Natural products produced by Streptomyces species underpin many industrially and medically important compounds. However, the majority of the ∼30 biosynthetic pathways harbored by an average species are not expressed in the laboratory. This unrevealed biochemical diversity is believed to comprise an untapped resource for natural product drug discovery. Major roadblocks preventing the exploitation of unexpressed biosynthetic pathways are a lack of insight into their regulation and limited technology for activating their expression. Our findings reveal that the abundance of σAntA, which is the cluster-situated regulator of antimycin biosynthesis, is controlled by the ClpXP protease. These data link proteolysis to the regulation of natural product biosynthesis for the first time to our knowledge, and we anticipate that this will emerge as a major strategy by which actinobacteria regulate production of their natural products. Further study of this process will advance understanding of how expression of secondary metabolism is controlled and will aid pursuit of activating unexpressed biosynthetic pathways.
KEYWORDS: ClpXP, ECF sigma factors, Streptomyces, antimycin, proteolysis, regulation of secondary metabolism
ABSTRACT
The survival of any microbe relies on its ability to respond to environmental change. Use of extracytoplasmic function (ECF) RNA polymerase sigma (σ) factors is a major strategy enabling dynamic responses to extracellular signals. Streptomyces species harbor a large number of ECF σ factors, nearly all of which are uncharacterized, but those that have been characterized generally regulate genes required for morphological differentiation and/or response to environmental stress, except for σAntA, which regulates starter-unit biosynthesis in the production of antimycin, an anticancer compound. Unlike a canonical ECF σ factor, whose activity is regulated by a cognate anti-σ factor, σAntA is an orphan, raising intriguing questions about how its activity may be controlled. Here, we reconstituted in vitro ClpXP proteolysis of σAntA but not of a variant lacking a C-terminal di-alanine motif. Furthermore, we show that the abundance of σAntA in vivo was enhanced by removal of the ClpXP recognition sequence and that levels of the protein rose when cellular ClpXP protease activity was abolished. These data establish direct proteolysis as an alternative and, thus far, unique control strategy for an ECF RNA polymerase σ factor and expands the paradigmatic understanding of microbial signal transduction regulation.
IMPORTANCE Natural products produced by Streptomyces species underpin many industrially and medically important compounds. However, the majority of the ∼30 biosynthetic pathways harbored by an average species are not expressed in the laboratory. This unrevealed biochemical diversity is believed to comprise an untapped resource for natural product drug discovery. Major roadblocks preventing the exploitation of unexpressed biosynthetic pathways are a lack of insight into their regulation and limited technology for activating their expression. Our findings reveal that the abundance of σAntA, which is the cluster-situated regulator of antimycin biosynthesis, is controlled by the ClpXP protease. These data link proteolysis to the regulation of natural product biosynthesis for the first time to our knowledge, and we anticipate that this will emerge as a major strategy by which actinobacteria regulate production of their natural products. Further study of this process will advance understanding of how expression of secondary metabolism is controlled and will aid pursuit of activating unexpressed biosynthetic pathways.
INTRODUCTION
The survival of any organism relies on its ability to respond to environmental change. This feature is especially true of bacteria, which often live in hostile and fluctuating environments. Streptomyces bacteria thrive in soils. The success of this genus of filamentous, sporulating bacteria is linked to their complex life cycle and keen ability to sense and respond to the surroundings. Notably, a multitude of bioactive secondary or specialized metabolites are produced in response to environmental cues (1). More than half of all small-molecule therapeutics critical for human health and well-being are derived from or inspired by Streptomyces natural products (2).
Streptomyces species typically harbor a large number of biosynthetic pathways, but only a few of them are expressed under common laboratory conditions. The biochemical diversity encoded by these silent pathways is a tremendous untapped resource for discovery of new antibacterial agents and other therapeutics. All available data indicate that the production of natural products is controlled predominantly at the level of transcription. Although there are complex regulatory cascades that tightly control expression of biosynthetic genes, they are ultimately activated, repressed, or derepressed by so-called cluster-situated regulators—regulatory proteins encoded within the biosynthetic gene cluster (BGC) (3, 4). Major roadblocks preventing the exploitation of silent biosynthetic pathways are a lack of insight into their regulation and limited technology for activating their expression.
Antimycins have been known for 70 years and are the founding members of a large class of natural products widely produced by Streptomyces species (5, 6). Recently, antimycins were shown to be potent and selective inhibitors of the mitochondrial Bcl-2/Bcl-XL-related antiapoptotic proteins that are overproduced by cancer cells and confer resistance to chemotherapeutic agents whose mode of action is activation of apoptosis (7). The ∼25-kb antimycin (ant) BGC harbored by Streptomyces albus is composed of 15 genes organized into four polycistronic operons: antBA, antCDE, antGF, and antHIJKLMNO (Fig. 1) (8, 9). The regulation of the ant BGC is unusual compared to other secondary metabolites. Its expression is regulated by FscRI, a cluster-situated LuxR-family regulator of candicidin biosynthesis; FscRI activates expression of antBA and antCDE (10). Importantly, antA is a cluster-situated regulator that encodes an extracytoplasmic function (ECF) RNA polymerase σ factor (σAntA) that activates expression of the remaining operons: antGF and antHIJKLMNO (Fig. 1) (9).
FIG 1.
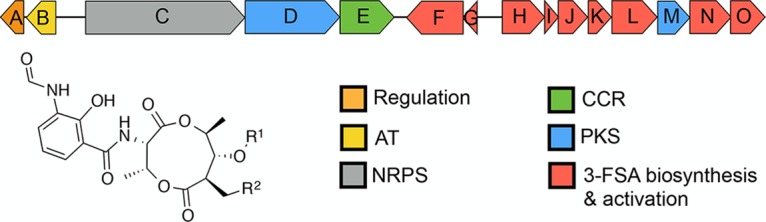
Schematic representation of the antimycin (ant) biosynthetic gene cluster. AT, acyltransferase; NRPS, nonribosomal peptide synthetase; PKS, polyketide synthase; CCR, crotonyl coenzyme A (crotonyl-CoA) carboxylase/reductase; 3-FSA, 3-formamidosalicylate. Antimycins: antimycin A1, R1= COCH(CH3)CH2CH3, R2= (CH2)4CH3; antimycin A2, R1=COCH(CH3)2, R2= (CH2)4CH3; antimycin A3, R1= COCH2CH(CH3)2, R2= (CH2)2CH3; antimycin A4, R1= COCH(CH3)2, R2= (CH2)2CH3.
σAntA, like all ECF σ factors, is similar to members of the housekeeping σ70 family but possesses only two of the four highly characteristic sigma domains: domains σ2 and σ4. The σ2 and σ4 regions of sigma factors bind the −10 and −35 promoter elements, respectively, and are sufficient for recruitment of RNA polymerase (11). Genes encoding ECF σ factors are almost always cotranscribed with their cognate anti-σ factor (12). Anti-σ factors are generally transmembrane proteins, but well-characterized examples of cytoplasmic anti-σ factors are known (13–15). Anti-σ factors selectively bind to and sequester a partner σ factor until its release is stimulated, usually by an exogenous signal (12, 16). After the σ factor is released, it recruits RNA polymerase to express a defined regulon that usually includes the σ factor–anti-σ factor operon itself, which thus establishes a positive auto-feedback loop in the presence of the inducing stimulus. Even when an ECF σ factor does have a cognate anti-σ factor, an additional mechanism of control can also exist, for instance, σRʹ in S. coelicolor is processed by the Clp-protease system (17). Streptomyces species encode a large number of ECF σ factors (>30 per strain), and the small number of these that have been characterized regulate genes required for morphological differentiation and/or responses to environmental stress and are not dedicated regulators of one biosynthetic pathway. Indeed, cluster-situated ECF σ factors have been observed previously only in the biosynthesis of lantibiotics produced by so-called rare actinomycetes. In Microbispora corallina, MibR and σMibX regulate microbisporicin biosynthesis and σMibX is controlled by the anti-σ factor MibW (18); in Planomonospora alba, PspR and σPspX regulate planosporicin production and σPspX is controlled by the anti-σ factor PspW (19). Interestingly, unlike the canonical ECF σ factors σMibX and σPspX, whose activities are controlled by cognate anti-σ factors, σAntA lacks an identifiable anti-σ factor partner and as a consequence has created curiosity about how its activity is controlled.
The Clp-protease system is essential for normal bacterial proteostasis and is best characterized in Escherichia coli (20, 21). The Clp protease is a multienzyme complex composed of a barrel-shaped peptidase, ClpP, and a regulatory enzyme, either ClpA or ClpX (or ClpC in some organisms). ClpA and ClpX (and ClpC) are AAA+-family protein unfoldases that recognize an N-terminal and/or C-terminal recognition signal (degron) and utilize ATP to unfold and translocate proteins to the peptidase chamber, where they are degraded into short peptides (22). In Streptomyces species, the peptidase is specified by two genes instead of one and is redundantly encoded (23). The primary peptidase is encoded by clpP1P2, whose corresponding proteins form a complex with ClpX or ClpA to facilitate normal proteostasis; the second peptidase is encoded by clpP3P4, but its expression occurs only when the primary system is compromised (24, 25). The best-understood degron is the SsrA tag from E. coli ( AANDENYALAA), which is added cotranslationally to polypeptides stalled on ribosomes (26, 27). The E. coli SsrA tag has been comprehensively studied, and Ala-Ala-COO-, at the C-terminal region of this motif, is essential for proteolysis by ClpXP (28). Intriguingly, the C terminus of σAntA harbors the sequence Ala-Ala-COO-, which previously led us to speculate that ClpXP may modulate its level/activity (9).
Here, we reconstituted ClpXP proteolysis of σAntA in vitro and showed that it is dependent upon the C-terminal Ala-Ala. We also found that the abundance of σAntA in vivo was higher when Ala-Ala was changed to Asp-Asp and that the abundance of σAntA was elevated in the absence of genes encoding the primary peptidase, ClpP, and its unfoldase, ClpX. These data establish direct proteolysis as an alternative, and thus far unique, control strategy of ECF RNA polymerase σ factors, expanding the paradigmatic understanding of microbial signal transduction regulation.
RESULTS AND DISCUSSION
σAntA orthologues are a new subfamily of ECF σ factors that regulate production of the antimycin biosynthetic starter unit.
Since its initial discovery 6 years ago, more than 70 ant BGCs have been identified within the members of Actinobacteria, including Actinospica, Saccharopolyspora, Streptacidiphilus, and Streptomyces (5). Each of these BGCs harbors a single regulator, σAntA (53% to 100% shared amino acid identity across all orthologues), which lacks a cognate anti-σ factor partner (5, 9). Our previous work with S. albus S4 established that σAntA orthologues comprise a new subfamily of ECF σ factors (9, 29). We demonstrated that σAntA is required for expression of antGF and antHIJKLMNO, which encode a standalone ketoreductase (AntM) and proteins required for the production/activation of the starter unit, 3-formamidosalicylate (3-FSA) (Fig. 1). We also mapped the transcriptional start sites and identified conserved promoter sequences for these operons in all ant BGCs known at the time (9). The conservation of σAntA and target promoters within ant BGCs from taxonomically diverse species suggests that σAntA-mediated regulation of these genes is direct. To verify this hypothesis, we performed chromatin immunoprecipitation sequencing (ChIP-seq) with a S. albus S4 ΔantA mutant complemented with a version of σAntA with a 3xFLAG tag at its N terminus, which we demonstrated restored antimycin production (see Fig. S2 in the supplemental material). Analysis of the resulting data revealed only one ChIP-seq peak across the whole chromosome for which the number of mapped reads was enriched for both biological replicates of ΔantA/3xFLAG-antA compared to that of the wild-type mock-immunoprecipitated control. This region corresponded to the intergenic space (297 bp) between antG and antH, which upon inspection revealed a prominent peak for the closely spaced and divergent σAntA-target promoters antGp and antHp and a second, albeit smaller peak corresponding to the 5′ end of the antH coding sequence (Fig. 2). Taken together, these data are consistent with the hypothesis that σAntA is a cluster-situated regulator that directly activates expression of genes for the production of 3-FSA during antimycin biosynthesis.
FIG 2.

3xFLAG-σAntA binds to the antGF and antHIJKLMNO promoters in vivo. Shown is a graphical representation of normalized sequence reads mapped to the intergenic region of antG-antH (shown at the bottom). The 1,242-bp genomic window depicts nucleotides 34,430 to 35,671 of contig CADY01000091.1 of the S. albus S4 genome. Each double slash denotes that genome window presented does not contain the entire antF or antH coding sequence. WT, wild type; IP, immunoprecipitation.
σAntA is degraded by the ClpXP protease in vitro.
The activities of almost all characterized ECF σ factors are modulated by a cognate anti-σ factor, which is typically a protein coencoded within the same operon. Intriguingly, σAntA lacks an anti-σ factor and is therefore an orphan, indicating that a unique mechanism is likely at work to control σAntA activity. An inspection of σAntA amino acid sequences revealed a C-terminal Ala-Ala in 67 of the 71 orthologues (Fig. S1). A C-terminal Ala-Ala is an important component of a common class of degrons for the ClpXP protease (28). This observation led us to hypothesize that the activity of σAntA might be modulated by proteolysis instead of by an anti-σ factor. To test this hypothesis, we performed in vitro proteolysis. Previous work indicated that S. albus S4 σAntA was insoluble when overproduced by E. coli, so we pursued the overproduction and purification of the orthologue from Streptomyces ambofaciens ATCC 23877, which has been experimentally demonstrated to be a producer of antimycins (30). S. ambofaciens σAntA (75% amino acid identity with S. albus S4 σAntA, including 13 of 15 amino acid residues at the C terminus) was purified as an N-terminal (His)6-SUMO-fusion protein. The (His)6-SUMO tag increases solubility and eases purification of putative substrates, without altering recognition of C-terminal degrons by ClpXP. ClpX orthologues from E. coli and S. ambofaciens possess 60% shared amino acid identity and therefore likely recognize similar substrates for degradation. Thus, ClpXP from E. coli was purified (Fig. S2) and its ability to degrade (His)6-SUMO-σAntA was assessed. Degradation of (His)6-SUMO-σAntA was apparent as early as 2.5 min after addition of ATP, and all of the sample was degraded by 15 min (Fig. 3). Substrates of ClpXP become resistant to proteolysis by specific alterations of the C-terminal Ala-Ala (28). Therefore, to investigate degradation specificity in the experiment described above, we constructed and tested a variant of S. ambofaciens σAntA in which the C-terminal Ala-Ala was mutated to Asp-Asp [(His)6-SUMO-σAntA-DD]. Strikingly, the Asp-Asp variant was stable against ClpXP degradation over the lifetime of the assay (Fig. 3). Thus, the degradation of (His)6-SUMO-σAntA and the characteristic resistance afforded by the Ala-Ala-to-Asp-Asp mutation demonstrated that σAntA is a substrate of ClpXP in vitro.
FIG 3.
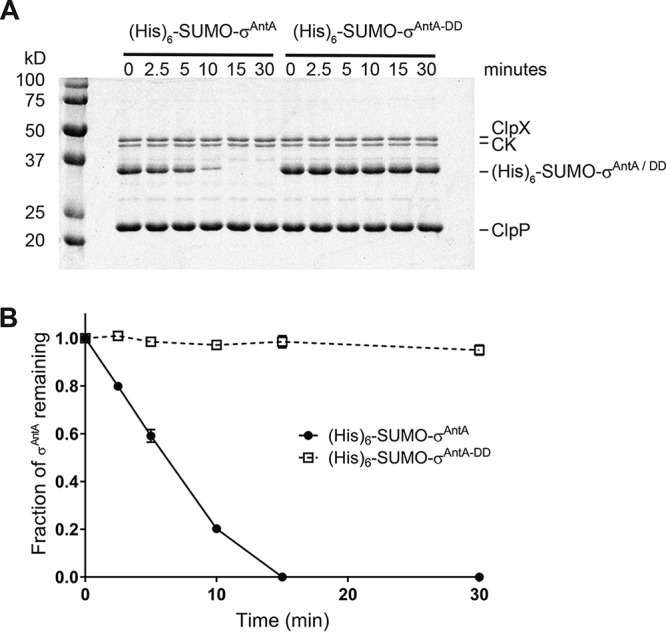
Proteolysis of S. ambofaciens σAntA by ClpXP in vitro. (A) SDS-PAGE analysis of proteolysis reaction mixtures containing 37 pmol (His)6SUMO-σAntA or (His)6SUMO-σAntA-DD. (B) Densitometry analysis SDS-PAGE images for three independent proteolysis experiments. The mean is plotted, and error bars illustrate the standard error of the mean (±1 SEM).
MUSCLE alignment of 71 σAntA orthologues. Only alignments of amino acid residues 160 to 180 are shown. Red asterisks indicate the C-terminal Ala-Ala motif conserved in 67 of 71 orthologues. Blue shading indicates the names of those taxa that do not possess the motif. The orthologue from Streptomyces sp. strain NRRL B-2790 terminated in Ala-Ala-Tyr; the inclusion of a terminal Tyr residue likely represents an artefact caused by poor genome sequence quality (>4,000 contigs). The remaining three orthologues lacking the di-alanine motif are Streptacidiphilus albus strains (terminating in Val-Ala) and Streptomyces sp. strain URHA-0041 (Tyr-Gly). Download FIG S1, GIF file, 0.8 MB (841.6KB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LC-HRMS analysis of ΔantA/pAU3-45-3xFLAG-antA strains. The m/z values corresponding to the [M + H]+ ions derived from antimycins A1 to A4 are shown. Download FIG S2, GIF file, 0.3 MB (263.4KB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
σAntA is degraded by the ClpXP protease in vivo.
To investigate if the in vitro degradation of σAntA demonstrated above is relevant to its regulation in vivo, we deleted the operon consisting of the clpX, clpP1, and clpP2 genes from S. albus S4. The resulting ΔclpXclpP1clpP2 mutant still harbored the second Clp peptidase encoded by clpP3clpP4 and underwent a normal developmental cycle, albeit sporulation was less robust, which is consistent with growth characteristics reported for mutation of equivalent genes in S. coelicolor (Fig. S3) (31). Next, genes encoding the 3xFLAG-σAntA or 3xFLAG-σAntA-DD fusion proteins were generated and introduced into the parental strain and the ΔclpXclpP1clpP2 mutant so the abundance of these proteins could be assessed over a developmental time course by Western blotting with anti-FLAG antisera. This experiment was initially performed with the σAntA fusions integrated on the chromosome under the control of the native promoter. However, a reliable signal could not be detected for 3xFLAG-σAntA and only a trace amount of the Asp-Asp variant was observed, presumably indicating that the cellular level of σAntA is normally low because the native promoter is relatively weak. The experiment was therefore repeated with 3xFLAG-σAntA and 3xFLAG-σAntA-DD expression driven by a stronger, constitutive promoter, ermE* (32). Analysis of the resulting immunoblot revealed that 3xFLAG-σAntA-DD was more abundant than 3xFLAG-σAntA in vegetative mycelium of the parent and ΔclpXclpP1clpP2 strains (Fig. 4; see also Fig. S5). Strikingly, in the later stages of development after aerial mycelium had formed (24 h and 30 h), 3xFLAG-σAntA and 3xFLAG-σAntA-DD were detected only in the ΔclpXclpP1clpP2 strain and not the parent; the Asp-Asp variant was also present in greater relative abundance (Fig. 4), which was consistent with our previous experiments that showed that the ant BGC is downregulated at the level of transcription upon the onset of aerial growth (9). Interestingly, the conspicuous absence of 3xFLAG-σAntA and the presence of 3xFLAG-σAntA-DD in protein samples prepared from the latest time point sampled suggest the potential involvement of an additional degradative factor(s). Taken together, these data support the hypothesis that the levels of σAntA, and thus its ability to activate gene expression of antFGHIJKLMNO, are modulated by the ClpXP protease.
FIG 4.
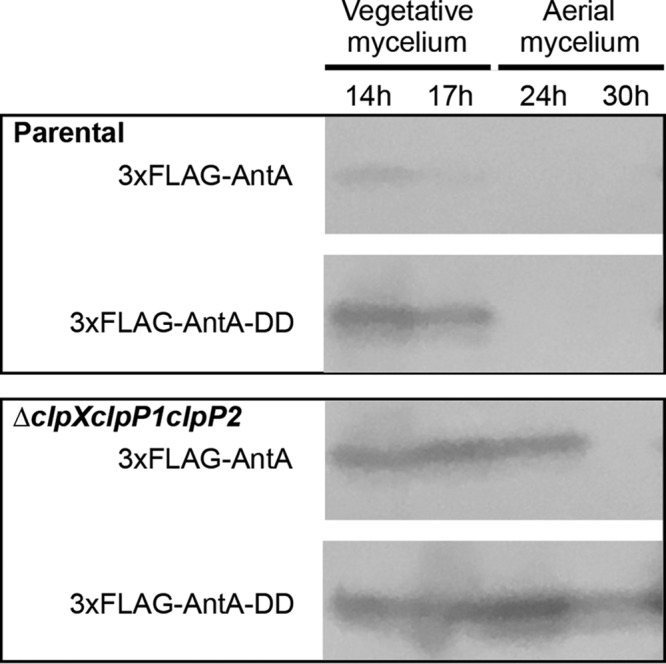
The abundance of σAntA is enhanced in the absence of the ClpXP in vivo. Cells from the indicated strains were cultivated over a developmental time course atop cellophane discs on agar media. Protein was isolated from 100 mg of either vegetative mycelium (14 and 17 h) or aerial mycelium (24 and 30 h). Thirty-microgram volumes of total protein were analyzed by Western blotting with anti-FLAG antisera. The images shown are derived from uncropped original images shown in Fig. S4 in the supplemental material, and the corresponding densitometry analysis is shown in Fig. S6.
Gel filtration of purified (His)6-SUMO-σAntA proteins. Gel filtration chromatograms show the elution profiles of purified (His)6-SUMO-σAntA and (His)6-SUMO-σAntA-DD proteins (upper panel) and results of associated SDS-PAGE analyses (lower panels). The chromatogram of (His)6-SUMO-σAntA differs slightly from that of (His)6-SUMO-σAntA-DD because gel filtration of (His)6-SUMO-σAntA protein was performed in a reaction mixture containing 50 mM HEPES-KOH (pH 7.5), 150 mM KCl, 10% glycerol, and 1 mM DTT, which was later dialyzed into the storage buffer, whereas gel filtration of (His)6-SUMO-σAntA-DD was performed in the storage buffer. Download FIG S3, GIF file, 1.3 MB (1.3MB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sporulation of S. albus S4 clp mutants. Photographs were taken after 6 days of growth on the indicated medium. The ΔclpXclpP1ΔclpP2 strain underwent a normal developmental cycle; however, sporulation was less robust on MS agar media than was seen with the parental strain. Download FIG S4, GIF file, 0.2 MB (226.9KB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Uncropped Western blotting images. Shown are the original Western blots depicted in Fig. 4. Red boxes indicate cropped sections for the strains indicated. The molecular weight (Mw) of the protein marker (M) is shown. Plus and minus symbols on each blot indicate positive-testing strains (mutant strain ΔclpXclpP1clpP2/pPDD) and negative controls (parental strain Δantall), respectively. Download FIG S5, GIF file, 1.2 MB (1.2MB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Densitometry analysis of Western blotting results. The mean intensities (in pixels) of relevant bands from the Western blots shown in Fig. 4 are displayed for the indicated strains and time points. The mean pixel intensity was determined using Photoshop v20.0.6, and values were normalized to the intensity of the ΔclpXclpP1clpP2/pPDD sample contained on each membrane. Download FIG S6, GIF file, 0.08 MB (85KB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antimycins are not overproduced in the absence of ClpXP.
The results of the experiments described above indicate that the cellular level of σAntA was more abundant in the absence of the ClpXP protease. In order to determine if an increased level of this transcription factor ultimately influenced the final production titer of antimycins, we used liquid chromatography–high-resolution mass spectrometry (LC-HRMS) to assess the abundance of antimycins in chemical extracts generated from the ΔclpXclpP1clpP2 and parental strains grown atop a cellophane disk on mannitol-soya flour (MS) agar in triplicate. The extracted ion chromatograms representing antimycin A1, A2, A3, and A4 were used to determine the peak area for each compound, which was subsequently normalized based on the wet mycelium weight of the sample. Interestingly, the results indicated that the total levels of antimycin production by the ΔclpXclpP1clpP2 mutant (15.57 arbitrary units [AU] ± 2.86) and the parental strain (16.59 AU ± 1.12) were not statistically significantly different (P value, 0.59) (see Table S1 in the supplemental material). This result is consistent with a previous experiment where overexpression of antA did not increase the titer of antimycins, because that experiment showed that it resulted in overexpression of only antGF and antHIJKLMNO (genes encoding the production of the AntG-S-3-formamidosalicylate starter unit) and not the remaining genes (antABCDE) in the BGC (9). This also presumably indicates that starter unit biosynthesis is not rate limiting for antimycin production.
LCMS quantification of antimycin production. Download Table S1, DOCX file, 0.02 MB (19.6KB, docx) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Model for the regulation of antimycin biosynthesis.
Our model for the regulation of antimycin biosynthesis is depicted in Fig. 5. Expression of the ant BGC is cross-activated by FscRI, a LuxR-family regulator, from the candicidin BGC, which activates expression of antBA and antCDE (10). This regulation in turn enables direct activation of the 3-FSA biosynthetic operons (antGF and antHIJKLMNO) by σAntA. The expression of antBA and antCDE is downregulated following the onset of morphological differentiation, presumably because the ligand sensed by the FscRI PAS domain is no longer available (9, 10). The cellular level of σAntA is antagonized by the ClpXP protease, for which it is a direct target, and is ultimately responsible for clearing residual σAntA when FscRI is inactivated following the onset of morphological differentiation (10). While ClpXP proteolytic control of transcription factor activity, and in particular that of ECF σ factor/anti-σ factors, has been shown previously (33–41), it has thus far not been directly linked to the control of cluster-situated regulators of natural product biosynthesis. This finding provides a new lens through which to examine microbial signal transduction and the regulation of natural product biosynthesis in Streptomyces species. Understanding the diversity of regulatory strategies controlling the expression of these pathways is critical for the development of new tools for exploiting the “silent majority” of biosynthetic pathways harbored by these organisms.
FIG 5.
Model for the regulation of antimycin biosynthesis. The upper panel displays the relative locations of the antimycin and candicidin BGCs in the S. albus S4 chromosome. As shown in the lower panel, FscRI, a LuxR-family regulator, from the candicidin BGC, activates expression of antBA and antCDE. This in turn enables direct activation of the 3-FSA biosynthetic operons (antGF and antHIJKLMNO) by σAntA. The cellular level of σAntA is antagonized by the ClpXP-protease system, for which it is a direct target and is ultimately responsible for clearing residual σAntA when FscRI is inactivated following the onset of differentiation.
MATERIALS AND METHODS
Growth media, strains, cosmids, plasmids, and other reagents.
Escherichia coli strains were propagated on Lennox agar (LA) or broth (LB) (42, 43), and Streptomyces albus S4 strains were cultivated using LA, LB, and mannitol-soya flour (MS) agar or broth (42). Development of clp mutants was assessed on MS and ISP2 medium (42). Culture medium was supplemented with antibiotics as required at the following concentrations: apramycin, 50 μg ml−1; carbenicillin, 100 μg ml−1; chloramphenicol, 25 μg ml−1; hygromycin, 50 μg ml−1; kanamycin, 50 μg ml−1; nalidixic acid, 25 μg ml−1. Streptomyces strains were constructed by conjugal mating with E. coli ET12567 as previously described (42). Enzymes were purchased from New England Biolabs unless otherwise stated, and oligonucleotides were purchased from Integrated DNA Technologies, Inc. All of the strains, cosmids, and plasmids used in this study are described in Table S2 in the supplemental material, and all of the oligonucleotides used are provided in Table S3.
Bacterial strains, cosmids, and plasmids used in this study. Download Table S2, DOCX file, 0.02 MB (25.6KB, docx) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in this study. Download Table S3, DOCX file, 0.02 MB (20.7KB, docx) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of plasmids.
The insertion for each plasmid generated in this study was prepared by PCR amplification with Q5 High-Fidelity DNA polymerase and oligonucleotides containing restriction sites. PCR-amplified insertions were restricted and cloned into the relevant plasmids cut with the same enzymes by standard molecular biology procedures. All clones were sequenced to verify the integrity of insertion DNA. The names of the restriction sites used for cloning are provided with the plasmid descriptions in Table S2.
ChIP-seq and bioinformatics analyses.
The antA coding sequence was amplified with RFS629 and RFS630, which contain KpnI and EcoRI restriction sites, respectively. The restricted PCR product was cloned into pSETNFLAG (10) digested with the same enzymes. The resulting plasmid was then restricted with NotI and EcoRI to release ermE*p-3xFLAG-antA, which was subsequently cloned into pAU3-45 (44) digested with the same enzymes. pAU3-45-3xFLAG-antA was mobilized to an apramycin-marked ΔantA strain (9). Cultivations of the wild-type and ΔantA/pAUNFLAG-antA strains for ChIP-seq were performed exactly as described previously (10). Pure DNA resulting from immunoprecipitates from two biological replicates of the wild-type and ΔantA/pAUNFLAG-antA strains and nonimmunoprecipitated chromosomal DNA were sequenced with a Illumina HiSeq 3000 platform with 150-nucleotide (nt) paired-end reads by the University of Leeds Next Generation Sequencing Facility at the St. James Teaching Hospital NHS Trust. The resulting reads were analyzed exactly as described previously (10). The graphic in Fig. 2 was generated using DeepTools computeMatrix in scale-regions mode with a bin size of 54 and plotProfile functions (45).
Construction of the S. albus S4 ΔclpXclpP1clpP2 mutant strain.
Deletion of clpXclpP1clpP2 was carried out using RecET recombineering in E. coli as follows. The clpXclpP1clpP2-containing cosmid, cos117, was obtained by screening a previously constructed S. albus S4 Supercos1 cosmid library (8) by PCR using oligonucleotides PBB001 and PBB002. Cos117 was mutagenized as required using E. coli recombineering with strain GB05-red (46) and a deletion cassette. The deletion cassette was generated by PCR from paac-apr-oriT (47) and consisted of the aac(3)IV apramycin resistance gene and a conjugal origin of transfer (oriT), which was flanked by ΦC31-attL and ΦC31-attR sites for excision of the cassette. The oligonucleotides used to generate deletion cassettes included 39 nt of homology upstream or downstream of the target open reading frame(s) and are listed in Table S3. The resulting PCR product was digested with DpnI, gel purified, and electroporated into arabinose-induced E. coli GB05-red harboring cos117. Transformants were screened for the presence of mutagenized cosmid by PCR using oligonucleotides listed in Table S3, and the integrity of the locus was verified by DNA sequencing. The mutagenized cosmid was electroporated into E. coli ET12567/pUZ8002 and mobilized to a strain of S. albus S4 harboring an entire antimycin BGC deletion (Δantall) (48) by conjugation as described previously (42). Transconjugants were screened for apramycin resistance and kanamycin sensitivity. The integrity of an apramycin-marked mutant was verified by PCR using the oligonucleotides listed in Table S3. The apramycin deletion cassette was subsequently excised from the chromosome by conjugal introduction of pUWLint31, which is a replicative plasmid with a temperature-sensitive origin of replication that expresses the ΦC31 integrase required for removal of the cassette (47). Transconjugants were screened for loss of apramycin resistance, and excision of the cassette was verified by polymorphic shift PCR and DNA sequencing of the product.
Immunoblot analysis.
Spores of the parental strain and of S. albus Δantall and the ΔclpXclpP1clpP2 mutant harboring pPDA or pPDD were grown on MS agar (buffered with 50 mM TES, pH 7.2) covered with cellophane discs. Protein was isolated from mycelium collected during growth at the following regular intervals: 14 h, 17 h, 24 h, and 30 h for the Δantall and ΔclpXclpP1clpP2 mutants harboring 3xFLAG-AntA constructs and 17 h, 20 h, 23 h, and 30 h for the Δantall and ΔclpXclpP1clpP2 mutants harboring the 3xFLAG-FscRI construct. Protein samples were generated as follows: 100-mg volumes of cells were resuspended in 200 μl lysis buffer (50 mM sodium phosphate buffer [pH 7.0]; 150 mM sodium chloride; 10 mg ml−1 lysozyme; cOmplete, Mini, EDTA-free protease inhibitors [Roche]; 100 mg of 0.1-mm-diameter glass beads [PowerLyzer]) and lysed by vortex mixing for 30 min at 2,000 rpm and 37°C, with a subsequent incubation for another 30 min at 37°C. The obtained suspension was centrifuged for 20 min at 20,000 × g at 18°C. Each clarified protein sample (30 μg) was subjected to SDS-PAGE and then transferred to a nitrocellulose membrane (pore size, 0.2 μm) for Western blot analysis. The membrane was probed with mouse monoclonal anti-FLAG M2-horseradish peroxidase (HRP) antibody (Sigma) (1:10,000, and the signals were detected using Pierce 1-Step Ultra TMB blotting solution (Thermo Scientific).
Protein purification and in vitro ClpXP proteolysis assays.
The wild-type antA gene was PCR amplified and cloned into the AgeI and HindIII sites of the pET23b-SUMO vector, which harbors an N-terminal (His)6-SUMO tag (49). The plasmid for production of (His)6-SUMO-σAntA-DD was generated by site-directed mutagenesis (Agilent QuikChange) using primers listed in Table S3. (His)6-SUMO-σAntA and (His)6-SUMO-σAntA-DD were produced by E. coli Rosetta(DE3) (Novagen) grown in LB at 37°C until an optical density at 600 nm (OD600) of 0.5 was reached, followed by induction with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and growth at 18°C for 16 h. Cells were resuspended in 50 mM sodium phosphate (pH 8)–1 M NaCl–20 mM imidazole–10% glycerol–1 mM dithiothreitol (DTT) and lysed by the use of a French press at 28,000 lb/in2, followed by treatment with protease inhibitor cocktail set III (EDTA-free) (Calbiochem), and benzonase (Millipore Sigma). (His)6-SUMO-σAntA and (His)6-SUMO-σAntA-DD proteins were purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography and Superdex-75 gel filtration and stored in 50 mM potassium phosphate (pH 6.8)–850 mM KCl–10% glycerol–1 mM DTT. E. coli ClpX and ClpP proteins were purified as described previously (50, 51).
In vitro ClpXP proteolysis assays were performed at 30°C by preincubating 0.3 μM ClpX6 and 0.8 μM ClpP14 with an ATP regeneration system (4 mM ATP, 50 μg ml−1 creatine kinase, 5 mM creatine phosphate) in 25 mM HEPES-KOH (pH 7.5)–20 mM KCl–5 mM MgCl2–10% glycerol–0.032% NP-40–0.2 mM DTT and adding substrate to initiate the reactions. Samples of each reaction mixture were taken at specific time points, and the reactions were stopped by addition of SDS-PAGE loading dye and boiling at 100°C before loading on Tris-Glycine-SDS gels. Bands were visualized by staining with colloidal Coomassie G-250 and quantified by ImageQuant (GE Healthcare) after scanning by Typhoon FLA 9500 (GE Healthcare). The (His)6-SUMO-σAntA fraction remaining was calculated by dividing the (His)6-SUMO-σAntA density at a given time point by the density at time zero and normalizing by ClpX density.
Chemical analysis.
S. albus S4 strains were cultivated atop a cellophane disc on MS agar at 30°C for 7 days in triplicate. At the time of harvest, the cellophane disc containing mycelium was removed and the quantity of biomass was determined. Bacterial metabolites were extracted from both the mycelium and the “spent” agar for 1 h using 50 ml of ethyl acetate. Thirty-milliliter volumes of ethyl acetate were evaporated to dryness under conditions of reduced pressure, and the resulting residue was resuspended in 100% methanol (300 μl). Immediately prior to LC-HRMS analysis, methanolic extracts were centrifuged at 16,000 × g in a microcentrifuge tube for 5 min to remove insoluble material. Only the supernatant (3 μl) was injected into a Bruker Maxis Impact time of flight (TOF) mass spectrometer equipped with a Dionext Ultimate 3000 HPLC system as previously described (52). The peak area associated with the extracted ion chromatograms for antimycin A1, A2, A3, and A4 present in agar and mycelium extracts was determined and used to calculate the total level of antimycins produced for each replicate. These values were subsequently used to determine the arithmetic mean for total antimycin production for each strain. Statistical significance was assessed in Microsoft Excel by a homoscedastic Student's t test with a two-tailed distribution.
Data availability.
The next-generation-sequencing data obtained in this study are available under ArrayExpress accession numbers E-MTAB-7700 and E-MTAB-5122.
ACKNOWLEDGMENTS
We thank Matt Hutchings, Paul Hoskisson, Kenneth McDowall, and Alex O’Neill for helpful discussion concerning the manuscript.
B.B., S.K., and A.F. performed experiments, interpreted data, and wrote sections of the manuscript; T.A.B. wrote sections of the manuscript; R.F.S. designed the study, performed experiments, and wrote the manuscript.
B.B. was supported by a grant from the Biotechnology and Biological Sciences Research Council (BB/N007980/1) awarded to R.F.S. A.F. was funded by a Ph.D. studentship funded by the University of Leeds. S.K. was supported by a National Science Foundation Graduate Research Fellowship and the Howard Hughes Medical Foundation; T.A.B. is an employee of the Howard Hughes Medical Foundation.
REFERENCES
- 1.Zhu H, Sandiford SK, van Wezel GP. 2014. Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol 41:371–386. doi: 10.1007/s10295-013-1309-z. [DOI] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM. 2012. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Wezel GP, McDowall KJ. 2011. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep 28:1311–1323. doi: 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- 4.van der Heul HU, Bilyk BL, McDowall KJ, Seipke RF, van Wezel GP. 2018. Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat Prod Rep 35:575–604. doi: 10.1039/C8NP00012C. [DOI] [PubMed] [Google Scholar]
- 5.Joynt R, Seipke RF. 2018. A phylogenetic and evolutionary analysis of antimycin biosynthesis. Microbiology 164:28–39. doi: 10.1099/mic.0.000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunshee BR, Leben C, Keitt GW, Strong FM. 1949. The isolation and properties of antimycin A. J Am Chem Soc 71:2436–2437. doi: 10.1021/ja01175a057. [DOI] [Google Scholar]
- 7.Tzung S-P, Kim KM, Basañez G, Giedt CD, Simon J, Zimmerberg J, Zhang KYJ, Hockenbery DM. 2001. Antimycin A mimics a cell-death-inducing Bcl-2 homology domain 3. Nat Cell Biol 3:183–191. doi: 10.1038/35055095. [DOI] [PubMed] [Google Scholar]
- 8.Seipke RF, Barke J, Brearley C, Hill L, Yu DW, Goss RJM, Hutchings MI. 2011. A single Streptomyces symbiont makes multiple antifungals to support the fungus farming ant Acromyrmex octospinosus. PLoS One 6:e22028–8. doi: 10.1371/journal.pone.0022028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seipke RF, Patrick E, Hutchings MI. 2014. Regulation of antimycin biosynthesis by the orphan ECF RNA polymerase sigma factor σ (AntA.). PeerJ 2:e253. doi: 10.7717/peerj.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLean TC, Hoskisson PA, Seipke RF. 2016. Coordinate regulation of antimycin and candicidin biosynthesis. mSphere 1:e00305-16. doi: 10.1128/mSphere.00305-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmann JD. 2002. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 46:47–110. [DOI] [PubMed] [Google Scholar]
- 12.Staroń A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) σ factor protein family. Mol Microbiol 74:557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 13.Campbell EA, Greenwell R, Anthony JR, Wang S, Lim L, Das K, Sofia HJ, Donohue TJ, Darst SA. 2007. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol Cell 27:793–805. doi: 10.1016/j.molcel.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang JG, Paget MS, Seok YJ, Hahn MY, Bae JB, Hahn JS, Kleanthous C, Buttner MJ, Roe JH. 1999. RsrA, an anti-sigma factor regulated by redox change. EMBO J 18:4292–4298. doi: 10.1093/emboj/18.15.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajasekar KV, Zdanowski K, Yan J, Hopper JTS, Francis M-L, Seepersad C, Sharp C, Pecqueur L, Werner J, Robinson CV, Mohammed S, Potts JR, Kleanthous C. 2016. The anti-sigma factor RsrA responds to oxidative stress by reburying its hydrophobic core. Nat Commun 7:1–14. doi: 10.1038/ncomms12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paget M. 2015. Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 5:1245–1265. doi: 10.3390/biom5031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim M-S, Hahn M-Y, Cho Y, Cho S-N, Roe J-H. 2009. Positive and negative feedback regulatory loops of thiol-oxidative stress response mediated by an unstable isoform of σR in actinomycetes. Mol Microbiol 73:815–825. doi: 10.1111/j.1365-2958.2009.06824.x. [DOI] [PubMed] [Google Scholar]
- 18.Foulston LC, Bibb MJ. 2010. Microbisporicin gene cluster reveals unusual features of lantibiotic biosynthesis in actinomycetes. Proc Natl Acad Sci U S A 107:13461–13466. doi: 10.1073/pnas.1008285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherwood EJ, Bibb MJ. 2013. The antibiotic planosporicin coordinates its own production in the actinomycete Planomonospora alba. Proc Natl Acad Sci U S A 110:E2500–E2509. doi: 10.1073/pnas.1305392110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gur E, Biran D, Ron EZ. 2011. Regulated proteolysis in Gram-negative bacteria–how and when? Nat Rev Microbiol 9:839–848. doi: 10.1038/nrmicro2669. [DOI] [PubMed] [Google Scholar]
- 21.Baker TA, Sauer RT. 2012. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta 1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivares AO, Baker TA, Sauer RT. 2016. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol 14:33–44. doi: 10.1038/nrmicro.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Crécy-Lagard V, Servant-Moisson P, Viala J, Grandvalet C, Mazodier P. 1999. Alteration of the synthesis of the Clp ATP-dependent protease affects morphological and physiological differentiation in Streptomyces. Mol Microbiol 32:505–517. doi: 10.1046/j.1365-2958.1999.01364.x. [DOI] [PubMed] [Google Scholar]
- 24.Viala J, Rapoport G, Mazodier P. 2000. The clpP multigenic family in Streptomyces lividans: conditional expression of the clpP3 clpP4 operon is controlled by PopR, a novel transcriptional activator. Mol Microbiol 38:602–612. doi: 10.1046/j.1365-2958.2000.02155.x. [DOI] [PubMed] [Google Scholar]
- 25.Viala J, Mazodier P. 2002. ClpP-dependent degradation of PopR allows tightly regulated expression of the clpP3 clpP4 operon in Streptomyces lividans. Mol Microbiol 44:633–643. doi: 10.1046/j.1365-2958.2002.02907.x. [DOI] [PubMed] [Google Scholar]
- 26.Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 27.Gottesman S, Roche E, Zhou Y, Sauer RT. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev 12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. 2001. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci U S A 98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seipke RF, Hutchings MI, Hutchings MI. 2013. The regulation and biosynthesis of antimycins. Beilstein J Org Chem 9:2556–2563. doi: 10.3762/bjoc.9.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoenian I, Paetz C, Dickschat JS, Aigle B, Leblond P, Spiteller D. 2012. An unprecedented 1,2-shift in the biosynthesis of the 3-aminosalicylate moiety of antimycins. ChemBioChem 13:769–773. doi: 10.1002/cbic.201200033. [DOI] [PubMed] [Google Scholar]
- 31.Viala J, Mazodier P. 2003. The ATPase ClpX is conditionally involved in the morphological differentiation of Streptomyces lividans. Mol Gen Genomics 268:563–569. doi: 10.1007/s00438-002-0783-1. [DOI] [PubMed] [Google Scholar]
- 32.Luo Y, Zhang L, Barton KW, Zhao H. 2015. Systematic identification of a panel of strong constitutive promoters from Streptomyces albus. ACS Synth Biol 4:1001–1010. doi: 10.1021/acssynbio.5b00016. [DOI] [PubMed] [Google Scholar]
- 33.Mika F, Hengge R. 2005. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Genes Dev 19:2770–2781. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao X-M, Sun N, Wang F, Luo S, Zhou Z, Feng W-H, Huang F-L, Li Y-Q. 2013. Dual positive feedback regulation of protein degradation of an extra-cytoplasmic function σ factor for cell differentiation in Streptomyces coelicolor. J Biol Chem 288:31217–31228. doi: 10.1074/jbc.M113.491498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ades SE, Connolly LE, Alba BM, Gross CA. 1999. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev 13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev 16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flynn JM, Levchenko I, Sauer RT, Baker TA. 2004. Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev 18:2292–2301. doi: 10.1101/gad.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zellmeier S, Schumann W, Wiegert T. 2006. Involvement of Clp protease activity in modulating the Bacillus subtilis sigma W stress response. Mol Microbiol 61:1569–1582. doi: 10.1111/j.1365-2958.2006.05323.x. [DOI] [PubMed] [Google Scholar]
- 39.Bellier A, Mazodier P. 2004. ClgR, a novel regulator of clp and lon expression in Streptomyces. J Bacteriol 186:3238–3248. doi: 10.1128/JB.186.10.3238-3248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellier A. 2006. Post-translational control of the Streptomyces lividans ClgR regulon by ClpP. Microbiology 152:1021–1027. doi: 10.1099/mic.0.28564-0. [DOI] [PubMed] [Google Scholar]
- 41.Mettert EL, Kiley PJ. 2005. ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe–4S] cluster. J Mol Biol 354:220–232. doi: 10.1016/j.jmb.2005.09.066. [DOI] [PubMed] [Google Scholar]
- 42.Skyrud W, Liu J, Thankachan D, Cabrera M, Seipke RF, Zhang W. 2018. Biosynthesis of the 15-membered ring depsipeptide neoantimycin. ACS Chem Biol 13:1398–1406. doi: 10.1021/acschembio.8b00298. [DOI] [PubMed] [Google Scholar]
- 43.Seipke RF, Grüschow S, Goss RJM, Hutchings MI. 2012. Isolating antifungals from fungus-growing ant symbionts using a genome-guided chemistry approach. Methods Enzymol 517:47–70. [DOI] [PubMed] [Google Scholar]
- 44.Bignell DRD, Tahlan K, Colvin KR, Jensen SE, Leskiw BK. 2005. Expression of ccaR, encoding the positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus, is dependent on bldG. Antimicrob Agents Chemother 49:1529–1541. doi: 10.1128/AAC.49.4.1529-1541.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Müller R, Stewart AF, Zhang Y. 2012. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol 30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 47.Myronovskyi M, Rosenkränzer B, Luzhetskyy A. 2014. Iterative marker excision system. Appl Microbiol Biotechnol 98:4557–4570. doi: 10.1007/s00253-014-5523-z. [DOI] [PubMed] [Google Scholar]
- 48.Fazal A, Thankachan D, Harris E, Seipke RF. 2020. A chromatogram-simplified Streptomyces albus host for heterologous production of natural products. Antonie Van Leeuwenhoek 113:511–510. doi: 10.1007/s10482-019-01360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang KH, Sauer RT, Baker TA. 2007. ClpS modulates but is not essential for bacterial N-end rule degradation. Genes Dev 21:403–408. doi: 10.1101/gad.1511907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neher SB, Sauer RT, Baker TA. 2003. Distinct peptide signals in the UmuD and UmuD’ subunits of UmuD/D’ mediate tethering and substrate processing by the ClpXP protease. Proc Natl Acad Sci U S A 100:13219–13224. doi: 10.1073/pnas.2235804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. 2000. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol Cell 5:639–648. doi: 10.1016/S1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- 52.Liu J, Zhu X, Seipke RF, Zhang W. 2015. Biosynthesis of antimycins with a reconstituted 3-formamidosalicylate pharmacophore in Escherichia coli. ACS Synth Biol 4:559–565. doi: 10.1021/sb5003136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MUSCLE alignment of 71 σAntA orthologues. Only alignments of amino acid residues 160 to 180 are shown. Red asterisks indicate the C-terminal Ala-Ala motif conserved in 67 of 71 orthologues. Blue shading indicates the names of those taxa that do not possess the motif. The orthologue from Streptomyces sp. strain NRRL B-2790 terminated in Ala-Ala-Tyr; the inclusion of a terminal Tyr residue likely represents an artefact caused by poor genome sequence quality (>4,000 contigs). The remaining three orthologues lacking the di-alanine motif are Streptacidiphilus albus strains (terminating in Val-Ala) and Streptomyces sp. strain URHA-0041 (Tyr-Gly). Download FIG S1, GIF file, 0.8 MB (841.6KB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LC-HRMS analysis of ΔantA/pAU3-45-3xFLAG-antA strains. The m/z values corresponding to the [M + H]+ ions derived from antimycins A1 to A4 are shown. Download FIG S2, GIF file, 0.3 MB (263.4KB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gel filtration of purified (His)6-SUMO-σAntA proteins. Gel filtration chromatograms show the elution profiles of purified (His)6-SUMO-σAntA and (His)6-SUMO-σAntA-DD proteins (upper panel) and results of associated SDS-PAGE analyses (lower panels). The chromatogram of (His)6-SUMO-σAntA differs slightly from that of (His)6-SUMO-σAntA-DD because gel filtration of (His)6-SUMO-σAntA protein was performed in a reaction mixture containing 50 mM HEPES-KOH (pH 7.5), 150 mM KCl, 10% glycerol, and 1 mM DTT, which was later dialyzed into the storage buffer, whereas gel filtration of (His)6-SUMO-σAntA-DD was performed in the storage buffer. Download FIG S3, GIF file, 1.3 MB (1.3MB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sporulation of S. albus S4 clp mutants. Photographs were taken after 6 days of growth on the indicated medium. The ΔclpXclpP1ΔclpP2 strain underwent a normal developmental cycle; however, sporulation was less robust on MS agar media than was seen with the parental strain. Download FIG S4, GIF file, 0.2 MB (226.9KB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Uncropped Western blotting images. Shown are the original Western blots depicted in Fig. 4. Red boxes indicate cropped sections for the strains indicated. The molecular weight (Mw) of the protein marker (M) is shown. Plus and minus symbols on each blot indicate positive-testing strains (mutant strain ΔclpXclpP1clpP2/pPDD) and negative controls (parental strain Δantall), respectively. Download FIG S5, GIF file, 1.2 MB (1.2MB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Densitometry analysis of Western blotting results. The mean intensities (in pixels) of relevant bands from the Western blots shown in Fig. 4 are displayed for the indicated strains and time points. The mean pixel intensity was determined using Photoshop v20.0.6, and values were normalized to the intensity of the ΔclpXclpP1clpP2/pPDD sample contained on each membrane. Download FIG S6, GIF file, 0.08 MB (85KB, gif) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LCMS quantification of antimycin production. Download Table S1, DOCX file, 0.02 MB (19.6KB, docx) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains, cosmids, and plasmids used in this study. Download Table S2, DOCX file, 0.02 MB (25.6KB, docx) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in this study. Download Table S3, DOCX file, 0.02 MB (20.7KB, docx) .
Copyright © 2020 Bilyk et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The next-generation-sequencing data obtained in this study are available under ArrayExpress accession numbers E-MTAB-7700 and E-MTAB-5122.



