Abstract
Summary
Sequence logos are visually compelling ways of illustrating the biological properties of DNA, RNA and protein sequences, yet it is currently difficult to generate and customize such logos within the Python programming environment. Here we introduce Logomaker, a Python API for creating publication-quality sequence logos. Logomaker can produce both standard and highly customized logos from either a matrix-like array of numbers or a multiple-sequence alignment. Logos are rendered as native matplotlib objects that are easy to stylize and incorporate into multi-panel figures.
Availability and implementation
Logomaker can be installed using the pip package manager and is compatible with both Python 2.7 and Python 3.6. Documentation is provided at http://logomaker.readthedocs.io; source code is available at http://github.com/jbkinney/logomaker.
1 Introduction
Sequence logos provide evocative graphical representations of the functional properties of DNA, RNA and protein sequences. Logos consist of characters stacked upon one another at a series of integer-valued positions, with the height of each character conveying some type of information about its biological importance. This graphical representation was introduced by Schneider and Stephens (1990) for illustrating statistical properties of multiple-sequence alignments. Although the specific representation they advocated is still widely used, sequence logos have since evolved into a general data visualization strategy that can be used to illustrate many different kinds of biological information (Kinney and McCandlish, 2019). For example, logos can be used to illustrate base-pair-specific contributions to protein–DNA binding energy (Foat et al., 2006), the effects of mutations in massively parallel selection experiments, and attribution method visualizations of deep neural networks (Jaganathan et al., 2019; Shrikumar et al., 2017).
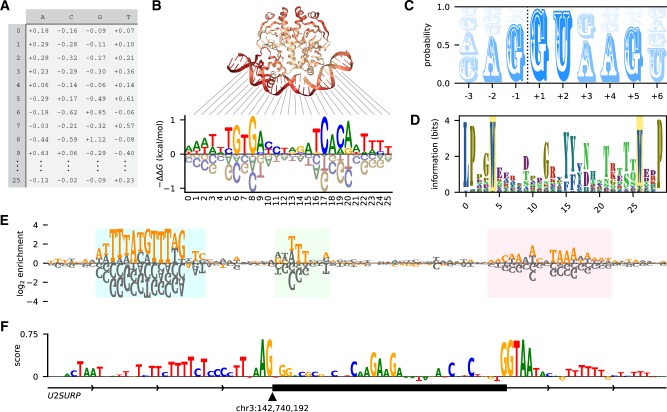
A substantial number of software tools for generating sequence logos have been described (Bailey et al., 2009; Colaert et al., 2009; Crooks et al., 2004; Gorodkin et al., 1997; Maddelein et al., 2015; Menzel et al., 2012; Nettling et al., 2015; Olsen et al., 2013; O’Shea et al., 2013; Ou et al., 2018; Rapin et al., 2010; Schuster-Böckler et al., 2004; Sharma et al., 2012; Thomsen and Nielsen, 2012; Waese et al., 2016; Wheeler et al., 2014; Workman et al., 2005; Wu and Bartel, 2017; Ye et al., 2017; Yu et al., 2015). However, each of these tools substantially limits the kinds of logos that one can make and the ways in which those logos can be styled. For example, WebLogo (Crooks et al., 2004) was one of the first logo-generating tools to be described and is still perhaps the most widely used. WebLogo allows users to create two standard types of sequence logos (information logos and probability logos) from a list of input sequences. However, it does not allow one to generate logos from arbitrary matrices of character heights. This capability is needed for illustrating the ΔΔG values of energy matrix models (Fig. 1B), the log-enrichment values obtained in high-throughput selection experiments (Fig. 1E) or importance scores that describe the predictions of deep neural networks (Fig. 1F). Moreover, although WebLogo is available as a Python package, the graphics it generates are written directly to file. This prevents logos from being customized using the matplotlib routines familiar to most Python users, or automatically incorporated into multi-panel figures.
Fig. 1.
Logomaker logos can represent diverse types of data. (A) Example input to Logomaker. Shown is an energy matrix for the transcription factor CRP; the elements of this pandas DataFrame represent - ΔΔG values contributed by each possible base (columns) at each nucleotide position (rows). Data are from Kinney et al. (2010). (B) An energy logo for CRP created by passing the DataFrame in panel A to Logomaker. The structural context of each nucleotide position is indicated [PDB 1CGP (Parkinson et al., 1996)]. (C) A probability logo computed from all annotated 5 splices sites in the human genome (Frankish et al., 2019). The dashed line indicates the exon/intron boundary. (D) An information logo computed from a multiple alignment of WW domain sequences [PFAM RP15 (Finn et al., 2014)], with the eponymous positions of this domain highlighted. (E) An enrichment logo representing the effects of mutations within the ARS1 replication origin of S.cerevisiae. Orange characters indicate the ARS1 wild-type sequence; highlighted regions correspond (from left to right) to the A, B1 and B2 elements of this sequence (Rao and Stillman, 1995). Data (unpublished; collected by J.B.K.) are from a mutARS-seq experiment analogous to the one reported by Liachko et al. (2013). (F) A masked logo (Shrikumar et al., 2017) representing the importance scores of nucleotides in the vicinity of U2SURP exon 9, as predicted by a deep neural network model of splice site selection. Logo adapted (with permission) from Fig. 1D of Jaganathan et al. (2019). The script used to make this figure is posted on the Logomaker GitHub page at logomaker/examples/figure.ipynb
In contrast to WebLogo and the other tools described above, ggseqlogo (Wagih, 2017) enables the creation of sequence logos within the R programming environment from arbitrary user-provided data. Importantly, ggseqlogo renders logos using native vector graphics, which facilitates post-hoc styling and the incorporation of logos into multi-panel figures. However, similar software is not yet available in Python. Because many biological data analysis pipelines are written in Python, there is a clear need for such logo-generating capabilities. Here we describe Logomaker, a Python package that addresses this need.
2 Implementation
Logomaker is a flexible Python API for creating sequence logos. Logomaker takes a pandas DataFrame as input, one in which columns represent characters, rows represent positions and values represent character heights (Fig. 1A). This enables the creation of logos for any type of data that are amenable to such a representation. The resulting logo is drawn using vector graphics embedded within a standard matplotlib Axes object, thus facilitating a high level of customization as well as incorporation into complex figures. Indeed, the logos in Figure 1 were generated as part of a single multi-panel matplotlib figure. Logomaker provides a variety of options for styling the characters within a logo, including the choice of font, color scheme, vertical and horizontal padding, etc. Logomaker also enables the highlighting of specific sequences within a logo (Fig. 1E), as well as the use of value-specific transparency in logos that illustrate probabilities (Fig. 1C). If desired, users can further customize individual characters within any rendered logo.
Because sequence logos are still commonly used to represent the statistics of multiple-sequence alignments, Logomaker provides methods for processing such alignments into matrices that can then be used to generate logos. Multiple types of matrices can be generated in this way, including matrices that represent probabilities (Fig. 1C), log odds ratios (Fig. 1E) or the information values described by Schneider and Stephens (1990) (Fig. 1D). Methods for transforming between these types of matrices are also provided. Finally, Logomaker supports the creation of masked matrices and logos that, e.g., represent deep neural network importance scores (Shrikumar et al., 2017), as in Figure 1F.
3 Conclusion
Logomaker thus fills a major need in the Python community for flexible logo-generating software. Indeed, Logomaker has already been used to generate logos for multiple preprints and publications (Belliveau et al., 2018; Barnes et al., 2019; Forcier et al., 2018; Kinney and McCandlish, 2019; Mason et al., 2019; Nguyen et al., 2019; Wong et al., 2018). Logomaker is thoroughly tested, has minimal dependencies and can be installed from PyPI by executing ‘pip install logomaker’ at the command line. A step-by-step tutorial on how to use Logomaker, as well as comprehensive documentation, is available at http://logomaker.readthedocs.io.
Acknowledgements
We thank William Ireland, David McCandlish and Bruce Stillman for helpful discussions. We also thank Kyle Farh and Kishore Jaganathan for providing data for the input-masked importance score logo in Figure 1F.
Funding
This work was supported by the National Institutes of Health [1R35GM133777, 5P30CA045508]; and the Cold Spring Harbor Laboratory/Northwell Health Alliance.
Conflict of Interest: none declared.
References
- Bailey T.L. et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes S.L. et al. (2019) Mapping DNA sequence to transcription factor binding energy in vivo. PLoS Comput. Biol., 15, e1006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliveau N.M. et al. (2018) Systematic approach for dissecting the molecular mechanisms of transcriptional regulation in bacteria. Proc. Natl. Acad. Sci. USA, 115, E4796–E4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaert N. et al. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods, 6, 786–787. [DOI] [PubMed] [Google Scholar]
- Crooks G.E. et al. (2004) WebLogo: a sequence logo generator. Genome Res., 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D. et al. (2014) Pfam: the protein families database. Nucleic Acids Res., 42, D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foat B. et al. (2006) Statistical mechanical modeling of genome-wide transcription factor occupancy data by MatrixREDUCE. Bioinformatics, 22, e141–9. [DOI] [PubMed] [Google Scholar]
- Forcier T.L. et al. (2018) Measuring cis-regulatory energetics in living cells using allelic manifolds. eLife, 7, e40618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A. et al. (2019) GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res., 47, D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodkin J. et al. (1997) Displaying the information contents of structural RNA alignments: the structure logos. Comput. Appl. Biosci., 13, 583–586. [DOI] [PubMed] [Google Scholar]
- Jaganathan K. et al. (2019) Predicting Splicing from Primary Sequence with Deep Learning. Cell, 176, 535–548.e24. [DOI] [PubMed] [Google Scholar]
- Kinney J.B., McCandlish D.M. (2019) Massively parallel assays and quantitative sequence-function relationships. Annu. Rev. Genom. Hum. Genet., 20, 99–127. [DOI] [PubMed] [Google Scholar]
- Kinney J.B. et al. (2010) Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc. Natl. Acad. Sci. USA, 107, 9158–9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I. et al. (2013) High-resolution mapping, characterization, and optimization of autonomously replicating sequences in yeast. Genome Res., 23, 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddelein D. et al. (2015) The iceLogo web server and SOAP service for determining protein consensus sequences. Nucleic Acids Res., 43, W543–W546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D.M. et al. (2019) Deep learning enables therapeutic antibody optimization in mammalian cells by deciphering high-dimensional protein sequence space. bioRxiv, doi:10.1101/617860, 1–26. [Google Scholar]
- Menzel P. et al. (2012) RILogo: visualizing RNA-RNA interactions. Bioinformatics, 28, 2523–2526. [DOI] [PubMed] [Google Scholar]
- Nettling M. et al. (2015) DiffLogo: a comparative visualization of sequence motifs. BMC Bioinformatics, 16, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.Q. et al. (2019) Quantitative mapping of protein-peptide affinity landscapes using spectrally encoded beads. eLife, 8, e40499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L.R. et al. (2013) BlockLogo: visualization of peptide and sequence motif conservation. J. Immunol. Methods, 400–401, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea J.P. et al. (2013) pLogo: a probabilistic approach to visualizing sequence motifs. Nat. Methods, 10, 1211–1212. [DOI] [PubMed] [Google Scholar]
- Ou J. et al. (2018) motifStack for the analysis of transcription factor binding site evolution. Nat. Methods, 15, 8–9. [DOI] [PubMed] [Google Scholar]
- Parkinson G. et al. (1996) Structure of the CAP-DNA complex at 2.5 angstroms resolution: a complete picture of the protein–DNA interface. J. Mol. Biol., 260, 395–408. [DOI] [PubMed] [Google Scholar]
- Rao H., Stillman B. (1995) The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl. Acad. Sci. USA, 92, 2224–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapin N. et al. (2010) The MHC motif viewer: a visualization tool for MHC binding motifs. Curr. Protoc. Immunol., Chapter 18, Unit 18.17. [DOI] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res., 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Böckler B. et al. (2004) HMM logos for visualization of protein families. BMC Bioinformatics, 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V. et al. (2012) CodonLogo: a sequence logo-based viewer for codon patterns. Bioinformatics, 28, 1935–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrikumar A. et al. (2017) Learning important features through propagating activation differences. In: Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, Vol. 70.
- Thomsen M.C.F., Nielsen M. (2012) Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res., 40, W281–W287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waese J. et al. (2016) Gene Slider: sequence logo interactive data-visualization for education and research. Bioinformatics, 32, 3670–3672. [DOI] [PubMed] [Google Scholar]
- Wagih O. (2017) ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics, 33, 3645–3647. [DOI] [PubMed] [Google Scholar]
- Wheeler T.J. et al. (2014) Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics, 15, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M.S. et al. (2018) Quantitative activity profile and context dependence of all human 5’ splice sites. Mol. Cell, 71, 1012–1026.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C.T. et al. (2005) enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res., 33, W389–W392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Bartel D.P. (2017) kpLogo: positional k-mer analysis reveals hidden specificity in biological sequences. Nucleic Acids Res., 45, W534–W538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z. et al. (2017) CircularLogo: a lightweight web application to visualize intra-motif dependencies. BMC Bioinformatics, 18, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.-K. et al. (2015) Log-odds sequence logos. Bioinformatics, 31, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]