Abstract
The vasopressin system has been implicated in the regulation of social behavior and cognition in humans, nonhuman primates and other social mammals. In chimpanzees, polymorphisms in the vasopressin V1a receptor gene (AVPR1A) have been associated with social dimensions of personality, as well as to responses to sociocommunicative cues and mirror self-recognition. Despite evidence of this association with social cognition and behavior, there is little research on the neuroanatomical correlates of AVPR1A variation. In the current study, we tested the association between AVPR1A polymorphisms in the RS3 promotor region and gray matter covariation in chimpanzees using magnetic resonance imaging and source-based morphometry. The analysis identified 13 independent brain components, three of which differed significantly in covariation between the two AVPR1A genotypes (DupB−/− and DupB+/−; P < .05). DupB+/− chimpanzees showed greater covariation in gray matter in the premotor and prefrontal cortex, basal forebrain, lunate and cingulate cortex, and lesser gray matter covariation in the superior temporal sulcus and postcentral sulcus. Some of these regions were previously found to differ in vasopressin and oxytocin neural fibers between nonhuman primates, and in AVPR1A gene expression in humans with different RS3 alleles. This is the first report of an association between AVPR1A and gray matter covariation in nonhuman primates, and specifically links an AVPR1A polymorphism to structural variation in the social brain network. These results further affirm the value of chimpanzees as a model species for investigating the relationship between genetic variation, brain structure and social cognition with relevance to psychiatric disorders, including autism.
Keywords: AVPR1A, brain structure, chimpanzee, gray matter, vasopressin
1 |. INTRODUCTION
Vasopressin is a neuropeptide implicated in the development and maintenance of social relationships, social cognition and social communication in mammals.1–4 For example, in voles, species differences and individual variation in brain regions specific to vasopressin receptor gene (AVPR1A) expression are associated with pair bonding, social monogamy and other dimensions of social behavior.3,5–9 Less is known about the functional role of vasopressin in social behavior and cognition of primates, including humans. Like voles, there is some evidence of taxonomic differences in AVPR1A polymorphisms among primate species10 but, unlike voles, they do not appear to be as closely related to pair bonding and mating systems (ie, social monogomy1,11–13).
Links between vasopressin and other aspects of social behavior, on the other hand, have been demonstrated in both apes and monkeys. In rhesus macaques, variation in sociability is associated with measures of vasopressin in cerebral spinal fluid,14 despite no significant associations between AVPR1A alleles and social behaviors, such as grooming, passive contact, approaches and aggression.15 In chimpanzees, several studies have shown that polymorphisms in AVPR1A are associated with individual variation in different dimensions of personality, including social “smarts” (using coalitions and engaging in reciprocal grooming and play) and sociability.16–20 Additional studies have shown that chimpanzee AVPR1A polymorphisms are associated with responses to sociocommunicative cues,21,22 videos and sounds of unfamiliar individuals23 and mirror self-recognition.24 Also, there is one report of an association between AVPR1A variation and the neuroticism dimension of personality in marmosets.25
In humans, AVPR1A polymorphic variation in the microsatellite element RS3 (a repetitive sequence element in the 5′ flanking region) is linked to variation in social behavior, including empathy and altruistic behavior, as well as pair bonding in males but not females.26–30 AVPR1A has also been proposed as a candidate susceptibility gene for autism spectrum disorder (ASD), a neurodevelopmental disorder that is marked by atypical development of social relationships and sociocognitive abilities.31 Indeed, several studies have reported that AVPR1A polymorphic variation is associated with scores on ASD diagnostic questionnaires, such as the Autism Diagnostic Observation Scale-Generic, Autism Diagnostic Interview-Revised32 and Autism Quotient.33
Despite the evidence implicating polymorphisms in the AVPR1A gene in different dimensions of social behavior and cognition, there is remarkably little data on the neuroanatomical or neurofunctional correlates that may link genetic variation and behavior in primates. In humans, variation in RS3 is related to activation of the amygdala during a face recognition task,34,35 an observation that supports the amygdala theory of autism.36,37 Immunohistochemistry analyses have revealed species similarities and differences in both oxytocin and vasopressin fibers in regions of the human, chimpanzee and rhesus macaque cortex.38 With respect to vasopressin, fibers were found in the anterior olfactory nucleus, primary olfactory cortex and subgenual region of the anterior cingulate cortex of all three species. However, vasopressin fibers were present in the agranular insula cortex and orbitofrontal cortex of humans and chimpanzees, but were absent in rhesus monkeys, and were found in the dysgranular insula and frontal operculum in humans only.38 Species differences have also been found in AVPR1A expression, with receptor binding in the amygdala, stria terminalis, lateral septum, hypothalamus and brainstem of rhesus macaques (similar to rodents), but spread throughout the cortex of titi monkeys (particularly in the insula, cingulate and occipital cortex).39–41
Here, we tested for associations between polymorphic variation in the RS3 portion of AVPR1A and gray matter covariation in chimpanzees (Pan troglodytes). RS3 is composed of two related repetitive sequence elements referred to as DupA and DupB.10 Interestingly, chimpanzees are polymorphic at the same AVPR1A locus as humans; however, chimpanzee alleles include either both DupA and DupB in RS3 microsatellite element in the 5′ flanking region (DupB+), like humans, or completely lack the DupB (DupB-).10 This creates two populations of chimpanzees that are either (a) homozygous for alleles lacking DupB (DupB−/−) or (b) heterozygous with one allele having both repetitive sequences (DupB+/−), as very few chimpanzees are DupB+/+. In the current study, we compared gray matter covariation between these two populations of chimpanzees, using source-based morphometry (SBM).42 SBM is a relatively new method that characterizes gray and white matter structural covariation in samples of magnetic resonance imaging (MRI) scans.43,44 Unlike univariate analytic methods, such as voxel-based morphometry (VBM), SBM is a multivariate, data-driven analytic approach that utilizes information about relationships among voxels and groups those carrying similar information across the brain. Without requiring prior determination of regions of interest, the resulting components, or sources, are identified based on the spatial information between voxels grouped in a natural manner and represent similar covariation networks between subjects. This approach has been described as a multivariate version of VBM.45 At the individual level, SBM produces a weighted score that reflects each subject’s relative contribution to the creation of each spatial component. Therefore, we compared DupB+/− and DupB−/− chimpanzees on these weighted scores for each of the components derived from the SBM analysis. Structures having high covariation scores vary together more strongly and the gray matter volume of one region can be used to predict the other. We hypothesized that DupB+/− and DupB−/− chimpanzees would differ on one or more of the SBM components. Given the function of vasopressin in social relationships and cognition, we further hypothesized that any differences in component scores between DupB+/− and Dup−/− chimpanzees would include brain regions comprising the social brain network (eg, prefrontal cortex, insula, cingulate and superior temporal sulcus).46–48
2 |. MATERIALS AND METHODS
2.1 |. Subjects
In the current study, we used 142 structural T1-weighted MRI scans obtained from captive chimpanzees (82 females and 60 males). The subjects were socially housed at either the Yerkes National Primate Research Center (YNPRC, n = 54) or the National Center for Chimpanzee Care at The University of Texas MD Anderson Cancer Center (NCCC, n = 88). The chimpanzees ranged in age from 12 to 51 years (M = 26.32, SD = 9.70). We used a matched-subjects design in this study. For this analysis, we included 71 DupB+/− and 71 DupB−/− chimpanzees (see below) that were matched on scanner magnet (1.5 and 3.0 Tesla), sex, rearing history and age. All work was carried out in accordance with the guidelines laid out by the National Institutes of Health in the USA, and were approved by the Institutional Animal Care and Use Committees at both NCCC and YNPRC.
2.2 |. DNA extraction, sequencing and genotyping
Prior to this study, all subjects were previously genotyped for the AVPR1A gene (see detailed methods21), and were classified as either homozygous (DupB−/−) or heterozygous (DupB+/−) for the RS3 microsatellite deletion. Briefly, DNA samples were isolated from buccal swabs or blood samples using Puregene DNA Purification system (Gentra, Minneapolis, MN) and stored at −80 °C until genotyping. Each individual was genotyped for the AVPR1A DupA/B polymorphism by polymerase chain reaction (PCR) using the extracted DNA and a forward primer 5′ GCATGGTAGCCTCTCTTTAAT and a reverse primer of 5′ CATACACATGGAAAGCACCTAA with an annealing temperature of 57°C for 30 cycles: 95°C, 5 minutes; 30 × (95°C, 30 seconds; 57°C, 30 seconds; 72°C, 3 minutes; 72°C, 10 minutes; 4°C, hold). PCR amplification was performed using the Epicenter Failsafe kit using premix H (Illumina Inc., Madison, WI) according to the manufacturer’s instructions. PCR products (20 μL) were resolved on a 2% agarose gel (SeaKem Agarose LE, Lonza, Basel, Switzerland) at 100 V for 45 minutes with a 100 bp DNA ladder (New England Biolabs, Ipswich, MA) in tris-borate-EDTA (TBE) buffer. The DupB+ allele resulted in a band of ~900 bp, while the DupB– allele was ~570 bp long, and genotypes were visually assigned. All genotyping was run in duplicate and confirmed in both analyses.
2.3 |. MRI acquisition
We obtained MRI scans using techniques previously described.44,49 Briefly, subjects were initially immobilized by injection of ketamine (10 mg/kg) or telazol (2–6 mg/kg) and subsequently anesthetized with propofol (40–60 mg/kg) per standard institutional guidelines. The subjects were then transported to the imaging facility and remained anesthetized for the duration of the scan (40–60 minutes depending on brain size), as well as during transportation back to a recovery cage (approximately 75–120 minutes in total). The subjects were placed in the scanner chamber in a supine position with their head inside the human-head coil. The NCCC chimpanzees (n = 88) were scanned using a 1.5-Tesla scanner (Phillips, Model 51), whereas the YNPRC individuals (n = 54) were scanned using a 3-Tesla scanner (Siemens Trio, Siemens Medical Solutions USA, Inc., Malvern, Pennsylvania). For the NCCC chimpanzees, T1-weighted images were collected in the transverse plane using a gradient echo protocol (pulse repetition = 19 milliseconds, echo time = 8.5 milliseconds, number of signals averaged = 8, and a 256 × 256 matrix). For the YNPRC chimpanzees, T1-weighted images were collected using a 3D gradient echo sequence (pulse repetition = 2300 milliseconds, echo time = 4.4 milliseconds, number of signals averaged = 3, matrix size = 320 × 320). After completing the MRI scan collection, the subjects were temporarily housed in a single cage for 6–12 hours to allow them to recover from the anesthesia, after which they were returned to their home group.
2.4 |. Postimage processing and analyses
Following acquisition, the MRI data were processed using a Macintosh computer. The procedures, detailed below, were completed twice: once for the images acquired from the 1.5-Tesla scanner and again for those acquired from the 3.0-Tesla scanner. First, we imported the raw DICOM files into 3D Slicer 4 (www.3Dslicer.org) and converted each into NIfTI format.50,51 Next we used the brain extraction tool (BET) function in FMRIB software library (FSL) (Analysis Group, FMRIB, Oxford, UK) for skull-stripping, using fractional intensity thresholds ranging between 0.35 and 0.80.52,53 The skull-stripped brains were then imported into 3D Slicer for N4ITK bias correction (spline distance = 50, bias field = 0.15, convergent threshold = 0.00154–58). Using the DWI Denoising Package for MATLAB (R2015b; Mathworks, Natick, Massachusetts), the scans were then denoised using an optimized nonlocal means denoising filter.59 The scans were then resampled at 0.625 isotropic voxels and aligned in radiological space in Analyze 11.0 (AnalyzeDirect, Overland Park, Kansas), using a capsule placed during the imaging process as a left-right orientation marker. Finally, we used FLIRT in FSL to perform a 12-parameter affine linear registration60,61 of the processed scan to a chimpanzee template brain.62
We used the FSL-VBM pipeline (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSLVBM) to process each preprocessed scan. The FSL-VBM pipeline included (a) segmentation of each scan into gray and white matter, (b) linear registration of each scan to a standard chimpanzee template,62 (c) creation of a study-specific gray matter template,63–65 (d) nonlinear registration of each subject’s gray matter images to the study-specific template, (e) modulation of the gray matter volume by use of a Jacobian warp to correct for local expansion or contraction of gray matter within each voxel, and (f) smoothing with an isotropic Gaussian kernel with a sigma of 2 mm.
2.5 |. SBM and statistical analyses
SBM analysis was performed using the software GIFT (http://icatb.sourceforge.net).45 See Xu et al45 for a detailed description of the computational methods used to develop the SBM analysis. We imported the smoothed gray matter volumes for each subject and allowed the software to estimate the number of components based on the independent components analysis using a neural network algorithm. This initial SBM analysis revealed a total of 13 components (see below). At the individual level, SBM produces a weighted score that reflects each subject’s relative contribution to the creation of each spatial component. These individual weighted scores for each component were analyzed using a multivariate analysis of covariance (MANCOVA), with weighted scores as the dependent variables and AVPR1A genotype as the independent variable. Many of the chimpanzees within each population (YNPRC and NCCC) were related to one another; therefore, we used relatedness as a covariate in the analysis. Relatedness coefficients were based on the entire pedigree66 and calculated using ENDOG v4.8.67 This program uses the dam and sire information from the entire pedigree to calculate average relatedness (or the probability that a randomly chosen allele belongs to a particular individual in the population). After identifying which SBM components differed between the AVPR1A genotypes, we visualized and identified significant regions within each component. To do so, we registered the component maps (scaled to SD units and z scores) to the standard chimpanzee template brain, and set the z-score threshold at |z| ≥ 3.00 as in previous studies in humans and chimpanzees.42,45 All brain regions reaching this threshold were considered significant. The volume of each region was then measured using the region-of-interest tool in Analyze 11.0.
3 |. RESULTS
The SBM analysis identified 13 independent components and calculated an individual weighted score reflecting each subject’s contribution to each component. The mean weighted score for the DupB +/− and DupB−/− chimpanzees for each component are shown in Table 1 (calculated by averaging each subject’s relative contribution to the creation of the components). The overall MANCOVA, examining the differences in these weighted scores between AVPR1A genotypes while controlling for relatedness, revealed a significant effect of genotype (F13,127 = 2.44, P = .006). The subsequent univariate F tests revealed significant differences between the DupB+/− and DupB−/− chimpanzees on component 4 (F1,139 = 5.80, P = .017), component 5 (F1,139 = 5.78, P = .018) and component 6 (F1,139 = 11.02, P = .001). For components 4 and 5, DupB+/− chimpanzees had higher gray matter covariation values than DupB−/− apes, whereas the opposite pattern was found for component 6. Higher covariation values imply that the regions within a single component are strongly correlated and, within the component, gray matter volume in one region can predict gray matter volume in other regions.
TABLE 1.
Mean (SE) weighted score for the DupB+/− and DupB −/− chimpanzees for each component (adjusted for relatedness)
| Component | AVPR1A genotype | |
|---|---|---|
| DupB+/− | DupB−/− | |
| 1 | 0.011 (0.116) | −0.011 (0.116) |
| 2 | −0.078 (0.095) | 0.078 (0.095) |
| 3 | −0.052 (0.109) | 0.052 (0.109) |
| 4 | 0.179 (0.105)* | −0.179 (0.105) |
| 5 | 0.190 (0.112)* | −0.190 (0.112) |
| 6 | −0.242 (0.103) | 0.242 (0.103)* |
| 7 | 0.022 (0.107) | −0.022 (0.107) |
| 8 | 0.005 (0.107) | −0.005 (0.107) |
| 9 | 0.009 (0.098) | −0.009 (0.098) |
| 10 | 0.024 (0.096) | −0.024 (0.096) |
| 11 | 0.022 (0.101) | −0.022 (0.101) |
| 12 | 0.038 (0.100) | −0.038 (0.100) |
| 13 | −0.009 (0.113) | 0.009 (0.113) |
Significantly greater gray matter covariation (P < .05).
The specific brain regions comprising components 4–6 are shown in Figure 1, and the volumes corresponding to each brain region are provided in Table 2. Component 4 was comprised of eight regions, largely in the premotor and prefrontal cortex, including dorsal prefrontal cortex (bilateral), superior medial prefrontal cortex (bilateral), precentral inferior sulcus (bilateral), middle central sulcus and precentral gyrus (both left), anterior insula (left), superior parietal sulcus (bilateral) and the inferior frontal sulcus (right). Component 5 was comprised of 14 brain regions including basal forebrain (bilateral), lunate sulcus (bilateral), motor hand area of the precentral gyrus (bilateral), cingulate gyrus (bilateral), ventral portion of the inferior frontal sulcus (bilateral), postcentral sulcus (bilateral), lateral occipital sulcus (bilateral), superior frontal gyrus, primary visual cortex (right), superior frontal orbital sulcus (left), inferior precentral gyrus (left) and hippocampus (left). Component 6 was comprised of 12 brain regions including superior temporal sulcus (bilateral), postcentral sulcus (bilateral), inferior portion of the lunate sulcus (bilateral), inferior frontal gyrus (bilateral), posterior cingulate gyrus (bilateral), lateral occipital sulcus (left), angular gyrus (left), anterior portion of the inferior temporal sulcus (left), inferior and middle temporal sulcus (right), intraparietal sulcus (right), medial parietal lobe (right), inferior occipital lobe (right) and Heschl’s gyrus (left).
FIGURE 1.
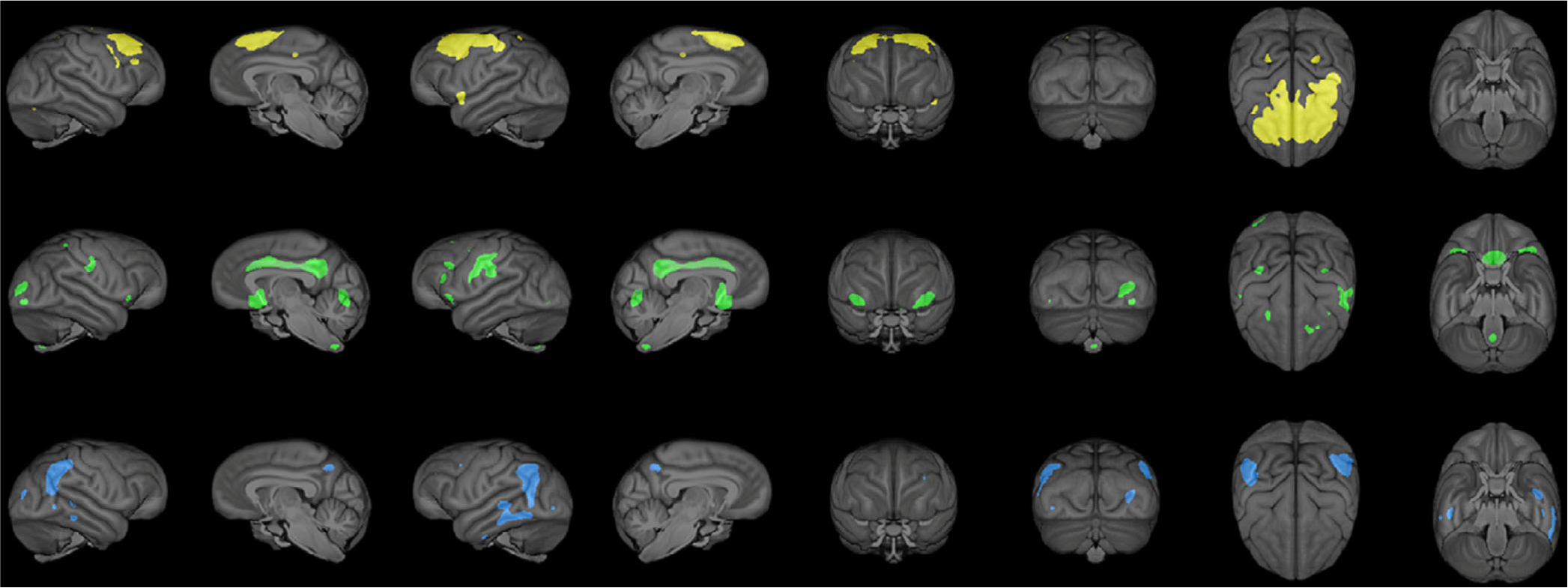
A 3D rendering of the surface area for components 4 (yellow), 5 (green) and 6 (blue). Yellow and green (top and middle) indicate regions where chimpanzees with the DupB+/− AVPR1A genotype had greater gray matter covariation values than DupB−/− chimpanzees, while blue (bottom) indicates regions where chimpanzees with the DupB−/− genotype had greater gray matter covariation values
TABLE 2.
Regions and corresponding volumes (mm3) of the three significant components
| Region | Total volume | L/R volume |
|---|---|---|
| Component 4 | ||
| Dorsal lateral prefrontal cortex | 15 033.00 | 8437.11/6595.89 |
| Superior medial prefrontal cortex | 747.05 | 313.50/433.55 |
| Precentral inferior | 653.07 | 527.88/125.19 |
| Middle central sulcus | 553.60 | 553.60/- |
| Anterior insula | 278.52 | 278.52/- |
| Middle precentral gyms | 269.60 | 269.60/- |
| Superior parietal | 161.90 | 95.70/66.20 |
| Inferior frontal sulcus | 62.43 | -/62.43 |
| Component 5 | ||
| Basal forebrain | 1426.88 | 827.32/599.56 |
| Lunate | 1299.97 | 641.41/658.56 |
| Motor hand area of precentral gyrus | 1194.32 | 900.37/293.95 |
| Posterior cingulate | 1095.54 | 488.09/607.45 |
| Inferior frontal orbital sulcus | 947.36 | 540.22/407.14 |
| Postcentral sulcus | 644.15 | 315.56/328.59 |
| Anterior and midcingulate | 570.75 | 219.18/351.57 |
| Primary visual cortex | 349.52 | -/349.52 |
| Posterior superior frontal | 286.75 | 190.02/96.73 |
| Inferior frontal sulcus | 284.69 | 284.69/- |
| Inferior precentral gyrus | 261.37 | 261.37/- |
| Superior frontal orbital | 113.53 | 113.53/- |
| Lateral occipital | 108.73 | 54.88/53.85 |
| Hippocampus | 81.63 | 81.63/- |
| Component 6 | ||
| Superior temporal sulcus | 5831.69 | 3235.52/2596.17 |
| Postcentral sulcus | 652.04 | 140.97/511.07 |
| Inferior lunate | 531.64 | 186.93/344.71 |
| Lateral occipital | 488.77 | 488.77/- |
| Inferior frontal gyrus | 249.02 | 179.05/69.97 |
| Inferior occipital gyrus | 228.44 | -/228.44 |
| Posterior cingulate (dorsal) | 139.94 | 85.75/54.19 |
| Angular gyrus | 137.54 | 137.54/- |
| Anterior inferior temporal sulcus | 97.41 | 97.41/- |
| Medial parietal | 96.04 | -/96.04 |
| Inferior temporal sulcus | 74.77 | -/74.77 |
| Heschel’s gyrus | 65.86 | 65.86/- |
| Middle temporal sulcus | 53.85 | -/53.85 |
| Intraparietal | 50.76 | -/50.76 |
4 |. DISCUSSION
The findings of this study are straightforward; chimpanzees with indel polymorphisms in the 5′ flanking region of the AVPR1A promoter exhibit significant differences in covariation in gray matter. Chimpanzees that had both RS3 alleles (DupB+/−) showed greater covariation in gray matter in brain regions within components 4 and 5, which included the premotor and prefrontal cortex, basal forebrain, lunate and the cingulate cortex, compared with those that lack DupB positive allele of RS3 (DupB−/−). In contrast, the DupB+/− chimpanzees show lesser gray matter covariation than DupB−/− chimpanzees in regions comprising component 6, including the superior temporal sulcus and the postcentral sulcus. These regional differences overlap with brain regions previously found to differ in vasopressin and oxytocin neural fibers between nonhuman primates, and in AVPR1A gene expression in humans with different RS3 alleles (cingulate, insula and the hippocampus).28,38,68 Moreover, several of the larger regions identified in components 4–6 overlap with those that comprise the social brain network (prefrontal cortex, insula, cingulate and superior temporal sulcus),46–48 which is consistent with our hypothesis that these polymorphisms affect brain structures inolved in regulation of social cognition and behavior.
Several of the brain regions that differed between DupB+/− and DupB−/− chimpanzees have also been implicated in various forms of social cognition in humans and chimpanzees, in particular, the anterior cingulate cortex and superior temporal gyrus/sulcus.69 For instance, Mundy and Newell70 have proposed that the initiation of joint attention is mediated in brain regions in the anterior portion of the cortex, including area 9, the anterior cingulate and orbital frontal cortex. In contrast, Mundy and Newell70 have suggested that responding to joint attention cues is mediated by posterior brain regions, including the superior temporal gyrus, supramarginal gyrus and superior parietal cortex. Previous studies have shown that, in chimpanzees, poor receptive joint attention skills are linked to reversed asymmetries in the superior temporal gyrus and that DupB−/− males perform significantly worse than DupB+/− males on receptive joint attention.22 Furthermore, chimpanzees that are poor at initiating joint attention (or making a behavioral request) have also been shown to differ in gray matter volume within the anterior cingulate cortex compared with chimpanzees who reliably initiate joint attention.71 In short, some of the brain regions from components 4–6 that differ between DupB +/− and DupB−/− chimpanzees are associated with measures of joint attention in chimpanzees.
In summary, this is the first report of an association between the V1a vasopressin receptor (AVPR1A) and gray matter covariation in primates, including humans. The results suggest that AVPR1A polymorphisms are related to structural changes within the social brain network which may have implications for social behavior and cognition. Clearly, additional studies are needed to evaluate the role of polymorphisms in AVPR1A on gray matter covariation in other species of primates as a means of understanding the evolution of the vasopressin system and its effect on brain and behavior. In addition, given the role of different polymorphisms in AVPR1A on social cognition in chimpanzees, further studies on its expression in brain tissue would be important for understanding how its effect on the brain may mediate behavioral variability. Lastly, because AVPR1A has been suggested to be a candidate gene for ASD, we believe the results reported here further affirm the value of chimpanzees as a model species for understanding neurogenetic processes relevant to psychiatric disorders such as ASD in humans. Indeed, as we have shown, many behavioral phenotypes linked to ASD can be readily and uniquely studied in chimpanzees, including joint attention, theory-of-mind, or empathy, just to name a few. Future research should expand the behavioral ASD-related phenotypes to include measures of mutual eye gaze and social motivation, as well as scores on nonhuman primate equivalents of autism diagnostic measures (eg, Autism Diagnostic Observation Scale, Autism Quotient, etc.). Future studies should also examine other peptide systems involved in social cognition, such as the oxytocin system.
ACKNOWLEDGMENTS
This research was supported by NIH grants NS-42867 and NS-73134 to WDH, and U42-OD-011197 to the NCCC and P51OD11132 to YNPRC. We would like to thank the veterinary staff of both the National Center for Chimpanzee Care and the Yerkes National Primate Research Center for assistance during MRI.
REFERENCES
- 1.Donaldson ZR, Young LJ. Oxytocin, vasopressin, and the neurogenetics of sociality. Science. 2008;322(5903):900–904. [DOI] [PubMed] [Google Scholar]
- 2.Goodson JL, Bass AH. Social behavior functions and related anatomical characteristics of vasotocin/vasopressin systems in vertebrates. Brain Res Rev. 2001;35:246–265. [DOI] [PubMed] [Google Scholar]
- 3.Hammock EA, Young LJ. Micosatellite instability generates diversity in brain and sociobehavioral traits. Science. 2005;308:1630–1634. [DOI] [PubMed] [Google Scholar]
- 4.Meyer-Lindenberg A, Domes G, Kirsch P, Heinrichs M. Oxytocin and vasopressin in the human brain: social neuropeptides for translational medicine. Nat Rev Neurosci. 2011;12(9):524–538. [DOI] [PubMed] [Google Scholar]
- 5.Donaldson ZR, Young LJ. The relative contribution of proximal 5′ flanking sequence and microsatellite variation on brain vasopressin 1a receptor (AVPR1A) gene expression and behavior. PLoS Genet. 2013; 9(8):e1003729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammock EA, Young LJ. Functional microsatellite polymorphisms associated with divergent social structure in vole species. Mol Biol Evol. 2004;21:1057–1063. [DOI] [PubMed] [Google Scholar]
- 7.Lim MM, Nair HP, Young LJ. Species and sex differences in brain distribution of CRF receptor subtypes 1 and 2 in monogomous and promiscuous vole species. J Comp Neurol. 2005;487:75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim MM, Wang Z, Olazabel DE, Ren X, Terwillinger EF, Young LJ. Enhanced partner prefernce in a promiscuous species by manipulating the expression of a single gene. Nature. 2004;429:754–757. [DOI] [PubMed] [Google Scholar]
- 9.Walum H, Young LJ. The neural mechanisms and circuitry of the pair bond. Nat Rev Neurosci. 2018;1:643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donaldson ZR, Bai Y, Kondrashov FA, Stoinski TL, Hammock EAD, Young LJ. Evolution of a behavior-linked microsatellite-containing element of the 5′ flanking region of the primate AVPR1A gene. BMC Evol Biol. 2008;8:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French JA, Taylor JH, Mustoe AC, Cavanaugh J. Neuropeptide diversity and the regulation of social behavior in New World primates. Front Neuroendocrinol. 2016;42:18–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosso L, Keller L, Kaessmann H, Hammond RL. Mating systems and AVPR1A promoter variation in primates. Biol Lett. 2008;4:375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staes N, Stevens JM, Helsen P, Hillyer M, Korody M, Eens M. Oxytocin and vasopressin receptor gene variation as a proximate base for inter- and intraspecific behavioral differences in bonobos and chimpanzees. PLoS One. 2014;9(11):e113364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker KJ, Garner JP, Oztan O, et al. Arginine vasopressin in cerebro-spinal fluid is a marker of sociality in nonhuman primates. Science Translational Medicine. 2018;10(439):643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madlon-Kay S, Montague MJ, Brent LJN, et al. Weak effects of common genetic variation in oxytocin and vasopressin receptor genes on rhesus macaque social behavior. Am J Primatol. 2018;80:e22873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anestis SF, Webster TH, Kamilar JM, Fontenot MB, Watts DP, Bradley BJ. AVPR1A variation in chimpanzees (Pan troglodytes): population differences and association with Behavioral style. Int J Primatol. 2014;35(1):305–324. [Google Scholar]
- 17.Latzman RD, Hopkins WD, Keebaugh AC, Young LJ. Personality in chimpanzees (Pan troglodytes): exploring the hierarchical structure and associations with the vasopressin V1A receptor gene. PLoS One. 2014;9(4):e95741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latzman RD, Schapiro SJ, Hopkins WD. Triarchic psychopathy dimensions in chimpanzees (Pan troglodytes): investigating associations with genetic variation in the vasopressin receptor 1A gene. Front Neurosci. 2017;11:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staes N, Koski SE, Helsen P, Fransen E, Eens M, Stevens JM. Chimpanzee sociability is associated with vasopressin (AVPR1A) but not oxytocin receptor gene (OXTR) variation. Horm Behav. 2015;75: 84–90. [DOI] [PubMed] [Google Scholar]
- 20.Wilson VA, Weiss A, Humle T, et al. Chimpanzee personality and the arginine vasopressin receptor 1A genotype. Behav Genet. 2017;47(2): 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopkins WD, Donaldson ZR, Young LJ. A polymorphic indel containing the RS3 microsatellite in the 5′ flanking region of the vasopressin V1a receptor gene is associated with chimpanzee (Pan troglodytes) personality. Genes Brain Behav. 2012;11(5):552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopkins WD, Keebaugh AC, Reamer LA, Schaeffer J, Schapiro SJ, Young LJ. Genetic influences on receptive joint attention in chimpanzees (Pan troglodytes). Sci Rep. 2014;4(3774):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latzman RD, Young LJ, Hopkins WD. Displacement behaviors in chimpanzees (Pan troglodytes): a neurogenomics investigation of the RDoC negative valence systems domain. Psychophysiology. 2016;53 (3):355–363. [DOI] [PubMed] [Google Scholar]
- 24.Mahovetz LM, Young LJ, Hopkins WD. The influence of AVPR1A genotype on individual differencs in behaviors during a mirror self-recognition tasks in chimpanzees (Pan troglodytes). Genes Brain Behav. 2016;15:445–452. [DOI] [PubMed] [Google Scholar]
- 25.Inoue-Murayama M, Yokoyama C, Yamanashi Y, Weiss A. Common marmoset (Callithrix jacchus) personality, subjective well-being, hair cortisol level and AVPR1A, OPRM1, and DAT genotypes. Sci Rep. 2018;8(1):10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avinum R, Israel S, Shalev I, et al. AVPR1A variant associated with preschoolers’ lower altruistic behavior. PLoS One. 2011;6(9):e25274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebstein RP, Israel S, Lerer E, et al. Arginine vasopressin and oxytocin modulate human social behavior. Ann N Y Acad Sci. 2009;1167: 87–102. [DOI] [PubMed] [Google Scholar]
- 28.Knafo A, Israel S, Darvasi A, et al. Individual differences in allocation of funds in the dictator game and post-mortem hippocampal mRNA levels are correlated with length of the arginine vasopressin 1 a receptor (AVPR1A) RS3 promotor region repeat. Genes Brain Behav. 2008;7:266–275. [DOI] [PubMed] [Google Scholar]
- 29.Uzefovsky F, Shalev I, Israel S, et al. Oxytocin receptor and vasopressin receptor 1a genes are respectively associated with emotional and cognitive empathy. Horm Behav. 2015;67:60–65. [DOI] [PubMed] [Google Scholar]
- 30.Walum H, Westberg L, Henningsson S, et al. Genetic variation in the vasopressin receptor 1a gene (AVPR1A) associates with pair-bonding behavior in humans. Proc Natl Acad Sci. 2008;105(37):14153–14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melke J Autism: which genes are involved? Clin Neuropsychiatry. 2008;5(1):63–69. [Google Scholar]
- 32.Yirmiya N, Rosenberg C, Levi S, et al. Association between the arginine vasopressin 1a receptor (AVPR1A) gene and autism in a family-based study: mediation by socialization skills. Mol Psychiatry. 2006;11 (5):488–494. [DOI] [PubMed] [Google Scholar]
- 33.Procyshyn TL, Hurd PL, Crespi BJ. Association testing of vasopressin receptor 1a microsatellite polymorphisms in non-clinical autism spectrum phenotypes. Autism Res. 2017;10(5):750–756. [DOI] [PubMed] [Google Scholar]
- 34.Haznedar MM, Buchsbaum MS, Wei TC, et al. Limbic circuitry in patients with autism spectrum disorders studied with positron emission tomography and magnetic resonance imaging. Am J Psychiatry. 2000;157:1994–2001. [DOI] [PubMed] [Google Scholar]
- 35.Meyer-Lindenberg A, Kolachana B, Gold B, et al. Genetic variants in AVPR1A linked to autism predict amygdala activation and personality traits in healthy humans. Mol Psychiatry. 2009;14(10):968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bachevalier J, Loveland KA. The orbitofrontal-amygdala circuit and self-regulation of social-emotional behavior in autism. Neurosci Biobehav Rev. 2006;30(1):97–117. [DOI] [PubMed] [Google Scholar]
- 37.Baron-Cohen S The cognitive neuroscience of autism. J Neurol Neurosurg Psychiatry. 2004;75:945–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers CN, Ross AP, Sahu SP, et al. Oxytocin- and arginine vasopressin-containing fibers in the cortex of humans, chimpanzees, and rhesus macaques. Am J Primatol. 2018;80: e22875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freeman SM, Walum H, Inoue K, et al. Neuroanatomical distribution of oxytocin and vasopressin 1a receptors in the socially monogamous coppery titi monkey (Callicebus cupreus). Neuroscience. 2014;273:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freeman SM, Young LJ. Comparative perspectives on oxytocin and vasopressin receptor research in rodents and primates: translational implications. J Neuroendocrinol. 2016;28(4):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young LJ, Toloczko D, Insel TR. Localization of vasopressin (V1a) receptor binding and mRNA in the rhesus monkey brain. J Neuroendocrinol. 1999;11(4):291–297. [DOI] [PubMed] [Google Scholar]
- 42.Hopkins WD, Latzman RD, Mareno MC, Schapiro SJ, Gómez-Robles A, Sherwood CC. Heritability of gray matter structural covariation and tool use skills in chimpanzees (Pan troglodytes): a source-based morphometry and quantitative genetic analysis. Cereb Cortex. 2018:3702–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexander-Bloch A, Giedd JN, Bullmore E. Imaging structural covariance between human brain regions. Nat Rev Neurosci. 2013;14(5): 322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bard KA, Hopkins WD. Early socioemotional intervention mediates long-term effects of atypical rearing on structural covariation in gray matter in adult chimpanzees. Psychol Sci. 2018;29(4):594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu L, Groth KM, Pearlson G, Schreffen DJ, Calhoun VD. Source-based morphometry: the use of independent component analysis to identify gray matter differences with application to schizophrenia. Hum Brain Mapp. 2009;30(3):711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adolphs R The social brain: neural basis of social knowledge. Annu Rev Psychol. 2009;60:693–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewis PA, Rezaie R, Brown R, Roberts N, Dunbar RI. Ventromedial prefrontal volume predicts understanding of others and social network size. Neuroimage. 2011;57(4):1624–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sliwa J, Freiwald WA. A dedicated network for social interaction processing in the primate brain. Science. 2017;356(6339):745–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogart SL, Bennett AJ, Schapiro SJ, Reamer LA, Hopkins WD. Different early rearing experiences have long-term effects on cortical organization in captive chimpanzees (Pan troglodytes). Dev Sci. 2014;17(2): 161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kikinis R, Pieper SD, Vosburgh KG. 3D slicer: a platform for subject-specific image analysis, visualization, and clinical support Intraoperative Imaging and Image-Guided Therapy. New York: Springer; 2014:277–289. [Google Scholar]
- 51.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30(9):1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17(3):143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jenkinson M, Pechaud M, Smith S. BET2: MR-based estimation of brain, skull and scalp surfaces Paper presented at: Eleventh annual meeting of the organization for human brain mapping 2005; Toronto. [Google Scholar]
- 54.Tustison N, Gee J. N4ITK: Nick’s N3 ITK implementation for MRI bias field correction. Insight J. 2009;9:1–8. [Google Scholar]
- 55.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tustison N, Avants BB, Cook PA, Gee JC. N4itk: Improved n3 bias correction with robust b-spline approximation. Paper presented at: Proceedings of the ISBI2010. [Google Scholar]
- 57.Boyes RG, Gunter JL, Frost C, et al. Intensity non-uniformity correction using N3 on 3-T scanners with multichannel phased array coils. Neuroimage. 2008;39(4):1752–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tustison NJ, Cook PA, Klein A, et al. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. Neuroimage. 2014; 99:166–179. [DOI] [PubMed] [Google Scholar]
- 59.Coupe P, Yger P, Prima S, Hellier P, Kervrann C, Barillot C. An optimized blockwise nonlocal means denoising filter for 3-D magnetic resonance images. IEEE Trans Med Imaging. 2008;27(4):425–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5(2): 143–156. [DOI] [PubMed] [Google Scholar]
- 61.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. [DOI] [PubMed] [Google Scholar]
- 62.Hopkins WD, Avants BB. Regional and hemispheric variation in cortical thickness in chimpanzees (Pan troglodytes). J Neurosci. 2013;33: 5241–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Douaud G, Smith S, Jenkinson M, et al. Anatomically related grey and white matter abnormalities in adolescent-onset schizophrenia. Brain. 2007;130(9):2375–2386. [DOI] [PubMed] [Google Scholar]
- 64.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. [DOI] [PubMed] [Google Scholar]
- 65.Andersson JL, Jenkinson M, Smith S. Non-linear Registration Aka Spatial Normalisation (FMRIB Technial Report TR07JA2). Oxford: FMRIB Analysis Group; 2007:1–21. [Google Scholar]
- 66.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gutiérrez JA, Goyache F. A note on ENDOG: a computer program for analysing pedigree information. J Anim Breed Genet. 2005;122(3):172–176. [DOI] [PubMed] [Google Scholar]
- 68.Tansey KE, Hill MJ, Cochrane LE, Gill M, Anney RJL, Gallagher L. Functionality of promoter microsatellites of arginine vasopressin receptor 1A (AVPR1A): implications for autism. Mol Autism. 2011;2(3):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mundy P A review of joint attention and social-cognitive brain systems in typical development and autism spectrum disorder. Eur J Neurosci. 2017;47(6):497–514. [DOI] [PubMed] [Google Scholar]
- 70.Mundy P, Newell L. Attention, joint attention, and social cognition. Curr Direct Psychol Sci. 2007;16(5):269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hopkins WD, Taglialatela JP. Initiation of joint attention is associated with morphometric variation in the anterior cingulate cortex of chimpanzees (Pan troglodytes). Am J Primatol. 2013;75(5):441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]


