Abstract
Extracellular cold-inducible RNA-binding protein (eCIRP) is a recently discovered damage-associated molecular pattern. Understanding the precise mechanism by which it exacerbates inflammation is essential. Here we identified that eCIRP is a new biologically active endogenous ligand of triggering receptor expressed on myeloid cells-1 (TREM-1), fueling inflammation in sepsis. Surface plasmon resonance revealed a strong binding affinity between eCIRP and TREM-1, and fluorescence resonance energy transfer assay confirmed eCIRP’s interaction with TREM-1 in macrophages. Targeting TREM-1 by its siRNA or a decoy peptide, LP17, or by using TREM-1–/– mice dramatically reduced eCIRP-induced inflammation. We developed a potentially novel 7-aa peptide derived from human eCIRP, M3, which blocked the interaction of TREM-1 and eCIRP. M3 suppressed inflammation induced by eCIRP or agonist TREM-1 antibody cross-linking in murine macrophages or human peripheral blood monocytes. M3 also inhibited eCIRP-induced systemic inflammation and tissue injury. Treatment with M3 further protected mice from sepsis, improved acute lung injury, and increased survival. Thus, we have discovered a potentially novel TREM-1 ligand and developed a new peptide, M3, to block eCIRP–TREM-1 interaction and improve outcomes in sepsis.
Keywords: Immunology, Inflammation
Keywords: Bacterial infections, Innate immunity, Macrophages
Extracellular cold-inducible RNA-binding protein is a biologically active endogenous ligand of triggering receptor expressed on myeloid cells-1 that fuels inflammation in sepsis.
Introduction
Pathogen-associated molecular patterns and damage-associated molecular patterns (DAMPs) play leading roles in fueling inflammation in sepsis (1–3). Cold-inducible RNA-binding protein (CIRP) is an 18-kDa RNA chaperone protein (4). In addition to its passive release due to necrotic cell death, CIRP can be released extracellularly during sepsis, hemorrhage, or ischemia/reperfusion injury, translocating from the nucleus to cytoplasmic stress granules before being released to the extracellular space (5). Extracellular CIRP (eCIRP) acts as a DAMP, causing inflammation and organ injury (5, 6). Elevated plasma levels of eCIRP have been correlated with a poor prognosis in patients with sepsis (5, 7). As a relatively new DAMP, a more thorough understanding of eCIRP’s pathobiology in inflammatory diseases is required to develop novel therapeutics.
Similar to other DAMPs, eCIRP recognizes Toll-like receptor 4 (TLR4) (5, 8) expressed in macrophages, lymphocytes, and neutrophils to increase cytokine production (5), T cell polarization (9), and neutrophil activation (10). Healthy mice injected with recombinant murine CIRP (rmCIRP) develop acute lung injury (ALI) via activation of macrophages, neutrophils, and endothelial cells and display increased vascular permeability in the lungs (11, 12). Conversely, CIRP–/– mice are protected from sepsis and ALI (5, 6). Consistent with these findings, treatment of septic animals with a neutralizing antibody against eCIRP attenuated organ injury and prolonged survival (5, 6). Collectively, these findings indicate that eCIRP is a major contributing factor in the pathogenesis of sepsis and that targeting eCIRP is a valid strategy to mitigate sepsis severity.
Triggering receptor expressed on myeloid cells-1 (TREM-1) is an innate immune receptor expressed primarily on neutrophils and macrophages (13). TREM-1 activation triggers inflammation independently (14), as well as by synergizing with the TLR4 pathways (14, 15). TREM-1 activation leads to DNAX-activating protein of 12 kDa (DAP12) phosphorylation (13, 15), which promotes activation of the tyrosine kinase Syk (16), resulting in the production of cytokines and chemokines (15, 16). The ligands for TREM-1 remain elusive, with only high mobility group box 1 (HMGB1) (17), extracellular actin (18), and peptidoglycan recognition protein 1 (PGLYRP1) (19) identified thus far. The nature of the TREM-1 ligand(s) and mechanisms of TREM-1 signaling are not yet thoroughly explored. Identification of a new natural endogenous TREM-1 ligand will not only help improve our understanding of the pathophysiology of inflammatory diseases but also discover new therapeutic avenues against those diseases.
Both eCIRP and TREM-1 are upregulated in sepsis to serve as mediators of inflammation (5, 20), but their interaction has not been studied. In sepsis, both eCIRP and TREM-1 reside extracellularly (5, 20), giving rise to the hypothesis that eCIRP could be a novel endogenous ligand of TREM-1. In this study, we have discovered that eCIRP is a new biologically active endogenous TREM-1 ligand and their interaction fuels inflammation. We have also developed a unique human eCIRP-derived ligand-dependent 7-aa peptide (RGFFRGG) to serve as an antagonist of TREM-1, named M3, and implemented M3 as a therapeutic in a preclinical model of sepsis.
Results
Identification of eCIRP as a potentially new TREM-1 ligand to promote inflammation.
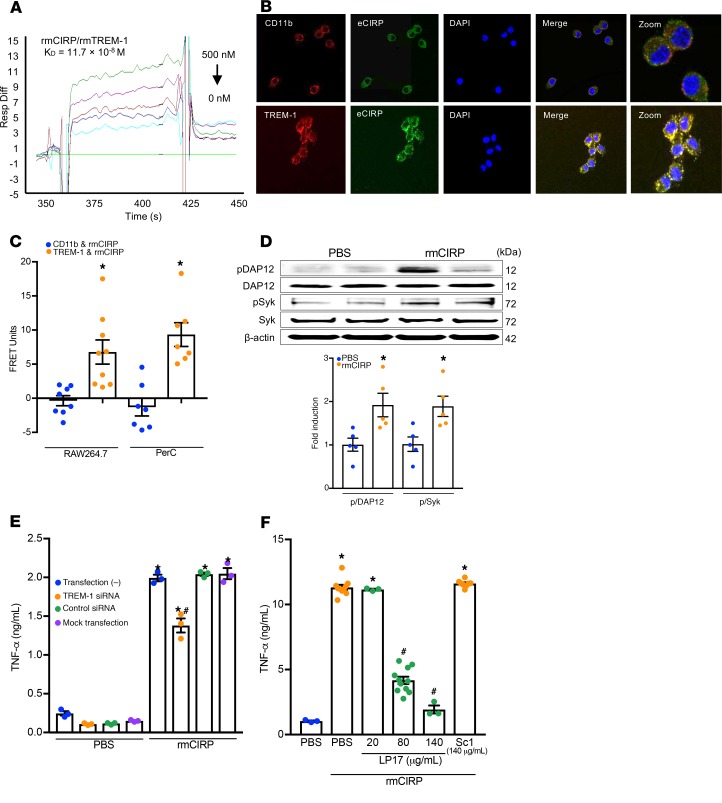
To study the direct interaction between eCIRP and TREM-1, we performed a surface plasmon resonance (SPR) assay, which demonstrated strong binding between rmCIRP and rmTREM-1 with a KD of 11.7 × 10–8 M (Figure 1A). An immunofluorescence study was performed to show the colocalization of eCIRP and TREM-1 in macrophages after rmCIRP stimulation. It clearly demonstrated the colocalization of rmCIRP and TREM-1, as indicated by the yellow color in the merged image (Figure 1B). Conversely, rmCIRP did not colocalize with a negative control, the pan-macrophage marker CD11b (Figure 1B). We next performed FRET analysis to quantitatively determine rmCIRP’s association with TREM-1. FRET analysis revealed a clear association between rmCIRP and TREM-1 with an increase in FRET units of nearly 7-fold compared with rmCIRP’s interaction with negative control CD11b in RAW264.7 macrophages (Figure 1C) and nearly 10-fold in WT peritoneal macrophages (Figure 1C). These findings imply that eCIRP is a novel TREM-1 ligand. We then studied the activation of downstream molecules DAP12 and Syk in macrophages treated with rmCIRP and found a significant increase in the phosphorylation of DAP12 and Syk at 10 minutes after rmCIRP stimulation (Figure 1D and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.134172DS1). We next confirmed the functional role of TREM-1 in eCIRP-mediated inflammation. We found that the siRNA-treated macrophages showed significant inhibition of TNF-α production following rmCIRP stimulation (Figure 1E). Similarly, the treatment of macrophages with LP17, an inhibitor of TREM-1 (21), dose-dependently inhibited rmCIRP-induced TNF-α production in RAW264.7 cells (Figure 1F). Conversely, the scramble peptide for LP17 did not show any inhibition of TNF-α production (Figure 1F). Collectively, these data show that eCIRP specifically binds to TREM-1 in macrophages and induces TNF-α production. TREM-1 expression in macrophages is increased in sepsis (15). To explore the role of eCIRP in this increase, RAW264.7 cells and murine primary peritoneal macrophages were stimulated with rmCIRP. TREM-1 mRNA levels were increased 2.5-fold in rmCIRP-treated RAW264.7 cells as compared with PBS control (Supplemental Figure 2A). The protein levels of TREM-1 expression on the cell surface of both RAW264.7 cells and primary murine peritoneal macrophages treated with rmCIRP were significantly increased by 4.3- and 1.6-fold, respectively, compared with PBS control (Supplemental Figure 2, B and C).
Figure 1. eCIRP binds TREM-1 to promote inflammation.
(A) SPR between rmCIRP and rmTREM-1. Anti-his antibody was used to capture rmCIRP-his. rmTREM-1 was injected as an analyte in concentrations of 0 to 500 nM. (B) RAW264.7 cells were treated with rmCIRP (5 μg/mL) at 4°C for 10 minutes, fixed in a nonpermeabilized fashion, and stained with primary antibodies against CIRP, TREM-1, and CD-11b as well as fluorescently labeled secondary antibodies. Confocal microscopy images were obtained with a 63× objective. Colocalization is indicated by the yellow color. (C) After the staining protocol described in B, cell-associated fluorescence was measured. The transfer of fluorescence was calculated as FRET units. Data are expressed as mean ± SEM obtained from 3 independent experiments; n = 8–9/group. Groups compared by unpaired t test (*P < 0.01 vs. CD11b). (D) RAW264.7 cells were stimulated with rmCIRP (1 μg/mL) for 10 minutes. Extracted proteins were immunoprecipitated by using anti-DAP12 antibody, followed by Western blotting using phospho-Tyr (p-Tyr; 4G10) and DAP12 antibody. Extracted total proteins obtained from RAW264.7 cells stimulated with rmCIRP (1 μg/mL) for 10 minutes were subjected to Western blotting using p-Syk, Syk, and β-actin antibodies. Representative Western blots for phosphotyrosine (4G10), DAP12, p-Syk, Syk, and β-actin are shown. Phosphotyrosine (p-DAP12) and p-Syk expression in each sample was normalized to DAP12 or Syk or β-actin expression and the mean values of 0 minutes of rmCIRP-treated groups were standardized as one for comparison. Data are expressed as mean ± SEM (n = 5 samples/group). The groups were compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. PBS). (E) RAW264.7 cells were transfected as shown. Cells were stimulated with PBS control or 1 μg/mL rmCIRP. After 6 hours, TNF-α in the supernatant was analyzed by ELISA. Data are expressed as mean ± SEM (n = 3 samples/group). Multiple groups were compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. respective PBS group; #P < 0.01 vs. rmCIRP-treated nontransfected cells). (F) RAW264.7 cells were stimulated with PBS or rmCIRP (1 μg/mL). Simultaneously cells were treated with various doses of LP17 or LP17-Sc1. After 24 hours, TNF-α in culture supernatants was measured by ELISA. Data are expressed as mean ± SEM obtained from 5 independent experiments (n = 3–10 wells/group). The groups were compared by Kruskal-Wallis test with Dunn’s method (*P < 0.05 vs. PBS; #P < 0.05 vs. rmCIRP). FRET, fluorescence resonance energy transfer; PerC, peritoneal cavity; ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline.
Pharmacological inhibition of TREM-1 attenuates eCIRP-induced systemic inflammation and lung injury in mice.
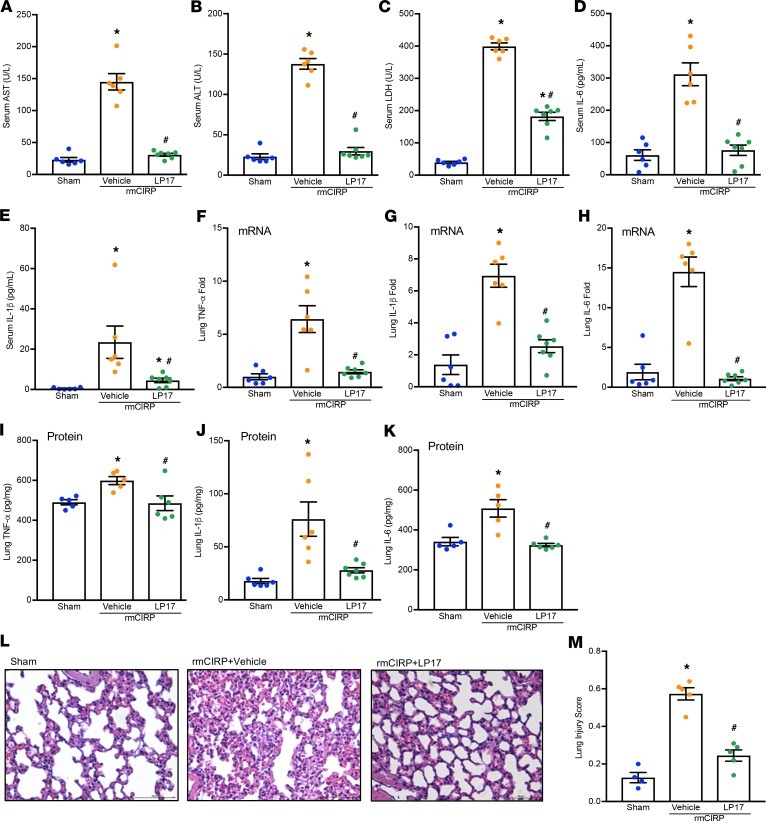
To further verify the interaction between TREM-1 and eCIRP in vivo, we exposed mice to rmCIRP and LP17, a TREM-1 decoy receptor, which functions to inhibit TREM-1–mediated signaling (21). We showed that LP17 attenuated rmCIRP-induced systemic inflammation and lung injury in mice (Figure 2). Treatment of mice with LP17 dramatically reduced the levels of AST, ALT, and LDH in the serum of rmCIRP-injected mice compared with vehicle-treated rmCIRP-injected mice (Figure 2, A–C). The levels of IL-6 and IL-1β in the serum were significantly decreased in LP17-treated mice (Figure 2, D and E). LP17 treatment significantly attenuated TNF-α, IL-1β, and IL-6 mRNA (Figure 2, F–H) and protein in the lungs (Figure 2, I–K). Similarly, treatment with LP17 significantly reduced the expression of chemokines macrophage inflammatory protein 2 (MIP-2) and keratinocyte chemoattractant (KC) and adhesion molecules intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) mRNA in the lungs (Supplemental Figure 3). Histological images of lung tissue showed increased levels of alveolar congestion, exudate, interstitial and alveolar cellular infiltrates, intra-alveolar capillary hemorrhages, and damage of epithelial architecture in rmCIRP-injected mice compared with sham mice (Figure 2L). LP17 treatment improved these histological injury parameters in rmCIRP-injected mice (Figure 2L). These histological changes were reflected in a significant decrease in lung tissue injury score in LP17-treated mice (Figure 2M).
Figure 2. LP17 inhibits eCIRP-induced inflammation in vivo.
Adult C57BL/6 mice were randomly assigned to sham, vehicle (PBS), or treatment group. rmCIRP at a dose of 5 mg/kg BW or equivalent volume of normal saline was administered i.v. via retro-orbital injection. LP17 at a dose of 5 mg/kg BW or vehicle was given i.p. at the time of rmCIRP injection. At 5 hours after rmCIRP injection, mice were euthanized, and blood and tissue were collected for analysis. (A) AST, (B) ALT, and (C) LDH were determined using specific colorimetric enzymatic assays. Serum (D) IL-6 and (E) IL-1β were measured by ELISA. Lung mRNA levels of (F) TNF-α, (G) IL-1β, and (H) IL-6 were measured by real-time PCR (RT-PCR). Equal amounts of total lung protein (250–350 μg) were loaded into respective ELISA wells for assessment of lung protein levels of (I) TNF-α, (J) IL-1β, and (K) IL-6. (L) Representative images of H&E-stained lung tissue at original magnification ×200. (M) Lung injury score calculated at original magnification ×400. n = 5 high-powered fields/group. Data are expressed as mean ± SEM. n = 5–7 mice/group. The groups were compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. sham, and #P < 0.05 vs. vehicle mice). ALT, alanine aminotransferase; AST, aspartate aminotransferase; LDH, lactate dehydrogenase.
TREM-1–/– macrophages and mice produce decreased levels of proinflammatory cytokines upon eCIRP exposure.
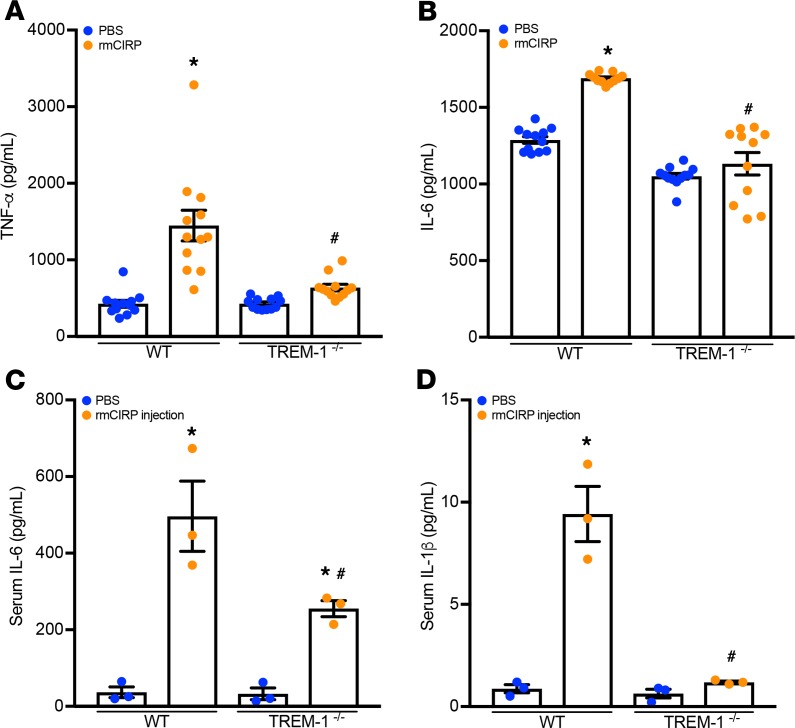
To definitively confirm the functional interaction between eCIRP and TREM-1, we performed both in vitro and in vivo studies using TREM-1–/– macrophages and mice. Primary peritoneal macrophages were isolated from both WT and TREM-1–/– mice and exposed to rmCIRP. IL-6 and TNF-α production after rmCIRP stimulation were significantly decreased in supernatant of TREM-1–/– macrophages as compared with WT macrophages (Figure 3, A and B). Similarly, WT mice exposed to rmCIRP increased the serum levels of IL-6 and IL-1β by 2.3- and 9.1-fold as compared with their TREM-1–/– counterparts, respectively (Figure 3, C and D). These data suggest that genetic ablation of TREM-1 controls eCIRP-induced inflammation.
Figure 3. TREM-1 deficiency ameliorates eCIRP-mediated inflammation.
Primary peritoneal macrophages were isolated from WT and TREM-1–/– mice and were stimulated with PBS or rmCIRP (1 μg/mL). After 24 hours, (A) TNF-α and (B) IL-6 in culture supernatants were measured by ELISA. Data are expressed as mean ± SEM; n = 12 wells/group. Multiple groups were compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. respective PBS group; #P < 0.001 vs. rmCIRP-treated WT cells). Adult C57BL/6 WT and TREM-1–/– mice were given rmCIRP at a dose of 5 mg/kg BW or equivalent volume PBS. After 5 hours, serum (C) IL-6 and (D) IL-1β were measured by ELISA. Data are expressed as mean ± SEM. n = 3 mice/group. Multiple groups were compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. respective PBS group; #P < 0.05 vs. rmCIRP-treated WT mice).
An eCIRP-derived antagonist, M3, inhibits the eCIRP–TREM-1 interaction.
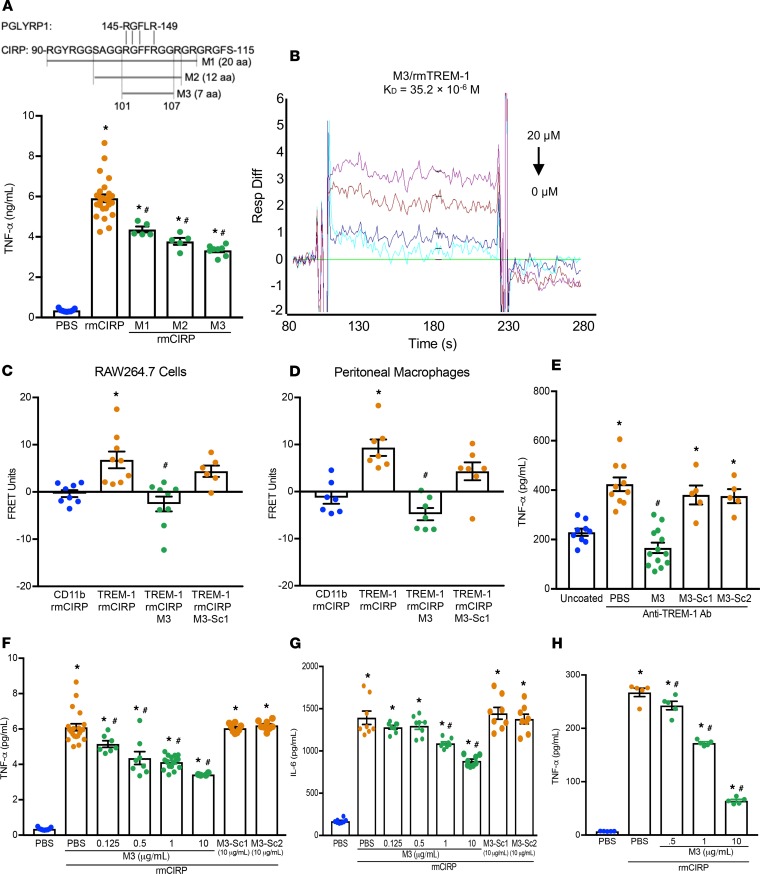
After establishing eCIRP as a novel TREM-1 ligand, we sought to identify a small molecule that inhibits the eCIRP–TREM-1 interaction. Using the Protein Model Portal, part of the Protein Structure Initiative KnowledgeBase, and PEP-FOLD3 (22), we compared structural images of a known TREM-1 ligand, murine PGLYRP1, and murine CIRP. We identified a section of CIRP with a similar structural form and associated similar aa sequences (Supplemental Figure 4, A and B). We then synthesized a series of small peptides (M1, M2, M3) from this region of CIRP and tested their ability to inhibit TNF-α secretion in RAW264.7 cells stimulated with rmCIRP (Figure 4A). We found that M3, a 7-aa peptide, consisting of the aa sequence of murine CIRP from 101 to 107 (RGFFRGG), which has 100% homology with human CIRP, demonstrated the greatest inhibitory effect on TNF-α production by the macrophages following rmCIRP treatment (Figure 4A). Using SPR, we identified a considerable binding affinity between M3 and rmTREM-1, with a KD of 35.2 × 10–6 M (Figure 4B). A FRET assay was performed between rmCIRP and TREM-1 in both RAW264.7 cells and murine primary peritoneal macrophages in the presence or absence of M3. Interestingly, we found that M3 was able to dramatically abrogate rmCIRP’s binding to TREM-1 (Figure 4, C and D). The agonist anti–TREM-1 Ab has been shown to induce TNF-α production through TREM-1 cross-linking (13). By inhibiting TREM-1–mediated inflammation caused solely by Ab-induced receptor activation, in the absence of any other stimuli, we have demonstrated the independent role of M3 on TREM-1–mediated inflammation. We found that M3 demonstrated significant inhibition of TNF-α production by the macrophages treated with an agonist anti–TREM-1 Ab, indicating M3 specifically worked on TREM-1 (Figure 4E). On the other hand, the scramble peptides, M3-Sc1 and M3-Sc2, did not demonstrate any inhibition (Figure 4E). We next validated M3’s inhibitory effect on controlling rmCIRP-treated inflammation in macrophages. We found that M3 inhibited rmCIRP-mediated TNF-α and IL-6 production by the RAW264.7 cells in a dose-dependent manner while the scramble peptides, M3-Sc1 and M3-Sc2, were not able to inhibit rmCIRP-induced inflammation (Figure 4, F and G). Similarly, M3 treatment was also able to significantly inhibit the production of TNF-α in rmCIRP-stimulated primary human PBMCs obtained from healthy volunteers, where the highest inhibition of 76% in TNF-α production occurred at 10 μg/mL of M3 treatment (Figure 4H). Collectively, we have discovered a potentially novel small peptide, M3, that blocks the eCIRP–TREM-1 interaction and inhibits eCIRP-mediated inflammation.
Figure 4. M3, a small CIRP-derived peptide, inhibits eCIRP and TREM-1 interaction.
(A) Partial aa sequence of CIRP highlighting an area of similarity with PGLYRP1. RAW264.7 cells were treated with 10 μg/mL of peptides M1, M2, or M3 for 30 minutes, then stimulated with rmCIRP (1 μg/mL). After 24 hours, TNF-α in culture supernatants was measured. Data are expressed as mean ± SEM obtained from 2 independent experiments (n = 5–7 wells/peptide group) and were compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. unstimulated cells, and #P < 0.05 vs. rmCIRP-treated cells). (B) SPR between rmTREM-1 and M3. (C) RAW264.7 cells and (D) primary peritoneal macrophages were treated with M3 or M3-Sc1 (both 10 μg/mL) for 30 minutes. Cells were then stimulated with PBS or rmCIRP (5 μg/mL), and FRET analysis was performed as described in Figure 1C. Data are expressed as mean ± SEM obtained from 3 independent experiments; n = 7–9/group, compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. CD11b + rmCIRP, and #P < 0.05 vs. TREM-1 + rmCIRP). (E) To activate RAW264.7 cells through TREM-1, 96-well plates were precoated with 20 μg/mL of an agonist anti–TREM-1 mAb. Then, 5 × 104 cells/well were premixed with PBS control or M3 (10 μg/mL) or scramble peptide (10 μg/mL) for 30 minutes, then plated. TNF-α production was measured in the culture supernatants after an additional 24 hours of incubation. The experiment was performed 2 independent times with n = 5–6 wells per group. Scramble peptide groups, n = 5/group. Data are expressed as mean ± SEM and were compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. uncoated; #P < 0.05 vs. TREM-1 Ab + PBS). (F and G) RAW264.7 cells were treated with M3 or scramble peptide for 30 minutes, then stimulated with PBS or rmCIRP (1 μg/mL). After 24 hours, (F) TNF-α and (G) IL-6 in culture supernatants were assessed. Data are expressed as mean ± SEM obtained from 3 independent experiments and compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. PBS-treated cells; #P < 0.05 vs. rmCIRP + PBS). (H) Human macrophages were treated with M3 for 30 minutes, then stimulated with PBS or rmCIRP (1 μg/mL). After 24 hours, TNF-α in culture supernatants was measured. Data are expressed as mean ± SEM (n = 5 wells/group) and were compared by Kruskal-Wallis test with Dunn’s method (*P < 0.05 vs. PBS-treated cells; #P < 0.05 vs. rmCIRP + PBS). Ab, antibody.
M3 inhibits eCIRP- and LPS-induced inflammation in mice.
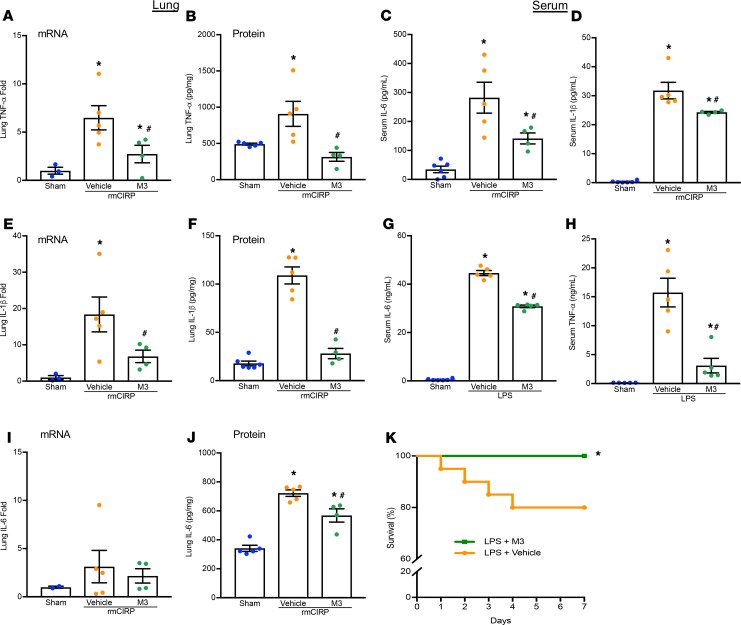
Having established M3 as an antagonist of eCIRP and TREM-1 binding, we sought to evaluate its efficacy in reducing pulmonary and systemic inflammation in vivo. Administration of rmCIRP in healthy mice dramatically increased lung mRNA and protein levels of TNF-α, IL-1β, and IL-6 while M3 treatment reduced their expression (Figure 5, A, B, E, F, I, and J). rmCIRP administration also increased serum levels of IL-6 and IL-1β, and M3 treatment decreased these levels (Figure 5, C and D). LPS is known to stimulate eCIRP release by macrophages (5). Using an endotoxemia model, we demonstrated that M3 treatment was able to reduce serum levels of IL-6 and TNF-α (Figure 5, G and H). Administration of M3 during endotoxemia increased the 7-day survival from 70% to 100% (Figure 5K). Therefore, TREM-1 is involved in eCIRP-mediated inflammation and tissue injury, and blockade of the eCIRP–TREM-1 interaction attenuates inflammation.
Figure 5. M3 inhibits eCIRP- and LPS-mediated inflammation.
Adult C57BL/6 mice were randomly assigned to sham, vehicle (PBS), or treatment group. rmCIRP at a dose of 5 mg/kg BW or equivalent volume normal saline was administered. M3 at a dose of 10 mg/kg BW or vehicle was given i.p. at the time of rmCIRP injection. At 5 hours after rmCIRP injection, mice were euthanized, and blood and tissue were collected for analysis. Lung mRNA and protein levels of (A and B) TNF-α, (E and F) IL-1β, and (I and J) IL-6 were measured by RT-PCR and ELISA. Serum levels of (C) IL-6 and (D) IL-1β were measured by ELISA. Data are expressed as mean ± SEM. The groups were compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. sham, and #P < 0.05 vs. vehicle mice). Adult C57BL/6 mice were i.p. injected with LPS at a dose of 15 mg/kg BW or equivalent volume normal saline (sham). M3 at a dose of 10 mg/kg BW or vehicle (PBS) was given simultaneously. After 90 minutes, serum was collected, and ELISA was used to measure (G) IL-6 and (H) TNF-α. Data are expressed as mean ± SEM. n = 4–5 mice/group. The groups were compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. sham, and #P < 0.05 vs. vehicle-treated mice). C57BL/6 mice were injected i.p. with 15 mg/kg BW LPS and simultaneously given M3 i.p. at a dose of 10 mg/kg BW or equivalent volume vehicle (PBS). (K) Mice were monitored for survival for 7 days. n = 20 mice/group; *P < 0.05 vs. LPS + vehicle (PBS), log-rank (Mantel-Cox) test.
M3 protects mice from polymicrobial sepsis.
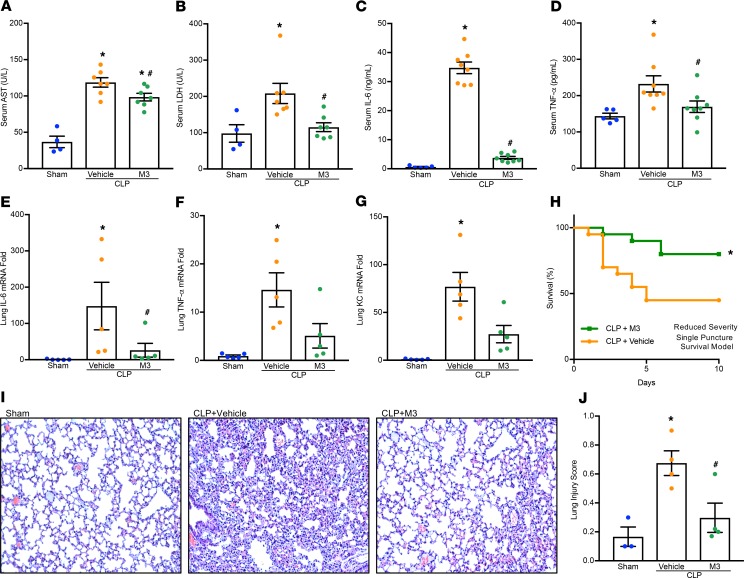
We next evaluated the efficacy of M3 in a more clinically relevant model of sepsis using cecal ligation and puncture (CLP) in mice. A double-puncture, high-mortality CLP resulted in increases in organ injury markers AST and LDH in the serum at 20 hours, while the mice treated with M3 simultaneously had a significant decrease in their levels (Figure 6, A and B). Similarly, serum levels of IL-6 and TNF-α were elevated by CLP; however, M3 treatment significantly reduced these levels (Figure 6, C and D). Expression of the proinflammatory cytokine IL-6 and TNF-α and chemokine KC mRNA in lung tissues was increased in CLP-induced sepsis, and were reduced with M3 treatment (Figure 6, E–G). Histological images of lung tissue in CLP mice displayed significant damage, with increased levels of alveolar congestion, proteinaceous debris, interstitial and alveolar neutrophil infiltration, intra-alveolar capillary hemorrhages, and damage of epithelial architecture (Figure 6I). M3 treatment improved these histological injury parameters in septic mice (Figure 6I). These histological changes were reflected in a significant decrease in lung tissue injury score in M3-treated mice compared with vehicle mice (Figure 6J). To determine if M3 was able to improve survival in sepsis, mice were subjected to a reduced-severity, single-puncture model of CLP and randomized to simultaneous treatment with M3 or vehicle. We found that M3 treatment increased the survival rate from 45% to 80% at day 10 after CLP (Figure 6H). In addition, in order to test the efficacy of M3 when given in a delayed manner after sepsis induction, mice were subjected to an increased-severity, but still single-puncture, CLP. M3 injection was given i.p. 90 minutes after CLP. M3 was able to improve 10-day survival from 10% to 25% (P = 0.02) (Supplemental Figure 5). In summary, the eCIRP released in sepsis binds to TREM-1 and potentiates proinflammatory signaling. Administration of M3, by inhibiting the eCIRP–TREM-1 interaction, exhibits excellent therapeutic potential against murine polymicrobial sepsis.
Figure 6. M3 protects mice from polymicrobial sepsis.
Adult C57BL/6 mice were randomly assigned to sham laparotomy, CLP plus vehicle (PBS), or CLP plus treatment group. At the time of CLP treatment, mice received an i.p. instillation of 10 mg/kg BW M3 before abdominal closure. Vehicle groups received an equivalent volume of normal saline. Analysis of serum and tissue in graphs (A–G and I–J) were obtained 20 hours after double-puncture CLP. (A) AST and (B) LDH were determined using specific colorimetric enzymatic assays. Serum (C) IL-6 and (D) TNF-α were measured by ELISA. Lung mRNA levels of (E) IL-6, (F) TNF-α, and (G) KC were measured by RT-PCR. Data are expressed as mean ± SEM (n = 5–8 mice/group) and were compared by 1-way ANOVA and Tukey’s method (A, B, D, and F) or Kruskal-Wallis test with Dunn’s method (C, E, and G) (*P < 0.05 vs. sham, and #P < 0.05 vs. vehicle-treated mice). (H) Kaplan-Meier survival curve generated from treatment (M3) and vehicle CLP mice during the 10-day monitoring period after reduced-severity CLP with simultaneous M3 treatment is shown. n = 20 mice in each group; *P < 0.05 vs. vehicle, determined by log-rank test. (I) Representative images of H&E-stained lung tissue at original magnification ×200. (J) Lung injury score calculated at original magnification ×400. n = 4 high-powered fields/group. Data are expressed as mean ± SEM and compared by 1-way ANOVA and Tukey’s method (*P < 0.05 vs. sham, and #P < 0.05 vs. vehicle mice).
Discussion
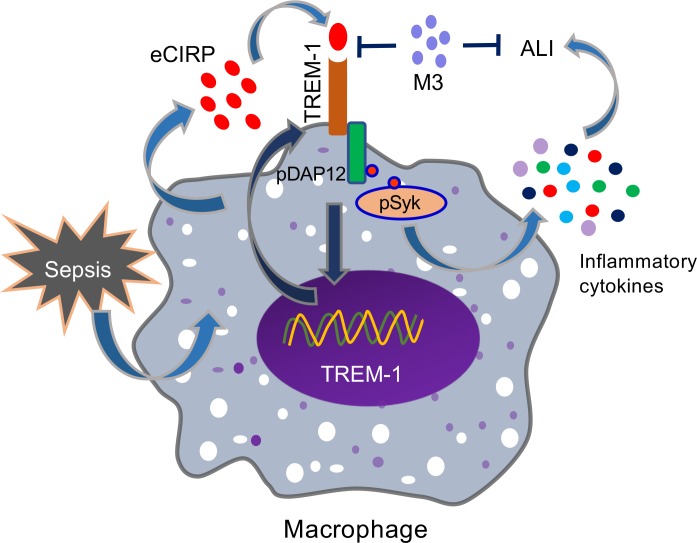
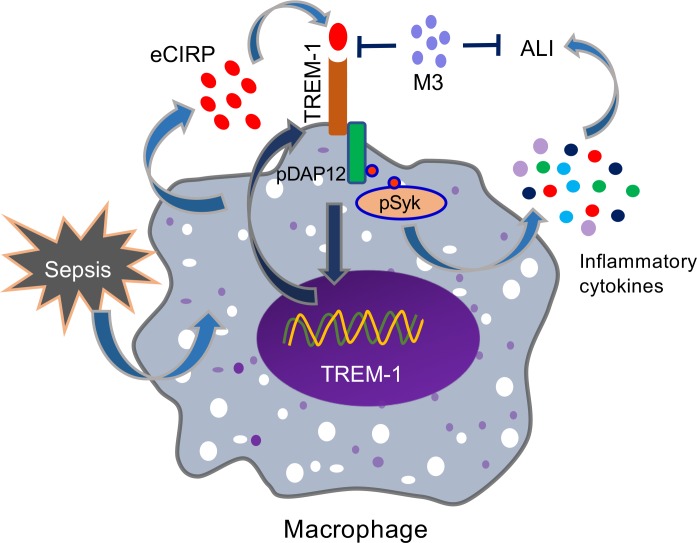
eCIRP has been recently identified as a DAMP that is released during sepsis, shock, and ischemia/reperfusion injury (5, 6). However, its molecular mechanism to induce inflammation remains enigmatic. Despite identification of the TREM-1 receptor nearly 2 decades ago (13), its ligands are not well elucidated. Our study fills a significant previous knowledge gap by establishing a potentially novel link between eCIRP and TREM-1 demonstrated by in vitro studies using murine and primary human macrophages and in several in vivo models. We summarized the overall findings in Figure 7, which demonstrates that eCIRP released during sepsis binds to TREM-1, serving as a novel biologically active endogenous TREM-1 ligand. This binding leads to the activation of intracellular DAP12 and Syk and increased production of inflammatory mediators to cause hyperinflammation and tissue injury. Targeting the interaction between eCIRP and TREM-1 by a small peptide, M3, derived from human eCIRP is protective in sepsis.
Figure 7. Summary of findings.
Sepsis causes an increased release of eCIRP. The endogenous ligand eCIRP recognizes TREM-1 and activates intracellular signaling molecules DAP12 and Syk, leading to increased expression of proinflammatory mediators that cause excessive inflammation and remote tissue injury. eCIRP increases TREM-1 expression, possibly via positive feedback induction. A small peptide, M3, derived from human eCIRP abrogates eCIRP–TREM-1 interaction, thereby leading to decreased inflammation and attenuated ALI.
We have, for the first time to our knowledge, identified eCIRP as an endogenous ligand for TREM-1. In other words, TREM-1 is a new receptor for eCIRP. Previously, TLR4 was the only identified receptor for eCIRP (5, 6). We hypothesized that, like most DAMPs, eCIRP has several receptors for signal transduction. HMGB1, for example, is known to potentiate signals through TLR2, TLR4, receptor for advanced glycation end product (RAGE), TREM-1, and CD163 (23–25). Using SPR, we found a strong KD of 11.7 × 10–8 M of binding between eCIRP and TREM-1, which is comparable to the dissociation constant of the previously identified receptor for eCIRP, TLR4 (5). Of the known ligands for TREM-1, PGLYRP1 (19), extracellular actin (18), and HMGB1 (17), SPR has been used between TREM-1 and PGLYRP1 and HMGB1. Surprisingly, the KD between eCIRP and TREM-1 (11.7 × 10–8 M) was more than 2-fold lower than the KD between HMGB1 and TREM-1 (35.4 × 10–6 M) (17), indicating higher binding affinity of eCIRP to TREM-1 than between HMGB1 and TREM-1. In addition to SPR, several approaches were taken to confirm the physical interaction of eCIRP and TREM-1, including confocal microscopy and FRET analysis.
After demonstrating eCIRP’s binding to TREM-1, the focus was shifted to determining whether eCIRP’s binding to TREM-1 resulted in fueling inflammation and causing tissue injury. This was a crucial step because we previously demonstrated that although SPR displays binding between eCIRP and TLR2 as well as RAGE, there are no functional outcomes of this association (5). Inhibition of TREM-1 by siRNA showed some, but not complete, reduction in the production of proinflammatory cytokines by the macrophages treated with eCIRP. This could be due to off-target effects of TREM-1 siRNA on other TREMs, although this remains speculative. The TREM family contains TREM-1, TREM-2, and in mice, TREM-3 (26). Although TREM-1 contains an immunoreceptor tyrosine-based activation motif, additional TREM family members contain an immunoreceptor tyrosine-based inhibitory motif, and thus they could play an antiinflammatory role (27, 28). In addition, macrophages deficient in TREM-2 are hyperresponsive to TLR ligands (29). We therefore used another approach to inhibit TREM-1 by LP17, which abrogated eCIRP-induced inflammation and ALI, confirming the notion that the eCIRP–TREM-1 interaction is functionally dynamic. LP17 is a 17-aa peptide whose sequence is taken from TREM-1; it is thought to function as a decoy receptor (21). We attempted to determine the interaction between LP17 and eCIRP using SPR and demonstrated a KD of 48 × 10–6 M between these 2 molecules (data not shown). This could be artificially elevated because of conditions present in vivo that we cannot reproduce artificially, such as involvement of other biological cofactors, temperature, and molecular stability. The TREM-1 antagonists function primarily as decoy receptors and to prevent receptor dimerization (21, 30). Additionally, Sigalov et al. have developed a ligand-independent inhibitor of TREM-1 (31, 32). Although use of these peptides in animal models of inflammatory diseases has resulted in decreased inflammation and improved survival, it is not clear which ligand of TREM-1 these decoy peptides neutralize, rendering their use somewhat less specific. Here, we have developed a ligand-dependent TREM-1 inhibitor, a potentially novel short peptide M3. As demonstrated with FRET analysis and functionally in vitro and in vivo, M3 is able to prevent eCIRP’s interaction with TREM-1, thereby impeding eCIRP’s proinflammatory role.
Both the specificity to the eCIRP and TREM-1 interaction and the small size of M3 may prove advantageous in future therapeutic applications of M3. M3 has a molecular weight of 795.9 Da. Most peptides in clinical use are injected subcutaneously. Peptides under 1 kDa can be absorbed directly into the systemic circulation via blood capillaries, whereas larger peptides are absorbed initially through the lymphatic system or in a combination of both blood and lymphatic absorption (33, 34). Additionally, because patient compliance is increased with noninvasive administration, small peptides have an advantage over larger peptides because they can be administered through mucosal absorption. In the current study, we demonstrated M3 had an excellent inhibitory role in mitigating inflammation not only in macrophage cells but also in bacterial sepsis. Prior to use of M3 in large animal models or human clinical trials, further studies on the stability, half-life, and safety would be required. However, at the doses used here, M3 did not demonstrate immunogenicity or tissue injury, as demonstrated by cytokine levels, organ injury markers, or histological analysis. In addition, cell proliferation assays in the presence of 100 μg/mL of M3 peptide — a dose 10 times higher than doses used in vitro in this study — demonstrated no toxicity (Supplemental Figure 6).
In addition to eCIRP’s interaction with TREM-1, we found that the expression of TREM-1 in macrophages was increased following treatment with eCIRP. Several studies have established the connection between TLR4 and TREM-1 in immune cells (14, 35, 36). The cellular consequences of TREM-1 activation following treatment with anti–TREM-1 Ab were examined in terms of global gene expression and compared with cells receiving LPS and cells receiving a combined treatment of anti–TREM-1 plus LPS (14). The results indicated that besides having the crosstalk with TLR4, TREM-1 signaling independently promoted the expression of some unique genes (14), implying the direct impact of the TREM-1 pathway in inflammation. As previously discussed, like other DAMPs, eCIRP has been shown to recognize TLR4 (5). TLR4 activation leads to upregulation of TREM-1 expression, which is MyD88 dependent and involves transcription factors NF-κB, PU.1, and AP1 (37). Additionally, simultaneous activation of TREM-1 and TLR4 leads to synergistic production of proinflammatory mediators through common signaling pathway activation, including PI3K, ERK1/2, IL-1 receptor associated kinase 1, and NF-κB (13–16). Thus, the upregulation of TREM-1 expression in macrophages after treatment with eCIRP could be promoted through both TLR4- and TREM-1-dependent pathways. The connection between TLR4 and TREM-1 has been shown in neutrophils: following LPS stimulation of neutrophils, TREM-1 was found to be colocalized with TLR4 (36). Genetic deletion of TREM-1 downregulates the expression of several genes implicated in the TLR4 pathway, including MyD88, CD14, and IκBα (35). Our data demonstrating the eCIRP-induced expression of proinflammatory cytokines by the macrophages also supports the synergistic effects of TREM-1 and TLR4. However, to demonstrate the independent proinflammatory role of TREM-1 in eCIRP-mediated inflammation, we activated TREM-1 using agonist Ab cross-linking (13, 14). TREM-1 activation resulted in increased levels of TNF-α; however, TNF-α production was diminished in cells treated with M3. M3 interfered with the cross-linking effect, demonstrating a TLR4-independent interaction between eCIRP and TREM-1.
In summary, we have discovered that eCIRP is a new ligand of TREM-1 in macrophages. We have developed a small peptide, M3, which successfully reduces systemic inflammation and improves survival in murine models of infectious inflammation. M3 was able to successfully inhibit inflammation in a human cell line. Future work should focus on additional immune cell types and other models of inflammatory diseases. Identification of eCIRP as a new biologically active endogenous TREM-1 ligand has allowed additional insight into the pathobiology of eCIRP-mediated inflammation in sepsis.
Methods
Experimental animals.
C57BL/6 male mice were purchased from Charles River Laboratories. The TREM-1–/– mouse, Trem1tm1(KOMP)Vlcg, was generated by the trans-NIH Knockout Mouse Project (KOMP) and obtained from the KOMP Repository, University of California, Davis. Age-matched (8–12 weeks) healthy mice were used in all experiments. Animals were randomly assigned to sham, vehicle, or treatment group. Every attempt was made to limit the number of animals used per experiment.
Only male mice were used in this study. Studies have found sex-specific differences in sepsis (38). It has been reported that male and female sex steroids exhibit diverse immune-modulating functions in both humoral and cell-mediated immune responses under normal conditions and various disease processes (38). Male sex hormones have been shown to suppress cell-mediated immune responses and cardiovascular function while female sex hormones have been shown to be protective. Survival rates after CLP have been shown to be greatly increased in female mice as compared with male mice (39). Given the impact of sex on sepsis pathogenesis, to generate reliable and consistent findings, only male mice were used.
Peptides.
LP17 (LQVTDSGLYRCVIYHPP), LP17-scramble control (TDSRCVIGLYHPPLQVY), M3 (RGFFRGG), and M3-scramble controls, M3-Sc1 (FGRGFRG) and M3-Sc2 (GFFGRGR), were synthesized by GenScript.
Animal model of polymicrobial sepsis.
Mice were anesthetized with inhaled isoflurane and placed in supine position. CLP was performed through a midline laparotomy (40, 41). Briefly, the abdomen was shaved and disinfected. A 2-cm incision was created; the cecum was exposed and ligated with a 4-0 silk suture 1 cm proximal from the distal cecal extremity. For 20-hour experiments the cecum was punctured twice with a 22-gauge needle. A small amount of cecal content was extruded, and the ligated cecum was returned to the peritoneal cavity. The wound was closed in layers. For 10-day CLP experiments the cecum was punctured only once with the same sized needle. However, the proximity of cecal ligation and amount of stool extruded varied between the reduced severity and the more severe model. Mice were allocated to the treatment or vehicle group. Treatment mice received an i.p. injection of 10 mg/kg BW M3 during abdominal closure in both the 20-hour experiments and the simultaneous treatment survival study. Vehicle groups received an equivalent volume of normal saline. Sham group animals underwent a laparotomy without cecal ligation or puncture. After closure, the mice received a subcutaneous injection of 1 mL of normal saline to avoid surgery-induced dehydration. Animals in the 20-hour experiment were not given antibiotics so that mice would develop severe sepsis and early mortality. Animals in the 10-day survival experiments were given 500 μL of the antibiotic imipenem (0.5 μg/kg, Merck) and 500 μL of normal saline subcutaneously at the time of laparotomy. For the 10-day experiment, mice were evaluated twice daily for their survival status.
Analgesics and sedatives to mitigate pain and discomfort in septic mice can modulate immune responses in sepsis. Studies have shown that the levels of proinflammatory cytokines are reduced by fentanyl and/or midazolam, which may contribute to the beneficial effects of these medications in septic mice. Additionally, analgesia can reduce inflammatory responses in septic mice, resulting in immunosedation and contributing to improved outcomes and mortality rates in murine sepsis (42). While we did use antibiotics for the survival studies, we did not use antibiotics for the short-term, 20-hour experiments. The inflammatory response in sepsis depends, at least in part, on bacterial load. Because the use of antibiotics reduces bacterial contents, and therefore inflammation, antibiotics were not used in the short-term experiments to allow for a rapid, robust inflammatory response. All experiments were in compliance with IACUC guidelines. Additional environmental enrichment and comfort was provided to all mice in the form of pressed cotton squares (43) and Shepard Shacks (44).
Animal model of endotoxemia.
Mice were injected i.p. with 15 mg/kg BW of LPS (Escherichia coli serotype O55:B5; MilliporeSigma). Mice were allocated to the treatment or vehicle group. Treatment mice received an i.p. injection of 10 mg/kg BW M3 at the time of LPS injection. The vehicle group received an equivalent volume of PBS. After 90 minutes, mice were sacrificed, and serum was collected for analysis. To study survival rates, mice were treated with 15 mg/kg BW LPS with or without 10 mg/kg BW M3 and were evaluated twice daily for 7 days.
In vivo administration of rmCIRP, LP17, and M3.
rmCIRP was produced in our lab as previously described (5). rmCIRP at a dose of 5 mg/kg BW or normal saline was administered i.v. via retro-orbital injection using a 29G × 1/2″ U-100 insulin syringe (Terumo Medical Corporation). LP17 at a dose of 5 mg/kg BW or M3 (10 mg/kg BW) or vehicle (PBS) was given i.p. at the time of rmCIRP injection. At 5 hours after rmCIRP injection, WT and TREM-1–/– mice were anesthetized, and blood and tissue were collected for analysis. A section of lung tissue was preserved in 10% formalin for histopathological analysis, and the rest was frozen in liquid nitrogen and stored at −80°C for quantitative PCR analysis.
Cell culture and isolation of peritoneal macrophages.
Mouse macrophage RAW264.7 cells were obtained from ATCC. Macrophages from healthy human donors were obtained from Hemacare (PBMACC-MON-2). Murine peritoneal macrophages were isolated from WT adult male mice (45). Briefly, 1 mL of 4% thioglycolate (MilliporeSigma) was injected i.p. Four days later, mice were euthanized using CO2 asphyxiation. Peritoneal fluid and cells were isolated using peritoneal lavage with PBS. Total peritoneal cells were isolated by centrifugation at 200 g for 10 minutes and subsequently cultured in DMEM (Thermo Fisher Scientific). After 4 hours nonadherent cells were removed, and adherent cells, primarily macrophages, were cultured overnight prior to use. For rmCIRP stimulation studies, murine peritoneal macrophages were isolated from WT and TREM-1–/– adult male mice. Mice were euthanized using CO2 asphyxiation. Peritoneal fluid and cells were isolated using peritoneal lavage with PBS. Total peritoneal cells were isolated by centrifugation at 200 g for 10 minutes and subsequently cultured in DMEM. After 4 hours nonadherent cells were removed, and adherent cells, primarily macrophages, were cultured overnight before use. All cultured medium was supplemented with 10% heat-inactivated fetal bovine serum (FBS, MP Biomedicals), 1% penicillin-streptomycin, and 2 mM glutamine. Cells were maintained in a humidified incubator with 5% CO2 at 37 °C.
Determination of organ injury markers.
Serum levels of LDH, AST, ALT, and lactate were determined using specific colorimetric enzymatic assays (Pointe Scientific) according to the manufacturer’s instructions.
ELISA.
Supernatant or serum was analyzed by ELISA kits specific for IL-6, TNF-α, IFN-γ (all from BD Biosciences), and IL-1β (Invitrogen, Thermo Fisher Scientific) according to the manufacturers’ instructions. The lung tissue was crushed in liquid nitrogen, and equal weights of powdered tissues (~50 mg) were dissolved in 500 μL of lysis buffer (10 mM HEPES at pH 7.4, 5 mM MgCl2, 1 mM DTT, 1% Triton X-100, and 2 mM each of EDTA and EGTA) and subjected to sonication on ice. Protein concentration was determined by the Bio-Rad protein assay reagent. Equal amounts of proteins (250–500 μg) were loaded into respective ELISA wells for the assessment of TNF-α, IL-6, and IL-1β.
Lung histopathology.
Lung tissues were fixed in 10% formalin before being embedded in paraffin. Tissues were cut into 5-μm sections and stained with H&E. Slides were evaluated under light microscopy to evaluate the degree of lung injury. Scoring was done using a system created by the American Thoracic Society (46). Scores ranged from 0 to 1 and were based on the presence of proteinaceous debris in the airspaces, the degree of septal thickening, and neutrophil infiltration in the alveolar and interstitial spaces. The average score per field was calculated at ×400 original magnification.
Assessment of TREM-1 expression in macrophages by flow cytometry.
To detect TREM-1 expression on the surface of macrophages, a total of 1 × 106 RAW264.7 or primary peritoneal macrophages were washed with FACS buffer containing PBS with 2% FBS and stained with APC anti-mouse TREM-1 Ab (clone 174021, R&D Systems, Bio-Techne). Unstained cells were used as a negative control to establish the flow cytometer voltage setting. Acquisition was performed on 10,000 events using a BD LSRFortessa flow cytometer (BD Biosciences), and data were analyzed with FlowJo software (Tree Star).
Real-time quantitative reverse transcription PCR.
Total RNA was extracted from tissue using TRIzol reagent (Invitrogen, Thermo Fisher Scientific). cDNA was synthesized using MLV reverse transcriptase (Applied Biosystems, Thermo Fisher Scientific). PCR reactions were carried out in 20 μL of a final volume of 0.08 μM of each forward and reverse primer (Supplemental Table 1), cDNA, water, and SYBR Green master mix (Applied Biosystems, Thermo Fisher Scientific). Amplification and analysis were conducted in a Step One Plus real-time PCR machine (Applied Biosystems, Thermo Fisher Scientific). Mouse β-actin mRNA was used as an internal control for amplification, and relative gene expression levels were calculated using the ΔΔCT method. Relative expression of mRNA was expressed as fold change in comparison with sham tissues.
Immunoprecipitation and Western blotting.
For immunoprecipitation assay of DAP12 and phosphotyrosine, RAW264.7 cells (1 × 106) were stimulated with rmCIRP (1 μg/mL). At different time points, cells were harvested and proteins were extracted using extraction buffer containing 25 mM Tris, 0.15 M NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, 2 mM Na3VO4, and protease inhibitor cocktail (Roche Diagnostics), pH 7.4. Equal amounts of protein were subjected to immunoprecipitation using anti-DAP12 Ab (Cell Signaling Technology) and protein A/G plus agarose beads (Pierce Classic IP Kit, Thermo Fisher Scientific). Following immunoprecipitation, samples were eluted and run on 4%–12% gradient polyacrylamide gels for electrophoresis. Gels were transferred into nitrocellulose membranes, blocked with 3% BSA, and finally reacted with primary Abs against phosphotyrosine (catalog 05-321; clone 4G10; MilliporeSigma) and DAP12 (Cell Signaling Technology). For the analysis of p-Syk, Syk, and β-actin, equal amounts of protein were subjected to Western blotting using 4%–12% gradient polyacrylamide gels. The blots were reacted with p-Syk (Abcam), Syk (Cell Signaling Technology), and β-actin (MilliporeSigma) primary Abs, followed by reaction with fluorescence-labeled secondary Abs (anti–mouse IgG, catalog 926-68070; anti–rabbit IgG, catalog 926-32211; Li-Cor Biosciences) and detection by Odyssey FC Dual-Mode Imaging system (Li-Cor Biosciences). The intensities of the bands were measured by using Image Studio 5.2 (Li-Cor Biosciences).
Stimulation of cytokine production by TREM-1 engagement.
To activate RAW264.7 cells through TREM-1, 96-well flat-bottom plates were precoated with 20 μg/mL of an agonist anti–TREM-1 mAb (clone 174031; R&D Systems, Bio-Techne) overnight at 37°C, similar to previously described protocols (13, 15). The wells were washed with sterile PBS and 5 × 104 cells/well were plated. Cells treated with M3 or scramble M3-Sc1 were premixed with the peptide for 30 minutes before adding to the well. TNF-α production was measured in the culture supernatants after an additional 24 hours of incubation.
SPR.
To examine the direct interaction between eCIRP and TREM-1, SPR, using Biacore technology (3000 instrument, GE Healthcare), was performed between rmCIRP and rmTREM-1. Anti-his Ab (His Capture Kit, GE Healthcare Life Sciences, catalog 28995056) was used to capture rmCIRP-his. TREM-1 (recombinant mouse TREM-1–Fc Chimera, R&D Systems, Bio-Techne, catalog 1187-TR-025) was injected as an analyte in concentrations of 0 to 500 nM. Binding reactions were performed in 1× HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.05% P20, pH 7.4.) The CM5 dextran chip (flow cell-2) was first activated by injection with a mix of 35 μL of 0.1 M N-ethyl-N′-[3-diethylaminopropyl]-carbodiimide and 0.1 M N-hydroxysuccinimide. An aliquot of 200 μL of 5 μg/mL of the ligand diluted in 10 mM sodium acetate (pH 4.5) was injected into flow cell-2 of the CM5 chip for immobilization. Next, 35 μL of 1 M ethanolamine (pH 8.2) was injected to block the remaining active sites. The flow cell-1 was treated with 35 μL of 0.1 M N-ethyl-N′-[3-diethylaminopropyl]-carbodiimide and 0.1 M N-hydroxysuccinimide and blocked with 1 M ethanolamine (pH 8.2) without the ligand and used as a control to evaluate nonspecific binding. The binding analyses were performed at a flow rate of 30 μL/min at 25°C. To evaluate the binding, the analyte (0.5 μM for the yes-or-no binding analysis and ranging from 31.25 nM to 0.5 μM for the kinetics analysis) was injected into flow cell-1 and flow cell-2, and the association of analyte and ligand was recorded by SPR. The signal from the blank channel (flow cell-1) was subtracted from the channel coated with the ligand (flow cell-2). Data were analyzed by the Biacore 3000 Evaluation software. For all samples, a blank injection with buffer alone was subtracted from the resulting reaction surface data. Data were globally fitted to the Langmuir model for 1:1 binding.
Immunofluorescence assay.
RAW264.7 cells were treated with rmCIRP (5 μg/mL) at 4°C for 10 minutes and then fixed in a nonpermeabilizing fashion using 4% paraformaldehyde. Immunofluorescence staining was performed using primary Abs against TREM-1, CD11b, and CIRP and fluorescently tagged secondary Abs. The fixed cells were washed in PBS and blocked with 5% normal horse serum for 1 hour. Primary Abs were diluted in 1% horse serum and incubated with cells for 2 hours at room temperature or overnight at 4°C. After washing with PBS, cells were incubated in the dark with diluted secondary Abs for 1 hour. After an additional washing step, slides were mounted immediately on Vectashield mounting medium with DAPI (Vector Laboratories). Antibodies used were as follows: rabbit anti-mouse CIRP Ab (catalog 10209-2-AP; ProteinTech), goat anti-mouse TREM-1 Ab (catalog AF1187; R&D Systems, Bio-Techne), goat anti-CD11b Ab (catalog MBS420973; MyBioSource), Cy5-conjugated AffiniPure donkey anti–goat IgG (code 705-175-147; Jackson ImmunoResearch Laboratories) and Cy3-conjugated AffiniPure F(ab′)2 donkey anti–rabbit IgG (code 711-166-152; Jackson ImmunoResearch Laboratories). Confocal microscopy images were obtained using a Zeiss LSM880 confocal microscope equipped with a 63× objective. Images were analyzed and quantified using ZenBlue software (Zeiss).
FRET analyses.
FRET analysis was performed as described previously (47). RAW264.7 cells and peritoneal macrophages from WT adult male mice were treated with rmCIRP at 4°C for 10 minutes with or without M3 or scramble peptide, fixed in a nonpermeabilized fashion, and stained using the Abs and protocol listed under Immunofluorescence assay in Methods. Cell-associated fluorescence was measured on a Biotek Synergy Neo2 at 566 nm upon excitation at 488 nm (E1), at 681 nm after excitation at 630 nm (E2), and at 681 nm after excitation at 488 nm (E3). The transfer of fluorescence was calculated as FRET units. FRET unit = (E3both − E3none) − ([E3Cy5 − E3none] × [E2both/E2Cy5]) − ([E3Cy3 − E3none] × [E1both/E1Cy3]).
Cell proliferation assay.
To confirm that there were no cytotoxic effects from LP17 or M3, a cell proliferation assay (CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay, Promega) was performed according to the manufacturer’s instructions. Briefly, RAW264.7 cells were plated in 96-well plates and treated with PBS, 1 μg/mL CIRP, or 1 μg/mL CIRP in addition to various doses of the inhibitory peptide. Cell viability was determined by colorimetric analysis at 490 nm.
Statistics.
Data represented in the figures are expressed as mean ± SEM. All data have been tested for normality using the Kolmogorov-Smirnov test of normality. Normally distributed data were analyzed using 1-way ANOVA for comparison among multiple groups, and the significance of differences between individual groups was determined by Tukey’s method. The 2-tailed t test was applied for 2-group comparisons. Nonparametric 1-way comparison among multiple groups was performed using the Kruskal-Wallis test with Dunn’s multiple-comparisons test. The specific tests used for each graph are identified in the figure legends. Significance was considered for P ≤ 0.05 between study groups. Data analyses were carried out using GraphPad Prism graphing and statistical software (GraphPad Software).
Study approval.
All experiments were performed in accordance with the guidelines for the use of experimental animals by the NIH and were approved by the IACUC of the Feinstein Institutes for Medical Research. Approval was not needed for the use of human PBMCs because these cells from deidentified human samples were obtained from a commercial vendor as primary cells.
Author contributions
NLD and MA designed the experiments. NLD performed animal and in vitro experiments. AM and SDG performed in vitro mechanistic studies. MO performed animal survival studies. NLD, MA, and JMP analyzed the data. NLD, MA, and PW prepared the figures and wrote the manuscript. PW revised and edited the manuscript. PW conceived the idea and supervised the project.
Supplementary Material
Acknowledgments
We thank Michael Bloch and Ying Zhang of the University of Maryland and Archna Sharma of the Center for Immunology and Inflammation (CII), the Feinstein Institutes for Medical Research (FIMR), for Biacore assays; Amanda Chan of Light Microscopy Core Facility (FIMR) for microscopic study assistance; Betsy Barnes, Center for Autoimmune, Musculoskeletal and Hematopoietic Diseases (FIMR), for the FRET assay help; and Max Brenner of CII (FIMR) for critical review. This study was supported by NIH grants R35GM118337 (to PW) and R01GM129633 (to MA).
Version 1. 02/06/2020
In-Press Preview
Version 2. 03/12/2020
Electronic publication
Funding Statement
This study was supported by the National Institutes of Health (NIH) grants R35GM118337 (P.W.) and R01GM129633 (M.A.).
Footnotes
Conflict of interest: PW is an inventor of patent applications (WO/2010/120726 and 61/881.798) covering the fundamental concept of targeting cold-inducible RNA-binding protein for the treatment of inflammatory diseases, licensed by TheraSource LLC. PW is a cofounder of TheraSource LLC.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(5):e134172.https://doi.org/10.1172/jci.insight.134172.
Contributor Information
Naomi-Liza Denning, Email: Ndenning@northwell.edu.
Monowar Aziz, Email: maziz1@nshs.edu.
Atsushi Murao, Email: amurao@northwell.edu.
Steven D. Gurien, Email: sgurien@northwell.edu.
Mahendar Ochani, Email: mochani@northwell.edu.
Jose M. Prince, Email: JPrince@northwell.edu.
Ping Wang, Email: pwang@northwell.edu.
References
- 1.Rittirsch D, Flierl MA, Ward PA. Harmful molecular mechanisms in sepsis. Nat Rev Immunol. 2008;8(10):776–787. doi: 10.1038/nri2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aziz M, Jacob A, Yang WL, Matsuda A, Wang P. Current trends in inflammatory and immunomodulatory mediators in sepsis. J Leukoc Biol. 2013;93(3):329–342. doi: 10.1189/jlb.0912437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denning NL, Aziz M, Gurien SD, Wang P. DAMPs and NETs in sepsis. Front Immunol. 2019;10:2536. doi: 10.3389/fimmu.2019.02536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishiyama H, Itoh K, Kaneko Y, Kishishita M, Yoshida O, Fujita J. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137(4):899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiang X, et al. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nat Med. 2013;19(11):1489–1495. doi: 10.1038/nm.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aziz M, Brenner M, Wang P. Extracellular CIRP (eCIRP) and inflammation. J Leukoc Biol. 2019;106(1):133–146. doi: 10.1002/JLB.3MIR1118-443R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y, Dong H, Zhong Y, Huang J, Lv J, Li J. The cold-inducible rna-binding protein (CIRP) level in peripheral blood predicts sepsis outcome. PLoS ONE. 2015;10(9):e0137721. doi: 10.1371/journal.pone.0137721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, et al. Cold-inducible RNA-binding protein through TLR4 signaling induces mitochondrial DNA fragmentation and regulates macrophage cell death after trauma. Cell Death Dis. 2017;8(5):e2775. doi: 10.1038/cddis.2017.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolognese AC, Sharma A, Yang WL, Nicastro J, Coppa GF, Wang P. Cold-inducible RNA-binding protein activates splenic T cells during sepsis in a TLR4-dependent manner. Cell Mol Immunol. 2018;15(1):38–47. doi: 10.1038/cmi.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ode Y, Aziz M, Jin H, Arif A, Nicastro JG, Wang P. Cold-inducible RNA-binding protein induces neutrophil extracellular traps in the lungs during sepsis. Sci Rep. 2019;9(1):6252. doi: 10.1038/s41598-019-42762-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang WL, Sharma A, Wang Z, Li Z, Fan J, Wang P. Cold-inducible RNA-binding protein causes endothelial dysfunction via activation of Nlrp3 inflammasome. Sci Rep. 2016;6:26571. doi: 10.1038/srep26571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ode Y, Aziz M, Wang P. CIRP increases ICAM-1+ phenotype of neutrophils exhibiting elevated iNOS and NETs in sepsis. J Leukoc Biol. 2018;103(4):693–707. doi: 10.1002/JLB.3A0817-327RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 2000;164(10):4991–4995. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 14.Dower K, Ellis DK, Saraf K, Jelinsky SA, Lin LL. Innate immune responses to TREM-1 activation: overlap, divergence, and positive and negative cross-talk with bacterial lipopolysaccharide. J Immunol. 2008;180(5):3520–3534. doi: 10.4049/jimmunol.180.5.3520. [DOI] [PubMed] [Google Scholar]
- 15.Bouchon A, Facchetti F, Weigand MA, Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410(6832):1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- 16.Arts RJ, Joosten LA, van der Meer JW, Netea MG. TREM-1: intracellular signaling pathways and interaction with pattern recognition receptors. J Leukoc Biol. 2013;93(2):209–215. doi: 10.1189/jlb.0312145. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Li J, Salcedo R, Mivechi NF, Trinchieri G, Horuzsko A. The proinflammatory myeloid cell receptor TREM-1 controls Kupffer cell activation and development of hepatocellular carcinoma. Cancer Res. 2012;72(16):3977–3986. doi: 10.1158/0008-5472.CAN-12-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu L, et al. Identification of extracellular actin as a ligand for triggering receptor expressed on myeloid cells-1 signaling. Front Immunol. 2017;8:917. doi: 10.3389/fimmu.2017.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Read CB, et al. Cutting Edge: identification of neutrophil PGLYRP1 as a ligand for TREM-1. J Immunol. 2015;194(4):1417–1421. doi: 10.4049/jimmunol.1402303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibot S, et al. Surface and soluble triggering receptor expressed on myeloid cells-1: expression patterns in murine sepsis. Crit Care Med. 2005;33(8):1787–1793. doi: 10.1097/01.CCM.0000172614.36571.75. [DOI] [PubMed] [Google Scholar]
- 21.Gibot S, et al. A soluble form of the triggering receptor expressed on myeloid cells-1 modulates the inflammatory response in murine sepsis. J Exp Med. 2004;200(11):1419–1426. doi: 10.1084/jem.20040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016;44(W1):W449–W454. doi: 10.1093/nar/gkw329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park JS, et al. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem. 2004;279(9):7370–7377. doi: 10.1074/jbc.M306793200. [DOI] [PubMed] [Google Scholar]
- 24.Yang H, et al. Identification of CD163 as an antiinflammatory receptor for HMGB1-haptoglobin complexes. JCI Insight. 2016;1(7):85375. doi: 10.1172/jci.insight.85375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol. 2011;29:139–162. doi: 10.1146/annurev-immunol-030409-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colonna M. TREMs in the immune system and beyond. Nat Rev Immunol. 2003;3(6):445–453. doi: 10.1038/nri1106. [DOI] [PubMed] [Google Scholar]
- 27.Underhill DM, Goodridge HS. The many faces of ITAMs. Trends Immunol. 2007;28(2):66–73. doi: 10.1016/j.it.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi K, Rochford CD, Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med. 2005;201(4):647–657. doi: 10.1084/jem.20041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turnbull IR, et al. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol. 2006;177(6):3520–3524. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- 30.Tammaro A, et al. Effect of TREM-1 blockade and single nucleotide variants in experimental renal injury and kidney transplantation. Sci Rep. 2016;6:38275. doi: 10.1038/srep38275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sigalov AB. Immune cell signaling: a novel mechanistic model reveals new therapeutic targets. Trends Pharmacol Sci. 2006;27(10):518–524. doi: 10.1016/j.tips.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Sigalov AB. A novel ligand-independent peptide inhibitor of TREM-1 suppresses tumor growth in human lung cancer xenografts and prolongs survival of mice with lipopolysaccharide-induced septic shock. Int Immunopharmacol. 2014;21(1):208–219. doi: 10.1016/j.intimp.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovalainen M, et al. Novel delivery systems for improving the clinical use of peptides. Pharmacol Rev. 2015;67(3):541–561. doi: 10.1124/pr.113.008367. [DOI] [PubMed] [Google Scholar]
- 34.Lundquist P, Artursson P. Oral absorption of peptides and nanoparticles across the human intestine: opportunities, limitations and studies in human tissues. Adv Drug Deliv Rev. 2016;106(Pt B):256–276. doi: 10.1016/j.addr.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Ornatowska M, et al. Functional genomics of silencing TREM-1 on TLR4 signaling in macrophages. Am J Physiol Lung Cell Mol Physiol. 2007;293(6):L1377–L1384. doi: 10.1152/ajplung.00140.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fortin CF, Lesur O, Fulop T. Effects of TREM-1 activation in human neutrophils: activation of signaling pathways, recruitment into lipid rafts and association with TLR4. Int Immunol. 2007;19(1):41–50. doi: 10.1093/intimm/dxl119. [DOI] [PubMed] [Google Scholar]
- 37.Zeng H, Ornatowska M, Joo MS, Sadikot RT. TREM-1 expression in macrophages is regulated at transcriptional level by NF-kappaB and PU.1. Eur J Immunol. 2007;37(8):2300–2308. doi: 10.1002/eji.200737270. [DOI] [PubMed] [Google Scholar]
- 38.Angele MK, Pratschke S, Hubbard WJ, Chaudry IH. Gender differences in sepsis: cardiovascular and immunological aspects. Virulence. 2014;5(1):12–19. doi: 10.4161/viru.26982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zellweger R, Wichmann MW, Ayala A, Stein S, DeMaso CM, Chaudry IH. Females in proestrus state maintain splenic immune functions and tolerate sepsis better than males. Crit Care Med. 1997;25(1):106–110. doi: 10.1097/00003246-199701000-00021. [DOI] [PubMed] [Google Scholar]
- 40.Baker CC, Chaudry IH, Gaines HO, Baue AE. Evaluation of factors affecting mortality rate after sepsis in a murine cecal ligation and puncture model. Surgery. 1983;94(2):331–335. [PubMed] [Google Scholar]
- 41.Rittirsch D, Huber-Lang MS, Flierl MA, Ward PA. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc. 2009;4(1):31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao D, et al. Effects of fentanyl, midazolam, and their combination on immune function and mortality in mice with sepsis. Exp Ther Med. 2015(4):1494–500. doi: 10.3892/etm.2015.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deacon RM. Assessing nest building in mice. Nat Protoc. 2006;1(3):1117–1119. doi: 10.1038/nprot.2006.170. [DOI] [PubMed] [Google Scholar]
- 44.Van Loo PL, Blom HJ, Meijer MK, Baumans V. Assessment of the use of 2 commercially available environmental enrichments by laboratory mice by preference testing. Lab Anim. 2005;39(1):58–67. doi: 10.1258/0023677052886501. [DOI] [PubMed] [Google Scholar]
- 45.Aziz M, Holodick NE, Rothstein TL, Wang P. B-1a cells protect mice from sepsis: critical role of CREB. J Immunol. 2017;199(2):750–760. doi: 10.4049/jimmunol.1602056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matute-Bello G, et al. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am J Respir Cell Mol Biol. 2011;44(5):725–738. doi: 10.1165/rcmb.2009-0210ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doucey MA, et al. CD3 delta establishes a functional link between the T cell receptor and CD8. J Biol Chem. 2003;278(5):3257–3264. doi: 10.1074/jbc.M208119200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.