv/atv lines. This thus demonstrated the existence of a strong synergy in the effects of the two genes, as initially hypothesized on the basis of the analyses of the phenotypes and of the initial molecular data (Povero et al., 2011).
An insertion in the SlMYB-ATV gene underlies the phenotype of the atv plants
The atv tomato line, derived from a spontaneous cross between S. lycopersicum and Solanum cheesmaniae (Rick et al., 1968), shows a general high anthocyanin abundance in the plant, which results in purple leaves, veins, branches, and stems (Fig. 4; Colanero et al., 2018). The phenotype was initially associated with a differential sensitivity to red/far red light, suggesting the involvement of a differential phytochrome-light perception and the classification of the line as a photomorphogenic mutant (Kendrick et al., 1997).
Figure 4.
Breeding anthocyanin-enriched tomato lines for the production of high-anthocyanin tomato fruits. The main regulation of the anthocyanin pathway is summarized under the picture of each tomato line. In the wild-type tomato fruit (top), the R2R3 MYB TF encoding SlAN2like gene is activated by an environmental signal (e.g. high amount of light), but, due to an alternative splicing mutation, its corresponding protein is truncated and nonfunctional. As a consequence, it cannot interact with a bHLH-partner protein and thus participate in the MBW complex that activates the transcription of different biosynthetic genes in the flavonoid pathway, including the LBGs, which are essential for the anthocyanin production. Anthocyanins are thus not produced. In the fruits of the atv tomato lines (left), the same situation described above is observed. However, in other organs of the atv plant, for example in the vegetative tissues, anthocyanin synthesis (activated by a suitable stimulus on the promoter of an R2R3 MYB TF encoding gene—e.g. SlAN2) is higher than in the wild-type plants because a repressor of the pathway, an R3 MYB factor encoded by the SlMYB-ATV gene, is mutated. The corresponding protein (slmyb-atv) is truncated and no longer able to repress the action of the MBW complex on the LBGs. Anthocyanin accumulation, therefore, is higher because it is not limited by the feedback repression mechanism exerted by the R3 MYB protein. In the fruits of the Aft line (right), the R2R3 MYB TF encoding gene SlAN2like is activated by an environmental signal (e.g. high amount of light) and produces a functional protein that interacts with the bHLH and WDR partners of the MBW complex, thus inducing anthocyanin synthesis. This is counterbalanced by the action of the repressor SlMYB-ATV factor, which, inhibiting the action of the MBW complex on the LBG promoters, prevents excessive accumulation of the purple pigments. In the fruits of the Aft/Aft atv/atv tomato genotype (bottom), the same activation mechanism mediated by the SlAN2like TF described for Aft takes place. In this case, however, the SlMYB-ATV gene is mutated as in atv plants. Therefore, once anthocyanin synthesis has been activated, it cannot be repressed by the R3 MYB protein—thus leading to a large accumulation of anthocyanins that can produce homogeneous purple pigmentation.
The first genetic mapping of the trait was carried out in the 1960s through the use of genetic markers, which highlighted chromosome 7 as being possibly linked with the phenotype (Rick et al., 1968; Clayberg, 1972). However, this genomic localization has only recently been confirmed. In 2017, through fine-mapping of chromosome 7, Cao et al. (2017) reached a 5-kb resolution of a region linked with the atv phenotype. This region harbors the gene Solyc07g052490 (Table 1) encoding an R3 MYB protein that has been considered as putatively responsible for the trait of study and thus named SlMYB-ATV. On the other hand, through genome sequencing of the atv line, Colanero et al. (2018) identified the exact location of the region introgressed from S. cheesmaniae in chromosome 7 and inside that region identified the same Solyc07g052490 gene as being responsible for the anthocyanin accumulation.
Table 1. Genes encoding the main regulators of the tomato anthocyanin biosynthetic pathway.
| Gene Code | Name | Function | References |
|---|---|---|---|
| Solyc10g086250 | SlAN2, SlMYB75 | R2R3 MYB | Povero et al., 2011; Myers, 2012; Kiferle et al., 2015 |
| Solyc10g086260 | SlANT1, anthocyanin1 | R2R3 MYB | Mathews et al., 2003; Sapir et al., 2008; Povero et al., 2011; Schreiber et al., 2012; Kiferle et al., 2015 |
| Solyc10g086270 | SlANT1like, SlMYB28 | R2R3 MYB | Kiferle et al., 2015 |
| Solyc10g086290 | SlAN2like, SlMYB114 | R2R3 MYB | Kiferle et al., 2015; Cao et al., 2017; Qiu et al., 2019; Yan et al., 2019; Colanero et al., 2020; Sun et al., 2020 |
| Solyc09g065100 | SlAN1, AH, SlTT8 | bHLH | Kiferle et al., 2015; Qiu et al., 2016 |
| Solyc08g081140 | SlJAF13, SlGL3 | bHLH | Nukumizu et al., 2013; Kiferle et al., 2015 |
| Solyc03g097340 | SlAN11 | WDR | Kiferle et al., 2015; Gao et al., 2018 |
| Solyc07g052490 | SlMYB-ATV | R3 MYB | Cao et al., 2017; Colanero et al., 2018; Yan et al., 2019; Sun et al., 2020 |
SlMYB-ATV is mutated and nonfunctional in the atv genotype, due to a 4-bp insertion in the second exon of the gene that leads to a frameshift and the onset of a premature stop codon. This produces a short truncated Slmyb-atv protein, with loss of the R3 domain (Cao et al., 2017; Colanero et al., 2018). In many plant species R3 MYB proteins act as negative regulators of various processes connected with cell fate determination (Wang and Chen, 2014), including the downregulation of anthocyanin biosynthesis (Zhu et al., 2009; Dubos et al., 2010). SlMYB-ATV is very similar in its structure to other R3 MYB proteins acting as repressors in the anthocyanin biosynthetic pathway, and particularly PhMYBx of petunia (Albert et al., 2014; Colanero et al., 2018).
R3 MYB proteins harbor in their R3 domain the conserved amino acid sequence responsible for the interaction with bHLH proteins (Zimmermann et al., 2004). It is thus believed that R3 MYBs downregulate anthocyanin synthesis by removing bHLH factors from the MBW complexes through competition with the activator R2R3 MYB TFs (Koes et al., 2005; Albert et al., 2014). This was demonstrated for SlMYB-ATV: the mutated Slmyb-atv protein cannot bind bHLH factors due to the loss of the R3 domain and thus cannot disrupt the MBW complex (Colanero et al., 2018; Sun et al., 2020). Furthermore, the transcriptional activation of the SlMYB-ATV gene by the same MBW complex, as previously postulated in the model of the regulation of the anthocyanin pathway (Albert et al., 2014), has been demonstrated (Colanero et al., 2018). Therefore, whereas in wild-type plants SlMYB-ATV acts as a repressor of the MBW complex in a negative feedback loop that fine-tunes anthocyanin production to avoid excessive pigment accumulation, in atv plants the lack of a functional R3 MYB repressor is responsible for the misregulated accumulation of anthocyanins wherever their production is induced (Fig. 4).
Aft encodes a functional R2R3 MYB TF that induces anthocyanin pigmentation in tomato fruits
The Aft accession derives from an interspecific cross between S. lycopersicum and S. chilense (Dunal) Reiche (Georgiev, 1972). The fruits of this wild species have purple cloves or spots in their peel, a trait that requires high light activation. This trait in Aft becomes a diluted phenotype that consists of spotted pigmentation (Davies et al., 2012) of the skin (Box 1) after exposure to intense light (Fig. 4). Similar to the identification of the atv mutation in chromosome 7, the region introgressed from S. chilense that contains the genes responsible for the phenotype was identified through genetic mapping (de Jong et al., 2004; Mes et al., 2008; Sapir et al., 2008). This region is located in the distal part of the long arm of chromosome 10, as finally confirmed by two recent studies (Yan et al., 2019; Sun et al., 2020). A genomic region associated with the production of anthocyanins was also mapped on chromosome 10 in eggplant and pepper, showing the existence of a strong synteny for this trait in various domesticated Solanaceae (Doganlar et al., 2002; Wang et al., 2018). Also the Abg locus, inherited from S. lycopersicoides, was mapped on the same chromosome region (Rick et al., 1994); however. it is still not clear if Abg is an allele of Aft, and it has not as yet been characterized.
Before the final identification of the gene responsible for the phenotype, several R2R3 MYB-encoding genes were identified in the Aft introgressed region: SlAN2, SlANT1, SlANT1like, and SlAN2like (Table 1; Sapir et al., 2008; Schreiber et al., 2012; Kiferle et al., 2015; Cao et al., 2017; Qiu et al., 2019). They are similar to the orthologous factors involved in anthocyanin regulation in other species (Mathews et al., 2003; Schreiber et al., 2012; Kiferle et al., 2015; Cao, et al., 2017). SlANT1 and SlAN2 were considered to be the main genes responsible for the anthocyanin-fruit phenotype (Sapir et al., 2008; Povero et al., 2011; Myers, 2012; Schreiber et al., 2012). In fact, when overexpressed under the constitutive cauliflower mosaic virus 35S promoter, both of these genes can cause a strong anthocyanin pigmentation in all of the main plant organs including the fruits (Kiferle et al., 2015). Therefore, they are functional activators of the pathway. Whereas SlANT1 has not yet been proven to play a physiological role in the plant, SlAN2 has been demonstrated to represent the main R2R3 MYB TF participating in the MBW complex in the vegetative tissues of tomato (Kiferle et al., 2015). However, later studies revealed very low expression of both genes not only in wild-type fruit peel, but also in Aft fruits (Cao et al., 2017; Qiu et al., 2019), thus weakening their possible role in the induction of fruit anthocyanin pigmentation.
Finally, three independent studies were recently published simultaneously, revealing the real genetic basis of the Aft trait, consisting of another R2R3 MYB encoding gene, SlAN2like, which is closely associated with the previous ones and with SlANT1like on the long arm of chromosome 10 (Yan et al., 2019; Colanero et al., 2020; Sun et al., 2020). Interestingly, the identification of SlAN2like with Aft was obtained through different and complementary approaches. Yan et al. (2019) fine-mapped the Aft locus to an interval of ∼145-kb on chromosome 10, a strategy that enabled them to exclude SlAN2, SlANT1, and SlANT1like from the candidates. Through CRISPR/Cas9-mediated silencing of SlAN2like, they then obtained a much lower accumulation of anthocyanins in the fruits, which was associated with the downregulation of the most important structural genes of the anthocyanin pathway.
Colanero et al. (2020) identified SlAN2like as Aft primarily through a functional approach. This revealed how, among all the wild-type and Aft versions of the four R2R3 MYB TFs encoded by the genes included in the Aft introgressed region, only the wild-type version of SlAN2like was nonfunctional. The Aft counterpart of the same protein is, on the contrary, fully functional in the activation of the anthocyanin biosynthetic pathway and also expressed at a much higher level in Aft fruit peel compared to all the other R2R3 MYB factors. Colanero et al. (2020) identified splicing mutations during the maturation of the precursor mRNA of the wild-type SlAN2like gene leading to aberrant mature SlAN2like mRNAs. The mutations in the wild-type SlAN2like create premature stop codons, which produce short proteins lacking most of the R2R3 domains and all of the C-terminal part of the TF. These proteins were found to be unable to bind the bHLH partners of the MBW anthocyanin complex, thus explaining their inefficacy in the induction of fruit pigmentation.
Sun et al. (2020) focused on the Aft/Aft atv/atv ‘Indigo Rose’ (InR) tomato line. They identified SlAN2like through a bulk-population–sequencing approach. Many of the results of the previous articles were confirmed in Sun’s study and additional pieces of the puzzle were found. They demonstrated that SlAN2like activates the transcription of both SlAN1 (Table 1), encoding the bHLH protein that takes part in the key MBW complex of tomato, and of the repressor SlMYB-ATV. Sun et al. (2020) also highlighted the competition of SlAN2like and SlMYB-ATV in binding the same bHLH partner SlAN1. They also found the exact location of the splicing mutation in SlAN2like that prevents wild-type plants from assembling a correct mature mRNA. Finally, through genetic engineering, Sun et al. (2020) reconstructed all of the network that induces anthocyanin synthesis and accumulation in the fruits of the double mutant Aft/Aft atv/atv InR (Fig. 4). They found that: CRISPR/Cas9-mediated silencing of all four R2R3 MYB genes SlAN2, SlANT1, SlANT1like, and SlAN2like in InR fruits proved that only SlAN2like is necessary to have the anthocyanin-enriched phenotype; and the overexpression of SlAN2like in wild-type plants induces a purple anthocyanin pigmentation in the fruit peel. They also revealed that the expression of the wild-type SlMYB-ATV gene in InR fruits reverts their phenotype to Aft; and, on the other hand, the knock-out of SlMYB-ATV in Aft fruits increases the pigmentation to that of InR. The essential role of SlAN2like in the induction of the anthocyanin synthesis in the fruits of tomato was thus definitively proved.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Purple tomato fruits are now one of the many variants of color offered on the market to consumers. Because tomato is one of the most consumed vegetables all over the world, anthocyanin-enriched tomatoes can provide our diet with the beneficial amount of antioxidants that preventive medicine recommends. Breeding has mostly exploited the characteristics of Aft and atv which, combined, produce the maximum amount of pigments in the fruit peel, the tissue that is genetically programmed to synthesize flavonoids. High concentrations of anthocyanins can thus be achieved without negatively impacting other metabolites, enhancing the nutraceutical properties of these tomatoes (Box 2).
Recent results have not ended research in this field (see Outstanding Questions). As shown by some genetically engineered purple tomatoes, anthocyanins can also be synthesized in the pulp of the fruit, provided the biosynthetic pathway is appropriately activated. Further studies may thus lead to the identification of new TFs that differentially modulate (either inducing more or repressing less) the pathway in the same peel or in other parts of the fruit. The accumulation of anthocyanins in the flesh would mean that purple tomatoes would not just be consumed fresh but could also be used for processed products, such as pastes, sauces, and juices. The characterization of the Abg mutation, stronger than Aft in inducing fruit pigmentation, certainly represents an interesting new step in this direction.
The uniformity of pigmentation could also be improved. In fact, the Aft/Aft atv/atv purple tomatoes, irrespectively of the background variety where the two alleles were introduced, show a light-dependent and cold-dependent phenotype (Povero et al., 2011). This often restricts the uniform purple coloration, which is so decisive in attracting consumers. Genome editing technologies, which can target the promoter regions of the regulatory genes, could contribute significantly to achieving such uniform coloration.
Acknowledgments
We apologize to those whose work we were unable to cite owing to space limitations.
Footnotes
Articles can be viewed without a subscription.
References
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26: 962–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Lindhout P (2007) Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Ann Bot 100: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassolino L, Zhang Y, Schoonbeek HJ, Kiferle C, Perata P, Martin C (2013) Accumulation of anthocyanins in tomato skin extends shelf life. New Phytol 200: 650–655 [DOI] [PubMed] [Google Scholar]
- Bedinger PA, Chetelat RT, McClure B, Moyle LC, Rose JK, Stack SM, van der Knaap E, Baek YS, Lopez-Casado G, Covey PA, et al. (2011) Interspecific reproductive barriers in the tomato clade: Opportunities to decipher mechanisms of reproductive isolation. Sex Plant Reprod 24: 171–187 [DOI] [PubMed] [Google Scholar]
- Blando F, Berland H, Maiorano G, Durante M, Mazzucato A, Picarella ME, Nicoletti I, Gerardi C, Mita G, Andersen Ø (2019) Nutraceutical characterization of anthocyanin-rich fruits produced by “Sun Black” tomato line. Front Nutr 6: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy A, de Vos R, Kemper M, Schijlen E, Almenar Pertejo M, Muir S, Collins G, Robinson S, Verhoeyen M, Hughes S, et al. (2002) High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy A, Schijlen E, Hall RD (2007) Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): The potential for metabolomics. Metabolomics 3: 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EG, Hall RD, Bovy AG, Luo J, et al. (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Cao X, Qiu Z, Wang X, Van Giang T, Liu X, Wang J, Wang X, Gao J, Guo Y, Du Y, Wang G, Huang Z (2017) A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit. J Exp Bot 68: 5745–5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy A, Mukamal KJ, Liu L, Franz M, Eliassen AH, Rimm EB (2013) High anthocyanin intake is associated with a reduced risk of myocardial infarction in young and middle-aged women. Circulation 127: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker-Scott L. (1999) Environmental significance of anthocyanins in plant stress responses. Photochem Photobiol 70: 1–9 [Google Scholar]
- Clayberg CD. (1972) Preliminary mapping of three chromosome 7 genes. Rep Tomato Genet Coop 22: 4 [Google Scholar]
- Colanero S, Perata P, Gonzali S (2018) The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front Plant Sci 9: 830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanero S, Tagliani A, Perata P, Gonzali S (2020) Alternative splicing in the Anthocyanin fruit gene encoding an R2R3 MYB transcription factor affects anthocyanin biosynthesis in tomato fruits. Plant Communications 1: 100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Albert NW, Schwinn KE (2012) From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct Plant Biol 39: 619–638 [DOI] [PubMed] [Google Scholar]
- de Jong WS, Eannetta NT, de Jong DM, Bodis M (2004) Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor Appl Genet 108: 423–432 [DOI] [PubMed] [Google Scholar]
- Doganlar S, Frary A, Daunay MC, Lester RN, Tanksley SD (2002) Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics 161: 1713–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Gao Y, Liu J, Chen Y, Tang H, Wang Y, He Y, Ou Y, Sun X, Wang S, Yao Y (2018) Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic Res 5: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev C. (1972) Anthocyanin fruit (Aft). Rep Tomato Genet Coop 22: 10 [Google Scholar]
- Gonzali S, Mazzucato A, Perata P (2009) Purple as a tomato: Towards high anthocyanin tomatoes. Trends Plant Sci 14: 237–241 [DOI] [PubMed] [Google Scholar]
- Gould KS. (2004) Nature’s Swiss Army knife: The diverse protective roles of anthocyanins in leaves. J Biomed Biotechnol 2004: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochem 55: 481–504 [DOI] [PubMed] [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62: 2465–2483 [DOI] [PubMed] [Google Scholar]
- Hoballah ME, Gübitz T, Stuurman J, Broger L, Barone M, Mandel T, Dell’Olivo A, Arnold M, Kuhlemeier C (2007) Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell 19: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch WA, Zeldin EL, McCown BH (2001) Physiological significance of anthocyanins during autumnal leaf senescence. Tree Physiol 21: 1–8 [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ET, Ryu S, Yi H, Shin B, Cheong H, Choi G (2001) Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J 25: 325–333 [DOI] [PubMed] [Google Scholar]
- Kendrick RE, Kerckhoffs LHJ, van Tuinen A, Koornneef M (1997) Photomorphogenic mutants of tomato. Plant Cell Environ 20: 746–751 [Google Scholar]
- Kiferle C, Fantini E, Bassolino L, Povero G, Spelt C, Buti S, Giuliano G, Quattrocchio F, Koes R, Perata P, et al. (2015) Tomato R2R3-MYB proteins SlANT1 and SlAN2: Same protein activity, different roles. PLoS One 10: e0136365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Krga I, Milenkovic D (2019) Anthocyanins: From sources and bioavailability to cardiovascular-health Benefits and molecular mechanisms of action. J Agric Food Chem 67: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Li XN, Lin J, Xia J, Qin L, Zhu SY, Li JL (2017) Lycopene mitigates atrazine-induced cardiac inflammation via blocking the NF-kB pathway and NO production. J Funct Foods 29: 208–216 [Google Scholar]
- Liu C, Zhu L, Fukuda K, Ouyang S, Chen X, Wang C, Zhang CJ, Martin B, Gu C, Qin L, et al. (2017) The flavonoid cyanidin blocks binding of the cytokine interleukin-17A to the IL-17RA subunit to alleviate inflammation in vivo. Sci Signal 10: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8: 689–708 [DOI] [PubMed] [Google Scholar]
- Liu Y, Tikunov Y, Schouten RE, Marcelis LFM, Visser RGF, Bovy A (2018) Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front Chem 6: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Butelli E, Hill L, Parr A, Niggeweg R, Bailey P, Weisshaar B, Martin C (2008) AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: Expression in fruit results in very high levels of both types of polyphenol. Plant J 56: 316–326 [DOI] [PubMed] [Google Scholar]
- Martí R, Roselló S, Cebolla-Cornejo J (2016) Tomato as a source of carotenoids and polyphenols targeted to cancer prevention. Cancers (Basel) 8: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Butelli E, Petroni K, Tonelli C (2011) How can research on plants contribute to promoting human health? Plant Cell 23: 1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín J, Navas MJ, Jiménez-Moreno AM, Asuero AG (2017) Anthocyanin Pigments: Importance, Sample Preparation and Extraction. Intech Open, Rijeka, Croatia, pp 117–152 [Google Scholar]
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, et al. (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15: 1689–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauray A, Felgines C, Morand C, Mazur A, Scalbert A, Milenkovic D (2010) Nutrigenomic analysis of the protective effects of bilberry anthocyanin-rich extract in apo E-deficient mice. Genes Nutr 5: 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucato A, Willems D, Bernini R, Picarella ME, Santangelo E, Ruiu F, Tilesi F, Soressi GP (2013) Novel phenotypes related to the breeding of purple-fruited tomatoes and effect of peel extracts on human cancer cell proliferation. Plant Physiol Biochem 72: 125–133 [DOI] [PubMed] [Google Scholar]
- Mes PJ, Boches P, Myers JR (2008) Characterization of tomatoes expressing anthocyanin in the fruit. J Am Soc Hortic Sci 133: 262–269 [Google Scholar]
- Mink PJ, Scrafford CG, Barraj LM, Harnack L, Hong CP, Nettleton JA, Jacobs DR Jr. (2007) Flavonoid intake and cardiovascular disease mortality: A prospective study in postmenopausal women. Am J Clin Nutr 85: 895–909 [DOI] [PubMed] [Google Scholar]
- Montefiori M, Brendolise C, Dare AP, Lin-Wang K, Davies KM, Hellens RP, Allan AC (2015) In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J Exp Bot 66: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozos I, Stoian D, Caraba A, Malainer C, Horbańczuk JO, Atanasov AG (2018) Lycopene and vascular health. Front Pharmacol 9: 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir SR, Collins GJ, Robinson S, Hughes S, Bovy A, Ric de Vos CH, van Tunen AJ, Verhoeyen ME (2001) Overexpression of Petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat Biotechnol 19: 470–474 [DOI] [PubMed] [Google Scholar]
- Muraki I, Imamura F, Manson JE, Hu FB, Willett WC, van Dam RM, Sun Q (2013) Fruit consumption and risk of type 2 diabetes: Results from three prospective longitudinal cohort studies. BMJ 347: f5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J. (2012) Breeding tomatoes for increased flavonoids. In Proceedings of the 6th Organic Seed Growers Conference. Organic Seed Alliance, Port Townsend, WA, pp 50–51 [Google Scholar]
- Nukumizu Y, Wada T, Tominaga-Wada R (2013) Tomato (Solanum lycopersicum) homologs of TRIPTYCHON (SlTRY) and GLABRA3 (SlGL3) are involved in anthocyanin accumulation. Plant Signal Behav 8: e24575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta IE, Spooner DM (2007) History, origin and early cultivation of tomato (Solanaceae). In Genetic Improvement of Solanaceous Crops, Vol. 2: Tomato. CRC Press, Boca Raton, Florida [Google Scholar]
- Povero G, Gonzali S, Bassolino L, Mazzucato A, Perata P (2011) Transcriptional analysis in high-anthocyanin tomatoes reveals synergistic effect of Aft and atv genes. J Plant Physiol 168: 270–279 [DOI] [PubMed] [Google Scholar]
- Qiu Z, Wang H, Li D, Yu B, Hui Q, Yan S, Huang Z, Cui X, Cao B (2019) Identification of candidate HY5-dependent and -independent regulators of anthocyanin biosynthesis in tomato. Plant Cell Physiol 60: 643–656 [DOI] [PubMed] [Google Scholar]
- Qiu Z, Wang X, Gao J, Guo Y, Huang Z, Du Y (2016) The Tomato Hoffman’s anthocyaninless gene encodes a bHLH transcription factor involved in anthocyanin biosynthesis that is developmentally regulated and induced by low temperatures. PLoS One 11: e0151067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Dixon RA (1999) Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11: 1537–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick CM, Cisneros P, Chetelat RT, Deverona JW (1994) Abg, a gene on chromosome 10 for purple fruit derived from S. lycopersicoides. Rep Tomato Genet Coop 44: 29–30 [Google Scholar]
- Rick CM, Reeves AF, Zobel RW (1968) Inheritance and linkage relations of four new mutants. Rep Tomato Genet Coop 18: 34–35 [Google Scholar]
- Robinson GM, Robinson R (1931) A survey of anthocyanins. I. Biochem J 25: 1687–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir M, Oren-Shamir M, Ovadia R, Reuveni M, Evenor D, Tadmor Y, Nahon S, Shlomo H, Chen L, Meir A, et al. (2008) Molecular aspects of Anthocyanin fruit tomato in relation to high pigment-1. J Hered 99: 292–303 [DOI] [PubMed] [Google Scholar]
- Sasaki N, Nishizaki Y, Ozeki Y, Miyahara T (2014) The role of acyl-glucose in anthocyanin modifications. Molecules 19: 18747–18766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarano A, Butelli E, de Santis S, Cavalcanti E, Hill L, de Angelis M, Giovinazzo G, Chieppa M, Martin C, Santino A (2018) Combined dietary anthocyanins, flavonols, and stilbenoids alleviate inflammatory bowel disease symptoms in mice. Front Nutr 4: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schijlen E, Ric de Vos CH, Jonker H, van den Broeck H, Molthoff J, van Tunen A, Martens S, Bovy A (2006) Pathway engineering for healthy phytochemicals leading to the production of novel flavonoids in tomato fruit. Plant Biotechnol J 4: 433–444 [DOI] [PubMed] [Google Scholar]
- Schreiber G, Reuveni M, Evenor D, Oren-Shamir M, Ovadia R, Sapir-Mir M, Bootbool-Man A, Nahon S, Shlomo H, Chen L, et al. (2012) ANTHOCYANIN1 from Solanum chilense is more efficient in accumulating anthocyanin metabolites than its Solanum lycopersicum counterpart in association with the Anthocyanin fruit phenotype of tomato. Theor Appl Genet 124: 295–307 [DOI] [PubMed] [Google Scholar]
- Sharma A, Shahzad B, Rehman A, Bhardwaj R, Landi M, Zheng B (2019) Response of pathway and the role of polyphenols in plants under abiotic stress. Molecules 24: 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata K, Shibata Y, Kasiwagi I (1919) Studies on anthocyanins: Color variation in anthocyanins. J Am Chem Soc 41: 208–220 [Google Scholar]
- Springob K, Nakajima J, Yamazaki M, Saito K (2003) Recent advances in the biosynthesis and accumulation of anthocyanins. Nat Prod Rep 20: 288–303 [DOI] [PubMed] [Google Scholar]
- Sun C, Deng L, Du M, Zhao J, Chen Q, Huang T, Jiang H, Li C-B, Li C (2020) A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol Plant 13: 42–58 [DOI] [PubMed] [Google Scholar]
- Tanksley SD. (2004) The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16(Suppl): S181–S189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D, Zhu G, Resende MFR Jr., Lin T, Nguyen C, Bies D, Rambla JL, Beltran KSO, Taylor M, Zhang B, et al. (2017) A chemical genetic roadmap to improved tomato flavor. Science 355: 391–394 [DOI] [PubMed] [Google Scholar]
- Tohge T, Zhang Y, Peterek S, Matros A, Rallapalli G, Tandròn Y-A, Butelli E, Kallam K, Hertkorn N, Mock H-P, et al. (2015) Ectopic expression of snapdragon transcription factors facilitates the identification of genes encoding enzymes of anthocyanin decoration in tomato. Plant J 83: 686–704 [DOI] [PubMed] [Google Scholar]
- Wang C, Yatsuya H, Tamakoshi K, Uemura M, Li Y, Wada K, Yamashita K, Kawaguchi L, Toyoshima H, Aoyama A (2013) Positive association between high-sensitivity C-reactive protein and incidence of type 2 diabetes mellitus in Japanese workers: 6-year follow-up. Diabetes Metab Res Rev 29: 398–405 [DOI] [PubMed] [Google Scholar]
- Wang D, Seymour GB (2017) Tomato flavor: Lost and found? Mol Plant 10: 782–784 [DOI] [PubMed] [Google Scholar]
- Wang D, Xia M, Yan X, Li D, Wang L, Xu Y, Jin T, Ling W (2012b) Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing miRNA-10b. Circ Res 111: 967–981 [DOI] [PubMed] [Google Scholar]
- Wang G, Chen B, Du H, Zhang F, Zhang H, Wang Y, He H, Geng S, Zhang X (2018) Genetic mapping of anthocyanin accumulation-related genes in pepper fruits using a combination of SLAF-seq and BSA. PLoS One 13: e0204690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ma C, Rong W, Jing H, Hu X, Liu X, Jiang L, Wei F, Liu Z (2012a) Bog bilberry anthocyanin extract improves motor functional recovery by multifaceted effects in spinal cord injury. Neurochem Res 37: 2814–2825 [DOI] [PubMed] [Google Scholar]
- Wang S, Chen JG (2014) Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front Plant Sci 5: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20: 176–185 [DOI] [PubMed] [Google Scholar]
- Yan S, Chen N, Huang Z, Li D, Zhi J, Yu B, Liu X, Cao B, Qiu Z (2019) Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol 18 [DOI] [PubMed] [Google Scholar]
- Youdim KA, Shukitt-Hale B, Joseph JA (2004) Flavonoids and the brain: Interactions at the blood-brain barrier and their physiological effects on the central nervous system. Free Radic Biol Med 37: 1683–1693 [DOI] [PubMed] [Google Scholar]
- Zhang B, Chopra D, Schrader A, Hülskamp M (2019) Evolutionary comparison of competitive protein-complex formation of MYB, bHLH, and WDR proteins in plants. J Exp Bot 70: 3197–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, Alseekh S, Tohge T, Rallapalli G, Luo J, Kawar PG, Hill L, Santino A, Fernie AR, et al. (2015) Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat Commun 6: 8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, de Stefano R, Schoonbeek HJ, Magusin A, Pagliarani C, Wellner N, Hill L, Orzaez D, Granell A, et al. (2013) Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr Biol 23: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HF, Fitzsimmons K, Khandelwal A, Kranz RG (2009) CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol Plant 2: 790–802 [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40: 22–34 [DOI] [PubMed] [Google Scholar]
- Zuluaga DL, Gonzali S, Loreti E, Pucciariello C, Degl’Innocenti E, Guidi L, Alpi A, Perata P (2008) Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct Plant Biol 35: 606–618 [DOI] [PubMed] [Google Scholar]