The drought tolerance of the cyanobacterium Nostoc flagelliforme is enhanced by a dehydration-induced chaperone pair that is involved in photosystem II repair under stress conditions.
Abstract
Maintaining the structural integrity of the photosynthetic apparatus during dehydration is critical for effective recovery of photosynthetic activity upon rehydration in a variety of desiccation-tolerant plants, but the underlying molecular mechanism is largely unclear. The subaerial cyanobacterium Nostoc flagelliforme can survive extreme dehydration conditions and quickly recovers its photosynthetic activity upon rehydration. In this study, we found that the expression of the molecular chaperone NfDnaK2 was substantially induced by dehydration, and NfDnaK2 proteins were primarily localized in the thylakoid membrane. NfDnaJ9 was identified to be the cochaperone partner of NfDnaK2, and their encoding genes shared similar transcriptional responses to dehydration. NfDnaJ9 interacted with the NfFtsH2 protease involved in the degradation of damaged D1 protein. Heterologous expression of NfdnaK2 enhanced PSII repair and drought tolerance in transgenic Nostoc sp. PCC 7120. Furthermore, the nitrate reduction (NarL)/nitrogen fixation (FixJ) family transcription factors response regulator (NfRre1) and photosynthetic electron transport-dependent regulator (NfPedR) were identified as putative positive regulators capable of binding to the promoter region of NfdnaK2 and they may mediate dehydration-induced expression of NfdnaK2 in N. flagelliforme. Our findings provide novel insights into the molecular mechanism of desiccation tolerance in some xerotolerant microorganisms, which could facilitate future synthetic approaches to the creation of extremophiles in microorganisms and plants.
Drought is one of the most deleterious environmental conditions in that it suppresses growth, development, and yield of plants (Ceccarelli and Grando, 1996; Huang et al., 2016). Drought stress results in water deficit and causes severe inhibition of photosynthesis due to reduced diffusion and metabolism of carbon dioxide in the chloroplast. Compromised photosynthesis under drought conditions leads to photoinhibition and further photodamage by the absorbed energy that failed to transmit through the photosynthetic electron transport chain (Pinheiro and Chaves, 2011; Wieners et al., 2012). Plants have evolved adaptation mechanisms to cope with drought stress by transcriptional reprogramming of gene expression and subsequent physiological adjustment, which is often initiated by a subset of differentially activated or repressed transcription factors (Skirycz and Inzé, 2010; Nakashima et al., 2014; Song et al., 2016; Li et al., 2019). Although most plants are unable to grow in drylands, some plant species, such as resurrection plants and certain subaerial cyanobacteria, can survive prolonged periods of desiccation by entering a dormant state called anhydrobiosis (Goyal et al., 2005). These organisms evolved some specific protective mechanisms to minimize damage, maintain the integrity of critical cellular and biochemical components during dehydration, and effectively repair the damage upon rehydration (Bewley, 1979; Oliver et al., 2000; Challabathula et al., 2018). However, the molecular basis of these protective and repairing mechanisms during dehydration and rehydration is still largely unknown.
Oxygenic photosynthesis resulting in oxygen accumulation in the atmosphere and metabolic evolvement in organisms is the most significant evolutionary event in the earth’s history (Fischer et al., 2016). Photosynthetic electron transport components include electron carriers such as plastoquinone and plastocyanin, as well as PSII, cytochrome b6f complex, and PSI, all of which are associated with the thylakoid membrane (Eberhard et al., 2008). PSII is a membrane-integral and multisubunit complex that initiates electron flow in oxygenic photosynthesis, and the core of PSII is conserved from cyanobacteria to vascular plants (Nickelsen and Rengstl, 2013). Functional PSII is formed by two different assembly pathways: one is de novo synthesis of all subunits and cofactors and the other is replacement of the damaged PSII proteins (primarily the D1 protein) with newly synthesized proteins following partial disassembly of the PSII complex (Aro et al., 2005; Allen et al., 2011). The highly coordinated PSII assembly inevitably involves protein synthesis, folding, assembly, degradation, and protein-protein interactions (Nixon et al., 2010). These biochemical processes are thought to be guided by molecular chaperones in the native cellular environment (Kampinga and Craig, 2010), and heat shock protein 70 (Hsp70) is one of the most ubiquitous chaperone proteins involved in some of these processes (Hartl and Hayer-Hartl, 2009).
Members of the Hsp70 family exist in almost all organisms from archaebacteria to eukaryotes and represent one of the most conserved protein families involved in a variety of cellular processes (Boorstein et al., 1994; Mayer and Bukau, 2005). One member of 40 kD heat shock protein family (Hsp40) is associated with Hsp70 and stimulates the ATP hydrolysis of Hsp70 by 1,000-fold (Laufen et al., 1999). The homologs of Hsp70 and Hsp40 are called DnaK and DnaJ, respectively, in prokaryotes. DnaJ defines the substrate specificity of its DnaK partner, and the complex is involved in a variety of essential cellular processes (Mayer and Bukau, 2005; Acebrón et al., 2008). Recent transcriptome analyses have indicated that Hsp70/DnaK transcripts increased in abundance following dehydration in the desert biological crust cyanobacterium Microcoleus vaginatus (Rajeev et al., 2013) and in leaves of the resurrection plants Craterostigma plantagineum (Costa et al., 2016) and Sporobolus stapfianus (Costa et al., 2016; Yobi et al., 2017). However, the mode of protection afforded by these dehydration-inducible Hsp70/DnaK proteins to desiccation-tolerant plants is still not defined.
Several studies have suggested the involvement of Hsp70 and Hsp40 in photosynthesis. Deletion of one of three chloroplastic DnaJ proteins (AtJ8, AtJ11, or AtJ20) in Arabidopsis (Arabidopsis thaliana) results in a decrease in photosynthetic efficiency, destabilization of PSII complexes, and imbalance of the redox reactions in chloroplasts (Chen et al., 2010). Overexpression of CrHSP70B enhances tolerance of PSII to photoinhibition in Chlamydomonas reinhardtii and causes a slower degradation rate of D1, CP43, and oxygen evolution complex proteins during high light treatment, but the underlying mechanism is still not clear (Schroda et al., 1999, 2001). In the green alga Dunaliella salina, HSP70B is upregulated upon high light exposure and is associated with D1, D2, and CP47 proteins forming a 320 kD PSII repair intermediate, suggesting a function of HSP70B in stabilization of the PSII holocomplex or removal of inactive reaction center proteins (Yokthongwattana et al., 2001). Liu et al. (2005) reported that the J-domain proteins CDJ2 and HSP70B are a chaperone pair that interacts with the Vesicle-Inducing Protein in Plastids1 (VIPP1), suggesting that HSP70B-CDJ2 plays a role in maintenance or biogenesis of the thylakoid membrane by modulating the assembly of VIPP1. Although these studies suggest a function of the Hsp70-Hsp40 chaperone in photosynthesis, more details remain to be revealed. Meanwhile, the specific role of Hsp70 and Hsp40 in the persistence of photosynthesis under various stress conditions remains largely unclear. PSII is one of the major desiccation-sensitive sites (Canaani et al., 1986), and drought has been reported to enhance the turnover of D1 protein and induce structural reorganization of PSII complex (Giardi et al., 1996; Challabathula et al., 2018). Nevertheless, the molecular mechanism of chaperones in photosynthesis protection under drought stress is still poorly understood.
The subaerial cyanobacterium Nostoc flagelliforme is distributed in arid or semiarid areas (Gao, 1998). It exhibits strong drought tolerance resembling that observed in resurrection plants and desert biological crust organisms (Xiao et al., 2015; Costa et al., 2016; Yobi et al., 2017; Oren et al., 2017, 2019). It can survive long periods in an almost completely dry state and recover photosynthetic activity after rehydration by absorbing dew at early morning or during rainfall (Gao, 1998; Shang et al., 2019). The photosynthetic performance of N. flagelliforme during dehydration and rehydration is similar to homoiochlorophyllous resurrection plants (Challabathula et al., 2018). Its chlorophyll and thylakoid membranes are well preserved during dehydration, which facilitates a rapid recovery of photosynthetic activity upon rehydration (Qiu et al., 2004a). The photosynthetic recovery of N. flagelliforme requires exogenous addition of potassium (Qiu and Gao, 1999; Qiu et al., 2004b), and extracellular carbonic anhydrase is activated to facilitate inorganic carbon acquisition in the process of rehydration (Ye et al., 2008). Weak red light plays an important role in triggering oxygen evolution complex photoactivation in N. flagelliforme upon rehydration (Xu et al., 2019). Cyanobacteria are generally considered to be the ancestors of chloroplasts (Rodríguez-Ezpeleta et al., 2005), and the major components of photosynthetic machinery are conserved among most oxygenic photosynthetic organisms (Nelson and Yocum, 2006; Shen, 2015). Therefore, N. flagelliforme serves as an excellent model for studying photosynthetic adaptation to drought stress. However, the mechanism of photosystem protection and regeneration during dehydration and rehydration in this cyanobacterium still require extensive study, and the involvement of NfDnaK-NfDnaJ in these processes could provide novel insights into photosystem protection in resurrection plants.
In this study, we found that the transcriptional level of NfdnaK2 was clearly induced by dehydration, and NfDnaK2 proteins were primarily localized in the thylakoid membrane. NfDnaJ9 was identified as a cochaperone partner of NfDnaK2 and interacted with NfFtsH2 (COO91_03537) protease, which suggests a role for the NfDnaK2-NfDnaJ9 chaperone pair in the PSII repair cycle under stress conditions. Heterologous expression of NfdnaK2 in Nostoc sp. PCC 7120 remarkably enhanced drought tolerance, which is likely mediated by enhanced D1 degradation under stress conditions. Furthermore, two transcription factors, COO91_04806 (NfPedR) and COO91_05451 (NfRre1), were found to bind to the promoter region of NfdnaK2, which could be the regulator for the adaptation of subaerial cyanobacteria to extreme arid environments. This study describes a molecular mechanism of action of the NfDnaK2-NfDnaJ9 chaperone pair during PSII repair under drought stress in Nostoc species.
RESULTS
NfDnaK2 Is Induced by Dehydration and Primarily Localizes in the Thylakoid Membrane
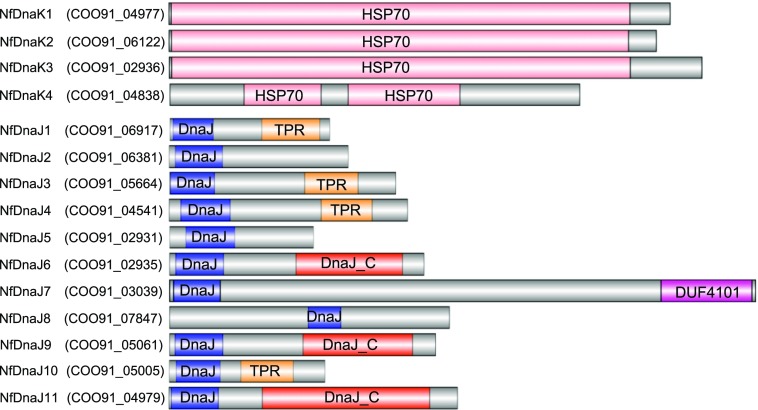
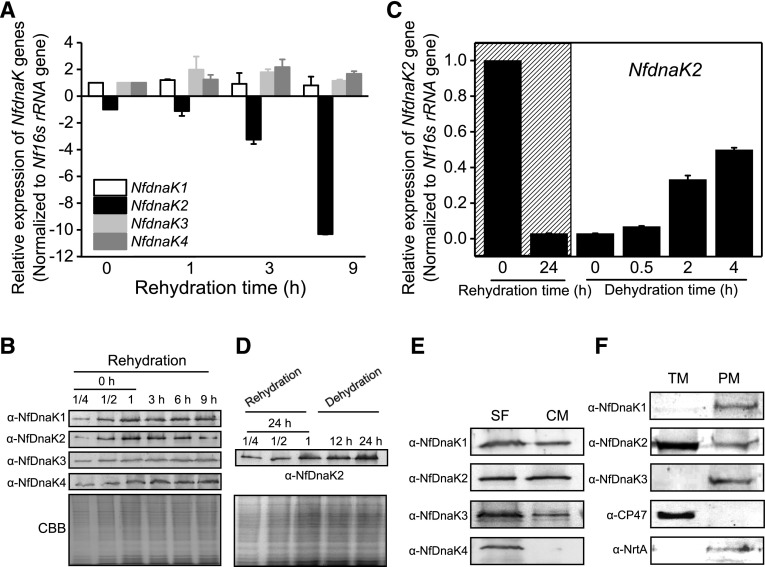
By using the BLAST search tool (BioEdit software version 7.0.5.3) against the genome of N. flagelliforme, four highly conserved NfdnaK genes were found, and the Hsp70 domain architectures were predicited by Pfam database as shown in Figure 1. The expression of NfDnaKs in response to dehydration and rehydration in N. flagelliforme was analyzed at both the transcriptional and protein levels (Fig. 2, A and B). The transcript abundances of NfdnaK3 and NfdnaK4 showed a slight increase, that of NfdnaK1 was unchanged, while that of NfdnaK2 was sharply decreased in response to rehydration. The NfdnaK2 transcripts decreased to 0.90-, 0.30-, and 0.09-fold of the value at the time zero point after rehydrating for 1, 3, and 9 h, respectively (Fig. 2A). Consistent with the changes in transcription levels, the protein level of NfDnaK2 was high in dry samples and was gradually decreased to about 0.5-fold of the value at the time zero point after 9 h rehydration, while the levels of the other three NfDnaK proteins showed a slight increase (Fig. 2B). In contrast to rehydration, dehydration substantially increased NfdnaK2 expression at both the transcriptional and protein levels (Fig. 2, C and D), while the transcriptional levels of the other three NfdnaKs did not show substantial changes in response to dehydration (Supplemental Fig. S1). In addition, the transcripts of NfdnaK2 were found to be the most abundant among the four NfdnaKs, since the transcriptional reads per kilobase of transcript per million value of NfdnaK2 accounted for 94% of the reads of all four NfdnaKs in dry N. flagelliforme according to the transcriptome analysis (Shang et al., 2019). These results indicated that NfDnaK2 is a dehydration-upregulated but rehydration-downregulated gene that may play a role in drought resistance in N. flagelliforme.
Figure 1.
Domain architectures of NfDnaK and NfDnaJ homologs predicted by the Pfam database. The gray color represents the area where the domain cannot be predicted. DnaJ_C, DnaJ C-terminal domain; DUF 4101, domain of unknown function; TPR, Tetratricopeptide repeat.
Figure 2.
The expression pattern of NfDnaK homologs during rehydration and dehydration and their subcellular localizations in field-collected N. flagelliforme. A, Relative transcriptional levels of NfdnaKs during rehydration. The dried field-collected N. flagelliforme samples were rehydrated for 0, 1, 6, and 9 h, and three independent replicates were performed for each time point. Total RNA was extracted as described in the “Materials and Methods”. Data are shown as the mean ± sd of three independent replicates. B, Protein levels of four NfDnaKs during rehydration. The dried field-collected N. flagelliforme samples were rehydrated as described in A. Total protein was extracted as described in the “Materials and Methods”. The total protein of the “0 h” sample (with series of dilution for protein loading, 7.5 μg [one-quarter dilution], 15 μg [half dilution], and 30 μg [1, without dilution], respectively) and other samples (30 μg) were loaded in each lane. Each NfDnaK was indicated by the corresponding specific antibodies (ɑ-NfDnaK1–ɑ-NfDnaK4). The corresponding Coomassie brilliant blue (CBB) stained gels beneath the western blotting results were used as loading controls. C, Relative transcriptional levels of NfdnaK2 during rehydration and subsequent dehydration. The dried field-collected N. flagelliforme samples were rehydrated (marked by hatching) for 0 h (dried field samples), 24 h (fully rehydrated), and subsequently dehydrated for 0.5 h (∼20% water loss), 2 h (∼50% water loss), and 4 h (∼90% water loss). Each treatment was repeated three times independently, and data are shown as the mean ± sd of three independent replicates. D, Changes in expression of the NfDnaK2 protein during rehydration and subsequent dehydration. The dried field-collected N. flagelliforme samples were rehydrated for 24 h and subsequently dehydrated for 12 h and 24 h. The total protein of the “24 h” sample (with series of dilution for protein loading, 7.5 μg [one-quarter dilution], 15 μg [half dilution], and 30 μg [1, without dilution], respectively) and other samples (30 μg) were loaded in each lane. The corresponding CBB stained gels beneath the western blotting results were used as loading controls. E and F, Subcellular localizations of NfDnaKs. The total proteins extracted from dried field N. flagelliforme samples were fractionated into soluble fractions (SF) and crude membranes (CM), and 30 μg protein was loaded in each lane (E). Crude membranes were subsequently separated into thylakoid membranes (TM) and plasma membranes (pm) by Suc density gradient centrifugation, and 30 μg protein was loaded in each lane (F).
We further investigated the subcellular location of NfDnaKs by using western blotting analysis of the fractionated plasma membrane, thylakoid membrane, and soluble proteins (Huang et al., 2002). The proteins of NfDnaK1, NfDnaK2, and NfDnaK3 were detected in both soluble and membrane fractions, while NfDnaK4 was only detected in soluble fractions (Fig. 2E). Moreover, NfDnaK2 primarily localized in the thylakoid membrane, and a lower level of NfDnaK2 was also detected in the plasma membrane, while NfDnaK1 and NfDnaK3 were only detected in the plasma membrane. The purity of the membrane fractions was assessed using the marker proteins NrtA and CP47 for the plasma membrane and thylakoid membrane, respectively (Fig. 2F). Localization of NfDnaK2 in the thylakoid membrane suggested a role for NfDnaK2 in the photosynthetic process.
NfDnaJ9 Is a Cochaperone Protein of NfDnaK2
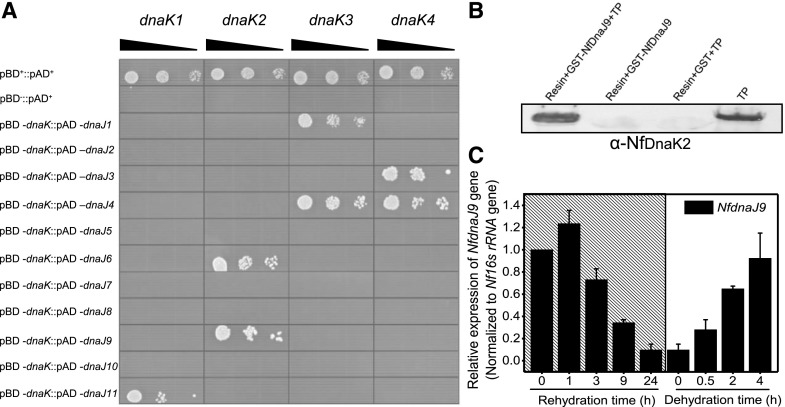
The DnaKs are regulated by several accessory partners, particularly the DnaJ family, and the DnaJ domain in DnaJ and DnaJ-like proteins is believed to be the major binding site for the interaction with DnaKs (Hennessy et al., 2005). By searching the genome of N. flagelliforme, 11 NfDnaJ proteins containing the DnaJ domain were found and named sequentially NfDnaJ1 to NfDnaJ11 (Fig. 1). To determine the interaction between NfDnaKs and NfDnaJs, yeast two-hybrid (Y2H) analyses were performed. The yeast Saccharomyces cerevisiae AH109 cotransformed with the plasmids pBD-NfDnaK2 and pAD-NfDnaJ6 or pAD-NfDnaJ9 could grow on synthetic defined (SD) medium lacking Trp, Leu, His and adenine (SD/-Trp-Leu-His-Ade), suggesting an interaction between NfDnaK2 and NfDnaJ6 or NfDnaJ9 (Fig. 3A). In addition, interactions between other NfDnaK and NfDnaJ proteins (pBD-NfDnaK1 and pAD-NfDnaJ11, pBD-NfDnaK3 and pAD-NfDnaJ1 or pAD-NfDnaJ4, pBD-NfDnaK4 and pAD-NfDnaJ3 or pAD-NfDnaJ4) were also identified. These results indicate that each member of the NfDnaK family has specific interaction partners in the family of NfDnaJs.
Figure 3.
Interaction of NfDnaK and NfDnaJ in N. flagelliforme, and the relative transcriptional levels of NfdnaJ9 during rehydration and subsequent dehydration. A, In vitro protein-protein interaction analyses of NfDnaK and NfDnaJ by Y2H assay. The yeast transformants (10 μL each of cell suspensions diluted from OD600 0.1 to 0.001, as indicated by the slope of the black triangles from left to right) expressing positive control plasmids (pAD+ and pBD+), negative control plasmids (pAD+ and pBD−), and plasmids for testing protein interaction (pAD-NfdnaJ and pBD-NfdnaK) were grown on SD/-Trp-Leu-His-Ade agar plates for 3 d. B, In vivo pull-down analysis of the interaction between NfDnaK2 and NfDnaJ9. The total proteins (TP) extracted from dried field N. flagelliforme samples were incubated with GST-binding resin and GST-tagged NfDnaJ9 (Resin+GST-NfDnaJ9+TP), and the coeluted products were detected with the specific antibody against NfDnaK2. The eluted products of the incubation mixture of resin and GST-tagged NfDnaJ9 (Resin+GST-NfDnaJ9),or resin, GST, and TP (Resin+GST+TP) were used as negative controls to exclude nonspecific interference, and TP extraction was used as the positive control. C, Relative transcriptional levels of NfdnaJ9 during rehydration and subsequent dehydration. The dried field-collected N. flagelliforme samples were rehydrated (hatch marks) for 0, 1, 3, 9, and 24 h and subsequently dehydrated for 0.5, 2, and 4 h. Each treatment was repeated three times independently, and data are shown as the mean ± sd of three independent replicates.
Transcriptome analysis of N. flagelliforme (Shang et al., 2019) revealed that the transcript level of NfdnaJ9 was higher than that of other NfdnaJs in dry samples, and that NfdnaJ9 transcripts account for 48% of the abundance of all 11 NfdnaJ transcripts, while NfdnaJ6 transcripts only account for 1.3%. Furthermore, the proportion of NfdnaJ9 decreased to 14.2% after 15 h rehydration. These results suggest that NfDnaJ9 may play an important role in drought tolerance. We therefore focused on the investigation of NfDnaJ9 among other NfDnaJs. The interaction between NfDnaK2 and NfDnaJ9 was verified by an in vivo pull-down assay (Fig. 3B), and in vivo copurified NfDnaK2 with glutathione S-transferase (GST)-tagged NfDnaJ9 was also confirmed by liquid chromatography tandem mass spectrometry analysis (Supplemental Table S1). Furthermore, the transcript levels of NfdnaJ9 increased in response to dehydration but decreased in the process of rehydration (Fig. 3C), which coincides with the expressing pattern of NfdnaK2. Altogether, these observations implied that the NfDnaK2-NfDnaJ9 chaperone could be important for drought resistance in N. flagelliforme.
NfFtsH2 Is the Target Substrate Protein of NfDnaJ9
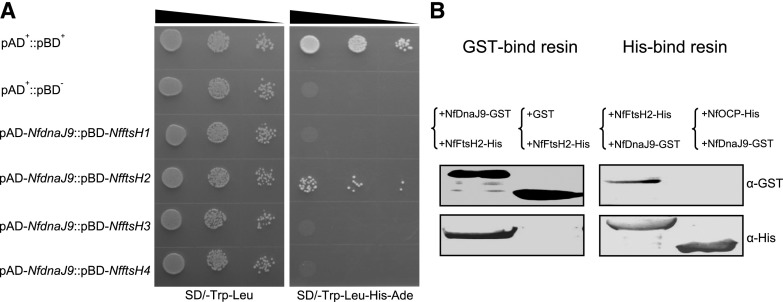
To explore the function of the NfDnaK2-NfDnaJ9 chaperone in drought tolerance of N. flagelliforme, a pull-down assay was used to identify possible target substrate proteins of NfDnaJ9. The recombinant GST-tagged NfDnaJ9 was incubated with the total proteins of N. flagelliforme, and the copurified proteins with NfDnaJ9 were isolated by GST-Bind resin and subjected to mass spectrometry (MS) analysis. The results showed that the bait NfDnaJ9 was ranked first, and its interacting NfDnaK2 and GroL chaperones were also at the top of the identified protein list (Supplemental Table S1). Neither NfDnaJ9 nor NfDnaK2 was detected in the negative control (only GST-tag pull down). Many putative NfDnaJ9-interacting proteins were identified by MS analysis, and FtsH protease, which is involved in the PSII repair cycle, appeared in all three replicates but was absent in the negative controls, and thus was chosen for further analysis. There are four conserved FtsH proteins in N. flagelliforme, which were named NfFtsH1–NfFtsH4 (COO91_03160, COO91_03537, COO91_03727, and COO91_00363, respectively). The interactions between NfDnaJ9 and NfFtsHs were tested by Y2H assays, and the yeast S. cerevisiae AH109 cotransformed with the plasmids pAD-NfDnaJ9 and pBD-NfFtsH2 could grow on SD/-Trp-Leu-His-Ade medium, indicating a specific interaction between NfDnaJ9 and NfFtsH2 (Fig. 4A). This interaction was further confirmed by in vitro pull-down assay. As shown in Figure 4B, GST-tagged NfDnaJ9 and His-tagged NfFtsH2 could be coeluted through either GST-Bind or His-Bind resin affinity purification but could not be coeluted with negative control proteins (His-tagged NfOCP and GST). This result further confirmed that NfDnaJ9 physically interacts with NfFtsH2. Since FtsH2 protease is involved in the D1 protein turnover by degrading damaged D1 protein, these results suggest that the upregulated NfDnaK2-NfDnaJ9 chaperone plays important roles in PSII protection in the process of dehydration in N. flagelliforme.
Figure 4.
In vitro interaction between NfDnaJ9 and NfFtsH2. A, Y2H assay of the interaction between NfDnaJ9 and NfFtsH. The yeast transformants (10 μL each of cell suspensions diluted from OD600 0.1 to 0.001, as indicated by the slope of the black triangles from left to right) expressing positive control plasmids (pAD+ and pBD+), negative control plasmids (pAD+ and pBD−) and plasmids for testing protein interaction (pAD-NfdnaJ9 and pBD-NfftsH) were grown on a SD/-Trp-Leu or SD/-Trp-Leu-His-Ade agar plate for 3 d. The yeast strain that grew on the SD/-Trp-Leu plate indicated that two plasmids (pAD and pGB) were cotransformed into S. cerevisiae AH109, and the yeast strain that grew on the SD/-Trp-Leu-His-Ade plate indicated that the two tested proteins interacted in S. cerevisiae AH109. B, Pull-down assay of the interaction between NfDnaJ9 and NfFtsH2. The supernatants of the extracts from GST-tagged NfDnaJ9 or His-tagged NfFtsH2 overexpressed E. coli were mixed at 4°C for overnight, and then were copurified through a GST-binding (left, +NfDnaJ9-GST+NfFtsH2-His) or His-binding resin (right, +NfFtsH2-His+NfDnaJ9-GST). The GST-tag and His-tag signals of copurified proteins were detected with the specific antibodies against His-Tag and GST-Tag, respectively. The samples that GST and His-tagged NfFtsH2 mixture and copurification through a GST-bind resin (left, +GST+NfFtsH2-His), and nonrelative His-tagged NfOCP and GST-tagged NfDnaJ9 mixture and copurification through a His-binding resin (right, +NfOCP-His+NfDnaJ9-GST) were used as negative controls to exclude the nonspecific interactions.
Heterologous Expression of NfDnaK2 Enhances PSII Repair
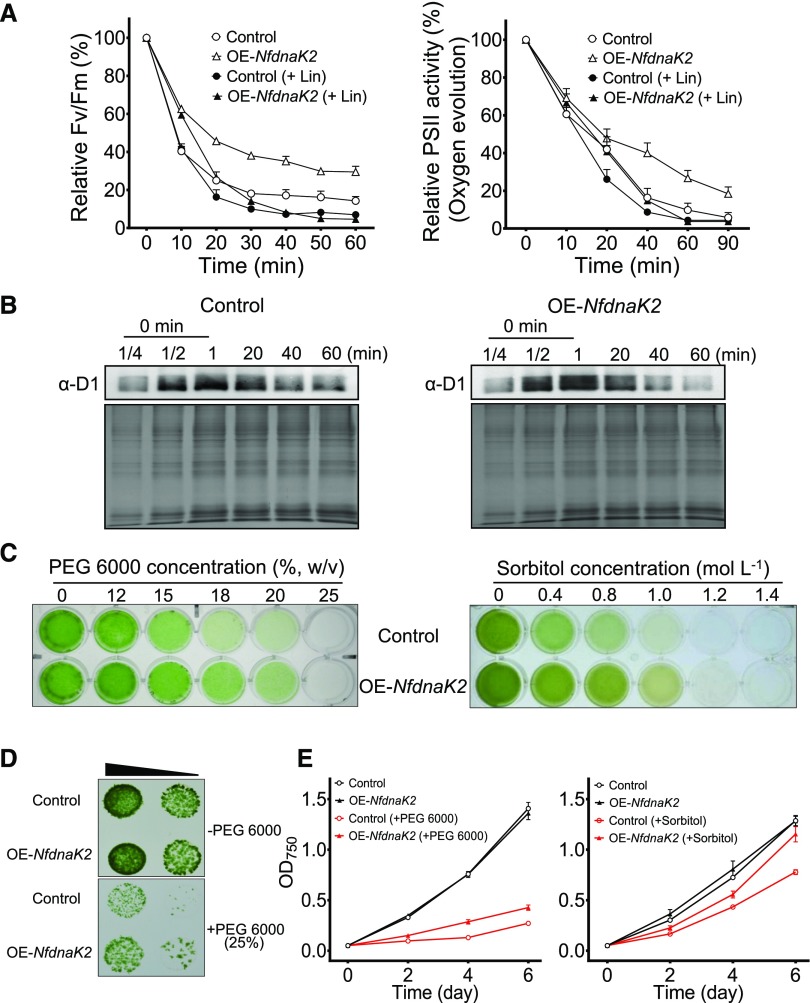
Since genetic manipulation is difficult in N. flagelliforme at present, NfdnaK2 of N. flagelliforme was transformed into Nostoc sp. PCC 7120 to investigate its function in PSII repair or protection. The inhibitory effect of high light on PSII activity was analyzed with and without inhibition of PSII repair by lincomycin (Lin), and the difference between PSII activity in the absence and that in the presence of Lin represents the contribution of PSII repair. As shown in Figure 5A, in the absence of Lin, the maximum potential quantum efficiency of PSII (Fv/Fm values) for both strains decreased rapidly during the initial 30 min exposure to high light (400 µmol photons m−2 s−1), and then gradually reached steady state with prolonged treatment time. However, the extent of the decrease of Fv/Fm values was much less in the transgenic strain (OE-NfdnaK2) than in wild-type Nostoc sp. PCC 7120. In the presence of Lin, high light led to a similar decrease in Fv/Fm values in these two strains. The difference in relative Fv/Fm values in the absence and presence of Lin in transgenic strains was larger than that in wild-type strains (Fig. 5A), indicating that the expression of NfDnaK2 in Nostoc sp. PCC 7120 enhanced PSII repair. PSII activity probed by the O2 evolution rate also supported enhanced PSII repair in the OE-NfdnaK2 strain when compared with the wild-type strain (Fig. 5A).
Figure 5.
Heterologous expression of NfdnaK2 enhanced PSII repair and drought tolerance in Nostoc sp. PCC 7120. A, Effect of high light (400 μmol photons m−2 s−1) on Fv/Fm and PSII oxygen evolution of wild-type and NfdnaK2-overexpressing (OE-NfdnaK2) Nostoc sp. PCC 7120 strains in the absence (open symbols) or presence (solid symbols) of 100 mg mL−1 Lin. Exponential growth cultures with OD750 0.4 were exposed to high light and withdrawn at the indicated times for Fv/Fm and oxygen evolution measurements. Wild-type Nostoc sp. PCC 7120 was used as a control to compare with the OE-NfdnaK2 strain. Each treatment was carried out for four independent replicates, and the 100% values corresponded to the values at time point zero of two strains. Data are shown as the means ± sd of four independent replicates. B, Comparison of D1 degradation between wild-type (Control) and OE-NfdnaK2 Nostoc sp. PCC 7120 strains. The samples were treated with high light, as shown in A, in the presence of Lin, and collected at the indicated times. The thylakoid membrane proteins of different samples (30 μg) were loaded into each lane (except for the series of dilution for the “0 min” sample protein loading, 7.5 μg [one-quarter dilution], 15 μg [half dilution], and 30 μg [1, without dilution], respectively) and separated by 15% (v/v) SDS-PAGE and subjected to western blotting analyses. The corresponding Coomassie brilliant blue stained gels beneath the western blotting results were used as protein loading controls. C to E, Growth comparison of wild-type (Control) and transgenic strains upon PEG 6000 (C–E) or sorbitol treatment (C and E). Cells were grown in culture plates for 5 d under treatment with various concentrations of PEG 6000 or sorbitol (C) or on BG11 agar plates (10 µL drops of cell culture with OD750 0.4 and 0.04, as indicated by the slope of the black triangle from left to right) with or without 25% (w/v) PEG infusion (D), or grown in flasks in the presence or absence of 18% (w/v) PEG 6000 or 1 mol L−1 sorbitol (E). Each treatment was carried out for three independent replicates for two strains, and data are shown as the mean ± sd of three independent replicates.
The repair of damaged PSII proceeds in two steps: damaged D1 protein is removed first (e.g. by FtsH and Deg), and newly synthesized D1 protein is then integrated into its place (Nixon et al., 2010; Nickelsen and Rengstl, 2013). Since NfFtsH2 was identified as the target protein of the NfDnaK2-NfDnaJ9 chaperone, the D1 degradation rates were compared between wild-type and transgenic strains exposed to high light (400 µmol photons m−2 s−1) with the addition of protein synthesis inhibitor Lin. As shown in Figure 5B, the degradation of D1 protein started within 20 min of high-light treatment in both strains, while the degradation proceeded much faster in the transgenic strain than in the wild-type strain. The D1 signal was very weak after exposure to high light for 60 min in the transgenic strain, while there was still a clearly detectable amount of damaged D1 in the wild-type Nostoc sp. PCC 7120 (Fig. 5B). MS analysis showed that the content of FtsH2 was significantly increased, by 65%, in the membrane fractions of OE-NfdnaK2 Nostoc sp. PCC 7120 when compared with that of the wild-type strain (P < 0.05, Student’s t test; Supplemental Fig. S2). Both physiological and biochemical results indicated that NfDnaK2 is involved in PSII repair through mediating the degradation of damaged D1 proteins.
Polyethylene glycol (PEG) and sorbitol are chemicals commonly used to create low water potentials mimicking the effect of drought stress on plants (Verslues et al., 2006; Gopal and Iwama, 2007). To determine whether NfdnaK2 could confer tolerance to water deficit, the growth of wild-type and transgenic Nostoc sp. PCC 7120 in response to PEG 6000 and sorbitol was analyzed. The growth of both strains was apparently inhibited by increased concentrations of PEG 6000 and sorbitol, although the wild-type strain appeared to be more inhibited by high concentrations of PEG 6000 and sorbitol than the transgenic Nostoc sp. PCC 7120 (Fig. 5C). The growth of transgenic Nostoc sp. PCC 7120 was less inhibited than that of the wild-type strain on the 25% PEG-infused BG11 agar plate (Fig. 5D). The growth rate of the OE-NfdnaK2 strain was 0.35 ± 0.01 d−1 in the presence of 18% (w/v) PEG 6000 and 0.52 ± 0.01 d−1 in the presence of 1 mol L−1 sorbitol, which was significantly higher than that of the wild-type strain (0.28 ± 0.01 and 0.45 ± 0.01 d−1; Student’s t test, P < 0.05; Fig. 5E). These results indicate that NfDnaK2 confers on cyanobacterium the tolerance of low water availability and drought stress.
NfRre1 and NfPedR Are the Transcription Factors for NfdnaK2 Response to Drought Stress
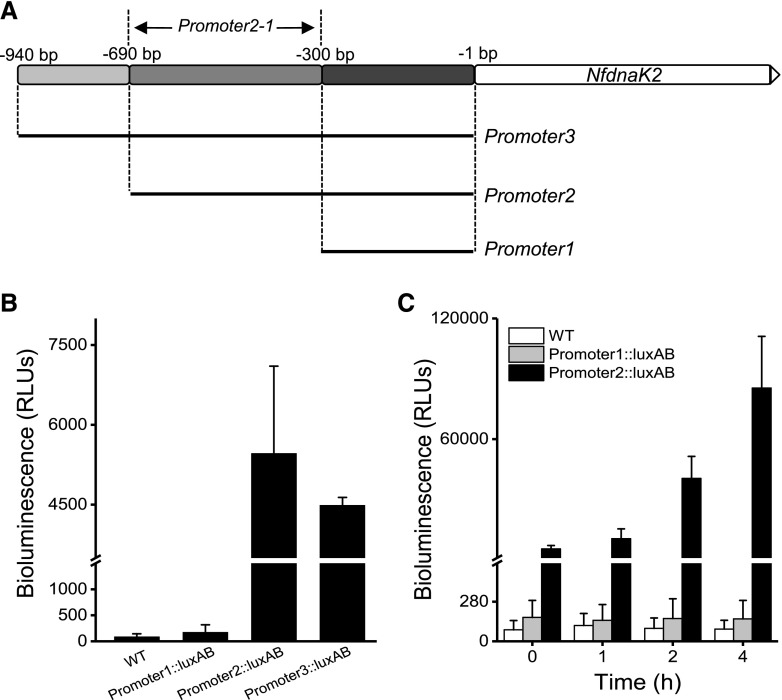
Previous studies suggested that the transcription of molecular chaperones is negatively regulated by HrcA in cyanobacteria (Rajaram and Apte, 2010), and the Hik34-Rre1 module directly activates heat-stress-inducible transcription of the major chaperones (Kobayashi et al., 2017). However, how these transcriptional regulations respond to drought stress is unknown. To determine the active promoter region of NfdnaK2, different promoter elements, promoter1–promoter3, were fused with the luxAB reporter gene (Fig. 6A). Expression of the luxAB reporter gene driven by the promoter elements was analyzed by detecting bioluminescence intensity. The strains carrying the LuxAB reporter gene fused with promoter2 (Promoter2::luxAB strain) or promoter3 (Promoter3::luxAB strain) showed high bioluminescence signals, and the Promoter1::luxAB strain exhibited no activity. This result indicated that promoter2 and promoter3 contain potential binding sites for transcription factors (Fig. 6B). Since the Promoter2::luxAB and Promoter3::luxAB produced similar levels of bioluminescence and Promoter1::luxAB did not show activity (Fig. 6B), the active promoter of NfdnaK2 is likely to be in the region from −690 bp to −300 bp, which was named promoter2-1 (Fig. 6A). Moreover, the bioluminescence intensity driven by promoter2 increased to about 15-fold within 4 h dehydration compared with that without dehydration treatment (Fig. 6C), which suggests that the promoter2 segment contains the cis-element for induced expression of NfdnaK2 in response to drought stress.
Figure 6.
Analysis of the promoter activity of NfdnaK2. A, Schematic of a nested series of NfdnaK2 promoter regions. Promoter1 (−300 to −1 bp region), Promoter2 (−690 to −1 bp region), Promoter3 (−940 to −1 bp region), and Promoter2-1 (−690 to −300 bp region) for promoter activity analysis. B, Measurements of luminescence driven by the NfdnaK2 promoter regions in the transgenic Nostoc sp. PCC 7120 carrying the LuxAB reporter fused with different promoter segments of the NfdnaK2 gene. Three individual cultures of wild-type (WT) and different transgenic Nostoc sp. PCC 7120 strains were incubated under grown condition (25°C and 30 µmol photons m−2 s−1), and the values of relative bioluminescence intensities (in relative luminescence units [RLUs]) are shown as the mean ± sd of three independent replicates. C, Time course of luminescence intensity in the transgenic Nostoc sp. PCC 7120 carrying the luxAB reporter gene fused with the Promoter1 and Promoter2 segments of the NfdnaK2 promoter in the process of dehydration. Cells of three strains were harvested on nylon membranes by filtration and put on the solid BG11 medium containing 2% (w/v) agar in the incubator for dehydration treatment for different periods of time. Each treatment was repeated independently three times, and data are shown as the mean ± sd of three independent replicates. The wild-type Nostoc sp. PCC 7120 strain was used as a negative control for luminescence measurements.
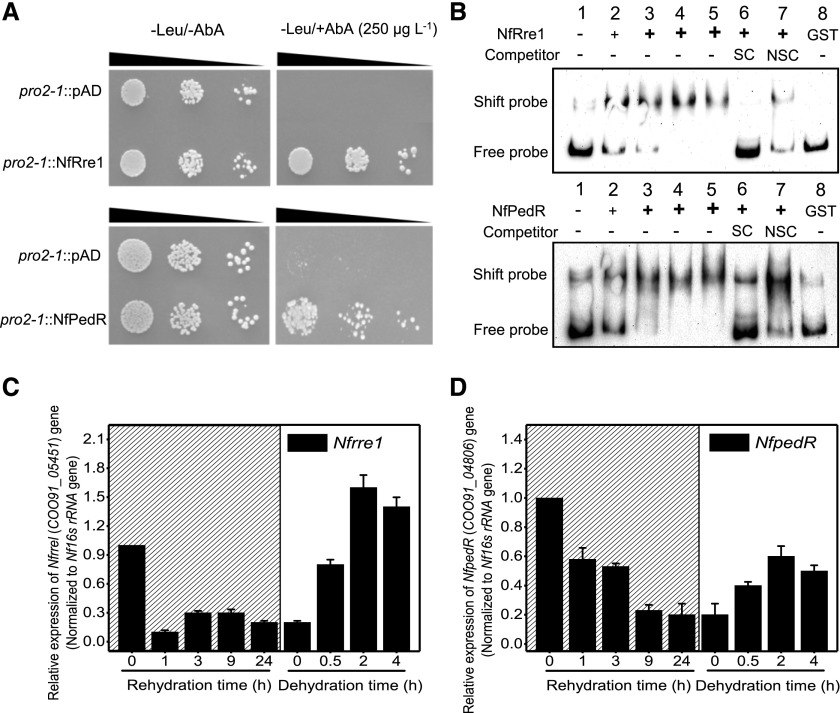
Yeast one-hybrid (Y1H) screening has been widely used for identification of transcription factors interacting with specific DNA fragments (Mitsuda et al., 2010). We used the promoter2-1 region as a bait and screened a Y1H transcription factor library of N. flagelliforme. Two transcription factors capable of binding to promoter2-1, NfRre1 (COO91_05451) and NfPedR (COO91_04806), were identified as putative regulators of NfdnaK2 (Fig. 7A). The interactions between these two candidate transcription factors and the NfdnaK2 promoter2-1 fragment were verified by electrophoretic mobility shift assay (EMSA). As shown in Figure 7B, retarded DNA bands were observed for 5 ng biotin-labeled promoter2-1 after addition of recombinant GST-tagged NfRre1 or NfPedR. Furthermore, the amounts of retarded DNA bands increased with the increase of GST-tagged NfRre1 or NfPedR concentrations (Fig. 7B, lanes 1–5). In addition, the shifted band was largely reduced by the addition of unlabeled promoter2-1 (specific competitor; Fig. 7B, lane 6), while addition of poly(deoxyinosinic-deoxycytidylic) acid [poly(dI-dC)] as a nonspecific competitor (NSC) did not affect the mobility shift of the bands (Fig. 7B, lane 7). GST as a negative control had no effect on the mobility of biotin-labeled promoter2-1 in the gel (Fig. 7B, lane 8). These results indicate that NfRre1 and NfPedR could physically bind to the promoter2-1 region in the promoter of NfdnaK2. Both NfRre1 and NfPedR contain a N-terminal receiver domain and a C-terminal DNA-binding domain and belong to the NarL/FixJ family. In N. flagelliforme, the pattern of transcriptional response of these two transcription factors to drought, especially that of NfRre1, coincided with that of NfdnaK2 (Fig. 7, C and D), which suggests that NfRre1 and NfPedR act as positive regulators of the expression of NfdnaK2 in response to drought stress in N. flagelliforme.
Figure 7.
Identification of transcriptional regulators binding to the promoter of NfdnaK2. A, Verification of interactions between the cis-acting element Promoter2-1 (pro2-1) and putative transcription factors NfRre1 or NfPedR by Y1H assay. The transformed yeast cells (10 μL) with DNA fragments and regulator proteins at OD600 0.1, 0.01, and 0.001 (indicated by the slope of the black triangles from left to right) were tested on SD/-Leu medium with or without 250 μg L−1 AbA. B, Interaction analyses between Promoter2-1 and NfRre1 or NfPedR by EMSA. Biotin-labeled Promoter2-1 (5 ng) was incubated with increasing concentrations of purified GST-tagged NfRre1 or NfPedR (lane 1, 0 ng; lane 2, 100 ng; lane 3, 200 ng; lane 4, 400 ng; and lane 5, 600 ng). The unlabeled Promoter2-1 (lane 6, 500 ng) and poly(dI-dC) (lane 7, 2,000 ng) were used as specific competitor (SC) and nonspecific competitor (NSC) inhibitors, respectively. GST-tag (lane 8, 5 ng Biotin-labeled Promoter2-1 was incubated with 200 ng GST protein, which was the equal amount as GST-tagged NfRre1 or NfPedR used in lanes 6 and 7) was used as a negative control to exclude the nonspecific binding of NfRre1/NfPedR with promoter2-1. The shift probe indicates the biotin-labeled promoter2-1 interacting with NfRre1 or NfPedR, and the free probe is the biotin-labeled promoter2-1 without interacting with NfRre1 or NfPedR. C and D, Relative transcriptional levels of Nfrre1 (C) or NfpedR (D) during rehydration and subsequent dehydration. The dried field-collected N. flagelliforme samples were rehydrated (marked by hatching) for 0, 1, 3, 9, and 24 h and subsequently dehydrated for 0.5, 2, and 4 h. Each treatment was repeated three times independently, and data are shown as the mean ± sd of three independent replicates.
DISCUSSION
Hsp70 proteins are ubiquitous molecular chaperones that serve as central components of the cellular network for protein folding and other critical biochemical processes (Mayer and Bukau, 2005). The genes encoding DnaK and its cochaperone partner DnaJ exist as multicopies in all sequenced cyanobacteria and are possibly involved in their adaptation to various environmental stresses (Rupprecht et al., 2007; Sato et al., 2007). In the desiccation-tolerant cyanobacterium N. flagelliforme, the DnaK-DnaJ chaperone pair could help in coping with harsh conditions such as extreme water deficit in the terrestrial habitat. The photosystems, especially PSII, are sensitive to drought, and normal function of PSII directly affects growth and even survival of plants under stress conditions (Pinheiro and Chaves, 2011). Previous study has demonstrated that photoinhibition of cyanobacteria at exceedingly high light intensities in desert biological crust is limited owing to a fast rate of PSII repair (Harel et al., 2004). N. flagelliforme is habituated to arid or semiarid areas with daily high light stress (Gao, 1998). This cyanobacterial species contains up to six copies of psbA encoding the PSII core subunit D1 protein, suggesting an abundant de novo biogenesis of D1 for efficient PSII repair in N. flagelliforme (Shang et al., 2019). We found that expression of NfdnaK2 is highly induced by dehydration, while the other three NfdnaKs show a negligible response to water loss (Supplemental Fig. S1). Meanwhile, NfDnaK2 is primarily localized in the thylakoid membrane and its heterologous expression in Nostoc sp. PCC 7120 enhanced tolerance to high light and drought stress, indicating that NfDnaK2 could be involved in photoprotection and adaptation of N. flagelliforme to subaerial habitat.
Previous studies showed that DnaK2 homologs are essential in cyanobacteria, and they could not be completely knocked out in Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942 (Nimura et al., 2001; Rupprecht et al., 2007). The NfDnaK2 homolog in Chlamydomonas reinhardtii is the chloroplast-located HSP70B, which is involved in protecting PSII subunits against photodamage (Schroda et al., 1999). In Dunaliella salina, HSP70B was identified as a component in the photodamage-repairing intermediate (Yokthongwattana et al., 2001). Consistently, our results showed that NfDnaK2 plays an important role in maintaining PSII activity under dehydration in N. flagelliforme (Figs. 4 and 5). The substrate specificity of Hsp70 is thought to be mediated mainly by its cochaperones, of which the J-domain cochaperones represent important Hsp70 partners (Acebrón et al., 2008). Liu et al. (2005) reported that J-domain protein CDJ2 and HSP70B form a plastidic chaperone pair and interact with VIPP1, suggesting a role of HSP70B-CDJ2 in maintenance or biogenesis of the thylakoid membrane. In tomato, the chloroplast-targeted DnaJ protein LeCDJ1 contributes to maintaining PSII activity under chilling stress, and another DnaJ protein SlCDJ2 protects Rubisco against heat stress (Kong et al., 2014; Wang et al., 2015). Both DnaJs interact with the cpHsp70 (chloroplast Hsp70), which indicates an involvement of HSP70 in stabilizing photosynthesis in tomato (Kong et al., 2014; Wang et al., 2015). Thus, the association of Hsp70 with its interaction partner DnaJ-like proteins to specify its functions seems to be conserved in both cyanobacteria and vascular plants.
In this study, NfDnaJ9 was identified as a specific cochaperone of NfDnaK2 in N. flagelliforme (Fig. 3, A and B), and the similar expression pattern of NfDnaJ9 and NfDnaK2 in response to dehydration provided further evidence for their functional association (Fig. 3C). NfDnaJ9 contains a N-terminal J domain and is classified as type II DnaJ according to the homology alignment with Synechocystis sp. PCC 6803 (Düppre et al., 2011). The NfDnaJ9 homolog in S. elongatus PCC 7942 is crucial for heat and high light stress response because it protects psbAII transcripts from RNase degradation (Sato et al., 2007; Watanabe et al., 2007). Phylogenetic analyses suggest that NfDnaJ9 is most closely related to the chloroplast-targeted AtJ20 in Arabidopsis, which is involved in optimization of photosynthetic reactions (Chen et al., 2010). Strikingly, NfFtsH2 protease was identified as interacting with NfDnaJ9 in our study (Fig. 4), and FtsH2 content was significantly increased, by 65%, in the membrane fractions of the OE-NfdnaK2 Nostoc sp. PCC 7120 compared to that in the control strain (Supplemental Fig. S2). FtsH proteases play critical roles in both cyanobacteria and plants, and NfFtsH2 homologs in Synechocystis sp. PCC 6803 (Slr0228) and Arabidopsis (AT2G30950) are involved in D1 turnover by degrading damaged D1 protein (Bailey et al., 2002; Nixon et al., 2005; Kato and Sakamoto, 2009; Boehm et al., 2012). Since chaperones function in folding and assembly of newly synthesized proteins (Mayer and Bukau, 2005), the NfDnaK2-NfDnaJ9 chaperone pair may facilitate the folding, maturation, and accumulation of NfFtsH2 protease in the thylakoid membrane. Therefore, the NfDnaK2-NfDnaJ9 chaperone in N. flagelliforme is likely to function in PSII repair through FtsH2 protease promoting degradation of damaged D1 in the process of dehydration. When exposed to high light, the D1 degradation rate was much faster in the NfdnaK2 transgenic Nostoc sp. PCC 7120 than in the wild-type strain, indicating that NfDnaK2-NfDnaJ9 chaperone-enhanced FtsH protease degrades D1 and protects PSII under high light conditions (Fig. 5B). Altogether, the findings presented here provide direct evidence linking DnaK-DnaJ chaperones with the degradation of D1 proteins during the PSII repair cycle.
Rajeev et al. (2013) have shown that one dnaK gene of the desert biological crust cyanobacterium M. vaginatus (WP_006633368.1), highly conserved to NfdnaK2, was induced during dehydration by transcriptome analysis. Consistently, the expression of NfdnaK2 was substantially decreased in response to rehydration but highly induced by dehydration (Fig. 2). This indicates that the expression of NfdnaK2 is regulated by the intracellular water status and is likely to play a role in protecting cells against dehydration damage. Previous studies showed that most Hsp genes are either regulated positively by σ32 (RpoH) or negatively by HrcA in bacteria (Yura and Nakahigashi, 1999), yet HrcA is only present in cyanobacteria and heat shock induction of the Hsp genes was observed even in a hrcA knockout mutant of Synechocystis sp. PCC 6803 (Nakamoto et al., 2003; Singh et al., 2006; Rajaram and Apte, 2010). This suggests that HrcA is not a key factor for Hsp gene transcription and that some other factors are involved. Rre1 is a transcription factor that was recently reported to directly regulate dnaK2 expression in S. elongatus PCC 7942 (Kobayashi et al., 2017). In this study, two transcription factors, NfRre1 (COO91_05451) and NfPedR (COO91_04806) were identified as able to bind to the cis-acting element of NfdnaK2 (Fig. 7). NfRre1 homologs are highly conserved and essential in all sequenced cyanobacterial strains (Rubin et al., 2015). The Hik34-Rre1 module controls the transcriptional activation of a subset of salt and hyperosmotic stress-responsive genes (Paithoonrangsarid et al., 2004; Shoumskaya et al., 2005) and positively regulates the major chaperone dnaK2 and other genes in S. elongatus PCC 7942 (Kobayashi et al., 2017). Whereas there is only one copy of Rre1 in S. elongatus PCC 7942, there are three NfRre1 homologous genes in N. flagelliforme, and only COO91_05451 can bind to the NfdnaK2 promoter in the yeast one-hybrid assay (Supplemental Fig. S3). These results suggest that each of the NfRre1 genes could participate in different signal transduction pathways, and COO91_05451 may specifically regulate the expression of NfdnaK2. In addition, the similar transcriptional patterns of COO91_05451 and NfdnaK2 in response to water status (Fig. 7C) suggest that this transcription factor acts as a positive regulator to control the expression of NfdnaK2.
The homolog of NfPedR (COO91_04806) in Synechocystis sp. PCC 6803, called PedR, is a small LuxR-type transcriptional regulator and is involved in photon-flux-density-dependent transcriptional regulation by interacting with thioredoxin (Nakamura and Hihara, 2006; Horiuchi et al., 2010). NfPedR showed a similar transcriptional pattern with NfdnaK2 (Fig. 7D), and thus it could also positively regulate NfdnaK2 expression in response to dehydration. The redox state of the PedR Cys residue is determined by thioredoxin, which drains electrons from the photosynthetic electron transport chain, resulting in a switch of the inactive (reduced) and active (oxidized) states of PedR to control the expression of its target genes (Nakamura and Hihara, 2006; Horiuchi et al., 2010). Based on this model, we propose that NfPedR is maintained in an active state to induce the expression of NfdnaK2 in N. flagelliforme upon dehydration due to decreased photosynthetic electron transport, and thus, reduction power is insufficient to reduce the PedR Cys residue. Moreover, upregulated expression of NfPedR during dehydration could further positively regulate the expression of NfdnaK2, which facilitates the maintenance of PSII integrity and activity.
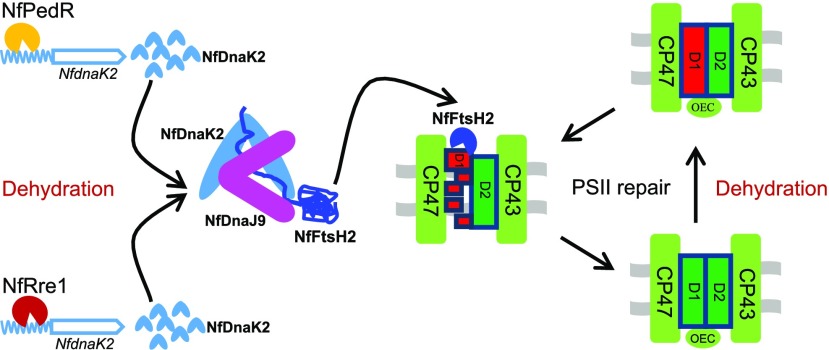
In conclusion, dnaKs and dnaJs are multicopy, conserved, and widely distributed chaperones in almost all organisms (Mayer and Bukau, 2005) and play important roles in stress tolerance of subaerial cyanobacteria. The NfDnaK2-NfDnaJ9 chaperone associates with the protease NfFtsH2 and is involved in PSII repair by promoting the degradation of damaged D1 protein in N. flagelliforme. NfRre1 and NfPedR transcription factors are putative positive regulators controlling the expression of NfdnaK2 in response to drought. The transcriptional regulation and involvement of the NfDnaK2-NfDnaJ9 chaperone in drought tolerance is summarized in Figure 8. Highly efficient PSII repair mediated by the NfDnaK2-NfDnaJ9 chaperone facilitates maintenance of PSII integrity, thus minimizing energy consumption for reconstruction of the photosynthetic machinery during rehydration (Klotz et al., 2016), which promotes a rapid recovery of photosynthetic activity to provide an energy source for the resuscitation of vegetative cells. Our findings provide novel insights into the protection of photosystems in desiccation-tolerant cyanobacteria, which could facilitate future synthetic approaches to engineer extremophiles via plant biotechnology.
Figure 8.
A working model of the transcriptional regulation and function of NfdnaK2 in protecting PSII against drought stress in the subaerial cyanobacterium N. flagelliforme. The steel blue wavy symbol represents the upstream region of the NfdnaK2 open reading frame (steel blue arrow region), and other symbols are as labeled. CP47, CP43, D1, and D2 proteins are the PSII core subunits, and green and red rectangles labeled by D1 protein represent functional and damaged D1 protein respectively. OEC, Oxygen evolution complex of PSII.
MATERIALS AND METHODS
Samples and Culture Conditions
The dry Nostoc flagelliforme was collected at Sunitezuoqi, Inner Mongolia, and stored under dry and dark conditions at room temperature until experimental use. Samples were rinsed three times using distilled water to remove surface dust and then rehydrated in BG11 medium at 25°C under 30 µmol photons m−2 s−1 for different time periods (0, 1, 3, and 9 h). The dehydration process was carried out at 25°C under 30 µmol photons m−2 s−1 in petri dishes by removing water drops on the surface of samples with filter paper. The dehydrated samples were collected when water loss (WL) had increased to 20%, 50%, and 90% at the corresponding dehydration time points 0.5 h, 2 h, and 4 h, respectively. WL was calculated according to the formula (Qiu and Gao, 2001) WL = (Ww − Wt)/(Ww − Wd) × 100%, where Ww is the initial full-rehydrated wet weight; Wt is the weight measured at a certain time interval; and Wd is the weight of the completely dry sample. The dry samples collected in the field were preserved in a desiccator for at least one year until use. The strain Nostoc sp. PCC 7120 was obtained from the Freshwater Algae Culture Collection of the Institute of Hydrobiology, Chinese Academy of Sciences, cultured in BG11 medium at 25°C, and illuminated with cool-white fluorescent lamps at an intensity of 30 μmol photons m−2 s−1 (Shang et al., 2018).
Total RNA Extraction and RT-qPCR Analyses
Total RNA isolation of N. flagelliforme was performed as described previously (Liu et al., 2010). The sample (100 mg) was ground in liquid nitrogen and homogenized in 1 mL Trizol reagent (Invitrogen). The genomic DNA digestion and complementary DNA synthesis from 1 μg of total RNA was performed by using the PrimeScript reagent gDNA eraser kit (Takara) according to the manufacturer’s instructions. Transcript levels of NfdnaK and NfdnaJ genes were quantified by RT-qPCR using the CFX96 Touch Real-Time PCR detection system (Bio-Rad), and 16s rDNA was used as the reference control. Amplifications were performed using SYBR Green Real-Time PCR Master Mix (Toyobo) and gene-specific primers are listed in Supplemental Table S2. One-step cycling was performed for amplification, with an initial preheating step of 3 min at 95°C followed by 39 cycles of 10 s at 95°C and 30 s at 58°C. Relative quantification of transcripts was estimated using the 2−∆∆CT method described by Livak and Schmittgen (2001). The transcript level of samples at time point zero was set to 100%. Three independent experiments were performed.
Antibody Production against NfDnaKs
The genomic DNA of N. flagelliforme CCNUN1 was extracted as described in Shang et al. (2019) and used as a DNA template to perform PCR amplification of four NfdnaKs (NfdnaK1, COO91_04977; NfdnaK2, COO91_06122; NfdnaK3, COO91_02936; NfdnaK4, COO91_04838) using the high-fidelity enzyme Pfu DNA polymerase (Promega). The primers used are listed in Supplemental Table S2. The amplified fragments were first cloned into the pMD18-T vector and sequenced. The resultant plasmids were digested with NdeI/XhoI to obtain the fragments, which were then cloned into pET41a vector (Novagen) and transformed into Escherichia coli strain BL21 for protein production. The NfDnaK proteins were purified by His-bind resin (Novagen) and used to generate polyclonal antibodies in rabbits, which were further tested for their recognition of NfDnaKs in N. flagelliforme.
Western Blotting and Subcellular Localization
Dry N. flagelliforme samples (0.5 g) were ground with liquid nitrogen and then resuspended in 20 mm potassium phosphate (pH 7.8) with 1 mm phenylmethylsulfonyl fluoride. Subsequently, the samples were homogenized by ultrasonication (Soniprep 150, Sanyo) on ice for 30 s per time at 15 μm amplitude, followed by a 30-s interval of cooling on ice. This process was repeated 20 times and the total time for cell disruption was 10 min. The cell debris was removed by centrifugation at 4,000g and 4°C for 10 min, and the supernatant was the total protein fraction. Total proteins were loaded and separated by 12% (w/v) SDS-PAGE, then transferred to polyvinylidene fluoride membranes (Millipore). NfDnaKs on these membranes were recognized with corresponding specific antibodies and visualized with goat antirabbit alkaline phosphatase antibody (Invitrogen) by staining with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (Amresco) as the substrates.
To analyze the subcellular location of NfDnaK proteins in N. flagelliforme, the total protein was extracted as described above. The total protein fraction was centrifuged at 100,000g and 4°C for 1 h to isolate soluble and membrane proteins (pellets). The membranes were washed three times with 20 mm potassium phosphate (pH 7.8) and called the “crude membrane”, which contains the thylakoid membrane and the plasma membrane. The separation of these two membrane components was performed according to the procedures described by Huang et al. (2002). The subcellular location of NfDnaK proteins was determined by western blotting. NrtA and CP47 were used as marker proteins for the plasma membrane and thylakoid membrane, respectively.
Y2H Assays
For Y2H assays, the protein-protein interactions of NfDnaKs and NfDnaJs were detected by a Matchmaker GAL4 Two-Hybrid System 3 (Clontech). Four NfdnaK genes and 11 NfdnaJ genes were obtained by PCR amplification with a N. flagelliforme CCNUN1 genomic DNA template using the high-fidelity enzyme Pfu DNA polymerase (Promega) and the primers listed in Supplemental Table S2. The amplified fragments were first cloned into the pMD18-T vector and sequenced. NfdnaJ and NfdnaK fragments were obtained by digestion with NdeI/XhoI (except for NfdnaJ11, which was digested with NdeI/SmaI) and NdeI/PstI, respectively, from the resultant plasmid, and then were cloned into pGADT7 and pGBKT7 vectors (Clontech), respectively. The two resultant plasmids were cotransformed into Saccharomyces cerevisiae AH109, and the transformants were isolated from colonies grown on synthetic defined plates lacking Trp and Leu (SD/-Trp-Leu). The transformants were then grown on SD plates lacking Trp, Leu, His, and adenine (SD/-Trp-Leu-His-Ade) for 3 d. The interactions between NfDnaJ9 (COO91_05061) and NfFtsHs (COO91_03160, COO91_03537, COO91_03727, and COO91_00363) by Y2H assay were performed as described above in this section.
Pull-Down Assays
To verify the interaction of NfDnaK2 and NfDnaJ9, an in vivo pull-down assay was carried out. Dry N. flagelliforme samples (0.5 g) were washed three times with distilled water, and total proteins were extracted in 30 mL phosphate-buffered saline (10 mm Na2HPO4, 1.8 mm KH2PO4, 140 mm NaCl, and 2.7 mm KCl, pH 7.3). After centrifuging at 16,000g for 30 min to separate the soluble proteins and membranes, 100 μL membranes (1 μg μL−1 chlorophyll a) were resuspended with TM buffer (50 mm MES-NaOH, 10 mm MgCl2, 5 mm CaCl2, and 25% [w/v] glycerol, pH 6.5), solubilized with 100 μL 2% (w/v) n-dodecyl-β-maltoside in TM buffer containing 1 mm phenylmethylsulfonyl fluoride for 30 min at 4°C. The solubilized membrane proteins were then collected by centrifugation at 16,000g for 30 min. The mixture of the solubilized membrane proteins and the soluble proteins was incubated with 1 mL GST-Bind resin (Novagen) and 30 mg total proteins of the GST-tagged NfDnaJ9 overexpressed E. coli strain BL21 after induction with 0.3 mm isopropylthio-β-d-galactoside for 6 h. After 6 h incubation with slight constant shaking at 4°C, the affinity proteins on GST-Bind resin were eluted with elution buffer (50 mm Tris-HCl and 10 mm glutathione, pH 8.0) and these coeluted proteins with GST-tagged NfDnaJ9 were then resolved by SDS-PAGE and western blotting performed with the specific antibody against NfDnaK2.
For the in vitro pull-down assay, NfdnaJ9 and NfftsH2 (COO91_03537), fragments were generated by PCR using N. flagelliforme CCNUN1 genomic DNA as template, and the primers are listed in Supplemental Table S2. The PCR products of NfdnaJ9 and NfftsH2 were digested with SpeI/XhoI and NdeI/XhoI, respectively. The resultant fragments were cloned into the pET41a vector and transformed into E. coli strain BL21. After induction with 0.4 mm isopropylthio-β-d-galactoside for 6 h, the cells were harvested by centrifugation and resuspended in phosphate-buffered saline (10 mm Na2HPO4, 1.8 mm KH2PO4, 140 mm NaCl, and 2.7 mm KCl, pH 7.3). Subsequently, cells were disrupted by sonication and centrifuged to obtain the supernatant. The supernatants from two strains of overexpressed E. coli (GST-tagged NfDnaJ9 and His-tagged NfFtsH2) were mixed and incubated overnight with slow constant shaking at 4°C, and then were copurified through a GST-Bind resin matrix. The His-tag and GST-tag signals of copurified proteins were detected with the specific antibodies (ABclonal) against His-Tag and GST-Tag, respectively. In addition, these two overexpressed E. coli strains were resuspended in His-binding buffer (20 mm Tris-HCl, 5 mm imidazole, and 500 mm NaCl, pH 7.9) to further verify the interaction. After cell lysis and centrifugation, the two generated supernatants were mixed and incubated overnight at 4°C and then copurified through a His-Bind resin matrix. The detection method was as described above. GST and nonrelative His-tagged NfOCP (COO91_07282) were used as negative controls to exclude the nonspecific interactions.
Construction of NfdnaK2 Transgenic Nostoc sp. PCC 7120 and Stress Treatments
The NfdnaK2 gene was cloned into pRL25C under control of the Nfrbcl promoter. Ribosome binding site of NfdnaK2 was fused to the 5̕′ end of the upstream primer (Supplemental Table S2). The constructed plasmid (pRL25C-Omega-Prbcl-NfdnaK2) was transformed into Nostoc sp. PCC 7120 to generate OE-NfdnaK2 Nostoc sp. PCC 7120 by the conjugal transfer method, as described previously (Wolk et al., 1984). The Nostoc sp. PCC 7120 transformants were selected on 1% agar BG11 plates containing 30 μg mL−1 spectinomycin under 30 µmol photons m−2 s−1, and the positive single clones were confirmed by PCR (Supplemental Fig. S4) and sequencing. The primers used are shown in Supplemental Table S2. To compare the PSII repair between transgenic strain and wild-type Nostoc sp. PCC 7120, the cells were grown in BG11 medium to an optical density (OD750) of 0.4 and then treated with high light (400 μmol photons m−2 s−1) in glass tubes. The Fv/Fm was monitored every 10 min by Handy Plant Efficiency Analysis (Hansatech Instruments; Strasser et al., 1995). The oxygen evolution rate of PSII was measured with a Clark-type oxygen electrode (Chlorolab 2, Hansatech Instruments), with water as the electron donor in the presence of an artificial electron acceptor, p-benzoquinone (p-BQ, 2 mmol L−1), and potassium ferricyanide (FeKCN, 2 mmol L−1; Xu et al., 2019). To distinguish the damage and repair of PSII, 100 μg mL−1 Lin was used to prevent the PSII repair cycle (Dai et al., 2014; Xu et al., 2019). In addition, the samples at different time points (0, 20, 40, and 60 min) were collected for D1 protein detection using western blotting.
Exponential growth cultures (adjusting the OD750 value to 0.05) were dispensed into cell culture plates (2 mL per well) and treated with various concentrations of PEG 6000 (0, 12%, 15%, 18%, 21%, and 25% [w/v]) or sorbitol (0, 0.4, 0.8, 1.0, 1.2, and 1.4 mol L−1) to simulate long-term drought stress treatment. Cells were incubated under 30 μmol photons m−2 s−1, resuspended three times per day in the clean bench, and photographed at day 5. For the growth curve measurement of wild-type and transgenic strains, samples were diluted to an OD750 of 0.05 and incubated in 100 mL Erlenmeyer flasks filled with 50 mL cultures with 18% (w/v) PEG 6000 or 1 mol L−1 sorbitol, and OD750 values were measured every 2 d. The growth rate (in microns) was calculated according to the equation μ = (ln X6 − ln X0)/6, where X6 and X0 are OD750 values from day 6 and the initial inoculums, respectively. The PEG plate system is ideal for studies of dehydration tolerance (van der Weele et al., 2000), and the preparation of PEG-infused agar plates with different water potential was as described in Verslues et al. (2006). Cells were adjusted to an OD750 of 0.4 and diluted 10 times. Then, 10 μL of each sample (OD750 of 0.4 and 0.04) was dropped onto a BG11 agar plate with or without 25% (w/v) PEG infusion and photographed at day 10.
Luciferase Reporter Assays
The −940 bp upstream region of NfdnaK2 orf was dissected into three segments (promoter1–promoter3) as shown in Figure 6A, and fused with the luxAB reporter fragment (Dai et al., 2014). The resulting DNA fragments were cloned into pRL25C to obtain the recombinant pRL25C-Omega-Promoter-luxAB plasmid, which was transformed into Nostoc sp. PCC 7120 as described in the previous section. The Nostoc sp. PCC 7120 transformants carrying different promoter segments fused with the luxAB reporter gene were confirmed by PCR amplification, as shown in Supplemental Fig. S4. The activity of promoter elements was determined by the relative bioluminescence intensity of luciferase in Nostoc sp. PCC 7120, as described previously (Shang et al., 2018). An aliquot of 0.2 mL cultures containing the same amount of chlorophyll a (1.5 mg L−1) was transferred into a 96-well plate and 5 μL of 0.1 mm decanal dissolved in dimethyl sulphoxide was added. The bioluminescence was measured at room temperature with a Synergy 2 microplate (BioTek), and the wild-type Nostoc sp. PCC 7120 was used as a negative control. All measurements were performed with three independent biological replicates.
Y1H Assays
For Y1H assays, the active promoter region (promoter2-1 fragment; Fig. 6A) was recombined into the yeast reporter vector pAbAi digested by KpnI/SalI, and the resulting construct was transformed into the Y1HGold yeast bait strain (Shang et al., 2018). The selective concentration of aureobasidin A (AbA) on SD plates minus uracil was determined by toxic effects of serial concentrations (150, 200, 250, and 300 μg L−1) of AbA, which repressed the growth of yeast cells with pAbAi-promoter elements. An efficient Y1H library composed only of transcription factors in N. flagelliforme (H.F. Xu, G.Z. Dai, and B.S Qiu, unpublished data) was transformed into the yeast bait Y1H Gold strain carrying the pAbAi-promoter2-1, which was used to screen for potential transcription factors that could interact with the promoter2-1 segment. The pGADT7 empty vector was used as a negative control. Growth of yeast cells on the SD plates minus Leu with 250 μg L−1 AbA indicates potential promoter2-1 interacting transcriptional factors in the cell of the yeast bait Y1HGold strain. Through PCR and sequencing of these clones, the genes encoding putative promoter2-1 binding proteins were identified. Finally, point-to-point Y1H verifications were carried out to verify the identified transcription factors.
EMSA
EMSA was performed as described by Song et al. (2018). DNA fragments composed of biotin-labeled promoter2-1, nonbiotin-labeled promoter2-1, and a nonspecific competitor (NSC), were prepared by PCR with the primers shown in Supplemental Table S2. The procedures for purifying recombinant GST-tagged NfRre1 (COO91_05451) and NfPedR (COO91_04806), both cloned into the pGEX-4T-1 vector, from E. coli cells were as described in the "Pull-Down Assays" section. The binding reactions between DNA and proteins were performed in the EMSA-binding buffer (10 mm Tris-HCl, 50 mm KCl, 1 mm dithiothreitol, 0.1% [w/v] bovine serum albumin, and 5% [w/v] glycerol, pH 7.5) for 30 min at 30°C. The unlabeled DNA and poly(dI-dC) were added to exclude unspecific binding. The reaction mixtures were separated on 6% (w/v) polyacrylamide gels at 60 V for 1 h and 80 V for 1 h in Tris-borate/EDTA buffer (45 mm Tris, 45 mm boric acid, and 1 mm EDTA, pH 8.3) at 4°C. After electrophoresis, the gels were soaked with 0.5× Tris-borate/EDTA buffer and then transferred to nylon membranes (Millipore) at 20 V for 1 h using Transblot Semi-dry (Bio-Rad). After UV cross-linking for 10 min, the nylon membranes were soaked with 5% dried skimmed milk for 30 min and incubated with horseradish peroxidase-conjugated streptavidin for 30 min. Then the samples were visualized with ECL solution (Yeasen) using FluorChem M (Protein Simple).
Statistical Analyses
All independent experiments were repeated three times, and statistical calculation was performed using Origin 6.1 (Originlab Corporation). Student’s t test was conducted to verify the statistical significance of the differences between means of two independent groups.
Accession Numbers
Sequence data from this article can be found in the KEGG database (https://www.kegg.jp/) with the accession numbers mentioned in the text.
Supplemental Data
The following supplemental information is available.
Supplemental Figure S1. Relative transcriptional levels of NfdnaKs during rehydration and subsequent dehydration.
Supplemental Figure S2. The expression levels of FtsH2, DnaK2, PsaA, and PsaB proteins in the membrane fractions of the control and OE-NfdnaK2 Nostoc sp. PCC 7120 strains.
Supplemental Figure S3. Interaction between Promoter2-1 and NfRre1 homologs.
Supplemental Figure S4. Identification of transgenic NfdnaK2 (OE-NfdnaK2) and transgenic Promoter1/2/3::luxAB Nostoc sp. PCC 7120 strains by PCR analysis.
Supplemental Table S1. Identification of proteins coeluted from GST-NfDnaJ9 pull down using a liquid chromatography tandem mass spectrophotometer and protein database searching.
Supplemental Table S2. Primers used in this study.
Supplemental Table S3. Strains used in this study.
Acknowledgments
We thank Dr. Hai-Bo Jiang, Dr. Xiang Gao, Dr. Zhong-Chun Zhang and Dr. Yi-Wen Yang (Central China Normal University) for their valuable advice, and Da Huang, Jian Wan, and Chen Xu (Central China Normal University) for assisting in the liquid chromatography tandem MS data collection and analysis. We also thank Xu-Dong Xu (Institute of Hydrobiology, Chinese Academy of Sciences) for kindly providing the pHB1347 plasmid and E. coli HB101.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31670332 and 31600190), Fundamental Research Funds for the Central Universities (CCNU18ZDPY05 and CCNU19QD013), and the Hubei Provincial Natural Science Foundation (grant no. 2013CFB211).
References
- Acebrón SP, Fernández-Sáiz V, Taneva SG, Moro F, Muga A (2008) DnaJ recruits DnaK to protein aggregates. J Biol Chem 283: 1381–1390 [DOI] [PubMed] [Google Scholar]
- Allen JF, de Paula WB, Puthiyaveetil S, Nield J (2011) A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci 16: 645–655 [DOI] [PubMed] [Google Scholar]
- Aro EM, Suorsa M, Rokka A, Allahverdiyeva Y, Paakkarinen V, Saleem A, Battchikova N, Rintamäki E (2005) Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J Exp Bot 56: 347–356 [DOI] [PubMed] [Google Scholar]
- Bailey S, Thompson E, Nixon PJ, Horton P, Mullineaux CW, Robinson C, Mann NH (2002) A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J Biol Chem 277: 2006–2011 [DOI] [PubMed] [Google Scholar]
- Bewley JD. (1979) Physiological aspects of desiccation tolerance. Annu Rev Plant Biol 30: 195–238 [Google Scholar]
- Boehm M, Yu J, Krynicka V, Barker M, Tichy M, Komenda J, Nixon PJ, Nield J (2012) Subunit organization of a synechocystis hetero-oligomeric thylakoid FtsH complex involved in photosystem II repair. Plant Cell 24: 3669–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA (1994) Molecular evolution of the HSP70 multigene family. J Mol Evol 38: 1–17 [DOI] [PubMed] [Google Scholar]
- Canaani O, Havaux M, Malkin S (1986) Hydroxylamine, hydrazine and methylamine donate electrons to the photooxidizing side of Photosystem II in leaves inhibited in oxygen evolution due to water stress. Biochim Biophys Acta 851: 151–155 [Google Scholar]
- Ceccarelli S, Grando S (1996) Drought as a challenge for the plant breeder. Plant Growth Regul 20: 149–155 [Google Scholar]
- Challabathula D, Zhang Q, Bartels D (2018) Protection of photosynthesis in desiccation-tolerant resurrection plants. J Plant Physiol 227: 84–92 [DOI] [PubMed] [Google Scholar]
- Chen KM, Holmström M, Raksajit W, Suorsa M, Piippo M, Aro EM (2010) Small chloroplast-targeted DnaJ proteins are involved in optimization of photosynthetic reactions in Arabidopsis thaliana. BMC Plant Biol 10: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MCD, Farrant JM, Oliver MJ, Ligterink W, Buitink J, Hilhorst HMW (2016) Key genes involved in desiccation tolerance and dormancy across life forms. Plant Sci 251: 162–168 [DOI] [PubMed] [Google Scholar]
- Dai GZ, Qiu BS, Forchhammer K (2014) Ammonium tolerance in the cyanobacterium Synechocystis sp. strain PCC 6803 and the role of the psbA multigene family. Plant Cell Environ 37: 840–851 [DOI] [PubMed] [Google Scholar]
- Düppre E, Rupprecht E, Schneider D (2011) Specific and promiscuous functions of multiple DnaJ proteins in Synechocystis sp. PCC 6803. Microbiology 157: 1269–1278 [DOI] [PubMed] [Google Scholar]
- Eberhard S, Finazzi G, Wollman FA (2008) The dynamics of photosynthesis. Annu Rev Genet 42: 463–515 [DOI] [PubMed] [Google Scholar]
- Fischer WW, Hemp J, Johnson JE (2016) Evolution of oxygenic photosynthesis. Annu Rev Earth Planet Sci 44: 647–683 [Google Scholar]
- Gao KS. (1998) Chinese studies on the edible blue-green alga, Nostoc flagelliforme: A review. J Appl Phycol 10: 37–49 [Google Scholar]
- Giardi MT, Cona A, Geiken B, Kučera T, Masojidek J, Mattoo AK (1996) Long-term drought stress induces structural and functional reorganization of photosystem II. Planta 199: 118–125 [Google Scholar]
- Gopal J, Iwama K (2007) In vitro screening of potato against water-stress mediated through sorbitol and polyethylene glycol. Plant Cell Rep 26: 693–700 [DOI] [PubMed] [Google Scholar]
- Goyal K, Walton LJ, Browne JA, Burnell AM, Tunnacliffe A (2005) Molecular anhydrobiology: Identifying molecules implicated in invertebrate anhydrobiosis. Integr Comp Biol 45: 702–709 [DOI] [PubMed] [Google Scholar]
- Harel Y, Ohad I, Kaplan A (2004) Activation of photosynthesis and resistance to photoinhibition in cyanobacteria within biological desert crust. Plant Physiol 136: 3070–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2009) Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 16: 574–581 [DOI] [PubMed] [Google Scholar]
- Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL (2005) Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci 14: 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi M, Nakamura K, Kojima K, Nishiyama Y, Hatakeyama W, Hisabori T, Hihara Y (2010) The PedR transcriptional regulator interacts with thioredoxin to connect photosynthesis with gene expression in cyanobacteria. Biochem J 431: 135–140 [DOI] [PubMed] [Google Scholar]
- Huang F, Parmryd I, Nilsson F, Persson AL, Pakrasi HB, Andersson B, Norling B (2002) Proteomics of Synechocystis sp. strain PCC 6803: Identification of plasma membrane proteins. Mol Cell Proteomics 1: 956–966 [DOI] [PubMed] [Google Scholar]
- Huang J, Yu H, Guan X, Wang G, Guo R (2016) Accelerated dryland expansion under climate change. Nat Clim Chang 6: 166–171 [Google Scholar]
- Kampinga HH, Craig EA (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W (2009) Protein quality control in chloroplasts: A current model of D1 protein degradation in the photosystem II repair cycle. J Biochem 146: 463–469 [DOI] [PubMed] [Google Scholar]
- Klotz A, Georg J, Bučinská L, Watanabe S, Reimann V, Januszewski W, Sobotka R, Jendrossek D, Hess WR, Forchhammer K (2016) Awakening of a dormant cyanobacterium from nitrogen chlorosis reveals a genetically determined program. Curr Biol 26: 2862–2872 [DOI] [PubMed] [Google Scholar]
- Kobayashi I, Watanabe S, Kanesaki Y, Shimada T, Yoshikawa H, Tanaka K (2017) Conserved two-component Hik34-Rre1 module directly activates heat-stress inducible transcription of major chaperone and other genes in Synechococcus elongatus PCC 7942. Mol Microbiol 104: 260–277 [DOI] [PubMed] [Google Scholar]
- Kong F, Deng Y, Zhou B, Wang G, Wang Y, Meng Q (2014) A chloroplast-targeted DnaJ protein contributes to maintenance of photosystem II under chilling stress. J Exp Bot 65: 143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B (1999) Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA 96: 5452–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lin YJ, Wang P, Zhang B, Li M, Chen S, Shi R, Tunlaya-Anukit S, Liu X, Wang Z, et al. (2019) The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell 31: 663–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Willmund F, Whitelegge JP, Hawat S, Knapp B, Lodha M, Schroda M (2005) J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol Biol Cell 16: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YH, Yu L, Ke WT, Gao X, Qiu BS (2010) Photosynthetic recovery of Nostoc flagelliforme (Cyanophyceae) upon rehydration after 2 years and 8 years dry storage. Phycologia 49: 429–437 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B (2005) Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Ikeda M, Takada S, Takiguchi Y, Kondou Y, Yoshizumi T, Fujita M, Shinozaki K, Matsui M, Ohme-Takagi M (2010) Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol 51: 2145–2151 [DOI] [PubMed] [Google Scholar]
- Nakamoto H, Suzuki M, Kojima K (2003) Targeted inactivation of the hrcA repressor gene in cyanobacteria. FEBS Lett 549: 57–62 [DOI] [PubMed] [Google Scholar]
- Nakamura K, Hihara Y (2006) Photon flux density-dependent gene expression in Synechocystis sp. PCC 6803 is regulated by a small, redox-responsive, LuxR-type regulator. J Biol Chem 281: 36758–36766 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 5: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N, Yocum CF (2006) Structure and function of photosystems I and II. Annu Rev Plant Biol 57: 521–565 [DOI] [PubMed] [Google Scholar]
- Nickelsen J, Rengstl B (2013) Photosystem II assembly: From cyanobacteria to plants. Annu Rev Plant Biol 64: 609–635 [DOI] [PubMed] [Google Scholar]
- Nimura K, Takahashi H, Yoshikawa H (2001) Characterization of the dnaK multigene family in the Cyanobacterium Synechococcus sp. strain PCC7942. J Bacteriol 183: 1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon PJ, Barker M, Boehm M, de Vries R, Komenda J (2005) FtsH-mediated repair of the photosystem II complex in response to light stress. J Exp Bot 56: 357–363 [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver MJ, Velten J, Wood AJ (2000) Bryophytes as experimental models for the study of environmental stress tolerance: Tortula ruralis and desiccation-tolerance in mosses. Plant Ecol 151: 73–84 [Google Scholar]
- Oren N, Raanan H, Kedem I, Turjeman A, Bronstein M, Kaplan A, Murik O (2019) Desert cyanobacteria prepare in advance for dehydration and rewetting: The role of light and temperature sensing. Mol Ecol 28: 2305–2320 [DOI] [PubMed] [Google Scholar]
- Oren N, Raanan H, Murik O, Keren N, Kaplan A (2017) Dawn illumination prepares desert cyanobacteria for dehydration. Curr Biol 27: R1056–R1057 [DOI] [PubMed] [Google Scholar]
- Paithoonrangsarid K, Shoumskaya MA, Kanesaki Y, Satoh S, Tabata S, Los DA, Zinchenko VV, Hayashi H, Tanticharoen M, Suzuki I, Murata N (2004) Five histidine kinases perceive osmotic stress and regulate distinct sets of genes in Synechocystis. J Biol Chem 279: 53078–53086 [DOI] [PubMed] [Google Scholar]
- Pinheiro C, Chaves MM (2011) Photosynthesis and drought: Can we make metabolic connections from available data? J Exp Bot 62: 869–882 [DOI] [PubMed] [Google Scholar]
- Qiu BS, Gao KS (1999) Dried field populations of Nostoc flagelliforme (Cyanophyceae) require exogenous nutrients for their photosynthetic recovery. J Appl Phycol 11: 535–541 [Google Scholar]
- Qiu BS, Gao KS (2001) Photosynthetic characteristics of the terrestrial blue-green alga, Nostoc flagelliforme. Eur J Phycol 36: 147–156 [Google Scholar]
- Qiu BS, Zhang AH, Liu ZL, Gao KS (2004a) Studies on the photosynthesis of the terrestrial cyanobacterium Nostoc flagelliforme subjected to desiccation and subsequent rehydration. Phycologia 43: 521–528 [Google Scholar]
- Qiu BS, Zhang AH, Zhou WB, Wei JM, Dong H, Liu ZL (2004b) Effects of potassium on the photosynthetic recovery of the terrestrial cyanobacterium, Nostoc flagelliforme (Cyanophyceae) during rehydration. J Phycol 40: 323–332 [Google Scholar]
- Rajaram H, Apte SK (2010) Differential regulation of groESL operon expression in response to heat and light in Anabaena. Arch Microbiol 192: 729–738 [DOI] [PubMed] [Google Scholar]
- Rajeev L, da Rocha UN, Klitgord N, Luning EG, Fortney J, Axen SD, Shih PM, Bouskill NJ, Bowen BP, Kerfeld CA, Garcia-Pichel F, Brodie EL, et al. (2013) Dynamic cyanobacterial response to hydration and dehydration in a desert biological soil crust. ISME J 7: 2178–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Löffelhardt W, Bohnert HJ, Philippe H, Lang BF (2005) Monophyly of primary photosynthetic eukaryotes: Green plants, red algae, and glaucophytes. Curr Biol 15: 1325–1330 [DOI] [PubMed] [Google Scholar]
- Rubin BE, Wetmore KM, Price MN, Diamond S, Shultzaberger RK, Lowe LC, Curtin G, Arkin AP, Deutschbauer A, Golden SS (2015) The essential gene set of a photosynthetic organism. Proc Natl Acad Sci USA 112: E6634–E6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupprecht E, Gathmann S, Fuhrmann E, Schneider D (2007) Three different DnaK proteins are functionally expressed in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 153: 1828–1841 [DOI] [PubMed] [Google Scholar]
- Sato M, Yamahata H, Watanabe S, Nimura-Matsune K, Yoshikawa H (2007) Characterization of dnaJ multigene family in the cyanobacterium Synechococcus elongatus PCC 7942. Biosci Biotechnol Biochem 71: 1021–1027 [DOI] [PubMed] [Google Scholar]
- Schroda M, Kropat J, Oster U, Rüdiger W, Vallon O, Wollman FA, Beck CF (2001) Possible role for molecular chaperones in assembly and repair of photosystem II. Biochem Soc Trans 29: 413–418 [DOI] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Wollman FA, Beck CF (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang JL, Chen M, Hou S, Li T, Yang YW, Li Q, Jiang HB, Dai GZ, Zhang ZC, Hess WR, et al. (2019) Genomic and transcriptomic insights into the survival of the subaerial cyanobacterium Nostoc flagelliforme in arid and exposed habitats. Environ Microbiol 21: 845–863 [DOI] [PubMed] [Google Scholar]
- Shang JL, Zhang ZC, Yin XY, Chen M, Hao FH, Wang K, Feng JL, Xu HF, Yin YC, Tang HR, et al. (2018) UV-B induced biosynthesis of a novel sunscreen compound in solar radiation and desiccation tolerant cyanobacteria. Environ Microbiol 20: 200–213 [DOI] [PubMed] [Google Scholar]
- Shen JR. (2015) The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu Rev Plant Biol 66: 23–48 [DOI] [PubMed] [Google Scholar]
- Shoumskaya MA, Paithoonrangsarid K, Kanesaki Y, Los DA, Zinchenko VV, Tanticharoen M, Suzuki I, Murata N (2005) Identical Hik-Rre systems are involved in perception and transduction of salt signals and hyperosmotic signals but regulate the expression of individual genes to different extents in synechocystis. J Biol Chem 280: 21531–21538 [DOI] [PubMed] [Google Scholar]
- Singh AK, Summerfield TC, Li H, Sherman LA (2006) The heat shock response in the cyanobacterium Synechocystis sp. Strain PCC 6803 and regulation of gene expression by HrcA and SigB. Arch Microbiol 186: 273–286 [DOI] [PubMed] [Google Scholar]
- Skirycz A, Inzé D (2010) More from less: Plant growth under limited water. Curr Opin Biotechnol 21: 197–203 [DOI] [PubMed] [Google Scholar]
- Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR (2016) A transcription factor hierarchy defines an environmental stress response network. Science 354: aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Zang SS, Li ZK, Dai GZ, Liu K, Chen M, Qiu BS (2018) Sycrp2 is essential for twitching motility in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 200: e00436-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weele CM, Spollen WG, Sharp RE, Baskin TI (2000) Growth of Arabidopsis thaliana seedlings under water deficit studied by control of water potential in nutrient-agar media. J Exp Bot 51: 1555–1562 [DOI] [PubMed] [Google Scholar]
- Strasser RJ, Srivastava A, Govindjee (1995) Polyphasic chlorophyll a fluorescence transient in plants and cyanobacteria. Photochem Photobiol 61: 32–42 [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Wang G, Kong F, Zhang S, Meng X, Wang Y, Meng Q (2015) A tomato chloroplast-targeted DnaJ protein protects Rubisco activity under heat stress. J Exp Bot 66: 3027–3040 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Sato M, Nimura-Matsune K, Chibazakura T, Yoshikawa H (2007) Protection of psbAII transcript from ribonuclease degradation in vitro by DnaK2 and DnaJ2 chaperones of the cyanobacterium Synechococcus elongatus PCC 7942. Biosci Biotechnol Biochem 71: 279–282 [DOI] [PubMed] [Google Scholar]
- Wieners PC, Mudimu O, Bilger W (2012) Desiccation-induced non-radiative dissipation in isolated green lichen algae. Photosynth Res 113: 239–247 [DOI] [PubMed] [Google Scholar]
- Wolk CP, Vonshak A, Kehoe P, Elhai J (1984) Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc Natl Acad Sci USA 81: 1561–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Yang G, Zhang L, Yang X, Zhao S, Ji Z, Zhou Q, Hu M, Wang Y, Chen M, et al. (2015) The resurrection genome of Boea hygrometrica: A blueprint for survival of dehydration. Proc Natl Acad Sci USA 112: 5833–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HF, Dai GZ, Qiu BS (2019) Weak red light plays an important role in awakening the photosynthetic machinery following desiccation in the subaerial cyanobacterium Nostoc flagelliforme. Environ Microbiol 21: 2261–2272 [DOI] [PubMed] [Google Scholar]
- Ye C, Gao K, Giordano M (2008) The odd behaviour of carbonic anhydrase in the terrestrial cyanobacterium Nostoc flagelliforme during hydration-dehydration cycles. Environ Microbiol 10: 1018–1023 [DOI] [PubMed] [Google Scholar]
- Yobi A, Schlauch KA, Tillett RL, Yim WC, Espinoza C, Wone BWM, Cushman JC, Oliver MJ (2017) Sporobolus stapfianus: Insights into desiccation tolerance in the resurrection grasses from linking transcriptomics to metabolomics. BMC Plant Biol 17: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokthongwattana K, Chrost B, Behrman S, Casper-Lindley C, Melis A (2001) Photosystem II damage and repair cycle in the green alga Dunaliella salina: Involvement of a chloroplast-localized HSP70. Plant Cell Physiol 42: 1389–1397 [DOI] [PubMed] [Google Scholar]
- Yura T, Nakahigashi K (1999) Regulation of the heat-shock response. Curr Opin Microbiol 2: 153–158 [DOI] [PubMed] [Google Scholar]