TCP transcription factors participate in stamen filament elongation in response to the plant hormone gibberellin through direct modulation of a family of genes involved in cell expansion.
Abstract
In autogamous plants like Arabidopsis (Arabidopsis thaliana), stamen filament elongation must be finely regulated to ensure that anthers reach the pistil at the correct developmental stage. In this work, we studied the roles of Arabidopsis TEOSINTE BRANCHED1, CYCLOIDEA, PCF15 (TCP15), and related class-I TCP transcription factors in stamen filament elongation. Plants with decreased expression of class-I TCPs and plants that express a fusion of TCP15 to a repressor domain (pTCP15::TCP15-EAR) had shorter stamens, indicating that class-I TCPs stimulate filament growth. These plants also showed reduced expression of several SMALL AUXIN UP RNA (SAUR)63 subfamily genes, which contain TCP target motifs in their promoters. Mutational analysis indicated that the TCP target motif in the SAUR63 promoter is required for expression of SAUR63 in stamen filaments. Moreover, TCP15 directly binds to the SAUR63 promoter region that contains the TCP target motif in vivo, highlighting the role of the TCPs in this process. Class-I TCPs are also required for the induction of SAUR63 subfamily genes by gibberellins (GAs). In addition, overexpression of SAUR63 restores filament growth in pTCP15::TCP15-EAR plants, whereas overexpression of TCP15 rescues the short stamen phenotype of GA-deficient plants. The results indicate that TCP15 and related class-I TCPs modulate GA-dependent stamen filament elongation by direct activation of SAUR63 subfamily genes through conserved target sites in their promoters. This work provides insight into GA-dependent stamen filament elongation.
Stamen filament elongation is particularly important in self-pollinating, autogamous species, like Arabidopsis (Arabidopsis thaliana). In these plants, the pollen fertilizes ovules from the same flower, usually before the flower opens. Stamens must elongate for anthers to reach the top of the gynoecium at the correct developmental stage (i.e. when the stigma becomes receptive and anthers dehisce). Incorrect stamen filament elongation may severely affect successful reproduction. Not surprisingly, the elongation of stamen filaments is a strictly controlled process and occurs during specific stages of flower development (Tashiro et al., 2009; Cardarelli and Cecchetti, 2014). Stamen filaments are first recognizable from the anther at flower stage 7. At stage 9, the filaments represent only 20% of the length of stamens. A phase of filament elongation, known as pre-anthesis growth, takes place between stages 10 and 13 and is mainly due to cell expansion. During this phase, the stamens reach the length of the pistil at stage 13, when the flower opens. Postanthesis growth occurs until stage 14, when the stigma becomes receptive for pollination.
Several hormones, including auxin, gibberellins (GAs), and jasmonic acid (JA), are involved in stamen filament elongation (Song et al., 2013). Plants defective in auxin biosynthesis, transport, or perception and double mutants in the AUXIN RESPONSE FACTOR genes ARF6 and ARF8 show defects in stamen filament elongation and anther maturation (Nagpal et al., 2005; Cecchetti et al., 2008; Tashiro et al., 2009; Tabata et al., 2010; Reeves et al., 2012). Particularly, a splice variant of ARF8 (ARF8.4) is required for stamen filament elongation and the correct expression of the auxin inducible gene AUX/IAA19 (Ghelli et al., 2018). Auxin transport from the tapetum through the middle layer and toward the filament is required to coordinate anther maturation with filament growth (Cecchetti et al., 2017). Mutations in JA biosynthesis genes, or in certain components of the JA signaling pathway, also affect filament elongation (Xie et al., 1998; Stintzi and Browse, 2000; Ishiguro et al., 2001; Park et al., 2002). The response to JA in stamens is mediated by two JA-inducible MYB transcription factors, MYB21 and MYB24 (Mandaokar et al., 2006), which are targeted by Jasmonate-ZIM domain proteins (Song et al., 2011). It has been reported that ARF6 and ARF8 induce the expression of JA biosynthesis genes during late stages of stamen development, indicating that auxin acts upstream of JA (Nagpal et al., 2005; Tabata et al., 2010; Reeves et al., 2012). However, the fact that stamen filament elongation is not rescued by JA treatment of arf6 arf8 mutants (Nagpal et al., 2005) suggests that additional pathways are involved. Among the genes downregulated in arf6 arf8 mutant flowers, there are several SMALL AUXIN UP RNA (SAUR) genes from the SAUR63 subfamily (Nagpal et al., 2005). SAUR proteins promote cell expansion by activating plasma membrane H+-ATPases (Spartz et al., 2014) and the overexpression of SAUR63 subfamily members stimulates stamen filament elongation (Chae et al., 2012). Thus, induction of SAUR genes by ARF6 and ARF8 may be required, in addition to JA biosynthesis, to stimulate filament elongation.
Plants defective in GA biosynthesis or perception also show defects in stamen filament elongation (Cheng et al., 2004; Tyler et al., 2004; Rieu et al., 2008). GAs induce the synthesis of JA and the expression of MYB transcription factors to modulate stamen development; however, the short stamen phenotype of GA-deficient plants cannot be rescued by exogenous JA, suggesting that other GA-dependent, JA-independent pathways are required for correct stamen filament elongation (Cheng et al., 2009). Notably, analysis of available microarray data indicates that several SAUR63 subfamily genes are also induced by GAs (Bai et al., 2012; Ren and Gray, 2015), suggesting that GA-dependent stamen filament elongation may involve the induction of SAUR genes. However, the mechanism involved in this process is largely unknown.
Teosinte branched1, cycloidea, PCF (TCP) transcription factors regulate several aspects of plant development, including plant architecture, leaf morphogenesis and maturation, inflorescence stem growth, and floral organ development (Martín-Trillo and Cubas, 2010; Manassero et al., 2013). Twenty-four TCP proteins (TCPs), assigned to either class I (13 proteins) or class II (11 proteins), are encoded in the Arabidopsis genome. Class-I proteins show a high degree of functional redundancy, and thus developmental phenotypes are usually observed only in higher-order mutants or plants that express fusions of the TCPs to the EAR domain (Kieffer et al., 2011; Uberti-Manassero et al., 2012; Aguilar-Martínez and Sinha, 2013). Fusions to the EAR domain convert transcription factors into strong dominant repressor forms (Hiratsu et al., 2003). This strategy is useful in cases of genetic redundancy and has been widely used to study the role of transcription factors, including those of the TCP family (Koyama et al., 2007, 2010; Kieffer et al., 2011; Li et al., 2012; Uberti-Manassero et al., 2012; Aguilar-Martínez and Sinha, 2013). This type of analysis revealed that class-I TCPs either positively or negatively modulate cell proliferation and expansion depending on the organ/tissue involved (Kieffer et al., 2011). Interplay of TCPs with hormone action was also described (Nicolas and Cubas, 2016). As an example, TCP14 and TCP15 negatively modulate both auxin biosynthesis genes during gynoecium development and the expression of the auxin reporter DR5:GUS in vegetative and reproductive tissues (Lucero et al., 2015). TCP14 and TCP15 also participate in GA-dependent germination, flowering, and inflorescence stem elongation (Davière et al., 2014; Resentini et al., 2015; Lucero et al., 2017). TCP20 and TCP9, more distantly related class-I TCPs, inhibit JA biosynthesis through the repression of the JA biosynthesis gene LIPOXYGENASE2 (Danisman et al., 2012).
In this work, we investigated the role of TCP15 and related class-I TCPs in stamen filament elongation. We report that class-I TCPs participate in GA-dependent stamen filament elongation by directly inducing the expression of SAUR63 subfamily genes. These results contribute to a more comprehensive understanding of the molecular pathway between GA action and stamen filament elongation.
RESULTS
TCP15 and Related Class-I TCPs Affect Stamen Filament Elongation
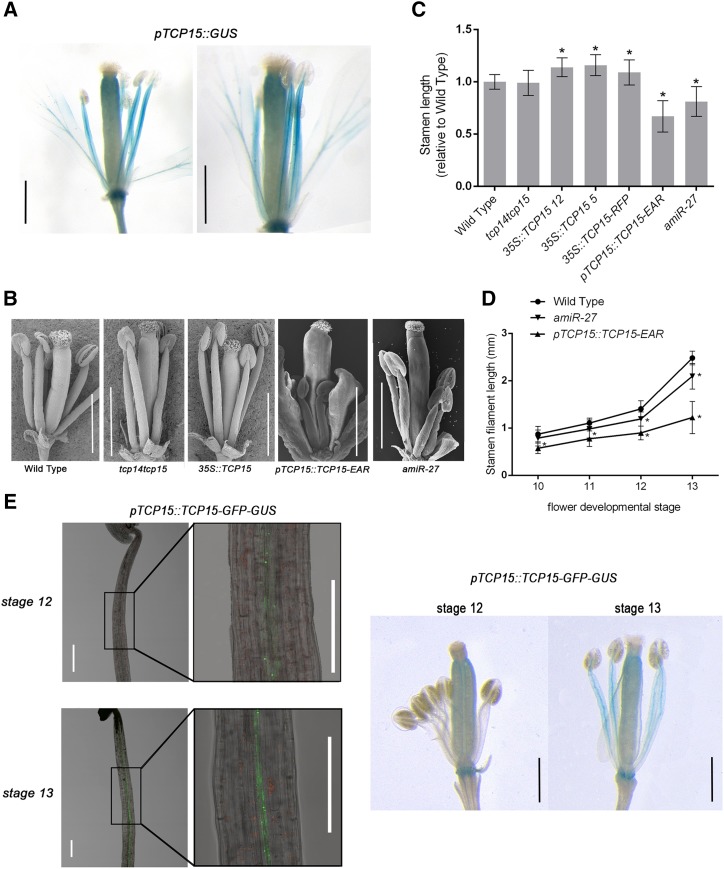
The analysis of plants bearing the TCP15 promoter region controlling the expression of the GUS reporter gene revealed that the TCP15 promoter is active in stamen filaments (Fig. 1A). Expression of the reporter gene was mainly observed in the vasculature and was extended to peripheral tissues. To evaluate a possible role of TCP15 in this organ, we analyzed stamen length in plants expressing a fusion of TCP15 to the EAR repressor domain under the control of the TCP15 promoter (pTCP15::TCP15-EAR plants). In this case, the assumption is that expressing TCP15-EAR within the expression domain of the TCP15 promoter will cause the downregulation of TCP15 target genes. Through this strategy, we expected to uncover a possible role of TCP15 in stamen filaments even in the presence of genetic redundancy with other class-I TCPs. We observed that the stamen length of pTCP15::TCP15-EAR plants was reduced by ∼30% to 40% relative to wild type (Fig. 1, B and C). As a complementary approach, we used plants that express TCP15 from the 35S Cauliflower mosaic virus (CaMV) promoter, either in its native form or fused to the red fluorescent protein (35S::TCP15 and 35S::TCP15-RFP plants, respectively), to analyze the effect of increased TCP15 expression. Plants of both lines exhibited longer filaments than wild-type plants (Fig. 1, B and C). This result suggests that TCP15 stimulates stamen filament elongation. Because the expression of a repressor form (TCP15-EAR) causes the opposite effect of overexpressing the native protein, it can be concluded that TCP15 most likely acts as an activator during this process. If TCP15 stimulates filament elongation, it is expected that loss-of-function of TCP15 will affect stamen length. We then analyzed filament length in a line carrying mutations in TCP15 and the closely related gene TCP14, but found that this double mutant showed no significant difference with wild type (Fig. 1, B and C). We speculate that this is likely due to functional redundancy with other class-I proteins, as observed in other processes (Kieffer et al., 2011; Uberti-Manassero et al., 2012; Aguilar-Martínez and Sinha, 2013). Analysis of expression levels of class-I TCPs in stamen filaments using data available in the Transcriptome Variation Analysis database (http://travadb.org/; Klepikova et al., 2016) indicated that several TCP genes are expressed in this organ, including TCP7, TCP8, TCP14, TCP15, TCP21, and TCP22 (Supplemental Table S1). Among these, TCP7 is the most highly expressed, accounting for 29% of all class-I TCP transcripts, similar to the sum of those of TCP14 and TCP15 (27%). Whereas available tcp7 insertional mutants do not show reduced transcript levels (Aguilar-Martínez and Sinha, 2013), plants that express an artificial micro RNA targeting the class-I TCP genes TCP8 and TCP22 in a tcp14 tcp15 mutant background were described before (Davière et al., 2014). We measured TCP8 and TCP22 transcript levels in stamen filaments of these plants (amiR-27 plants) and found that they were reduced by ∼35% and 70%, respectively, whereas the expression levels of TCP7 and TCP21 were not significantly affected (Supplemental Fig. S1). Analysis of amiR-27 plants indicated that they showed shorter stamen filaments than wild type (Fig. 1, B and C), leading to the conclusion that downregulated expression of TCP8 and TCP22, in addition to TCP14 and TCP15, affects filament elongation. Thus, our results indicate that several TCP15-related class-I TCPs redundantly participate in stamen filament elongation. In the case of pTCP15::TCP15-EAR, no changes in expression were observed for class-I TCP genes other than for TCP15 (Supplemental Fig. S1). TCP15 transcript levels were ∼2.5-fold higher in these plants that in wild-type plants (Supplemental Fig. S1). Because this represents the sum of transcripts arising from the endogenous gene and the TCP15-EAR transgene, it can be concluded that the transgene was expressed at roughly similar levels as the endogenous TCP15 gene in these plants. Altogether, our results suggest that the short stamen phenotype is due to a direct effect of the repressor form of TCP15 and not caused by changes in the expression of the other class-I TCPs.
Figure 1.
TCP15 and related class-I TCPs participate in stamen filament elongation. A, GUS expression pattern in flowers of plants that contain a fusion of the TCP15 promoter region to the gus reporter gene. Scale bars = 1 mm. B, SEM images of stage 13–15 flowers from wild-type (Col-0) plants and plants with altered TCP function, as indicated. Organs from the outer two whorls were removed to allow visualization of the stamens. Scale bars = 1 mm. C, Stamen filament length, relative to wild type, in flowers at stage 13 of different plant lines with altered TCP function. Bars indicate the mean ± se (n = 19–43 stamens, depending on the line). D, Stamen filament length of wild-type, pTCP15::TCP15-EAR, and amiR-27 plants at stages 10–13 of flower development. Bars indicate the mean ± sd (n = 10–54 stamens, depending on the stage). Asterisks in (C) and (D) indicate significant differences with wild-type plants (P < 0.05; Student’s t test). E, GFP (left; scale bars = 0.2 mm) and GUS expression (right; scale bars = 1 mm) in stamen filaments and flowers of plants that contain a fusion of the TCP15 promoter region to the TCP15, GFP, and GUS coding regions (pTCP15::TCP15-GFP-GUS). Stamens from flowers at different stages are shown.
Measurement of stamen filament length at different stages of flower development indicated that amiR plants show shorter filaments than wild type at stages 12 and 13 (before and at anthesis, respectively; Fig. 1D), suggesting that the TCPs are particularly important during the late stages of pre-anthesis growth, when faster elongation occurs (Cecchetti et al., 2008). By contrast, pTCP15::TCP15-EAR flowers showed shorter filaments already by stage 10 (Fig. 1D). This implies that TCP15 and other TCPs may also have a role during early stages of filament growth. Nevertheless, the growth rate was more markedly decreased between stages 12 and 13, again pointing to a particular role of the TCPs during these stages. Accordingly, analysis of GFP and GUS reporters indicated that the TCP15 promoter is active in stamen filaments by stage 12 and its activity increases at stage 13, mainly in vascular tissues (Fig. 1E). Expression of the reporters was not detected at earlier stages.
Class-I TCPs Modulate the Expression of SAUR63 Subfamily Genes
To gain insight into the action mechanism of TCP15, we analyzed an available microarray experiment of gene expression changes in pTCP15::TCP15-EAR plants (Lucero et al., 2015). Among the genes that were previously related to stamen filament elongation, consistent expression changes were not evident for those related to JA metabolism or for JA-regulated MYB genes (Table 1). ARF8 expression was also not significantly affected, whereas ARF6 showed only an ∼40% decrease in expression relative to wild type (Table 1). Because the microarray experiment of pTCP15::TCP15-EAR plants was performed with RNA prepared from rosettes, we confirmed this result by measuring ARF6 and ARF8 transcript levels in pTCP15::TCP15-EAR flowers by reverse-transcription quantitative PCR (RT-qPCR; Supplemental Fig. S2). We also found that most members of the SAUR63 gene subfamily (composed of SAUR61-68 and SAUR75; Ren and Gray, 2015) were repressed in pTCP15::TCP15-EAR plants (Table 1). The only exception was SAUR68, whose expression was below background levels in the microarray. Because decreased expression of SAUR63 subfamily genes causes reduced stamen filament elongation (Chae et al., 2012), this may explain the shorter filament phenotype of pTCP15::TCP15-EAR plants.
Table 1. Expression levels of genes related to stamen filament elongation in pTCP15::TCP15-EAR plants.
Expression data were obtained from GEO datasets GSE57742, GSE57743 and GSE57744 (Lucero et al., 2015). AGI, Arabidopsis Genome Initiative Locus Code.
| Probe ID | AGI | Name | Fold-Changea | Adjusted P Value |
|---|---|---|---|---|
| JA synthesis genes | ||||
| A_84_P18289 | AT2G44810 | DAD1 | 0.34 | 0.3335 |
| A_84_P15574 | AT3G45140 | LOX2 | 0.28 | 0.6584 |
| A_84_P13078 | AT5G42650 | AOS | −0.38 | 0.4058 |
| A_84_P853664 | AT2G06050 | OPR3 | −0.26 | 0.8151 |
| JA-regulated MYB genes | ||||
| A_84_P19845 | AT3G27810 | MYB21 | 0.09 | 0.6923 |
| A_84_P825353 | AT5G40350 | MYB24 | −0.13 | 0.8349 |
| A_84_P767898 | AT5G06100 | MYB33 | 0.03 | 0.7715 |
| A_84_P15465 | AT3G01530 | MYB57 | 0.20 | 0.4211 |
| A_84_P16408 | AT3G11440 | MYB65 | 0.00 | 0.9881 |
| Auxin response factor genes | ||||
| A_84_P21873 | AT1G30330 | ARF6 | −0.70 | 0.0442 |
| A_84_P18731 | AT5G37020 | ARF8 | −0.20 | 0.4085 |
| SAUR63 subfamily genes | ||||
| A_84_P537376 | AT1G29420 | SAUR61 | −1.19 | 0.0395 |
| A_84_P10257 | AT1G29430 | SAUR62 | −1.73 | 0.0136 |
| A_84_P279980 | AT1G29440 | SAUR63 | −1.97 | 0.0076 |
| A_84_P19713 | AT1G29450 | SAUR64 | −2.29 | 0.0077 |
| A_84_P15930 | AT1G29460 | SAUR65 | −1.91 | 0.0085 |
| A_84_P11207 | AT1G29500 | SAUR66 | −2.06 | 0.0085 |
| A_84_P22555 | AT1G29510 | SAUR67 | −2.08 | 0.0076 |
| A_84_P563710 | AT5G27780 | SAUR75 | −1.40 | 0.0488 |
Log2 (pTCP15::TCP15-EAR/wild-type).
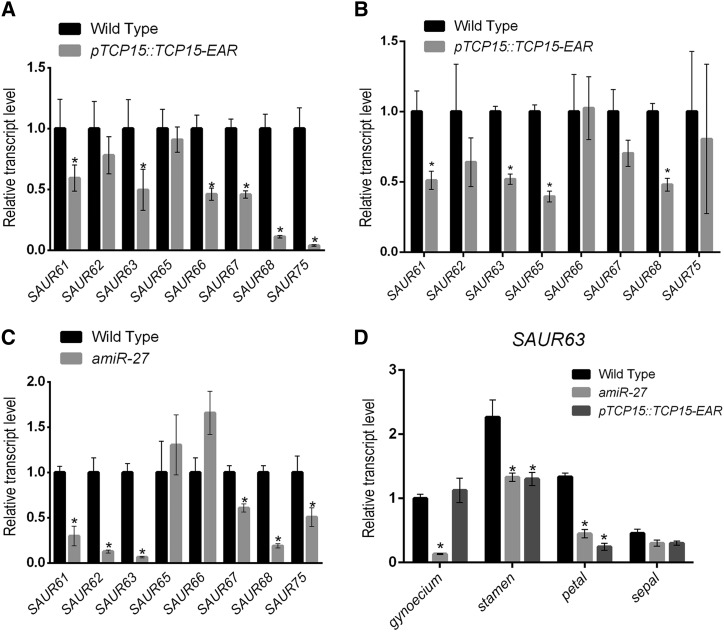
To confirm the repression of SAUR63 subfamily genes in floral organs of pTCP15::TCP15-EAR plants, we performed RT-qPCR analyses using RNA prepared from flowers. Decreased expression of most SAUR63 subfamily genes, with the exception of SAUR62 and SAUR65, was observed in these samples (Fig. 2A). Repression of several SAUR63 subfamily genes in pTCP15::TCP15-EAR plants was also evident when RNA isolated from stamens was tested (Fig. 2B). Most SAUR63 subfamily genes, except SAUR65 and SAUR66, also showed reduced expression in flowers of amiR-27 plants (Fig. 2C), indicating that class-I TCPs are required for the correct expression of these genes. The differences in expression observed in pTCP15::TCP15-EAR and amiR-27 plants probably reflect changes in the preference of class-I TCPs for distinct SAUR genes. Notably, SAUR63 was repressed in all the samples analyzed (Fig. 2, A–C).
Figure 2.
Transcript levels of SAUR63 subfamily genes in plants with altered TCP function. A and B, Transcript levels, determined by RT-qPCR, in flowers and stamens, respectively, of pTCP15::TCP15-EAR plants. C, Transcript levels, determined by RT-qPCR, in flowers of amiR-27 plants. D, SAUR63 transcript levels, determined by RT-qPCR, in different organs of flowers from wild-type, pTCP15::TCP15-EAR, and amiR-27 plants. Flowers at stage 13 were used. The bars indicate the mean ± se of three biological replicates. Asterisks indicate significant differences with wild-type plants (P < 0.05; Student’s t test).
We then took SAUR63 as a representative gene of this group and measured its expression in different flower organs. The highest expression of SAUR63 was observed in stamens of wild-type plants (Fig. 2D). A significant decrease in SAUR63 transcript levels was evident in stamens and petals of both pTCP15::TCP15-EAR and amiR-27 plants (Fig. 2D). For amiR-27 plants, but not for pTCP15::TCP15-EAR plants, significantly decreased SAUR63 expression was also observed in the gynoecium (Fig. 2D), indicating that class-I TCPs other than TCP15 likely regulate SAUR63 expression in this organ. These results show that the regulation of SAUR63 by class-I TCPs probably extends to other flower organs, in addition to stamens.
Overexpression of SAUR63 Restores Stamen Filament Elongation in pTCP15::TCP15-EAR Plants
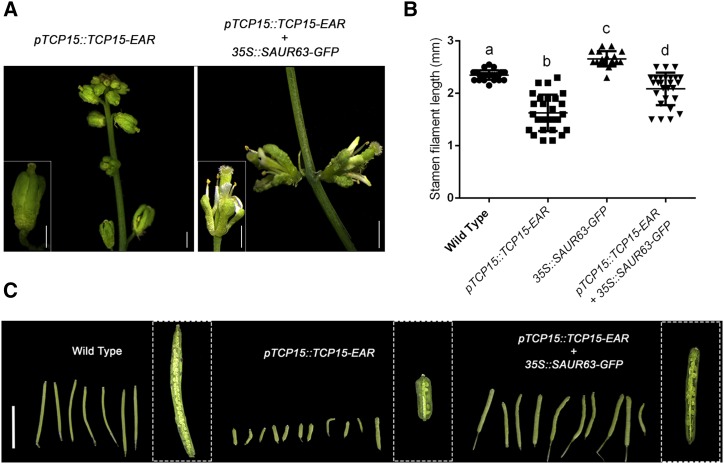
To evaluate if reduced expression of SAUR63 subfamily genes is the cause of the short stamen phenotype observed in pTCP15::TCP15-EAR plants, we generated and analyzed pTCP15::TCP15-EAR plants expressing SAUR63 under the control of the 35SCaMV promoter. Because pTCP15::TCP15-EAR plants are largely infertile, we first obtained plants transformed with the 35S::SAUR63-GFP construct that contain flowers with elongated stamens (Supplemental Fig. S3). It is worth noting that the fusion to protein tags is necessary to stably express SAUR proteins, which are otherwise rapidly degraded (Chae et al., 2012; Spartz et al., 2012). We then transformed wild-type and 35S::SAUR63-GFP plants in parallel with the pTCP15::TCP15-EAR construct and analyzed the phenotype of at least 10 independent plants from the T1 population in each background. In the wild-type background, the expression of TCP15-EAR caused defects in rosette and inflorescence development, similar to those previously reported by Kieffer et al. (2011) and Uberti-Manassero et al. (2012). Particularly, the inflorescence showed a decrease in stem and pedicel lengths, flowers showed shorter petals and stamens, sepals were curved inwards, and the protrusion of stigmatic tissue from the replum was observed in the gynoecium (Fig. 3A, left). Similar phenotypes were observed in the 35S::SAUR63-GFP background, except for stamens, which were considerably longer, protruding from sepals and almost reaching the top of the gynoecium (Fig. 3A, right). Petals were also elongated in comparison with transformants in the wild-type background (Fig. 3A, right). Increased elongation of stamens and petals in pTCP15::TCP15-EAR plants that express SAUR63-GFP agrees with the observation that SAUR63 transcript levels are reduced in both organs in pTCP15::TCP15-EAR plants (Fig. 2D).
Figure 3.
Overexpression of SAUR63 rescues the short stamen and infertility phenotypes of pTCP15::TCP15-EAR plants. A, Phenotype of pTCP15::TCP15-EAR plants in wild-type (Col-0) and 35S::SAUR63-GFP backgrounds. A representative image of 10 independent transformants analyzed for each background is shown. Scale bars = 1 mm. B, Stamen filament length in flowers at stage 13 of wild-type plants, 35S::SAUR63-GFP plants, and plants that express TCP15-EAR under the control of the TCP15 promoter in either a wild-type or a 35S::SAUR63-GFP background. The bars indicate the mean ± se (n = 18–27 stamens, depending on the line). Different letters indicate significant differences (P < 0.05; one-way ANOVA for multiple comparisons). C, Images of siliques obtained from wild-type plants, pTCP15::TCP15-EAR plants, and pTCP15::TCP15-EAR plants that express SAUR63-GFP from the 35SCaMV promoter. Scale bar = 1 cm.
Measurement of stamen filament length indicated that the effect of expressing TCP15-EAR was more pronounced in a wild-type than in a 35S::SAUR63-GFP background (Fig. 3B). Indeed, the median of stamen filament length for the population of plants analyzed was decreased by only 15% in the 35S::SAUR63-GFP background and by 36% in the wild-type background after TCP15-EAR expression. SAUR63-GFP expression also partially restored fertility, as judged from silique growth and seed production, which were considerably affected in pTCP15::TCP15-EAR plants (Fig. 3C). Thus, our results suggest that reduced stamen filament elongation in pTCP15::TCP15-EAR plants is largely due to repression of SAUR63 and related genes by TCP15-EAR.
SAUR63 is a direct TCP15 target
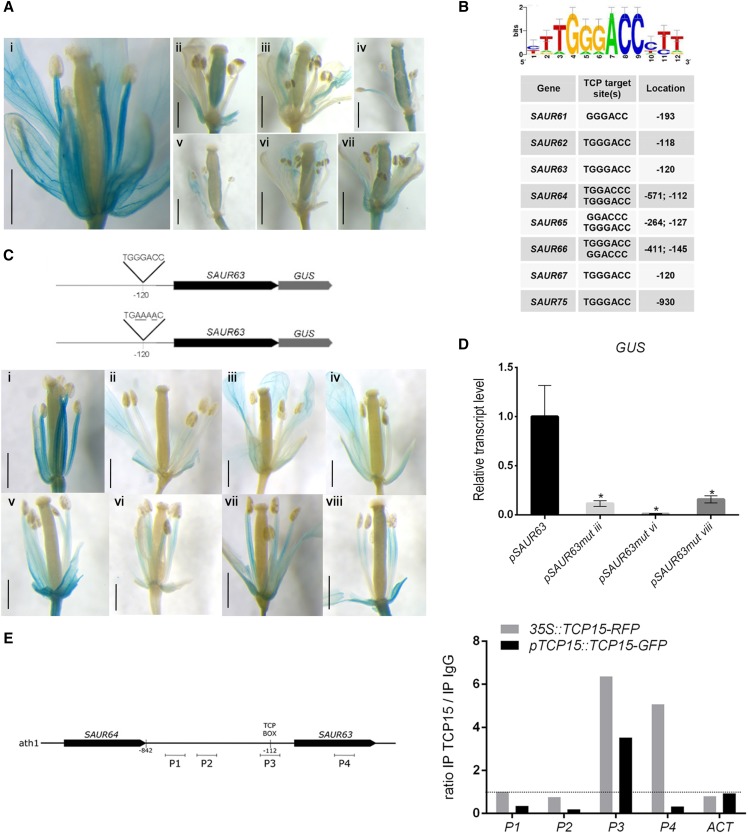
To analyze in more detail the regulation of SAUR63 subfamily genes by TCP15, we focused on SAUR63 itself as a representative member of the subfamily. We obtained plants that express a translational fusion of SAUR63 with the GUS reporter gene under the control of the SAUR63 promoter. The analysis of independent lines revealed strong GUS activity in stamen filaments, as well as in petals (Fig. 4A, left), as previously reported by Chae et al. (2012). We then transformed one of these lines with the pTCP15::TCP15-EAR construct. A strong decrease in GUS expression was evident in the stamen filaments of the transformed lines (Fig. 4A), indicating that TCP15-EAR represses the activity of the SAUR63 promoter in filaments.
Figure 4.
SAUR63 is a direct TCP15 target. A, GUS expression in a representative plant line that contains a fusion of SAUR63 to gus under the control of the SAUR63 promoter, either in a wild-type background (i) or after transformation of this line with the pTCP15::TCP15-EAR construct (ii–vii are different independent transformants). Scale bars = 1 mm. B, Enrichment of sequences bound by TCPs in the promoter regions of SAUR63 subfamily genes. A sequence logo of a sequence overrepresented in the promoters of SAUR63 subfamily genes, obtained using the Regulatory Sequence Analysis Tool platform (http://rsat.eead.csic.es/plants/), together with a list of the sequences and their locations in the respective promoters, is shown. Sequences are from the coding strand and numbers are relative to the putative transcription start site, except for SAUR75, which corresponds to the complementary strand and is relative to the translation start site. C, Expression of SAUR63-GUS under the control of the native SAUR63 promoter (i; a representative line is shown), or under the control of a mutated version of the promoter in which the sequence TGGGACC located at −120 was mutagenized (ii–viii are different independent lines). Scale bars = 1 mm. Schematics of the constructs used for transformation are shown above. D, Quantification by RT-qPCR of GUS transcript levels in flowers of representative lines from those shown in (C). The bars indicate the mean ± se of three biological replicates. Asterisks indicate significant differences (P < 0.05; Student’s t test). E, ChIP analysis of the binding of TCP15-GFP to the SAUR63 promoter region. The results of two independent experiments, one with 35S::TCP15-RFP plants and the other with plants that express TCP15-GFP under the control of the native TCP15 promoter, are shown. Primer pairs for the amplification of different regions of the SAUR63 gene (P1–P4) and of ACT2 and ACT8 genes (ACT; control) were used. A schematic of the genomic region analyzed, indicating the location of the different fragments, is shown to the left. The results are expressed as the ratio of the signal obtained after immunoprecipitation with specific antibodies and with anti-IgG (control).
Remarkably, the promoter regions of all SAUR63 subfamily genes, except SAUR68, contain sequences that closely match the consensus target site bound by TCP transcription factors (GGGNCC; Fig. 4B; Kosugi and Ohashi, 2002; Viola et al., 2012). In fact, motif enrichment analysis in SAUR63 subfamily gene promoters using the Regulatory Sequence Analysis Tool platform (http://rsat.eead.csic.es/plants/) yielded the sequence TGGGACC as an enriched motif in these promoters (Fig. 4B; Supplemental Table S2), which is recognized by TCP15 and other class-I TCPs in vitro (Kosugi and Ohashi, 2002; Viola et al., 2011) and in vivo (Lucero et al., 2017). This suggests that SAUR63 and related genes may be direct targets of the TCPs. To evaluate the role of the putative TCP target sequence in the SAUR63 promoter, we mutated the TGGGACC sequence to TGAAAAC and obtained plants that express SAUR63-GUS under the control of the mutated promoter. Several lines of these plants (eight out of 10 independent lines analyzed) showed a strong decrease in GUS expression in stamen filaments when compared with lines that express SAUR63-GUS under the control of the native promoter (Fig. 4C). Notably, expression in petals was less significantly affected, whereas strong expression in rosettes was observed in all the lines (Supplemental Fig. S4). Reduced GUS expression in plants expressing SAUR63-GUS from the mutated promoter was also observed by RT-qPCR (Fig. 4D). The results indicate that TGGGACC is an active cis-acting element in the promoter region of SAUR63, driving expression in stamen filaments. Furthermore, this result suggests that SAUR63 may be a direct target of TCP15 and other class-I TCPs.
Direct binding of TCP15 to the SAUR63 promoter in vivo was analyzed by chromatin immunoprecipitation (ChIP) in 35S::TCP15-RFP and pTCP15::TCP15-GFP plants. ChIP-qPCR resulted in a relative enrichment of fragment P3 of the SAUR63 promoter, including the TGGGACC site, in both lines analyzed (Fig. 4E). By comparison, two fragments located further upstream on the SAUR63 promoter and ACTIN genes showed no enrichment, whereas a fragment located downstream, within the SAUR63 coding region, was enriched only in samples from 35S::TCP15-RFP plants (Fig. 4E). Therefore, SAUR63 can be considered a direct in vivo target of TCP15.
Genes Induced By GAs and ARF6/8, But Not JA-Induced Genes, Are Repressed in pTCP15::TCP15-EAR Plants
It was previously shown that mutants with deficiencies in GA synthesis or signaling show defects in stamen filament elongation (Cheng et al., 2004; Tyler et al., 2004; Rieu et al., 2008). We thus analyzed available microarray data for GA-regulated genes in flowers (Cao et al., 2006) and compared these with those genes exhibiting modified expression in the microarray of pTCP15::TCP15-EAR plants. As shown in Supplemental Figure S5, a significant overlap was observed between genes downregulated in pTCP15::TCP15-EAR plants and genes induced by GA or repressed by DELLA proteins, which are negative regulators of GA responses. This, together with previous reports on the inhibition of class-I TCPs by interaction with DELLAs (Davière et al., 2014; Resentini et al., 2015), suggests that class-I TCPs may be involved in mediating GA responses in flowers. It was also shown that GAs promote JA synthesis during stamen filament elongation, but the short stamen phenotype of GA-deficient plants cannot be rescued by JA treatment (Cheng et al., 2009), leading to the suggestion that GA-dependent pathways that are independent of JA exist. In agreement with the induction of JA synthesis by GAs, there is a significant overlap between genes induced by GAs, or repressed by DELLAs, and JA-upregulated genes (Nemhauser et al., 2006; Supplemental Fig. S5). However, there is no significant overlap between genes repressed in pTCP15::TCP15-EAR plants and genes induced by JA (Supplemental Fig. S5). In fact, several genes repressed by JA are also repressed in pTCP15::TCP15-EAR plants and those induced by JA are induced in these plants (Supplemental Fig. S5), suggesting that TCP15 affects stamen filament elongation independently from the JA-related pathway.
JA-dependent filament elongation was shown to be mediated by MYB21, MYB24, and MYB57 (Cheng et al., 2009; Reeves et al., 2012). As for JA-responsive genes, most (331 out of 345; listed in Supplemental Table S3) genes upregulated by MYB21 and MYB24 (e.g. genes with reduced expression in myb21 myb24 mutant flowers; Reeves et al., 2012) were not repressed in pTCP15::TCP15-EAR plants (Supplemental Fig. S5), which was further confirmed by RT-qPCR analysis of a subset of five genes with reduced expression in myb21 myb24 mutants using RNA prepared specifically from flowers of pTCP15::TCP15-EAR plants. None of these genes showed reduced expression in pTCP15::TCP15-EAR flowers and even some of them were upregulated (Supplemental Fig. S6). Thus, the function of TCP15 and related class-I TCPs is probably not related to JA- and/or MYB21/24-dependent stamen filament elongation.
We also found a significant overlap between genes repressed in pTCP15::TCP15-EAR plants and those induced by ARF6 and ARF8 (i.e. genes with reduced expression in arf6 arf8 flowers; Reeves et al., 2012; Supplemental Fig. S5; genes listed in Supplemental Table S4). We confirmed this result by measuring the expression of four genes selected from this group in pTCP15::TCP15-EAR flowers (Supplemental Fig. S6). Three of these genes (IAA19, IAA3, and EXP8) are preferentially expressed in stamen filaments (Supplemental Fig. S7), suggesting that the observed expression changes in flowers are related to changes in this organ. In addition, IAA19 expression levels are related to stamen filament elongation (Tashiro et al., 2009; Cecchetti et al., 2017; Ghelli et al., 2018). Most of the aforementioned SAUR63 subfamily genes also belong to this group of genes with reduced expression in pTCP15::TCP15-EAR and arf6 arf8 flowers accompanied by preferential expression in stamen filaments (Supplemental Fig. S7). These results raise the possibility that class-I TCPs also participate in auxin-dependent responses. On the other hand, none of the class-I TCP genes show significant expression changes in arf6 arf8 flowers (Reeves et al., 2012). Many of the genes induced by ARF6 and ARF8 are also induced by GAs or repressed by DELLAs (Supplemental Fig. S5). Based on this analysis, it can be speculated that TCP15 and related class-I TCPs participate in GA-dependent and/or auxin-dependent stamen filament elongation pathways. The action of class-I TCPs may be at least partially related to the induction of SAUR63 subfamily genes, which are direct targets of TCP15 and probably other TCPs. However, it is conceivable that other genes either directly or indirectly regulated by the TCPs are also involved.
Overexpression of TCP15 Restores Stamen Filament Elongation in a GA-Deficient Background
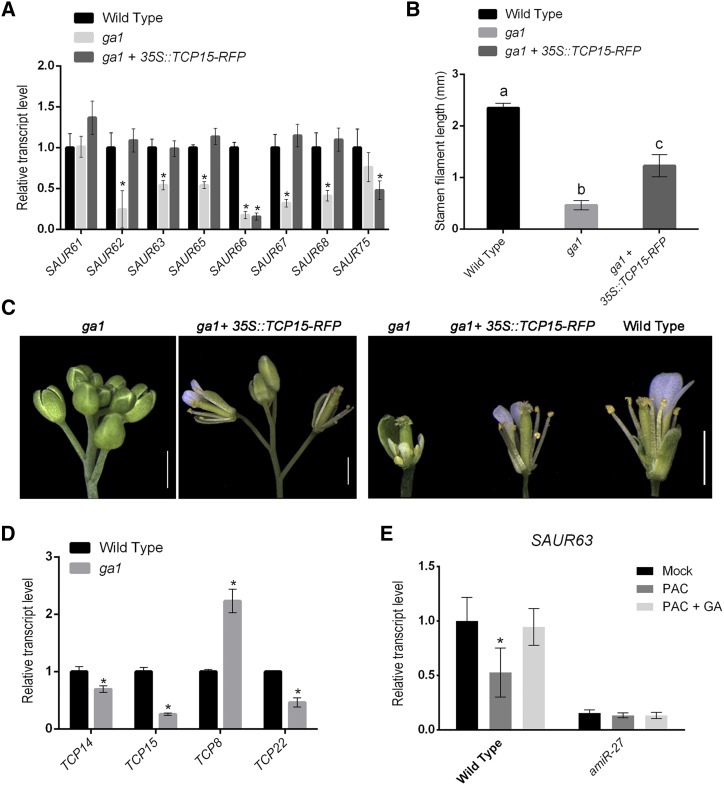
To analyze a possible role of class-I TCPs in GA-dependent expression of SAUR63 subfamily genes, we analyzed transcript levels of these genes in flowers of ga1-mutant plants, deficient in an early step of GA biosynthesis. Most SAUR63 subfamily genes, except SAUR61 and SAUR75, showed reduced expression in GA-deficient plants (Fig. 5A), indicating that they are under GA regulation. If reduced expression of SAUR63 subfamily genes in GA-deficient plants is related to decreased activity of the TCPs, overexpression of TCP15 would restore the expression levels of these genes. As shown in Figure 5A, expression of TCP15 from the 35SCaMV promoter restored the expression of most SAUR63 subfamily genes that were repressed in the GA-deficient background.
Figure 5.
Class-I TCPs participate in GA-dependent stamen filament elongation and SAUR63 subfamily gene expression. A, Transcript levels of SAUR63 subfamily genes in flowers from wild-type, ga1 plants, and ga1 plants transformed with the 35S::TCP15-RFP construct. The results are expressed relative to wild type. Values represent the mean ± se of three biological replicates. Asterisks indicate significant differences with wild-type plants (P < 0.05; Student’s t test). B, Stamen filament length in flowers at stage 13 of wild-type plants, ga1 plants, and ga1 plants that express TCP15-RFP under the control of the 35SCaMV promoter. The bars indicate the mean ± sd (n = 11–23 stamens, depending on the line). Different letters indicate significant differences (P < 0.05; one-way ANOVA for multiple comparisons). C, Phenotype of ga1 plants before and after transformation with a 35S::TCP15-RFP construct. A representative image of five independent lines is shown. A flower of a wild-type plant is included for comparison. Scale bars = 1 mm. D, Transcript levels of class-I TCP genes in flowers from wild-type and ga1 plants. The results are expressed relative to wild type. Values represent the mean ± se of three biological replicates. Asterisks indicate significant differences with wild-type plants (P < 0.05; Student’s t test). E, SAUR63 transcript levels in wild-type and amiR-27 plants 4 h after a single treatment with 10 μM of PAC (PAC) or 10 µM of PAC plus 100 μM of GA3 (PAC + GA). Control plants (Mock) were mock-treated with solvent solution. Values are expressed as relative to wild type under control conditions and represent the mean ± se of three biological replicates. Asterisks indicate significant differences between control and treated plants of the same genetic background (P < 0.05; Student’s t test).
In addition, TCP15 overexpression increased stamen elongation of ga1-mutant plants (Fig. 5, B and C), indicating that decreased activity of the TCPs is a likely cause of decreased filament elongation in these plants. Notably, flowers from ga1 plants that were overexpressing TCP15 remained smaller than wild-type flowers, but their stamen filaments reached the top of the gynoecium, unlike those of ga1 plants (Fig. 5C). This suggests that other GA-dependent pathways, not related to TCP15, are also important to determine the size of flower organs. Despite that stamens reached the top of the gynoecium, we did not observe restoration of fertility in TCP15-overexpressing ga1 plants. This may be due to the fact that ga1 plants show male and female infertility (Plackett et al., 2012). In addition to stamens, the length of petals was also increased upon TCP15 overexpression (Fig. 5C), suggesting that TCP15 may also be involved in promoting the elongation of these organs.
Measurement of TCP15 transcript levels indicated that TCP15 and TCP22 are also repressed in the ga1 mutant (Fig. 5D). For TCP15, this agrees with results of a microarray experiment with ga1 flowers (Cao et al., 2006). A slight repression was also observed for TCP14, whereas TCP8 was induced in the mutant (Fig. 5D). Thus, decreased expression of class-I TCPs in the GA-deficient background may partly explain the repression of SAUR63 subfamily genes observed in these plants. The reported inhibition of class-I TCPs by DELLA proteins (Davière et al., 2014), which would be more pronounced in the GA-deficient background, is likely another factor playing a key role in this regulation.
We also analyzed the response of SAUR63 to GA in wild-type plants. For this purpose, we treated inflorescences with the GA-synthesis inhibitor paclobutrazol (PAC). The expression of SAUR63 was significantly reduced in wild-type plants 4 h after a single PAC treatment (Fig. 5E), most likely due to a decrease in endogenous GA levels. In agreement, treatment with GA, in addition to PAC, restored normal SAUR63 transcript levels (Fig. 5E). To analyze if class-I TCPs are required for the correct expression of SAUR63 in response to GA, we also treated amiR-27 plants with PAC and PAC + GA. Contrary to the wild type, SAUR63 expression was not significantly modified by the treatments in amiR-27 plants (Fig. 5E), indicating that TCP15 and related class-I TCPs participate in GA-dependent modulation of SAUR63 gene expression. We also measured the expression of other SAUR63 subfamily genes with reduced expression in amiR-27 plants. For SAUR61 and SAUR75, we observed a similar behavior to that of SAUR63 (Supplemental Fig. S8). Notably, these two genes did not show reduced expression in ga1 plants (Fig. 5A). For SAUR62 and SAUR68, we did not observe a significant effect of the PAC treatment on expression, although SAUR68 was induced by GA in wild-type plants and not in amiR-27 plants (Supplemental Fig. S8). The differences observed between ga1 and wild-type plants treated with PAC may reflect differences in the response of SAUR genes to constitutive versus transient GA deprivation. Nevertheless, the results indicate that most SAUR63 subfamily genes undergo regulation by GA levels in flowers, and that TCP15 and related class-I TCPs participate in this regulation.
Induction of SAUR63 Subfamily Genes By Auxin Is Not Compromised in Class-I TCP-Deficient Plants
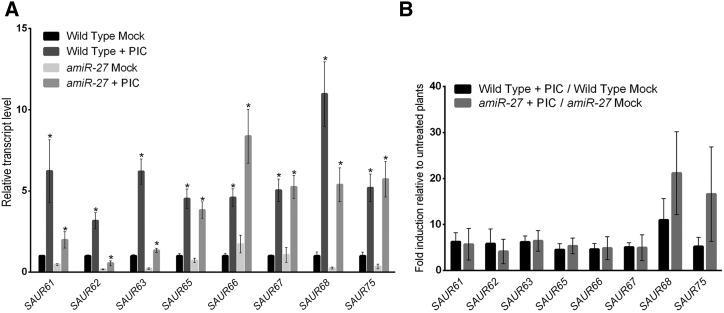
SAUR genes were discovered based on their rapid response to auxin (McClure and Guilfoyle, 1987). Despite later studies showing that not all SAUR genes are induced by auxin, those belonging to the SAUR63 subfamily are indeed reported as auxin-inducible (Nagpal et al., 2005; Chapman et al., 2012) and targets of ARF6 (Oh et al., 2014). In addition, most members of the subfamily show reduced expression in flowers of arf6 arf8 mutants (Nagpal et al., 2005; Reeves et al., 2012). We thus analyzed the effect of auxin treatment on SAUR63 subfamily gene expression in wild-type and amiR-27 flowers, the latter deficient in several class-I TCPs. A significant induction by auxin was observed for all tested SAUR63 subfamily genes in wild-type plants and amiR-27 plants (Fig. 6A). Particularly for SAUR61-63 and SAUR68, transcript levels after auxin treatment were lower in amiR-27 plants than in wild-type plants (Fig. 6A). However, the relative induction of SAUR63 subfamily genes by auxin was similar (or even higher in the case of SAUR68 and SAUR75) in amiR-27 and wild-type plants when compared with mock-treated plants (Fig. 6B).
Figure 6.
Induction of SAUR63 subfamily genes by auxin is not compromised in class-I TCP-deficient plants. A, Transcript levels of SAUR63 subfamily genes in wild-type and amiR-27 plants 4 h after a single treatment with 100 μM of the synthetic auxin picloram (+ PIC). Control plants (Mock) were mock-treated with solvent solution. Values are expressed as relative to wild type under control conditions and represent the mean ± se of three biological replicates. Asterisks indicate significant differences between control and treated plants of the same genetic background (P < 0.05; Student’s t test). B, Relative induction of SAUR63 subfamily genes by auxin in wild-type and amiR-27 plants. The fold induction of each gene, relative to mock-treated plants, is indicated.
These results indicate that the response of SAUR63 subfamily genes to auxin is not significantly impaired by a deficiency in the class-I TCPs studied here, suggesting that these TCPs do not participate in the induction of these genes by auxin. The results also suggest that, at least for SAUR61-63 and SAUR68, auxin cannot fully replace the lack of class-I TCPs, indicating that the TCPs are required, in addition to auxin, for maximal expression of these genes.
DISCUSSION
In this work, we report that plants affected in the function of class-I TCPs show defects in stamen filament elongation. This was observed for plants that express a fusion of TCP15 to the EAR repression domain under the control of the TCP15 promoter (pTCP15::TCP15-EAR plants), but not in loss-of-function mutants in TCP15 and the related class-I gene TCP14. Because TCP15-EAR most likely represses the expression of genes that are under direct control of TCP15, the results suggest that TCP15 is involved in the regulation of genes that affect stamen filament elongation. This is also supported by the fact that overexpression of TCP15 yielded plants with longer filaments. The opposite effects on filament elongation observed after expressing the native and repressor forms of TCP15 indicates that TCP15 acts as an activator in this process. The lack of appreciable changes in filament length after loss-of-function of TCP15 is probably due to redundancy of TCP15 with other class-I TCPs, which was previously observed among members of this class in different developmental contexts (Kieffer et al., 2011; Uberti-Manassero et al., 2012; Aguilar-Martínez and Sinha, 2013). In agreement, plants affected in TCP8 and TCP22, in addition to TCP14 and TCP15 (amiR-27 plants), show shorter filaments.
Members of the SAUR63 subfamily of SAUR genes are required for correct stamen filament elongation (Chae et al., 2012), a process mainly governed by cell expansion (Tashiro et al., 2009). The fact that several members of the SAUR63 subfamily show reduced expression in pTCP15::TCP15-EAR and amiR-27 plants and that overexpression of SAUR63 rescues the short stamen and infertility phenotypes of pTCP15::TCP15-EAR plants suggest that reduced expression of SAUR63 subfamily genes is responsible for defective stamen elongation in these plants. Interestingly, overexpression of SAUR63 also promotes the elongation of petals, which are shorter than wild type in pTCP15::TCP15-EAR plants. This is consistent with the fact that SAUR63 subfamily genes are also required for petal growth (Chae et al., 2012) and suggests that the regulation of SAUR63 subfamily genes by class-I TCPs described here may also be important for this process. Indeed, reduced expression of SAUR63 was observed in both petals and stamens of pTCP15::TCP15-EAR and amiR-27 plants. Notably, reporter gene assays indicated that TCP15 is mainly expressed in the vasculature of stamen filaments, whereas organ elongation is thought to be limited mainly by epidermal cell expansion (Savaldi-Goldstein and Chory, 2008). Chae et al. (2012) observed that SAUR63 is also more prominently expressed in filament vascular tissues and hypothesized that it may have a role in redirecting auxin flux to peripheral tissues. The similar expression characteristics of TCP15 and SAUR63 within stamen filaments supports a role for TCP15 in regulating SAUR63 gene expression.
Analysis of the proximal promoter regions of different members of the SAUR63 subfamily revealed the existence of putative TCP binding motifs. At least for SAUR63, mutation of this motif leads to considerably reduced expression in flowers, and particularly in stamen filaments, as deduced from GUS histochemical assays. This, together with the observed binding of TCP15 to the promoter region containing this motif, strongly indicates that SAUR63 subfamily genes are direct targets of TCP15 and related class-I TCPs. It is noteworthy that several members of the SAUR63 subfamily were also uncovered as target genes of other transcription factors, namely BZR1 (Sun et al., 2010), PIF4 (Oh et al., 2012), and ARF6 (Oh et al., 2014). Thus, class-I TCPs can be added to this set of transcription factors that directly impinge on SAUR63 subfamily gene expression to regulate cell growth. We show here that the response of members of the SAUR63 subfamily to auxin treatment of flowers is not significantly compromised in plants defective in TCP15 and related class-I TCPs, suggesting that the TCPs are not essential for the response to auxin, which is most likely mediated by the ARFs. Previous reports indicated that 35S::TCP15 and pTCP15::TCP15-EAR plants show lower and higher expression, respectively, of the auxin reporter DR5::GUS (Uberti-Manassero et al., 2012; Lucero et al., 2015; Viola et al., 2016), indicating that TCP15 has a negative impact on expression from auxin response elements, to which ARF transcription factors bind. In fact, a higher response to auxin could be observed for some members of the SAUR63 subfamily, considering that basal expression levels were lower in TCP-deficient plants. Thus, auxin-dependent expression of SAUR63 subfamily genes would not be mediated by class-I TCPs. However, the TCPs, as well as ARFs, seem to be required for correct expression of SAUR63 subfamily genes during stamen filament elongation, most likely directly acting through their respective target sites in the promoters of these genes.
Previous reports showed that GAs promote the synthesis of JA during late stages of stamen development (Cheng et al., 2009). JA, in turn, induces the expression of three MYB genes required for filament growth and anther maturation (Mandaokar et al., 2006). Further analysis indicated that induction of these MYB genes after JA treatment did not restore normal stamen growth in a GA-deficient background (Cheng et al., 2009), leading to the conclusion that alternative GA-dependent pathways exist. In agreement with this, the short stamen phenotype of plants deficient in the GA transporter GTR1 can be rescued by GA treatment, but not by JA, whereas the anther maturation defects of JA-deficient plants can be rescued by JA, but not by GA (Saito et al., 2015). Based on this, Saito et al. (2015) proposed that GA and JA pathways act in parallel and coordinate each other to modulate stamen development, and that anther maturation is primarily controlled by JA, whereas filament elongation is under GA control. The fact that overexpression of TCP15 rescues the short stamen phenotype of GA-deficient plants suggests that TCP15 and related class-I proteins are major players in a GA-dependent pathway controlling stamen filament elongation. Furthermore, we showed that the expression of TCP15 is reduced in GA-deficient plants. However, the observed repression of TCP15 would not be enough to explain the short stamen phenotype of GA-deficient plants, because the single tcp15 mutant shows normal stamens. The reported interaction of class-I TCPs with DELLA proteins (Davière et al., 2014; Resentini et al., 2015) provides an additional mechanism for the regulation of TCP action by GAs. In addition, many of the genes induced by GAs in flowers show reduced expression in pTCP15::TCP15-EAR plants, suggesting that they may be under the control of TCP15 and related class-I TCPs. Specifically, our experiments showing that the changes in expression of SAUR63 and related genes observed after PAC or GA treatment are attenuated in TCP-deficient plants indicate that the TCPs are required for the induction of these genes by GA. Moreover, the restoration of filament growth in GA-deficient plants brought about by TCP15 overexpression was accompanied by an increase in the expression of most SAUR63 subfamily genes. Thus, one of the mechanisms involved in GA-dependent stamen filament elongation most likely involves the activation of class-I TCPs to induce growth through changes in the expression of SAUR63 subfamily genes.
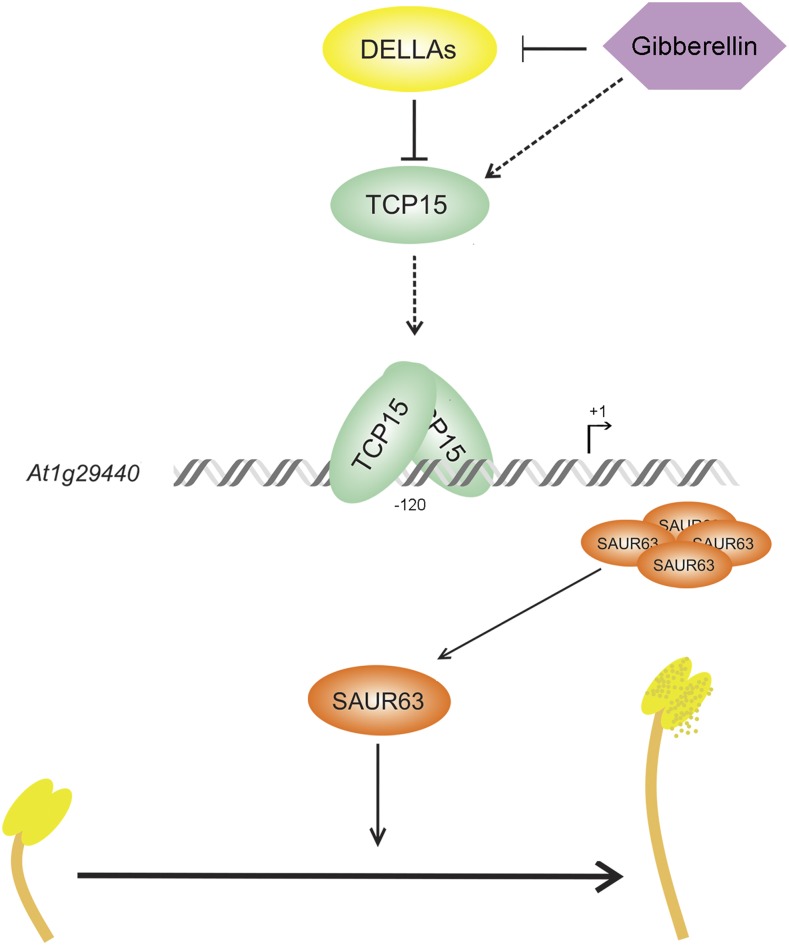
In conclusion, our results show that TCP15 and related class-I TCPs link GA action to stamen filament elongation by direct transcriptional activation of SAUR63 subfamily genes through target sites present in their promoters (Fig. 7). The results presented here contribute to a more comprehensive understanding of the pathway behind GA-dependent stamen filament elongation.
Figure 7.
Proposed model for the involvement of TCP15 and other class-I TCPs in the stimulation of stamen filament elongation by GAs. GAs induce expression of TCP15 and activate class-I TCP proteins through GA-dependent DELLA degradation. Class-I TCP proteins induce SAUR63 and related genes by direct interaction with TCP target sites present in their promoters. SAUR63 proteins stimulate stamen filament elongation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) lines used in this study were in the Columbia (Col-0) ecotype. Lines that express TCP15 and TCP15-RFP from the 35SCaMV promoter (35S::TCP15 and 35S::TCP15-RFP) were described in Lucero et al. (2015) and Viola et al. (2016), as well as lines that express GUS or TCP15-EAR under the control of the TCP15 promoter (Uberti-Manassero et al., 2012). The tcp14-4 tcp15-3 mutant was described by Kieffer et al. (2011) and provided by Dr. Simona Masiero (Universitá degli Studi di Milano), whereas amiR-27 plants, which express an artificial micro RNA directed against TCP8 and TCP22 in the tcp14-4 tcp15-3 background (Davière et al., 2014), were kindly sent by Drs. Jean-Michel Davière and Patrick Achard (Institut de Biologie Moléculaire des Plantes). A homozygous ga1 (SALK_109115) T-DNA insertion mutant was provided by Dr. Stephen Thomas (Rothamsted Research). Plants were grown in pots filled with soil under long-day conditions (16-h light/8-h dark) at an intensity of 100 μmol m−2 s−1 and a temperature of 22°C to 24°C. For auxin, GA, and PAC treatments, the inflorescences of plants were sprayed with 100 μm of picloram, 100 μm of GA3, and 10 μm of PAC (dissolved in ethanol) and samples were harvested 4 h later for expression analysis. Controls were mock-treated with the respective solutions without the reagent.
DNA Constructs and Plant Transformation
For expression of TCP15 or SAUR63 fused to GFP, the respective coding regions, without the stop codons, were amplified with specific primers (Supplemental Table S5) and cloned into entry vector pENTR3C (Thermo Fisher Scientific). For expression driven by the 35SCaMV promoter, the inserts were then recombined into the pGreen-based binary vector pFK248 (Hellens et al., 2000) using the Gateway cloning system. For expression of TCP15-GFP from the TCP15 promoter, an ∼1,500-bp sequence located upstream of the translation start codon used previously to analyze TCP15 expression (Uberti-Manassero et al., 2012) was amplified (Supplemental Table S5) and cloned into the SalI and KpnI sites of the pENTR3C construct that contained the TCP15 coding region between KpnI and XhoI. The fragment containing the TCP15 promoter and coding region was then recombined into vector pFAST-R07 (Shimada et al., 2010). The pTCP15::TCP15-GFP-GUS reporter construct was generated by recombining the fragment into vector pKGWFS7. The constructs expressing SAUR63-GUS under the control of the native or mutated SAUR63 promoters were obtained by amplification of a 1,256-bp fragment comprising sequences located upstream of the SAUR63 translation stop codon (Supplemental Table S5), followed by cloning into the SalI and BamHI sites of pBluescript SK−. After mutagenesis of the SAUR63 promoter through the QuikChange method (Xia et al., 2015) using Phusion DNA polymerase (Thermo Fisher Scientific), the fragments were cloned into pBI101.3. All constructs were verified by DNA sequencing. The pTCP15::TCP15-EAR construct, used to express a fusion of TCP15 to the EAR repressor domain, was previously described by Uberti-Manassero et al. (2012). Plants were transformed using floral dip (Clough and Bent, 1998).
Phenotypic Analysis
Stamen lengths shown in Figure 1C were measured with a caliper under a stereoscopic microscope. Alternatively, flowers at different stages were photographed using a model no. MZ10F stereomicroscope (Leica) equipped with a digital camera and filament length was determined using the software ImageJ (Schindelin et al., 2012). Because wild-type Arabidopsis flowers have four long and two short stamens, only the long stamens in each flower were considered. For SEM analysis, flowers were fixed in formalin/acetyl alcohol/acetic anhydride/water (10:50:5:35 v/v) during 24 h, transferred to 70% (v/v) ethanol, and dissected under an SMZ-10 stereomicroscope (Nikon). After dehydration with successive rounds of ethanol solutions (80%, 96%, and 100% [v/v]), samples were transferred to acetone for desiccation with CO2 in an Emitech K850 critical point drier (UK-RAS Network). Finally, they were coated with gold-palladium and photographed with either an FEI Quanta 200 (Thermo Fisher Scientific) or a model no. XL30 TMP New Look SEM (Philips) at Centro Científico Tecnológico Consejo Nacional de Investigaciones Científicas y Técnicas, Rosario (Argentina) or Museo Bernardino Rivadavia, respectively.
Analysis of Gene Expression
Transcript levels were measured through RT-qPCR. RNA was prepared from inflorescences (complete bolts) or isolated organs from flowers at stage 13 using TRIzol reagent followed by LiCl precipitation. Reverse transcription was performed with an oligo(dT)18 primer and MMLV reverse transcriptase (Promega) on 1.5–2.0 μg of RNA. An aliquot of the complementary DNA was used for qPCR with specific primers for the genes of interest (Supplemental Table S5). Amplified products were monitored by SYBR Green detection in an Applied Biosystems StepOnePlus apparatus (Thermo-Fisher Scientific). A comparative Ct method, with ACT2 and ACT8 actin genes for normalization (Charrier et al., 2002), was used to calculate relative transcript levels. For histochemical analysis of GUS expression, flowers were incubated in 1 mm of 5-bromo-4-chloro-3-indolyl-beta-d-GlcA, 50 mm of sodium phosphate (pH 7.0), and 0.1% (v/v) Triton X-100. Vacuum was applied for 5 min and reactions were incubated at 37°C until satisfactory staining was observed. When comparing different lines, incubation times were the same for all lines analyzed. GFP fluorescence in stamens at different stages was detected using a model no. TCS SP8 confocal microscope (Leica) with excitation at 488 nm and detection at 498–531 nm.
ChIP
ChIP was performed on inflorescences of plants that express TCP15-RFP or TCP15-GFP under the control of the 35SCaMV promoter or the TCP15 promoter, respectively, with anti-RFP (632496; Clontech), anti-GFP (ab290; Abcam), and anti-IgG (ab6702; Abcam) antibodies, mainly as described by Ariel et al. (2014). After cross linking and extraction, chromatin was sonicated in a Bioruptor Pico water bath (Diagenode; 10 cycles of 30 s on-/30 s off-pulses at high intensity, using Bioruptor microtubes). For immunoprecipitation, samples were incubated for 12 h at 4°C with Protein A Dynabeads (Invitrogen) precoated with the corresponding antibodies. Immunoprecipitated DNA was recovered using phenol/chloroform/isoamyl alcohol mix (25:24:1) and ethanol precipitation. The amount of specific genomic regions in the immunoprecipitated DNA was analyzed by qPCR using primers listed in Supplemental Table S5. Untreated sonicated chromatin was processed in parallel and considered the input sample.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers: TCP8 (At1g58100), TCP22 (At1g72010), TCP14 (At3g47620), and TCP15 (At1g69690). Accession numbers for other genes are listed in Table 1 and Supplemental Tables S3 and S4.
Supplemental Data
The following supplemental information is available.
Supplemental Figure S1. Transcript levels of class-I TCPs in stamen filaments of amiR-27 and pTCP15::TCP15-EAR plants.
Supplemental Figure S2. ARF6 and ARF8 transcript levels in flowers of pTCP15::TCP15-EAR plants.
Supplemental Figure S3. Stamen length of 35S::SAUR63-GFP plants.
Supplemental Figure S4. Expression in rosettes of SAUR63-GUS under the control of the native or mutated versions of the SAUR63 promoter.
Supplemental Figure S5. Comparison of genes regulated by TCP15, GAs, ARF6/8, MYB21/24, and JA.
Supplemental Figure S6. Expression of genes regulated by MYB21/24 and ARF6/8 in flowers of pTCP15::TCP15-EAR plants.
Supplemental Figure S7. Relative expression of genes repressed in arf6 arf8 flowers and pTCP15::TCP15-EAR plants in different flower organs.
Supplemental Figure S8. Transcript levels of SAUR genes in wild-type and amiR-27 plants after treatment with PAC or PAC plus GA.
Supplemental Table S1. Expression levels of class-I TCPs in stamen filaments.
Supplemental Table S2. Motif enrichment analysis of the promoter regions of SAUR63 subfamily genes.
Supplemental Table S3. List of genes with reduced expression in myb21 myb24 plants but not in pTCP15::TCP15-EAR plants.
Supplemental Table S4. List of genes with reduced expression in arf6 arf8 and pTCP15::TCP15-EAR plants.
Supplemental Table S5. List of primers used.
Acknowledgments
We gratefully acknowledge the Arabidopsis Biological Resource Center, Jean-Michel Davière, Patrick Achard, Martin Kieffer, Simona Masiero, and Stephen Thomas for sending seeds of lines used in this study. We also thank Claudia Studdert for advice with the mutagenesis procedure.
Footnotes
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica, Argentina (PICT-2016-0655).
Articles can be viewed without a subscription.
References
- Aguilar-Martínez JA, Sinha N (2013) Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front Plant Sci 4: 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M (2014) Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell 55: 383–396 [DOI] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Cheng H, Wu W, Soo HM, Peng J (2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarelli M, Cecchetti V (2014) Auxin polar transport in stamen formation and development: How many actors? Front Plant Sci 5: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M (2008) Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchetti V, Celebrin D, Napoli N, Ghelli R, Brunetti P, Costantino P, Cardarelli M (2017) An auxin maximum in the middle layer controls stamen development and pollen maturation in Arabidopsis. New Phytol 213: 1194–1207 [DOI] [PubMed] [Google Scholar]
- Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71: 684–697 [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Greenham K, Castillejo C, Sartor R, Bialy A, Sun TP, Estelle M (2012) Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS One 7: e36210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130: 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J (2009) Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk AD, Muino JM, Cutri L, Dornelas MC, et al. (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159: 1511–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P (2014) Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol 24: 1923–1928 [DOI] [PubMed] [Google Scholar]
- Ghelli R, Brunetti P, Napoli N, de Paolis A, Cecchetti V, Tsuge T, Serino G, Matsui M, Mele G, Rinaldi G, et al. (2018) A newly identified flower-specific splice variant of AUXIN RESPONSE FACTOR8 regulates stamen elongation and endothecium lignification in Arabidopsis. Plant Cell 30: 620–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J 88: 1058–1070 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Li B, Dong AW (2012) The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol Plant 5: 270–280 [DOI] [PubMed] [Google Scholar]
- Lucero LE, Manavella PA, Gras DE, Ariel FD, Gonzalez DH (2017) Class I and class II TCP transcription factors modulate SOC1-dependent flowering at multiple levels. Mol Plant 10: 1571–1574 [DOI] [PubMed] [Google Scholar]
- Lucero LE, Uberti-Manassero NG, Arce AL, Colombatti F, Alemano SG, Gonzalez DH (2015) TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J 84: 267–282 [DOI] [PubMed] [Google Scholar]
- Manassero NG, Viola IL, Welchen E, Gonzalez DH (2013) TCP transcription factors: Architectures of plant form. Biomol Concepts 4: 111–127 [DOI] [PubMed] [Google Scholar]
- Mandaokar A, Thines B, Shin B, Lange BM, Choi G, Koo YJ, Yoo YJ, Choi YD, Choi G, Browse J (2006) Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J 46: 984–1008 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: A family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T (1987) Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol 9: 611–623 [DOI] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, et al. (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nicolas M, Cubas P (2016) TCP factors: New kids on the signaling block. Curr Opin Plant Biol 33: 33–41 [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Plackett AR, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, Jikumaru Y, Benlloch R, Nilsson O, Ruiz-Rivero O, et al. (2012) Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24: 941–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PH, Ellis CM, Ploense SE, Wu MF, Yadav V, Tholl D, Chételat A, Haupt I, Kennerley BJ, Hodgens C, et al. (2012) A regulatory network for coordinated flower maturation. PLoS Genet 8: e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Gray WM (2015) SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant 8: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resentini F, Felipo-Benavent A, Colombo L, Blázquez MA, Alabadí D, Masiero S (2015) TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol Plant 8: 482–485 [DOI] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG, et al. (2008) The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53: 488–504 [DOI] [PubMed] [Google Scholar]
- Saito H, Oikawa T, Hamamoto S, Ishimaru Y, Kanamori-Sato M, Sasaki-Sekimoto Y, Utsumi T, Chen J, Kanno Y, Masuda S, et al. (2015) The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat Commun 6: 6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Chory J (2008) Growth coordination and the shoot epidermis. Curr Opin Plant Biol 11: 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Hara-Nishimura I (2010) A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J 61: 519–528 [DOI] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D (2011) The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Xie D (2013) Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol Plant 6: 1065–1073 [DOI] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM (2014) SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Ikezaki M, Fujibe T, Aida M, Tian CE, Ueno Y, Yamamoto KT, Machida Y, Nakamura K, Ishiguro S (2010) Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol 51: 164–175 [DOI] [PubMed] [Google Scholar]
- Tashiro S, Tian CE, Watahiki MK, Yamamoto KT (2009) Changes in growth kinetics of stamen filaments cause inefficient pollination in massugu2, an auxin-insensitive, dominant mutant of Arabidopsis thaliana. Physiol Plant 137: 175–187 [DOI] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP (2004) Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberti-Manassero NG, Lucero LE, Viola IL, Vegetti AC, Gonzalez DH (2012) The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J Exp Bot 63: 809–823 [DOI] [PubMed] [Google Scholar]
- Viola IL, Camoirano A, Gonzalez DH (2016) Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol 170: 74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola IL, Reinheimer R, Ripoll R, Manassero NG, Gonzalez DH (2012) Determinants of the DNA binding specificity of class I and class II TCP transcription factors. J Biol Chem 287: 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola IL, Uberti Manassero NG, Ripoll R, Gonzalez DH (2011) The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem J 435: 143–155 [DOI] [PubMed] [Google Scholar]
- Xia Y, Chu W, Qi Q, Xun L (2015) New insights into the QuikChangeTM process guide the use of Phusion DNA polymerase for site-directed mutagenesis. Nucleic Acids Res 43: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]