Correction to: Scientific Reports 10.1038/srep11666, published online 02 July 2015
This Article contains an error, where the protein name corresponding to 4IM4 (pdbid) is incorrectly given as “Lic26A-Cel5E”, and should read “CelE”.
In addition, three references were omitted from Table 2, and are given below as References 1–3.
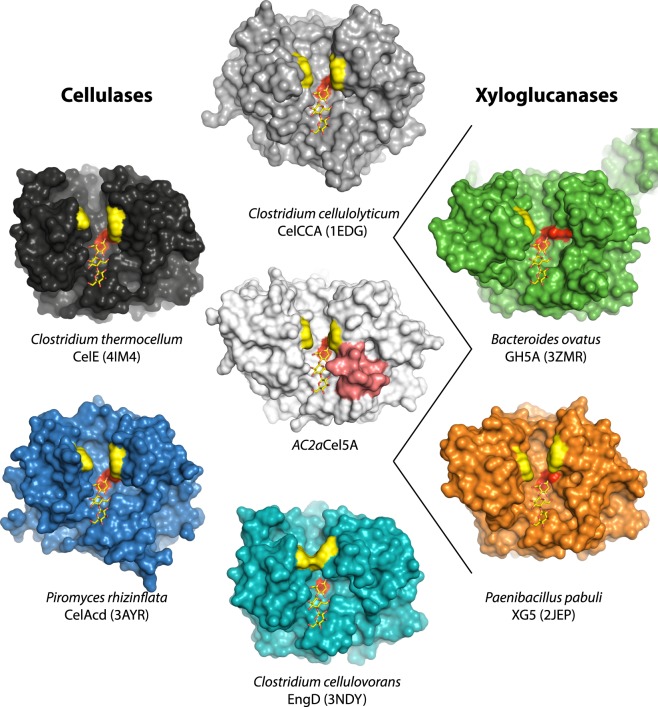
The correct Table 2 and Figure 4 are given below as Table 1 and Figure 1 respectively.
Table 1.
Structural comparison of AC2aCel5A with its six closest structural homologues identified using the DALI server17, along with reported enzyme activities.
| Structural comparison | Enzymatic activities | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DALI Z-score | RMSD (Å) | % id | CMC (U/mg) | β-glucan (U/mg) | Filter paper (U/mg) | Avicel (U/mg) | Lichenan (U/mg) | Xylan (U/mg) | Xyloglucan (U/mg) | ||
| Bacteroidetes AC2a Cellulase Cel5A | — | — | — | 216.8 | 1471.4 | 0.152a | 0.115a | 839.9 | nd | trace | This study and7 |
| Paenibacillus pabuli Xyloglucanase XG5 (2JEP) | 49.2 | 1.6 | 34 | nd | nd | — | nd | nd | nd | 8700 | 33 |
| Clostridium cellulovorans Cellulase EngD (3NDY) | 47.0 | 1.7 | 33 | 15 | 42 | — | 0.017 | — | 0.5 | 36 | 15 |
| Bacteroides ovatus Xyloglucanase BoGH5A (3ZMR) | 46.6 | 1.9 | 32 | nd | nd | — | — | nd | — | 514.2 | 44 |
| Clostridium thermocellum Cellulase CelE (4IM4) | 46.0 | 1.6 | 32 | Active | 75.4 | Active | — | — | Active | — | 1–3, b |
| Clostridium cellulolyticum Cellulase CelCCA (1EDG) | 45.9 | 1.9 | 32 | 101.3 | 104.3 | — | 0.028 (0.124)d | 79.2 | 10.1 | — | 45,46, c |
| Piromyces rhizinflata Cellulase CelAcd (3AYR) | 43.2 | 1.8 | 30 | 344.9 | 576 | 0.64 | 1.39 | 542.5 | 106.2 | — | 47 |
RMSD; root-mean-square deviation of C-alpha atoms. The structural homologues are sorted based on the Z-score obtained in the DALI search. One Unit of enzyme activity was defined as the amount of enzyme releasing 1 µmol of reducing sugar equivalents per minute. “nd” means not detected, whereas a hyphen, “—”, indicates “not tested”.
aµmol reducing sugar equivalents calculated as µmol cellotriose + µmol cellobiose + µmol glucose, quantified by HPAEC-PAD7.
bGH5 domain of CelE.
cU/mg calculated from U/µmol reported in45.
dFor this enzyme data for both the full length protein (in parentheses), and the catalytic domain only were published.
Figure 1.
.
Finally, the updated Supplementary Information file, containing the correct protein name in Figure S1, is given below.
Supplementary information
Contributor Information
A. E. Naas, Email: adrian.naas@nmbu.no
P. B. Pope, Email: phil.pope@nmbu.no
Supplementary information
is available for this paper at 10.1038/s41598-020-62786-2.
References
- 1.Hall J, Hazlewood GP, Barker PJ, Gilbert HJ. Conserved reiterated domains in Clostridium thermocellum endoglucanases. Gene. 1988;69:29–38. doi: 10.1016/0378-1119(88)90375-7. [DOI] [PubMed] [Google Scholar]
- 2.Durrant AJ, Hall J, Hazlewoodt GP, Gilbert HJ. The non-catalytic C-terminal region of endoglucanase E from Clostridium thermocellum contains a cellulose-binding domain. Biochem. J. 1991;273:289–293. doi: 10.1042/bj2730289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takasuka TE, et al. Cell-free translation of biofuel enzymes. Methods Mol. Biol. 2014;1118:71–95. doi: 10.1007/978-1-62703-782-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.