Abstract
The long noncoding RNA (lncRNA), HOX antisense intergenic RNA myeloid 1 (HOTAIRM1), has been shown to act as a tumor suppressor in various human cancers. However, the overall biological roles and clinical significance of HOTAIRM1 in papillary thyroid cancer (PTC) have not been investigated. In this study, we used quantitative reverse transcription PCR (qRT-PCR) to show that HOTAIRM1 was significantly downregulated in PTC tissues and low HOTAIRM1 expression levels were associated with lymph node metastasis and advanced TNM stage. We performed Cell Counting Kit-8, plate colony-formation, flow cytometric apoptosis, transwell, and scratch wound healing assays. Overexpression of HOTAIRM1 was found to inhibit PTC cell proliferation, invasion, and migration in vitro. Additionally, we identified miR-107 as a target of HOTAIRM1 using online bioinformatics tools. Dual-luciferase reporter gene and RNA immunoprecipitation assays were used to confirm that HOTAIRM1 acted as a competing endogenous RNA of miR-107. Furthermore, enhancement of miR-107 could potentially reverse the effects of HOTAIRM1 overexpression in vitro. Inhibition of miR-107 suppressed PTC cell proliferation, invasion, and migration in vitro. HOTAIRM1 overexpression and miR-107 inhibition impaired tumorigenesis in vivo in mouse xenografts. Bioinformatics prediction and a dual-luciferase reporter gene assay demonstrated the binding between miR-107 and the 3′-untranslated region of TDG. The results of qRT-PCR and western blotting assays suggested that HOTAIRM1 could regulate the expression of TDG in an miR-107-meditated manner. In conclusion, we validated HOTAIRM1 as a novel tumor-suppressor lncRNA in PTC and proposed that the HOTAIRM1/miR-107/TDG axis may serve as a therapeutic target for PTC.
Subject terms: Thyroid cancer, Long non-coding RNAs
Introduction
Papillary thyroid carcinoma (PTC) is the most common thyroid malignancy1. In recent years, an increasing number of new PTC cases have been reported each year and patients are being diagnosed with PTC at a younger age. Although it is usually accompanied by long-term and disease-specific survival, recurrence, metastases, and cancer-related deaths may occur in 10–15% of PTC patients. Thus, understanding the pathological metastasis or progression of PTC is required.
Long noncoding RNAs (lncRNAs), which are more than 200 nucleotides in length, used to be very poorly understood. However, in recent years, various functions of lncRNAs have been identified, including participating in the autophagy pathway2,3, controlling cell differentiation4, and acting as competing endogenous RNAs (ceRNAs) for microRNAs2,5. The role of some lncRNAs as ceRNAs of miRNAs has been demonstrated in cancer metastasis and invasion6–9. For example, Lian et al. reported that lncRNA AFAP1-AS1 acted as a ceRNA of miR-432-5p, to promote metastasis in nasopharyngeal carcinoma. In PTC, an increasing number of lncRNAs are being identified, including the isoform 2 of NEAT1 (NEAT1_2)10, SNHG15 11, and n384546 12.
HOX antisense intergenic RNA myeloid 1 (HOTAIRM1) is a lncRNA with a length of 1052 bp. It was recently found to participate in various cancers, such as gastrointestinal malignancies13–16, breast cancer17, lung cancer18, glioblastoma multiforme19, and acute myeloid leukemia20,21. Of note, HOTAIRM1 is under-expressed in colorectal cancer13 and gastric cancer14. A previous study has also shown that HOTAIRM1 may impair the development of head and neck tumors by acting as a ceRNA and sponging miRNA-148a 9. These previous findings suggest that HOTAIRM1 may play a crucial role as a tumor suppressor. However, to date, the role of HOTAIRM1 in thyroid cancer has not been investigated.
On the basis of target prediction using bioinformatics analyses, HOTAIRM1 may serve as a ceRNA for miR-107. Previous studies have demonstrated that miR-107 is engaged in numerous biological processes, including cell differentiation22, response to chemotherapy23, insulin resistance24, and metastasis25,26. Accumulating evidence has shown that high levels of miR-107 may be a risk factor in the prognostic monitoring of malignant diseases, such as gastric cancer27, oropharyngeal cancer28, colorectal cancer29, and breast cancer30. However, the association between miR-107 and HOTAIRM1 in the mechanism of PTC metastasis remains unknown.
In the present study, we performed quantitative reverse transcription PCR (qRT-PCR) to measure the expression of HOTAIRM1 in PTC tissues and adjacent normal tissues and we found that HOTAIRM1 was significantly downregulated in tumor tissues. Under-expression of HOTAIRM1 was significantly correlated with the clinicopathological features of PTC patients, including TNM staging and lymph node metastasis. In vitro experiments showed that the overexpression of HOTAIRM1 inhibited PTC cell proliferation, migration, and invasion and promoted apoptosis. In vivo experiments confirmed that tumor growth was suppressed after HOTAIRM1 overexpression. In addition, high levels of HOTAIRM1 were found to have a tumor-suppressor effect by sponging miR-107 and regulating the expression of TDG. Taken together, the results of the current study indicated that the lncRNA, HOTAIRM1, might be a therapeutic target for PTC.
Results
HOTAIRM1 levels were decreased in papillary thyroid cancer
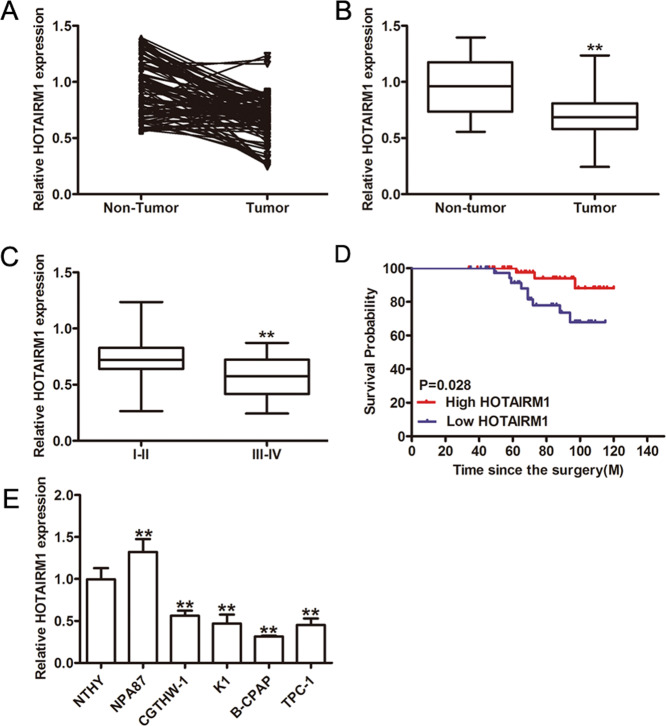
The expression levels of HOTAIRM1 in PTC samples were downregulated compared to the matched adjacent normal thyroid tissues (p < 0.001; Fig. 1a, b). Subgroup analyses on clinical features revealed that low HOTAIRM1 expression levels were significantly correlated with advanced stage (TIII and TIV; p = 0.0497; Fig. 1c, Supplementary Table 1) and lymph node metastasis (p < 0.01, Supplementary Table 1). Thus, Kaplan-Meier survival analysis was performed, which confirmed that PTC patients with low HOTAIRM1 levels (n = 40) had poorer overall survival than patients with high HOTAIRM1 levels (n = 56; p = 0.028; Fig. 1d). Therefore, we performed an in vitro study to determine the function of HOTAIRM1 in PTC. We investigated the expression of HOTAIRM1 in Nthy-ori 3-1, NPA87, CGTHW-1, K1, B-CPAP, and TPC-1 cells and confirmed the low HOTAIRM1 expression levels in the latter four PTC cell lines compared to the normal thyroid cell line, Nthy-ori 3-1 (Fig. 1e).
Fig. 1. Decreased expression of lncRNA HOTAIRM1 in human PTC tissues and cells.
a, b Relative expression levels of lncRNA HOTAIRM1 in human PTC tissues (n = 96) and corresponding adjacent non-tumor tissues (n = 96) by qRT-PCR (**p < 0.001). c HOTAIRM1 expression levels were lower in patients with an advanced tumor stage (TIII–TIV, n = 32) than that in those with an early stage (TI–TII, n = 64, **p < 0.001). d Kaplan-Meier survival analysis showed that patients with low HOTAIRM1 expression levels (n = 40) had poorer overall survival than patients with high HOTAIRM1 levels (n = 56, p = 0.028). e Relative expression levels in PTC cell lines (K1, B-CPAP, CGTHW-1, NPA87, and TPC-1) compared in a pairwise manner with expression levels in a normal human thyroid follicular epithelial cell line (Nthy-ori 3-1, n = 3, **p < 0.001).
HOTAIRM1 inhibited the proliferation of PTC cells in vitro and in vivo
Since HOTAIRM1 expression levels were relatively low in B-CPAP and TPC-1 cell lines, we studied the effect of HOTAIRM1 overexpression on the proliferation and invasion of these two PTC cell lines. B-CPAP and TPC-1 cells were divided into negative control vector (cells transfected with pcDNA3.1 plasmid vectors) and HOTAIRM1 (cells transfected with pcDNA3.1-HOTAIRM1) groups. A high efficiency of HOTAIRM1 overexpression was achieved in B-CPAP and TPC-1 cell lines using pcDNA3.1-HOTAIRM1, as illustrated in Fig. 2a.
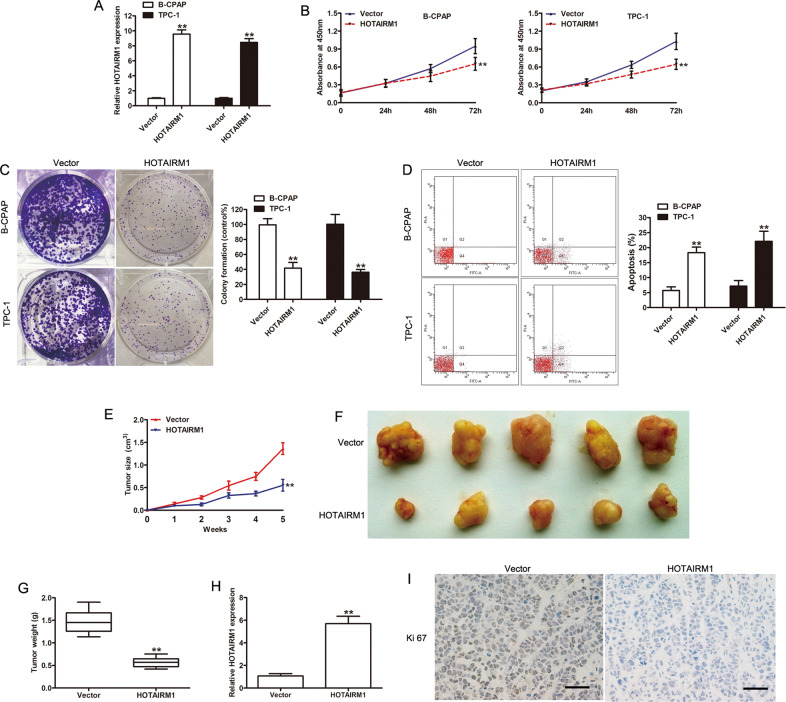
Fig. 2. Overexpression of HOTAIRM1 inhibited tumor growth in vitro and in vivo.
a High levels of HOTAIRM1 expression in B-CPAP and TPC-1 cells, achieved by transfection with pcDNA3.1-HOTAIRM1, were measured by qRT-PCR (n = 3, **p < 0.001). b, c The effect of HOTAIRM1 on PTC cell proliferation determined by CCK-8 and colony-formation assay (n = 3, **p < 0.001); d Apoptosis was evaluated by flow cytometry (n = 3, **p < 0.001). e The tumor growth curve of xenograft tumors formed from B-CPAP cells, with or without HOTAIRM1 overexpression, in the indicated week (n = 5, **p < 0.001). f Photographs of xenograft tumors after sacrifice. g The weight of xenograft tumors after sacrifice (n = 5, **p < 0.001). h Expression of HOTAIRM1 in xenograft tumors, assessed by qRT-PCR (n = 5, **p < 0.001). i Representative photographs of immunohistochemical staining of Ki-67 in xenograft tumors. Scale bar: 50 μm.
The effects of HOTAIRM1 on PTC cell proliferation in vitro were measured by a Cell Counting Kit-8 (CCK-8) assay, a colony-formation assay, and flow cytometry. The CCK-8 assay showed that HOTAIRM1 overexpression caused a decrease in the proliferation of B-CPAP and TPC-1 cells compared with the vector group (p < 0.001, Fig. 2b). The colony-formation assay demonstrated that HOTAIRM1 overexpression attenuated the proliferation of these cells (p < 0.001, Fig. 2c). The results of flow cytometry indicated that HOTAIRM1 increased apoptosis (p < 0.001, Fig. 2d).
A mouse tumor xenograft model was established to examine the effects of HOTAIRM1 on PTC cell proliferation in vivo. B-CPAP cells from the vector or HOTAIRM1 group were injected subcutaneously on the back of each nude mouse (n = 5 per group). Tumor volume was measured every 7 days after the injection. B-CPAP cells with high levels of HOTAIRM1 expression formed smaller tumors at each indicated time point, compared to vector-transfected cells (p < 0.001, Fig. 2e). At the termination of the experiment (the 35th day), mice were sacrificed and the entire tumors were excised. The resected tumors in the HOTAIRM1 group were significantly smaller and weighed less than the resected tumors in the vector group (p < 0.001, Fig. 2e–g). Further, qRT-PCR results confirmed the upregulation of HOTAIRM1 in xenograft tumors in the HOTAIRM1 group compared with the vector group (p < 0.001, Fig. 2h). In addition, immunohistochemistry (IHC) assays of the proliferation indicator, Ki-67, were performed. The percentage of Ki-67-positive cells in xenograft tumors was lower in the HOTAIRM1 group than the vector group (Fig. 2i). Taken together, these results showed that HOTAIRM1 overexpression efficiently impaired the proliferation of PTC cells in vitro and in vivo.
HOTAIRM1 suppressed cell migration and invasion in vitro
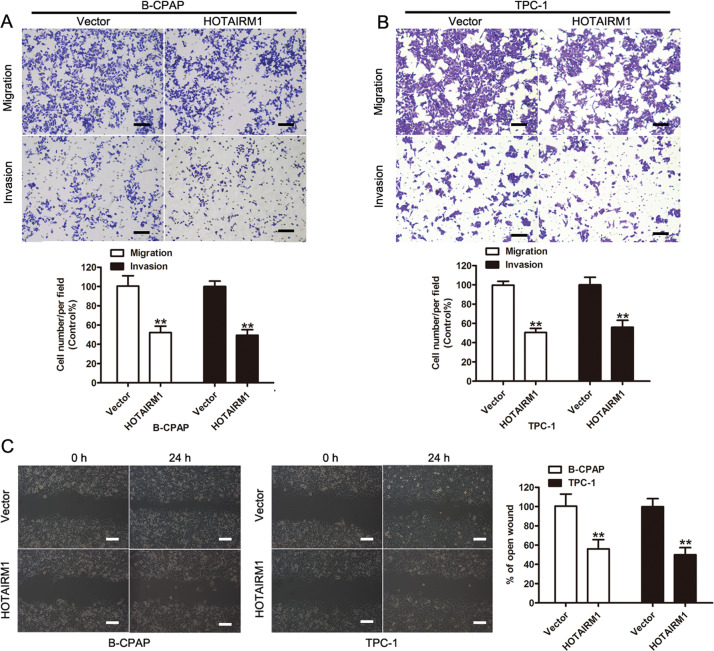
Transwell and scratch wound healing assays were performed to evaluate the involvement of HOTAIRM1 in the migration and invasion of B-CPAP and TPC-1 cells. The results of the transwell assay showed that HOTAIRM1 overexpression significantly suppressed the migration and invasion of PTC cells (p < 0.001, Fig. 3a, b). The results of the scratch wound healing assay confirmed that HOTAIRM1 overexpression inhibited the migration of these cells (p < 0.001, Fig. 3c). The above results demonstrated the function of HOTAIRM1 in PTC cells and that overexpression of HOTAIRM1 could significantly inhibit PTC cell migration and invasion in vitro.
Fig. 3. Overexpression of HOTAIRM1 suppressed PTC cell migration and invasion in vitro.
a, b Transwell assays were performed to evaluate the involvement of HOTAIRM1 in the invasion of B-CPAP (a) and TPC-1 cells (b) (n = 3, **p < 0.001). Scale bar: 100 μm. c Scratch wound healing assays were performed to assess the migration ability of B-CPAP and TPC-1 cells (n = 3, **p < 0.001). Scale bar: 100 μm.
HOTAIRM1 acted as a sponge for miR-107 in PTC cells
We further investigated the underlying molecular mechanism by which HOTAIRM1 affected PTC cell proliferation and invasion. Since there is increasing evidence showing that lncRNAs serve as ceRNAs to modulate the function of miRNAs5, we utilized an online bioinformatics database (miRcode31) and identified 13 microRNAs as potential competing targets of HOTAIRM1. Moreover, in a previous study of prognosis-related lncRNAs in ovarian cancer tissues, HOTAIRM1 was found to regulate hub genes through seven miRNAs, including miR-107, miR-103a-3p, miR-129-5p, miR-152-3p, miR-148a-3p, and miR-148b-3p 32. Thus, among the candidate targets identified, we focused on miR-107, since high expression levels of this miRNA are considered a prognostic risk factor in several cancers27–30.
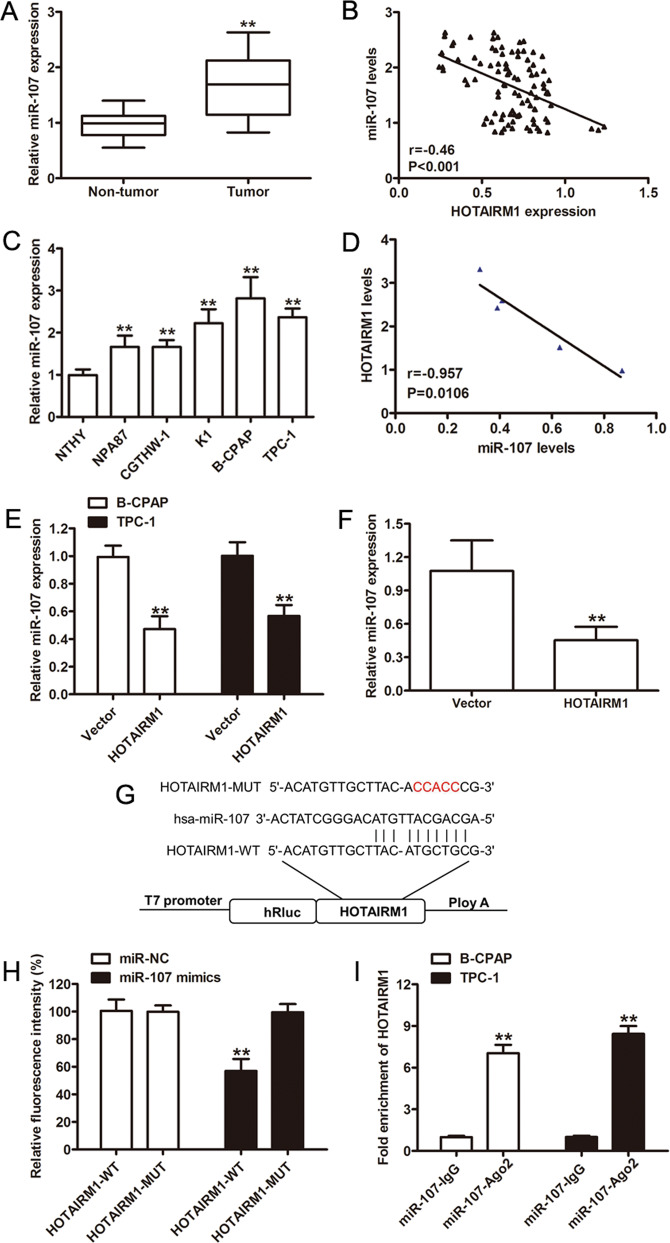
To investigate the potential relationship between miR-107 and HOTAIRM1 expression in PTC, we analyzed miR-107 expression using qRT-PCR. High expression levels of miR-107 were found in PTC tissues compared with patient-matched non-tumor tissues (n = 96, p < 0.001, Fig. 4a). Since we previously found decreased levels of HOTAIRM1 in PTC by qRT-PCR, a negative correlation between miR-107 and HOTAIRM1 levels was observed (r = −0.46, p < 0.001, Fig. 4b). In addition, we investigated the expression of miR-107 in Nthy-ori 3-1, NPA87, CGTHW-1, K1, B-CPAP, and TPC-1 cells and confirmed the high miR-107 expression levels in the latter four PTC cell lines compared to the normal thyroid cell line, Nthy-ori 3-1 (Fig. 4c). There were negative correlations between HOTAIRM1 and miR-107 were found in the same panel of cell lines (Fig. 4d). Meanwhile, we verified that miR-107 levels were reduced in HOTAIRM1-overexpressing PTC cells (p < 0.001; Fig. 4e, f).
Fig. 4. HOTAIRM1 regulated the expression of miR-107.
a Relative miR-107 expression levels determined by qRT-PCR in PTC samples (n = 96, **p < 0.001). b Negative correlation between HOTAIRM1 and miR-107 levels in 96 paired PTC tissues. c, d Relative expression levels of miR-107 in PTC cell lines (NPA87, CGTHW-1, K1, B-CPAP, and TPC-1) compared in a pairwise manner with expression levels in a normal human thyroid follicular epithelial cell line (Nthy-ori 3-1, n = 3, **p < 0.001). Negative correlations between HOTAIRM1 and miR-107 in the same panel of cell lines. e, f Relatively low miR-107 expression levels detected by qRT-PCR in HOTAIRM1-overexpressing PTC cells (n = 3, **p < 0.001) and in HOTAIRM1-overexpressing xenograft tumor tissues (n = 5, **p < 0.001). g Online bioinformatics software tools predicted a putative binding site between HOTAIRM1 and miR-107 and the binding site was mutated for dual-luciferase reporter assays. h A dual-luciferase reporter gene assay showed that miR-107 decreased the luciferase activity in the HOTAIRM1-WT group (n = 3, **p < 0.001). i RNA immunoprecipitation assay using an anti-Ago2 antibody and an IgG control showed a high degree of HOTAIRM1 enrichment (n = 3, **p < 0.001).
To determine whether HOTAIRM1 binds to miR-107, we performed dual-luciferase reporter and RNA immunoprecipitation (RIP) assays. The predicted miR-107-binding site of HOTAIRM1 was mutated, as illustrated in Fig. 4g. The results of the dual-luciferase reporter assay demonstrated that miR-107 mimics significantly suppressed luciferase activity in the HOTAIRM1-WT group; however, the miR-107 mimics were not able to bind to the mutant construct, HOTAIRM1-MUT and HOTAIRM1-WT was not able to bind to miR-NC without the seed region (p < 0.001, Fig. 4h). An anti-argonaute2 (Ago2) antibody was then used to capture mature miRNAs33. Data from the Ago2-RIP assay revealed that HOTAIRM1 was enriched by the anti-Ago2 antibody, compared with the negative control (p < 0.001, Fig. 4i). Taken together, these results indicated that HOTAIRM1 may act as a ceRNA and sponge miR-107 in PTC cells.
Inhibition of miR-107 induced the suppression of PTC cell proliferation, migration, and invasion
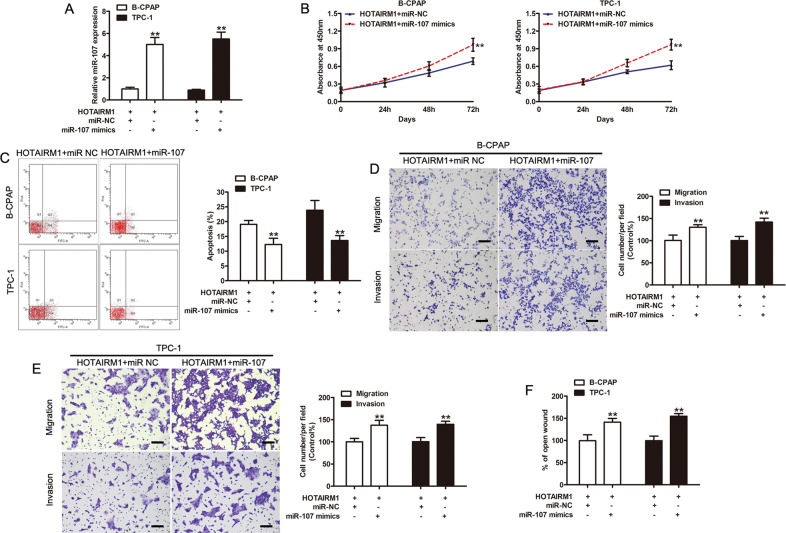
To investigate the role of miR-107 in HOTAIRM1-mediated regulation, we further investigated whether miR-107 could affect biologic activity in HOTAIRM1-overexpressing PTC cells. A stable increase in miR-107 expression, compared to cells transfected with the negative control, miR-NC, was achieved in HOTAIRM1-overexpressing PTC cell lines using miR-107 mimics (p < 0.001, Fig. 5a).
Fig. 5. miRNA-107 reversed the effects of HOTAIRM1 overexpression.
a Expression levels of miR-107 in HOTAIRM1-overexpressing B-CPAP and TPC-1 cells transfected with miR-NC or miR-107 mimics, assessed by qRT-PCR (n = 3, **p < 0.001). b Micro-RNA-107 mimics reversed the HOTAIRM1-induced inhibition of cell proliferation (n = 3, **p < 0.001). c miRNA-107 mimics reversed the HOTAIRM1-induced cell apoptosis (N = 3, **p < 0.001). d, e miRNA-107 mimics restored the migration and invasion ability impaired by HOTAIRM1 in B-CPAP (D) and TPC-1 (E) cells (n = 3, **p < 0.001). Scale bar: 100 μm. f Migration was evaluated by wound healing assays in B-CPAP and TPC-1 cells stably overexpressing HOTAIRM1 and transfected with miR-NC or miR-107 mimics (n = 3, **p < 0.001).
CCK-8 and flow cytometry assays demonstrated that miR-107 promoted cell proliferation and increased apoptosis (p < 0.001, Fig. 5b, c). The results of transwell and scratch wound healing assays showed that miR-107 significantly increased cell migration and invasion in HOTAIRM1-overexpressing PTC cell lines (p < 0.001, Fig. 5d–f). These results indicated that increased levels of miR-107 could potentially reverse the effects of HOTAIRM1 overexpression in vitro.
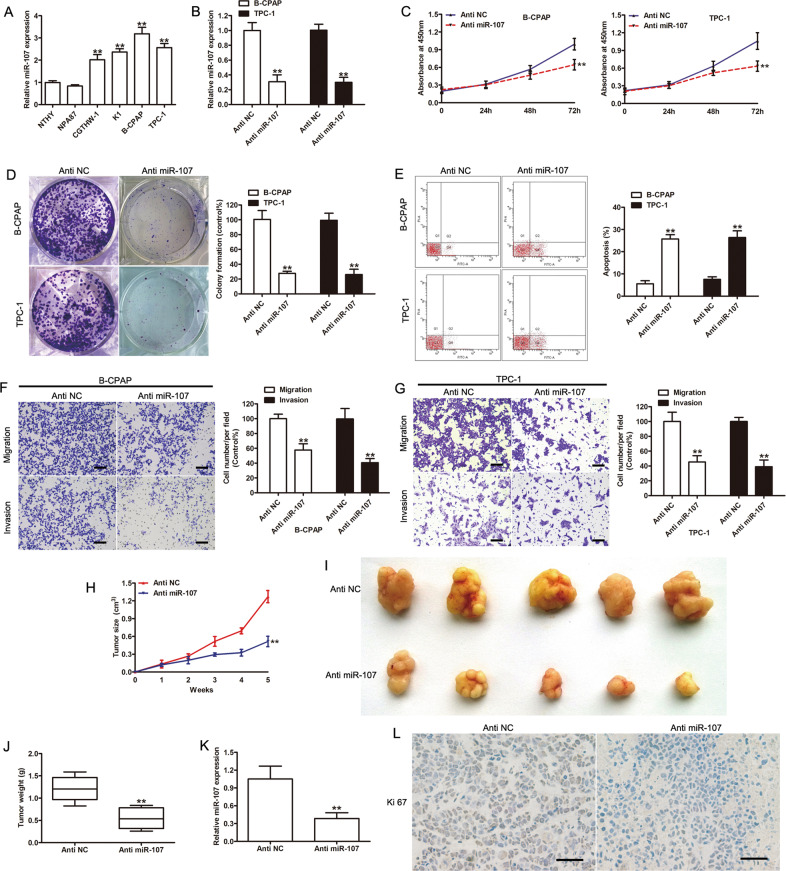
Based on the qRT-PCR results, PTC cells have high levels of miR-107 expression (p < 0.001, Fig. 6a), which was consistent with the negative correlation found between HOTAIRM1 and miR-107 in PTC tissues (Fig. 4b). To further verify the function of miR-107 in PTC, we inhibited miR-107 in B-CPAP and TPC-1 cell lines using anti-miR-107 (p < 0.001 versus the negative control, anti-NC; Fig. 6b). The results of the CCK-8, colony formation, and flow cytometry assays indicated that the inhibition of miR-107 significantly suppressed the proliferation, migration, and invasion of B-CPAP and TPC-1 cells in vitro, compared with the negative control group (p < 0.001, Fig. 6c–g). The tumorigenesis of miR-107-inhibited B-CPAP cells was also evaluated in a xenograft model. The inhibition of miR-107 significantly attenuated B-CPAP cell proliferation in vivo, based on the observed decrease in tumor size and weight (p < 0.001, Fig. 6h–j). Further, qRT-PCR and Ki-67 IHC results confirmed that miR-107 expression was downregulated and cell proliferation was inhibited in xenograft tumor tissues of the anti-miR-107 group (p < 0.001, Fig. 6k–l). These results demonstrated that tumorigenesis was inhibited once the function of miR-107 was impaired.
Fig. 6. The effects of miR-107 on PTC cell proliferation and invasion.
a The expression levels of miR-107 in PTC cell lines and normal thyroid follicular epithelial cells (n = 3, **p < 0.001). b The expression levels of miR-107 were determined in miR-107-knockdown B-CPAP and TPC-1 cells by qRT-PCR (n = 3, **p < 0.001). c, d Cell proliferation was evaluated by CCK-8 (n = 3, **p < 0.001) (c) and colony-formation assays (n = 3, **p < 0.001) (d) in B-CPAP and TPC-1 cells transfected with anti-miR-107. e Apoptosis was analyzed by flow cytometry (n = 3, **p < 0.001). f, g Knockdown of miR-107 inhibited the invasion of B-CPAP (f) and TPC-1 (g) cells, as determined by transwell assays (n = 3, **p < 0.001). Scale bar: 100 μm. h Tumor growth curve of xenograft tumors formed from B-CPAP cells transfected with anti-NC or anti-miR-107 (n = 5, **p < 0.001). i Photographs of xenograft tumors. j Weight of xenograft tumors (n = 5, **p < 0.001). k The expression levels of miR-107 in xenograft tumor tissues assessed by qRT-PCR (n = 5, **p < 0.001). l Representative photographs of immunohistochemical staining of Ki-67 in xenograft tumors. Scale bar: 50 μm.
TDG was an miR-107 target gene and was indirectly regulated by HOTAIRM1
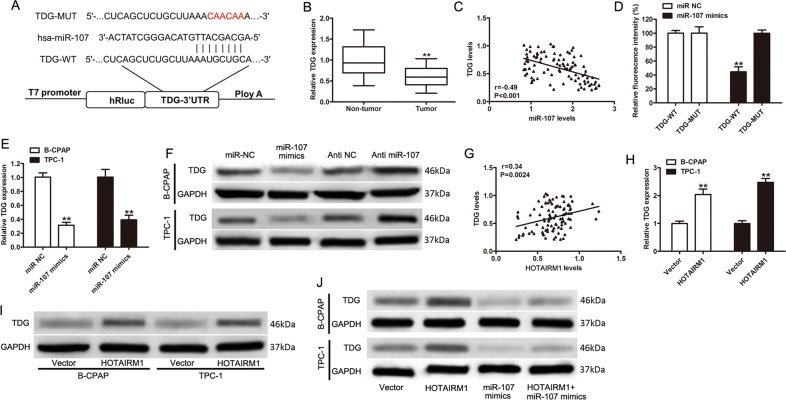
In view of these results, we assumed that the target genes of miR-107 might function directly in the pathogenesis of PTC. Therefore, we performed bioinformatics analysis (TargetScanHuman34) and found that the 3′-untranslated region (UTR) of TDG was one of the potential binding sites of miR-107 (Fig. 7a). Since TDG has been shown to be involved in active DNA demethylation35,36, we investigated it further. We found that the expression level of TDG was lower in tumor tissues compared with non-tumor tissues (n = 96, Fig. 7b, p < 0.001). Moreover, there was a negative correlation between miR-107 and TDG levels in PTC tissues (r = −0.49, p < 0.001, Fig. 7c). A dual-luciferase activity assay was performed to confirm that TDG was an miR-107 target. The results indicated that miR-107 mimics reduced luciferase activity in the TDG-WT group but not in TDG-MUT group (Fig. 7d, p < 0.001). In both B-CPAP and TPC-1 cell lines, decreased TDG mRNA and protein expression levels were detected after increasing the expression level of miR-107 (p < 0.001, Fig. 7e, f) and western blotting results indicated that TDG protein levels increased after the inhibition of miR-107 (Fig. 7f). These results suggested that the 3′-UTR of TDG was a target of miR-107. The functions of TDG were then investigated in PTC cells. Endogenous TDG was significantly transiently overexpressed in B-CPAP and TPC-1 cells and the efficiencies of interference were confirmed by western blotting (Supplementary Fig. 1a). A CCK-8 assay showed that TDG overexpression significantly inhibited cell proliferation (Supplementary Fig. 1b). Simultaneously, flow cytometric analysis indicated that the overexpression of TDG markedly increased the apoptosis of PTC cells (Supplementary Fig. 1c). Transwell assays demonstrated that TDG overexpression significantly decreased PTC cell migration and invasion (Supplementary Fig. 1d). To further assess the metastatic effect of TDG in vivo, TPC-1 cells with transient overexpression of TDG were injected into the tail vein of nude mice. Thus, overexpressed TDG significantly inhibited lung metastasis in vivo (Supplementary Fig. 1e). These findings strongly suggested that TDG suppressed PTC progression in vitro and in vivo.
Fig. 7. TDG was a target of miR-107 and was increased by HOTAIRM1 overexpression.
a Online bioinformatics software tools predicted a putative binding site between miR-107 and the 3′-UTR of TDG. This binding site was mutated for dual-luciferase reporter assays. b The mRNA levels of TDG in tissues from PTC patients (n = 96, **p < 0.001). c Association analysis of miR-107 and TDG mRNA levels in PTC tissues. d The dual-luciferase reporter gene assay showed that miR-107 decreased luciferase activity in the 3′-UTR-TDG-WT group (n = 3, **p < 0.001). e, f Relative mRNA and protein levels of TDG in B-CPAP and TPC-1 cells transfected as indicated (n = 3, **p < 0.001). g Positive correlation between HOTAIRM1 and TDG expression levels in 96 paired PTC tissues. h, i The mRNA and protein levels of TDG in cells with stable overexpression of HOTAIRM1 (n = 3, **p < 0.001). j TDG protein levels of in B-CPAP and TPC-1 cells following overexpression of HOTAIRM1 and/or enhancement of miR-107.
Since a positive correlation between HOTAIRM1 and TDG expression levels was observed in tumor tissues (r = 0.34, p = 0.0024, Fig. 7g), we sought to determine whether the regulation of TDG expression was influenced by HOTAIRM1. The results of qRT-PCR and western blotting indicated that the overexpression of HOTAIRM1 increased the mRNA and protein levels of TDG (Fig. 7h–i). However, TDG protein remained under-expressed when miR-107 expression was increased, despite the overexpression HOTAIRM1, in B-CPAP and TPC-1 cell lines (Fig. 7i). These results suggested that TDG expression might be enhanced by HOTAIRM1, in the presence of relatively low miR-107 expression levels.
Discussion
Proliferation, migration, and invasion are the three main characteristics of malignant tumor cells. Our present work demonstrated that HOTAIRM1 was significantly under-expressed in PTC tissues, while PTC patients with lymph node metastasis or higher TNM stage showed lower HOTAIRM1 expression levels than patients with no lymph node metastases or with a lower TNM stage. This suggested that HOTAIRM1 expression is involved in the development of PTC. Further investigation verified that overexpression of HOTAIRM1 inhibited the proliferation, migration, and invasion of PTC cell lines in vitro, and inhibited cell proliferation in vivo, as evidenced by decreased tumor size in a mouse model of thyroid cancer. Our findings suggested the role of HOTAIRM1 as a negative regulator of PTC progression.
Recently, HOTAIRM1 has been reported to be involved in many malignant diseases, acting as either a positive or a negative regulator in various cancers. Here, we showed that the expression levels of HOTAIRM1 were significantly lower in PTC tissues than in adjacent normal tissues and that the expression levels of HOTAIRM1 were lower in advanced-stage tumors than in early-stage tumors. The low levels of HOTAIRM1 expression observed in advanced-stage PTC was consistent with the previously identified role of HOTAIRM1 as a negative regulator of cancer progression. A study of myeloid leukemia showed that HOTAIRM1 regulates autophagy and oncoprotein degradation during the process of myeloid cell differentiation blockade20. Zheng et al. reported that the overexpression of HOTAIRM1 suppresses the proliferation, apoptosis, migration, and invasion of prehuman hypopharyngeal tumor cells in vitro, and suppresses human hypopharyngeal tumor cell growth in vivo9. The role of HOTAIRM1 as a potential tumor suppressor has also been observed in colorectal cancer13 and gastric cancer cell lines14. Subsequently, our in vitro and in vivo results also supported the role of HOTAIRM1 in inhibiting the growth, metastasis, and invasion of PTC tumors. These findings provided further evidence that HOTAIRM1 may have a role in suppressing tumor progression.
In the past 10 years, various studies have shown that lncRNAs inhibit the expression of microRNAs through a sponge-like effect, by binding to specific sites on the microRNA and thus, interfering with the binding of the microRNA to its target gene. In the present study, the binding of HOTAIRM1 and miR-107 was verified by dual-luciferase reporter and RIP assays. Moreover, HOTAIRM1 was found to negatively regulate miR-107 in TPC-1 and B-CPAP cell lines. We also confirmed the function of miR-107 in promoting cell growth, metastasis, and invasion in vitro and in vivo. Previous studies have shown that miR-107 is involved in metastasis25,26 and may serve as a prognostic risk factor in various cancers27–30. In view of these findings, the regulation of miR-107 by HOTAIRM1 may be a feasible strategy to inhibit PTC metastasis. However, the endogenous relationship between HOTAIRM1 and miR-107 is not exclusive, as indicated by the prediction of target microRNAs for HOTAIRM1. Therefore, the repression of HOTAIRM1 may affect miRNAs other than miR-107. A previous study to identify prognosis-related lncRNAs in ovarian cancer tissues, showed that HOTAIRM1 regulates hub genes through several miRNAs, including miR-107, miR-103a-3p, miR-129-5p, miR-152-3p, miR-148a-3p, and miR-148b-3p 32. In vitro and in vivo studies demonstrated that HOTAIRM1 may play a negative role in the development of head and neck tumors through the HOTAIRM1/microRNA-148a axis9. HOTAIRM1 is also regulated by miR-17-5p in 5-fluorouracil-resistant colorectal cancer cells37 and gastric cancer14. Moreover, studies have shown that various lncRNAs, such as lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1)38, lncRNA tissue differentiation-inducing non-protein coding (TINCR)39, and lncRNA long intervening noncoding 00467 (LINC00467)40, modulate miR-107 by acting as endogenous sponges. These results indicated that HOTAIRM1 has a relatively extensive inhibitory effect on microRNAs and it may target other genes in PTC, which may have a similar impact as the interaction between miR-107 and HOTAIRM1. Despite the limitations of our study, the present data may contribute to further studies of the role of HOTAIRM1 in the progression and metastasis of PTC. The role of miR-107 and whether it has a major or minor effect on the regulation of metastasis-related processes in PTC remains undetermined and thus, requires further investigation.
To further clarify the impact of HOTAIRM1 and miR-107 on PTC, the downstream regulatory mechanism of miR-107 in PTC progression was investigated. Using bioinformatics tools, the 3′-UTR of TDG was predicted as one of the direct binding targets of miR-107. TDG encodes thymine DNA glycosylase, which participates in active DNA demethylation in the mammalian genome35,36. Therefore, decreased TDG levels may induce the accumulation of 5-hydroxymethylcytosine, 5-carboxylcytosine, and 5-formylcytosine35,36, which may lead to genomic instability and the occurrence of malignancy41. For instance, previous studies have reported that low levels of TDG expression are associated with poor prognosis in breast cancer (HR = 2.178, 95% confidence interval: 1.140–4.163, p = 0.018)42. Previously, some retrospective studies have suggested that TDG mutations are associated with the occurrence of tumors43–45; however, there is no direct evidence suggesting that post-transcriptional regulation by microRNAs, such as microRNA-29a46, microRNA-29b47, and microRNA-26a48, is associated with tumors. In our study, the targeting of TDG by miR-107 was verified. Moreover, we observed a negative correlation between TDG mRNA levels and miR-107 in PTC tissues, but a positive correlation between TDG and HOTAIRM1 levels. These data suggested a potential involvement of the HOTAIRM1/miR-107/TDG axis in PTC, which would be a novel pathway of the microRNA-mediated regulation of TDG in cancers. Since the present finding was only based on bioinformatics prediction and a dual-luciferase reporter assay, the evidence is not yet comprehensive and further studies are required to confirm these findings. The proposed HOTAIRM1/miR-107/TDG axis in PTC remains to be further explored.
In conclusion, we demonstrated that HOTAIRM1 functioned as an oncogene in PTC. HOTAIRM1 promotes PTC progression and acted as a ceRNA to exert malignant effects via miR-107/TDG axis. We propose a model that highlights the function of HOTAIRM1 in regulating cell proliferation and invasion during PTC progression (Fig. 8). Collectively, our results showed that the HOTAIRM1/ miR-107/TDG axis has a critical role in PTC progression and is thus, a promising target for PTC therapy.
Fig. 8. Structure chart of the role of the HOTAIRM1/miR-107/TDG axis in PTC progression.
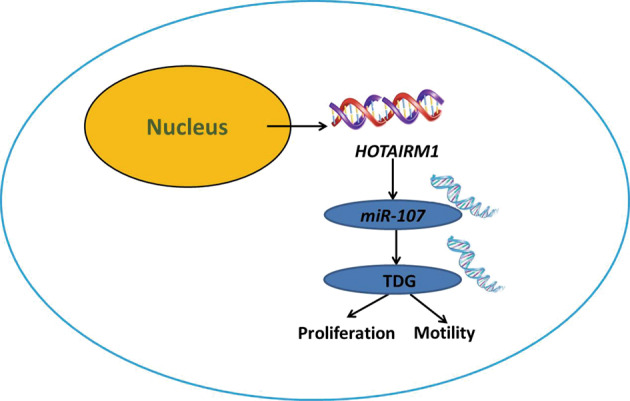
Our combined data indicated a model whereby HOTAIRM1 inhibited PTC cell proliferation and invasion through sponging miR-107 to decrease the expression of TDG, a key DNA repair and replication proteins associated with critical aspects of cellular function.
Methods
Sample collection
The sampling and experimental processes were performed with the approval of the Institutional Review Board and Ethics Committee of Shanghai Tenth People’s Hospital and informed written consent was obtained from 96 PTC patients who were admitted to Shanghai Tenth People’s Hospital. PTC tissues and corresponding adjacent normal thyroid tissues from all patients were stored at −80 °C. Prior to thyroidectomy, none of the patients had received chemotherapy or radical treatment.
Cell culture
A normal human thyroid follicular epithelial cell line (Nthy-ori 3-1) and human thyroid cancer cell lines (NPA87, CGTHW-1, K1, B-CPAP, and TPC-1) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). NPA87 cells were cultivated in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS), K1 cells were cultivated in DMEM supplemented with 10% FBS, and the other cell lines were cultivated in RPMI-1640 medium supplemented with 10% FBS. All cells were cultured at 37 °C under a humidified atmosphere with 5% CO2.
Plasmid construction and lentiviral transfection
To overexpress HOTAIRM1, a cDNA encoding HOTAIRM1 was amplified by PCR and subcloned into the pCDH-CMV-MCS-EF1-Puro vector (System Biosciences, Mountain View, CA, USA). To overexpress TDG, a cDNA encoding TDG was amplified by PCR and subcloned into the pcDNA3.1(+) vector (Invitrogen Thermo Fisher, Shanghai, China). miR-107 mimics and a negative control miRNA (miR-NC) and miR-107 inhibitors (anti-miR-107) and an inhibitor control (anti-NC) were purchased from Invitrogen (Carlsbad, CA, USA). B-CPAP and TPC-1 cells were seeded in 6-well plates the day before lentivirus transduction. Lentiviruses were transduced into cells at a suitable multiplicity of infection with polybrene (8 mg/mL). After incubation for 24 h, the medium was replaced with fresh medium.
RNA extraction and qRT-PCR
Total RNA was isolated from tissues or cells using TRIzol™ (Invitrogen) or GenElute™ Total RNA Purification Kit (Sigma-Aldrich, St. Louis, MO, USA), following the instructions of the manufacturers. Reverse transcription was performed using the PrimeScript™ RT Master Mix (Takara Biomedical Technology Co., Ltd, Beijing, China) in a S1000™ Thermal Cycler (Bio-Rad, Hercules, CA, USA). Real-time PCR was performed using the KAPA SYBR® FAST qPCR Master Mix (2×) Kit (Kapa Biosystem, Wilmington, MA, USA) in a CFX96™ Real-Time System (Bio-Rad). All the primers used in these experiments are listed in Supplementary Table 2. Either GAPDH or U6 was used as an endogenous reference and the 2−ΔΔCt method was used to calculate expression levels.
Western blotting
Protein extracts were boiled in RIPA buffer (Beyotime, Shanghai, China) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Subsequently, the proteins were transferred to polyvinylidene fluoride membranes (0.45 μm pore diameters). Membranes were blocked in phosphate- buffered saline with 0.05% Tween-20 (PBS-T), containing 5% non-fat milk for 1 h and then incubated at 4 °C overnight with the following primary antibodies: rabbit anti-human TDG (13370-1-AP, 1:800, Proteintech, Wuhan, Hubei, China) and rabbit anti-human GAPDH (ab181602, 1:800, Abcam, Cambridge, UK). After washing with PBS-T, the membranes were hybridized with horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (ab6721, 1:2,000, Abcam, Cambridge, UK) for 1 h. Signal detection was performed using an ECL system (Amersham Pharmacia, Piscataway, NJ, USA).
Cell counting kit-8 assay
A CCK-8 assay (Dojindo, Kumamoto, Japan) was used to determine the effect of miR-107 and HOTAIRM1 on cell proliferation. Briefly, cells were seeded in 96-well plates at a concentration of 2 × 103 cells per well and incubated for 24, 48, 72, or 96 h. At the indicated time point, 10 μL of CCK-8 assay reagent was added and the plates were incubated for another 4 h. A SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA, USA) was used to measure the absorbance at 450 nm.
Plate colony-formation assay
Briefly, after 24 h of transfection, B-CPAP and TPC-1 cells were initially seeded into 3.5 cm culture dishes at a density of 800 cells per dish and maintained in medium containing 10% FBS, which was refreshed every 2 days. After the cells had been incubated for approximately 2 weeks at 37 °C in 5% CO2, the resulting colonies were visible to the naked eye. The cells were fixed with 4% paraformaldehyde for 15 min before staining with 0.1% crystal violet for 15 min and then counted. The colony numbers were counted using ImageJ software (National Institutes of Health, Bethesda, MD, USA) and manually counted from three randomly chosen fields. Experiments were tested in triplicate.
Flow cytometric analysis of cell apoptosis
A fluorescein-conjugated annexin V (annexin V-FITC)/propidium iodide (PI) staining kit (BD Biosciences, San Jose, CA, USA) was used to detect apoptosis, following the manufacturer’s instructions. A FACS Calibur FCM instrument (BD Biosciences) was used to observe apoptosis. In brief, 48 h after transfection, cells were suspended in 100 μL of binding buffer at a concentration of 1 × 105 cells/mL, after which 5 μL of FITC-annexin V and 5 μL of PI were added to the solution. After incubating for 15 min at room temperature protected from light, 400 μL of binding buffer was added and apoptosis was assessed within 1 h. Experiments were performed in triplicate to help reduce errors. FACS Diva software (BD Biosciences) was used for data analysis.
Cell invasion and migration assays
For transwell migration assays, transfected B-CPAP and TPC-1 cells transfected cells (4 × 105) were plated in the top chamber and a noncoated membrane (24-well insert; pore size, 8 μm; BD Biosciences) was polymerized in transwell inserts for 45 min at 37 °C. In both assays, cells were plated in the top chamber in medium without serum and the lower chamber was filled with 20% FBS (GIBCO BRL, Grand Island, NY, USA) as a chemoattractant. Cells were incubated for 24 h and those that did not migrate or invade through the pores were removed with a cotton swab. Cells that migrated to the lower surface of the membrane were fixed and stained with 0.1% crystal violet. The cells on the bottom of the membrane were counted from five different microscopic fields and the average number was calculated.
Scratches extending the length of each well were made on the cellular surface of each well of six-well plates containing B-CPAP and TPC-1 cells, using standard 200 μL pipette tips. Cells within the wound area were washed with PBS and the images were photographed under an inverted microscope (Leica Microsystems) 24 h later.
Animal experiments
All animal studies were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Animal Care and Use Committee of Shanghai Tenth People’s Hospital. Six-week-old nude mice (n = 20) were purchased from SLAC Laboratory Animal Co., Ltd (Shanghai, China) and were randomly divided into four groups and no blinding was done. Serum-free suspensions (1 × 107/mL) of B-CPAP cells transfected with pcDNA3.1-HOTAIRM1, pcDNA3.1vector (as negative control), anti-NC, or anti-miR-107 were injected subcutaneously on the back of each mouse (0.2 mL). When the tumor grew to approximately 100-200 mm3, the tumor volume was calculated using the following formula: 1/2 × L2 × W, where L is the length (mm) and W is the width (mm) of the tumor. The average volume of the tumor was measured three times every 7 days. At the termination of the experiment (the 35th day), mice were sacrificed and the tumors were excised for volume and weight measurements. Total RNA was isolated and the expression level of HOTAIRM1 was determined by qRT-PCR.
To establish an in vivo lung metastasis model, 1 × 106 cells were intravenously injected into the lateral tail vein of nude mice (n = 5 per group). The mice were measured using a bioluminescence system (Titertek Berthold, Pforzheim, Germany) and at week 8, the mice were sacrificed. Thereafter, to analyze the presence of metastatic nodules, the lungs were fixed, photographed, preserved, and stained with hematoxylin and eosin.
Immunohistochemistry assay
A Ki-67 cell proliferation kit (Sangon Biotech, Shanghai, China) was used to evaluate cell proliferation in xenograft tumors, following the manufacturer’s instructions.
Dual-luciferase activity assay
Luciferase plasmids containing wild-type (pmirGLO-HOTAIRM1-WT) or mutated (pmirGLO-HOTAIRM1-MUT) putative HOTAIRM1 binding sites used to target miR-107 were generated. Luciferase plasmids containing wild-type (pmirGLO-TDG-WT) or mutated (pmirGLO-TDG-MUT) putative miR-107-binding sites from the 3′-UTR of TDG were also generated. All plasmids were obtained from Genepharma (Shanghai, China).
To detect binding between HOTAIRM1 and miR-107, pmirGLO-HOTAIRM1-WT or pmirGLO-HOTAIRM1-MUT were co-transfected with miR-107 mimics or miR-NC (Invitrogen) into HEK-293T cells using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Cells were harvested 48 h after transfection and luciferase activity was measured as chemiluminescence using a luminometer (PerkinElmer Life Sciences, Boston, MA, USA) and a dual-luciferase reporter assay system (Promega, Madison, WI, USA), according to the manufacturer’s protocol. The detection of binding between the 3′-UTR of TDG and miR-107 was performed in the same way.
RNA immunoprecipitation assay
RIP assays were performed, using B-CPAP and TPC-1 cell lines, to investigate the binding of miR-107 to HOTAIRM1. An Imprint RIP kit was used according to the manufacturer’s instructions (Sigma-Aldrich), with an anti-Ago2 antibody (Sigma-Aldrich). Total RNA was isolated using a GenElute™ Total RNA Purification Kit (Sigma-Aldrich) and the final analysis was performed using qRT-PCR, as described above.
Bioinformatics analyses
The prediction of candidate target microRNAs of HOTAIRM1 and the prediction of potential binding sites were performed using the online tool, miRcode31 (http://www.mircode.org/). Results were retrieved using the gene symbol, “HOTAIRM1.” The site conservation parameter was set as “most primates” and the other parameters were set as default.
The prediction of putative target genes for miR-107 was performed using the online tool, TargetScanHuman34 (http://www.targetscan.org/vert_71/). Results were retrieved using the microRNA name, ‘miR-107’ and the other parameters were set as default.
Statistical analysis
Statistical analyses were performed using SPSS statistics 22 software (IBM, Armonk, NY, USA) and GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA). All data are presented as the mean ± standard deviation and all in vitro experiments were performed in triplicate. The expression data for HOTAIRM1, miR-107, and TDG conformed to a normal distribution and the differences in expression levels between tumor and paired normal tissues were evaluated by a paired Student’s t-test. The analysis of correlation between lncRNA HOTAIRM1 expression and clinicopathological features of PTC patients was performed by Chi-square test. Pearson’s correlation analysis was performed to assess the correlation between HOTAIRM1/mir-107, mir-107/TDG, and HOTAIRM1/TDG. P-values less than 0.05 were considered statistically significant.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant number 81501505).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by E. Candi
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41419-024-06556-2
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Dan Li, Li Chai, Xiaqing Yu
Change history
2/21/2024
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41419-024-06556-2
Contributor Information
Lihong Fan, Email: fanlih@aliyun.com.
Zhongwei Lv, Email: lzwkxy@163.com.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-2416-1).
References
- 1.Kitahara CM, Sosa JA. The changing incidence of thyroid cancer. Nat. Rev. Endocrinol. 2016;12:646–653. doi: 10.1038/nrendo.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao X, Su L, He X, Zhao B, Miao J. Long noncoding RNA CA7-4 promotes autophagy and apoptosis via sponging MIR877-3P and MIR5680 in high glucose-induced vascular endothelial cells. Autophagy. 2019;16:70–85. doi: 10.1080/15548627.2019.1598750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang S, et al. A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy. 2015;11:2172–2183. doi: 10.1080/15548627.2015.1106663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez-Mejias A, Tay Y. Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J. Hematol. Oncol. 2015;8:30. doi: 10.1186/s13045-015-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lian Y, et al. Long noncoding RNA AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the Rho/Rac pathway. J. Exp. Clin. Cancer Res. 2018;37:253. doi: 10.1186/s13046-018-0918-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, et al. MEG3, as a competing endogenous RNA, binds with miR-27a to promote PHLPP2 protein translation and impairs bladder cancer invasion. Mol. Ther. Nucleic Acids. 2019;16:51–62. doi: 10.1016/j.omtn.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng M, Liu X, Zhou Q, Liu G. HOTAIRM1 competed endogenously with miR-148a to regulate DLGAP1 in head and neck tumor cells. Cancer Med. 2018;7:3143–3156. doi: 10.1002/cam4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun W, et al. NEAT1_2 functions as a competing endogenous RNA to regulate ATAD2 expression by sponging microRNA-106b-5p in papillary thyroid cancer. Cell Death Dis. 2018;9:380. doi: 10.1038/s41419-018-0418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu DM, et al. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis. 2018;9:947. doi: 10.1038/s41419-018-0975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng J, et al. A novel lncRNA n384546 promotes thyroid papillary cancer progression and metastasis by acting as a competing endogenous RNA of miR-145-5p to regulate AKT3. Cell Death Dis. 2019;10:433. doi: 10.1038/s41419-019-1637-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan L, et al. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J. Cell Mol. Med. 2016;20:2036–2044. doi: 10.1111/jcmm.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R, et al. Long noncoding RNA HOTAIRM1 inhibits cell progression by regulating miR-17-5p/ PTEN axis in gastric cancer. J. Cell Biochem. 2019;120:4952–4965. doi: 10.1002/jcb.27770. [DOI] [PubMed] [Google Scholar]
- 15.Luo Y, et al. High expression of long noncoding RNA HOTAIRM1 is associated with the proliferation and migration in pancreatic ductal adenocarcinoma. Pathol. Oncol. Res. 2019;25:1567–1577. doi: 10.1007/s12253-018-00570-4. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, et al. Microarray expression profile analysis of long non-coding RNAs in pancreatic ductal adenocarcinoma. Int. J. Oncol. 2016;48:670–680. doi: 10.3892/ijo.2015.3292. [DOI] [PubMed] [Google Scholar]
- 17.Su X, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5:9864–9876. doi: 10.18632/oncotarget.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian X, et al. Long non-coding RNA HOXA transcript antisense RNA myeloid-specific 1-HOXA1 axis downregulates the immunosuppressive activity of myeloid-derived suppressor cells in lung cancer. Front. Immunol. 2018;9:473. doi: 10.3389/fimmu.2018.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, Dong C, Cui J, Wang Y, Hong X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J. Exp. Clin. Cancer Res. 2018;37:265. doi: 10.1186/s13046-018-0941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen ZH, et al. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24:212–224. doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Beya M, et al. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget. 2015;6:31613–31627. doi: 10.18632/oncotarget.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahonen MA, et al. miR-107 inhibits CDK6 expression, differentiation, and lipid storage in human adipocytes. Mol. Cell Endocrinol. 2019;479:110–116. doi: 10.1016/j.mce.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Su PF, Song SQ. Regulation of mTOR by miR-107 to facilitate glioma cell apoptosis and to enhance cisplatin sensitivity. Eur. Rev. Med. Pharm. Sci. 2018;22:6864–6872. doi: 10.26355/eurrev_201810_16155. [DOI] [PubMed] [Google Scholar]
- 24.Foley NH, O’Neill L. A. miR-107: a toll-like receptor-regulated miRNA dysregulated in obesity and type II diabetes. J. Leukoc. Biol. 2012;92:521–527. doi: 10.1189/jlb.0312160. [DOI] [PubMed] [Google Scholar]
- 25.Xia H, Li Y, Lv X. MicroRNA-107 inhibits tumor growth and metastasis by targeting the BDNF-mediated PI3K/AKT pathway in human non-small lung cancer. Int. J. Oncol. 2016;49:1325–1333. doi: 10.3892/ijo.2016.3628. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Xiong J, et al. Deregulated expression of miR-107 inhibits metastasis of PDAC through inhibition PI3K/Akt signaling via caveolin-1 and PTEN. Exp. Cell Res. 2017;361:316–323. doi: 10.1016/j.yexcr.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 27.Inoue T, Iinuma H, Ogawa E, Inaba T, Fukushima R. Clinicopathological and prognostic significance of microRNA-107 and its relationship to DICER1 mRNA expression in gastric cancer. Oncol. Rep. 2012;27:1759–1764. doi: 10.3892/or.2012.1709. [DOI] [PubMed] [Google Scholar]
- 28.Hui AB, et al. Potentially prognostic miRNAs in HPV-associated oropharyngeal carcinoma. Clin. Cancer Res. 2013;19:2154–2162. doi: 10.1158/1078-0432.CCR-12-3572. [DOI] [PubMed] [Google Scholar]
- 29.Chen HY, et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72:3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 30.Kleivi Sahlberg K, et al. A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin. Cancer Res. 2015;21:1207–1214. doi: 10.1158/1078-0432.CCR-14-2011. [DOI] [PubMed] [Google Scholar]
- 31.Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li N, Zhan X. Identification of clinical trait-related lncRNA and mRNA biomarkers with weighted gene co-expression network analysis as useful tool for personalized medicine in ovarian cancer. EPMA J. 2019;10:273–290. doi: 10.1007/s13167-019-00175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, Y., Chen, B. & Huang, S. Identification of circRNAs for miRNA targets by argonaute2 RNA immunoprecipitation and luciferase screening assays. Methods Mol. Biol. 1724, 209–218 (2018). [DOI] [PubMed]
- 34.Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. eLife4, e05005 (2015). [DOI] [PMC free article] [PubMed]
- 35.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen L, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren T, et al. The long non-coding RNA HOTAIRM1 suppresses cell progression via sponging endogenous miR-17-5p/ B-cell translocation gene 3 (BTG3) axis in 5-fluorouracil resistant colorectal cancer cells. Biomed. Pharmacother. 2019;117:109171. doi: 10.1016/j.biopha.2019.109171. [DOI] [PubMed] [Google Scholar]
- 38.Zhen Y, et al. Knockdown of NEAT1 repressed the malignant progression of glioma through sponging miR-107 and inhibiting CDK14. J. Cell Physiol. 2018;234:10671–10679. doi: 10.1002/jcp.27727. [DOI] [PubMed] [Google Scholar]
- 39.Gao YW, et al. Sp1-induced upregulation of the long noncoding RNA TINCR inhibits cell migration and invasion by regulating miR-107/miR-1286 in lung adenocarcinoma. Am. J. Transl. Res. 2019;11:4761–4775. [PMC free article] [PubMed] [Google Scholar]
- 40.Li GC, Xin L, Wang YS, Chen Y. Long intervening noncoding 00467 RNA contributes to tumorigenesis by acting as a competing endogenous RNA against miR-107 in cervical cancer cells. Am. J. Pathol. 2019;189:2293–2310. doi: 10.1016/j.ajpath.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 41.Xu X, Watt DS, Liu C. Multifaceted roles for thymine DNA glycosylase in embryonic development and human carcinogenesis. Acta Biochim. Biophys. Sin. 2016;48:82–89. doi: 10.1093/abbs/gmv083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang L, Yu SJ, Hong Q, Yang Y, Shao ZM. Reduced expression of TET1, TET2, TET3 and TDG mRNAs are associated with poor prognosis of patients with early breast cancer. PLoS ONE. 2015;10:e0133896. doi: 10.1371/journal.pone.0133896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broderick P, et al. Evaluation of NTHL1, NEIL1, NEIL2, MPG, TDG, UNG and SMUG1 genes in familial colorectal cancer predisposition. BMC Cancer. 2006;6:243. doi: 10.1186/1471-2407-6-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruczinski I, et al. A population-based study of DNA repair gene variants in relation to non-melanoma skin cancer as a marker of a cancer-prone phenotype. Carcinogenesis. 2012;33:1692–1698. doi: 10.1093/carcin/bgs170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li WQ, et al. Genetic variants in DNA repair pathway genes and risk of esophageal squamous cell carcinoma and gastric adenocarcinoma in a Chinese population. Carcinogenesis. 2013;34:1536–1542. doi: 10.1093/carcin/bgt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang P, Huang B, Xu X, Sessa WC. Ten-eleven translocation (Tet) and thymine DNA glycosylase (TDG), components of the demethylation pathway, are direct targets of miRNA-29a. Biochem. Biophys. Res. Commun. 2013;437:368–373. doi: 10.1016/j.bbrc.2013.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, et al. miR-29b targets LPL and TDG genes and regulates apoptosis and triglyceride production in MECs. DNA Cell Biol. 2016;35:758–765. doi: 10.1089/dna.2016.3443. [DOI] [PubMed] [Google Scholar]
- 48.Fu X, et al. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc. Natl Acad. Sci. USA. 2013;110:17892–17897. doi: 10.1073/pnas.1317397110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.