Abstract
A critical component of wound healing is the transition from the inflammatory phase to the proliferation phase to initiate healing and remodeling of the wound. Macrophages are critical for the initiation and resolution of the inflammatory phase during wound repair. In diabetes, macrophages display a sustained inflammatory phenotype in late wound healing characterized by elevated production of inflammatory cytokines, such as TNF-α. Previous studies have shown that an altered epigenetic program directs diabetic macrophages toward a proinflammatory phenotype, contributing to a sustained inflammatory phase. Males absent on the first (MOF) is a histone acetyltransferase (HAT) that has been shown be a coactivator of TNF-α signaling and promote NF-κB–mediated gene transcription in prostate cancer cell lines. Based on MOF’s role in TNF-α/NF-κB–mediated gene expression, we hypothesized that MOF influences macrophage-mediated inflammation during wound repair. We used myeloid-specific Mof-knockout (Lyz2Cre Moffl/fl) and diet-induced obese (DIO) mice to determine the function of MOF in diabetic wound healing. MOF-deficient mice exhibited reduced inflammatory cytokine gene expression. Furthermore, we found that wound macrophages from DIO mice had elevated MOF levels and higher levels of acetylated histone H4K16, MOF’s primary substrate of HAT activity, on the promoters of inflammatory genes. We further identified that MOF expression could be stimulated by TNF-α and that treatment with etanercept, an FDA-approved TNF-α inhibitor, reduced MOF levels and improved wound healing in DIO mice. This report is the first to our knowledge to define an important role for MOF in regulating macrophage-mediated inflammation in wound repair and identifies TNF-α inhibition as a potential therapy for the treatment of chronic inflammation in diabetic wounds.
Keywords: Immunology, Inflammation
Keywords: Diabetes, Macrophages
TNF-α drives expression of the histone acetyltransferase MOF to promote inflammation in macrophages in nonhealing diabetic wounds.
Introduction
The innate immune system plays a fundamental role in the resolution of wounds after injury. Wound healing occurs through a series of phases of hemostasis/clotting, inflammation, proliferation, and resolution/remodeling (1). Following hemostasis, the inflammatory phase is initiated when peripheral monocytes are recruited to the wound site, where they initially differentiate into proinflammatory macrophages secreting inflammatory cytokines (e.g., IL-1β, TNF-α, and IL-6) (2–4). This phase is critical to clear the wound site of microbes and cellular debris. In order for the wound to heal, these proinflammatory macrophages must transition to an antiinflammatory phenotype to promote tissue repair (2–6). Sustained inflammation due to the inability of wound macrophages to transition to the regeneration stage necessary for normal skin repair is a hallmark of impaired wound healing associated with diseases such as type 2 diabetes (T2D) (6–11). The mechanisms regulating this failed macrophage transition are not completely clear, but recent evidence suggests that epigenetic signatures can drive prolonged inflammation in diabetic wounds (12–16). Our group and others have identified that diabetic wound macrophages exhibit changes in histone methylation that promote a proinflammatory phenotype (12, 14, 16). Specifically, we have shown that MLL1 and JMJD3; a histone methyltransferase and histone demethylase, respectively, are altered in T2D macrophages, drive production of proinflammatory cytokines, and establish a proinflammatory feedback loop leading to uncontrolled inflammation (12, 14, 16).
The histone acetyltransferase (HAT) males absent on the first (MOF) (also called MYST1 or KAT8) is a MYST family HAT that is known to specifically acetylate lysine 16 on the histone H4 tail (H4K16) and promote gene transcription (17–19). MOF has been well studied in a wide variety of cellular processes (17, 20–31), and recently, MOF was reported to regulate macrophage function in response to viral infection, where it acetylates interferon regulatory factor 3 (IRF3) and reduces type I interferon (IFN-I) production (32). Despite this, its role in regulating both normal and diabetic wound macrophages following cutaneous injury remains unknown. Given that MOF may regulate macrophages in response to viruses, we sought to investigate the role of MOF in directing macrophage-mediated inflammation during both normal and pathological wound healing.
Here, we report that TNF-α drives MOF expression in wound macrophages, and this alters macrophage phenotype during normal and diabetic wound healing. Using a myeloid-specific MOF-knockout (Lyz2Cre Moffl/fl) model, we demonstrate that MOF directed inflammatory cytokine production in wound macrophages during the inflammatory phase of wound repair. We show that MOF was overexpressed in wound macrophages from both diet-induced obese (DIO) mouse and human T2D wounds and drove expression of inflammatory cytokines by acetylation of histone H4K16. Importantly, local treatment with an FDA-approved TNF-α inhibitor improved wound healing in DIO mice and reduced MOF levels in DIO wound macrophages. Given the recently FDA-approved anti–TNF-α therapy, these findings allow for a local targeted therapy to reverse the inflammatory macrophage phenotype in diabetic wounds.
Results
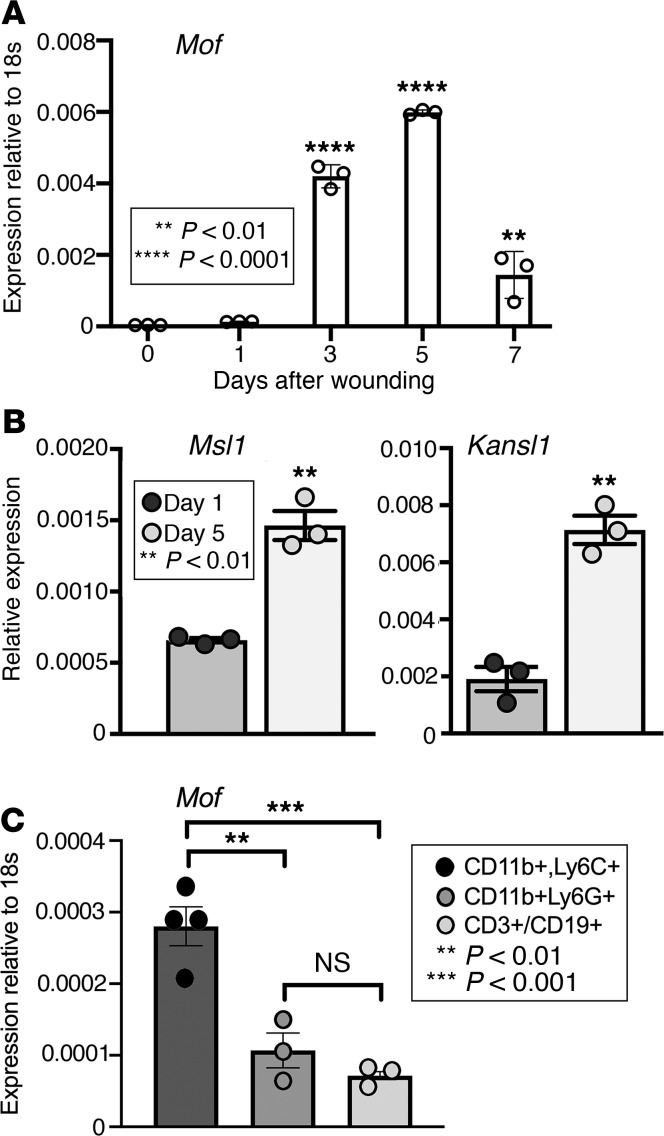
MOF is dynamic during the inflammatory phase of wound healing.
Wound repair requires progression from the inflammatory to the proliferative phase of healing (5, 6, 9). Because macrophages transition from an inflammatory to reparative phenotype to initiate the proliferative phase and previous studies have found that histone acetylation is involved in regulating macrophage function (32), we examined the role of MOF on macrophage function following tissue injury. To test this, C57BL/6J mice were subjected to 6-mm full-thickness wounds as previously described, (12) and myeloid cells (CD11b+CD3–CD19–Ly6G–) were isolated from wounds at multiple time points (0, 1, 3, 5, and 7 days) and analyzed for Mof expression. We found that Mof was upregulated at both days 3 and 5 after injury during a key transition where macrophages shift from a proinflammatory to an antiinflammatory phenotype (Figure 1A). MOF is the catalytic subunit of 2 complexes: the male-specific lethal (MSL) and nonspecific lethal (NSL), which govern its target specificity and subcellular localization (33, 34). Although the MOF-MSL complex is highly specific for H4K16 acetylation (19, 33–35), the MOF-NSL complex is more promiscuous, acetylating lysines 5, 8 (K5, K8), and 16 on the H4 tail (33), and allows MOF to target nonhistone proteins and traffic to mitochondria (22, 23, 34, 36, 37). To better understand MOF’s role in macrophages in wound repair, we also examined levels of Msl1 and Kansl1, integral components of the MSL and NSL complexes, respectively. We found that both Msl1 and Kansl1 were upregulated in wound macrophages at day 5 after injury, suggesting that MOF may play an important role in the transition to an antiinflammatory phenotype in wounds (Figure 1B).
Figure 1. MOF regulates macrophage function during wound healing.
(A) Representative figure showing Mof expression in wounds over time. Three wounds were created using a 6-mm punch on the backs of WT C57BL/6J mice. Wound macrophages (CD11b+CD3–CD19–Ly6G–) were isolated at baseline (day 0) and on days 1, 3, 5, and 7 after injury, and Mof gene expression was assessed (n = 3 × 3 mice pooled/replicate/time point; repeated twice). Data were analyzed for normality and 1-way ANOVA was performed followed by Tukey’s multiple-comparisons test. (B) Representative figures of Msl1 and Kansl1 expression in day 5 wounds. Wound macrophages (CD11b+CD3–CD19–Ly6G–) were isolated on days 1 and 5 after injury. Msl1 and Kansl1 gene expression were measured by quantitative PCR (qPCR) (n = 3 × 3 mice pooled/replicate/time point; repeated twice). Data were analyzed for normality and 2-tailed Student’s t test was performed. (C) Representative figure showing divergence in Mof expression between cell types in day 5 wounds. Macrophages (CD11b+CD3–CD19–Ly6G–, n = 4), neutrophils (CD11b+CD3–CD19–Ly6G+, n = 3), and combined T/B cells (CD3+/CD19+CD11b–Ly6G–, n = 3) were isolated on 5 after wounding. Mof gene expression was measured by qPCR (n = 3 mice pooled/replicate/time point; repeated twice). Data were analyzed using 1-way ANOVA followed by Tukey’s test for multiple comparisons.
Because wound healing is a complex process involving contributions from many cells, we examined whether the increase in Mof we observed is intrinsic to macrophages. Here, we isolated monocytes/macrophages (CD11b+Ly6C+CD3–CD19–Ly6G–), neutrophils (CD11b+CD3–CD19–Ly6G+), and a combined population of T and B cells (CD3+/CD19+CD11b–Ly6G–Ly6C–) from wounds on day 5 after injury and analyzed Mof expression. Mof was significantly elevated in the monocyte/macrophage population compared with all other cell fractions (Figure 1C). Taken together, these data suggest that MOF may regulate macrophage-specific functions during the transition from the inflammatory to the reparative phenotype during wound repair.
MOF regulates NF-κB–mediated inflammatory cytokine expression in wound macrophages.
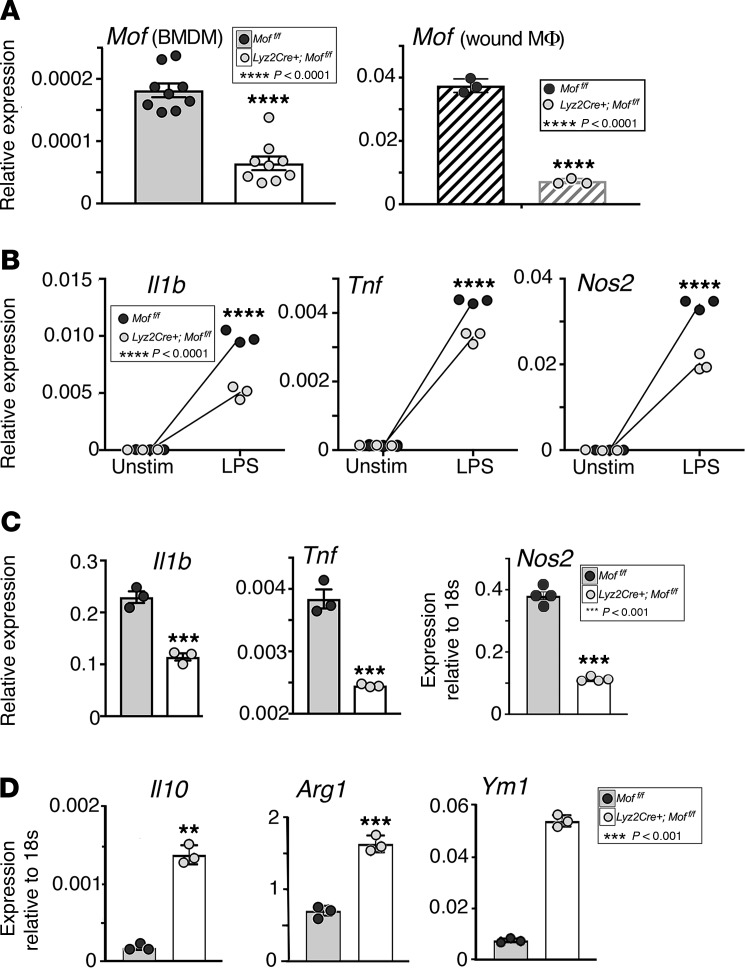
Because MOF has been shown to promote NF-κB–mediated gene transcription in cancer cell lines (38), we examined its role in NF-κB–mediated inflammatory cytokine expression in upregulation in wound macrophages during the macrophage phenotype transition. To test this, we generated a mouse line with a myeloid-specific Mof deletion by crossing Lyz2Cre mice that express Cre recombinase in a myeloid-specific manner to mice containing loxP sites flanking exons 4–6 of Mof (28), hereafter referred to as Lyz2Cre Moffl/fl. Myeloid-specific deletion of MOF was confirmed by examining Mof expression in bone marrow–derived macrophages (BMDMs) and wound macrophages, B cells, and T cells generated from Lyz2Cre Moffl/fl mice and Moffl/fl control mice lacking Cre expression (Figure 2A and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.132306DS1). To determine the effect of MOF on inflammatory cytokine expression, we activated BMDMs with lipopolysaccharide (LPS) (100 ng/mL for 8 hours) and analyzed NF-κB–mediated inflammatory cytokine expression. We found that canonical proinflammatory cytokines, Il1b, Tnf, and Nos2, which are highly relevant for wound repair, were significantly decreased in Lyz2Cre Moffl/fl BMDMs compared with controls (Figure 2B). We then sought to determine whether the reduction in proinflammatory expression associated with MOF deletion in vitro persisted in vivo in wound macrophages. We measured inflammatory cytokine expression in day 5 wound macrophages isolated from Lyz2Cre Moffl/fl and Moffl/fl control mice and found that expression of Il1b, Tnf, and Nos2 was significantly decreased in Lyz2Cre Moffl/fl wound macrophages (Figure 2C).
Figure 2. Macrophages from myeloid-deficient MOF mice demonstrate decreased NF-κB inflammatory genes.
(A) Representative figures showing Mof deletion in BMDMs and wound macrophages from Lyz2Cre Moffl/fl mice. BMDMs were derived from Lyz2Cre Moffl/fl or Moffl/fl mice. Wound macrophages were sorted by magnetic-activated cell sorting (MACS) from Lyz2Cre Moffl/fl or Moffl/fl mice on day 5 after wounding. Mof gene expression was measured by qPCR (n = 3 × 4 mice pooled/replicate, repeated twice). (B) Representative figures showing inflammatory cytokine gene expression in LPS-stimulated BMDMs. BMDMs were isolated from Lyz2Cre Moffl/fl or Moffl/fl mice and treated with LPS or vehicle control for 8 hours. Il1b, Tnf, and Nos2 gene expression was measured by qPCR (n = 3, repeated twice). (C) Representative figures showing baseline inflammatory cytokine gene expression in wound macrophages. Wound macrophages (CD11b+CD3–CD19–Ly6G–) were isolated from Lyz2Cre Moffl/fl or Moffl/fl mice on day 5 after wounding. Il1b (n = 3), Tnf (n = 3), and Nos2 (n = 4) expression was measured by qPCR (n = 3 mice pooled/replicate, repeated twice). (D) Representative figures showing antiinflammatory gene expression in wound macrophages. Wound macrophages (CD11b+CD3–CD19–Ly6G–) were isolated from Lyz2Cre Moffl/fl or Moffl/fl mice on day 5 after wounding. Il10, Arg1, and Ym1 expression was measured by qPCR (n = 3 × 3 mice pooled/replicate, repeated twice). A, C, and D were analyzed using a 2-tailed Student’s t test. B was analyzed using 2-way ANOVA followed by Holm-Šídák test for multiple comparisons.
Because Mof expression was elevated at day 5 after wounding (Figure 1A), we hypothesized that MOF may regulate expression of genes involved with the shift in macrophage phenotype observed during this transitional phase. To test this, we measured expression of canonical antiinflammatory genes in Lyz2Cre Moffl/fl and control wound macrophages. We found that the antiinflammatory genes Il10, Ym1, and Arg1 were more highly expressed in Lyz2Cre Moffl/fl wound macrophages compared with control wound macrophages from Moffl/fl (Figure 2D). Taken together, these data demonstrate that MOF regulates NF-κB–mediated inflammation in wound macrophages and may play a role in regulating macrophage phenotype during wound repair.
MOF is upregulated in diabetic wound macrophages and regulates NF-κB–mediated inflammation via H4K16 acetylation.
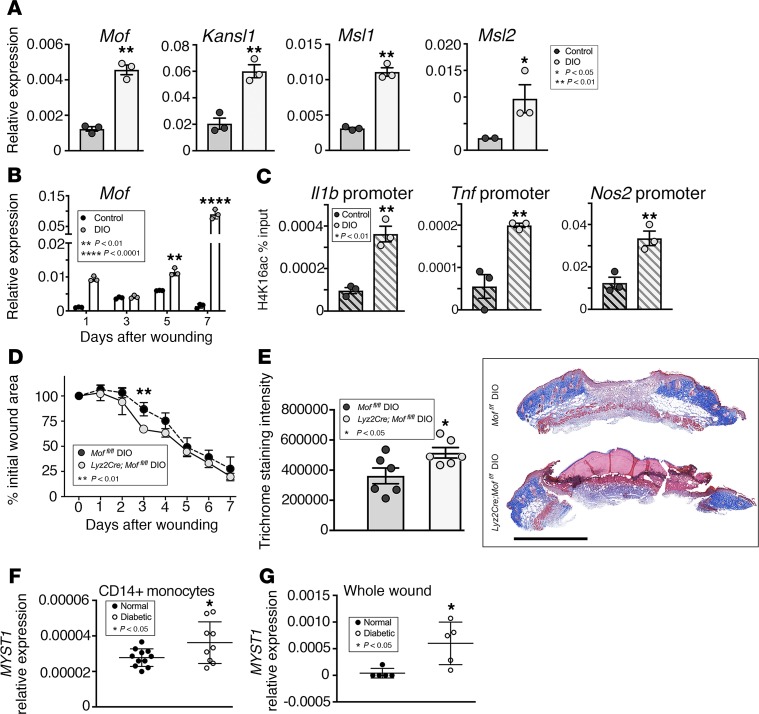
Diabetic wounds are characterized by an inflammatory phase where macrophages fail to transition from an inflammatory to a reparative phenotype (6, 9, 11). These diabetic wound macrophages express high levels of NF-κB–mediated inflammatory cytokines that promote chronic inflammation in the wound and prevent transition to the proliferative phase (39, 40). Because we found that Mof regulates NF-κB–mediated inflammatory cytokines in wound macrophages and was upregulated in wound macrophages during their transition to an antiinflammatory phenotype, we examined whether MOF is relevant in diabetic wound macrophages. To examine this, a physiological murine model of T2D, the high-fat diet (60% saturated fat) DIO mouse, was used because of its lack of effect on the innate immune system (41, 42). DIO and normal diet (13.5% saturated fat) control mice were wounded, and wound macrophages were isolated on day 5 and analyzed for Mof, Msl1, Msl2, and Kansl1 expression. We found that Mof, Msl1, Msl2, and Kansl1 were all significantly elevated in DIO wound macrophages compared with wound macrophages isolated from normal diet controls (Figure 3A). To determine whether Mof upregulation was specific to the DIO mouse model, we examined another established model of T2D, the db/db murine model, which lacks a functional leptin receptor (43). We measured Mof expression in wound macrophages isolated from db/db and db/wt controls at day 5 after injury and found that Mof expression was significantly higher in db/db wound macrophages compared with wound macrophages isolated from db/wt controls (Supplemental Figure 2A). To determine the kinetics of Mof expression in diabetic macrophages following injury, we isolated wound macrophages at several time points (days 1, 3, 5, and 7) during repair. We found that Mof continued to increase over the course of wound healing and was highest at day 7, a late time point when transition of macrophages to a reparative phenotype should have occurred (Figure 3B).
Figure 3. MOF is elevated in diabetic wound macrophages at late time points.
(A) Representative figures showing Mof complex member expression in day 5 wounds from control and DIO wounds. Three wounds were created using a 6-mm punch on the backs of WT C57BL/6J mice on a normal (13.5% saturated fat) or high-fat (DIO, 60% saturated fat) diet. Wound macrophages (CD11b+CD3–CD19–Ly6G–) were isolated on day 5 after injury. Mof, Msl1, Msl2, and Kansl1 expression was examined by qPCR (n = 3 × 3 mice pooled/replicate; repeated twice). (B) Representative figure showing changes in Mof expression in control and DIO wounds over time. Wound macrophages (CD11b+CD3–CD19–Ly6G–) were isolated on days 1, 3, 5, and 7 after injury and assessed for Mof expression by qPCR (n = 3 × 3 mice pooled/replicate/time point; repeated twice). (C) Representative figures showing H4K16ac deposition on inflammatory cytokine gene promoters in day 5 wounds. DIO and control wound macrophages (CD11b+CD3–CD19–Ly6G–) were isolated on day 5 after wounding. ChIP analysis was performed for H4K16ac at the NF-κB binding site on the promoters of Il1b, Tnf, and Nos2 promoters (n = 3 × 4 mice pooled/replicate, repeated twice). (D) Representative figure showing wound closure in Lyz2Cre Moffl/fl mice on high-fat diet. Two wounds were created using 6-mm punch biopsies on the backs of DIO Lyz2Cre Moffl/fl or DIO Moffl/fl mice. Change in wound area was analyzed daily using ImageJ software (NIH) (n = 6 mice/group; repeated twice). (E) Representative figures showing collagen deposition in day 5 Lyz2Cre Moffl/fl DIO wounds. Wounds from DIO Lyz2Cre Moffl/fl or DIO Moffl/fl mice were harvested on day 5 after injury and processed for histology. Trichrome staining was performed and calculated using ImageJ software (n = 6 mice/group). Scale bar: 2 mm. (F) Representative figure showing MYST1 expression in diabetic and control human PBMCs. Human PBMCs (CD14+) were isolated from T2D or non-T2D control patients. MYST1 expression was measured using qPCR (n = 11 normal, 9 diabetic). (G) Representative figure showing MYST1 expression in diabetic and control human wound tissue. Wounds from T2D or non-T2D control patients were examined for MYST1 expression by qPCR (n = 5). A and C were analyzed using a 2-tailed Student’s t test. B and D were analyzed by multiple t tests (1 per time point). E, F, and G were analyzed using a 1-tailed Student’s t test with Welch’s correction.
Because of MOF’s well-established role in promoting transcription by histone H4K16 acetylation, we examined whether MOF promotes expression of NF-κB–mediated inflammatory genes via an H4K16-mediated mechanism. To examine this, we performed chromatin immunoprecipitation (ChIP) for acetylated H4K16 (H4K16ac) on the NF-κB binding sites of promoters of inflammatory genes Il1b, Tnf, and Nos2 in wound macrophages isolated from DIO and normal diet controls. We found that H4K16ac was increased on the promoters of Il1b, Tnf, and Nos2 in the DIO wound macrophages compared with control wound macrophages (Figure 3C). Further, we performed H4K16ac ChIP in BMDMs from db/db and db/wt controls and found that H4K16ac deposition was higher on the NF-κB binding sites of the promoters of Il1b and Tnf in db/db wound macrophages compared with controls (Supplemental Figure 2B). These data suggest that MOF regulates inflammatory gene expression in diabetic wound macrophages via acetylation of histone H4K16.
Because MOF was marked elevated in diabetic wound macrophages, we examined the effects of myeloid-specific MOF deletion on wound repair. Lyz2Cre Moffl/fl mice and Moffl/fl control mice were wounded, and wound closure was monitored during healing. We found that MOF deletion conferred an improvement during the very early stages of wound healing compared with controls (Supplemental Figure 3). We then examined whether MOF deletion could improve healing in diabetic wounds. Lyz2Cre Moffl/fl mice and Moffl/fl control mice were placed on a high-fat diet for 10 weeks to create a glucose-intolerant, insulin-resistant, prediabetic murine model (Lyz2Cre Moffl/fl DIO). The mice were then wounded, and wound closure was monitored and measured over the course of healing. We identified a modest but significant improvement in wound closure in Lyz2Cre Moffl/fl DIO mice during the inflammatory phase of healing compared with Moffl/fl DIO control mice (Figure 3D). Histological examination showed a moderate increase in collagen deposition at the wound edge in Lyz2Cre Moffl/fl DIO mice compared with Moffl/fl DIO control (Figure 3E and Supplemental Figure 4). Taken together, these results suggest that modulation of myeloid-specific MOF expression improves diabetic wound healing.
To translate our findings to human disease, we isolated human CD14+ peripheral blood monocytes from T2D and matched control patients and assessed expression of MYST1, MOF’s human ortholog. We found that MYST1 was significantly elevated in T2D monocytes compared with controls (Figure 3F). Further, we measured MYST1 expression in human wound tissue from patients with T2D and non-T2D controls and found that MYST1 was significantly increased in wounds from T2D patients compared with wounds from non-T2D controls (Figure 3G). Taken together, these data suggest that MOF’s elevated expression at late time points in diabetic wound macrophages/monocytes prevents resolution of inflammation in wounds in both mice and humans and that myeloid-specific reduction in MOF may improve diabetic wound repair.
TNF-α regulates MOF expression in macrophages.
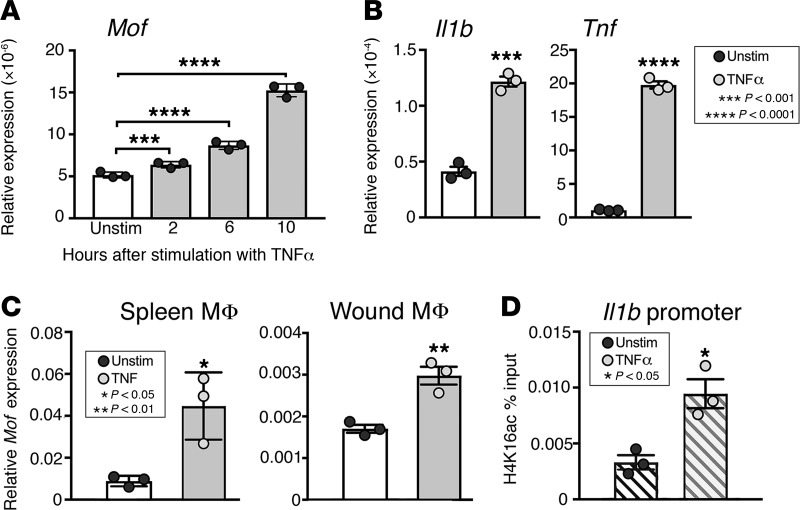
TNF-α has been shown to possibly regulate NF-κB transcriptional activation via MOF in prostate cancer cell lines (38); however, the regulation of MOF in macrophages is unknown. TNF-α is a critical cytokine involved in wound repair and is known to be significantly elevated in diabetic wounds (6); hence, we examined whether TNF-α directs Mof expression in macrophages. To first determine whether TNF-α directly activates Mof expression, we isolated BMDMs and treated them with TNF-α (25 ng/mL) for 2, 6, and 10 hours. We found that Mof expression was upregulated upon stimulation with TNF-α (Figure 4A). Further, we found that TNF-α stimulation significantly upregulated inflammatory gene expression in BMDMs, particularly of Il1b and Tnf (Figure 4B). To determine whether TNF-α increases MOF in vivo in blood monocytes/macrophages or peripheral wound macrophages, we isolated in vivo splenic (CD11b+CD3–CD19–Ly6G–NK1.1–Ter119–) and wound macrophages (CD11b+CD3–CD19–Ly6G–), stimulated them ex vivo with TNF-α, and analyzed for Mof expression. We found that TNF-α promotes Mof expression in vivo in both blood and wound monocytes/macrophages (Figure 4C). These data suggest that the increased MOF seen in diabetic macrophages may be regulated by the increased TNF-α known to be present in high levels in diabetic tissue. To determine whether the TNF-α–dependent increase in MOF is driving the inflammatory phenotype in diabetic wound macrophages, we performed ChIP for H4K16ac on NF-κB binding sites of the Il1b promoter. We found that wound macrophages stimulated with TNF-α demonstrate an increase in H4K16ac deposition on the Il1b promoter (Figure 4D). Hence, in diabetic tissues, elevated TNF-α levels may promote MOF expression in wound macrophages and alter macrophage function/phenotype by driving prolonged expression of inflammatory genes.
Figure 4. TNF-α promotes MOF expression in vitro and in wound macrophages.
(A) Representative figure showing induction of Mof expression by TNF-α in BMDMs. BMDMs were isolated from C57BL6/J mice and stimulated in culture with 25 ng/mL TNF-α for 0, 2, 6, and 10 hours. Mof expression was measured by qPCR (n = 3/time point, repeated twice). (B) Representative figures showing induction of inflammatory cytokine gene expression by TNF-α. BMDMs were isolated from C57BL6/J mice and stimulated in culture with 25 ng/mL TNF-α for 12 hours. Il1b and Tnf expression were measured by qPCR (n = 3, repeated twice). (C) Representative figures showing induction of Mof expression by TNF-α in splenic and wound macrophages. Monocyte/macrophage (CD11b+CD3–CD11c–CD19–Ly6G–NK1.1–) single-cell suspensions were MACS sorted from C57BL/6J mouse spleens and stimulated ex vivo with 25 ng/mL TNF-α for 12 hours. Wound macrophages (CD11b+CD3–CD19–Ly6G–) were MACS sorted at day 5 after injury from C57BL/6J mice and stimulated ex vivo with 25 ng/mL TNF-α for 12 hours. Mof expression was measured by qPCR in both splenic and wound monocytes/macrophages (n = 3 mice pooled/replicate, repeated twice). (D) Representative figure showing H4K16ac deposition on Il1b promoter in response to TNF-α stimulation. Wound macrophages (CD11b+CD3–CD19–Ly6G–) were isolated on day 5 after wounding from C57BL/6J mice, and ChIP analysis was performed for H4K16ac at the NF-κB binding site on the Il1b promoter (n = 6 mice, repeated three times). A was analyzed by 1-way ANOVA followed by Tukey’s test for multiple comparisons. B–D were analyzed using a 2-tailed Student’s t test.
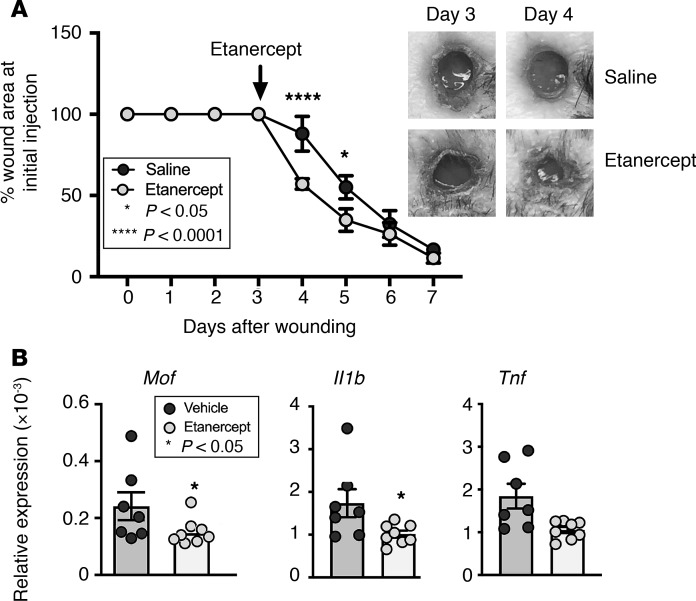
TNF-α inhibition reduces MOF in diabetic wound macrophages and improves diabetic wound healing.
Given that Mof expression was increased by TNF-α stimulation in wound macrophages, we examined whether TNF-α inhibition reduces MOF expression and modulates inflammation in wound macrophages. To examine this, DIO and control mice were wounded and administered a single local injection of the FDA-approved TNF-α inhibitor etanercept (5 mg/kg) or vehicle saline (controls), starting on day 3 after the initial inflammatory phase necessary for repair. We found that DIO mice treated with etanercept demonstrated significantly improved wound repair compared with DIO mice with saline control (Figure 5A). Further, we found that expression of Mof and the inflammatory cytokines Il1b, Tnf, and Nos2 was reduced in macrophages isolated from mice given etanercept compared with saline controls (Figure 5B and Supplemental Figure 5). Because we saw increased expression of antiinflammatory genes in wound macrophages from Lyz2Cre Moffl/fl mice, we examined antiinflammatory gene expression in DIO wound macrophages treated ex vivo with etanercept. Unlike the result in MOF-deleted wound macrophages, etanercept-treated wound macrophages did not show a difference in expression of Il10, Arg1, and Ym1 (Supplemental Figure 6). Thus, TNF-α inhibition was likely having a broader effect in the wound. Regardless, these results suggest that Mof expression in diabetic wound macrophages is driven, at least in part, by TNF-α and that TNF-α inhibition is sufficient to reduce MOF expression and inflammatory cytokine production in these cells, leading to improved diabetic wound repair.
Figure 5. Local TNF-α inhibition improves diabetic wound healing by modulating MOF in wound macrophages.
(A) Representative figure showing improved wound closure after treatment with etanercept. Two wounds were created using 6-mm punch biopsies on the backs of DIO mice. On day 3 after injury, the mice were injected locally with 5 mg/kg etanercept or vehicle control, and change in wound area was analyzed daily using ImageJ software (n = 5 mice/group; repeated twice). Data were analyzed using multiple t tests, 1 per time point. (B) Representative figures showing reduced Mof and inflammatory cytokine expression in response to TNF-α. Wound macrophages (CD11b+CD3–CD19–Ly6G–) were isolated on day 5 after injury from untreated DIO mice and stimulated ex vivo with 250 mg/mL etanercept or vehicle control for 12 hours, and Mof, Il1b, and Tnf expression was measured by qPCR. Data were analyzed using a 1-way Student’s t test with Welch’s correction.
Discussion
During the inflammatory state of wound repair, macrophages transition from an inflammatory to a reparative phenotype. This switch is significantly impaired in obesity/T2D and results in chronic inflammation and nonhealing wounds; however, the etiology is unclear. We and others have previously identified that chronic inflammation in diabetic wounds is governed by epigenetic regulation of macrophages (12–16). In this study, we identify that the HAT MOF regulates inflammation in macrophages during normal and diabetic wound repair. Here, we demonstrate that MOF regulates NF-κB–mediated inflammatory gene transcription in wound macrophages via an H4K16ac mechanism in increased inflammatory cytokine production. Further, MOF is pathologically upregulated in diabetic wound macrophages and corresponds to increased macrophage-mediated inflammation. Functionally, we demonstrate that Mof expression in wound macrophages is regulated by TNF-α and that Mof can be controlled in wound macrophages by TNF-α inhibition. Importantly, local TNF-α inhibition decreased MOF and inflammation in diabetic wound macrophages and improved healing in diabetic mice. Hence, these findings define a potential therapy to decrease inflammatory macrophages in diabetic wounds.
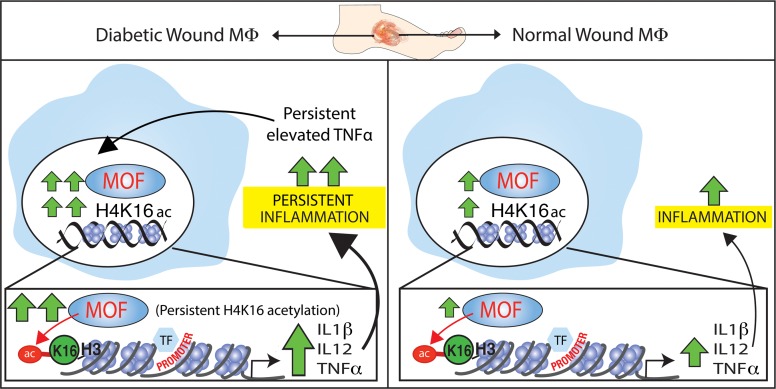
Our findings suggest a model whereby MOF is required for normal inflammatory cytokine expression during wound healing but is downregulated in the later stages, while in the context of diabetic wound healing, MOF expression increases unrestrained over time and contributes to the proinflammatory nature of diabetic wounds (Figure 6). Our data align with studies showing that activation of sirtuin 1 (SIRT1), a known H4K16 deacetylase, is protective against metabolic pathologies and that SIRT1 regulates inflammation and insulin sensitivity in macrophages (44–46). Additionally, mechanistic studies have shown that MOF and SIRT1 act in an opposing manner to regulate the NoRC chromatin remodeling complex (36). Thus, it is possible that SIRT1 and MOF act antagonistically in macrophages to regulate inflammation in an H4K16-dependent manner. SIRT1 has also been shown to deacetylate MOF directly in its autoacetylation site, although the impacts of this on MOF function are not fully clear (47). Whether there is interplay between SIRT1 and MOF in macrophage function remains uncertain.
Figure 6. Schematic showing TNF-α drives MOF-mediated inflammatory gene expression in diabetic wound macrophages via H4K16 acetylation.
The Lyz2Cre mouse model used in our study may also affect Mof in neutrophils, which are extremely important during early wound healing. Because we observed significant increases in Mof expression in normal wound macrophages only at day 5 (Figure 1C and Supplemental Figure 1), we did not specifically investigate neutrophils; thus, this is a limitation of our study. Further, the improvement in wound closure that we observed in the TNF-α inhibition study (Figure 5A) was more pronounced than in Lyz2Cre Moffl/fl mice on the high-fat diet (Figure 3B). Hence, TNF-α is likely acting on cell types other than macrophages and may potentially be affecting MOF function, or other functions, in these cells.
Another potential limitation of the Lyz2Cre Moffl/fl mouse model is that deletion of MOF in the bone marrow prior to macrophage differentiation may affect early events that require low-level expression of MOF. Because we show that MOF promotes inflammatory cytokine expression, and inflammatory macrophages are required in the early inflammatory phase of wound repair to promote bactericidal functions and clean up apoptotic debris (1, 2), it is feasible that proinflammatory macrophages are hampered during the early stages of wound repair. However, we saw that MOF deletion improved wound closure very early in the wound healing process (Supplemental Figure 4). Whether this is due to effects on proinflammatory macrophages or some other cell type will require deeper investigation into MOF’s complex regulation. Additionally, because histone deacetylase inhibition promotes keratinocyte proliferation in the wound and accelerates wound repair (48, 49), global MOF deletion or inhibition may affect keratinocyte and fibroblast signaling that promotes collagen deposition and re-epithelialization (50, 51). Thus, further investigation is necessary to determine the effect of early MOF deletion on wound repair.
In addition to facilitating transcription via H4K16 acetylation, MOF has been shown to play decisive roles in nonhistone protein acetylation, autophagy, and mitochondrial function (22, 24, 34, 52, 53). MOF function is governed by the NSL and MSL complexes that determine its target specificity and subcellular localization. For example, the MSL complex imparts high specificity toward histone H4K16 (33). On the other hand, the NSL complex determines MOF’s ability to acetylate nonhistone proteins, e.g., p53, and localize to mitochondria (22, 34, 36, 37). In the NSL complex, affinity for histone H4K16 is less specific because MOF acetylates additional lysine residues on the H4 tail (i.e., H4K5, H4K8) (33). In our study, we tested key complex members of the MSL and NSL complexes to examine regulation of MOF function in wound macrophages. Because we saw that components of both complexes were increased in diabetic wound macrophages, we concluded that MOF is acting through acetylation of histone H4K16. Indeed, our ChIP data support a role for MOF in promoting inflammatory gene expression in an H4K16ac-dependent manner. Although MSL and NSL have been identified in developmental disorders and malignancies, very little is known about their roles in regulating inflammation. Because MOF activity is defined by the complex it resides in and we saw increased Msl1 and Kansl1 expression at day 5 in diabetic wound macrophages, it is possible that MOF alters wound repair through other mechanisms not examined here. In view of our data here, further study into the mechanisms by which the MOF/MSL/NSL functional axis regulates inflammation during normal and diabetic wound healing is merited.
Recently, MOF was shown to reduce antiviral immunity in macrophages by acetylating IRF3 and, in turn, reducing IFN-I production (32). Our group has recently reported that IFN-I mediates inflammation in wound macrophages by driving expression of the histone methyltransferase SETDB2, which silences inflammatory cytokine gene expression (54). Thus, MOF may play an indirect role in wound healing by regulating IFN-I signaling. In the current study, we observed a direct effect of MOF by increased H4K16ac on NF-κB–mediated inflammatory gene promoters. Because members of both the MSL and NSL complexes increased similarly in diabetic wound macrophages, we did not investigate acetylation of nonhistone proteins. However, we used an MOF-knockout model, which could affect any component of the MOF complex. Thus, MOF may also affect wound macrophage phenotype via mechanisms independent of H4K16 acetylation.
In summary, we identified that MOF promotes NF-κB–mediated inflammatory gene expression via H4K16 acetylation in wound macrophages and that this process is increased in diabetic wound macrophages in a TNF-α–dependent manner. We also found that TNF inhibition reduced MOF expression in wound macrophages and improved wound healing in a murine model of diabetes. Thus, with the recent FDA approval of TNF inhibitor therapies, targeting MOF via TNF-α could be a viable therapeutic strategy to regulate the sustained inflammation in diabetic wounds.
Methods
Mouse models.
C57BL/6J mice were purchased from The Jackson Laboratory (stock number 000664) at 6–8 weeks. Mice with myeloid-specific deletion of Mof were generated by mating Moffl/fl mice (a gift from Yali Dou, Department of Pathology, University of Michigan, Ann Arbor, Michigan, USA) (28) with Lyz2Cre mice (stock number 004781, The Jackson Laboratory) and backcrossed for 8 or more generations. C57BL/6J and Lyz2Cre Moffl/fl mice were maintained on either a normal chow diet (13.5% kcal fat; LabDiet) or a high-fat diet (60% saturated fat; LabDiet) for 12 weeks to induce the DIO murine model of T2D as previously described (55). Animals underwent all procedures at 20–24 weeks of age.
Wound healing assessment.
Prior to wounding, mice were anesthetized and depilated with Veet (Reckitt Benckiser), and skin was cleaned with sterile water. Full-thickness back wounds were created using a 4-mm punch biopsy as previously described (23, 24). Initial wound surface area was recorded. Digital photographs were obtained daily using an 8-megapixel iPad camera with an internal scale to standardize measurements. Wound area was quantified using ImageJ software (56) and was expressed as the percentage of original wound size over time.
Wound histology.
Whole wounds were excised at day 5 using a 6-mm punch biopsy and fixed in 10% formalin overnight before embedding in paraffin. We stained 5-μm sections with Masson’s trichrome stain for collagen deposition. Images were captured using an Olympus BX43 microscope and Olympus cell Sens Dimension software. Collagen deposition was calculated using ImageJ software (56).
Wound digestion.
Mice were euthanized according to guidelines approved by the University of Michigan Animal Care and Use program. Wounds were excised from the backs of the mice postmortem containing full-thickness dermis with a 1- to 2-mm margin to ensure collection of granulation tissue and placed in 1 mL RPMI (12-167F, Lonza). Wounds were then carefully minced with sharp scissors and digested by incubating in a solution of 50 mg/mL Liberase TM (Roche) and 20 U/mL DNase I (MilliporeSigma). Because the final cell number is limited, wounds from the same genotype/treatment were pooled to maximize cell yield, as previously published (11, 12, 14, 16, 54, 57–59). Wound cell suspensions were then gently plunged and filtered through a 100-μm filter to yield a single-cell suspension. Cells were then sorted by MACS for CD11b+CD3–CD19–Ly6G– cells (described below) or by FACS for specific cell populations or cultured ex vivo.
Wound myeloid cell isolation and MACS.
Wounds were digested as described above. Single-cell suspensions were incubated with FITC-labeled anti-CD3, anti-CD19, and anti-Ly6G antibodies (BioLegend) followed by anti-FITC microbeads (Miltenyi Biotec). Flow-through was then incubated with anti-CD11b microbeads (Miltenyi Biotec) to isolate the non-neutrophil, nonlymphocyte, CD11b+ cells. Cells were then lysed in TRIzol (Invitrogen, Thermo Fisher Scientific) for quantitative reverse transcription PCR (RT-PCR) analyses or fixed with 10% formalin for ChIP assay.
FACS.
Cells were digested as described above. Single-cell suspensions were washed with FACS buffer (Dulbecco’s PBS + 2% FBS + 0.2% EDTA). Cell suspensions were then stained with antibodies against CD3 (clone 145-2C11), CD19 (clone 1D3/CD19), CD11b (clone M1/70), Ly6C (clone HK1.4), and Ly6G (clone 1A8) (all from BioLegend) for 30 minutes. Cells were washed twice with FACS buffer and then stained with DAPI. Cell suspensions were then sorted using an Aria III flow cytometry–assisted cell sorter (BD) for CD11b+Ly6C+, CD11b+Ly6G+, and CD3+/CD19+ cell populations.
Gene expression analysis.
Total RNA was extracted using TRIzol and precipitated using isopropanol according to the manufacturer’s instructions. RNA was then normalized and reversed-transcribed to cDNA using Superscript III (Invitrogen, Thermo Fisher Scientific). qPCR was performed in triplicate with 2X TaqMan PCR mix using the 7500 Real-Time PCR System and primers for Il1b (Mm00434228_m10), Tnfa (Mm00443258_m1), Mof (Mm01137921_g1), Msl1 (Mm00511921_m1), Msl2 (Mm01235702_m1), Kansl1 (Mm1246891_m1), Il10 (Mm01288386_m1), Arg1 (Mm00475988_m1), Ym1 (Mm00657889_mH) (all from Thermo Fisher Scientific), and 18s as the internal control. Data were analyzed relative to 18S ribosomal RNA (2-ΔCt). Data were compiled in Microsoft Excel and the 2-ΔCt values were plotted and analyzed using Prism software (GraphPad).
ChIP.
ChIP assay was performed as described previously (26). Briefly, cells were fixed in 1% paraformaldehyde and lysed and sonicated using a Bioruptor Pico (Diagenode) to generate 300- to 500-bp fragments. Samples were then incubated overnight in anti-H4K16ac antibody (39167, Active Motif) or isotype control (rabbit polyclonal IgG) (ab171870, Abcam) in parallel followed by addition of protein A-Sepharose beads (Thermo Fisher Scientific). Beads were washed and bound; DNA was eluted and purified using phenol/chloroform/isoamyl alcohol extraction followed by ethanol precipitation. H4K16ac deposition was measured by qPCR using 2X SYBR PCR mix (Invitrogen, Thermo Fisher Scientific) and primers targeting NF-κB–binding sites in the Il1b, Tnf, and Nos2 promoters described previously (60).
Cell culture and cytokine analysis.
Bone marrow cells were collected by flushing mouse femurs and tibias with RPMI. BMDMs were cultured as previously detailed (26). At 6 days in culture, the cells were replated and, after resting for 24 hours, were incubated with or without recombinant TNF-α (25 ng/mL; MilliporeSigma, 2880, purified by phenol extraction to <3% impurities) for 2–10 hours, after which cells were placed in TRIzol for RNA analysis.
Isolation of macrophages from human wounds.
All experiments using human samples were approved by the IRB at the University of Michigan (protocols HUM00060733 and HUM00098915) and were conducted in accordance with the principles in the Declaration of Helsinki. For a detailed description of the patient cohort, see Supplemental Table 1. Briefly, wounds were isolated from age-matched patients with or without T2D who were undergoing amputation for medical reasons. Comorbid conditions were not statistically different between the groups. Wounds were obtained from the lateral edge of wound specimens using an 8-mm punch biopsy tool. Wounds were then processed for RT-PCR as described for the murine wounds. RNA with RNA integrity number scores of greater than 8 were used, and all values were done with comparison to 28S/18S ratios and other housekeeping genes.
Isolation of human monocytes.
For human monocyte isolation, peripheral blood was collected and subjected to RBC lysis and Ficoll-Hypaque separation (GE Healthcare). Cell suspensions were then treated with anti–human CD14 microbeads and purified by MACS as described above. For a detailed description of the patient cohort, see Supplemental Table 1.
Statistics.
Data were analyzed using GraphPad Prism software version 8. Data are expressed as mean ± SEM. Data were tested for normal distribution and variance. Differences between 2 normally distributed groups were analyzed using 2-tailed Student’s t test for groups with equal variance or 1-tailed Student’s t test with Welch’s correction for groups with unequal variances. A nonparametric Mann-Whitney U test was used to test differences between 2 groups with non-normal distributions. Differences between more than 2 groups were evaluated by either 1-way or 2-way ANOVA followed by post hoc analysis for multiple comparisons for data that passed tests for normality and equal variance (Bartlett’s test); otherwise, a nonparametric Kruskal-Wallis test followed by Dunn’s post hoc analysis was used. Time course experiments between 2 groups were analyzed using multiple t tests (1 per time point) between the groups. P values less than 0.05 were considered significant.
Study approval.
Mice were maintained in the University of Michigan pathogen-free animal facility, and all protocols were approved by and in accordance with the guidelines established by the Institutional Animal Care and Use Committee at the University of Michigan. Approval for obtaining human peripheral blood was approved by the IRB (protocol HUM00098915). Patient consent for obtaining human wound tissue was deemed exempt by the IRB because tissue was obtained from discarded surgical material (protocol HUM00060733).
Author contributions
ADD and KAG designed the study, conducted experiments, acquired and analyzed data, and wrote the manuscript. FMD, ADJ, RA, JL, BN, JK, and DN conducted experiments and reviewed and edited the manuscript. SJW, JB, BBM, and SLK reviewed and edited the manuscript. JEG acquired and analyzed data and reviewed and edited the manuscript. ADD and KAG are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Supplementary Material
Acknowledgments
The authors thank Yali Dou, professor in the Department of Pathology, University of Michigan, for the gift of the Moffl/fl mice and Robin G. Kunkel, research associate in the Department of Pathology, University of Michigan, for the artwork. This work was supported in part by NIH grant R01 HL137919 (to KG) and Doris Duke Clinical Scientist Development Award (to KG).
Version 1. 02/18/2020
In-Press Preview
Version 2. 03/12/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(5):e132306.https://doi.org/10.1172/jci.insight.132306.
Contributor Information
Aaron D. denDekker, Email: adendekk@med.umich.edu.
Frank M. Davis, Email: davisfr@umich.edu.
Amrita D. Joshi, Email: ajoshi@med.umich.edu.
Sonya J. Wolf, Email: sonwolf@med.umich.edu.
Ronald Allen, Email: allenrm@med.umich.edu.
Jay Lipinski, Email: jaylip@med.umich.edu.
Brenda Nguyen, Email: nbrenda@med.umich.edu.
Joseph Kirma, Email: jokirma@med.umich.edu.
Dylan Nycz, Email: nyczd@med.umich.edu.
Bethany B. Moore, Email: Bmoore@umich.edu.
Johann E. Gudjonsson, Email: johanng@med.umich.edu.
Steven L. Kunkel, Email: slkunkel@med.umich.edu.
Katherine A. Gallagher, Email: kgallag@med.umich.edu.
References
- 1.Diegelmann RF, Evans MC. Wound healing: an overview of acute, fibrotic and delayed healing. Front Biosci. 2004;9:283–289. doi: 10.2741/1184. [DOI] [PubMed] [Google Scholar]
- 2.Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44(3):450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front Immunol. 2014;5:514. doi: 10.3389/fimmu.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucas T, et al. Differential roles of macrophages in diverse phases of skin repair. J Immunol. 2010;184(7):3964–3977. doi: 10.4049/jimmunol.0903356. [DOI] [PubMed] [Google Scholar]
- 5.Dal-Secco D, et al. A dynamic spectrum of monocytes arising from the in situ reprogramming of CCR2+ monocytes at a site of sterile injury. J Exp Med. 2015;212(4):447–456. doi: 10.1084/jem.20141539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boniakowski AE, Kimball AS, Jacobs BN, Kunkel SL, Gallagher KA. Macrophage-mediated inflammation in normal and diabetic wound healing. J Immunol. 2017;199(1):17–24. doi: 10.4049/jimmunol.1700223. [DOI] [PubMed] [Google Scholar]
- 7.Mirza R, Koh TJ. Dysregulation of monocyte/macrophage phenotype in wounds of diabetic mice. Cytokine. 2011;56(2):256–264. doi: 10.1016/j.cyto.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirza RE, Fang MM, Ennis WJ, Koh TJ. Blocking interleukin-1β induces a healing-associated wound macrophage phenotype and improves healing in type 2 diabetes. Diabetes. 2013;62(7):2579–2587. doi: 10.2337/db12-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirza RE, Fang MM, Weinheimer-Haus EM, Ennis WJ, Koh TJ. Sustained inflammasome activity in macrophages impairs wound healing in type 2 diabetic humans and mice. Diabetes. 2014;63(3):1103–1114. doi: 10.2337/db13-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimball A, et al. Ly6CHi blood monocyte/macrophage drive chronic inflammation and impair wound healing in diabetes mellitus. Arterioscler Thromb Vasc Biol. 2018;38(5):1102–1114. doi: 10.1161/ATVBAHA.118.310703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher KA, et al. Epigenetic changes in bone marrow progenitor cells influence the inflammatory phenotype and alter wound healing in type 2 diabetes. Diabetes. 2015;64(4):1420–1430. doi: 10.2337/db14-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Cao Q, Yu L, Shi H, Xue B, Shi H. Epigenetic regulation of macrophage polarization and inflammation by DNA methylation in obesity. JCI Insight. 2016;1(19):e87748. doi: 10.1172/jci.insight.87748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimball AS, et al. The histone methyltransferase MLL1 directs macrophage-mediated inflammation in wound healing and is altered in a murine model of obesity and type 2 diabetes. Diabetes. 2017;66(9):2459–2471. doi: 10.2337/db17-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan J, et al. Diabetes impairs wound healing by Dnmt1-dependent dysregulation of hematopoietic stem cells differentiation towards macrophages. Nat Commun. 2018;9(1):33. doi: 10.1038/s41467-017-02425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis FM, et al. Histone methylation directs myeloid TLR4 expression and regulates wound healing following cutaneous tissue injury. J Immunol. 2019;202(6):1777–1785. doi: 10.4049/jimmunol.1801258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5(2):367–375. doi: 10.1016/S1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 18.Rea S, Xouri G, Akhtar A. Males absent on the first (MOF): from flies to humans. Oncogene. 2007;26(37):5385–5394. doi: 10.1038/sj.onc.1210607. [DOI] [PubMed] [Google Scholar]
- 19.Taipale M, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25(15):6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buscaino A, et al. MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol Cell. 2003;11(5):1265–1277. doi: 10.1016/S1097-2765(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 21.Chelmicki T, et al. MOF-associated complexes ensure stem cell identity and Xist repression. Elife. 2014;3:e02024. doi: 10.7554/eLife.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee A, et al. MOF acetyl transferase regulates transcription and respiration in mitochondria. Cell. 2016;167(3):722–738.e23. doi: 10.1016/j.cell.2016.09.052. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, et al. The histone acetylranseferase hMOF acetylates Nrf2 and regulates anti-drug responses in human non-small cell lung cancer. Br J Pharmacol. 2014;171(13):3196–3211. doi: 10.1111/bph.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Füllgrabe J, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500(7463):468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta A, et al. MOF phosphorylation by ATM regulates 53BP1-mediated double-strand break repair pathway choice. Cell Rep. 2014;8(1):177–189. doi: 10.1016/j.celrep.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, et al. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 2010;30(22):5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfister S, et al. The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma. Int J Cancer. 2008;122(6):1207–1213. doi: 10.1002/ijc.23283. [DOI] [PubMed] [Google Scholar]
- 28.Li X, et al. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11(2):163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma GG, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30(14):3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas T, Dixon MP, Kueh AJ, Voss AK. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol. 2008;28(16):5093–5105. doi: 10.1128/MCB.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 32.Huai W, et al. KAT8 selectively inhibits antiviral immunity by acetylating IRF3. J Exp Med. 2019;216(4):772–785. doi: 10.1084/jem.20181773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai Y, et al. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem. 2010;285(7):4268–4272. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell. 2009;36(2):290–301. doi: 10.1016/j.molcel.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morales V, Straub T, Neumann MF, Mengus G, Akhtar A, Becker PB. Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J. 2004;23(11):2258–2268. doi: 10.1038/sj.emboj.7600235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y, Schmitz KM, Mayer C, Yuan X, Akhtar A, Grummt I. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat Cell Biol. 2009;11(8):1010–1016. doi: 10.1038/ncb1914. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Dou Y. New perspectives for the regulation of acetyltransferase MOF. Epigenetics. 2010;5(3):185–188. doi: 10.4161/epi.5.3.11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaganathan A, et al. Coactivator MYST1 regulates nuclear factor-κB and androgen receptor functions during proliferation of prostate cancer cells. Mol Endocrinol. 2014;28(6):872–885. doi: 10.1210/me.2014-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1(4):a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dixit VD, et al. Ghrelin inhibits leptin- and activation-induced proinflammatory cytokine expression by human monocytes and T cells. J Clin Invest. 2004;114(1):57–66. doi: 10.1172/JCI21134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lam QL, Lu L. Role of leptin in immunity. Cell Mol Immunol. 2007;4(1):1–13. [PubMed] [Google Scholar]
- 43.Tartaglia LA, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83(7):1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 44.Hui X, et al. Adipocyte SIRT1 controls systemic insulin sensitivity by modulating macrophages in adipose tissue. EMBO Rep. 2017;18(4):645–657. doi: 10.15252/embr.201643184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshizaki T, et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;298(3):E419–E428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillum MP, et al. SirT1 regulates adipose tissue inflammation. Diabetes. 2011;60(12):3235–3245. doi: 10.2337/db11-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu L, Li L, Lv X, Wu XS, Liu DP, Liang CC. Modulations of hMOF autoacetylation by SIRT1 regulate hMOF recruitment and activities on the chromatin. Cell Res. 2011;21(8):1182–1195. doi: 10.1038/cr.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spallotta F, et al. Enhancement of lysine acetylation accelerates wound repair. Commun Integr Biol. 2013;6(5):e25466. doi: 10.4161/cib.25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spallotta F, et al. A nitric oxide-dependent cross-talk between class I and III histone deacetylases accelerates skin repair. J Biol Chem. 2013;288(16):11004–11012. doi: 10.1074/jbc.M112.441816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Usui ML, Mansbridge JN, Carter WG, Fujita M, Olerud JE. Keratinocyte migration, proliferation, and differentiation in chronic ulcers from patients with diabetes and normal wounds. J Histochem Cytochem. 2008;56(7):687–696. doi: 10.1369/jhc.2008.951194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Werner S, Krieg T, Smola H. Keratinocyte-fibroblast interactions in wound healing. J Invest Dermatol. 2007;127(5):998–1008. doi: 10.1038/sj.jid.5700786. [DOI] [PubMed] [Google Scholar]
- 52.Luo H, et al. MOF Acetylates the histone demethylase LSD1 to suppress epithelial-to-mesenchymal transition. Cell Rep. 2016;15(12):2665–2678. doi: 10.1016/j.celrep.2016.05.050. [DOI] [PubMed] [Google Scholar]
- 53.Sykes SM, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24(6):841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kimball AS, et al. The histone methyltransferase Setdb2 modulates macrophage phenotype and uric acid production in diabetic wound repair. Immunity. 2019;51(2):258–271.e5. doi: 10.1016/j.immuni.2019.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Surwit RS, Kuhn CM, Cochrane C, McCubbin JA, Feinglos MN. Diet-induced type II diabetes in C57BL/6J mice. Diabetes. 1988;37(9):1163–1167. doi: 10.2337/diab.37.9.1163. [DOI] [PubMed] [Google Scholar]
- 56.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boniakowski AM, et al. SIRT3 regulates macrophage-mediated inflammation in diabetic wound repair. J Invest Dermatol. 2019;139(12):2528–2537.e2. doi: 10.1016/j.jid.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boniakowski AE, et al. Murine macrophage chemokine receptor CCR2 plays a crucial role in macrophage recruitment and regulated inflammation in wound healing. Eur J Immunol. 2018;48(9):1445–1455. doi: 10.1002/eji.201747400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimball AS, et al. Notch regulates macrophage-mediated inflammation in diabetic wound healing. Front Immunol. 2017;8:635. doi: 10.3389/fimmu.2017.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishii M, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114(15):3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.