Abstract
Gliomas are the most commonly occurring tumors of the central nervous system. Glioblastoma multiforme (GBM) is the most malignant and aggressive brain cancer in adults. Further understanding of the mechanisms underlying the aggressive nature of GBM is urgently needed. Here we identified homeobox B8 (HOXB8), a member of the homeobox family, as a crucial contributor to the aggressiveness of GBM. Data mining of publicly accessible RNA sequence datasets and our patient cohorts confirmed a higher expression of HOXB8 in the tumor tissue of GBM patients, and a strong positive correlation between the expression level and pathological grading of tumors and a negative correlation between the expression level and the overall survival rate. We next showed that HOXB8 promotes the proliferation and migration of glioblastoma cells and is crucial for the activation of the PI3K/AKT pathway and expression of epithelial–mesenchymal transition-related genes, possibly through direct binding to the promoter of SAMD9 (Sterile Alpha Motif Domain-Containing Protein 9) and activating its transcription. Collectively, we identified HOXB8 as a critical contributor to the aggressiveness of GBM, which provides insights into a potential therapeutic target for GBM and opens new avenues for improving its treatment outcome.
Electronic supplementary material
The online version of this article (10.1007/s12264-019-00436-y) contains supplementary material, which is available to authorized users.
Keywords: HOXB8, Glioma, Aggressiveness, SAMD9, Treatment
Introduction
Glioma is a set of primary brain tumors which account for almost 80% of all malignant primary tumors of the brain [1]. They are classified according to their presumed cell of origin and include astrocytic tumors, oligodendrogliomas, ependymomas, and mixed gliomas [2–5]. The World Health Organization classifies gliomas pathologically as grades I to IV based on the level of malignancy as determined by histopathological criteria [6, 7]. Glioblastoma multiforme (GBM) is the most aggressive type of tumor and has been designated Grade IV [8]. The global incidence of GBM is < 10 per 100,000 people. However, the poor prognosis and survival rate after diagnosis makes it a crucial public health issue [9–11]. Currently, the etiology of gliomas remains poorly understood and the aggressiveness of GBM needs urgent investigation in order to develop more effective treatment strategies to improve the poor prognosis of patients with this high-grade glioma.
Hox genes are a group of related genes, each of which contains a well-conserved DNA sequence known as the homeobox [12]. The protein product of each Hox gene is a transcription factor. In mammals, 39 Hox genes have been identified that are clustered in four genomic loci, the HoxA, HoxB, HoxC and HoxD complexes, which control the body plan of the embryo along the head-tail axis [13, 14]. In addition, Hox proteins regulate numerous other processes including apoptosis, receptor signaling, differentiation, motility, and angiogenesis [15, 16]. Aberrations in Hox gene expression have been reported in malignancy, indicating that altered expression of Hox genes could be important for both oncogenesis and tumor suppression, depending on context [17, 18].
HOXB8 is located at 17q21.32, is composed of 5 exons, and encodes 243 amino-acids. In addition to its critical role in the development of the central nervous system, accumulating evidence suggests a significant role of HOXB8 in colorectal cancer [19], liver cancer [20], gastric cancer [21], and ovarian cancer [22]. However, its role in the development and progression of glioma is unclear.
Therefore, in this study, we set out to determine the role of HOXB8 in the development of glioma.
Materials and Methods
Ethics Statement
Human brain tissue microarrays were provided by Shanghai Outdo Biotech Co. Ltd., comprising 3 from normal brain tissue and 87 from glioma patients (3 with grade I, 16 with grade II, 26 with grade III, and 42 with grade IV) (Cat No. HBraG090PG01, Lot No. XT16-017).
Tissues from 26 glioma cases (13 with grade III and 13 with grade IV) were provided by Beijing Tiantan Hospital and individual informed consent was given by all patients. Normal brain tissues were acquired from patients who underwent brain surgery for unrelated causes. We confirm that the procedures involving experiments on human subjects met the ethical standards of the Helsinki Declaration in 1975. This study was approved by the Ethics Committee of Peking University Health Science Center.
Bioinformatics Analysis of HOXB8 Expression
HOXB8 mRNA expression data from gliomas were downloaded and extracted from three datasets [European Bioinformatics Institute (EBI) ArrayExpress data, Chinese Glioma Genome Atlas (CGGA) data, and Gene Expression Omnibus (GEO) data]. The Kruskal-Wallis rank sum test was performed to assess the difference in HOXB8 expression between non-tumor brain tissue and gliomas, and between low-grade and high-grade gliomas. Gene Set Enrichment Analysis (GSEA) was also used to find significantly enriched gene sets and pathways. The relationship between HOXB8 mRNA expression levels and tumor patient survival was analyzed online using Oncolnc (http://www.oncolnc.org/), which linked the survival data to mRNAs, miRNAs, and lncRNAs from The Cancer Genome Atlas.
Cell Culture
Human glioma cell lines U251 (Sigma-Aldrich, 0906301, Porton Down, UK), U87 (American Type Culture Collection, HTB-14, Manassas, VA), A172 (American Type Culture Collection, CRL-1620), GOS-3 (German Collection of Microorganisms and Cell Cultures, ACC 408, Braunschweig, Germany), and rat C6 glioma cells (American Type Culture Collection, CCL-107) were cultured in RPMI 1640 medium (pH 7.4) supplemented with 10% fetal bovine serum (FBS) and 100 IU/mL penicillin and streptomycin (all from Gibco, Thermo Scientific, NY, USA) in a humidified atmosphere of 5% CO2 at 37°C.
Plasmids and Antibodies
Full-length HOXB8 was cloned into the pcDNA3.1 vector. Anti-HOXB8 antibody (Abcam, ab125727, WB 1:1000, ICC 1:200) and anti-SAMD9 antibody (Abcam, ab13603, WB 1:1000, ICC 1:100) were from Abcam. MMP2 (#13132, D8N9Y), MMP3 (#14351, D7F5B), MMP-9 (#13667, D6O3H), Slug (#9585, C19G7), Snail (#3879, C15D3), PI3K kinase p85 (#4942), phospho-PI3 kinase p85 (#4228,1:1000,Tyr458)/p55 (Tyr199), E-cadherin (#3195, 24E10), N-cadherin (#13116,D4R1H), p44/42 MAPK (#4695, Erk1/2) (137F5), phospho-p44/42 MAPK (#9101, Erk1/2) (Thr202/Tyr204), Akt (#4691, pan) (C67E7), phospho-Akt (#4060, Ser473) (D9E), vimentin (#5741, D21H3), β-actin (#3700, 8H10D10), and GAPDH (#8884, D16H11) were from Cell Signaling Technology (Massachusetts, USA). Mouse anti-rabbit IgG-HRP (BE0107-100, Shenzhen, China) was from EASYBIO and KI67 rabbit polyclonal antibody was from Proteintech (27309-1-AP, Rosemont, USA).
si-RNA Sequences
The small interfering (si)-RNA sequences were designed and synthesized by GenePharma (Shanghai, China). U251 and U87 cells were transfected with Smart pool siRNAs against HOXB8 or SAMD9 (20 nmol/L) using Lipofectamine 3000 (Thermo Fisher Scientific Transfection, Waltham, MA). The sequences were as follows: siRNA HOXB8 1, 5′-CCUAUUUAAUCCCUAUCUGTT-3′; siRNA HOXB8 2, 5′-UCAACUCACUGUUCUCCAATT-3′; siRNA SAMD9, 5′-GUGCAUUCGAGAGCCAAGAUU-3′; siNC, 5′-GUGCAUUCAAGAGCCAAGAUU-3′.
Lentivirus Infection
Lentiviruses carrying short hairpin (sh)-RNA targeting human HOXB8 were constructed by GeneChem (GIDL0158539, Shanghai, China). Cells were infected by the viruses for 48 h, then they were cultured in medium containing puromycin for selecting stable HOXB8-knockdown cells. The knockdown efficiency was verified by Western blotting. The shRNA sequences were as follows: HOXB8 5′-GCAAATCCAGGAGTTCTAC-3′, non-targeting control: 5′-TTCTCCGAACGTGTCACGT -3′.
Cell Survival Assay
Cells were assayed using Cell Counting Kit-8 (CCK-8) (Beijing Solarbio Science & Technology Co., Ltd, CA1210). The cells were plated in 96-well plates (100 µL cell suspensions, 3 × 103 cells), 10 μL of CCK8 per well was added at 0, 12, 24, 36, 48, and 72 h, incubated for 2–4 h in a 37°C cell incubator, then the absorbance (OD) at 450 nm was measured using a microplate reader (Thermo, Waltham, MA).
Colony Formation
Approximately 200 stably-transfected cells were plated into a 12-well plate. Two weeks later, visible colonies were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet (Selleck Chemicals, S1917, Houston, TX). All experiments were performed in triplicate wells.
Cell Migration Assays
Migration assays were conducted by seeding 1 × 104 U251 or U87 glioma cells in the upper chamber of a Transwell plate (Corning, NY, USA). The glioma cells were cultured without serum for 12 h before each experiment; a single cell suspension (300 μL) in serum-free medium was added to the upper chamber, then 700 μL of complete medium containing 10% FBS was added to the lower level. After incubation at 37°C for 24 h, the cells in the lower chamber were fixed in 4% formalin and stained with 0.1% crystal violet. The migrated cells were counted in three randomly-selected fields from each membrane and each experiment was performed three times.
Cell Invasion Assay
Invasion assays were performed by seeding 1×104 U251 or U87 glioma cells in the upper chamber of a Transwell plate (Corning). Matrigel (0.5%) in a coating buffer solution (BD Biosciences, NJ, USA) was added to the upper chamber prior to seeding the cells. The glioma cells were cultured without serum for 12 h before each experiment and an appropriate amount (300 μL) of a single-cell suspension in serum-free medium was added to the upper chamber, and complete medium containing 10% FBS (700 μL) was then added to the lower layer. After incubation at 37°C for 24 h, the cells in the lower chamber were fixed in 4% formalin and stained with 0.1% crystal violet. The migrated cells were counted in three randomly-selected fields from each membrane; each experiment was performed three times.
Western Blotting
Glioma cells were harvested and lysed in RIPA buffer (Beyotime, P0013C, Beijing, China), containing complete mini protease inhibitor cocktail (Roche, 04693124001, Basel, Switzerland). Proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes (GE, PVDF 0.45 μmol/L, 10600023). Non-specific binding was blocked using 5% non-fat milk and the primary antibody was incubated at 4°C overnight; then the secondary antibody was incubated at room temperature for 1 h. Signals were visualized by chemiluminescence (Millipore Corp., Billerica, MA).
RNA Sequencing and Target Gene Search by qPCR
U251 cells were treated with HOXB8 siRNA and siRNA NC for 48 h, each with 3 technical replicates. RNA was extracted using TRIzol (Invitrogen, 15596-026, Grand Island, NY) according to the manufacturer’s protocol. Total RNA was reverse transcribed using an RT-PCR kit (Tiangen, KR103-03, Beijing, China) according to the manufacturer’s protocol. The sequencing reads were generated using the BGISEQ-500 platform following the manufacturer’s recommendations. The paired-end clean reads were aligned to the reference human genome (UCSC version hg19) using TopHat v2.0.12. HTSeq v0.6.1 was used to count the read numbers mapped to each gene and the gene expression levels were calculated with RSEM version v1.2.31. Fragments per kilobase of transcript per million mapped reads for each gene were calculated based on the length of the gene and the read-count mapped to that gene, all these were provided by the Beijing Genomics Institute (Beijing, China). We used MA, volcano, scatter, and heatmap plots to show the distributions of differentially-expressed genes. A Holm’s corrected P-value of 0.005 and log2 (fold-change) of 1 were set as the threshold for significant differential expression. Then functional enrichment analysis was performed on Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathways. Subsequently, qPCR was performed using Go Taq qPCR Master Mix (Cat No. A6001, Promega Corp., Madison, WI). Cycling conditions were as follows: 5 min at 95°C followed by 45 cycles each consisting of 10 s at 95°C, 20 s at 60°C, and 30 s at 72°C. The relative concentrations of genes were normalized to β-actin. Fold-change was calculated using the 2−∆∆Ct method. The primers are listed in Table S1.
Semi-quantitative Immunohistochemistry
Paraffin sections were made as previously described [23–25]. After deparaffinization, sections were immersed in 100% ethanol (2×), 96% ethanol (2×), and 70% ethanol (2×) followed by heat-induced antigen retrieval at 120°C for 10 min in citrate buffer (pH 6), then allowed to cool to room temperature. After blocking endogenous enzyme activity and non-specific protein binding sites, each slide was incubated overnight at 4°C with the primary antibody followed by incubation with the secondary antibody for 1 h. Color development was produced using 3, 3-diaminobenzidine tetrahydrochloride (Beijing Solarbio Science & Technology Co., Ltd, DA1010-10) for 5 min, and counterstained with hematoxylin (Selleck Chemicals, S2384).
HOXB8 expression was evaluated independently by two experienced pathologists using the following method. A: Cell staining intensity (at 10×20 magnification, 5 different fields of view were selected randomly and observed under the microscope; the average of the combined counts of 5 fields was calculated): negative staining, 0 point; weakly positive staining, 1 point; positive staining but with light brown background, 2 points; positive staining without background, 3 points. B: Area staining intensity (at 10 × 4 magnification the total positive area was observed and evaluated): positive area = 0%, 0 point; positive area = 1%–25%, 1 point; positive area = 26%–50%, 2 points; positive area = 51%–75%, 3 points; positive area > 75%, 4 points. C: The degree of positive staining for each section was determined by multiplication of the values for A and B: 1–3 was classified as weakly positive (+); 4–6 as positive (++); and 7–12 as strongly positive (+++).
Nuclear Protein Extraction and Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were isolated from shNC U251 and shHOXB8 U251 cells with Nuclear and Cytoplasmic Extraction Reagents (Pierce, Cat: 78833, NY, USA) according to the manufacturer’s instructions. Protein concentrations were determined with a BCA protein assay kit (Pierce, Cat: 23227). Nuclear extracts were stored at − 80°C until use. We predicted HOXB8 binding sites in the SAMD9 promoter and designed three biotin end-labeled probes (probe 1 sequence, AAGAGTAATTAAGTTA, location − 778 to − 794 bp; probe 2 sequence, CTCTGCAATAAATGAA, location − 488 to − 478 bp; probe 3 sequence, ACCCTTAAAGGCCAGT, − 370 to − 386 bp), synthesized by Sangon Biotech (Shanghai, China). EMSA was carried out using the LightShift® Chemiluminescent EMSA Kit (Pierce, Cat: 89880, NY, USA) according to the manufacturer’s instructions. DNA binding reactions were detected in 20-μL volumes containing biotin-labeled oligonucleotides and nuclear extracts. Unlabeled oligonucleotides were added as competition controls. The reaction products were incubated at 4°C for 20 min, then the reaction mixtures were separated by electrophoresis, transferred to a nylon membrane (Roche) and detected by chemiluminescence (Millipore Corp.).
Statistical Analysis
Quantitative data are presented as the mean ± SD. Differences in the mean of two samples were analyzed using Student’s t-test. Correlation between HOXB8 and SAMD9 expression levels was analyzed using the Spearman rank correlation test. Correlations between HOXB8 expression and various clinical pathological variables were analyzed using the Kruskal-Wallis test. Overall survival rates were determined using the Kaplan-Meier method. All statistical tests were two-tailed exact tests with a P < 0.05 considered significant. All statistical analyses were performed using SPSS software (version 15.0)/R version 3.4.4 (http://www.cran.r-project.org).
Results
HOXB8 is Involved in the Pathogenesis of Gliomas
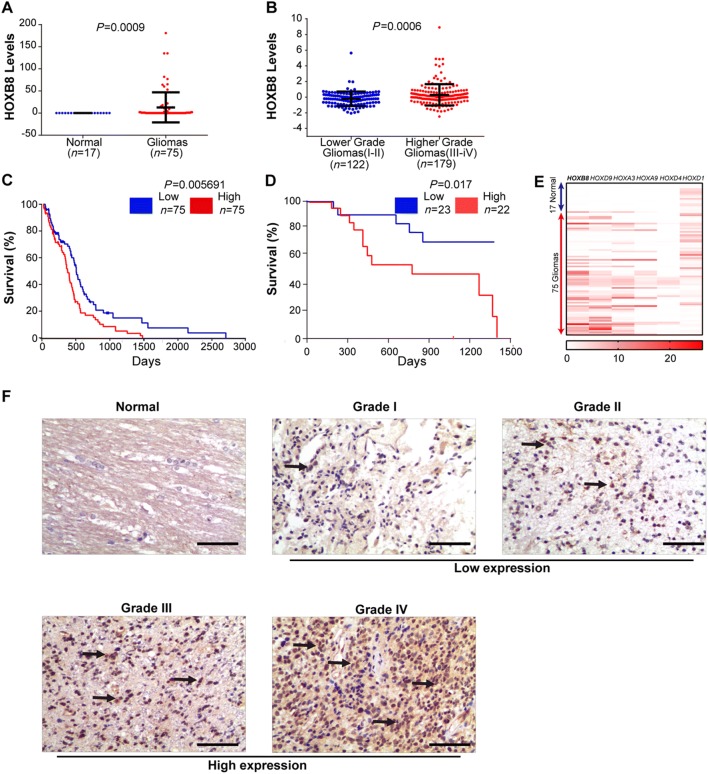
We first turned to publicly-accessible databases for evidence of involvement of HOXB8 in the pathogenesis of gliomas. We found that the expression level of HOXB8 was significantly higher in gliomas than in non-tumor tissue (Fig. 1A). To further explore the clinical significance of HOXB8 in glioma progression, we took advantage of data from the CGGA database and found a strong positive correlation between the expression level of HOXB8 and the pathological grading of tumors (Fig. 1B). These findings were supported by data from a cohort of glioma patients from the GEO database (Table S2). A strong negative correlation was found between the expression level of HOXB8 and patients’ overall survival rate (Fig. 1C), which was confirmed in our cohort of glioma patients (23 low grade and 22 high grade) (Fig. 1D). It is noteworthy that, according to data derived from the GSEA database (75 gliomas and 17 non-tumor controls), many HOX family members were strongly expressed in gliomas, including HOXB8 (Fig. 1E). We confirmed the stronger expression of HOXB8 in gliomas in our cohort of patients by immunohistochemistry staining (IHC) (Fig. 1F). Consistent with the above data-mining results, HOXB8 expression in our cohort was also correlated with pathological grading (Table 1).
Fig. 1.
HOXB8 expression in gliomas in relation to their grades. A HOXB8 expression in gliomas and non-tumor brain tissue from EBI ArrayExpress data (17 non-tumor brain tissues and 75 gliomas). ***P < 0.001. B HOXB8 expression in low-grade and high-grade gliomas from CGGA data (122 low-grade and 179 high-grade). ***P < 0.001. C Overall survival of glioma patients with high and low expression of HOXB8. Data from OncoLnc (http://www.oncolnc.org/). *P < 0.05. D Overall survival of glioma patients with high and low expression of HOXB8. Data from our cohort of patients. *P < 0.05. E Expression of HOX family members including HOXB8 in gliomas from the GSEA database (17 non-tumor controls and 75 gliomas). F Immunohistochemical analysis of HOXB8 expression in gliomas of different pathological grades. Data are presented as the mean ± SD. Scale bars, 50 μm.
Table 1.
HOXB8-related factor analysis in grade I–IV gliomas
| Variable | Total | (−) | (+) | (++) | (+++) | P value |
|---|---|---|---|---|---|---|
| Age (years) | ||||||
| > 50 | 26 | 6 | 4 | 7 | 9 | 0.432788 |
| ≤ 50 | 50 | 18 | 10 | 7 | 15 | |
| Gender | ||||||
| Male | 38 | 9 | 8 | 8 | 16 | 0.146729 |
| Female | 38 | 15 | 6 | 9 | 8 | |
| Glioma grade | ||||||
| I–II | 19 | 11 | 4 | 3 | 1 | 0.005443 |
| III–IV | 57 | 13 | 10 | 11 | 23 | |
HOXB8 Promotes Proliferation of Glioma Cells
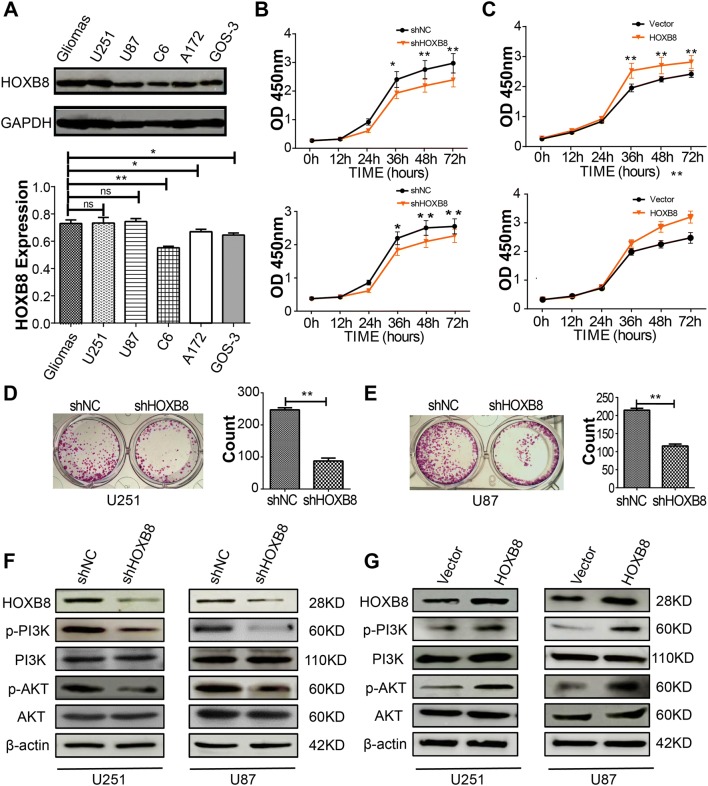
We screened several glioma cell lines with grade IV glioma tissue as a positive control. Consequently, we selected U251 and U87 glioma cells for subsequent experiments based on the abundant expression of HOXB8 in these two cell lines (Fig. 2A). After transient knockdown of HOXB8 in the glioma cells, proliferation experiments showed that the knockdown of HOXB8 reduced their ability to proliferate (Fig. S1A, B). The knockdown efficiency in these cells was verified by western blotting (Fig. S2A). Stable silencing of HOXB8 in U251 and U87 glioma cells was achieved using a lentivirus-mediated knockdown system. Silencing HOXB8 significantly reduced the proliferative capacity (Fig. 2B). Consistently, over-expression of HOXB8 promoted the proliferation of glioma cells (Fig. 2C). Silencing HOXB8 also significantly reduced the colony-forming ability (Fig. 2D, E). To explore the mechanism underlying the decreased proliferation of glioma cells following knockdown of HOXB8, we investigated the activation of the AKT pathway and found that it was significantly decreased (Fig. 2F). Conversely, overexpression of HOXB8 increased the activation of the PI3K/AKT pathway (Fig. 2G). We also examined the ERK pathway and found no notable difference (Fig. S2C).
Fig. 2.
HOXB8 promotes proliferation of glioma cells and affects activation of the AKT/PI3K pathway. A Expression of HOXB8 assessed by western blot in glioma samples. Upper panel, blots of U251, U87, C6, A172, GOS-3 cells; lower panel, statistical analysis of grayscale values. B Proliferation assays in U251 cells (upper) and U87 (lower) with stable expression of shHOXB8. C Proliferation assays in U251 (upper) and U87 (lower) cells with overexpression of HOXB8. D Colony formation assays in U251 cells (left) with stable expression of shHOXB8 and statistical results (right; 3 independent experiments). E Colony formation assays in U87 cells (left) with stable expression of shHOXB8, and statistical results (right: 3 independent experiments). F, G Western blots of PI3K and AKT, and phosphorylation of these proteins in U251 and U87 cells with stable expression of shHOXB8 (F) and overexpression of HOXB8 (G). Data are shown as the mean ± SD and were pooled from 3 independent experiments. *P < 0.05, **P < 0.01.
HOXB8 Promotes Migration of Glioma Cells
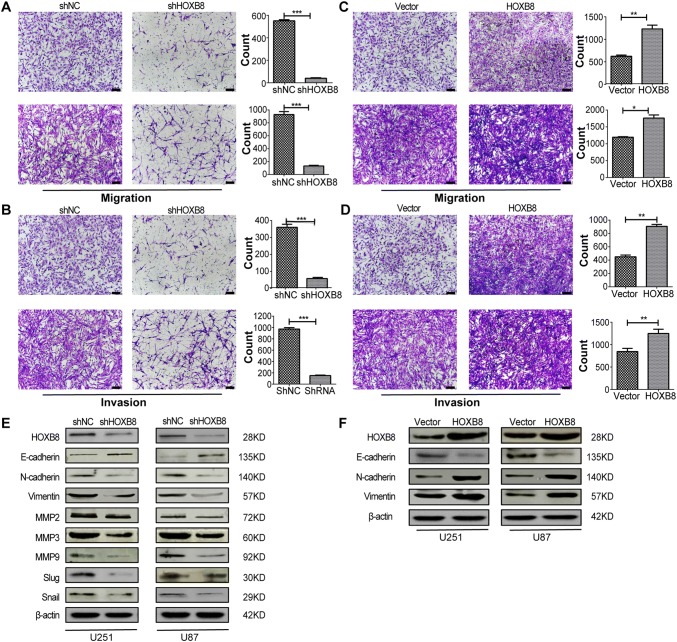
Transient silencing of HOXB8 in glioma cells reduced their migration and invasion ability (Fig. S1C, D). Consistently, stable silencing of HOXB8 produced the same results (Fig. 3A, B). Epithelial-mesenchymal transition (EMT)-related proteins were examined to reveal the relationship between HOXB8 and the EMT process. We detected upregulation of an epithelial cell marker (E-cadherin) and downregulation of mesenchymal cell markers (N-cadherin and vimentin) after transient knockdown of HOXB8 (Fig. S2A, B). Similar results were obtained in stably-transfected cells (Fig. 3E). Consistently, overexpression of HOXB8 significantly decreased the epithelial cell markers and increased the mesenchymal cell markers (Fig. 3C, D, F). These results indicated that HOXB8 is involved in the process of epithelial-to-mesenchymal transition.
Fig. 3.
HOXB8 promotes migration of glioma cells and upregulates EMT-related genes. A, B Migration (A) and invasion (B) assays in U251 (upper) and U87 (lower) cells with stable expression of shHOXB8. C, D Migration (C) and invasion (D) assays in U251 (upper) and U87 (lower) cells with overexpressed HOXB8. E Western blots of EMT-related proteins (E-cadherin, N-cadherin, vimentin, MMP2, MMP3, MMP9, Slug, and Snail) in U251 and U87 cells with stable expression of shHOXB8. F Western blots of EMT-related proteins E-cadherin, N-cadherin and vimentin in U251 cells with HOXB8 overexpression. Data are shown as the mean ± SD and were pooled from three independent experiments. Scale bars, 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
SAMD9 is a Potential Target of HOXB8 and Induces Proliferation, Migration, and Invasion of Glioma Cells
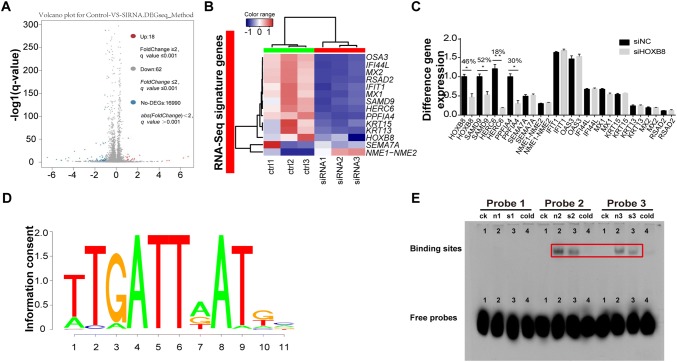
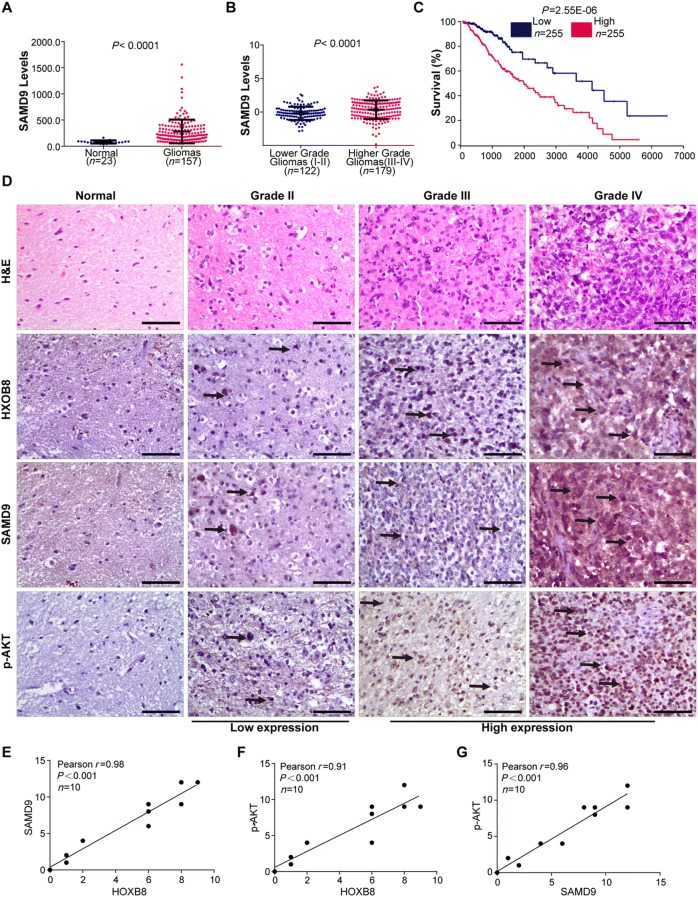
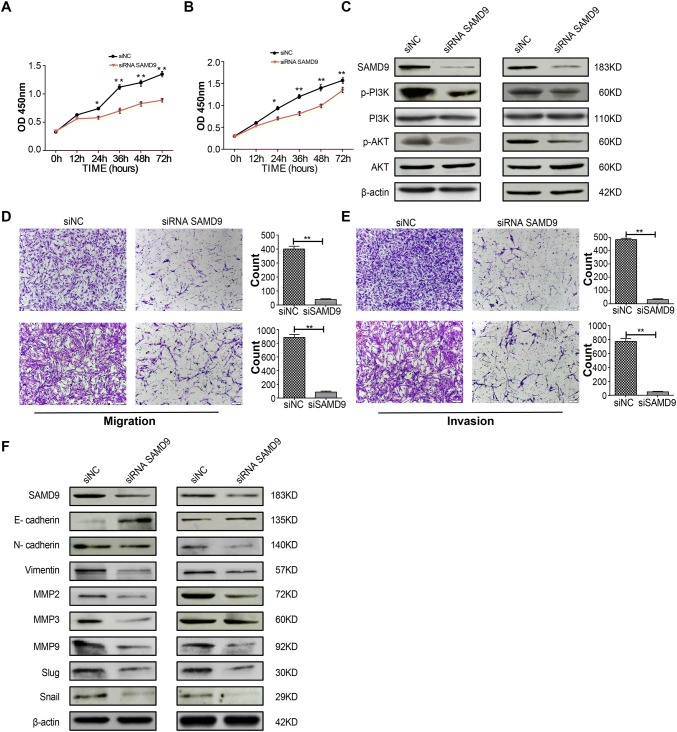
To gain better insight into the molecular mechanisms involving HOXB8 during the proliferation, migration, and invasion of glioma cells, we performed whole genome RNA sequencing in U251 cells transfected with control siRNA and siRNA against HOXB8. We found down-regulation of 62 genes and up-regulation of 18 genes among the differentially-expressed genes (Fig. 4A). We analyzed numerous differentially-expressed genes, including HOXB8, SAMD9, OSA3, IFI44L, RSAD2, IFIT1, HERC6, PPFIA4, KRT15, KRT13, SEMA7A, NME1-NME2, MX1, and MX2 (Fig. 4B). Based on the role of HOXB8 in promoting the proliferation, migration, and invasion of tumor cells, 14 genes associated with metastasis were identified from the top genes with a 20-fold change (Table S3). Three out of 14 genes were down-regulated in siRNA HOXB8-treated U251 cells by qPCR analysis (Fig. 4C). Among these genes, SAMD9 is closely associated with tumorigenesis. To test if HOXB8 binds directly to the promoter of SAMD9, we designed three probes in the SAMD9 promotor region and analyzed the possible protein motif for HOXB8 (Fig. 4D). We performed EMSA experiments and showed that HOXB8 directly binds to the promoter of SAMD9 and transactivates SAMD9 (Fig. 4E). More importantly, SAMD9 was also elevated in gliomas compared to non-tumor brain tissue (Fig. 5A). Similar to HOXB8, we found that the expression level of SAMD9 had a positive correlation with the glioma clinical pathology data (Fig. 5B) and a negative correlation with the overall survival rate in the CGGA database (Fig. 5C). These findings were further supported by data from a cohort of glioma patients from the GEO database (Table S4). Using semi-quantitative IHC analysis of 1 normal brain and 9 gliomas (Fig. 5D), we found that HOXB8 expression correlated with SAMD9 expression (Fig. 5E) and p-AKT expression (Fig. 5F); we also found that SAMD9 expression correlated with p-AKT expression (Fig. 5G). Transient knockdown of SAMD9 in glioma cells reduced their ability to proliferate (Fig. 6A, B) and decreased the activation of the AKT pathway (Fig. 6C). Silencing of SAMD9 in glioma cells reduced their migration and invasion ability (Fig. 6D, E). We revealed upregulation of an epithelial cell marker (E-cadherin) and downregulation of mesenchymal cell markers (N-cadherin and vimentin) after transient knockdown of SAMD9 (Fig. 6F). Taken together, these data suggest the working model that HOXB8/SAMD9 drives the development of glioma cells through the PI3K/AKT signaling pathway (Fig. 7).
Fig. 4.
SAMD9 is one of the mediators of HOXB8 in glioma cells. A Volcano plot depicting a log transformation plot of the fold-change of different genes by whole genome RNA sequencing in U251 cells transfected with control siRNA and HOXB8 siRNA. B Top differentially-expressed genes in RNA-seq results. C Top differentially-expressed genes in RNA sequencing confirmed in U251 cells by qPCR. *P < 0.05, **P < 0.01. DDe novo motifs recovered by the MD module from the HOXB8 binding sites with a RefSeq gene within 2 kb. E EMSA of biotinylated SMAD9 promoter probes in nuclear extracts from siNC-treated and HOXB8 siRNA-treated U251 cells. Biotinylated probes were detected with HRP-streptavidin. “ck” and “cold” indicate competitor and unlabeled probe negative controls, respectively. Data are shown as the mean ± SD and were pooled from three independent experiments.
Fig. 5.
SAMD9 expression in glioma cells and its correlation with HOXB8. A SAMD9 expression in gliomas from GEO data (23 non-tumor brain tissue samples and 157 gliomas). ***P < 0.001. B SAMD9 expression levels in higher and lower grade gliomas from CGGA data (122 low-grade and 179 high-grade). ***P < 0.001. C Overall survival of glioma patients with high and low expression of SAMD9. Data from OncoLnc (http://www.oncolnc.org/). ***P < 0.001. D One normal brain sample and 9 gliomas stained with hematoxylin and eosin (H&E) and for HOXB8, SAMD9, and p-AKT. The arrows show the positive protein expression. E Correlation analysis of HOXB8 and SAMD9 immunohistochemical scores. X-axis, HOXB8 staining score; y-axis, SAMD9 staining score; correlation coefficient r = 0.96; ***P < 0.001. F Correlation analysis of HOXB8 and p-AKT immunohistochemical scores. X-axis, HOXB8 staining score; y-axis, p-AKT staining score; correlation coefficient r = 0.91; ***P < 0.001. G Correlation analysis of SAMD9 and p-AKT immunohistochemical scores. X-axis, SAMD9 staining score; y-axis, p-AKT staining score; correlation coefficient r = 0.96; ***P < 0.001. Data are shown as the mean ± SD. Scale bars, 50 μm.
Fig. 6.
SAMD9 mediates the proliferation, migration, and invasion of glioma cells and affects the activation of the AKT/PI3K pathway and expression of EMT-related genes. A, B Transient silencing of SAMD9 in U251 cells (A) and U87 cells (B) decreased the rate of proliferation. C Western blots of phosphorylation of PI3K and AKT in U251 and U87 cells with transiently silenced SAMD9. D Transient silencing of HOXB8 in U251 (upper) and U87 (lower) cells decreased the rate of migration. E Transient silencing of SAMD9 in U251 (upper) and U87 (lower) cells decreased the rate of invasion. F Western blots of EMT-related proteins (E-cadherin, N-cadherin, vimentin, MMP2, MMP3, MMP9, Slug, and Snail) in U251 and U87 cells with transiently silenced SAMD9. Data are shown as the mean ± SD and were pooled from three independent experiments. Scale bars, 100 μm. *P < 0.05, **P < 0.01.
Fig. 7.
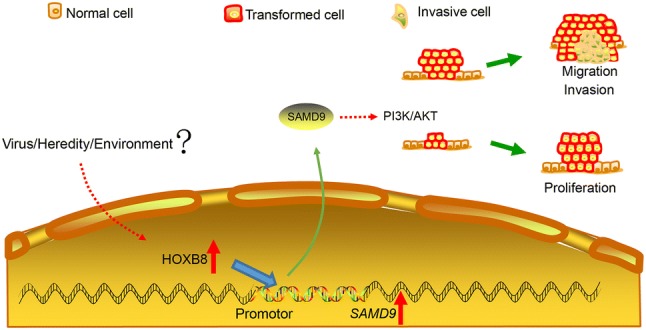
Working model. Schematic showing the speculation that HOXB8/SAMD9 drives the proliferation, migration, and invasion of glioma cells and involves the PI3K/AKT signaling pathway.
Discussion
Increased expression of HOXB8 is associated with a wide variety of cancers. Higher expression of HOXB8 promotes gastric cancer cell migration, invasion, and the EMT, possibly through interacting with ZEB2 [21]. HOXB8 knockdown inhibits the proliferation and invasion of colon cancer cells in vitro as well as carcinogenesis and metastasis in vivo. HOXB8 also induces the EMT in colon cancer cells, and this is characterized by the down-regulation of AKT phosphorylation [26]. Higher HOXB8 expression in effusions is associated with shorter overall and progression-free survival in serous ovarian carcinoma [22]. Consistent with these findings, we found higher expression of HOXB8 in human gliomas than in non-tumor tissue (Fig. 1A). Furthermore, we found that the expression level of HOXB8 was strongly correlated with tumor pathological grading (Fig. 1B) and negatively correlated with the overall survival rate (Fig. 1C, D). We also demonstrated that strong expression of HOXB8 was critical for the proliferation (Fig. 2B), migration, and invasion (Fig. 3A, B) of glioblastoma cells and that HOXB8 might also induce the EMT in glioblastoma cells. The EMT of tumor cells is a major step towards invasion and metastasis. After knockdown of HOXB8, the epithelial marker E-cadherin was up-regulated, while Snail was down-regulated (Fig. 3E). Snail plays an important role in tumor metastasis by transcriptionally regulating a variety of factors involved in the EMT process. Vimentin was also down-regulated (Fig. 3E). Vimentin is an intermediate microfilament protein specifically expressed in mesenchymal cells and its presence indicates that cancer cells have changed from an epithelial phenotype to a mesenchymal phenotype. Nevertheless, whether the dysregulation of EMT-related proteins is directly responsible for the increased proliferation, migration, and invasion of glioma cells by HOXB8 overexpression needs further investigation.
Activation of the PI3K-AKT pathway plays an important role in the malignant proliferation of tumor cells. This pathway is considered to be one of the most basic survival signaling pathways in tumors. It is abnormally activated in many tumor types including glioma and is closely associated with the aggressiveness of glial tumors. PI3K/AKT further activates its downstream target mTOR through the TSC1/2 complex. In head and neck tumors, PI3K/AKT activation up-regulates the expression of MMP9, which degrades E-cadherin on the cell surface and promotes cell invasion and migration. We identified SAMD9 as a downstream target gene of HOXB8 in glioma cells (Fig. 4E). SAMD9 is involved in the control of cell proliferation and functions as a tumor suppressor in some cancers. Deleterious mutations in the SAMD9 gene are known to cause normophosphatemic familial tumoral carcinosis, a rare autosomal recessive disease [27]. In malignant glioma, SAMD9 is involved in death signaling in response to inactivated Sendai virus particles (HVJ-E) or type I interferon treatment. When SAMD9 expression is knocked down by RNA interference, the apoptotic cell death induced by HVJ-E is blocked in U251 cells [28]. Studies have shown that SAMD9 is closely associated with invasive fibromatosis [29]. Overexpression of SAMD9 inhibits the proliferation and invasion of non-small-cell lung carcinoma cells [30]. SAMD9 is significantly up-regulated in highly metastatic esophageal carcinoma, suggesting that it plays an important role in tumor metastasis [31]. Bioinformatics analysis found that SAMD9 was higher in gliomas than normal brain tissue (Fig. 5A), and its expression was positively correlated with glioma pathological grading (Fig. 5B). SAMD9 was highly expressed in gliomas and positively correlated with pathological grades from the GEO database (Table S4). Subsequent overexpression and knockdown assays showed a similar phenotype for HOXB8. However, whether dysregulated PI3K/AKT activation plays a direct role downstream of both HOXB8 and SAMD9 in the proliferation, migration, and invasion of glioma cells is not clear. In addition, further investigation is needed to fully establish SAMD9 is a direct effector gene for the HOXB8-mediated proliferation, migration, and invasion of glioma cells. Nevertheless, similar to other neural disorders [32], there is a critical need for biomarkers for diagnosis and prognosis in glioma treatment. Currently, one clinical trial is recruiting patients to evaluate and identify effective prognostic biomarkers for malignant glioma [33, 34]. HOXB8 expression evaluation in initial diagnostic biopsy sections could provide a valuable indicator in terms of prognosis.
In conclusion, our study identified HOXB8 as a crucial contributor to the aggressiveness of GBM. With further detailed mechanistic investigation, HOXB8 may prove to be an interesting prognostic biomarker and therapeutic target, blockade of which increases the sensitivity of glioma cells to current treatment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to give special thanks to Dr. Wangshu Xu for her helpful editing improvements and Dr. Hongqiang Du for his contributions to trial management. This work was supported by the National Natural Science Foundation of China (31571298).
Conflict of interest
The authors declare no competing interests.
Contributor Information
Youtao Yu, Email: yuyoutao@126.com.
Haifeng Yang, Email: 429810560@qq.com.
Hongshan Zhao, Email: hongshan@bjmu.edu.cn.
References
- 1.Ostrom QT, Gittleman H, Stetson L, Virk SM, Barnholtz-Sloan JS. Epidemiology of gliomas. Cancer Treat Res. 2015;163:1–14. doi: 10.1007/978-3-319-12048-5_1. [DOI] [PubMed] [Google Scholar]
- 2.Schwab DE, Lepski G, Borchers C, Trautmann K, Paulsen F, Schittenhelm J. Immunohistochemical comparative analysis of GFAP, MAP-2, NOGO-A, OLIG-2 and WT-1 expression in WHO 2016 classified neuroepithelial tumours and their prognostic value. Pathol Res Pract. 2018;214:15–24. doi: 10.1016/j.prp.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 3.DeWeerdt S. The genomics of brain cancer. Nature. 2018;561:S54–S55. doi: 10.1038/d41586-018-06711-8. [DOI] [PubMed] [Google Scholar]
- 4.Xie H, Wang M, Bonaldo Mde F, Rajaram V, Stellpflug W, Smith C, et al. Epigenomic analysis of Alu repeats in human ependymomas. Proc Natl Acad Sci U S A. 2010;107:6952–6957. doi: 10.1073/pnas.0913836107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 2017, 355. [DOI] [PMC free article] [PubMed]
- 6.Perry A, Wesseling P. Histologic classification of gliomas. Handb Clin Neurol. 2016;134:71–95. doi: 10.1016/B978-0-12-802997-8.00005-0. [DOI] [PubMed] [Google Scholar]
- 7.Birk HS, Han SJ, Butowski NA. Treatment options for recurrent high-grade gliomas. CNS Oncol. 2017;6:61–70. doi: 10.2217/cns-2016-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sehmer EA, Hall GJ, Greenberg DC, O’Hara C, Wallingford SC, Wright KA, et al. Incidence of glioma in a northwestern region of England, 2006-2010. Neuro Oncol. 2014;16:971–974. doi: 10.1093/neuonc/not301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu VM, Welby JP, Mahajan A, Laack NN, Daniels DJ. Reirradiation for diffuse intrinsic pontine glioma: a systematic review and meta-analysis. Childs Nerv Syst. 2019;35:739–746. doi: 10.1007/s00381-019-04118-y. [DOI] [PubMed] [Google Scholar]
- 11.Kieran MW, Goumnerova L, Manley P, Chi SN, Marcus KJ, Manzanera AG, et al. Phase I study of gene-mediated cytotoxic immunotherapy with AdV-tk as adjuvant to surgery and radiation for pediatric malignant glioma and recurrent ependymoma. Neuro Oncol. 2019;21:537–546. doi: 10.1093/neuonc/noy202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 13.Quinonez SC, Innis JW. Human HOX gene disorders. Mol Genet Metab. 2014;111:4–15. doi: 10.1016/j.ymgme.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Kutejova E, Engist B, Self M, Oliver G, Kirilenko P, Bobola N. Six2 functions redundantly immediately downstream of Hoxa2. Development. 2008;135:1463–1470. doi: 10.1242/dev.017624. [DOI] [PubMed] [Google Scholar]
- 15.Domsch K, Papagiannouli F, Lohmann I. The HOX-apoptosis regulatory interplay in development and disease. Curr Top Dev Biol. 2015;114:121–158. doi: 10.1016/bs.ctdb.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 16.Bakalenko NI, Novikova EL, Nesterenko AY, Kulakova MA. Hox gene expression during postlarval development of the polychaete Alitta virens. Evodevo. 2013;4:13. doi: 10.1186/2041-9139-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pastori C, Kapranov P, Penas C, Peschansky V, Volmar CH, Sarkaria JN, et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci U S A. 2015;112:8326–8331. doi: 10.1073/pnas.1424220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutlet B, Theys N, Coste C, Ahn MT, Doshishti-Agolli K, Lizen B, et al. Systematic expression analysis of Hox genes at adulthood reveals novel patterns in the central nervous system. Brain Struct Funct. 2016;221:1223–1243. doi: 10.1007/s00429-014-0965-8. [DOI] [PubMed] [Google Scholar]
- 19.Vider BZ, Zimber A, Hirsch D, Estlein D, Chastre E, Prevot S, et al. Human colorectal carcinogenesis is associated with deregulation of homeobox gene expression. Biochem Biophys Res Commun. 1997;232:742–748. doi: 10.1006/bbrc.1997.6364. [DOI] [PubMed] [Google Scholar]
- 20.Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112:532–538. doi: 10.1038/bjc.2014.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding WJ, Zhou M, Chen MM, Qu CY. HOXB8 promotes tumor metastasis and the epithelial-mesenchymal transition via ZEB2 targets in gastric cancer. J Cancer Res Clin Oncol. 2017;143:385–397. doi: 10.1007/s00432-016-2283-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stavnes HT, Holth A, Don T, Kaern J, Vaksman O, Reich R, et al. HOXB8 expression in ovarian serous carcinoma effusions is associated with shorter survival. Gynecol Oncol. 2013;129:358–363. doi: 10.1016/j.ygyno.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 23.Kelppe J, Thoren H, Ristimaki A, Haglund C, Sorsa T, Hagstrom J. BRAF V600E expression in ameloblastomas-A 36-patient cohort from Helsinki University Hospital. Oral Dis. 2019;25:1169–1174. doi: 10.1111/odi.13072. [DOI] [PubMed] [Google Scholar]
- 24.Nishida H, Kashima K, Yano S, Daa T, Arakane M, Oyama Y, et al. A biotin tagging immunoelectron microscopy for paraffin-embedded sections. Appl Immunohistochem Mol Morphol. 2019 doi: 10.1097/PAI.0000000000000735. [DOI] [PubMed] [Google Scholar]
- 25.Sousa DA, Silva K, Cascon CM, Silva FBF, Mello MFV, Leite JDS, et al. Epidermal growth factor receptor 2 immunoexpression in gastric cells of domestic cats with H. heilmannii infection. Acta Histochem. 2019;121:413–418. doi: 10.1016/j.acthis.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, et al. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089–2096. doi: 10.3748/wjg.15.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hershkovitz D, Gross Y, Nahum S, Yehezkel S, Sarig O, Uitto J, et al. Functional characterization of SAMD9, a protein deficient in normophosphatemic familial tumoral calcinosis. J Invest Dermatol. 2011;131:662–669. doi: 10.1038/jid.2010.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka M, Shimbo T, Kikuchi Y, Matsuda M, Kaneda Y. Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int J Cancer. 2010;126:1982–1991. doi: 10.1002/ijc.24965. [DOI] [PubMed] [Google Scholar]
- 29.Li CF, MacDonald JR, Wei RY, Ray J, Lau K, Kandel C, et al. Human sterile alpha motif domain 9, a novel gene identified as down-regulated in aggressive fibromatosis, is absent in the mouse. BMC Genomics. 2007;8:92. doi: 10.1186/1471-2164-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Q, Yu T, Ren YY, Gong T, Zhong DS. Overexpression of SAMD9 suppresses tumorigenesis and progression during non small cell lung cancer. Biochem Biophys Res Commun. 2014;454:157–161. doi: 10.1016/j.bbrc.2014.10.054. [DOI] [PubMed] [Google Scholar]
- 31.Tang S, Zheng X, He L, Qiao L, Jing D, Ni Z, et al. Significance of SAMD9 expression in esophageal squamous cell carcinoma. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2014;30:411–413. [PubMed] [Google Scholar]
- 32.Yan X, Mai L, Lin C, He W, Yin G, Yu J, et al. CSF-based analysis for identification of potential serum biomarkers of neural tube defects. Neurosci Bull. 2017;33:436–444. doi: 10.1007/s12264-017-0154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paech D, Dreher C, Regnery S, Meissner JE, Goerke S, Windschuh J, et al. Relaxation-compensated amide proton transfer (APT) MRI signal intensity is associated with survival and progression in high-grade glioma patients. Eur Radiol. 2019;29:4957–4967. doi: 10.1007/s00330-019-06066-2. [DOI] [PubMed] [Google Scholar]
- 34.Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.