Abstract
Normal cellular physiology and biochemical processes require undamaged RNA molecules. However, RNAs are frequently subjected to oxidative damage. Overproduction of reactive oxygen species (ROS) leads to RNA oxidation and disturbs redox (oxidation-reduction reaction) homeostasis. When oxidation damage affects RNA carrying protein-coding information, this may result in the synthesis of aberrant proteins as well as a lower efficiency of translation. Both of these, as well as imbalanced redox homeostasis, may lead to numerous human diseases. The number of studies on the effects of RNA oxidative damage in mammals is increasing by year due to the understanding that this oxidation fundamentally leads to numerous human diseases. To enable researchers in this field to explore information relevant to RNA oxidation and effects on human diseases, we developed DES-ROD, an online knowledgebase that contains processed information from 298,603 relevant documents that consist of PubMed abstracts and PubMed Central full-text articles. The system utilizes concepts/terms from 38 curated thematic dictionaries mapped to the analyzed documents. Researchers can explore enriched concepts, as well as enriched pairs of putatively associated concepts. In this way, one can explore mutual relationships between any combinations of two concepts from used dictionaries. Dictionaries cover a wide range of biomedical topics, such as human genes and proteins, pathways, Gene Ontology categories, mutations, noncoding RNAs, enzymes, toxins, metabolites, and diseases. This makes insights into different facets of the effects of RNA oxidation and the control of this process possible. The usefulness of the DES-ROD system is demonstrated by case studies on some known information, as well as potentially novel information involving RNA oxidation and diseases. DES-ROD is the first knowledgebase based on text and data mining that focused on the exploration of RNA oxidation and human diseases.
1. Background
Oxidative damage induced by reactive oxygen species (ROS) to the cellular elements such as proteins, lipids, and DNA has proven to be deleterious to organisms as a whole. Until recently, RNA damage was not recognized and explored as one such component of ROS effects on cellular elements. RNA oxidation was believed to be primarily a consequence of a dying cell until it was shown that changes in RNA structure are early events in the development of aging-related disorders such as Alzheimer's disease (AD), Parkinson's disease (PD), amyotrophic lateral sclerosis (ALS), and cardiovascular diseases (CVD) [1–6]. These findings are strengthened by the notion that RNA species rRNA and tRNA are abundantly present in the cell and are not readily degraded during cell growth. These features have recognized RNA oxidation to be a great challenge to cell function and to cell surveillance mechanisms that control oxidative-reductive stress. Alterations of these processes may advance the development of pathologies in various diseases [1, 6–8].
RNA undergoes oxidative damage more often than DNA, owing to the RNA's location in the cytosol, where they are in closer proximity to the mitochondria where oxidative stress is generated and because there are no protective histones in single-stranded RNA structure [1]. These oxidative modifications of RNA affect translation and synthesis of proteins, as well as the self-regulatory processes of transcription repression by various miRNAs [9, 10]. Thus, curbing the accumulation of oxidatively damaged RNA aids the maintenance of cellular health and prevents disease development. Several RNA oxidation surveillance mechanisms prevent such accumulation. That is, oxidized RNAs appear to be targeted for degradation [11] in a process that involves the ribosome [12, 13]. Another RNA oxidation surveillance mechanism is via RNA-binding proteins such as YB-1, which was shown to bind to 8-oxoguanosine (8-oxoG) with high affinity [14]. This action of YB-1 and its known role in mRNA stability associated with helping the winding of RNA duplexes suggest that this protein may be functioning as an RNA chaperone that targets oxidized RNA for degradation [14]. Another mechanism of RNA quality control is promoted by proteins such as MTH1, MTH2, and NUDT5. These proteins can hydrolyze oxidatively damaged RNA (such as 8-oxoG), thereby eliminating them from the RNA precursor pool [15]. Many such research findings showed the complexity and importance of the RNA oxidation-related processes. However, research related to RNA oxidation mechanisms and its role in different diseases is scattered in a large volume of scientific literature. For example, indexed in the Web of Science (All Databases) (https://clarivate.com/), specifically focused on the RNA oxidations in human diseases, there are 50,905 and 273,633 scientific articles published in 2018 and the 2014-2018 period, respectively, while in the most strict selection of the Web of Science Core Collection, there are 21,578 and 100,016 articles published in 2018 and the 2014-2018 period, respectively. This volume of literature makes it infeasible to efficiently search for RNA oxidation-related information or track significant developments manually. Such bottlenecks are not new to specialized domains; thus, several groups have been looking for ways to simplify the search for useful information.
2. Exploring Voluminous Information
It has been acknowledged that automated systems are needed to search for and retrieve useful information from such voluminous data. Thus, several automated systems have been developed using text mining (TM) and/or natural language processing (NLP) for over 30 years [16–23]. Moreover, TM and NLP methods have been combined with different approaches for knowledge extraction from free text. For example, ontologies provide a systematic representation of interrelationships between terms in a specific domain [24, 25]. Various ontology-based frameworks have been developed [26] such as Aber-OWL [27]. Additionally, ontology-based systems have different purposes, for example, identifying pathways using pharmacogenomics data [28] and selecting gene candidates [29]. Other methods are based on network analysis [30] and biological knowledge graphs [31]. In addition, TM has been combined with methods from bioinformatics. For example, position weight matrices have been used for text representation and feature generation in a TM system to extract associations between methylated genes and diseases [32, 33]. Another study combined TM and bioinformatics approaches for the interpretation of mutations in protein kinases [34].
Several generalized automated systems were designed to facilitate extracting information from biomedical literature [35]. For example, iHOP uses a text mining approach wherein genes and proteins are used as hyperlinks between sentences and PubMed abstracts and then uses the text-mined information to produce network representations that users can browse [36]. Other tools include Twister that is aimed at reducing the screening time of systematic literature reviews [37]; SWIFT-Review, which is a workbench for systematic review based on NLP [38]; SparkText, which is a big data framework for mining biomedical literature [39]; and GIS, which is an NLP-based framework for gene discovery from scientific literature [40]. In addition to these tools, several frameworks for mining biomedical literature have been developed [41–47].
Automated extraction of relevant and necessary information helps improve our understanding and knowledge of specific domains, propose hypotheses, and potentially discover new knowledge. For example, TM and NLP systems have been used to identify new candidate compounds for drug repurposing [48, 49], analyze relationships between proteostasis protein factors and cancer [50], prioritize cancer genes and pathways [51], predict protein functions [52], and extract disease-related biomarkers [53], as well as find associations between TFs [54]. Additionally, the text has been used as features to represent protein structures and subsequently predict their characteristics computationally [55]. Other useful applications of TM and NLP have been reported in the literature [56–61].
Moreover, various domain-specific knowledgebases (KB) exist. For example, CRAB is a KB implemented to support chemical health risk assessment through literature TM [61]. Another KB is ERIC, developed to support research focused on molecular mechanisms of bacterial enteropathogens using TM of PubMed abstracts [62]. Also, CNVdigest created using TM assists geneticists or physicians to find rare CNVs and the original literature context for more detailed information [63]. Other tools include CHAT (Cancer Hallmark Analytics Tool), developed to organize and evaluate cancer-related scientific literature [64]; FamPlex, designed for exploring associations between human protein families and complexes in the scientific literature [65]; and PPInterFinder and PIminer, implemented for mining protein-protein interactions from biomedical text [66, 67], while [54] presents a tool for context-specific protein interaction networks based on TM. In addition to these tools, several tools have been developed for specialized domains [68].
Here, we develop DES-ROD, the KB focused on RNA oxidation-related research, and demonstrate its utility in this domain, focusing on the role of RNA oxidation in the development of AD, CVD, and obesity.
3. The DES-ROD Exploration System
We developed DES-ROD using the DES V3.0 framework on 26 November 2018. DES is a text mining and data mining system that allows the exploration of text through enriched concepts and enriched pairs of concepts in topic-specific literature. We used the DES framework to create several topic-specific KBs [32, 33, 54, 67, 69–82]. The underlying systems, workflow, and concept enrichment process used in the current version of DES have been described in [69]. The user manual is provided at https://des-documentation.readthedocs.io/en/des-rod/.
Specific to this DES-ROD, our local MongoDB repository (updated September 03, 2018) hosting PubMed and PubMed Central articles was used to retrieve all topic-specific articles using the following query: “(human OR mouse OR rat OR mammal∗) AND (“RNA damage” OR Fenton OR PNPase OR hPNPase OR APE1 OR “apyrmidinic endonuclease 1” OR “apurinic endonuclease 1” OR nucleophosmin∗OR NPM1 OR “purine nucleoside phosphorylase” OR PNP OR “oxidative demethylase” OR “tRNA nucleotidyl transferase” OR “Y box binding protein” OR “Ro autoantigen” OR “8-hydroxyguanine” OR “8-oxoG” OR “8-hydroxyguanosine” OR “8-oxo-deoxyguanosine triphosphate” OR “8-oxodGTP” OR “8-oxo-guanosine triphosphate” OR “8-oxo-GTP” OR “nucleoside-diphosphate kinase” OR NDK OR “adenosine-diphosphate kinase” OR ADK OR Lipoxygenase∗OR LOs OR “4-hydroxy-2,3-nonenal” OR HNE OR “4-oxo-2-nonenal” OR acrolein OR “reductive stress” OR radical∗OR peroxide∗OR ROS OR “reactive oxygen species” OR RNS OR “reactive nitrogen species” OR redox OR “reduction-oxidation reaction” OR oxidat∗OR nitrosat∗OR peroxide∗OR superoxide∗OR detoxifi∗OR antioxid∗OR “polyunsaturated fatty acids” OR “arachidonic acid” OR “linoleic acid” OR hydroperoxide∗OR “hypochlorous acid” OR peroxynitrit∗OR flavoprot∗OR oxidase∗OR “cytochromes P450” OR catalase∗OR sulfiredoxin∗OR peroxiredoxin∗) AND (clinic∗OR disease∗OR diabet∗OR obes∗OR syndrome∗OR neuro∗OR heart OR cardi∗OR cancer∗). The query retrieved 286,370 articles used as the literature corpus. This literature corpus was indexed using 38 dictionaries: 28 dictionaries from the preexisting DES v2.0 vocabularies (used to develop other KBs) and 10 newly compiled topic-relevant dictionaries (see Table 1).
Table 1.
Dictionaries used in DES-ROD with data source references.
| Dictionary | Enriched unique terms in the KB | Source |
|---|---|---|
| Chemicals/compounds | ||
| Chemical Entities of Biological Interest (ChEBI) [83] | 19,298 | Preexisting in DES |
| Toxins (T3DB) [84] | 2,193 | Preexisting in DES |
| Lipids (lipid maps) [85,86] | 3,099 | Preexisting in DES |
| Amyloids (Human and Mouse), compiled in-house | 394 | Newly compiled |
| Functional annotation | ||
| Biological Process (GO) [87] | 5,868 | Preexisting in DES |
| Cellular Component (GO) [87] | 1,284 | Preexisting in DES |
| Molecular Function (GO) [87] | 1,963 | Preexisting in DES |
| Pathways (KEGG [88], Reactome [89], UniPathway [90], and PANTHER [91]) | 1,584 | Preexisting in DES |
| Diseases | ||
| DOID Ontology (BioPortal) Human Disease Ontology [92] | 3,637 | Preexisting in DES |
| ADO Ontology (BioPortal) Alzheimer's Disease Ontology [93] | 937 | Newly compiled |
| DMTO Ontology (BioPortal) Diabetes Mellitus Treatment Ontology [94] | 1,980 | Newly compiled |
| HFO Ontology (BioPortal) Heart Failure Ontology [95] | 1,002 | Newly compiled |
| CVDO Ontology (BioPortal) Cardiovascular Disease Ontology [96] | 49 | Newly compiled |
| HP Ontology (BioPortal) Human Phenotype Ontology [97] | 3,306 | Preexisting in DES |
| UBERON Ontology (BioPortal) Uber Anatomy Ontology [98] | 6,657 | Newly compiled |
| ICD9 Ontology (BioPortal) International Classification of Diseases, Version 9-Clinical Modification [99] | 719 | Preexisting in DES |
| Drugs | ||
| Drugs (DrugBank) [100] | 4,025 | Preexisting in DES |
| ATC Ontology (BioPortal) Anatomical Therapeutic Chemical Classification [101] | 2,008 | Newly compiled |
| CSSO Ontology (BioPortal) Clinical Signs and Symptoms Ontology | 206 | Newly compiled |
| SIDER (Drug Indications and Side Effects) [102] | 3,203 | Preexisting in DES |
| Human | ||
| Human Genes and Proteins (EntrezGene) [103] | 22,896 | Preexisting in DES |
| Human Transcription Factors [104] | 1,565 | Preexisting in DES |
| Human Transcription Cofactors (TcoF-DB) [104] | 388 | Preexisting in DES |
| Human microRNAs (HGNC [105] and EntrezGene) [106] | 2,088 | Updated |
| Human Long Noncoding RNAs (HGNC) [105] | 527 | Preexisting in DES |
| Mutations (tmVar) [107] | 15,852 | Preexisting in DES |
| Human Anatomy (in-house compiled) | 2,569 | Preexisting in DES |
| OMIT Ontology (BioPortal) Ontology for MicroRNA Target [19] | 695 | Newly compiled |
To integrate these newly compiled dictionaries into DES-ROD, redundant dictionary concepts are unified and concepts are normalized to ensure that a single concept represents synonymous symbols and names. Then, initial indexing is performed to identify and remove promiscuous or ambiguous concepts. After this dictionary cleaning, the literature corpus is reindexed to calculate and ensure the accuracy of concepts' enrichment estimates.
Concepts are recognized as enriched, if their occurrence in the DES-ROD literature corpus is proportionally higher than its occurrence in the complete set of PubMed and PubMed Central articles in our local repository and has a false discovery rate (FDR) < 0.05. A total of 131,741 concepts were determined to be statistically enriched in DES-ROD (see Table 1). Also, 10,846,802 pairs of concepts were determined to be statistically enriched. Concepts are regarded as cooccurring based on their cooccurrence in the text within a 200-character distance from each other. The resulting network of concept pairs was also embedded in a high-dimensional semantic space, enabling the computation of semantic similarity between concepts. The literature corpus, 38 dictionaries, enriched concepts, enriched pairs of concepts, and semantic similarities were integrated to create DES-ROD.
4. Knowledgebase Utilities
DES-ROD allows RNA oxidation-related literature to be easily explored using concepts found to be statistically enriched in the topic-specific literature. The KB is designed to provide users with multiple means to explore the literature with topic-relevant concepts (determined through concept enrichment estimates). Users are provided with multiple views, including “Enriched Concepts”, “Enriched Pairs”, “Semantic Similarity”, and “Literature”. Briefly, individual-enriched concepts can be explored on the “Enriched Concepts” page where their mentions in the text are highlighted on the right-hand side annotation pane, enriched cooccurring concepts on the “Enriched Pairs” page are also linked to their cooccurrence context in the literature, and concepts with semantic similarity to a chosen enriched concept are displayed on the “Semantic Similarity” page. The “Semantic Similarity” link is new in this version of DES. Using these utilities, users can view all enriched concepts, search for their concept of interest, or select a specific dictionary. Furthermore, provided is a “Column visibility” tab that allows viewing the enriched concepts using several ranking options, including false discovery rate (FDR), KB frequency, background frequency, or density. Moreover, highlighting the concept or concept pair of interest allows the user to view the literature from where the indexing was retrieved. Also, concepts are highlighted, making them easily identifiable in the literature, as well as color-coded to indicate in which dictionary the concept is located. Each concept is also linked to a right-click menu which allows users to generate a “Network” view or “Term Co-occurrences” table. The literature in DES-ROD can also be explored via the “Literature” view. Case study examples are given below to demonstrate the utility of DES-ROD.
5. Case Studies that Demonstrate the Use of DES-ROD as a Research Supporting System
Example 1 . —
Hypothesis derived through the use of DES-ROD.
Hypothesis: Let-7b may be preventing RNA oxidation through suppression of OGG1, and this may be the cause of dopaminergic neuron death and Alzheimer's disease.
Only recently was it reported that ROS could oxidatively modify miRNAs. Wang et al. [107] demonstrated that oxidatively modified miR-184 associates with the 3′ UTRs of some mRNAs (Bcl-xL and Bcl-w that are known to initiate apoptosis) that are not the usual targets of this miRNA. In this manner, oxidized miR-184 promotes apoptosis via suppression of Bcl-xL and Bcl-w. Also, miR-205/let-7/miR-184 is highly expressed in the nondiseased brain, and miR-205 directly inhibits LRKK2 [108]. In line with this, dopamine neurons were shown to be devoid of LRRK2 mRNA [109]. Moreover, the other miRNAs, miR-184 and let-7, repress E2F1 and DP, respectively, and downregulation of E2F1 and DP suppresses the death of dopaminergic neurons [110]. Also, inhibition of both let-7 and miR-184 is sufficient to phenocopy pathogenic LRRK2 in wild-type animal models, and both miRNAs regulate dopaminergic survival and activity [110]. This finding is interesting as the death of dopaminergic neurons is being looked at as the possible leading cause of both Alzheimer's disease (AD) and Parkinson's diseases (PD), and oxidized miR-184 not binding to its usual mRNA targets suggests that oxidized miR-184 might not be providing protection against the death of dopaminergic neurons. This reveals the complexity and necessity of oxidation research.
Also, RNA oxidation was shown to be significantly elevated in early preclinical stages of AD, and this increase is observed with a compensatory increase in 8-oxoguanine glycosylase (OGG1) levels [111, 112]. OGG1 is the primary enzyme responsible for the excision of 8-oxoguanine (8-oxoG), a mutagenic base byproduct of reactive oxygen species (ROS) that may be responsible for the RNA oxidation. Knowing that a single miRNA can modulate multiple genes and that multiple miRNAs are usually involved in a single disease or physiological phenotype, discerning the overall intricacies of these complex networks is needed. Thus, we here use DES-ROD to explore miRNA associated with RNA oxidation in AD.
In search of novel insights, we looked at AD concepts associated with OGG1. Thus, we explored DES-ROD by clicking on the “Enriched Concepts” link. In the search bar, we typed the concept of interest “OGG1” and then used the concepts' right-click menu to generate a network (Figure 1, Step 1). On the “Network” page, we selected the “ADO Ontology (BioPortal) Alzheimer's Disease Ontology” dictionary; then, the “OGG1” node was highlighted (Figure 1, Step 2) and expanded with the top ten enriched associated terms from the selected dictionary. This process was repeated by selecting the “Human microRNAs” dictionary only and then expanding the “inflammation” node with these concepts, as oxidative stress generally leads to inflammation. We then selected the “ADO Ontology (BioPortal) Alzheimer's Disease Ontology” dictionary only and then expanded all the microRNA nodes with concepts from this dictionary. All nodes with a single edge were removed and were nonspecific nodes such as “things related to severe stage”, “micro RNA”, “Chi-Square test”, “in vivo model”, and “In silico thing” (see Figure 1, Step 3).
Figure 1.
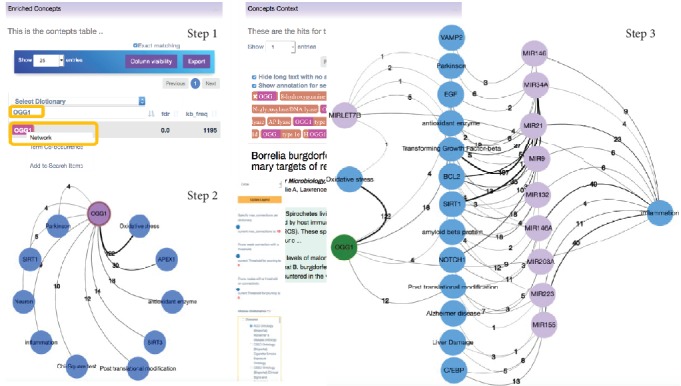
Step-by-step illustration of how DES-ROD can be used to identify relationships between the concepts. The blue circles represent nodes from the “ADO Ontology (BioPortal) Alzheimer's Disease Ontology” dictionary, the green circles represent the nodes from the “Human Genes and Proteins (EntrezGene)” dictionary, and the light purple circles represent the nodes from the “Human microRNAs” dictionary. The edge color is distributed across a color spectrum from black (strong association) to grey (weaker association) based on the frequency of cooccurrence. The number of publications that link the associated nodes is displayed on each edge. Note that the generated networks were exported from DES-ROD and manually adjusted in Cytoscape for better visibility.
Of the miRNAs retrieved, only “MIRLET7B” (referred to in the text as Let-7b) was associated with oxidative stress. Elevated levels of Let-7b have been detected in AD patients [113], and it was further identified as a blood-based molecular biomarker signature in AD [114]. However, we found no literature connecting Let-7b and OGG1 despite this indirect association depicted by the network generated by DES-ROD. Consequently, we used miRDB for microRNA target prediction [115]. This tool retrieved several predicted targets of “MIRLET7B” including OGG1. This finding indicates that Let-7b might have a direct role in RNA oxidation surveillance that protects against the development of AD.
Example 2 . —
Finding the relevant concepts and potentially new knowledge derived through the use of DES-ROD: focused on the association between type 2 diabetes and heart failure.
Finding ROS-induced DNA damage in atherosclerosis led Martinet et al. [116] to assess whether oxidative stress-induced RNA damage occurs in human atherosclerotic plaques. They reported that 11 of 20 atherosclerotic plaques assessed showed significant loss of RNA integrity and strong staining for the oxidative damage marker 8-oxoG, compared to 20 nonatherosclerotic mammary arteries. Moreover, they showed that plaque pretreated with RNase A diminished in cytoplasmic 8-oxoG staining, which suggests RNA damage [116]. Also, in the mouse model of myocardial injury, oxidative modification of miR-184 results in decreased levels of Bcl-xL and Bcl-w, which are essential for apoptosis of cells [107]. On the other hand, different types of miRNA that are present in the cardiomyocytes, such as miR-1, miR-499, and miR-208, are not affected by RNA oxidation. This finding raises the possibility to suspect the presence of specific sequences that could be subjected to RNA oxidation and shows that RNA oxidation plays a role in the development of cardiovascular diseases.
Also, type 2 diabetes mellitus (T2DM) patients usually have high urinary levels of 8-oxo-7,8-dihydroguanosine (8-oxoGuo) and are at risk of cardiovascular mortality. Consequently, Kjaer et al. [117] set out to determine if 8-oxoGuo is associated with this cardiovascular mortality risk. They conducted a five-year follow-up clinical study on 1,863 patients with T2DM wherein they measured the level of 8-oxoGuo. It was concluded that in patients with type 2 diabetes, high RNA oxidation is associated with cardiovascular mortality risk [117].
Here, we attempt to search for novel insights into the association found between type 2 diabetes and cardiovascular risk, focused on oxidative stress. To do this, we start exploring DES-ROD by clicking on the “Enriched Pairs” link. In the search bars, we typed the concepts of interest “Type II diabetes” and “OGG1”, to check if this association was enriched in DES-ROD. Then, we used “OGG1” concepts' right-click menu to generate a network (Step 1). On the “Network” page, we selected the “HFO Ontology (BioPortal) Heart Failure Ontology” and the “HP Ontology (BioPortal) Human Phenotype Ontology” dictionaries; then, the “OGG1” node was highlighted and expanded with the top ten enriched associated terms from the selected dictionaries. To restrict our search to the T2DM and cardiovascular risk association, we removed all retrieved associations except “Type II diabetes mellitus” and “Cardiac Hypertrophy” (Step 2). This process was repeated by selecting the “Human Genes and Proteins (EntrezGene)”, “Human Long Non-Coding RNAs”, and “Human microRNAs” dictionaries to individually expand “OGG1”, “Type II diabetes mellitus”, and “Cardiac Hypertrophy” and then adjust the “current Threshold for pruning is: 1” (Step 3). Now, we had 4 additional nodes “MTOR”, “PGR-AS1”, “SOD2-OT1”, and “MIR21” that were similarly expanded with same dictionaries used in Step 3; then, the threshold was again adjusted “current Threshold for pruning is: 1” (see Figure 2(a), Step 4). We used the DIANA tool TarBase v.8 [118] to search if any of the miRNAs retrieved through DES-ROD target OGG1. This tool provides a collection of experimentally supported miRNA-gene interactions. Figure 2(b) shows that this tool retrieves results for mir-155, mir-17, and mir-34, but only mir-17 interacts with OGG1.
Figure 2.
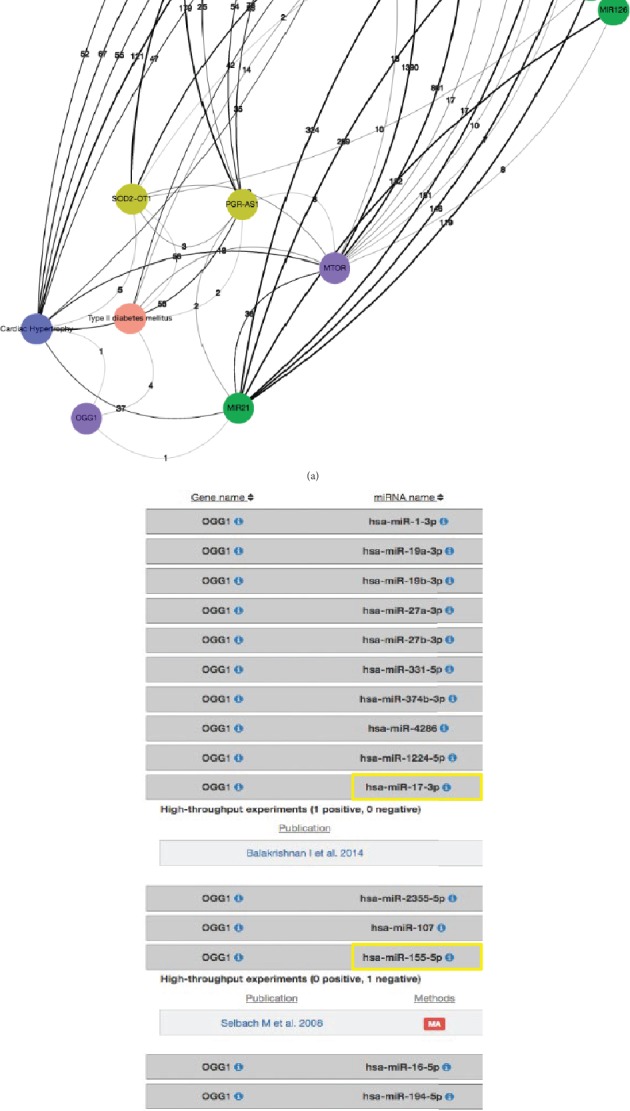
(a) An illustration of the concepts that link “Type 2 diabetes mellitus”, “Cardiac Hypertrophy”, and “OGG1”. The blue circles represent nodes from the “HFO Ontology (BioPortal) Heart Failure Ontology” dictionary, the peach circles represent the nodes from the “HP Ontology (BioPortal) Human Phenotype Ontology” dictionary, the purple circles represent the nodes from the “Human Genes and Proteins (EntrezGene)” dictionary, the greenish-yellow circles represent the nodes from the “Human Long Non-Coding RNAs, and the green circles represent the nodes from the “Human microRNAs” dictionary. The edge color is distributed across a color spectrum from black (strong association) to grey (weaker association) based on the frequency of cooccurrence. The number of publications that link the associated nodes is displayed on each edge. (b) Experimentally supported miRNA-gene interactions retrieved from the DIANA tool TarBase v.8.
However, Ikitimur et al. conducted a study to determine the miRNAs involved in heart failure (HF) using blood samples of 42 HF patients and 15 healthy controls [119]. They found that 29 showed miRNAs with significant dysregulation, which included upregulated miRNA-155. Moreover, miRNA-155 was positively correlated with the left ventricular mass index [119]. Marques et al. consistently demonstrated upregulated miRNA-155 in HF patients [120]. Also, He et al. confirmed the role of miRNA-155 in pathological cardiac remodeling that causes HF. They demonstrated that loss of miRNA-155 in fibroblasts protects left ventricular function after experimental acute myocardial infarction [121]. This is interesting as Corral-Fernandez et al. reported a significant correlation between the basal expression of miR-155 and miR-146a with HbA1c, glucose, and BMI [122]. This altered distribution of miR-155 and miR-146a expression related to HbA1c, glucose, and BMI was also detected using the analysis of a three-dimensional association of variables in the group of T2DM patients. Based on these findings, this group further suggested that downregulated levels of miR-155 could play an essential role in the pathogenesis of T2DM [122].
Taken together, this study demonstrates that the retrieval of miR-155 is a relevant concept to both T2DM and cardiovascular risk and serves as potentially new knowledge as to answering why T2DM and cardiovascular risk are associated.
6. Discussion and Limitations
DES-ROD provides users with over 10 million statistically enriched (FDR < 0.05) cooccurring concepts (with cooccurrence based on a distance up to 200 characters), compared to the documents in the background set. The cooccurring concepts or associations that are of interest to the user can be evaluated through the text from where the associations are derived; this makes it easier for users to find meaningful associations than can be used to develop novel hypotheses. However, to find meaningful associations, users should have some domain-specific knowledge. Users can also explore over 10 billion associations between any of the individual statistically enriched concepts that are semantically similar. However, this number of associations is a bit misleading; as such, associations appear to be most meaningful when the similarity between concepts is sufficiently high, i.e., >0.75.
Furthermore, DES-ROD carries all shortcomings as other text mining approaches. (1) Information extraction is restricted to electronically available documents; (2) information extraction is restricted to what the author chose to mention in the text of the manuscript, such as biomarkers, whereas the complete gene set is placed in a depository or supplementary material that DES does not analyze; (3) peer-reviewed literature contains errors that may cause literature to be omitted; (4) completeness of the concept set extracted depends on the quality and completeness of the dictionaries used and availability of synonyms of a concept; (5) some concepts are “promiscuous” and thus do not retrieve the correct information pertaining to the concept of interest; and (6) cooccurrence of terms does not necessarily imply meaningful association/link between paired terms.
Given the constraints, DES-ROD is useful as most initiated studies start with the review of literature, which DES-ROD can provide comprehensively and visually in minutes, and knowledge of this literature and summarized information extracted from it help not only with developing hypotheses but also with the interpretation of the data. Nonetheless, users should acknowledge the limitation of this system and consequently use it to draw attention to linked concepts or new emerging concepts in the field and to provide a bird's eye view on the topic of interest.
7. Concluding Remarks
DES-ROD rapidly and comprehensively sifts through 298,603 topic-specific publications and extracts relevant topic-specific concepts that may be known or novel. This type of information is not at all available or not easily found in other related databases. The current release comprises 131,741 statistically enriched concepts from 38 topic-relevant dictionaries, together with 10,846,802 statistically enriched pairs of concepts.
DES-ROD provides a user-friendly interface and instructional material to facilitate navigation through the KB. DES-ROD has various tools that enable users to explore enriched concepts, enriched concept pairs, or enriched associated terms based on semantic similarity between these terms, as well as the literature from which terms are derived. Users are further provided with a network viewer to visualize the associations of concepts of interests based on user-selected dictionaries, providing a flexible information exploration experience.
To our knowledge, DES-ROD is the first KB focused on RNA oxidation in human disease discoveries through literature mining and data mining. It will be updated every six months to ensure that the KB remains current. We hope that users find DES-ROD to be a useful tool for supporting RNA oxidation in human disease-related research questions.
Acknowledgments
This work is part of the collaboration between the Laboratory of Radiobiology and Molecular Genetics, Institute of Nuclear Sciences Vinca, University of Belgrade, Belgrade, Serbia, and King Abdullah University of Science and Technology (KAUST), Computational Bioscience Research Center (CBRC), Thuwal, Saudi Arabia. This work has been supported by grants 173033 (EI) and 173034 (VPB) from the Ministry of Education, Science and Technological Development, Republic of Serbia, and by the KAUST Office of Sponsored Research (OSR) grant OSR#4129 (to EI and VPB). VPB has been supported by the KAUST Base Research Fund (BAS/1/1606-01-01), while ME has been supported by KAUST OSR grant no. FCC/1/1976-17-01. TG has been supported by the King Abdullah University of Science and Technology (KAUST) Base Research Fund (BAS/1/1059-01-01). This article is dedicated to the memory of our coauthor, colleague, and world-class leader and researcher in his field, Professor Vladimir Bajic, who passed away after a valiant battle against lymphatic cancer on 31 October 2019.
Abbreviations
- ALS:
Amyotrophic lateral sclerosis
- AD:
Alzheimer's disease
- CVD:
Cardiovascular disease
- FDR:
False discovery rate
- KB:
Knowledgebase
- NLP:
Natural language processing
- miRNA:
MicroRNA
- OGG1:
8-Oxoguanine glycosylase
- OS:
Oxidative stress
- PD:
Parkinson's disease
- ROS:
Reactive oxygen species
- T2DM:
Type 2 diabetes mellitus
- TM:
Text mining
- 8-oxoG:
8-Oxoguanosine.
Contributor Information
Magbubah Essack, Email: magbubah.essack@kaust.edu.sa.
Esma Isenovic, Email: isenovic@yahoo.com.
Data Availability
The DES-ROD portal is free for academic and nonprofit users and can be accessed at http://cbrc.kaust.edu.sa/des-rod/.
Conflicts of Interest
All authors declare no competing interests.
Authors' Contributions
M.E., V.P.B., and V.B.B. conceived and designed the study. A.S. conducted the main technical development. F.T., C.V.N., A.H., and M.U. contributed to technical implementation. M.E., A.B.R., C.V.N., B.Z., S.Z., E.R.I., T.G., V.B.B., and V.P.B. wrote the paper. Magbubah Essack, Adil Salhi, and Christophe Van Neste contributed equally to this work.
References
- 1.Kong Q., Lin C. L. Oxidative damage to RNA: mechanisms, consequences, and diseases. Cellular and Molecular Life Sciences. 2010;67(11):1817–1829. doi: 10.1007/s00018-010-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nunomura A., Lee H. G., Zhu X., Perry G. Consequences of RNA oxidation on protein synthesis rate and fidelity: implications for the pathophysiology of neuropsychiatric disorders. Biochemical Society Transactions. 2017;45(5):1053–1066. doi: 10.1042/BST20160433. [DOI] [PubMed] [Google Scholar]
- 3.Nunomura A., Hofer T., Moreira P. I., Castellani R. J., Smith M. A., Perry G. RNA oxidation in Alzheimer disease and related neurodegenerative disorders. Acta Neuropathologica. 2009;118(1):151–166. doi: 10.1007/s00401-009-0508-1. [DOI] [PubMed] [Google Scholar]
- 4.Nunomura A., Moreira P. I., Castellani R. J., et al. Oxidative damage to RNA in aging and neurodegenerative disorders. Neurotoxicity Research. 2012;22(3):231–248. doi: 10.1007/s12640-012-9331-x. [DOI] [PubMed] [Google Scholar]
- 5.Nunomura A., Tamaoki T., Motohashi N., et al. The earliest stage of cognitive impairment in transition from normal aging to Alzheimer disease is marked by prominent RNA oxidation in vulnerable neurons. Journal of Neuropathology and Experimental Neurology. 2012;71(3):233–241. doi: 10.1097/NEN.0b013e318248e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madamanchi N. R., Vendrov A., Runge M. S. Oxidative stress and vascular disease. Arteriosclerosis, Thrombosis, and Vascular Biology. 2005;25(1):29–38. doi: 10.1161/01.ATV.0000150649.39934.13. [DOI] [PubMed] [Google Scholar]
- 7.Islam M. T. Oxidative stress and mitochondrial dysfunction-linked neurodegenerative disorders. Neurological Research. 2017;39(1):73–82. doi: 10.1080/01616412.2016.1251711. [DOI] [PubMed] [Google Scholar]
- 8.Li Z., Deutscher M. P. Exoribonucleases and endoribonucleases. EcoSal Plus. 2004;1(1):p. 3. doi: 10.1128/ecosalplus.4.6.3. [DOI] [PubMed] [Google Scholar]
- 9.Bregeon D., Sarasin A. Hypothetical role of RNA damage avoidance in preventing human disease. Mutation Research. 2005;577(1-2):293–302. doi: 10.1016/j.mrfmmm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Shan X., Chang Y., Lin C. L. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. The FASEB Journal. 2007;21(11):2753–2764. doi: 10.1096/fj.07-8200com. [DOI] [PubMed] [Google Scholar]
- 11.Hofer T., Badouard C., Bajak E., Ravanat J. L., Mattsson A., Cotgreave I. A. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biological Chemistry. 2005;386(4):333–337. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- 12.Simms C. L., Hudson B. H., Mosior J. W., Rangwala A. S., Zaher H. S. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Reports. 2014;9(4):1256–1264. doi: 10.1016/j.celrep.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegde V., Wang M., Deutsch W. A. Characterization of human ribosomal protein S3 binding to 7,8-dihydro-8-oxoguanine and abasic sites by surface plasmon resonance. DNA Repair. 2004;3(2):121–126. doi: 10.1016/j.dnarep.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Hayakawa H., Uchiumi T., Fukuda T., et al. Binding capacity of human YB-1 protein for RNA containing 8-oxoguanine. Biochemistry. 2002;41(42):12739–12744. doi: 10.1021/bi0201872. [DOI] [PubMed] [Google Scholar]
- 15.Ishibashi T., Hayakawa H., Ito R., Miyazawa M., Yamagata Y., Sekiguchi M. Mammalian enzymes for preventing transcriptional errors caused by oxidative damage. Nucleic Acids Research. 2005;33(12):3779–3784. doi: 10.1093/nar/gki682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saffer J. D., Burnett V. L. Introduction to biomedical literature text mining: context and objectives. In: Kumar V., Tipney H., editors. Biomedical Literature Mining Methods in Molecular Biology. Vol. 1159. Springer; 2014. [DOI] [PubMed] [Google Scholar]
- 17.Zeng Z., Shi H., Wu Y., Hong Z. Survey of natural language processing techniques in bioinformatics. Computational and Mathematical Methods in Medicine. 2015;2015:10. doi: 10.1155/2015/674296.674296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jovanović J., Bagheri E. Semantic annotation in biomedicine: the current landscape. Journal of Biomedical Semantics. 2017;8(1):p. 44. doi: 10.1186/s13326-017-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C. C., Lu Z. Community challenges in biomedical text mining over 10 years: success, failure and the future. Briefings in Bioinformatics. 2016;17(1):132–144. doi: 10.1093/bib/bbv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilicoglu H. Biomedical text mining for research rigor and integrity: tasks, challenges, directions. Briefings in Bioinformatics. 2018;19(6):1400–1414. doi: 10.1093/bib/bbx057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fluck J., Hofmann-Apitius M. Text mining for systems biology. Drug Discovery Today. 2014;19(2):140–144. doi: 10.1016/j.drudis.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 22.Cohen A. M., Hersh W. R. A survey of current work in biomedical text mining. Briefings in Bioinformatics. 2005;6(1):57–71. doi: 10.1093/bib/6.1.57. [DOI] [PubMed] [Google Scholar]
- 23.Shatkay H., Feldman R. Mining the biomedical literature in the genomic era: an overview. Journal of Computational Biology. 2003;10(6):821–855. doi: 10.1089/106652703322756104. [DOI] [PubMed] [Google Scholar]
- 24.Ruch P. The Gene Ontology Handbook. Springer; 2017. Text mining to support gene ontology curation and vice versa; pp. 69–84. (Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- 25.Bada M. Biomedical Literature Mining. Springer; 2014. Mapping of biomedical text to concepts of lexicons, terminologies, and ontologies; pp. 33–45. (Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- 26.Hoehndorf R., Schofield P. N., Gkoutos G. V. The role of ontologies in biological and biomedical research: a functional perspective. Briefings in Bioinformatics. 2015;16(6):1069–1080. doi: 10.1093/bib/bbv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoehndorf R., Slater L., Schofield P. N., Gkoutos G. V. Aber-OWL: a framework for ontology-based data access in biology. BMC Bioinformatics. 2015;16(1) doi: 10.1186/s12859-015-0456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoehndorf R., Dumontier M., Gkoutos G. V. Identifying aberrant pathways through integrated analysis of knowledge in pharmacogenomics. Bioinformatics. 2012;28(16):2169–2175. doi: 10.1093/bioinformatics/bts350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiffin N., Kelso J. F., Powell A. R., Pan H., Bajic V. B., Hide W. A. Integration of text- and data-mining using ontologies successfully selects disease gene candidates. Nucleic Acids Research. 2005;33(5):1544–1552. doi: 10.1093/nar/gki296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon O. S., Kim J., Choi K. H., Ryu Y., Park J. E. Trends in deqi research: a text mining and network analysis. Integrative Medicine Research. 2018;7(3):231–237. doi: 10.1016/j.imr.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alshahrani M., Khan M. A., Maddouri O., Kinjo A. R., Queralt-Rosinach N., Hoehndorf R. Neuro-symbolic representation learning on biological knowledge graphs. Bioinformatics. 2017;33(17):2723–2730. doi: 10.1093/bioinformatics/btx275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bin Raies A., Mansour H., Incitti R., Bajic V. B. Combining position weight matrices and document-term matrix for efficient extraction of associations of methylated genes and diseases from free text. PLoS One. 2013;8(10, article e77848) doi: 10.1371/journal.pone.0077848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raies A. B., Mansour H., Incitti R., Bajic V. B. DDMGD: the database of text-mined associations between genes methylated in diseases from different species. Nucleic Acids Research. 2015;43(D1):D879–D886. doi: 10.1093/nar/gku1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izarzugaza J. M. G., Krallinger M., Valencia A. Interpretation of the consequences of mutations in protein kinases: combined use of bioinformatics and text mining. Frontiers in Physiology. 2012;3 doi: 10.3389/fphys.2012.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neves M., Leser U. A survey on annotation tools for the biomedical literature. Briefings in Bioinformatics. 2014;15(2):327–340. doi: 10.1093/bib/bbs084. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann R., Valencia A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics. 2005;21(Supplement 2):ii252–ii258. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- 37.Kreiner K., Hayn D., Schreier G. Twister: a tool for reducing screening time in systematic literature reviews. Studies in Health Technology and Informatics. 2018;255:5–9. [PubMed] [Google Scholar]
- 38.Howard B. E., Phillips J., Miller K., et al. SWIFT-Review: a text-mining workbench for systematic review. Systematic Reviews. 2016;5(1) doi: 10.1186/s13643-016-0263-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye Z., Tafti A. P., He K. Y., Wang K., He M. M. SparkText: biomedical text mining on big data framework. PLoS One. 2016;11(9, article e0162721) doi: 10.1371/journal.pone.0162721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang J. H., Yu H. C., Hsu H. J. GIS: a biomedical text-mining system for gene information discovery. Bioinformatics. 2004;20(1):120–121. doi: 10.1093/bioinformatics/btg369. [DOI] [PubMed] [Google Scholar]
- 41.Chung D., Lawson A., Zheng W. J. A statistical framework for biomedical literature mining. Statistics in Medicine. 2017;36(22):3461–3474. doi: 10.1002/sim.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paynter R., Bañez L. L., Erinoff E., Lege-Matsuura J., Potter S. Commentary on EPC methods: an exploration of the use of text-mining software in systematic reviews. Journal of Clinical Epidemiology. 2017;84:33–36. doi: 10.1016/j.jclinepi.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 43.Jacome A. G., Fdez-Riverola F., Lourenco A. BIOMedical Search Engine Framework: lightweight and customized implementation of domain-specific biomedical search engines. Computer Methods and Programs in Biomedicine. 2016;131:63–77. doi: 10.1016/j.cmpb.2016.03.030. [DOI] [PubMed] [Google Scholar]
- 44.Khare R., Wei C. H., Mao Y., Leaman R., Lu Z. tmBioC: improving interoperability of text-mining tools with BioC. Database. 2014;2014 doi: 10.1093/database/bau073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H., Christiansen T., Baumgartner W. A., Jr., Verspoor K. BioLemmatizer: a lemmatization tool for morphological processing of biomedical text. Journal of Biomedical Semantics. 2012;3(1) doi: 10.1186/2041-1480-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roeder C., Jonquet C., Shah N. H., Baumgartner W. A., Jr., Verspoor K., Hunter L. A UIMA wrapper for the NCBO annotator. Bioinformatics. 2010;26(14):1800–1801. doi: 10.1093/bioinformatics/btq250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baumgartner W. A., Jr., Cohen K. B., Hunter L. An open-source framework for large-scale, flexible evaluation of biomedical text mining systems. Journal of Biomedical Discovery and Collaboration. 2008;3(1):p. 1. doi: 10.1186/1747-5333-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang H. T., Ju J. H., Wong Y. T., Shmulevich I., Chiang J. H. Literature-based discovery of new candidates for drug repurposing. Briefings in Bioinformatics. 2017;18(3):488–497. doi: 10.1093/bib/bbw030. [DOI] [PubMed] [Google Scholar]
- 49.Tari L. B., Patel J. H. Systematic drug repurposing through text mining. Methods in Molecular Biology. 2014;1159:253–267. doi: 10.1007/978-1-4939-0709-0_14. [DOI] [PubMed] [Google Scholar]
- 50.Carvalho A. S., Rodriguez M. S., Matthiesen R. Review and literature mining on proteostasis factors and cancer. Methods in Molecular Biology. 2016;1449:71–84. doi: 10.1007/978-1-4939-3756-1_2. [DOI] [PubMed] [Google Scholar]
- 51.Luo Y., Riedlinger G., Szolovits P. Text mining in cancer gene and pathway prioritization. Cancer Informatics. 2014;13(Suppl 1):69–79. doi: 10.4137/CIN.S13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verspoor K. M. Roles for text mining in protein function prediction. Methods in Molecular Biology. 2014;1159:95–108. doi: 10.1007/978-1-4939-0709-0_6. [DOI] [PubMed] [Google Scholar]
- 53.Bravo A., Cases M., Queralt-Rosinach N., Sanz F., Furlong L. I. A knowledge-driven approach to extract disease-related biomarkers from the literature. BioMed Research International. 2014;2014:11. doi: 10.1155/2014/253128.253128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan H., Zuo L., Choudhary V., et al. Dragon TF association miner: a system for exploring transcription factor associations through text-mining. Nucleic Acids Research. 2004;32(Web Server):W230–W234. doi: 10.1093/nar/gkh484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shatkay H., Brady S., Wong A. Text as data: using text-based features for proteins representation and for computational prediction of their characteristics. Methods. 2015;74:54–64. doi: 10.1016/j.ymeth.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 56.Park S. H., Hwang M. S., Park H. J., Shin H. K., Baek J. U., Choi B. T. Herbal prescriptions and medicinal herbs for Parkinson-related rigidity in Korean medicine: identification of candidates using text mining. Journal of Alternative and Complementary Medicine. 2018;24(7):733–740. doi: 10.1089/acm.2017.0387. [DOI] [PubMed] [Google Scholar]
- 57.Xiao F., Li C., Sun J., Zhang L. Knowledge domain and emerging trends in organic photovoltaic technology: a scientometric review based on CiteSpace analysis. Frontiers in Chemistry. 2017;5 doi: 10.3389/fchem.2017.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abbe A., Grouin C., Zweigenbaum P., Falissard B. Text mining applications in psychiatry: a systematic literature review. International Journal of Methods in Psychiatric Research. 2016;25(2):86–100. doi: 10.1002/mpr.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spasic I., Livsey J., Keane J. A., Nenadic G. Text mining of cancer-related information: review of current status and future directions. International Journal of Medical Informatics. 2014;83(9):605–623. doi: 10.1016/j.ijmedinf.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 60.Piedra D., Ferrer A., Gea J. Text mining and medicine: usefulness in respiratory diseases. Archivos de Bronconeumología. 2014;50(3):113–119. doi: 10.1016/j.arbres.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 61.Korhonen A., Ó Séaghdha D., Silins I., Sun L., Högberg J., Stenius U. Text mining for literature review and knowledge discovery in cancer risk assessment and research. PLoS One. 2012;7(4):p. e33427. doi: 10.1371/journal.pone.0033427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zaremba S., Ramos-Santacruz M., Hampton T., et al. Text-mining of PubMed abstracts by natural language processing to create a public knowledge base on molecular mechanisms of bacterial enteropathogens. BMC Bioinformatics. 2009;10(1):1471–2105. doi: 10.1186/1471-2105-10-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang X., Song Z., Wu C., et al. Constructing a database for the relations between CNV and human genetic diseases via systematic text mining. BMC Bioinformatics. 2018;19(S19):528–2526. doi: 10.1186/s12859-018-2526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baker S., Ali I., Silins I., et al. Cancer Hallmarks Analytics Tool (CHAT): a text mining approach to organize and evaluate scientific literature on cancer. Bioinformatics. 2017;33(24):3973–3981. doi: 10.1093/bioinformatics/btx454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bachman J. A., Gyori B. M., Sorger P. K. FamPlex: a resource for entity recognition and relationship resolution of human protein families and complexes in biomedical text mining. BMC Bioinformatics. 2018;19(1):248–2211. doi: 10.1186/s12859-018-2211-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raja K., Subramani S., Natarajan J. PPInterFinder—a mining tool for extracting causal relations on human proteins from literature. Database. 2013;2013 doi: 10.1093/database/bas052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chowdhary R., Zhang J., Tan S. L., Osborne D. E., Bajic V. B., Liu J. S. PIMiner: a web tool for extraction of protein interactions from biomedical literature. International Journal of Data Mining and Bioinformatics. 2013;7(4):450–462. doi: 10.1504/ijdmb.2013.054232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.French L., Liu P., Marais O., et al. Text mining for neuroanatomy using WhiteText with an updated corpus and a new web application. Frontiers in Neuroinformatics. 2015;9 doi: 10.3389/fninf.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kordopati V., Salhi A., Razali R., et al. DES-mutation: system for exploring links of mutations and diseases. Scientific Reports. 2018;8(1):p. 13359. doi: 10.1038/s41598-018-31439-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salhi A., Negrão S., Essack M., et al. DES-TOMATO: a knowledge exploration system focused on tomato species. Scientific Reports. 2017;7(1):p. 5968. doi: 10.1038/s41598-017-05448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salhi A., Essack M., Alam T., et al. DES-ncRNA: a knowledgebase for exploring information about human micro and long noncoding RNAs based on literature-mining. RNA Biology. 2017;14(7):963–971. doi: 10.1080/15476286.2017.1312243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dawe A. S., Radovanovic A., Kaur M., et al. DESTAF: a database of text-mined associations for reproductive toxins potentially affecting human fertility. Reproductive Toxicology. 2012;33(1):99–105. doi: 10.1016/j.reprotox.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 73.Essack M., Radovanovic A., Bajic V. B. Information exploration system for sickle cell disease and repurposing of hydroxyfasudil. PLoS One. 2013;8(6):p. e65190. doi: 10.1371/journal.pone.0065190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Essack M., Radovanovic A., Schaefer U., et al. DDEC: dragon database of genes implicated in esophageal cancer. BMC Cancer. 2009;9(1) doi: 10.1186/1471-2407-9-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwofie S. K., Radovanovic A., Sundararajan V. S., Maqungo M., Christoffels A., Bajic V. B. Dragon exploratory system on hepatitis C virus (DESHCV) Infection, Genetics and Evolution. 2011;11(4):734–739. doi: 10.1016/j.meegid.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 76.Maqungo M., Kaur M., Kwofie S. K., et al. DDPC: dragon database of genes associated with prostate cancer. Nucleic Acids Research. 2011;39(Database):D980–D985. doi: 10.1093/nar/gkq849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sagar S., Kaur M., Dawe A., et al. DDESC: dragon database for exploration of sodium channels in human. BMC Genomics. 2008;9(1):p. 622. doi: 10.1186/1471-2164-9-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sagar S., Kaur M., Radovanovic A., Bajic V. B. Dragon exploration system on marine sponge compounds interactions. Journal of Cheminformatics. 2013;5(1) doi: 10.1186/1758-2946-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Salhi A., Essack M., Radovanovic A., et al. DESM: portal for microbial knowledge exploration systems. Nucleic Acids Research. 2016;44(D1):D624–D633. doi: 10.1093/nar/gkv1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bajic V. B., Veronika M., Veladandi P. S., et al. Dragon Plant Biology Explorer. A text-mining tool for integrating associations between genetic and biochemical entities with genome annotation and biochemical terms lists. Plant Physiology. 2005;138(4):1914–1925. doi: 10.1104/pp.105.060863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chowdhary R., Tan S. L., Zhang J., Karnik S., Bajic V. B., Liu J. S. Context-specific protein network miner–an online system for exploring context-specific protein interaction networks from the literature. PLoS One. 2012;7(4, article e34480) doi: 10.1371/journal.pone.0034480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaur M., Radovanovic A., Essack M., et al. Database for exploration of functional context of genes implicated in ovarian cancer. Nucleic Acids Research. 2009;37(Database):D820–D823. doi: 10.1093/nar/gkn593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hastings J., de Matos P., Dekker A., et al. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Research. 2013;41(Database issue):D456–D463. doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wishart D., Arndt D., Pon A., et al. T3DB: the toxic exposome database. Nucleic Acids Research. 2015;43(D1):D928–D934. doi: 10.1093/nar/gku1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cotter D., Maer A., Guda C., Saunders B., Subramaniam S. LMPD: LIPID MAPS proteome database. Nucleic Acids Research. 2006;34(Database issue):D507–D510. doi: 10.1093/nar/gkj122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sud M., Fahy E., Cotter D., et al. LMSD: LIPID MAPS structure database. Nucleic Acids Research. 2007;35(Database issue):D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.The Gene Ontology Consortium. Gene ontology consortium: going forward. Nucleic Acids Research. 2015;43(D1):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fabregat A., Sidiropoulos K., Garapati P., et al. The reactome pathway knowledgebase. Nucleic Acids Research. 2016;44(D1):D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morgat A., Coissac E., Coudert E., et al. UniPathway: a resource for the exploration and annotation of metabolic pathways. Nucleic Acids Research. 2012;40(D1):D761–D769. doi: 10.1093/nar/gkr1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mi H., Lazareva-Ulitsky B., Loo R., et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Research. 2005;33(Database issue):D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kibbe W. A., Arze C., Felix V., et al. Disease ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Research. 2015;43(Database issue):D1071–D1078. doi: 10.1093/nar/gku1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Malhotra A., Younesi E., Gündel M., Müller B., Heneka M. T., Hofmann-Apitius M. ADO: a disease ontology representing the domain knowledge specific to Alzheimer's disease. Alzheimers Dement. 2014;10(2):238–246. doi: 10.1016/j.jalz.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 94.El-Sappagh S., Kwak D., Ali F., Kwak K. S. DMTO: a realistic ontology for standard diabetes mellitus treatment. Journal of Biomedical Semantics. 2018;9(1):p. 8. doi: 10.1186/s13326-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang L., Bray B. E., Shi J., Del Fiol G., Haug P. J. A method for the development of disease-specific reference standards vocabularies from textual biomedical literature resources. Artificial Intelligence in Medicine. 2016;68:47–57. doi: 10.1016/j.artmed.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arguello Casteleiro M., Demetriou G., Read W., et al. Deep learning meets ontologies: experiments to anchor the cardiovascular disease ontology in the biomedical literature. Journal of Biomedical Semantics. 2018;9(1):p. 13. doi: 10.1186/s13326-018-0181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Köhler S., Vasilevsky N. A., Engelstad M., et al. The human phenotype ontology in 2017. Nucleic Acids Research. 2017;45(D1):D865–D876. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mungall C. J., Torniai C., Gkoutos G. V., Lewis S. E., Haendel M. A. Uberon, an integrative multi-species anatomy ontology. Genome Biology. 2012;13(1):p. R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Slee V. N. The international classification of diseases: ninth revision (ICD-9) Annals of Internal Medicine. 1978;88(3):424–426. doi: 10.7326/0003-4819-88-3-424. [DOI] [PubMed] [Google Scholar]
- 100.Wishart D. S., Feunang Y. D., Guo A. C., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Research. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen L., Zeng W. M., Cai Y. D., Feng K. Y., Chou K. C. Predicting Anatomical Therapeutic Chemical (ATC) classification of drugs by integrating chemical-chemical interactions and similarities. PLoS One. 2012;7(4, article e35254) doi: 10.1371/journal.pone.0035254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kuhn M., Letunic I., Jensen L. J., Bork P. The SIDER database of drugs and side effects. Nucleic Acids Research. 2016;44(D1):D1075–D1079. doi: 10.1093/nar/gkv1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmeier S., Alam T., Essack M., Bajic V. B. TcoF-DB v2: update of the database of human and mouse transcription co-factors and transcription factor interactions. Nucleic Acids Research. 2017;45(D1):D145–D150. doi: 10.1093/nar/gkw1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yates B., Braschi B., Gray K. A., Seal R. L., Tweedie S., Bruford E. A. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Research. 2017;45(D1):D619–D625. doi: 10.1093/nar/gkw1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maglott D., Ostell J., Pruitt K. D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Research. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wei C.-H., Harris B. R., Kao H. Y., Lu Z. tmVar: a text mining approach for extracting sequence variants in biomedical literature. Bioinformatics. 2013;29(11):1433–1439. doi: 10.1093/bioinformatics/btt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang J. X., Gao J., Ding S. L., et al. Oxidative modification of miR-184 enables it to target Bcl-xL and Bcl-w. Molecular Cell. 2015;59(1):50–61. doi: 10.1016/j.molcel.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 108.Cho H. J., Liu G., Jin S. M., et al. MicroRNA-205 regulates the expression of Parkinson's disease-related leucine-rich repeat kinase 2 protein. Human Molecular Genetics. 2013;22(3):608–620. doi: 10.1093/hmg/dds470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Galter D., Westerlund M., Carmine A., Lindqvist E., Sydow O., Olson L. LRRK2 expression linked to dopamine-innervated areas. Annals of Neurology. 2006;59(4):714–719. doi: 10.1002/ana.20808. [DOI] [PubMed] [Google Scholar]
- 110.Gehrke S., Imai Y., Sokol N., Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466(7306):637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang W. X., Huang Q., Hu Y., Stromberg A. J., Nelson P. T. Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta Neuropathologica. 2011;121(2):193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lovell M. A., Soman S., Bradley M. A. Oxidatively modified nucleic acids in preclinical Alzheimer's disease (PCAD) brain. Mechanisms of Ageing and Development. 2011;132(8-9):443–448. doi: 10.1016/j.mad.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Grasso M., Piscopo P., Confaloni A., Denti M. A. Circulating miRNAs as biomarkers for neurodegenerative disorders. Molecules. 2014;19(5):6891–6910. doi: 10.3390/molecules19056891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rahman M. R., Islam T., Zaman T., et al. Identification of molecular signatures and pathways to identify novel therapeutic targets in Alzheimer's disease: Insights from a systems biomedicine perspective. Genomics. 2020;(2):1290–1299. doi: 10.1016/j.ygeno.2019.07.018. [DOI] [PubMed] [Google Scholar]
- 115.Wong N., Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Research. 2015;43(D1):D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Martinet W., de Meyer G. R., Herman A. G., Kockx M. M. Reactive oxygen species induce RNA damage in human atherosclerosis. European Journal of Clinical Investigation. 2004;34(5):323–327. doi: 10.1111/j.1365-2362.2004.01343.x. [DOI] [PubMed] [Google Scholar]
- 117.Kjær L. K., Cejvanovic V., Henriksen T., et al. Cardiovascular and all-cause mortality risk associated with urinary excretion of 8-oxoGuo, a biomarker for RNA oxidation, in patients with type 2 diabetes: a prospective cohort study. Diabetes Care. 2017;40(12):1771–1778. doi: 10.2337/dc17-1150. [DOI] [PubMed] [Google Scholar]
- 118.Karagkouni D., Paraskevopoulou M. D., Chatzopoulos S., et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Research. 2018;46(D1):D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ikitimur B., Cakmak H. A., Coskunpinar E., Barman H. A., Vural V. A. The relationship between circulating microRNAs and left ventricular mass in symptomatic heart failure patients with systolic dysfunction. Kardiologia Polska. 2015;73(9):740–746. doi: 10.5603/KP.a2015.0082. [DOI] [PubMed] [Google Scholar]
- 120.Marques F. Z., Vizi D., Khammy O., Mariani J. A., Kaye D. M. The transcardiac gradient of cardio-microRNAs in the failing heart. European Journal of Heart Failure. 2016;18(8):1000–1008. doi: 10.1002/ejhf.517. [DOI] [PubMed] [Google Scholar]
- 121.He W., Huang H., Xie Q., et al. MiR-155 knockout in fibroblasts improves cardiac remodeling by targeting tumor protein p53-inducible nuclear protein 1. Journal of Cardiovascular Pharmacology and Therapeutics. 2016;21(4):423–435. doi: 10.1177/1074248415616188. [DOI] [PubMed] [Google Scholar]
- 122.Corral-Fernández N., Salgado-Bustamante M., Martínez-Leija M., et al. Dysregulated miR-155 expression in peripheral blood mononuclear cells from patients with type 2 diabetes. Experimental and Clinical Endocrinology & Diabetes. 2013;121(6):347–353. doi: 10.1055/s-0033-1341516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The DES-ROD portal is free for academic and nonprofit users and can be accessed at http://cbrc.kaust.edu.sa/des-rod/.


