Abstract
The gut microbiota produce hundreds of molecules that are present at high concentrations in the host circulation. Unraveling the contribution of each molecule to host biology remains difficult. We developed a system for constructing clean deletions in Clostridium spp., the source of many molecules from the gut microbiome. By applying this method to the model commensal organism Clostridium sporogenes, we knocked out genes for 10 C. sporogenes–derived molecules that accumulate in host tissues. In mice colonized by a C. sporogenes for which the production of branched short-chain fatty acids was knocked out, we discovered that these microbial products have immunoglobulin A–modulatory activity.
Gut bacteria produce hundreds of diffusible molecules that are notable for four reasons: (i) Most have no host source, so their levels are determined predominantly or exclusively by the microbiome. (ii) Many get into the bloodstream, so they can access peripheral tissues. (iii) They often reach concentrations that approach or exceed what a typical drug reaches, and the concentration range can be large—more than an order of magnitude in many cases—so they have the potential to cause biological differences among humans. (iv) Several of these molecules are known to be ligands for key host receptors; additional compounds from this category are candidate ligands for, for example, G protein–coupled receptors (GPCRs) and nuclear hormone receptors that play an important role in the host immune and metabolic systems (1). Thus, the gut microbiome is a prolific endocrine organ, but its output is not well understood.
The biological activities of most of these molecules remain unknown. One reason is that there has not been a general method for “toggling” one or more of them on or off in the host, akin to a gene knockout experiment in a model organism. Such a method would open the door to interrogating—and ultimately controlling—one of the most concrete contributions gut bacteria make to host biology.
Previous efforts that have sought to study an individual microbiome-derived molecule in the setting of host colonization have used two main strategies: First, a compound is administered by injection or gavage, which can offer insights into mechanism of action but suffers from the lack of a clean background (i.e., existing physiologic levels of the molecule of interest) and the possible effects of differences in route and timing of administration relative to the native context of gut bacterial production. Second, a bacterial species that produces the molecule is added or removed, which has the advantage of a more native-like context but makes it difficult to distinguish between molecule-induced phenotypes and other biological activities of the organism (2–4).
The most precise format for interrogating a microbiome-derived molecule is to compare two organisms that differ only in its production. Such an experiment has two threshold requirements: (i) knowledge of the metabolic genes for the molecule of interest and (ii) the ability to perform genetics in a robustly producing strain. This approach has been successful in Bacteroides (5, 6), Escherichia coli (7), and Lactobacillus (8), but a key technical barrier limiting its generalization is that many of the known highly abundant gut-derived molecules are produced by Clostridium and its relatives, which have been difficult to manipulate genetically. We recently reported the use of a group II intron, a ribozyme-encoding mobile element (9), to mutate a single pathway in Clostridium sporogenes (10), but this insertional mutagenesis system performs unpredictably and cannot be used to make strains that carry multiple mutations. Here, we address these challenges by developing a highly efficient CRISPR-Cas9–based system for constructing single and multiple mutants in a model gut-resident Clostridium species.
Selection of C. sporogenes as a model gut Clostridium
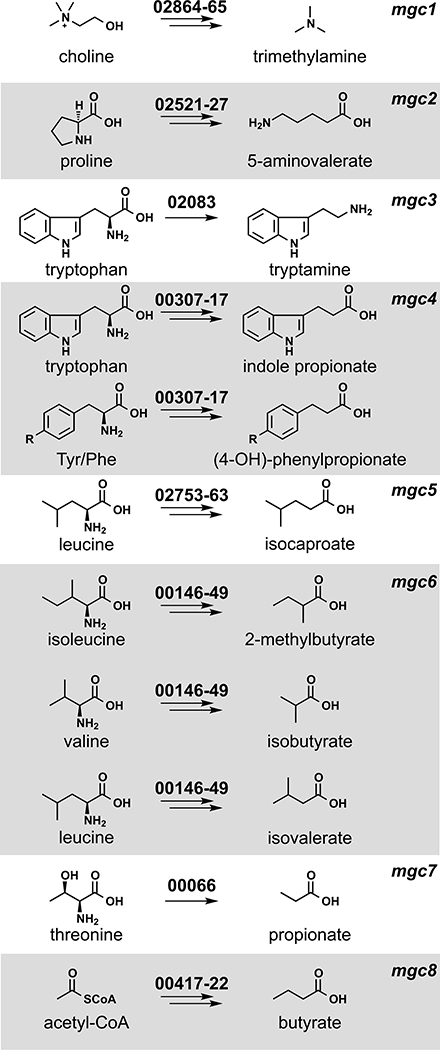
We chose C. sporogenes ATCC 15579 (hereafter C. sporogenes) as a model gut commensal from the anaerobic Firmicutes for three reasons: This strain has long been known as a robust producer of highly abundant small molecules (11–13), it stably colonizes germ-free mice (10), and it is a commensal or mutualist (i.e., neither a pathogen nor a pathobiont). We began by performing metabolic profiling experiments to systematically determine the set of highly abundant small molecules produced by this strain in vitro. As shown in Fig. 1, C. sporogenes produces 10 molecules that are highly abundant and either primarily or exclusively derived from the gut microbiota: 5-aminovalerate, trimethylamine, tryptamine, indole propionate and other aryl propionates, isobutyrate, 2-methylbutyrate, isovalerate, isocaproate, propionate, and butyrate, confirming previous studies (10–13).
Fig. 1. Metabolic pathways from C. sporogenes ATCC 15579 examined in this study.
Each pathway generates a microbiome-derived metabolite present at high abundance in the host’s circulation. The prefix “mgc” stands for “metabolic gene cluster.” Genes that comprise each pathway are shown above the corresponding arrows; the numbers indicate a locus tag suffix for C. sporogenes, where the prefix is “CLOSPO_.”
Prediction and computational analysis of metabolic pathways
Next, we sought to predict the genes responsible for producing each molecule. Metabolic genes for trimethylamine (14), tryptamine (13), and indole propionate (10) have been discovered using genetics in C. sporogenes or another gut bacterium. Pathways for the remaining seven molecules had not been validated genetically in the gut microbiota. We made predictions for each one on the basis of three sources of evidence (Fig. 1): (i) pathways validated in nonmicrobiome organisms [e.g., the pathway for 5-aminovalerate production from the terrestrial isolate Clostridium sticklandii (15)]; (ii) biochemical studies that implicate an enzyme superfamily, which enabled us to search for orthologs in C. sporogenes [e.g., 2-hydroxyisocaproate dehydratases (16), which led us to a predicted cluster for isocaproate]; and (iii) a metagenomic analysis of butyrate gene clusters (17), which yielded a predicted gene cluster for butyrate production in C. sporogenes.
We determined whether the metabolic genes we predicted are widely distributed in the human gut microbiome and actively transcribed during colonization. We used MetaQuery (18) to measure the abundance of the key genes in each pathway (colored red in fig. S1) in >2000 publicly available human gut metagenomes. Every gene, except tdcA, was present in >95% of the stool metagenomes, indicating that the predicted pathways are cosmopolitan (minimum abundance = 1 copy per 1000 cells) (fig. S2). Because the mere presence of a gene in a metagenome does not imply that it is transcribed, we determined the transcript abundance of the key metabolic genes (including close homologs from non–C. sporogenes genomes) by recruiting reads from nine publicly available RNA-sequencing datasets derived from stool samples of healthy subjects (19). This analysis revealed that multiple homologs of each pathway are highly transcribed in the host (fig. S2). For example, porA and its homologs—ALIPUT_00387 in Alistipes putredinis DSM 17216, BACSTE_01839 from Bacteroides stercoris ATCC 43183, and BVU_2313 from Bacteroides vulgatus ATCC 8482—are transcribed in at least one sample. Taken together, these data suggest that the predicted pathways are widely distributed and actively transcribed in the host.
Development of a CRISPR-Cas9–based genetic system for C. sporogenes
Next, we tested our metabolic pathway predictions by constructing mutants in each of them, along with the known pathways in C. sporogenes for tryptamine (13), indole propionate (10), and trimethylamine (14) (Fig. 1). The genetic system we used previously (10), which is based on a group II intron, had two important limitations: Because the intron’s targeting mechanism is not well understood, generating one insertional mutant typically requires testing several targeting sequences; moreover, this process regularly fails to yield a mutant. After multiple attempts, we were not able to recycle the antibiotic resistance marker using ClosTron in order to create strains with multiple mutations; our experience is consistent with a previous report in the literature (9).
Reasoning that a dependable, markerless, recyclable genetic system for Clostridium would open the door to more systematic studies of microbiome metabolism, we developed a CRISPR-Cas9–based genome-editing system for C. sporogenes. Clostridium species have been notoriously difficult to modify genetically, in part because of inefficient homologous recombination (9). We postulated that a Cas9-induced double-strand break would help select for a rare homologous recombination event. To this end, we constructed a single vector that includes all the essential components of a bacterial CRISPR-Cas9 system—the Cas9 gene, a guide RNA (gRNA), and a 1.5- to 2.5-kb repair template—and transferred it by conjugation into C. sporogenes. However, we did not obtain viable colonies, even after multiple rounds of vector design modifications (see supplementary text for more detail).
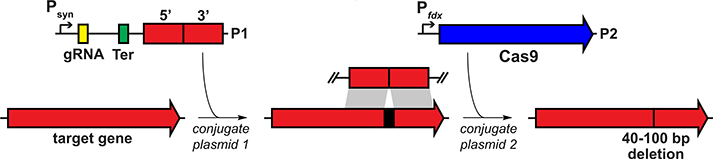
Having observed that the conjugation efficiency for C. sporogenes is greatly diminished for plasmids >10 kb, we redesigned the system by splitting its components into two separate vectors: one that contains the gRNA and repair template and another that harbors Cas9 under the control of a ferredoxin promoter (Fig. 2 and fig. S3). When we introduced these plasmids sequentially, we failed to get any viable colonies after introducing the second vector that harbors the Cas9 coding sequence. Reasoning that the efficiency of the second conjugation step could be the source of failure, we lengthened the donor-acceptor cocultivation step of the second conjugal transfer from 24 to 72 hours; this optimized protocol yielded reproducible, high-efficiency mutations at multiple test loci (see supplementary text for a more detailed description of the method’s development).
Fig. 2. Development of a CRISPR-Cas9–based genetic system for C. sporogenes.
Schematic view of the genetic system. In the first step, plasmid P1 is introduced by conjugation into C. sporogenes. P1 contains a gRNA expressed under the control of Psyn, a synthetic promoter generated using PePPER (31); Ter, a terminator sequence from the C. sporogenes 16S ribosomal RNA gene; and a ~1.5- to 2.0-kb repair template. In the second step, plasmid P2 is introduced by conjugation. P2 consists of the Cas9 gene from Streptococcus pyogenes expressed under the control of Pfdx, the promoter from the C. sporogenes ferredoxin gene. Key steps in the development of the method were to introduce the genome-editing components (Cas9, gRNA, and repair template) sequentially on two plasmids and to lengthen the donor-acceptor cocultivation step of the second conjugal transfer from 24 to 72 hours.
Constructing mutants to validate C. sporogenes pathways in vitro
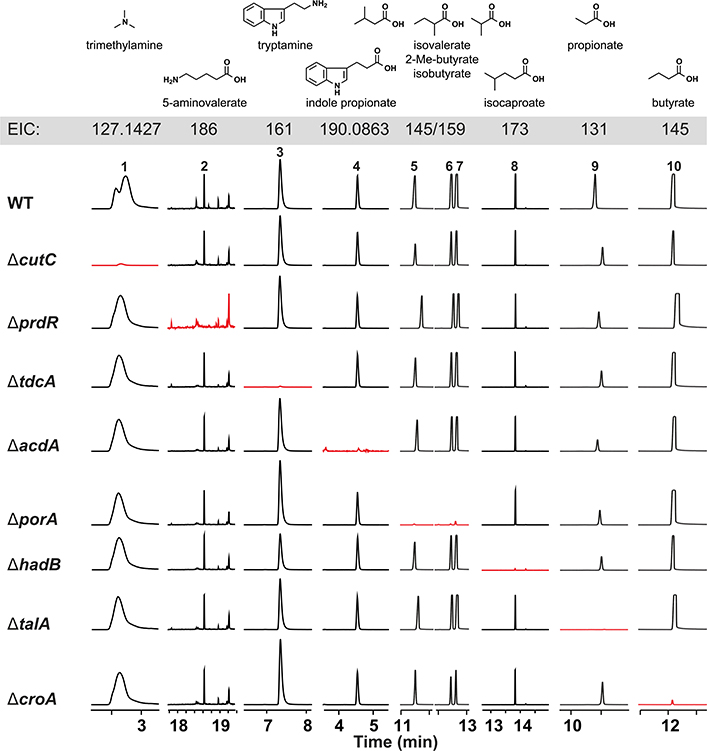
We used the Cas9-based genetic system to construct deletion mutants in each of the 10 pathways. In each case, our repair template effected the removal of an 80– to 150–base pair (bp) portion of the targeted gene in the C. sporogenes chromosome (Fig. 2 and fig. S4); for simplicity, the resulting mutants [e.g., ΔporA (coding sequence 330–409)] are referred to simply as ΔporA (see table S4 for a list of deleted regions). We cultured each strain in vitro and analyzed culture extracts by liquid chromatography–mass spectrometry (LC-MS) or gas chromatography (GC)–MS (depending on the analyte), yielding the following conclusions: (i) The production of each of the 10 molecules is blocked by the corresponding pathway mutant (Fig. 3 and fig. S5), validating our prediction set; an important exception is detailed below in (iii). (ii) Deleting one pathway does not appreciably alter the production of other molecules nor does it affect growth rate (fig. S6), indicating that these pathways are dispensable and function independently in vitro. (iii) Our predicted genetic locus for the branched short-chain fatty acids (SCFAs) isobutyrate, 2-methylbutyrate, and isovalerate was incorrect. In other organisms [e.g., Streptomyces avermitilis (20)], the branched-chain α-ketoacid dehydrogenase complex (BCKDH) converts branched-chain amino acids (BCAAs) into branched SCFAs. To our surprise, deletion of the BCKDH gene CLOSPO_03305 did not affect branched SCFA production. The ΔporA mutant—a gene we previously showed to be involved in the oxidative catabolism of aromatic amino acids (10)—was deficient in the production of all three branched SCFAs (Fig. 3). PorA is a member of the pyruvate:ferredoxin oxidoreductase (PFOR) superfamily; like the BCKDH, PFOR enzymes are thiamine-pyrophosphate dependent, but they harbor an array of iron-sulfur clusters for electron transfer in place of lipoate and flavin and reduce ferredoxin or flavodoxin instead of nicotinamide adenine dinucleotide (oxidized form) (NAD+) (21). Although PFOR superfamily members are known to use pyruvate in anaerobic bacteria (generating acetate), their use of branched-chain a-keto acids derived from BCAAs constitutes a noncanonical pathway for branched SCFA production, having only been previously observed in Archaea (22, 23). We found multiple porA homologs in the genomes of other human gut isolates that are highly transcribed in human stool metatranscriptomes, suggesting that this pathway could be a substantial contributor to the host branched SCFA pool (Figs. 1 and 3 and fig. S2).
Fig. 3. C. sporogenes mutants exhibit specific loss of metabolite production in vitro.
WT and mutant strains of C. sporogenes were cultured individually with the pathway substrates, and metabolites were assayed by LC-MS and GC-MS. Extracted ion chromatogram windows corresponding to each of the pathway products are shown to compare the metabolic output of each strain; ion counts ( y axis) are on the same scale within a column. Full traces are displayed in fig. S5. Traces in red show the metabolite whose production is blocked by the mutation indicated at the beginning of each row. Each mutant is deficient in the production of the corresponding pathway product but proficient in the production of all other pathway products. EIC, extracted-ion chromatogram.
In vivo modulation of C. sporogenes–derived molecules
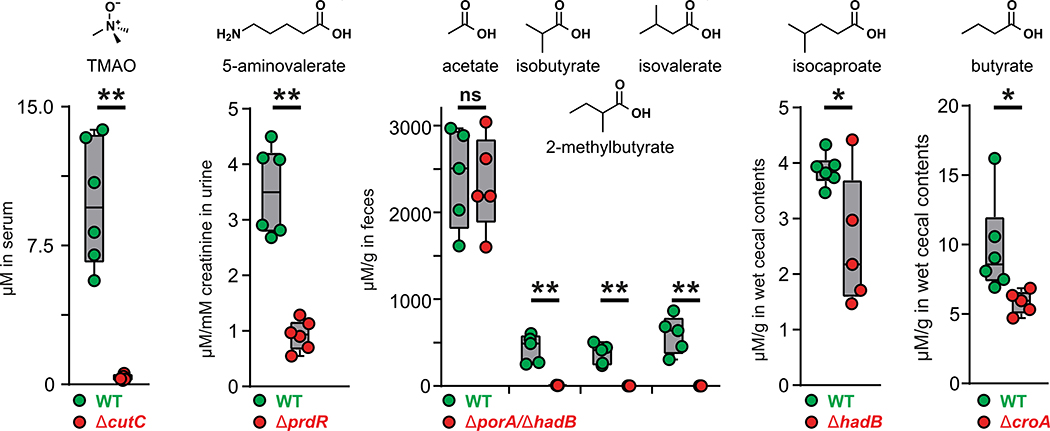
Having validated our target pathways in vitro, we set out to determine whether we could use these mutants to knock out the production of each pathway product in the host. For this experiment, we used a subset of five mutants. The first four were ΔcutC, ΔprdR, ΔcroA, and ΔhadB; for the fifth, we took advantage of the markerless nature of our CRISPR-based genetic system to construct a ΔporA/ΔhadB double mutant, with the goal of eliminating the production of all four branched SCFAs (isobutyrate, 2-methylbutyrate, and isovalerate via ΔporA and isocaproate via ΔhadB). We monocolonized germ-free mice with wild-type (WT) C. sporogenes and the five mutants (Fig. 4); after 4 weeks, we sacrificed the mice and measured the concentration of each molecule in serum, urine, cecal contents, and fecal pellets. WT C. sporogenes and the mutant strains colonized germ-free mice at comparable levels (fig. S7). We drew three observations from these data: (i) Each metabolite (or its host metabolic product) was substantially reduced in the corresponding mutant (Fig. 4). (ii) The WT and mutant differences were smaller for isocaproate and butyrate as a result of two factors: low production by WT C. sporogenes relative to the native level of each molecule (usually mM) and a background level of the metabolite in mutant-colonized mice, possibly owing to a source of contaminating molecule in the chow. (iii) The WT and mutant difference was especially large for the branched SCFAs isobutyrate, 2-methylbutyrate, and isovalerate, which form a pool of >2 mM in the cecal contents of WT C. sporogenes–associated mice that falls to near baseline in the ΔporA/ΔhadB–associated animals. Overall, these data validate the utility and generality of using C. sporogenes mutants to deplete microbiome-derived molecules in the host, and they highlight that the presence of a pathway in an organism can lead to production levels that vary from native to orders of magnitude below native. Thus, the choice of a producer organism that can support the biosynthesis of native metabolite levels depends on unknown factors beyond the mere presence of a corresponding pathway.
Fig. 4. Genetic depletion of metabolites in vivo by colonization with WT versus mutant strains of C. sporogenes.
Germ-free mice were monocolonized with WT C. sporogenes or the ΔcutC, ΔprdR, ΔporA/ΔhadB, ΔhadB, or ΔcroA mutants (n ≥ 5 mice per group). Metabolite levels were altered in the host by mutation of the corresponding pathway. Error bars indicate standard deviation. *P < 0.05, **P < 0.01, and ns is not significant.
Immune modulatory properties of branched SCFAs
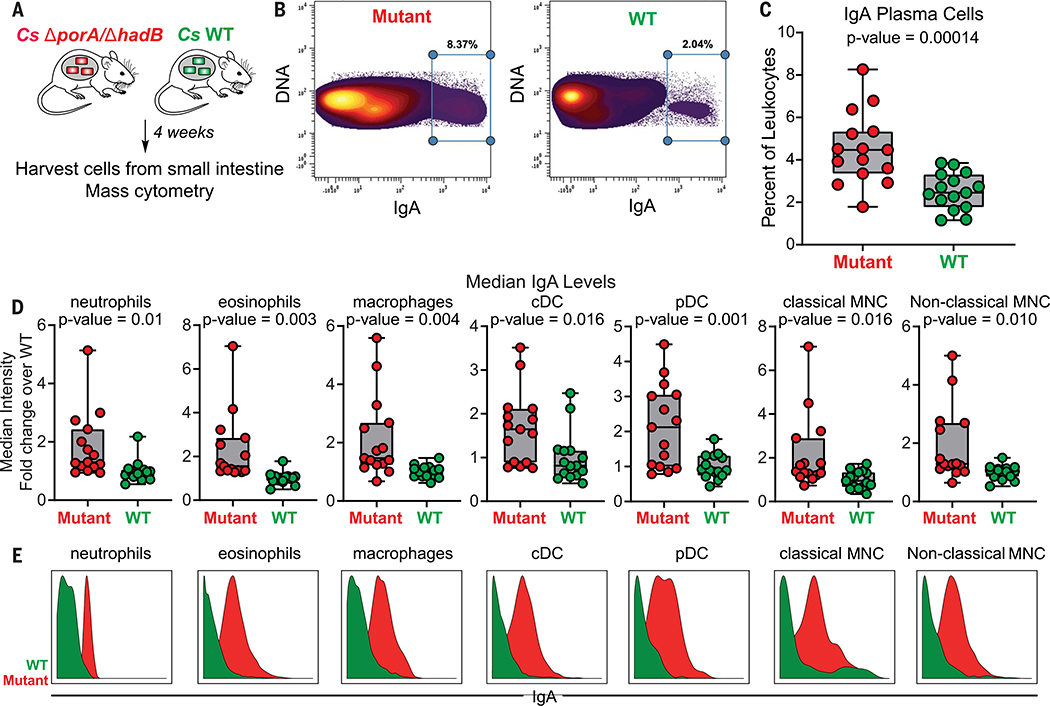
In light of our ability to knock out the production of the C. sporogenes–derived small molecules in vivo, we returned to our original motivation for establishing the new genetic system: to study the role of microbiome-derived molecules in mediating microbe-host interactions. We chose the branched SCFAs for two reasons: (i) Like the conventional SCFAs acetate, propionate, and butyrate, they are highly abundant in the cecum and colon (concentrations in the mM range) (24); but unlike the SCFAs, which are known to modulate the host immune response via GPCRs GPR41 and 43 (1), very little is known about the biology of the branched SCFAs. (ii) C. sporogenes produces them robustly; they constitute a pool of >2 mM (Fig. 4). Hypothesizing that the branched SCFAs—like conventional SCFAs—might modulate the host immune response, we colonized germ-free mice with WT C. sporogenes and the ΔporA/ΔhadB mutant (Fig. 5A). After 5 weeks, we sacrificed the mice, isolated immune cells from the small intestine and mesenteric lymph nodes, and analyzed them by mass cytometry. WT- and ΔporA/ΔhadB-colonized mice were similar by broad metrics of immune function (e.g., total numbers of CD4+ and CD8+ T cells, B cells, and innate immune cells; percentages of effector cell subpopulations; and cytokine levels). However, the ΔporA/ΔhadB-colonized mice had an increased number of immunoglobulin A (IgA)–producing plasma cells (Fig. 5, B and C, and figs. S8 and S9) and increased levels of IgA bound to the surface of a variety of innate immune cells: neutrophils, eosinophils, macrophages, plasmacytoid and conventional dendritic cells, and classical and nonclassical monocytes (Fig. 5, D and E). These results suggest a previously unrecognized role for the branched SCFAs in modulating IgA-related immune cells. There is no significant difference in the levels of free IgA and Clostridium-specific IgA in fecal pellets from WT-colonized versus ΔporA/ΔhadB-colonized mice (fig. S10). Thus, the branched SCFAs regulate the frequency of IgA+ plasma cells in the small intestine without resulting in the accumulation of IgA in feces, in contrast to another C. sporogenes–derived metabolite, indole propionic acid, which affects IgA levels in the lumen (10). These data highlight the compartmentalized nature of the IgA response and suggest multiple points of microbiomemediated control.
Fig. 5. In vivo modulation of branched SCFAs reveal their IgA-related modulatory activity.
(A) Schematic of germ-free mouse monocolonization experiment. Cs, C. sporogenes. (B to E) Small intestinal lamina propria cells were analyzed by mass cytometry (n = 15 mice per group; three biological replicates of n = 5 mice per group). The frequency of IgAhi cells of total live immune cells are shown in (B). In (C), IgA plasma cells were quantified as a percent of live immune cells (CD45+) after excluding cells expressing common markers of other lineages (Ly6G, Siglec-F, B220,CD3, F4/80, CD64,CD11c, CD90, CD115, NK1.1,CD49b, and FcεR1α). In (D) and (E), levels of surface-bound IgA were quantified on the indicated immune cell populations. Statistical analysis was performed using a Wilcoxon rank-sum test for all comparisons. Error bars indicate standard deviation. cDC, conventional dendritic cells; pDC, plasmacytoid dendritic cells; MNC, monocytes.
Discussion
The hundreds of microbiome-derived molecules that accumulate in circulation represent one of the most concrete modes of communication between the host and its microbiota. Little progress has been made in systematically studying their effects on host biology, owing in part to the absence of a method that enables the selective depletion of one member of this pool. One approach to addressing this challenge is to colonize germ-free mice with a WT gut bacterial species versus a metabolite-deficient mutant. Given the outsize role of Clostridium and related anaerobic Firmicutes in generating this pool of highly abundant metabolites, the lack of a reliable genetic system for commensal strains of Clostridium has been a key impediment to generalizing this approach. The system we introduce here is a first step toward that goal; it validates that genetics can be performed in a microbiome-derived Clostridium species rapidly, reliably, and without the need for a marker, and it demonstrates the utility of WT-mutant pairs for interrogating the host effects of microbiome-derived molecules. Given the versatility of Cas9, the key factors for generalizing this system to other Clostridium species are the ability to get DNA into a strain and the availability of replication origins for the plasmids that deliver the mutagenesis and repair elements. An alternative strategy would be to deliver Cas9 and the gRNA as a purified ribonucleoprotein, forgoing the need for plasmid-borne elements. Both strategies could expand the scope of Cas9-mediated mutagenesis to important but previously inaccessible Firmicutes, such as butyrate producers from Clostridium clusters IV and XIVa (e.g., Faecalibacterium prausnitzii) (25), the bile acid metabolizers Clostridium scindens and Clostridium hylemonae (26), and the leanness-associated bacterium Christensenella minuta (27).
C. sporogenes is a prolific producer of amino acid metabolites, and our in vitro studies show that we can predictably block each pathway. However, one of the most notable observations is the difference in the concentration of each pathway product in the context of host colonization. 5-Aminovalerate, indole propionate, isobutyrate, 2-methylbutyrate, isovalerate, propionate, and trimethylamine-N-oxide (TMAO) are produced robustly in vivo, whereas isocaproate and butyrate are generated at 10- to 100-fold below the native concentration range. By highlighting that the mere presence of a pathway does not imply that it will function robustly in the host, our data raise questions about computational approaches that predict metabolic function on the basis of gene or transcript levels (28, 29), and they suggest the importance of understanding pathway regulation and substrate availability under conditions of colonization by a complex native community.
The conventional SCFAs acetate, propionate, and butyrate modulate host immune function and induce regulatory T cells via a pair of GPCRs, GPR41 and 43. Although they are also predominantly microbiome-derived and present at high concentration (24), much less is known about the branched SCFAs isobutyrate, 2-methylbutyrate, and isovalerate, which are produced by a distinct pathway from a different set of precursors, BCAAs. Isovalerate was recently shown to be a ligand for Olfr558, a GPCR in enterochromaffin cells that controls the secretion of serotonin (30). Our data uncover a role for the branched SCFA pathway in modulating IgA-related immune cells.
Materials and methods summary
Constructing C. sporogenes mutants
The coding sequence of Cas9 was assembled with pMTL83153 using Instant Sticky-end Ligase (NEB). The gRNA and repair templates were assembled with pMTL82254. The repair template consists of two 700- to 1200-bp sequences flanking the ~100-bp C. sporogenes genome sequence targeted for Cas9 excision. To construct a C. sporogenes mutant, pMTL82254 harboring the gRNA and repair template was introduced into C. sporogenes via E. coli conjugation. Next, a second conjugation (~72 hours) was performed to introduce pMTL83153 harboring the Cas9 coding sequence. C. sporogenes colonies typically appeared after 36 to 48 hours. Genomic DNA was isolated from at least 16 colonies, and mutations were verified by sequencing.
Quantification of C. sporogenes metabolites using LC-MS or GC-MS
A single colony of WT or mutant C. sporogenes was inoculated in TYG (tryptone–yeast extract– glucose) broth at 37°C for 48 hours in an anaerobic chamber. The conversion of choline to trimethylamine (TMA) was quantified using a previously published protocol (14). The serum TMAO level was quantified using Agilent 6570 LC-QQQ by comparing its area under the curve (AUC) with that of d9-TMAO. The metabolites 5-aminovalerate, indole propionate, propionate, butyrate, isovalerate, 2-methylbutyrate, isobutyrate, and isocaproate were detected and quantified using previously published protocols (3, 10). The creatinine concentration of each urine sample was measured using a Creatinine Assay Kit (ab204537).
Germ-free mouse experiments
All experiments were performed using germfree Swiss Webster mice (male, 6 to 10 weeks of age, n = 5 or 6 mice per group) originally from Taconic Biosciences. Germ-free mice were maintained in gnotobiotic isolators and monocolonized with an overnight culture of either WT or mutant C. sporogenes in TYG medium [200 μl; ~1 × 107 colony forming units (CFU)]. For quantifying C. sporogenes–derived small molecules in vivo, mice were maintained on standard chow (LabDiet 5K67). After 4 weeks of colonization, mice were euthanized humanely by CO2 asphyxiation. Blood was collected by cardiac puncture, and serum was prepared using serum separator tubes from BD (cat. #365967). For mass cytometry analysis, mice were maintained on a high-protein chow (Teklad TD.90018).
Preparation of single-cell suspensions from the small intestine for mass cytometry
Small intestines were harvested from animals and placed in cold RPMI (Roswell Park Memorial Institute) medium. Intestines were filleted open and washed two times in phosphate-buffered saline (PBS). Tissues were cut, washed, and digested in complete RPMI supplemented with Liberase (Roche) and DNAse I (Sigma-Aldrich). Digestion was quenched with complete RPMI, and cells were washed with PBS and labeled with cisplatin viability dye for mass cytometry analysis. The viability stain was quenched, and the cells were washed twice before fixation with 1.5% paraformaldehyde (Electron Microscopy Sciences). Cells were then washed twice and stored at −80°C for subsequent mass cytometry analysis. A summary of all mass cytometry antibodies, reporter isotopes, and concentrations used for analysis can be found in table S3, and a detailed protocol for mass cytometry analysis is available in the supplementary materials.
Supplementary Material
ACKNOWLEDGMENTS
We are deeply indebted to members of the Fischbach group for helpful suggestions and comments on the manuscript.
Funding: This work was supported by NSF Graduate Research Fellowship DGE-114747 (W.V.T.). This work was also supported by an HHMI-Simons Faculty Scholar Award (M.A.F.); a Fellowship for Science and Engineering from the David and Lucile Packard Foundation (M.A.F.); an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Foundation (M.A.F.); an award from BASF (M.A.F.); an award from the Leducq Foundation (M.A.F.); NIH grants DK110174 (M.A.F.), DK113598 (M.A.F.), DK101674 (M.A.F. and J.L.S.), OD023056 (M.H.S.), OD018040 (UCSF, to acquire the mass cytometer used in this study), DK110335 (D.D.), DK085025 (J.L.S.), and 1DP2HD101401-01 (C.-J.G.); the Stanford Microbiome Therapies Initiative (M.A.F., J.L.S., and D.D.); the Chan Zuckerberg Biohub (M.A.F., M.H.S., and J.L.S.); Stanford’s ChEM-H Institute (C.R.F.); the Parker Institute for Cancer Immunotherapy (M.H.S.); the Human Frontier Science Program LT000493/2018-L (K.N.); and the Fellowship of Astellas Foundation for Research on Metabolic Disorders (K.N.).
Footnotes
Competing interests: M.A.F. is a co-founder and director of Federation Bio. J.L.S. is a co-founder of Novome Biotechnologies and of January, Inc. He serves on the scientific advisory board of Second Genome and Kaleido Biosciences.
Data and materials availability: Data described in the paper are present in the main text or archived in the supplementary materials.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Rooks MG, Garrett WS, Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352 (2016). doi: 10.1038/nri.2016.42; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith PM et al. , The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013). doi: 10.1126/science.1241165; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furusawa Y et al. , Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). doi: 10.1038/nature12721; [DOI] [PubMed] [Google Scholar]
- 4.Zhao L et al. , Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156 (2018). doi: 10.1126/science.aao5774; [DOI] [PubMed] [Google Scholar]
- 5.Devlin AS et al. , Modulation of a circulating uremic solute via rational genetic manipulation of the gut microbiota. Cell Host Microbe 20, 709–715 (2016). doi: 10.1016/j.chom.2016.10.021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL, An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). doi: 10.1016/j.cell.2005.05.007; [DOI] [PubMed] [Google Scholar]
- 7.Romano KA et al. , Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe 22, 279–290.e7 (2017). doi: 10.1016/j.chom.2017.07.021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas CM et al. , Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLOS ONE 7, e31951 (2012). doi: 10.1371/journal.pone.0031951; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP, The ClosTron: A universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70, 452–464 (2007). doi: 10.1016/j.mimet.2007.05.021; [DOI] [PubMed] [Google Scholar]
- 10.Dodd D et al. , A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652 (2017). doi: 10.1038/nature24661; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsden SR, Hilton MG, Waller JM, The end products of the metabolism of aromatic amino acids by clostridia. Arch. Microbiol. 107, 283–288 (1976). doi: 10.1007/BF00425340; [DOI] [PubMed] [Google Scholar]
- 12.Elsden SR, Hilton MG, Volatile acid production from threonine, valine, leucine and isoleucine by clostridia. Arch. Microbiol. 117, 165–172 (1978). doi: 10.1007/BF00402304; [DOI] [PubMed] [Google Scholar]
- 13.Williams BB et al. , Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 16, 495–503 (2014). doi: 10.1016/j.chom.2014.09.001; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-del Campo A et al. , Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. mBio 6, e00042–15 (2015). doi: 10.1128/mBio.00042-15; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabisch UC et al. , Identification of D-proline reductase from Clostridium sticklandii as a selenoenzyme and indications for a catalytically active pyruvoyl group derived from a cysteine residue by cleavage of a proprotein. J. Biol. Chem. 274, 8445–8454 (1999). doi: 10.1074/jbc.274.13.8445; [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Hetzel M, Boiangiu CD, Buckel W, Dehydration of (R)-2-hydroxyacyl-CoA to enoyl-CoA in the fermentation of α-amino acids by anaerobic bacteria. FEMS Microbiol. Rev. 28, 455–468 (2004). doi: 10.1016/j.femsre.2004.03.001; [DOI] [PubMed] [Google Scholar]
- 17.Vital M, Howe AC, Tiedje JM, Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 5, e00889 (2014). doi: 10.1128/mBio.00889-14; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nayfach S, Fischbach MA, Pollard KS, MetaQuery: A webserver for rapid annotation and quantitative analysis of specific genes in the human gut microbiome. Bioinformatics 31, 3368–3370 (2015). doi: 10.1093/bioinformatics/btv382; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David LA et al. , Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). doi: 10.1038/nature12820; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denoya CD et al. , A second branched-chain alpha-keto acid dehydrogenase gene cluster (bkdFGH) from Streptomyces avermitilis: Its relationship to avermectin biosynthesis and the construction of a bkdF mutant suitable for the production of novel antiparasitic avermectins. J. Bacteriol. 177, 3504–3511 (1995). doi: 10.1128/jb.177.12.3504-3511.1995; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chabrière E et al. , Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat. Struct. Biol. 6, 182–190 (1999). doi: 10.1038/5870; [DOI] [PubMed] [Google Scholar]
- 22.Heider J, Mai X, Adams MW, Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new and reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J. Bacteriol. 178, 780–787 (1996). doi: 10.1128/jb.178.3.780-787.1996; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma K, Hutchins A, Sung SJ, Adams MW, Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase. Proc. Natl. Acad. Sci. U.S.A. 94, 9608–9613 (1997). doi: 10.1073/pnas.94.18.9608; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT, Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28, 1221–1227 (1987). doi: 10.1136/gut.28.10.1221; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ,The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217, 133–139 (2002). doi: 10.1111/j.1574-6968.2002.tb11467.x; [DOI] [PubMed] [Google Scholar]
- 26.Ridlon JM, Harris SC, Bhowmik S, Kang D-J,Hylemon PB, Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39 (2016). doi: 10.1080/19490976.2015.1127483; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodrich JK et al. , Human genetics shape the gut microbiome. Cell 159, 789–799 (2014). doi: 10.1016/j.cell.2014.09.053; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langille MGI et al. , Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013). doi: 10.1038/nbt.2676; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaminski J et al. , High-specificity targeted functional profiling in microbial communities with ShortBRED. PLOS Comput. Biol. 11, e1004557 (2015). doi: 10.1371/journal.pcbi.1004557; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellono NW et al. , Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell 170, 185–198.e16 (2017). doi: 10.1016/j.cell.2017.05.034; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Jong A, Pietersma H, Cordes M, Kuipers OP, Kok J, PePPER: a webserver for prediction of prokaryote promoter elements and regulons. BMC Genomics 13, 299 (2012). doi: 10.1186/1471-2164-13-299; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.