Abstract
Microorganisms in soil are known to be a source and a sink of volatile organic compounds (VOCs). The role of the microbial VOCs on soil ecosystem regulation has been increasingly demonstrated in the recent years. Nevertheless, little is known about the influence of the microbial soil community structure and diversity on VOC emissions. This novel study analyzed the effect of reduced microbial diversity in soil on VOC emissions. We found that reduced levels of microbial diversity in soil increased VOC emissions from soils, while the number of different VOCs emitted decreased. Furthermore, we found that Proteobacteria, Bacteroidetes and fungi phyla were positively correlated to VOC emissions, and other prokaryotic phyla were either negatively correlated or very slightly positively correlated to VOCs emissions. Our interpretation is that Proteobacteria, Bacteroidetes and fungi were VOC producers while the other prokaryotic phyla were consumers. Finally, we discussed the possible role of VOCs as mediators of microbial interactions in soil.
Subject terms: Environmental chemistry, Environmental impact
Introduction
Recent studies have been focused on biogenic volatile organic compounds (bVOCs) released by aboveground biomass, because of their contribution to atmospheric chemistry1–3. Often, VOC fluxes by soil, and in particular by microorganisms, are not directly considered. A recent study showed that VOCs from vegetation could be overestimated compared to soil emissions by reporting comparable fluxes of methanol from bare soil and canopy in a maize crop4.
VOC emissions by soils may be boosted by organic amendment, due to soil organic matter (SOM) content and soil microbial biomass increase, compared to soil receiving mineral fertilization5. Among the different organic amendments, organic waste products (OWPs) resulting from human activities (i.e., sewage sludge, municipal solid waste composts, farmyard manure) are being increasingly used since they facilitate the recycling of nutrients and improve soil fertility. OWPs are also responsible for changing the microbial structure and increasing soil microbial diversity in soil6–10 due to the incorporation of exogenous microorganisms colonizing the organic amendment11. Microbial diversity in soil ensures several soil ecosystems functions such as (i) carbon (C) balance between organic C sequestration and C mineralization12, (ii) organic matter break down13, and more generally, (iii) nutrient recycling. Microorganisms are also directly involved in the production of VOCs emitted by soil14. Moreover, VOCs play a pivotal role in several processes such as the communication between organisms15, alterations of microbial nutrient cycles16 and growth rate of microorganisms colonizing the ecosystem17. Recently, it has been demonstrated that several microbial VOCs are involved in plant seed germination,plants growth18 and plant pathogens suppression19. The microbial community is not only a source of VOCs, but also a sink that microorganisms use as a source of energy20. Thanks to this characteristic, microorganisms are ideal to be used for biofiltration21,22. Biofiltration mechanisms consist in maintaining a mixed microbial community into biofilters in order to absorb and break down multiple VOCs23. The biofilters performance is particularly affected by the structure of the microbial community, the diversity and the interaction between microorganisms24, showing that VOC absorption depends the microbial community structure. VOC emissions can also depend on the taxa25,26 and thus, on the structure of the microbial community in soil. To our knowledge, no studies have reported the measurement of VOC emissions in response to soil microbial diversity loss and organic waste amendment in the soil. Hence, the effect of microbial diversity loss on VOC emissions is still unknown. Since microorganisms play a central role in soil VOC emissions, we hypothesized that the manipulation of the microbial community in soil might lead to some variations in the quantity and quality of VOC emissions.
The manipulation of the microbial diversity was performed by inoculating 30 g of sterile soils amended with 4 different organic waste products (OWPs): municipal solid waste compost derived from the composting of residual solid wastes after removing dry and clean packaging (MWS), farmyard manure (FYM), a compost derived from the co-composting of green wastes with sewage sludge (GWS) and the bio-waste compost derived from the co-composting of green wastes and source-separated organic fractions of municipal solid wastes (BIOW). All the samples were compared to a control (CN) with no OWPs. The inoculation for each soil was performed with three dilutions of a soil suspension taken from the same soil sampled before sterilization. After six weeks of incubation at 20 °C, VOC emissions were measured with a dynamic chamber method using a highly sensitive proton transfer, quadrupole injection, time of flight mass spectrometer (PTR-Qi-TOF-MS, Ionicon). The VOC concentrations were measured every second for 3 min for each sample (Samples = 45, replicates = 3). The mass spectra were analyzed using PTR-MS viewer-3 software in order to integrate each ion peak. The microbial diversity was measured after VOC emission measurements by a high throughput sequencing approach targeting 16S and 18S ribosomal genes.
Results
Soil microbial biomass
After six weeks of incubation, the microbial biomass was not significantly different among microbial dilution levels for each type of amended treatment except for FYM (soil amended with farmyard manure), where the highest dilution level had the highest microbial biomass (Table 1). A normalization of the VOC emissions by the microbial biomass levels showed no significant difference in the variation of VOC emissions with dilution compared to non-normalized data. We could attribute the variation of VOC emissions measured in the different dilution treatments to microbial diversity and not to microbial biomass quantity. Whatever the dilution levels considered, the microbial biomass was statistically higher in MWS treatment, FYM and GWS than in BIOW and CN.
Table 1.
Effect of different microbial diversity dilution on soil prokaryotic and fungal diversity indices (Shannon and Richness) and density of the genes copies of fungi and prokatyiotes (18S and 16S genes copies).
| Organic Waste Product | Level of diversity | Biomass µg DNA g sol−1 ± sd | Indices of diversity | Density | F/P ratio | ||||
|---|---|---|---|---|---|---|---|---|---|
| Shannon (Prokaryotes) | Richness (Prokaryotes) | Shannon (Fungi) | Richness (Fungi) | 16S gene copies per g of dry soil × 109 ± sd × 109 | 18S gene copies per g of dry soil × 108 ± sd × 108 | ||||
| Control without organic input | D0 | 13.52 ± 2.41 a | 4.44 ± 0.45a | 1308 ± 247.67a | 3.30 ± 0.16a | 553.00 ± 111.53a | 3.59 ± 1.56a | 2.34 ± 1.31a | 0,065 |
| D1 | 12.48 ± 2.09 a | 4.06 ± 0.41ab | 1083.67 ± 189.74ab | 3.16 ± 0.51a | 506.67 ± 152.75a | 3.54 ± 0.641a | 1.45 ± 0.332ab | 0,041 | |
| D2 | 12.69 ± 2.30 a | 3.47 ± 0.16b | 876.67 ± 88.29b | 2.95 ± 0.84a | 708.67 ± 207.61a | 4.24 ± 1.73a | 0.346 ± 0.281b | 0,008 | |
| Biowaste | D0 | 11.24 ± 1.07 a | 4.16 ± 1.31a | 1751 ± 450.85a | 3.27 ± 0.31b | 533.33 ± 77.02b | 2.97 ± 0.548a | 2.49 ± 0.695a | 0,084 |
| D1 | 15.83 ± 1.33 a | 4.03 ± 0.35a | 1346 ± 224.31ab | 2.93 ± 0.42b | 467.33 ± 93.50b | 5.96 ± 0.148a | 1.67 ± 0.929ab | 0,028 | |
| D2 | 17.38 ± 2.77 a | 3.5 ± 0.30a | 931 ± 3.5b | 4.62 ± 0.26a | 1184.33 ± 106.33a | 6.87 ± 2.40a | 0.129 ± 0.0548b | 0,002 | |
| Farmyard manure | D0 | 16.25 ± 4.35 a | 4.50 ± 0.13a | 1462.67 ± 49.56a | 3.83 ± 0.22a | 746.33 ± 82.31a | 3.78 ± 1.30b | 1.93 ± 0.172b | 0,051 |
| D1 | 21.22 ± 2.20 ab | 4.11 ± 0.19a | 1049.67 ± 97.03b | 3.03 ± 0.14b | 423.00 ± 40.01b | 5.99 ± 1.67b | 3.75 ± 0.294a | 0,063 | |
| D2 | 28.52 ± 3.48 b | 3.24 ± 0.15b | 718.67 ± 31.5c | 4.58 ± 0.25a | 1018.67 ± 173.37a | 13.90 ± 2.93a | 0.187 ± 0.0476c | 0,001 | |
| Green waste and sludge | D0 | 18.30 ± 1.18 a | 4.35 ± 0.10a | 1442 ± 90.7a | 2.93 ± 0.20ab | 508.67 ± 28.75ab | 5.02 ± 1.19b | 6.51 ± 1.94a | 0,129 |
| D1 | 20.26 ± 4.78 a | 4.08 ± 0.12a | 1153.67 ± 91.36ab | 2.85 ± 0.22b | 400.00 ± 53.07b | 5.60 ± 0.746b | 2.69 ± 0.812b | 0,048 | |
| D2 | 21.99 ± 3.77 a | 3.42 ± 0.05b | 961.33 ± 156.67b | 4.67 ± 0.66a | 1123.67 ± 325.92a | 10.50 ± 2.23a | 0.140 ± 0.0186c | 0,001 | |
| Municipal solid waste | D0 | 22.31 ± 2.12 a | 4.56 ± 0.13a | 1579.00 ± 131.19a | 2.86 ± 0.34a | 514.67 ± 112.21a | 6.70 ± 0.895b | 7.92 ± 2.44a | 0,118 |
| D1 | 21.52 ± 1.19 a | 4.00 ± 0.13b | 1114.00 ± 67.02b | 2.68 ± 0.37a | 380.67 ± 96.84a | 6.93 ± 0.735b | 3.70 ± 1.57a | 0,053 | |
| D2 | 23.97 ± 1.39 a | 3.39 ± 0.24c | 798.33 ± 39.80c | 3.02 ± 1.90a | 729.33 ± 379.96a | 11.00 ± 2.06a | 2.20 ± 3.44a | 0,02 | |
Letters indicate significant differences according to the Tukey test with p.value > 0.05. F/P ratio represent the fungi-to-prokaryotic ratio.
Microbial diversity
The Shannon index is a measure of microbial diversity and it has been calculated for both fungi and prokaryotic community in soil. The prokaryotic Shannon index shows a significant decrease of prokaryotic diversity with increasing dilution (p.value < 0.05) in our samples (Fig. 1). These results confirm the efficiency of the prokaryotic diversity manipulation. The dilution of the prokaryotic diversity was also efficient for the FYM samples, even though a larger biomass was found for the highest dilution (D2, Table 1). For fungi, the Shannon index revealed a decrease of fungal diversity in the higher dilution level (Fig. 1). In our microcosms, the fungi-to-prokaryotes (F/P) ratio was smaller (between 0.05 and 0.20) compared to the F/P ratio previously observed in similar studies27 where it was always higher than 0.20. Even if the portion of fungi in our microcosms seems to be negligible compared to prokaryotes, if we consider the density of gene copies of 18S, which represent the density of fungi, we notice that their density is between 3.75 × 108 and 1.4 × 107 (Table 1). This density might be consistent in all soils and might influence the VOC emissions, and the reason why we did not ignore this part of the microcosms.
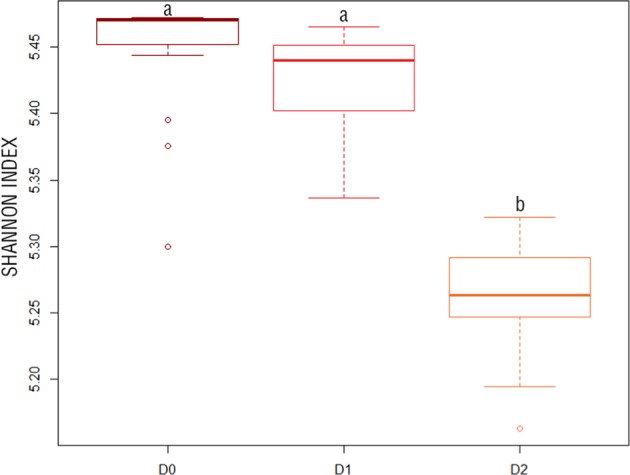
Figure 1.
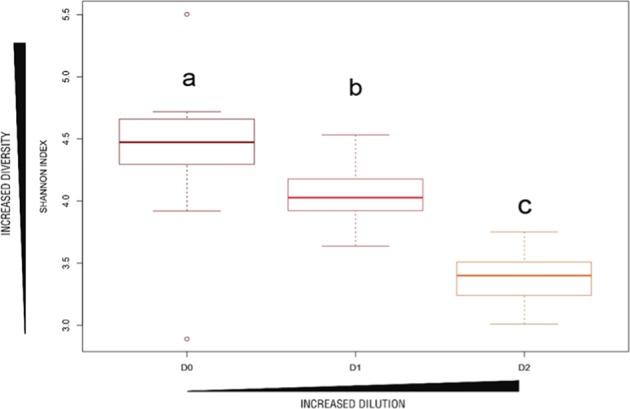
Shannon index for prokaryotes in the soil as a function of dilution levels. D0: no dilution of the microbial diversity. D1: microbial diversity dilution equal to 10−3. D2: microbial diversity dilution equal to 10−5. Bold line = median, boxes = interquartile, whiskers = minimum and maximum. Point = Shannon Index of each sample. Letters indicate significant differences according to the Tukey test with p-value < 0.05.
A decrease of F/P ratio throughout the diversity gradient was also observed (Table 1) suggesting that while the microbial dilution was higher, the fungal community was disadvantaged compared to prokaryotic community.
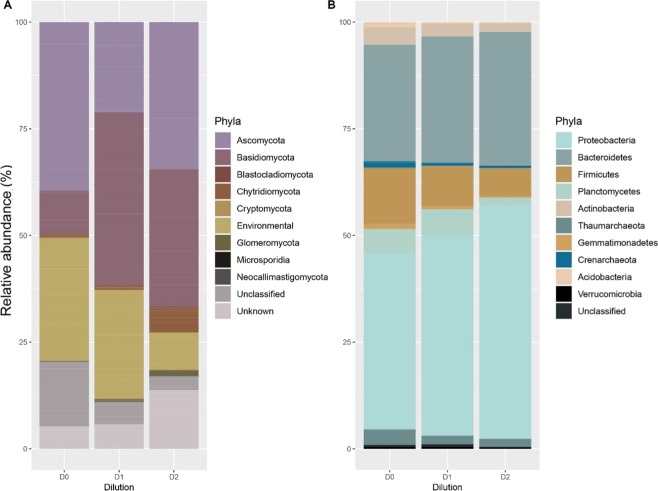
We observed the opposite pattern for fungi: Basidiomycota, Chytridiomycota, Glomeromycota, and Neoclimastigomycota were more abundant in the highest dilution levels (D2 samples) than in the lowest ones (D0 samples) although they were not the most abundant fungi in D0 (Fig. 2b). The opposite trend was observed for Ascomycota and Blastoclamidiomycota, which were the most abundant in D0.
Figure 2.
Relative abundances of the phyla. (a) Relative abundances of the prokaryotic phyla. (b) Relative abundances of the fungal phyla. D0: no dilution equal of the microbial diversity. D1: microbial diversity dilution equal to 10−3. D2: microbial diversity dilution equal to 10−5.
As expected, the diversity manipulation tended to strengthen the presence of the most abundant prokaryotic phyla and reduce the presence of the less abundant ones. This was indeed observed with Proteobacteria and Bacteroides phyla which were the most abundant in the lowest dilution samples and showed an increased abundance in the highest dilution samples. Firmicutes, Crenarchaeota, Actinobacteria, Thaumarchaeota, Planctomycetes, Gemmatimonadetes, and Acidobacteria showed the opposite behavior (Fig. 2a).
VOC emissions from manipulated soils
We detected 754 ion peaks, spanning m/z 15 to m/z 510. VOC emissions summed among all detected ion peaks increased while the microbial diversity in the soil was lower (Fig. 3).
Figure 3.
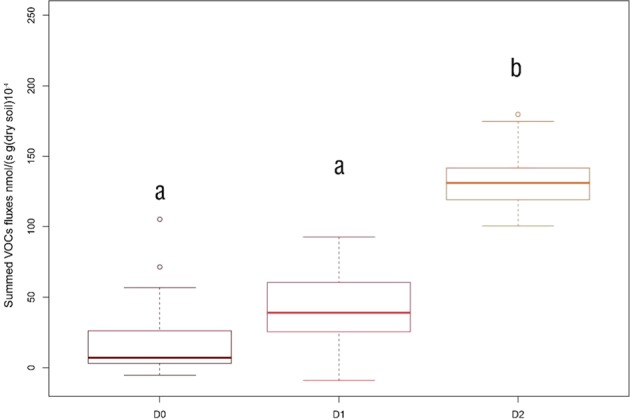
Summed VOCs fluxes rate as a function of microbial dilution in soils. Bold line = median, boxes = interquartile, whiskers = minimum and maximum, colored circles = outliers, grey dots = individual values. Letters indicate significant differences according to the Tukey test with p.value < 0.05. D0: no dilution equal of the microbial diversity. D1: microbial diversity dilution equal to 10−3. D2: microbial diversity dilution equal to 10−5.
The lower microbial diversity level showed a summed VOC flux between 0.5 and 3 times higher than the flux at the higher microbial diversity levels. This was true for all the considered soils. This means that, in our experiment, the effect of microbial dilution in driving soil VOC emissions overpassed the effect of OWPs amendment (Figs. S1 and S2, Tukey test on OWPs). The diversity in VOCs (Shannon index on VOCs emissions) was lowest in the lowest microbial diversity level, denoting that the larger summed emission rates were combined with a lower number of VOCs emitted (Fig. 4).
Figure 4.
Shannon index for VOCs emissions in soil. D0 = microbial diversity pure or 100, D1 = microbial dilution diversity equal to 10−3, D2 = microbial dilution diversity equal to 10−5. Letters indicate significant differences according to the Tukey test with p.value < 0.05.
The diversity of the VOCs emitted (Table 1) also showed that the microbial dilution levels D0 and D1 had similar VOC profiles while the lowest microbial diversity level (D2) had a different profile (Fig. 5). Several VOC compounds explain the differences in VOC profiles between microbial dilutions. Tentative identification of the compounds explaining the variance in Fig. 5 are reported in Table 2, together with the 50 most emitted compounds in all dilution levels. These 65 compounds contribute almost 99% of the total emissions rate. The most emitted compounds were m/z 121.097 (tentatively identified as Propylbenzene, isopropylbenzene or 1,3,5-trimethylbenzene, Phenilacetaldheyde), m/z 135.113 (p-cymene) and m/z 73.062 (Butanone, MEK). Together, these compounds represented 70% of the summed VOCs emission rate (Table S2). Acetaldehyde, butanone, and acetoin were emitted between 10 and 20 times more in samples with the highest dilution (D2) compared to samples with the lowest dilution (D0).
Figure 5.
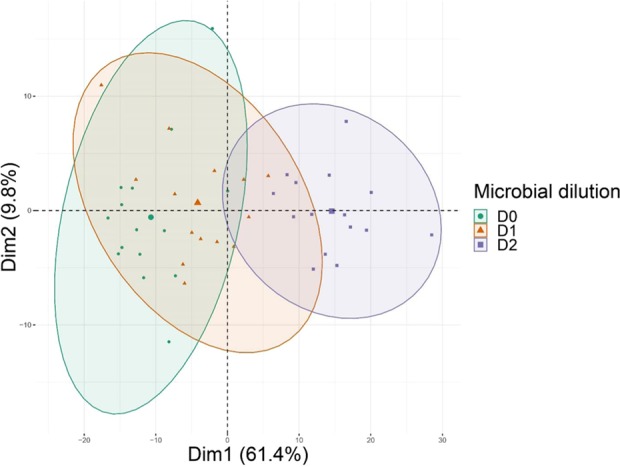
Effect of the different microbial dilutions on VOCs emissions by soil. The m/z of the 20 compounds that are explaining the variance in the two first components are shown on the graph. The percentage of the variance explained by the 2 first components is shown on each axis. The ellipses represent the similarity between samples. Samples in the same ellipsis are more similar than samples displayed in two different ellipses. D0: no dilution equal of the microbial diversity. D1: microbial diversity dilution equal to 10−3. D2: microbial diversity dilution equal to 10−5.
Table 2.
Most emitted compounds for the different microbial dilution levels D0, D1 and D2 as well as compounds explaining most the variance in VOC emissions between dilution levels and those known as fungistatic VOCs.
| m/z | Most likely formula | Tentative identification | Compounds class | Fungistatic compounds |
Compounds most explaining the variance (PCA analysis) | Mean emission ± sd (nmol s−1 g−1(DW)) × 104 |
Mean emission rates ± sd (nmol s−1 g−1(DW)) × 104 |
Percentage contribution (% mol) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D0 | D1 | D2 | D0 | D1 | D2 | |||||||
| m121,097 | C9H12 |
Propylbenzene cumene mesytilene 1,3,5-trimethylbenzene, Phenilacetaldheyde |
Aromatic | Phenilac-etaldheyde66 | 7,83 ± 13,04 | 14,34 ± 11,04 | 49,59 ± 8,2 | 23,92 ± 17,11 | 52,37 | 49,41 | 54,95 | |
| m135,113 | C10H14 | p-cymene | Aromatic | x | 2,04 ± 2,77 | 4,39 ± 2,44 | 10,45 ± 2,35 | 5,63 ± 3,22 | 13,65 | 15,13 | 11,58 | |
| m73.062 | C4H8O | Butanone, MEK | Carbonyl- Ketone | 0,82 ± 1,02 | 0,96 ± 1,49 | 2,49 ± 1,36 | 1,42 ± 0,71 | 5,49 | 3,32 | 2,75 | ||
| m122.100 | C8H11N | Benzenamine/2,6-Xylidine | Nitrogen compounds | 0,75 ± 1,31 | 1,37 ± 1,12 | 5,23 ± 1,09 | 2,45 ± 1,85 | 5,04 | 4,72 | 5,79 | ||
| m57.068 | C4H8 | Butene | Hydrocarbon - Alkene | 0,51 ± 0,77 | 2 ± 0,81 | 3,11 ± 0,55 | 1,87 ± 0,91 | 3,43 | 6,89 | 3,44 | ||
| m41.037 | C3H4 | Propyne | Hydrocarbon - Alkyine | 0,35 ± 0,37 | 0,94 ± 0,55 | 2,19 ± 0,47 | 1,16 ± 0,69 | 2,32 | 3,25 | 2,43 | ||
| m119.078 | C8H6O | Oxygenated compound | 0,31 ± 0,48 | 0,61 ± 0,42 | 1,88 ± 0,39 | 0,93 ± 0,63 | 2,07 | 2,09 | 2,08 | |||
| m120.090 | C8H9N | Nitrogen compounds | 0,29 ± 0,53 | 0,52 ± 0,45 | 2,03 ± 0,53 | 0,95 ± 0,72 | 1,94 | 1,81 | 2,25 | |||
| m105.066 | C5H12S | Butane 1-methylthio, styrene | Hydrocarbon - Alkane | 0,26 ± 0,43 | 0,47 ± 0,35 | 1,66 ± 0,34 | 0,8 ± 0,57 | 1,71 | 1,63 | 1,84 | ||
| m136.021 | C7H5NS | Benzothiazole | Aromatic heterocyclic compound | x66 | 0,23 ± 0,07 | 0,3 ± 0,15 | 0,4 ± 0,12 | 0,31 ± 0,06 | 1,52 | 1,05 | 0,44 | |
| m91.072 | C4H10S | Diethyl sulphide | Organosulfur | x66 | 0,19 ± 0,31 | 0,33 ± 0,27 | 0,88 ± 0,25 | 0,47 ± 0,28 | 1,26 | 1,14 | 0,97 | |
| m136.118 | C10H15 | Hydrocarbon | x | 0,16 ± 0,23 | 0,35 ± 0,2 | 0,87 ± 0,21 | 0,46 ± 0,27 | 1,08 | 1,20 | 0,97 | ||
| m94.070 | 0,14 ± 0,21 | 0,27 ± 0,18 | 0,89 ± 0,17 | 0,43 ± 0,3 | 0,93 | 0,93 | 0,98 | |||||
| m79.071 | C6H6 | Benzene | Aromatic | x | 0,12 ± 0,16 | 0,27 ± 0,18 | 0,9 ± 0,15 | 0,43 ± 0,31 | 0,83 | 0,93 | 1,00 | |
| m108.084 | C7H9N | Dimethyl pyridine | Nitrogen compounds | 0,1 ± 0,15 | 0,18 ± 0,12 | 0,66 ± 0,14 | 0,31 ± 0,23 | 0,66 | 0,61 | 0,73 | ||
| m33.033 | CH4O | Methanol | Alcohol | 0,06 ± 0,04 | 0,08 ± 0,05 | 0,18 ± 0,12 | 0,1 ± 0,05 | 0,37 | 0,29 | 0,19 | ||
| m134.108 | C9H11N | Amine | 0,05 ± 0,08 | 0,12 ± 0,07 | 0,28 ± 0,08 | 0,15 ± 0,08 | 0,37 | 0,41 | 0,31 | |||
| m55.051 | C4H6 | 1,3 butadiene | Hydrocarbon - Diene | x | 0,05 ± 0,07 | 0,08 ± 0,13 | 0,27 ± 0,14 | 0,13 ± 0,09 | 0,35 | 0,27 | 0,30 | |
| m106.073 | C8H9 | Hydrocarbon | 0,05 ± 0,08 | 0,08 ± 0,06 | 0,32 ± 0,07 | 0,15 ± 0,11 | 0,31 | 0,28 | 0,36 | |||
| m133.108 | C10H13/C5H12N2O2 | Hydrocarbon/Amine | 0,04 ± 0,05 | 0,09 ± 0,04 | 0,17 ± 0,03 | 0,1 ± 0,05 | 0,30 | 0,30 | 0,19 | |||
| m122.064 | C7H8NO | Amide | 0,04 ± 0,06 | 0,08 ± 0,05 | 0,14 ± 0,02 | 0,08 ± 0,03 | 0,25 | 0,27 | 0,15 | |||
| m95.052 | C6H6O | Phenol | Oxygenated Aromatic | x | 0,03 ± 0,04 | 0,06 ± 0,04 | 0,2 ± 0,03 | 0,1 ± 0,07 | 0,23 | 0,22 | 0,22 | |
| m74.065 | C3H7NO | Dimethylformamide/Propanamide | Amide | x | 0,03 ± 0,04 | 0,04 ± 0,06 | 0,1 ± 0,05 | 0,05 ± 0,03 | 0,21 | 0,13 | 0,11 | |
| m95.043 | C2H6S2 | Dimethyl disulfide | Organosulfur | x66,67 | 0,03 ± 0,04 | 0,06 ± 0,04 | 0,18 ± 0,03 | 0,09 ± 0,06 | 0,21 | 0,21 | 0,20 | |
| m123.073 | C8H11O | Oxygenated compound | 0,03 ± 0,04 | 0,05 ± 0,04 | 0,17 ± 0,02 | 0,08 ± 0,06 | 0,20 | 0,18 | 0,18 | |||
| m45.033 | C2H4O | Acetaldehyde | Carbonyl - Aldheyde | 0,03 ± 0,29 | 0,06 ± 0,47 | 0,42 ± 0,45 | 0,17 ± 0,17 | 0,19 | 0,19 | 0,46 | ||
| m122.062 | C8H9O | Acetophenone | Aromatic ketone | 0,03 ± 0,04 | 0,06 ± 0,03 | 0,1 ± 0,02 | 0,06 ± 0,02 | 0,17 | 0,20 | 0,11 | ||
| m92.056 | C6H5N | Heterocyclic compound | 0,03 ± 0,04 | 0,05 ± 0,04 | 0,14 ± 0,03 | 0,07 ± 0,04 | 0,17 | 0,19 | 0,15 | |||
| m58.071 | C4H9 | Hydrocarbon | 0,02 ± 0,03 | 0,08 ± 0,03 | 0,12 ± 0,02 | 0,07 ± 0,03 | 0,14 | 0,27 | 0,13 | |||
| m83.047 | C5H7O | Oxygenated compound | 0,02 ± 0,01 | 0,03 ± 0,02 | 0,17 ± 0,12 | 0,07 ± 0,07 | 0,12 | 0,10 | 0,19 | |||
| m158.162 | C9H19NO | Amide | 0,02 ± 0,02 | 0,02 ± 0,02 | 0,02 ± 0,02 | 0,02 ± 0 | 0,12 | 0,07 | 0,02 | |||
| m109.099 | C8H6O | Benzofuran | Oxygenated Aromatic | 0,01 ± 0,01 | 0,02 ± 0,01 | 0,05 ± 0,01 | 0,03 ± 0,01 | 0,10 | 0,07 | 0,05 | ||
| m149.130 | C6H12O4/C10H12O | Oxygenated compound | 0,01 ± 0,01 | 0,03 ± 0,01 | 0,05 ± 0,01 | 0,03 ± 0,01 | 0,09 | 0,10 | 0,05 | |||
| m95.083 | C5H6N2 | Methyl pyrazine | Hydrocarbon | x66 | 0,01 ± 0,01 | 0,02 ± 0,01 | 0,06 ± 0,01 | 0,03 ± 0,02 | 0,09 | 0,07 | 0,06 | |
| m111.041 | C7H10O | 2,3,5-trimethyl- Furan | Heterocyclic compound | 0,01 ± 0,01 | 0,02 ± 0,01 | 0,02 ± 0,01 | 0,02 ± 0 | 0,09 | 0,06 | 0,02 | ||
| m95.028 | C2H6O2S | Organosulfur | 0,01 ± 0,01 | 0,03 ± 0,02 | 0,1 ± 0,02 | 0,05 ± 0,03 | 0,09 | 0,10 | 0,11 | |||
| m108.953 | 0,01 ± 0 | 0,01 ± 0 | 0,01 ± 0 | 0,01 ± 0 | 0,08 | 0,04 | 0,01 | |||||
| m117.084 | C6H12O2 | Ethyl Butyrate | Oxygenated compound | 0,01 ± 0,01 | 0,02 ± 0,01 | 0,06 ± 0,01 | 0,03 ± 0,02 | 0,08 | 0,08 | 0,07 | ||
| m61.049 | C2H4O2 | Acetic acid | Carboxylic acid | 0,01 ± 0,01 | 0,01 ± 0,02 | 0,05 ± 0,05 | 0,02 ± 0,02 | 0,08 | 0,04 | 0,05 | ||
| m80.048 | 0,01 ± 0,02 | 0,03 ± 0,02 | 0,09 ± 0,02 | 0,04 ± 0,03 | 0,08 | 0,09 | 0,09 | |||||
| m105.031 | C7H4O | Heterocyclic compound | 0,01 ± 0,02 | 0,02 ± 0,01 | 0,06 ± 0,01 | 0,03 ± 0,02 | 0,08 | 0,08 | 0,07 | |||
| m53.037 | C4H4 | Hydrocarbons | 0,01 ± 0,02 | 0,03 ± 0,02 | 0,1 ± 0,02 | 0,04 ± 0,03 | 0,08 | 0,09 | 0,11 | |||
| m89.056 | C4H8O2 | Ethyl acetate/Butanoic acid/Acetoin | Acetate ester/Carboxylic acid | 0,01 ± 0,02 | 0,08 ± 0,07 | 1,96 ± 1,68 | 0,68 ± 0,85 | 0,06 | 0,26 | 2,17 | ||
| m118.089 | C5H11NO2 | Valine | Amino acid | 0,01 ± 0,01 | 0,02 ± 0,01 | 0,05 ± 0,01 | 0,03 ± 0,02 | 0,06 | 0,06 | 0,06 | ||
The correlation between the summed VOC emissions from soil and the prokaryotic diversity, richness and the F/P ratio showed a significant negative correlation (Fig. 6) with a Spearman coefficient always lower than −0.71. Non-significant correlations were found for all other combinations (VOC diversity and fungal richness, fungal diversity and VOC summed emissions, and fungal richness and fungal diversity, Fig. S3. The variance partitioning showed that Prokaryotic and fungal richness explained 33.35% of the variability of summed VOC emission rates (adjusted R2; P < 0.01).
Figure 6.
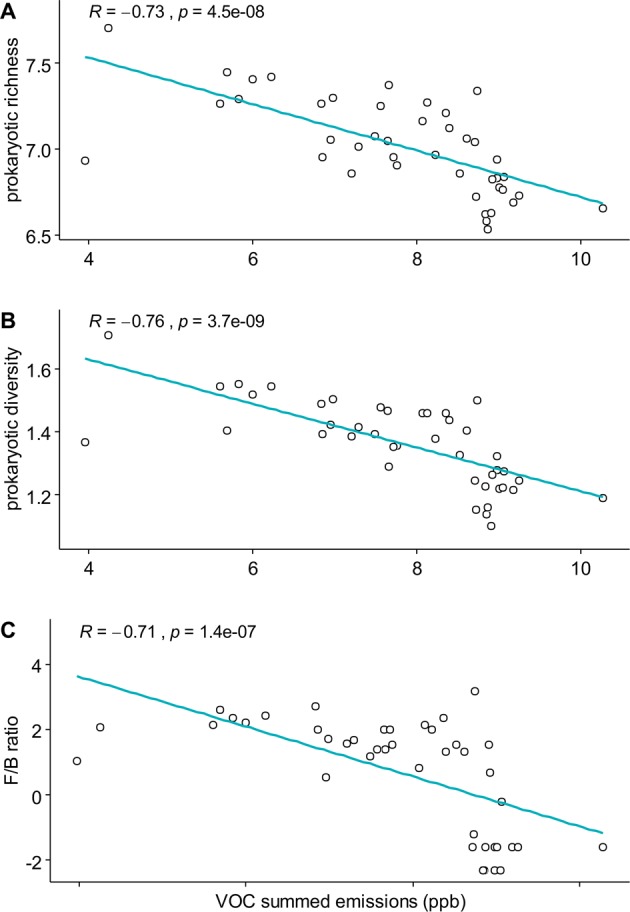
Correlation between log (VOC summed emissions) and (a) log (prokaryotic richness), (b) log (prokaryotic diversity), (c) log (F/P ratio). The Spearman correlation coefficient (R) and the p.value (p) are also displayed for all correlations.
Finally, we found that individual VOC emission rates were mostly negatively correlated with prokaryotic phyla abundance, except for Bacteroidetes and Proteobacteria whose abundances were positively correlated with almost all VOCs (Fig. 7). An opposite behavior was observed for the fungal phyla, where most had positive correlations with VOCs except for Cryptomicota.
Figure 7.
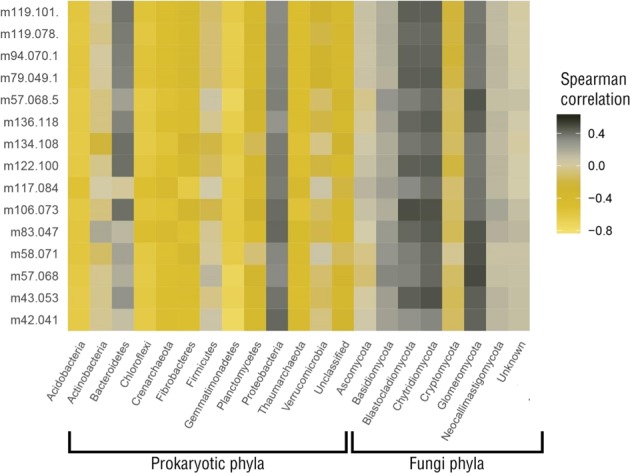
Correlation coefficients between VOCs emission rates and phyla abundance. Pearson correlation between log (emission rate + c) and log (phyla abundance + 1). The constant c was tuned to get always positive values. VOCs are named as ion mass to charge ratios (m/z). VOCs for which at least one correlation coefficient was larger than 0.6 with phyla are displayed.
Discussion
Microbial diversity
Tukey tests shows that the decrease of prokaryotic diversity index (Shannon index) was effective in all D2 samples compared to D0 (Fig. 1). This decrease can be explained as the result of a competition between slow-growing organisms such as Acidobacteria, Actinobacteria, Planctomycetes, and Gemmatimonadetes28, and fast-growing organisms, such as Proteobacteria, Bacteroidetes and Firmicutes28. Indeed, in reduced competition conditions29, fast-growing organisms (pioneer species) can colonize microcosms faster than others (slow-growing ones). The larger biomass observed in the highest dilution level (D2) in the FYM microcosms may be attributed to specific conditions of that soil treatment promoting even more the pioneering species.
Microbial diversity affects VOCs total emission rates
Although significantly higher VOC emission rates from BIOW treatments have been reported in one of our previous studies30, in this study we noticed that the effect of the substrate on VOC emissions was negligible compared to the effect of the prokaryotic diversity (Figs. S1–S2). In our previous study30 we used the same initial soil samples without performing the microbial diversity manipulation and it was found that the most emitted compounds were acetone, butanone, and acetaldehyde. In this study we found that the masses m/z 121.097 (tentatively identified as propylbenzene, isopropylbenzene or 1,3,5-trimethylbenzene), m/z 135.113 (p-cymene) and m/z 73.062 (Butanone, MEK) were the most emitted ones. Acetone was not in the list of the 50 most emitted compounds which means that its contribution to the summed VOC emissions was lower than 0.01%. Acetaldehyde and butanone were emitted 100 and 10 times less, respectively, compared to our preview study30. The reason why the most emitted compound were this different it could be due to the different sample treatment. The samples used in our first study30 were not sterilized and thus the characteristics of the soils played a stronger role in the VOC emissions. For instance, acetone is one of the most emitted compound by agricultural soils31, especially during the degradation of plant material32, in this study all the plant residues have been removed. Because of these results, we can hypothesize that microbial diversity in soil has a greater impact than OWP amendment or the structure and composition of the soils on even the most highly emitted VOCs. Differences in VOC emissions, however, may also be due to different sample treatment between two studies.
The two most emitted compounds reported in this study are aromatic compounds. Aromatic compounds are released from the shikimate pathway leading to the production of aromatic aminoacids33,34. In particular, this pathway is used by microorganisms in order to produce amino acids like phenylalanine, tyrosine, and tryptophan which are used to build proteins.
It has been shown by several studies that microorganisms are a source and a sink of VOCs35,36. For instance, in vitro soil incubation studies reported that when the concentration of VOCs increased the respiration rate was higher, suggesting that VOCs were used as a nutrient source for some bacteria20. In our study we counted more than 250 VOCs having a detectable flux and we found that VOC emissions rates increased with decreasing bacterial diversity.
We further show that VOC emissions were positively correlated with only two prokaryotic phyla (Proteobacteria and Bacteriodetes), which increased in abundance with the decreasing of the diversity. These data can be interpreted in two non-exclusive ways: (1) either increased VOC emissions are due to the two phyla that are more present than the others, or (2) decreased VOC absorption by the other prokaryotes, which are more abundant. The two interpretations lead to the observed correlation pattern. If the first interpretation is correct, it could be explained by an increased VOCs release by secondary metabolisms sources37 which is triggered with the colonization of the space within the microcosms38. The second interpretation may be explained by an increased colonization rate of Proteobacteria and Bacteroidetes which were boosted by the higher concentrations of VOCs in the higher dilution levels where absorbing bacteria take up fewer VOCs39.
Studies on biofilters reported that fungi biofilters have a higher efficiency in removing VOCs than bacterial filters40. Our results show that fungi that are most present in the most diverse environment are indeed negatively correlated with VOC emissions suggesting they are good VOCs absorbers. These fungi are however not clearly identified in our study as they belong to the “environmental and unclassified” phyla. Other studies have reported that mix of bacteria were however found to be more efficient for absorbing and converting specifics VOCs such as: methanol23 and styrene41. Our study tends to demonstrate that bacterial diversity decreases emissions, meaning an increased absorption by most of the bacteria phyla, which is in line with these studies. In view of all these we can hypothesize that a complex microbial structure with a high diversity level might increase the efficiency on converting VOCs also in soil
Interactions between prokaryotic and fungi diversity
The decrease in density for some fungi phyla and the lower fungi diversity observed at the highest dilution level (D2) might also be the consequence of the competition between fungi and prokaryotes. Probably, fungi have been negatively impacted by the pioneer prokaryotic species. Indeed, prokaryotes secondary metabolisms produced fungistatic VOCs limiting the colonization of the microcosms42,43. More specifically, some VOCs were only found in strongly fungistatic soils, and others VOCs had higher concentrations in fungistatic soils44. The prokaryotes which emit fungistatic volatiles span a wide phylogenetic spectrum44. It has been also reported that several prokaryotes from the Firmicutes phylum inhibited the growth of common fungal species45. Furthermore, Phenilacetaldheyde, Benzothaziole, Diehtyl sulfide, Dimethyl disulfide and methyl pyrazine, produced by pseudomonas sp (Proteobacteria phylum), were one of the inhibitors of the fungi mycelia growth and of the spore germination46,47. Our results reported a higher relative abundance of Proteobacteria in the lowest dilution level (D0) compared to D2 (Fig. 2) and we found that all the fungistatic compounds were emitted around 2 and 10 times more in D2 microcosms than in the D0 (Table 2). Finally, our hypothesis is that the increase of fungistatic VOCs might be related with the increase of the Proteobacteria phylum in the highest dilution level (D2).
Prokaryotic and fungi VOCs emissions profiles
As mentioned above, prokaryotes and fungi are both capable of producing and consuming VOCs. The heatmap (Fig. 7) shows that some phyla are mostly positively correlated with VOC emissions while others are mostly negatively correlated with VOC emissions. A previous study reported that the attribution of a unique VOC emission pattern to a phylum was not possible48. For this reason, the aim of this study was not to find common intraspecific VOC patterns. However, it is interesting to underline that data showing increased VOC emission positively correlated with microorganism abundance was observed much less than high VOC emissions associated with low microbial abundance. Other studies found, as in the present study, positive and negative correlations between microorganisms and specific VOCs, and hypothesized two possible VOC-prokaryote interactions for both the positive and negative correlations49.
A positive correlation may result from the emission of a given VOC by a given microorganism or from the stimulated growth of this microorganism when that VOC concentration increases. For instance, this correlation could result from the VOCs linked to the quorum sensing like the molecule dimethyl disulfide38. Quorum sensing is a system used by microorganisms for intra-specific communication. Quorum sensing microorganisms release specific VOCs as a signal to monitor the cell density50. The concentration of these VOCs increases as a function of microorganism growth. In this study, the dimethyl disulfide was positively correlated with the Proteobacteria and Bacteroidetes phyla and was higher when the colonization of those phyla was higher (D2 samples). An example of microbial VOCs promoting the growth of other microorganisms was given by Collimonas pratensis and S. plymuthica (Proteobacteria phylum) species. Those two species induced the growth of P. fluorecens (Proteobcteria phylum) with the collaborative purpose of increasing their chances of survival against other microorganisms in soil51.
A negative correlation may result from the absorption of a given VOC by a given microorganism as a carbon source, or from the inhibition of this microorganism when that VOC concentration increases. For the first case it was demonstrated that in carbon-restricted substrate, the growth of Ascomycota phylum was increased from 22% to 45% while exposed to microbial VOCs emission52. Whereas an example of inhibition mechanisms was the interaction between Debaryomycetes sp. (Ascomycota phylum) and B. theobromae fungi. When B. theobromae were exposed to the VOCs emitted from Debaryomycetes sp their colonization of the substrate decreased53. The same happened to S. pithyophila (Basidiomycota phylum) when exposed to the VOCs emitted from an unidentified mix of bacteria53.
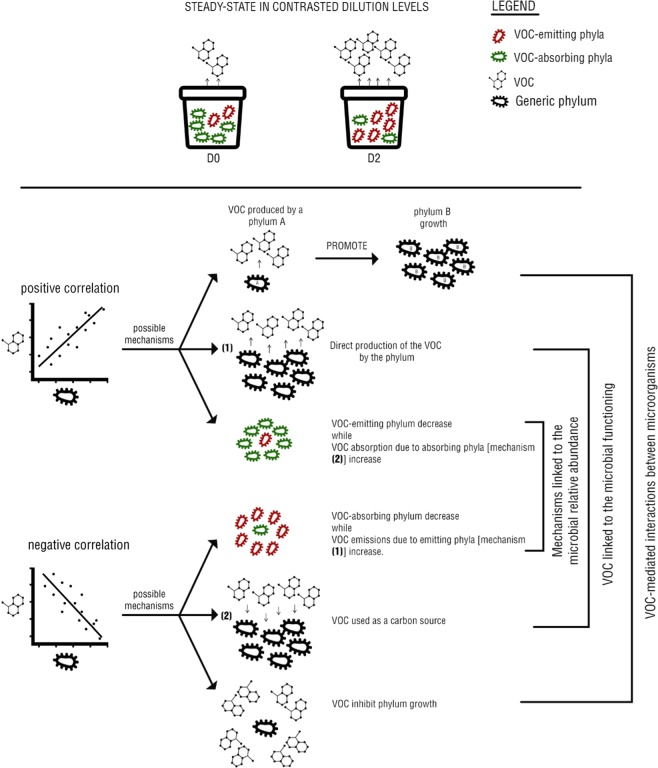
These mechanisms can hence imply some VOC-mediated interactions between the microorganisms since the further decrease of VOC concentrations in the soil may be itself linked with the microorganism metabolisms. The positive and the negative correlation between VOC emission and microorganism abundance might also result from a decrease in VOC-absorbing phyla, while VOC-emitting phyla increased and vice versa. For instance, samples D2 reported an increase of the Proteobacteria and Bacteroidetes phyla which were mostly correlated to the VOC emissions. At the same time, the VOC absorbing phyla in D2 samples decrease. Hence, in the D2 samples, Proteobacteria and Bacteroidetes phyla were positively correlated to VOC emission levels, because of the decrease of the absorbing phyla in the microcosms. On the other hand, absorbing phyla were negatively correlated to the VOC emissions, because of the increase of the Proteobacteria and Bacteroidetes phyla. In this case, VOCs are not directly involved in the correlation, but still, be correlated to the phylum. These possible mechanisms are summarized in Fig. 8.
Figure 8.
Observation of the steady statement given from the performed analysis (VOCs detection and DNA extraction) of the D0 and D2 samples. The pathways explaining the possible mechanisms behind the positive and the negative correlations are displayed. We hypothesized three different type of pathways (i) Mechanisms linked to the microbial relative abundance: VOCs are not the direct players in the correlation since absorbing phyla decrease while emissions increase due to the increase of the emitting phyla in the microcosms and vice versa; (ii) VOC might be linked to the microbial functioning since we hypothesized a direct absorption or production of the VOC by the phylum; (iii) The interaction between microorganisms such as competition and synergy.
VOCs mediating interaction between phyla
VOCs were shown to be essential intermediates in the interactions between prokaryotes for nutrients36. In particular, some VOCs produced by prokaryotes, that have access to nutrients, are shown to stimulate but suppress the growth of starved bacteria36. Compounds tentatively identified in this study as 1,3-butadiene are known to be used as biocontrol agents against other bacteria. In particular, Pseudomonas sp. which form part of the Proteobacteria phylum can inhibit the growth of Bacillus sp, which form part of the Firmicutes phylum52. In this study, we observed an increase in emission of 1,3-butadiene with dilution levels, while we measured an increase of Proteobacteria phylum abundance and a decrease of Firmicutes phylum abundance, giving indirect evidence of the effect of 1,3-butadiene in the interaction between these two phyla.
Another interesting compound detected was acetoin (3-hydroxy-2-butanone, m/z 89.056). We detected acetoin in the three dilution levels with an increased relative contribution in the summed VOC with dilution levels. The acetoin is derived from the pyruvate fermentation under anaerobic conditions53, and was shown to increase the virulence factors of Proteobacteria in the colonization process54,55. Other studies have found a positive correlation between the bacteria cell numbers of Salmonella enterica (Proteobacteria phylum)55 with the signal of an unidentified signal from the mass 89, which corresponds to the mass of protonated acetoin. We hypothesize that acetoin could have been a vector promoting Proteobacteria growth in reduced microbial diversity conditions. It is interesting to further note that acetoin is a promoter of plant growth inducing systemic resistance in plants44,53,56. We can hypothesize that the effect of acetoin on plants may be indirectly linked to promotion of Proteobacteria growth.
Our results also showed a doubled emission rate of acetaldehyde and butanone in the most diluted levels compared to the lowest ones. A positive correlation between the number count of Shigella flexneri and Salmonella enterica (Proteobacteria phylum) cells and the emissions of acetaldehyde and butanone have been reported55. We can therefore also hypothesize that the increase in acetaldehyde and butanone emissions in D2 might be due to Proteobacteria phylum growth in that dilution level.
Conclusions
This work shows that reduced microbial diversity in soil increased the VOC emissions and decreased the number of VOCs emitted. This work also strongly suggests that there are more bacteria absorbing VOCs than emitting them in soil. Several microbial processes involved in the production of VOCs within the microcosms have been hypothesized, underlining the possibility of primary and secondary metabolism production pathways. Furthermore, our study suggests that Proteobacteria and Bacteroidetes phyla would be a source of VOCs in soils while other phyla may be mostly sinks. Our results further suggest that positive and negative VOC-mediated interactions between microorganisms may be an important soil colonization process.
Methods
Sampling and site description
Samples were collected in the QualiAgro site, a field station taking part of the SOERE-PRO-network (https://www6.inra.fr/qualiagro_eng/Nos-partenaires/The-SOERE-PRO-network). The QualiAgro agronomic set up which is described in this study started in September 1998. The QualiAgro site is located at Feucherolles in northwestern France (35 km west of Paris; 48°52′N, 1°57′E, alt 150 m) on a silt loam textured soil. The soil is classified as a hortic glossic Luvisol (IUSS Working Group WRB, 2014), representative of the Parisian Basin. The main characteristics of these soils are represented by the lack of clay, a silt-loam texture (15.0% clay, 78.3% slit) and an initial pH of 6.9 in the surface horizon (0–30 cm) and good drainage. Moreover, the QualiAgro field experiment is in a cropland dominated region, which leads to low organic carbon and low organic matter concentrations (initial content of 1.1%)57.
The experiment was a randomized block design with 4 replicates comparing 4 organic waste products: BIOW (bio-waste compost derived from the co-composting of green wastes and source-separated organic fractions of municipal solid wastes), GWS (compost derived from the co-composting of green wastes with sewage sludge), FYM (farmyard manure) and MSW (municipal solid waste compost derived from the composting of residual solid wastes after removing dry and clean packaging); plus a control without organic input (CN). Samples were collected in 5 blocks of the site amended with mineral N in order to reach optimal N application. Since 1998, the organic waste products (OWPs) have been applied at a rate of ~ 4 t C ha−1 every two years on the wheat stubbles in September after harvesting. In each plot, 5 soil cores were randomly sampled at 0–30 cm depth using a core drill and stored in a cold chamber at 4 °C prior to analysis. The sampling was performed in early September 2016, one year after the last amendment of OWPs.
Microcosms and experimental setup
Soil samples were sieved at 4 mm and homogenized before gamma-ray sterilization (35 kGy; Conservatome, Dagneux, France). The sterility of the irradiated soil was verified by spreading serial dilutions of the soil onto nutrient agar plates. After the sterilization process, soils were inoculated with a diluted soil suspension prepared with the same soil before sterilization58. The soil suspension was created by mixing 30 g of soil with 90 mL of sterilized water. From this soil suspension, we used a pure sample (100) D0, and two levels of dilution were prepared with a water ratio of 1:103 (D1) and 1:105 (D2). The second step was the re-inoculation of the sterilized soil with the three different soil suspensions (pure or 100, 10−3 and 10−5). In order to create the microcosms, 30 g of each sterilized soil sample were transferred to a flask, and we added 50% of the water necessary to reach 60% of the water holding capacity (WHC). We completed the microcosms by adding one of the three soil suspension until 60% of the WHC. Soil samples consisted of soils amended with 4 different OWPs and a control without organic input. Three replicates of each combination of soil and microbial diversity level were prepared, resulting in 45 microcosms in total (n = 45, replicates = 3). Microcosms were sealed hermetically and pre-incubated at 20 °C in the dark. Once a week during six weeks the microcosms have been aerated and the water content was adjusted to maintain constant soil moisture at 60% of the WHC.
One week before the measurement of VOC emissions with the PTR-QiTOF-MS, the silicone flask plugs were substituted with Teflon coated plugs. This was necessary in order to reduce the influence of the emissions of VOCs released from the silicone plugs. Teflon is an inert material reducing VOC emissions that would have been released from the silicone plug. After the six weeks incubation, the microcosms were connected to the PTR-QiTOF-MS in order to measure VOC emissions.
VOC emissions measurements with the PTR-QiTOF-MS
PTR-QiTOF-MS setup
The VOCs were analyzed with a PTR-QiTOF-MS (PTR-Quadrupole Ion guide-TOF, Ionicon, Analytik GmbH, AU). The analyzer setup is described in details by Abis et al.30 and is only briefly described here. In this study, ionization was carried out with H3O+ as proton donor. In the drift tube, the pressure was tuned to 4 mbar, the temperature to 80 °C, and the drift voltage to 1000 V. The E/N ratio (Electric field/density of natural particles) was 132 Td where 1 Td is 10−17 V cm2. The setup of the time of flight timing was: TOF extraction period 40000 ns, pulse width 2000 ns, trigger delay 100 ns. The number of channels was 240.000. This gave a mass spectrum measurement up to 510 m/z. The measurement period was set to 1 s, which means that each sample corresponded to 60 acquisitions of 25000 individual spectra. Raw PTR-ToF-MS data were recorded by TofDaq software (Tofwerk AG, Switzerland).
Flask sampling method and flux calculations
Each flask plug was equipped with two PEEK tubes, one allowing the connection with the PTR-QiTOF-MS and the other one was connected with a bottle of dry synthetic air (Alphagaz 1 Air: 80% nitrogen, 20% oxygen, 99.9999%, Air Liquide®). The flasks had a volume of 88 cm3. The detection of the VOC emissions from the microcosms was performed during 180 s for every sample. For each sample, an empty flask was used as a reference for zero emissions, using the same Teflon plug as the sample. An air flow () of 0.3 L min−1 (equivalent volumetric flow at 0 °C and 1 atm) of dry synthetic air was passed through a hydrocarbons and humidity filter (Filter for fuel gas, final purity = 99.999%, Restek®) and a Hydrocarbon Trap (Supelco, Supelpure® HC) prior to injection in the flask. A mass flowmeter (Bronkhorst® model F-201CV, accuracy: standard 0.5% Rd plus 0.1% FS, range: 0.2 L min−1 to 5 L min−1 air) was used to control the synthetic air flow rate. Air was sampled at the chamber outlet into a PTR-QiToF-MS with a 0.05 L min−1 flow rate with a 2 m long, 1 mm internal diameter PEEK tube, heated at 80 °C. A measurement cycle consisted in measuring the VOC mixing ratio at the outlet of the flask containing the microcosms ( in ppb) for 180 s. Then air was sampled for 180 s on the empty chamber with the same Teflon plug sealing to determine (ppb). Only the last 60 s of each measurement were kept to calculate averaged mixing ratios in order to ensure a stable VOCs mixing ratio. The VOC emission ( in nmol g−1 s−1 dry soil) was calculated as:
| 1 |
where is the air molar volume at standard temperature and pressure (22.4 L mol−1 at 0 °C and 1 atm), and 30 g. After each measurement, the flask was cleaned and the soil transferred in a −40 °C chamber before the DNA extraction.
VOCs data analysis
The analysis of the ion peaks in the mass spectra measured with the PTR-QiTOF-MS, the mass calibration, and the processing of the mass table with all the compounds detected were performed using the Spectra Analyser of the PTR viewer 3.1.0.29 software (Ionicon, Analytik GmbH) following the protocol published in Abis et al.30. Likely isotopes and fragments were identified using a correlation coefficient of 0.99. Therefore, the ions having a time-correlation coefficient higher than 0.99 were considered as either isotopes or fragments depending on the m/z difference. Furthermore, correlated ion peaks that were closer than the resolution of the used PTR-QiTOF-MS were considered a single ion and only counted once.
Microbial analysis
DNA extraction
The DNA extraction has been performed for all microcosms following the protocol developed by GenoSol platform (INRA, Dijon, France, www.dijon.inra.fr/plateforme_genosol) (Terrat et al., 2012) for application in large-scale soil survey (Terrat et al., n.d.). The protocol consist of mixing in a 15 mL Falcon tube 1 g of each soil sample with 2 g of 100 mm diameter silica beads, 2.5 g of 1.4 mm diameter ceramic beads and 4 glass bead of 4 mm diameter and 5 mL of a solution containing 100 mMTris (pH 8.0), 100 mMEDTA (pH 8.0), 100 mM NaCl, and 2% (wt/vol) sodium dodecyl sulfate. Then, we proceeded with homogenizing the samples in a FastPrep-24 (MP-Biomedicals, Santa Ana, CA, USA) during 90 s and incubated for 30 min at 70 °C before centrifugation at 7000 g for 5 min at 20 °C. The deproteination was performed by collecting 1 mL of the supernatant and incubating for 10 min on ice with 1/10 volume of 3 M potassium acetate (pH 5.5) and centrifuged at 14.000 g during 5 min. The precipitation of the proteins was performed with one volume of ice-cold isopropanol. The last step of the extraction consisted of washing the nucleic acid with 70% ethanol. DNA concentrations of crude extracts were determined by electrophoresis in 1% agarose gel stained with ethidium bromide using a calf thymus DNA standard curve, and used as estimates of microbial biomass (Dequiedt et al., 2011). After quantification, nucleic acids were separated from the residual impurities, particularly humic substances, by centrifuging through two types of minicolumn. After quantification, 100 µl of crude DNA extract were separated from the residual impurities, particularly humic substances, by using only the purification steps of Nucleospin Soil kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany). Purified DNA concentrations were finally measured using the Quantifluor (Promega, Lyon, France) staining kit, according to the manufacturer’s instructions.
Quantitative PCR (qPCR)
Quantitative real-time PCR was performed on extracted DNA to quantify 16S and 18S rRNA gene sequences59,60, which led to the estimation of the F/P ratio. Prokaryotic and fungal quantitative PCR assays were performed using a StepONE (Applied Biosystems, Courtaboeuf, France) with a SYBRGreen® detection system. Purified DNA extract was amplified in a total reaction volume of 20 µl, containing 500 ng of T4 gene 32 protein (MP Biomedicals, France) and 10 µl of Takyon Rox Sybr 2X Mastermix dTTP blue (Eurogentec, Liège, Belgium).
For prokaryotic quantification, the reaction mixtures contained 1 µM of each primer (341 F: 5′ - CCT ACG GGA GGC AGC AG - 3′ and 515 R: 5′ - ATT ACC GCG GCT GCT GGC A - 3′) (López-Gutiérrez et al. 2004) and 2 ng of template DNA. The PCR conditions consisted of an initial step of 15 min at 95 °C then 35 cycles of 15 s at 95 °C, 30 s at 60 °C, 30 s at 72 °C and 20 s at 80 °C. The 16S rDNA gene from a pure culture of Pseudomonas aeruginosa PAO was used as a standard for the prokaryotic quantitative PCR assay.
Soil fungi were quantified using 1.25 µM of each primer (FR1: 5′-AIC CAT TCA ATC GGT AIT-3′, and FF390: 5′-CGA TAA CGA ACG AGA CCT-3′)61, and 2 ng of template DNA. The PCR conditions were: an initial step of 10 min at 95 °C for activation; followed by 35 cycles of 15 s at 95 °C, 30 s at 50 °C and 60 s at 70 °C. Amplified DNA from a pure culture of Fusarium oxysporum 47 was used as a fungal standard. These measures were used to estimate the prokaryotic and fungal densities in the samples.
High throughput sequencing of 16S and 18S rRNA gene sequences
Prokaryotic diversity was obtained by amplifying a 440-base 16S rRNA from each DNA samples. The corresponding primers were: F479 (5’-CAG CMG CYG CNG TAA NAC-3’) and R888 (5’-CCG YCA ATT CMT TTR AGT-3’) (Tardy et al., 2014). The amplification of the DNA was performed during a 25 µL PCR (with 5 ng of DNA for each sample) under the following set up conditions: 94 °C for 2 min, 35 cycles of 30 s at 94 °C, 52 °C for 30 s and 72 °C for 1 min, followed by 7 min at 72 °C.
For fungal diversity, a 350-base 18S rRNA fragment was amplified from each DNA sample (5 ng) with the corresponding primers: FF390 (5’-CGA TAA CGA ACG AGA CCT-3’) and FR1 (5’-ANC CAT TCA ATC GGT ANT-3’)61. For each sample, 5 ng of DNA were used for a 25 µL PCR conducted under the following conditions: 94 °C for 3 min, 35 cycles of 30 s at 94 °C, 52 °C for 1 min and 72 °C for 1 min, followed by 5 min at 72 °C.
The purification of the PCR products was performed using the Agencourt® AMPure® XP kit (Beckman Coulter, Italy, Milano) and quantified with the Quantifluor (Promega, Lyon, France) staining kit according to the manufacturer’s instructions. Purified PCR products (7.5 ng of DNA for prokaryotes and 5 ng of DNA for fungi) were amplified twice in order to integrate to the 5′ end of the primers a 10-bp multiplex identifiers allowing the specific identification of the samples and the prevention of PCR biases. For prokaryotes, the second PCR conditions were the same as previously described but with only seven cycles. For fungi, the second PCR conditions were optimized, with the number of cycles being reduced to seven and the denaturation step processed at 94 °C during 1 min. PCR products were purified with the MinElute PCR purification kit (Qiagen NV) and quantified with the Quantifluor (Promega, Lyon, France) staining kit according to the manufacturer’s instructions. Equal amounts of each sample were pooled and then cleaned with the SPRI (Solid Phase Reverse Immobilization Method) using the Agencourt® AMPure® XP kit (Beckman Coulter, Italy, Milano). The pool was finally sequenced with a MiSeq Illumina instrument (Illumina Inc, San Diego, CA) operating with V3 chemistry and producing 250 bp paired-reads.
Bioinformatic analysis of 16S and 18S rRNA gene sequences
The bioinformatics analyses were performed using the GnS-PIPE developed by the Genosol platform (INRA, Dijon, France) (Terrat et al., 2012). At first, all the 16S and 18S raw reads were organized according to the multiplex identifier sequences. All raw sequences were checked and discarded if: (i) they contained any ambiguous base (Ns), (ii) if their length was less than 350 nucleotides for 16S reads or 300 nucleotides for 18S reads, (iii) if the exact primer sequences were not found (for the distal primer, the sequence can be shorter than the complete primer sequence, but without ambiguities). A PERL program was then applied for rigorous dereplication (i.e. clustering of strictly identical sequences). The dereplicated reads were then aligned using Infernal alignment62, and clustered into operational taxonomic units (OTU) using a PERL program that groups rare reads to abundant ones, and does not count differences in homopolymer lengths with a threshold of 95% of sequence similarity. A filtering step was then carried out to check all single-singletons (reads detected only once and not clustered, which might be artifacts, such as PCR chimeras) based on the quality of their taxonomic assignments. Finally, in order to compare the datasets efficiently and avoid biased community comparisons, the reads retained were homogenized by random selection (23 700 and 11900 reads for 16S and 18S rRNA gene sequences, respectively). The retained high-quality reads were used for: (i) taxonomy-independent analyses, determining the Shannon index (ii) taxonomy-based analysis using similarity approaches against dedicated reference databases from SILVA63. The raw datasets are available in the EBI database system under project accession number PRJEB29286.
Statistical analysis
Microbial biomass, microbial diversity Index (Shannon) and the relative abundance of prokaryotes, and fungal phyla in the microbial composition were processed by the ANOVA test. With the ANOVA test, we also analyzed the effect of the organic waste product on the microbial community for the different dilution level The dataset before the statistical analysis was made of 754 variables (number of peaks detected) and 45 samples comprising the 3 replicate for each sample. In order to select the most representing variables of the dataset, several statistical tests have been performed using the R software (Version 1.0.153 – © 2009–2017 RStudio). At first, the normality test (Shapiro-Wilk test, W > 0.9) has been applied to verify that the mean mixing ratios were normally distributed for each VOC. Next, the homogeneity of the variances was verified for each treatment using the Levene test. Once the normality and the homogeneity of the variances were validated, a t-test was performed in order to see if the VOC flux was significantly higher than 0. Correlated masses that were closer than the resolution of the used PTR-QiTOF-MS were removed in order to not count twice the same peak. Finally, an ANOVA test was performed. The final selected dataset comprised of 45 samples and 239 variables meaning that only 32% of the dataset was kept for further analysis. The ANOVA tests were followed by the Tukey post hoc test.
A principal component analysis (PCA, Package Ade4 Version 1.0.153 – © 2009–2017 R Studio) was performed to see the different VOCs compounds differentiating the three dilution levels. The Shannon index of diversity (Figs. 1 and S4) was calculated with the diversity function of the vegan package (version 2.4-3) in the R software (version 3.2.3). The diversity index was calculated as , where the sum is over all VOCs recorded in the mass table. The correlation matrix between VOCs and microorganisms has been created selecting the VOC having a R2 of the correlation microorganisms/VOC larger than 0.6, the final number of compounds displayed was 107.
Microbial biomass, microbial diversity Index (Shannon) and the relative abundance of prokaryotic, and fungal phyla in the microbial composition were processed by the ANOVA test. With the ANOVA test, we also analyzed the effect of the organic waste product on the microbial community for the different dilution levels. All significant effects were assessed by Tukey’s Honestly Significant Difference (HSD) post hoc test (P < 0.05).
The correlation between the summed VOC emissions and VOC diversity and the different microbiological factors has been calculated using the basic package STAT of R studio. We choose the Spearman correlation as a method since the distribution of the dataset was not normal.
To visualize the relationship between VOCs and soil physico-chemical and microbiological factors, redundancy analysis (RDA) and variance partitioning were carried out. All statistical analyses were performed with the Vegan package (v.2.0-8)64 in R software version 3.0.1. RDA was applied to VOCs variables (diversity and total emission rates) using soil physico-chemical (Corg, pH, C/N) and microbial community parameters (molecular microbial biomass, prokaryotic and fungal richness, prokaryotic and fungal diversity, F/P ratio) as explanatory variables. At first, a global variance partitioning was applied to quantify the respective effect of soil physico-chemistry and soil biology (rda followed by anova.cca function). Then, a detailed variance partitioning provided the pure effect of each exploratory variable was proceeded (ordiR2step followed by anova function)65. The interactions between all explanatory variables were also included in the model. The significance of the model and of each explanatory variable included in the model was tested using 10000 permutations.
Supplementary information
Acknowledgements
We gratefully acknowledge ANAEE-France that provided the PTR-QiTOF-MS, ADEME for partially funding this study through the projects COV3ER (n°1562C0032) and DICOV (n°1662C0020). The Qualiagro site conducted in partnership with Veolia, forms part of the SOERE-PRO integrated as a service of the infrastructure AnaEE-France, overseen by the French National Research Agency (ANR-11-INBS-0001). The QUALIAGRO field experiment forms part of the SOERE-PRO (network of long-term experiments dedicated to the study of impacts of organic waste product recycling) certified by ALLENVI (Alliance Nationale de Recherche pour l’Environnement).
Author contributions
L. Abis wrote the article, carried out the experiment, and performed the statistical analysis with B. Loubet. B. Loubet and S. Sadet-Bourgeteau conceived the experiment and supervised the redaction and the statistical analysis of the manuscript. R. Ciuraru revised the manuscript and performed the tentative identification of the compounds. Pierre-Alain Maron conceived the experiment. S. Houot managed the long-term field experiment. F. Lafouge, V. Nowak and J. Tripied carried out the experiment. S. Dequiedt performed the bioinformatics analysis.
Data availability
Data are available in data.inra.fr website (10.15454/4W9DM2).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-63091-8.
References
- 1.Karl M, Guenther A, Köble R, Leip A, Seufert G. A new European plant-specific emission inventory of biogenic volatile organic compounds for use in atmospheric transport models. Biogeosciences. 2009;6:1059–1087. doi: 10.5194/bg-6-1059-2009. [DOI] [Google Scholar]
- 2.Lindfors V, Laurila T. Biogenic volatile organic compound (VOC) emissions from forests in Finland. Boreal Environ. Res. 2000;5:95–113. [Google Scholar]
- 3.Zemankova K, Brechler J. Emissions of biogenic VOC from forest ecosystems in central Europe: Estimation and comparison with anthropogenic emission inventory. Environ. Pollut. 2010;158:462–469. doi: 10.1016/j.envpol.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 4.Bachy A, et al. Are BVOC exchanges in agricultural ecosystems overestimated? Insights from fluxes measured in a maize field over a whole growing season. Atmospheric Chem. Phys. 2016;16:5343–5356. doi: 10.5194/acp-16-5343-2016. [DOI] [Google Scholar]
- 5.Girvan MS, Bullimore J, Ball AS, Pretty JN, Osborn AM. Responses of Active Bacterial and Fungal Communities in Soils under Winter Wheat to Different Fertilizer and Pesticide Regimens. Appl Env. Microbiol. 2004;70:2692–2701. doi: 10.1128/AEM.70.5.2692-2701.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadet-Bourgeteau S, et al. Lasting effect of repeated application of organic waste products on microbial communities in arable soils. Appl. Soil Ecol. 2018;125:278–287. doi: 10.1016/j.apsoil.2018.02.006. [DOI] [Google Scholar]
- 7.Poulsen PHB, et al. Effects of fertilization with urban and agricultural organic wastes in a field trial – Prokaryotic diversity investigated by pyrosequencing. Soil Biol. Biochem. 2013;57:784–793. doi: 10.1016/j.soilbio.2011.12.023. [DOI] [Google Scholar]
- 8.Hartmann M, Frey B, Mayer J, Mäder P, Widmer F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015;9:1177–1194. doi: 10.1038/ismej.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, et al. Microbial communities of an arable soil treated for 8 years with organic and inorganic fertilizers. Biol. Fertil. Soils. 2016;52:455–467. doi: 10.1007/s00374-016-1089-5. [DOI] [Google Scholar]
- 10.Francioli D, et al. Mineral vs. Organic Amendments: Microbial Community Structure, Activity and Abundance of Agriculturally Relevant Microbes Are Driven by Long-Term Fertilization Strategies. Front. Microbiol. 2016;7:1446. doi: 10.3389/fmicb.2016.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Gil JC, Plaza C, Senesi N, Brunetti G, Polo A. Effects of Long-Term Sewage Sludge Amendment on the Composition, Structure and Proton Binding Activity of Soil Fulvic Acids. CLEAN – Soil Air Water. 2007;35:480–487. doi: 10.1002/clen.200720013. [DOI] [Google Scholar]
- 12.Orgill SE, et al. Sensitivity of soil organic carbon to grazing management in the semi-arid rangelands of south-eastern Australia. Rangel. J. 2017;39:153. doi: 10.1071/RJ16020. [DOI] [Google Scholar]
- 13.Baumann K, et al. Soil microbial diversity affects soil organic matter decomposition in a silty grassland soil. Biogeochemistry. 2013;114:201–212. doi: 10.1007/s10533-012-9800-6. [DOI] [Google Scholar]
- 14.Stahl PD, Parkin TB. Microbial production of volatile organic compounds in soil microcosms. Soil Sci. Soc. Am. J. 1996;60:821–828. doi: 10.2136/sssaj1996.03615995006000030020x. [DOI] [Google Scholar]
- 15.Wenke K, Kai M, Piechulla B. Belowground volatiles facilitate interactions between plant roots and soil organisms. Planta. 2010;231:499–506. doi: 10.1007/s00425-009-1076-2. [DOI] [PubMed] [Google Scholar]
- 16.Bending GD, Lincoln SD. Inhibition of soil nitrifying bacteria communities and their activities by glucosinolate hydrolysis products. Soil Biol. Amp Biochem. 2000;32:1261–1269. doi: 10.1016/S0038-0717(00)00043-2. [DOI] [Google Scholar]
- 17.Stotzky G, Schenck S. Volatile organic compounds and microorganisms. CRC Crit. Rev. Microbiol. 1976;4:333–382. doi: 10.3109/10408417609102303. [DOI] [PubMed] [Google Scholar]
- 18.Lee S, Behringer G, Hung R, Bennett J. Effects of fungal volatile organic compounds on Arabidopsis thaliana growth and gene expression. Fungal Ecol. 2019;37:1–9. doi: 10.1016/j.funeco.2018.08.004. [DOI] [Google Scholar]
- 19.de Boer, W., Li, X., Meisner, A. & Garbeva, P. Pathogen suppression by microbial volatile organic compounds in soils. FEMS Microbiol. Ecol. 95 (2019). [DOI] [PubMed]
- 20.Paavolainen L, Kitunen V, Smolander A. Inhibition of nitrification in forest soil by monoterpenes. Plant Soil. 1998;205:147–154. doi: 10.1023/A:1004335419358. [DOI] [Google Scholar]
- 21.Cheng Y, et al. Challenges and solutions for biofiltration of hydrophobic volatile organic compounds. Biotechnol. Adv. 2016;34:1091–1102. doi: 10.1016/j.biotechadv.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Yang C, et al. Simultaneous Removal of Multicomponent VOCs in Biofilters. Trends Biotechnol. 2018;36:673–685. doi: 10.1016/j.tibtech.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Zehraoui A, Hassan AA, Sorial GA. Biological treatment of n-hexane and methanol in trickle bed air biofilters under acidic conditions. Biochem. Eng. J. 2013;77:129–135. doi: 10.1016/j.bej.2013.06.001. [DOI] [Google Scholar]
- 24.Cabrol L, Malhautier L, Poly F, Lepeuple A-S, Fanlo J-L. Bacterial dynamics in steady-state biofilters: beyond functional stability. FEMS Microbiol. Ecol. 2012;79:260–271. doi: 10.1111/j.1574-6941.2011.01213.x. [DOI] [PubMed] [Google Scholar]
- 25.Misztal PK, et al. Emission Factors of Microbial Volatile Organic Compounds from Environmental Bacteria and Fungi. Environ. Sci. Technol. 2018;52:8272–8282. doi: 10.1021/acs.est.8b00806. [DOI] [PubMed] [Google Scholar]
- 26.Isidorov V, Jdanova M. Volatile organic compounds from leaves litter. Chemosphere. 2002;48:975–979. doi: 10.1016/S0045-6535(02)00074-7. [DOI] [PubMed] [Google Scholar]
- 27.Maron, P.-A. et al. High microbial diversity promotes soil ecosystem functioning. Appl Env. Microbiol AEM.02738-17, 10.1128/AEM.02738-17 (2018). [DOI] [PMC free article] [PubMed]
- 28.Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 29.Bevivino, A. et al. Soil Bacterial Community Response to Differences in Agricultural Management along with Seasonal Changes in a Mediterranean Region. PLoS ONE9 (2014). [DOI] [PMC free article] [PubMed]
- 30.Abis L, et al. Profiles of volatile organic compound emissions from soils amended with organic waste products. Sci. Total Environ. 2018;636:1333–1343. doi: 10.1016/j.scitotenv.2018.04.232. [DOI] [PubMed] [Google Scholar]
- 31.Bachy A, et al. Methanol exchange dynamics between a temperate cropland soil and the atmosphere. Atmos. Environ. 2018;176:229–239. doi: 10.1016/j.atmosenv.2017.12.016. [DOI] [Google Scholar]
- 32.Warneke C, et al. Acetone, methanol, and other partially oxidized volatile organic emissions from dead plant matter by abiological processes: Significance for atmospheric HOx chemistry. Glob. Biogeochem. Cycles. 1999;13:9–17. doi: 10.1029/98GB02428. [DOI] [Google Scholar]
- 33.Gosset G. Production of aromatic compounds in bacteria. Curr. Opin. Biotechnol. 2009;20:651–658. doi: 10.1016/j.copbio.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt R, Cordovez V, de Boer W, Raaijmakers J, Garbeva P. Volatile affairs in microbial interactions. ISME J. 2015;9:2329–2335. doi: 10.1038/ismej.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller A, et al. Volatile profiles of fungi – Chemotyping of species and ecological functions. Fungal Genet. Biol. 2013;54:25–33. doi: 10.1016/j.fgb.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Schulz-Bohm K, Zweers H, de Boer W, Garbeva P. A fragrant neighborhood: volatile mediated bacterial interactions in soil. Front. Microbiol. 2015;6:1212. doi: 10.3389/fmicb.2015.01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Citron CA, Gleitzmann J, Laurenzano G, Pukall R, Dickschat JS. Terpenoids are Widespread in Actinomycetes: A Correlation of Secondary Metabolism and Genome Data. Chembiochem. 2012;13:202–214. doi: 10.1002/cbic.201100641. [DOI] [PubMed] [Google Scholar]
- 38.Chernin L, et al. Quorum-sensing quenching by rhizobacterial volatiles. Environ. Microbiol. Rep. 2011;3:698–704. doi: 10.1111/j.1758-2229.2011.00284.x. [DOI] [PubMed] [Google Scholar]
- 39.Yuan J, et al. Microbial volatile compounds alter the soil microbial community. Environ. Sci. Pollut. Res. Int. 2017;24:22485–22493. doi: 10.1007/s11356-017-9839-y. [DOI] [PubMed] [Google Scholar]
- 40.Estrada JM, Hernández S, Muñoz R, Revah S. A comparative study of fungal and bacterial biofiltration treating a VOC mixture. J. Hazard. Mater. 2013;250–251:190–197. doi: 10.1016/j.jhazmat.2013.01.064. [DOI] [PubMed] [Google Scholar]
- 41.Chang K, Lu C. Biofiltration of toluene and acetone mixtures by a trickle-bed air biofilter. World J. Microbiol. Biotechnol. 2003;19:791–798. doi: 10.1023/A:1026065206638. [DOI] [Google Scholar]
- 42.de Boer W, Verheggen P, Klein Gunnewiek PJA, Kowalchuk GA, van Veen JA. Microbial Community Composition Affects Soil Fungistasis. Appl. Environ. Microbiol. 2003;69:835–844. doi: 10.1128/AEM.69.2.835-844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garbeva P, Hol WHG, Termorshuizen AJ, Kowalchuk GA, de Boer W. Fungistasis and general soil biostasis - A new synthesis. Soil Biol. Biochem. 2011;43:469–477. doi: 10.1016/j.soilbio.2010.11.020. [DOI] [Google Scholar]
- 44.Chuankun X, Minghe M, Leming Z, Keqin Z. Soil volatile fungistasis and volatile fungistatic compounds. Soil Biol. Biochem. 2004;36:1997–2004. doi: 10.1016/j.soilbio.2004.07.020. [DOI] [Google Scholar]
- 45.Campos VP, Pinho RSCde, Freire ES. Volatiles produced by interacting microorganisms potentially useful for the control of plant pathogens. Ciênc. E Agrotecnologia. 2010;34:525–535. doi: 10.1590/S1413-70542010000300001. [DOI] [Google Scholar]
- 46.Zou C-S, Mo M-H, Gu Y-Q, Zhou J-P, Zhang K-Q. Possible contributions of volatile-producing bacteria to soil fungistasis. Soil Biol. Biochem. 2007;39:2371–2379. doi: 10.1016/j.soilbio.2007.04.009. [DOI] [Google Scholar]
- 47.Kai M, et al. Bacterial volatiles and their action potential. Appl. Microbiol. Biotechnol. 2009;81:1001–1012. doi: 10.1007/s00253-008-1760-3. [DOI] [PubMed] [Google Scholar]
- 48.Bennett, J. W., Hung, R., Lee, S. & Padhi, S. 18 Fungal and Bacterial Volatile Organic Compounds: An Overview and Their Role as Ecological Signaling Agents. In Fungal Associations (ed. Hock, B.) 373–393, 10.1007/978-3-642-30826-0_18 (Springer, 2012).
- 49.Garbeva P, Hordijk C, Gerards S, de Boer W. Volatile-mediated interactions between phylogenetically different soil bacteria. Front. Microbiol. 2014;5:289. doi: 10.3389/fmicb.2014.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cale JA, et al. Fungal Volatiles Can Act as Carbon Sources and Semiochemicals to Mediate Interspecific Interactions Among Bark Beetle-Associated Fungal Symbionts. PLOS ONE. 2016;11:e0162197. doi: 10.1371/journal.pone.0162197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruce A, Stewart D, Verrall S, Wheatley RE. Effect of volatiles from bacteria and yeast on the growth and pigmentation of sapstain fungi. Int. Biodeterior. Biodegrad. 2003;51:101–108. doi: 10.1016/S0964-8305(02)00088-4. [DOI] [Google Scholar]
- 52.Tahir HAS, et al. Bacillus volatiles adversely affect the physiology and ultra-structure of Ralstonia solanacearum and induce systemic resistance in tobacco against bacterial wilt. Sci. Rep. 2017;7:40481. doi: 10.1038/srep40481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryu C-M, et al. Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2003;100:4927–4932. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Audrain B, Farag MA, Ryu C-M, Ghigo J-M. Role of bacterial volatile compounds in bacterial biology. FEMS Microbiol. Rev. 2015;39:222–233. doi: 10.1093/femsre/fuu013. [DOI] [PubMed] [Google Scholar]
- 55.Bunge M, et al. On-Line Monitoring of Microbial Volatile Metabolites by Proton Transfer Reaction-Mass Spectrometry. Appl. Environ. Microbiol. 2008;74:2179–2186. doi: 10.1128/AEM.02069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farag MA, Zhang H, Ryu C-M. Dynamic Chemical Communication between Plants and Bacteria through Airborne Signals: Induced Resistance by Bacterial Volatiles. J. Chem. Ecol. 2013;39:1007–1018. doi: 10.1007/s10886-013-0317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meersmans J, et al. A high resolution map of French soil organic carbon. Agron. Sustain. Dev. 2012;32:841–851. doi: 10.1007/s13593-012-0086-9. [DOI] [Google Scholar]
- 58.Wertz S, et al. Maintenance of soil functioning following erosion of microbial diversity. Environ. Microbiol. 2006;8:2162–2169. doi: 10.1111/j.1462-2920.2006.01098.x. [DOI] [PubMed] [Google Scholar]
- 59.Plassart P, et al. Evaluation of the ISO Standard 11063 DNA Extraction Procedure for Assessing Soil Microbial Abundance and Community Structure. PLOS ONE. 2012;7:e44279. doi: 10.1371/journal.pone.0044279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lienhard P, et al. Pyrosequencing evidences the impact of cropping on soil bacterial and fungal diversity in Laos tropical grassland. Agron. Sustain. Dev. 2014;34:525–533. doi: 10.1007/s13593-013-0162-9. [DOI] [Google Scholar]
- 61.Prévost-Bouré NC, et al. Validation and Application of a PCR Primer Set to Quantify Fungal Communities in the Soil Environment by Real-Time Quantitative PCR. PLOS ONE. 2011;6:e24166. doi: 10.1371/journal.pone.0024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cole JR, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927-930–4. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 65.Peres-Neto PR, Legendre P, Dray S, Borcard D. Variation Partitioning of Species Data Matrices: Estimation and Comparison of Fractions. Ecology. 2006;87:2614–2625. doi: 10.1890/0012-9658(2006)87[2614:VPOSDM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 66.Zou C-S, Mo M-H, Gu Y-Q, Zhou J-P, Zhang K-Q. Possible contributions of volatile-producing bacteria to soil fungistasis. Soil Biol. Biochem. 2007;39:2371–2379. doi: 10.1016/j.soilbio.2007.04.009. [DOI] [Google Scholar]
- 67.Chuankun X, Minghe M, Leming Z, Keqin Z. Soil volatile fungistasis and volatile fungistatic compounds. Soil Biol. Biochem. 2004;36:1997–2004. doi: 10.1016/j.soilbio.2004.07.020. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in data.inra.fr website (10.15454/4W9DM2).