U.S. university students are at a higher risk of invasive meningococcal disease than the general population. The responsible pathogen, Neisseria meningitidis, can be carried asymptomatically in the oropharynx; the dynamics of meningococcal carriage and the genetic features that distinguish carriage versus disease states are not completely understood. Through our analyses, we aimed to provide data to address these topics. We whole-genome sequenced 1,514 meningococcal carriage isolates from individuals at three U.S. universities, two of which underwent mass vaccination campaigns following recent meningococcal outbreaks. We describe the within-host genetic changes among individuals carrying a strain with the same molecular type over time, the primary strains being carried in this population, and the genetic differences between closely related outbreak and carriage strains. Our results provide detailed information on the dynamics of meningococcal carriage and the genetic differences in carriage and outbreak strains, which can inform future efforts to reduce the incidence of invasive meningococcal disease.
KEYWORDS: Neisseria meningitidis, United States, WGS, carriage, comparative genomics, meningococcal, university, whole-genome sequencing
ABSTRACT
In 2015 and 2016, meningococcal carriage evaluations were conducted at two universities in the United States following mass vaccination campaigns in response to Neisseria meningitidis serogroup B (NmB) disease outbreaks. A simultaneous carriage evaluation was also conducted at a university near one of the outbreaks, where no NmB cases were reported and no mass vaccination occurred. A total of ten cross-sectional carriage evaluation rounds were conducted, resulting in 1,514 meningococcal carriage isolates collected from 7,001 unique participants; 1,587 individuals were swabbed at multiple time points (repeat participants). All isolates underwent whole-genome sequencing. The most frequently observed clonal complexes (CC) were CC198 (27.3%), followed by CC1157 (17.4%), CC41/44 (9.8%), CC35 (7.4%), and CC32 (5.6%). Phylogenetic analysis identified carriage isolates that were highly similar to the NmB outbreak strains; comparative genomics between these outbreak and carriage isolates revealed genetic changes in virulence genes. Among repeat participants, 348 individuals carried meningococcal bacteria during at least one carriage evaluation round; 50.3% retained N. meningitidis carriage of a strain with the same sequence type (ST) and CC across rounds, 44.3% only carried N. meningitidis in one round, and 5.4% acquired a new N. meningitidis strain between rounds. Recombination, point mutations, deletions, and simple sequence repeats were the most frequent genetic mechanisms found in isolates collected from hosts carrying a strain of the same ST and CC across rounds. Our findings provide insight on the dynamics of meningococcal carriage among a population that is at higher risk for invasive meningococcal disease than the general population.
IMPORTANCE U.S. university students are at a higher risk of invasive meningococcal disease than the general population. The responsible pathogen, Neisseria meningitidis, can be carried asymptomatically in the oropharynx; the dynamics of meningococcal carriage and the genetic features that distinguish carriage versus disease states are not completely understood. Through our analyses, we aimed to provide data to address these topics. We whole-genome sequenced 1,514 meningococcal carriage isolates from individuals at three U.S. universities, two of which underwent mass vaccination campaigns following recent meningococcal outbreaks. We describe the within-host genetic changes among individuals carrying a strain with the same molecular type over time, the primary strains being carried in this population, and the genetic differences between closely related outbreak and carriage strains. Our results provide detailed information on the dynamics of meningococcal carriage and the genetic differences in carriage and outbreak strains, which can inform future efforts to reduce the incidence of invasive meningococcal disease.
INTRODUCTION
Invasive meningococcal disease (IMD) is a severe bacterial infection. Although overall incidence of IMD has declined in the United States to historically low levels (1), college students are at increased risk for serogroup B meningococcal disease compared to that of other adolescents (2). Between 2013 and 2018, 11 university-associated outbreaks of IMD were reported (3) (CDC, unpublished data). The responsible organism, Neisseria meningitidis, can colonize the human oropharynx asymptomatically. Approximately 8% to 25% of the healthy human population are asymptomatic carriers of N. meningitidis (4), with higher prevalence of carriage in teenagers and young adults (5). The highest carriage prevalence has been reported in populations in semiclosed settings or with extensive close contact, such as universities (6–8) and military barracks (9, 10). Studies have shown that meningococcal carriage is highly dynamic and may be chronic, intermittent, or transient (11–15).
The meningococcal polysaccharide capsule is a major virulence factor that protects the bacteria against the host’s immune system. Six encapsulated serogroups (A, B, C, W, X, and Y) are responsible for the majority of IMD worldwide, while nonencapsulated meningococci are mostly associated with carriage and rarely cause IMD (11). Most of the IMD cases in the United States are caused by strains belonging to a few hyperinvasive lineages, namely, clonal complex (CC) 32, CC41/44, CC11, and CC23 (16–18). These hyperinvasive lineages are rarely found among healthy carriers, and carriage isolates are primarily associated with sequence types (STs) or CCs that seldom cause disease (12, 19, 20).
In response to two separate N. meningitidis serogroup B (NmB) outbreaks at two U.S. universities in Oregon (OR) and Rhode Island (RI-1) between January and May 2015, mass vaccination campaigns with serogroup B meningococcal vaccines (predominantly MenB-FHbp [Trumenba] at both universities and some MenB-4C [Bexsero] at OR) were launched (7, 8). Meningococcal carriage evaluations were conducted at both universities during 2015 and 2016 to evaluate meningococcal carriage before and after completion of the vaccination campaigns (6, 21); in both evaluations, carriage rates remained stable, suggesting the vaccination did not rapidly reduce meningococcal carriage or prevent acquisition. Additional work is under way to analyze the possible impact of mass vaccination on the carriage and acquisition of strains potentially covered by the vaccine, and this work will be published at a later date. A third carriage evaluation was conducted at another university in Rhode Island (RI-2), where no IMD outbreak or cases were reported (22) and no mass vaccination occurred. We sequenced isolates from these three carriage evaluations in order to describe within-host genetic changes in individuals carrying N. meningitidis at multiple time points, elucidate the meningococcal carriage population structure, and compare closely related carriage and outbreak isolates.
RESULTS
Population structure of N. meningitidis carriage isolates.
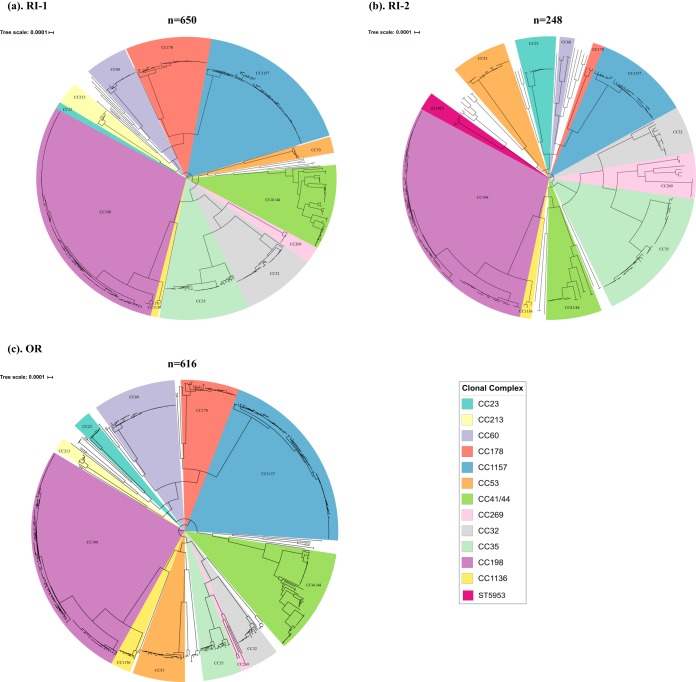
A total of 1,514 meningococcal carriage isolates from the three universities were sequenced and analyzed. Sequencing information, multilocus sequence typing (MLST), and serogroup information about these isolates are provided in Table S1 in the supplemental material. The vast majority of these isolates were nongroupable (92.1% [1,395/1,514]), followed by serogroup B (4.8% [74/1,514]) and serogroup E (3.0% [45/1,514]). Fewer than five isolates belonging to serogroups Y, X, W, C, and Z were identified. Phylogenetic analysis revealed that these isolates clustered into CC-specific phylogenetic clades with 12 major clades (≥20 isolates per clade) and 28 minor clades (<20 isolates per clade) across the three universities (Fig. 1). The most frequent CCs were CC198 (414/1,514 [27.3%]), followed by CC1157 (264/1,514 [17.4%]), CC41/44 (148/1,514 [9.8%]), CC35 (112/1,514 [7.4%]), and CC32 (85/1,514 [5.6%]). A total of 20 STs were found in all three universities, while the majority (n = 69/89 [77.5%]) of STs were only detected in a single university (Table 1).
FIG 1.
Whole-genome phylogenetic relationship of carriage isolates from 3 U.S. universities. (a) The phylogenetic relationship among carriage isolates identified at RI-1. (b) The phylogenetic relationship among carriage isolates identified at RI-2. (c) The phylogenetic relationship among carriage isolates identified at OR. CC/ST lineages are highlighted and named according to the corresponding CC or ST.
TABLE 1.
Sequence types identified within the 12 major CCs across all the 3 universitiesa
| Clonal complex | ST |
|||
|---|---|---|---|---|
| OR, RI-1, RI-2 | OR | RI-1 | RI-2 | |
| CC41/44 | 136, 2578 | 44, 414, 1489, 5881, 10982, 11737, 11857, 12632, 12918, 12920 | 43, 437, 2578, 4682, 4971, 7574, 11525, 11541, 11737, 13079 | 8052 |
| CC32 | 32, 1130, 11395 | 2506 | ||
| CC35 | 35, 278 | 12908 | 11735, 11740 | 1679, 3085, 11530 |
| CC178 | 178 | 11550, 12933, 13038 | 11733 | 12927 |
| CC1157 | 1157, 1649, 12475 | 11580 | ||
| CC23 | 23 | 2533, 3582 | 5733, 6800 | |
| CC60 | 60, 11739 | 12912 | 2476, 13042 | |
| CC213 | 3496, 11852 | 3123, 11528 | 9413 | |
| CC269 | 467 | 3091, 11294, 12910, 12919 | 269 | 4221, 5329, 12753 |
| CC53 | 53 | 2647, 12909 | ||
| CC198 | 198, 823, 2384 | 2236, 10148, 12916, 12710 | 12716, 12755 | 10042, 12754, 12756, 12937, 12932 |
| CC1136 | 1136 | 11459, 12917 | ||
STs identified uniquely in each of the three universities and those identified at all three universities are shown.
Isolate information and sequencing statistics. Download Table S1, XLSX file, 0.1 MB (105.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Stability of population structure across rounds.
The stability of the most frequently observed CCs at each university was assessed across rounds (see Table S2). Overall, the observed counts of these CCs at each university were not significantly different across rounds, indicating that they remained stable throughout the course of our study within each respective university. Specific CCs, such as CC198, CC1157, and CC41/44 were among the most frequently observed CCs across each university.
Stability of the most observed CCs per round at each university. Download Table S2, DOCX file, 0.1 MB (16.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genomic comparison between the meningococcal outbreak strain and carriage isolates.
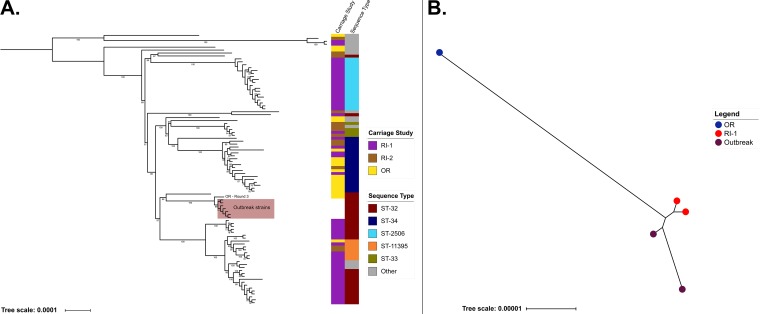
Whole-genome sequences of 7 CC32/ST-32 IMD isolates from the 2015 meningococcal outbreak at OR that precipitated the carriage study (21) were compared with those of 86 CC32 carriage isolates from the three universities (Fig. 2A). Phylogenetic analysis demonstrated that these 93 CC32 carriage and outbreak isolates clustered into five main clades, each associated with a specific ST. Clades 1 to 4 contained only carriage isolates, while clade 5 (ST-32) contained a subclade that included the seven outbreak isolates and one carriage isolate collected from OR during round three (Fig. 2A).
FIG 2.
Whole-genome phylogenetic relationships for CC32 and ST-9069 carriage and outbreak isolates. (A) The phylogenetic relationship among CC32 carriage isolates and outbreak strains collected from OR. Major carriage clades and STs are indicated in the figure. The OR carriage isolate from round 3 that was most closely related to the outbreak isolates is noted. Bootstrap values of >50 are included in the phylogeny. The “other” ST category consists of those STs with less than or equal to 3 isolates included in the phylogeny and consisted of ST-1130, ST-11527, ST-12936, ST-12914, ST-11397, ST-11278, ST-8758, ST-12924, ST-10875, and ST-12750. (B) The phylogenetic relationship among ST-9069 carriage isolates and outbreak isolates from RI-1.
ST-9069 caused the outbreak at RI-1 in 2015, and whole-genome sequences of the two outbreak isolates were used for comparison against those from carriage isolates. Three ST-9069 carriage isolates were identified, with two obtained from a single participant at RI-1 during rounds two and three and one from a participant at OR in round four. The two RI-1 ST-9069 carriage isolates were more closely related to the two RI-1 outbreak isolates than to the OR carriage isolate (Fig. 2B).
Comparative genomic analysis revealed genetic changes in 30 genes between the round three ST-32 OR carriage isolate and the most closely related outbreak isolate and in 72 genes between the ST-9069 RI-1 carriage isolates and the outbreak strain. Genetic changes were found in known N. meningitidis virulence genes such as those involved in adherence to the human epithelium and tissue tropism (type IV pili genes pilC, pilF, and pilU), host invasion (opacity protein genes such as opa), csb (capsular polymerase gene), lgtA, lgtG (N. meningitidis adhesion genes), and modB (DNA methylation) as well as in other genes not known to be associated with N. meningitidis virulence (see Table S3).
List of genes that showed genetic variation in the CC32 and ST-9069 carriage isolates and outbreak isolates. Download Table S3, XLSX file, 0.1 MB (45.1KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
N. meningitidis carriage dynamics among repeat participants.
Among 1,587 individuals who participated in multiple rounds of carriage evaluations (repeat participants), 348 were identified to be N. meningitidis carriers in at least one of the rounds and 194 were carriers of N. meningitidis in two or more rounds (repeat carriers) (Table 2). RI-1 had the highest number of repeat participants (n = 615), followed by OR (n = 613) and RI-2 (n = 359). Among the 348 repeat participants with N. meningitidis carriage in at least one round, 50.3% (175/348) maintained carriage of N. meningitidis with the same ST and CC across rounds (see Table S4), 44.3% (154/348) carried N. meningitidis in only one round of the study, and 5.4% (19/348) acquired a new N. meningitidis strain between rounds.
TABLE 2.
Number of repeat carriers that were positive for N. meningitidis carriage in each combination of rounds
| Round | No. of carriersa
|
|||
|---|---|---|---|---|
| RI-1 | RI-2 | OR | Total | |
| Two carriage rounds | ||||
| 1 + 2 | 36 | 29 | 19 | 84 |
| 1 + 3 | 9 | 0 | 1 | 10 |
| 1 + 4 | 10 | 0 | 2 | 12 |
| 2 + 3 | 19 | 0 | 8 | 27 |
| 2 + 4 | 3 | 0 | 4 | 7 |
| 3 + 4 | 9 | 0 | 9 | 18 |
| 1, 2 different ST | 1 | 1 | 3 | 5 |
| 2, 3 different ST | 3 | 0 | 0 | 3 |
| 1, 4 different ST | 2 | 0 | 0 | 2 |
| 1, 3 different ST | 0 | 0 | 0 | 0 |
| 2, 4 different ST | 2 | 0 | 1 | 3 |
| 3, 4 different ST | 1 | 0 | 2 | 3 |
| Total | 95 | 30 | 49 | 174 |
| Three carriage rounds | ||||
| 1 + 2 + 3 | 8 | 0 | 2 | 10 |
| 1 + 2 + 4 | 3 | 0 | 0 | 3 |
| 1 + 3 + 4 | 1 | 0 | 2 | 3 |
| 2 + 3 + 4 | 1 | 0 | 0 | 1 |
| 1 + 2, 4 different ST | 3 | 0 | 0 | 3 |
| Total | 16 | 0 | 4 | 20 |
The elapsed days from round 1 to subsequent rounds for each university were as follows. RI-1: round 2 (56 days), round 3 (185 days), round 4 (380 days). RI-2: round 2 (41 days); OR, round 2 (69 days), round 3 (218 days), round 4 (350 days).
Counts of repeat carriers that maintained the same ST and CC across rounds. Download Table S4, XLSX file, 0.1 MB (11.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The numbers of repeat participants who carried N. meningitidis in only one round of the study were 70/615 (11.4%), 15/359 (4.2%), and 69/613 (11.3%) for RI-1, RI-2, and OR, respectively. The number of repeat carriers among repeat participants was 111/615 (18.0%) at RI-1, 30/359 (8.3%) at RI-2, and 53/613 (8.6%) at OR. Among the repeat carriers from the three universities, 174/194 (89.7%) were carriers in two rounds (Table 2). Excluding repeat carriers from RI-2 (as it contained only two carriage rounds), 20/164 (12.2%) were carriers in three rounds; no repeat carriers were identified to be carriers in all four rounds. Eight repeat carriers tested negative for N. meningitidis carriage after testing positive in a previous round, and regained N. meningitidis carriage in a subsequent round; two regained carriage of a strain with the same CC and ST, and six gained carriage of a new strain.
Acquisition of new N. meningitidis strains in repeat carriers.
The 19 new strain acquisition events which were observed (Table 3) among repeat carriers involved the carrier either acquiring a different ST within the same CC or acquiring an entirely different CC. The only instance of new strain acquisition from a different ST within the same CC was detected in an individual from OR who was carrying an ST-178 (CC178) strain during round three and a ST-11550 (CC178) strain in round four. We did not observe any patterns or trends of certain STs or CCs being preferentially gained between any of the carriage evaluation rounds.
TABLE 3.
Within-host new strain (with a new CC/ST) acquisition events during the carriage evaluation time period in all three U.S. universitiesa
| Evaluation | CC (ST) or outcome |
|||
|---|---|---|---|---|
| Round 1 | Round 2 | Round 3 | Round 4 | |
| RI-1 | ||||
| A | CC32 (ST-11527) | CC32 (ST-11527) | Not swabbed | CC35 (ST-35) |
| B | CC41/44 (ST-409) | Not swabbed | Not swabbed | CC198 (ST-823) |
| C | CC198 (ST-2384) | CC198 (ST-2384) | Not swabbed | CC269 (ST-269) |
| E | CC1136 (ST-1136) | CC1136 (ST-1136) | Not swabbed | CC35 (ST-11740) |
| F | Swabbed (negb ) | CC32 (ST-1130) | CC198 (ST-6321) | Not swabbed |
| G | Not swabbed | CC198 (ST-823) | Swabbed (neg) | CC41/44 (ST-7574) |
| H | Not swabbed | CC1157 (ST-1157) | CC60 (ST-60) | Not swabbed |
| I | CC1157 (ST-1157) | CC198 (ST-823) | Swabbed (neg) | Not swabbed |
| J | Swabbed (neg) | CC60 (ST-11739) | CC35 (ST-11740) | Not swabbed |
| K | Swabbed (neg) | CC60 (ST- 60) | Swabbed (neg) | CC32 (ST-2506) |
| L | CC32 (ST-34) | Not swabbed | Not swabbed | CC41/44 (ST-409) |
| M | Not swabbed | Not swabbed | CC41/44 (ST-409) | CC60 (ST-60) |
| RI-2 | ||||
| A | CC53 (ST-53) | CC35 (ST-3085) | Not swabbed | Not swabbed |
| OR | ||||
| A | Not swabbed | Not swabbed | CC178 (ST-178) | CC178 (ST-11550) |
| B | Not swabbed | CC35 (ST-35) | Not swabbed | CC53 (ST-12930) |
| C | CC60 (ST-60) | CC1157 (ST-1649) | Not swabbed | Not swabbed |
| D | CC198 (ST 823) | CC41/44 (ST 12632) | Not swabbed | Not swabbed |
| E | CC60 (ST 60) | CC35 (ST 278) | Not swabbed | Not swabbed |
| F | Not swabbed | Not swabbed | CC 1157 (ST 1649) | CC53 (ST 53) |
A new strain was defined as identification of a strain with a different CC or ST than was previously identified for a given carrier.
neg, negative for N. meningitidis.
Within-host genetic diversity and variations of N. meningitidis isolates over time.
To assess if the 175 repeat carriers who maintained carriage of N. meningitidis with the same ST and CC across rounds were each carrying the same strain, the single nucleotide polymorphism (SNP) differences among these isolates, along with the SNP differences from the carried isolates among nonrepeat participants of the same CC, were compared (see Table S5). Within each of the CCs, the median SNP difference between isolates carried by repeat carriers was significantly different than the median SNP difference for the rest of the carried population, indicating that these repeat carriers were likely carrying the same strain across rounds. The pairwise SNP differences between isolates collected from these repeat carriers were compared against the elapsed time between rounds. In general, a weak correlation (Pearson’s r = 0.21) (Fig. S2) was observed between SNPs and days among the isolates collected from these repeat carriers.
SNP differences within clonal complexes from repeat carriers who maintained the same ST and CC across rounds and nonrepeat carriers. Download Table S5, DOCX file, 0.1 MB (15.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Among the isolates carried by these 175 repeat carriers, genetic variations were detected in 782 genes over time (see Table S6), with 16 of these genes being common among the strains harbored by >20% (30/175) of the repeat carriers. The most frequently observed gene with genetic variations was pilC1, which had undergone genetic variations in strains harbored by 72.6% (127/175) of the repeat carriers. The next most frequently changed genes were pilE (54.3%), pilC2 (53.14%), hpuA (39.4%), and pglA (37.7%).
List of genes and counts of genetic changes in the N. meningitidis carriage isolates from repeat participants who maintained carriage of a strain with the same ST and CC across rounds. Download Table S6, XLSX file, 0.3 MB (317.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Recombination was the most common mechanism behind the genetic differences observed between isolates obtained from repeat carriers with the same ST and CC and presented as multiple nucleotide differences along with insertions or deletions in the gene. Point mutations, deletions, and short sequence repeats (SSRs) were the next most frequently observed mechanisms responsible for genetic variations in isolates obtained from these repeat carriers. The genes that most frequently sustained SSRs in the isolates were NEIS1156 (unknown function), NEIS1750 (S-adenosylmethionine-dependent methyltransferase), lgtG (O-3 linked glucosyltransferase), NEIS3112 (unknown function), pglI (O-acetyltransferase), and pilC1 (pilus-associated protein). Previously characterized as phase variations induced by SSR, (CTTCT)n and poly(G/C) (23), were also found in opaA and pilC2 genes. Two SSRs, (CCTTGG)3 in the NEIS1987 gene and (CTTCAG)7 in the IbpB gene, were only identified in CC198 isolates.
DISCUSSION
To better understand the dynamics of U.S. meningococcal carriage in university and outbreak settings, we analyzed whole-genome sequencing (WGS) data for 1,514 N. meningitidis carriage isolates collected from ten cross-sectional carriage evaluation rounds conducted at three U.S. universities. While the predominant CCs were the same at the three universities, these three student populations were largely differentiated by the carried meningococcal ST distributions found at each, as many STs were observed only within one university during the evaluation time period. Carriage of N. meningitidis belonging to ST-32/CC32 and ST-9069, responsible for outbreaks at OR and RI-1, respectively, were detected. N. meningitidis carriage among U.S. university students appeared to be dynamic, and new strain acquisition events among repeat carriers primarily consisted of acquisition of a CC or ST which was also observed among other carriers within the same university population. Recombination, point mutations, and simple sequence repeats were the most frequent mechanisms for the genetic variation observed among carriage isolates that were detected within the hosts who maintained carriage of a strain with the same ST and CC for multiple rounds.
In this evaluation, CC198, CC1157, CC41/44, CC32, and CC35 were observed to be the five predominantly carried CCs among study participants from all three universities. Similarly, a previous carriage evaluation study among U.S. high school students in Georgia and Maryland during the academic year 2006 to 2007 also identified CC198 as the predominant CC, followed by CC23 and CC41/44, while carriage isolates associated within CC1157 and CC35 were the 5th and 6th most common CCs detected (24). In general, the most observed CCs identified at each university remained stable across evaluation rounds. CC198, CC1157, and CC41/44 were frequently observed in all 3 universities across all rounds, suggesting that strains belonging to these CCs were commonly carried in these populations.
Phylogenetic comparison indicated that the two ST-9069 carriage isolates from RI-1 were genetically similar to the RI-1 outbreak isolates, while the carriage ST-9069 isolate detected from OR was genetically different. ST-9069 is newly identified in the United States, with the only previous case reported before the RI-1 outbreak being a single carriage isolate from Ireland in 2009. As such, it is likely that the full diversity of this virulent lineage has not yet been captured. While ST-32 (CC32), which caused the outbreak in OR, is common among invasive disease cases in the United States, only two carriage isolates from this ST were detected at OR. One of these two isolates was highly similar to the outbreak strains. Overall, the low number of carriage isolates which were similar to the outbreak strains suggests that these strains were not commonly carried in the student population.
N. meningitidis carriage among U.S. university students appeared to be highly dynamic; nearly half of repeat participants with N. meningitidis carriage in at least one round retained carriage of a strain with the same ST and CC across rounds, and the other 44.3% carried N. meningitidis in only one round, and 5.4% acquired a new N. meningitidis strain. In a previous meningococcal carriage study among European teenagers, researchers found that among repeat participants who carried N. meningitidis in at least one round, 54.4% (147/270) retained the same strain across rounds, 37.4% (101/270) carried N. meningitidis in only one round, and 8.1% (22/270) acquired a new strain at some point over a 23-week time period (25). While these results were similar to ours, more data are needed to be able to produce generalized conclusions about carriage dynamics and strain acquisition events among the general population.
Given the extent of recombination and horizontal gene transfer observed in meningococci, it is likely that simultaneous carriage of multiple meningococcal strains occurs (26). Since only a single colony was selected for sequencing from the culture plates, this study does not consider whether strain heterogeneity was present within an individual or even on the collected swab. However, it was previously shown that colonization of the throat by multiple meningococcal clones is rare (27) and that only a small minority of swabs yield more than one simultaneous phenotypically distinct meningococcal isolate (28). Nevertheless, it is not possible to conclude from this study whether the new strain acquisition events observed were actual replacement of the existing strain or due to mixed carriage in which one strain was sampled during one round and the other in a subsequent round. As culturing several isolates from a single throat swab is labor intensive, metagenomics approaches for answering these questions should be considered for future carriage studies, such as sequencing bacteria directly from swabs. While expensive, this methodology would provide higher resolution and confidence in identifying novel strain acquisition events.
Recent studies investigating within-host meningococcal evolution by comparing genome sequences of throat-blood strain pairs from a limited number of patients with IMD (29), and within-host genetic changes in paired carriage isolates (30), identified genetic changes predominantly affecting the meningococcal type IV pilus genes. These genes are involved in the adherence to the human epithelium and tissue tropism. Similarly, our analyses comparing within-host genetic changes across rounds and outbreak isolates to their closest carriage isolate also found genetic changes in the genes involved in N. meningitidis adherence and virulence, as well as other genes associated with N. meningitidis invasion, escape from host immune defenses, iron uptake, and proteases. These results can inform future studies focused on identifying the significant genetic variations between carried and invasive meningococci.
Exposure to meningococcal bacteria can lead to either asymptomatic carriage, or less commonly, to IMD. Asymptomatic carriage may be an immunizing event, thereby helping to protect an individual against future IMD. In cases where IMD occurs, the time between exposure to the bacteria and development of IMD may be anywhere from 1 day to approximately 2 weeks after the acquisition of the bacterium (4, 31). The pathophysiological factors that lead to meningococci invading the bloodstream are not fully known. It has been hypothesized that this fast-track from exposure to the invasive phenotype is related to the short time required by SSRs to modulate gene expression and promote phenotypic variation (32). During this process, a random reassortment of proteins relevant to the host-pathogen interaction may produce a pathogenic variant capable of crossing the nasopharyngeal epithelium, resulting in IMD (33). The data collected during this carriage evaluation were not sufficient to confirm this hypothesis mainly because, thankfully, no participant developed IMD during this evaluation. Additionally, no preceding carriage isolates were available for the patients who developed meningococcal disease during the outbreaks. Nevertheless, future carriage studies will help to address questions about carriage acquisition, transmission, risk factors, and duration of carriage. Knowledge gained from such studies will be important for future research efforts working toward disrupting the transmission of meningococcal carriage isolates in the population, thereby potentially preventing future cases of IMD.
MATERIALS AND METHODS
Data collection.
From February 2015 to March 2016, a total of ten rounds of cross-sectional carriage surveys were conducted at three U.S. universities (RI-1, RI-2, an OR) using standardized methods, as previously described (6, 21, 22). A total of 1,514 N. meningitidis carriage isolates were obtained from 8,905 swabs collected from 7,001 unique participants. A total of 1,587 individuals participated in multiple survey rounds (repeat participants). N. meningitidis carriage isolates included 650 from four RI-1 carriage rounds (February 2015, April 2015, September 2015, and March 2016), 616 from four OR rounds (March 2015, May 2015, October 2015, and February 2016), and 248 from two RI-2 rounds (March 2015 and April 2015) (see Fig. S1 in the supplemental material). The serogroup for all isolates was determined using slide agglutination (SASG).
Flowchart detailing participants and isolates collected across the three universities. Download FIG S1, PDF file, 0.1 MB (95.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Scatter plot showing the comparison of SNP differences and days elapsed among isolates obtained from repeat carriers carrying the same strain across rounds. Pearson’s correlation r = 0.24. Download FIG S2, DOCX file, 0.02 MB (25.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Whole-genome sequencing and molecular typing.
All N. meningitidis carriage isolates were characterized with whole-genome sequencing (WGS). Genomic DNA was prepared for sequencing using the 5Prime ArchivePure DNA Purification kit (5Prime, Gaithersburg, MD) or Gentra Puregene yeast/bacteria DNA extraction kit (Qiagen, Germantown, MD) as previously described (34) for sequencing on Pacific Biosystems (PacBio, Menlo Park, CA, USA) and/or Illumina (HiSeq2500 or MiSeq; Illumina, San Diego, CA, USA) platforms; genomic DNA was further prepared for Illumina sequencing with the dual-index NEBNext Ultra DNA library preparation kits (New England BioLabs Inc., Ipswich, MA). Genomes generated using the PacBio platform were processed as previously described (35). The Illumina reads were trimmed with Cutadapt (36), and de novo short read assembly was carried out using SPAdes 3.7.0 (37) with the “careful” option. Assemblies were compared with BLAST (38) against the PubMLST Neisseria allele collection (39) to identify MLST alleles. The MLST genes were compared against the PubMLST MLST scheme collection to identify the N. meningitidis ST and CC.
Phylogenetic and comparative analyses.
The phylogenetic trees were constructed on carriage isolates from each university using a whole-genome alignment (WGA) produced by Parsnp (Harvest suite, version 1.2) (40). WGA was used to infer the maximum likelihood (ML) phylogeny using RAxML with a general time-reversible nucleotide model (41). The recombination corrected phylogenetic tree was generated using ClonalFrameML (42). The final phylogenetic tree was annotated in Interactive Tree of Life (43).
For the CC32 and ST-9069 phylogenetic analysis and comparative genomic analysis of carriage and outbreak isolates and the calculations of SNP differences between repeat and nonrepeat participants, trimmed Illumina sequencing reads were aligned to a completed reference genome of each CC/ST generated using PacBio sequencing and curated Illumina reads. Whole-genome alignments were generated using Snippy (https://github.com/tseemann/snippy), which identifies variants using Freebayes v1.0.2 (44) with a minimum 10× read coverage and 90% read concordance at a locus for each SNP. Putative regions of recombination were predicted and masked using Gubbins (45). The resulting SNP alignment was used to calculate the SNP differences among repeat and nonrepeat participants, and for CC32 and ST-9069, the alignment was used as input for RAxML in order to infer the ML phylogenetic tree using the general time reversible (GTR) model and 100 bootstrap replicates. The genomes involved in the CC32 and ST-9069 comparative genome analysis were annotated using the PubMLST Neisseria allele collection using custom scripts with BLAST (38). The genes harboring genetic changes between the carriage and invasive isolates were identified by comparing the alleles present for each locus. Pairwise SNP differences between isolates obtained from repeat carriers harboring a strain with the same ST and CC across rounds were obtained by generating a recombination corrected whole-genome alignment using Snippy and Gubbins, as described above, against a same-ST reference.
To identify genes with frequent genetic variations, as well as the types of variations, among carried isolates from repeat carriers with the same ST and CC across rounds, genetic variations were identified through mapping sequencing reads of carriage isolates against a common reference genome of the same CC to identify variant locations. Variation type was defined as point mutation if only a single nucleotide difference was detected in a gene, homologous recombination if multiple nucleotide variations (SNPs, multinucleotide polymorphisms [MNPs], and complex) and insertions were observed in the same gene, simple sequence repeats if repeated nucleotides with various length were inserted into a gene, and indels if random sequences were inserted or deleted in the gene. These variant locations were mapped against gene locations determined by annotating each reference genome using the PubMLST Neisseria allele database, through the use of custom python scripts, to identify the genes harboring these variations. The significance of the median SNP differences between repeat and nonrepeat participants was calculated using the Wilcoxon’s rank sum test (46). The stability of the most frequently observed CCs per university across each round was assessed with a chi-square statistical analysis using SAS 9.4.
Data availability.
Sequence reads for all isolates used in this analysis are available in BioProject PRJNA533315.
ACKNOWLEDGMENTS
Sandeep J. Joseph, Nadav Topaz, How-Yi Chang, Fang Hu, and Lorraine D. Rodriguez-Rivera are contractors assigned to the Meningitis and Vaccine Preventable Diseases Branch, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
We thank the CDC’s Biotechnology Core Facility for their assistance in whole-genome sequencing of isolates included in this work. We also thank members of the CDC Meningitis and Vaccine Preventable Diseases Branch for their support and feedback.
This work was supported by the Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Mbaeyi SA, Blain A, Whaley MJ, Wang X, Cohn AC, MacNeil JR. 2018. Epidemiology of meningococcal disease outbreaks in the United States, 2009–2013. Clin Infect Dis 68:580–585. doi: 10.1093/cid/ciy548. [DOI] [PubMed] [Google Scholar]
- 2.Mbaeyi SA, Joseph SJ, Blain A, Wang X, Hariri S, MacNeil JR. 2019. Meningococcal disease among college-aged young adults: 2014–2016. Pediatrics 143:e20182130. doi: 10.1542/peds.2018-2130. [DOI] [PubMed] [Google Scholar]
- 3.Soeters HM, McNamara LA, Blain AE, Whaley M, MacNeil JR, Hariri S, Mbaeyi S. 2019. University-based outbreaks of meningococcal disease caused by serogroup B, United States, 2013–2018. Emerg Infect Dis 25:434–440. doi: 10.3201/eid2503.181574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephens DS, Greenwood B, Brandtzaeg P. 2007. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369:2196–2210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- 5.Claus H, Maiden MC, Wilson DJ, McCarthy ND, Jolley KA, Urwin R, Hessler F, Frosch M, Vogel U. 2005. Genetic analysis of meningococci carried by children and young adults. J Infect Dis 191:1263–1271. doi: 10.1086/428590. [DOI] [PubMed] [Google Scholar]
- 6.Soeters HM, Whaley M, Alexander-Scott N, Kanadanian KV, MacNeil JR, Martin SW, McNamara LA, Sicard K, Vanner C, Vuong J, Wang X, Bandy U, Patel M, Rhode Island Meningococcal Carriage Evaluation Team. 2017. Meningococcal carriage evaluation in response to a serogroup B meningococcal disease outbreak and mass vaccination campaign at a college-Rhode Island, 2015-2016. Clin Infect Dis 64:1115–1122. doi: 10.1093/cid/cix091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soeters HM, McNamara LA, Whaley M, Wang X, Alexander-Scott N, Kanadanian KV, Kelleher CM, MacNeil J, Martin SW, Raines N, Sears S, Vanner C, Vuong J, Bandy U, Sicard K, Patel M, Centers for Disease Control (CDC). 2015. Serogroup B meningococcal disease outbreak and carriage evaluation at a college-Rhode Island, 2015. MMWR Morb Mortal Wkly Rep 64:606–607. [PMC free article] [PubMed] [Google Scholar]
- 8.McNamara LA, Shumate AM, Johnsen P, MacNeil JR, Patel M, Bhavsar T, Cohn AC, Dinitz-Sklar J, Duffy J, Finnie J. 2015. First use of a serogroup B meningococcal vaccine in the US in response to a university outbreak. Pediatrics 135:794–804. doi: 10.1542/peds.2014-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caugant D, Høiby E, Rosenqvist E, Frøholm L, Selander R. 1992. Transmission of Neisseria meningitidis among asymptomatic military recruits and antibody analysis. Epidemiol Infect 109:241–253. doi: 10.1017/s0950268800050196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jounio U, Saukkoriipi A, Bratcher HB, Bloigu A, Juvonen R, Silvennoinen-Kassinen S, Peitso A, Harju T, Vainio O, Kuusi M, Maiden MCJ, Leinonen M, Käyhty H, Toropainen M. 2012. Genotypic and phenotypic characterization of carriage and invasive disease isolates of Neisseria meningitidis in Finland. J Clin Microbiol 50:264–273. doi: 10.1128/JCM.05385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caugant DA, Maiden MC. 2009. Meningococcal carriage and disease—population biology and evolution. Vaccine 27:B64–B70. doi: 10.1016/j.vaccine.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ala'Aldeen DA, Neal KR, Ait-Tahar K, Nguyen-Van-Tam JS, English A, Falla TJ, Hawkey PM, Slack RC. 2000. Dynamics of meningococcal long-term carriage among university students and their implications for mass vaccination. J Clin Microbiol 38:2311–2316. doi: 10.1128/JCM.38.6.2311-2316.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson ME, Mile R, Li Y, Nair H, Kyaw MH. 2018. Meningococcal carriage in high-risk settings: a systematic review. Int J Infect Dis 73:109–117. doi: 10.1016/j.ijid.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 14.De Wals P, Gilquin C, De Maeyer S, Bouckaert A, Noël A, Lechat M, Lafontaine A. 1983. Longitudinal study of asymptomatic meningococcal carriage in two Belgian populations of schoolchildren. J Infect 6:147–156. doi: 10.1016/s0163-4453(83)92756-1. [DOI] [PubMed] [Google Scholar]
- 15.Ali O, Aseffa A, Omer AB, Lema T, Demissie TM, Tekletsion Y, Worku A, Xabher HG, Yamuah L, Boukary RM, Collard J-M, Dan Dano I, Habiboulaye I, Issaka B, Jusot J-F, Ousmane S, Rabe I, Dauglaz DM, Gami JP, Gamougam K, Mbainadji L, Naibei N, Narbé M, Toralta J, Berthe A, Diallo K, Keita M, Coulibaly A, Onwuchekwa U, Sow SO, Tamboura B, Traore A, Toure A, Clark T, Mayer L, Amodu M, Beida O, Gadzama G, Omotara B, Zailani S, Yahya S, Chandramohan D, Greenwood BM, Hassan-King M, Manigart O, Nascimento M, M Stuart J, Woukeu A, Basta NE, Bai X, et al. 2016. Household transmission of Neisseria meningitidis in the African meningitis belt: a longitudinal cohort study. Lancet Glob Health 4:e989–e995. doi: 10.1016/S2214-109X(16)30244-3. [DOI] [PubMed] [Google Scholar]
- 16.Harrison LH, Shutt KA, Schmink SE, Marsh JW, Harcourt BH, Wang X, Whitney AM, Stephens DS, Cohn AA, Messonnier NE, Mayer LW. 2010. Population structure and capsular switching of invasive Neisseria meningitidis isolates in the pre-meningococcal conjugate vaccine era—United States, 2000–2005. J Infect Dis 201:1208–1224. doi: 10.1086/651505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Cohn A, Comanducci M, Andrew L, Zhao X, MacNeil JR, Schmink S, Muzzi A, Bambini S, Rappuoli R, Pizza M, Murphy E, Hoiseth SK, Jansen KU, Anderson AS, Harrison LH, Clark TA, Messonnier NE, Mayer LW. 2011. Prevalence and genetic diversity of candidate vaccine antigens among invasive Neisseria meningitidis isolates in the United States. Vaccine 29:4739–4744. doi: 10.1016/j.vaccine.2011.04.092. [DOI] [PubMed] [Google Scholar]
- 18.Potts CC, Joseph SJ, Chang H-Y, Chen A, Vuong J, Hu F, Jenkins LT, Schmink S, Blain A, MacNeil JR, Harrison LH, Wang X. 2018. Population structure of invasive Neisseria meningitidis in the United States, 2011–15. J Infect 77:427–434. doi: 10.1016/j.jinf.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoen C, Blom J, Claus H, Schramm-Glück A, Brandt P, Müller T, Goesmann A, Joseph B, Konietzny S, Kurzai O, Schmitt C, Friedrich T, Linke B, Vogel U, Frosch M. 2008. Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc Natl Acad Sci U S A 105:3473–3478. doi: 10.1073/pnas.0800151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yazdankhah SP, Kriz P, Tzanakaki G, Kremastinou J, Kalmusova J, Musilek M, Alvestad T, Jolley KA, Wilson DJ, McCarthy ND, Caugant DA, Maiden MCJ. 2004. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J Clin Microbiol 42:5146–5153. doi: 10.1128/JCM.42.11.5146-5153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNamara LA, Thomas JD, MacNeil J, Chang HY, Day M, Fisher E, Martin S, Poissant T, Schmink SE, Steward-Clark E, Jenkins LT, Wang X, Acosta A, Oregon Meningococcal Carriage Team. 2017. Meningococcal carriage following a vaccination campaign with MenB-4C and MenB-FHbp in response to a university serogroup B meningococcal disease outbreak-Oregon, 2015-2016. J Infect Dis 216:1130–1140. doi: 10.1093/infdis/jix446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breakwell L, Whaley M, Khan UI, Bandy U, Alexander-Scott N, Dupont L, Vanner C, Chang H-Y, Vuong JT, Martin S, MacNeil JR, Wang X, Meyer SA. 2018. Meningococcal carriage among a university student population–United States, 2015. Vaccine 36:29–35. doi: 10.1016/j.vaccine.2017.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saunders NJ, Jeffries AC, Peden JF, Hood DW, Tettelin H, Rappuoli R, Moxon ER. 2000. Repeat‐associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol Microbiol 37:207–215. doi: 10.1046/j.1365-2958.2000.02000.x. [DOI] [PubMed] [Google Scholar]
- 24.Harrison LH, Shutt KA, Arnold KE, Stern EJ, Pondo T, Kiehlbauch JA, Myers RA, Hollick RA, Schmink S, Vello M, Stephens DS, Messonnier NE, Mayer LW, Clark TA. 2015. Meningococcal carriage among Georgia and Maryland high school students. J Infect Dis 211:1761–1768. doi: 10.1093/infdis/jiu679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glitza I, Ehrhard I, Müller-Pebody B, Reintjes R, Breuer T, Ammon A, Sonntag H-G. 2008. Longitudinal study of meningococcal carrier rates in teenagers. Int J Hyg Environ Health 211:263–272. doi: 10.1016/j.ijheh.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Trotter CL, Maiden MC. 2016. Carriage and transmission of Neisseria meningitidis, p 15–23. In Feavers I, Pollard AJ, Sadarangani M. (eds), Handbook of meningococcal disease management. Springer, Cham, Switzerland. [Google Scholar]
- 27.Caugant DA, Tzanakaki G, Kriz P. 2007. Lessons from meningococcal carriage studies. FEMS Microbiol Rev 31:52–63. doi: 10.1111/j.1574-6976.2006.00052.x. [DOI] [PubMed] [Google Scholar]
- 28.Andersen J, Berthelsen L, Bech Jensen B, Lind I. 1998. Dynamics of the meningococcal carrier state and characteristics of the carrier strains: a longitudinal study within three cohorts of military recruits. Epidemiol Infect 121:85–94. doi: 10.1017/s0950268898008930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klughammer J, Dittrich M, Blom J, Mitesser V, Vogel U, Frosch M, Goesmann A, Müller T, Schoen C. 2017. Comparative genome sequencing reveals within-host genetic changes in Neisseria meningitidis during invasive disease. PLoS One 12:e0169892. doi: 10.1371/journal.pone.0169892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bårnes GK, Brynildsrud OB, Børud B, Workalemahu B, Kristiansen PA, Beyene D, Aseffa A, Caugant DA. 2017. Whole genome sequencing reveals within-host genetic changes in paired meningococcal carriage isolates from Ethiopia. BMC Genomics 18:407. doi: 10.1186/s12864-017-3806-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzeng Y-L, Stephens DS. 2000. Epidemiology and pathogenesis of Neisseria meningitidis. Microbes Infect 2:687–700. doi: 10.1016/s1286-4579(00)00356-7. [DOI] [PubMed] [Google Scholar]
- 32.Siena E, D'Aurizio R, Riley D, Tettelin H, Guidotti S, Torricelli G, Moxon ER, Medini D. 2016. In-silico prediction and deep-DNA sequencing validation indicate phase variation in 115 Neisseria meningitidis genes. BMC Genomics 17:843. doi: 10.1186/s12864-016-3185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siena E, Bodini M, Medini D. 2018. Interplay between virulence and variability factors as a potential driver of invasive meningococcal disease. Comput Struct Biotechnol J 16:61–69. doi: 10.1016/j.csbj.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Retchless AC, Hu F, Ouédraogo A-S, Diarra S, Knipe K, Sheth M, Rowe LA, Sangaré L, Ky Ba A, Ouangraoua S, Batra D, Novak RT, Ouédraogo Traoré R, Wang X. 2016. The establishment and diversification of epidemic-associated serogroup W meningococcus in the African meningitis belt, 1994 to 2012. mSphere 1:e00201-16. doi: 10.1128/mSphere.00201-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kretz CB, Retchless AC, Sidikou F, Issaka B, Ousmane S, Schwartz S, Tate AH, Pana A, Njanpop-Lafourcade B-M, Nzeyimana I, Nse RO, Deghmane A-E, Hong E, Brynildsrud OB, Novak RT, Meyer SA, Oukem-Boyer OOM, Ronveaux O, Caugant DA, Taha M-K, Wang X, Niger Response Team. 2016. Whole-genome characterization of epidemic Neisseria meningitidis serogroup C and resurgence of serogroup W, Niger, 2015. Emerg Infect Dis 22:1762–1768. doi: 10.3201/eid2210.160468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 37.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907 [q-bio.GN] https://arxiv.org/abs/1207.3907.
- 45.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peto R, Peto J. 1972. Asymptotically efficient rank invariant test procedures. J R Stat Soc Ser A 135:185–198. doi: 10.2307/2344317. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Isolate information and sequencing statistics. Download Table S1, XLSX file, 0.1 MB (105.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Stability of the most observed CCs per round at each university. Download Table S2, DOCX file, 0.1 MB (16.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of genes that showed genetic variation in the CC32 and ST-9069 carriage isolates and outbreak isolates. Download Table S3, XLSX file, 0.1 MB (45.1KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Counts of repeat carriers that maintained the same ST and CC across rounds. Download Table S4, XLSX file, 0.1 MB (11.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
SNP differences within clonal complexes from repeat carriers who maintained the same ST and CC across rounds and nonrepeat carriers. Download Table S5, DOCX file, 0.1 MB (15.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of genes and counts of genetic changes in the N. meningitidis carriage isolates from repeat participants who maintained carriage of a strain with the same ST and CC across rounds. Download Table S6, XLSX file, 0.3 MB (317.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Flowchart detailing participants and isolates collected across the three universities. Download FIG S1, PDF file, 0.1 MB (95.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Scatter plot showing the comparison of SNP differences and days elapsed among isolates obtained from repeat carriers carrying the same strain across rounds. Pearson’s correlation r = 0.24. Download FIG S2, DOCX file, 0.02 MB (25.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Sequence reads for all isolates used in this analysis are available in BioProject PRJNA533315.