Abstract
Biochemical and biophysical cues directly affect cell morphology, adhesion, proliferation, and phenotype, as well as differentiation; thus, they have been commonly utilized for designing and developing biomaterial systems for tissue engineering applications. To bioengineer skeletal muscle tissues, the efficient and stable formation of aligned fibrous multinucleated myotubes is essential. To achieve this goal, we employed a decellularized extracellular matrix (dECM) as a biochemical component and a modified three-dimensional (3D) cell-printing process to produce an in situ uniaxially aligned/micro-topographical structure. The dECM was derived from the decellularization of porcine skeletal muscles and chemically modified by methacrylate process to enhance mechanical stability. By using this ECM-based material and the 3D printing capability, we were able to produce a cell-laden dECM-based structure with unique topographical cues. The myoblasts (C2C12 cell line) laden in the printed structure were aligned and differentiated with a high degree of myotube formation, owing to the synergistic effect of the skeletal muscle-specific biochemical and topographical cues. In particular, the increase of the gene-expression levels of the dECM structure with topographical cues was approximately 1.5–1.8-fold compared with those of a gelatin methacrylate (GelMA)-based structure with the same topographical cues and a dECM-based structure without topographical cues. According to these in vitro cellular responses, the 3D printed dECM-based structures with topographical cues have the potential for bioengineering functional skeletal muscle tissues, and this strategy can be extended for many musculoskeletal tissues, such as tendons and ligaments and utilized for developing in vitro tissue-on-a-chip models in drug screening and development.
Keywords: Decellularization, skeletal muscle, extracellular matrix, bioink, 3D bioprinting
Introduction
Although skeletal muscle damaged in a small region can be healed with inherent regeneration capability, the critically sized defect of muscle cannot be recovered via general physiological procedures; thus, alternative clinical treatment options are needed to avoid the scar formation and muscular atrophy that can result in skeletal muscle dysfunction [1]. Generally, the regeneration of muscle requires the fusion of myoblasts to form fully aligned multinucleated myotubes with diameters ranging from 20 to 100 μm [2–4]. Thus, to bioengineer skeletal muscle tissues, biomaterial scaffolding system should not only have topographical cues for aligning the myoblasts, but also provide biochemical components to ensure efficient myogenic differentiation and maturation, including thick and elongated myotube formation [5].
Several studies have been performed to examine myogenic differentiation/maturation with various extracellular matrix (ECM) components [6–8], because the interaction between cells and ECMs can provoke numerous cellular activities. Especially, decellularized ECM (dECM)-derived hydrogels have been widely investigated as a 3D cell culture system due to their tissue-specific biochemical components, including growth factors, cytokines, proteoglycans, and structural and adhesive proteins [9,10]. Although many attempts have utilized tissue-specific dECM hydrogels for tissue engineering applications [11–13], the structural integrity/stability of dECM hydrogel-based tissue constructs is significantly limited to build complex structures of tissue architecture [14].
Three-dimensional (3D) bioprinting or additive manufacturing strategy has been extensively employed for bioengineering tissue-specific constructs due to the reproducibility, suitability, and accuracy [15–17]. Because the layer-by-layer manufacturing system allows the fabrication of controllable construct geometry, including pore size and structure, this has recently been applied for the cell-based bioprinting process to fabricate cellularized tissue constructs with various cell types using cell-laden bioinks [18–21]. Thus, this cell-based bioprinting strategy has provided new insights to bioengineer various tissues or organs.
In order to bioengineer skeletal muscle tissues, the topographical cues to guide cellular orientation have been considered as a major requirement for facilitating proficient in vitro myogenic differentiation and maturation. However, the extrusion-based bioprinting method, which is the most commonly used bioprinting system, can only produce a few hundred size resolution [14,22]. To overcome this limitation, we previously established a novel printing method to fabricate a fibrous (a few micron scale) poly(ε-caprolactone) bundle and a micropatterned collagen structure for skeletal muscle tissue engineering [23,24]. Using the extrusion bioprinting method, fibrillated poly(vinyl alcohol) (PVA) as a sacrificial material was aligned by a controllable wall shear stress within a microsized nozzle. This novel method was able to create a uniaxially aligned fibrillated printed construct that could induce myoblast orientation, resulting in accelerated myogenic differentiation.
Herein, we utilized this novel printing method with a skeletal muscle-specific dECM to bioengineer biochemically and topographically mimicked skeletal muscle constructs. To achieve this goal, a dECM methacrylate (dECM-MA) derived from porcine skeletal muscles (as a bioink) was combined with fibrillated PVA to fabricate a uniaxially oriented dECM-MA patterned structure. We hypothesized that the combinations of tissue-specific biochemical cues from dECM-MA and topographical cues generated by PVA-fibrillation could provide a synergistic effect on the cell growth/orientation, myotube formation, and myogenic differentiation for skeletal muscle tissue engineering. The in vitro biological evaluations to validate this printed skeletal muscle construct were performed with two controls: (1) a dECM-MA-based cell-laden structure without topographical cues and (2) a gelatin methacrylate (GelMA)-based cell-laden structure with topographical cues.
Experimental Procedures
Decellularization of porcine skeletal muscle tissue
The decellularization process for the skeletal muscle tissue was performed via slightly modified methods based on the previously described method of Christman Et al [25]. Skeletal muscle tissues were isolated from the lower limbs of an adult Yorkshire Porcine (female, 10–15 months old) in accordance with the guidelines of Wake Forest University Institutional Animal Care and Use Committee (IACUC). The outer layer of the tissue (epimysium) was removed, and the tissue was cut into a cube (8 × 8 × 3 mm3). The chopped tissues were rinsed three times with Dulbecco’s phosphate-buffered saline (DPBS) and immersed in a 1% (w/v) sodium dodecyl sulfate (SDS) solution in sterile glass bottle for 3 d. The SDS solution was changed three times until the tissues became transparent. Then, the transparent tissues were washed with deionized water and treated with 1% Triton X-100 for 2 d. After treatment, the tissues were immersed in DPBS containing 1% antibiotic/antimycotic for 30 min and incubated in a DNase I solution for 2 h. To remove the remaining chemical materials, the decellularized skeletal muscle ECM (dECM) was rinsed three times with DPBS and lyophilized in a freeze-dryer (Labconco, USA) for 3 d.
The freeze-dried dECM was digested with 0.1% (w/v) pepsin in a 0.5 M acetic acid solution at a ratio of 10 mg of dECM per 1 mL of pepsin solution at room temperature for 2 d. For dECM precipitation, sodium chloride was added to the solution, followed by centrifugation (3000 rpm) for 15 min. The dECM precipitation was dialyzed in deionized water using dialysis tubing (molecular cutoff of 1000 kDa; Spectrum Labs, Inc., USA) at 40°C. for 2 d and then lyophilized. The freeze-dried dECM was stored at –80°C until use. All chemical reagents were obtained from Millipore Sigma (St. Louis, MO, USA) unless stated otherwise.
Characterization of dECM
To observe the decellularized structure, the native and decellularized tissues were fixed with 10% neutral-buffered formalin (Leica Biosystem, Germany) at room temperature for 3 d. The samples were washed twice using DPBS (HyClone™, USA) and dehydrated with ethanol series (50%, 60%, 70%, 80%, 90%, and 100%). The dehydrated tissues were immersed in xylene and embedded in paraffin blocks. A microtome (Leica Microsystems, Germany) was used to cut the paraffinized tissues for obtaining a 5-μm thin section. The sections were stained by hematoxylin and eosin (H&E), and the stained sections were visualized using a bright-field/fluorescent microscope (Leica DM4000B; Leica Microsystems, Germany).
For visualizing the longitude cross-sectional morphology and for DNA, collagen, glycosaminoglycans (GAGs), and elastin quantification, the dehydrated samples were lyophilized in a freeze-dryer (Labconco, USA) for 3 d. The freeze-dried tissues were observed using scanning electron microscopy (SEM) (Flex SEM 1000; Hitachi, Japan) and optical microscopy (Leica DM300; Leica Microsystems, Germany).
For the quantification of the DNA content, the DNA in the tissues was extracted using Qiagen a DNeasy Blood & Tissue kit (Qiagen Inc., USA). The DNA quantification was performed using a Quant-iT PicoGreen dsDNA Assay kit (ThermoFisher Scientific) according to the protocol of the manufacturer. Additionally, collagen, GAGs, and elastin were examined using the decellularized tissue and native tissue. For each component, 20 mg of freeze-dried tissues was solubilized chemically, as follows: pepsin mixed with 0.5 M acetic acid (0.1 mg of pepsin per 1 mL of acetic acid) for a collagen assay; a papain extraction solution for a GAG assay; and 0.25 M oxalic acid for an elastin assay. The analyses were performed using Sircol™ Soluble Collagen, Blycan™ Sulfated Glycosaminoglycans, and Fastin™ Elastin assay kits (Biocolor Life Sciences Assays, UK) according to the protocols of the manufacturer.
To evaluate the intactness of the ECM after the decellularization process, the collagen type-I, elastin, fibronectin, and laminin of the tissues were visualized. The sectioned tissues were treated with a serum-free blocking agent (Dako, USA) for 2 h and permeabilized using 2% Triton X-100 for 30 min. The permeabilized samples were then incubated with an anti-collagen type-I antibody (5 μg mL−1), an anti-elastin antibody (1:50 in DPBS), a fluorescein-conjugated anti-fibronectin antibody (1:50 in DPBS; Santa Cruz Biotechnology, Inc., USA), and an anti-laminin antibody (5 μg mL−1) overnight at 4°C. The anti-collagen type-I antibody, anti-elastin antibody, and anti-laminin antibody-treated samples were washed with DPBS and stained with an Alexa Fluor 488 (1:50 in DPBS; Invitrogen, USA) conjugated secondary antibody for 1 h at room temperature. The stained tissues were counterstained with 5 μM diamidino-2-phenylindole (DAPI) and visualized using a fluorescence microscope (Leica DM4000B; Leica Microsystem, Germany).
Methacrylation process of dECM
A methacrylation process was performed using dECM to obtain a photo-crosslinkable bioink. Briefly, digested dECM was dissolved in 0.5 M acetic acid at a density of 3.75 mg mL−1 until the solution became transparent (adjusted to pH 8–9 using 1 M NaOH) at 4°C. Under continuous stirring at 4°C, methacrylic anhydride at various densities (207, 414, and 621 mg per 600 mg of dECM) was added to the solution for 2 d for chemical modification of the dECM methacrylate (dECM-MA). After the chemical reaction, the dECM-MA solution was dialyzed and freeze-dried.
Characterization of dECM-Ma
To analyze the maintenance of ECM proteins after the dECM-MA fabrication process, the protein composition of dECM-MA was evaluated via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and compared with native tissue and gelatin methacrylate (GelMA). A Tris-HCl, 12% polyacrylamide gel (Bio-Rad Laboratories, Inc., USA) in a Tris/glycine/SDS buffer (ThermoFisher Scientific, Inc., USA) was used to characterize the protein fragment size according to the previously reported protocol of K. L. Christman Et al [26]. In addition, collagen, GAGs, and elastin were examined using Sircol™ Soluble Collagen, Blycan™ Sulfated Glycosaminoglycans, and Fastin™ Elastin assay kits (Biocolor Life Sciences Assays, UK) according to the protocols of the manufacturer.
The biodegradation rate of the dECM-MA was evaluated using Dulbecco’s modified Eagle’s medium (DMEM/high glucose; HyClone™, USA) and collagenase. Following measurement of the initial weight (M0) of the freeze-dried dECM-MA structures, printed using 3 wt% dECM-MA solution in DPBS onto the 38 oC printing plate with fixed fabricating conditions (nozzle inner diameter = 150 μm, moving speed = 10 mm s−1, pneumatic pressure = 20 kPa, and barrel temperature = 20°C, UV light strength = 300 mW/cm2), the structures were incubated in the DMEM/high glucose supplemented with fetal bovine serum (FBS; 10%), and penicillin/streptomycin (PS; 1%), and collagenase solution (0.1, and 20 U/mL in DPBS) for 0.5, 1, 3, 5, 6, 7, 10, 14, 21, and 28 d. The incubated samples were washed using deionized water, freeze-dried, and weighed at each timepoint (Md). The mass loss was calculated by the following equation:
The calculated Mass loss values are shown as the mean ± SD of five samples.
To confirm the rate of methacrylation, a 2,4,6-trinitrobenzene sulfonic acid (TNBS) assay kit (ThermoFisher Scientific, USA) was used. For the TNBS assay, the lyophilized dECM and dECM-MA were dissolved in a 0.1 M sodium bicarbonate solution (pH 8.5) at a density of 0.2 mg mL−1, and 250 μL of TNBS was added [27]. After 2 h of incubation, 125 mL of 1 M hydrochloric acid (HCl) and 250 μL of 10% SDS were added to stop the reaction. The optical density (OD) of the solution was measured using a spectrophotometer (SpectraMax M5 Microplate Reader; Molecular Devices, USA) at 335 nm. The degree of methacrylation was calculated using the following equation:
Preparation of dECM-MA-based bioink
For bioink formulation, a dECM-MA/PVA solution and a dECM-MA solution were prepared, respectively. Briefly, 6 wt% dECM-MA was dissolved in DPBS, and the solution was mixed with 20 wt% PVA (in DPBS) at a volume ratio of 1:1. The final weight fractions of the dECM-MA and PVA were 3 and 10 wt%, respectively. Also, 3 wt% dECM-MA was dissolved in DPBS. To prepare the bioink, the dECM-MA/PVA solution and the dECM-MA solution were mixed with 1 × 107 cells/mL myoblasts (C2C12; American-type Culture Collection, USA) and 3 mg/mL of photoinitiator [2-hydroxy-4’-(2-hydroxyethoxy)-2-methylpropiophenone (Irgacure 2959)].
Rheological properties of dECM-MA-based bioinks
To evaluate the rheological properties of the dECM-MA and dECM-MA/PVA bioinks, including the storage modulus (G′) and complex viscosity (η*), a rotational rheometer (Discovery HR2; TA Instruments, Inc., USA) with a cone-and-plate geometry (1° angle, 40 mm dia., 150 μm gap) was used. A time sweep was conducted on 3 wt% dECM-MA bioinks with different ratios of methacrylation [30% (dECM-MA-30), 50% (dECM-MA-50), and 80% (dECM-MA-80)] for 300 s at a frequency of 1 Hz, 1% strain, and 20°C. After 1 min, the bioinks were exposed to a UV source with different intensities (181, 300, 410 mW/cm2) for 60 s. To identify the gelation temperature of the 3 wt% dECM-MA-80 bioink, a temperature sweep (from 10 to 50°C) was conducted at a frequency of 1 Hz and 1% strain. Additionally, a frequency sweep (0.1 to 10 Hz) was conducted on the 3 wt% dECM-MA-80 bioink with 1% strain before UV crosslinking at 20°C and 38°C and on dECM-MA-80 (3 wt%)/PVA bioinks with various weight fractions of PVA (5, 10, and 15 wt%) without UV crosslinking at 20°C. All the values are shown as the mean ± SD of five samples.
Fabrication of cell-laden micropatterned dECM-Ma scaffold
To fabricate the aligned cell-laden dECM-MA structure (dE-AS), the dECM (3 wt%)/PVA (10 wt%) bioink was printed onto a sterilized PET film on a 38°C working plate using a printing system (DTR3–2210 T-SG; DASA Robot, South Korea) supplemented with a dispensing system (AD-3000C; Ugin-tech, South Korea) and a 30G single nozzle (inner diameter = 150 μm). The nozzle moving speed (10 mm s−1), pneumatic pressure (200 kPa), and barrel temperature (20°C) were fixed. In the printing process, the dispensed bioink was exposed to 300 mW/cm2 UV light to crosslink the dECM-MA. The printing procedures were progressed in a closed chamber with humidifier. To remove the PVA component and remnant photoinitiator, the printed structures were placed in a 6-well cell culture plate and incubated in a culture medium for 1 d at 37°C and washed thrice with pre-warmed DPBS (37°C). The films were removed after placing the structures in the cell culture plate.
For evaluating the effect of dECM-MA on the cellular activities, we used a cell-laden aligned GelMA structure (CON-1) as a control. GelMA was synthesized by a modified protocol based on the previous publication of H. Bergmans Et al [28]. For a GelMA/PVA bioink, GelMA (6 wt%) was dissolved in PBS and mixed with a PVA solution (20 wt%) at a volume ratio of 1:1. The final weight fractions of GelMA and PVA were 3 and 10 wt%, respectively. Then, the C2C12 cells (1 × 107 cells/mL) and the photoinitiator (3 mg/mL) were mixed with the GelMA/PVA solution. The printing and crosslinking processes were the same as described above for the aligned cell-laden dECM-MA structure, except for the temperature of the barrel (15°C) and the working plate (15°C) [29,30].
A dECM-MA structure without PVA fibrillation (CON-2) was printed using the 3 wt% dECM-MA bioink for assessing the effect of topographical cues in the printed structure. The printing and crosslinking processes were the same as described above for the aligned cell-laden dECM-MA structure, except for the pneumatic pressure (20 kPa).
In vitro cell culture
The printed C2C12-laden structures (8 × 2 × 1 mm3), placed in the 6-well plates, were cultured in Dulbecco’s modified Eagle’s medium (DMEM/high glucose; HyClone™, USA) supplemented with 1% penicillin/streptomycin (PS) (Antimycotic; ThermoFisher Scientific, USA) and 10% fetal bovine serum (FBS, Gemini Bio-Products, USA) with 5% CO2 at 37°C. The medium was changed every 2 d.
In vitro Cellular activities
The C2C12 cells in the printed structures were stained using 0.15 mM calcein AM and 2 mM ethidium homodimer-1 at 37°C for 1 h to visualize the live (green) and dead (red) cells. Images of the stained cells were obtained using a fluorescence microscope (Leica DM4000B; Leica, Germany). The cell viability and circularity were calculated using the ImageJ software (National Institutes of Health, USA).
To assess the proliferation of C2C12 cells in the structures, an AlamarBlue assay (Alamar BioScience, USA) was performed. The samples were incubated in DMEM/high glucose containing 1% PS, 10% FBS, and 10% AlamarBlue indicator at 37°C for 4 h. The absorbance of the medium was measured at 570 and 600 nm using a microplate reader (Spectra Max M5; Molecular Devices, USA). A known number of C2C12 cells reacting with the AlamarBlue indicator was used to generate the standard curve, and then all of the levels were normalized by the level of CON-2 scaffold at 1 d. The cell proliferation was evaluated using nine samples (n = 9, *P < 0.05).
The cultured C2C12 cells in the printed structures were stained using diamidino-2-phenylindole (DAPI; blue; 1:100 in DPBS; Invitrogen, USA) and fluorescein phalloidin (green; 1:100 in DPBS; Invitrogen) to visualize the nuclei and cytoskeleton of the C2C12 cells. A fluorescence microscope was used to observe the stained nuclei and cytoskeleton of the cells. The density and circularity of the nuclei and F-actin coverage and alignment were calculated using the ImageJ software.
The quantitative analyses using live/dead and DAPI/phalloidin images were evaluated using nine samples (n = 9, *P < 0.05).
Immunofluorescence
The printed cell-laden structures were washed with DPBS and fixed using 10% formalin for 60 min at room temperature. The specimens were then blocked with a serum-free blocking agent (X090930–1; Dako, USA) for 2 h and permeabilized with 2% Triton X-100 for 30 min. Then, the structures were treated with an anti-MF20 primary antibody (5 μg mL−1; Developmental Studies Hybridoma Bank, USA), anti-vinculin primary antibody (5 μg mL−1; EMD Millipore Corp., USA), and anti-integrin-β1 primary antibody (5 μg mL−1; Abcam, USA), anti-laminin primary antibody (5 μg mL−1; Abcam, USA), and anti-sarcomeric α-actinin primary antibody (5 μg mL−1; Invitrogen, USA) for the cells, and anti-collagen type-I primary antibody (reacts with porcine) (5 μg mL−1; Sigma-Aldrich, USA) for the dECM-MA overnight at 4°C. Subsequently, the samples were washed with DPBS and stained with Alexa Fluor 488 (1:50 in DPBS; Invitrogen, USA) and Alexa Fluor 594 (1:50 in DPBS; Invitrogen, USA) conjugated secondary antibodies for 1 h. The stained cells were counterstained with 5 μM DAPI. For the imaging analysis, we captured the fluorescence images using a confocal microscope (Leica DM4000B; Leica, Germany) with z-stacking mode to obtain the combined images. Additionally, the ImageJ software was used to calculate the development and orientation of the myotubes, and relative expression area of vinculin. The quantification of the myotube maturity was calculated by measuring the values of each myotubes (area, length, and diameter) or normalizing the values with the number of DAPI to calculate the fusion index and maturation rate. The quantitative analyses using immunofluorescence images were evaluated using nine samples (8 × 50 random measurements per image, n = 9, *P < 0.05).
Real-time polymerase chain reaction (RT-PCR)
To measure the expression levels of the myogenic markers of the C2C12 cells, including myogenic differentiation 1 (Myod1), myosin heavy chain 2 (Myh2), and Myogenin (Myog), an RT-PCR was conducted after 14 and 21 d of culturing. The total RNA from the cultured cells was isolated using a TRI reagent (Millipore Sigma). A spectrophotometer (SpectraMax M5 Microplate Reader; Molecular Devices, USA) was used to examine the concentration and purity of the isolated RNA. A complementary DNA (cDNA) synthesis from the RNase-free DNase-treated total RNA was conducted with a reverse transcription system using a QuantiTect Reverse Transcription Kit (Qiagen, Germany). To conduct a quantitative RT-PCR, a QuantiStudio™ 3 Real-Time PCR System (Applied Biosystems, USA), Power SYBR® Green PCR Master Mix (Quiagen) was used to amplify the cDNA. The gene-specific primers were listed as follows: mouse beta-actin (Actb) (forward: 5’-AAG GAA GGC TGG AAA AGA GC-3’, reverse: 5’-GCT ACA GCT TCA CCA CCA CA-3’), mouse Myod1 (forward: 5’-CGG CTA CCC AAG GTG GAG AT-3’, reverse: 5’-ACC TTC GAT GTA GCG GAT GG-3’), mouse Myh2 (forward: 5’-AGC AGA CG GAGA GGA GCA GGA AG-3’, reverse: 5’-CTT CAG CTC CTC CGC CAT CAT G-3’), mouse Myog (forward: 5’-CTG ACC CTA CAG ACG CCC AC-3’, reverse: 5’-TGT CCA CGA TGG ACG TAA GG-3’).
Statistical analyses
For evaluating statistical analyses, the SPSS software (SPSS, Inc., USA) was used. A single-factor analysis of variance and post-hoc analysis, Tukey’s HSD test, was performed, and *P < 0.05 was considered statistically significant. In case of the DNA, collagen, GAGs, and elastin contents, t-test was performed, and *P < 0.05 and **P < 0.01 were considered statistically significant.
Results and discussion
Preparation of decellularized ECM and its reaction with methacrylate
Because dECM possesses biochemical and physiological cues expressing the native microcellular environment, it has been considered as one of the most effective functionalized biomaterials [11–13, 31–33]. Because ECM consists of various bioactive components (collagen, elastin, laminin, fibronectin, proteoglycans, GAG, and complex molecules), the decellularization process (Fig. 1(a)) should not only conserve tissue-specific bioactive constituents safely, but also effectively remove the cell components [34].
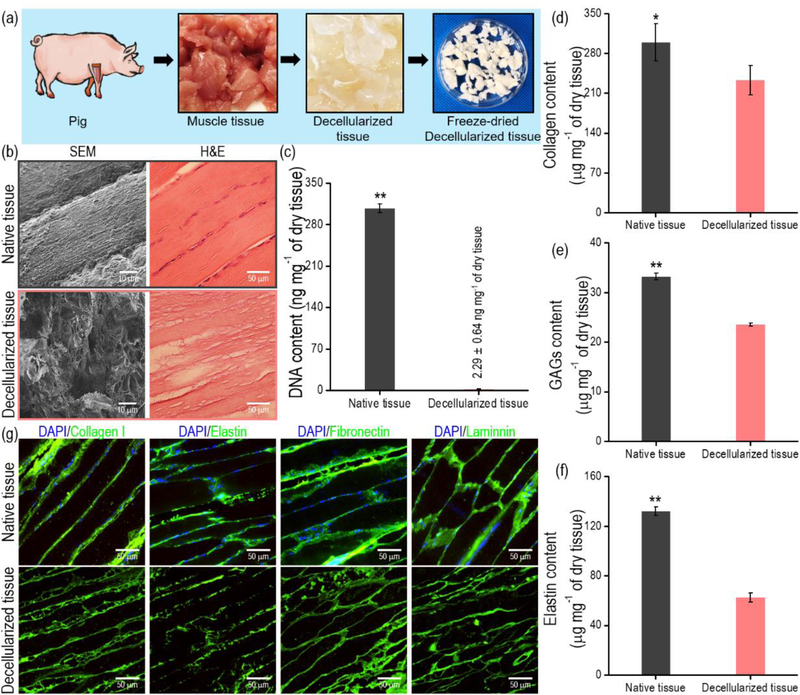
Figure 1.
(a) Schematic of the decellularization process using muscle tissue. (b) SEM and histomorphological images of native and decellularized muscle tissue. (c) DNA, (d) collagen, (e) GAG, and (f) elastin contents before and after decellularization (n = 5, *P < 0.05, **P < 0.01). (g) Immunofluorescence images of native and decellularized muscle tissue.
In this study, dECM was obtained from porcine skeletal muscle because it has similar biochemical components to human tissue [35,36]. As indicated by the histomorphological results in Fig. 1(b), the cellular components of the ECM were fully removed by the decellularization procedure. To quantitatively observe the remnant cell components after the decellularization, the relative DNA contents of the decellularized muscle were measured, and the value was significantly decreased below 3 ng/mg dry weight (Fig. 1(c)). The decellularization process was sufficient to remove the cell components because <50 ng/mg dry weight is an acceptable range for the DNA amount [35]. Additionally, collagen, GAGs, and elastin contents before and after the decellularization were measured, and although the absolute amount of the components was decreased considerably, the components remained in the decellularized matrix (dECM) (Figs. 1(d)–(f)). The obtained dECM and native tissues were stained to visualize the ECM components, including collagen type-I, elastin, fibronectin, and laminin (Fig. 1(g)). As shown in the immunofluorescence images, these proteins were well expressed in both the dECM and the native tissue.
As indicated by several studies, the dECM can provide outstanding bioactive components, but the low structural integrity and stability of the dECM using an extrusion-based 3D printer has been a significant issue [14,37]. Recently, to enhance the structural integrity and stability of the dECM bioink, a photo-crosslinkable process using the methacrylate reaction in various hydrogels has been proposed, e.g., GelMA, methacrylated hyaluronic acid, methacrylated collagen, and kidney-dECM-based hydrogel [29,30,38,39]. We also used a method of reacting the photo-crosslinkable methacrylate in the dECM derived from porcine muscle (dECM-MA) (Figs. 2(a) and (b)). The optical images in Fig. 2(c) show the dECM-MA solution (3 wt% in PBS) before and after UV crosslinking for 1 min. A viscosity increase was qualitatively observed for the UV-treated dECM-MA solution.
Figure 2.
(a) Optical images of the fabrication of a decellularized muscle tissue ECM (dECM-MA) and (b) schematics illustrating a photo-crosslinkable dECM-MA. (c) Optical images of dECM-MA before and after UV crosslinking. (d) SDS-PAGE separation of the dECM-Ma, native skeletal muscle, and GelMa. (e) Degree of methacrylation for various weights of methacrylic anhydride (n = 5, *P < 0.05).
Using SDS-PAGE, the relative biochemical components, including proteins, peptides, and polysaccharides, of the dECM-MA were compared with those in the native skeletal muscle and GelMA (Fig. 2(d)). As indicated by the results, the dECM-MA conserved the biochemical compounds after the various sample preparation processes, although it was not completely conserved for the biocomponents of the native muscle tissue. The quantitative results of the collagen, GAGs, and elastin contents also verified that the ECM components remained after the methacrylate procedures (Supplementary Fig. S1(a)).
Additionally, the substitution degree of the amino group from the dECM was measured using a TNBS assay. Fig. 2(e) shows the percentages of methacrylation in the dECM (600 mg) reacted with various methacrylic anhydride contents. As indicated by the results, the degree of the methacrylation in the dECM-MA could be manipulated. According to the results, the methacrylate procedure is efficient for fabricating a photo-crosslinkable skeletal muscle-specific material (dECM-MA). Moreover, after the reaction process, we measured the various growth factors of the skeletal muscle, dECM, and dECM-MA-80 (methacrylation: 80%) using a Quantibody Human Growth Factor Array (RayBiotech, Norcross, GA). As shown in Table 1, several growth factors and cytokines were resided in the dECM-MA through the decellularization and methacrylation processes. The remained biochemical components, such as vascular endothelial growth factor (VEGF), transforming growth factor beta-1 (TGF β1), insulin-like growth factor 1 (IGF-1), growth hormone 1 (GH-1), and bone morphogenic protein 7 (BMP7), in the dECM-MA could upregulate the proliferation and myogenesis of the skeletal muscle cells and promotes the growth of muscle fibers [40–44].
Table 1.
Growth factors and cytokines of porcine native skeletal muscle, dECM, and dECM-MA-80.
| [pg mL−1] | Skeletal muscle [n = 4] | dECM [n = 4] | dECM/MA [n = 4] | LOD* |
|---|---|---|---|---|
| Amphiregulin | 77.9 ± 18.3 | 102.8 ± 25.9 | 144.9 ± 11.4 | 8.3 |
| BDNF | 2.2 ± 0.8 | 1.4 ± 0.3 | 0.0 ± 0.0 | 1.1 |
| bFGF | 106.5 ± 15.8 | 4.9 ± 1.3 | 1.0 ± 0.4 | 20.7 |
| BMP-4 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 88.6 |
| BMP-5 | 612.6 ± 39.7 | 525.3 ± 88.6 | 877.7 ± 93.1 | 4,365.3 |
| BMP-7 | 1,024.8 ± 16.4 | 976.1 ± 96.6 | 1,405.8 ± 297.5 | 57.1 |
| β-NGF | 1.4 ± 0.2 | 0.0 ± 0.0 | 0.0 ± 0.0 | 1.4 |
| EGF | 0.1 ± 0.04 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 |
| EGF receptor | 8.0 ± 4.7 | 1.5 ± 0.2 | 10.6 ± 3.8 | 0.6 |
| EG-VEGF | 11.0 ± 2.1 | 11.1 ± 1.2 | 4.2 ± 1.2 | 4.9 |
| FGF-4 | 69.5 ± 10.1 | 73.0 ± 4.3 | 96.1 ± 9.3 | 132.0 |
| FGF-7 | 10.1 ± 0.9 | 0.0 ± 0.0 | 0.0 | 34.5 |
| GDF-15 | 1.4 ± 0.4 | 0.7 ± 0.3 | 0.2 ± 0.1 | 1.0 |
| GDNF | 1.1 ± 0.1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 53.7 |
| GH1 | 49.8 ± 9.2 | 60.1 ± 13.3 | 61.4 ± 6.1 | 3.7 |
| HB-EGF | 64.4 ± 5.1 | 30.4 ± 6.1 | 46.7 ± 2.5 | 4.7 |
| HGF | 5.3 ± 0.9 | 1.4 ± 0.3 | 1.1 ± 0.3 | 34.2 |
| IGFBP-1 | 6.5 ± 2.2 | 5.5 ± 1.2 | 7.5 ± 0.4 | 14.7 |
| IGFBP-2 | 23.1 ± 2.9 | 6.9 ± 1.7 | 30.8 ± 9.3 | 28.1 |
| IGFBP-3 | 967.3 ± 28.7 | 730.7 ± 59.7 | 102.8 ± 16.1 | 281.3 |
| IGFBP-4 | 8,938.1 ± 524.9 | 9,091.0 ± 706.8 | 1,754.4 ± 233.7 | 866.6 |
| IGFBP-6 | 219.3 ± 18.3 | 130.2 ± 16.7 | 187.2 ± 10.2 | 35.6 |
| IGF-1 | 59.6 ± 15.1 | 31.4 ± 4.9 | 41.0 ± 9.9 | 7.7 |
| Insulin | 2.1 ± 0.5 | 0.0 ± 0.0 | 8.0 ± 2.9 | 10.8 |
| MCSF R | 3.6 ± 0.4 | 1.4 ± 0.6 | 6.0 ± 1.8 | 38.1 |
| NGF R | 7.4 ± 0.8 | 7.9 ± 0.8 | 0.0 ± 0.0 | 13.6 |
| NT-3 | 6.7 ± 1.1 | 6.4 ± 0.5 | 8.4 ± 1.5 | 42.8 |
| NT-4 | 10.8 ± 2.6 | 9.0 ± 4.2 | 8.9 ± 2.5 | 20.2 |
| Osteoprotegerin | 12.4 ± 3.1 | 10.3 ± 3.5 | 0.9 ± 0.3 | 12.8 |
| PDGF-AA | 8.3 ± 1.3 | 12.6 ± 3.1 | 0.5 ± 0.2 | 2.6 |
| PIGF | 1.1 ± 0.2 | 0.0 ± 0.0 | 0.5 ± 0.1 | 4.7 |
| SCF | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.1 | 8.3 |
| SCF R | 2.8 ± 0.3 | 0.0 ± 0.0 | 0.0 ± 0.0 | 15.4 |
| TGFα | 0.0 | 0.0 ± 0.0 | 0.0 | 0.1 |
| TGF β1 | 2,464.5 ± 339.1 | 2,096.3 ± 160.8 | 2,503.3 ± 312.8 | 1,099.2 |
| TGF β3 | 6.2 ± 1.1 | 0.0 ± 0.0 | 0.0 | 13.0 |
| VEGF | 34.5 ± 9.7 | 8.2 ± 2.2 | 25.3 ± 6.7 | 7.2 |
| VEGF receptor 2 | 1.7 ± 0.2 | 0.0 ± 0.0 | 0.0 | 49.8 |
| VEGF receptor 3 | 3.3 ± 0.5 | 1.1 ± 0.3 | 0.0 | 17.9 |
| VEGF-D | 1.5 ± 0.1 | 0.0 ± 0.0 | 0.2 ± 0.1 | 4.4 |
LOD: limit of detection.
BDNF: Brain-derived neurotrophic factor, bFGF: Basic fibroblast growth factor, BMP: Bone morphogenetic protein, β-NGF: Beta-nerve growth factor, EGF: Epidermal growth factor, EG-VEGF: Endocrine gland-derived vascular endothelial growth factor, FGF: Fibroblast growth factor, GDF-15: Growth/differentiation factor 15, GDNF: Glial cell line-derived neurotrophic factor, GH1: Growth hormone 1, HB-EGF: Heparin-binding epidermal growth factor-like growth factor, HGF: Hepatocyte growth factor, IGFBP: Insulin-like growth factor-binding protein, IGF-1: Insulin growth factor 1, MCSF R: Macrophage colony-stimulating factor 1 receptor, NGF R: Nerve growth factor receptor, NT-3: Neurotrophin-3, NT-4: Neurotrophin-4, PDGF-AA: Platelet-derived growth factor A chain, PIGF: Placenta growth factor, SCF: Stem cell factor, SCF R: Stem cell factor receptor, TGF α: Transforming growth factor alpha, TGF β1: Transforming growth factor beta-1, TGF β3: Transforming growth factor beta-3, VEGF: Vascular endothelial growth factor
To provide more proper microenvironments to the myoblasts, the remained biochemical molecules should be released from the dECM-MA structures. M. T. Wolf Et al. [6] reviewed that the biochemical components could be released from the ECM-based biomaterials during the degradation events. Furthermore, when the degradable ECM-based scaffolds had been implanted, the degraded structures could be replaced by remodeled native ECM, recruited perivascular cells and remodeled vessels [6,45]. To evaluate the degradation of the dECM-MA, we incubated the dECM-MA structures in the growth medium and collagenase solution with low and high concentration (0.1 and 20 U/mL in DPBS). The structures were fully degraded after 1 day in the collagenase solution with high concentration (20 U/mL), while the structures in the growth medium and 0.1 U/mL of collagenase were not (Supplementary Fig. S1(b)). At 30 days, the volumetric dECM-MA structure was degraded around 80% in the collagenase solution with low concentration (0.1 U/mL) and completely collapsed, while the structure was degraded around 40% in culture medium and (Supplementary Fig. S1(b)). By the results, we can carefully estimate that the scaffolds fabricated using the dECM-MA bioinks can be utilized effectively in muscle regeneration by providing proper biodegradable microenvironmental condition for the skeletal muscle tissues.
Viscoelastic properties of dECM-MA solution and proper UV-exposure condition
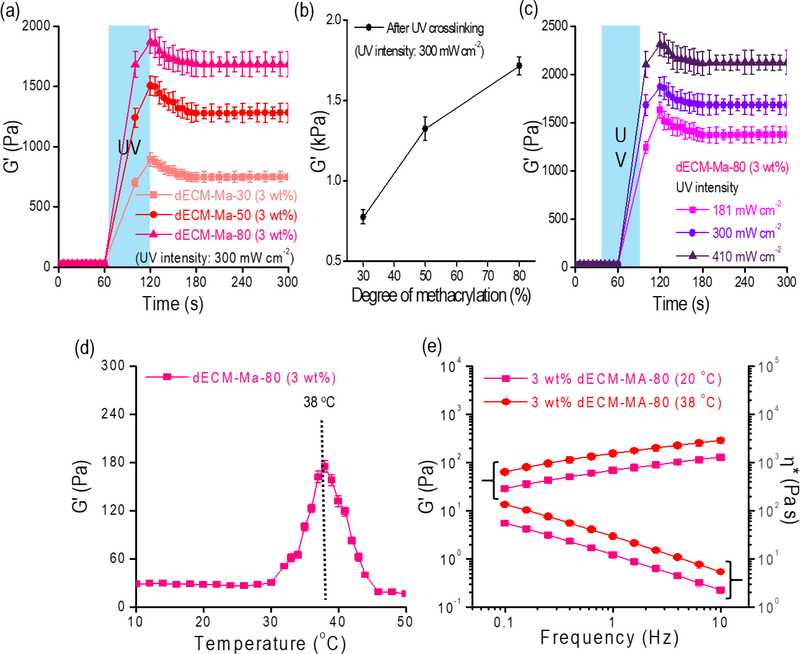
To measure the viscoelastic properties of the 3 wt% dECM-MA solutions with different methacrylation (dECM-MA-30, dECM-MA-50, and dECM-MA-80, as shown in Fig. 2(e)), the storage modulus (G’) of the solution was assessed with a time sweep before and after the UV exposure (300 mW/cm2 and 60 s) (Fig. 3(a)). As indicated by the results, the modulus after the UV exposure was significantly increased, and also the modulus increased linearly with respect to the increasing degree of the methacrylation (Fig. 3(b)). Additionally, to evaluate the effect of the UV power on the rheological properties of dECM-MA-80, we applied three different UV powers (181, 300, 410 mW/cm2) for 60 s (Fig. 3(c)). As expected, the G’ of the dECM-MA solution before the UV treatment was significantly low, but with an increase in the UV power, the modulus of the dECM-MA solution gradually increased.
Figure 3.
(a, b) Storage modulus (G’) of dECM-based bioinks (3 wt%) with different degrees of methacrylation (30%, 50%, and 80%) before and after the UV exposure (300 mW/cm2 and 60 s) (n = 5, *P < 0.05). (c) Storage modulus of dECM-MA-80 for different UV intensities (181, 300, and 410 mW/cm2) (n = 5, *P < 0.05). (d) Gelation temperature of dECM-MA-80 (not crosslinked with UV) tested in the temperature-sweep mode (n = 5, *P < 0.05). (e) Rheological properties (G’ and complex viscosity (η*)) of dECM-MA-80 at 20°C and 38°C before UV exposure (n = 5, *P < 0.05).
The rheological properties of dECM-MA can depend on the applied temperature because collagen, which is one of the main components of the dECM, has a gelation temperature of 37–38°C; thus, we measured the rheological properties of the dECM-MA-80, which was not treated with UV, with a temperature sweep from 10–50°C (Fig. 3(d)). As expected, the dECM-MA-80 exhibited an increased modulus (six-fold) near the collagen gelation temperature (38°C), and the frequency-sweep test at different temperatures (20 and 38°C) indicated that the processing temperature of 38°C is more favorable for fabricating dECM-MA with a stable structure (Fig. 3(e)).
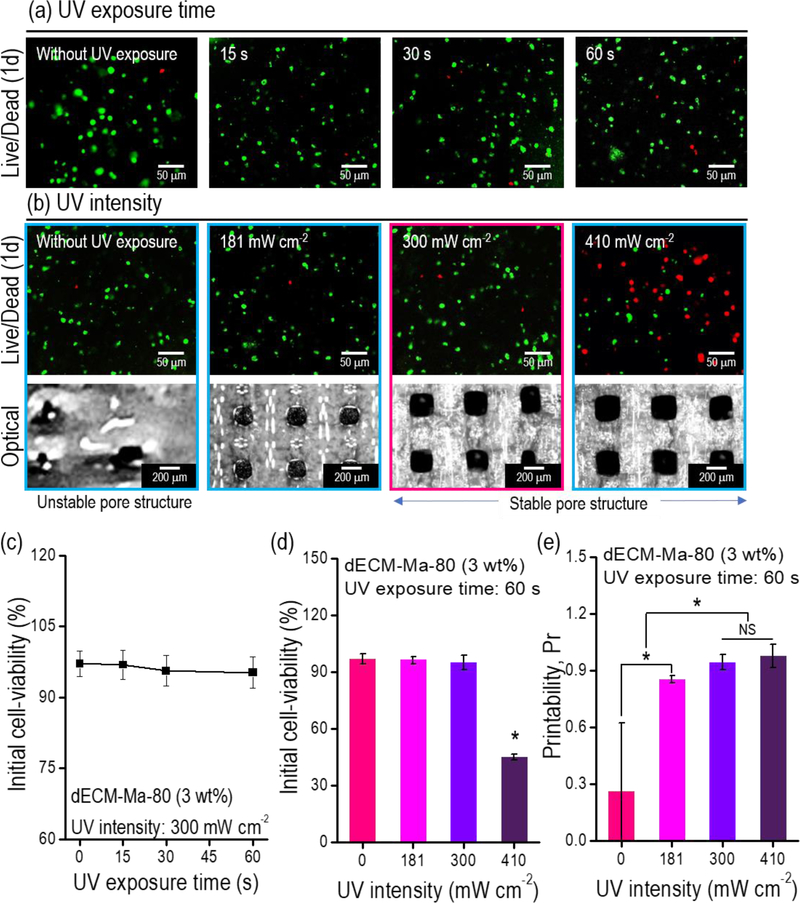
UV treatment can damage cells, depending on the conditions, e.g., the intensity and exposure time. To select a reasonable UV condition for the laden cells (1 × 107 C2C12 cells/mL in dECM-MA-80 bioink), various UV exposure times and intensities were applied to the printed structures, and the structures were cultured for 1 d. Figs. 4(a) and (b) show images of the live (green) and dead (red) cells. As indicated by the images, UV exposure for <60 s (at an intensity of 300 mW/cm2) did not affect the cell viability (Fig. 4(c)), and UV intensities of <300 mW/cm2 (with a constant exposure time of 60 s) did not damage the cells, while 410 mW/cm2 affected the cell viability significantly (Fig. 4(d)). Additionally, the printability (Pr) defined by the equation, Pr = L2 /16A, where A and L represent the area and perimeter of the pore, respectively [46], was measured for the applied UV intensities, as shown in the optical image of Fig. 4(b). As indicated by the results, with an increase in the UV intensity, the printability gradually increased, and at a UV intensity of 300 mW/cm2, the printability of the structure was maximized (Fig. 4(e)).
Figure 4.
(a) Live (green)/dead (red) images for various UV exposure times under a fixed intensity (300 mW/cm2). (b) Live/dead and optical images of the printed mesh structure (8 × 8 × 0.5 mm3) for various UV intensities at a fixed exposure time (60 s). Measured initial cell viability with respect to the (c) UV exposure time and (d) UV intensity (n = 9, *P < 0.05). (e) Printability calculated using the pore size of the printed mesh structure (8 × 8 × 0.5 mm3) for various UV intensities (n = 9, *P < 0.05).
According to the results for the rheological property and UV conditions of the dECM-MA bioink, we selected the degree of methacrylation (dECM-MA-80) of the dECM-based bioink, the temperature (38°C) of a printing stage to support the printed shape, and the UV conditions (power: 300 mW/cm2; exposure time: 60 s).
In situ micropatterning of printed dECM-MA/PVA structure using 3D printing
Previously, we developed a fibrillation process of PVA to obtain uniaxially aligned/micropatterned collagen struts processed with a low-temperature 3D printer (processing temperature of the working stage: −20 to −10°C) [24]. After the mixture (PVA/collagen) was printed using the low-temperature working stage and a freeze-drying process was conducted, the fibrillated PVA phase in the 3D-printed collagen/PVA struts was dissolved; thus, the remnant collagen structure was crosslinked with 1-ethyl-(3,3-(dimethylamino)propyl) carbodiimide hydrochloride for 30 min, and the topographical pattern on the collagen surface was well preserved [24]. However, although the myoblasts cultured on the uniaxially patterned collagen surface exhibited higher cellular activities, including myotube formation, than the non-patterned collagen struts, the complex and harsh processes, such as the low-temperature printing process (approximately −10°C) and the usage of a toxic chemical to crosslink the patterned collagen, were significant obstacles to directly laden cells in the functional collagen struts [24].
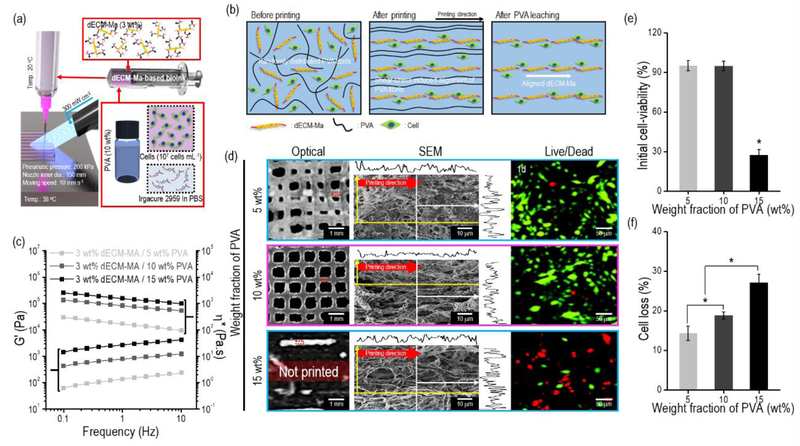
To overcome the limitations for fabricating cell-laden structures with an in situ topographical cues, we proposed the bioink consisting of cell-laden dECM-MA-80, which is photo-crosslinkable with UV during the printing process, and a sacrificial fibrillated PVA component in which hydroxyl groups can be bound with water and the fibrillated PVA molecules can be attained [24,47]. Through a microsize-nozzle (ID: 150 μm) in the 3D printer, the component of fibrillated PVA in the bioink could be aligned owing to the wall shear stresses at the printing temperature (20°C), which is not the gelation temperature of the collagen in the dECM. Additionally, the temperature (38°C) of the working stage was set to obtain a mechanically stable dECM-MA strut using the gelation temperature of the dECM bioink (Fig. 5(a)). After the printing, the aligned PVA phase from the cell-laden structure was leached, and finally, a uniaxial topographical cue could be achieved on the dECM-MA based bioink, as shown in the schematic of Fig. 5(b).
Figure 5.
(a) Schematic of the 3D cell printing process using the dECM-MA-based bioink and UV crosslinking process. (b) Schematics describing the alignment and leaching of PVA fibrils during the processes. (c) Rheological properties (G’ and complex viscosity (η*)) of dECM-MA-based bioinks (3 wt%) mixed with three different weight fractions of PVA (5, 10, and 15 wt%) (n = 5, *P < 0.05). (d) Optical, SEM, and live (green)/dead (red) images of the structures (8 × 8 × 0.5 mm3) printed using the bioinks at 1 d. (e) Initial cell viability and (f) cell loss of the three different bioinks at 1 d after the removal of the PVA components (n = 9, *P < 0.05).
Because the PVA weight fraction in the dECM-MA-80 (3 wt%)/PVA bioink can directly affect the cell viability for the printing process owing to the induced wall shear stress and can also influence the formation of the uniaxially aligned topographical pattern on a matrix material [24], we had to select a proper PVA concentration using various weight fractions of PVA (5–15 wt%).
To observe the viscoelastic properties of the dECM-MA/PVA bioinks, we measured the complex viscosity (η*) and G’ for the frequency sweep at the printing temperature (20°C). Figure 5(c) indicates that the G’ and η* of the bioinks gradually increased with the increasing weight fraction of PVA. Three different C2C12-laden bioinks (density: 1 × 107 cells/mL) with different weight fractions (5, 10, and 15 wt%) of PVA were printed using similar processing conditions (pneumatic pressure = 200 kPa; nozzle moving speed = 10 mm s−1; printing temperature = 20°C) (Fig. 5(d)). The printed cell-laden struts with different PVA concentrations were examined via optical images. The strut was stable at the PVA concentrations of 5 and 10 wt%, whereas for the PVA concentration of 15 wt%, the printed strut was unstable, with a discontinuous shape, due to the high viscosity of the bioink.
In addition, the PVA concentration is an important parameter for aligned fibrillated PVA molecules because water can penetrate between the PVA molecules and ultimately influence the alignment of the molecules [48]. Therefore, the concentration of PVA in the dECM-MA bioink should be considered from the viewpoint of the alignment of the fabricated structure. As shown in the SEM images of Fig. 5(d), a relatively well-developed anisotropic pore shape on the strut surface was observed in the PVA-concentration range of 10–15 wt%, whereas at a low concentration (5 wt%) the pore shape on the printed surface was round owing to the insufficient development of fibrillated PVA molecules [47].
As indicated by the live/dead results in Figs. 5(d) and (e), the bioinks printed at the PVA concentrations of 5 and 10 wt% had high cell viability (>94%), while the relatively low cell viability (27.2 ± 4.1%) of the bioink at 15 wt% PVA was observed owing to the relatively high viscosity of the bioink, as evidenced by the rheological data.
As a sacrificial material was used as a supplementary processing agent for the bioinks, the cell loss from the cell-laden struts during culture period should be considered, because the released cells from the struts cannot participate the important cell-to-cell function in the structure. As shown in Fig. 5(f), the cell loss from the three bioinks using the PVA was measured 1 d after fabrication. As indicated by the results, with the increasing weight fraction of the PVA component from the printed cell-laden strut, the cell loss was gradually increased.
Although all the best conditions were not satisfied, i.e., the cell viability, topographical shape, and cell loss of the printed cell-laden strut, we selected the optimal PVA concentration as 10 wt% in the dECM-MA bioink.
Effect of pneumatic pressure on cell alignment in dECM-MA structure
The pneumatic pressure in the 3D-printing process significantly affects the alignment of the fibrillated PVA component [24], eventually inducing the orientation of laden cells. To observe the effect of the pneumatic pressure on the alignment of cells, various pneumatic pressures were applied with fixed UV conditions and the following printing conditions: nozzle ID = 150 μm; nozzle moving speed = 10 mm s−1. Following the printing, the cell viability was measured after 1 d in cell culture, and the cell alignment was observed after 7 d of cell culturing.
Figure 6(a) shows the optical, SEM, live/dead (1 d), and DAPI/phalloidin (7 d) images of the cell-printed structures with three pneumatic pressures (100, 200, and 300 kPa). In the optical images, the relatively low pneumatic pressure (100 kPa) yielded an unstable strut formation owing to the insufficient printing amount, whereas for the high pressure (300 kPa), the uniaxial topographical structure on the strut surface was not formed, owing to the excessive extrudate-swell phenomenon, which disturbed the alignment of the fibrillated PVA component in the dECM-MA/PVA bioink. However, as shown in the optical and F-actin images, the formation of a stable structure and a highly aligned cell/ECM were observed in the cell-laden structure fabricated using 200 kPa.
Figure 6.
(a) Optical, SEM, live (green)/dead (red), and DAPI (blue)/phalloidin (green) images, along with the measured FWHM, for various pneumatic pressures (100, 200, and 300 kPa). (b) Initial cell viability calculated using the live/dead images at 1 d for the scaffolds (8 × 8 × 0.5 mm3) printed using the various pneumatic pressures (n = 9, *P < 0.05). (c) Circularity of the DAPI and (d) orientation factor of the F-actin calculated using the DAPI/phalloidin images of the structures (n = 9, *P < 0.05).
The initial cell viability at 1 d, circularity of the nucleus (4 × π × area/perimeter2), and orientation factor, ((90-φo)/90), based on the full width at half maximum (FWHM, φo) of F-actin at 7 d were measured, as shown in Figs. 6(b)–(d), respectively. The initial cell viability was significantly reduced at 300 kPa owing to the relatively high wall shear stress. For the alignment of the cell orientation, 100 and 200 kPa were appropriate; however, for the stable formation of the cell-laden structure, the appropriate pneumatic pressure in the printing process was 200 kPa.
In vitro cellular activities of the cell-laden structures
To observe the effect of the biochemical properties of dECM-MA, we used a control that was fabricated using GelMA with a similar topographical pattern (CON-1). CON-1 was fabricated using the same weight fraction of the fibrillated PVA sacrificial component. Furthermore, to assess the effect of the uniaxial topographical pattern of the dECM-MA laden with C2C12 cells on the in vitro cell-responses, cell-laden dECM-MA struts without a uniaxial topographical pattern (control; CON-2) were fabricated.
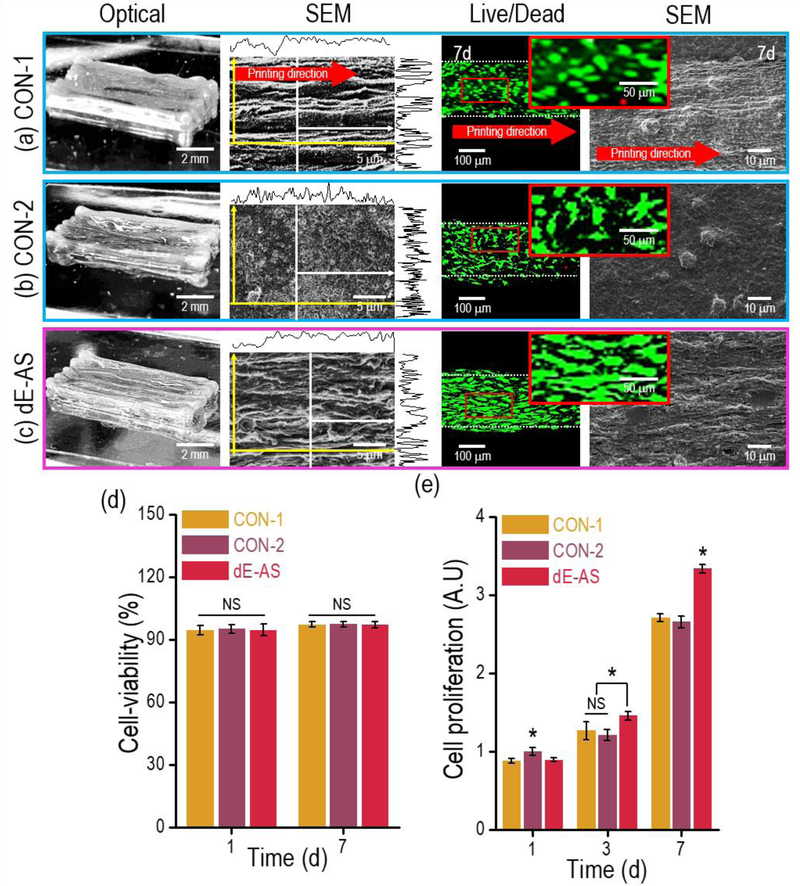
Figure 7(a)–(c) shows optical, SEM, and fluorescence images of the fabricated cell-laden structures, i.e., CON-1, CON-2, and the experimental group (dECM-MA having the aligned structure; dE-AS), taken at 1 and 7 d. As shown in the SEM images, a flat surface on CON-2 was observed, whereas, for the CON-1 and dE-AS structures, uniaxially grooved surface patterns were observed. The myoblasts cultured for 7 d in the CON-2 and dE-AS structures were spread over a wider range, exhibiting a spindle shape, than those cultured in the CON-1 structure.
Figure 7.
Optical, SEM, and live (green)/dead (red) images for the (a) CON-1, (b) CON-2, and (c) dE-AS structures (8 × 2 × 1 mm3). (d) Cell viability for the C2C12 cells in the printed structures (CON-1, CON-2, and dE-AS) calculated using the live/dead images at 7 d (n = 9, *P < 0.05). (e) Cell proliferation of the cells in the structures, measured using an AlamarBlue staining assay (n = 9, *P < 0.05).
The cell viability of the printed structures was measured using the live/dead images taken at 1 and 7 d (Figs. 7(a)–(c)). The cell viability was >90% for all structures (Fig. 7(d)). The cell proliferation (determined using an AlamarBlue assay) of dE-AS was significantly higher at 7 d compared to the others owing to the synergistic effect of the muscle-specific cell-growth components of dECM-MA and the microgrooved patterned surface, which induced more effectual cell-to-cell interactions (Fig. 7(e)) [49].
The morphologies of the cells in the structures after 7 and 14 d were observed using DAPI (blue) and phalloidin (green) (Figs. 8(a)–(c)). As expected, the nucleus density and the area fraction of the actin fibers were significantly higher in the dE-AS structure (Figs. 8(d) and (e)). Furthermore, the dE-AS structure exhibited far lower circularity of the nuclei (Fig. 8(f)) and higher alignment of the actin fibers after 7 and 14 d (based on the FWHM and orientation factor, Fig. 8(g)–(j)) than the CON-1 and CON-2 structures. Thus, the results indicate that the dE-AS structure effectively induced myoblast alignment with the synergistic effect of contact guidance and the biochemical component of the dECM.
Figure 8.
DAPI (blue)/phalloidin (green) images of C2C12 cells in the (a) CON-1, (b) CON-2, and (c) dE-AS structures (8 × 2 × 1 mm3) at 7 and 14 d. (d) Cell nucleus density, (e) F-actin area, and (f) circularity at 7 and 14 d calculated using the images (n = 9, *P < 0.05). Distribution of the orientation of the F-actin of (g) CON-1, (h) CON-2, and (i) dE-AS at 14 d. (j) Orientation factor for the F-actin of the cultured cells in the structures at 7 and 14 d (n = 9, *P < 0.05).
In vitro myotube formation and gene expression of cell-laden structures
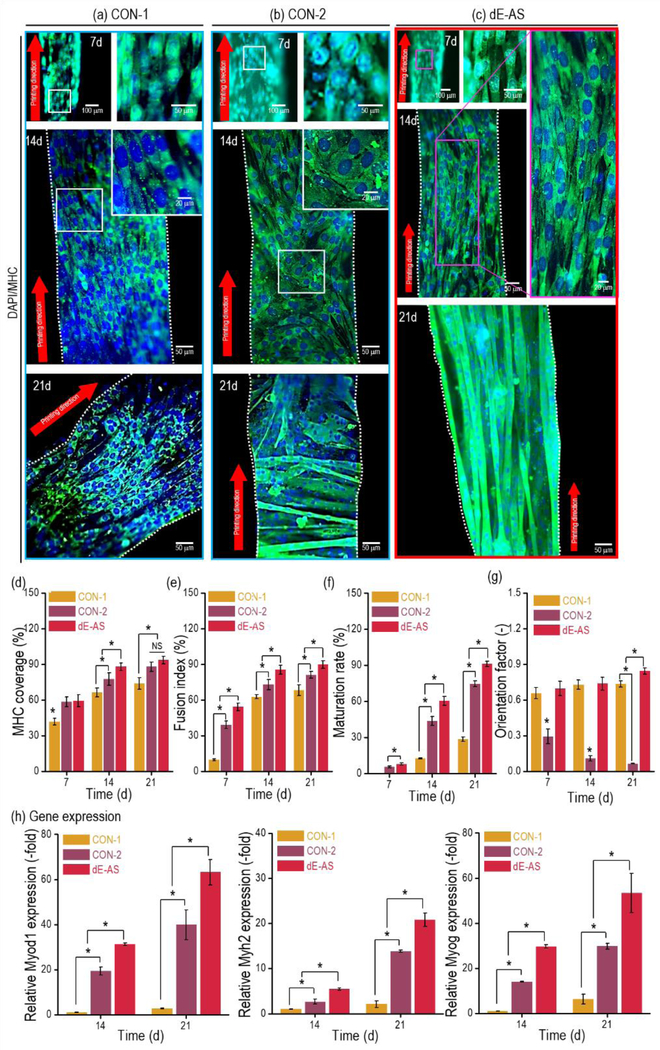
Immunofluorescence images showing the nuclei/myosin heavy chain (MHC, green), which is a marker of maturation of the skeletal muscle tissue, for the CON-1, CON-2, and dE-AS structures were taken after 7, 14, and 21 d (Figs. 9(a)–(c)). As indicated by the images, the dE-AS structure resulted in significantly higher myoblast alignment and even multinucleated myotube formation compared with those of the controls. In particular, for the CON-2 structure using the same dECM-MA bioink (but, without the micropattern), although the multinucleated myotubes were formed, a random distribution of the aligned myotubes was observed.
Figure 9.
DAPI (blue) and MHC (green) images of C2C12 cells in the (a) CON-1, (b) CON-2, and (c) dE-AS structures (8 × 2 × 1 mm3) at 7, 14, and 21 d. Quantitative analysis of the MHC: (d) MHC coverage, (e) fusion index, (f) maturation rate, and (g) orientation factor at 7, 14, and 21 d, calculated using the MHC images (n = 9, *P < 0.05). (h) Gene expression (Myod1, Myh2, and Myog), which was compared with that of Actb and then normalized by the level of cells in the CON-1 at 7 d (n = 9, *P < 0.05).
By analyzing the fluorescence images in four aspects—(1) the stained MHC area (MHC coverage); (2) the fusion index, i.e., the ratio of the number of nuclei in the MHC area to the total number of nuclei; (3) the rate of maturation, i.e., the ratio of the number of myotubes with more than five nuclei to the total number of myotubes; and (4) the orientation factor of the MHC—we examined the quantitative degree of myotube formation for several culture days. According to the results, although CON-1 exhibited a high alignment of MHC (but low myotube formation) and CON-2 exhibited high myotube formation (but a low MHC orientation), the dE-AS structure provided a far more cell-affiliated microcellular environment than the controls (CON-1 and CON-2) owing to the synergistic effect of the biochemical components derived from the dECM and the biophysical property induced by the uniaxially aligned topographical cue (Figs. 9(d)–(g)).
To observe the interaction between the C2C12 cells and matrix materials, focal adhesion kinase (FAK; vinculin), integrin-β1, MHC, and type-I collagen, which can react with porcine, were used (Supplementary Fig. S2) [50,51]. In the dECM-MA structures (CON-2 and dE-AS), high expression of vinculin was detected compared to that in the GelMa-based structure. However, much higher vinculin expression in the dE-AS at 14 d was measured than that of the CON-2 (Supplementary Figs. S2(a,b)). In addition, since the interaction of integrin-β1 and type-I collagen can upregulate the cellular activities [52], we believe that the type-I collagen can interact with the integrin-β1 receptor on the cell-membrane and even may affect extracellular signals into cells (Supplementary Fig. S2(d)). The results can be an evidence that the C2C12 cells interacted with the dECM-MA, and the guidance of the matrix material could affect the alignment and myotube formation of the myoblasts.
The cultured C2C12 cells in the CON-1, CON-2, and dE-AS structures were stained using laminin and sarcomeric α-actinin (Supplementary Figs. S3(a,b)). The cells in the structures well secreted laminin, abundant in the basement membrane of skeletal muscle tissue [53,54], to form a basement membrane-like structure. In addition, the expression of sarcomeric α-actinin, a muscle contraction unit, was observed in the scaffolds. While striation pattern of sarcomere was not observed in the CON-1, the striated sarcomeres were detected clearly in the CON-2 and dE-AS scaffolds at 28 d (Supplementary Fig. S3(b)). Furthermore, the patterned myotubes were uniaxially aligned to the printed direction in the dE-AS scaffold. The results showed that the biochemical and topographical cues of the dE-AS scaffolds provided much efficient microenvironment to the skeletal muscle cells for inducing myogenic differentiation.
The levels of myogenic genes (Myod1, Myh2, and Myog) in the structures cultured for 14 and 21 d were observed using an RT-PCR. The expression was normalized with the Actb, and the value of CON-1 at 7 d was set as 1, as shown in Fig. 9(h). As indicated by the immunofluorescence images, the expression levels of dE-AS were significantly greater than those of the controls, because the biochemical and topographical cues clearly induced highly efficient myoblast differentiation.
Conclusions
We successfully developed the biochemically and topographically mimicked skeletal muscle constructs combined by the skeletal muscle tissue-derived dECM-MA bioink and the PVA fibrillation/leaching by the modified 3D bioprinting process. By optimizing bioink compositions and 3D printing parameters, we were able to fabricate cellularized skeletal muscle-like tissue constructs having biochemical and topographical cues. We demonstrated that these printed constructs accelerated the myogenic differentiation by the dECM components, while the cellular alignment was achieved by topographical cues. This study suggests that this innovative dECM-based cell-based 3D bioprinting may lead to efficient functional recovery of damaged or diseased skeletal muscle tissues.
Supplementary Material
Acknowledgments
This study was supported by the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology (MEST) (NRF-2018R1A2B2005263) and National Institutes of Health (1P41EB023833-346 01).
References
- [1].Jarvinen TA, Jarvinen TL, Kaariainen M, Aarimaa V, Vaittinen S, Kalimo H, Jarvinen M, Muscle Injuries: Optimising Recovery, Best Pract. Res. Clin. Rheumatol 21 (2007) 317–331. [DOI] [PubMed] [Google Scholar]
- [2].Lagrange M, Boulade-Ladame C, Mailly L, Weiss E, Orfanoudakis G, Deryckere F, Intracellular scFvs against the viral E6 oncoprotein provoke apoptosis in human papillomavirus-positive cancer cells, Biochem. Bioph. Res. Co 361 (2007) 487–492. [DOI] [PubMed] [Google Scholar]
- [3].Huang YC, Dennis RG, Larkin L, Baar K, Rapid formation of functional muscle in vitro using fibrin gels, J. Appl. Physiol. 98 (2005) 706–713. [DOI] [PubMed] [Google Scholar]
- [4].Jana S, Leung M, Chang J, Zhang M, Effect of nano- and micro-scale topological features on alignment of muscle cells and commitment of myogenic differentiation, Biofabrication 6 (2014) 035012. [DOI] [PubMed] [Google Scholar]
- [5].Koning M, Harmsen MC, van Luyn MJA, Werker PMN, Current opportunities and challenges in skeletal muscle tissue engineering,. J Tissue Eng. Regen. Med. 3 (2009) 407–415. [DOI] [PubMed] [Google Scholar]
- [6].Wolf MT, Dearth CL, Sonnenberg SB, Loboa EG, Badylak SF, Naturally derived and synthetic scaffolds for skeletal muscle reconstruction, Adv. Drug Deliv. Rev. 84 (2015) 208–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hosseini V, Ahadian S, Ostrovidov S, Camci-Unal G, Chen S, Kaji H, Ramalingam M, Khademhosseini A, Engineered Contractile Skeletal Muscle Tissue on a Microgrooved Methacrylated Gelatin Substrate, Tissue Eng. Part A 18 (2012) 2453–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Duffy RM, Sun Y, Feinberg AW, Understanding the Role of ECM Protein Composition and Geometric Micropatterning for Engineering Human Skeletal Muscle, Ann. Biomed. Eng. 44 (2016) 2076–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Badylak SF, Freytes DO, Gilbert TW, Extracellular matrix as a biological scaffold material: Structure and function, Acta Biomater. 5 (2009) 1–13. [DOI] [PubMed] [Google Scholar]
- [10].Badylak SF, Brown BN, Gilbert TW, Daly KA, Huber A, Biologic scaffolds for constructive tissue remodeling, Biomaterials 32 (2011) 316–319. [DOI] [PubMed] [Google Scholar]
- [11].Noor N, Shapira A, Edri R, Gal I, Wertheim L, Dvir T, 3D Printing of Personalized Thick and Perfusable Cardiac Patches and Hearts, Adv. Sci (2019) 1900344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Choi YJ, Jun YJ, Kim DY, Yi HG, Chae SH, Kang J, Lee J, Gao G, Kong JS, Jang J, Chung WK, Rhile JW, Cho DW, A 3D cell printed muscle construct with tissue-derived bioink for the treatment of volumetric muscle loss, Biomaterials 206 (2019) 160. [DOI] [PubMed] [Google Scholar]
- [13].Choudhury D, Tun HW, Wang T, Naing MW, Organ-Derived Decellularized Extracellular Matrix: A Game Changer for Bioink Manufacturing?, Trends Biotechnol. 36 (2018) 787–805. [DOI] [PubMed] [Google Scholar]
- [14].Visser J, Levett PA, te Moller NCR, Besems J, Boere KWM, van Rijen MHP, de Grauw JC, Dhert WJA, van Weeren PR, Malda J, Crosslinkable Hydrogels Derived from Cartilage, Meniscus, and Tendon Tissue, Tissue Eng. Part A 21 (2015) 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Murphy SV, Atala A, 3D bioprinting of tissues and organs, Nature Biotechnol. 32 (2014) 773–785. [DOI] [PubMed] [Google Scholar]
- [16].Derby B, Printing and Prototyping of Tissues and Scaffolds, Science 338 (2012) 921–926. [DOI] [PubMed] [Google Scholar]
- [17].Leong KF, Cheah CM, Chua CK, Solid freeform fabrication of three-dimensional scaffolds for engineering replacement tissues and organs, Biomaterials 24 (2003) 2363–2378. [DOI] [PubMed] [Google Scholar]
- [18].Ouyang L, Highley CB, Sun W, Burdick JA, A Generalizable Strategy for the 3D Bioprinting of Hydrogels from Nonviscous Photo-crosslinkable Inks, Adv. Mater. 29 (2017) 1604983. [DOI] [PubMed] [Google Scholar]
- [19].Kim YB, Lee H, Kim GH, Strategy to Achieve Highly Porous/Biocompatible Macroscale Cell Blocks, Using a Collagen/Genipin-bioink and an Optimal 3D Printing Process, ACS Appl. Mater. Interfaces 8 (2016) 32230–32240. [DOI] [PubMed] [Google Scholar]
- [20].Kang HW, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, A 3D bioprinting system to produce human-scale tissue constructs with structural integrity, Nature Biotechnol. 34 (2016) 312–319. [DOI] [PubMed] [Google Scholar]
- [21].Rutz AL, Hyland KE, Jakus AE, Burghardt WR, Shah RN, A Multimaterial Bioink Method for 3D Printing Tunable, Cell-Compatible Hydrogels, Adv. Mater. 27 (2015) 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Benders KE, van Weeren PR, Badylak SF, Saris DB, Dhert WJ, Malda J, Extracellular matrix scaffolds for cartilage and bone regeneration, Trends Biotechnol. 31 (2013) 169–176. [DOI] [PubMed] [Google Scholar]
- [23].Kim W, Kim M, Kim GH, 3D-Printed Biomimetic Scaffold Simulating Microfibril Muscle Structure, Adv. Funct. Mater. 28 (2018) 1800405. [Google Scholar]
- [24].Kim M, Kim W, Kim GH, Topologically Micropatterned Collagen and Poly(ε-caprolactone) Struts Fabricated Using the Poly(vinyl alcohol) Fibrillation/Leaching Process To Develop Efficiently Engineered Skeletal Muscle Tissue, ACS Appl. Mater. interfaces 9 (2017) 43459–43469. [DOI] [PubMed] [Google Scholar]
- [25].Johnson TD, DeQuach JA, Gaetani R, Ungerleider J, Elhag D, Nigam V, Behfar A, Christman KL, Human versus porcine tissue sourcing for an injectable myocardial matrix hydrogel, Biomater. Sci 2 (2014) 735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Singelyn JM, DeQuach JA, Seif-Naraghi SB, Littlefield RB, Schup-Magoffin PJ, Christman KL, Naturally derived myocardial matrix as an injectable scaffold for cardiac tissue engineering, Biomaterials 30 (2009) 5409–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Habeeb AFSA, Determination of free amino groups in proteins by trinitrobenzenesulfonic acid, Anal. Biochem. 14 (1966) 328–336 [DOI] [PubMed] [Google Scholar]
- [28].Van Den Bulcke AI, Bogdanov B, De Rooze N, Schacht EH, Cornelissen M, Berghmans H, Structural and Rheological Properties of Methacrylamide Modified Gelatin Hydrogels, Biomacromolecules 1 (2000) 31–38. [DOI] [PubMed] [Google Scholar]
- [29].Yin J, Yan M, Wang Y, Fu J, Suo H, 3D Bioprinting of Low-Concentration Cell-Laden Gelatin Methacrylate (GelMA) Bioinks with a Two-Step Cross-linking Strategy, ACS Appl. Mater. Interfaces 10 (2018) 6849–6857. [DOI] [PubMed] [Google Scholar]
- [30].Ying GL, Jiang N, Maharjan S, Yin YX, Chai RR, Cao X, Yang JZ, Miri AK, Hassan S, Zhang YS, Aqueous Two-Phase Emulsion Bioink-Enabled 3D Bioprinting of Porous Hydrogels, Adv. Mater. 30 (2018) 1805460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Skardal A, Devarasetty M, Kang HW, Mead I, Bishop C, Shupe T, Lee SJ, Jackson J, Yoo JJ, Soker S, Atala A, A hydrogel bioink toolkit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs, Acta Biomater. 25 (2015) 24–34. [DOI] [PubMed] [Google Scholar]
- [32].Badylack SF, The extracellular matrix as a biologic scaffold material, Biomatrials 28 (2007) 3587. [DOI] [PubMed] [Google Scholar]
- [33].Pati1 F, Jang J, Ha DH, Kim SW, Rhie JW, Shim JH, Kim DH, Cho DW, Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink, Nat. Comm 5 (2014) 3587–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rijal G, The decellularized extracellular matrix in regenerative medicine, Regen. Med. 12 (2017) 475–477. [DOI] [PubMed] [Google Scholar]
- [35].Crapo PM, Gilbert TW, Badylak SF, An overview of tissue and whole organ decellularization processes, Biomaterials 32 (2011) 3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Choi YJ, Park SJ, Yi HG, Lee H, Kim DS, Cho DW, Muscle-derived extracellular matrix on sinusoidal wavy surfaces synergistically promotes myogenic differentiation and maturation, J. Mater. Chem. B 6 (2018) 5530–553. [DOI] [PubMed] [Google Scholar]
- [37].Toprakhisar B, Nadernezhad A, Bakirci E, Khani N, Skvortsov GA, Koc B, Development of Bioink from Decellularized Tendon Extracellular Matrix for 3D Bioprinting, Macromol. Biosci. 18 (2018) 1800024. [DOI] [PubMed] [Google Scholar]
- [38].Komez A, Baran ET, Erdem U, Hasirici N, Hasirici V, Construction of a patterned hydrogel—fibrous mat bilayer structure to mimic choroid and Bruch’s membrane layers of retina, J. Biomed. Mater. Res. A 104 (2016) 2166–2177. [DOI] [PubMed] [Google Scholar]
- [39].Ali M, Kumar PR A, Yoo JJ, Zahran F, Atala A, Lee SJ, A Photo-Crosslinkable Kidney ECM-Derived Bioink Accelerates Renal Tissue Formation, Adv. Healthcare Mater. (2019) 1800992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Arsic N, Zacchigna S, Zentilin L, Ramirez-Correa G, Pattarini L, Salvi A, Sinagra G, Giacca Mauro., Vascular endothelial growth factor stimulates skeletal muscle regeneration in vivo, Mole. Ther. 10 (2004) 844–854. [DOI] [PubMed] [Google Scholar]
- [41].Cannon JG, St BA. Pierre, Cytokines in exertion-induced skeletal muscle injury, Mol. Cell Biochem. 175 (1998) 159–167. [DOI] [PubMed] [Google Scholar]
- [42].Tonkin J, Temmerman L, Sampson RD, Gallego-Colon E, Barberi L, Bilbao D, Schneider MD, Musarò A, Rosenthal N, Monocyte/macrophage-derived IGF-1 orchestrates murine skeletal muscle regeneration and modulates autocrine polarization, Mol. Ther. 23 (2015) 1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].McCusker RH, Campion DR, Effect of growth hormone-secreting tumours on skeletal muscle cellularity in the rat, J. Endocr. 111 (1986) 279–285. [DOI] [PubMed] [Google Scholar]
- [44].Winbanks CE, Chen JL, Qian H, Liu Y, Bernardo BC, Beyer C, Watt KI, Thomson RE, Connor T, Turner BJ, McMullen JR, Larsson L, McGee SL, Harrison CA, Gregorevic P, The bone morphogenetic protein axis is a positive regulator of skeletal muscle mass, J. Cell Biol. 203 (2013) 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Serbo JV, Gerecht S, Vascular tissue engineering: biodegradable scaffold platforms to promote angiogenesis, Stem Cell Res. Ther. 4 (2013) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ouyang L, Yao R, Zhao Y, Sun W, Effect of bioink properties on printability and cell viability for 3D bioplotting of embryonic stem cells, Biofabrication 8 (2016) 035020. [DOI] [PubMed] [Google Scholar]
- [47].Lyoo WS, Kim JH, Choi JH, Kim BC, Blackwell J, Role of Degree of Saponification in the Shear-Induced Molecular Orientation of Syndiotacticity-Rich Ultrahigh Molecular Weight Poly(vinyl alcohol), Macromolecules 34 (2001) 3982–3987. [Google Scholar]
- [48].Lyoo WS, Ha WS, In situ fibrillation of poly(vinyl alcohol) during saponification of poly(vinyl ester) (1). Chemorheological and morphological investigations of in situ fibrillation, Polymer 40 (1999) 497–505. [Google Scholar]
- [49].Lee JS, Shin J, Park HM, Kim YG, Kim BG, Oh JW, Cho SW, Liver Extracellular Matrix Providing Dual Functions of Two-Dimensional Substrate Coating and Three-Dimensional Injectable Hydrogel Platform for Liver Tissue Engineering, Biomacromolecules 15 (2013) 206–218. [DOI] [PubMed] [Google Scholar]
- [50].Zhou Q, Zhao Z, Zhou z., Zhang G, Chiechi RC, van Rijn P, directing mesenchymal stem cells with gold nanowire arrays, Adv. Mater. Interfaces 5 (2018) 1800334. [Google Scholar]
- [51].Navaei A, Saini H, Christenson W, Sullivan RT, Ros R, Nikkhah M, Gold nanorod-incorporated gelatin-based conductive hydrogels for engineering cardiac tissue constructs, Acta Biomater. 41 (2016) 133–146. [DOI] [PubMed] [Google Scholar]
- [52].Maller O, Hansen KC, Lyons TR, Acerbi I, Weaver VM, Prekeris R, Tan AC, Schedin P, Collagen architecture in pregnancy-induced protection from breast cancer, J. Cell Sci. 126 (2013) 4108–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sane JR, The basement membrane/basal lamina of skeletal muscle, J. Biol. Chem. 278 (2003) 12601–12604. [DOI] [PubMed] [Google Scholar]
- [54].Reinhard JR, Lin S, McKee KK, Meinen S, Crosson S. c., Sury M, Hobbs S, Maier G, Yurchenco PD, Rüegg MA, Linker proteins restore basement membrane and correct LAMA2-related muscular dystrophy in mice, Sci. Transl. Med 9 (2017) eaal4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.