Abstract
Hepatocyte growth factor (HGF) is a multifunctional protein that signals through the MET receptor. HGF stimulates cell proliferation, cell dispersion, neuronal survival, and wound healing. In the inner ear, levels of HGF must be fine-tuned for normal hearing. In mice, a deficiency of HGF expression limited to the auditory system, or an overexpression of HGF, causes neurosensory deafness. In humans, noncoding variants in HGF are associated with nonsyndromic deafness DFNB39. However, the mechanism by which these noncoding variants causes deafness was unknown. Here, we reveal the cause of this deafness using a mouse model engineered with a noncoding intronic 10 bp deletion (del10) in Hgf. Male and female mice homozygous for del10 exhibit moderate-to-profound hearing loss at 4 weeks of age as measured by tone burst auditory brainstem responses. The wild type (WT) 80 mV endocochlear potential was significantly reduced in homozygous del10 mice compared with WT littermates. In normal cochlea, endocochlear potentials are dependent on ion homeostasis mediated by the stria vascularis (SV). Previous studies showed that developmental incorporation of neural crest cells into the SV depends on signaling from HGF/MET. We show by immunohistochemistry that, in del10 homozygotes, neural crest cells fail to infiltrate the developing SV intermediate layer. Phenotyping and RNAseq analyses reveal no other significant abnormalities in other tissues. We conclude that, in the inner ear, the noncoding del10 mutation in Hgf leads to developmental defects of the SV and consequently dysfunctional ion homeostasis and a reduction in the EP, recapitulating human DFNB39 nonsyndromic deafness.
SIGNIFICANCE STATEMENT Hereditary deafness is a common, clinically and genetically heterogeneous neurosensory disorder. Previously, we reported that human deafness DFNB39 is associated with noncoding variants in the 3′UTR of a short isoform of HGF encoding hepatocyte growth factor. For normal hearing, HGF levels must be fine-tuned as an excess or deficiency of HGF cause deafness in mouse. Using a Hgf mutant mouse with a small 10 bp deletion recapitulating a human DFNB39 noncoding variant, we demonstrate that neural crest cells fail to migrate into the stria vascularis intermediate layer, resulting in a significantly reduced endocochlear potential, the driving force for sound transduction by inner ear hair cells. HGF-associated deafness is a neurocristopathy but, unlike many other neurocristopathies, it is not syndromic.
Keywords: deafness, DFNB39, hepatocyte growth factor, HGF, neural crest cells, stria vascularis
Introduction
Hepatocyte growth factor (HGF) is an activator of mitosis and identical to “scatter factor,” which stimulates epithelial cells to disperse in culture (Stoker et al., 1987; Nakamura, 1989). HGF is also implicated in branching morphogenesis (Y. Zhang and Vande Woude, 2003), tumorigenesis (Y. Zhang et al., 2018), immune cell regulation (Papaccio et al., 2018), neuronal survival (Thompson et al., 2004; Nakano et al., 2017), wound healing (Miyagi et al., 2018), and neuronal differentiation, synapse formation, and maturation (Matsumoto et al., 2014). There are numerous studies of HGF splice isoforms and HGF protein structure, domains, and diverse functions (Comoglio et al., 2003; Matsumoto et al., 2014). Inactive pre-pro-HGF is secreted and proteolytically processed into a functional α and β disulfide-linked heterodimer (Fig. 1A). The α-chain contains an N-terminal hairpin loop followed by four kringle domains (Fig. 1A). Alternate splice transcripts of HGF give rise to shorter protein isoforms, called HGF/NK1 and HGF/NK2, depending on the number of kringle domains encoded.
Figure 1.
A, Schematic structure of HGF, which binds to and activates the MET receptor. The N-terminus signal peptide (SP) of HGF is removed by a signal peptidase. HGF is proteolytically cleaved extracellularly into a heavy alpha-chain that has a hairpin loop (HL), four kringle domains (K1-K4), each characterized by three disulfide bonds, and a light beta-chain with sequence similarity to serine proteases but catalytically inoperative. B, HGF-MET signaling pathways (abridged). Protein designations in uppercase are the official human nomenclature (HGNC) and mouse nomenclature protein acronyms (Mouse Genome Informatics) as of August 2019. For example, the previous METTL13 is a synonym for EEF1AKNMT, and SHP-2 is a synonym for PTPN11. Proteins are noted whose genes are associated with human and mouse hereditary deafness. C, Targeting construct used to generate a mouse 10 bp deletion mutation that models human deafness DFNB39. The targeting construct has a 10 bp deletion engineered into intron 5 of mouse Hgf. The NotI, XhoI, and BamHI restriction endonuclease sites were introduced into the targeting construct. The BamHI site downstream of exon 6 is present endogenously in mouse genomic DNA. The Neomycin (Neo) cassette is flanked by loxP sites (left pointing black triangles). Depicted below the targeting construct are the alternate transcripts of the Hgf locus. RefSeq NM_001289458 encodes the 728 amino acid canonical longest isoform (HGF/SF), whereas NM_001289461 encodes an isoform using an alternate exon 6 acceptor site, and is 5 amino acids shorter (HGF723). A short isoform Hgf/NK0.5 of mouse Hgf (AK142159.1) is orthologous to an annotated human short HGF isoform (ENST00000643024.1). Hgf/NK0.5 encodes approximately one-half (35 residues) of the first of the four kringle domains (K1; 80 residues) and would differ from the well-studied short HGF/NK1 Hgf isoform that has an entire K1 of 80 residues. The protein coding sequence of Hgf/NK0.5 ends with sequence in exon 5 plus one intronic nucleotide to complete a conserved arginine codon, which is followed by a conserved UAA translation termination codon and regions of conserved sequence of the 3′UTR. D, The deleted 10 bp sequence in intron 5 (green underline) is also part of 3′UTR sequence of HGF/NK0.5. The deleted 10 bp is 100% identical to human sequence and to many mammals, including chimp, rhesus monkey, dog, and opossum. In human, a 10 bp sequence is tandemly duplicated (two boxes); one copy is deleted in some subjects with DFNB39 deafness. By convention, the deletion is annotated as the second copy (http://varnomen.hgvs.org). The location of the 3 bp deletion (TGA, short red underline) is the first unambiguous sequence deleted and is also a recessive variant associated with DFNB39 nonsyndromic deafness segregating in numerous human families (Schultz et al., 2009; Richard et al., 2019). DNA Sanger sequence traces are shown for a WT and a Hgfdel10Neo mouse. E, Schematic gene structures of mouse Hgf/SF, Hgf/NK1, and Hgf/NK0.5 showing the locations of RT-PCR primers. Each primer pair was designed to detect unique sequence of Hgf/SF, Hgf/NK1, or Hgf/NK0.5. Exon 6 has two acceptor splice sites, such that the encoded sequence of exons 6a and 6b differs in length by five evolutionarily conserved residues (SFLFS). F, RT-PCR analysis of portions of three Hgf isoforms. Hgf/SF (180 bp), Hgf/NK1 (181 bp), and Hgf/NK0.5 (191 and 2229 bp) are all expressed in the P1 mouse cochlea. G, Developmental expression in WT mouse cochlea using ddPCR (digital droplet) analyses. Expression levels in the cochlea of the Hgf/SF, Hgf/NK1, and Hgf/NK0.5 isoforms of the gene encoding hepatocyte growth factor (Hgf) relative to Gusb expression. Levels of expression of these three isoforms decrease from E16.5 to P7. Error bars indicate mean ± SD for three independent biological ddPCR determinations of cDNA synthesized three different times from mRNA isolated each time from four cochlea of 2 mice. Each ddPCR point is the mean of three technical replicates of cDNAs from mice at E16.5, P1 and P7.
HGF activates the MET receptor tyrosine kinase, which mediates diverse downstream pathways involved in epithelial-mesenchymal transition and the development of neural crest-derived lineages (Fig. 1B) (Sonnenberg et al., 1993; Birchmeier et al., 2003). When MET is active, several effector molecules are recruited that trigger signaling cascades involved in cell survival, transformation, motility and invasion, proliferation, and cell cycle progression (Fig. 1B) as well as MET autoregulation (Organ and Tsao, 2011). HGF is also necessary for normal hearing in human and mouse. Sensorineural deafness segregating as a recessive trait in several families was genetically mapped to human chromosome 7q11.22-q21.12 and designated as the DFNB39 locus (Wajid et al., 2003). Sanger sequencing of all genes in the smallest obligate genetic linkage interval revealed 3 and 10 bp deletions in intron 4 of HGF in numerous families segregating nonsyndromic deafness (Schultz et al., 2009; Richard et al., 2019) (Fig. 1C,D). The intronic deletions are predicted to occur in the 3′UTR of an alternative splice isoformat the HGF locus (referred to here as HGF/NK0.5; RefSeq NM_001010933); distinct from and smaller than HGF/NK1 (Fig. 1A) (Cioce et al., 1996).
Homozygous KO of mouse Hgf results in embryonic lethality (Kato, 2017). However, we reported that a conditional deficiency in HGF in the ear results in viable deaf mice with thinning of the stria vascularis (SV) in the cochlea (Schultz et al., 2009). Additionally, a Hgf transgenic mouse constitutively overexpressing HGF is also deaf, suggesting that normal cochlear development is sensitive to the amount of HGF (Takayama et al., 1996; Schultz et al., 2009). Variants of MET are also associated with human deafness DFNB97 (Mujtaba et al., 2015; Alabdullatif et al., 2017). In light of these observations, cochlear epithelial-specific Hgf and Met KO mouse models were engineered to study HGF-MET signaling during development of the SV (Shibata et al., 2016). The SV generates the 80 mV endocochlear potential (EP) necessary for hair cell mechano-transduction (Wangemann, 2002). Mutations in genes expressed by SV cell types can cause a reduction or loss of EP, resulting in deafness (Steel and Barkway, 1989; Tachibana, 1999). Deficits of either HGF or MET resulted in a failure of neural crest cells to incorporate into the intermediate cell layer of the SV (Shibata et al., 2016), providing an explanation for the abnormally thin SV in Hgf conditional KO mice (Schultz et al., 2009).
Despite evidence related to the roles of HGF in hearing, the question remained as to whether the noncoding variants of human HGF described by Schultz et al. (2009) are benign but are merely closely linked to the actual deafness-causing variants. Alternatively, are the noncoding variants the direct cause of cochlear pathology in DFNB39 deafness? If the latter, then what is the mechanism? To answer these questions, a mouse model was engineered with a 10 bp deletion in the homologous region in mouse (Fig. 1D). The phenotype of this mouse recapitulates human DFNB39 deafness and reveals the pathophysiological mechanism of neurosensory deafness.
Materials and Methods
Mouse model of DFNB39 human deafness
A targeting construct comprising 5028 bp (left arm) and 3648 bp (right arm) of mouse genomic DNA from the Hgf locus was cloned into a targeting vector based on pPNT-loxP-Neo (Fig. 1C). The right and left arms spanned exons 4, 5, and 6. A 10 bp deletion in intron 5 was introduced by site-directed mutagenesis. In human DFNB39 families, homozygous 3 and 10 bp deletions occur in a highly conserved region of intron 4 (Schultz et al., 2009). We deleted a 10 bp sequence that is 90% identical between mouse and human and overlaps the region of identity corresponding to both the 3 bp (c.482 + 1986_1988delTGA; RefSeq NM_000601) and 10 bp (c.482 + 1991_2000delGATGATGAAA) deletions in humans (Fig. 1D). In humans, this 10 bp sequence is the first of two identical tandem 10 bp sequences (Fig. 1D). A 1.8 kb neomycin (neo) selection cassette was introduced into intron 5. Targeting constructs corresponding to both mutant and WT sequences were provided to the University of Michigan Transgenic Animal Model Core for electroporation into Bruce4 ES cells. Mutant-containing ES cells were recovered, and two independent lines, designated 11224 and 11225, were established with the targeted mutation of Hgf. The 11225 line was crossed with a ZP3-cre mouse (de Vries et al., 2000) and the Neo cassette was excised, while a separate 11225 line with the Neo cassette intact was maintained. The JAX mouse nomenclature committee designated the lines B6.Cg-Hgf Tm1.1Tbf (MGI:6294040, founder line with neo cassette) and B6.Cg-HgfTm1Tbf (MGI:6294042, neo cassette removed), referred to here as Hgfdel10Neo and Hgfdel10, respectively. Back-crossing of each line to C57BL/6J continued for at least six generations before crosses between heterozygotes were performed to generate mice for auditory evaluations.
Evaluation of B6.Cg-HgfTm1.1Tbf mice by the National Institutes of Health phenotyping service
Founder males for B6.Cg-HgfTm1.1Tbf (Hgfdel10Neo) were backcrossed to C57BL/6J, and heterozygous pups were crossed to generate mice for evaluation by the Mouse Phenotyping Service, Division of Veterinary Resources at the National Institutes of Health. Eight WT, 6 heterozygous (HET), and 4 homozygous knock-in (KI; i.e., Hgfdel10Neo/del10Neo) mice, for a total of 18 mice, were evaluated at the age of 3 months. There were equal numbers of males and females of each genotype. The phenotype assessment was a comprehensive evaluation of major organ weights, hematology, serum chemistries, and gross and microscopic organ evaluation.
Generation of Hgf conditional KO mice
Hgf conditional KO mice were generated as previously described (Shibata et al., 2016). Briefly, Pax2-Cre (RRID:MMRRC_010569-UNC; CD1 background) and Hgf-floxed (RRID:MMRRC_000423-UNC; B6/129 hybrid) were used to generate cochlear epithelium-specific deletions of Hgf.
Immunohistochemistry and measurements of strial thickness
For immunohistochemistry of cochlear sections, fixed adult mouse inner ears were decalcified in 150 mm EDTA for 5-7 d, transferred to 30% sucrose, and then embedded and frozen in SCEM tissue embedding medium (C-EM001, Section-Lab). Adhesive film (C-FUF303, Section-Lab) was fastened to the cut surface of the sample in order to support the section and cut slowly with a blade to obtain 10-µm-thickness sections. The adhesive film with sections attached was submerged for 60 s in 100% ethanol, then transferred to distilled water. The adhesive film prevents specimen shrinkage and detachment. This methodology allows for high-quality anatomic preservation of the specimen and sectioning at a thickness of 0.5 µm. Mid-modiolar sections were obtained from each cochlea where an EP recording had been performed.
Fluorescence immunohistochemistry for known SV cell-type markers was performed as follows. Mid-modiolar sections were washed in PBS, then permeabilized and blocked for 1 h at room temperature in PBS with 0.2% Triton X-100 (PBS-T) with 10% FBS (A3840001, Thermo Fisher Scientific). Samples were then incubated in the appropriate primary antibodies in PBS-T with 10% FBS, followed by three rinses in PBS-T and labeling with AlexaFluor-conjugated secondary antibodies (1:250, Invitrogen) in PBS-T for 1 h at room temperature. Where indicated, DAPI (1:10,000, Invitrogen) was included with the secondary antibodies to detect nuclei. Organs were washed in PBS three times and mounted in SlowFade Gold (S36937, Invitrogen, Thermo Fisher Scientific). Specimens were imaged using a confocal microscope (Carl Zeiss). Sections were mounted with SCEM tissue embedding medium (C-EM001, Section-Lab). Primary antibodies used included rabbit anti-KCNJ10 (RRID:AB_2040120, Alomone Labs, APC-035, polyclonal, dilution 1:200), rabbit anti-CLDN11 (RRID:AB_2533259, Invitrogen, 364500, polyclonal, dilution 1:200), goat anti-SLC12A2 (RRID:AB_2188633, Santa Cruz Biotechnology, sc-21 545, polyclonal, dilution 1:200), goat anti-KCNQ1 (RRID:AB_2131554, Santa Cruz Biotechnology, sc-10 646, polyclonal, dilution 1:200), and Phalloidin AlexaFluor-647 (RRID:AB_2620155, Invitrogen, A22287, dilution 1:250).
ISH with digoxigenin-labeled antisense riboprobes
ISH on postnatal day 0 (P0) cochlear cross sections was performed as previously described (Shibata et al., 2016). Briefly, P0 mice heads were fixed in 4% PFA in PBS overnight at 4°C, sunk in 30% sucrose in PBS at 4°C, incubated in Tissue-Tek O.C.T. compound (Sakura Finetek) at room temperature for 10 min, and frozen on dry ice. Sections, 14 μm thick, were cut using a 3050S cryostat (Leica Microsystems). Digoxigenin-labeled antisense riboprobes were synthesized using standard protocols (Stern, 1998). The following probes were used: Hgf, Aldh1a2 (gift from U. Dräger, University of Massachusetts Medical School, Worcester, MA), Cldn11 and Dct (gift from A. Kispert, Hannover Medical School, Hannover, Germany). The ISH procedure was modified from a published protocol (Henrique et al., 1995).
Single-molecule FISH (smFISH) using RNAscope probes
ISHs were performed using RNAscope Probe-Mm-Hgf-No-XHs (target region: 6-2185 nucleotides, NM_001289458.1) and Probe-Mm-Hgf-C3 (target region: 1203-2113 nucleotides, NM_001289458.1, that is equal to 1120-2030 nucleotides of NM_010427.4) (Table 1). RNAscope probes from Advanced Cell Diagnostics were used with sections of cochleae from C57BL/6J WT mice at embryonic ages E14.5, E18.5, and P30. Embryonic cochleae with the brain hemisected were fixed overnight at 4°C in 4% PFA in 1× PBS. Cochleae were then cryopreserved overnight in 15% and then overnight in 30% sucrose at 4°C. Adult cochleae were dissected from the head and fixed overnight at 4°C in 4% PFA in 1× PBS. Fixed adult mouse inner ears were decalcified in 150 mm EDTA for 5-7 d, transferred to 30% sucrose, and then embedded and frozen in SCEM tissue embedding medium (Section-Lab). Adhesive film (Section-Lab) was fastened to the cut surface of the sample in order to support the section and cut slowly with a blade to obtain thin mid-modiolar sections. The adhesive film with section attached was submerged in 100% EtOH for 60 s, then transferred to distilled water. This methodology allows for high-quality anatomic preservation of the specimen. Frozen tissues were sectioned (10 µm thickness) with a CM3050S cryostat microtome (Leica Microsystems). Sections were mounted with SCMM mounting medium (Section-Lab) and imaged using a 1.4 NA objective.
Table 1.
RNAScope probes used for smFISH
| Probe name | Gene | Target region | Accession no. | Catalog no. | Manufacturer |
|---|---|---|---|---|---|
| Probe-Mm-Hgf-No-XHs | Hgf | 6-2185 | NM_001289458.1 | 441431 | Advanced Cell Diagnostics |
| Probe-Mm-Hgf-C3 | Hgf | 1203-2113a | NM_001289458.1a | 315631-C3 | Advanced Cell Diagnostics |
aThat is equal to 1120-2030 nucleotides of NM_010427.4.
SV measurements and fluorescence intensity quantifications of strial cell type markers
ImageJ was used to calculate the cross-sectional area and thickness of the SV in mid-modiolar sections of both WT and homozygous KI mice in both the Hgfdel10Neo and Hgfdel10 mouse lines at postnatal day 60 (P60). Fluorescence intensity quantification was performed in ImageJ by calculating the fluorescence intensity of the outlined region of the SV. Fluorescence intensity was normalized by comparing the SV fluorescence intensity to that of a corresponding region in the scala media. The number of mice used with each mouse serving as a biological replicate are as follows: Hgfdel10Neo/+ (N = 8 mice), Hgfdel10Neo/del10Neo (N = 7 mice), Hgfdel10/+ (N = 8 mice), and Hgfdel10/del10 (N = 6 mice). Measurements for the upper (apical), middle, and lower (basal) turns of the cochlea were obtained. These measurements were obtained for known SV cell types, including intermediate cells (KCNJ10), marginal cells (SLC12A2, KCNQ1), and basal cells (CLDN11). KCNJ10 fluorescence intensity in the spiral ganglion neurons served as a control for immunofluorescence signal intensity measurements. Spiral ganglion fluorescence intensity was unchanged between KI and WT mice with no statistically significant difference between fluorescence intensity measurements (data not shown).
Auditory and vestibular testing
Auditory brainstem responses (ABRs) were measured at ages 4, 8, and 25 weeks after birth. Mice were anesthetized by an intraperitoneal injection of ketamine (56 mg/kg) and dexdomitor (0.375 mg/kg) and placed on a heating pad connected to a temperature controller (T-1000 or T-2000, World Precision Instruments) inside a sound-proofed booth (Acoustic Systems). A rectal probe was used to monitor body temperature, and a heating pad was used to maintain body temperature near 37°C. ABRs were obtained using Tucker-Davis Technologies hardware (RZ6 Processor) and software (BioSigRZ, version 5.1; RRID:SCR_014820).
For ABR testing, subdermal needle electrodes (Rhythmlink) were placed at the vertex, under the test ear, and under the contralateral ear (ground). Blackman-gated tone burst stimuli (3 ms, 29.9/s, alternating polarity) were presented to the test ear at 8, 16, 32, and 40 kHz via a closed-field Tucker-Davis Technologies MF-1 speaker. Responses were amplified (20×), filtered (0.3-3 kHz), and digitized (25 kHz) with 512-1024 artifact-free responses per waveform. For each frequency, testing began at 80 dB SPL and decreased in 10 dB steps until the ABR waveform was no longer discernable. If no response was obtained at 80 dB SPL, testing was performed at a maximum level of 90 dB SPL. Once the response was lost, testing continued in 5 dB steps with a minimum of two waveforms per stimulus level to verify repeatability of ABR waves. ABR thresholds were determined by visual inspection of stacked waveforms for the lowest stimulus level that yielded repeatable waves. Distortion-product otoacoustic emissions (DPOAEs) were measured in the right ear using Tucker-Davis Technologies hardware (RZ6 Multi I/O processor, MF-1 speakers) and software (BioSigRz, version 5.1).
Vestibular function of Hgfdel10Neo/+ (N = 4 mice) andHgfdel10Neo/del10Neo (N = 3 mice) mice was assessed at P140 with vestibular sensory-evoked potentials (VsEP) responses as previously described (Tona et al., 2019). Briefly, mice were anesthetized as described above for ABR analyses. VsEP responses were measured on the left side using Tucker-Davis Technologies hardware (RZ6 Multi I/O processor, Medusa preamplifier, and a headstage) and BioSigRz, version 5.7.2 software.
EP measurements
Methods for EP measurement have been described (Wangemann et al., 2004, 2007). Here, mice were anesthetized with 2,2,2-tribromoethanol (Sigma Millipore) at a dose of 0.35 mg/g body weight. EP measurements were made using glass microelectrodes inserted into the round window and through the basilar membrane of the first turn of the cochlea. Induction of anoxia, allowing measurement of anoxic-state EP, was accomplished by intramuscular injection of succinylcholine chloride (0.1 µg/g, NDC-0409-6629-02, Pfizer) after establishment of deep anesthesia followed by additional injection of 2,2,2-tribromoethanol (Sigma Millipore). Anoxic-state EP provides an indicator of the lowest EP and sensory hair cell function. In the presence of functional hair cells, the anoxic-state EP is negative, whereas the EP is 0 if the hair cells are not functional. Data were recorded digitally (Digidata 1440A and AxoScope 10; Axon Instruments) and analyzed using Clampfit10 (RRID:SCR_011323, Molecular Devices). For EP measurements, the following number of mice were used with each mouse serving as a biological replicate: Hgfdel10Neo/+ (N = 8 mice), Hgfdel10Neo/del10Neo (N = 7 mice), Hgfdel10/+ (N = 8 mice), and Hgfdel10/del10 (N = 6 mice).
Spiral ganglion region cell nuclei counts
From the same cochlea in which EPs were recorded, counts of cell nuclei in the spiral ganglion region in mid-modiolar cross sections were performed for P30 WT and KI mice from both mouse lines. The following number of mice were analyzed: Hgfdel10Neo/+ (N = 8 mice), Hgfdel10Neo/del10Neo (N = 7 mice), Hgfdel10/+ (N = 8 mice), and Hgfdel10/del10 (N = 5 mice). Similarly, H&E mid-modiolar cross sections of P90 WT (N = 4 mice) and KI mice (N = 4 mice) from the Hgfdel10Neo mouse line were examined for differences in spiral ganglion region cell nuclei counts.
Hair cell counts
Cochlear whole mounts from P30 WT (N = 4 cochleae) and Hgfdel10Neo/del10Neo (N = 4 cochleae) mice were immunostained with anti-MYO7A antibody (Axxora) for hair cells and DAPI (Invitrogen) for cell nuclei. Apical, medial, and basal turns were microdissected and mounted on slides. Inner and outer hair cell (OHC) counts were performed from two 210-μm-length regions from each cochlear turn (apical, medial, basal), and the mean hair cell count was determined.
Transcriptomics
KI and WT littermate mice at 5-27 weeks of age were killed via CO2 asphyxiation, and dissections of the inner ear, including cochlea and vestibule, lung, and kidney, were immediately frozen in liquid nitrogen. Tissue was pulverized with a Covaris CPO2 CryoPrep Automated Dry Pulverizer (Covaris), and RNA was extracted with Trizol (Invitrogen). For RNA-seq, total RNA from cochleae was reverse-transcribed with random primers after ribo-depletion using a TruSeq library kit (15031048 Rev. C, Illumina), and 2 × 93 bp sequenced on an Illumina HiSeq1500 instrument. The reads were mapped to the mouse genome (GRCm38.vM11) using STAR (Dobin et al., 2013). Differentially expressed genes (DEGs) were determined by DeSeq2 (Love et al., 2014). A total of 19 DEGs were found to be downregulated, and 14 DEGs upregulated, in Hgfdel10Neo/del10Neo cochleae (GEO Accession ID: GSE137721). The list of DEGs was analyzed by EnrichR for gene ontology analyses (RRID:SCR_001575; http://amp.pharm.mssm.edu/Enrichr/) as previously described (Chen et al., 2013; Kuleshov et al., 2016; Pazhouhandeh et al., 2017). Enrichr is an integrated web-based application that includes updated gene-set libraries, alternative approaches to ranking enriched terms, and a variety of interactive visualization approaches to display the enrichment results. Enrichr uses three approaches to compute enrichment as previously described (Jagannathan et al., 2017). The combined score approach where enrichment was calculated from the combination of the p value computed using the Fisher exact test and the z score was used. RT-PCR was performed using C57BL/6J WT P1 mouse cochlea cDNA, and PCR products were Sanger sequenced. qRT-PCR was performed using ddPCR (digital droplet) Supermix for Probes (186-3024, Bio-Rad) on a QX200 droplet digital PCR system (Bio-Rad). Each experiment was repeated three times, and expression levels were calculated as ratio of positive Hgf cDNA droplets to droplet positive for the housekeeping gene Gusb.qRT-PCR was performed using TaqMan Gene Expression Master Mix (4369514, Thermo Fisher Scientific) on a ViiA7 Realtime PCR instrument (Applied Biosystems). TaqMan probes for Hgf either spanned the exon 3-exon 4 junction, which should recognize all isoforms of Hgf, or were specific to the exon 5-exon6a and exon 5-exon 6b junctions, which are downstream of the 10 bp deletion. Expression levels of Hgf probe 3-4 were calculated as the delta Cq between the target probe and the geometric mean of reference probes Actb and Gusb. The 6a versus 6b alternative splice acceptor site usage was calculated as the Log2 fold difference for each sample. Details for primers and TaqMan probes used are listed in Table 2.
Table 2.
Primer and probe sequences
| Target | Sequence | |
|---|---|---|
| Hgf/SF | Forward 1 (exon 6)a | 5′-GAGCTATCGCGGTAAAGACCT-3′ |
| Reverse 1 (exon 7) | 5′-TGATCCATGGGACCTCTGTAG-3′ | |
| Probe 1 | 5′-/56-FAM/TTGAATGCA/ZEN/TGACCTGCAAT/3IABkFQ/-3′ | |
| Hgf/NK1 | Forward 2 (junction of exon 5–6)a | 5′-AACACAGCTTTTTGCCTTCG-3′ |
| Reverse 2 (exon 6 3′ UTR)a | 5′-AGCACCCCATGTGATATTCAG-3′ | |
| Probe 2 | 5′-/56-FAM/CGCGGTAAA/ZEN/GACCTACAGGA/3IABkFQ/-3′ | |
| Hgf/NK0.5 | Forward 3 (exon 4)a | 5′-GGTTTGGCCATGAATTTGAC-3′ |
| Reverse 3 (exon 5 3′ UTR)a | 5′-GTAAGGTTGACTGACCCTCTTT-3′ | |
| Probe 3 | 5′-/56-FAM/ATCCCCCAT/ZEN/GAACACAGGTA/3IABkFQ/-3′ | |
| Reverse 4 (exon 5 3′ UTR)b | 5′-AGGCCAGTGAAGGGTATAGA-3′ | |
| Hgf/SF, NK1, NK0.5 | Forward 5 (exon 3) | 5′-AGTGTGCCAACAGGTGTATC-3′ |
| Reverse 5 (exon 4) | 5′-TGAAAGGATACCAGTAGCATCG-3′ | |
| Probe 5 | 5′-/56-FAM/TGCAAGTGA/ZEN/ACGTAAAGCCCCTGT/3IABkFQ/-3′ | |
| Hgf/SF, exon 6a | Forward 6 (exon 5) | 5′-GCAGCTATAAAGGGACGGTATC-3′ |
| Reverse 6 (exon 6) | 5′-CTCGAGGATTTCGACAGTAGTTT-3′ | |
| Probe 6a | 5′-/56-FAM/TTTGCCTTC/ZEN/GAGCTATCGCGGT/3IABkFQ/-3′ | |
| Hgf/SF, exon 6b | Forward 6 (exon 5) | 5′-GCAGCTATAAAGGGACGGTATC-3′ |
| Reverse 6 (exon 6) | 5′-CTCGAGGATTTCGACAGTAGTTT-3′ | |
| Probe 6b | 5′-/56-FAM/TCTTTACCG/ZEN/CGATAGCTGTGTTC/3IABkFQ/-3′ | |
| Gusb | Forward 7 (exon 10) | 5′-GAGAACTGGTATAAGACGCATCA-3′ |
| Reverse 7 (exon11) | 5′-GAACAGCCTTCTGGTACTCC-3′ | |
| Probe 7 | 5′-/5HEX/CGAGTATGG/ZEN/AGCAGACGCAATCCC/3IABkFQ/-3′ |
aForward 1-3 and Reverse 1-3 primers were also used for RT-PCR in Figure 1E,F.
bReverse 4 primer was only used for RT-PCR.
Statistical analyses
For pairwise comparisons between mutants and WT littermates, an unpaired two-tailed Student's t test was used. ANOVA was used for comparisons between genotypes involving multipletissues. Differences between genotypes within tissues were calculated using Sidak's multiple comparisons test. All statistical analyses were performed using GraphPad Prism version 6.0 (RRID:SCR_002798; GraphPad) for PC. For measurements of strial thickness, cross-sectional area, and fluorescent intensity, all values are mean ± SD. Both males and females were tested, and there was no evidence of a significant effect of sex on any measures. Therefore, the data displayed in all graphs are from males and females combined.
Results
Overall phenotype of 10 bp deletion homozygotes
WT, HET, and homozygous KI (Hgfdel10Neo/del10Neo) mice were evaluated at age P90 (3 months) by the Mouse Phenotyping Service at the National Institutes of Health Division of Veterinary Resources. This comprises a comprehensive analysis of gross and histopathologic evaluations, including organ weights, serum chemistries, hematology, and histology. There were no obvious differences between genotypes in body size, weight, or coat pigmentation. Gross inspection of the major organs and histopathology of organs, including those that normally show high expression of Hgf (lung, liver, kidney), showed no abnormalities in Hgfdel10Neo/del10Neo mice. A schematic of the organ of Corti and the SV, which reside in the cochlea, is shown in Figure 2A. The organ of Corti is composed of one row of inner hair cells (IHCs) and three rows of OHCs surrounded by supporting cells. The SV is composed of three cellular layers, which consist predominantly of marginal, intermediate, and basal cells, respectively. Despite the lack of abnormalities in other organs, the inner ears of Hgfdel10Neo homozygous KI mice exhibited gross defects, including thin and detached SV, missing or pyknotic hair cells, and supporting cells along with general atrophy of the organ of Corti and degeneration of the spiral ligament, among other malformations (Fig. 2B,C). These defects occurred in an otherwise normally developed and patterned cochlea. The organ of Corti developed with apparently all cell types present and morphologically identifiable, although OHCs were reduced in numbers or showed signs of degeneration. Circling behavior and head-bobbing were not noticed in KI mice. Vestibular function was quantitatively evaluated by measuring VsEPs in the Hgfdel10Neo. No significant difference was observed between homozygous KI mice and heterozygous mice (Fig. 3A). These results were not significantly different from WT mice tested (N = 11 mice), with a mean VsEP threshold of −12.95 ± 2.62 dB (range, −10.5 to −16.5 dB). This finding is consistent with the absence of detectable Hgf expression as shown by smFISH in the vestibule at E18.5 mice (Fig. 3B).
Figure 2.
A, Schematic of the adult cochlea, including the organ of Corti and the SV. The organ of Corti is housed within the endolymph-containing scala media and is composed of IHCs and OHCs surrounded by supporting cells. The SV is composed of three cellular layers consisting of marginal, intermediate, and basal cells. The SV generates the 80 mV EP. FC, Fibrocyte; SP, spiral prominence. B, C, H&E staining of adult P90 homozygous KI mice demonstrates changes in cochlear structure, including loss of OHCs, strial atrophy, and spiral ligament atrophy. B, Representative H&E-stained section of P90 WT littermate from Hgfdel10Neo mouse line. Arrows indicate SV. Arrowheads indicate IHCs. Brackets indicate OHCs. C, Representative H&E-stained section of P90 homozygous KI mouse Hgfdel10Neo mouse line. Note the detachment and thinning of the SV (arrow) from spiral ligament, spiral ligament atrophy, and loss of OHCs (bracket) in the Hgfdel10Neo homozygous KI mouse. Scale bar, 200 μm. Arrowhead indicates IHCs. Brackets indicate the OHCs. Arrow points to the SV. A similar pathology was noted between Hgfdel10Neo and Hgfdel10 KI mouse lines at P90. Estimated cell counts in the spiral ganglion region between WT and KI mice at P30 in both mouse lines and at P90 in the Hgfdel10Neo mouse line are provided along with representative images (see Fig. 7).
Figure 3.
Expression of Hgf mRNA in the vestibular system and VsEP evaluation of vestibular function in Hgfdel10Neo/+ and Hgfdel10Neo/del10Neo mice. A, Mean VsEP thresholds of Hgfdel10Neo/+ (HET) and Hgfdel10Neo/del10Neo (KI) at P140. Hgfdel10Neo/del10Neo mice reveal normal vestibular function. Error bars indicate mean ± SD. These results were not significantly different from WT mice tested (N = 11 mice), which had a mean VsEP threshold of −12.95 ± 2.62 dB (range, −10.5 to −16.5 dB). B, Expression of Hgf mRNA at E18.5 in a WT mouse inner ear using two smFISH probes. A representative image of the SV colabeled with smFISH probes Mm-Hgf-No-XHs (red) and Mm-Hgf-C3 (blue). In an adjacent area of the same section of tissue that included the SV as a positive control, no labeling was observed in the saccule (S). The targeted region of the two smFISH probes is shown in Figure 10. Scale bars: 200 µm; insets, 20 µm.
Homozygous KI mice exhibit hearing loss
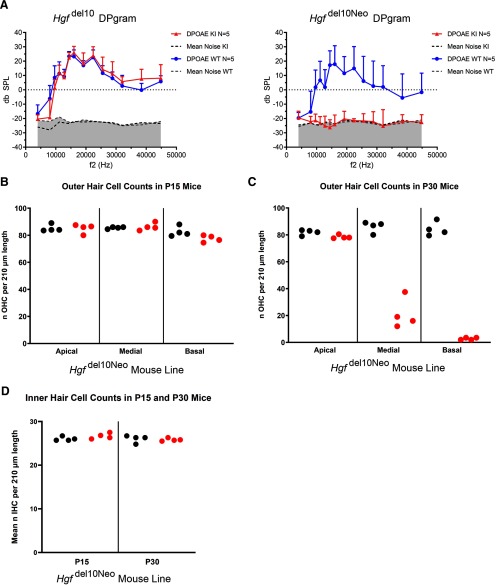
Homozygous KI mice in both lines (Hgfdel10Neo and Hgfdel10) by measuring ABRs to pure-tone stimuli and DPOAEs: homozygous Hgfdel10Neo KI mice displayed profound hearing loss at 4 weeks of age (Fig. 4A), which was unchanged at 8 and 25 weeks. No significant differences in ABR thresholds were seen between heterozygotes or WT littermates. Homozygous Hgfdel10Neo KI mice had significantly reduced DPOAEs at 8 weeks, indicating pathology likely involving OHC loss (Fig. 5A). In the Hgfdel10 mouse line, homozygous KI mice had moderate-to-severe hearing loss at age 4 weeks (Fig. 4A) that was unchanged at 8 and 25 weeks. The DPOAEs of homozygous Hgfdel10 KI mice were normal (Fig. 5A), indicating normal OHC function.
Figure 4.
Hearing loss phenotyping and cochlear structural observations in Hgfdel10Neo/del10Neo mice and Hgfdel10/del10 mice at P60. A, ABR for Hgfdel10Neo KI (top) and Hgfdel10 KI (bottom) mouse lines at 4 weeks. Tone burst stimuli were delivered, and responses were measured at 8, 16, 32, and 40 kHz as described in Materials and Methods. B, Representative mid-modiolar cross sections of P60 organ of Corti from Hgfdel10Neo/del10Neo mice (top) and Hgfdel10/del10 mice (bottom). Sections are stained with MYO7A for hair cells (red), Phalloidin (green), acetylated tubulin for supporting cells (AT, blue), and DAPI for nuclei (white). Note the IHCs and OHCs in red labeled with anti-MYO7A antibody. Scale bars, 20 μm. Label and arrow indicate IHC. Label and bracket indicate OHCs. DPOAE measurements at P60 for both mouse lines as well as IHC and OHC counts in the Hgfdel10Neo mouse line at P15 and P30 were performed (see Fig. 5). C, Strial thickness and strial cross-sectional area measurements in both Hgfdel10Neo (top) and Hgfdel10 (bottom) homozygous KI mice compared with WT littermate controls. ***p < 0.0001.
Figure 5.
Assessment of hair cell function and number. A, Right ear DPOAEs for Hgfdel10 (left) and Hgfdel10Neo (right). Reduction of DPOAEs is noted in the KI compared with WT mice from the Hgfdel10Neo mouse line, whereas DPOAEs are comparable between KI and WT mice from the Hgfdel10 mouse line. Five adult mice (age P30–P60) were tested for each genotype in each mouse line. B, OHC counts comparing P15 WT (n = 3 mice) and KI (n = 2 mice) mice from the Hgfdel10Neo mouse line. There are comparable OHC numbers between WT and KI mice. C, OHC counts comparing P30 WT (n = 2 mice) and KI (n = 2 mice) mice from the Hgfdel10Neo mouse line. Counts broken down by apical, medial, and basal turns. There is apical-to-basal gradient of OHC loss starting in the medial turn and increasing toward the basal turn of the cochlea. D, IHC counts at P15 and P30 are unchanged between WT and KI mice in the Hgfdel10Neo mouse line.
Histologic evaluation of homozygous KI mice indicates that hearing loss originates from defects in the SV
For both Hgfdel10Neo and Hgfdel10 KI lines at P60, examination of the organ of Corti with fluorescent immunohistochemistry suggests the possibility of normal morphology and complement of IHCs and OHCs and supporting cells (Fig. 4B), but by P90 there was a loss of OHCs in homozygous Hgfdel10Neo KI mice (Fig. 2C). Closer examination of hair cell counts between P30 WT and KI mice from the Hgfdel10Neo mouse line reveals a gradient of OHC loss from apex to base with normal OHC numbers in the apex and increasing hair cell loss from medial to basal turns of the cochlea (Fig. 5B). Examination of OHC counts between P15 WT and KI mice from the Hgfdel10Neo mouse line showed no difference between WT and KI OHC numbers (Fig. 5C). IHC counts in homozygous KI mice from the Hgfdel10Neo mouse line did not significantly differ from WT mice at both P15 and P30 (Fig. 5D).
Strial thinning (Fig. 4) and reduced KCNJ10 expression were observed at P60 (Fig. 6). Examination of the spiral ganglion region revealed no statistically significant difference in the number of cell nuclei (including both glia and neurons) between KI and WT mice from both mouse lines at P30 (Fig. 7A). Hgfdel10Neo/del10Neo mice at P90 have no statistically significant difference in cell number in the spiral ganglion region between KI and WT mice (Fig. 7B). While there was a small decrease in cell nuclei at P30 in the KI mice from both mouse lines, there was only a minimal difference at P90 between the KI and WT mice from the Hgfdel10Neo line. Representative spiral ganglion images are shown for P30 (Fig. 7C), whereas images for P90 mice are provided (Fig. 7D). After a gross WT developmental patterning of the cochlea, we sought to identify the primary defect causing the hearing loss in the homozygous Hgfdel10Neo and Hgfdel10 KI lines.
Figure 6.
Physiologic and histologic evaluation of the Hgfdel10Neo and Hgfdel10 lines demonstrate deficits in SV melanocytes. A, EP measurements in the Hgfdel10Neo (left) and Hgfdel10 (right) mouse lines demonstrate significant differences between homozygous KI and WT littermates (unpaired two-tailed Student's t test, p < 0.001). B, Representative immunostaining for KCNJ10 in the Hgfdel10Neo and Hgfdel10 lines demonstrates visually apparent decrease in KCNJ10 immunostaining in the homozygous KI mice from both mouse lines. Top, WT mice. Bottom, homozygous KI mice. Sections are stained with KCNJ10 for intermediate cells (red), Phalloidin for tight junctions (blue), and DAPI for nuclei (white). C, Representative immunostaining for SLC12A2 in the Hgfdel10Neo and Hgfdel10 lines demonstrates SLC12A2 immunostaining in the homozygous KI mice from both mouse lines. Differences in immunostaining are not readily apparent. Top, WT mice. Bottom, Homozygous KI mice. Sections are stained with SLC12A2 for marginal cells (red), Phalloidin for tight junctions (blue), and DAPI for nuclei (white). D, Representative immunostaining for CLDN11 in the Hgfdel10Neo and Hgfdel10 lines demonstrates CLDN11 immunostaining in the homozygous KI mice from both mouse lines. No differences are apparent in CLDN11 immunostaining. Top, WT mice. Bottom, Homozygous KI mice. Sections are stained with CLDN11 for basal cells (red), Phalloidin for tight junctions (blue), and DAPI for nuclei (white). E, Quantitative intensity analysis of KCNJ10, SLC12A2, and CLDN11 immunostaining in the Hgfdel10Neo and Hgfdel10 lines (n ≥ 6 animals per line). A significant decrease in KCNJ10 intensity in homozygous KI mice is shown for both mouse lines. For SLC12A2 immunostaining, only the Hgfdel10Neo mouse line demonstrates significant increase in SLC12A2 intensity in homozygous KI mice, whereas no significant change is apparent between homozygous KI and WT mice in the Hgfdel10 mouse line. Finally, no significant change in CLDN11 intensity is seen in either mouse line. *p < 0.05; ***p < 0.0001.
Figure 7.
Estimation of cell number in the spiral ganglion region of the cochlea from mid-modiolar cochlear cross sections. A, DAPI-labeled nuclei were counted in the spiral ganglion region of P30 WT and KI mice from the Hgfdel10Neo (WT = 8 mice, KI = 7 mice) and Hgfdel10 (WT = 8 mice, KI = 5 mice) mouse lines. The number of DAPI-labeled nuclei did not demonstrate a statistically significant difference between WT and KI mice from either mouse line (p = NS). B, Cells in the spiral ganglion region were counted from H&E-stained mid-modiolar cross sections of P90 WT (n = 4 mice) and KI (n = 4 mice) mice from the Hgfdel10Neo mouse line. The number of cells in the spiral ganglion region between WT and KI mice did not differ significantly (p = NS). C, Representative images of DAPI-labeled mid-modiolar cross sections depicting the spiral ganglion region of both P30 WT and KI mice from both the Hgfdel10Neo and Hgfdel10 mouse lines. No visually apparent differences were noted between WT and KI mice. Scale bars, 50 μm. D, Representative images of H&E-stained mid-modiolar cross sections depicting the spiral ganglion region of both P90 WT and KI mice from the Hgfdel10Neo mouse line. No visually apparent difference was noted between WT and KI mice. Scale bars, 50 μm.
The thinning and sometimes detachment of the SV seen in the KI mice was reported in a mouse with a conditional deletion of Hgf exon 6 and in a constitutively overexpressing Hgf mouse (MH19) (Takayama et al., 1996; Schultz et al., 2009). This was especially interesting given that expression of Hgf and Met, encoding the HGF receptor tyrosine kinase, is expressed in the early developing SV, and HGF-MET signaling has been shown to be crucial to the incorporation of melanocytes into the SV (Shibata et al., 2016). Therefore, we made detailed measurements of strial thickness and total area at three defined locations in the cochlea, comparing P60 WT with Hgfdel10Neo and Hgfdel10 lines (Fig. 4C). Significant reductions in both the Hgfdel10Neo and Hgfdel10 lines compared with WT in thickness and total strial area were noted at the base, mid-turn, and apex of the cochlea (unpaired two-tailed Student's t test, p < 0.001) (Fig. 4C).
Given the possibility of a SV defect as the primary site of pathology associated with hearing loss and the relationship between SV function and ion homeostasis in the inner ear (Hibino et al., 2010; Patuzzi, 2011), we measured EP in P60 mice as a direct measure of SV function. The EP has been shown to be directly proportional to strial volume (Schulte and Schmiedt, 1992). Homozygous Hgfdel10Neo and Hgfdel10 lines showed statistically significant reduction in their EPs compared with their WT littermates (Fig. 6A; unpaired two-tailed Student's t tests, p < 0.001).
The SV is composed of three cellular layers: marginal, intermediate, and basal cells (Fig. 2A). Examination of the SV with immunohistochemistry using antibodies specific for each layer (Fig. 6B–D) indicates that intermediate cells are deficient, as indicated by the reduced expression of the intermediate cell-specific marker, KCNJ10, in homozygous KI mice compared with WT mice (Fig. 6B,E) (p < 0.0001). However, notably, KCNJ10 immunostaining is not absent in homozygous KI mice. By comparison, markers for the other two strial cell types are not reduced in homozygous KI mice (Fig. 6C,D). Immunostaining intensities for SLC12A2, a marker for marginal cells (Fig. 6C,E), are slightly increased in the apical and basal turns of the Hgfdel10Neo homozygous KI mice compared with the SLC12A2 intensities in WT littermates, whereas they are unchanged in the Hgfdel10 homozygous KI mice (p < 0.01) (Fig. 6C,E). Expression intensities for CLDN11, a tight junction protein and marker for basal cells, are indistinguishable for KI and WT littermates from both mouse lines (Fig. 6D,E). This finding was confirmed and extended by ISH experiments with probes for Cldn11 (basal cells), Aldh1a2 (marginal cells), and Dct (intermediate cells) (Fig. 8A–L) in Hgf-conditional KO (CKO) mice at P0. The expression of Aldh1a2 and Cldn11 is unchanged between WT, heterozygotes, and homozygous Hgf-CKO mice. The images for Dct are particularly illuminating since this gene encodes dopachrome tautomerase, which is expressed by melanocytes, the cells that constitute the future SV intermediate cell layer. The Dct signal is markedly reduced, but not absent, at P0 in Hgf-CKO mice compared with WT (Fig. 8A–F). These data are consistent with previous observations (Shibata et al., 2016) and reminiscent of the reduced KCNJ10 immunostaining seen in homozygous KI (Hgfdel10/del10) mice in our mouse model. Together, the data imply that the primary defect in Hgfdel10/del10 mice is a significant reduction in the number of neural crest-derived melanocytes that infiltrate the developing SV.
Figure 8.
Observations in Hgf-CKO mice at P0 confirm a reduction in future strial intermediate cells. A–F, ISH for Dct demonstrates a reduction of Dct signal in homozygous CKO mice versus WT and homozygous CKO mice. Low-magnification imaging of Dct signal in mid-modiolar cross sections of WT (A), homozygous (B), and heterozygous (C) cochleae from Hgf-CKO mice. Scale bars, 100 μm. High magnification of representative SV from WT (D), homozygous (E), and heterozygous (F) Hgf-CKO mice. Dct signal identifies intermediate cell layer. G–I, ISH for Cldn11 demonstrates unchanged signal in WT (G), homozygous (H), and heterozygous (I) Hgf-CKO mice. Cldn11 signal identifies basal cell layer. J–L, ISH for Aldh1a2 demonstrates unchanged signal in WT (J), homozygous (K), and heterozygous (L) Hgf-CKO mice. Scale bars, 100 μm. Aldh1a2 signal identifies marginal cell layer. Each figure panel represents the results from at least 3 animals.
The del10 intronic mutation results in altered Hgf expression in the inner ear
We evaluated Hgf mRNA levels in the cochleae, kidneys, and lungs of KI mice and their WT littermates using TaqMan assays and qRT-PCR. Using TaqMan assays that span the constitutively expressed exons 3 and 4 (probe 3-4), which recognize all known splice isoforms, the expression level of Hgf was ∼70% lower in cochlea of KI mice compared with WT (p = 0.0004 and p = 0.03, respectively) in both Hgfdel10Neo and Hgfdel10 lines (Fig. 9A,B). In kidney and lung, there were no expression differences between WT and KI mice (p = 0.11 and p = 0.79 for kidney, and p = 0.64 and p = 0.99 for lung). Probes specific to the junction using the exon 6a or exon 6b alternative splice acceptor sites were used to evaluate the relative ratio of 6a to 6b isoform usage among Hgf transcripts. The 6a/6b ratios were reduced in KI compared with WT adult mice in the cochleae of both lines (p = 0.03 and p = 0.01, respectively), and these changes were not observed in the kidney and lung (all p values > 0.3 for kidney and lung, both lines) (Fig. 9B). These data suggest that the noncoding 10 bp deletion in Hgf intron 5 may alter Hgf expression exclusively in the adult cochlea.
Figure 9.
The del10 intronic Hgf mutation results in an inner ear alteration in Hgf expression. A, B, Cochlear Hgf expression as determined by qRT-PCR using TaqMan probe that spans exon 3-exon 4 (detects all isoforms) is significantly lower in KI compared with WT mice in both the Hgfdel10Neo (A) and Hgfdel10 (B) lines (p = 0.0004 and p = 0.03, respectively). Graph represents relative quantification of Hgf probe 3-4 versus the geometric mean of two reference genes (Actb and Gusb). More negative numbers indicate larger target probe Cq relative to stable reference gene Cq, and thus lower expression levels. Expression of Hgf in the mouse kidney and lungs is unchanged KI compared with WT mice (p = 0.11 and p = 0.79 for kidney, and p = 0.64 and p = 0.99 for lung). Sidak's multiple comparisons test was used to determine significance between KI and WT values after one-way ANOVA. *p < 0.05. **p < 0.001. C, D, Use of TaqMan probes specific for exon 6a or exon 6b splice acceptor sites demonstrates significant, but slight, change in the relative ratio of 6a versus 6b alternative splice acceptor usage between WT and KI mice in the cochlea in both the Hgfdel10Neo (C) and Hgfdel10 (D) lines (p = 0.03 and p = 0.01, respectively), whereas no change in this ratio was noted in the kidney and lung (all p values > 0.03 for kidney and lung, both lines). E, RNA-Seq comparing transcriptome profiles of whole cochlea from Hgfdel10Neo homozygous KI (KI.S1, KI.S2, KI.S3) to transcriptome profiles of whole cochlea from WT littermates (WT.S1, WT.S2, WT.S3) demonstrate downregulation of genes consistent with a loss of SV intermediate cells. Heatmap illustrates the 33 top DEGs, including 19 genes that were downregulated and 14 genes that wereupregulated in the KI transcriptomes. Higher red intensity represents higher relative expression; higher blue intensity represents lower relative expression. Histogram color bar is shown for reference.
Differential gene expression analysis revealed 34 significantly DEGs, of which 19 genes were upregulated and 14 genes were downregulated (Fig. 9C). Among the few downregulated DEGs are two genes, Dct and Tyr, that are specific to the intermediate cells in the SV and two genes, Slc45a2 and Mlana, which are known to be expressed by melanocytes. Analysis with Enrichr reveals that the downregulated genes are involved in Mitf signaling (TRRUST Transcription Factors 2019, Transcription Factors PPI). Gene ontology (GO) analysis (GO Biological Process,GO Molecular Function, GO Cellular Component) reveals that these genes, such as tyrosinase, are involved in the melanin synthesis pathway. Melanin may have a protective role, possibly as an ROS scavenger (Bustamante et al., 1993). These RNA-Seq analyses provide independent confirmation of a loss of intermediate cells in the setting of the Hgfdel10 mutation.
The preferential reduction of Hgf in the cochlea in the case of the del10 mutation coupled with the confinement of DEGs to melanocyte-specific genes and pathways also suggests that the effect of the Hgfdel10 mutation may be confined to the cochlea. Thus, this mutation appears to represent a unique situation, since neurocristopathies most often result in syndromes involving multiple organ systems (Bolande, 1974; Vega-Lopez et al., 2018; Ritter and Martin, 2019).
Hgf RNA expression is present in the developing SV
smFISH using two RNAscope probes was used to detect Hgf transcript expression in the SV during development and adulthood. One probe (Mm-Hgf-No-XHs) detected the full-length sequence of mouse Hgf and the other probe (Mm-Hgf-C3) detected the Hgf RNA sequence toward the C-terminus (Fig. 10A). Both probes detected the full-length Hgf mRNA in the SV of E14.5, E18.5, and P30 WT mice, providing dual confirmation of Hgf mRNA in the SV (Fig. 10B,C). smFISH labeling demonstrates the presence of both Mm-Hgf-No-XHs (red) and Mm-Hgf-C3 (blue) in marginal cells at E18.5 WT mice (Fig. 10B). In adult WT mice, Hgf mRNA expression is present as measured by both smFISH probes, Mm-Hgf-No-XHs and Mm-Hgf-C3, in marginal cells and, to a lesser extent, in the spiral ligament and Reissner's membrane (Fig. 10C).
Figure 10.
Hgf mRNA expression in the developing and adult SV. A, Schematic showing targeted regions for smFISH RNAscope probes for Hgf mRNA. Mm-Hgf-No-XHs (red) corresponds to the full-length sequence of Hgf mRNA, whereas Mm-Hgf-C3 (blue) targets the sequence toward the C-terminus of Hgf mRNA. In effect, these probes enable dual methods of detecting the full-length form of Hgf. B, Expression of Hgf mRNA in the perinatal SV at E14.5 and E18.5. Representative images of E14.5 (n = 2 mouse cochleae) and E18.5 (n = 2 mouse cochleae) SV colabeled with smFISH probes for Mm-Hgf-No-XHs (red) and Mm-Hgf-C3 (blue). The full-length form of Hgf is detected by both probes at E14.5 and E18.5. Scale bars: first row, 50 µm; other rows, 10 µm. Two WT cochleae from different mice were used for cryosectioning for each stage (E14.5, E18.5), and ISH was performed 3 times using RNAscope probes. C, Expression of Hgf mRNA in the adult SV (P30) (n = 3 adult mice). Representative images of P30 SV colabeled with smFISH probes for Mm-Hgf-No-XHs (red) and Mm-Hgf-C3 (turquoise) demonstrate expression of Hgf RNA in the adult SV. The full-length form of Hgf is detected by both probes at P30. Scale bars, 10 µm. Artifactual labeling of capillaries noted.
Discussion
The ionic composition of cochlear endolymph within the inner ear has a notably high potassium concentration of 150 mm, which is necessary for hair cell mechano-electrical transduction of sound. In this study, we demonstrate a mechanistic link between dysfunctional ion homeostasis in the inner ear and a 10 bp deletion (del10) of conserved intronic sequence of Hgf, which accounts for the neurosensory deafness of Hgfdel10Neo/del10Neo and Hgfdel10/del10 mutant mice and, by analogy, is the likely reason for human DFNB39 deafness. Homozygosity for the del10 mutation results in a failure of neural crest-derived melanocytes to incorporate into the SV during development, leading to a reduced intermediate cell layer and, consequently, compromised EP, deafness, and subsequent hair cell loss. Thus, DFNB39 qualifies as a neurocristopathy that is surprisingly nonsyndromic (Bolande, 1974; Vega-Lopez et al., 2018; Ritter and Martin, 2019). Mouse models described here provide an opportunity to study the role of HGF-MET signaling in neural crest cell incorporation into the SV.
SV intermediate cells express the KCNJ10 inward-rectifying potassium channel, which is critical for EP generation (Tachibana, 1999; Marcus et al., 2002; Wangemann et al., 2004). The EP reduction in Hgfdel10Neo and Hgfdel10 homozygous KI mice suggests that the initial tissue affected is the SV. Cell type-specific protein expression for markers of marginal (SLC12A2), intermediate (KCNJ10), and basal (CLDN11) cells reveals a reduction in KCNJ10 protein expression in Hgfdel10Neo and Hgfdel10 homozygous KI mice. This is consistent with the observed reduction in future intermediate cells derived from Dct-expressing neural crest cells seen in the Hgf-CKO mouse line (Fig. 8) (Shibata et al., 2016), and with the depletion of intermediate cell specific transcripts in homozygous mutant cochleae (Fig. 9E). These results link the hearing loss phenotype to an intermediate cell reduction, supporting the contention that neural crest-derived intermediate cells play a significant role in EP generation by the SV.
While SLC12A2 and CLDN11 protein levels remain unchanged between homozygous KI and WT Hgfdel10/del10 littermates, SLC12A2 demonstrates a slight, significant increase in expression in the Hgfdel10Neo line. Because marginal cells use SLC12A2 to transport potassium from the intrastrial space to maintain low intrastrial potassium essential for EP generation (Takeuchi et al., 2000), the hearing loss severity in Hgfdel10Neo mice may result in the induction of a compensatory mechanism heralded by an increase in SLC12A2 protein expression to account for the diminished intermediate cell population's ability to generate the high potassium concentration necessary for the EP. However, a change in expression of certain proteins, such as SLC12A2, may upset the fine balance necessary for normal function and result in dysfunction instead of compensation. For these reasons, the effect of the Neo cassette may be a tool to explore the response of marginal cells to reduction of SV intermediate cells.
The auditory phenotype of the Hgfdel10Neo line is more severe than the Hgfdel10 line. While ABR thresholds are elevated in both the homozygous Hgfdel10Neo/del10Neo and Hgfdel10/del10 KI mice, DPOAEs at P60 are reduced only in the homozygous Hgfdel10Neo/del10Neo KI mice but are normal in homozygous Hgfdel10/del10 KI mice (Fig. 5A). These data suggest that OHCs are dysfunctional in the KI mice from the Hgfdel10Neo mouse line, while they are functional in the Hgfdel10 mouse line at P60. OHC counts are normal, however, as late as P15 in the Hgfdel10Neo mouse line (Fig. 5B) with demonstrated OHC degeneration at P30 (Fig. 5C). IHC counts are normal at both P15 and P30 in the Hgfdel10Neo mouse line (Fig. 5D).
The unifying feature of both lines is the significant reduction in EP and evidence of reduced neural crest-derived intermediate cells in the SV. A reduced EP in the presence of normal DPOAEs in homozygous KI mice from the Hgfdel10 mouse line and a normally developed organ of Corti in P15 homozygous KI mice from the Hgfdel10Neo mouse line support the contention that a strial deficit precedes pathogenesis of the organ of Corti, likely due to dysfunctional ion homeostasis in the cochlear endolymph (Liu et al., 2016; Huebner et al., 2019). The timeline and relationship between EP reduction and hair cell loss have not been definitively established in this study or by others. The reduction, but not absence, of KCNJ10 combined with the thinning of the SV seen in homozygous Hgfdel10Neo and Hgfdel10 mice suggests the possibility of two different populations of KCNJ10-expressing cells within the SV. Light and dark cells have been reported as two distinct intermediate cell types in the SV (Cable et al., 1992). Light intermediate cells are dendritic with electron-lucent cytoplasm containing numerous cell organelles indicative of synthetic activity, whereas dark intermediate cells possess numerous melanin granules. Another possibility is that the remaining KCNJ10-expressing cells in the homozygous KI mouse from both Hgfdel10Neo and Hgfdel10 lines may be perivascular-resident macrophage-like melanocytes as described by W. Zhang et al. (2012). A third nonmutually exclusive possibility is that Hgfdel10 incompletely damages the process of neural crest cells integration into the SV.
HGF expression during inner ear development must be fine-tuned, with a deficit or an excess resulting in hearing loss (Takayama et al., 1996; Schultz et al., 2009). When there is a deficit of HGF, neural crest cells fail to migrate sufficiently into the strial intermediate cell layer. The molecular mechanism by which excess HGF causes deafness is not understood (Schultz et al., 2009). An understanding of the role of HGF in cochlear development and homeostasis will require a comprehensive survey of the spatial and temporal expression of the splice isoforms of Hgf. The data in this paper contribute to such a study. We show by ISH that Hgf is expressed at E14.5 and E18.5 in the SV as well as during adulthood at P30 (Fig. 10). With qRT-PCR, we show the temporal expression of alternative transcripts HGF, HGF/NK1, and HGF/NK0.5 (AK142159.1) in the developing inner ear (Fig. 1G). The latter is of particular interest since its 3′UTR encompasses the 10 bp deletion. The HGF/NK0.5 isoform is conserved in human and terminates with only 35 of 80 residues of the kringle domain K1 (Fig. 1A). The function of HGF/NK0.5 is unknown and will be a challenge to study in vivo. Because an arginine residue at the C-terminus of HGF/NK0.5 is the only difference from this sequence in the full-length HGF protein, obtaining a specific HGF/NK0.5 antibody is unlikely. The HGF/NK1 isoform includes the entire first kringle domain and can interact with the MET receptor, as can HGF/NK2, a competitive antagonist of HGF mitogenicity (Cioce et al., 1996). This isoform diversity is further complicated by the existence of an alternate splice acceptor site in exon 6, which either removes or retains 5 amino acids from the first kringle domain. Coexpression of the resulting shorter protein, HGF723, along with the canonical long isoform, HGF728, has been shown to potentiate HGF's angiogenic effects (Pyun et al., 2010; Hahn et al., 2011). We demonstrate that homozygosity for the 10 bp deletion in the adult results in reduced usage of the exon 6a splice acceptor, leaving exon 6b levels unaltered. The reduction of 6a acceptor usage was seen only in the cochlea, and not in kidney or lung (Fig. 9C,D). A future study will concentrate on the diversity and functional roles of each HGF isoform using mouse models that permit tissue-specific, isoform-specific, and quantitative regulation of Hgf expression, including the NK0.5 isoform, as well as the 6a and 6b splice acceptor sites during the developmental period when neural crest cells migrate to become SV intermediate cells.
Although we have focused on the developmental component of the mutant phenotype, the persistent expression of MET in adult SV intermediate cells suggests an ongoing requirement for HGF/MET signaling after SV development is complete (Shibata et al., 2016). In the adult, the ototoxic effects of aminoglycoside antibiotics can be ameliorated by exogenous HGF or an HGF mimetic, preventing hair cell loss (Kikkawa et al., 2009; Uribe et al., 2015). Specifically, exogenous HGF appears to be protective against neomycin in cochlear explants (Kikkawa et al., 2009). Dihexa, a HGF mimetic, crosses the blood-brain barrier and protects against acute aminoglycoside ototoxicity by upregulating MET in hair cells in zebrafish neuromasts (Uribe et al., 2015). These studies suggest that, when MET expression is altered, increasing HGF expression may prevent hair cell death.
In conclusion, we describe a mouse model of human deafness DFNB39 and demonstrate a functional link between a 10 bp deletion in a highly conserved intronic sequence of Hgf and hereditary hearing loss. The del10 mutation diminishes Hgf expression in the cochlea, leading to failure of neural crest-derived melanocytes to infiltrate the SV during development. Consequently, the intermediate cell layer is reduced and compromised, leading to reduced EPs, hearing loss, and, ultimately, hair cell loss. The Hgfdel10Neo and Hgfdel10 lines will be invaluable resources in further studies of HGF expression and function in the auditory system.
Footnotes
This work was supported in part by National Institute on Deafness and Other Communication Disorders/National Institutes of Health Intramural Research Program DC000048 to T.B.F., DC000086 to R.J.M., DC000080 to T.F, DC000088 to M.H., and National Institute on Deafness and Other Communication Disorders/National Institutes of Health Extramural Award R01 DC012085 to T.O.
The authors declare no competing financial interests.
References
- Alabdullatif MA, Al Dhaibani MA, Khassawneh MY, El-Hattab AW (2017) Chromosomal microarray in a highly consanguineous population: diagnostic yield, utility of regions of homozygosity, and novel mutations. Clin Genet 91:616–622. 10.1111/cge.12872 [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF (2003) Met, metastasis, motility and more. Nat Rev Mol Cell Biol 4:915–925. 10.1038/nrm1261 [DOI] [PubMed] [Google Scholar]
- Bolande RP. (1974) The neurocristopathies: a unifying concept of disease arising in neural crest maldevelopment. Hum Pathol 5:409–429. 10.1016/S0046-8177(74)80021-3 [DOI] [PubMed] [Google Scholar]
- Bustamante J, Bredeston L, Malanga G, Mordoh J (1993) Role of melanin as a scavenger of active oxygen species. Pigment Cell Res 6:348–353. 10.1111/j.1600-0749.1993.tb00612.x [DOI] [PubMed] [Google Scholar]
- Cable J, Barkway C, Steel KP (1992) Characteristics of stria vascularis melanocytes of viable dominant spotting (Wv/Wv) mouse mutants. Hear Res 64:6–20. 10.1016/0378-5955(92)90164-i [DOI] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce V, Csaky KG, Chan AM, Bottaro DP, Taylor WG, Jensen R, Aaronson SA, Rubin JS (1996) Hepatocyte growth factor (HGF)/NK1 is a naturally occurring HGF/scatter factor variant with partial agonist/antagonist activity. J Biol Chem 271:13110–13115. 10.1074/jbc.271.22.13110 [DOI] [PubMed] [Google Scholar]
- Comoglio PM, Boccaccio C, Trusolino L (2003) Interactions between growth factor receptors and adhesion molecules: breaking the rules. Curr Opin Cell Biol 15:565–571. 10.1016/s0955-0674(03)00096-6 [DOI] [PubMed] [Google Scholar]
- de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB (2000) Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis 26:110–112. [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn W, Pyun WB, Kim DS, Yoo WS, Lee SD, Won JH, Shin GJ, Kim JM, Kim S (2011) Enhanced cardioprotective effects by coexpression of two isoforms of hepatocyte growth factor from naked plasmid DNA in a rat ischemic heart disease model. J Gene Med 13:549–555. 10.1002/jgm.1603 [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D (1995) Expression of a Delta homologue in prospective neurons in the chick. Nature 375:787–790. 10.1038/375787a0 [DOI] [PubMed] [Google Scholar]
- Hibino H, Nin F, Tsuzuki C, Kurachi Y (2010) How is the highly positive endocochlear potential formed? The specific architecture of the stria vascularis and the roles of the ion-transport apparatus. Pflugers Arch 459:521–533. 10.1007/s00424-009-0754-z [DOI] [PubMed] [Google Scholar]
- Huebner AK, Maier H, Maul A, Nietzsche S, Herrmann T, Praetorius J, Hubner CA (2019) Early hearing loss upon disruption of Slc4a10 in C57BL/6 mice. J Assoc Res Otolaryngol 20:233–245. 10.1007/s10162-019-00719-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan R, Seixas A, St-Jules D, Jagannathan L, Rogers A, Hu L, Jean-Louis G, Sevick MA (2017) Systems biology genetic approach identifies serotonin pathway as a possible target for obstructive sleep apnea: results from a literature search review. Sleep Disord 2017:6768323. 10.1155/2017/6768323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T. (2017) Biological roles of hepatocyte growth factor-Met signaling from genetically modified animals. Biomed Rep 7:495–503. 10.3892/br.2017.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa YS, Nakagawa T, Tsubouchi H, Ido A, Inaoka T, Ono K, Ito J (2009) Hepatocyte growth factor protects auditory hair cells from aminoglycosides. Laryngoscope 119:2027–2031. 10.1002/lary.20602 [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li Y, Chen L, Zhang Q, Pan N, Nichols DH, Zhang WJ, Fritzsch B, He DZ (2016) Organ of Corti and stria vascularis: is there an interdependence for survival? PLoS One 11:e0168953. 10.1371/journal.pone.0168953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DC, Wu T, Wangemann P, Kofuji P (2002) KCNJ10 (Kir4.1) potassium channel knockout abolishes endocochlear potential. Am J Physiol Cell Physiol 282:C403–C407. 10.1152/ajpcell.00312.2001 [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Funakoshi H, Takahashi H, Sakai K (2014) HGF-Met pathway in regeneration and drug discovery. Biomedicines 2:275–300. 10.3390/biomedicines2040275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi H, Thomasy SM, Russell P, Murphy CJ (2018) The role of hepatocyte growth factor in corneal wound healing. Exp Eye Res 166:49–55. 10.1016/j.exer.2017.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba G, Schultz JM, Imtiaz A, Morell RJ, Friedman TB, Naz S (2015) A mutation of MET, encoding hepatocyte growth factor receptor, is associated with human DFNB97 hearing loss. J Med Genet 52:548–552. 10.1136/jmedgenet-2015-103023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. (1989) Molecular characterization of hepatocyte growth factor (HGF). Seikagaku 61:1243–1247. [PubMed] [Google Scholar]
- Nakano M, Tamura Y, Yamato M, Kume S, Eguchi A, Takata K, Watanabe Y, Kataoka Y (2017) NG2 glial cells regulate neuroimmunological responses to maintain neuronal function and survival. Sci Rep 7:42041. 10.1038/srep42041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ SL, Tsao MS (2011) An overview of the c-MET signaling pathway. Ther Adv Med Oncol 3:S7–S19. 10.1177/1758834011422556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaccio F, Della Corte CM, Viscardi G, Di Liello R, Esposito G, Sparano F, Ciardiello F, Morgillo F (2018) HGF/MET and the immune system: relevance for cancer immunotherapy. Int J Mol Sci 19:E3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patuzzi R. (2011) Ion flow in stria vascularis and the production and regulation of cochlear endolymph and the endolymphatic potential. Hear Res 277:4–19. 10.1016/j.heares.2011.01.010 [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh M, Samiee F, Boniadi T, Khedmat AF, Vahedi E, Mirdamadi M, Sigari N, Siadat SD, Vaziri F, Fateh A, Ajorloo F, Tafsiri E, Ghanei M, Mahboudi F, Rahimi Jamnani F (2017) Comparative network analysis of patients with non-small cell lung cancer and smokers for representing potential therapeutic targets. Sci Rep 7:13812. 10.1038/s41598-017-14195-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyun WB, Hahn W, Kim DS, Yoo WS, Lee SD, Won JH, Rho BS, Park ZY, Kim JM, Kim S (2010) Naked DNA expressing two isoforms of hepatocyte growth factor induces collateral artery augmentation in a rabbit model of limb ischemia. Gene Ther 17:1442–1452. 10.1038/gt.2010.101 [DOI] [PubMed] [Google Scholar]
- Richard EM, Santos-Cortez RL, Faridi R, Rehman AU, Lee K, Shahzad M, Acharya A, Khan AA, Imtiaz A, Chakchouk I, Takla C, Abbe I, Rafeeq M, Liaqat K, Chaudhry T, Bamshad MJ, Nickerson DA, Schrauwen I, Khan SN, Morell RJ, et al. (2019) Global genetic insight contributed by consanguineous Pakistani families segregating hearing loss. Hum Mutat 40:53–72. 10.1002/humu.23666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter KE, Martin DM (2019) Neural crest contributions to the ear: implications for congenital hearing disorders. Hear Res 376:22–32. 10.1016/j.heares.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte BA, Schmiedt RA (1992) Lateral wall Na,K-ATPase and endocochlear potentials decline with age in quiet-reared gerbils. Hear Res 61:35–46. 10.1016/0378-5955(92)90034-K [DOI] [PubMed] [Google Scholar]
- Schultz JM, Khan SN, Ahmed ZM, Riazuddin S, Waryah AM, Chhatre D, Starost MF, Ploplis B, Buckley S, Velásquez D, Kabra M, Lee K, Hassan MJ, Ali G, Ansar M, Ghosh M, Wilcox ER, Ahmad W, Merlino G, Leal SM, et al. (2009) Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. Am J Hum Genet 85:25–39. 10.1016/j.ajhg.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Miwa T, Wu HH, Levitt P, Ohyama T (2016) Hepatocyte growth factor-c-MET signaling mediates the development of nonsensory structures of the mammalian cochlea and hearing. J Neurosci 36:8200–8209. 10.1523/JNEUROSCI.4410-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg E, Weidner KM, Birchmeier C (1993) Expression of the met-receptor and its ligand, HGF-SF during mouse embryogenesis. EXS 65:381–394. [PubMed] [Google Scholar]
- Steel KP, Barkway C (1989) Another role for melanocytes: their importance for normal stria vascularis development in the mammalian inner ear. Development 107:453–463. [DOI] [PubMed] [Google Scholar]
- Stern CD. (1998) Detection of multiple gene products simultaneously by in situ hybridization and immunohistochemistry in whole mounts of avian embryos. Curr Top Dev Biol 36:223–243. 10.1016/s0070-2153(08)60505-0 [DOI] [PubMed] [Google Scholar]
- Stoker M, Gherardi E, Perryman M, Gray J (1987) Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature 327:239–242. 10.1038/327239a0 [DOI] [PubMed] [Google Scholar]
- Tachibana M. (1999) Sound needs sound melanocytes to be heard. Pigment Cell Res 12:344–354. 10.1111/j.1600-0749.1999.tb00518.x [DOI] [PubMed] [Google Scholar]
- Takayama H, La Rochelle WJ, Anver M, Bockman DE, Merlino G (1996) Scatter factor/hepatocyte growth factor as a regulator of skeletal muscle and neural crest development. Proc Natl Acad Sci USA 93:5866–5871. 10.1073/pnas.93.12.5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi S, Ando M, Kakigi A (2000) Mechanism generating endocochlear potential: role played by intermediate cells in stria vascularis. Biophys J 79:2572–2582. 10.1016/S0006-3495(00)76497-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Dolcet X, Hilton M, Tolcos M, Davies AM (2004) HGF promotes survival and growth of maturing sympathetic neurons by PI-3 kinase- and MAP kinase-dependent mechanisms. Mol Cell Neurosci 27:441–452. 10.1016/j.mcn.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Tona R, Chen W, Nakano Y, Reyes LD, Petralia RS, Wang YX, Starost MF, Wafa TT, Morell RJ, Cravedi KD, du Hoffmann J, Miyoshi T, Munasinghe JP, Fitzgerald TS, Chudasama Y, Omori K, Pierpaoli C, Banfi B, Dong L, Belyantseva IA, et al. (2019) The phenotypic landscape of a Tbc1d24 mutant mouse includes convulsive seizures resembling human early infantile epileptic encephalopathy. Hum Mol Genet 28:1530–1547. 10.1093/hmg/ddy445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe PM, Kawas LH, Harding JW, Coffin AB (2015) Hepatocyte growth factor mimetic protects lateral line hair cells from aminoglycoside exposure. Front Cell Neurosci 9:3. 10.3389/fncel.2015.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Lopez GA, Cerrizuela S, Tribulo C, Aybar MJ (2018) Neurocristopathies: new insights 150 years after the neural crest discovery. Dev Biol 444:S110–S143. 10.1016/j.ydbio.2018.05.013 [DOI] [PubMed] [Google Scholar]
- Wajid M, Abbasi AA, Ansar M, Pham TL, Yan K, Haque S, Ahmad W, Leal SM (2003) DFNB39, a recessive form of sensorineural hearing impairment, maps to chromosome 7q11.22-q21.12. Eur J Hum Genet 11:812–815. 10.1038/sj.ejhg.5201041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangemann P. (2002) K+ cycling and the endocochlear potential. Hear Res 165:1–9. 10.1016/s0378-5955(02)00279-4 [DOI] [PubMed] [Google Scholar]
- Wangemann P, Nakaya K, Wu T, Maganti RJ, Itza EM, Sanneman JD, Harbidge DG, Billings S, Marcus DC (2007) Loss of cochlear HCO3– secretion causes deafness via endolymphatic acidification and inhibition of Ca2+ reabsorption in a Pendred syndrome mouse model. Am J Physiol Renal Physiol 292:F1345–F1353. 10.1152/ajprenal.00487.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangemann P, Itza EM, Albrecht B, Wu T, Jabba SV, Maganti RJ, Lee JH, Everett LA, Wall SM, Royaux IE, Green ED, Marcus DC (2004) Loss of KCNJ10 protein expression abolishes endocochlear potential and causes deafness in Pendred syndrome mouse model. BMC Med 2:30. 10.1186/1741-7015-2-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Dai M, Fridberger A, Hassan A, Degagne J, Neng L, Zhang F, He W, Ren T, Trune D, Auer M, Shi X (2012) Perivascular-resident macrophage-like melanocytes in the inner ear are essential for the integrity of the intrastrial fluid-blood barrier. Proc Natl Acad Sci USA 109:10388–10393. 10.1073/pnas.1205210109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Vande Woude GF (2003) HGF/SF-met signaling in the control of branching morphogenesis and invasion. J Cell Biochem 88:408–417. 10.1002/jcb.10358 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xia M, Jin K, Wang S, Wei H, Fan C, Wu Y, Li X, Li X, Li G, Zeng Z, Xiong W (2018) Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer 17:45. 10.1186/s12943-018-0796-y [DOI] [PMC free article] [PubMed] [Google Scholar]