Abstract
Purpose
To provide a quantitative clinical-regulatory insight into the status of FDA orphan drug designations for compounds intended to treat lysosomal storage disorders (LSDs).
Methods
Assessment of the drug pipeline through analysis of the FDA database for orphan drug designations with descriptive and comparative statistics.
Results
Between 1983 and 2019, 124 orphan drug designations were granted by the FDA for compounds intended to treat 28 lysosomal storage diseases. Orphan drug designations focused on Gaucher disease (N = 16), Pompe disease (N = 16), Fabry disease (N = 10), MPS II (N = 10), MPS I (N = 9), and MPS IIIA (N = 9), and included enzyme replacement therapies, gene therapies, and small molecules, and others. Twenty-three orphan drugs were approved for the treatment of 11 LSDs. Gaucher disease (N = 6), cystinosis (N = 5), Pompe disease (N = 3), and Fabry disease (N = 2) had multiple approvals, CLN2, LAL-D, MPS I, II, IVA, VI, and VII one approval each. This is an increase of nine more approved drugs and four more treatable LSDs (CLN2, MPS VII, LAL-D, and MPS IVA) since 2013. Mean time between orphan drug designation and FDA approval was 89.7 SD 55.00 (range 8–203, N = 23) months.
Conclusions
The drug development pipeline for LSDs is growing and evolving, with increased focus on diverse small-molecule targets and gene therapy. CLN2 was the first and only LSD with an approved therapy directly targeted to the brain. Newly approved products included “me-too”–enzymes and innovative compounds such as the first pharmacological chaperone for the treatment of Fabry disease.
Introduction
Lysosomal storage disorders (LSDs) are a group of more than 50 inherited, multisystemic, progressive conditions caused by a genetic defect that results in the progressive accumulation of complex non-metabolized substrates in the lysosomes of cells, tissues and organs, inducing distinct but heterogeneous somatic and neurological disease phenotypes [1–7]. In general, lysosomal storage disorders lead to significant morbidity and decreased life expectancy. Reported prevalences of LSDs in industrialized countries range between 7.6 per 100,000 live births (= 1 in 13,158) and 25 per 100,000 live births (= 1 in 4000) [8–11]. Some LSDs are treatable and the drug development in the field has traditionally been very active and dynamic after the successful development of enzyme replacement therapy in Gaucher disease which seeded further innovation [1, 12, 13]. The development of new compounds and new concepts of treatment for lysosomal storage disorders has been very dynamic. Therefore, the purpose of the present paper is to precisely analyze the most recent advances and novel trends in orphan drug development for lysosomal storage diseases as documented in the FDA Orphan Drug Product designation database.
Methods
The FDA orphan drug database was accessed over the internet at the following address http://www.accessdata.fda.gov/scripts/opdlisting/oopd/. Search criteria were “all designations” from 1 January 1983 until 10 May 2019, i.e., all data entries until 10 May 2019 were taken into account (N = 4979). The output format was an excel file which was downloaded on a local computer. Orphan designations for lysosomal storage diseases were extracted with pertinent keywords (N = 124) [1]. STROBE criteria (S1 Checklist) were respected [14].
Definitions
Pharmacological compounds were categorized based on their chemical structures into the following classes, listed in alphabetical order: “enzyme”, “enzyme/small molecule combination”, “gene therapy”, “polymer”, “protein (other than enzyme)”, and “small molecule” [1]. A small molecule was defined as a compound with a molecular weight below 900 Da [15]. In addition, compounds were further grouped into functionally meaningful subtypes based on their biochemical properties, molecular mechanisms of action, or gene therapy platforms, i.e., (in alphabetical order): “AAV vector”, “adjunctive therapy”, “anaplerotic”, “anti-inflammatory/neuroprotective”, “anti-inflammatory/pro-chondrogenic”, “anti-inflammatory/TPP1 enhancing”, “anti-inflammatory/TPP1 enhancing/vitamin combination”, “pharmacological chaperone”, “cytochrome P450 rescue”, “enzyme replacement therapy”, “enzyme replacement therapy–pharmacological chaperone co-administration”, “lentiviral vector”, “membrane stabilization”, “lysosomal cholesterol redistributor”, “replacement therapy with a modified enzyme”, “nonviral vector directing transgene integration”, “receptor amplification”, “retroviral vector”, “small molecule facilitating intracellular substrate transport”, “stem cells”, “stop-codon read-through”, and “substrate reduction”.
Time to FDA approval was defined as the time period from orphan drug designation until approval by the FDA [1]. Drug approval rates were defined as the proportion of orphan drug designations approved out of overall orphan drug designations granted. Missing data were not imputed.
Statistical analysis
Standard techniques of descriptive statistics were applied: continuous variables were summarized with mean, standard deviation, median, minimum and maximum values. Categorical variables were summarized with frequencies and percentages.
Comparative statistics were performed with the appropriate parametric test for data with Gaussian distribution. Differences in mean times to approval (defined as time between orphan drug designation and FDA approval) between drug compound subtypes were analyzed with ANOVA. Differences between frequency counts for approval rates of lysosomal orphan drugs versus approval rates for non-lysosomal orphan drugs were compared with the chi-square test. A two-sided p-value < 0.05 was considered statistically significant.
The following groups were analyzed:
Orphan drug designations and approvals by the FDA (overall and for compounds intended to treat lysosomal storage diseases) by year
- Orphan drug designations by the FDA for compounds intended to treat lysosomal storage diseases by year and
- by disease
- by pharmacological technology platform (as specified in the definitions section)
Withdrawn FDA orphan drug designations for compounds intended to treat lysosomal storage diseases
- FDA approved therapies for lysosomal storage disorders
- by disease with time periods from orphan drug designation until approval and market exclusivity
- by year by pharmacological technology platform (as specified in the definitions section)
All statistical analyses were performed using SAS Enterprise guide 7.13 HF4, SAS Institute Inc., Cary, NC, USA. Graphs were generated with R [16] and GraphPad Prism 5.04, GraphPad Software, Inc., San Diego, CA, USA.
Results
The drug development pipeline: Orphan drug designations granted by the FDA
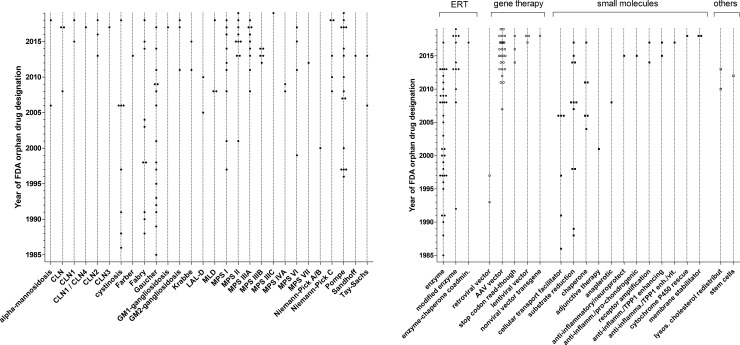
Between 1 January 1983 and 10 May 2019, 124 orphan drug designations were granted by the FDA for compounds intended to treat 28 lysosomal storage diseases (Fig 1A). For comparison, in the same time period, the FDA granted 4979 orphan drug designations overall, out of which 783 were approved (Fig 1B). Twenty lysosomal conditions had multiple orphan drug designations. Most orphan drug designations were granted for Gaucher disease (N = 16), Pompe disease (N = 16), Fabry disease (N = 10), MPS II (N = 10), MPS I (N = 9), and MPS IIIA (N = 9), followed by 14 other diseases depicted in Fig 2A. Eight lysosomal conditions had one orphan drug designation each. Enzyme replacement therapies, gene therapies, small molecules, and other technology platform classes were designated as orphan drugs intended to treat lysosomal storage diseases (Fig 2B). Nine granted orphan drug designations were subsequently withdrawn (Table 1). The reason for withdrawal is not specified in the FDA orphan drug database. The approval rates of lysosomal orphan drugs (18.5%) did not differ from approval rates for non-lysosomal orphan drugs (15.7%, p = 0.38, chi-square)
Fig 1.
A: Number of orphan drug designations (open bars) and FDA approvals (full bars) for compounds intended to treat lysosomal storage diseases by year. * indicates close of database: 10 May 2019. B: Overall number of orphan drug designations (open bars) and FDA approvals (full bars) by year. * indicates close of database: 10 May 2019.
Fig 2.
A: Orphan drug designations granted by the FDA for compounds intended to treat lysosomal storage disorders by year and specific disease. B: Orphan drug designations granted by the FDA for compounds intended to treat lysosomal storage disorders by year and pharmacological technology platform.
Table 1. Withdrawn orphan drug designations.
Reasons for and time of withdrawal were not specified in the FDA database.
| Compound | Pharmacological subtype | Year of orphan drug designation | Indication under development |
|---|---|---|---|
| Ataluren | Stop-codon read-through | 2014 | Treatment of mucopolysaccharidosis type I |
| Recombinant human alpha-N-acetylglucosaminidase | Enzyme | 2013 | Treatment of mucopolysaccharidosis IIIB (Sanfilippo B syndrome) |
| Recombinant human arylsulphatase A | Enzyme | 2008 | Treatment of metachromatic leukodystrophy (MLD) |
| Miglustat | Substrate reduction | 2008 | Treatment of the neurological manifestations of Niemann-Pick disease, type C |
| Duvoglustat hydrochloride | Substrate reduction | 2007 | Treatment of Pompe disease |
| Isofagomine tartrate | Chaperone | 2006 | Treatment of Gaucher disease |
| Retroviral vector, R-GC and GC gene 1750 | Retroviral vector | 1997 | Treatment of Gaucher disease |
| Human acid precursor alpha-glucosidase, recombinant | Enzyme | 1996 | Treatment of glycogen storage disease type II |
| Phosphocysteamine | Substrate reduction | 1988 | Treatment of cystinosis. |
Lysosomal storage disorders with FDA approved therapies
Twenty-three orphan drugs were approved for the treatment of 11 lysosomal storage diseases. Four diseases had multiple therapeutics approved, i.e. Gaucher disease (N = 6), cystinosis (N = 5), Pompe disease (N = 3), and Fabry disease (N = 2), (Fig 3A). The remaining seven diseases had one compound each approved by the FDA (i.e., CLN2, LAL-D, MPS I, II, IVA, VI, VII). CLN2 was the only neuronopathic lysosomal storage disease with an FDA approved therapy directly targeting the brain; all the other therapies address systemic non-neurological manifestations. FDA approved therapies included enzyme replacement therapies (N = 15) and small molecules (N = 8), but no other class of drugs (Fig 3B, Table 2). Approved treatments for cystinosis included three different formulations and three different age groups. Enzyme replacement therapies with alglucosidase alfa for Pompe disease were manufactured in two different bioreactor systems and approved for pediatric and adult age groups.
Fig 3. FDA approved compounds for the treatment of lysosomal storage disorders (depicted as compound #disease), development times and market exclusivity.
A: Grey bars indicate drug development times, i.e. time from orphan drug designation to orphan drug approval by the FDA. Black bars indicate, if applicable, market exclusivity periods. (1)–systemic administration, immediate release (IR). (2)—ophthalmic solution (OS). (3)–systemic administration, delayed release (DR), adults. (4)–systemic administration, delayed release (DR), age 2 to 6 years. (5)—systemic administration, delayed release (DR), age 1 to less than 2 years. (6)—bioreactor 160 L. (7)—bioreactor 4000 L, 8 years and older.(8)—bioreactor 4000 L, all ages. B: FDA approved therapies for the treatment of lysosomal storage disorders by year of approval and pharmacological technology platform.
Table 2. Mechanism of action of FDA approved small molecules (*) and small molecules in development, intended to treat a lysosomal storage disorder.
| Mechanism of action | Compound | Disease with FDA orphan drug designation |
|---|---|---|
| Targeting the affected gene: Stop-codon read through in missense mutations [36] | ||
| 6'-(R)-methyl-5-O-(5-amino-5,6-dideoxy-alpha-L-talofuranosyl)-paromamine sulfate | MPS I Cystinosis | |
| Targeting the affected enzyme: TPP1 enhancer, chaperone, enzyme/chaperone co-administration [37–42] | ||
| Gemfibrozil | CLN | |
| N-t-butylhydroxylamine | CLN1 | |
| Modified cholera toxin¶ | Gaucher disease | |
| Pyrimethamine | GM2-gangiosidosis (Tay-Sachs and Sandhoff disease) | |
| Ambroxol | Gaucher disease | |
| N-acetyl-glucosamine thiazoline | Adult Tay-Sachs disease | |
| Migalastat hydrochloride* | Fabry disease | |
| Recombinant human acid a-glucosidase/miglustat | Pompe disease | |
| Targeting storage: Substrate reduction and subcellular storage redistribution [43–45] | ||
| Odiparcil | MPS VI | |
| Lucerastat | Fabry disease | |
| Venglustat | Fabry disease | |
| (3S)-1-azabicyclo[2.2.2]oct-3-yl {2-[2-(4-fluorophenyl)-1,3-thiazol-4-yl]propan-2-yl}carbamate | Gaucher disease | |
| 2-hydroxypropyl-B-cyclodextrin§ | Niemann-Pick disease type C | |
| Hydroxy-Propyl-Beta-Cyclodextrin§ | Niemann-Pick disease type C | |
| Miglustat* | Gaucher disease* | |
| Eliglustat* | Gaucher disease type I* | |
| Cysteamine* | Cystinosis* NCL (Batten disease) | |
| 1,5-(Butylimino)-1,5 dideoxy,D-glucitol | Fabry disease | |
| L-cycloserine | Gaucher disease | |
| Targeting cellular uptake of therapeutic enzymes: Receptor amplification [46] | ||
| Clenbuterol | Pompe disease | |
| Mitigation of cellular damage (Antiinflammatory, pro chondrogenic, neuroprotective cytochrome P450 rescue) or anaplerotic [20, 37, 47–49] | ||
| Ursodeoxycholic acid | Niemann-Pick C | |
| Gemfibrozil and vitamin A | CLN | |
| Ibudilast | Krabbe disease | |
| Pentosan polysulfate sodium | MPS VI | |
| Triheptanoin | Pompe disease |
¶protein acting as a chaperone
§polymer
Regulatory drug development timelines
Overall mean time to approval, defined as time between orphan drug designation and FDA approval was 89.7 SD 55.00 (range 8–203, N = 23) months. Stratified by drug compound subtypes, mean time to approval for enzyme replacement therapies was 81.2 SD 56.42 (range 8–203, N = 15) months, mean time to approval for small molecules facilitating subcellular transport was 107.8 SD 52,96 (range 40–181, N = 5) months, and mean time to approval for substrate reduction therapies was 66.5 SD 6.36 (range 62 to 71, N = 2) months. Time to approval for the pharmacological chaperone therapy was 173 months. Differences between the above groups were not statistically significant (p = 0.33, ANOVA). The drug development timelines and market exclusivity periods, an incentive granted by the FDA to stimulate orphan drug development [13], are illustrated in Fig 3A.
Discussion
By 10 May 2019, 23 orphan drugs were approved by the FDA for the treatment of 11 lysosomal storage disorders. This is an increase of nine more approved orphan drugs and four more treatable lysosomal disease (i.e. CLN2, MPS VII, LAL-D, and MPS IVA) compared to 2013 [1].
While alglucerase for Gaucher disease was the first orphan drug approved for a lysosomal storage disease in 1991, intrathecally administered cerliponase alfa for CLN2, FDA approved in 2017, is the first orphan drug for a lysosomal storage disorder to directly treat the brain, which is a significant therapeutic innovation [17, 18]. Since 2013, 54 more orphan drug designations were granted. In addition, diseases such as CLN1, CLN3, CLN4, Farber disease, and GM1-gangliosidosis did not have orphan drug designations in 2013. This indicates that drug development in lysosomal storage disorders is now being driven into mainly neuronopathic conditions (Fig 2A). The overall growth curve of orphan drug designations appears to accelerate over time and may become exponential (Fig 1A), possibly following a global trend (Fig 1B). Interestingly, the drug approval rate in lysosomal orphan drug development and non-lysosomal orphan drug development did not differ. Technology is evolving: while enzyme replacement therapies had initially set the trend, more modified enzymes, including fusion proteins, and an enzyme-chaperone co-administration entered the development pipeline. This may be a reaction to the increasing recognition in the field that, in general, systemically administered enzyme replacement therapy with conventional enzymes can easily access organs such as liver and spleen but have little impact on bone and CNS manifestations. Four small molecules have been approved by the FDA for the treatment of a lysosomal storage disease. Their mechanisms of action target the facilitation of subcellular transport (e.g., cysteamine for cystinosis, approved in 1994) and the reduction of storage (miglustat, approved in 2003, and eligustat, approved in 2014, both for the treatment of Gaucher disease) [1]. In 2018, migalastat, which stabilizes the misfolded enzyme alpha-galactosidase A, was approved as a first-of-its kind pharmacological chaperone by the FDA for the treatment of Fabry disease [19], (Table 2). Mechanisms of action for small molecules, either approved or in drug development, encompassed the broad spectrum of underlying pathophysiology and aimed at 1) targeting the affected gene, 2) targeting the affected enzyme, 3) targeting storage, 4) targeting cellular uptake of therapeutic enzymes, and 4) mitigating the cellular damage or anaplerotic (Table 2). It is possible and likely that not all mechanistically meaningful approaches lead to clinical benefit in patients [20]. The plethora of innovative ideas for pharmacological approaches is laudable but should not lead a treating physician to engage in off-label use but rather encourage international collaboration aimed to generate the highest standard of evidence-based knowledge by respecting excellence clinical research [21].
Gene therapy now plays a larger role in the drug development pipeline compared to the situation in our last analysis [22]. This may again be a reaction to the increased recognition of enzyme replacement therapies’ substantial limitations, as described in detail above. The technical approach towards gene therapy is evolving as illustrated in Fig 2B. Gene therapies rely, at least in principle, on the assumption that a single treatment may result in a sustained, potentially curative clinical benefit for the patients. The first molecular tools enabling efficient non-toxic gene transfer into human somatic cells were recombinant replication-deficient vectors [23]. Among those, retroviral and adeno-associated viral (AAV) vectors have been the most widely used in particular for ex vivo T cell engineering or genetically engineered hematopoietic stem cells (HSCs) for the treatment of primarily hematologic or oncologic conditions such as pediatric ALL, β-thalassemia or adenosine deaminase deficiency [24–26]. In contrast, while the first two orphan drug designations for gene therapy for lysosomal storage diseases in 1993 and 1997 (both for Gaucher disease) relied on retroviral vectors, this platform was subsequently abandoned. This is likely due to the emergence of serious toxicities related to high gene transfer including insertional genotoxicity, immune destruction of genetically modified cells, and immune reactions related to the application of certain vectors [27, 28]. The next step in gene technology was the introduction of AAV (designated for Pompe disease in 2007), followed by stop-codon read-through (designated in 2014 and 2016 for MPS I, and in 2018 for cystinosis). Moreover, lentiviral vector (designated in 2018 for Fabry disease and MLD), and nonviral vector directing transgene integration (designated in 2018 for MPS I) technologies are being considered, all of which have to prove their safety and efficacy in the future. More sophisticated genome editing technologies that enable a variety of therapeutic genome modifications (gene addition, gene ablation or “gene correction”) consist of the administration of transcription activator-like effector nucleases (TALENs) and or CRISPR-Cas 9 system to efficiently cleave and modify DNA at sites of interest [29–33]. Those approaches are currently limited to applications in basic research, but transfer into clinical trials can be expected in the near future [34, 35]. Until close of database no gene therapy was approved for the treatment of lysosomal storage disorders (Fig 3B). If proven successful in registration trials—which would in all likelihood be small clinical trials of a limited duration—it is of particular interest, how long the therapeutic effect of gene therapy can be sustained during a patients’ lifetime, and, if this time is limited, whether it would be safe and feasible to repeat the administration of a gene therapy multiple times in a single patient. It is anticipated that novel therapies will be costly. Important topics for future investigations will include patient selection, starting and stopping criteria.
For the correct contextual interpretation it is important to be aware of important limitations of the present work as pointed out previously [1]. Orphan drug designation was considered as the expressed intent to develop a drug. This may be influenced by strategic and patent related considerations. Not all manufacturers may choose such a publicly visible pathway at an early stage. Therefore, time to approval as presented here may be biased by the intellectual property strategy of the respective drug development program. Orphan drug development outputs in jurisdictions other than the FDA were, as in our previous analysis, not taken into account because this analysis was by definition focused on the impact of the US orphan drug act [1, 13]. As drug development in lysosomal storage disorders is, in general, a global enterprise we consider the present findings generalizable within the context of their limitations.
Conclusions
Activities in orphan drug development for lysosomal storage disorders are steadily increasing, which follows a global trend in orphan drug development overall. Newly approved products included “me-too”–enzymes, and also innovative compounds such as the first ERT targeting the brain in CLN2 and the first-of-its-kind pharmacological chaperone for the treatment of Fabry disease. The drug development pipeline for LSDs is growing and evolving, with increased focus on diverse small-molecule targets and gene therapy.
Supporting information
(DOC)
Acknowledgments
We thank Lorna Stimson, PhD, for language editing.
Data Availability
All data are in the paper and supporting information.
Funding Statement
SphinCS GmbH provided support in the form of salary for EM, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of this author are articulated in the ‘author contributions’ section.
References
- 1.Mechler K, Mountford WK, Hoffmann GF, Ries M. Pressure for drug development in lysosomal storage disorders—a quantitative analysis thirty years beyond the US orphan drug act. Orphanet journal of rare diseases. 2015;10:46 10.1186/s13023-015-0262-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zielonka M, Garbade SF, Kölker S, Hoffmann GF, Ries M. Quantitative clinical characteristics of 53 patients with MPS VII: a cross-sectional analysis. Genetics in medicine: official journal of the American College of Medical Genetics. 2017;19(9):983–8. 10.1038/gim.2017.10 . [DOI] [PubMed] [Google Scholar]
- 3.Zielonka M, Garbade SF, Kölker S, Hoffmann GF, Ries M. A cross-sectional quantitative analysis of the natural history of Farber disease: an ultra-orphan condition with rheumatologic and neurological cardinal disease features. Genetics in medicine: official journal of the American College of Medical Genetics. 2018;20(5):524–30. 10.1038/gim.2017.133 . [DOI] [PubMed] [Google Scholar]
- 4.Zielonka M, Garbade SF, Kölker S, Hoffmann GF, Ries M. A cross-sectional quantitative analysis of the natural history of free sialic acid storage disease-an ultra-orphan multisystemic lysosomal storage disorder. Genetics in medicine: official journal of the American College of Medical Genetics. 2018. 10.1038/s41436-018-0051-3 . [DOI] [PubMed] [Google Scholar]
- 5.Sláma T, Garbade SF, Kölker S, Hoffmann GF, Ries M. Quantitative natural history characterization in a cohort of 142 published cases of patients with galactosialidosis-A cross-sectional study. Journal of inherited metabolic disease. 2019;42(2):295–302. 10.1002/jimd.12010 . [DOI] [PubMed] [Google Scholar]
- 6.Komatsuzaki S, Zielonka M, Mountford WK, Kölker S, Hoffmann GF, Garbade SF, et al. Clinical characteristics of 248 patients with Krabbe disease: quantitative natural history modeling based on published cases. Genetics in medicine: official journal of the American College of Medical Genetics. 2019. 10.1038/s41436-019-0480-7 . [DOI] [PubMed] [Google Scholar]
- 7.Zielonka M, Garbade SF, Kölker S, Hoffmann GF, Ries M. Ultra-orphan lysosomal storage diseases: A cross-sectional quantitative analysis of the natural history of alpha-mannosidosis. Journal of inherited metabolic disease. 2019. 10.1002/jimd.12138 . [DOI] [PubMed] [Google Scholar]
- 8.Meikle PJ, Hopwood JJ, Clague AE, Carey WF. Prevalence of lysosomal storage disorders. Jama. 1999;281(3):249–54. 10.1001/jama.281.3.249 . [DOI] [PubMed] [Google Scholar]
- 9.Poorthuis BJ, Wevers RA, Kleijer WJ, Groener JE, de Jong JG, van Weely S, et al. The frequency of lysosomal storage diseases in The Netherlands. Human genetics. 1999;105(1–2):151–6. 10.1007/s004399900075 . [DOI] [PubMed] [Google Scholar]
- 10.Applegarth DA, Toone JR, Lowry RB. Incidence of inborn errors of metabolism in British Columbia, 1969–1996. Pediatrics. 2000;105(1):e10 10.1542/peds.105.1.e10 . [DOI] [PubMed] [Google Scholar]
- 11.Pinto R, Caseiro C, Lemos M, Lopes L, Fontes A, Ribeiro H, et al. Prevalence of lysosomal storage diseases in Portugal. European journal of human genetics: EJHG. 2004;12(2):87–92. 10.1038/sj.ejhg.5201044 . [DOI] [PubMed] [Google Scholar]
- 12.Ries M. Enzyme replacement therapy and beyond-in memoriam Roscoe O. Brady, M.D. (1923–2016). Journal of inherited metabolic disease. 2017;40(3):343–56. 10.1007/s10545-017-0032-8 . [DOI] [PubMed] [Google Scholar]
- 13.FDA. Public Law 97–414, 97th Congress, An Act To amend the Federal Food, Drig, and Cosmetic Act to facilitate the development of drugs for rare diseases and conditions, and for other purposes 1983 [accessed 29 May 2019]. Available from: https://www.fda.gov/media/99546/download.
- 14.Vandenbroucke JP, von Elm E, Altman DG, Gotzsche PC, Mulrow CD, Pocock SJ, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. PLoS Med. 2007;4(10):e297 10.1371/journal.pmed.0040297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macielag MJ. Chemical properties of antibacterials and their uniqueness. In: Dougherty TJ, Pucci MJ, editors. Antibiotic Discovery and Development. New York Springer Science+Business Media, LLC; 2012. p. 793–820. [Google Scholar]
- 16.R core team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria 2019. [accessed 29 May 2019]. Available from: https://www.R-project.org/ [Google Scholar]
- 17.Barton NW, Brady RO, Dambrosia JM, Di Bisceglie AM, Doppelt SH, Hill SC, et al. Replacement therapy for inherited enzyme deficiency—macrophage-targeted glucocerebrosidase for Gaucher's disease. The New England journal of medicine. 1991;324(21):1464–70. 10.1056/NEJM199105233242104 . [DOI] [PubMed] [Google Scholar]
- 18.Schulz A, Ajayi T, Specchio N, de Los Reyes E, Gissen P, Ballon D, et al. Study of Intraventricular Cerliponase Alfa for CLN2 Disease. The New England journal of medicine. 2018;378(20):1898–907. 10.1056/NEJMoa1712649 . [DOI] [PubMed] [Google Scholar]
- 19.Germain DP, Hughes DA, Nicholls K, Bichet DG, Giugliani R, Wilcox WR, et al. Treatment of Fabry's Disease with the Pharmacologic Chaperone Migalastat. The New England journal of medicine. 2016;375(6):545–55. 10.1056/NEJMoa1510198 . [DOI] [PubMed] [Google Scholar]
- 20.Schiffmann R, Wallace ME, Rinaldi D, Ledoux I, Luton MP, Coleman S, et al. A double-blind, placebo-controlled trial of triheptanoin in adult polyglucosan body disease and open-label, long-term outcome. Journal of inherited metabolic disease. 2018;41(5):877–83. 10.1007/s10545-017-0103-x . [DOI] [PubMed] [Google Scholar]
- 21.Lampert A, Hoffmann GF, Ries M. Ten Years after the International Committee of Medical Journal Editors' Clinical Trial Registration Initiative, One Quarter of Phase 3 Pediatric Epilepsy Clinical Trials Still Remain Unpublished: A Cross Sectional Analysis. PloS one. 2016;11(1):e0144973 10.1371/journal.pone.0144973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagree MS, Scalia S, McKillop WM, Medin JA. An update on gene therapy for lysosomal storage disorders. Expert opinion on biological therapy. 2019;19(7):655–70. 10.1080/14712598.2019.1607837 . [DOI] [PubMed] [Google Scholar]
- 23.Kotterman MA, Chalberg TW, Schaffer DV. Viral Vectors for Gene Therapy: Translational and Clinical Outlook. Annu Rev Biomed Eng. 2015;17:63–89. 10.1146/annurev-bioeng-071813-104938 . [DOI] [PubMed] [Google Scholar]
- 24.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. The New England journal of medicine. 2014;371(16):1507–17. 10.1056/NEJMoa1407222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467(7313):318–22. 10.1038/nature09328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. The New England journal of medicine. 2009;360(5):447–58. 10.1056/NEJMoa0805817 . [DOI] [PubMed] [Google Scholar]
- 27.Jenks S. Gene therapy death—"everyone has to share in the guilt". J Natl Cancer Inst. 2000;92(2):98–100. 10.1093/jnci/92.2.98 . [DOI] [PubMed] [Google Scholar]
- 28.Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Molecular therapy: the journal of the American Society of Gene Therapy. 2006;13(6):1031–49. 10.1016/j.ymthe.2006.03.001 . [DOI] [PubMed] [Google Scholar]
- 29.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, et al. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39(1):359–72. 10.1093/nar/gkq704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Port F, Bullock SL. Creating Heritable Mutations in Drosophila with CRISPR-Cas9. Methods Mol Biol. 2016;1478:145–60. 10.1007/978-1-4939-6371-3_7 . [DOI] [PubMed] [Google Scholar]
- 31.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30(5):460–5. 10.1038/nbt.2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096 10.1126/science.1258096 . [DOI] [PubMed] [Google Scholar]
- 34.Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14(1):49–55. 10.1038/nrm3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168(1–2):20–36. 10.1016/j.cell.2016.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leubitz A, Frydman-Marom A, Sharpe N, van Duzer J, Campbell KCM, Vanhoutte F. Safety, Tolerability, and Pharmacokinetics of Single Ascending Doses of ELX-02, a Potential Treatment for Genetic Disorders Caused by Nonsense Mutations, in Healthy Volunteers. Clinical pharmacology in drug development. 2019. 10.1002/cpdd.647 . [DOI] [PubMed] [Google Scholar]
- 37.Kim K, Kleinman HK, Lee HJ, Pahan K. Safety and potential efficacy of gemfibrozil as a supportive treatment for children with late infantile neuronal ceroid lipofuscinosis and other lipid storage disorders. Orphanet journal of rare diseases. 2017;12(1):113 10.1186/s13023-017-0663-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adnan H, Zhang Z, Park HJ, Tailor C, Che C, Kamani M, et al. Endoplasmic Reticulum-Targeted Subunit Toxins Provide a New Approach to Rescue Misfolded Mutant Proteins and Revert Cell Models of Genetic Diseases. PloS one. 2016;11(12):e0166948 10.1371/journal.pone.0166948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parenti G, Fecarotta S, la Marca G, Rossi B, Ascione S, Donati MA, et al. A chaperone enhances blood alpha-glucosidase activity in Pompe disease patients treated with enzyme replacement therapy. Molecular therapy: the journal of the American Society of Gene Therapy. 2014;22(11):2004–12. 10.1038/mt.2014.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enshaei H, Molina BG, Del Valle LJ, Estrany F, Arnan C, Puiggali J, et al. Scaffolds for Sustained Release of Ambroxol Hydrochloride, a Pharmacological Chaperone That Increases the Activity of Misfolded beta-Glucocerebrosidase. Macromolecular bioscience. 2019:e1900130 10.1002/mabi.201900130 . [DOI] [PubMed] [Google Scholar]
- 41.Osher E, Fattal-Valevski A, Sagie L, Urshanski N, Sagiv N, Peleg L, et al. Effect of cyclic, low dose pyrimethamine treatment in patients with Late Onset Tay Sachs: an open label, extended pilot study. Orphanet journal of rare diseases. 2015;10:45 10.1186/s13023-015-0260-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valenzano KJ, Khanna R, Powe AC, Boyd R, Lee G, Flanagan JJ, et al. Identification and characterization of pharmacological chaperones to correct enzyme deficiencies in lysosomal storage disorders. Assay and drug development technologies. 2011;9(3):213–35. 10.1089/adt.2011.0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guerard N, Oder D, Nordbeck P, Zwingelstein C, Morand O, Welford RWD, et al. Lucerastat, an Iminosugar for Substrate Reduction Therapy: Tolerability, Pharmacodynamics, and Pharmacokinetics in Patients With Fabry Disease on Enzyme Replacement. Clinical pharmacology and therapeutics. 2018;103(4):703–11. 10.1002/cpt.790 . [DOI] [PubMed] [Google Scholar]
- 44.Entchev E, Jantzen I, Gawronski X, Feraille L, Luccarini J, Abitbol J, et al. Odiparcil is a promising substrate reduction therapy in MPS VI murine model. Mol Genet Metab. 2018;123(2):S42–S3. [Google Scholar]
- 45.Ory DS, Ottinger EA, Farhat NY, King KA, Jiang X, Weissfeld L, et al. Intrathecal 2-hydroxypropyl-beta-cyclodextrin decreases neurological disease progression in Niemann-Pick disease, type C1: a non-randomised, open-label, phase 1–2 trial. Lancet. 2017;390(10104):1758–68. 10.1016/S0140-6736(17)31465-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koeberl DD, Case LE, Smith EC, Walters C, Han SO, Li Y, et al. Correction of Biochemical Abnormalities and Improved Muscle Function in a Phase I/II Clinical Trial of Clenbuterol in Pompe Disease. Molecular therapy: the journal of the American Society of Gene Therapy. 2018;26(9):2304–14. 10.1016/j.ymthe.2018.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bongarzone ER, Escolar ML, Gray SJ, Kafri T, Vite CH, Sands MS. Insights into the Pathogenesis and Treatment of Krabbe Disease. Pediatric endocrinology reviews: PER. 2016;13 Suppl 1:689–96. . [PubMed] [Google Scholar]
- 48.Schuchman EH, Ge Y, Lai A, Borisov Y, Faillace M, Eliyahu E, et al. Pentosan polysulfate: a novel therapy for the mucopolysaccharidoses. PloS one. 2013;8(1):e54459 10.1371/journal.pone.0054459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roe CR, Mochel F. Anaplerotic diet therapy in inherited metabolic disease: therapeutic potential. Journal of inherited metabolic disease. 2006;29(2–3):332–40. 10.1007/s10545-006-0290-3 . [DOI] [PubMed] [Google Scholar]