Abstract
Maintaining cellular proteostasis is essential for oligodendrocyte viability and function; however, its underlying mechanisms remain unexplored. Unfolded protein response (UPR), which comprises 3 parallel branches, inositol requiring enzyme 1 (IRE1), pancreatic ER kinase (PERK), and activating transcription factor 6α (ATF6α), is a major mechanism that maintains cellular proteostasis by facilitating protein folding, attenuating protein translation, and enhancing autophagy and ER-associated degradation. Here we report that impaired UPR in oligodendrocytes via deletion of PERK and ATF6α did not affect developmental myelination but caused late-onset mature oligodendrocyte dysfunction and death in young adult mice. The detrimental effects of the impaired UPR on mature oligodendrocytes were accompanied by autophagy impairment and intracellular proteolipid protein (PLP) accumulation and were rescued by PLP deletion. Data indicate that PLP was degraded by autophagy and that intracellular PLP accumulation was cytotoxic to oligodendrocytes. Thus, these findings imply that the UPR is required for maintaining cellular proteostasis and the viability and function of mature oligodendrocytes in adults by regulating autophagy of PLP.
Keywords: Cell Biology, Neuroscience
Keywords: Cell stress, Demyelinating disorders
Impaired unfolded protein response in oligodendrocytes causes late-onset demyelination in young adult mice via autophagy impairment and intracellular proteolipid protein accumulation.
Introduction
The endoplasmic reticulum (ER) is the site in eukaryotic cells in which modification and folding of membrane and secretory proteins take place (1, 2). Properly folded proteins are transported to their destinations through the cellular secretory pathway. Perturbations in protein modification or folding lead to accumulation of unfolded or misfolded proteins in the ER and result in ER stress and activation of the unfolded protein response (UPR), which comprises 3 parallel branches: inositol requiring enzyme 1 (IRE1), pancreatic ER kinase (PERK), and activating transcription factor 6α (ATF6α) (3–5). IRE1 activation increases the expression of genes that enhance protein folding and protein degradation by splicing X-box binding protein 1 (XBP1) mRNA. PERK activation inhibits protein translation but stimulates the expression of genes related to macroautophagy (a bulk protein degradation process, hereafter referred to as autophagy) and genes related to ER-associated degradation (ERAD) by phosphorylating eukaryotic translation initiation factor 2α. ATF6α activation enhances the expression of ER chaperones, autophagy-related genes, and ERAD-related genes. The UPR is a major mechanism that maintains ER protein homeostasis and a major component of the cellular proteostasis network that preserves cellular proteostasis (4, 6, 7).
Myelin is an enormous membrane structure that wraps and insulates axons. Oligodendrocytes, the myelin-producing cells in the central nervous system (CNS), must produce a vast number of membrane proteins via the ER to assemble and maintain the structure of myelin sheaths (2, 8, 9). The current dogma is that maintaining ER protein homeostasis is essential for the production of myelin proteins and the myelinating function of oligodendrocytes (2, 10, 11). It is well documented that oligodendrocytes are highly sensitive to disruption of ER protein homeostasis and that this increased susceptibility contributes to the pathogenesis of various myelin disorders (2, 10–12). A number of studies have demonstrated that the PERK branch of the UPR is a major player in regulating the viability and function of oligodendrocytes in myelin disorders (13–15). A recent study also suggests cytoprotective effects of the ATF6α branch on oligodendrocytes in immune-mediated demyelinating diseases (16). Nevertheless, it remains unknown whether the UPR is involved in maintaining ER protein homeostasis and cellular proteostasis in oligodendrocytes, or their viability and function, under physiological conditions.
Although the IRE1 branch is regarded as the most conserved branch of the UPR, there is no evidence that this branch plays a major role in oligodendrocytes (12, 17, 18). Conversely, evidence suggests that PERK and ATF6α are activated in oligodendrocytes under normal and disease conditions (12, 18, 19). However, deletion of either PERK or ATF6α has no effect on oligodendrocytes in mice under physiological conditions (15, 16). The minimal effect of PERK deletion or ATF6α deletion on oligodendrocytes is likely due to the functional redundancy of these 2 branches, as both of them enhance autophagy and ERAD. Thus, in this study, we determined the physiological role of the UPR in oligodendrocytes by generating a mouse model that allows for inactivation of both PERK and ATF6α in oligodendrocytes. Intriguingly, we found that inactivation of PERK and ATF6α in oligodendrocytes led to late-onset demyelination in the CNS of young adult mice. We also found that inactivation of PERK and ATF6α caused impairment of the autophagy/lysosome pathway and soma retention of proteolipid protein (PLP) in mature oligodendrocytes of young adult mice. It is known that soma retention of PLP in oligodendrocytes is a major contributor to dysmyelination or late-onset demyelination in the CNS in patients with Pelizaeus-Merzbacher disease (PMD) and its animal models that carry increased copies of Plp1 gene and that the autophagy/lysosome pathway is one of the major mechanisms responsible for PLP degradation in oligodendrocytes (20–23). Importantly, we showed that PLP deficiency attenuated myelin abnormalities in the CNS of young adult mice with double deletion of PERK and ATF6α in oligodendrocytes. Collectively, our results suggest that impaired UPR leads to impairment of the autophagy/lysosome pathway and soma retention of PLP in mature oligodendrocytes, resulting in late-onset demyelination in the CNS of young adult mice.
Results
Inactivation of PERK and ATF6α in oligodendrocytes caused late-onset demyelination in young adult mice.
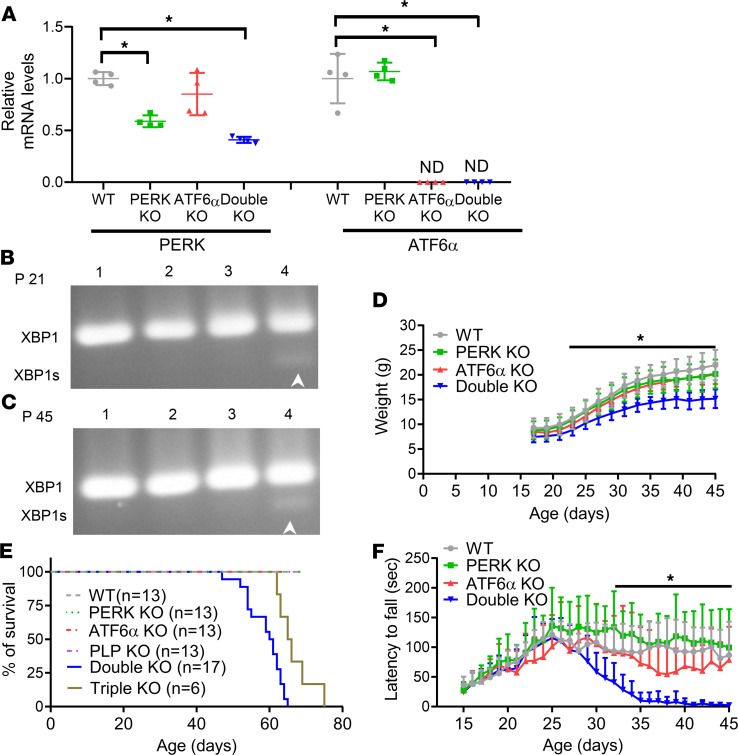
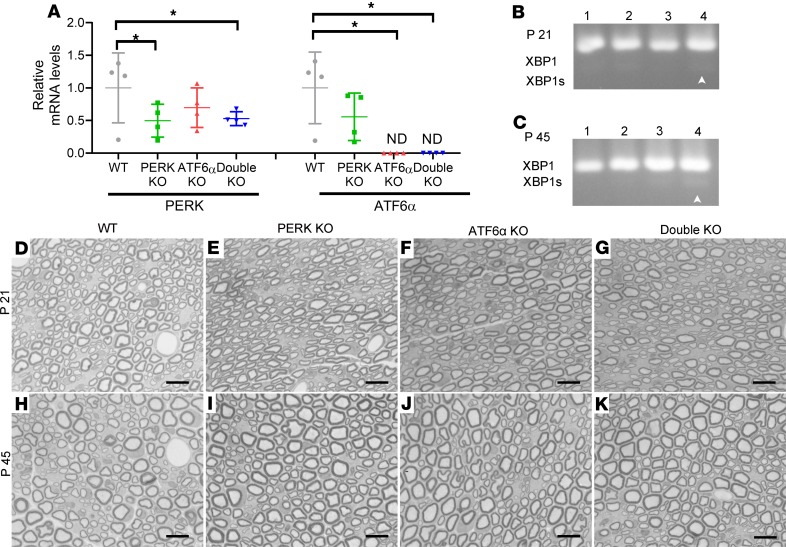
Active developmental myelination occurs in the rodent CNS between P7 and P21 (24). Currently available data indicate that actively myelinating oligodendrocytes during this stage have a much higher rate of myelin protein production and are more susceptible to disruption of ER protein homeostasis than mature oligodendrocytes in adults (2, 8, 10). Several studies show that ATF6α homozygous-knockout (ATF6α-KO) mice appear healthy and exhibit normal myelination in the CNS (16, 25). A previous study demonstrated that PERK is deleted specifically in oligodendrocytes in the CNS of PERKloxP/loxP CNP/Cre (PERK-KO) mice and that PERK inactivation has no effect on oligodendrocytes or myelin in these mice (15). To determine the physiological role of the UPR in oligodendrocytes, we crossed ATF6α-KO mice with PERK-KO mice to generate mice with double deletion of PERK and ATF6α in oligodendrocytes (double-KO mice), PERK-KO mice, ATF6α-KO mice, and corresponding control mice (WT mice). We performed real-time PCR to verify the deletion of PERK and ATF6α in oligodendrocytes of double-KO mice using the optic nerve because approximately 60% of cells in the optic nerve are oligodendrocytes (26, 27). As expected, we found that the mRNA levels of PERK and ATF6α were significantly reduced in the optic nerve of double-KO mice compared with WT mice (Figure 1A). Moreover, we determined whether inactivation of PERK and ATF6α disrupts ER protein homeostasis in oligodendrocytes in young developing and/or adult mice. PCR analysis showed that the spliced XBP1 (XBP1s) mRNA was undetectable in the spinal cord of WT mice, PERK-KO mice, and ATF6α-KO mice but became moderately detectable in double-KO mice at P21 and P45 (Figure 1, B and C). Moderate splicing of XBP1 mRNA suggests moderate disruption of ER protein homeostasis and modest activation of ER stress in oligodendrocytes of double-KO mice. Thus, these results suggest that inactivation of PERK and ATF6α moderately disrupts ER protein homeostasis in both actively myelinating oligodendrocytes in young developing mice and in mature oligodendrocytes in young adult mice.
Figure 1. Mice with double deletion of PERK and ATF6α in oligodendrocytes developed a late-onset and progressive tremoring phenotype.
(A) Real-time PCR analysis showed that the mRNA level of PERK was significantly reduced in the optic nerve of PERK-KO mice and double-KO mice compared with WT mice and ATF6α-KO mice and that ATF6α mRNA was undetectable in the optic nerve of ATF6α-KO mice and double-KO mice (N = 4 animals). (B and C) PCR analysis showed that XBP1s was undetectable in the spinal cord of WT mice (lane 1), PERK-KO mice (lane 2), and ATF6α-KO mice (lane 3) at P21 and P45 but became moderately detectable in double-KO mice (lane 4) at P21 and P45 (N = 4 animals). (D) Double-KO mice started to show lighter body weight at P23 compared with WT mice, PERK-KO mice, and ATF6α-KO mice (N = 10 animals). (E) Mouse survival curve. (F) Latency to fall in Rotarod test (N = 10 animals). ND, not detected. Error bars represent mean ± SD. Statistical analyses were done with a 1-way ANOVA with Tukey’s posttest (A) or a 2- way ANOVA with Tukey’s posttest (D and F); *P < 0.05.
As expected, both PERK-KO mice and ATF6α-KO mice looked healthy and survived well. Surprisingly, double-KO mice were asymptomatic and their body weight was comparable to WT mice, PERK-KO mice, and ATF6α-KO mice during the active developmental myelination stage. Interestingly, double-KO mice gained body weight at a reduced rate compared with WT mice, PERK-KO mice, and ATF6α-KO mice starting at P23 (Figure 1D) and displayed a tremoring phenotype starting around P28. All double-KO mice showed severe tremor by P45 and died by P65 (Figure 1E). Similarly, Rotarod test showed that double-KO mice displayed normal motor coordination compared with WT mice, PERK-KO mice, and ATF6α-KO mice before P28; however, motor coordination of double-KO mice gradually declined after P28 and became severely impaired after P35 (Figure 1F). These results demonstrate that inactivation of PERK and ATF6α in oligodendrocytes induces a late-onset and progressive tremoring phenotype in mice.
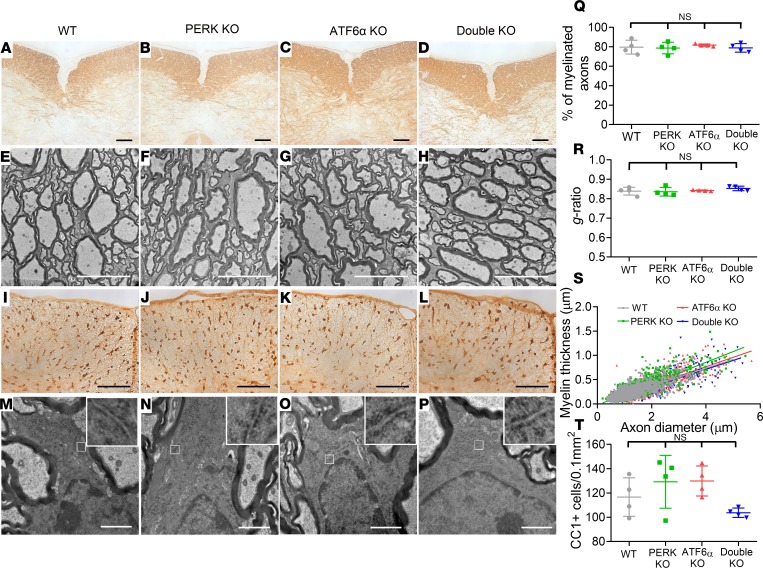
A late-onset tremoring phenotype displayed by double-KO mice implies myelin abnormalities in the CNS of these mice. We first determined whether inactivation of PERK and ATF6α in oligodendrocytes affects developmental myelination. Myelin basic protein (MBP) immunohistochemistry (IHC), CC1 (an oligodendrocyte marker) IHC, and electron microscopy (EM) analysis showed a comparable degree of myelination and a comparable number of oligodendrocytes in the CNS of WT mice, PERK-KO mice, and ATF6α-KO mice at P21 (Figure 2, A–C, E–G, I–K, Q–T; and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.132364DS1). Unexpectedly, MBP IHC showed that inactivation of PERK and ATF6α in oligodendrocytes did not alter the degree of myelination in the CNS at P21 (Figure 2, A–D; and Supplemental Figure 1). EM analysis confirmed that inactivation of PERK and ATF6α in oligodendrocytes did not alter the percentage of myelinated axons, the thickness of myelin, or the g-ratio in the CNS at P21 (Figure 2, E–H and Q–S). CC1 IHC showed that inactivation of PERK and ATF6α in oligodendrocytes did not change oligodendrocyte numbers in the CNS at P21 (Figure 2, I–L and T; and Supplemental Figure 1). Moreover, EM analysis showed that both the morphology of oligodendrocytes and the morphology of the ER in these cells in double-KO mice were indistinguishable from those of WT mice, PERK-KO mice, and ATF6α-KO mice at P21 (Figure 2, M–P). Taken together, these data suggest that the inactivation of PERK and ATF6α has a minimal effect on the viability and function of actively myelinating oligodendrocytes during developmental myelination.
Figure 2. Inactivation of PERK and ATF6α in oligodendrocytes did not affect developmental myelination in the CNS (P21).
(A–D) MBP IHC showed that the degree of myelination in the spinal cord of double-KO mice was comparable to WT mice, PERK-KO mice, and ATF6α-KO mice at P21. (E–H and Q–S) EM analysis showed that the percentage of myelinated axons, the thickness of myelin, and the g-ratio were not altered in the spinal cord of double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice at P21. (I–L and T) CC1 IHC showed a comparable number of oligodendrocytes in the spinal cord of WT mice, PERK-KO mice, ATF6α-KO mice, and double-KO mice at P21. (M–P) EM analysis showed that the morphology of oligodendrocytes and the ER morphology (insets) in oligodendrocytes appeared normal in double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice at P21. Scale bars: 100 μm (A–D, I–L), 5 μm (E–H), 1 μm (M–P). N = 4 animals. Error bars represent mean ± SD. Statistical analyses were done with a 1-way ANOVA with Tukey’s posttest. NS, not significant.
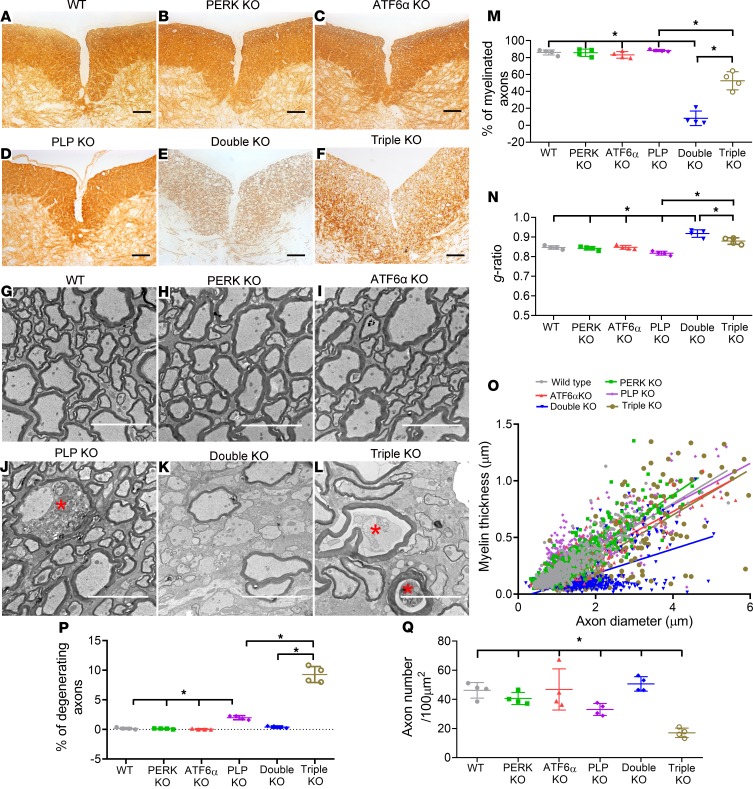
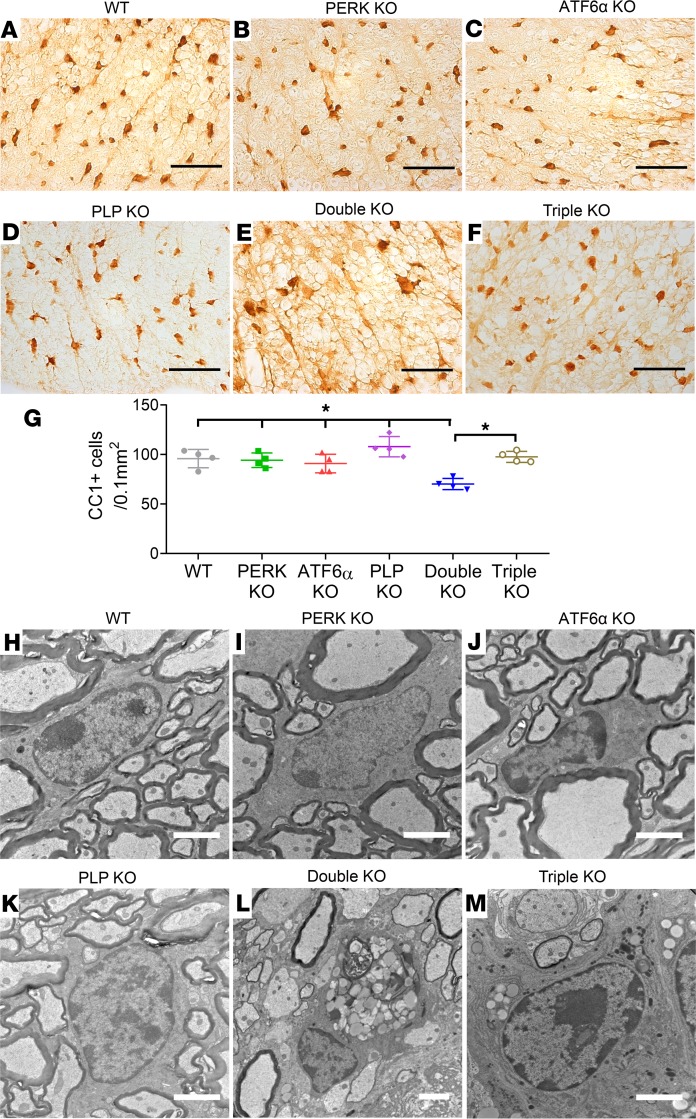
We further determined whether inactivation of PERK and ATF6α in oligodendrocytes leads to late-onset demyelination in the CNS. MBP IHC, CC1 IHC, and EM analysis showed that the degree of myelination and the number of oligodendrocytes were comparable in the CNS of WT mice, PERK-KO mice, and ATF6α-KO mice at P45 (Figure 3, A–C, G–I, and M–O; Figure 4, A–C and G; and Supplemental Figure 2). Importantly, MBP IHC showed severe myelin loss in the CNS of double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice at P45 (Figure 3, A–C and E; and Supplemental Figure 2). EM analysis confirmed that inactivation of PERK and ATF6α in oligodendrocytes significantly reduced the percentage of myelinated axons and the thickness of myelin and increased the g-ratio in the CNS at P45 (Figure 3, G–I, K, and M–O). CC1 IHC showed that the number of oligodendrocytes was significantly reduced in the CNS of double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice at P45 (Figure 4, A–C, E, and G; and Supplemental Figure 2). Notably, the soma of oligodendrocytes in double-KO mice appeared markedly larger than WT mice, PERK-KO mice, and ATF6α-KO mice at P45 (Figure 4, A–C and E; and Supplemental Figure 2). Moreover, CC1 and cleaved caspase-3 (a marker of apoptosis) double immunostaining showed that none of CC1-positive oligodendrocytes was positive for cleaved caspase-3 in the spinal cord of WT mice, PERK-KO mice, and ATF6α-KO mice at P45; however, there were a few cleaved caspase-3–positive oligodendrocytes in double-KO mice (Supplemental Figure 3). Interestingly, EM analysis showed accumulation of vacuolar structures that are morphologically reminiscent of autophagic vacuoles in oligodendrocytes of double-KO mice at P45, which were absent from WT mice, PERK-KO mice, and ATF6α-KO mice (Figure 4, H–J and L), suggesting that the enlarged soma of oligodendrocytes results from the accumulation of autophagic vacuoles in these cells. Thus, these data demonstrate that inactivation of PERK and ATF6α leads to dysfunction and death of mature oligodendrocytes in young adult mice.
Figure 3. Inactivation of PERK and ATF6α in oligodendrocytes led to late-onset demyelination in the CNS, and PLP deficiency attenuated myelin loss in the CNS of double-KO mice (P45).
(A–F) MBP IHC revealed severe myelin loss in the spinal cord of double-KO mice compared with WT mice, PERK-KO mice, ATF6α-KO mice, and PLP-KO mice at P45. Interestingly, myelin loss was noticeably attenuated in the spinal cord of triple-KO mice compared with double-KO mice at P45. (G–O) EM analysis revealed a dramatic reduction in myelinated axons, a significant increase in the g-ratio, and a significant reduction in myelin thickness in the spinal cord of double-KO mice compared with WT mice, PERK-KO mice, ATF6α-KO mice, and PLP-KO mice at P45. Interestingly, there was a significant increase in myelinated axons, a significant reduction in the g-ratio, and a significant increase in myelin thickness in the spinal cord of triple-KO mice compared with double-KO mice at P45. (G–L, P, and Q) EM analysis revealed a few degenerating axons (red asterisk) in the spinal cord of PLP-KO mice at P45 compared with WT mice, PERK-KO mice, and ATF6α-KO mice at P45. Importantly, there was a significant increase in degenerating axons and a significant reduction in healthy-looking axons in the spinal cord of triple-KO mice compared with PLP-KO mice and double-KO mice at P45. Scale bars: 100 μm (A–F), 5 μm (G–L); N = 4 animals. Error bars represent mean ± SD. Statistical analyses were done with a 1-way ANOVA with Tukey’s posttest; *P < 0.05.
Figure 4. Inactivation of PERK and ATF6α in oligodendrocytes led to late-onset oligodendrocyte loss in the CNS, and PLP deficiency attenuated oligodendrocyte loss in the CNS of double-KO mice (P45).
(A–G) CC1 IHC showed that the number of oligodendrocytes was significantly reduced in the spinal cord of double-KO mice compared with WT mice, PERK-KO mice, ATF6α-KO mice, and PLP-KO mice at P45; however, the soma of oligodendrocytes was markedly enlarged in double-KO mice. Interestingly, oligodendrocyte loss was significantly attenuated in the spinal cord of triple-KO mice compared with double-KO mice at P45. (H–M) EM analysis revealed accumulation of autophagic vacuoles in oligodendrocytes in the spinal cord of double-KO mice that were absent in WT mice, PERK-KO mice, ATF6α-KO mice, and PLP-KO mice at P45. There was still accumulation of autophagic vacuoles in oligodendrocytes of triple-KO mice at P45. Scale bars: 50 μm (A–F), 2 μm (H–M). N = 4 animals. Error bars represent mean ± SD. Statistical analyses were done with a 1-way ANOVA with Tukey’s posttest; *P < 0.05.
It is known that the promoter of the 2′,3′ cyclic nucleotide 3′-phosphodiesterase (CNP) gene drives Cre recombinase expression in both oligodendrocytes in the CNS and in Schwann cells in the peripheral nervous system (PNS) in CNP/Cre mice starting at early developmental myelination (28, 29). Next, we assessed myelin integrity in the PNS of double-KO mice. Real-time PCR analysis showed that the mRNA levels of PERK and ATF6α were significantly reduced in the sciatic nerve of double-KO mice compared with WT mice (Figure 5A). PCR analysis showed that the XBP1s mRNA was undetectable in the sciatic nerve of WT mice, PERK-KO mice, and ATF6α-KO mice but became modestly detectable in double-KO mice at P21 and P45 (Figure 5, B and C). Nevertheless, toluidine blue staining showed that the degree of myelination in the sciatic nerve of double-KO mice was comparable to WT mice, PERK-KO mice, and ATF6α-KO mice at P21 and P45 (Figure 5, D–K). Thus, these data demonstrate that inactivation of PERK and ATF6α modestly disrupts ER protein homeostasis in Schwann cells but does not affect their viability or function under normal conditions.
Figure 5. Mice with double deletion of PERK and ATF6α in Schwann cells showed normal myelination in the PNS.
(A) Real-time PCR analysis showed that the mRNA level of PERK was significantly reduced in the sciatic nerve of PERK-KO mice and double-KO mice compared with WT mice and that ATF6α mRNA was undetectable in the sciatic nerve of ATF6α-KO mice and double-KO mice. (B and C) PCR analysis showed that XBP1s was undetectable in the sciatic nerve of WT mice (lane 1), PERK-KO mice (lane 2), and ATF6α-KO mice (lane 3) at P21 and P45 but became modestly detectable in double-KO mice (lane 4) at P21 and P45. (D–G) Toluidine blue staining showed normal myelination in the sciatic nerve of 21-day-old double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice. (H–K) Toluidine blue staining showed normal myelination in the sciatic nerve of 45-day-old double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice. Scale bars: 10 μm. N = 4 animals. ND, not detected. Error bars represent mean ± SD. Statistical analyses were done with a 1-way ANOVA with Tukey’s posttest; *P < 0.05.
Inactivation of PERK and ATF6α led to impairment of the autophagy/lysosome pathway in mature oligodendrocytes of young adult mice.
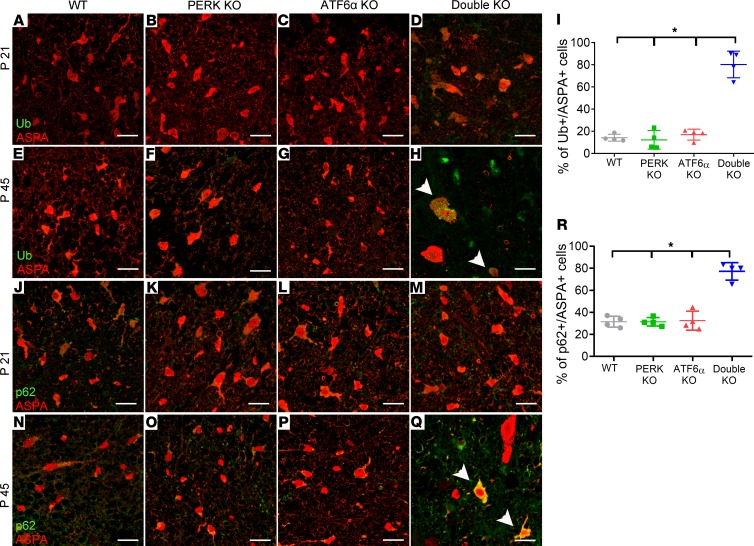
Evidence suggests that inactivation of PERK or ATF6α causes autophagy defects in ER-stressed cells (30–32). Accumulation of autophagic vacuoles in mature oligodendrocytes of 45-day-old double-KO mice indicates that inactivation of PERK and ATF6α caused defects in the autophagy/lysosome pathway in these cells. It is known that defects of autophagic flux cause intracellular accumulation of p62 (sequestosome-1, SQSTM1) and ubiquitinated proteins (33–35). An autophagic cargo adapter, p62 can bind to ubiquitinated proteins as well as autophagosome membrane proteins and deliver ubiquitinated protein aggregates for autophagy (36). p62 itself is also degraded by autophagy. Ubiquitin and aspartoacylase (ASPA, an oligodendrocyte marker) double immunostaining showed that the immunoreactivity of ubiquitin was barely detectable in oligodendrocytes of 21- or 45-day-old WT mice, PERK-KO mice, and ATF6α-KO mice (Figure 6, A–C and E–G). Similarly, p62 and ASPA double immunostaining showed that the immunoreactivity of p62 was barely detectable in oligodendrocytes of 21- or 45-day-old WT mice, PERK-KO mice, and ATF6α-KO mice (Figure 6, J–L and N–P). There was no accumulation of ubiquitinated proteins or p62 in oligodendrocytes of 21-day-old double-KO mice (Figure 6, A–D and J–M). Importantly, ubiquitin and ASPA double immunostaining showed that the immunoreactivity of ubiquitin was markedly increased in oligodendrocytes in the CNS of 45-day-old double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice (Figure 6, E–I). Similarly, p62 and ASPA double immunostaining showed that the immunoreactivity of p62 was markedly increased in oligodendrocytes in the CNS of 45-day-old double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice (Figure 6, N–R). The elevated levels of p62 and ubiquitinated proteins in oligodendrocytes of 45-day-old double-KO mice provide further evidence that inactivation of PERK and ATF6α impairs autophagic flux in mature oligodendrocytes of young adult mice.
Figure 6. Inactivation of PERK and ATF6α led to accumulation of ubiquitinated proteins and p62 in mature oligodendrocytes at P45.
(A–D) ASPA and ubiquitin (Ub) double immunostaining revealed little immunoreactivity of ubiquitin in oligodendrocytes in the spinal cord of WT mice, PERK-KO mice, ATF6α-KO mice, and double-KO mice at P21. (E–I) ASPA and ubiquitin double immunostaining showed that both the intensity of ubiquitin immunoreactivity in oligodendrocytes (arrowheads) and the number of oligodendrocytes positive for ubiquitin were significantly increased in the spinal cord of double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice at P45. (J–M) ASPA and p62 double immunostaining revealed little immunoreactivity of p62 in the oligodendrocytes in the spinal cord of WT mice, PERK-KO mice, ATF6α-KO mice, and double-KO mice at P21. (N–R) ASPA and p62 double immunostaining showed that both the intensity of p62 immunoreactivity in oligodendrocytes (arrowheads) and the number of oligodendrocytes positive for p62 were significantly increased in the spinal cord of double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice at P45. Scale bars: 20 μm. N = 4 animals. Error bars represent mean ± SD. Statistical analyses were done with a 1-way ANOVA with Tukey’s posttest; *P < 0.05.
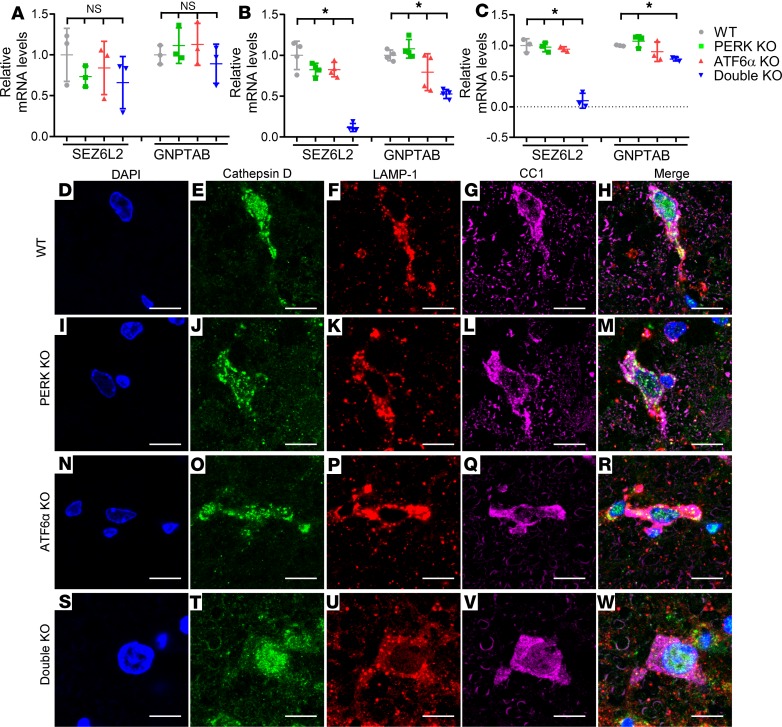
Data indicate that activation of PERK or ATF6α enhances the expression of genes related to the autophagy/lysosome pathway (37–39). Interestingly, real-time PCR analysis showed that inactivation of PERK and ATF6α reduced the expression of 2 genes related to the autophagy/lysosome pathway, SEZ6L2 (seizure related 6 homolog like 2) and GNPTAB (encoding the α and β subunits of GlcNAc-1-phosphotransferase). The mRNA levels of SEZ6L2 were significantly decreased in the optic nerve of double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice at P28 and P45 but not at P21 (Figure 7, A–C). Furthermore, the mRNA levels of GNPTAB were significantly decreased in the optic nerve of double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice at P28, and in the optic nerve of double-KO mice compared with WT mice and PERK-KO mice at P45, but were not decreased at P21. Both SEZ6L2 and GNPTAB participate in the transport of the lysosomal hydrolase cathepsin D from the trans-Golgi network to the lysosome (40–43). We further assessed the subcellular localization of cathepsin D in oligodendrocytes of these mice. As expected, CC1, lysosome-associated membrane protein 1 (LAMP1), and cathepsin D triple immunostaining showed that a substantial amount of cathepsin D was colocalized with the lysosome marker LAMP1 in CC1-positive oligodendrocytes of 45-day-old WT mice, PERK-KO mice, and ATF6α-KO mice (Figure 7, D–R). In contrast, the amount of cathepsin D that was colocalized with LAMP1 in mature oligodendrocytes of 45-day-old double-KO mice was minimal (Figure 7, S–W). These results suggest that the attenuated expression of SEZ612 and GNPTAB disrupts the transport of cathepsin D from the trans-Golgi network to the lysosome and results in mislocalization of cathepsin D in mature oligodendrocytes of young adult double-KO mice. Cathepsin D is a major lysosomal protease responsible for degradation of proteins delivered to the lysosomes via autophagy (41, 44). It is well documented that inactivation or mislocalization of cathepsin D causes lysosome dysfunction and impairment of autophagic flux (44–46). Taken together, these data suggest that inactivation of PERK and ATF6α leads to delocalization of cathepsin D from the lysosome by attenuating the expression of SEZ6L2 and GNPTAB and subsequently results in impairment of the autophagy/lysosome pathway in mature oligodendrocytes of young adult mice.
Figure 7. Inactivation of PERK and ATF6α caused delocalization of cathepsin D from the lysosome in mature oligodendrocytes at P45.
(A) Real-time PCR analysis showed that the levels of SEZ6L2 and GNPTAB were comparable in the optic nerve of WT mice, PERK-KO mice, ATF6α-KO mice, and double-KO mice at P21. N = 3 animals. (B) Real-time PCR analysis showed that the levels of SEZ6L2 and GNPTAB were significantly reduced in the optic nerve of double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice at P28. N = 4 animals. (C) Real-time PCR analysis showed that the level of SEZ6L2 was significantly reduced in the optic nerve of double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice and that the level of GNPTAB was significantly reduced in the optic nerve of double-KO mice compared with WT mice and PERK-KO mice at P45. N = 4 animals. (D–W) CC1, LAMP1, and cathepsin D triple labeling revealed significant colocalization of LAMP1 and cathepsin D in oligodendrocytes of WT mice, PERK-KO mice, and ATF6α-KO mice at P45. Conversely, there was little to no colocalization of LAMP1 and cathepsin D in oligodendrocytes of double-KO mice. N = 4 animals. Scale bars: 10 μm. Error bars represent mean ± SD. Statistical analyses were done with a 1-way ANOVA with Tukey’s posttest; *P < 0.05.
Inactivation of PERK and ATF6α led to soma retention of PLP in mature oligodendrocytes of young adult mice.
PLP, the most abundant myelin protein in the CNS, is synthesized in the ER and transported to myelin sheaths (21). The autophagy/lysosome pathway is a major mechanism responsible for PLP degradation in oligodendrocytes (20, 22). It is well documented that overexpression of PLP induced by increased copies of the Plp1 gene causes accumulation of this protein in the soma of oligodendrocytes and results in dysmyelination or late-onset demyelination in the CNS of human patients with PMD and its animal models (20–23). Interestingly, a number of studies show that overexpression of PLP leads to accumulation of this protein in autophagic vacuoles in oligodendrocytes of mice carrying increased copies of the Plp1 gene (20, 22, 47). We showed here that inactivation of PERK and ATF6α led to impairment of autophagic flux in mature oligodendrocytes and late-onset demyelination in young adult mice. Thus, these data raise the possibility that inactivation of PERK and ATF6α attenuates PLP degradation through impairment of the autophagy/lysosome pathway, resulting in soma retention of PLP in mature oligodendrocytes and subsequent late-onset demyelination in the CNS of young adult mice.
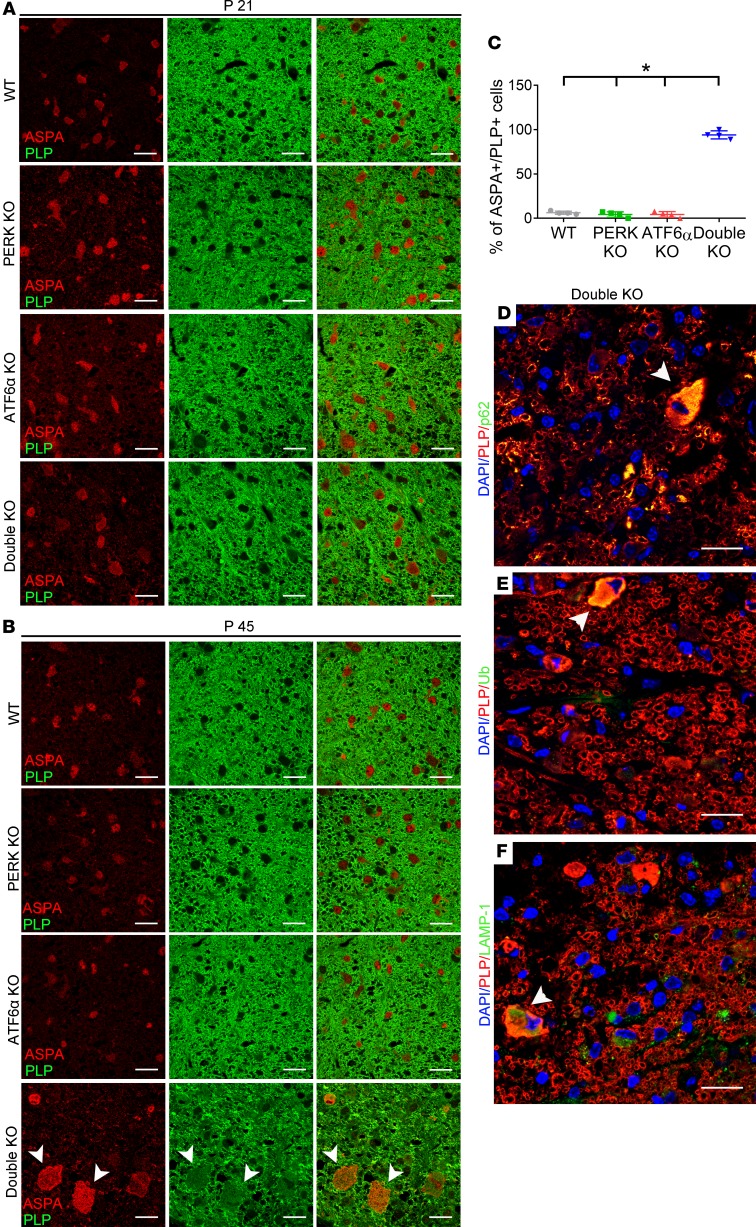
We first determined whether inactivation of PERK and ATF6α leads to soma retention of PLP in oligodendrocytes. PLP and ASPA double immunostaining showed strong PLP immunoreactivity in myelinating tracts and little PLP immunoreactivity in the soma of oligodendrocytes in the CNS of 21- or 45-day-old WT mice, PERK-KO mice, and ATF6α-KO mice (Figure 8, A and B). The distribution pattern of PLP in the CNS of 21-day-old double-KO mice was comparable to WT mice, PERK-KO mice, and ATF6α-KO mice (Figure 8A). Conversely, we found that immunoreactivity of PLP was noticeably decreased in myelinated tracts but noticeably increased in the soma of oligodendrocytes in the CNS of 45-day-old double-KO mice compared with WT mice, PERK-KO mice, and ATF6α-KO mice (Figure 8, B and C). These results suggest that inactivation of PERK and ATF6α does not affect PLP homeostasis in actively myelinating oligodendrocytes of young developing mice but leads to soma retention of PLP in mature oligodendrocytes of young adult mice.
Figure 8. Inactivation of PERK and ATF6α led to soma retention of PLP in mature oligodendrocytes at P45.
(A) ASPA and PLP double immunostaining showed that PLP immunoreactivity was undetectable in the soma of oligodendrocytes in the spinal cord of WT mice, PERK mice-KO, ATF6α-KO mice, and double-KO mice at P21. (B and C) ASPA and PLP double immunostaining showed that PLP immunoreactivity was undetectable in the soma of the majority of oligodendrocytes in the spinal cord of WT mice, PERK-KO mice, and ATF6α-KO mice at P45 but became detectable in the soma of the majority of oligodendrocytes (arrowheads) in double-KO mice. (D) PLP and p62 double labeling revealed colocalization of PLP and p62 in oligodendrocytes (arrowhead) of double-KO mice at P45. (E) PLP and ubiquitin double labeling revealed colocalization of PLP and ubiquitin in oligodendrocytes (arrowhead) of double-KO mice at P45. (F) PLP and LAMP1 double labeling revealed colocalization of PLP and LAMP1 in oligodendrocytes (arrowhead) of double-KO mice at P45. Scale bars: 20 μm. N = 4 animals. Error bars represent mean ± SD. Statistical analyses were done with a 1-way ANOVA with Tukey’s posttest; *P < 0.05.
We further determined the subcellular localization of PLP in the soma of mature oligodendrocytes in 45-day-old double-KO mice. PLP and p62 double immunostaining showed colocalization of PLP and p62 in the soma of mature oligodendrocytes in 45-day-old double-KO mice (Figure 8D). PLP and ubiquitin double immunostaining also showed colocalization of PLP and ubiquitin in the soma of mature oligodendrocytes of 45-day-old double-KO mice (Figure 8E). Moreover, PLP and LAMP1 double immunostaining showed colocalization of PLP and LAMP1 in the soma of mature oligodendrocytes of 45-day-old double-KO mice (Figure 8F). Colocalization of PLP with p62, ubiquitin, and LAMP1 suggests accumulation of PLP in autophagic vacuoles in mature oligodendrocytes. As mentioned above, EM analysis showed accumulation of autophagic vacuoles in mature oligodendrocytes of 45-day-old double-KO mice. Taken together, these data suggest that impaired autophagic flux induced by inactivation of PERK and ATF6α leads to PLP accumulation in autophagic vacuoles in mature oligodendrocytes of young adult mice.
PLP deficiency attenuated late-onset demyelination in mice with double deletion of PERK and ATF6α in oligodendrocytes.
The Plp1 gene lies on the X chromosome. PLP-KO mice, including female PLP homozygous-KO (PLP–/–) mice and male hemizygous-KO (PLP–/Y) mice, develop normally and are phenotypically normal at a young age; however, aged mice show signs of spasticity (48–50). Importantly, PLP deficiency does not cause gross oligodendrocyte loss or obvious myelin abnormalities in young developing or young adult mice but induces axonal swellings and degeneration starting at the age of 6–8 weeks (48–50). Thus, we examined the involvement of the soma retention of PLP in the detrimental effects of inactivation of PERK and ATF6α on mature oligodendrocytes in young adults using PLP-KO mice. PLP-KO mice were crossed with double-KO mice to generate PERKloxP/loxP CNP/Cre ATF6α–/– PLP–/– mice or PERKloxP/loxP CNP/Cre ATF6α–/– PLP–/Y mice (triple-KO mice). Similar to double-KO mice, triple-KO mice were asymptomatic during the active developmental myelination stage and displayed a tremoring phenotype starting around P28. Importantly, the survival of triple-KO mice was significantly prolonged compared with double-KO mice (Figure 1E).
As expected, MBP IHC, CC1 IHC, and EM analysis showed that PLP deficiency alone did not significantly alter the degree of myelination or the number and morphology of oligodendrocytes but caused mild axon degeneration in the CNS at P45 (Figure 3, A, D, G, J, and M–Q; Figure 4, A, D, G, H, and K; and Supplemental Figure 2). Interestingly, MBP IHC showed that PLP deficiency noticeably attenuated myelin loss in the CNS of mice with double deletion of PERK and ATF6α in oligodendrocytes at P45 (Figure 3, E and F; and Supplemental Figure 2). EM analysis confirmed that the percentage of myelinated axons and the thickness of myelin were significantly increased in the CNS of triple-KO mice compared with double-KO mice at P45 and that the g-ratio was significantly decreased in the CNS of triple-KO mice compared with double-KO mice at P45 (Figure 3, K–O). Nevertheless, there were a significantly increased number of degenerating axons and a significantly decreased number of healthy-looking axons in the CNS of triple-KO mice compared with double-KO mice at P45 (Figure 3, K, L, P, and Q). While CC1 IHC showed that the number of oligodendrocytes was significantly increased in the CNS of triple-KO mice compared with double-KO mice at P45 (Figure 4, E–G; and Supplemental Figure 2), EM analysis showed that there was still accumulation of autophagic vacuoles in oligodendrocytes of triple-KO mice at P45 (Figure 4, L and M). Thus, these data demonstrate that PLP deficiency attenuates late-onset oligodendrocyte death and demyelination in the CNS of double-KO mice, resulting in prolonged survival of these mice. Collectively, these results provide direct evidence that soma retention of PLP contributes to the detrimental effects of PERK and ATF6α’s inactivation on mature oligodendrocytes in young adult mice.
Discussion
To assemble the myelin sheaths, actively myelinating oligodendrocytes must produce vast amounts of myelin proteins via the ER during developmental myelination. Mature oligodendrocytes in adults must also produce large amounts of myelin proteins via the ER to maintain myelin structure. It was thought that actively myelinating oligodendrocytes are more sensitive to disruption of ER protein homeostasis than mature oligodendrocytes because of the rate of production of myelin proteins (2, 8, 10). The UPR is a major mechanism that maintains ER protein homeostasis and cellular proteostasis (3, 4, 6, 7). As expected, we found that the impaired UPR in oligodendrocytes via inactivation of PERK and ATF6α induced moderate disruption of ER protein homeostasis in these cells, as evidenced by moderate XBP1 mRNA splicing and the normal morphology of the ER in oligodendrocytes in the CNS of double-KO mice. Surprisingly, inactivation of PERK and ATF6α in oligodendrocytes did not affect the viability or function of actively myelinating oligodendrocytes during developmental myelination but led to late-onset mature oligodendrocyte dysfunction and death in young adult mice. Thus, these data suggest that the UPR is not a major player in regulating ER protein homeostasis or cellular proteostasis in actively myelinating oligodendrocytes, or in maintaining their viability and function during developmental myelination.
Cathepsin D, a ubiquitously expressed aspartic-type protease, is a major lysosomal protease responsible for degradation of proteins delivered to the lysosomes via autophagy (41,44). Inactivation or mislocalization of cathepsin D causes lysosome dysfunction and accumulation of autophagic vacuoles in cells (44–46). Cathepsin D is synthesized in the ER and transported to the lysosome via the trans-Golgi network through the mannose 6-phosphate–dependent (M6P-dependent) pathway and M6P-independent pathways (41, 42, 51). GlcNAc-1-phosphotransferase catalyzes the synthesis of the M6P residues on cathepsin D in the trans-Golgi network, which allows cathepsin D to be recognized by M6P receptors (MPRs), which initiate protease packaging into clathrin-coated vesicles that travel to prelysosomal compartments (40, 43, 51). Moreover, data indicate that SEZ6L2 participates in the trafficking of cathepsin D to prelysosomal compartments via an M6P-independent pathway (42). Deficiency of either GNPTAB (encoding the α and β subunits of GlcNAc-1-phosphotransferase) or SEZ6L2 leads to delocalization of cathepsin D from the lysosome in cells (40, 42, 43). Interestingly, we showed here that inactivation of PERK and ATF6α attenuated the expression of both GNPTAB and SEZ6L2, induced delocalization of cathepsin D from the lysosome, and led to accumulation of autophagic vacuoles, p62, and ubiquitinated proteins in mature oligodendrocytes of young adult mice. Collectively, these results suggest that inactivation of PERK and ATF6α diminishes cathepsin D activity in the lysosome through disruption of its transport by attenuating the expression of GNPTAB and SEZ6L2 and results in lysosome dysfunction and impaired autophagic flux in mature oligodendrocytes of young adult mice.
A very recent study suggests that autophagy is an important player in regulating oligodendrocyte viability and function (52). Evidence suggests that autophagy is one of the major mechanisms responsible for degradation of PLP (20, 22). Several studies show that cathepsin D deficiency causes dysmyelination in the CNS of mice, accompanied by intracellular PLP accumulation and colocalization of PLP with LAMP1 (a lysosome marker) in oligodendrocytes (53, 54). Herein, we showed that inactivation of PERK and ATF6α led to intracellular PLP accumulation and colocalization of PLP with ubiquitin, p62, and LAMP1 in mature oligodendrocytes of young adult mice. Moreover, previous reports show that overexpression of PLP induced by increased copies of the Plp1 gene leads to accumulation of this protein in autophagic vacuoles in oligodendrocytes of mice (20, 22). Thus, these data suggest that inactivation of PERK and ATF6α attenuates autophagy-mediated PLP degradation by impairing cathepsin D activity in the lysosomes and results in PLP accumulation in autophagic vacuoles in mature oligodendrocytes of young adult mice.
It is believed that overexpression of PLP induced by increased copies of the Plp1 gene causes intracellular PLP accumulation in oligodendrocytes and subsequently results in dysmyelination or late-onset demyelination in humans with PMD and its animal models, although the mechanisms responsible for the cytotoxic effects of intracellular PLP accumulation on oligodendrocytes remain unknown (20–23). Moreover, a number of studies show that PLP is accumulated in autophagic vacuoles in oligodendrocytes of mice that carry increased copies of the Plp1 gene (20, 22, 47). Thus, we tested the possibility that PLP accumulation in autophagic vacuoles in oligodendrocytes contributes to the late-onset demyelination in young adult double-KO mice. We crossed double-KO mice with PLP-KO mice to obtain mice with deletion of PERK, ATF6α, and PLP in oligodendrocytes (triple-KO mice). Importantly, we showed that PLP deficiency significantly extended the life span (Figure 1E) and attenuated the abnormalities of oligodendrocytes and myelin in the CNS of double-KO mice. These results demonstrate a causal relationship between the intracellular accumulation of PLP and the dysfunction and death of mature oligodendrocytes in young adult mice with double deletion of PERK and ATF6α in oligodendrocytes. Nevertheless, triple-KO mice still exhibited not only substantial myelin loss but also exacerbated axon degeneration in the CNS and died prematurely. Several lines of evidence have suggested that PLP deficient mice display subtle myelin defects, including condensed intraperiod lines and persistence of short stretches of cytoplasm in myelin lamellae but develop progressive axonal degeneration starting as early as 6 weeks (48–50). On the other hand, recent studies show that oligodendrocytes support axon integrity through both myelin-dependent and myelin-independent mechanisms (55, 56). Thus, it is possible that oligodendrocyte dysfunction caused by inactivation of PERK and ATF6α facilitates PLP deficiency–induced axon degeneration in the CNS of triple-KO mice and that exacerbated axon degeneration leads to premature death of triple-KO mice. Alternatively, because triple-KO mice lack ATF6α in all cell types, there is a possibility that ATF6α inactivation in neurons enhanced PLP deficiency–induced axon degeneration in triple-KO mice. Our data support the notion that the partial rescuing effects of PLP deficiency on double-KO mice result from its inherently deleterious effects on axons and myelin. Additionally, there was still accumulation of autophagic vacuoles in mature oligodendrocytes of triple-KO mice. Therefore, an alternative, but not mutually exclusive, possibility is that inactivation of PERK and ATF6α in oligodendrocytes causes accumulation of not only PLP but also other cytotoxic proteins in the soma. Intracellular accumulation of other cytotoxic proteins leads to mature oligodendrocyte dysfunction and death in triple-KO mice.
Data indicate that endocytic sorting and recycling of PLP are involved in myelin sheath assembly and remodeling (57). There is evidence that PLP is also accumulated in the late endosomes/endolysosome in oligodendrocytes of mice carrying increased copies of the Plp1 gene (58). Interestingly, a previous report shows that feeding these mice a high-cholesterol diet reduces PLP accumulation in the late endosomes/endolysosome by facilitating incorporation of PLP into myelin sheaths and prevents disease progression (59). To explore the possibility of accumulation of PLP in the late endosomes/endolysosome of mature oligodendrocytes in double-KO mice, we fed these mice the high-cholesterol diet as described in the previous report (59). Nevertheless, we found that high-cholesterol treatment did not affect the disease progression or life span of double-KO mice (Supplemental Figure 4). Thus, it is unlikely that inactivation of PERK and ATF6α induces PLP accumulation in the late endosomes/endolyososome in mature oligodendrocytes of young adult mice.
Paradoxically, there was no evidence that inactivation of PERK and ATF6α led to impairment of autophagic flux or intracellular PLP accumulation in actively myelinating oligodendrocytes during developmental myelination. These data suggest that the role of the UPR in regulating lysosome function, autophagic flux, and PLP homeostasis in oligodendrocytes is dependent on the differentiation stages of the cells. It is unclear how the UPR is required for maintaining cellular proteostasis in mature oligodendrocytes of young adult mice but not in actively myelinating oligodendrocytes of young developing mice. Additionally, we showed here that inactivation of PERK and ATF6α had no effect on Schwann cells in young developing or adult mice, suggesting that the UPR is not a major player in maintaining ER protein homeostasis or cellular proteostasis in Schwann cells. Clearly, the mechanisms by which actively myelinating oligodendrocytes and Schwann cells maintain their ER protein homeostasis and/or cellular proteostasis warrant further investigation.
In summary, we showed here that inactivation of PERK and ATF6α in oligodendrocytes had a minimal effect on actively myelinating oligodendrocytes during developmental myelination but led to late-onset mature oligodendrocyte dysfunction and death in young adult mice by inducing intracellular accumulation of PLP through impairment of the autophagy/lysosome pathway. These findings imply that the UPR is required for maintaining cellular proteostasis and the viability and function of mature oligodendrocytes in adults by regulating autophagy of PLP.
Methods
Mice.
ATF6α homozygous-KO (ATF6α–/–) mice (25), PERKloxP mice (60) (The Jackson Laboratory stock 023066), CNP/Cre mice (28), and PLP heterozygous-KO (PLP+/–) mice (48) were on the C57BL/6J background. All mice were specific pathogen free and maintained on a 12-hour light/12-hour dark cycle. PERKloxP mice were crossed with ATF6α–/– mice to obtain PERKloxP ATF6α+/– mice, which were then crossed with CNP/Cre mice to obtain PERKloxP CNP/Cre ATF6α+/– mice. PERKloxP CNP/Cre ATF6α+/– mice were crossed with PERKloxP ATF6α+/– mice to obtain PERKloxP/loxP CNP/Cre ATF6α–/– mice (double-KO mice), PERKloxP/loxP CNP/Cre mice (PERK-KO mice), ATF6α–/– mice (ATF6α-KO mice), and control mice (WT mice), including PERKloxP/loxP mice, CNP/Cre mice, and WT mice. Moreover, we crossed PERKloxP CNP/Cre ATF6α+/– mice with PLP+/– mice to generate PERKloxP CNP/Cre ATF6α+/– PLP+/– mice and PERKloxP ATF6α+/– PLP+/– mice. PERKloxP CNP/Cre ATF6α+/– PLP+/– mice were crossed with PERKloxP ATF6α+/– PLP+/– mice to obtain PERKloxP/loxP CNP/Cre ATF6α–/– PLP–/– mice or PERKloxP/loxP CNP/Cre ATF6α–/– PLP–/Y mice (triple-KO mice), double-KO mice, PLP–/– or PLP–/Y mice (PLP-KO mice), and WT mice. Genotypes were determined by PCR from DNA extracted from tail tips as described in previous papers (25, 28, 48, 60). Both male and female mice were used in this study and no sex-based differences were observed.
Rotarod test.
The Rotarod apparatus (Panlab/Harvard Apparatus) with a spindle diameter of 3 cm was used to test motor coordination in mice. Mice were trained by placing them on the Rotarod at a constant speed (4 rpm) for 60 seconds a day before testing. For the trials, mice were placed on the Rotarod at 4 rpm, and then the Rotarod was set to accelerate continuously from 4 to 40 rpm over 300 seconds, and the latency to fall was recorded (falls were detected with a pressure-sensitive lever). Trials were repeated 3 times/d and the time of the fall was averaged. The maximum time for the trial was 300 seconds. Mice that reached this were recorded as 300 seconds and removed from the Rotarod.
Real-time PCR and XBP1 splicing assay.
Optic nerves and sciatic nerves harvested from mice were rinsed in ice-cold PBS and snap-frozen with dry ice. RNA was isolated from optic nerves or sciatic nerves using TRIzol reagent (Invitrogen, Thermo Fisher Scientific) or RNeasy Plus Mini Kit (QIAGEN), then treated with DNase I (Invitrogen, Thermo Fisher Scientific) to eliminate genomic DNA. Reverse transcription was performed using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories). Real-time PCR was performed with iQ Supermix (Bio-Rad Laboratories) on the LightCycler 480 System (Roche Diagnostics Corporation) as described in our previous papers (13, 16, 29). PCR for XBP1 was performed to detect XBP1s mRNA using Taq DNA Polymerase (QIAGEN). PCR products were separated by electrophoresis on a 3% agarose gel as described in our previous paper (16).
IHC.
Anesthetized mice were perfused through the left cardiac ventricle with 4% paraformaldehyde in PBS and the tissues were removed. Half sagittal brain and the cephalic half of the cervical spinal cord were postfixed in 4% paraformaldehyde for 2 hours, cryoprotected in 30% sucrose for 48 hours, embedded in optimum cutting temperature compound, and frozen on dry ice. Frozen sections were cut using a cryostat at a thickness of 10 μm. The other half sagittal brain and the caudal half of the cervical spinal cord were postfixed in 4% paraformaldehyde for 72 hours, dehydrated through graded alcohols, and embedded in paraffin wax. Paraffin sections were cut using a microtome at a thickness of 5 μm. Fluorescein (1:100, Vector Laboratories, anti-mouse catalog FI-2000, anti-rabbit catalog FI-1000), Cy3 (1:500, MilliporeSigma, anti-mouse catalog AP124C, anti-rat catalog AP136C, anti-rabbit catalog AP132C), Cy5 (1:200, Jackson ImmunoResearch Labs, anti-mouse catalog 115-175-146), or enzyme-labeled secondary antibodies (1:200, Vector Laboratories, anti-mouse catalog PI-2000) were used for detection. Immunohistochemical detection of CC1 (anti-APC, 1:50; MilliporeSigma, catalog OP80), MBP (1:1000; Covance, catalog SMI-99P-100), ASPA (1:1000, provided by M.A. Aryan Namboodiri at Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA), ubiquitin (1:100; Santa Cruz Biotechnology, catalog sc-8017), sequestosome 1 (p62, 1:15,000; Abcam, catalog ab56416), LAMP1 (1:100, FITC conjugate, LifeSpan BioSciences, catalog LS-B4246), LAMP1 (1:100, unconjugated, Thermo Fisher Scientific, catalog 14-1071-82), cleaved caspase-3 (1:100, Cell Signaling Technology, catalog 9664), cathepsin D (1:200, Abcam, catalog ab75852; provided by Michael K. Lee, University of Minnesota, Minneapolis, Minnesota, USA), PLP (AA3, 1:100; provided by Alexander Gow, Wayne State University, Detroit, Michigan, USA), and PLP (1:100; Santa Cruz Biotechnology, catalog sc-73336) was performed as described in previous papers (16, 29). Fluorescence stained sections were mounted with Vectashield mounting medium with DAPI (Vector Laboratories) and visualized with a Zeiss Axioskop 2 fluorescence microscope or an Olympus FV1000 confocal microscope. Whole-brain sections were scanned by a TissueScope LE120 slide scanner (Huron Digital Pathology). Immunopositive cells in the CNS were quantified as described in our previous papers (14, 16, 27, 29).
Toluidine blue staining and EM analysis.
Mice were anesthetized and perfused with PBS containing 4% paraformaldehyde and 2.5% glutaraldehyde. The sciatic nerve and cervical spinal cord were processed and embedded as previously described (13, 16, 27, 29). One-micrometer sections were cut, stained with toluidine blue, visualized with a Nikon C2 upright confocal microscope, and analyzed as described previously (13, 16, 27, 19). Ultrathin sections were cut, stained with uranyl acetate and lead citrate, and analyzed as described previously (13, 16, 27, 29). We counted the total number of axons, the number of degenerating axons, the number of healthy-looking axons, and the number of myelinated axons and calculated the percentage of myelinated axons as described previously (13, 16, 27, 29). Additionally, the diameter of axon as well as the diameter of axon and myelin were measured, and the g-ratio was calculated as diameter of axon/diameter of axon and myelin as described previously (14, 16, 27).
Statistics.
The sample size for each individual experiment is listed in the corresponding figure legend. Data are expressed as mean ± SD. Comparisons between 2 groups were statistically evaluated by 2-tailed t test (Supplemental Figure 4) using GraphPad Prism 6 (GraphPad Software RRID:SCR_002798). Multiple comparisons were statistically evaluated by a 1-way ANOVA with Tukey’s posttest or a 2-way ANOVA with Tukey’s posttest using GraphPad Prism 6. P values less than 0.05 were considered significant.
Study approval.
All animal procedures were conducted in complete compliance with the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and were approved by the Institutional Animal Care and Use Committee of the University of Minnesota.
Author contributions
WL designed the experiments, analyzed the data, and wrote the manuscript. SS designed the experiments, performed the experiments, analyzed the data, and wrote the manuscript. SW performed the experiments, analyzed the data, and wrote the manuscript. KAN provided the CNP/Cre mice and PLP-KO mice and reviewed the manuscript.
Supplementary Material
Acknowledgments
We thank Kazutoshi Mori (Department of Biophysics, Graduate School of Science, Kyoto University, Kyoto, Japan) for providing the ATF6α-KO mice. We thank Wendy Macklin (University of Colorado School of Medicine, Aurora, Colorado, USA) for sending us the PLP-KO mice. We thank M.A. Aryan Namboodiri (Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA) for providing the antibody against ASPA. We thank Alexander Gow (Wayne State University, Detroit, Michigan, USA) for providing the antibody against PLP (AA3 antibody). We thank Michael K. Lee (University of Minnesota) for purchasing and providing the antibodies against LAMP1 and cathepsin D. This study was supported by grants from the NIH (NS094151 and NS105689) to WL.
Version 1. 02/13/2020
In-Press Preview
Version 2. 03/12/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(5):e132364.https://doi.org/10.1172/jci.insight.132364.
Contributor Information
Sarrabeth Stone, Email: sarrabeth.stone@gmail.com.
Shuangchan Wu, Email: wuxx1763@umn.edu.
Klaus-Armin Nave, Email: nave@em.mpg.de.
Wensheng Lin, Email: linw@umn.edu.
References
- 1.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13(10):1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 2.Lin W, Popko B. Endoplasmic reticulum stress in disorders of myelinating cells. Nat Neurosci. 2009;12(4):379–385. doi: 10.1038/nn.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86(4):1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 4.Hetz C, Chevet E, Oakes SA. Proteostasis control by the unfolded protein response. Nat Cell Biol. 2015;17(7):829–838. doi: 10.1038/ncb3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529(7586):326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 6.Martínez G, Duran-Aniotz C, Cabral-Miranda F, Vivar JP, Hetz C. Endoplasmic reticulum proteostasis impairment in aging. Aging Cell. 2017;16(4):615–623. doi: 10.1111/acel.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klaips CL, Jayaraj GG, Hartl FU. Pathways of cellular proteostasis in aging and disease. J Cell Biol. 2018;217(1):51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeiffer SE, Warrington AE, Bansal R. The oligodendrocyte and its many cellular processes. Trends Cell Biol. 1993;3(6):191–197. doi: 10.1016/0962-8924(93)90213-K. [DOI] [PubMed] [Google Scholar]
- 9.Anitei M, Pfeiffer SE. Myelin biogenesis: sorting out protein trafficking. Curr Biol. 2006;16(11):R418–R421. doi: 10.1016/j.cub.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Clayton BLL, Popko B. Endoplasmic reticulum stress and the unfolded protein response in disorders of myelinating glia. Brain Res. 2016;1648(pt B):594–602. doi: 10.1016/j.brainres.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volpi VG, Touvier T, D’Antonio M. Endoplasmic reticulum protein quality control failure in myelin disorders. Front Mol Neurosci. 2016;9:162. doi: 10.3389/fnmol.2016.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin W, Stone S. Unfolded protein response in myelin disorders. Neural Regen Res. 2020;15(4):636–645. doi: 10.4103/1673-5374.266903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin W, et al. Oligodendrocyte-specific activation of PERK signaling protects mice against experimental autoimmune encephalomyelitis. J Neurosci. 2013;33(14):5980–5991. doi: 10.1523/JNEUROSCI.1636-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y, et al. PERK activation preserves the viability and function of remyelinating oligodendrocytes in immune-mediated demyelinating diseases. Am J Pathol. 2014;184(2):507–519. doi: 10.1016/j.ajpath.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussien Y, Cavener DR, Popko B. Genetic inactivation of PERK signaling in mouse oligodendrocytes: normal developmental myelination with increased susceptibility to inflammatory demyelination. Glia. 2014;62(5):680–691. doi: 10.1002/glia.22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone S, Wu S, Jamison S, Durose W, Pallais JP, Lin W. Activating transcription factor 6α deficiency exacerbates oligodendrocyte death and myelin damage in immune-mediated demyelinating diseases. Glia. 2018;66(7):1331–1345. doi: 10.1002/glia.23307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussien Y, Podojil JR, Robinson AP, Lee AS, Miller SD, Popko B. ER Chaperone BiP/GRP78 is required for myelinating cell survival and provides protection during experimental autoimmune encephalomyelitis. J Neurosci. 2015;35(48):15921–15933. doi: 10.1523/JNEUROSCI.0693-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stone S, Lin W. The unfolded protein response in multiple sclerosis. Front Neurosci. 2015;9:264. doi: 10.3389/fnins.2015.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naughton MC, McMahon JM, FitzGerald U. Differential activation of ER stress pathways in myelinating cerebellar tracts. Int J Dev Neurosci. 2015;47(pt B):347–360. doi: 10.1016/j.ijdevneu.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths IR, Schneider A, Anderson J, Nave KA. Transgenic and natural mouse models of proteolipid protein (PLP)-related dysmyelination and demyelination. Brain Pathol. 1995;5(3):275–281. doi: 10.1111/j.1750-3639.1995.tb00604.x. [DOI] [PubMed] [Google Scholar]
- 21.Woodward KJ. The molecular and cellular defects underlying Pelizaeus-Merzbacher disease. Expert Rev Mol Med. 2008;10:e14. doi: 10.1017/S1462399408000677. [DOI] [PubMed] [Google Scholar]
- 22.Karim SA, et al. PLP/DM20 expression and turnover in a transgenic mouse model of Pelizaeus-Merzbacher disease. Glia. 2010;58(14):1727–1738. doi: 10.1002/glia.21043. [DOI] [PubMed] [Google Scholar]
- 23.Mayer JA, Larsen EC, Kondo Y, Duncan ID. Characterization of a PLP-overexpressing transgenic rat, a model for the connatal form of Pelizaeus-Merzbacher disease. Neurobiol Dis. 2011;44(2):231–238. doi: 10.1016/j.nbd.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81(2):871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13(3):365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Vaughn JE. An electron microscopic analysis of gliogenesis in rat optic nerves. Z Zellforsch Mikrosk Anat. 1969;94(3):293–324. doi: 10.1007/BF00319179. [DOI] [PubMed] [Google Scholar]
- 27.Lin Y, et al. Impaired eukaryotic translation initiation factor 2B activity specifically in oligodendrocytes reproduces the pathology of vanishing white matter disease in mice. J Neurosci. 2014;34(36):12182–12191. doi: 10.1523/JNEUROSCI.1373-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lappe-Siefke C, et al. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33(3):366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- 29.Stone S, Jamison S, Yue Y, Durose W, Schmidt-Ullrich R, Lin W. NF-κB activation protects oligodendrocytes against inflammation. J Neurosci. 2017;37(38):9332–9344. doi: 10.1523/JNEUROSCI.1608-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dey S, Tameire F, Koumenis C. PERK-ing up autophagy during MYC-induced tumorigenesis. Autophagy. 2013;9(4):612–614. doi: 10.4161/auto.23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou L, et al. Avian metapneumovirus subgroup C induces autophagy through the ATF6 UPR pathway. Autophagy. 2017;13(10):1709–1721. doi: 10.1080/15548627.2017.1356950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song S, Tan J, Miao Y, Zhang Q. Crosstalk of ER stress-mediated autophagy and ER-phagy: Involvement of UPR and the core autophagy machinery. J Cell Physiol. 2018;233(5):3867–3874. doi: 10.1002/jcp.26137. [DOI] [PubMed] [Google Scholar]
- 33.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 34.Inoue K, et al. Macroautophagy deficiency mediates age-dependent neurodegeneration through a phospho-tau pathway. Mol Neurodegener. 2012;7:48. doi: 10.1186/1750-1326-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menzies FM, et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93(5):1015–1034. doi: 10.1016/j.neuron.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 36.Lamark T, Svenning S, Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017;61(6):609–624. doi: 10.1042/EBC20170035. [DOI] [PubMed] [Google Scholar]
- 37.Martina JA, Diab HI, Brady OA, Puertollano R. TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 2016;35(5):479–495. doi: 10.15252/embj.201593428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Z, et al. Activation of the ATF6 branch of the unfolded protein response in neurons improves stroke outcome. J Cereb Blood Flow Metab. 2017;37(3):1069–1079. doi: 10.1177/0271678X16650218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luhr M, et al. The kinase PERK and the transcription factor ATF4 play distinct and essential roles in autophagy resulting from tunicamycin-induced ER stress. J Biol Chem. 2019;294(20):8197–8217. doi: 10.1074/jbc.RA118.002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee WS, Payne BJ, Gelfman CM, Vogel P, Kornfeld S. Murine UDP-GlcNAc:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase lacking the gamma-subunit retains substantial activity toward acid hydrolases. J Biol Chem. 2007;282(37):27198–27203. doi: 10.1074/jbc.M704067200. [DOI] [PubMed] [Google Scholar]
- 41.Zaidi N, Maurer A, Nieke S, Kalbacher H. Cathepsin D: a cellular roadmap. Biochem Biophys Res Commun. 2008;376(1):5–9. doi: 10.1016/j.bbrc.2008.08.099. [DOI] [PubMed] [Google Scholar]
- 42.Boonen M, Staudt C, Gilis F, Oorschot V, Klumperman J, Jadot M. Cathepsin D and its newly identified transport receptor SEZ6L2 can modulate neurite outgrowth. J Cell Sci. 2016;129(3):557–568. doi: 10.1242/jcs.179374. [DOI] [PubMed] [Google Scholar]
- 43.Velho RV, et al. The lysosomal storage disorders mucolipidosis type II, type III alpha/beta, and type III gamma: update on GNPTAB and GNPTG mutations. Hum Mutat. 2019;40(7):842–864. doi: 10.1002/humu.23748. [DOI] [PubMed] [Google Scholar]
- 44.Vidoni C, Follo C, Savino M, Melone MA, Isidoro C. The role of cathepsin D in the pathogenesis of human neurodegenerative disorders. Med Res Rev. 2016;36(5):845–870. doi: 10.1002/med.21394. [DOI] [PubMed] [Google Scholar]
- 45.Koike M, et al. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci. 2000;20(18):6898–6906. doi: 10.1523/JNEUROSCI.20-18-06898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koike M, et al. Purkinje cells are more vulnerable to the specific depletion of cathepsin D than to that of Atg7. Am J Pathol. 2017;187(7):1586–1600. doi: 10.1016/j.ajpath.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 47.Karim SA, et al. PLP overexpression perturbs myelin protein composition and myelination in a mouse model of Pelizaeus-Merzbacher disease. Glia. 2007;55(4):341–351. doi: 10.1002/glia.20465. [DOI] [PubMed] [Google Scholar]
- 48.Klugmann M, et al. Assembly of CNS myelin in the absence of proteolipid protein. Neuron. 1997;18(1):59–70. doi: 10.1016/S0896-6273(01)80046-5. [DOI] [PubMed] [Google Scholar]
- 49.Griffiths I, et al. Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science. 1998;280(5369):1610–1613. doi: 10.1126/science.280.5369.1610. [DOI] [PubMed] [Google Scholar]
- 50.Rosenbluth J, Nave KA, Mierzwa A, Schiff R. Subtle myelin defects in PLP-null mice. Glia. 2006;54(3):172–182. doi: 10.1002/glia.20370. [DOI] [PubMed] [Google Scholar]
- 51.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10(9):623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 52.Bankston AN, et al. Autophagy is essential for oligodendrocyte differentiation, survival, and proper myelination. Glia. 2019;67(9):1745–1759. doi: 10.1002/glia.23646. [DOI] [PubMed] [Google Scholar]
- 53.Mutka AL, et al. Murine cathepsin D deficiency is associated with dysmyelination/myelin disruption and accumulation of cholesteryl esters in the brain. J Neurochem. 2010;112(1):193–203. doi: 10.1111/j.1471-4159.2009.06440.x. [DOI] [PubMed] [Google Scholar]
- 54.Guo DZ, et al. Cathepsin D deficiency delays central nervous system myelination by inhibiting proteolipid protein trafficking from late endosome/lysosome to plasma membrane. Exp Mol Med. 2018;50(3):e457. doi: 10.1038/emm.2017.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bankston AN, Mandler MD, Feng Y. Oligodendroglia and neurotrophic factors in neurodegeneration. Neurosci Bull. 2013;29(2):216–228. doi: 10.1007/s12264-013-1321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saab AS, Tzvetanova ID, Nave KA. The role of myelin and oligodendrocytes in axonal energy metabolism. Curr Opin Neurobiol. 2013;23(6):1065–1072. doi: 10.1016/j.conb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 57.Winterstein C, Trotter J, Krämer-Albers EM. Distinct endocytic recycling of myelin proteins promotes oligodendroglial membrane remodeling. J Cell Sci. 2008;121(pt 6):834–842. doi: 10.1242/jcs.022731. [DOI] [PubMed] [Google Scholar]
- 58.Simons M, et al. Overexpression of the myelin proteolipid protein leads to accumulation of cholesterol and proteolipid protein in endosomes/lysosomes: implications for Pelizaeus-Merzbacher disease. J Cell Biol. 2002;157(2):327–336. doi: 10.1083/jcb.200110138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saher G, et al. Therapy of Pelizaeus-Merzbacher disease in mice by feeding a cholesterol-enriched diet. Nat Med. 2012;18(7):1130–1135. doi: 10.1038/nm.2833. [DOI] [PubMed] [Google Scholar]
- 60.Zhang P, et al. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22(11):3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.