Abstract
Plants interact with a great variety of microorganisms that inhabit the rhizosphere or the epiphytic and endophytic phyllosphere and that play critical roles in plant growth as well as the biocontrol of phytopathogens and insect pests. Avocado fruit damage caused by the thrips species Scirtothrips perseae leads to economic losses of 12–51% in many countries. In this study, a screening of bacteria associated with the rhizosphere or endophytic phyllosphere of avocado roots was performed to identify bacterial isolates with plant growth-promoting activity in vitro assays with Arabidopsis seedlings and to assess the biocontrol activity of the isolates against Scirtothrips perseae. The isolates with beneficial, pathogenic and/or neutral effects on Arabidopsis seedlings were identified. The plant growth-promoting bacteria were clustered in two different groups (G1 and G3B) based on their effects on root architecture and auxin responses, particularly bacteria of the Pseudomonas genus (MRf4-2, MRf4-4 and TRf2-7) and one Serratia sp. (TS3-6). Twenty strains were selected based on their plant growth promotion characteristics to evaluate their potential as thrips biocontrol agents. Analyzing the biocontrol activity of S. perseae, it was identified that Chryseobacterium sp. shows an entomopathogenic effect on avocado thrips survival. Through the metabolic profiling of compounds produced by bacteria with plant growth promotion activity, bioactive cyclodipeptides (CDPs) that could be responsible for the plant growth-promoting activity in Arabidopsis were identified in Pseudomonas, Serratia and Stenotrophomonas. This study unravels the diversity of bacteria from the avocado rhizosphere and highlights the potential of a unique isolate to achieve the biocontrol of S. perseae.
Introduction
Bacteria inhabiting the roots of trees have been scarcely investigated. These microbial populations may include plant growth promoters and biological control agents [1]. In model plants, an increase in plant biomass correlated with changes in root architecture such as, increased lateral root formation and root hair development or changes in the main root growth and angle the of branching structures [2,3]. In addition, other factors, including nutrient acquisition [4,5], phytohormones production (e.g., auxins or cytokinins) or host physiological alterations may account for the probiotic properties of certain bacteria [6–9],. On the other hand, the biocontrol of pathogens and insect pests that challenge plants may be exerted by rhizobacteria through competition for nutrients, the production of antimicrobial and antifungal compounds, or the activation of plant immune responses [10–12].
The identification of rhizobacteria for their use as probiotics includes the exploration of the rhizospheres of wild and crop plants from various ecosystems. In Mexico, the growth of avocado trees in the temperate zone in Michoacán state is a growing industry for which effective and environmentally friendly management are urgently needed. Despite the importance of avocado around the world, there are just a few studies regarding its rhizosphere biology. Carzola et al. [13] reported that four strains of Bacillus subtilis were associated with avocado antifungal activity and showed apparent enzymatic and antibiotic action against phytopathogenic fungi, whereas Guevara-Avendaño et al. [14] identified Bacillus strains with the ability to control the phytopathogenic fungi Fusarium euwallaceae and Graphium sp. Méndez-Bravo et al. [15] described the effects of Bacillus, Pseudomonas and Arthrobacter isolated from the avocado rhizosphere against Phytophthora cinnamomi through the production of volatile organic compounds.
A major goal is to identify biological strategies to combat insect pests such as thrips, which are currently controlled by the application of highly toxic insecticides that are responsible for the decline in the populations of pollinators such as bees. Thrips diminish the fruit quality as well as avocado production rates [16]. In the United States, the avocado industry in California has estimated annual losses attributable to the avocado thrips Scirtothrips perseae Nakahara at $8.51 million in the short term [17]. Considering that the production of Hass avocado hybrids has grown significantly in Mexico, making it the leading producer worldwide, the severity of this pest can translate into losses of millions of dollars. In 2015, avocado production reached a total of 1,624,000 tons in Mexico [18]. Some thrips species with phytosanitary importance have been reported in Mexico, including: Frankliniella bruneri Watson, Heliothrips haemorrhoidalis Bouché, Scirtothrips perseae Nakahara, and Pseudophilothrips perseae Watson [19].
Certain bacterial genera, such as Pseudomonas, Bacillus, Burkholderia, Serratia, Paenibacillus, and Streptomyces, include species with entomopathogenic activity against different insect pests [20,21], but there is limited information regarding the biological control of avocado thrips. Therefore, searching for rhizobacteria that could be used for the biocontrol of this avocado crop pest is relevant. In this report, we isolated and identified bacteria associated with avocado trees. Since avocado is a fruit crop whose optimal yield begins five years after planting, we characterized the plant growth-promoting effects of isolated bacteria on Arabidopsis thaliana as well as evaluating their direct entomopathogenic activity against avocado thrips Scirtothrips perseae. The results obtained are relevant for the development of biotechnological applications to reduce the use of fertilizers and chemical pesticides to sustainably improve avocado growth and health.
Material and methods
Sampling, bacterial isolation and morphotype groups
The study/sampling collection was carried out on private land, owned by different members of APEAM (Asociación de Productores y Empacadores Exportadores de Aguacate de México), who provided us the permission to conduct the study/sampling in all locations. Soil and root samples were collected from avocado plants (Persea americana Miller) in the orchards of La Meza and La Meza San Angel in Taretan, El Camino 3 in Tingambato and Los Diamantes in San Juan Nuevo between October 2016 and March 2017. All these places are in Michoacán, Mexico.
The bacterial isolates were obtained from soil, rhizosphere soil, and roots (endophytic bacteria) by serial dilutions. The growth conditions were 30°C for 24–48 h on Luria-Bertani agar (LB) medium. To confirm their purity, all strains were streaked at least three times and analyzed with gram stain. The colonial macroscopic data were used to form groups by morphotype. At least 20% of each group with similar morphological characteristics and 100% of the unique strains were selected for the plant growth promotion assay with the Arabidopsis-bacteria coculture system.
In-vitro plant growth promotion assay
For the Arabidopsis-bacteria coculture system, A. thaliana Col-0 seedlings, strains of Pseudomonas aeruginosa PAO1 (wild type strain, negative control), P. aeruginosa ΔlasI (quorum-sensing (QS) mutant strain, positive control for plant growth promotion) and selected isolated strains were used [7,22]. Arabidopsis seeds were surface disinfected and germinated on 0.2x Murashige and Skoog (MS) medium [23] and grown in a plant growth chamber (CARON, model 7300–50) according to conditions described by Ortiz-Castro et al. [7]. Seven-day-old A. thaliana seedlings (8 seedlings per plate [n = 24]) were coinoculated in direct contact with each bacterial isolate. The A. thaliana seedlings were grown for a further 7-d period by placing the plates in the growth chamber in a completely randomized design, according to the method reported by Ortiz-Castro et al. [22]. After the growth period, the primary root length, lateral root number, root and shoot fresh weight, and anthocyanin concentrations were measured according to the method in a previous report [24]. The lateral root density was calculated by dividing the lateral root number by the primary root length. All experiments were replicated at least twice.
Molecular characterization and phylogenetic analysis of isolates
Genomic DNA extraction was performed by the protocol of Nicholson et al. [25], Cenis, [26] and Ogram et al. [27]. The DNA was diluted with ultrapure water (1:10). To perform molecular characterization of the isolates, the molecular markers used were the hypervariable region V3-V5 from 16S rDNA for all the bacterial isolates and the 16S rDNA gene for those isolates selected for metabolic profiling. In the PCR, the hypervariable region was amplified using the primers: V3-V5 AFRW: TACGGRAGGCAGCAG; V3-V5 BREV: CCGTCAATTCMTTTGAGTTT, while the 16S rDNA gene was amplified using the primers: 27F: AGAGTTTGATCMTGGCTCAG and R1494: CTACGGRTACCTTGTTACGAC. Both markers were amplified using the protocol described by Espinosa Asuar [28]. Each sample of extracted DNA was amplified in triplicate and purified using the GeneAll® ExpinTM PCR SV minikit according to the manufacturer's protocol. Finally, the samples were sent to Macrogen Inc. for sequencing.
The obtained sequences were compared against nucleotide databases using the NCBI BLAST algorithm at http://www.ncbi.n1m.nih.gov/blast/Blast.cgi. Triplicate sequences were edited with Applied Biosystems Sequence Scanner Software v2.0 from Thermo Fisher Scientific and processed in EGassember [29] to obtain the consensus sequence. The alignment of consensus sequences and phylogenetic analysis by maximum likelihood using bootstraping with 1,000 replicates were performed in MEGA7 [30]. 16S rDNA sequences obtained from the selected isolates for metabolic profiling were deposited in GenBank (Accession Numbers: MN098850-MN098867).
Auxin-responsive gene expression analysis
To evaluate auxin-responsive gene expression, 5-day-old A. thaliana DR5::uidA [31] transgenic seedlings were coinoculated with different bacterial strains with plant growth-promoting activity and grown for an additional 6-day period. For the histochemical analysis of β-glucuronidase (GUS) activity, A. thaliana seedlings were incubated at 37°C in a GUS reaction buffer (0.5 mg/mL 5-bromo-4-chloro-3-indolyl-BD-glucuronide in 100 mM sodium phosphate, pH 7). The seedlings were clarified according to Malamy and Benfey [32]. At least 10 transgenic seedlings were analyzed for GUS activity by using Leica S8 APO stereoscopic microscope (Leica, Microsystem). A qualitative scale from -4 to 4 was established to compare the GUS activity of uninoculated and cocultivated Arabidopsis seedlings. A score of 4 represented four times more expression than the control.
Phosphate solubilization assay
To estimate the phosphate solubilization capacity of the bacterial isolates with plant growth promotion activity, bacteria were cultured in LB medium overnight and then cultured on Pikovskaya-bromophenol blue (PKV-BMP) medium at 0.025 OD600 (n = 3). The PKV-BMP medium contained 10 g/L dextrose, 5 g/L Ca3(PO4)2, 0.5 g/L yeast extract, 0.5 g/L (NH4)2SO4, 0.2 g/L KCl, 0.1 g/L MgSO4 7H2O, 0.0001 g/L MnSO4, 0.0001 g/L FeSO4, and 0.025 g/L BPB. The bacterial isolates were incubated at 30° C, 180 rpm for 48 h. The cultures were harvested by centrifugation at 8,000 rpm for 5 min; the obtained supernatant was measured at 590 nm by using an Epoch2 (Biotek) spectrophotometer for the quantitative assay. The experiment was repeated twice per triplicate for each bacterial isolated evaluated.
Entomopathogenic activity assay
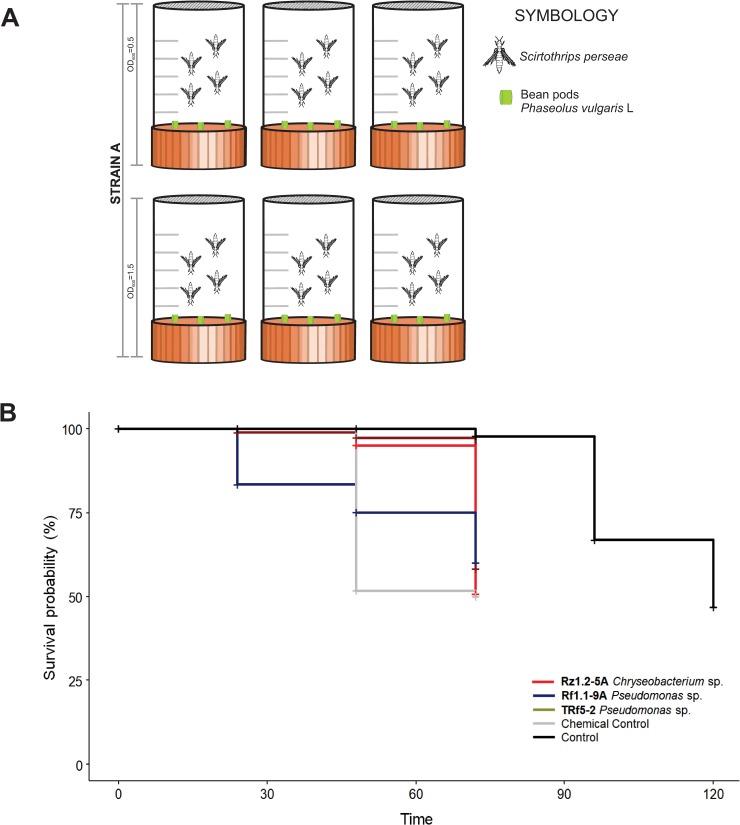
We assessed the entomopathogenic activity of a selected group of bacterial strains with plant growth promotion activity with a thrips oral toxicity test. The thrips used in the test were laboratory-reared Scirtothrips perseae individuals reared and maintained in plastic cages with disinfected fragments of bean pods (Phaseolus vulgaris L.) as food and oviposition sources, at 23–25°C, relative humidity (RH) 70–75% and 12 h light. For this assay, the 20 strains were cultured in LB broth for 48 h at 28°C and adjusted by dilution until they reached 0.5 and 1.5 OD600. The chambers used in the assay were made with 50 mL Falcon® tubes cut in half and sealed with antithrips mesh (Fig 4A). The chambers were autoclaved before use. Disinfected fragments of bean pods (length <5 mm) were impregnated by immersion in bacterial solutions and were introduced into the chambers as a source of food and oviposition sites. Due to limitations on the number of laboratory-reared thrips, the 20 strains were subjected to a first screening with only four adult thrips in each chamber. In the first screening, uninoculated bean pod fragments were used as controls. The chambers were maintained at 23–25°C, 70–75% RH and 12 h light, and the number of living insects was recorded at 24, 48, 72 and 96 h. All strains were tested in triplicate. The bacterial treatments with fewer recorded live insects were selected for a second oral toxicity test. The second test was performed by placing 10 adult thrips in each chamber, using all selected strains at 1.5 OD600 with an insecticide mix of imidacloprid plus lamba-cyhalothrine (Turner SC® Allister) at the commercial dose as a positive control and uninoculated bean pod fragments as negative control. The selected strains were tested in triplicate, the chambers were maintained under the previously described conditions and the number of live insects was recorded at 24, 48, 72, 96 and 120 h.
Fig 4. Entomopathogenic activity of plant growth-promoting rhizobacteria from avocado.
(A) Representative diagram of the chamber design of the in-vitro entomopathogenic activity assay per strain. The image shows the number of repetitions per concentration of bacterial strain analyzed, as well as the number of insects used per chamber. (B) Oral toxicity assay of seven of the twenty strains evaluated with S. perseae. Thrips were exposed to the strains for 96 h in a bioassay chamber. Bean pod slices impregnated with two bacterial concentrations were used as food. Data for the commercial control and the control with no inoculation are presented in red and black, respectively. The median is shown as a bar inside the boxes, and the whiskers indicate to the range of the data.
Chemical profiling of plant growth-promoting activity strains
A selected group of bacterial strains with plant growth promotion activity in A. thaliana were cultured in 300 mL of LB broth with 0.5 OD600 and grown for 48 h at 30°C and 150 rpm. The cultures were centrifuged at 12,000 rpm and the supernatant was lyophilized (Labconco FreeZone™ Freeze-Dry Systems 4.5L). Extractions were performed by sonication for 3 periods of 5 min each using methanol (1:10 v/v) in an ultrasonic bath (CPXH series) at 20°C. The extracts were centrifuged at 6500 rpm for 5 min at 20°C. The supernatant was collected, and the methanol was removed under vacuum pressure through a rotaevaporator (Büchi® Rotavapor® RII). The samples were analyzed in a Waters ultrahigh-performance liquid chromatographic (UHPLC) class I system coupled to a Synapt HDMi mass spectrometer (see Supplementary information). The data were acquired and processed with MassLynx (WatersTM, version 4.1) and MarkerLynx (WatersTM, version 4.1) software to determine the distinct chemical biomarkers through orthogonal partial least squares discriminant analysis (OPLS-DA).
Data analysis
Data were analyzed with the R programming language [33]. For the plant growth promotion assay, a cluster analysis of the primary root length, lateral root number, root and shoot fresh weight, and anthocyanin concentration was performed with all the assessed bacterial isolates. The data were normalized (Scale) and grouped with the K-means method in three blocks. For the dendrogram construction, a distance matrix was calculated using the "Euclidian" method based on the grouped data and graphed with the "complete" method of the cluster package [34]. Group formation was evaluated statistically through similarity analysis (ANOSIM) using the vegan package [35]. Chemical profiling data were visualized with a heatmap graph, and for the proposed candidate compounds, the METLIN metabolomics database (https://metlin.scripps.edu) was consulted. The mass/charge (m/z) ratio of the quasi-molecular ions and the fragmentation pattern were compared with the Metlin database, and the maximum error allowed was five (Δppm). The results of the entomopathogenic bioassay with the thrips were analyzed through survival analysis with the log-rank test from the survival package [36,37]. The multiple paired comparison of the curves was performed with the survminer package [38]. Graphs were constructed using the ggplot [39] and ggpubr [40] packages.
Results
Bacteria associated with avocado trees show plant growth-promoting activity in Arabidopsis thaliana
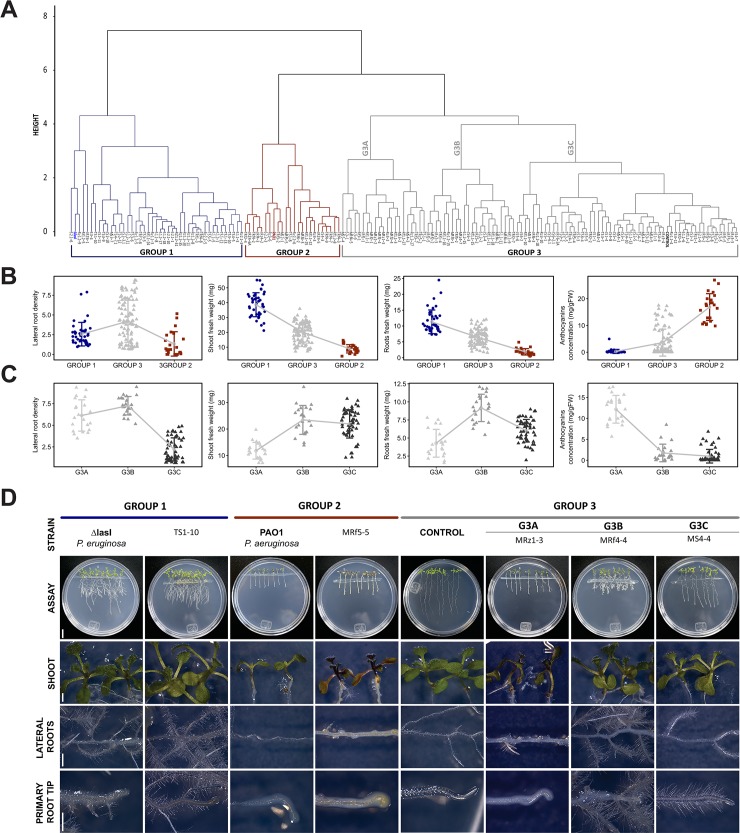
A total of 446 bacterial isolates were obtained from the different soil, rhizosphere and root samples. We evaluated the in vitro effect of the 162 selected strains by using an Arabidopsis-bacteria coculture system to screen the bacterial growth promotion activity on plants. The dendrogram in Fig 1A was obtained from cluster analysis and differentiated the treatments into three groups: Group 1 (G1) contained 42 isolates and the positive control P. aeruginosa ΔlasI, Group 2 (G2) contained 23 isolates and the negative control P. aeruginosa PAO1 and Group 3 (G3) contained 97 isolates and the uninoculated control. Statistically significant differences were found among groups through an analysis of similarity (ANOSIM, p<0.05, R = 0.67).
Fig 1. Avocado rhizosphere bacteria with plant growth-promoting activity in Arabidopsis thaliana.
Cluster analysis of bacterial isolates associated with Persea americana Miller evaluated in the Arabidopsis-bacteria coculture system, ANOSIM, p<0.05, R = 0.67. (A) Dendrogram resulting from the cluster analysis of the 162 selected isolates. (B) Analysis of the grouping pattern of Groups 1, 2 and 3 considering the arithmetic means per group for each variable. (C) Grouping patterns of subgroups G3A, G3B, and G3C. The points represent each of the observations, and the bars represent the standard deviation. (D) Representative images of the growth of Arabidopsis seedlings inoculated with strains associated with the different groups and subgroups. Control plants are shown as uninoculated or inoculated with P. aeruginosa PAO1 (wild-type strain, pathogenic) and P. aeruginosa ΔlasI (QS mutant strain, promoter). Scale bar: Petri dishes = 1 cm; plant zones = 3 mm.
When plotting the group means of the variables measured in the three groups (Fig 1B), some patterns were observed. The isolates associated with P. aeruginosa ΔlasI (G1) induced the highest shoot and root fresh weight values and a lower anthocyanin concentration in the inoculated seedlings while the isolates grouped with P. aeruginosa PAO1 in G2 showed the opposite pattern (Fig 1B). Plant growth promotion by G1 isolates is represented by TS1-10 (Fig 1D), whereas isolates from G2 with a deleterious effect are shown by MRf5-5 (Fig 1D).
G3 isolates were associated with control as "neutral" strains. As shown in Fig 1C, we found three types of bacteria in G3. The first type, isolates grouped in G3A, caused some type of damage in the plant (necrosis, highest mean for anthocyanin concentration), but induced many lateral roots and an inhibition in the growth of the primary root tip. These effects are shown by the Arabidopsis seedlings coinoculated with the MRz1-3 strains (Fig 1D). The second group, G3B strains, in addition to producing a short primary root length, stimulated lateral root formation and enhanced shoot fresh weight without anthocyanin induction, for example, MRf4-4 (Fig 1D). The third group, G3C, were the isolates most similar to the uninoculated control. Representative photographs of the growth of Arabidopsis coinoculated with the MS4-4 strains from this subgroup were compared with photographs of the other subgroups (Fig 1D).
The following experiments were focused on the strains in the G1 and G3B groups because of their clearly observed plant growth-promoting effects.
Molecular characterization of bacteria with plant growth promoting activity
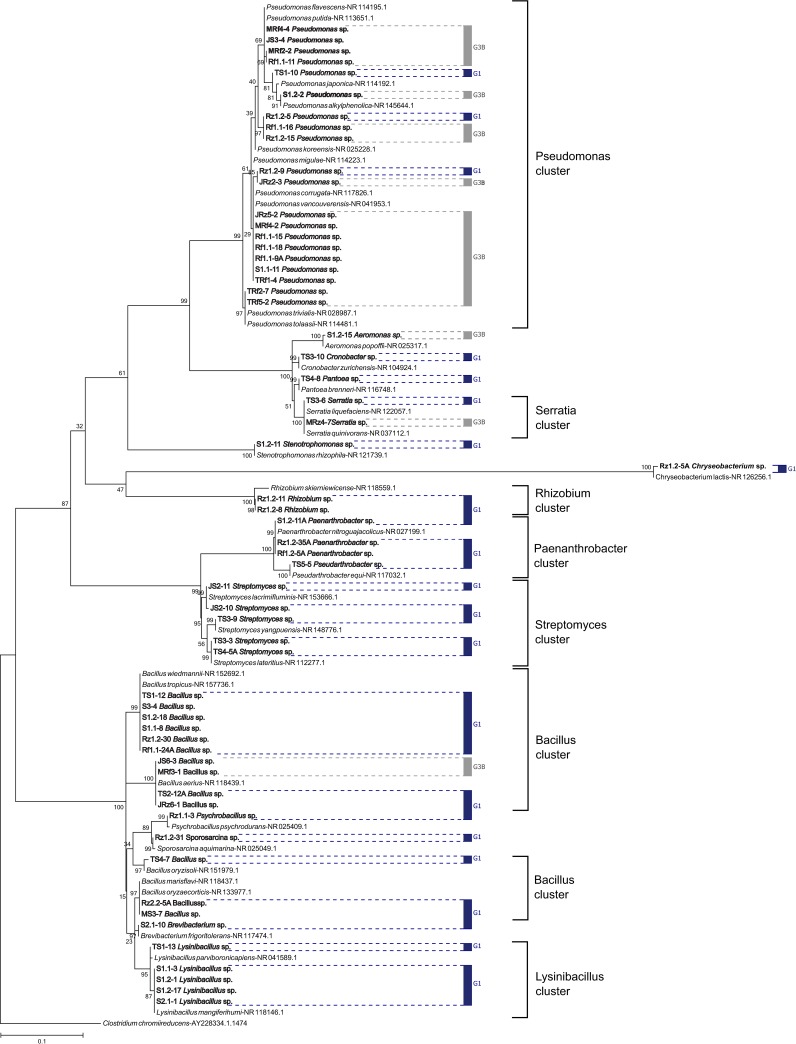
The bacterial isolates were molecularly identified by analyzing the hypervariable region V3-V4 from 16S rDNA. Sixteen genera were identified from the G1 and G3B groups. The G1 Bacillus group comprised 10 strains, while G3B mostly consisted of Pseudomonas. In general, a greater richness of bacterial genera was found in the G1 group than in the G3B group. The phylogenetic tree presented in Fig 2 shows 88% G1 and 100% G3B strains. Notably, although the dominant genera were different for each group, there was not a strict taxonomic separation between the two groups analyzed. The genera Bacillus, Pseudomonas and Serratia were found in both groups.
Fig 2. Phylogenetic relationships among G1 and G3B strains, including NCHB references, were inferred using the maximum likelihood method.
The percentage of the replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 runs) is shown next to the branches. The tree is drawn to scale. G1 is indicated with blue lines, and G3B is indicated with gray lines.
Although this study focused on bacterial isolates with plant growth-promoting activity (PGPB), at least 15% of the isolates from the other groups were sequenced. The phylogenetic relationships among the G2, G3A and G3C groups are shown in S1 Fig.
Plant growth-promoting activity is regulated through phosphate solubilization and the modulation of auxin-related gene expression in Arabidopsis
One indirect mechanism of action of PGPB is inorganic phosphate solubilization, allowing P uptake in the plant. Therefore, we analyzed the capability of strains with plant growth promotion activity to solubilize tricalcium phosphate by using Pikovskaya-bromophenol blue (PKV-BMP) medium. Interestingly, in comparison with the control, five strains from Pseudomonas sp. (Rf1.1-9A, TRf5-2, TS1.10, TRf2-7, JRz5-2) and one strain from Brevibacterium sp. (S2.1–10) showed the highest phosphate solubilization activity, while strains Lysinibacillus sp. S1.2–17, Pseudomonas sp. Rz1.2–5, and Streptomyces sp. JS2-10 showed low phosphate solubilization activity (S2 Fig).
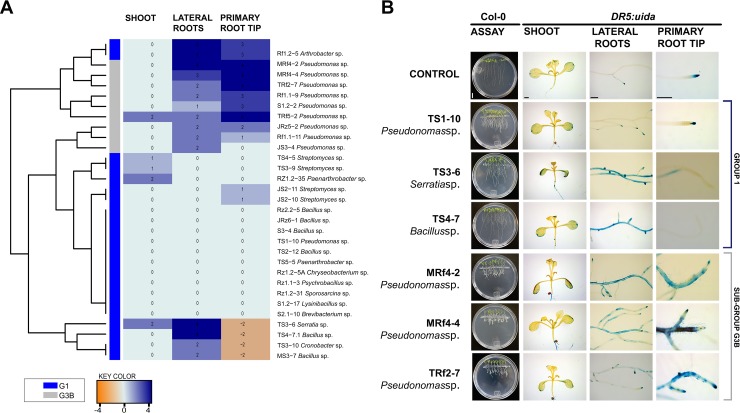
Several effects shown by the PGPB from the G1 and G3B groups are related to auxin-like activity. Arabidopsis transgenic lines of DR5::uidA were inoculated with 21 selected strains, and uidA gene expression was analyzed (Fig 3). The heat map constructed showed a clear differentiation between strains G1 and G3B (Fig 3A) on the DR5::uidA gene marker. Ten strains (Serratia sp. TS3-6; Bacillus sp. TS4-7.1; Pseudomonas sp. MRF4-2, MRF4-4, Rz1.2–5, Rf1.1–9, S1.2–2, TRf5-2 and TRF2-7; Arthrobacter sp. Rf1.2–5) induced the highest production of auxin in lateral root zone and/or in the primary root meristem (Fig 3A and 3B). Interestingly, Serratia sp. TS3-6, Bacillus sp. TS4-7 and Cronobacter sp. TS3-10 had an unusual lack of auxinic gene expression in the primary root tip, in contrast with the high expression in the lateral root zone.
Fig 3. Effect of some bacterial isolates from G1 and G3B on the DR5::uidA reporter gene for auxin responses in Arabidopsis thaliana.
(A) Qualitative analysis of uidA gene expression in A. thaliana transgenic lines inoculated with bacterial isolates. Values were obtained by comparing the treatments with the control without inoculation. A scale with a range of 4 to -4 was established for this analysis. Higher values and a more intense blue color indicate higher expression compared to that in the control. The orange color indicates an absence of gene expression (NE). (B) Representative photographs of the shoot, lateral root zone and meristem of the primary root of transgenic DR5::uidA seedlings inoculated with bacterial strains. Scale bar: Petri dishes = 1 cm, plant zones = 500 μm.
Plant growth-promoting bacteria associated with avocado show entomopathogenic activity
Twenty bacterial strains with the best plant growth promotion characteristics were evaluated in an entomopathogenic bioassay with laboratory-reared avocado thrips Scirtothrips perseae. As a first approach, the 20 strains were used in bacterial solutions at 0.5 and 1.5 OD600 to test their oral toxicity on S. perseae for four days (96 h, Fig 4A). To compare the number of dead insects in the different treatments, a survival analysis with a log-rank test was performed. Statistically significant differences were found both when comparing the curves with respect to the OD concentrations [X2 = 83.5, p<0.05] and when only the strains were considered [X2 = 70.9, p<0.05]. In the multiple paired comparisons of the curves with p values adjusted with the BH method, only Chryseobacterium sp. Rz1.2-5A presented a difference at the margin of statistical significance (p = 0.058) compared with the control. Although no significant differences were recorded between the strains and the control, some tendencies were observed. After 48 h, no live insects were counted in six of the 20 strains analyzed (S3 Fig). In the chambers where the insects were exposed to Chryseobacterium sp. Rz1.2-5A, regardless to the concentration, the insects died at 24 h, while in the control, live insects were recorded until 72 h (S3 Fig). From this first screening, the three bacterial strains with the smallest numbers of live thrips recorded were selected for a second oral toxicity test with 120 h of evaluation. In the second test, a higher number of thrips and an insecticide mix of imidacloprid plus lamda-cyhalotrine (chemical control) were used. The selected strains were Chryseobacterium sp. Rz1.2-5A, Pseudomonas sp. Rf1.1-9A and Pseudomonas sp. TRf5-2, in which <1 live thrips were recorded after 24 h at OD600 1.5 (S3 Fig). In the second test, dead thrips were recorded at 24 h in Pseudomonas sp. Rf1.1-9A treatment, even earlier than in the chemical control, and the survival probability was reduced to <80%. Interestingly, at 72 h in Chryseobacterium sp. Rz1.2-5A treatment the thrips survival probability was reduced to 50%, the same level observed in the chemical control (Fig 4B). To compare the number of dead thrips in the different treatments, a survival analysis with a log-rank test comparison was performed, and significant differences were detected between the control and all treatments, including the chemical control (X2 = 65.9, df = 4, p<0.0001; Fig 4B). After a multiple pairwise comparisons of the curves, Chryseobacterium sp. Rz1.2-5A showed a similar survival curve to the chemical control (p = 0.655), suggesting the same effectiveness level.
Chemical profiling of plant growth-promoting bacteria
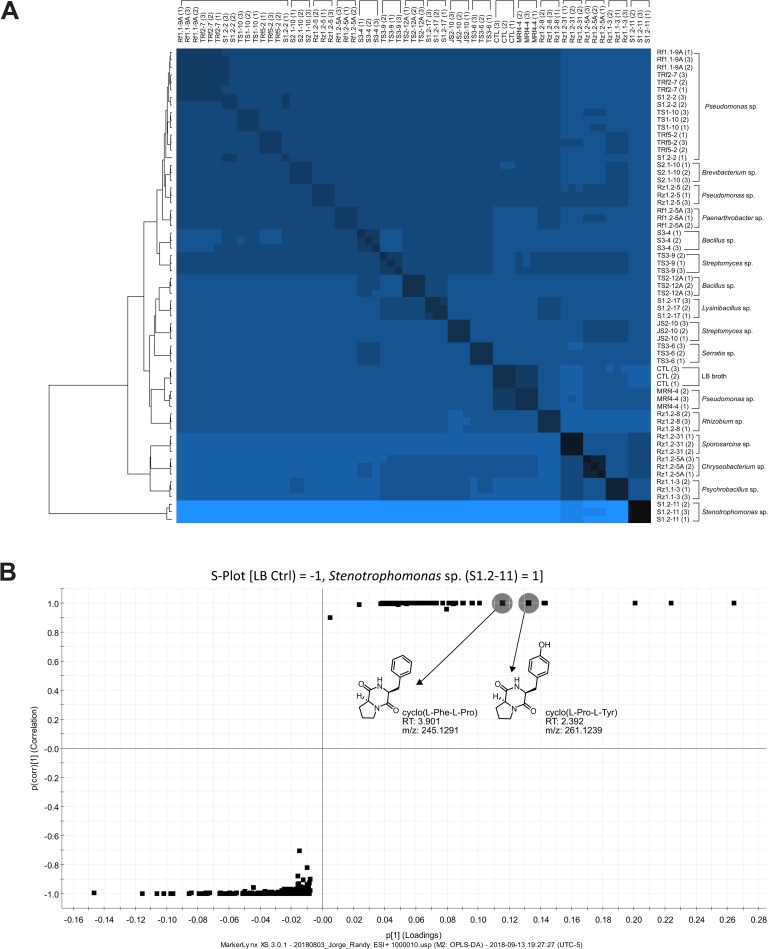
Considering the chemical natures of the compounds produced by different bacteria, chemical profiling of 20 bacterial strains was performed to determine whether their plant growth-promoting characteristics could be reflected at the metabolic level. The 20 bacterial strains subjected to chemical profiling were chosen based on their plant growth promotion characteristics and their potential as thrips biocontrol agents. The heat map in Fig 5A shows the chemical profile for each of the selected strains, and it is possible to differentiate some bands or groups. Interestingly, the bands at the ends (top and bottom) of the heat map showed differences compared with the control. The strains inserted in the middle band seem to be similar to the LB broth profile (CTL). At the bottom, the Stenotrophomonas sp. S1.2–11 band presents a completely different chemical profile compared to all others, while at the other end a Pseudomonas sp. (Rf1.1-9A, Rz1.2–5, TRf2-7, S1.2–2, TS1-10, TRf5-2) cluster is located. Of the seven strains of Pseudomonas included in the analysis, only Pseudomonas sp. MRf4-4 is separate from this group (Fig 5A).
Fig 5. Chemical profile of avocado rhizosphere bacteria with plant growth-promoting activity.
(A) Heat map representation of the chemical profile of the twenty plant growth promoting rhizobacteria strain extracts. Cluster relationships are presented through the dendrogram on the left side of the heat map. This graph was constructed considering analytical triplicates. LB broth was used as the culture medium and is indicated in the graph as CTL. (B) S-plot analysis of the differential compounds produced between Pseudomonas MRf4-4 and a set of 6 other Pseudomonas strains.
Exploring the chromatograms and spectrometric fingerprints allowed the tentative identification of a group of cyclodipeptides (CDPs, Table 1), compounds with auxin-like activity [7]. From the Pseudomonas cluster we identified cyclo (L-His-L-Leu). This same CDP was found for Serratia sp. TS3-6 as well (Table 1). Regarding Stenotrophomonas sp. S1.2–11, differential analysis with orthogonal partial least squares discriminant analysis (OPLS-DA) was performed to compare this unique strain to the LB broth (control). The S-plot of this analysis is presented in Fig 5B, where points located at negative values represent the differential compounds or fragments of the LB broth, while points at positive values indicate the differential compounds or fragments of the Stenotrophomonas sp. strain. This analysis identified two CDPs: cyclo(L-Pro-L-Tyr) and cyclo(L-Phe-L-Pro). The fragment pattern matches the databases for these three CDPs (mass spectra in S4 Fig). These results suggest the possible role of cyclodipeptides in plant growth promotion and entomopathogenic activity.
Table 1. Identification of cyclodipeptides from bacteria with plant growth promoting activity.
| STRAIN | ID | GROUP | m/z | RT (min) | QMI | ERROR (ppm) |
CDP |
|---|---|---|---|---|---|---|---|
| Pseudomonas sp. | S1.2–2 | G3B | 251.15 | 1.343 | [M+H]+ | 4.4 | 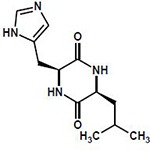 |
| Rf1.1-9A | G3B | 251.15 | 1.343 | [M+H]+ | 2.8 | ||
| TRf2-7 | G3B | 251.15 | 1.326 | [M+H]+ | 1.2 | ||
| TRf5-2 | G3B | 251.15 | 1.343 | [M+H]+ | 1.2 | ||
| Rz1.2–5 | G1 | 251.15 | 1.363 | [M+H]+ | 1.9 | cyclo(L-His-L-Leu) | |
| TS1-10 | G1 | 251.15 | 1.363 | [M+H]+ | 2.3 | C12H18N4O2 | |
| Serratia sp. | TS3-6 | G1 | 251.15 | 1.380 | [M+H]+ | 2.8 | M: 250.14297 |
| Stenotrophomonas sp. | S1.2–11 | G1 | 261.12 | 2.392 | [M+H]+ | 1.9 | 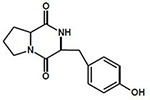 |
| cyclo(L-Pro-L-Tyr) | |||||||
| C14H16N2O3 | |||||||
| M: 260.116089 | |||||||
| 245.12 | 3.901 | [M+H]+ | 0.8 | 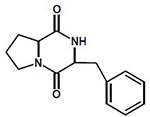 |
|||
| cyclo(L-Phe-L-Pro) | |||||||
| C14H16N2O2 | |||||||
| M: 244.121185 |
ID: code name, m/z: mass-charge ratio, RT: retention time, QMI: quasi-molecular ion, CDP: cyclodipeptide.
Discussion
Several studies have used the Arabidopsis thaliana plant model to evaluate the beneficial, pathogenic or neutral effects of microorganisms associated with plants and as a rapid system for identifying or characterizing plant growth-promoting bacteria [6–8,15,22,41,44,63]. Avocados are large forest trees, often over 20 m tall, that exhibit rhythmic growth with two or more flushes of shoot growth per year, alternating with periods of rest, and that reach optimal yields until five years after planting [42], while A. thaliana completes its life cycle in 8–12 weeks from germination to harvesting. In this study, we used the A. thaliana model to evaluate the plant growth-promoting effects of bacteria isolated from avocado trees in a shorter time. The notable high biomass of A. thaliana seedlings after inoculation with strains from Group 1 is a characteristic commonly attributed to several plant growth-promoting rhizobacteria (PGPR) of Bacillus and Pseudomonas strains in Phaseolus vulgaris [41], Solanum lycopersicum L., Amaranthus sp. [43] and Arabidopsis [44]. This suggest that G1 could be considered plant growth-promoting bacteria.
In Group 2, the high anthocyanin concentration it was a defining characteristic. The role of anthocyanins as indicators of stress in plant-microbe interactions is well understood. Specifically, transcriptomic studies have shown an increased expression of genes related to anthocyanin biosynthesis during pathogen attack [45,46]. Himemo et al. [47] showed the correlation of this type of compound with a defense mechanism to prevent cell death in the shoots of infected plants. The effects observed in plants inoculated with strains associated with negative control P. aeruginosa PAO1 in G2 reflect phytopathogenic or opportunistic bacteria causing some kind of damage or stress in the plant and compromising foliar and radicular development, since the plants with the lowest lateral root number and shoot weight were recorded in this group.
In this sense, the high anthocyanin concentrations recorded with G3A strains denote a pathogenic effect in Arabidopsis seedlings; therefore, the high lateral root number values for these strains imply an opposite effect. Dovana et al. [48] described how some endophytic pathogenic fungi might reduce cell elongation by producing more cells per unit volume when the increase in dryness is not related to fresh weight and noted the involvement of IAA in this effect. This could explain how G3A strains showed high values for lateral root density and low shoot and root fresh weights. Thus, we propose that the G3A group contains phytopathogenic strains. In the case of the strains grouped in G3B, the effects observed in Arabidopsis plants are similar to those in G1, except for a primary root shortening, which can be explained by the concentration of the inoculum. Shi et al. [49] and Kwon et al. [50], who worked with Serratia marcescens and Paenibacillus polymyxa, respectively, indicated that low concentrations of inoculum cause a growth promotion effect, while a high concentration inhibits the elongation of the primary root. Therefore, the strains of the G3B group can be considered growth promoters, and a larger primary root could be achieved by regulating the inoculum concentration. Finally, the G3C group strains associated with the uninoculated control can be considered neutral strains that have a null or reduced effect on the inoculated plants within the framework of the assay performed.
We molecularly identified the strains with plant growth-promoting characteristics from the G1 and G3B groups. The identified taxonomic positions have been reported in other studies describing the microbial community of avocado tree soil [51] and the rhizospheres of other Lauraceae species [52]. Recently, Méndez-Bravo et al. [15] described seven bacterial isolates of the genera Bacillus, Pseudomonas and Arthrobacter (the old name of several Paenarthrobacter species) with PGPR activity through the production of volatile organic compounds in Arabidopsis seedlings. All these genera were identified in our study. Bacillus and Pseudomonas were the two main genera we found, with Pseudomonas being better represented in group G3B and Bacillus in group G1. In general, all the genera identified include at least one species that has been reported in other studies as s plant growth promoter [53–60].
The presence of Pseudomonas in all groups represents an interesting point not only for the versatility of its effects in many plant systems, but also because it can be identified as a good auxin gene expression promoter and phosphate solubilizer when it is related to plant growth promotion activity. Most of the Pseudomonas strains that induced high DR5::uidA gene expression for auxin biosynthesis belonged to the subgroup G3B, so the expression of this particular gene could be directly associated with short primary root length, the main characteristic that distinguishes this group from G1. Strain concentration has been noted to directly affect the primary root length as in the G3B strains. Some authors attribute this effect to indole-3-acetic acid (IAA), a phytohormone that acts as a repressor of the elongation of the primary root at high concentrations [61,62]. We found this relationship between reduced primary root elongation and high auxin expression in G3B group.
The modulation of auxin expression by plant growth promoter strains from G1 is more heterogeneous than that of strains of G3B, which seems to be related to the higher diversity of the bacterial strains in G1. Moreover, some authors have described nutritional pathways such as phosphate solubilization, nitrogen fixation and siderophore production [63] as well as auxin-independent mechanisms [41]. In the same way, 2,3-butanediol, acetoin and other volatile organic compounds from rhizobacteria are reported to enhance plant growth through auxin-independent hormonal pathways [64].
Considering the plant growth-promoting characteristics described for the strains in the G1 and G3B groups, we studied the potential of a select group of strains to act as biocontrol agents through an entomopathogenic bioassay against S. perseae, one of the most harmful avocado pests. Notably, Chryseobacterium sp. Rz1.2-5A had an entomopathogenic effect on the thrips, showing a similar efficiency level as the chemical control. In a detailed search of the literature, we did not find publications about bacteria in the Chryseobacterium genus showing entomopathogenic activity. The closest report was one published by Yi et al. [65], who described a strain of the genus Flavobacterium, which is phylogenetically close to Chryseobacterium, showing entomopathogenic activity against the larvae of Spodoptera exigua, or commonly known as the settled worm, which is a pest of multiple crops.
Interestingly, in exploring the chemical profiles presented in the heatmap we identified the same presumed CDPs for almost all strains of Pseudomonas, except MRf4-4, the only strain in the heatmap not grouped with its congeners. The similarity found in the heatmap between the MRf4-4 and LB medium profiles (control), indicates the need to explore longer incubation times or larger extraction volumes to corroborate the capacity of this strain to produce the CDPs identified. Two others presumed CDPs were identified in Stenotrophomonas sp. and located as differential metabolites compared to those in the control. In a recent study, Song et al. [66] evaluated plant immune activation through seed defense biopriming (SDB) by using metabolites from root-associated Bacillus spp. isolates and found that B. gaemokensis strain PB69 increased the mortality of insect the pest Spodoptera litura fed with cucumber or pepper tissues previously treated with B. gaemokensis. They identified cyclo(L-Leu-L-Pro) as the compound responsible for SDB activity. Previously, Ortiz-Castro et al., [7] described three CDPs, cyclo(L-Pro-L-Tyr), cyclo(L-Pro-L-Phe) and cyclo(L-Pro-L-Val), from Pseudomonas aeruginosa that acted as auxin-like compounds responsible for plant growth promotion in Arabidopsis seedlings. This is relevant not only for understanding the possible participation of these compounds in plant growth promotion and pest biocontrol but could also be related to other important roles of the bacterial strains during plant-microbe interactions. The current study provides a basis for the potential application of rhizobacterial strains in avocado.
Supporting information
The percentage of the replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 runs) is shown next to the branches. The tree is drawn to scale. G2 is indicated with red lines, while G3A and G3C are indicated with gray lines.
(EPS)
Bacterial isolates were cultured on Pikosvskaya-bromophenol (PKV-BMP) medium with a 0.025 OD600 at 30°C, 180 rpm for 48 h. The data represent the means ± SD (n = 3). The experiment was replicated two times with similar results.
(TIFF)
Thrips were exposed to the strains for 96 h in a bioassay chamber. Bean pod slices impregnated with two bacterial concentrations were used as food. Data for the commercial control and the control with no inoculation are presented in red and black, respectively. The median is shown as a bar inside the boxes and the whiskers indicate to the range of the data.
(EPS)
(A) Compound with RT = 3.944 and m/z = 245.1295 obtained from the methanolic extract of Stenotrophomonas sp. S1.2–11. The fragmentation pattern corresponds to cyclo(L-Phe-L-Pro). (B) Compound with RT = 2.40 and m/z = 261.1243 obtained from the methanolic extract of Stenotrophomonas sp. S1.2–11. The fragmentation pattern corresponds to cyclo(L-Pro-L-Tyr). (C) Compound with RT = 1.343 and m/z = 251.1515 obtained from the methanolic extract of Pseudomonas sp. Rf1.1-9A. The fragmentation pattern corresponds to cyclo(L-His-L-Leu).
(EPS)
(DOCX)
Acknowledgments
We thank Dr. José López-Bucio for kindly providing seeds of the WT and transgenic lines. We thank the Asociación de Productores y Empacadores Exportadores de Aguacate de México A.C. (APEAM) and the Consejo Nacional de Ciencia y Tecnología (CONACyT). J.A. T.-I. thanks CONACyT for his master’s fellowship.
Data Availability
16S rDNA sequences obtained from the selected isolates for metabolic profile were deposited in GenBank (Accession Numbers: MN098850-MN098867).
Funding Statement
This work was supported by grants from the Asociación de Productores y Empacadores Exportadores de Aguacate de México A.C. (APEAM)-Instituto de Ecología A.C. (INECOL) (grant no. 42002); Consejo Nacional de Ciencia y Tecnología (CONACyT, México, grant no. PDCPN-2015-882). J.A. T.-I. thanks to CONACyT for his Master fellowship.
References
- 1.Besset-Manzoni Y, Rieusset L, Joly P, Comte G, Prigent-Combaret C. Exploiting rhizosphere microbial cooperation for developing sustainable agriculture strategies. Environ. Sci. Pollut. R. 2018; 25:29953–29970. [DOI] [PubMed] [Google Scholar]
- 2.Delaplace P, Delory BM, Baudson C, de Cazenave MMS, Spaepen S, Varin S, et al. Influence of rhizobacterial volatiles on the root system architecture and the production and allocation of biomass in the model grass Brachypodium distachyon (L.) P. Beauv. BMC Plant Biology. 2015; 15:1–15. 10.1186/s12870-014-0410-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pérez-Flores P, Valencia-Cantero E, Altamirano-Hernández J, Pelagio-Flores R, López-Bucio J, García-Juárez P,et al. Bacillus methylotrophicus M4-96 isolated from maize (Zea mays) rhizoplane increases growth and auxin content in Arabidopsis thaliana via emission of volatiles. Protoplasma. 2017; 254:2201–2213. 10.1007/s00709-017-1109-9 [DOI] [PubMed] [Google Scholar]
- 4.Hernández-Rodríguez A, Heydrich-Pérez M, Acebo-Guerrero Y, Velazquez-del Valle MG, Hernández-Lauzardo AN. Antagonistic activity of Cuban native rhizobacteria against Fusarium verticillioides (Sacc.) Nirenb. in maize (Zea mays L.). Appl. Soil Ecol. 2008; 39:180–186. [Google Scholar]
- 5.Molina-Romero D, Bustillos-Cristales MR, Rodríguez-Andrade O, Morales-García EY, Santiago-Saenz Y, Castañeda-Lucio M, et al. Mecanismos de fitoestimulación por rizobacterias, aislamientos en América y potencial biotecnológico. Revista de la DES Ciencias Biológico Agropecuarias. 2015; 17:24–34. [Google Scholar]
- 6.Ortiz-Castro R, Valencia-Canteros E, López-Bucio J. Plant growth promotion by Bacillus megaterium involves cytokinin signaling. Plant Signal. Behav. 2008; 3:263–265. 10.4161/psb.3.4.5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortiz-Castro R, Díaz-Pérez C, Martínez-Trujillo M, del Rio RE, Campos-García J, López-Bucio J. Transkingdom signaling based on bacterial cyclodipeptides with auxin activity in plants. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:7253–7258. 10.1073/pnas.1006740108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spaepen S, Bossuyt S, Engelen K, Marchal K, Vanderleyden J. Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 2013; 201:850–861. 10.1111/nph.12590 [DOI] [PubMed] [Google Scholar]
- 9.Rosier A, Medeiros FHV, Bais HP. Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil. 2018; 428:35–55. [Google Scholar]
- 10.Lee KJ, Oh BT, Seralathan KK. Advances in plant growth promoting rhizobacteria for biological control of plant diseases In: Bacteria in Agrobiology: Disease Management. Berlin, Heidelberg: Springer Berlin Heidelberg, 2013. pp. 1–13. [Google Scholar]
- 11.Islam S, Akanda AM, Prova A, Islam MT, Hossain MM. Isolation and identification of plant growth promoting rhizobacteria from Cucumber rhizosphere and their effect on plant growth promotion and disease suppression. Front. Microbiol. 2015; 6:1360 10.3389/fmicb.2015.01360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naseem M, Kaltdorf M, Dandekar T. The nexus between growth and defence signalling: auxin and cytokinin modulate plant immune response pathways. J. Exp. Bot. 2015; 66:4885–4896. 10.1093/jxb/erv297 [DOI] [PubMed] [Google Scholar]
- 13.Cazorla FM, Romero D, Pérez-García A, Lugtenberg BJ, Vicente A, Bloemberg G. Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity. J. Appl. Microbiol. 2007; 103:1950–1959. 10.1111/j.1365-2672.2007.03433.x [DOI] [PubMed] [Google Scholar]
- 14.Guevara-Avendaño E, Carrillo JD, Ndinga-Muniania C, Moreno K, Méndez-Bravo A, Guerrero-Analco JA, et al. Antifungal activity of avocado rhizobacteria against Fusarium euwallaceae and Graphium spp., associated with Euwallacea spp. nr. fornicatus, and Phytophthora cinnamomi. Anton. Leeuw. Int. J. G. 2018; 111:563–572. [DOI] [PubMed] [Google Scholar]
- 15.Méndez-Bravo A, Cortazar-Murillo EM, Guevara-Avendaño E, Ceballos-Luna O, Rodríguez-Haas B, Kiel-Martínez AL, et al. Plant growth-promoting rhizobacteria associated with avocado display antagonistic activity against Phytophthora cinnamomi through volatile emissions. PLoS ONE. 2018; 13:e0194665 10.1371/journal.pone.0194665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yee WL, Phillips PA, Faber BA, Rodgers JL. Relationships between Scirtothrips perseae (Thysanoptera: Thripidae) populations on avocado leaves, fruit, and scarring damage on fruit. Environ. Entomol. 2001; 30:932–938. [Google Scholar]
- 17.Hoddle MS, Jetter KM, Morse JG. The economic impact of Scirtothrips perseae Nakahara (Thysanoptera: Thripidae) on California avocado production. Crop Prot. 2003; 22:485–493. [Google Scholar]
- 18.Moral Barrera LE, Murillo Villanueva B, Moral BLE, Murillo VB. Producción y precio del aguacate en México, 2011-2016.II. Revista Paradigma Económico. 2016; 9:3–7. [Google Scholar]
- 19.Johansen R, Mojica-Guzmán A, Ascención-Betanzos G. Introducción al conocimiento de los insectos tisanopteros mexicanos, en el aguacatero (Persea americana Miller). Revista Chapingo Serie Horticultura. 1999; 5:279–285. [Google Scholar]
- 20.Narayanasamy P., Detection and Identification of Bacterial Biological Control Agents In: Biological Management of Diseases of Crops. Dordrecht: Springer Netherlands; 2013. pp. 201–293. [Google Scholar]
- 21.Ruiu L. Insect pathogenic bacteria in integrated pest management. Insects. 2015; 6:352–367. 10.3390/insects6020352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortiz-Castro R, García-Campos J, Bucio-López J. Rapid identification of plant-growth-promoting rhizobacteria using an agar plate cocultivation system with Arabidopsis In: Molecular Microbial Ecology of the Rhizosphere. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2013. pp. 345–353. [Google Scholar]
- 23.Murashige T, Skoog F. A revised medium for rapid growth and bio-assays with tobacco tissue culture. Physiol. Plantarum. 1962; 15:473–497. [Google Scholar]
- 24.Strack D, Wray V. Anthocyanins In: Methods in Plant Biochemistry. Harborne J. B. (Ed). Volume 1 Elsevier; 1989. pp. 325–356. [Google Scholar]
- 25.Nicholson TP, Rudd BAM, Dawson M, Lazarus M, Simpson TJ, Cox RJ. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem. Biol. 2001; 8:157–178. 10.1016/s1074-5521(00)90064-4 [DOI] [PubMed] [Google Scholar]
- 26.Cenis JL. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992; 20:2380 10.1093/nar/20.9.2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogram A, Sayler GS, Barkay T. The extraction and purification of microbial DNA from sediments. J. Microbiol. Meth. 1987; 7:57–66. [Google Scholar]
- 28.Espinosa Asuar L. Guía práctica sobre la técnica de PCR In: Eguiarte LE, Souza V, Aguirre X, eds. Ecología molecular. Secretaría de Medio Ambiente y Recursos Naturales, Instituto Nacional de Ecología, Universidad Nacional Autónoma de México, Comisión Nacional para el Conocimiento y Uso de la Biodiversidad; 2007. pp. 517–540. [Google Scholar]
- 29.Masoudi-Nejad A, Tonomura K, Kawashima S, Moriya Y, Suzuki M, Itoh M, et al. EGassembler: Online bioinformatics service for large-scale processing, clustering and assembling ESTs and genomic DNA fragments. Nucleic Acids Res. 2006; 34:W459–W462. 10.1093/nar/gkl066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016; 33:1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulmasov D, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 (1997) 1963–1971. 10.1105/tpc.9.11.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malamy JE, Benfey PN, Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997; 124:33–44. [DOI] [PubMed] [Google Scholar]
- 33.R.C. Team, R: A Language and Environment for Statistical Computing. (2017) https://www.R-project.org/
- 34.Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. Cluster: Cluster Analysis Basics and Extensions. (2017) https://cran.r-project.org/web/packages/cluster/citation.html [Google Scholar]
- 35.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: Community Ecology Package. R Package Version 2.2–0; 2017. http://CRAN.Rproject.org/package=vegan. [Google Scholar]
- 36.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the {C}ox Model. Springer, New York, 2000, ISBN:0-387-98784-3. [Google Scholar]
- 37.Therneau TM. A Package for Survival Analysis in S; 2015, https://CRAN.R-project.org/package=survival [Google Scholar]
- 38.Kassambara A, Kosinski M. Survminer: Drawing Survival Curves using ‘ggplot2’; 2018, https://rpkgs.datanovia.com/survminer/index.html [Google Scholar]
- 39.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: ISBN: 978-0-387-98140-6. 2009; http://www.springer.com/978-0-387-98140-6. [Google Scholar]
- 40.Kassambara A. ggpubr: ‘ggplot2’ Based Publication Ready Plots, 2018, https://github.com/kassambara/ggpubr
- 41.López-Bucio J, Campos-Cuevas JC, Hernández-Calderón E, Velásquez-Becerra C, Farías-Rodríguez R, Macías-Rodríguez LI, et al. Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an auxin- and ethylene-independent signaling mechanism in Arabidopsis thaliana. Mol. Plant-Microbe In. 2007; 20:207–217. [DOI] [PubMed] [Google Scholar]
- 42.Alcaraz ML, Thorp TG, and Hormaza JI. Phenological growth stages of avocado (Persea americana) according to the BBCH scale. Scientia Horticulturae. 2013; 164: 434–439. [Google Scholar]
- 43.Adesemoye AO, Obini M, Ugoji EO. Comparison of plant growth-promotion with Pseudomonas aeruginosa and Bacillus subtilis in three vegetables. Braz. J. Microbiol. 2008; 39:423–426. 10.1590/S1517-83822008000300003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Kim MS, Krishnamachari V, Payton P, Sun Y, Grimson M, et al. Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226(2007) 839–851. 10.1007/s00425-007-0530-2 [DOI] [PubMed] [Google Scholar]
- 45.Miranda M, Ralph SG, Mellway R, White R, Heath MC, Bohlmann J, et al. The transcriptional response of hybrid poplar (Populus trichocarpa x P. deltoides) to infection by Melampsora medusae leaf rust involves induction of flavonoid pathway genes leading to the accumulation of proanthocyanidins. Mol. Plant-Microbe In. 2007; 20:816–831. [DOI] [PubMed] [Google Scholar]
- 46.Hren M, Nikolić P, Rotter A, Blejec A, Terrier N, Ravnikar M, et al. ‘Bois noir’ phytoplasma induces significant reprogramming of the leaf transcriptome in the field grown grapevine. BMC Genomics. 2009; 10:460 10.1186/1471-2164-10-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Himeno M, Kitazawa Y, Yoshida T, Maejima K, Yamaji Y, Oshima K, et al. Purple top symptoms are associated with reduction of leaf cell death in phytoplasma-infected plants. Scientific Reports. 2014; 4:4111 10.1038/srep04111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dovana F, Mucciarelli M, Mascarello M, Fusconi A. In vitro morphogenesis of Arabidopsis to search for novel endophytic fungi modulating plant growth. PLOS ONE. 2015; 10:e0143353 10.1371/journal.pone.0143353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi CL, Park HB, Lee JS, Ryu S, Ryu CM. Inhibition of primary roots and stimulation of lateral root development in Arabidopsis thaliana by the rhizobacterium Serratia marcescens 90–166 is through both auxin-dependent and -independent signaling pathways. Mol. Cells. 2010; 29:251–258. 10.1007/s10059-010-0032-0 [DOI] [PubMed] [Google Scholar]
- 50.Kwon YS, Lee DY, Rakwal R, Baek SB, Lee JH, Kwak YS, et al. Proteomic analyses of the interaction between the plant-growth promoting rhizobacterium Paenibacillus polymyxa E681 and Arabidopsis thaliana. Proteomics. 2016; 16:122–135. 10.1002/pmic.201500196 [DOI] [PubMed] [Google Scholar]
- 51.Vida C, Bonilla N, de Vicente A, Cazorla FM. Microbial profiling of a suppressiveness-induced agricultural soil amended with composted almond shells. Front. Microbiol. 2016; 7:4 10.3389/fmicb.2016.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lambais MR, Lucheta AR, Crowley DE. Bacterial community assemblages associated with the phyllosphere, dermosphere, and rhizosphere of tree species of the Atlantic forest are host taxon dependent. Microb. Ecol. 2014; 68:567–574. 10.1007/s00248-014-0433-2 [DOI] [PubMed] [Google Scholar]
- 53.Janarthine SRS, Eganathan P. Plant growth promoting of endophytic Sporosarcina aquimarina SjAM16103 isolated from the pneumatophores of Avicennia marina L. Int. J. Microbiol. 2012; 2012:532060 10.1155/2012/532060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vejan P, Abdullah R, Khadiran T, Ismail S, Nasrulhaq Boyce A. Role of plant growth promoting rhizobacteria in agricultural sustainability-A review. Molecules. 2016; 21: 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naureen Z, Ur Rehman N, Hussain H, Hussain J, Gilani SA, Al Housni SK, et al. Exploring the potentials of Lysinibacillus sphaericus ZA9 for plant growth promotion and biocontrol activities against phytopathogenic fungi. Front. Microbiol. 2017; 8:1477 10.3389/fmicb.2017.01477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rahmoune B, Morsli A, Khelifi-Slaoui M, Khelifi L, Strueh A, Erban A,et al. Isolation and characterization of three new PGPR and their effects on the growth of Arabidopsis and Datura plants. J. Plant Interact. 2017; 12:1–6. [Google Scholar]
- 57.Ullah A, Mushtaq H, Fahad S, Shah A, Chaudhary HJ. Plant growth promoting potential of bacterial endophytes in novel association with Olea ferruginea and Withania coagulans. Microbiology. 2017; 86:119–127. [Google Scholar]
- 58.Gouda S, Kerry RG, Das G, Paramithiotis S, Shin H, Patra JK. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 2018; 206:131–140. 10.1016/j.micres.2017.08.016 [DOI] [PubMed] [Google Scholar]
- 59.Xu J, Kloepper JW, Huang P, McInroy JA, Hu CH. Isolation and characterization of N2 -fixing bacteria from giant reed and switchgrass for plant growth promotion and nutrient uptake. J. Basic Microb. 2018; 58:459–471. [DOI] [PubMed] [Google Scholar]
- 60.Etesami H, Maheshwari DK. Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotox. Environ. Safe. 2018; 156:225–246. [DOI] [PubMed] [Google Scholar]
- 61.Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007; 31:425–448. 10.1111/j.1574-6976.2007.00072.x [DOI] [PubMed] [Google Scholar]
- 62.Remans R, Beebe S, Blair M, Manrique G, Tovar E, Rao I, et al. Physiological and genetic analysis of root responsiveness to auxin-producing plant growth-promoting bacteria in common bean (Phaseolus vulgaris L.). Plant Soil. 2008; 302:149–161. [Google Scholar]
- 63.Martínez-Viveros O, Jorquera M, Crowley D, Gajardo G, Mora M. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci. Plant Nut. 2010. 10:293–319. [Google Scholar]
- 64.Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Pare PW, et al. Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2003; 100:4927–4932. 10.1073/pnas.0730845100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yi YK, Park HW, Shrestha S, Seo J, Kim YO, Shin CS, et al. Identification of two entomopathogenic bacteria from a nematode pathogenic to the Oriental beetle, Blitopertha orientalis. J. Microbiol Biotechn. 2007; 17:968–978. [PubMed] [Google Scholar]
- 66.Song GC, Choi HK, Kim YS, Choi J, Ryu C-M. Seed defense biopriming with bacterial cyclodipeptides triggers immunity in cucumber and pepper. Scientific Reports. 2017; 7:14209 10.1038/s41598-017-14155-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The percentage of the replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 runs) is shown next to the branches. The tree is drawn to scale. G2 is indicated with red lines, while G3A and G3C are indicated with gray lines.
(EPS)
Bacterial isolates were cultured on Pikosvskaya-bromophenol (PKV-BMP) medium with a 0.025 OD600 at 30°C, 180 rpm for 48 h. The data represent the means ± SD (n = 3). The experiment was replicated two times with similar results.
(TIFF)
Thrips were exposed to the strains for 96 h in a bioassay chamber. Bean pod slices impregnated with two bacterial concentrations were used as food. Data for the commercial control and the control with no inoculation are presented in red and black, respectively. The median is shown as a bar inside the boxes and the whiskers indicate to the range of the data.
(EPS)
(A) Compound with RT = 3.944 and m/z = 245.1295 obtained from the methanolic extract of Stenotrophomonas sp. S1.2–11. The fragmentation pattern corresponds to cyclo(L-Phe-L-Pro). (B) Compound with RT = 2.40 and m/z = 261.1243 obtained from the methanolic extract of Stenotrophomonas sp. S1.2–11. The fragmentation pattern corresponds to cyclo(L-Pro-L-Tyr). (C) Compound with RT = 1.343 and m/z = 251.1515 obtained from the methanolic extract of Pseudomonas sp. Rf1.1-9A. The fragmentation pattern corresponds to cyclo(L-His-L-Leu).
(EPS)
(DOCX)
Data Availability Statement
16S rDNA sequences obtained from the selected isolates for metabolic profile were deposited in GenBank (Accession Numbers: MN098850-MN098867).