Increasing multidrug resistance, including vancomycin resistance, in enterococci is a major concern in clinical settings. Horizontal gene transfer, such as via plasmids, has been shown to play a crucial role in the acquisition of vancomycin resistance. Among vancomycin resistance types, the VanA type is one of the most prevalent, and outbreaks caused by VanA-type vancomycin-resistant enterococci (VRE) have occurred worldwide. Here, we describe an enterococcal linear plasmid responsible for multispecies local spread of VanA-type VRE. Such a study is important because although hospital outbreaks caused by mixed enterococcal species have been reported, this particular spread indicates plasmid transfer across species. This is a crucial finding because the high risk for such a spread of antimicrobial resistance calls for regular monitoring and surveillance.
KEYWORDS: vancomycin-resistant enterococci, conjugative linear plasmid, local spread, interspecies transmission
ABSTRACT
Vancomycin-resistant enterococci pose a threat in the clinical setting and have been linked to hospital outbreaks worldwide. In 2017, a local spread of VanA-type vancomycin-resistant enterococci (VRE) occurred in Japan, and 25 enterococcal isolates, including 14 Enterococcus faecium, 8 E. raffinosus, and 3 E. casseliflavus isolates, were identified from four inpatients. Molecular analysis of the multispecies of VanA-type VRE revealed the involvement of both the dissemination of clonally related VRE strains between patients and the horizontal transfer of plasmids harboring the vanA gene cluster between Enterococcus spp. Pulsed-field gel electrophoresis showed that the plasmid DNAs without S1 nuclease treatment were able to migrate into the gel, suggesting that the topology of the plasmid was linear. Whole-genome sequencing revealed that this plasmid, designated pELF2, was 108,102 bp long and encoded multiple antimicrobial resistance genes, including ermA and ant(9). The amino acid sequences of putative replication- and transfer-related genes were highly conserved between pELF2 and pELF1, the latter of which was the first identified enterococcal conjugative linear plasmid. On comparing the genomic structure, pELF2 showed the presence of a backbone similar to that of pELF1, especially with respect to the nucleotide sequences of both terminal ends, indicating a hybrid-type linear plasmid, possessing two different terminal structures. pELF2 possessed a broad host range and high conjugation frequencies for enterococci. The easy transfer of pELF2 to different Enterococcus spp. in vitro might explain this local spread of multiple species, highlighting the clinical threat from the spread of antimicrobial resistance by an enterococcal linear plasmid.
IMPORTANCE Increasing multidrug resistance, including vancomycin resistance, in enterococci is a major concern in clinical settings. Horizontal gene transfer, such as via plasmids, has been shown to play a crucial role in the acquisition of vancomycin resistance. Among vancomycin resistance types, the VanA type is one of the most prevalent, and outbreaks caused by VanA-type vancomycin-resistant enterococci (VRE) have occurred worldwide. Here, we describe an enterococcal linear plasmid responsible for multispecies local spread of VanA-type VRE. Such a study is important because although hospital outbreaks caused by mixed enterococcal species have been reported, this particular spread indicates plasmid transfer across species. This is a crucial finding because the high risk for such a spread of antimicrobial resistance calls for regular monitoring and surveillance.
INTRODUCTION
Enterococci are normal inhabitants of the human intestinal tract and are typical nosocomial pathogens, with more than 40 species having been described thus far (1). Among the various Enterococcus spp., clinical infections are mainly caused by Enterococcus faecalis and E. faecium (2). In adult health care-associated infections in the United States, E. faecalis and E. faecium are the fifth and eighth most commonly reported pathogens, respectively (3). Other Enterococcus spp., including E. raffinosus and E. casseliflavus, are considered low-virulence organisms; however, with regard to severe enterococcal infections, the incidence of non-faecium and non-faecalis enterococcal bacteremia has been gradually increasing (2, 4). These clinical isolates have been reported to be resistant to clinical drugs, including penicillin, aminoglycosides, and glycopeptides (5). The Japan Nosocomial Infections Surveillance (JANIS) program showed that the rates of resistance to penicillin G, ampicillin, erythromycin, and levofloxacin were more than 80% among E. faecium isolates (https://janis.mhlw.go.jp/english/index.asp). Vancomycin has been among the antibiotic drugs most commonly used against severe Gram-positive bacterial infections (6, 7). For over 30 years after the clinical use of vancomycin was approved, vancomycin resistance was not a concern; however, since the first isolation of vancomycin-resistant enterococci (VRE), the number of VRE strains has been increasing (8–10). This is alarming because of the limited treatment options for VRE infections. As of 2018, the proportion of VRE isolates identified in Japan was reported to be less than 1% (JANIS; https://janis.mhlw.go.jp/english/index.asp); however, sporadic outbreaks have been occurring every year.
Most VRE outbreaks worldwide have been caused by E. faecium and E. faecalis and occurred because of the dissemination of clonally related VRE strains between patients or the horizontal transfer of glycopeptide resistance genes mediated by mobile genetic elements, such as transposons or plasmids, between Enterococcus spp. (5, 11–13). Of these, plasmids are considered to have the more significant impact because they are more amenable to intraspecies or interspecies transmission with Enterococcus spp. (14). In general, interspecies transfer of plasmids of Enterococcus spp. is considered to be much less frequent than intraspecies transfer (15). Recently, we discovered a transferrable linear plasmid harboring the vanA and vanM gene clusters from E. faecium in Japan (16). This plasmid conferring vancomycin resistance was reported to show interspecies transferability in enterococci.
In 2017, local spread of VanA-type VRE strains occurred in a community hospital in Japan. Here, we describe this VanA-type VRE spread and attempt to identify the plasmid underlying this event.
RESULTS AND DISCUSSION
Description of the local spread of VanA-type VRE and patient characteristics.
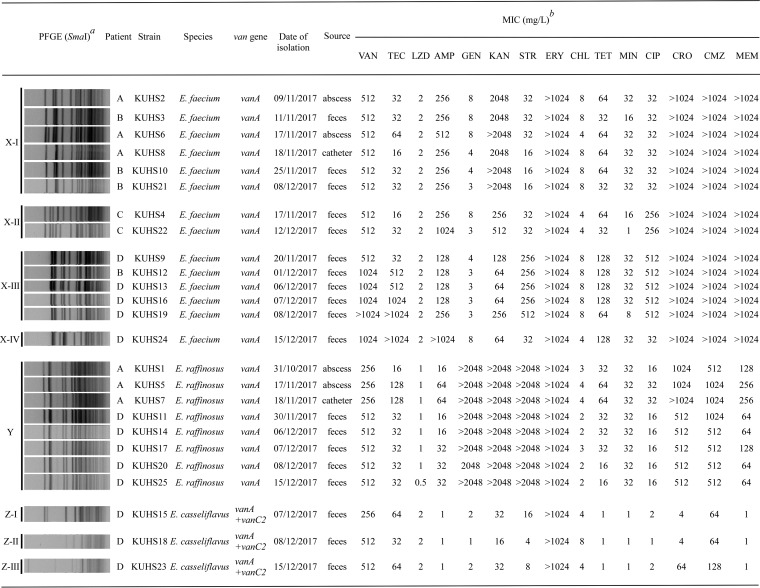
Index patient A was admitted to the hospital for the treatment of pneumonia. On day 31 of admission, a bloodstream infection with Staphylococcus caprae occurred for which vancomycin was administered intravenously for 18 days. The first vancomycin-resistant E. raffinosus isolate, designated KUHS1, was recovered from a sacral abscess on day 54 (see Fig. S1 in the supplemental material). Vancomycin-resistant E. faecium (KUHS2) was isolated from the same sacral abscess after a further 9 days; vancomycin-resistant E. faecium (KUHS6) and E. raffinosus (KUHS5) were also isolated after a further 8 days. In addition, vancomycin-resistant E. raffinosus (KUHS7) and E. faecium (KUHS8) were isolated from the central venous catheter. During this period, all patients present in the same room were screened using rectal swabs. The screening results revealed that among six patients, three (patients B to D) carried the vancomycin-resistant E. faecium strain in their feces. In the case of patient D, the presence of vancomycin-resistant E. raffinosus and E. casseliflavus was accompanied by that of vancomycin-resistant E. faecium. Regardless of frequent screening tests, vancomycin-resistant E. casseliflavus was isolated only from patient D. Unlike patient A, who exhibited the invasive VRE infection, no clinical signs of infection were detected in patients B to D, indicating that they were carriers. In total, 25 VRE isolates (14 E. faecium, 8 E. raffinosus, and 3 E. casseliflavus) were collected from four inpatients during a period of 2 months. All these isolates were determined to harbor the vanA gene by the use of a multiplex PCR assay (17). Data from JANIS revealed that the number of isolates of VRE in the prefecture, where the hospital is located, was low (vancomycin-resistant E. faecium, 1.7% [n = 75/4,420] in 2017; vancomycin-resistant E. faecalis, 0.0% [n = 3/9,499] in the same year). Furthermore, VRE isolates had not been identified from other inpatients for over 5 years before the isolation of KUHS1 in this hospital. These findings suggested the local spread of multispecies of VRE in the hospital during a short period of time. The MICs of antibiotics are shown in Fig. 1. Analyses of these MICs revealed that all the isolates exhibited high-level resistance to vancomycin. In addition, all of them showed resistance to erythromycin.
FIG 1.
Bacterial strains used in this study and their pulsed-field gel electrophoresis (PFGE) patterns and drug susceptibilities. The superscript italic “a” indicates results of PFGE analysis of SmaI-digested DNA isolated from the locally spread strains. Pulse time varied from 5.3 to 34.9 s during the 20.0 h of electrophoresis. The superscript italic “b” indicates the following abbreviations and definitions: VAN, vancomycin; TEC, teicoplanin; LZD, linezolid; AMP, ampicillin; GEN, gentamicin; KAN, kanamycin; STR, streptomycin; ERY, erythromycin; CHL, chloramphenicol; TET, tetracycline; MIN, minocycline; CIP, ciprofloxacin; CRO, ceftriaxone; CMZ, cefmetazole; MEM, meropenem. To determine the MICs, E. raffinosus strains were grown for 48 h because their growth rate was low.
Time course of isolation of multispecies vancomycin-resistant enterococci (VREs) from four patients. Dates of isolation of multispecies VREs from patients A to D are shown. Characters by themselves represent Enterococcus faecium isolates, characters on gray background represent Enterococcus raffinosus isolates, and characters in boxes represent Enterococcus casseliflavus isolates. Characters in brackets represent SmaI-PFGE patterns. N.D., not detected. Download FIG S1, TIF file, 0.05 MB (48.6KB, tif) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pulsed-field gel electrophoresis analyses of multispecies of VanA-type VRE.
To analyze the genetic correlations between these isolates, pulsed-field gel electrophoresis (PFGE) was performed (Fig. 1; see also Fig. S2) (16). On the basis of the patterns of SmaI-digested chromosomal DNA, isolates were categorized into three major groups. Accordingly, 14 E. faecium isolates were shown to belong to group X, which was divided into four subgroups (four subtypes, including X-I to X-IV). All E. raffinosus isolates belonged to the same group, the Y group, and three E. casseliflavus isolates belonged to group Z, which was divided into three subgroups (subtypes I to III). According to a guideline proposed previously by Tenover (18), the PFGE results showed that some of these isolates were indistinguishable, suggesting the occurrence of clonal dissemination of the VRE strains between the different patients. For example, regarding E. raffinosus in group Y, clonal isolates were identified in the comparisons between patients A and D. In addition, among the E. faecium strains in group X-I and group X-III, clonal VRE isolates were detected between patients A and B and patients B and D, respectively. However, the coexistence of isolates showing different PFGE patterns was also determined among E. faecium and E. casseliflavus isolates. For patient D, the number of isolated species of VRE increased over time during that short period (Fig. S1). Therefore, these findings implicating the involvement of genetically unrelated isolates indicated the horizontal transfer of glycopeptide resistance genes.
Pulse-field gel electrophoresis (PFGE) analysis of SmaI-digested DNA of the locally spread strains. PFGE analysis of SmaI-digested DNA isolated from the strains was performed. Pulse times ranged from 5.3 to 34.9 s during the 20.0 h of electrophoresis. Download FIG S2, TIF file, 0.2 MB (197.6KB, tif) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Molecular analyses of the plasmid carrying the vanA gene cluster.
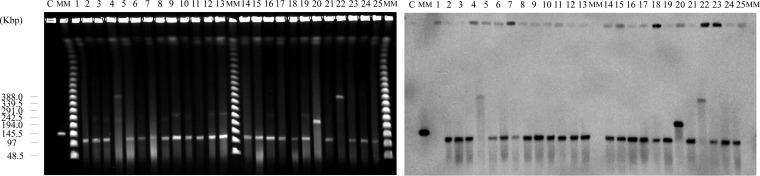
Because clonal dissemination of VRE strains alone cannot fully explain the PFGE results, we proceeded to confirm the plasmid content of these isolates. According to the results of PFGE performed with S1 nuclease as previously described (16), all these isolates carried several plasmids (Fig. S3). In particular, 22 of 25 isolates harbored an ∼110-kb plasmid, with the exceptions being KUHS4, KUHS20, and KUHS22. To confirm the plasmid topology, PFGE without S1 nuclease was performed (Fig. 2). The levels of electrophoretic mobility of these plasmids seen with or without S1 nuclease treatment were not different, indicating that they corresponded in a linear manner (Fig. 2; see also Fig. S3) (19). In contrast to the 22 isolates that seemed to carry the same linear plasmid, the size of each plasmid of KUHS4, KUHS20, and KUHS22 was observed to be larger and not consistent with that of each of the others; however, these plasmids also migrated into the PFGE gel without S1 nuclease treatment, suggesting that their topology was also linear (Fig. 2). Southern blotting using a vanA gene probe revealed that the vanA gene cluster was located on these linear plasmids (Fig. 2) (20), indicating that linear plasmid-mediated horizontal transfer had occurred between Enterococcus spp.
FIG 2.
Pulsed-field gel electrophoresis (PFGE) analysis of S1 nuclease-untreated DNA and Southern blotting of the locally spread strains. PFGE analysis of S1 nuclease-untreated DNA (left) and Southern hybridization with a vanA gene probe (right). Lanes: C, AA708, a control strain harboring the linear plasmid pELF1; MM, Lambda Ladder PFG Marker (New England BioLabs); 1, KUHS1; 2, KUHS2; 3, KUHS3; 4, KUHS4; 5, KUHS5; 6, KUHS6; 7, KUHS7; 8, KUHS8; 9, KUHS9; 10, KUHS10; 11, KUHS11; 12, KUHS12; 13, KUHS13; 14, KUHS14; 15, KUHS15; 16, KUHS16; 17, KUHS17; 18, KUHS18; 19, KUHS19; 20, KUHS20; 21, KUHS21; 22, KUHS22; 23, KUHS23; 24, KUHS24; 25, KUHS25.
Pulse-field gel electrophoresis (PFGE) analysis of S1 nuclease-treated DNA of the locally spread strains. PFGE analysis of S1 nuclease-treated DNA isolated from the strains was performed. Pulse times ranged from 5.3 to 66 s during the 19.5 h of electrophoresis. Lanes: MM, Lambda ladder PFG marker (New England BioLabs); 1, KUHS1; 2, KUHS2; 3, KUHS3; 4, KUHS4; 5, KUHS5; 6, KUHS6; 7, KUHS7; 8, KUHS8; 9, KUHS9; 10, KUHS10; 11, KUHS11; 12, KUHS12; 13, KUHS13; 14, KUHS14; 15, KUHS15; 16, KUHS16; 17, KUHS17; 18, KUHS18; 19, KUHS19; 20, KUHS20; 21, KUHS21; 22, KUHS22; 23, KUHS23; 24, KUHS24; 25, KUHS25; C, AA708, a control strain harboring linear plasmid pELF1. Download FIG S3, TIF file, 0.6 MB (634.1KB, tif) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Whole-genome-sequencing-based analyses of the linear plasmid.
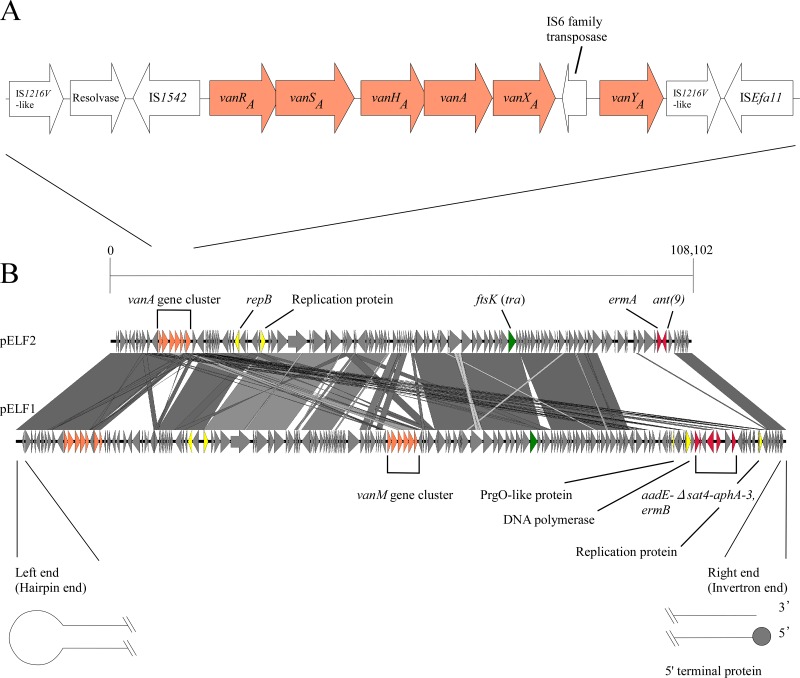
To investigate the genetic structure, whole-genome sequencing (WGS) of the E. faecium KUHS13 isolate was performed (see Table S1 in the supplemental material) using short-read Illumina MiniSeq (Illumina, San Diego, CA, USA) and long-read Nanopore MinION (Oxford Nanopore Technologies, Oxford, United Kingdom). The complete sequence of KUHS13 included the chromosome and three plasmids (Table S2). Two of the plasmids were circular and 253,148 and 6,258 bp long, respectively, whereas the third one (pELF2) was presumed to be linear and 108,102 bp long. According to the annotation, the pELF2 plasmid carried the vanA gene cluster, consistent with the results of our Southern blotting. The vanA gene cluster was carried on the Tn1546-like family. In the vanA gene cluster, a lack of the vanZ gene was identified; however, all other genes comprising the vanA gene cluster were intact (Fig. 3A). In addition, pELF2 also harbored drug resistance genes, including ermA and ant(9). Interestingly, compared to pELF1, the first identified enterococcal conjugative linear plasmid, pELF2, was found to harbor a similar genomic backbone (Fig. 3B) (21). Notably, the nucleotide sequences of both ends were shown to be highly conserved between pELF2 and pELF1, suggesting that pELF2 was a linear plasmid possessing two different terminal structures (Fig. S4) (16). The left end of pELF2 was presumed to be a hairpin structure harboring a 5′-TATA-3′ loop. In contrast, the right end contained abundant palindromic sequences forming secondary loop structures as shown by mfold analysis (Fig. S5) (22). Putative repB and ftsK genes were also carried on pELF2 (Fig. 3B). Notably, FtsK was considered to be a translocator protein involved in plasmid transfer (23, 24). The amino acid sequences of RepB and FtsK from pELF2 displayed 92.2% and 98% identities with the corresponding proteins from pELF1, respectively, indicating that the replication and transfer mechanisms of pELF2 were highly similar to those of pELF1.
FIG 3.
Schematics of the genetic structure of the vanA gene cluster (A) and pELF2 (B). The panels show the genetic structure of the vanA gene cluster carried on pELF2 (A) and a genomic comparison between pELF2 and pELF1 generated by Easyfig (B) (21). Gray lines connecting the nucleotide sequences of the two plasmids represent nucleotide identity of >67%. Arrows represent vancomycin resistance genes (orange), other drug resistance genes (red), putative replication-related genes (yellow), and putative transfer-related genes (green), respectively.
Nucleotide sequence alignment of each end of the pELF2 and pELF1 linear plasmids. The nucleotide sequence alignments of the left (A) and right (B) ends of pELF2 and pELF1 are shown. The box indicates putative hairpin sequences of the left end (5′-TATA-3′). Black boxes indicate single nucleotide variations between pELF2 and pELF1. Download FIG S4, TIF file, 0.7 MB (734.9KB, tif) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Secondary structure of the right-end nucleotide sequence of pELF2. (A) The secondary structure of the 280-nucleotide sequence of the 3′ right end of pELF2 was predicted using mfold software (ΔG = −52.31 kcal/mol) (A) (22). (B) The 280-nucleotide sequence of the 3′ right end of pELF2 is shown. Arrows and Roman numerals indicate palindromic sequences. Boxes indicate predicted loop-structure sequences of each palindromic sequence. Download FIG S5, TIF file, 0.3 MB (337.8KB, tif) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Whole-genome sequence statistics. The superscript italic “a” indicates reads that were checked using Nanoplot (ver. 1.25.0) (36). The superscript italic “b” indicates reads that were checked using FastQC (ver. 0.11.8) (37). Download Table S1, XLSX file, 0.01 MB (8.9KB, xlsx) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed information of WGS of KUHS13. The superscript italic “a” indicates that WGS was performed using Illumina Miniseq and Nanopore Minion and data were assembled using Canu (v1.8) and refined with Racon (v1.3.1.1) and Pilon (v1.20.1) (30–32). Download Table S2, XLSX file, 0.01 MB (8.7KB, xlsx) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transferability of linear plasmid pELF2 to different Enterococcus spp.
To confirm the horizontal plasmid transfer in vitro, conjugation experiments were performed. The pELF2 plasmid was easily transferred to E. faecium, E. faecalis, E. hirae, and E. casseliflavus by both filter and broth mating. The observed frequencies were 10−3 to 10−5 per donor cell in filter mating and 10−6 per donor cell in broth mating (Table S3). The transfer frequency seemed to be relatively high (15). The resulting transconjugants exhibited resistance to glycopeptides and erythromycin, consistent with the results of our WGS analysis (Fig. 3; see also Table S4). To investigate the possibility of the presence of an existing plasmid-like prophage, we prepared and collected the supernatant from a mitomycin C-treated KUHS13 culture (25). However, transfer of pELF2 could not be detected in recipient strains incubated with this supernatant.
Transfer frequency of pELF2 between KUHS13 and recipients strains. The superscript italic “a” indicates the frequencies were estimated from the CFU ratio of transconjugant strains to donor strains. The values represent means of three independent experiments with standard errors. Download Table S3, XLSX file, 0.01 MB (9KB, xlsx) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MICs of various antibiotic drugs. The superscript italic “a” indicates the following abbreviations and definitions: VAN, vancomycin; TEC, teicoplanin; LZD, linezolid; AMP, ampicillin; GEN, gentamicin; STR, streptomycin; KAN, kanamycin; ERY, erythromycin; CHL, chloramphenicol; TET, tetracycline; MIN, minocycline; CIP, ciprofloxacin; CRO, ceftriaxone; CMZ, cefmetazole; MEM, meropenem. The superscript italic “b” indicates the corresponding transconjugants that were obtained by filter mating with the donor KUHS13 strain. The superscript italic “c” indicates instances in which the strain was used as a recipient strain for the conjugation experiment. Download Table S4, XLSX file, 0.01 MB (9.5KB, xlsx) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conclusion.
Here, we described the interspecies parallel dissemination of clonal VRE strains and the linear plasmid carrying the vanA gene cluster during a local spread in a single hospital in Japan. To our knowledge, this is the first report of a local spread of VRE as a result of the transmission of a linear plasmid.
Hospital outbreaks caused by mixed enterococcal species have already been reported, but this kind of local spread seems to be rare (13, 26) and might explainable partially by the presence of plasmid-transfer barriers across species. In general, the rate of plasmid transfer between different enterococcal species was low (15); however, our PFGE results showed interspecies transfer of pELF2, consistent with results of conjugation experiments performed in vitro. Our results highlight the clinical importance of the enterococcal conjugative linear plasmid. The linear plasmids of KUHS4, KUHS20, and KUHS22 were found to be longer than the pELF2 plasmid. This finding suggests concurrent evolution of enterococcal linear plasmids during the local spread.
The clinical threat of non-faecium and non-faecalis enterococci has been often underestimated because they do not cause frequent infections (27, 28); however, non-faecium and non-faecalis enterococci have the potential to cause outbreaks of infection (29). Moreover, they might become important reservoirs of vancomycin resistance genes (13). As described above, pELF2 appears to possess a broad host range in enterococci. Our study also emphasized the importance of the surveillance of non-faecium and non-faecalis VRE. Because of the lack of information and epidemiological data concerning enterococcal linear plasmids and the absence of knowledge regarding the molecular mechanisms of plasmid transfer and plasmid replication, further intensive studies will be needed to clarify these points.
MATERIALS AND METHODS
Strains and culture media.
Bacterial strains isolated and used in this study are shown in Fig. 1. Screened stool cultures were inoculated onto BD vancomycin-resistant enterococcal selective agar (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) at 35°C. After purification was performed at least twice, these strains were subjected to further investigations. Enterococci were grown in Todd-Hewitt broth (THB; Difco, Detroit, MI, USA) at 37°C.
Antimicrobial susceptibility test.
After overnight culture, each strain was grown in Mueller-Hinton broth (MHB; Nissui, Tokyo, Japan), and the cultures were diluted 1:100 with fresh MHB. Approximately 5 × 105 cells were spotted onto a series of Mueller-Hinton agar plates (Eiken, Tokyo, Japan) containing the appropriate test drugs. The MICs of antibiotics were determined using the agar-dilution method according to the CLSI guidelines (http://clsi.org/). To determine MICs, E. raffinosus strains were grown for 48 h owing to their low growth rate.
Pulsed-field gel electrophoresis.
PFGE analysis was performed as previously described (16). Briefly, 1% agarose plugs containing embedded enterococcal strains were digested with lysozyme solution (Roche Diagnostics K.K., Minneapolis, MN, USA) (10 mg/ml) at 37°C for 6 h, followed by digestion of proteinase K (Merck Millipore, Darmstadt, Germany) solution (60 mAnson units/ml) at 50°C for 48 h. For analysis of genetic relatedness, enterococcal DNA embedded in agarose plugs was digested using SmaI (New England Biolabs, Ipswich, MA, USA) at 25°C for 12 h and then subjected to PFGE using a Chef Mapper system (Bio-Rad, Hercules, CA, USA). Pulse times ranged from 5.3 to 34.9 s during the 20.0 h of electrophoresis. For analysis of plasmid contents, enterococcal DNA embedded in agarose plugs was digested using S1 nuclease (Promega, San Luis Obispo, CA, USA) at 37°C for 20 min, and then subjected to PFGE using CHEFF MAPPER. Pulse time varied from 5.3 to 66 s during the 19.5 h of electrophoresis.
Southern blot hybridization.
To determine the topology of plasmids and the location of vanA, enterococcal DNA without S1 nuclease digestion was prepared and subjected to PFGE, as described above. Southern blot hybridization was performed with a digoxigenin-based nonradioisotope system (Boehringer GmbH, Mannheim, Germany) according to the manufacturer’s protocol.
Whole-genome sequencing.
Hybrid assembly was performed using short-read Illumina MiniSeq (Illumina) and long-read Nanopore MinION (Oxford Nanopore Technologies). WGS of strain KUHS13 was performed using a MiniSeq system (Illumina) with a High Output reagent kit (300 cycles). The library for sequencing (insertion size, 500 to 900 bp) was prepared using a Nextera XT DNA library prep kit (Illumina). On the other hand, the DNA library for Nanopore MinION was prepared using a rapid barcoding kit (SQK RBK-004; Oxford Nanopore Technologies) from total DNAs extracted using a Qiagen Genomic-tip 20/G or Gentra Puregene yeast/bacteria kit (Qiagen, Hilden, Germany) and then sequenced on a MinION flow cell (R9.4.1). WGS statistics are shown in Table S1 in the supplemental material. Raw data sets from the Nanopore MinION assay were submitted to Porechop (v0.2.3). The reads were assembled de novo using Canu (v1.8) (30). After the data were trimmed from Canu, they were polished with Racon (v1.3.1.1) and Pilon (v1.20.1) (31, 32). The nucleotide sequence of the left end of the linear plasmid (pELF2) was further checked by Sanger sequencing. DFAST and RAST were used to obtain the annotation (33, 34).
Conjugation experiments and transfer frequency.
Conjugation experiments were performed as previously described (16, 35). KUHS13 was used as the donor strain, and FA2-2 (E. faecalis), BM4105RF (E. faecium), ATCC 9790RF (E. hirae), and KT06RF (E. casseliflavus) were used as the recipient strains (Fig. 1; see also Table S3). Briefly, for filter mating, the overnight cultures of donor and recipient strains were diluted 1:50 with fresh THB and then incubated at 37°C until the end of the exponential phase. Each 100 μl of the donor or the recipient strain was mixed with 5 ml THB. The mixture was collected on a 0.22-μm-pore-size nitrocellulose membrane filter (Merck) and then incubated on a THB plate for 5 h. After the membrane filter was washed with 1 ml of THB by vortex mixing, the mating mixture was plated on a selective THB plate containing vancomycin (12 mg/liter), rifampin (25 mg/liter), and fusidic acid (25 mg/liter) and was incubated at 37°C for 24 to 48 h.
For broth mating, the preculture was prepared as described above. Fifty microliters of the donor culture and 450 μl of the recipient culture were mixed in 4.5 ml of THB. The mixture of donor and recipient cells was incubated at 37°C with gentle shaking for 3 h and then plated on a selective THB plate, as described above. Each transfer frequency was estimated from the CFU ratio of transconjugant strains to donor strains.
Accession number(s).
The data set for this study (the assembled sequence of KUHS13 strain) can be found in the DNA Data Bank of Japan database (DDBJ) (https://www.ddbj.nig.ac.jp/) under accession numbers AP022341, AP022342, AP022343, and AP022344. The Illumina reads and Nanopore reads of KUHS13 strain have been deposited in DDBJ under accession number DRA009515.
ACKNOWLEDGMENTS
This work was supported by grants from the Japanese Ministry of Health, Labor and Welfare (the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development [AMED]) (19fk0108061h0502, H30-Shokuhin-Ippan-006) and the Japanese Ministry of Education, Culture, Sport, Science and Technology [Grant-in-Aid for Early-Career Scientists (19K17922), Kiban (C) 18K0710100].
We declare that we have no competing interests.
REFERENCES
- 1.Yuen GJ, Ausubel FM. 2014. Enterococcus infection biology: lessons from invertebrate host models. J Microbiol 52:200–210. doi: 10.1007/s12275-014-4011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monticelli J, Knezevich A, Luzzati R, Di Bella S. 2018. Clinical management of non-faecium non-faecalis vancomycin-resistant enterococci infection. Focus on Enterococcus gallinarum and Enterococcus casseliflavus/flavescens. J Infect Chemother 24:237–246. doi: 10.1016/j.jiac.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Weiner-Lastinger LM, Abner S, Edwards JR, Kallen AJ, Karlsson M, Magill SS, Pollock D, See I, Soe MM, Walters MS, Dudeck MA. 2020. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: summary of data reported to the National Healthcare Safety Network, 2015–2017. Infect Control Hosp Epidemiol 41:1–18. doi: 10.1017/ice.2019.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan CK, Lai CC, Wang JY, Lin SH, Liao CH, Huang YT, Wang CY, Lin HI, Hsueh PR. 2010. Bacteremia caused by non-faecalis and non-faecium enterococcus species at a medical center in Taiwan, 2000 to 2008 J Infect 61:34–43. doi: 10.1016/j.jinf.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Werner G, Coque TM, Franz CM, Grohmann E, Hegstad K, Jensen L, van Schaik W, Weaver K. 2013. Antibiotic resistant enterococci-tales of a drug resistance gene trafficker. Int J Med Microbiol 303:360–379. doi: 10.1016/j.ijmm.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Nagarajan R. 1991. Antibacterial activities and modes of action of vancomycin and related glycopeptides. Antimicrob Agents Chemother 35:605–609. doi: 10.1128/AAC.35.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estes KS, Derendorf H. 2010. Comparison of the pharmacokinetic properties of vancomycin, linezolid, tigecyclin, and daptomycin. Eur J Med Res 15:533–543. doi: 10.1186/2047-783X-15-12-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cattoir V, Leclercq R. 2013. Twenty-five years of shared life with vancomycin-resistant enterococci: is it time to divorce? J Antimicrob Chemother 68:731–742. doi: 10.1093/jac/dks469. [DOI] [PubMed] [Google Scholar]
- 9.Leclercq R, Derlot E, Duval J, Courvalin P. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med 319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 10.Uttley AH, Collins CH, Naidoo J, George RC. 1988. Vancomycin-resistant enterococci. Lancet 331:57–58. doi: 10.1016/S0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 11.Cetinkaya Y, Falk P, Mayhall CG. 2000. Vancomycin-resistant enterococci. Clin Microbiol Rev 13:686–707. doi: 10.1128/CMR.13.4.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollenbeck BL, Rice LB. 2012. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 3:421–433. doi: 10.4161/viru.21282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawalec M, Kedzierska J, Gajda A, Sadowy E, Wegrzyn J, Naser S, Skotnicki AB, Gniadkowski M, Hryniewicz W. 2007. Hospital outbreak of vancomycin-resistant enterococci caused by a single clone of Enterococcus raffinosus and several clones of Enterococcus faecium. Clin Microbiol Infect 13:893–901. doi: 10.1111/j.1469-0691.2007.01774.x. [DOI] [PubMed] [Google Scholar]
- 14.Jovanovic M, Tosic T, Jovanovic S, Stevanovic G, Drakulovic M, Stosovic R, Zervos M. 2017. Local spread of Tn1546-like element among three species of vancomycin resistant enterococci in an intensive care unit. J Infect Dev Ctries 11:684–690. doi: 10.3855/jidc.7959. [DOI] [PubMed] [Google Scholar]
- 15.Werner G, Freitas AR, Coque TM, Sollid JE, Lester C, Hammerum AM, Garcia-Migura L, Jensen LB, Francia MV, Witte W, Willems RJ, Sundsfjord A. 2011. Host range of enterococcal vanA plasmids among Gram-positive intestinal bacteria. J Antimicrob Chemother 66:273–282. doi: 10.1093/jac/dkq455. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto Y, Taniguchi M, Uesaka K, Nomura T, Hirakawa H, Tanimoto K, Tamai K, Ruan G, Zheng B, Tomita H. 2019. Novel multidrug-resistant enterococcal mobile linear plasmid pELF1 encoding vanA and vanM gene clusters from a Japanese vancomycin-resistant enterococci isolate. Front Microbiol 10:2568. doi: 10.3389/fmicb.2019.02568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nomura T, Hashimoto Y, Kurushima J, Hirakawa H, Tanimoto K, Zheng B, Ruan G, Xue F, Liu J, Hisatsune J, Sugai M, Tomita H. 2018. New colony multiplex PCR assays for the detection and discrimination of vancomycin-resistant enterococcal species. J Microbiol Methods 145:69–72. doi: 10.1016/j.mimet.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornell CR, Marasini D, Fakhr MK. 2018. Molecular characterization of plasmids harbored by Actinomycetes isolated from the Great Salt Plains of Oklahoma using PFGE and next generation whole genome sequencing. Front Microbiol 9:2282. doi: 10.3389/fmicb.2018.02282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng B, Tomita H, Xiao YH, Wang S, Li Y, Ike Y. 2007. Molecular characterization of vancomycin-resistant enterococcus faecium isolates from mainland China. J Clin Microbiol 45:2813–2818. doi: 10.1128/JCM.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grohmann E, Muth G, Espinosa M. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev 67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosted TJ, Wang T, Horan AC. 2004. Characterization of the Streptomyces lavendulae IMRU 3455 linear plasmid pSLV45. Microbiology 150:1819–1827. doi: 10.1099/mic.0.26994-0. [DOI] [PubMed] [Google Scholar]
- 25.Mazaheri Nezhad Fard R, Barton MD, Heuzenroeder MW. 2010. Novel bacteriophages in Enterococcus spp. Curr Microbiol 60:400–406. doi: 10.1007/s00284-009-9555-z. [DOI] [PubMed] [Google Scholar]
- 26.Hsueh PR, Teng LJ, Pan HJ, Chen YC, Wang LH, Chang SC, Ho SW, Luh KT. 1999. Emergence of vancomycin-resistant enterococci at a university hospital in Taiwan: persistence of multiple species and multiple clones. Infect Control Hosp Epidemiol 20:828–833. doi: 10.1086/501592. [DOI] [PubMed] [Google Scholar]
- 27.Yin M, Jiang Y, Qian C, Wu F, Ying Y, Wu C, Li P, Ying J, Li K, Xu T, Bao Q, Sun C. 2018. Molecular characteristics and comparative genomics analysis of a clinical Enterococcus casseliflavus with a resistance plasmid. Infect Drug Resist 11:2159–2167. doi: 10.2147/IDR.S180254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tschudin Sutter S, Frei R, Dangel M, Gratwohl A, Bonten M, Widmer AF. 2010. Not all patients with vancomycin-resistant enterococci need to be isolated. Clin Infect Dis 51:678–683. doi: 10.1086/655824. [DOI] [PubMed] [Google Scholar]
- 29.Samuel J, Coutinho H, Galloway A, Rennison C, Kaufmann ME, Woodford N. 2008. Glycopeptide-resistant Enterococcus raffinosus in a haematology unit: an unusual cause of a nosocomial outbreak. J Hosp Infect 70:294–296. doi: 10.1016/j.jhin.2008.06.029. [DOI] [PubMed] [Google Scholar]
- 30.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaser R, Sovic I, Nagarajan N, Sikic M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanizawa Y, Fujisawa T, Nakamura Y. 2018. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34:1037–1039. doi: 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA III, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanimoto K, Ike Y. 2002. Analysis of the conjugal transfer system of the pheromone-independent highly transferable Enterococcus plasmid pMG1: identification of a tra gene (traA) upregulated during conjugation. J Bacteriol 184:5800–5804. doi: 10.1128/JB.184.20.5800-5804.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrews S, Krueger F, Segonds-Pichon A, Biggins L, Krueger C, Wingett S. 2012. FastQC: a quality control tool for high throughput sequence data. Babraham Institute (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time course of isolation of multispecies vancomycin-resistant enterococci (VREs) from four patients. Dates of isolation of multispecies VREs from patients A to D are shown. Characters by themselves represent Enterococcus faecium isolates, characters on gray background represent Enterococcus raffinosus isolates, and characters in boxes represent Enterococcus casseliflavus isolates. Characters in brackets represent SmaI-PFGE patterns. N.D., not detected. Download FIG S1, TIF file, 0.05 MB (48.6KB, tif) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pulse-field gel electrophoresis (PFGE) analysis of SmaI-digested DNA of the locally spread strains. PFGE analysis of SmaI-digested DNA isolated from the strains was performed. Pulse times ranged from 5.3 to 34.9 s during the 20.0 h of electrophoresis. Download FIG S2, TIF file, 0.2 MB (197.6KB, tif) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pulse-field gel electrophoresis (PFGE) analysis of S1 nuclease-treated DNA of the locally spread strains. PFGE analysis of S1 nuclease-treated DNA isolated from the strains was performed. Pulse times ranged from 5.3 to 66 s during the 19.5 h of electrophoresis. Lanes: MM, Lambda ladder PFG marker (New England BioLabs); 1, KUHS1; 2, KUHS2; 3, KUHS3; 4, KUHS4; 5, KUHS5; 6, KUHS6; 7, KUHS7; 8, KUHS8; 9, KUHS9; 10, KUHS10; 11, KUHS11; 12, KUHS12; 13, KUHS13; 14, KUHS14; 15, KUHS15; 16, KUHS16; 17, KUHS17; 18, KUHS18; 19, KUHS19; 20, KUHS20; 21, KUHS21; 22, KUHS22; 23, KUHS23; 24, KUHS24; 25, KUHS25; C, AA708, a control strain harboring linear plasmid pELF1. Download FIG S3, TIF file, 0.6 MB (634.1KB, tif) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nucleotide sequence alignment of each end of the pELF2 and pELF1 linear plasmids. The nucleotide sequence alignments of the left (A) and right (B) ends of pELF2 and pELF1 are shown. The box indicates putative hairpin sequences of the left end (5′-TATA-3′). Black boxes indicate single nucleotide variations between pELF2 and pELF1. Download FIG S4, TIF file, 0.7 MB (734.9KB, tif) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Secondary structure of the right-end nucleotide sequence of pELF2. (A) The secondary structure of the 280-nucleotide sequence of the 3′ right end of pELF2 was predicted using mfold software (ΔG = −52.31 kcal/mol) (A) (22). (B) The 280-nucleotide sequence of the 3′ right end of pELF2 is shown. Arrows and Roman numerals indicate palindromic sequences. Boxes indicate predicted loop-structure sequences of each palindromic sequence. Download FIG S5, TIF file, 0.3 MB (337.8KB, tif) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Whole-genome sequence statistics. The superscript italic “a” indicates reads that were checked using Nanoplot (ver. 1.25.0) (36). The superscript italic “b” indicates reads that were checked using FastQC (ver. 0.11.8) (37). Download Table S1, XLSX file, 0.01 MB (8.9KB, xlsx) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed information of WGS of KUHS13. The superscript italic “a” indicates that WGS was performed using Illumina Miniseq and Nanopore Minion and data were assembled using Canu (v1.8) and refined with Racon (v1.3.1.1) and Pilon (v1.20.1) (30–32). Download Table S2, XLSX file, 0.01 MB (8.7KB, xlsx) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transfer frequency of pELF2 between KUHS13 and recipients strains. The superscript italic “a” indicates the frequencies were estimated from the CFU ratio of transconjugant strains to donor strains. The values represent means of three independent experiments with standard errors. Download Table S3, XLSX file, 0.01 MB (9KB, xlsx) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MICs of various antibiotic drugs. The superscript italic “a” indicates the following abbreviations and definitions: VAN, vancomycin; TEC, teicoplanin; LZD, linezolid; AMP, ampicillin; GEN, gentamicin; STR, streptomycin; KAN, kanamycin; ERY, erythromycin; CHL, chloramphenicol; TET, tetracycline; MIN, minocycline; CIP, ciprofloxacin; CRO, ceftriaxone; CMZ, cefmetazole; MEM, meropenem. The superscript italic “b” indicates the corresponding transconjugants that were obtained by filter mating with the donor KUHS13 strain. The superscript italic “c” indicates instances in which the strain was used as a recipient strain for the conjugation experiment. Download Table S4, XLSX file, 0.01 MB (9.5KB, xlsx) .
Copyright © 2020 Hashimoto et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.