Abstract
Cytidine triphosphate (CTP) synthetase 1 (CTPS1) deficiency is caused by a unique homozygous frameshift splice mutation (c.1692-1G>C, p.T566Dfs26X). CTPS1-deficient patients display severe bacterial and viral infections. CTPS1 is responsible for CTP nucleotide de novo production involved in DNA/RNA synthesis. Herein, we characterized in depth lymphocyte defects associated with CTPS1 deficiency. Immune phenotyping performed in 7 patients showed absence or low numbers of mucosal-associated T cells, invariant NKT cells, memory B cells, and NK cells, whereas other subsets were normal. Proliferation and IL-2 secretion by T cells in response to TCR activation were markedly decreased in all patients, while other T cell effector functions were preserved. The CTPS1T566Dfs26X mutant protein was found to be hypomorphic, resulting in 80%–90% reduction of protein expression and CTPS activity in cells of patients. Inactivation of CTPS1 in a T cell leukemia fully abolished cell proliferation. Expression of CTPS1T566Dfs26X failed to restore proliferation of CTPS1-deficient leukemia cells to normal, except when forcing its expression to a level comparable to that of WT CTPS1. This indicates that CTPS1T566Dfs26X retained normal CTPS activity, and thus the loss of function of CTPS1T566Dfs26X is completely attributable to protein instability. This study supports that CTPS1 represents an attractive therapeutic target to selectively inhibit pathological T cell proliferation, including lymphoma.
Keywords: Immunology
Keywords: Adaptive immunity, Monogenic diseases, T cells
A hypomorphic homozygous mutation that leads to reduced CTP synthetase 1 expression causes defects in T lymphocyte functions and differentiation.
Introduction
T lymphocytes need a very active metabolism to facilitate rapid cellular proliferation and the production of effector molecules in response to antigen challenge (1–3). The production of purine and pyrimidine nucleotides has a critical role in the synthesis of nucleic acids and lipids, and these pathways are thus necessary for cell cycle progression of activated T lymphocytes (4). Activation of T lymphocytes with mitogens or in response to TCR stimulation leads to increased purine and pyrimidine pools (5, 6). Cytidine triphosphate (CTP) synthetase is the limiting nucleotide in cells, and its synthesis depends on 2 pathways: a salvage pathway allowing production of CTP from cytidine derived from nucleic acid degradation and a de novo pathway that relies on CTP synthase (CTPS) activity (7). CTPS activity is encoded by 2 genes, CTPS1 and CTPS2, and their proteins share 80% identity and homology (8–10). CTPS catalyzes ATP-dependent amination of uridine triphosphate (UTP) to CTP with ammonia (NH3) transfer from hydrolyzed glutamine. In normal T cells, CTPS activity is increased only in the S phase of cell cycle progression, while in cancer cells, including in acute T cell lymphoblastic leukemia (T-ALL), CTPS activity is elevated irrespective of cell cycle progression (11–13). Thus, CTPS might be considered an attractive therapeutic target for the treatment of T-ALL (14).
Recently, we and others identified immunodeficient patients with a CTPS1 deficiency leading to mainly life-threatening herpes virus infections with varicella-zoster virus (VZV) and EBV. All patients characterized so far have been shown to carry the same homozygous mutation IVS18-1 G>C or NM_001905.3: c.1692-1G>C, p.T566Dfs26X (rs145092287) in the acceptor splice site at introns 17–18, with a likely common ancestor from northwest England (6, 15–17). In our initial study, we showed that CTPS1 is important for the expansion of activated T cells. In T lymphocytes, CTPS1 expression is upregulated in response to TCR stimulation. The capacity of activated CTPS1-deficient T cells to proliferate is impaired (6). However, in this previous study, analysis of immunological parameters and lymphocyte functions was carried out in only 1 patient available at that time, and no molecular and functional characterization of the mutation was done.
In the present work, a detailed analysis of immune parameters was performed in 7 CTPS1-deficient patients. The consequences of the CTPS1 mutation at the protein and enzymatic levels were also characterized in normal cells and derived cell lines from patients and in a T cell leukemia cell line in which CTPS1 was inactivated by CRISPR/Cas9.
Results
Identification of new patients with CTPS1 deficiency.
Since our original report (6), we have screened further patients originating from England and the United States with a clinical phenotype compatible with a CTPS1 deficiency that notably includes severe VZV and EBV infections (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.133880DS1). Five of them (P10 to P14) were found to carry the same homozygous mutation IVS18-1 G>C in CTPS1 (data not shown). In addition, a further 6 patients have been reported since our original report (6). Clinical manifestations appeared during the first 2 years of life in the majority of patients. Hematopoietic stem cell transplantation (HSCT) was performed in 15 of the 19 patients. Thirteen out of 15 are alive and well at the most recent follow-up (17). Two patients died from severe viral infection (P3.1 and P5) before HSCT could be performed, and 2 (P12 and P13) are alive and have yet to receive transplants. No extrahematopoietic manifestation (not related to their immunodeficiency or HSCT treatment) was reported in any of the 19 patients with a median follow-up extending to 14.6 years (with the oldest patient aged 24 years old). Routine laboratory investigations were carried out in the 5 newly identified patients (Supplemental Table 2 and Supplemental Table 3). Blood cell counts of the different subsets were within, or close to, normal ranges. IgGs were elevated but specific antibodies were low as well as T cell proliferation in response to phytohemagglutinin (PHA) and anti-CD3 (OKT3). These data were similar to those reported previously for CTPS1-deficient patients. (6, 15–17). Ranges and numbers of the different blood cell populations were compared to established healthy pediatric standards (18–21).
Blood leukocyte subset analysis in CTPS1-deficient individuals.
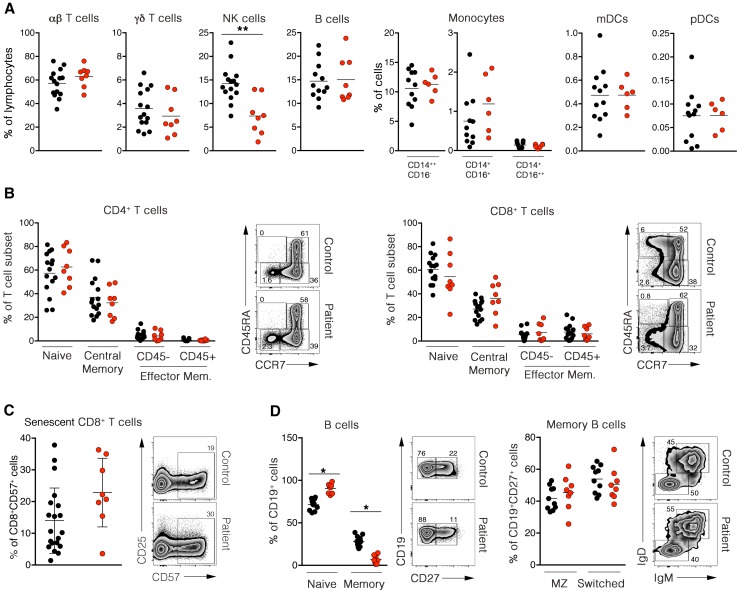
Immunological investigations were performed in depth in 7 CTPS1-deficient patients (P1.2, P7.1, P7.2, P10, P11, P12, P13, and P14). Frequencies of αβ and γδ T cells and B cells were in the normal ranges, whereas absolute numbers and frequency of NK cells were significantly diminished in all patients when compared with healthy controls (Figure 1A and Supplemental Table 3). Analysis of NK cell subsets in 1 patient showed reduced proportions of CD56dimCD16+ NK cells (Supplemental Figure 1A). Patients also exhibited normal absolute numbers and percentages of myeloid subsets (CD14++CD16–, CD14+CD16+, CD14+CD16++), mDCs (HLADR+ CD123loCD11c+), and pDCs (HLADR+CD123hiCD11c) (Figure 1A, Supplemental Figure 1B, and Supplemental Table 3). Analysis of T cell subsets showed no significant difference in the proportions of naive recent emigrant (CD31+CD45RA+CCR7+), central memory (CD45RA–CCR7+CD27+), effector memory (CD45RA–CCR7–CD27–), and terminal effector memory (CD45RA+CCR7–CD27–) in CD4+ and CD8+ T cells and CD8+ senescent T cells (CD57+) cells compared to controls (Figure 1, B and C). Total B cell numbers were normal, but the frequency of memory B cells (CD19+CD27+) was decreased compared with healthy controls (Supplemental Table 3 and Figure 1D) (22). Within the memory B cell compartment, the proportions of switched memory IgM–IgD– and marginal zone B cells were normal (Figure 1D).
Figure 1. Immunophenotyping of CTPS1-deficient patients.
(A) Percentages of αβ and γδ T cells, NK cells, B cells, monocytes, myeloid dendritic cells (mDCs), and plasmacytoid dendritic cells (pDCs) in PBMCs are represented in dot plot graphs. Data were obtained from FACS analysis after cell-specific staining. Reference ranges for age-matched controls (3–18 years) correspond to 21%–98%, 0.9%–17%, 4%–22 %, 6%–24%, 2%–21%, 0.1%–7.8%, 0%–4.5%, 0.1%–3.1%, and 0.8%–0.36% for αβ T cells, γδ T cells, NK cells, B cells, classical CD14++CD16–, intermediate CD14+CD16+, nonclassical CD14+CD16++ monocytes, mDCs, and pDCs, respectively. (B) Frequencies of naive (CD31+CD45RA+CCR7–), central memory (CD45RA–CCR7+CD27–), effector memory (CD45RA–CCR7+CD27–), and exhausted effector memory/TEMRA (CD45RA+CCR7+CD27–) compartments in CD4+ and CD8+ T cells are represented in the dot plot graph. Representative dot plot FACS analysis of naive and memory CD4+ cells (left) and CD8+ cells (right) are also depicted. (C) Frequencies of senescent cells (CD8+CD57+) gated on CD3+ cells are shown in the dot plot graph (left). Representative dot plot FACS analysis of senescent CD8+ cell populations are depicted at right . Data were obtained from FACS analysis of cell-specific staining. (D) Frequencies of naive (CD19+CD27–), memory (CD19+CD27+) (left), circulating marginal zone (MZ) (CD19+CD27+IgM+IgD+) and switched B cells (CD19+CD27+IgM–IgD–) (right) are shown in the dot plot graph. Representative dot plots FACS analysis of naive/memory (left) and marginal zone/switched (right) B cell populations are depicted. Data were obtained from FACS analysis after cell-specific staining. In A–D, black or red dots correspond to a healthy individual donor or a patient, respectively. Each circle represents an independent biological sample. Ten to 19 adult controls and 8 patients were analyzed. The horizontal bars represent the median. Data obtained from 6 independent experiments. Groups of values were compared 2 by 2 using Mann-Whitney U tests. *P < 0.05; **P < 0.01.
CD4+CD25+CD127loFOXP3+ regulatory T cells (Tregs) were detected at normal ranges (Figure 2A). In contrast, invariant T cell populations invariant natural killer T (iNKT) (CD3+Vα24+Vβ11+ CD161hi or Tet-CD1d+CD161hi) and mucosal-associated invariant T (MAIT) (CD3+Vα7+CD161hi or Tet-MR1+CD161hi) cells were absent or markedly reduced (Figure 2B and Supplemental Figure 1C). These defects did not result from impaired VDJ recombination, as percentages of mainstream T cells using Vα7.2 and Vα24 chains of the TCR (Figure 2B), Vα and Vβ gene segment usage, and PROMIDISα signature (23) were all comparable to controls (Supplemental Figure 2, A and B). In summary, CTPS1 deficiency is characterized by an almost complete absence of blood iNKT cell and MAIT cell subpopulations and decreased NK cells and memory B cells. This phenotypic analysis of 8 patients corroborates well the results of our first report (6).
Figure 2. Absence of invariant T cells in blood of CTPS1-deficient patients.
(A) Percentages of CD127loCD25hi regulatory T cells (Tregs) in blood are shown in the dot plot graph (upper left). Data were obtained from FACS analysis after cell-specific staining. Representative dot plot of FACS analysis of Treg population is depicted (lower). Representative dot plot of FACS analysis of Foxp3 expression in CD127hiCD25lo mainstream T cells (black cell population) and CD127loCD25hi Tregs (red cell population) are shown (upper right). (B) Percentages of Vα7.2+CD161– and Vα24+CD161– mainstream T cells, Vα7.2+CD161+ mucosal-associated invariant T (MAIT) and Vα24+CD161+ invariant NK T (iNKT) cells in blood are shown in the dot blot graph (upper). Representative dot plot FACS analysis of MAIT cells (lower left) and iNKT cells (lower right) in control and patient blood are depicted. In A–B, red and black dots represent independent biological samples of patients and controls, respectively. The horizontal bars represent the median. Data obtained from 3 independent experiments. Groups of values were compared 2 by 2 using Mann-Whitney U tests. ***P < 0.001.
Impaired TCR-induced IL-2 secretion and proliferation of CTPS1-deficient T lymphocytes.
In our first report we showed that CTPS1 was required for T cell proliferation but not for IFN-γ and TNF-α cytokine production in response to TCR activation (6). To extend these observations, PHA-expanded T cell blasts from 5 CTPS1-deficient patients and 10 healthy donors/controls were stimulated for 48 hours with anti-CD3 antibody, anti-CD3/CD28–coated beads, or PMA/ionomycin, and production of IL-2, IL-4, IL-17, IFN- γ, TNF-α, and granzyme B was analyzed in culture supernatants. IFN- γ, TNF-α, IL-17, granzyme B, and IL-4 release was comparable between patient and control cells (Figure 3A). In contrast, IL-2 production was significantly reduced. To exclude the possibility that IL-2 was consumed faster by CTPS1-deficient T cells, intracellular IL-2 production from 6 CTPS1-deficient patients and 10 controls was examined after 12 hours of stimulation (Figure 3B). Intracellular IL-2 expression in T cells of patients was reduced following anti-CD3/CD28 or PMA/ionomycin stimulation. In contrast, intracellular IFN-γ was slightly increased in T cells from patients (Figure 3B). Degranulation of cytotoxic granules by CD8+ T cells was also normal from 2 examined CTPS1-deficient patients (Supplemental Figure 2C). The proliferation of CTPS1-deficient T lymphocytes was impaired in response to TCR stimulation by an anti-CD3 antibody or anti-CD3/CD28–coated beads (Figure 3D). This was variably associated with increased activation-induced cell death (Supplemental Figure 2D). Remarkably, CTPS1-deficient T cells stopped dividing after a few divisions and expressed decreased levels of the activation marker CD25 (Figure 3, C and D). This defective proliferation was restored after addition of cytidine in the culture medium (Figure 3E). To investigate whether impaired autocrine IL-2 secretion was contributing to the defective proliferation, exogenous IL-2 was added to CTPS1-deficient T cells or control T cells treated with a specific inhibitor of CTPS1 (3-deazauridine; 3-DAZ) (24) (Figure 3F and Supplemental Figure 3A). Addition of IL-2 only slightly restored the proliferation of CTPS1-deficient cells or 3-DAZ–treated control T cells. Thus, we conclude that the defective capacity of CTPS1-deficient T cells to produce IL-2 did not contribute significantly to impaired proliferation. Interestingly, in contrast to CTPS1-deficient cells, 3-DAZ treatment did not impair IL-2 production while CD25 expression was decreased (Supplemental Figure 3, B and C), suggesting that CD25 expression but not IL-2 production is dependent on CTPS activity. These observations support the selective role of CTPS1 in promoting the expansion of human T cells following TCR-mediated activation, while T cell differentiation and most effector functions were spared and normal, with the notable exception of IL-2 production (which was significantly decreased, Figure 3A).
Figure 3. Functional studies of CTPS1-deficient T cells.
(A) Quantification of IFN-γ, TNF-α, IL-17A, granzyme B, IL-4, and IL-2 in supernatants of activated T cell blasts from 10 healthy donors and patients using coated beads array technology. T cell blasts were stimulated or not with 1 μg/mL of anti-CD3 (OKT3) antibody, anti-CD3/CD28 beads, or PMA plus ionomycin for 48 hours. Data were obtained from 4 independent experiments. (B) Flow cytometry analysis of IFN-γ and IL-2 intracellular detections gated on CD3+ T cell blasts from healthy donors and CTPS1-deficient patients. T cell blasts were stimulated or not with 1 μg/mL of anti-CD3 (OKT3) antibody, anti-CD3/CD28 beads or PMA plus ionomycin for 12 hours. Representative dot plots of FACS analyses corresponding to IFN-γ and IL-2 stainings after PMA stimulation of control and patient T cell blasts are depicted (lower left and right panels, respectively) with isotype staining (upper panels). Frequencies of IFN-γ– and IL-2–secreting T cell blasts are shown in the dot plot graphs (lower left and right panels, respectively). Data were normalized on isotype staining and obtained from 4 independent experiments. (C) Expression of CD25-based mean fluorescence intensity (MFI) calculated from FACS histograms following anti-CD3 antibody or anti-CD3/CD28 bead stimulation of control and patient T cell blasts for 96 hours. Graph from 5 independent experiments. (D) Index values of proliferation of CD3+ T cell blasts from controls and CTPS1-deficient patients after anti-CD3/CD28 beads (left) or coated anti-CD3 antibody (right) stimulation for 4 days. The index value was obtained from histogram analyses of cell trace violet staining of healthy donors and patient T cells using FlowJo software. Data were obtained from 1 experiment. (E) Same as D, except that stimulated CD3+ T cell blasts were cultured in medium supplemented or not with cytidine (200 μM). (F) Analysis of control (black histogram) and patient (red histogram) T cell blast proliferations using cell trace violet staining. Cells were stimulated with anti-CD3 antibody for 4 days and supplemented or not with IL-2 (100 U/mL) or cytidine (200 μM). Histogram analysis was performed using FlowJo software. Data shown are representative of 3 independent experiments. In A–E, red and black dots represent independent biological samples of patients and controls, respectively. The horizontal bars represent the median. Group of values were compared 2 by 2 using Mann-Whitney U tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Residual CTPS1 mutant protein expression in CTPS1-deficient patients.
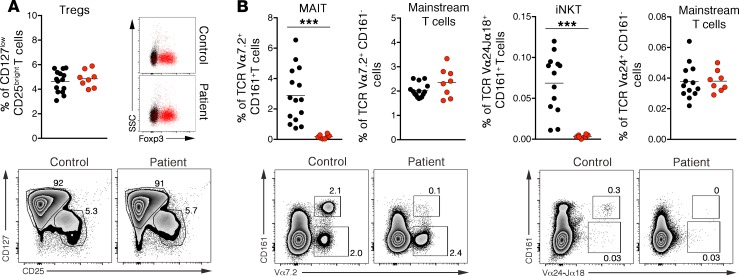
We previously showed that the mutation in CTPS1 was associated with no detectable expression of CTPS1 in cell lysates from patients, suggesting that the mutation behaved as an amorphic (null) mutation (6). Using a newly available anti-CTPS1 monoclonal antibody, which gives a more sensitive and specific signal than the polyclonal antibody previously used (Supplemental Figure 4), CTPS1 expression profile and the effect of the CTPS1 mutation were reanalyzed by Western blot and intracellular FACS staining in PBMCs, T cell blasts, lymphoblastoid cell lines (LCLs), primary fibroblasts, transformed/immortalized fibroblasts, and T cells activated with PMA or anti-CD3 antibody (Figure 4 and Supplemental Figure 5). PMA induced marked CTPS1 expression in T and B lymphocytes from healthy donors (Figure 4A). Among the different T cell subsets, iNKT and MAIT cells (without stimulation) expressed the highest levels of CTPS1, while the induction of CTPS1 was the strongest in naive T cells following anti-CD3 stimulation (Figure 4B). Expression of CTPS1 was also found to be high in cell lines such as the leukemia Jurkat cells, the LCLs, and SV-40–transformed cells compared with T cell blasts and primary fibroblasts (Figure 4C). CTPS2 protein expression was also readily detectable in these cell lines, except in Jurkat T cells.
Figure 4. Analysis of CTPS1 expression in lymphocytes subsets and activated T cells in control individuals.
(A) Dot plot graph representing the MFI of CTPS1 expression in αβ T cells (black circle), γδ T cells (blue circle), NK cells (green circle), B cells (red circle), and monocytes (gray circle). PBMCs were stimulated (right) or not (No stim) (left) with PMA plus ionomycin for 48 hours. (B) MFI of CTPS1 expression in T cell subpopulations: MAIT (red circle), iNKT (green circle), naive (circle), memory (square), CD45RA– (up triangle) and CD45RA+ (down triangle), effector memory CD4 (black symbols), or CD8 (blue symbols). T cells were stimulated (middle) or not (upper) with 1 μg/mL of coated anti-CD3 antibody for 48 hours. CTPS1 MFI ratios of unstimulated and anti-CD3–stimulated T cells are represented in the lower panel. (C) Immunoblots for CTPS1, CTPS2, and actin protein expression in Jurkat cells and control T cell blasts stimulated or not with anti-CD3/CD28–coated beads for 48 hours, control (Ctrl), LCLs, primary fibroblast cell lines, SV-40 T antigen–immortalized fibroblast cell lines (SV40 Fibro), and telomerized SV40-transformed fibroblast cell lines (Tel. SV40 Fibro). (D) Immunoblots (upper) and quantitative PCRs (qPCRs) (lower) for CTPS1, CTPS2, and actin protein expression in T cell blasts stimulated with anti-CD3/CD28 beads for different periods. In lower panels, the bar graphs depict relative CTPS1 (left) and CTPS2 (right) mRNA expression normalized on GAPDH mRNA expression. Data were obtained from 3 independent experiments. In A–B, all MFIs were calculated from FACS histogram analysis (data not shown). Each symbol corresponds to an independent biological sample of a healthy individual. The horizontal bars represent the median ± SEM.
After stimulation with anti-CD3, CTPS1 protein expression in normal T cells was upregulated from 24 hours and declined after 96 hours (Figure 4, C and D). CTPS2 protein expression was also strongly increased with a more rapid and transient kinetic, being detectable 12 hours after stimulation but diminishing after 24 hours (Figure 4D). Of note, CTPS2 detection resolved in 2 protein products in T cells that are differentially expressed after CD3 stimulation, possibly indicative of posttranslational modifications. As expected, CTPS1 and CTPS2 protein expression was preceded by a peak in CTPS1 and CTPS2 mRNA expression, respectively (Figure 4D). The upregulation of CTPS2 could not compensate for CTPS1 deficiency in patient cells.
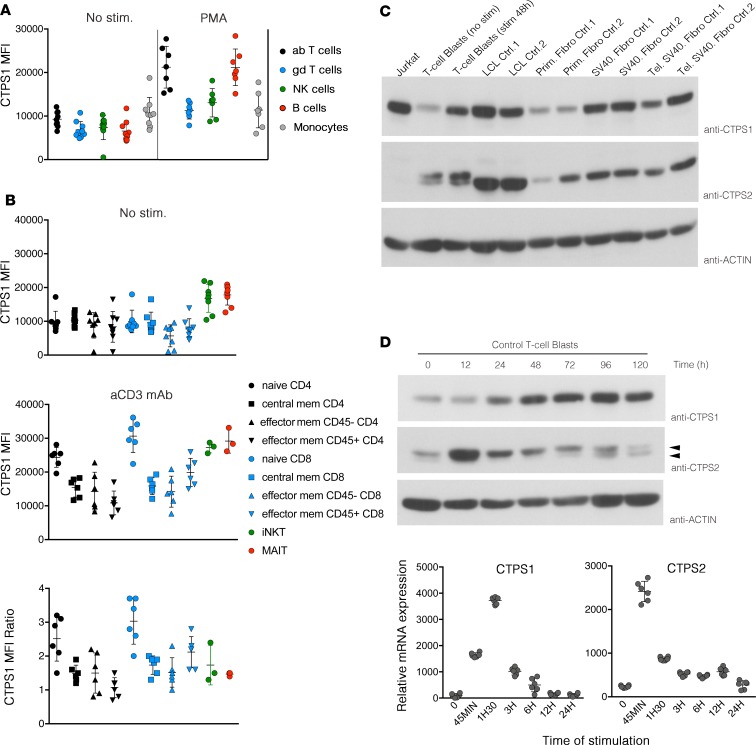
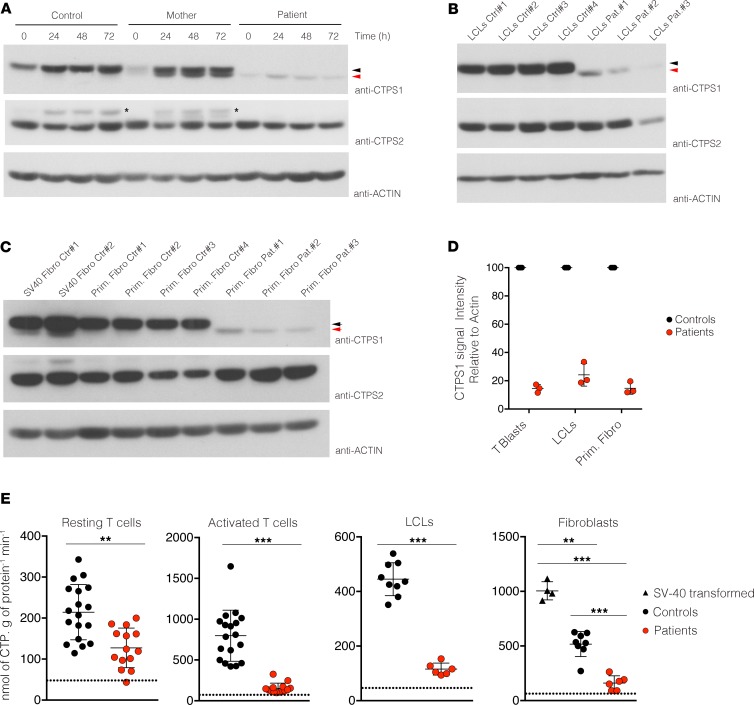
Importantly, by using this new monoclonal CTPS1 antibody, we detected in activated CTPS1-deficient T cells from patients a weak expression with an apparent lower molecular weight (Figure 5A). These findings are in agreement with our previous data showing that CTPS1 cDNA from patients in which exon 18 is lacking encodes a CTPS1 protein with a smaller size when transiently expressed (6). This shorter mutated CTPS1 protein, hereafter denoted as CTPS1Δ18, was also detectable in LCLs and primary fibroblasts derived from patients (Figure 5, B and C). The CTPS1Δ18 protein was also found at higher levels in activated T cells from 2 heterozygous maternal carriers of the mutation, but not in control cells (Figure 5A and Supplemental Figure 5). These results suggest that the presence of WT CTPS1 stabilizes the CTPS1Δ18. This hypothesis was supported by transfection of WT CTPS1 cDNA in LCLs derived from 1 patient. In these conditions, expression of WT CTPS1 increased the expression of CTPS1Δ18 (Supplemental Figure 5B). Analysis of CTPS1 mRNA expression by qPCR showed no significant difference between cells from controls and patients (Supplemental Figure 5C), indicating that the mutation had no impact on the expression or the stability of CTPS1 mRNA as initially reported (6). Quantification of CTPS1Δ18 protein in T cell blasts, LCLs, and fibroblasts of patients showed that levels were 15%, 25%, and 12%, respectively, of the WT CTPS1 level in control cells (Figure 5D), while CTPS2 protein expression in CTPS1-deficient cells from patients showed comparable levels to those of control cells (Figure 5, A–C). Treatment of activated T cell blasts from 1 patient with the proteasome inhibitor MG-132 enhanced expression of CTPS1Δ18 protein after 4 hours of incubation, while WT CTPS1 expression in control cells was unchanged (Supplemental Figure 5D). After 8 hours of treatment, both WT and mutant CTPS1 protein were decreased because of MG-132 toxicity that induced cell death (data not shown). However, these data indicate that CTPS1Δ18 protein is highly susceptible to proteasome degradation.
Figure 5. CTPS1 expression and CTPS activity in CTPS1-deficient cells from patients.
(A) Immunoblots for CTPS1, CTPS2, and actin protein expression in lysates of control (healthy donor), mother, and patient T cell blasts stimulated with anti-CD3/CD28 beads for different periods. Data are representative of 4 independent experiments. The asterisks indicate CTPS1 signals that were not removed by stripping. Black and red arrows indicate CTPS1 and mutated CTPS1Δ18 proteins, respectively. (B) Same as A with lysates from LCLs of 4 controls and 3 patients. Data are representative of 2 independent experiments. (C) Same as A with lysates from primary fibroblasts (Prim. Fibro) or SV-40–transformed fibroblasts (SV40 Fibro) of 4 controls and 3 patients. Data were obtained from 2 independent experiments. (D) Bar graph represents the quantification of the mean of signal intensity of CTPS1 protein expression from immunoblots of 3 independent experiments. CTPS1 signal was normalized to actin protein expression in each sample. CTPS1 signal intensity was fixed to 100% in control cells. Quantification was done using ImageJ software (NIH). (E) Dot plot graphs representing the quantification of CTPS activity in lysates of resting T cells and stimulated T cells with anti-CD3/CD28 beads for 48 hours from 7 patients and 8 to 15 controls and LCLs and fibroblast cell lines from 3 patients. Patients and controls correspond to red and black circles, respectively. Data from 7 independent experiments for T cells and 2 experiments for LCLs and fibroblasts. The horizontal bars represent the median ± SEM. Groups of values were compared 2 by 2 using Mann-Whitney U tests. **P < 0.01; *** P < 0.001. The dotted line represents the threshold of CTPS activity detection.
CTPS activity was then measured in protein lysates extracted from cells of controls and patients. CTPS activity was found to be markedly decreased but detectable in lysates from T cell blasts, LCLs, and fibroblasts derived from CTPS1-deficient patients as compared with control cells (Figure 5E). In control T cell blasts, CTPS activity was significantly increased following TCR-CD3 stimulation, while no or a weak increase in CTPS activity was detected in T cell blasts of patients. CTPS activity in CTPS1-deficient T cells was in the order of 20% of the activity found in control cells. These data demonstrate that expression of a mutant CTPS1 protein is detected, albeit at low levels, in CTPS1-deficient cells and is associated with a marked reduction of the CTPS activity.
Mutated CTPS1 protein is unstable but has a preserved CTPS activity.
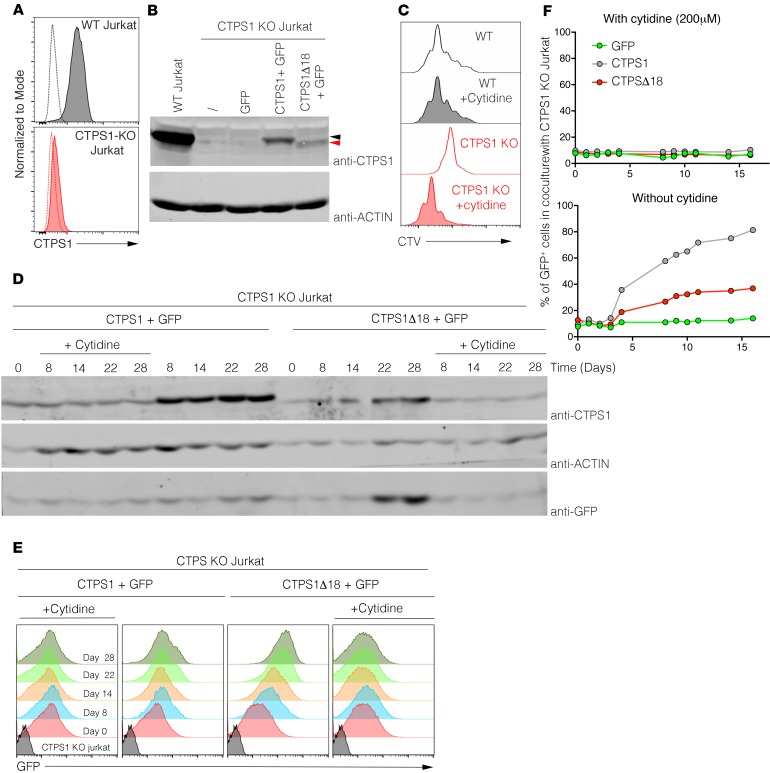
To determine whether the mutant CTPS1 protein was active and accounted for the residual CTPS activity observed in patients’ T cells, we tested its capacity to rescue the proliferation of CTPS1-deficient Jurkat cells that do not express CTPS2 (Figure 4A and data not shown). CTPS1-deficient Jurkat cells (CTPS1-KO Jurkat cells) were obtained by CRISPR/Cas9 technology through the introduction of deleterious mutations targeting exon 10 of CTPS1. In these cells, CTPS1 expression was abrogated (Figure 6, A and B) and proliferation profoundly impaired (Figure 6C). The addition of cytidine to the culture medium, allowing CTP production through the salvage pathway, rescued proliferation of the CTPS1-deficient cells.
Figure 6. Expression of CTPS1D18 mutant partially restores the proliferation of CTPS1-deficient Jurkat cells.
(A) Histograms representing flow cytometry analysis of CTPS1 expression in WT (gray) or CTPS1-KO (red) Jurkat cells. Isotype staining is represented by dotted lines. (B) Immunoblots for CTPS1 and actin protein expression in lysates of WT, CTPS1-KO, or complemented CTPS1-KO Jurkat cell lines. CTPS1-KO Jurkat cells were complemented with lentivirus expressing GFP, CTPS1 plus GFP, CTPS1Δ18 plus GFP, or the empty lentivirus (shown with a slash). Black and red arrows indicate CTPS1 and CTPS1Δ18 proteins, respectively. (C) Analysis of WT (gray histogram) and CTPS1-KO (red histogram) Jurkat cell proliferation using cell trace violet staining. Cells were treated or not with 200 μM of cytidine for 4 days. Data shown are representative of 2 independent experiments. (D) Immunoblots for CTPS1, actin, and GFP protein expressions in lysates of CTPS1 and CTPS1Δ18 complemented CTPS1-KO Jurkat cell lines at different times of culture in the presence or not of cytidine. (E) Same as D except that GFP protein expression was quantified by flow cytometry and represented in histograms. (F) Percentage of GFP+ cells in cocultures of CTPS1-KO Jurkat cells complemented with GFP alone (green symbols), CTPS1Δ18 plus GFP (red symbols), or WT CTPS1 plus GFP (gray symbols) with noncomplemented CTPS1-KO Jurkat cells at a 1:10 ratio for 16 days. Cells were cultured with cytidine (left) or not (right). Percentage values were obtained from FACS analysis. Data shown are representative of 2 independent experiments.
CTPS1-KO Jurkat cells were cultured with cytidine for their growth and then transduced with lentivirus to express WT CTPS1 or CTPS1Δ18 alongside GFP as a reporter gene (Figure 6B). Cells were sorted for GFP expression to purify. Expression of WT CTPS1 and GFP were then analyzed by Western blot and FACS. If cytidine was removed from the culture, expression of GFP and particularly WT CTPS1 rapidly increased in cell lines transduced with CTPS1 (Figure 6, D and E). In the cell lines transduced with CTPS1Δ18, the expression of the mutated protein was reduced compared with WT CTPS1 and relative to GFP (the expression of which was stronger than in cell lines transduced with CTPS1). These results suggest that in the absence of cytidine supplementation, the CTPS1Δ18-expressing cells that were selected integrated more copies of the lentivirus containing the transgene coding CTPSΔ18 and the GFP reporter (than WT CTPS1-expressing cells).
The proliferation of CTPS1- and CTPS1Δ18-expressing cells was examined in coculture experiments by mixing CTPS1-deficient cells with WT Jurkat cells at a 1:10 ratio (Figure 6F). When cytidine supplementation was maintained, no selective proliferation advantage was observed, indicating that the proliferative rates of the different cell lines were similar. In contrast, when cytidine was removed, a rapid accumulation of GFP+ cells expressing CTPS1 and to a lesser extent CTPS1Δ18 was observed (Figure 6F). The accumulation of GFP+ CTPS1Δ18-expressing cells was slower and plateaued at 30% compared with that of GFP+ CTPSΔ-expressing cells that reached 80% of the whole culture. Taken together, these data indicate that CTPS1Δ18 has a diminished capacity to rescue proliferation of CTPS1-KO cells likely because of its instability.
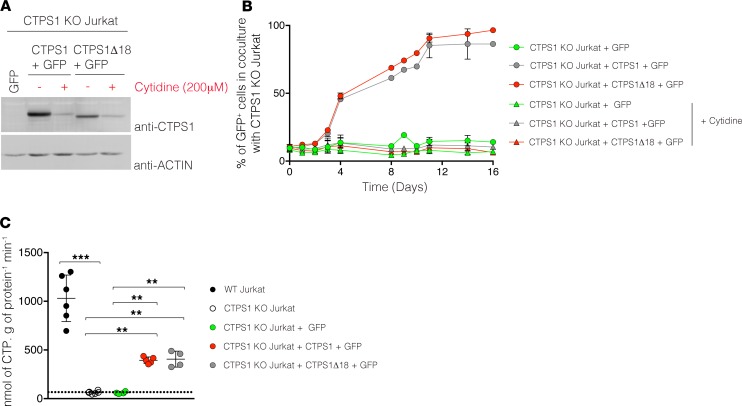
To further analyze CTPS1Δ18 enzymatic activity, CTPS1Δ18-expressing cell lines were maintained in culture without cytidine for 2 months, to select cells expressing higher amounts of CTPS1Δ18. Of note, however, CTPS1Δ18 expression remained lower than that of WT CTPS1 (Figure 7A). Proliferative capacity of these CTPS1Δ18-expressing cells was then tested in coculture experiments with CTPS1-KO Jurkat cells. In the absence of cytidine, there was a rapid expansion of GFP+ cells expressing CTPS1 or CTPS1Δ18 with similar kinetics, indicating that the level of CTPS1Δ18 in these cells was reached following selection, enabling a proliferation rate similar to that of WT CTPS1 (Figure 7B). Finally, to formally prove that the enzymatic activity of CTPSΔ18 is preserved, CTPS activity was quantified from cell extracts of CTPS1 and CTPS1Δ18-expressing cells. While CTPS activity was almost undetectable in extracts from CTPS1-KO Jurkat cells, in CTPS1- and CTPS1Δ18-expressing cell extracts, CTPS activity was comparable and much higher (Figure 7C). These results indicate that CTPSΔ18 has enzymatic activity comparable to that of CTPS1.
Figure 7. Preserved enzymatic activity of CTPS1D18 protein.
(A) Immunoblots for CTPS1 and actin protein expression in CTPS1-KO Jurkat cell lines complemented with GFP, CTPS1Δ18 plus GFP, or CTPS1 plus GFP and maintained in culture with or without 200 μM of cytidine. (B) Percentages of GFP+ cells of a coculture of CTPS1-KO Jurkat cells complemented with GFP (green symbols), CTPS1Δ18 plus GFP (red symbols), or CTPS1 plus GFP (gray symbols) with noncomplemented CTPS1-KO Jurkat cells at a 1:10 ratio for 16 days. Coculture was supplemented with 200 μM cytidine (triangle) or not (circle). Complemented cells were preselected for high CTPS1 expression by cytidine starvation before the coculture. Percentage values were obtained from FACS analysis. The values represent the mean ± SEM of 2–3 independent experiments. (C) Dot plot graph representing the quantification of CTPS activity in lysates of WT Jurkat cells (black circle), CTPS1-KO Jurkat cells (white circle), CTPS1-KO Jurkat cells complemented with GFP (green circle), CTPS1Δ18 plus GFP (red circle), or CTPS1 plus GFP (gray circle). Complemented cells were preselected for high CTPS1 expression by cytidine starvation. Data from 3–5 independent experiments. Each symbol corresponds to the CTPS activity of an independent biological replicate. The horizontal bars represent the median ± SEM. Values were compared 2 by 2 using Mann-Whitney U tests. **P < 0.01; ***P < 0.001.
In conclusion, these results demonstrate that the mutation in CTPS1 causing immunodeficiency in patients behaves as a hypomorphic mutation, characterized by 80%–90% lower expression of an unstable mutated CTPS1 protein (CTPSΔ18), which has close to normal enzymatic activity. This should be taken into account in the analysis of the immunodeficiency phenotype.
Discussion
This study provides an in-depth analysis of the CTPS1 deficiency phenotype and consequences of the mutation on protein expression and function based on data from 19 CTPS1 patients from 13 families. All patients exhibited a combined immunodeficiency phenotype that resulted in severe viral infections that developed early in life, justifying a requirement for HSCT in 15 patients. All patients carried the same mutation in CTPS1, which descends from a common ancestor in northwest England, as suggested by our previous studies (6). Patients from the United States all had ancestors who could be traced back to this geographical area.
The immunological phenotype was remarkably similar between patients. All patients showed a markedly reduced number of iNKT and MAIT cells, as well as reduced NK cells and memory B cells, whereas the other populations, such as naive and memory conventional αβ T cells, γδ T cells, Tregs, and myeloid populations (i.e., monocytes, mDCs, and pDCs), remained normal. CTPS1 was highly expressed in normal iNKT and MAIT cells, and therefore this enzyme may be critically important for the expansion of these innate lymphoid populations or their differentiation or maintenance. Of note, in contrast to conventional T cells, iNKT and MAIT cells undergo a proliferative boost in a late stage of development, when they acquire a memory and activated phenotype (25, 26). Until now, concomitant, profound defects in iNKT and MAIT cells have been reported in only 3 other primary immunodeficiencies (PIDs) caused by mutations in RORC, RASGRP1, and GINS1 (27–29). Interestingly, in RASGRP1 deficiency, CTPS1 expression was found to be strongly diminished in activated T cells, further supporting the concept that CTPS1 is necessary for the development of these invariant T cell populations (28). In RORC-deficient patients (and RORγt-KO mice), the absence of iNKT and MAIT cells is associated with a decrease in thymocytes’ survival at the double-positive stage, as shown by the underrepresentation of upstream Vα segments in the TCRα repertoire (27).
It may appear surprising that CTPS1 deficiency seems to have little or no effect on the development of conventional T cells, given the massive production of immature thymocytes that occurs in the thymus (30). It may be that the nucleoside salvage pathway compensates for this defect in CTP synthesis (31). Alternatively, residual de novo CTP synthesis may be sufficient to enable normal thymocyte proliferation and differentiation, which may be less intensive than that of activated T cells. Although conventional T cells developed normally in CTPS1-deficient patients, they exhibited poor proliferative capacities when activated by TCR stimulants. T cell effector responses, including degranulation and production of different cytokines, were unaffected, except for IL-2. Interestingly, inhibition of CTPS activity by deazauridine in control T cells completely blocked proliferation but did not impair IL-2 production, suggesting a role of CTPS1 in IL-2 production independent of its enzymatic activity. Memory B cells were lower in patients, but the proportions of switched memory cells were preserved, suggesting a role of CTPS1 in their expansion rather than in their function, like T cells. We have previously shown that, like in T cells, CTPS1 is required for B cell proliferation in response to B cell receptor stimulation (6).
Using newly available anti-CTPS1 and anti-CTPS2 monoclonal antibodies, we also showed that CTPS1 and CTPS2 expression are differentially regulated following TCR activation. CTPS2 expression is more rapidly upregulated after stimulation than CTPS1. CTPS1-deficient T cells can initiate cell division, but this is rapidly limited by the lack of CTP. Initial cell divisions might rely on CTPS2 activity, while CTPS1 takes over after 12–24 hours. Analysis of CTPS1/2 expression in different cell lines suggests a direct correlation between respective level of expression and CTPS activity and rate of proliferation. This is particularly clear in fibroblasts, in which transformation by SV-40 or telomerase ectopic expression is associated with increased CTPS1 expression and CTPS activity.
Importantly, we now demonstrate that the mutation causing CTPS1 deficiency behaves as a hypomorphic mutation. The mutation does not affect RNA stability but results in the expression of an unstable protein lacking the last exon (exon 18). Its expression is estimated to be 10%–20% of the WT CTPS1. In heterozygous carriers, the mutated CTPS1 protein is better expressed because the stability of the mutated CTPS1 is supported through its association with the WT CTPS1, known to form active tetramers (32). In complementation experiments, we showed that the activity of the mutated CTPS1 is similar to that of the WT, thereby accounting for most of the residual CTPS activity detected in cells of patients.
Of note, we recently obtained conditional Ctps1–/– KO mice that exhibit embryonic lethality (Claire Soudais and Sylvain Latour, unpublished observations). One may thus envisage that in humans too, null mutations in CTPS1 will be not viable and that the residual CTPS1 expression and activity found in patients may be sufficient to allow normal development (including lymphocyte development) and cellular function except for lymphocytes. None of the patients developed extrahematopoietic manifestations, the oldest patient having now reached the age of 20 years (following HSCT). However, we cannot exclude the possibility that CTPS1 and CTPS2 are nonredundant in mice while they may be in humans in nonlymphoid tissues. Alternatively, the pyrimidine salvage pathway may compensate for diminished CTPS activity in humans but not in the mouse.
Finally, we have generated a CTPS1-deficient Jurkat cell line obtained by CRISPR/Cas9 technology. The Jurkat cells have been derived from a patient with an acute T cell leukemia (33). We showed that Jurkat cells are completely dependent on CTPS1 for their proliferation. In the absence of CTP or cytidine supplementation, cells rapidly enter apoptosis (Norbert Minet and Sylvain Latour, unpublished data). We previously proposed that CTPS1 may represent an interesting target for selective therapy of pathologies caused by excessive T cell responses (6). The observation that CTPS1 is mandatory for this T cell leukemia cell proliferation indicates that such inhibitors of CTPS1 could also be used for the treatment of T cell lymphoma/leukemia. We previously showed that CTPS1 is involved in B cell proliferation (6); hence CTPS1 might also represent an interesting target for B cell leukemia/lymphoma. However, CTPS1 may be less important for proliferation in B cells than in T cells. Indeed, we showed that EBV-transformed tumor B cell lines expressed high levels of CTPS2 (Figure 4C).
In conclusion, the detailed description of the consequences of this hypomorphic CTPS1 mutation in the human immune system sheds light on a critical checkpoint of antigen-triggered adaptive immune responses supporting rapid cell amplification, which is critical for the control of viral infections, notably EBV and VZV.
Methods
Genomic DNA sequencing.
Genomic DNA was sequenced for CTPS1 as previously described (6). The diagnosis of patient P10 was made via whole-genome sequencing and confirmed by Sanger sequencing. Whole-genome sequencing data sets were created through the UK 100,000 Genomes Project main/pilot program, using Illumina X10 sequencing chemistry. Sequencing reads were aligned to build GRCh37 of the human reference genome using Issac (Aligner) and variants within the PID panel version analyzed. All collected sequences were analyzed using 4peaks software (version 1.8; https://nucleobytes.com/4peaks/index.html).
Gene expression analysis.
Total RNA was isolated from resting and activated T cell blasts, EBV-transformed LCLs, and fibroblasts of patients and control donors using the RNeasy Mini kit (QIAGEN). The samples were depleted of genomic DNA, and reverse transcription was performed using Superscript III First-Strand Synthesis System (Thermo Fisher Scientific). cDNAs were used as a template to quantify CTPS1, CTPS2, and GAPDH gene expressions by qPCR using universal reaction mixture (Thermo Fisher Scientific). Gene expression assays were performed with custom and primer Assays-on-Demand probes (CTPS1, Hs01041858; CTPS2, Hs00219845; GAPDH, Hs027558991) from Applied Biosystems, Thermo Fisher Scientific. All probes were labeled with dye (6-carboxy-fluorescein [FAM] or 2′-chloro-7′phenyl-1,4-dichloro-6-carboxy-fluoroscein [VIC]) and Quencher (Black Hole Quencher-1 [BHQ-1]). Real-time qPCRs were performed in triplicate using a LightCycler 1536 System (Roche). Expression levels were determined by relative quantification using the comparative threshold cycle method 2ΔΔCt, in which ΔΔCt is determined as follows: (Cttarget gene – Ctreference gene) target tissue − (Cttarget gene – Ctreference gene) calibrator tissue. The results were normalized to GAPDH gene expression and are presented as the relative value in gene expression.
Cell cultures.
PBMCs collected from patients and healthy donors were isolated by Ficoll-Paque density gradient (Lymphoprep, Proteogenix) from blood samples using standard procedures. Expansion of T cell blasts was obtained by incubating PBMCs for 72 hours with PHA (2.5 μg/mL, MilliporeSigma) in Panserin 401 (Pan Biotech) supplemented with 5% human AB serum (Bio West), penicillin (100 UI/mL), and streptomycin (100 μg/mL). After 3 days, dead cells were removed by Ficoll-Paque density gradient, and blasts were maintained in culture with IL-2 (100 UI/mL). EBV-transformed LCLs were obtained following procedures previously described (34). Primary fibroblasts, SV-40–transformed fibroblasts, Jurkat cells, and LCLs of patients and controls were cultured in RPMI 1640 (Life Technologies) supplemented with 10% heat-inactivated fetal calf serum (Gibco, Thermo Fisher Scientific), penicillin (100 UI/mL), and streptomycin (100 μg/mL).
Flow cytometry analysis.
Cell staining and the flow cytometry–based phenotypic analyses of PBMCs and cells were performed according to standard flow cytometry methods. The following monoclonal antibodies were conjugated to fluorescein isothiocyanate (FITC), R-phycoerythrin (PE), phycoerythrin-cyanin5 (PE-Cy5), phycoerythrin-cyanin5.5 (PE-Cy5.5), phycoerythrin-cyanin7 (PE-Cy7), peridinin-chlorophyll (PerCP), peridinin-chlorophyll-cyanin5.5 (PerCP-Cy5.5), allophycocyanin (APC), allophycocyanin-cyanin7 (APC-Cy7), allophycocyanin-Vio7 (APC-Vio7), Alexa Fluor 700, Brilliant Violet 421 (BV421), Brilliant Violet 510 (BV510), Brilliant Violet 650 (BV650), and Brilliant Violet 785 (BV785): anti-CD3 (catalog UCHT1), anti-CD4 (catalog OKT4), anti-CD8 (catalog RPA-T8), anti-CD11c (catalog 3.9), anti-CD14 (catalog M5E2), anti-CD16 (catalog 3G8), anti-CD19 (catalog HIB19), anti-CD25 (catalog BC96), anti-CD27 (catalog LG.3A10), anti-CD28 (catalog CD28.2), anti-CD31 (catalog WM59), anti-CD45RA (catalog HI100), anti-CD45RO (catalog UCHL1), anti-CD56 (catalog HCD56), anti-CD57 (catalog HNK-1), anti-CD70 (catalog 113-16), anti-CD161 (catalog HP-3G10), anti-CD197 (catalog G043H7), anti-CD303 (catalog BDCA-2), anti-IgM (catalog G20-127), anti-IgD (catalog IA6-2), anti-CD355 (catalog 29A1.4), anti-TCRαβ (catalog IP26), anti-TCRγδ (catalog B1), anti-IgM (catalog MHM-88), and anti-IgD (catalog IA6-2), all purchased from BioLegend.
iNKT cells were detected by staining with anti–Vα24-Jα18 (6B11, BioLegend) and anti-Vβ11 (C21, Beckman Coulter) or using α-GalCer–loaded CD1d tetramer (NIH, to be completed). MAIT cells were detected by staining with anti-Vα7.2 (3C10, BioLegend) and anti-CD161 (HP-3G10, BioLegend) or using 5-OP-RU–loaded MR1 tetramer (NIH Tetramer Core Facility, Atlanta, Georgia, USA).
For intracellular staining of CTPS1, cells were stimulated or not with PMA and ionomycin or anti-CD3/CD28 beads for 48 hours; then cells were fixed and permeabilized using Intraprep kit (Beckman Coulter) according to the manufacturer’s instructions. Cells were stained first with an anti-CTPS1 antibody [clone EPR8086(B), Abcam] or an isotype-matched antibody (rabbit IgG, clone EPR25A, Abcam) and then labeled with a PE-goat anti-rabbit secondary antibody. For detection of Treg cells, extracellular staining was performed using BV510–anti-CD4 (OKT4), PE-Cy7–anti-CD127 (A019D5), and APC–anti-CD25 (BC96) antibodies. Then, cells were fixed, permeabilized using transcription factor staining buffer set (eBioscience, Thermo Fisher Scientific), and stained with a PE–anti-Foxp3 (150D-E4) or an isotype-matched antibody. All data were collected on an LSRFortessa cytometer (BD Biosciences) and analyzed using FlowJo version 10.4.2 software (Tree Star).
Cytokine production assay.
For intracellular staining of cytokines, cells were stimulated overnight with PMA and ionomycin, anti-CD3/CD28 beads, or 1 μg/mL coated anti-CD3 (OKT3, eBioscience, Thermo Fisher Scientific) in the presence of brefeldin A (GolgiPlug, BD Biosciences). Cells were then fixed and permeabilized using the BD Cytofix/Cytoperm Plus kit (BD Pharmingen) according to the manufacturer’s instructions. Cells were labeled with PE–anti–IL-2 (rat IgG2a, MHQ1-17H12), APC–anti–IFN-γ (mouse IgG1, 4S.B3), and isotype-matched monoclonal antibodies purchased from BioLegend and analyzed by flow cytometry. For measurement of cytokine secretion, cells were stimulated with PMA and ionomycin or anti-CD3/CD28 beads or 1 μg/mL coated anti-CD3 (OKT3); supernatants were collected after 36 hours of culture. IFN-γ, TNF-α, IL-17A, IL-4, and IL-2 cytokines and granzyme B were simultaneously quantified in supernatants using coated beads array technology (BD Biosciences). All collected flow cytometry data were analyzed on FCAP Array Software (version 3.0, BD Biosciences).
Proliferation assay.
For proliferation analysis, T cell blasts were washed and cultured without IL-2 for 72 hours to synchronize the cells. Cell proliferation was monitored by labeling cells with cell-tracking violet dye (Tag-it Violet, BioLegend) before stimulation, according to the manufacturer’s instructions. Blasts or PBMCs were then cultured for 4 to 6 days in complete medium alone or in the presence of 1 μg/mL coated anti-CD3 antibody (clone OKT3, eBioscience) or anti-CD3/CD28–coated beads (Invitrogen, Thermo Fisher Scientific). PBMCs were incubated with 40 μM 3-DAZ (MilliporeSigma), 200 μM of cytidine, and 100 U/mL of IL-2 in combination or separately during the time of stimulation. Then, cells were harvested and cell-tracking violet dye dilution was assessed by flow cytometry with an LSRFortessa cytometer. The index of proliferation was calculated using FlowJo software and corresponds to the average number of cell divisions per cell including the undivided peak.
Degranulation assay.
T cell blasts were washed and stimulated for 3 hours with different concentrations of anti-CD3 antibody in the presence of anti-CD107a and anti-CD107b antibodies (H4A3 and H4B4, BioLegend). After 3 hours of stimulation, CD107a/b membrane protein expression was quantified by flow cytometry using an LSRFortessa cytometer.
Immunoblots.
The following antibodies were used for immunoblotting with standard procedures: rabbit polyclonal anti-CTPS1 (SAB111072, MilliporeSigma), rabbit monoclonal anti-CTPS1 [EPR8086(B)] and rabbit anti-CTPS2 (ab235109, both from Abcam), anti-GFP (C163, Life Technologies), and rabbit anti-actin (D18C11, Cell Signaling Technology).
CTPS activity assay.
The assay method for measuring CTPS activity is based on the transamidation of UTP by cellular extracts containing the enzyme and the simultaneous quantitative determination of UTP and CTP by liquid chromatography coupled to tandem mass spectrometry. Glutamine was added as a source of nitrogen and ATP and guanosine-5′-triphosphate (GTP) as positive regulators of the enzymatic reaction. Cell pellets were sonicated on ice and centrifuged for 30 minutes at 9300 g at 4°C. Protein concentration was determined on supernatants using the BCA protein quantitation kit (Interchim). Thirty micrograms of proteins were used for measuring CTPS activity in reaction mixture containing 1.7 mM EDTA, 13 mM MgCl2, 1.3 mM ATP, 0.2 mM GTP, 13 mM glutamine, 1.3 mM phosphoenolpyruvate, 10 mM NaF, 1.3 mM UTP, and 10 μM stable CTP isotope as an internal standard in 10 μL HEPES buffer, 70 mM, at pH 8.0. The reaction mixtures were incubated at 37°C for 90 minutes, and then the enzymatic reaction was stopped by the addition of 2 volumes of HClO4. Five microliters of sample was injected onto an Acquity HSS T3 column, 1.8-μm particle size, 2.1 × 100 mm (Waters), connected to an Acquity H-Class ultra-performance liquid chromatography interfaced with a Xevo TQ-S triple-quadrupole mass spectrometer (Waters), both controlled by MassLynx software (Waters). Aqueous mobile phase A was water at pH 6.5, and organic mobile phase B was acetonitrile, both containing 5 mM dibutylamine acetate ion pair. A programmed mobile phase gradient was used for 12-minute runs: 0 minutes, 11% B; 8 minutes, 13% B; 8.1 minutes, 100% B; 9.1 minutes, 100% B; 9.2 minutes, 10% B; and 12 minutes, 11% B. The CTP identification and detection were performed in the electrospray positive ion mode with multiple reaction monitoring mode (MRM). The most intense MRM transitions used for integration and quantification were m/z 484 > 112 and 484 > 208 for CTP and m/z 495 > 119 and 495 > 102 for the internal standard. Quantification was performed using TargetLynx software. The linearity and intraassay variability were determined for the technical validation of this assay and gave a correlation coefficient of the linear regression curves greater than 0.99 and a variation coefficient less than 15%. The threshold of CTPS activity detection corresponds to the mean of activity detected in Jurkat cells deficient for CTPS1 (that did not express CTPS2) from 4 independent experiments (51 ± 5 nmol of CTP/g protein/min).
CRISPR/Cas9 genome editing and infection.
The SpCas9(BB)-2A-EGFP (PX458) plasmid (Addgene plasmid 48138) was used for genome editing. All single-guide RNAs (sgRNAs) were designed following a previously reported procedure (34). SgRNA sequences targeting the CTPS1 gene were forward 5′-CACCGGCTTCGTGGTAGCGCAC and reverse 5′-AAACGTGCGCTACCACGAAGCC for exon 10. Pairs of synthesized oligonucleotides were annealed, phosphorylated, ligated to linearized PX458 plasmid, and transferred into Stabl3 bacteria (Thermo Fisher Scientific). sgRNA insertion in the plasmid was confirmed by Sanger sequencing using forward 5′-TTTCTTGGGTAGTTTGCAGTTTT-3′ and reverse 5′-ACGGGTACCTCTAGAGCCATT-3′ primers. Jurkat cells were transfected 2 times by electroporation using Nepa21 electroporator (Nepagene) with PX458 plasmids, sorted on EGFP expression, and subcloned. CTPS1 expression was analyzed by Western blot in clones. Clones lacking CTPS1 expression were sequenced for CTPS1 by Sanger sequencing using forward 5′-AAGTTGATCACTTTAAAGCCAGTATT-3′ and reverse 5′-TGGCACATGAAAACAAGAGG to determine the mutations that had been introduced by the CRISPR/Cas9 genome. CTPS1-KO Jurkat clones were maintained in complete medium supplemented with 200 μM of cytidine. All CTPS1-KO clones failed to proliferate in the absence of supplementation of cytidine. One clone was selected for complementation experiments.
Full-length CTPS1 and CTPS1Δ18 cDNAs were obtained by real-time PCR from donor and patient blasts, respectively, using the same forward 5′-CGGGATCCCACCATGAAGTATATTCTGGTT-3′ and different reverse 5′-CCGCTCGAGTCAGTCATGATTTATTGA-3′ (CTPS1) or 5′-CCGCTCGAGTTAAAGAAAGTCTCCAAAGC-3′ (CTPS1Δ18) primers. The cDNA was verified by sequencing and inserted into a bicistronic lentiviral expression vector encoding GFP as a reporter (pLenti7.3/V5-TOPO, Invitrogen, Thermo Fisher Scientific). Viral particles for infection were obtained by cotransfection of the lentiviral vector containing CTPS1 and the plasmids containing Gag-Pol, Rev, and the G protein of the vesicular stomatitis virus into HEK293T cells (ATCC). Viral supernatants were collected every 12 hours on 2 consecutive days, starting 48 hours after transfection, and viral particles were concentrated by ultracentrifugation at 49,000 g for 1.5 hours at 12°C. CTPS1-KO Jurkat cells were infected with viral particles at a minimal titer of 107 TU/mL, and 48 hours after infection, cells were sorted on GFP expression and starved in cytidine. The percentages of GFP+ cells were determined by flow cytometry.
Jurkat coculture assay.
All Jurkat cells were maintained in RPMI plus 10% fetal bovine serum plus 1% penicillin/streptomycin. CTPS1-KO Jurkat cells and CTPS1-KO Jurkat cells infected with a pLenti 7.3 lentivirus vector either empty or containing the CTPS1 or mutant CTPS1Δ18 sequences along with a GFP reporter gene were supplemented with 200 μM of cytidine. After an initial expansion, part of the CTPS1 or Δ18CTPS1-complemented cells were cytidine starved, leading to the loss of uninfected cells (GFP–). The number of GFP+ cells was assessed with FlowJo software using Super Enhanced Dmax. This quantification was used to adjust the number of cells needed to obtain a mix of 10% of GFP-expressing cells with 90% of WT Jurkat or CTPS1-KO Jurkat cells. The cells were then seeded at a density of 150 × 103 cells/mL with or without 200 μM of cytidine. The initial and subsequent percentages of GFP+ cells during the culture were assessed by flow cytometry.
Statistics.
P values were calculated with Mann-Whitney U test with a 2-tailed distribution. A P value less than 0.05 was considered significant.
Study approval.
Informed consent was obtained from donors, patients, and families of patients. The study and protocols conform to the 1975 Declaration of Helsinki as well as to local legislation and ethical guidelines from the Comité de Protection des Personnes de l’Ile de France II, Hôpital Necker-Enfants Malades, Paris, France. Study protocols were approved by the Comité de Protection des Personnes de l’Ile de France II.
Author contributions
EM and NM designed and performed experiments and analyzed the data. EM and SL wrote the manuscript. CP, JPV, NM, ACB, S Sobrino, S Sanquer, CL, and MLDM performed experiments and/or analyzed the data. SP, PDA, SMH, RFW, TAB, SH, and MGL identified the patients, provided and analyzed clinical information and data, and reviewed the manuscript draft. CS, TB, and AF interpreted data and reviewed manuscript draft. SL interpreted data, designed and supervised the research, and supervised the writing of the manuscript.
Supplementary Material
Acknowledgments
We acknowledge the patients, their families, and the healthy donors for cooperation and blood gifts. We also thank the members of the Latour lab for discussions. SL is a senior scientist at the Centre National de la Recherche Scientifique (CNRS, France). This work was supported by grants from Ligue Contre le Cancer-Equipe Labellisée (France) (to SL), Inserm (France), ANR-18-CE15-0025-01 (to SL) and ANR-10-IAHU-01 (Imagine Institute), the European Research Council ERC-2009-AdG_20090506 n°FP7-249816 (to AF) and Proof of Concept ERC-2015-PoC_Master/ERC-2015-PoC_680465_SAFEIMMUNOSUPPRESS (to AF, SL). NM is a fellowship recipient of the Association National de la Recherche Technologique under agreement with the industrial partner Step Pharma. ACB is supported by Imagine Institute. We acknowledge the NIH Tetramer Core Facility at Emory (Atlanta, Georgia, USA) for providing us the MR1 and CD1d tetramers. We thank Annick Lim (Institut Pasteur) for TCR repertoire analysis by Immunoscope. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health). The 100,000 Genomes Project is funded by the National Institute for Health Research and National Health Service England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support. See Supplemental Acknowledgments for Genomics England Research Consortium details.
Version 1. 03/12/2020
Electronic publication
Version 2. 03/13/2020
Updated author name
Footnotes
Conflict of interest: Step Pharma has had a partnership agreement with Imagine Institute to support the development of CTPS1 inhibitors since 2015 and has financed the PhD program of NM since 2017.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(5):e133880.https://doi.org/10.1172/jci.insight.133880.
Contributor Information
Emmanuel Martin, Email: emmanuel.martin@inserm.fr.
Norbert Minet, Email: norbert.minet@institutimagine.org.
Anne-Claire Boschat, Email: anneclaire.boschat@gmail.com.
Sylvia Sanquer, Email: sylvia.sanquer@gmail.com.
Steicy Sobrino, Email: steicy.sobrino@institutimagine.org.
Christelle Lenoir, Email: christelle.lenoir@inserm.fr.
Capucine Picard, Email: capucine.picard@inserm.fr.
Claire Soudais, Email: claire.soudais@inserm.fr.
Sophie Hambleton, Email: sophie.hambleton@newcastle.ac.uk.
Stephen M. Hughes, Email: Stephen.Hughes@cmft.nhs.uk.
Tracy A. Briggs, Email: Tracy.Briggs@manchester.ac.uk.
Monica G. Lawrence, Email: ML4NZ@hscmail.mcc.virginia.edu.
Alain Fischer, Email: alain.fischer@aphp.fr.
Peter D. Arkwright, Email: peter.arkwright@nhs.net.
Sylvain Latour, Email: sylvain.latour@inserm.fr.
References
- 1.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38(4):633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maciolek JA, Pasternak JA, Wilson HL. Metabolism of activated T lymphocytes. Curr Opin Immunol. 2014;27:60–74. doi: 10.1016/j.coi.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 3.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quéméneur L, Gerland LM, Flacher M, Ffrench M, Revillard JP, Genestier L. Differential control of cell cycle, proliferation, and survival of primary T lymphocytes by purine and pyrimidine nucleotides. J Immunol. 2003;170(10):4986–4995. doi: 10.4049/jimmunol.170.10.4986. [DOI] [PubMed] [Google Scholar]
- 5.Fairbanks LD, Bofill M, Ruckemann K, Simmonds HA. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans. Disproportionate expansion of pyrimidine pools and contrasting effects of de novo synthesis inhibitors. J Biol Chem. 1995;270(50):29682–29689. doi: 10.1074/jbc.270.50.29682. [DOI] [PubMed] [Google Scholar]
- 6.Martin E, et al. CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature. 2014;510(7504):288–292. doi: 10.1038/nature13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140(1):1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 8.van Kuilenburg AB, Meinsma R, Vreken P, Waterham HR, van Gennip AH. Isoforms of human CTP synthetase. Adv Exp Med Biol. 2000;486:257–261. doi: 10.1007/0-306-46843-3_50. [DOI] [PubMed] [Google Scholar]
- 9.van Kuilenburg AB, Meinsma R, Vreken P, Waterham HR, van Gennip AH. Identification of a cDNA encoding an isoform of human CTP synthetase. Biochim Biophys Acta. 2000;1492(2-3):548–552. doi: 10.1016/s0167-4781(00)00141-x. [DOI] [PubMed] [Google Scholar]
- 10.Kassel KM, Au da R, Higgins MJ, Hines M, Graves LM. Regulation of human cytidine triphosphate synthetase 2 by phosphorylation. J Biol Chem. 2010;285(44):33727–33736. doi: 10.1074/jbc.M110.178566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams JC, Kizaki H, Weber G, Morris HP. Increased CTP synthetase activity in cancer cells. Nature. 1978;271(5640):71–73. doi: 10.1038/271071a0. [DOI] [PubMed] [Google Scholar]
- 12.Verschuur AC, Van Gennip AH, Leen R, Meinsma R, Voute PA, van Kuilenburg AB. In vitro inhibition of cytidine triphosphate synthetase activity by cyclopentenyl cytosine in paediatric acute lymphocytic leukaemia. Br J Haematol. 2000;110(1):161–169. doi: 10.1046/j.1365-2141.2000.02136.x. [DOI] [PubMed] [Google Scholar]
- 13.van den Berg AA, van Lenthe H, Kipp JB, de Korte D, van Kuilenburg AB, van Gennip AH. Cytidine triphosphate (CTP) synthetase activity during cell cycle progression in normal and malignant T-lymphocytic cells. Eur J Cancer. 1995;31A(1):108–112. doi: 10.1016/0959-8049(94)00442-8. [DOI] [PubMed] [Google Scholar]
- 14.van den Berg AA, et al. Evidence for transformation-related increase in CTP synthetase activity in situ in human lymphoblastic leukemia. Eur J Biochem. 1993;216(1):161–167. doi: 10.1111/j.1432-1033.1993.tb18128.x. [DOI] [PubMed] [Google Scholar]
- 15.Trück J, et al. Variable phenotype and discrete alterations of immune phenotypes in CTP synthase 1 deficiency: report of 2 siblings. J Allergy Clin Immunol. 2016;138(6):1722–1725.e6. doi: 10.1016/j.jaci.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 16.Kucuk ZY, Zhang K, Filipovich L, Bleesing JJ. CTP synthase 1 deficiency in successfully transplanted siblings with combined immune deficiency and chronic active EBV infection. J Clin Immunol. 2016;36(8):750–753. doi: 10.1007/s10875-016-0332-z. [DOI] [PubMed] [Google Scholar]
- 17.Nademi Z, et al. Hematopoietic stem cell transplantation for cytidine triphosphate synthase 1 (CTPS1) deficiency. Bone Marrow Transplant. 2019;54(1):130–133. doi: 10.1038/s41409-018-0246-x. [DOI] [PubMed] [Google Scholar]
- 18.Damasceno D, et al. Distribution of subsets of blood monocytic cells throughout life. J Allergy Clin Immunol. 2019;144(1):320–323.e6. doi: 10.1016/j.jaci.2019.02.030. [DOI] [PubMed] [Google Scholar]
- 19.Duchamp M, et al. B-cell subpopulations in children: national reference values. Immun Inflamm Dis. 2014;2(3):131–140. doi: 10.1002/iid3.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shearer WT, et al. Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J Allergy Clin Immunol. 2003;112(5):973–980. doi: 10.1016/j.jaci.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 21.van der Burg M, et al. The EuroFlow PID orientation tube for flow cytometric diagnostic screening of primary immunodeficiencies of the lymphoid system. Front Immunol. 2019;10:246. doi: 10.3389/fimmu.2019.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piątosa B, Wolska-Kuśnierz B, Pac M, Siewiera K, Gałkowska E, Bernatowska E. B cell subsets in healthy children: reference values for evaluation of B cell maturation process in peripheral blood. Cytometry B Clin Cytom. 2010;78(6):372–381. doi: 10.1002/cyto.b.20536. [DOI] [PubMed] [Google Scholar]
- 23.Berland A, et al. PROMIDISα: a T-cell receptor α signature associated with immunodeficiencies caused by V(D)J recombination defects. J Allergy Clin Immunol. 2019;143(1):325–334.e2. doi: 10.1016/j.jaci.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 24.McPartland RP, Wang MC, Bloch A, Weinfeld H. Cytidine 5’-triphosphate synthetase as a target for inhibition by the antitumor agent 3-deazauridine. Cancer Res. 1974;34(11):3107–3111. [PubMed] [Google Scholar]
- 25.Martin E, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009;7(3):e54. doi: 10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 27.Okada S, et al. Immunodeficiencies. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 2015;349(6248):606–613. doi: 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winter S, et al. Loss of RASGRP1 in humans impairs T-cell expansion leading to Epstein-Barr virus susceptibility. EMBO Mol Med. 2018;10(2):188–199. doi: 10.15252/emmm.201708292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cottineau J, et al. Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J Clin Invest. 2017;127(5):1991–2006. doi: 10.1172/JCI90727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yates AJ. Theories and quantification of thymic selection. Front Immunol. 2014;5:13. doi: 10.3389/fimmu.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Austin WR, et al. Nucleoside salvage pathway kinases regulate hematopoiesis by linking nucleotide metabolism with replication stress. J Exp Med. 2012;209(12):2215–2228. doi: 10.1084/jem.20121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch EM, et al. Human CTP synthase filament structure reveals the active enzyme conformation. Nat Struct Mol Biol. 2017;24(6):507–514. doi: 10.1038/nsmb.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider U, Schwenk HU, Bornkamm G. Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int J Cancer. 1977;19(5):621–626. doi: 10.1002/ijc.2910190505. [DOI] [PubMed] [Google Scholar]
- 34.Izawa K, et al. Inherited CD70 deficiency in humans reveals a critical role for the CD70-CD27 pathway in immunity to Epstein-Barr virus infection. J Exp Med. 2017;214(1):73–89. doi: 10.1084/jem.20160784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.