Abstract
This Perspective explores the application of machine learning toward improved diagnosis and treatment. We outline a vision for how machine learning can transform three broad areas of biomedicine: clinical diagnostics, precision treatments, and health monitoring, where the goal is to maintain health through a range of diseases and the normal aging process. For each area, early instances of successful machine learning applications are discussed, as well as opportunities and challenges for machine learning. When these challenges are met, machine learning promises a future of rigorous, outcomes-based medicine with detection, diagnosis and treatment strategies that are continuously adapted to individual and environmental differences.
eToC blurb - Gray
Gray and colleagues provide an ambitious visión of how machine learning could impact healthcare, outlining the progress and challenges of applying machine learning approaches to clinical diagnostics, precision therapeutics, and health monitoring.
Introduction
Machine learning leverages sophisticated algorithms operating on large-scale, heterogeneous data sets to uncover useful patterns that would be difficult or impossible for even well-trained individuals to identify. There already are many applications of this approach throughout science and society ranging from game playing (Silver et al., 2018) to product recommendations (Batmaz et al., 2019) to controlling self-driving cars (Bojarski et al., 2016). In biomedicine, work in the human genome project (Venter et al., 2001), efforts in cancer omics (e.g., The Cancer Genome Atlas (Tomczak et al., 2015), the International Cancer Genome Consortium (Zhang et al., 2019), and the Clinical Proteomic Tumor Analysis Consortium (Ellis et al., 2013)), and numerous international machine learning competitions such as DREAM challenges (Saez-Rodriguez et al., 2016; Sage Bionetworks, 2020) and the Critical Assessment of Genome Interpretation (Andreoletti et al., 2019) have shown the power of this approach. The ability to collect and analyze large datasets related to medical treatments and outcomes promises to transform medicine into a data-driven, outcomes-oriented discipline with profound implications for disease detection, diagnosis and treatment. Collection of molecular and phenotypic data has become pervasive, and includes genomic testing for personalized treatment of cancer, high-resolution two-and three-dimensional anatomical imaging of organs, histological analyses of tissue biopsies, and smart watches that monitor heart rates and notify wearers of irregularities (Shilo et al., 2020). These and many other collected data provide the raw material for a future of early, more accurate diagnoses, personalized treatments, and ongoing monitoring in support of overall health.
Machine learning will help realize a future of improved health care by unlocking the potential of large biomedical and patient datasets. Early uses of machine learning in diagnosis and treatment have shown promise to diagnosis breast cancer from x-rays (McKinney et al., 2020; Wu et al., 2019), discover new antibiotics (Stokes et al., 2020), predict onset of gestational diabetes from electronic health records (Artzi et al., 2020), and identify clusters of patients that share a molecular signature of treatment response (Zitnik et al., 2019). Automated pattern recognition through machine learning is essential due to the enormity and complexity of biomedical data; manual analysis is both inefficient and untenable. Equally important, many human diseases involve a complex constellation of changes that occur dynamically and vary from patient to patient. Understanding this complexity requires analysis of large-scale heterogeneous data to identify novel patterns that, after rigorous evaluation, can be used for diagnosis and treatment. Machine learning, then, can assist biomedicai scientists and medical professionals by identifying and summarizing meaningful patterns from large datasets (Rajkomar et al., 2019). Careful evaluation of the patterns found and predictions made by machine learning applications in diagnosis and treatment is essential. “Ground truth” data, in which associations between data and outcome are known, can be used to rigorously evaluate the performance of novel algorithms. Such evaluation data may be quantitative, such as biomarker reduction on treatment, or more qualitative such as overall patient health. It is also important to appreciate that ground truth may change depending on individual characteristics such as age, gender, and environmental exposures.
Recognizing this, there are a growing number of research programs designed to collect and organize large scale datasets linking variables to health status, which can be used to train and evaluate machine learning approaches. Programs in cancer that aggregate molecular profiles from experimental model systems or patient samples together with diagnostic, prognostic, and therapeutic responses provide examples of these valuable data repositories. For example, the Cancer Dependency Map (Tsherniak et al., 2017) has collected multimodal molecular profiles, drug response, and genetic viability data on more than 1,000 cancer cell lines. The AACR Project GENIE (AACR Project GENIE Consortium, 2017) has collected genomic profiles and clinical data for more than 19,000 patients, and the ASCO CancerLinQ is building a similar database of hundreds of thousands of patients. Coupled with advanced algorithms, such programs have the potential to transform our understanding of diseases and improve our ability to predict disease outcomes.
Machine learning is a subdiscipline of artificial intelligence, and the main conceptual approaches in machine learning are summarized in Table 1. Whereas artificial intelligence includes all methods for enabling computers to display human-like understanding and intelligence, machine learning is focused specifically on developing algorithms to learn from data. General classes of machine learning methods include: (a) supervised learning in which data groups are associated with a specific outcome; categorical data (e.g., disease vs. normal) rely on classification methods whereas continuous values (e.g., strength of response to therapy) are used in regression methods, (b) unsupervised or semi-supervised methods to cluster data into discrete groups that can then be manually labeled and associated with outcome, (c) ensemble learning, where results from multiple computational models are combined to produce a final prediction, can lead to more accurate predictions by enabling models to generalize to new data better (d) deep learning, which uses artificial neural networks, a formalization modeled on the human brain, to recognize patterns or associations in the data, is especially useful when working with unstructured data such as images, speech, and text and (e) Bayesian learning, in which prior knowledge is encoded into the learning process, is especially useful in data-poor situations.
Table 1.
Key concepts in machine learning.
| Concepts | Definition |
|---|---|
| Supervised, unsupervised, and semi-supervised learning | Supervised learning predicts labels or classes on future data based on past data that includes labels/classes. Unsupervised learning identifies structure, usually clusters, amongst unlabeled data. Semi-supervised learning first performs unsupervised learning, and humans label structures found from unsupervised learning. |
| Classification and regression | Both are supervised learning methods. Classification predicts discreet categories such as normal vs. diseased while regression predicts real-valued outputs such as response to therapy. |
| Ensemble learning | Ensemble methods build many models and use the average of all models to produce predictions. Common ensemble approaches include random forests, gradient-boosting, and stacking/meta-ensembles. |
| Deep learning | Multi-layer artificial neural networks that can learn complex non-linear functions. Very useful for unstructured data such as images, speech, or text but typically do not provide insights in to the aspects of the data that are driving the functions. |
| Bayesian learning | Methods that combine prior knowledge in addition to data to perform machine learning. |
| Dimensionality reduction | Reduces the number of attributes or features of a dataset by selecting important features or combining features to capture variance in a dataset. Often used to improve performance of machine learning models and to aid visualization. |
| Federated learning | Approaches for incrementally learning from data distributed in multiple locations and which cannot be combined into a single dataset. Federated learning is useful when data are located in multiple clinical systems or when learning from sensitive personal data. |
There are two complementary approaches that can be used with any of these learning methods and are especially useful for biomedical applications. Many biomedical datasets have a large number of features (dimensions), and the number of features may exceed the number of data points. Dimensionality reduction can help improve the performance of machine learning approaches by selecting a subset of relevant attributes of a dataset or combining attributes into a smaller number that capture variability in a dataset. Reducing the dimensions of a dataset is also useful for visualizing data or model predictions. When data are distributed across multiple sites and cannot be moved to create a single dataset for machine learning, federated learning approaches are used to learn incrementally across all the data (Konecny et al., 2016; Yang et al., 2019). Federated learning is especially important in many biomedical applications where data contain sensitive or protected health information that cannot be easily shared. Most of these approaches are conceptually mature but are now finding increased use as structured biomedical data become available and as computer technology becomes sufficiently powerful to enable discovery of subtle but important patterns in large data sets. A recent review provides a brief tutorial on machine learning approaches in the life sciences (Camacho et al., 2018). The application goals and available data dictate appropriate machine learning methods to use. Table 2 lists prototypic examples of machine learning applications for medical diagnosis and treatment.
Table 2.
Example applications of machine learning for diagnosis and treatment.
| Dataset | Goals | Successes | Data type | ML method |
|---|---|---|---|---|
| Patient molecular patient profiles without clinical data | 1. Discover subtypes or stratify patients 2. Identify similarities among clustered patients |
Cancer subtyping (Curtis et al., 2012; Gao et al., 2019) | • High-dimensional, structured data • Unlabeled data |
Unsupervised clustering for cluster discovery; supervised learning and deep learning for subtype assignment. |
| Patient or laboratory molecular profiles with clinical data | Predict most efficacious therapies | Cancer cell line drug response prediction (Chiu et al., 2019b; Costello et al., 2014) | • High-dimensional, structured data • Unlabeled data |
Supervised learning, deep learning, and ensemble learning |
| Images and associated diagnoses | Automated diagnoses | Medical imaging diagnostics (Liu et al., 2019) | • Unstructured data • Labeled data |
Deep learning |
| EMR data + clinical outcomes | Predict clinical outcomes | Diagnosis of gestational diabetes (Artzi et al., 2020); patient similarity (Lee et al., 2018) | • Structured and unstructured data • Labeled data |
Traditional machine learning on structured data with labels. Deep learning/natural language processing to mine unstructured data. Federated learning. |
| Wearable and home device ambient data collection | Early diagnoses | Detection of atrial fibrillation (Bumgarner et al., 2018) and agonal breathing, an audible biomarker of cardiac arrest (Chan et al., 2019) | • Unstructured, longitudinal data • Labeled data |
Both supervised and deep learning approaches, with adjustments made for time-series analyses. |
| Deep longitudinal data | Ongoing health management | None yet due to lack of available datasets | • Structured and unstructured data • Labeled data |
Continuous learning |
We expect that applications of machine learning will have a profound impact on many aspects of health management as computers optimized for machine learning increase in power and as infrastructure for accurate data collection and curation becomes more widely deployed. Immediate biomedical opportunities summarized in the following sections include earlier and more accurate disease detection, better diagnosis, and more durable and tolerable treatments. Of course, the accuracy of the underlying “learned” relationships depends on the accuracy and magnitude of the data on which learning is based. This can be enhanced substantially by widely deploying standardized electronic medical record systems designed specifically to support machine learning and by supporting their widespread use. Acquisition of data “at home” using smartphones, commercial home assistant devices (e.g. Amazon Echo, Google Home), and other electronic devices will further enhance robust biomedical machine learning. Looking ahead, we envision these trends merging to enable outcomes-based personalized management of patient health (Figure 1) using algorithms that increase in accuracy as the quantity and quality of data grows.
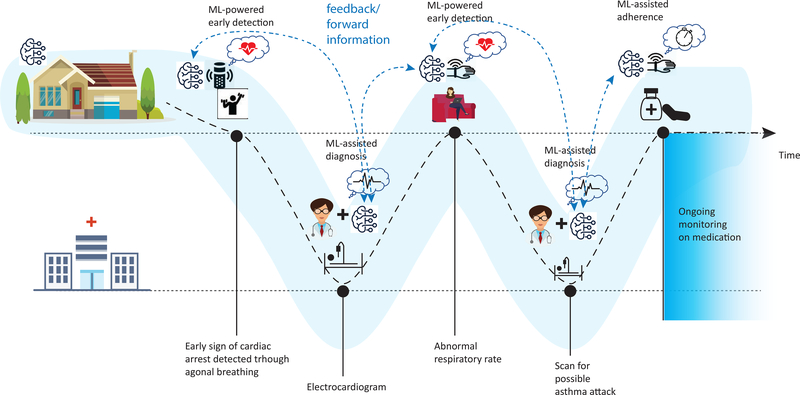
Figure 1.
How machine learning applications could help individuals maintain health. At home, machine learning may help in early detection of disease, monitoring response to treatment, and adherence to treatment regimens. In the clinic or hospital, machine learning may aid medical professionals to diagnosis and tune treatment for an individual patient. The dashed line shows how a patient moves between home and clinical settings and how machine learning can help at each step to maintain health.
In this Perspective, we outline a vision for how machine learning can be applied to make critical advances in biomedicine. We focus on three biomedical areas: improved clinical diagnostics, precision treatment, and health management and monitoring. For each area, we describe opportunities for machine learning applications to enable new insights or improve on current state-of-the-art approaches, discuss successful early applications of machine learning, and highlight unmet needs to be addressed. We conclude by identifying several cross-cutting challenges that, if solved, will help realize the full potential of machine learning in biomedicine.
Improved Diagnostics from Clinical Imaging and Molecular Tests
Technological advances in clinical testing are generating orders of magnitude more data than tests in the past. High-fidelity imaging tests now produce large two-, three or four-dimensional (the fourth dimension being time) images of tissue and organs, and molecular tests can provide assessment of hundreds or even thousands of genes and proteins. Machine learning is both essential and ubiquitous for automated analysis of diagnostic features in these data that are strongly associated with disease type, status or response to treatment.
The use of deep learning to extract meaning from biomedical images is one of the most active areas of current research. Several recent publications have shown that computer-aided detection (CAD) software using machine learning can interpret radiologie images on par with medical professionals indicating the power of this approach. For example, deep learning-based CAD software was able to detect diabetic retinopathy at high levels of accuracy (Gulshan et al., 2016) and to retrospectively identify invasive and in-situ breast cancer of all grades similar to radiologists (McKinney et al., 2020; Wu et al., 2019). A recent review found that deep learning based approaches performed as well as medical professionals across a range of medical imaging diagnostic tasks, though many of these studies are small and have yet to perform a prospective evaluation (Liu et al., 2019). Importantly, deep learning approaches benefit from large data sets and will increase continually in accuracy as the sizes of the training data sets grow.
Molecular assays can identify genetic mutations and quantify gene expression levels and protein abundance from a variety of samples, including blood, saliva, and tissue. Machine learning has the potential to increase the utility of these data by discovering complex sets of biomarkers associated with various disease states, which ultimately can inform patient outcome and identify effective treatment strategies. Some examples from cancer biology include using DNA methylation (Kang et al., 2017) and nucleosome positioning (Heitzer et al., 2019) from blood to predict tumor tissue of origin, quantifying cellular pathway activation levels in biopsies and other tissue samples (Way and Greene, 2019; Way et al., 2018), predicting genomic features of brain cancers using magnetic resonance images (Chang et al., 2018a), and forecasting cancer patient outcomes based on multi-omics (Chaudhary et al., 2018) or imaging-omics integrations (Mobadersany et al., 2018). Beyond cancer, machine learning has been used to identify individuals with sleep deprivation through analysis of mRNA in the blood, informing how sleep insufficiencies negatively affect health (Laing et al., 2018). Through integration of multiple data types and biomarkers, machine learning models are likely to be substantially more accurate than current practice, which is often limited to a few markers and reflects only a narrow view of complex diseases.
Joint human-computer diagnostic approaches such as those illustrated in Figure 1, are likely to become common because they take advantage of the strengths of both humans and computers. In this collaborative approach, physicians will make a final diagnosis by integrating all available information, including that provided by machine learning systems (Ahuja, 2019). Machine learning systems will have a key role by automating routine diagnosis, flagging challenging cases that require more human input, and providing additional information useful in making diagnoses (e.g., (Ardila et al., 2019)). Moreover, machine learning systems may use different features than medical professionals to make diagnoses, though care will be required to assess the biological utility of such features. As a result, approaches that integrate knowledge from both medical professionals and advanced algorithms will lead to improved diagnoses. Ensuring that machine learning software is transparent will be critical before widespread deployment and adoption. ‘Transparency’ in this context includes description of the optimized objectives, strengths, quantitative performance and limitations of a particular algorithm (Cai et al., 2019) as well as the procedures used to validate the algorithm. These attributes will help medical professionals decide when and how to use machine learning applications to obtain valid results and improve decision making. Applications that use machine learning can help build trust in the system and facilitate deeper understanding of the underlying biological mechanism of disease by explaining predictions, such as by highlighting the most important features used (Ching et al., 2018; Litjens et al., 2016).
As more advanced clinical testing technologies are coupled with machine learning, it will be important to consider tradeoffs between disease detection rates, patient outcomes, and other factors that impact patient health and quality of life. Disease detection rates may increase with the use of machine learning technologies, and disease-specific research will be needed to differentiate indolent versus fatal disease to avoid over-treatment and to identify disease subtypes in order to guide the selection of the most effective treatments for each subtype. Careful framing of clinical goals that can be connected to evaluation and validation metrics will ensure that machine learning improves patient care and overall health (Chen et al., 2019b).
Precision Treatment through Multiscale Modeling and Expert Guidance
One of the most promising application areas for machine learning is precision medicine, where a patient receives medical care and treatment tailored to their personal disease profile. Precision oncology, where the goal is to prescribe cancer treatments based on tumor molecular characteristics, is a prime example of the challenges and opportunities for machine learning in precision medicine. In current practice, individual molecular markers such as somatic mutations and gene expression levels are often used to inform treatment selection. However, responses are often highly variable between patients due to differences at other genomic and epigenomic loci as well as anatomic disease distribution (Brown et al., 2019; Kobayashi and Mitsudomi, 2016; Rotow and Bivona, 2017). Further complicating precision oncology is that there are hundreds of potential drugs and not every combination can be tested for every disease profile (Gerstung et al., 2017; Kurnit et al., 2018).
One way that machine learning can help overcome these challenges is through the development of multifactorial predictive models that are robust against individual diversity. For example, single-purpose models have been built to forecast the functional consequences of biological changes, such as how genetic mutations influence splicing and gene expression (Xiong et al., 2015) as well as transcription factor binding (Chen et al., 2019a). Machine learning models have also been built to predict drug response in cancer cell lines (Chang et al., 2018b), transfer predictions from cell lines to patient tumors (Chiu et al., 2019a), and forecast patient response to therapies based on clinical response data (Huang et al., 2018). Future advancements in modeling for precision therapeutics are likely to operate over multiple scales and serve multiple purposes. Multiscale modeling will use large biological datasets to investigate the growth and development of an organism across diverse temporal and spatial domains. Already there are computational models of human-virus interactions (Lasso et al., 2019), cell-cell interplay such tumor-immune cell interactions, and even whole cells (Metzcar et al., 2019; Rahman et al., 2017; Sakamoto et al., 2018). Eventually we anticipate that computational models of organs and entire individuals—so-called “digital twins” (Bjornsson et al., 2019)—will be developed. The goal of digital twins will be multifaceted, such as predicting the efficacy of different combination therapies that have never been used together and modeling the impact of disease on different organs.
While multiscale models may become accurate enough that their predictions can be used directly for treatment, we envision an intermediate stage in which machine learning approaches generate a ranked list of suggested therapies that can be used by expertly trained physicians to help guide treatment decisions. For instance, patient-derived laboratory models could be used to test predictions from computational models, with the best-performing predictions recommended for use in treatment. This hybrid approach has many advantages: machine learning models can dramatically reduce the space of potential treatment combinations to be considered and identify others that might otherwise be overlooked. An experimental validation step could be added to provide additional evidence that a predicted therapy is likely to be effective.
Precision medicine will also be advanced by using machine learning to automatically mine and search expert knowledge in published literature and patient databases (Rajkomar et al., 2019). Patient databases, usually in the form of electronic health records (EHRs), represent a rich source of information about diagnosis, treatment, and treatment response for large patient cohorts. Early efforts have attempted to use natural language processing algorithms to mine publications (Dong et al., 2018), EHRs (Shickel et al., 2018), and clinical reports (Kreimeyer et al., 2017; Pons et al., 2016) for useful knowledge, such as biomarker-therapy associations and biological pathways of interest. Other applications have used structured information from EHRs to predict disease onset (Artzi et al., 2020). Machine learning will help harness this information and make it useful for precision medicine through advanced approaches that address the unstructured nature of data and metadata in publications and EHRs. Of course, the EHR mining approach assumes that the information needed to establish a useful association is accurately and completely captured. Unfortunately, this is not always the case and future work will be needed to increase the utility of EHR analyses.
Health Management and Monitoring
We envision a shift in how complex diseases are treated, moving from the goal of a cure to one of disease management. This comprehensive health management approach will strive to maintain health through a range of diseases and the normal aging process. Health management is demanding as it requires ongoing monitoring of all aspects of health for potential disease, choosing treatments suited to individual patients, and adapting treatments based on patient response (Figure 2). Here machine learning has a key role to play, largely by integrating many of the ideas already discussed for diagnosis and treatment into a continuous learning approach.
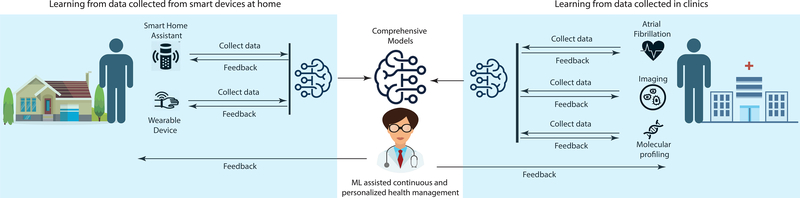
Figure 2.
Combining data collected from both home (left) and clinical settings (right), or combining predictive models built at home and in the clinic, has the potential to lead to comprehensive and integrated models that support personalized health management. Comprehensive models are more likely to perform well as they incorporate more information about an individual, and these models have the potential to be applied in the home, clinic, or wherever an individual may be.
Outside of clinical settings, wearable devices and at-home smart electronic devices provide a new avenue for health management. These devices can collect large amounts of fine-grained data on patient health status that can be used by machine learning applications to suggest one-time actions, changes in daily activities, or referral to a physician for assessment and testing. Wearable devices now include sensors for motion, pulse, respiratory rate, body temperature, blood pressure, oxygen levels, and other biometrics. Prototype applications show how data from wearables might be useful, including: diabetes management (Chang et al., 2016), detection of atrial fibrillation (Bumgarner et al., 2018), blood cholesterol monitoring (Fu and Guo, 2018), early detection of Parkinson’s disease (Lonini et al., 2018), self-adherence to medications (Car et al., 2017; Toh et al., 2016), and early warning of heart attack (Sahoo et al., 2017). Speech-driven home assistants have been used to detect agonal breathing, an audible biomarker that is an early sign of cardiac arrest (Chan et al., 2019). In the future, machine learning software is likely to be used to identify new biomarkers from wearable and audio sensor data, perhaps by integrating data across different types of devices. Both traditional supervised learning and deep learning are likely to play roles in developing models from wearable data.
Using machine learning together with data collected from smartphones provides new opportunities for diagnostics as well. Deep learning approaches have been applied to analyze pictures from smartphone cameras to identify different types of skin cancers (Esteva et al., 2017) and also to diagnose diabetic retinopathy (Micheletti et al., 2016). Recent studies have found that sensory data (e.g., voice, tapping, response time, and accelerometer data) collected from smartphones and processed using machine learning can be used to monitor symptoms and progression of Parkinson’s disease (Arora et al., 2015; Espay et al., 2016; Ginis et al., 2016; Pereira et al., 2016). These prototype applications suggest a role for machine learning where wearables, home devices, and smartphones are used to capture all kinds of data, including biometric measurements, photos, dietary intake, and even environmental information (i.e., the “exposome” (Vermeulen et al., 2020)). By connecting this information with diagnoses, machine learning will be used to identify patterns within the data that suggest a particular diagnosis.
The foundation of health management is the ongoing monitoring of individual behavior and body functioning through home and wearable devices together with readouts from routine blood sampling. Personalized models of baseline functions and activity will be built by customizing population-level models with data collected for each individual. A key advantage of this approach is that personal baselines can be established and deviations from baselines--which may indicate a change in health status--can be detected. Using personalized models, machine learning applications will monitor individuals for any changes from normal and notify individuals when a change requires consult with a medical professional. An interesting possibility along these lines is suggested by recent work showing that monitoring of individual internet symptom searches (in essence, self-reported health issues such as weight loss, bronchitis, cough, chest pain, etc.) coupled with machine-learned tendencies from many individuals can enable early detection of lung (White and Horvitz, 2017) and pancreatic (Paparrizos et al., 2016) cancers. This could lead to a physician or patient alert system that recommends medical attention when a more serious issue may explain the seemingly innocuous symptoms searched for. Of course, many issues regarding privacy would have to be overcome to make this possible.
Once in a clinical setting, high-fidelity imaging and molecular testing will be interpreted by medical professionals with the help of machine learning to identify noteworthy biomarkers and make a final diagnosis. Disease diagnoses that require treatment will use multiscale modeling and automated search results for similar patients to inform treatment selection.
After diagnosis and treatment, health management begins again with ongoing monitoring of individual health. This time, however, there are multiple goals that a machine learning system must meet: monitor how the individual is responding to treatment, watch for any adverse effects, and monitor overall health and changes from baseline not accounted for by treatment. Machine learning will help adapt the initial personalized model to include the new diagnosis and therapy information, creating an expected trajectory on treatment that will serve as the new baseline.
Health management across a person’s lifespan will require data integration and modeling at a level of sophistication and automation that is only possible with machine learning. Each step in health management—building personalized models and using them to monitor for and accurately detect anomalies, aiding physicians in diagnosis and treatment through automated processing of large datasets and patient databases, and updating individual models for new diagnoses and treatments—is data intensive and requires automated pattern recognition of complex datasets. Health management will also continuously learn as models will be updated with availability of new data. Two general approaches for continuous learning are to build new predictive models or to update existing models, and more work is needed to understand the strengths and limitations of these approaches for different applications.
Challenges and Concluding Thoughts
For machine learning to play a transformative role in diagnosis and treatment, it is necessary to develop high-quality, well-curated datasets. High-quality datasets have several important benefits: they improve the predictive power of machine learning methods while reducing the size of the data needed for training and the complexity of the learned representations. Famously, machine learning approaches for image recognition accelerated when ImageNet (Deng et al., 2009), a corpus of labelled and ontologically-linked images, was introduced. Similar efforts in biomedicine are needed across the variety the tasks where machine learning may be applied.
Creating high-quality datasets for machine learning applications in diagnosis and treatment will require addressing technical, legal, and economic issues that often result in siloed biomedical data that are not standardized. As discussed above, federated learning provides a technical solution for combining data amongst siloed systems because no actual data movement is necessary and individual privacy can be protected. Wearables and home devices provide a way to collect accurate data, and machine learning can be used as a preprocessing step to extract accurate analytic and clinical data from unstructured sources such as electronic health records and publications. Legal procedures must be developed for the secure management and analysis of Private Health Information (PHI) and community and legal standards that define the performance of these procedures must be established. Biomedical institutions and individuals must be incentivized to engage in data standardization and sharing. Similarly, insurers, the pharmaceutical industry, and agencies that support biomedical research must be willing to invest the infrastructure, data acquisition, and data curation required to generate high quality data.
Approaches and incentives for data sharing that promote diversity in the datasets used for learning are needed as well. This includes national and international data sharing standards that make it possible to obtain data from both major medical centers and community clinics. It is likely, for instance, that machine learning applications that improve patient treatment response in major medical centers may not perform well in community settings due to differences in overall care and patient populations. However, the ultimate goal of biomedical data collection for machine learning is to obtain suitable representative data from patient cohorts to develop accurate machine learning models that will generalize to diverse populations. Therefore there must be a concerted effort to also account for variables such as patient status prior to treatment, treatment regimes, age, gender, race, ethnicity, and environmental exposures.
Rigorous evaluation approaches are needed for biomedical machine learning applications, especially in settings where continuous learning is required. In our view, the performance of a machine learning system is best measured by the accuracy of its predictions in a prospective setting. We advocate for an iterative approach to machine learning that includes: training with retrospective data, algorithm lock-down and deployment, followed by assessment of the application’s accuracy based on predictions obtained during deployment. Data collected during deployment, coupled with additional or larger retrospective datasets, can then be used to retrain and optimize the algorithm, followed by a subsequent deployment-evaluation cycle. Evaluating continuous learning systems--such as those we envision for health monitoring that must adapt to changes in health status or habits--will likely require tightening this loop and use of data collected during the deployment phase to detect limitations or failures. Quantifying not only accuracy but also confidence intervals is critical, as some uses of machine learning will be more tolerant to inexact predictions than others and confidence intervals can be used by physicians to inform decision making. Iteratively training and deploying machine learning applications poses regulatory challenges as most diagnostic and therapeutic tests assume that models and data are fixed. When models are updated in response to new data or adapted for new diagnoses or treatments, ongoing evaluation is needed to ensure that predictions remain accurate. Real or simulated datasets that are multi-modal, expansive, and longitudinal will be needed to ensure robust evaluation of biomedical machine learning applications.
While the challenges outlined above are significant, we are optimistic that they can be overcome. Further, we believe the effort is worthwhile, as success promises a future of rigorous, outcomes-based medicine with detection, diagnosis, and treatment strategies that are continuously adapted via machine learning to individual and environmental differences and enable comprehensive health management.
Acknowledgements
J.G. and J.W.G. are supported by National Cancer Institute grant U2C CA233280. J.G. is supported by National Cancer Institute grant U24 CA231877. L.M.H. and J.W.G. are supported by the National Institutes of Health grant U54 HG008100 and National Cancer Institute grant U54 CA209988. J.G., L.M.H., and J.W.G. are supported by the Prospect Creek Foundation. J.W.G. is supported by the Susan G. Komen Foundation. L.M.H. is supported by the Breast Cancer Research Foundation and The Jayne Koskinas Ted Giovanis Foundation.
Declaration of Interests
J.W.G. receives research support from Micron and ThermoFisher and has stock in NVIDIA, Microsoft, Amazon and Google (Alphabet) and GE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AACR Project GENIE Consortium (2017). AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 7, 818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja AS (2019). The impact of artificial intelligence in medicine on the future role of the physician. In PeerJ, pp. e7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoletti G, Pal LR, Moult J, and Brenner SE (2019). Reports from the fifth edition of CAGI: The Critical Assessment of Genome Interpretation. Hum Mutat 40, 1197–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, et al. (2019). End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nature Medicine 25, 954–961. [DOI] [PubMed] [Google Scholar]
- Arora S, Venkataraman V, Zhan A, Donohue S, Biglan KM, Dorsey ER, and Little MA (2015). Detecting and monitoring the symptoms of Parkinson’s disease using smartphones: A pilot study. Parkinsonism & Related Disorders 21, 650–653. [DOI] [PubMed] [Google Scholar]
- Artzi NS, Shilo S, Hadar E, Rossman H, Barbash-Hazan S, Ben-Haroush A, Balicer RD, Feldman B, Wiznitzer A, and Segal E (2020). Prediction of gestational diabetes based on nationwide electronic health records. Nat Med 26, 71–76. [DOI] [PubMed] [Google Scholar]
- Batmaz Z, Yurekli A, Bilge A, and Kaleli C (2019). A review on deep learning for recommender systems: challenges and remedies. Artificial Intelligence Review 52, 1–37. [Google Scholar]
- Björnsson B, Borrebaeck C, Elander N, Gasslander T, Gawel DR, Gustafsson M, Jörnsten R, Lee EJ, Li X, Lilja S, et al. (2019). Digital twins to personalize medicine. Genome Medicine 12, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojarski M, Del Testa D, Dworakowski D, Firner B, Flepp B, Goyal P, Jackel LD, Monfort M, Muller U, Zhang J, et al. (2016). End to End Learning for Self-Driving Cars (arXiv). [Google Scholar]
- Brown BP, Zhang Y-K, Westover D, Yan Y, Qiao H, Huang V, Du Z, Smith JA, Ross JS, Miller VA, et al. (2019). On-target resistance to the mutant-selective EGFR inhibitor osimertinib can develop in an allele specific manner dependent on the original EGFR activating mutation. Clinical Cancer Research, clincanres.3829.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner JM, Lambert CT, Hussein AA, Cantillon DJ, Baranowski B, Wolski K, Lindsay BD, Wazni OM, and Tarakji KG (2018). Smartwatch Algorithm for Automated Detection of Atrial Fibrillation. Journal of the American College of Cardiology 71, 2381–2388. [DOI] [PubMed] [Google Scholar]
- Cai CJ, Winter S, Steiner D, Wilcox L, and Terry M (2019). “Hello AI”: Uncovering the Onboarding Needs of Medical Practitioners for Human-AI Collaborative Decision-Making. Proc ACM Hum-Comput Interact 3, Article 104. [Google Scholar]
- Camacho DM, Collins KM, Powers RK, Costello JC, and Collins JJ (2018). Next-Generation Machine Learning for Biological Networks. Cell 173, 1581–1592. [DOI] [PubMed] [Google Scholar]
- Car J, Tan WS, Huang Z, Sloot P, and Franklin BD (2017). eHealth in the future of medications management: personalisation, monitoring and adherence. BMC Medicine 15, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Rea T, Gollakota S, and Sunshine JE (2019). Contactless cardiac arrest detection using smart devices. npj Digital Medicine 2, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, Su M-Y, Cha S, Filippi CG, Bota D, et al. (2018a). Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. American Journal of Neuroradiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Chiang R, Wu S, and Chang W (2016). A Context-Aware, Interactive M-Health System for Diabetics. IT Professional 18, 14–22. [Google Scholar]
- Chang Y, Park H, Yang H-J, Lee S, Lee K-Y, Kim TS, Jung J, and Shin J-M (2018b). Cancer Drug Response Profile scan (CDRscan): A Deep Learning Model That Predicts Drug Effectiveness from Cancer Genomic Signature. Scientific Reports 8, 8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary K, Poirion OB, Lu L, and Garmire LX (2018). Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clinical Cancer Research 24, 1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KM, Cofer EM, Zhou J, and Troyanskaya OG (2019a). Selene: a PyTorch-based deep learning library for sequence data. Nature Methods 16, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P-HC, Liu Y, and Peng L (2019b). How to develop machine learning models for healthcare. Nature Materials 18, 410–414. [DOI] [PubMed] [Google Scholar]
- Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, Ferrero E, Agapow P-M, Zietz M, Hoffman MM, et al. (2018). Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface 15, 20170387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y-C, Chen H-IH, Zhang T, Zhang S, Gorthi A, Wang L-J, Huang Y, and Chen Y (2019a). Predicting drug response of tumors from integrated genomic profiles by deep neural networks. BMC Medical Genomics 12, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YC, Chen HH, Zhang T, Zhang S, Gorthi A, Wang LJ, Huang Y, and Chen Y (2019b). Predicting drug response of tumors from integrated genomic profiles by deep neural networks. BMC Med Genomics 12, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JC, Heiser LM, Georgii E, Gonen M, Menden MP, Wang NJ, Bansal M, Ammad-ud-din M, Hintsanen P, Khan SA, et al. (2014). A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol 32, 1202–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. (2012). The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Dong W, Socher R, Li L, Kai L, and Li F-F (2009). ImageNet: A large-scale hierarchical image database. Paper presented at: 2009 IEEE Conference on Computer Vision and Pattern Recognition. [Google Scholar]
- Dong W, Wang X, Xia Z, Zhang X, and Yang H (2018). A legacy of the “1% program” - The “Chinese Chapter” of the human genome reference sequence. J Genet Genomics 45, 565–568. [DOI] [PubMed] [Google Scholar]
- Ellis MJ, Gillette M, Carr SA, Paulovich AG, Smith RD, Rodland KK, Townsend RR, Kinsinger C, Mesri M, Rodriguez H, et al. (2013). Connecting genomic alterations to cancer biology with proteomics: the NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discov 3, 1108–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espay AJ, Bonato P, Nahab FB, Maetzler W, Dean JM, Klucken J, Eskofier BM, Merola A, Horak F, Lang AE, et al. (2016). Technology in Parkinson’s disease: Challenges and opportunities. Movement Disorders 31, 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, and Thrun S (2017). Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, and Guo J (2018). Blood Cholesterol Monitoring With Smartphone as Miniaturized Electrochemical Analyzer for Cardiovascular Disease Prevention. IEEE Transactions on Biomedical Circuits and Systems 12, 784–790. [DOI] [PubMed] [Google Scholar]
- Gao F, Wang W, Tan M, Zhu L, Zhang Y, Fessler E, Vermeulen L, and Wang X (2019). DeepCC: a novel deep learning-based framework for cancer molecular subtype classification. Oncogenesis 8, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstung M, Papaemmanuil E, Martincorena I, Bullinger L, Gaidzik VI, Paschka P, Heuser M, Thol F, Bolli N, Ganly P, et al. (2017). Precision oncology for acute myeloid leukemia using a knowledge bank approach. Nature Genetics 49, 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginis P, Nieuwboer A, Dorfman M, Ferrari A, Gazit E, Canning CG, Rocchi L, Chiari L, Hausdorff JM, and Mirelman A (2016). Feasibility and effects of home-based smartphone-delivered automated feedback training for gait in people with Parkinson’s disease: A pilot randomized controlled trial. Parkinsonism & related disorders 22, 28–34. [DOI] [PubMed] [Google Scholar]
- Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, et al. (2016). Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 316, 2402–2410. [DOI] [PubMed] [Google Scholar]
- Heitzer E, Haque IS, Roberts CES, and Speicher MR (2019). Current and future perspectives of liquid biopsies in genomics-driven oncology. Nature Reviews Genetics 20, 71–88. [DOI] [PubMed] [Google Scholar]
- Huang C, Clayton EA, Matyunina LV, McDonald LD, Benigno BB, Vannberg F, and McDonald JF (2018). Machine learning predicts individual cancer patient responses to therapeutic drugs with high accuracy. Scientific Reports 8, 16444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Li Q, Chen Q, Zhou Y, Park S, Lee G, Grimes B, Krysan K, Yu M, Wang W, et al. (2017). CancerLocator: non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biology 18, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, and Mitsudomi T (2016). Not all epidermal growth factor receptor mutations in lung cancer are created equal: Perspectives for individualizad treatment strategy. Cancer Science 107, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecny J, McMahan HB, Yu FX, Richtárik P, Suresh AT, and Bacon D (2016). Federated learning: Strategies for improving communication efficiency. arXiv preprint arXiv:161005492. [Google Scholar]
- Kreimeyer K, Foster M, Pandey A, Arya N, Halford G, Jones SF, Forshee R, Walderhaug M, and Botsis T (2017). Natural language processing systems for capturing and standardizing unstructured clinical information: A systematic review. Journal of Biomedical Informatics 73, 14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurnit KC, Ileana Dumbrava EE, Litzenburger BC, Khotskaya YB, Johnson A, Yap TA, Rodon J, Zeng J, Shufean MA, Bailey A, et al. (2018). PRECISION ONCOLOGY DECISION SUPPORT: CURRENT APPROACHES AND STRATEGIES FOR THE FUTURE. Clinical Cancer Research, clincanres.2494.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing EE, Moller-Levet CS, Dijk D-J, and Archer SN (2018). Identifying and validating blood mRNA biomarkers for acute and chronic insufficient sleep in humans: a machine learning approach. Sleep 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasso G, Mayer SV, Winkelmann ER, Chu T, Elliot O, Patino-Galindo JA, Park K, Rabadan R, Honig B, and Shapira SD (2019). A Structure-Informed Atlas of Human-Virus Interactions. Cell 178, 15261541 e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sun J, Wang F, Wang S, Jun C-H, and Jiang X (2018). Privacy-Preserving Patient Similarity Learning in a Federated Environment: Development and Analysis. JMIR Med Inform 6, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litjens G, Sánchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I, Hulsbergen - van de Kaa C, Bult P, van Ginneken B, and van der Laak J (2016). Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Scientific Reports 6, 26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Faes L, Kale AU, Wagner SK, Fu DJ, Bruynseels A, Mahendiran T, Moraes G, Shamdas M, Kern C, et al. (2019). A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. The Lancet Digital Health 1, e271–e297. [DOI] [PubMed] [Google Scholar]
- Lonini L, Dai A, Shawen N, Simuni T, Poon C, Shimanovich L, Daeschler M, Ghaffari R, Rogers JA, and Jayaraman A (2018). Wearable sensors for Parkinson’s disease: which data are worth collecting for training symptom detection models. npj Digital Medicine 1, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, Back T, Chesus M, Corrado GC, Darzi A, et al. (2020). International evaluation of an AI system for breast cancer screening. Nature 577, 89–94. [DOI] [PubMed] [Google Scholar]
- Metzcar J, Wang Y, Heiland R, and Macklin P (2019). A Review of Cell-Based Computational Modeling in Cancer Biology. JCO Clinical Cancer Informatics, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheletti JM, Hendrick AM, Khan FN, Ziemer DC, and Pasquel FJ (2016). Current and Next Generation Portable Screening Devices for Diabetic Retinopathy. Journal of Diabetes Science and Technology 10, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velázquez Vega JE, Brat DJ, and Cooper LAD (2018). Predicting cancer outcomes from histology and genomics using convolutional networks. Proceedings of the National Academy of Sciences 115, E2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paparrizos J, White RW, and Horvitz E (2016). Screening for Pancreatic Adenocarcinoma Using Signals From Web Search Logs: Feasibility Study and Results. J Oncol Pract 12, 737–744. [DOI] [PubMed] [Google Scholar]
- Pereira CR, Weber SAT, Hook C, Rosa GH, and Papa JP (2016). Deep Learning-Aided Parkinson’s Disease Diagnosis from Handwritten Dynamics. Paper presented at: 2016 29th SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI). [Google Scholar]
- Pons E, Braun LMM, Hunink MGM, and Kors JA (2016). Natural Language Processing in Radiology: A Systematic Review. Radiology 279, 329–343. [DOI] [PubMed] [Google Scholar]
- Rahman MM, Feng Y, Yankeelov TE, and Oden JT (2017). A fully coupled space-time multiscale modeling framework for predicting tumor growth. Computer Methods in Applied Mechanics and Engineering 320, 261–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkomar A, Dean J, and Kohane I (2019). Machine Learning in Medicine. New England Journal of Medicine 380, 1347–1358. [DOI] [PubMed] [Google Scholar]
- Rotow J, and Bivona TG (2017). Understanding and targeting resistance mechanisms in NSCLC. Nature Reviews Cancer 17, 637–658. [DOI] [PubMed] [Google Scholar]
- Saez-Rodriguez J, Costello JC, Friend SH, Kellen MR, Mangravite L, Meyer P, Norman T, and Stolovitzky G (2016). Crowdsourcing biomedical research: leveraging communities as innovation engines. Nat Rev Genet 17, 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage Bionetworks (2020). DREAM Challenges.
- Sahoo KP, Thakkar KH, and Lee M-Y (2017). A Cardiac Early Warning System with Multi Channel SCG and ECG Monitoring for Mobile Health. Sensors 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M, Ikeyama N, Yuki M, and Ohkuma M (2018). Draft Genome Sequence of Faecalimonas umbilicata JCM 30896(T), an Acetate-Producing Bacterium Isolated from Human Feces. Microbiol Resour Announc 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shickel B, Tighe PJ, Bihorac A, and Rashidi P (2018). Deep EHR: A Survey of Recent Advances in Deep Learning Techniques for Electronic Health Record (EHR) Analysis. IEEE Journal of Biomedical and Health Informatics 22, 1589–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilo S, Rossman H, and Segal E (2020). Axes of a revolution: challenges and promises of big data in healthcare. Nat Med 26, 29–38. [DOI] [PubMed] [Google Scholar]
- Silver D, Hubert T, Schrittwieser J, Antonoglou I, Lai M, Guez A, Lanctot M, Sifre L, Kumaran D, Graepel T, et al. (2018). A general reinforcement learning algorithm that masters chess, shogi, and Go through self-play. Science 362, 1140–1144. [DOI] [PubMed] [Google Scholar]
- Stokes JM, Yang K, Swanson K, Jin W, Cubillos-Ruiz A, Donghia NM, MacNair CR, French S, Carfrae LA, Bloom-Ackerman Z, et al. (2020). A Deep Learning Approach to Antibiotic Discovery. Cell 180, 688–702 e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh X, Tan H, Liang H, and Tan H (2016). Elderly medication adherence monitoring with the Internet of Things. Paper presented at: 2016 IEEE International Conference on Pervasive Computing and Communication Workshops (PerCom Workshops). [Google Scholar]
- Tomczak K, Czerwinska P, and Wiznerowicz M (2015). The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 19, A68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. (2017). Defining a Cancer Dependency Map. Cell 170, 564–576 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. (2001). The sequence of the human genome. Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- Vermeulen R, Schymanski EL, Barabási A-L, and Miller GW (2020). The exposome and health: Where chemistry meets biology. Science 367, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way GP, and Greene CS (2019). Discovering Pathway and Cell Type Signatures in Transcriptomic Compendia with Machine Learning. Annual Review of Biomedical Data Science 2, 1–17. [Google Scholar]
- Way GP, Sanchez-Vega F, La K, Armenia J, Chatila WK, Luna A, Sander C, Cherniack AD, Mina M, Ciriello G, et al. (2018). Machine Learning Detects Pan-cancer Ras Pathway Activation in The Cancer Genome Atlas. Cell Rep 23, 172–180.e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RW, and Horvitz E (2017). Evaluation of the Feasibility of Screening Patients for Early Signs of Lung Carcinoma in Web Search Logs. jAmA Oncol 3, 398–401. [DOI] [PubMed] [Google Scholar]
- Wu N, Phang J, Park J, Shen Y, Huang Z, Zorin M, Jastrzebski S, Fevry T, Katsnelson J, Kim E, et al. (2019). Deep Neural Networks Improve Radiologists’ Performance in Breast Cancer Screening. IEEE Trans Med Imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RKC, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR, et al. (2015). The human splicing code reveals new insights into the genetic determinants of disease. Science 347, 1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Liu Y, Chen T, and Tong Y (2019). Federated Machine Learning: Concept and Applications. ACM Trans Intell Syst Technol 10, Article 12. [Google Scholar]
- Zhang J, Bajari R, Andric D, Gerthoffert F, Lepsa A, Nahal-Bose H, Stein LD, and Ferretti V (2019). The International Cancer Genome Consortium Data Portal. Nat Biotechnol 37, 367–369. [DOI] [PubMed] [Google Scholar]
- Zitnik M, Nguyen F, Wang B, Leskovec J, Goldenberg A, and Hoffman MM (2019). Machine Learning for Integrating Data in Biology and Medicine: Principles, Practice, and Opportunities. Inf Fusion 50, 71–91. [DOI] [PMC free article] [PubMed] [Google Scholar]