During differentiation, oligodendrocyte precursor cells (OPCs) extend a network of processes that make contact with axons and initiate myelination. Recent studies revealed that actin polymerization is required for initiation of myelination whereas actin depolymerization promotes myelin wrapping.
Keywords: ArpC3, cytoskeleton, MBP, mTOR, myelination, oligodendrocyte
Abstract
During differentiation, oligodendrocyte precursor cells (OPCs) extend a network of processes that make contact with axons and initiate myelination. Recent studies revealed that actin polymerization is required for initiation of myelination whereas actin depolymerization promotes myelin wrapping. Here, we used primary OPCs in culture isolated from neonatal rat cortices of both sexes and young male and female mice with oligodendrocyte-specific deletion of mechanistic target of rapamycin (mTOR) to demonstrate that mTOR regulates expression of specific cytoskeletal targets and actin reorganization in oligodendrocytes during developmental myelination. Loss or inhibition of mTOR reduced expression of profilin2 and ARPC3, actin polymerizing factors, and elevated levels of active cofilin, which mediates actin depolymerization. The deficits in actin polymerization were revealed in reduced phalloidin and deficits in oligodendrocyte cellular branching complexity at the peak of morphologic differentiation and a delay in initiation of myelination. We further show a critical role for mTOR in expression and localization of myelin basic protein (Mbp) mRNA and MBP protein to the cellular processes where it is necessary at the myelin membrane for axon wrapping. Mbp mRNA transport deficits were confirmed by single molecule RNA FISH. Moreover, expression of the kinesin family member 1B, an Mbp mRNA transport protein, was reduced in CC1+ cells in the mTOR cKO and in mTOR inhibited oligodendrocytes undergoing differentiation in vitro. These data support the conclusion that mTOR regulates both initiation of myelination and axon wrapping by targeting cytoskeletal reorganization and MBP localization to oligodendrocyte processes.
SIGNIFICANCE STATEMENT Myelination is essential for normal CNS development and adult axon preservation and function. The mechanistic target of rapamycin (mTOR) signaling pathway has been implicated in promoting CNS myelination; however, there is a gap in our understanding of the mechanisms by which mTOR promotes developmental myelination through regulating specific downstream targets. Here, we present evidence that mTOR promotes the initiation of myelination through regulating specific cytoskeletal targets and cellular process expansion by oligodendrocyte precursor cells as well as expression and cellular localization of myelin basic protein.
Introduction
Progression through specific stages of oligodendrocyte differentiation has been well characterized and involves extensive morphologic changes in preparation for axon contact and initiation of myelination (for reviews, see Michalski and Kothary, 2015; Snaidero and Simons, 2017; Brown and Macklin, 2020). Oligodendrocyte precursor cells (OPCs) transition into immature oligodendrocytes characterized by increased number and length of cellular processes to contact axons and initiate myelination. Final maturation of oligodendrocytes requires the cell membrane to flatten out to wrap axons in the process of myelination (Nawaz et al., 2015; Zuchero et al., 2015). The inability of OPCs to differentiate contributes to impaired developmental myelination as well as to reduced myelin repair after demyelination in diseases such as multiple sclerosis (for reviews, see Franklin and Ffrench-Constant, 2017; Snaidero and Simons, 2017; Abu-Rub and Miller, 2018).
The evolving morphology of oligodendrocytes during cellular differentiation and myelination requires a dynamic cytoskeleton (for reviews, see Michalski and Kothary, 2015; Snaidero and Simons, 2017; Thomason et al., 2019; Brown and Macklin, 2020). The actin cytoskeleton exists in monomer and filament form; the balance between polymerization and depolymerization is tightly regulated and controls cytoskeletal reorganizations. Numerous laboratories have contributed to our current understanding of cytoskeletal dynamics in developing oligodendroglia. Prior reports support two phases of developmental myelination involving opposing cytoskeletal processes in differentiating oligodendrocytes; phase I requires actin polymerization for growth cone formation mediated process extension (Fox et al., 2006; Zuchero et al., 2015), whereas phase II requires actin depolymerization for proper axon wrapping and myelination (Nawaz et al., 2015; Zuchero et al., 2015). Actin binding proteins that regulate actin cytoskeletal reorganization include the actin polymerizing protein profilin, the ARP2/3 branched actin nucleator complex, and the depolymerizing protein family actin depolymerizing factor (ADF)/cofilin1. The ARP2/3 complex in oligodendroglia is known to regulate axon ensheathment (Zuchero et al., 2015). In contrast, ADF/cofilin1 regulates filamentous actin (F-actin) turnover for proper myelin wrapping; however, it is hypothesized that F-actin at the membrane leading edge provides the driving force needed for myelin growth (Nawaz et al., 2015). Moreover, the involvement of F-actin at the inner tongue of myelinating cells helps create the balance between myelin compaction and cytoplasmic channels (Snaidero and Simons, 2017). The presence of myelin basic protein (MBP) in the oligodendrocyte processes also has been proposed to be important in the wrapping phase of myelination to enable MBP to displace cofilin at the membrane (Zuchero et al., 2015). The membrane displacement of cofilin then may facilitate actin depolymerization necessary for wrapping (Zuchero et al., 2015).
The function of specific intracellular signaling pathways in the developmental progression of oligodendroglia has been the focus of numerous publications (Sperber et al., 2001; J. Zou et al., 2011; Ishii et al., 2012; Bercury et al., 2014; Dai et al., 2014; Wahl et al., 2014; Furusho et al., 2017). However, it is unknown how these pathways orchestrate the morphologic changes observed during differentiation and myelination. Moreover, the studies on actin dynamics in developing oligodendrocytes and myelination did not identify upstream regulators (Nawaz et al., 2015; Zuchero et al., 2015). Studies in a variety of other cell types including neutrophils, fibroblasts, and tumor cells demonstrate a role for the mechanistic target of rapamycin (mTOR) pathway in regulating the cytoskeleton (Jacinto et al., 2004; Sarbassov et al., 2004; Liu et al., 2008; He et al., 2013); however, these studies have provided little information on specific cytoskeletal targets downstream of mTOR. Our previous studies revealed that mTOR regulates oligodendrocyte differentiation, initiation of myelination and myelin thickness in the spinal cord (Tyler et al., 2009; Bercury et al., 2014; Wahl et al., 2014). Collectively, these findings led us to hypothesize that mTOR regulates cytoskeletal dynamics in oligodendrocytes necessary for initiation of myelination and for proper myelin wrapping.
The goal of the studies presented here was to determine the function of mTOR signaling in regulating cytoskeletal reorganization during oligodendrocyte development. We show that loss or inhibition of mTOR alters expression of cytoskeletal proteins involved in actin polymerization in differentiating oligodendrocytes in vitro and in vivo. Moreover, we demonstrate that mTOR in oligodendroglia regulates the localization of MBP to oligodendrocyte processes during myelination, through promoting transport of Mbp mRNAs.
Materials and Methods
Experimental animals
All mouse protocols were conducted in accordance with Rutgers University Institutional Animal Care and Use Committee and the National Institute of Health guidelines for care and use of laboratory animals. Mice were housed in a barrier facility with a 12 h light/dark cycle. The CNP-Cre/mTOR conditional knock-out (mTOR cKO) mouse line carrying CNP-Cre and floxed alleles for Mtor was described previously (Wahl et al., 2014).
Mice homozygous for Mtor floxed and heterozygous for CNP-Cre were used for breeding to generate Cre+ or Cre- littermates for experiments. The PLP-Cre/mTOR inducible cKO (icKO) line was established by crossing Mtor fl/fl mice (C. H. Lang et al., 2010; S. A. Lang et al., 2010; Carr et al., 2012; Wahl et al., 2014) with PLP-CreERT mice (The Jackson Laboratory, 005975; RRID:IMSR_JAX:005975), henceforth referred to as PLP-mTOR icKO mice. Mice homozygous for Mtor floxed and heterozygous for PLP-CreERT were used for breeding to generate Cre+ or Cre− littermates for experiments. Tamoxifen was injected intraperitoneally (60 mg/kg) for 4 consecutive days to induce recombination at P7. Tamoxifen was dissolved in a 9:1 ratio of sesame oil–100% ethanol. Both males and females were used in all analyses. All strains were on a C57BL/6 background.
All zebrafish experiments were approved by the Institutional Animal Care and Use Committee at the University of Colorado School of Medicine. Embryos were raised at 28.5°C in embryo media (EM) and staged according to hours postfertilization (hpf), days postfertilization (dpf), and morphologic criteria (Kimmel et al., 1995). Rapamycin (Tocris Bioscience) was dissolved in 100% DMSO at a concentration of 20 mm. Drugs were diluted in EM to make a working concentration of 5 mm with a final concentration of 1% DMSO. Control solutions contained 1% DMSO in EM. Tg(nkx2.2a:mEGFP) zebrafish embryos were collected following timed matings. Embryos were sorted for GFP, dechorionated and treated with rapamycin or DMSO control. Zebrafish drug treatments were initiated at 48 hpf until 56 hpf, when zebrafish were anesthetized using tricaine (MS-222). Embryos were mounted laterally in 1% low-melt agarose and tricaine and imaged directed above the yolk sac extension on a Leica DM-6000 confocal. Individual oligodendrocytes were analyzed using IMARIS image analysis software (Bitplane).
Preparation and isolation of primary oligodendrocytes
OPCs were purified from cortical mixed glial cultures isolated from postnatal days (P)0–P2 Sprague-Dawley rat pups by established methods and cultured as described previously (McCarthy and Vellis, 1980; Tyler et al., 2009). To initiate OPC differentiation, we followed an established mitogen withdrawal protocol in the presence of 30 ng/ml triiodothyronine (T3) and plus or minus the addition of rapamycin (15 nm) as for prior studies (Tyler et al., 2009). In some experiments, we initiated differentiation for 48 h prior to adding rapamycin. For all experiments, differentiation medium plus/minus rapamycin was replenished every 48 h except as noted for Figure 1.
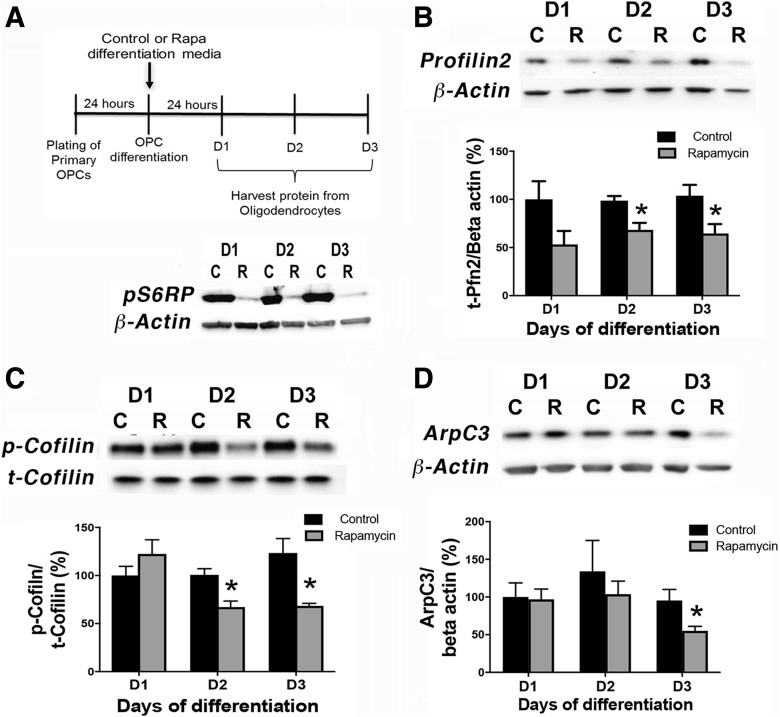
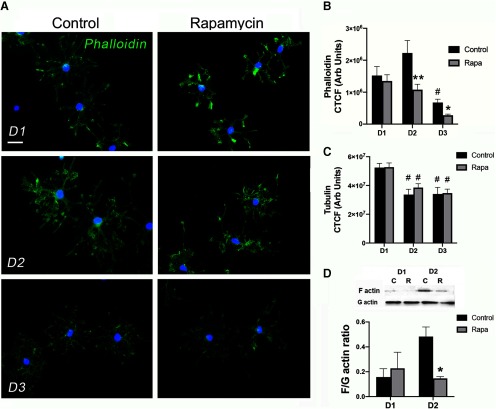
Figure 1.
mTOR inhibition downregulates expression of cytoskeletal targets in differentiating OPCs in vitro. A, Experimental paradigm of OPC differentiation and protein harvest from primary rat OPCs in vitro. Western blot shows reduction in pS6RP in the presence of rapamycin (R) versus control (C) from D1 to D3 of differentiation with rapamycin added at the onset of differentiation. B–D, Control (C) or rapamycin (R) treated OPC cultures were analyzed for levels of profilin2 (PFN2; B); n = 4, control versus rapamycin *p = 0.013 at D2; *p = 0.038 at D3; p/t-cofilin (C); n = 3, control versus rapamycin *p = 0.019 at D2, *p = 0.022 at D3; or ARPC3 (D); n= 4, control versus rapamycin *p = 0.044 at D3. Representative Western blots are presented in B–D. All values are expressed as ±SEM.
Protein isolation and Western immunoblotting
Plated cells from OPC isolation were harvested in TRIS-triton buffer (10 mm Tris, 100 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 10% glycerol, 0.1% SDS, 0.5% deoxycholate) supplemented with phosphatase and protease inhibitors (5 mm NaF, 1 mm PMSF, 1% protease inhibitor cocktail, 1 mm Na3V04), then sonicated. Protein concentrations were determined using the Bio-Rad protein assay and separated by SDS-PAGE. Primary antibodies were as follows: Rb-profilin2 (1:250; Novus Biologicals, NBP1-87 426; RRID:AB_11007824), Rb-p-cofilin (1:1000; Cell Signaling Technology, 3313; RRID:AB_2080597), Rb-total cofilin (1:1000; Cell Signaling Technology, 5175; RRID:AB_10622000), Rb-Kif1b (1:500; Abcam, ab69614; RRID:AB_1860749), Rb-ARPC3 (1:500; Novus Biologicals, NBP1-89 016; RRID:AB_11003588), Rb-MBP (1:1000; Abcam, ab40390; RRID:AB_1141521), and Ms-beta-actin (1:10 000; Sigma-Aldrich, A5441; RRID:AB_476744). Secondary antibodies were as follows: (Gt anti-Rb 1:2000 for all Rb antibodies, Jackson ImmunoResearch, 111-035-144; RRID:AB_ 2307391) or (Gt anti-Ms 1:5000 for beta-actin, Jackson ImmunoResearch, 115-035-003; RRID:AB_10015289).
For filamentous to globular (F/G) actin analysis, proteins were harvested from control or rapamycin treated cells following differentiation in T3 supplemented differentiation media as detailed in the previous section. F/G actin ratios were performed according to the manufacturer's instructions (Cytoskeleton, BK 037).
Tissue preparation and immunofluorescent staining
Animals at P7 and P14 were rapidly decapitated, and spinal cords were dissected and drop fixed in 3% PFA at 4°C, overnight. Tissues were then dehydrated in 30% sucrose and frozen in OCT for cryosectioning. Mounted cryosections (25 µm) were rinsed in 0.1% PBST and incubated in 50 mm ammonium chloride for 1 h at room temperature. Slides were washed in 0.1% PBST 3 × 10 min. Sections were then permeabilized and blocked in 10% BSA/5% NGS in 0.1% PBST for 1 h at room temperature. Primary and secondary antibodies were added to 1% BSA/5% NGS in 0.1% PBST. Sections were incubated in primary antibody in a humid chamber, overnight at 4°C. Primary antibodies were obtained as described in the previous section and used at the indicated dilutions: profilin2 (1:50), ARPC3 (1:100), Kif1b (1:200), MBP (1:200), CC1 (1:100; Millipore, OP80; RRID:AB_2057371), α-Tubulin (1:500; Sigma-Aldrich, T6199; RRID:AB_477583), Acti-Stain 488 Fluorescent Phalloidin (100 nm; Cytoskeleton, PHDG1), and mouse-neurofilament (NF) 200 (1:20; Sigma-Aldrich, N5389; RRID:AB_260781). Secondary antibodies purchased from Jackson ImmunoResearch were as follows: goat anti-rabbit 647 (1:500; 111-605-144; RRID:AB_2338078), mouse biotin (1:100; 200-002-211; RRID:AB_2339006), and streptavidin 405 (1:300; 016-470-084; RRID:AB_2337248).
Following primary antibody incubation, sections were washed in 0.1% PBST and incubated in the appropriate secondary antibody (described above) for 2 h at room temperature at the following dilutions: goat anti-rabbit 647 (1:500) or mouse biotin (1:100). Sections were washed in 0.1% PBST and incubated with streptavidin 405 (1:300) for 2 h at room temperature followed by washes in 0.1% PBST and PBS and coverslipped with either Fluorogel or Prolong Gold.
For cell staining in vitro, media was removed from cultured cells in well-slides. Slides were rinsed with PBS, fixed in 3% PFA for 15 min, washed with PBS and permeabilized with 0.3% triton X-100 for 15 min. Cells were blocked in 4% BSA/3% goat serum in PBS for 1 h and incubated overnight with primary antibody in blocking solution. Following washes in PBS, cells were incubated in secondary antibody for 1 h at room temperature. Cells were then washed in PBS and incubated in DAPI for 5 min, washed again in PBS and coverslipped with Fluorogel. For F-actin visualization, cells were incubated in 100 nm Acti-Stain 488 Fluorescent Phalloidin at room temperature in the dark for 30 min prior to DAPI incubation.
In situ hybridization
Mice were intracardially perfused with 4% PFA in PBS; spinal cords were dissected and postfixed with 4% PFA overnight, cryoprotected with 30% sucrose-PBS buffer overnight and frozen. Mounted cryosections were prepared at 20 µm thickness. In situ hybridization was performed as described previously (Hashimoto et al., 2016) with slight modifications. The following plasmids containing mouse cDNA were used to generate cRNA probes: Plp (full coding region; Harlow et al., 2014) and Mbp (nucleotides 683–1286 corresponding to NM_010777.3). Briefly, the sections were treated with proteinase K (2 μg/ml for 15 min at room temperature) and hybridized overnight at 63°C with DIG-labeled antisense riboprobes in a hybridization solution consisting of 50% formamide, 20 mm Tris-HCl, pH 7.5, 600 mm NaCl, 1 mm EDTA, 10% dextran sulfate, 200 μg/ml yeast tRNA and 1× Denhardt's solution. After the sections were washed in buffers with decreasing stringency, they were incubated with an alkaline phosphatase-conjugated anti-DIG antibody (1:5000; Roche Diagnostics). The color was developed in the presence of 4-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate (Roche Diagnostics) in the dark at room temperature.
Quantification of Plp+ cell numbers were performed using ImageJ (National Institutes of Health) on 63× control (n = 3) and mTOR cKO (n = 4) ventral white matter fields. Cells were counted on at least three sections per animal. Quantification of Mbp mRNA intensity in the ventral white matter was performed using ImageJ on 63× control (n = 3) and mTOR cKO (n = 4). The average intensity was measured in at least three sections per animal, and the results were expressed as arbitrary units.
FISH
A set of 50 (20 nt long) probes was designed to hybridize to the coding sequence of the Mbp mRNA using freely available software at LGC Biosearch Technologies as described previously (Batish et al., 2011). The oligos were ordered with the 3′ terminal nucleotide having an amino modification. All the modified oligos were pooled in equimolar concentration and coupled en mass with AlexaFluor 594 dye. The labeled probes were purified from unlabeled oligos and unbound dye using a C14 reverse phase hydrophobic HPLC column as described previously (Batish et al., 2011). Primary rat OPCs were grown on glass coverslips under differentiation conditions as described in the section, Preparation and isolation of primary oligodendrocytes, ±15 nm of rapamycin for 7 d with media change every 48 h. The cells were fixed using 4% paraformaldehyde for 10 min, permeabilized with 70% ethanol at 4°C and hybridized overnight at 37°C with the labeled probes. The coverslips were washed multiple times to remove any unbound probes, stained with DAPI and mounted using deoxygenated media (Batish et al., 2011; Markey et al., 2014). The coverslips were imaged using 100× oil objective in a Nikon TiE inverted automated fluorescence microscope equipped with cooled CCD Pixis 1024b camera. The z stacks of 3 μm with 0.2 μm apart were acquired using MetaMorph software and the images were analyzed using custom written programs in MATLAB (MathWorks; Batish et al., 2012; Markey et al., 2014). At least 100 cells for each treatment were quantified.
Isolation of O4-positive cells
Spinal cord tissue dissociation was performed according to manufacturer's instructions using a neural dissociation kit (Miltenyi Biotec, 130-092-628). The O4+ oligodendrocytes were isolated from spinal cords using O4 microbeads (Miltenyi Biotec, 130-094-543) and MS columns (Miltenyi Biotec, 130–042-201) and used for protein or RNA isolation.
RNA isolation and RT-PCR analysis
RNA isolation from O4-positive cells was completed using RNeasy Plus Micro Kit (Qiagen, 74 034) and the Quick-StartProtocol. RNA concentration was measured using a NanoDrop spectrophotometer (ThermoFisher Scientific); 50–80 ng of RNA was used to reverse transcribe cDNA using Superscript II (Invitrogen). For RT-PCR, 1.5–2 µl of cDNA per well was used in reactions containing 1× SYBR Green detection master mix and 1× QuantiTect primer mix to detect mRNA levels. Amplification was normalized to expression levels of β -actin (IDT) for each sample.
Data analysis and statistics
At least three animals per genotype were used to investigate cytoskeletal or lineage marker protein expression at all ages. Cells were counted on at least four nonadjacent rostral and caudal sections, with a minimum of eight sections per animal. Images (10×) were taken using an Olympus AX70 microscope of dorsal white matter (DWM) and ventral white matter (VWM) to count CC1+ cells. Absolute numbers of CC1+ cells in defined ROIs were normalized to number of cells/mm2. Images (20×) were taken of DWM and VWM to count percentage of cells expressing the protein-of-interest. A MATLAB software code described previously was used to count cells (Gould et al., 2018). Additional spinal cord samples from P14 were collected to address variability observed in analyses of CC1+ cells observed in original cohorts. At least three images of VWM regions were analyzed. Using ImageJ, the VWM was outlined and defined as the ROI. The “Adjust Threshold” function was used to highlight CC1+ cell bodies. The number of CC1+ cells was automatically quantified using the “Analyze Particles” function to define upper (infinity) and lower (10 µm) size limits.
Phalloidin and tubulin intensity measurements and Sholl analyses were performed on individual primary OPCs using Fiji/NIH Image. For intensity measurements, corrected total cell fluorescence was calculated by integrated density – (area of cell × mean fluorescence of background).
For analysis of confocal images of ARPC3 and NF staining, three z stacks ranging from the surface of the tissue to the end of NF antibody penetration in rostral VWM regions from each sample were taken with a 60× oil objective using a Nikon A1R confocal microscope. The maximum intensity projection of each z stack was used to analyze the number of interactions between ARPC3 and NF immunostaining in each image of VWM. An interaction was defined as an ARPC3+ soma or process in contact with ≥50% of the circumference of a NF+ axon. The absolute number of interactions was normalized to the area of the image, 12, 683 µm2.
In vitro experiments were performed on at least three preparations of rat primary OPCs representing biological replicates. Numbers of animals and in vitro biological replicates were determined based on our prior studies using control and mTOR cKO mice or primary rat OPCs to quantify changes in expression of RNA or proteins. In some cases when pilot studies indicated high variability, a power analysis was performed to determine the number of biological replicates necessary for statistical power. Student's unpaired t test was used to statistically compare groups for all two group comparisons. An unpaired t test with Welch's correction was performed for Sholl analyses in Figure 3, for cell area by tubulin staining in Figure 8, and for Mbp RNA analyses in Figure 10 because of unequal variances between groups. One-way or two-way analysis of variance assays (ANOVAs) were used for multigroup comparisons across time points where specified; adjusted p values were determined using Sidak's multiple-comparison test.
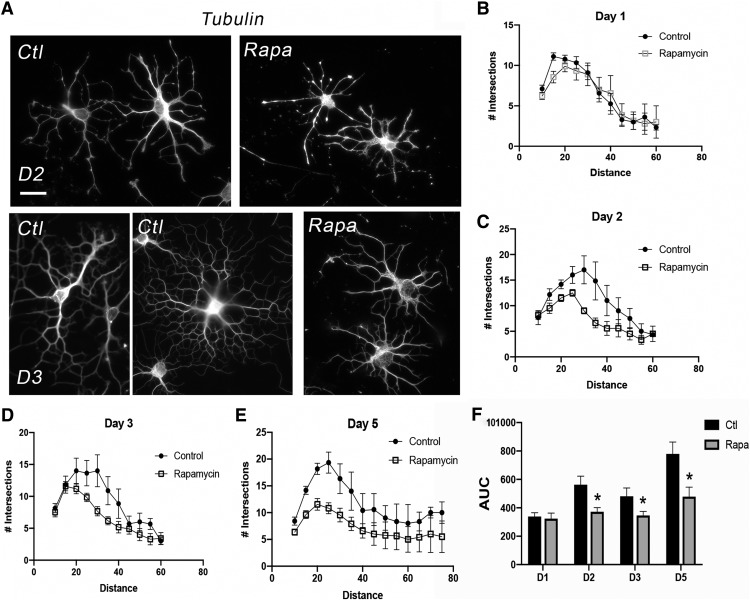
Figure 3.
mTOR inhibition during differentiation reduces morphologic complexity in differentiating OPCs in vitro. A, Images show representative primary rat oligodendroglia stained with tubulin at D2 or D3 of differentiation plus or minus rapamycin. Scale bar, 20 µm. B–E, Individual cells were analyzed using NIH ImageJ to quantify the number of Sholl intersections at D2, D3, and D5, error bars are SEM. F, Area under curve (arbitrary units): D1, Control (Ctl): 338.7 ± 26.49, Rapamycin (Rapa): 323.4 ± 39.76, p = 0.749; D2, Ctl: 563.3 ± 59.89, Rapa: 372.2 ± 28.72, *p= 0.005; D3, Ctl: 480.9 ± 58.66, Rapa: 346.2 ± 28.38, *p = 0.041; D5, Ctl: 779.2 ± 84.06, Rapa: 479.0 ± 66.07, *p = 0.006.
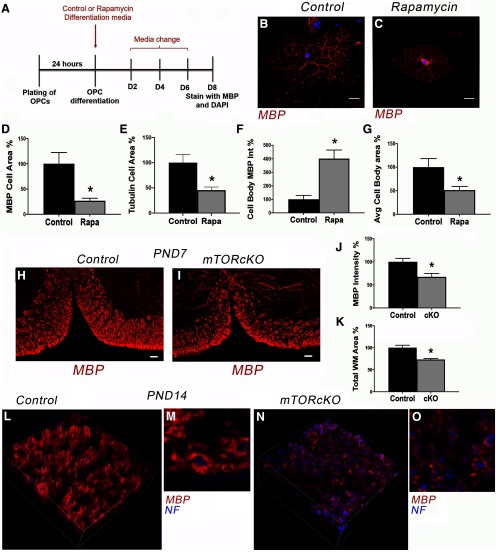
Figure 8.
Inhibiting mTOR signaling perturbs MBP localization and reduces area of membrane spreading in differentiating oligodendrocytes in vitro. A, In vitro experimental paradigm of OPC differentiation plus/minus rapamycin treatment. B, C, Micrographs showing representative control or rapamycin (Rapa)-treated oligodendrocytes after 8 d of differentiation. Scale bar, 10 µm. D–G, Quantification to determine total MBP+ cell area (D) n = 7, *p = 0.007, tubulin+ cell area at D5 (E), n = 23 Ctrl, n = 11 Rapa, *p = 0.004; MBP protein intensity in the cell body (F), n = 9, *p = 0.0007; and MBP+ cell body area (G) n = 9, *p = 0.028. Values are expressed as mean ± SEM. H, I, Representative micrographs showing MBP protein in control and mTOR cKO spinal cords at P7. Scale bar, 50 µm. J, K, Quantification of MBP intensity (J) and white matter area (K) in control versus mTOR cKO spinal cord VWM at P7. n = 4 control, n = 3 cKO; J, *p = 0.030; K, *p = 0.013. Values are expressed as ± SEM. L–O, Representative confocal z stack images showing persistent decrease in myelin thickness in mTOR cKO at P14.
Figure 10.
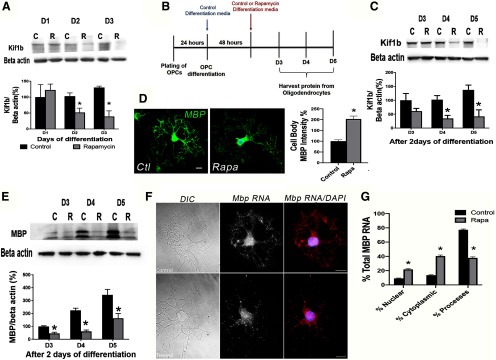
mTOR regulates KIF1B expression and Mbp mRNA transport in differentiating oligodendrocytes in vitro. A, Western blot analysis showing decreased KIF1B expression with mTOR inhibition during oligodendrocyte differentiation in vitro. Rapamycin was added at the initiation of differentiation as for prior experiments (Fig. 1A); n = 3; p= 0.041 at D2, p = 0.007 at D3. B, Experimental paradigm of OPC differentiation plus or minus rapamycin addition following 2 d of normal differentiation. C, Western blot analysis of KIF1B expression after delayed rapamycin (R) treatment in vitro; n = 5, p = 0.006 at D4, p = 0.011 at D5. D, Representative images and percentage of MBP protein localization restricted to the cell body in rapamycin-treated oligodendrocytes at D4 in delayed addition paradigm; n = 4 Ctrl, n = 3 Rapa, p = 0.0008. E, Western blot analysis of MBP expression after delayed rapamycin treatment in vitro; n = 3, p = 0.007 at D3; p = 0.001 at D4; p = 0.032 at D5. F, G, Representative images of DIC and Mbp mRNA fluorescence (F) and quantification of Mbp mRNA localization (G) following single-molecule FISH for Mbp mRNA on primary rat OPCs after 7 d of differentiation in the absence (F, top panels; G control) or presence (F, bottom panels; G Rapa) of rapamycin from the start of differentiation. Scale bar, 5 µm. Mbp mRNA is reduced in the cellular processes of rapamycin-treated OPCs with a corresponding accumulation in the cytoplasm and nucleus; n = 51 Ctrl, n = 53 Rapa. *p < 0.001 for Control versus Rapa in all compartments.
Results
mTOR signaling regulates actin polymerization during early oligodendrocyte morphologic differentiation in vitro
Actin binding proteins that regulate actin cytoskeleton reorganization include profilins that promote actin polymerization, the ARP2/3 complex that mediates branched actin nucleation, and the actin depolymerizing factor family (ADF)/cofilin. Profilin interaction with formin proteins regulates polymerization of long actin cables necessary for polarity (Evangelista et al., 2002; Romero et al., 2004; Winder and Ayscough, 2005; Kovar et al., 2006; Suarez et al., 2015). Profilin exchanges the ADP bound to G-actin with ATP for addition to the growing end of the filament. In contrast, the ADF/cofilin family regulates actin filament turnover by binding to ADP bound F-actin at the negative end of the filament to facilitate filament depolymerization (Kanellos and Frame, 2016). Phosphorylation of cofilin results in its inactivation, inhibiting it from binding to and depolymerizing actin filaments (Arber et al., 1998; Mizuno, 2013).
To determine the impact of mTOR signaling on actin polymerization during oligodendrocyte differentiation, we initially determined levels of profilin2 and of phospho/total cofilin in primary rat OPCs undergoing differentiation in the presence or absence of the mTOR inhibitor rapamycin. Profilin2 is highly expressed in the brain (Witke et al., 1998), and its RNA expression is induced transiently in newly formed oligodendrocytes (Zhang et al., 2014). For these in vitro experiments, we used a standard mitogen withdrawal protocol to induce differentiation of rat OPCs with or without rapamycin to inhibit mTOR signaling as determined by phosphorylation of S6-ribosomal protein (Fig. 1A). Using this paradigm, we found that profilin2 protein expression decreased by day (D)2 of differentiation in the presence of rapamycin (D1: p = 0.093, t = 1.99, df = 6; D2: p = 0.013, t = 3.48, df = 6; D3: p = 0.038, t = 2.64, df = 6; Fig. 1B). Moreover, mTOR inhibition also decreased p-cofilin, the inactive form of cofilin, whereas total cofilin expression levels were unchanged (p/t cofilin, D1: p = 0.271, t = 1.27, df = 4; D2: p = 0.019, t = 3.77, df = 4; D3: p = 0.022, t = 3.61, df = 4; Fig. 1C).
Whereas profilins have a major role in polymerization of linear actin filaments, the ARP2/3 complex is a branched nucleation factor that binds to the side of an existing actin mother filament and forms a nucleation core from which new daughter filaments are formed (Pollard et al., 2001; Rouiller et al., 2008). The ARPC3 subunit of the ARP2/3 complex is necessary for nucleation core formation (Yae et al., 2006). A study on oligodendrocyte cytoskeleton and myelination demonstrated that deletion of Arpc3 in oligodendroglia impaired initiation of myelination (Zuchero et al., 2015). Inhibiting mTOR significantly reduced ARPC3 expression by D3 of differentiation in vitro (D1: p = 0.898, t = 0.13, df = 6; D2: p = 0.520, t = 0.68, df = 6; D3: p = 0.043, t= 2.54, df = 6; Fig. 1D), later than the reduction in profilin2 and p-cofilin expression suggesting that mTOR regulates ARPC3 expression subsequent to initial actin polymerization.
To determine F-actin levels in early oligodendrocyte development, we quantified phalloidin intensity during OPC differentiation in the presence or absence of rapamycin (Fig. 2). Two-way ANOVA revealed a main effect of treatment and time (F(2,80) = 15.08, p < 0.0001 and F(2,80) = 9.35, p = 0.003, respectively). Phalloidin intensity was highest in control cells at D2 of differentiation and then decreased dramatically by D3 [adjusted (adj) p values: D1 vs D2, p = 0.101; D1 vs D3, p = 0.036; D2 vs D3, p < 0.0001; Fig. 2A,B]. Similarly, phalloidin decreased in rapamycin-treated cells at D3 (adj p values: D1 vs D2, p = 0.788; D1 vs D3, p = 0.049; D2 vs D3, p = 0.045; Fig. 2A,B). However, inhibition of mTOR significantly reduced phalloidin intensity at D2 (adj p values: D1, p = 0.940; D2, p = 0.002; D3, p = 0.526; Fig. 2A,B). In contrast with phalloidin, two-way ANOVA of tubulin intensity showed a main effect only of time (F(2118) = 14.74, p < 0.0001). Tubulin intensity decreased from D1 to D2 in both control and rapamycin-treated cells (adj p values: control D1 vs D2, p = 0.002; control D1 vs D3, p = 0.001; control D2 vs D3, p = 0.997; rapamycin D1 vs D2, p = 0.029; rapamycin D1 vs D2, p = 0.004; rapamycin D2 vs D3, p = 0.854; Fig. 2, but was unchanged by mTOR inhibition at any time point from D1 to D3 (adj p values: D1, p > 0.999; D2, p = 0.758; D3, p = 0.999; Fig. 2C). These data suggest that mTOR increases F-actin in early oligodendrocyte development by promoting actin polymerization and/or decreasing actin depolymerization.
Figure 2.
mTOR inhibition during differentiation reduces F-actin assembly in differentiating OPCs in vitro. A, Images show representative primary rat oligodendroglia stained with phalloidin from 1 to 3 d of differentiation plus or minus rapamycin. Scale bar, 20 µm. B, Quantification of phalloidin intensity in individual cells in control (black bars) and rapamycin (Rapa; gray bars) conditions from D1 to D3 of differentiation. n = 13–15 per group; #p = 0.036 control D1 versus D3 and p < 0.0001 control D2 versus D3; *p = 0.049 rapamycin D1 versus D3 and p = 0.045 rapamycin D2 versus D3; **p = 0.002 control versus rapamycin D2. C, Quantification of tubulin intensity in individual cells in control (black bars) and rapamycin (Rapa; gray bars) conditions from D1 to D3 of differentiation. n = 13-15 per group; #p = 0.002 control D1 versus D2; p = 0.001 control D1 versus D3; p = 0.029 rapamycin D1 versus D2; p = 0.004 rapamycin D1 versus D3. D, Representative Western blot and quantification of F/G actin ratio in control C, (black bars) and rapamycin-treated R, (Rapa; gray bars) cells from D1 to D2 of differentiation. n = 4, *p = 0.005 control versus rapamycin D2; All values are expressed as ± SEM.
We further analyzed actin dynamics by measuring cellular F/G actin ratios to determine whether the changes in phalloidin with mTOR inhibition were reflected in changes in cellular F/G actin. Rapamycin treatment reduced the F/G actin ratio at D2 of differentiation (D1: p = 0.643, t = 0.49, df = 6; D2: p = 0.005, t = 4.25, df = 6; Fig. 2D), indicating reduced actin polymerization. By D3 of differentiation, the F/G actin ratio was highly variable (data not shown) consistent with the rapid decrease in phalloidin from D2 to D3 (Fig. 2B). Together, these data support the conclusion that mTOR transiently induces F-actin linear polymerization that is critical for inducing early process outgrowth by increasing expression of profilin2 and decreasing active cofilin. mTOR signaling subsequently induces ARPC3 that is necessary for nucleating branched actin filaments. Our data also suggest that mTOR signaling is not a major regulator of tubulin expression although we cannot rule out an effect on microtubule branching or nucleation important for oligodendrocyte cellular process extension and myelination (Song et al., 2001; Fu et al., 2019; Lee and Hur, 2020).
Inhibiting mTOR reduces oligodendrocyte branching complexity
To directly determine whether mTOR inhibition impacts cellular branching during differentiation, we performed Sholl analyses on differentiating rat OPCs cultured with or without rapamycin in vitro (Fig. 3). We used tubulin immunostaining for measuring cellular branching complexity, because tubulin intensity was unchanged with mTOR inhibition (Figs. 2C, 3A). The number of Sholl intersections was equivalent between control and mTOR inhibited cells at D1 of differentiation (Fig. 3. By D2 the number of Sholl intersections was reduced in rapamycin-treated cells (Fig. 3A,C). The reduction in process complexity persisted through D3 and D5 and was particularly apparent in the regions proximal to the cell body (Fig. 3A,D,E). Quantification of the area under the curve of the Sholl intersection plots confirmed a significant reduction in complexity from D2 to D5 in mTOR inhibited cells (D1: p = 0.749, t = 0.32, df = 122.5; D2: p = 0.005, t = 2.88, df = 72.3; D3: p = 0.041, t = 2.07, df = 115.4; D5: p = 0.006, t = 2.81, df = 178.1; Fig. 3F). Interestingly, the Sholl analyses indicated that mTOR inhibition compromised cellular branching complexity in proximal regions but that linear extension of the longest processes was unaffected or even trended toward longer with rapamycin treatment at later time points (average radii length: D1 control = 53.9±13.2 µm, rapamycin = 52.5±13.9 µm, p = 0.835; D2 control = 61.7±13.7 µm, rapamycin = 50.0±13.1 µm, p = 0.131; D3 control = 44.6±17.4 µm, rapamycin = 58.8±17.9 µm, p = 0.062; D5 control = 53.8±26.0 µm, rapamycin = 60.5±18.9 µm, p = 0.491). These data suggest the possibility that another pathway may be compensating to promote linear actin polymerization in the mTOR inhibited cells. Studies in other cell types indicate there are multiple actin polymerization pathways; for example, there is evidence for a zyxin-dependent actin polymerization system that is independent of the ARP2/3/WASP polymerization pathway in HeLa cells (Fradelizi et al., 2001).
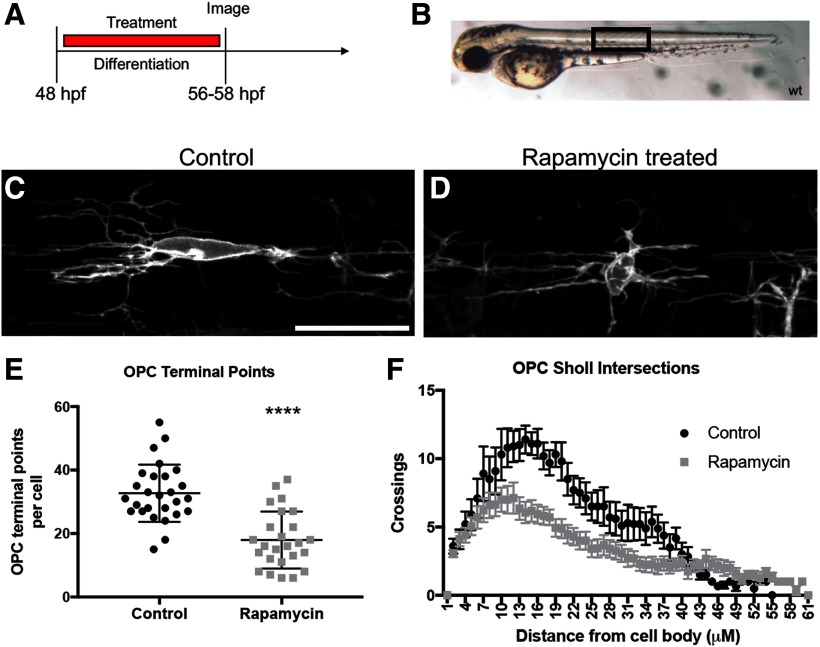
We next extended our analyses of the effect of mTOR on OPC branching complexity into zebrafish. The use of zebrafish for understanding cell developmental processes has been on the rise over the past decade. The quick developmental timeline and transparent exterior of zebrafish makes it an ideal in vivo model to investigate complex cellular morphology in real time. To further determine the function of mTOR in OPC branching morphology in an in vivo setting, we analyzed oligodendrocyte complexity in a membrane-tethered transgenic zebrafish line Tg(nkx2.2α:mEGFP) that labels OPCs with GFP (Fig. 4). Transgenic zebrafish embryos were treated with a vehicle control or with 5 μm of the mTOR inhibitor rapamycin from 48 to 56 hpf (Fig. 4A). This timeline targets dorsally migrating OPCs prior to differentiation and mimics our in vitro rat OPC differentiation paradigm (Fig. 1A). Analysis was completed above the yolk sac extension in the dorsal spinal cord of the zebrafish embryo (Fig. 4B). Zebrafish treated with 5 μm of rapamycin exhibited decreased numbers of OPC terminal points, indicating fewer OPC branches (p = 0.0001, t = 5.9, df = 50; Fig. 4C–E) as well as a decrease in Sholl intersections, indicating a decrease in overall OPC morphologic complexity (p = 0.0011, t = 3.694, df = 24; Fig. 4F). These data suggest that mTOR similarly regulates the actin cytoskeleton in rodent and zebrafish oligodendrocytes; however, no antibodies were available that detected profilin or ARPC3 in the zebrafish to quantify changes in the zebrafish proteins. Overall, these data support the conclusion that mTOR signaling is a positive regulator of OPC branching complexity and morphologic differentiation in both fish and rodents.
Figure 4.
mTOR inhibition decreases OPC morphologic complexity in zebrafish embryos. A, Individual OPCs were analyzed in Tg(nkx2.2α:mEGFP) zebrafish embryos treated with vehicle control or with 5 μm rapamycin from 48 to 56 hpf. B, Zebrafish were placed in live imaging media and mounted in low-melt agarose. All images are a lateral view taken above the yolk sac extension (black box). C, D, Control or rapamycin-treated cells displayed different morphologic complexities. Scale bar, 25 µm. E, F, Individual cells were analyzed using IMARIS software to quantify the number of OPC terminal points (E), a representation of the number of OPC branches, ****p < 0.0001, error bars are SD, or the number of Sholl intersections (F), error bars are SEM. Area under curve (arbitrary units): Control 298.2 ± 16.49, Rapamycin 200.8 ± 12.48, p = 0.0011.
mTOR signaling regulates expression of profilin2 and ARPC3 in vivo
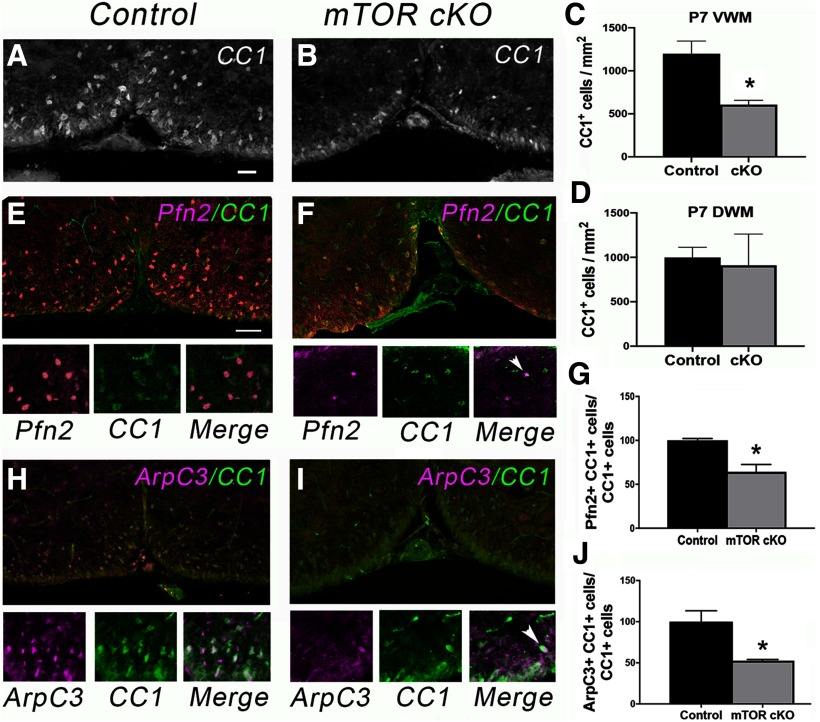
Loss of mTOR in developing OPCs in vivo results in a delay in initiation of myelination (i.e., reduced numbers of myelinated axons early in developmental myelination) as well as reduced myelin thickness in the spinal cord (Wahl et al., 2014). From these data and our in vitro findings, we proposed the hypothesis that the absence of mTOR in differentiating OPCs results in defects in process extension and initial axon contact. To test whether these defects involve alterations in cytoskeletal proteins similar to what we observed in vitro with mTOR inhibition, we determined levels of profilin2 and ARPC3 in CC1+ cells in developing mTOR cKO spinal cords. At P7, we observed a 50% reduction in the number of CC1+ cells in mTOR cKO spinal cords in VWM (p = 0.022, t = 3.27, df = 5; Fig. 5A–C), but a nonsignificant 10% reduction in DWM (p = 0.794, t = 0.28, df = 5; Fig. 5D). Regardless of region, the percentage of CC1+ cells that were profilin2+ was significantly reduced (p = 0.016, t = 3.54, df = 5; Fig. 5E–G). Similarly, we observed a significant reduction in the percentage of CC1+ cells that expressed ARPC3 in the mTOR cKO at P7 (p = 0.024, t = 3.54, df = 4; Fig. 5H–J). In contrast to profilin and ARPC3, there was no significant difference in CC1+ cells expressing p-cofilin at P7 (data not shown).
Figure 5.
Oligodendrocyte loss of mTOR reduces expression of profilin2 and ARPC3 at P7 in vivo. A, B, Representative images of immunostaining for CC1 in VWM during differentiation. Scale bar, 100 µm. C, D, Quantification of the number of CC1+ cells in VWM (C) or DWM (D) in control versus mTOR cKO spinal cords at P7; n = 3; *p = 0.022. E, F, Representative images of double staining for CC1 (green) with profilin 2 (Pfn2). G, Quantification of the percentage of CC1+ cells expressing profilin2, n = 3, *p = 0.016. H, I, Representative images of double staining for CC1 (green) with ARPC3 (pink). J, Quantification of the percentage of CC1+ cells expressing ARPC3; n = 3; *p = 0.024. I, Arrowhead indicates one double-positive cell in the field in contrast to H where most of the cells are double-positive. All values are expressed as ± SEM. Scale bars: (in E) E, F, H, I, 50 µm.
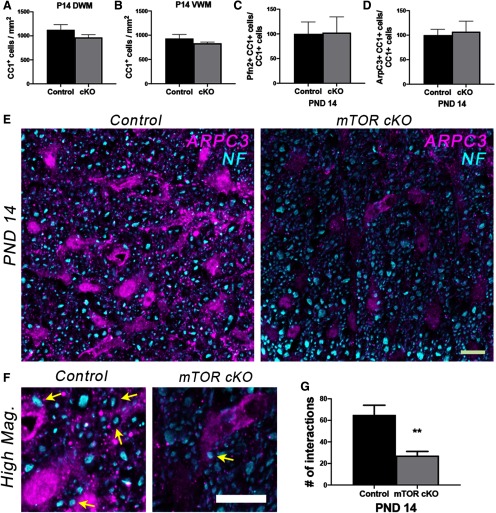
The number of CC1+ cells had largely recovered by P14 in the mTOR cKO VWM; we observed only a modest nonsignificant reduction in CC1+ cells in both DWM (14% decrease; p = 0.223, t = 1.39, df = 5; Fig. 6A) and VWM (11% decrease; p = 0.203, t = 1.34, df = 13; Fig. 6B at this time. Regardless of region, there were also no significant differences at P14 in the percentage of CC1+ cells that expressed profilin2 (p = 0.95, t = 0.05, df = 5; Fig. 6C), ARPC3 (p = 0.78, t = 0.28, df = 4; Fig. 6D) or p-cofilin (data not shown), indicating recovery in the number of morphologically differentiated cells. However, because the previous studies from Zuchero et al. (2015) showed the involvement of ARPC3 in axon ensheathment, we costained sections from P14 spinal cords for ARPC3 and NF. Confocal image analysis revealed that the number of interactions between ARPC3+ processes and NF+ axons was significantly reduced in the mTOR cKO spinal cord compared with controls (p = 0.004, t = 4.48, df = 6; Fig. 6E–G). These data support our previous observation that there are fewer myelinated axons present in mTOR cKO spinal cords at P14 (Wahl et al., 2014).
Figure 6.
Recovery in the number of morphologically differentiated cells but decreased ARPC3/NF interaction in mTOR cKO at P14 in vivo. A, B, Quantification of the number of CC1+ cells in DWM (A) or VWM (B) at P14 in control versus mTOR cKO spinal cords; n = 3–9. C, D, Percentage of CC1+ cells expressing profilin2 (C) or ARPC3 (D) at P14 in control versus mTOR cKO spinal cords; n = 3. E, Representative confocal maximum intensity images showing that despite the recovery of the number of CC1+ cells expressing ARPC3 by P14, there was decreased interaction between ARPC3+ (magenta) soma or processes with NF+ (cyan) in mTOR cKO VWM spinal cords. F, Higher-magnification confocal images from representative images in E, with yellow arrows pointing to ARPC3 and NF interactions. Scale bars: E, F, 10 µm. G, Quantification of the number of interactions between ARPC3+ soma or processes and NF+ axons in control and mTOR cKO spinal cords at P14 (see Materials and Methods); n = 3-5. Control = 65.0 ± 9.03, mTOR cKO = 27.2 ± 3.94, **p = 0.004. All values are expressed mean as ± SEM.
mTOR inhibition decreases Mbp mRNA expression and number of Plp-positive cells
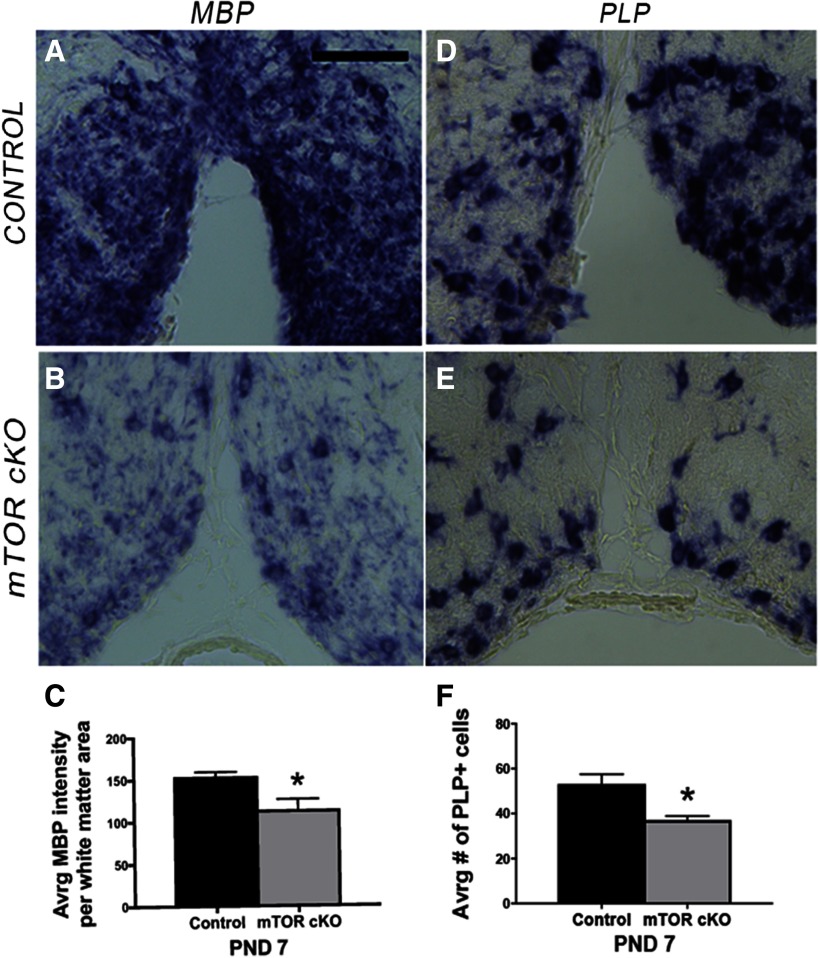
The results above collectively demonstrate that mTOR signaling regulates actin polymerization, cellular branching and axon ensheathment during early oligodendrocyte differentiation. To determine if mTOR also regulates the later process of myelin wrapping that is hypothesized to be modulated in part through expression and membrane localization of MBP, we initially performed in situ hybridization (ISH) to determine mRNA expression of the myelin proteins MBP and PLP in control and mTOR cKO spinal cords at P7 (Fig. 7). The intensity of Mbp mRNAs was reduced in the mTOR cKO spinal cords (p = 0.004, t =−5.08, df = 5; Fig. 7A–C), as well as the intensity of mRNA staining in the processes surrounding each cell body (Fig. 7A,B). However, because overall Mbp mRNA expression was reduced, it was not possible to accurately quantify expression in the processes versus cell bodies from these data. ISH for Plp mRNA revealed a reduction in number of Plp-positive cells (p = 0.002, t =−5.90, df = 5; Fig. 7D–F), consistent with the reduction in number of CC1+ cells in VWM at P7 (Fig. 5A–C).
Figure 7.
Loss of mTOR in oligodendrocytes decreases Mbp, and Plp mRNA expression in developing spinal cord oligodendrocytes in vivo. A, B, Representative images of ISH for Mbp RNA in ventral spinal cord sections from control (A) or mTOR cKO (B) mice at P7. Scale bar, 100 µm. C, Quantification of Mbp RNA expression in spinal cord white matter; WT, n = 3; cKO, n = 4. *p = 0.004. Error bars are SD. D, E, Representative images of ISH for Plp RNA in ventral spinal cord sections from control (D) or mTOR cKO (E) mice at P7. F, Quantification of number of Plp-positive cells in spinal cord white matter; WT, n = 3; mTOR cKO, n = 4. *p = 0.002. Error bars are SD.
Inhibiting mTOR signaling disrupts MBP localization and membrane expansion
To better analyze the effect of mTOR inhibition on myelin wrapping, we determined the extent of myelin membrane sheet expansion in primary OPCs differentiated for 8 d in vitro in the presence or absence of rapamycin (Fig. 8A). We observed a significant reduction in the area of MBP+ myelin membrane in rapamycin-treated cells (Fig. 8B–D; p = 0.007, t= 3.19, df = 12). We also confirmed the effect of mTOR inhibition on overall cell size by calculating cell area from the tubulin stained cells at D5 used for previous analyses (Fig. 3). These analyses demonstrated that rapamycin decreased cell size by more than twofold at D5 (p = 0.004, t = 3.142, df = 27; Fig. 8E). Even more striking was the accumulation of MBP protein in the cytoplasm and proximal processes in rapamycin-treated cells that was not observed in control cells (Fig. 8B,C,F; p = 0.0007, t = 4.29, df = 14). In addition, the cell bodies were significantly smaller with mTOR inhibition (p = 0.028, t = 2.44, df = 14; Fig. 8G) confirming the contribution of mTOR signaling to cell size documented in other cell types (Edinger and Thompson, 2002; Fingar et al., 2002; Kim et al., 2002; Fingar and Blenis, 2004; Lloyd, 2013).
We next examined MBP intensity in white matter in mTOR cKO and control mice at P7. As expected based on the reduction in CC1+ cells at this age in VWM, there was a significant reduction in MBP intensity (Fig. 8H–J; p = 0.030, t = 2.97, df = 5) and a decrease in white matter area in mTOR cKO mice (p = 0.013, t = 3.76, df = 5; Fig. 8K). However, despite the recovery in cytoskeletal targets and in CC1+ cells to 90% normal numbers at P14, the extent of MBP expression surrounding axons was reduced at P14 (Fig. 8L–O).
mTOR promotes Kif1b expression in oligodendrocytes in the spinal cord
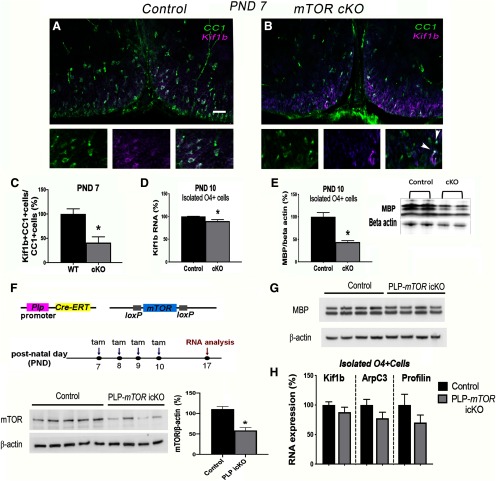
The cytoplasmic MBP accumulation with mTOR inhibition in vitro suggests a possible defect in Mbp mRNA transport to the processes where it is normally translated (Colman et al., 1982; Ainger et al., 1993). In zebrafish oligodendrocytes, Lyons et al. (2009) demonstrated that the kinesin family member 1B (KIF1B) transports Mbp mRNAs in a vesicle from the cytoplasm to the periphery where it is locally translated, and lack of KIF1B leads to Mbp mRNA translation in the cell body. We therefore investigated whether KIF1B is downstream of mTOR signaling in rodent oligodendrocytes. To determine whether mTOR deletion in differentiating oligodendrocytes in vivo impairs KIF1B expression, we analyzed KIF1B levels in CC1+ cells at P7 in mTOR cKO spinal cords. These analyses revealed a significant decrease in the percentage of CC1+ cells that express KIF1B (Fig. 9A–C; p = 0.010, t = 3.68, df = 6). By P14, the number of CC1+ cells expressing KIF1B through immunofluorescent staining was similar in the control and mTOR cKO spinal cords (data not shown) consistent with the prior data on recovery of cytoskeletal polymerization factors by P14 (Fig. 6). However, we observed reduced Kif1b RNA expression (p = 0.028, t = 3.34, df = 4; Fig. 9D) as well as decreased MBP protein expression (p = 0.004, t = 4.91, df = 5; Fig. 9E) in O4-positive cells acutely isolated from spinal cords of mTOR cKO versus WT mice at P10.
Figure 9.
mTOR regulates KIF1B expression in differentiating oligodendrocytes in vivo. A, B, Representative micrographs showing immunostaining for CC1 (green) and KIF1B (magenta) in control (A) or mTOR cKO (B) spinal cords at P7. Scale bar, 50 µm. Note, most CC1+ cells are also KIF1B+ in control section; arrowheads indicate only two double-positive cells in the mTOR cKO higher-power image. C, Quantification of the percentage of CC1+ cells expressing CC1 and KIF1B at P10; n = 4, *p = 0.010. D, QPCR for Kif1b mRNA expression in isolated O4+ cells from control and mTOR cKO spinal cords at P10; n = 7 control, n = 6 cKO, p = 0.028. E, Representative Western blot and quantification of MBP protein expression in isolated O4+ cells from control and mTOR cKO spinal cords at P10; n= 4 control, n = 3 cKO, *p = 0.004. F, Experimental paradigm for tamoxifen injections and sample collection for RNA analysis in PLP-mTOR icKO mouse model [tamoxifen (tam)]. mTOR protein is significantly reduced in isolated O4+ cells from PLP-mTOR icKO versus controls at P17, 10 d after the last tamoxifen injection, *p = 0.001. G, MBP protein is unchanged in isolated O4+ cells from PLP-mTOR icKO versus controls at P17. H, qPCR for Kif1b, Arpc3, and profilin2 in isolated O4+ cells shows no significant differences in mRNA expression between control and PLP-mTOR icKO at P17, n = 4 control, n = 5 PLP-mTOR icKO, p = 0.2938 for Kif1b, p = 0.1604 for Arpc3, and p = 0.2114 for profiin2. All values are expressed as ± SEM.
Previous observations from our laboratory showed stage-dependent effects of Tsc1 deletion on remyelination (McLane et al., 2017). TSC1 and TSC2 form the tuberous sclerosis complex that serves as an upstream negative regulator of mTORC1. Loss of Tsc1 from adult OPCs using inducible NG2-Cre deletion in mice improved remyelination efficiency in spinal cord in the lysolecithin model, whereas inducible deletion of Tsc1 in PLP+ pre-myelinating oligodendrocytes during remyelination led to decreased efficiency of remyelination in this model (McLane et al., 2017). Based on these and other published data showing the complex role of TSC/mTOR signaling depends on the specific developmental time of recombination (Y. Zou et al., 2014; Figlia et al., 2017), we tested how Mtor deletion in PLP+ oligodendrocytes affected targets we observed altered in early oligodendroglia in the CNP-Cre/mTOR cKO mouse model. We thus established a PLP-mTOR icKO mouse line using a PLP-CreERT line (Doerflinger et al., 2003) and induced recombination with tamoxifen from P7 to P10 to target the majority of myelinating oligodendrocytes (Fig. 9F). In our prior studies using the PLP-CreERT line to delete Tsc1, we observed that ∼ 60% of oligodendroglia showed recombination after tamoxifen injections resulting in a significant reduction in TSC1 protein in O4+ cells (McLane et al., 2017). To examine mTOR expression in the PLP-Mtor icKO line, we isolated O4+ cells from spinal cords of mice at P17 (n = 4–5 per genotype) 7 d after the last tamoxifen injection. MTOR protein expression was significantly reduced in O4+ cells isolated from PLP-mTOR icKO mice compared with Cre-negative O4+ cells (p = 0.001, t = 5.375, df = 7; Fig. 9F). However, in contrast to the CNP-Cre-driven deletion of Mtor, which occurs in the early progenitor stage, deletion of Mtor in PLP+ differentiated oligodendrocytes had no impact on MBP protein expression at P17 (Fig. 9G) or on Kif1b mRNA expression in O4+ cells at P17 (p = 0.2938, t = 1.135, df = 7; Fig. 9H). Similarly, the mRNA levels of Arpc3 and Pfn2 were unchanged between control and PLP-mTOR icKO O4+ cells at P17 (p = 0.1604, t = 1.570, df = 7 for Arpc3 and p = 0.2114, t = 1.375, df = 7 for Pfn2). Together, these data suggest that mTOR positively regulates cytoskeletal, Kif1b and MBP expression in early oligodendroglia in the spinal cord during initiation of myelination but is no longer required for expression of these targets in myelin-producing oligodendrocytes at a later stage of maturation.
mTOR is necessary for KIF1B expression and Mbp mRNA localization in differentiating OPCs
To confirm the downregulation of KIF1B after mTOR loss in OPCs in vitro, we used the same experimental paradigm shown in Figure 1A to analyze levels of KIF1B at D1–D3 of differentiation plus/minus rapamycin treatment to inhibit mTOR in primary OPCs. KIF1B expression was significantly reduced at D2 and D3 in the presence of rapamycin (D1: p= 0.633, t= 0.51, df= 4; D2: p= 0.041, t= 2.96, df= 4; D3: p = 0.007, t= 5.01, df = 4; Fig. 10A). These data suggest an explanation for mislocalized MBP protein in rapamycin-treated oligodendrocytes (Fig. 8).
Our recently published study demonstrated that mTOR signaling normally downregulates the bone morphogenetic pathway to promote the transition from the early OPC to immature oligodendrocyte (Ornelas et al., 2020). To determine whether mTOR signaling is required for KIF1B maintenance following the initial transition of the early bipolar OPCs to late progenitor cells, we used a modified in vitro differentiation paradigm where we allowed OPCs to differentiate normally for 48 h prior to inhibiting mTOR with rapamycin (Tyler et al., 2009; Fig. 10B). We then analyzed KIF1B expression at 24, 48, and 72 h after the start of treatment (equivalent to 3–5 d of differentiation). KIF1B protein was reduced within 2 d of exposure to rapamycin, and levels remained low with continued rapamycin treatment in the delayed mTOR inhibition paradigm (D1: p = 0.175, t = 1.48, df = 8; D2: p = 0.006, t = 3.62, df = 8; D3: p = 0.011, t = 3.23, df = 8; Fig. 10C). These data suggest that mTOR promotes normal expression of KIF1B in late progenitors/pre-myelinating oligodendrocytes even after its expression is initiated.
Similar to our prior observations where mTOR was inhibited from the start of differentiation, inhibiting mTOR for 48 h after cells were allowed to differentiate normally for 48 h resulted in MBP protein concentrated in the cell body (p = 0.0008, t = 7.25, df = 5; Fig. 10D) and decreased MBP protein expression (D1: p = 0.007, t = 5.03, df = 4; D2: p = 0.001, t = 7.84, df = 4; D3: p = 0.032, t = 3.22, df = 4; Fig. 10E).
Together, the mislocalization of MBP protein and downregulation of KIF1B with loss or inhibition of mTOR suggests an effect on Mbp mRNA transport. To directly determine if mTOR regulates Mbp mRNA localization in vitro, we performed single molecule FISH for Mbp mRNA on primary rat OPCs undergoing differentiation in the presence or absence of rapamycin treatment from the start of differentiation. Inhibiting mTOR resulted in reduced Mbp mRNA localization to the cellular processes of OPCs with a corresponding accumulation in the cytoplasm after 7 d of differentiation (nuclear: adj p < 0.001, t = 10.13, df = 77.78; cytoplasm: adj p < 0.001, t = 16.08, df = 77.13; processes: adj p < 0.001, t = 20.63, df = 96.23; Fig. 10F,G). These data support the conclusion that Mbp mRNA transport is regulated by mTOR in differentiating OPCs.
Discussion
In this study we defined a function for mTOR signaling in the regulation of actin binding proteins necessary for reorganization of oligodendrocyte cytoskeleton during development and show that mTOR promotes cellular branching complexity and axon ensheathment. We further demonstrate that mTOR regulates MBP expression as well as proper localization in the oligodendrocyte processes due to promoting Mbp mRNA transport likely, in part, via KIF1B. We distinguish the requirements for mTOR in proper morphologic differentiation and ultimately, myelination, into two phases, as discussed below.
mTOR induces ARPC3 and profilin2 that are important for phase I-dependent actin polymerization
Consistent with previous data showing that ARPC3 is important for cellular branching (Pollard and Borisy, 2003; Yae et al., 2006; Rouiller et al., 2008) and in oligodendrocytes for axon ensheathment (Zuchero et al., 2015), we observed reduced expression of ARPC3 in the mTOR cKO at P7; a time point when oligodendrocytes are differentiating and initiating myelination. Although the number of CC1+ cells expressing ARPC3 recovered at P14, we observed a significant decrease in ARPC3+ processes interacting with axons in mTOR cKO spinal cords. These findings provide the basis for the delay in initiation of myelination that we described previously in the mTOR cKO spinal cords (Wahl et al., 2014). However, unlike the results from complete deletion of Arpc3 reported by Zuchero et al. (2015), the number of myelinated axons recovered in the mTOR cKO spinal cords likely because of the partial and transient reduction in ARPC3 in CC1+ cells. We observed similar results with differentiating primary OPCs treated with rapamycin in vitro expressing reduced levels of ARPC3. The earlier response of profilin2 to mTOR inhibition in vitro suggests the importance of mTOR signaling in regulating actin polymerization and initial cellular process extension. Importantly, profilin is still required to facilitate addition of actin-GTP to the growing daughter actin filament even after actin branching is initiated by ARPC3 nucleation activity. Profilin is the major protein responsible for polymerization of actin; however, its regulation in oligodendrocytes leading to cytoskeletal reorganization was unknown. Similar to ARPC3, we found a decrease in the expression of profilin2 during oligodendrocyte differentiation in vivo with loss of mTOR in oligodendrocytes. The low levels of profilin2 during peak differentiation in the mTOR cKO likely results in a lack of adequate cellular processes during oligodendrocyte development. These data are supported by our in vitro findings showing low expression of profilin2 following rapamycin treatment of primary OPCs during differentiation. In addition, these data support and provide an explanation for our previously published studies showing impaired process extension with rapamycin treatment of rat OPCs during differentiation in vitro (Tyler et al., 2009) and deficits in OPC branching and morphologic complexity in vivo in fish treated with rapamycin (Fig. 4).
mTOR regulates proper MBP expression and localization necessary for phase II dependent actin depolymerization
Prior studies have shown that Mbp mRNA is transported from the nucleus to oligodendrocyte processes where it is locally translated (Colman et al., 1982) and that this transport requires microtubules, kinesin, and dynein/dynactin proteins that regulate anterograde and retrograde transport along the microtubules, respectively, and RNA granules containing heterogeneous nuclear ribonuclear protein A2 (Carson et al., 1997; Song et al., 2003; Lyons et al., 2009; White et al., 2012; Yang et al., 2015; Herbert et al., 2017). KIF1B is a specific kinesin shown to regulate the transport of Mbp mRNA to oligodendrocyte processes (Lyons et al., 2009). We found reduced expression of MBP as well as oligodendrocyte expression of KIF1B in the mTOR cKO spinal cords at P7 in vivo. To determine whether the lack of myelin in our mTOR cKO animals is a result of both lack of MBP protein expression and mislocalization of MBP, we determined localization of MBP protein in vitro in differentiating OPCs with or without mTOR inhibition. In mTOR-inhibited cells, MBP protein accumulated within the cytoplasm and main processes whereas in control cells, MBP protein expression appeared evenly distributed throughout the cell body, main branches and processes. In addition, mTOR inhibition in rat OPCs in culture or oligodendrocyte deletion of mTOR in vivo reduced expression of KIF1B. Our FISH data showing deficits in Mbp mRNA localization in rapamycin-treated oligodendrocytes confirm our conclusion that, in the absence of mTOR signaling, Mbp mRNA is not properly transported to the processes and thus is translated in the cell body. This is likely contributed to by the low expression of KIF1B with mTOR inhibition; however, our recent RNAseq data revealed reductions in several subunit proteins that form dyneins and kinesins that may also contribute to the Mbp RNA transport deficits with loss of mTOR function (Luipa Khandker, Marisa A. Jeffries, Yun-juan Chang, Marie Mather, Jennifer N. Bourne, Wendy B. Macklin, Teresa L. Wood, unpublished observations). Moreover, our prior study identified Fyn kinase in the mTOR regulated proteome in differentiating oligodendrocytes (Tyler et al., 2011) potentially linking mTOR to the Fyn regulation of hnRNPs involved in Mbp mRNA transport and translation (White et al., 2008, 2012).
A current model for myelin wrapping is based on MBP protein in the cell processes causing displacement of the actin severing proteins cofilin and gelsolin from PIP2 binding resulting in their activation to promote actin depolymerization necessary for myelin membrane wrapping (Zuchero et al., 2015). Thus, the low levels of MBP in the processes during myelination with loss of mTOR is expected to disrupt myelin wrapping. Interestingly, along with depolymerization, F-actin at the leading edge during myelin wrapping also is important for myelin wrapping (Nawaz et al., 2015). Thus, the function of mTOR in regulating actin polymerization and actin filament branching as well as expression and localization of MBP explain the observed deficits in myelin thickness in the mTOR cKO (Wahl et al., 2014). That the myelin stays thinner even into the adult spinal cord in the absence of mTOR suggests either that there is a critical developmental window for the wrapping phase during myelination and/or that the persistent deficit in MBP observed even at 8 weeks of age in the mTOR cKO prevents recovery to normal myelin thickness although the number of myelinated axons eventually recovers (Wahl et al., 2014). Multiple studies demonstrate that inhibiting mTOR in zebrafish reduces myelin sheath number and length, depending on the timing of inhibition (Mathews and Appel, 2016; Preston et al., 2019). Together with our current data demonstrating that mTOR regulates the actin cytoskeleton, this suggests mTOR may determine myelin production by modulating actin cytoskeletal regulators. Overall, our findings demonstrate critical roles for mTOR in regulating both phases of cytoskeletal reorganization necessary for axon ensheathment and myelination.
Footnotes
This work was supported by National Institute of Neurological Disorders and Stroke NS082203 to W.B.M. and T.L.W., National Multiple Sclerosis Society RG5371-A-4 to T.L.W. and NIH Director's Early Independence Award DP5 OD012160 to M.B. We thank Luke Fritzky for technical assistance and Luipa Khandker for critical reading of the paper.
The authors declare no competing financial interests.
References
- Abu-Rub M, Miller RH (2018) Emerging cellular and molecular strategies for enhancing central nervous system (CNS) remyelination. Brain Sci 8:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH (1993) Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol 123:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P (1998) Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393:805–809. 10.1038/31729 [DOI] [PubMed] [Google Scholar]
- Batish M, Raj A, Tyagi S (2011) Single molecule imaging of RNA in situ. Methods Mol Biol 714:3–13. 10.1007/978-1-61779-005-8_1 [DOI] [PubMed] [Google Scholar]
- Batish M, van den Bogaard P, Kramer FR, Tyagi S (2012) Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci U S A 109:4645–4650. 10.1073/pnas.1111226109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercury KK, Dai J, Sachs HH, Ahrendsen JT, Wood TL, Macklin WB (2014) Conditional ablation of raptor or rictor has differential impact on oligodendrocyte differentiation and CNS myelination. J Neurosci 34:4466–4480. 10.1523/JNEUROSCI.4314-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TL, Macklin WB (2020) The actin cytoskeleton in myelinating cells. Neurochem Res 45:684–693. 10.1007/s11064-019-02753-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr TD, DiGiovanni J, Lynch CJ, Shantz LM (2012) Inhibition of mTOR suppresses UVB-induced keratinocyte proliferation and survival. Cancer Prev Res (Phila) 5:1394–1404. 10.1158/1940-6207.CAPR-12-0272-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JH, Worboys K, Ainger K, Barbarese E (1997) Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil Cytoskeleton 38:318–328. [DOI] [PubMed] [Google Scholar]
- Colman DR, Kreibich G, Frey AB, Sabatini DD (1982) Synthesis and incorporation of myelin polypeptides into CNS myelin. J Cell Biol 95:598–608. 10.1083/jcb.95.2.598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Bercury KK, Macklin WB (2014) Interaction of mTOR and Erk1/2 signaling to regulate oligodendrocyte differentiation. Glia 62:2096–2109. 10.1002/glia.22729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger NH, Macklin WB, Popko B (2003) Inducible site-specific recombination in myelinating cells. Genesis 35:63–72. 10.1002/gene.10154 [DOI] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB (2002) Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell 13:2276–2288. 10.1091/mbc.01-12-0584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A (2002) Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol 4:260–269. 10.1038/ncb770 [DOI] [PubMed] [Google Scholar]
- Figlia G, Norrmen C, Pereira JA, Gerber D, Suter U (2017) Dual function of the PI3K-Akt-mTORC1 axis in myelination of the peripheral nervous system. eLife 6:e29241 10.7554/eLife.29241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar DC, Blenis J (2004) Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23:3151–3171. 10.1038/sj.onc.1207542 [DOI] [PubMed] [Google Scholar]
- Fingar DC, Salama S, Tsou C, Harlow E, Blenis J (2002) Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 16:1472–1487. 10.1101/gad.995802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MA, Afshari FS, Alexander JK, Colello RJ, Fuss B (2006) Growth conelike sensorimotor structures are characteristic features of postmigratory, premyelinating oligodendrocytes. Glia 53:563–566. 10.1002/glia.20293 [DOI] [PubMed] [Google Scholar]
- Fradelizi J, Noireaux V, Plastino J, Menichi B, Louvard D, Sykes C, Golsteyn RM, Friederich E (2001) ActA and human zyxin harbour Arp2/3-independent actin-polymerization activity. Nat Cell Biol 3:699–707. 10.1038/35087009 [DOI] [PubMed] [Google Scholar]
- Franklin RJM, Ffrench-Constant C (2017) Regenerating CNS myelin: from mechanisms to experimental medicines. Nat Rev Neurosci 18:753–769. 10.1038/nrn.2017.136 [DOI] [PubMed] [Google Scholar]
- Fu MM, McAlear TS, Nguyen H, Oses-Prieto JA, Valenzuela A, Shi RD, Perrino JJ, Huang TT, Burlingame AL, Bechstedt S, Barres BA (2019) The Golgi outpost protein TPPP nucleates microtubules and is critical for myelination. Cell 179:132–146.e14. 10.1016/j.cell.2019.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusho M, Ishii A, Bansal R (2017) Signaling by FGF receptor 2, not FGF receptor 1, regulates myelin thickness through activation of ERK1/2-MAPK, which promotes mTORC1 activity in an Akt-independent manner. J Neurosci 37:2931–2946. 10.1523/JNEUROSCI.3316-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould EA, Busquet N, Shepherd D, Dietz RM, Herson PS, Simoes de Souza FM, Li A, George NM, Restrepo D, Macklin WB (2018) Mild myelin disruption elicits early alteration in behavior and proliferation in the subventricular zone. eLife 7:e34783 10.7554/eLife.34783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow DE, Saul KE, Culp CM, Vesely EM, Macklin WB (2014) Expression of proteolipid protein gene in spinal cord stem cells and early oligodendrocyte progenitor cells is dispensable for normal cell migration and myelination. J Neurosci 34:1333–1343. 10.1523/JNEUROSCI.2477-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Ishino Y, Jiang W, Yoshimura T, Takeda-Uchimura Y, Uchimura K, Kadomatsu K, Ikenaka K (2016) Keratan sulfate regulates the switch from motor neuron to oligodendrocyte generation during development of the mouse spinal cord. Neurochem Res 41:450–462. 10.1007/s11064-016-1861-9 [DOI] [PubMed] [Google Scholar]
- He Y, Li D, Cook SL, Yoon MS, Kapoor A, Rao CV, Kenis PJ, Chen J, Wang F (2013) Mammalian target of rapamycin and Rictor control neutrophil chemotaxis by regulating Rac/Cdc42 activity and the actin cytoskeleton. Mol Biol Cell 24:3369–3380. 10.1091/mbc.EXIII-07-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert AL, Fu MM, Drerup CM, Gray RS, Harty BL, Ackerman SD, O'Reilly-Pol T, Johnson SL, Nechiporuk AV, Barres BA, Monk KR (2017) Dynein/dynactin is necessary for anterograde transport of Mbp mRNA in oligodendrocytes and for myelination in vivo. Proc Natl Acad Sci U S A 114:E9153–E9162. 10.1073/pnas.1711088114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A, Fyffe-Maricich SL, Furusho M, Miller RH, Bansal R (2012) ERK1/ERK2 MAPK signaling is required to increase myelin thickness independent of oligodendrocyte differentiation and initiation of myelination. J Neurosci 32:8855–8864. 10.1523/JNEUROSCI.0137-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6:1122–1128. 10.1038/ncb1183 [DOI] [PubMed] [Google Scholar]
- Kanellos G, Frame MC (2016) Cellular functions of the ADF/cofilin family at a glance. J Cell Sci 129:3211–3218. 10.1242/jcs.187849 [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–175. 10.1016/S0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD (2006) Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124:423–435. 10.1016/j.cell.2005.11.038 [DOI] [PubMed] [Google Scholar]
- Lang CH, Frost RA, Bronson SK, Lynch CJ, Vary TC (2010) Skeletal muscle protein balance in mTOR heterozygous mice in response to inflammation and leucine. Am J Physiol Endocrinol Metab 298:E1283–E1294. 10.1152/ajpendo.00676.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang SA, Hackl C, Moser C, Fichtner-Feigl S, Koehl GE, Schlitt HJ, Geissler EK, Stoeltzing O (2010) Implication of RICTOR in the mTOR inhibitor-mediated induction of insulin-like growth factor-I receptor (IGF-IR) and human epidermal growth factor receptor-2 (Her2) expression in gastrointestinal cancer cells. Biochim Biophys Acta 1803:435–442. 10.1016/j.bbamcr.2010.01.009 [DOI] [PubMed] [Google Scholar]
- Lee BY, Hur EM (2020) A role of microtubules in oligodendrocyte differentiation. Int J Mol Sci 21:1062 10.3390/ijms21031062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Chen L, Chung J, Huang S (2008) Rapamycin inhibits F-actin reorganization and phosphorylation of focal adhesion proteins. Oncogene 27:4998–5010. 10.1038/onc.2008.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AC. (2013) The regulation of cell size. Cell 154:1194–1205. 10.1016/j.cell.2013.08.053 [DOI] [PubMed] [Google Scholar]
- Lyons DA, Naylor SG, Scholze A, Talbot WS (2009) Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat Genet 41:854–858. 10.1038/ng.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markey FB, Ruezinsky W, Tyagi S, Batish M (2014) Fusion FISH imaging: single-molecule detection of gene fusion transcripts in situ. PloS one 9:e93488. 10.1371/journal.pone.0093488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews ES, Appel B (2016) Cholesterol biosynthesis supports myelin gene expression and axon ensheathment through modulation of P13K/Akt/mTor signaling. J Neurosci 36:7628–7639. 10.1523/JNEUROSCI.0726-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K, Vellis J (1980) Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol 85:890–902. 10.1083/jcb.85.3.890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane LE, Bourne JN, Evangelou AV, Khandker L, Macklin WB, Wood TL (2017) Loss of tuberous sclerosis complex1 in adult oligodendrocyte progenitor cells enhances axon remyelination and increases myelin thickness after a focal demyelination. J Neurosci 37:7534–7546. 10.1523/JNEUROSCI.3454-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski JP, Kothary R (2015) Oligodendrocytes in a nutshell. Front Cell Neurosci 9:340. 10.3389/fncel.2015.00340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K. (2013) Signaling mechanisms and functional roles of cofilin phosphorylation and dephosphorylation. Cell Signal 25:457–469. 10.1016/j.cellsig.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Nawaz S, Sánchez P, Schmitt S, Snaidero N, Mitkovski M, Velte C, Bruckner BR, Alexopoulos I, Czopka T, Jung SY, Rhee JS, Janshoff A, Witke W, Schaap IA, Lyons DA, Simons M (2015) Actin filament turnover drives leading edge growth during myelin sheath formation in the central nervous system. Dev Cell 34:139–151. 10.1016/j.devcel.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornelas IM, Khandker L, Wahl SE, Hashimoto H, Macklin WB, Wood TL (2020) The mechanistic target of rapamycin pathway downregulates bone morphogenetic protein signaling to promote oligodendrocyte differentiation. Glia. Advance online publication. Retrieved January 6, 2020. doi: 10.1002/glia.23776 10.1002/glia.23776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112:453–465. 10.1016/s0092-8674(03)00120-x [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD (2001) Actin dynamics. J Cell Sci 114:3–4. [DOI] [PubMed] [Google Scholar]
- Preston MA, Finseth LT, Bourne JN, Macklin WB (2019) A novel myelin protein zero transgenic zebrafish designed for rapid readout of in vivo myelination. Glia 67:650–667. 10.1002/glia.23559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF (2004) Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119:419–429. 10.1016/j.cell.2004.09.039 [DOI] [PubMed] [Google Scholar]
- Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D (2008) The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol 180:887–895. 10.1083/jcb.200709092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14:1296–1302. 10.1016/j.cub.2004.06.054 [DOI] [PubMed] [Google Scholar]
- Snaidero N, Simons M (2017) The logistics of myelin biogenesis in the central nervous system. Glia 65:1021–1031. 10.1002/glia.23116 [DOI] [PubMed] [Google Scholar]
- Song J, Goetz BD, Baas PW, Duncan ID (2001) Cytoskeletal reorganization during the formation of oligodendrocyte processes and branches. Mol Cell Neurosci 17:624–636. 10.1006/mcne.2001.0974 [DOI] [PubMed] [Google Scholar]
- Song J, Carson JH, Barbarese E, Li FY, Duncan ID (2003) RNA transport in oligodendrocytes from the taiep mutant rat. Mol Cell Neurosci 24:926–938. 10.1016/s1044-7431(03)00254-9 [DOI] [PubMed] [Google Scholar]
- Sperber BR, Boyle-Walsh EA, Engleka MJ, Gadue P, Peterson AC, Stein PL, Scherer SS, McMorris FA (2001) A unique role for Fyn in CNS myelination. J Neurosci 21:2039–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR (2015) Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev Cell 32:43–53. 10.1016/j.devcel.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason EJ, Escalante M, Osterhout DJ, Fuss B (2019) The oligodendrocyte growth cone and its actin cytoskeleton: a fundamental element for progenitor cell migration and CNS myelination. Glia. Advance online publication. Retrieved from November 7, 2019. doi: 10.1002/glia.23735 10.1002/glia.23735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler WA, Gangoli N, Gokina P, Kim HA, Covey M, Levison SW, Wood TL (2009) Activation of the mammalian target of rapamycin (mTOR) is essential for oligodendrocyte differentiation. J Neurosci 29:6367–6378. 10.1523/JNEUROSCI.0234-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler WA, Jain MR, Cifelli SE, Li Q, Ku L, Feng Y, Li H, Wood TL (2011) Proteomic identification of novel targets regulated by the mammalian target of rapamycin pathway during oligodendrocyte differentiation. Glia 59:1754–1769. 10.1002/glia.21221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl SE, McLane LE, Bercury KK, Macklin WB, Wood TL (2014) Mammalian target of rapamycin promotes oligodendrocyte differentiation, initiation and extent of CNS myelination. J Neurosci 34:4453–4465. 10.1523/JNEUROSCI.4311-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R, Gonsior C, Krämer-Albers EM, Stöhr N, Hüttelmaier S, Trotter J (2008) Activation of oligodendroglial Fyn kinase enhances translation of mRNAs transported in hnRNP A2-dependent RNA granules. J Cell Biol 181:579–586. 10.1083/jcb.200706164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R, Gonsior C, Bauer NM, Krämer-Albers EM, Luhmann HJ, Trotter J (2012) Heterogeneous nuclear ribonucleoprotein (hnRNP) F is a novel component of oligodendroglial RNA transport granules contributing to regulation of myelin basic protein (MBP) synthesis. J Biol Chem 287:1742–1754. 10.1074/jbc.M111.235010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder SJ, Ayscough KR (2005) Actin-binding proteins. J Cell Sci 118:651–654. 10.1242/jcs.01670 [DOI] [PubMed] [Google Scholar]
- Witke W, Podtelejnikov AV, Di Nardo A, Sutherland JD, Gurniak CB, Dotti C, Mann M (1998) In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J 17:967–976. 10.1093/emboj/17.4.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yae K, Keng VW, Koike M, Yusa K, Kouno M, Uno Y, Kondoh G, Gotow T, Uchiyama Y, Horie K, Takeda J (2006) Sleeping beauty transposon-based phenotypic analysis of mice: lack of Arpc3 results in defective trophoblast outgrowth. Mol Cell Biol 26:6185–6196. 10.1128/MCB.00018-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ML, Shin J, Kearns CA, Langworthy MM, Snell H, Walker MB, Appel B (2015) CNS myelination requires cytoplasmic dynein function. Dev Dyn 244:134–145. 10.1002/dvdy.24238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhou L, Du XX, Ji Y, Xu J, Tian J, Jiang W, Zou Y, Yu S, Gan L, Luo M, Yang Q, Cui Y, Yang W, Xia X, Chen M, Zhao X, Shen Y, Chen PY, Worley PF, Xiao B (2011) Rheb1 is required for mTORC1 and myelination in postnatal brain development. Dev Cell 20:97–108. 10.1016/j.devcel.2010.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Jiang W, Wang J, Li Z, Zhang J, Bu J, Zou J, Zhou L, Yu S, Cui Y, Yang W, Luo L, Lu QR, Liu Y, Chen M, Worley PF, Xiao B (2014) Oligodendrocyte precursor cell-intrinsic effect of Rheb1 controls differentiation and mediates mTORC1-dependent myelination in brain. J Neurosci 34:15764–15778. 10.1523/JNEUROSCI.2267-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Fu MM, Sloan SA, Ibrahim A, Olson A, Zaremba A, Dugas JC, Wienbar S, Caprariello AV, Kantor C, Leonoudakis D, Leonoudakus D, Lariosa-Willingham K, Kronenberg G, Gertz K, Soderling SH, Miller RH, Barres BA (2015) CNS myelin wrapping is driven by actin disassembly. Dev Cell 34:152–167. 10.1016/j.devcel.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]