Abstract
Guanylate binding proteins (GBPs) belongs to the interferons (IFNs) induced guanylate-binding protein family (Guanosine triphosphatases, GTPases) consisting of seven homologous members, termed GBP1 to GBP7. We used multidimensional survey ways to explore GBPs expression, regulation, mutations, immune infiltration and functional networks in head and neck squamous cell carcinoma (HNSCC) patient data based on various open databases. The study provides staggered evidence for the significance of GBPs in HNSCC and its potential role as a novel biomarker. Our results showed that over expressions of 7 GBPs members and multivariate analysis suggested that N-stage, high expressions of GBP1 and low expression of GBP6/7 were linked to shorter OS in HNSCC patients. In addition, B cells of immune infiltrates stimulant the prognosis and might have a medical prognostic significance linked to GBPs in HNSCC. We assume that GBPs play a synergistic role in the viral related HNSCC. Our results show that data mining efficiently reveals information about GBPs expression in HNSCC and more importance lays a foundation for further research on the role of GBPs in cancers.
Subject terms: Diagnostic markers, Risk factors
Introduction
Head and Neck Squamous Cell Carcinoma (HNSCC) is a common head and neck malignancy that originates from lips, mouth, paranasal sinuses, oropharynx, larynx, nasopharynx, and other pharyngeal cancers1. As the sixth most common type of malignant tumors, with more than 655,000 new cases and 90,000 deaths every year2. Currently, smoking, drinking, and human papillomavirus (HPV) infection are considered risk factors for HNSCC occurrence and prognosis3. Unfortunately, the 5-year survival rate is still below 50% due to the usual lack of early symptoms when HNSCC is detected early, while the survival rate is reduced to 35% due to local recurrence and metastasis4.
Once in advanced stages, treatment can notably effect organ function and harm the structures involved in speaking and swallowing, causing devastating results in patient’s quality of life5,6. Studies reported that lots of HNSCC not only has experienced epithelial-to-mesenchymal transition (EMT) but also presents a mesenchymal-like (ML) phenotype and thus effect the drug resistance, tumor migration or tumor growth7. The incidence and development of HNSCC is a complex process involving multiple molecules. Guan et al. found long non-coding RNA H19 and its mature miR-675 were significantly high level in two HNSCC cell lines as well as a cohort of 65 primary tumor samples8. Wu et al. reported that SUZ12 protein was particularly overexpression in primary HNSCC samples and is up-regulated significantly associated with cervical node metastasis, overall survival and disease-free survival9. Reed et al. believed that inactivation of the p16 tumor suppressor gene is a common event in HNSCC10. In spite of advances in combine chemotherapy, radiation, and surgery during the past decades have fundamentally improved survival rate of the HNSCC patients, many patients increase secondary tumors, recurrences or metastasis after treatment and thus becomes a challenging problem and leading to therapeutic failure11.
Guanylate binding proteins (GBPs) belongs to the interferons (IFNs) induced guanylate-binding protein family (Guanosine triphosphatases, GTPases) consisting of seven homologous members, termed GBP1 to GBP712. Until now, seven different GBPs and six in mouth have been identified and human GBPs show a high level of homology. GTPases are involved in process of signal transduction, cell proliferation, differentiation and intracellular protein transportation13,14. Inside host cells, GBPs quickly assemble into big antimicrobial defense complexes that fight various bacterial pathogens15. Studies found the immune roles of human GBPs in regulating not only pyroptosis, but also apoptosis16. Most previous studies the function of GBPs in bacterial pathogens, but more and more studies explore the role of GBPs in tumor mechanism. Study found that GBP1 acts specifically as a tumor suppressor in colorectal carcinoma and the reduction of GBP1 expression might imply cancer evasion from the IFN-γ-dominated Th1 immune response17. Nevertheless, the mechanism by which GBPs are depressed or activated still remained unclear in the development and progression of HNSCC. Until now, a comprehensive bioinformatics analysis has not yet to analysis the potential role of GBPs in HNSCC. we explored the mutations and expressions of various GBPs based on thousands of variations in copy numbers or gene expressions in patients whom with HNSCC in detail to provide new insights into the potential functions, expression patterns, molecular mechanisms and distinct prognostic underlying GBPs regulation.
Results
Over-expression of different GBPs family members in HNSCC
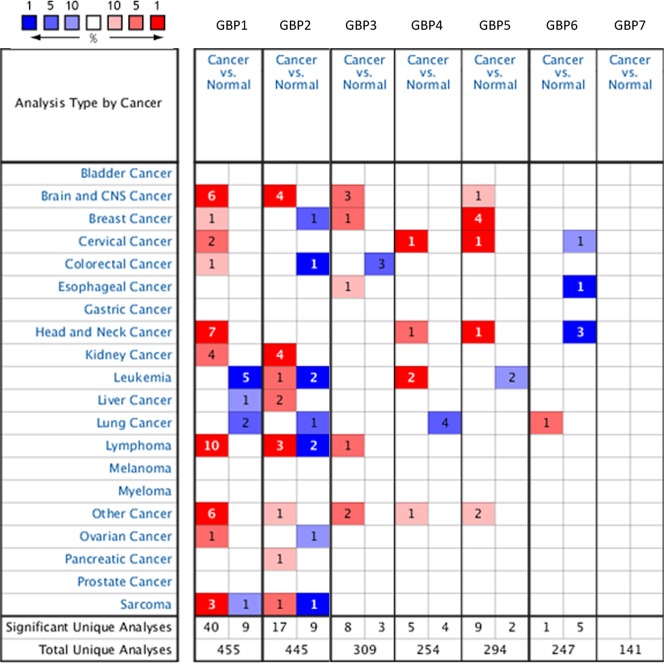
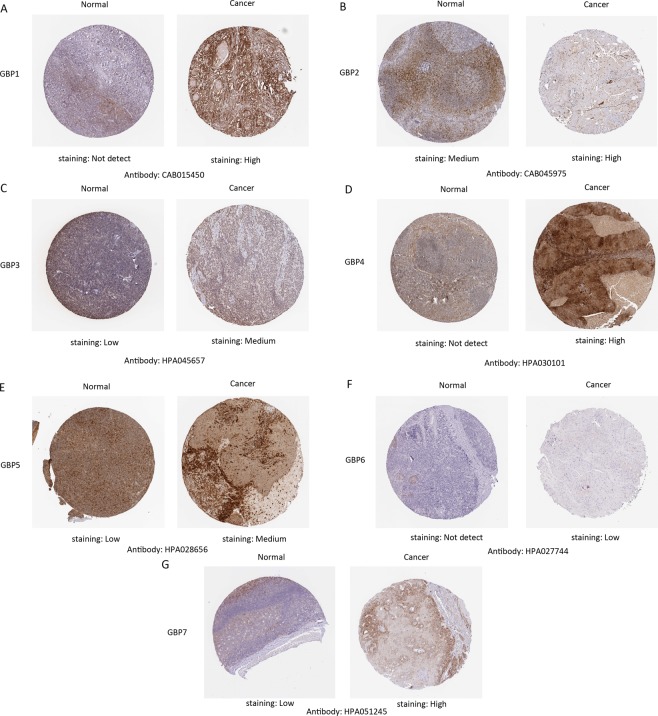
In order to explore the potential therapeutic value and distinct prognostic of different GBPs members in HNSCC patients, the expression GBPs were explored by ONCOMINE database, TCGA database and HPA database. We first contrast the transcriptional levels of GBPs in cancers and normal samples and significantly higher mRNA expressions of GBP1/4/5 were found in HNSCC tissues in multiple datasets (Fig. 1). In Peng Head-Neck statistics18, GBP1 over-expression was found in oral cavity squamous cell carcinoma tissues compared with normal tissues with a fold change of 7.854 (P = 4.15E-24), while Talbot19 observed 2.962-fold increase in GBP1 mRNA expression in tongue samples (P = 3.10E-12), Ginos20 found 5.397- fold increase in GBP1 mRNA expression in HNSCC tissues (P = 2.70E-13), Estilo21 found 4.286- fold increase in GBP1 mRNA expression in tongue tissues (P = 3.51E-9) and Ye22 found 4.450- fold increase in GBP1 mRNA expression in tongue tissues (P = 5.39E-5, Table 1). Significant up-regulation of GBP4 was also found in HNSCC tissues compared to normal tissues. In Peng Head-Neck statistics18, GBP4 over-expression was found in oral cavity squamous cell carcinoma tissues compared with normal tissues with a fold change of 3.256 (P = 2.06E-12). Similarly, In Peng Head-Neck statistics18, GBP5 over-expression was found in oral cavity squamous cell carcinoma tissues compared with normal tissues with a fold change of 14.065 (P = 3.61E-26), while Ye22 observed 3.376-fold increase in GBP5 mRNA expression in tongue samples (P = 1.53E-4). We then tried to analysis the protein expression patterns of GBPs in HNSCC by the HPA database. As the result shown GBP1/4/5 proteins were not expressed in normal head and neck tissues, whereas high and medium expressions of them were observed in HNSCC tissues. In addition, low protein expressions of GBP3/7 were expressed in normal tissues, while medium and high protein expressions of them were observed in tumor tissues. Moreover, medium protein expression of GBP2 was observed in normal tissues and high expression in HNSCC tissues (Fig. 2). However, no expression and low protein expression of GBP6 was observed in normal and tumor tissues. In total, our results suggested that transcriptional and proteinic expressions of GBPs were over-expressed in patients with HNSCC.
Figure 1.
The transcriptional levels of GBPs in cancers and normal samples and significantly higher mRNA expressions of GBP1/4/5 were found in HNSCC tissues in multiple datasets. Redder means higher expression and bluer means lower expression, the analysis which can give us an understanding of the expression of GBPs family in different cancer types. (ONCOMINE Database).
Table 1.
The Significant Changes of GBPs Expression in Transcription Level between Different Types of HNSCC and Normal Tissues (Oncomine database).
| GBPs | Type of Breast Cancer versus Normal Breast Tissue | Fold Change | P Value | t Test | Source and/or Reference |
|---|---|---|---|---|---|
| GBP1 | Oral Cavity Squamous Cell Carcinoma | 7.854 | 4.15E-24 | 16.990 | Peng Head-Neck statistics18 |
| Tongue Squamous Cell Carcinoma | 2.962 | 3.10E-12 | 9.029 | Talbot Lung statistics19 | |
| Head and Neck Squamous Cell Carcinoma | 5.397 | 2.70E-13 | 10.813 | Ginos Head-Neck statistics20 | |
| Tongue Squamous Cell Carcinoma | 4.286 | 3.51E-9 | 7.071 | Estilo Head-Neck statistics21 | |
| Tongue Squamous Cell Carcinoma | 4.450 | 5.39E-5 | 3.433 | Ye Head-Neck statistics22 | |
| GBP2 | Oral Cavity Squamous Cell Carcinoma | 1.636 | 3.53E-8 | 6.125 | Peng Head-Neck statistics18 |
| Oral Cavity Squamous Cell Carcinoma | 1.692 | 0.035 | 2.151 | Toruner Head-Neck Statistics | |
| GBP3 | Oral Cavity Squamous Cell Carcinoma | 1.792 | 4.54E-9 | 6.542 | Peng Head-Neck statistics18 |
| GBP4 | Oral Cavity Squamous Cell Carcinoma | 3.265 | 2.06E-12 | 8.363 | Peng Head-Neck statistics18 |
| GBP5 | Oral Cavity Squamous Cell Carcinoma | 14.065 | 3.61E-26 | 16.300 | Peng Head-Neck statistics18 |
| Tongue Squamous Cell Carcinoma | 3.376 | 1.53E-4 | 3.997 | Ye Head-Neck statistics22 | |
| GBP6 | NA | NA | NA | NA | NA |
| GBP7 | Oral Cavity Squamous Cell Carcinoma | 1.636 | 3.53E-8 | 6.125 | Peng Head-Neck statistics18 |
NA, not available; HNSCC: head and neck squamous cell carcinoma.
Figure 2.
Representative immunohistochemistry images of distinct GBPs family members in HNSCC tissues and normal tissues (Human Protein Atlas Database).
Association of mRNA expression of different GBPs family members with clinicopathological parameters of HNSCC patients
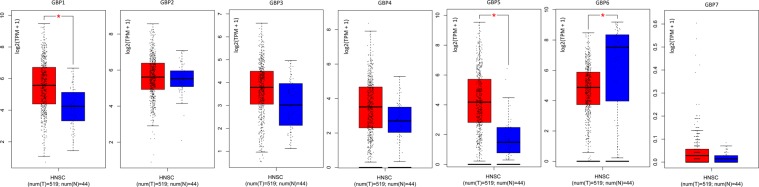
Using the GEPIA dataset, we first contrast the mRNA expression difference of GBPs between cancer and normal tissues. The results demonstrated that the expression of GBP1 and GBP5 were higher in HNSCC tissues, while the expression level of GBP6 was lower in cancer tissues (Fig. 3). Moreover, we also explored the expression of GBPs with cancer stage for HNSCC and we found GBP1/5/6 groups significantly varied, but GBP2/3/4/7 groups did not significantly differ (Fig. 4).
Figure 3.
The box plot Expression of GBPs in HNSCC. The box color of red indicates tumor and blue indicates normal. The method for differential analysis is one-way ANOVA, using disease state as variable for calculating differential expression and asterisk means statistically significant, with each dot representing a distinct tumor or normal sample. (GEPIA Database; TPM: Transcripts Per Million; T: Tumor; N: Normal).
Figure 4.
The expression of GBPs with cancer stage (Stage I~Stage IV) for HNSCC and we found GBP1/5/6 groups significantly varied, but GBP2/3/4/7 groups did not significantly differ. The method for differential gene expression analysis is one-way ANOVA, using pathological stage as variable for calculating differential expression. The larger F, the better the study fit and P-Value < 0.05 as statistically significant. (GEPIA Database).
Prognostic value of mRNA expression of GBPs in HNSCC
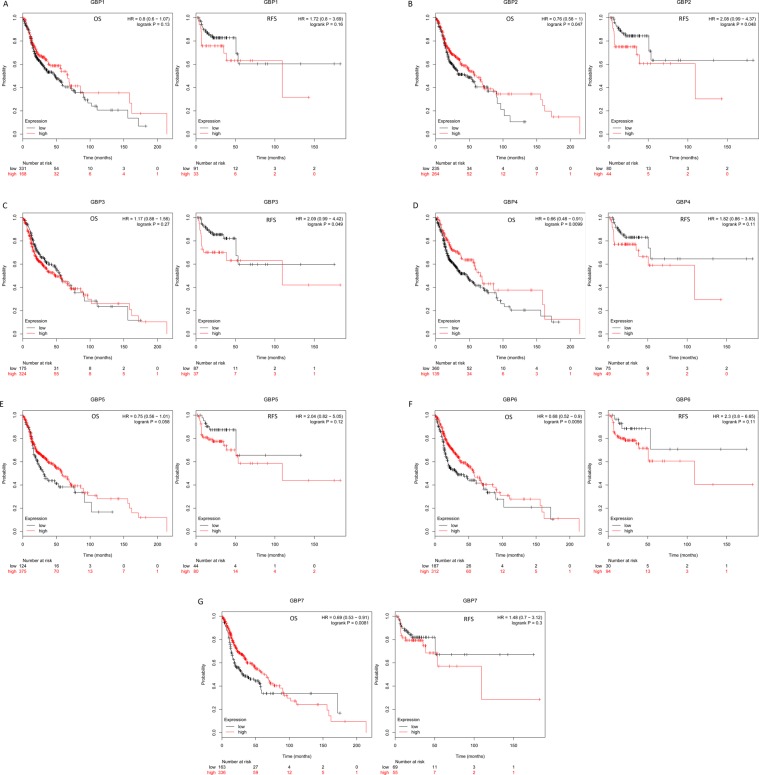
We next used Kaplan-Meier plotter to explored the prognostic survival values of the mRNA expression of GBPs in HNSCC patients by using publicly free online datasets. The Kaplan-Meier curve and log rank test analyses were shown in Fig. 5, higher mRNA expression of GBP2 (HR = 2.08, 95% CI: 0.99–4.37, P = 0.048) and GBP3 (HR = 2.09, 95% CI: 0.99–4.42, P = 0.049) were significantly associated with shorter relapse free survival (RFS) of HNSCC patients. Interestingly, higher mRNA expression of GBP2 (HR = 0.76, 95% CI: 0.58–1, P = 0.047), GBP4 (HR = 0.66, 95% CI: 0.48–0.91, P = 0.0099), GBP6 (HR = 0.68, 95% CI: 0.52–0.9, P = 0.0056) and GBP7 (HR = 0.69, 95% CI: 0.53–0.91, P = 0.0081) were significantly associated with longer overall survival (OS) of HNSCC patients. These results suggested that mRNA level of GBP2/3/4/6/7 plays an important role in cancer patients’ prognosis and they may be exploited as novel useful biomarkers for prediction of HNSCC patients’ survival. We then tried to evaluate the independent prognostic value of expression of GBPs involve in OS in HNSCC patients from TCGA database. Significant high expression of GBP1/3/4/5/6/7 in HNSCC tissues compared to normal tissues (Supplementary Fig.). In univariate analysis, we found that tumor stage (HR = 1.80, 95% CI: 1.23–2.66, P < 0.05), T-stage (HR = 1.36, 95% CI: 1.05–1.76, P < 0.05), N-stage (HR = 1.36, 95% CI: 1.10–1.68, P < 0.05), high expressions of GBP1/6/7 were linked to shorter OS of HNSCC patients. Multivariate analysis suggested that N-stage (HR = 1.33, 95% CI: 1.03–1.72, P < 0.05), high expressions of GBP1 and low expression of GBP6/7 were linked to shorter OS of cancer patients (Supplementary Table 1). These results showed that transcriptional expressions of GBP6/7 were independent prognostic factors for patients with HNSCC.
Figure 5.
The Prognostic Value of mRNA Level of GBP Factors in HNSCC Patients. The patient samples are split into two groups according to various quantile expressions of the proposed biomarker and gene expression data and relapse free survival (RFS) and overall survival (OS) information are downloaded from GEO, EGA and TCGA. (Kaplan-Meier Plotter Database).
Predicted Functions and Pathways of GBPs in HNSCC
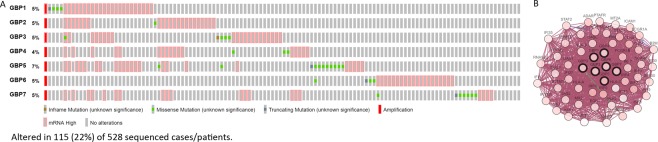
We next explored the GBPs alterations and networks via the cBioPortal online tool and we further analyzed 50 neighbor genes which were significantly related to GBPs mutations. In the 528 sequenced HNSCC patients, genetic alteration was found in 115 patients and the mutation rate was 22%, GBP5 ranked the highest genes with genetic alterations with the mutation rates were 7% and we also shown the network for GBPs and the 50 most frequently altered neighbor genes (Fig. 6). The top 10 type and frequency of GBPs neighbor gene alterations in HNSCC including ADAR, IRF6, IRF9, IRF2, CD44, STAT1, FCGR1A, NFATC2, ICAM1 and STAT2 (Table 2). In addition, functions and pathways of GBPs and their 50 frequently altered neighbor genes were analyzed by GO and KEGG in DAVID online database (Supplementary Table 3). GO analysis results showed that changes in biological processes (BP) were significantly enriched in cellular response to type I interferon, regulation of immune response, multi-organism process, response to virus and cytokine production et al. Changes in molecular function (MF) were mainly enriched in oligoadenylate synthetase activity, protein binding, regulatory region DNA binding and nucleic acid binding. Changes in cell component (CC) were mainly enriched in intracellular membrane-bounded organelle, cytoplasmic part, membrane-bounded organelle and MHC class I protein complex et al.
Figure 6.
GBPs Gene Expression and Mutation Analysis in HNSCC (cBioPortal Database). (A) GBPs gene expression and mutation analysis; (B) The protein- protein interaction network for GBPs and the 50 most frequently altered neighbor genes.
Table 2.
The top 10 type and frequency of GBPs neighbor gene alterations in HNSCC (cBioPortal).
| Gene Symbol | Amplification | Up-regulation | Down-regulation | Mutation | Total Alteration |
|---|---|---|---|---|---|
| ADAR | 1.3 | 6.2 | 0.4 | 1.5 | 9.1 |
| IRF6 | 0.9 | 6.6 | 0.4 | 0.9 | 8.7 |
| IRF9 | 0.6 | 6.0 | 1.5 | 0.4 | 8.7 |
| IRF2 | 0.4 | 2.1 | 2.8 | 1.5 | 8.4 |
| CD44 | 3.4 | 6.6 | 0.4 | 0.6 | 8.5 |
| STAT1 | 2.8 | 4.9 | 0 | 2.8 | 8.5 |
| FCGR1A | 2.1 | 4.2 | 0 | 0.2 | 7.9 |
| NFATC2 | 0.2 | 4.3 | 0 | 2.8 | 7.5 |
| ICAM1 | 0.9 | 4.0 | 0 | 1.1 | 7.0 |
| STAT2 | 0.4 | 5.1 | 0 | 0.9 | 6.4 |
KEGG were mainly enriched in antigen processing and presentation and viral infection.
Immune infiltrates in correlation with GBPs in HNSCC
Correlation between GBPs in HNSCC expression and abundance of immune infiltrates was statistically significant (P < 0.05, Supplementary Table. 2). Cumulative survive showed that B cells of immune infiltrates statistically significant (P < 0.05) of GBPs in HNSCC and it is worth further research and exploration (Supplementary Table 4).
Discussion
Although certain GBPs members have been shown to play a key role in cancer, the different roles of GBP family members in HNSCC remain to be elucidated. So, we first explored the mutations and expressions of various GBPs based on thousands of variations in copy numbers or gene expressions in patients whom with HNSCC. Our study showed that high expressions of mRNA in all the GBPs family members, and mRNA expression of GBPs was significantly related to patients’ individual cancer stages in HNSCC patients. Multivariate analysis suggested that N-stage (HR = 1.33, 95% CI: 1.03–1.72, P < 0.05), high expressions of GBP1 and low expression of GBP6/7 were linked to shorter OS of cancer patients.
The occurrence of HPV-related HNSCC has steadily rised over the past two decades and now indicate a majority of HNSCC cases23. Epstein–Barr virus (EBV) infection is found in 90% to 100% of nasopharyngeal carcinoma (NPC) cases in endemic regions24. GBPs belongs to the IFNs induced guanylate-binding protein family, while IFNs are the main antiviral factors released during the process of infecting host cells by viruses or other pathogens, and have cell surface receptors that bind to responsive cells, regulating host antiviral status, anti-malignant cell proliferation and immune response15. Functions and pathways of GBPs and their 50 frequently altered neighbor genes were analyzed by GO and KEGG demonstrated that changes in BP of DEGs were significantly enriched in regulation of immune response and virus and cytokine production, MF were mainly enriched in protein/region DNA/nucleic acid binding and CC were mainly enriched in intracellular membrane-bounded organelle and MHC class I protein complex et al. KEGG were mainly enriched in antigen processing and presentation and viral infection. So, we presume that GBPs play a synergistic role in the HPV-HNSCC or other viral related cancers.
GBP1 expression is excited not only by type I and type II interferons (IFN), including IFN-γ, and also by interleukin-1α, IL- 1β, and tumor necrosis factor-α25–28. In vivo, GBP1 is related to the existence of inflammation and has been adhere to in inflammatory bowel diseases (IBD), autoimmune diseases and cancers. GBP1 regulates the suppression of endothelial cells invasion and proliferation in reaction to inflammatory cytokines and exhibit antiviral activity27. Yu et al. reported that GBP1 positively regulating the invasion and migration of oral cavity squamous cell carcinoma cells29. A recent study showed that GBP1 may promote lung adenocarcinoma invasiveness by stimulate cell motility and via manage of GBP1 expression may development of new therapeutic ways for tumor30. GBP1 also found as a fetal T lymphocyte-induced protein that cause breast cancer cells to cross the blood-brain barrier31. Our results showed that higher mRNA expressions of GBP1 were found in HNSCC tissues, and that mRNA expression of GBP1 was significantly related with patients’ cancer stages and abundance of immune infiltrates. Moreover, previous studies focus on the role of T immune cell in cancers, while our cumulative survive showed that B cells significantly stimulant the prognosis and might have a medical prognostic significance linked to GBPs in HNSCC. GBP2 is strongly induced by IFN-γ. GBP2 is obviously higher in esophageal squamous cell carcinomas than in adjacent normal epithelium cells and is consistently increased in a p53-dependent manner and also followed by an rise in protein levels of IRF-132. Zhang et al.33 first found that GBP2 inhibits cell metastasis and mitochondrial fission in breast cancer cells not only in vitro but in vivo and another study found an interesting result that GBP2 related to a recently generally T cell signature, suggesting tumor infiltration with T cells in breast cancer34. Other study found GPB2/5 shown wide antiviral activity by suppressing the activation of the virus-dependency factor furin35. GBP2 was similarly relocalized in cells in which autophagy was impaired36. Our study results shown that higher mRNA expression of GBP2 were significantly related to longer OS and RFS of HNSCC patients. So, we guess GBP2 may play a critical role in cancer autophagy and needs more study to convince.
Xu et al.37 first found that GBP3 lead to the proliferation of glioma cells via SQSTM1-ERK1/2 pathway. GBP3 also has been reported to contributed to anti-influenza activity via suppressing viral transcription and replication38. GBP1 and GBP3 exhibit highest homology between the seven members (88% amino acid sequence)12 so GBP3 may be a novel proto-oncogene. Similarly, our results showed that higher mRNA expressions of GBP3 and related to shorter OS in HNSCC tissues. Study reported that GBP4 is a negative regulator of virus-induced IFN-I production resulting in impaired TRAF6-mediated IRF7 ubiquitination and it is deemed as a novel protein targeting IRF7 and suppressing its function39. Therefore, GBP4 may affect ubiquitination-mediated pathways, thereby inhibiting viral infection. IRF regulation of tumors is mainly through regulation of the cell cycle, apoptosis or tumor suppressor gene p53, but also regulation of anti-tumor immunity, or regulation of interferon signaling Pathway (eg IRF9)40,41. Our results found the 50 most frequently altered GBPs neighbor gene alterations in HNSCC including interferon regulatory factors 6 (IRF6), IRF9 and IRF2, indicated that IRFs and GBPs coordinate the development of tumors and requires more study. GBP5 have splice variants (GBP5a, GBP5b, GBP5ta) before 199242. Krapp et al.43 reported that GBP5 has a broad spectrum of antiretroviral capabilities. Wehner et al.44 reported that only the a/b splice variant is rise in healthy cells, but the truncated splice variant has been discovered in each melanoma and lots of lymphoma cell lines detected. Study also found GBP5 act as an only rheostat for NLRP3 inflammasome activation and thus alert the immune system to the exist of infection or tissue damage45. Our results showed that higher mRNA expressions of GBP5 in HNSCC tissues and may related with patients’ tumor immune microenvironment. Until now, about the function of GBP6/7 is rare. HuGBP-6 lacks a GAS or NF-κB site and therefore, would not be expected to be induced by IFNs46. In GEPIA database analysis we found high GBP6 expression in normal tissue and related with patients’ cancer stages, while in TCGA database analysis we found high expression in cancer tissue, in addition, in HPA databased we found no expression and low protein expression of GBP6 was observed in normal and tumor tissues. Both GBP6/7 significantly related to longer OS of HNSCC patients. GBP7 can triggered an IFIT5-IRF1/3-RSAD5 pathway in the DF-1 cells which potentially limited the viral replication cycle in the initial infection stage47. Thus, more researches should conduct to provide new ideas for the development of anti-tumor drugs and the treatment of human diseases.
There were some limitations in our study, one is that all the data analyzed in our study was retrieved from the online databases and cannot provides precise clinical data, larger sample sizes are needed to validate our findings, another is that we did not evaluate the potential diagnostic and therapeutic roles of GBPs and tumor markers can vary widely due to the histological type of HNSCC and multiple anatomical sites, so future related researches are required. Finally, due to insufficient data, we were unable to compare differences in function of GBPs between HPV-positive and HPV-negative HNSCC patients and we will do a lot of work in the future. In conclusion, our results showed that over expressions of 7 GBPs members and multivariate analysis suggested that N-stage, high expressions of GBP1 and low expression of GBP6/7 were linked to shorter OS in HNSCC patients. In addition, B-cell infiltration of immune cells affects prognosis and may be related to GBPs in HNSCC. We hypothesized that the GBPs plays a synergistic role in virus-associated HNSCC. This research uses online tools that rely on the most popular bioinformatics theories to perform target gene analysis of tumor data information from open databases, and authorizes large-scale HNSCC genomics research and follow-up functional exploration.
Materials and Methods
Ethics statement
As the work is a bioinformatics analysis article, so the ethical approval was not necessary and all the data were retrieved from the free online databases.
ONCOMINE database
The DNA copy number and mRNA expression of GBPs in HNSCC were investigated inside the Oncomine 4.5 database. Oncomine (www.oncomine.org) contains 715 gene expression data sets and 86,733 samples, is also the biggest oncogene chip database and incorporated data mining platform worldwide48. This analysis drew on a series of HNSCC studies and GBPs expression was involved in evaluated in HNSCC tissue in respect to its expression in normal tissue, and P < 0.05 as the cutoff criterion considered statistically significant.
Human protein atlas
The Human Protein Atlas (https://www.proteinatlas.org/about/licence) is a website that involve immunohistochemistry-based expression data for distribution and expression of 20 tumor tissues, 47 cell lines, 48 human normal tissues and 12 blood cells, the results are represented by at least 576 immunohistochemical staining maps, which have been read and indexed by professionals. These tissues were from 144 different individuals and 216 tumor tissues, which ensured that the staining results were fully representative49. In our study, direct contrast of protein expression of different GBPs family members between normal and HNSCC tissues was used by immunohistochemistry image.
GEPIA dataset
GEPIA50 is a newly created online interactive web server which enables users to exploring the RNA sequencing expression information of tumors/normal tissues or samples from the Genotype Tissue Expression (GTEx) projects and The Cancer Genome Atlas (TCGA), based on a criterion processing pipeline. GEPIA offer customizable functions such as profiling regarding to pathological stages, cancer types, differential expression analysis, survival analysis, correlation analysis and similar gene detection.
Kaplan-meier plotter
The prognostic significance of mRNA expression of different GBPs in HNSCC was evaluated by using Kaplan-Meier plotter (http://kmplot.com/analysis/)51, in which data about gene expression with survival of patients in 21 cancer types. The system includes gene chip and RNA-seq information - sources for the databases include TCGA, GEO and EGA. By parameter setting, we can analyze the prognosis of patients in different subgroups, different pathological parameters, different treatment modes, and different data sets. In Kaplan-Meier plotter, cancer patient samples were split into low and high expression group according to median values of mRNA expression and assessed by K-M survival plot.
TCGA database
TCGA uses large-scale genome sequencing to map out the genomic variation maps of all human cancers, and conduct systematic analysis to understand the mechanism of cancer cell occurrence and development, and on this basis, to obtain new diagnostic and therapeutic methods52. In our analysis, clinicopathological parameters of 512 HNSCC patients and mRNA expression of GBPs of 529 HNSCC patients were downloaded from the TCGA. The listwise deletion technique was utilized to deal with missing data, which excluded the entire sample from the investigation if any single value was absent. Cox analysis was utilized to evaluate the impact of GBPs expression on survival alongside other clinical attributes (such as age, gender, stage, distant metastasis).
cBioPortal
The cBioPortal (http://cbioportal.org)53 is an open-access asset gives visualization, analysis and download of substantial scale cancer genomics data sets. We utilized c-BioPortal to analyze GBPs alterations in the TCGA HNSCC samples and shows an overview of genetic alterations per test in GBPs. A tab biological interaction network of the GBPs and their co-expression genes was analyzed and neighboring genes with alteration frequencies were included. Functions of GBPs mutations and 50 genes significantly related to GBPs mutations were performed by GO and KEGG in the Database for Annotation, Visualization, and Integrated Discovery (DAVID) online tool. P-Value<0.05 as statistically significant.
TIMER analysis
TIMER (https://cistrome.shinyapps.io/timer/)54 is a comprehensive asset for systematical investigation of immune infiltrates over various malignancy types. The abundances of six immune infiltrates (CD8+ T cells, B cells, CD4+ T cells, Macrophages, Neutrphils and Dendritic cells) are assessed by our statistical method, which is approved using pathological estimations. Using Gene module to explore correlation between GBPs expression and abundance of immune infiltrates in HNSCC; Survival module to draw Kaplan-Meier plots for immune infiltrates and GBPs to picture the survival differences. P-Value < 0.05 as statistically significant.
Supplementary information
Author contributions
W.Z.H. designed and analyzed the research study; W.Z.H. and Z.Y. wrote and revised the manuscript, C.F.C. and W.Z.H. collected the data and all authors contributed to and approved the final version of manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zeng-Hong Wu and Fu-Cheng Cai.
Supplementary information
is available for this paper at 10.1038/s41598-020-63246-7.
References
- 1.Xiao-Nan F, et al. Comprehensive analysis of competitive endogenous RNAs network associated with head and neck squamous cell carcinoma. Scientific Reports. 2018;8(1):10544. doi: 10.1038/s41598-018-28957-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray, F. et al. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians (2018). [DOI] [PubMed]
- 3.Magnes T, Egle A, Greil R, Melchardt T. Update on squamous cell carcinoma of the head and neck: ASCO annual meeting. Memo. 2017;10:220–223. doi: 10.1007/s12254-017-0358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin D, et al. Novel markers for poor prognosis in head and neck cancer. Int J Cancer. 2005;113:789–797. doi: 10.1002/ijc.20608. [DOI] [PubMed] [Google Scholar]
- 5.Bressan V, et al. The effects of swallowing disorders, dysgeusia, oral mucositis and xerostomia on nutritional status, oral intake and weight loss in head and neck cancer patients: a systematic review. Cancer Treat Rev. 2016;45:105–119. doi: 10.1016/j.ctrv.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Coskun HH, et al. Current philosophy in the surgical management of neck metastases for head and neck squamous cell carcinoma. Head Neck. 2015;37(6):915–926. doi: 10.1002/hed.23689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavón M. A. et al. The combined use of EFS, GPX2, and SPRR1A expression could distinguish favorable from poor clinical outcome among epithelial-like head and neck carcinoma subtypes. Head Neck. Jun;41(6):1830-1845 (2019) [DOI] [PubMed]
- 8.Guo-Fang G, et al. Overexpression of lncRNA H19/miR-675 promotes tumorigenesis in head and neck squamous cell carcinoma. International Journal of Medical Sciences. 2016;13(12):914–922. doi: 10.7150/ijms.16571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y. et al. SUZ12 is a novel putative oncogene promoting tumorigenesis in head and neck squamous cell carcinoma. Journal of Cellular and Molecular Medicine (2018). [DOI] [PMC free article] [PubMed]
- 10.Reed AL, et al. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Research. 1996;56(16):3630–3633. [PubMed] [Google Scholar]
- 11.Wang L. et al. FSCN1 is upregulated by SNAI2 and promotes epithelial to mesenchymal transition in head and necksquamous cell carcinoma[J]. Cell Biology International (2017). [DOI] [PubMed]
- 12.Tripal PM, et al. Unique features of different members of the human guanylate-binding protein family. J. Interferon Cytokine Res. 2007;27:44e52. doi: 10.1089/jir.2007.0086. [DOI] [PubMed] [Google Scholar]
- 13.Boehm UL, et al. Two families of GTPases dominate the complex cellular response to IFN-gamma, J. Immunol. 1998;161:6715e6723. [PubMed] [Google Scholar]
- 14.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117e127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 15.Ji C, et al. Structural mechanism for guanylate-binding proteins (GBPs) targeting by the Shigella E3 ligase IpaH9.8. PLoS Pathog. 2019 Jun 19. 2019;15(6):e1007876. doi: 10.1371/journal.ppat.1007876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisch D, et al. Human GBP1 is a microbe-specific gatekeeper of macrophage apoptosis and pyroptosis. EMBO J. 1. 2019;38(13):e100926. doi: 10.15252/embj.2018100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Britzen-Laurent N, et al. GBP-1 acts as a tumor suppressor in colorectal cancer cells. Carcinogenesis. 2013;34(1):153–162. doi: 10.1093/carcin/bgs310. [DOI] [PubMed] [Google Scholar]
- 18.Peng C H. et al. A Novel Molecular Signature Identified by Systems Genetics Approach Predicts Prognosis in Oral Squamous Cell Carcinoma. PLOS ONE, 6 (2011). [DOI] [PMC free article] [PubMed]
- 19.Talbot SG, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Research. 2005;65(8):3063–71. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- 20.Ginos MA. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck[J] Cancer Research. 2004;64(1):55. doi: 10.1158/0008-5472.CAN-03-2144. [DOI] [PubMed] [Google Scholar]
- 21.Estilo CL, et al. Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9(1):11–0. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye H, et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9(1):69–0. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koneva L. A. et al. HPV Integration in HNSCC Correlates with Survival Outcomes, Immune Response Signatures, and Candidate Drivers. Molecular Cancer Research:molcanres.0153 (2017). [DOI] [PMC free article] [PubMed]
- 24.Ho Y, et al. STAT3 as a therapeutic target for epstein-barr virus (EBV): associated nasopharyngeal carcinoma. Cancer Lett. 2013;330(2):141–9. doi: 10.1016/j.canlet.2012.11.052. [DOI] [PubMed] [Google Scholar]
- 25.Naschberger E, et al. Nuclear factor-kappaB motif and interferon-alpha-stimulated response element co-operate in the activation of guanylate-binding protein-1 expression by inflammatory cytokines in endothelial cells. Biochem. J. 2004;379:409–420. doi: 10.1042/bj20031873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lubeseder-Martellato C, et al. Guanylate-binding protein-1 expression is selectively induced by inflammatory cytokines and is an activation marker of endothelial cells during inflammatory diseases. Am. J. Pathol. 2002;161:1749–1759. doi: 10.1016/S0002-9440(10)64452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guenzi E, et al. The helical domain of GBP-1 mediates the inhibition of endothelial cell proliferation by inflammatory cytokines. EMBO J. 2001;20:5568–5577. doi: 10.1093/emboj/20.20.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Decker T, Lew DJ, Darnell JE., Jr. Two distinct alpha-interferondependent signal transduction pathways may contribute to activation of transcription of the guanylate-binding protein gene. Mol. Cell. Biol. 1991;11:5147–5153. doi: 10.1128/MCB.11.10.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu CJ, et al. Identification of Guanylate-Binding Protein 1 as a Potential Oral Cancer Marker Involved in Cell Invasion Using Omics-Based Analysis. Journal of Proteome Research. 2011;10(8):3778–3788. doi: 10.1021/pr2004133. [DOI] [PubMed] [Google Scholar]
- 30.Yamakita I. et al. Guanylate binding protein 1 (GBP-1) promotes cell motility and invasiveness of lung adenocarcinoma. Biochem Biophys Res Commun. 14 (2019). [DOI] [PubMed]
- 31.Mustafa D. A. M. et al. T lymphocytes facilitate brain metastasis of breast cancer by inducing Guanylate-Binding Protein 1 expression. Acta Neuropathologica (2018). [DOI] [PMC free article] [PubMed]
- 32.Degrandi D, et al. Murine guanylate binding protein 2 (mGBP2) controls Toxoplasma gondii replication. Proc Natl Acad Sci USA. 2013;110(1):294–9. doi: 10.1073/pnas.1205635110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, et al. Guanylate-binding protein 2 regulates Drp1-mediated mitochondrial fission to suppress breast cancer cell invasion. Cell Death and Disease, 2017;8(10):e3151. doi: 10.1038/cddis.2017.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Godoy P, et al. Interferon-inducible guanylate binding protein (GBP2) is associated with better prognosis in breast cancer and indicates an efficient T cell response. Breast Cancer. 2014;21(4):491–499. doi: 10.1007/s12282-012-0404-8. [DOI] [PubMed] [Google Scholar]
- 35.Braun E, et al. Guanylate-Binding Proteins 2 and 5 Exert Broad Antiviral Activity by Inhibiting Furin-Mediated Processing of Viral Envelope Proteins. Cell Rep. 2019;27(7):2092–2104.e10. doi: 10.1016/j.celrep.2019.04.063. [DOI] [PubMed] [Google Scholar]
- 36.Traver MK, et al. Immunity-related GTPase M (IRGM) Proteins Influence the Localization of Guanylate-binding Protein 2 (GBP2) by Modulating Macroautophagy. Journal of Biological Chemistry. 2011;286(35):30471–30480. doi: 10.1074/jbc.M111.251967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H. et al. GBP3 promotes glioma cell proliferation via SQSTM1/p62-ERK1/2 axis. Biochemical and Biophysical Research Communications, S0006291X17322283 (2017). [DOI] [PubMed]
- 38.Nordmann LA, et al. A new splice variant of the human guanylate-binding protein 3 mediates anti-influenza activity through inhibition of viral transcription and replication, FASEB. J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2012;26:1290e1300. doi: 10.1096/fj.11-189886. [DOI] [PubMed] [Google Scholar]
- 39.Hu Y, et al. Guanylate Binding Protein 4 Negatively Regulates Virus-Induced Type I IFN and Antiviral Response by Targeting IFN Regulatory Factor 7. The Journal of Immunology. 2011;187(12):6456–6462. doi: 10.4049/jimmunol.1003691. [DOI] [PubMed] [Google Scholar]
- 40.Restivo G, et al. IRF6 is a mediator of Notch pro-differentiation and tumor suppressive function in keratinocytes. EMBO J. 2011;30(22):4571–4585. doi: 10.1038/emboj.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattei F, et al. IRF-8 controls melanoma progression by regulating the cross talk between cancer and immune cells within the tumor microenvironment. Neoplasia. 2012;14(12):1223–1235. doi: 10.1593/neo.121444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hallenberger S, et al. Inhibition of furin-mediated cleavage activation of HIV‐1 glycoprotein gp160. Nature. 1992;360(6402):358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 43.Krapp C, et al. Guanylate binding protein (GBP) 5 is an interferon-in‐ducible inhibitor of HIV‐1 infectivity. Cell Host Mi‐crobe. 2016;19(4):504–514. doi: 10.1016/j.chom.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 44.Wehner M, Herrmann C. Biochemical properties of the human guanylate binding protein 5 and a tumor-specific truncated splice variant. Febs Journal. 2010;277(7):1597–1605. doi: 10.1111/j.1742-4658.2010.07586.x. [DOI] [PubMed] [Google Scholar]
- 45.Shenoy A. R. et al. GBP5 Promotes NLRP3 Inflammasome Assembly and Immunity in Mammals. Science, 336 (2012). [DOI] [PubMed]
- 46.Olszewski MA, Gray J, Vestal DJ. In Silico Genomic Analysis of the Human and Murine Guanylate-Binding Protein (GBP) Gene Clusters. Journal of Interferon & Cytokine Research. 2006;26(5):328–352. doi: 10.1089/jir.2006.26.328. [DOI] [PubMed] [Google Scholar]
- 47.Hui RK, Leung FC, Yuntao W. Differential Expression Profile of Chicken Embryo Fibroblast DF-1 Cells Infected with Cell-Adapted Infectious Bursal Disease Virus. PLOS ONE. 2015;10(6):e0111771. doi: 10.1371/journal.pone.0111771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asplund A, Edqvist PH, Schwenk JM, Pontén F. Antibodies for profiling the human proteome-The Human Protein Atlas as a resource for cancer research. Proteomics. 2012;12:2067–77. doi: 10.1002/pmic.201100504. [DOI] [PubMed] [Google Scholar]
- 50.Tang Z, et al. Gepia: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagy A, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Scientific Reports. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19:A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao et al. Sci. Signal. 2013 & Cerami et al. Cancer Discov. 2012 when publishing results based on cBioPortal (2013).
- 54.Li T, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Research, 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.