Abstract
Parsing the relative importance of environmental (recent disturbances) and spatial factors (historical processes) in determining community structure is a core issue in ecology. The Bohai Bay is a typical semi‐enclosed bay located in the north of China, surrounding by the metropolitan area with anthropogenic disturbances made it a complex marine coastal system with pollution gradients, where the distributions and determinants of bacterioplankton communities remain unclear. In this study, we collected surface water samples from 19 sites across Bohai Bay at about 100 km scale to investigate the relative roles of local environments and regional spatial factors in shaping bacterioplankton community composition (BCC). The environmental parameters in the sampling region showed gradient change according to the geographic variation. Several abundant OTUs were significantly correlated with the pollution parameters in the studied area, and 16 OTUs of them showed distinct distribution pattern in different polluted regions with obvious geographic segmentation, which indicated the effects of pollution gradient and dispersal limitation on specific taxon. The BCCs did not show obviously clustering effect between different polluted regions, which indicated the complexity for explaining the BCC variation in the studied region. The partial Mantel test revealed stronger spatial effects on beta diversity than those of local environmental factors, which indicated that dispersal limitation accounted more for the beta diversity than environmental heterogeneity. Furthermore, variation partitioning analysis (VPA) conducted by combining the environmental variables, linear trends, and principal coordinates of the variables from neighbor matrices (PCNM) showed that it was the joint effects of environmental and spatial factors contributed to the explained variation of BCC in the studied area. Considering the special human geography characteristics of Bohai Bay, the unmeasured biotic/abiotic factors, stochastic factors, and anthropogenic disturbances may be responsible for the unexplained variation of the BCC.
Keywords: 16S rRNA V4 region gene, bacterioplankton community composition, Bohai Bay, marine environment, multivariate analysis, spatial factors
Parsing the relative importance of environmental and spatial factors in determining community structure in anthropogenic disturbed semi‐enclosed bay environment is rarely few. This study found that the joint effects of environmental and spatial factors contributed to the explained variation of bacterioplankton community composition in Bohai Bay (China), in which stronger spatial effects on bacterial beta diversity than those of local environmental factors were detected, which indicated that dispersal limitation accounted more for the bacterial community variation than environmental heterogeneity.

1. INTRODUCTION
Coastal marine ecosystems have long been a focused issue among microbial ecologists worldwide. Bacterioplankton has long been recognized as a key player in food web dynamics and biogeochemical cycling in the global ocean (Fuhrman, 2009). The structure and abundance of bacterioplankton involved in the marine microbial cycle (Farooq & Francesca, 2007) are canonical indicators of ecosystem status and the extent of anthropogenic impact on coastal waters (He et al., 2017; Zhang, Wang, et al., 2014). Several studies found that the distribution of bacterioplankton communities in coastal zones depends on both environmental and spatial processes, and most of the studies revealed that the microbial taxa exhibit distinct biogeographical distribution patterns in the ocean, which are correlated with varying environmental parameters (Delong & Karl, 2005; Seo, Kang, Yang, & Cho, 2017; Wang et al., 2015; Zhang, Zhao, Dai, Jiao, & Herndl, 2014). In the East China Sea, bacterial biogeography in the coastal waters of northern Zhejiang was found to be highly influenced by spatially structured environmental gradients (Wang et al., 2015). In the South Sea of Korea, the bacterial community structures showed statistically significant relationships to spatial characteristics and environmental factors (Seo et al., 2017). In the Baltic Sea, a strong correlation between salinity and bacterial beta diversity was observed when controlling for geographic distance (Herlemann et al., 2011). With the growing interest in biogeography, microbial ecologists are attempting to elucidate the mechanisms driving community structure, which is central to understanding the processes underlying the variation and maintenance of microbial communities (Van der Gucht et al., 2007). The environmental conditions (representing contemporary environmental disturbances, such as contemporary abiotic and biotic environmental variables that influence community composition) and spatial factors (representing legacies of historical events, such as dispersal‐related processes) are widely considered to be the major factors driving microbial distribution in multiple environments (Bokulich, Thorngate, Richardson, & Mills, 2014; Ge et al., 2008; Lindström & Langenheder, 2012; Martiny et al., 2006). The main aim of these studies was to illustrate how biodiversity scales with space, and the extent to which this variation is due to contemporary environmental factors or historical contingencies (Hanson, Fuhrman, Horner‐Devine, & Martiny, 2012).
Studies on parsing the relative importance of environmental or spatial factors for the local community structure had been conducted in several habitats across different spatial scales (Beisner, Peres‐Neto, Lindstrom, Barnett, & Longhi, 2006; Martiny et al., 2006; Martiny, Eisen, Penn, Allison, & Horner‐Devine, 2011; Van der Gucht et al., 2007). Most of these studies in aquatic systems were focused on the “island‐like” habitats from a diverse range, such as lakes or marsh sediments, which provided relatively constant environmental conditions (Beisner et al., 2006; Martiny et al., 2011). While there are relatively few studies in local and regional marine environments, a significant geographic gradient of marine bacterial plankton diversity was observed on the worldwide scale (Fuhrman et al., 2008), and many studies verified the spatial patterns of bacterial richness and evenness in the coastal marine environment (Du et al., 2013; Pommier et al., 2010; Wang et al., 2015; Zhang, Zhao, et al., 2014). However, there are few quantitative studies revealing the role of environmental and spatial processes in structuring bacterial community composition in the inner bay environment.
Anthropogenic factors have significant effects on the semi‐enclosed bay, which was one of the most studied coastal ecosystems over the last decades (Caddy, 2000; Peng et al., 2012; Qiao, Feng, Cui, & Zhu, 2017; Shi, Li, & Wang, 2011). Bohai Bay is a typical semi‐enclosed bay with a warm‐temperate marine monsoon climate situated in the western region of Bohai Sea in the north of China, less influenced by current trends and weakly exchanged with the open sea, surrounding by the metropolitan area of Tianjin, Hebei, and Shandong provinces. The increases in port construction, chemical and shipbuilding industries, and aquaculture development have accompanied the urbanization of the Tianjin coastal area over the last two decades (Qiao et al., 2017). It was reported that the seawater quality has deteriorated rapidly in recent years. Many studies of this area had focused either on the primary pollutants such as nitrogen and phosphate enrichment (Chen et al., 2016; Qiao et al., 2017), heavy metal pollution (Zhu, Xie, Li, Ma, & Xu, 2017) or on the aquatic plants and animals (Guo, Li, & Zhang, 2014; Qiao et al., 2017; Yang et al., 2018). By contrast, studies on microorganisms in the Bohai Bay are relatively few, this research focused on the region of Bohai Bay, which is relatively stable and suitable for biogeographical research.
Surface water samples ranged from nearshore to offshore areas across Bohai Bay from north to south and west to east on a scale of about 1–100 km were analyzed based on the sequences of the V4 region of the 16S rRNA gene, and we would like to know: (a) how the bacterioplankton community in such a complex coastal marine system distributed in the “local” bay environment; (b) how the bacterioplankton community composition is influenced by the local environment and spatial factors, since the relative roles of local environment versus. regio‐spatial influences have rarely been compared in the inner bay environment (Lear, Bellamy, Case, Lee, & Buckley, 2014).
2. MATERIALS AND METHODS
2.1. Sampling, DNA extraction, and measurement of environmental parameters
Nineteen sites were selected in Bohai Bay from a routine monitoring project of the Marine Environmental Monitoring Center of Tianjin, SOA, China, in the western part of Bohai Sea (Figure 1a and Figure A1). Surface water samples at 0.5 m depth were collected at each site during 17–19 August 2014 using a 2.5‐L Perspex bottle with an automatic switch in the bottom, and when the bottle is filled with water, the bottom will be closed automatically. Samples comprising 500 ml of seawater were filtered on board through a 0.2‐μm polycarbonate membrane (Millipore) with a diameter of 47 mm at a pressure of <0.03 MPa, and stored at −20°C prior to DNA extraction. Each site was sampled three times. All the sampling was undertaken during high‐tide conditions. Environmental DNA from each polycarbonate membrane was extracted using a PowerWater DNA Isolation Kit (MO BIO) according to the manufacturer's instructions. At the time of sampling, in situ environmental parameters including air temperature, humidity, and air pressure were measured with a Kestrel handheld weather station (Kestrel4500, NK, USA). The depth, water temperature, transparency, pH, salinity, and conductivity were measured using a probe (Multi 3630 portable multi‐parameter water quality automatic analyzer, WTW, Germany) which was calibrated according to the operating manual. The conductivity calibration was conducted using a 0.01 M KCl solution, and the pH was set using two‐point calibration with standard buffer solutions of pH = 7 and pH = 10. The oxygen concentrations were determined on board using the Winkler method (Carpenter, 1965). Nitrate (), nitrite (), ammonium (), phosphate (), and silicate () concentrations were quantified using a UV‐1800 spectrophotometer (SHIMADZU, Japan) according to standard methods (AQSIQ, 2007). The concentration of was determined using the molybdate‐blue method with a detection wavelength of 882 nm. The was determined by the phenol hypochlorite method with a detection wavelength of 640 nm. The concentration was determined using the molybdenum blue method with a detection wavelength of 812 nm. Nitrogen in the form of was determined using the sulfanilamide method with a detection wavelength of 543 nm, and was measured as NO2 ‐ after reduction in a Cd‐Cu column. The dissolved inorganic nitrogen (DIN) content was calculated as the sum of , , and . The geographic coordinates and chemical parameters of water samples are listed in Dataset S1.
Figure 1.
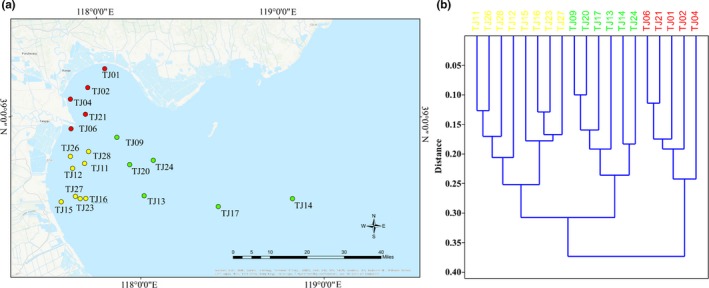
Location of the nineteen sampling sites (a) and clustering result (b) of the environmental factors sampled from the nineteen sites. Signals with red color represent sites which were located in the north of Bohai Bay (group 1); signals with yellow color represent sites which were located in the south of Bohai Bay (group 2); signals with green color represent sites which were located in the east of Bohai Bay (group 3)
2.2. Bacterial 16S rRNA gene amplification and Illumina MiSeq sequencing
The extracted DNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). The V4 region of the bacterial 16S rRNA genes was amplified using primers F515 (5′ ‐ GTG CCA GCM GCC GCG GTA/A3′) and R806 (5′ ‐ GGA CTA CHV GGG TWT CTA AT ‐ 3′) (Caporaso et al., 2011) with the reverse primer modified to contain a unique 12 nt barcode at the 5′ end. An aliquot comprising 12.5 ng of purified DNA template from each sample was amplified in triplicate using a 25 μl reaction system under the following conditions: denaturation at 95°C for 3 min, followed by 25 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, with a final extension at 72°C for 5 min. A sample comprising 1 µl of the PCR product was loaded onto a Bioanalyzer DNA 1000 chip to verify the size. Using the V4 primer pairs in the protocol, the expected size on a Bioanalyzer trace after the amplification PCR step was ~380 bp. Next, AMPure XP beads were used to purify the 16S V4 amplicon, removing free primers and primer dimers. The dual indices and Illumina sequencing adapters were attached to the above products using the Nextera XT Index Kit with a 50 μl reaction system under conditions of 95°C for 3 min, then 8 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, with a final extension at 72°C for 5 min. The polymerase chain reaction amplicons were purified using AMPure XP beads; the library was validated on a Bioanalyzer DNA 1000 chip to verify the size; and the expected size of the final library on a Bioanalyzer trace was ~460 bp. The DNA concentration was quantified using the Quant‐It Pico Green kit (Invitrogen) with a Qubit fluorometer (Life Technologies). An equimolar amount of each PCR amplicon was combined into one pooled sample and run on an Illumina MiSeq sequencing machine (Illumina, Inc.), producing 250 bp paired‐end reads from the reverse direction, containing 806R with the barcode.
2.3. Processing of the sequencing data
The paired reads were joined using FLASH (Magoc & Salzberg, 2011) with default settings. The joined pairs were then quality‐filtered and checked for chimeras using the QIIME workflow (Caporaso, Kuczynski, et al., 2010). Briefly, the bacterial reads whose lengths were outside the bounds of 200 and 450 bp and cases in which the homopolymer run exceeded 6 were removed using the NGSQC Toolkit v2.3 (Patel & Jain, 2012). The remaining sequences were analyzed and the chimeras removed using USEARCH (Edgar, Haas, Clemente, Quince, & Knight, 2011). Sequences with the same barcode were then assigned to the same sample (Caporaso, Kuczynski, et al., 2010). Bacterial phylotypes were identified using UCLUST (Edgar, 2010) and assigned to operational taxonomic units (OTUs) based on 97% similarity. The most abundant sequence of each OTU was selected as the representative sequence. The representative sequences were classified taxonomically according to the Greengenes database (Desantis et al., 2006) and aligned against a template alignment of the Greengenes database using PyNAST (Caporaso, Bittinger, et al., 2010). The singletons and sequences classified as Chloroplast, Mitochondria, Archaea, Eukaryota or unknown kingdom were removed. To avoid sample‐size‐based artifacts, we used a randomly selected subset of 3,000 sequences (corresponding to the smallest sequencing effort for any of the samples) per sample to calculate the diversities and distances between samples. The Biological Observation Matrix (BIOM) obtained was analyzed using the summarize_taxa.py script in order to obtain the relative abundance of each taxonomic group for all samples (Caporaso, Kuczynski, et al., 2010).
2.4. Statistical analysis
Cluster analysis was performed to evaluate the overall variation trend of environmental factors based on Gower distances (Flynn, Mirotchnick, Jain, Palmer, & Naeem, 2011). Before the cluster analysis, the raw environmental data were standardized using the equation SNDE = (x − mean of the raw data)/standard deviation of the raw data, where SNDE represents the standard normal deviate equivalents and x represents the raw data for one sample (Zhang, Wang, et al., 2014). The differences of each environmental factor among different groups were analyzed using one‐way ANOVA. Pearson's correlation coefficients were calculated to measure the relationships between the abundant OTUs and chemical variables. Principal coordinates analysis (PCoA) was performed to evaluate the overall differences in microbial community structure based on weighted UniFrac distances and Bray–Curtis distances. The analysis of similarities (ANOSIM) and permutational multivariate analysis of variance (PERMANOVA) were then employed to test the significance of differences among the BCCs using Vegan's function anosim () and adonis () (Anderson, 2001; Clarke, 1993; Oksanen et al., 2019). Alpha‐diversity estimates were calculated using QIIME with multiple indices (observed species, Shannon–Wiener index, and phylogenetic diversity).
2.4.1. Relationship between environmental factors and bacterial alpha diversity
The difference of bacterial alpha diversity among all sites and groups was analyzed using one‐way ANOVA. Pearson's correlation coefficients between geo‐physico‐chemical variables and bacterial alpha‐diversity indices were calculated to measure the relationships between them. To address how environmental and spatial factors affect the microbial alpha diversity in Bohai Bay, the partial least squares path modeling (PLS‐PM) algorithm (Hair, Sarstedt, Pieper, & Ringle, 2012) was used to inspect the effects between them. The path model was developed in a formative way. At first, four latent variables (LVs, an abstract concept) were defined as follows: geographic location, seawater environment, nutrient content, and microbial alpha diversity. Then, manifest variables (MVs, measured variables) for each LV were selected as follows: (a) longitude and latitude were selected as MVs of geographic location; (b) temperature, humidity, air pressure, depth, water temperature, transparency, salinity, and conductivity were selected as physical environmental variables; (c) DO, pH, COD, SRP, nitrite, nitrate, AN, DIN, and SRSi were selected as chemical environmental variables; and (d) phylogenetic diversity, observed species, and Shannon index were defined as bacterial alpha‐diversity variables. Each LV was considered a linear combination of its own MVs (Tenenhaus, Vinzi, Chatelin, & Lauro, 2005). During the modeling, all variables were tested to assess the indicator reliability and maximize the efficiency of PLS‐PM. Indicator reliability (the indicative ability of MV for its LV) is usually determined by the construct loadings, and we selected those MVs with loadings significant at the 0.05 level and above the recommended 0.7 parameter value for each LV. Loadings greater than 0.7 are considered acceptable (Sanchez, 2013). Model parameters (the path coefficients, the loadings, and communalities of the MVs) and fit indices (R 2) were validated by bootstrapping. The significance of the path coefficients was estimated at the 0.05 level. The PLS‐PM algorithm was implemented using the package “plspm” in the R environment (Sanchez, 2013).
2.4.2. Relationship between environmental or geospatial factors and bacterial beta diversity
When analyzing the relationship between beta diversity and environmental/spatial factors, the distance matrices based on Bray–Curtis and weighted UniFrac distances were averaged among the triplicate samples for each site before the correlation analysis. Simple and partial Mantel tests were used to test the correlations of geographic distance and environmental variables with bacterial beta diversity. Subsequently, a Mantel correlogram was used to examine the significance of distance effects on beta diversity across a subset of different geographic distance scales (classes) among the sampling stations (Wang et al., 2016). Pairwise geographic distances between sites were calculated using the distm () function from the geosphere R package (Robert, 2019). In order to investigate the relative contributions of spatial and environmental parameters to the variation of BCC, variance partitioning analysis (VPA) (Borcard, Legendre, & Drapeau, 1992) was conducted by combining the environmental factors, linear trends, and principle coordinates of neighbor matrices (PCNM). The PCNM analytical approach was able to indicate whether the variation of responsible variables was caused by undetected environmental factors with a similar spatial structure (Borcard, Gillet, & Legendre, 2011). Before we conducted this analysis, the raw environmental data were log‐transformed and the species data were center‐transformed using the Hellinger transformation (Legendre & Gallagher, 2001). Environmental factors, as well as spatial variables derived from geographic coordinates using both linear trends and PCNM procedure (Griffith & Peres‐Neto, 2006), were forward‐selected by sequentially adding one variable that improves the selection criterion (R 2) the most at each step, until no further improvement of R 2 was observed. In all models, the significance was tested through 999 permutations and all the R 2 values were calculated and adjusted as described in the literature (Peres‐Neto, Legendre, Dray, & Borcard, 2006).
3. RESULTS
3.1. Geo‐physico‐chemical characteristics of the seawater environment
The geographical coordinates and 17 environmental parameters of 19 seawater samples were summarized in Dataset S1. Clustering analysis based on all environmental factors of each site showed that the 19 sites can be classified into three groups (Figure 1b). The classified groups showed obvious geographic segmentation on the sampling map (Figure 1a). Almost all of the measured environmental variables (except salinity and soluble reactive silicate [SRSi]) were significantly different among the three clustered groups (Table A1). The dissolved inorganic nitrogen (DIN) concentrations in the north, south, and east region were, respectively, inferior to Category IV, Category III, and Category II seawater quality according to the Sea Water Quality Standard of China (Tables A1 and Table A2) (MEP, 1997) (the classification standards for clean water, relatively clean water, slightly polluted water, medium polluted water, and heavily polluted water were listed in Table A2). The environmental variability was significantly increased with geographic distance (Table A3).
3.2. Distribution of taxa and phylotypes
Across all the 19 seawater samples, a total of 652,037 high‐quality sequences were obtained (three replicates for one sample) and 3,066–30,493 sequences per sample (mean = 11,445). Two cases (one replicate of sample TJ17 and one replicate of sample TJ20) with less than 1,000 sequences, as well as three samples with biased community composition, were excluded from analysis (Figure A2). The dominant bacterial phyla (>5%) in Bohai Bay were Alphaproteobacteria (22.53%), Gammaproteobacteria (40.49%), and Bacteroidetes (19.94%) (Figure 2). At the family level, Rhodobacteraceae, Flavobacteriaceae, and Alteromonadaceae constituted the top three, followed by taxa mostly from the Gammaproteobacteria among the top 15 (Figure A2). Among the 19 sites, only TJ14 had Cyanobacteria as a dominant taxon, where they accounted for 29% of the relative abundance of all taxa, albeit mainly represented by Synechococcus at genus level (Figure A3). While they had relatively low abundance in other samples, the numbers of reads classified as Cyanobacteria among the total in TJ14 were 2837/7761 and 1577/7157 in the two replicates, respectively. In addition, there were 2,789 and 1,563 reads classified as Synechococcus, respectively.
Figure 2.
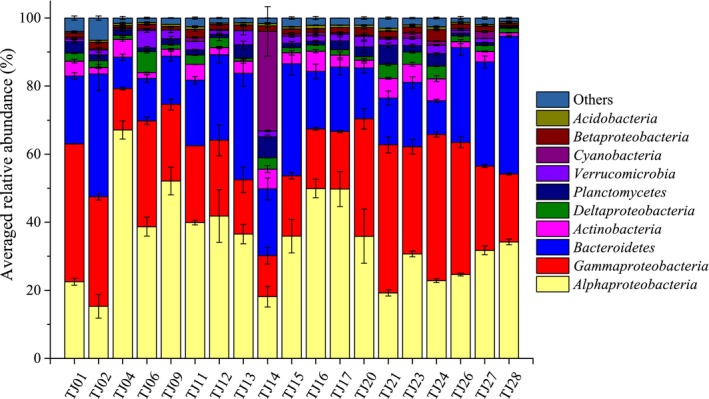
Relative abundances of the dominant taxa in 19 seawater samples of Bohai Bay at phylum level. Top 10 phyla were presented in the figure, and others represented the sum of the rest phyla. Taxa of Proteobacteria were grouped at the class level
3.3. Distribution of specific taxon and the correlation with pollution parameters
A total of 36 OTUs (with relative abundance >1%) were identified significantly correlated with the concentrations of DIN, nitrate, nitrite, ammonia nitrogen (AN), soluble reactive phosphate (SRP), and SRSi, and most of them showed different variation trends among the three clustered regions in Bohai Bay (Figure 3, Dataset S2, Table A4). The relative abundance of OTU65886 (member of Flammeovirga from Bacteroidetes) and OTU5954 (member of Flavobacteriaceae from Bacteroidetes) was significantly higher in the nearshore north region (group 1) than in the other two groups and significantly positively correlated with nitrogen nutrients and SRP. The relative abundance of OTU126053 (member of Fluviicola from Bacteroidetes), OTU85516 (member of Flavobacteriaceae from Bacteroidetes), OTU7191, and OTU84157 (members of OM60 from Gammaproteobacteria) was significantly higher in the nearshore south region (group 2) than in the other two groups and significantly negatively correlated with AN. The relative abundance of OTU104369 (member of OCS155 from Actinobacteria), OTU30515 (member of Flavobacteriaceae from Bacteroidetes), OTU82462 (member of Phyllobacteriaceae from Alphaproteobacteria), OTU73029 (member of Rhodobacteraceae from Alphaproteobacteria), and OTU12256 (member of Methylophilaceae from Betaproteobacteria) was significantly higher in the offshore east region (group 3) than in the other two groups and significantly negatively correlated with DIN. While the relative abundance of OTU3224 (member of Cryomorphaceae from Bacteroidetes), OTU92726 (member of Flavobacteriaceae from Bacteroidetes), OTU66225 (member of Flavobacterium from Bacteroidetes), and OTU32430 (member of Kiloniellales from Alphaproteobacteria) was significantly higher in the south and east region (groups 2 and 3) than in north region (group 1) and negatively correlated with nitrogen nutrients (Figure 3, Table A4). In addition, OTUs identified as Bacteroidaceae, Porphyromonadaceae, Prevotellaceae, Sphingobacteriaceae, Enterococcus, Ochrobactrum, Achromobacter, Arcobacter, and Enterobacteriaceae showed relatively high abundance only in TJ02 (Figure 3).
Figure 3.
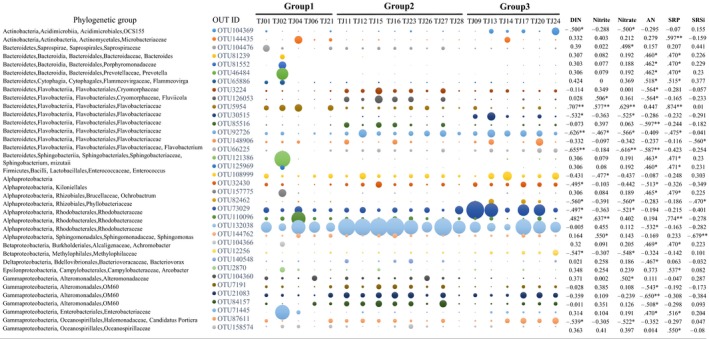
Taxonomic identities of the 36 OTUs serving as indicator taxa which were significantly correlated with the pollution parameters measured. The diameters of the circles are proportional to the abundances of the OTUs, with the size of the circle indicating the average abundance of each OTU at a given site. The relative abundance of each OTU was listed in Dataset S2. *p < .05; **p < .01; ***p < .001
3.4. Bacterial alpha diversity
The bacterial alpha diversity was calculated for each sample with three replicates, including phylogenetic diversity (PD), observed species (OTU richness), and Shannon–Wiener index (Shannon) (Table 1). Significant differences were tested between sites in the offshore (TJ13) and sites in the nearshore (TJ28, TJ06). However, there were no significant differences of alpha diversity among the three regions grouped according to the environmental clustering results (Table A5).
Table 1.
Values of OTU richness, phylogenetic diversity (PD), and Shannon diversity estimates for the bacterioplankton in different sampling sites of Bohai Bay
| Sample ID | OTU richness | PD | Shannon |
|---|---|---|---|
| TJ01 | 637.50 ± 20.97ab | 66.84 ± 2.66ab | 4.98 ± 0.07abcd |
| TJ02 | 568.07 ± 36.58b | 63.81 ± 1.39ab | 4.60 ± 0.37bcd |
| TJ04 | 648.97 ± 66.67ab | 57.70 ± 7.35cd | 4.36 ± 0.17cd |
| TJ06 | 582.90 ± 39.75b | 58.05 ± 4.80cd | 4.36 ± 0.13cd |
| TJ09 | 789.00 ± 110.23ab | 65.81 ± 1.93ab | 5.11 ± 0.35abc |
| TJ11 | 703.65 ± 46.15ab | 70.90 ± 5.25ab | 5.24 ± 0.16abc |
| TJ12 | 607.00 ± 30.43ab | 58.82 ± 3.40b | 4.56 ± 0.24bcd |
| TJ13 | 628.80 ± 13.30a | 63.19 ± 2.23a | 4.83 ± 0.11a |
| TJ14 | 603.05 ± 27.05ab | 64.57 ± 1.49ab | 4.92 ± 0.05abcd |
| TJ15 | 674.77 ± 85.89ab | 64.05 ± 3.44ab | 4.86 ± 0.40abcd |
| TJ16 | 718.30 ± 60.28ab | 69.92 ± 3.73ab | 5.09 ± 0.02abc |
| TJ17 | 827.85 ± 92.55ab | 72.22 ± 2.34ab | 5.43 ± 0.43abc |
| TJ20 | 659.70 ± 29.30ab | 67.26 ± 4.66ab | 4.75 ± 0.15abcd |
| TJ21 | 875.67 ± 84.06ab | 85.90 ± 3.93ab | 5.80 ± 0.28ab |
| TJ23 | 836.73 ± 125.36ab | 79.48 ± 8.22ab | 5.78 ± 0.28ab |
| TJ24 | 703.00 ± 63.10ab | 71.65 ± 5.74ab | 5.16 ± 0.23abc |
| TJ26 | 597.97 ± 63.90ab | 57.79 ± 7.90cd | 4.38 ± 0.16cd |
| TJ27 | 639.23 ± 46.51ab | 63.89 ± 4.82ab | 5.06 ± 0.09abcd |
| TJ28 | 531.70 ± 37.52b | 48.21 ± 3.51d | 3.81 ± 0.10d |
Data in the table are the means ± SD; the different normal letters in the same column indicate significant difference among sites at 0.05 level (n = 3) under Duncan test, except for TJ11, TJ13, TJ14, TJ17, and TJ20, which were tested through Kruskal–Wallis test since their replicate was <3.
3.5. Investigation of the relationship between bacterial alpha diversity and environmental/spatial factors
Bacterial alpha‐diversity estimates according to OTU richness and Shannon index were significantly negatively correlated with ammonia nitrogen (Table 2). The results showed that most of the environmental factors were not significantly correlated with alpha diversity of the BCC in the surface waters of Bohai Bay.
Table 2.
Pearson's correlation coefficients (r) between alpha diversity of bacterial community and biogeochemical characteristics of seawater samples
| Variables | Richness | PD | Shannon | n |
|---|---|---|---|---|
| Longitude | 0.192 | 0.244 | 0.18 | 19 |
| Latitude | −0.208 | −0.143 | −0.129 | 19 |
| pH | −0.091 | −0.176 | −0.086 | 19 |
| Air temperature | −0.195 | −0.178 | −0.118 | 19 |
| Humidity | −0.005 | 0.067 | 0.027 | 19 |
| DO | 0.029 | −0.11 | 0.028 | 19 |
| COD | 0.038 | −0.019 | 0.128 | 19 |
| Air pressure | −0.347 | −0.27 | −0.4 | 19 |
| Depth | −0.059 | −0.001 | 0.045 | 19 |
| Water temperature | −0.311 | −0.321 | −0.313 | 19 |
| Salinity | 0.203 | 0.19 | 0.223 | 19 |
| Transparency | 0.264 | 0.189 | 0.296 | 19 |
| Conductivity | 0.2 | 0.142 | 0.264 | 19 |
| SRP | −0.086 | −0.047 | −0.126 | 19 |
| Nitrite | 0.08 | 0.002 | −0.044 | 19 |
| Nitrate | −0.119 | −0.068 | −0.13 | 19 |
| AN | −0.461* | −0.366 | −0.503* | 19 |
| DIN | −0.221 | −0.172 | −0.272 | 19 |
| SRSi | −0.327 | −0.103 | −0.073 | 19 |
Data in bold indicate significant correlations, *p < .05, **p < .01, ***p < .001.
Abbreviations: AN, ammonia nitrogen; DIN, dissolved inorganic nitrogen; DO, dissolved oxygen; PD, phylogenetic diversity; Shannon, Shannon–Wiener index; SRP, soluble reactive phosphate; SRSi, soluble reactive silica.
The goodness of fit (GoF) of PLS‐PM analysis was 0.6046, which indicated that the prediction power of this model was close to 60%. The constructed path model (Figure 4) showed that the path coefficients between geographic location and chemical (0.7439, p = .005), as well as physical (0.7984, p < .0001), environmental variables were more significant and relatively higher than the coefficient with bacterial alpha diversity (0.2048, p = .123). The correlations between microbial alpha diversity and other LVs were not significant (the details of the PLS‐PM are shown in the Appendix, Table A6).
Figure 4.
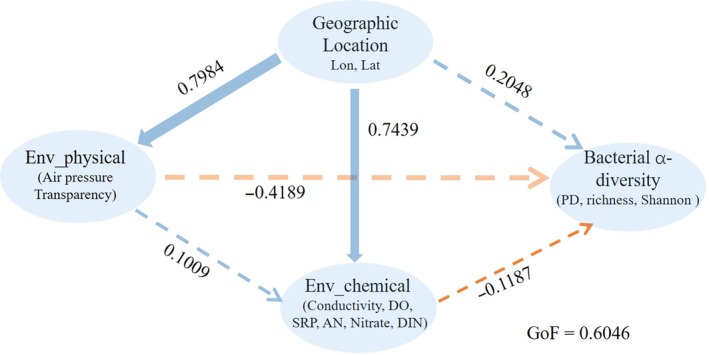
Path models examining how geographic location alters seawater environment properties and the nutrient contents and the microbial alpha diversity through direct and indirect pathways. Solid and dashed arrows indicate significant and no significant relationships between latent variables at the level p = .05, respectively. Red and blue arrows indicate negative and positive effects between latent variables. Numbers associated with pathways between variables represent standardized path coefficients. AN, ammonia nitrogen; DIN, dissolved inorganic nitrogen; DO, dissolved oxygen; Env_chemical, environmental chemical factors; Env_physical, environmental physical factors; GoF, the goodness of fit; Lat, latitude; Lon, longitude; PD, phylogenetic diversity; richness, OTU richness; Shannon, Shannon–Wiener index; SRP, soluble reactive phosphate
3.6. Bacterial beta diversity
The results of PCoA using the Bray–Curtis distance metric showed that the bacterial assemblages from the three groups clustered according to environmental factors and geographic locations were weakly separated, with the first coordinate explaining 17.73% of the variance and the second coordinate explaining 12.98% (Figure 5a). PCoA analysis based on the weighted UniFrac distance metric revealed an overlap between most of the sampling stations, except for bacterial communities in TJ02, TJ21, TJ14, and TJ24, with the first coordinate explaining 29.27% of the variance and the second coordinate explaining 15.62% (Figure 5b). Notably, significant differences were found among the three groups using the Bray–Curtis distance matrix (ANOSIM, R = .3751, p = .0001; ADONIS, R 2 = .05695, p = .049), but not the weighted UniFrac distance matrix (ANOSIM, R = −.006712, p = .528; ADONIS, R 2 = .04824, p = .228).
Figure 5.
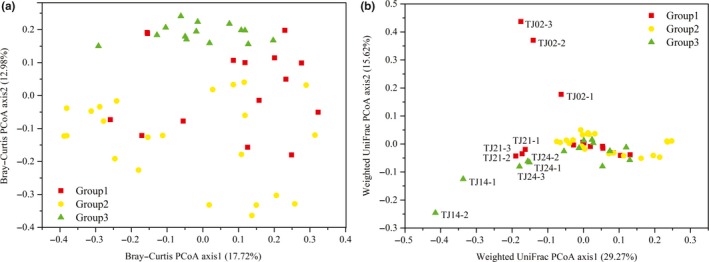
Principal coordinates analysis (PCoA) plot derived from the Bray–Curtis (a) and weighted UniFrac (b) distance among seawater samples based on the occurrences and abundances of operational taxonomic units (OTUs). The two principal coordinate axes and the distribution of the bacterial assemblages (designated with the sampling station names) in response to these axes are shown. Group 1 were comprised of sites TJ01, TJ02, TJ04, TJ06, and TJ21; group 2 were comprised of sites TJ11, TJ12, TJ15, TJ16, TJ23, TJ26, TJ27, and TJ28; and group 3 were comprised of sites TJ09, TJ13, TJ14, TJ17, TJ20, and TJ24
3.7. Investigation of the relationship between bacterial beta diversity and the environmental/spatial factors
We identified significant distance–decay relationships in the studied area. The weighted UniFrac and Bray–Curtis distances of bacterial communities (beta diversity) were both significantly correlated with geographic distances (Pearson correlation, N = 170, p < .001; Figure 6). The Mantel test based on the Bray–Curtis distances and weighted UniFrac distances both showed that geographic distance as well as environmental distance, were significant individual determinants of community composition. At the same time, stronger geographic distance effects on beta diversity were observed when controlling for environmental variations compared with environmental effects when controlling for geographic distance using either of the two distance matrix methods (Table 3). The Mantel correlogram showed a positive spatial correlation in beta diversity at short geographic distance scales of 0–7 km, and a negative spatial correlation at geographic distance scales of 19 km–31 km (Bray–Curtis similarity distance) and 79 km–91 km (weighted UniFrac distance) (Figure A4). However, on the rest of the spatial scales, the spatial correlation was not significant in the studied area of Bohai Bay.
Figure 6.
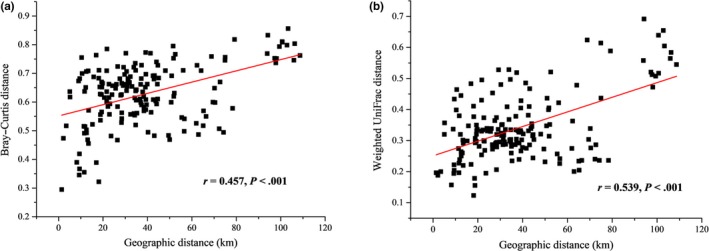
Correlations between community dissimilarity (a: Bray–Curtis measure; b: weighted UniFrac measure) with geographic distances between sites (all samples)
Table 3.
Simple Mantel test demonstrating the correlations of environmental variations (Euclidean distance) and geographic distance with the variation in bacterioplankton community composition (based on Bray–Curtis/weighted UniFrac distance, 999 permutations)
| Simple | Controlled by | Partial | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Bray–Curtis distance | Weighted UniFrac distance | Bray–Curtis distance | Weighted UniFrac distance | ||||||
| r | p | r | p | r | p | r | p | ||
| Geo_dist | .3566 | .008 | .5387 | .024 | Temp | .4776 | .002 | .5612 | .007 |
| Geo_dist | WT | .4870 | .001 | .5360 | .008 | ||||
| Geo_dist | Humidity | .4712 | .005 | .5394 | .010 | ||||
| Geo_dist | pH | .4684 | .005 | .5501 | .017 | ||||
| Geo_dist | COD | .4625 | .005 | .5501 | .020 | ||||
| Geo_dist | Salinity | .4605 | .002 | .5389 | .016 | ||||
| Geo_dist | DO | .4557 | .009 | .5351 | .017 | ||||
| Geo_dist | SRSi | .4572 | .006 | .5402 | .019 | ||||
| Geo_dist | SRP | .4529 | .005 | .5353 | .015 | ||||
| Geo_dist | AN | .4493 | .004 | .5326 | .023 | ||||
| Geo_dist | Nitrite | .4540 | .005 | .5671 | .008 | ||||
| Geo_dist | Air_press | .4430 | .006 | .5544 | .013 | ||||
| Geo_dist | Conduct | .4321 | .007 | .5206 | .031 | ||||
| Geo_dist | Nitrate | .3947 | .023 | .5430 | .023 | ||||
| Geo_dist | IN | .3795 | .025 | .5334 | .034 | ||||
| Geo_dist | Trans | .3614 | .010 | .4605 | .020 | ||||
| Geo_dist | Depth | .4585 | .005 | .5388 | .023 | ||||
| Geo_dist | Env_dist | .3630 | .034 | .3974 | .036 | ||||
| Env_dist | .2753 | .020 | .4621 | .015 | Geo_dist | .0615 | .324 | .2588 | .093 |
| Depth | .3196 | .021 | .0000 | .452 | Geo_dist | .1996 | .091 | .0123 | .374 |
| Conduct | .3398 | .021 | .2540 | .124 | Geo_dist | .3071 | .041 | .1982 | .183 |
| Trans | .3178 | .002 | .3374 | .022 | Geo_dist | .1737 | .058 | .1275 | .158 |
| DIN | .2854 | .007 | .1426 | .133 | Geo_dist | .1409 | .142 | −.1119 | .774 |
| AN | .2646 | .008 | .2117 | .091 | Geo_dist | .2471 | .022 | .1897 | .126 |
| Nitrate | .2024 | .018 | .1423 | .138 | Geo_dist | .0401 | .357 | −.1635 | .895 |
| SRP | .1447 | .229 | .1277 | .211 | Geo_dist | .1731 | .138 | .1062 | .236 |
| Salinity | .2466 | .074 | .0842 | .221 | Geo_dist | .1837 | .137 | .0861 | .209 |
| WT | .1156 | .238 | .2016 | .148 | Geo_dist | −.2236 | .942 | −.1916 | .861 |
| Air_press | .0354 | .327 | −.0107 | .489 | Geo_dist | .0354 | .335 | −.1560 | .932 |
| Nitrite | .0804 | .274 | .0178 | .353 | Geo_dist | −.0739 | .653 | −.2112 | .952 |
| Humidity | .0868 | .277 | .0836 | .276 | Geo_dist | −.1304 | .773 | −.0902 | .654 |
| DO | .0552 | .287 | .0762 | .247 | Geo_dist | −.0110 | .481 | .0186 | .371 |
| SRSi | −.0616 | .720 | −.1031 | .779 | Geo_dist | −.0592 | .659 | −.1135 | .798 |
| COD | .0053 | .439 | −.0434 | .505 | Geo_dist | −.0790 | .649 | −.1393 | .805 |
| Temp | −.0661 | .639 | −.1160 | .699 | Geo_dist | −.1902 | .907 | −.2187 | .933 |
| pH | −.1257 | .824 | −.1216 | .737 | Geo_dist | −.1710 | .898 | −.1788 | .880 |
Partial Mantel test demonstrating the correlation of geographic distance with variation in community composition controlled by significantly correlative environmental variables obtained from simple Mantle tests and those of environmental variables controlled by geographic distance. Refer to Table 2 for other variable abbreviations. Data in bold indicate significant correlations (p < .05).
Abbreviations: Air_press, air pressure; Conduct, conductivity; Env_dist, environmental distance (Euclidean distance); Geo_dist, geographic distance; Temp, temperature; Trans, transparency; WT, water temperature.
The VPA results performed on the OTU matrix showed that the spatial linear trend together with the spatial factors derived from the PCNM procedure contributed 9.9% to the variation of BCC; however, neither of their effects were significant (bootstrap test for trend fractions, p = .075; bootstrap test for PCNM fractions, p = .098) after controlling for other variables; the pure environmental factors accounted for 8.4% (bootstrap test for Env fractions, p = .012); and conductivity and nitrite were reserved in the environmental factors after the forward selection. Additionally, 9.1% could be attributed to the interaction between environmental variables and spatial parameters (Figure 7a), demonstrating the strong influence of the spatial scale and environmental variables. The results of VPA performed on the principal coordinates based on the weighted UniFrac distance matrix showed a weak contribution of pure environmental factors, trends, or PCNM on the BCC variation, but the interaction between environmental variables and spatial parameters accounted for 16.5% of the variation, which underscored the strong combined influence of spatial factors and environmental variables (Figure 7b). However, a large proportion of the variation (70.8%–72.6%) across all bacterial taxa could not be explained.
Figure 7.
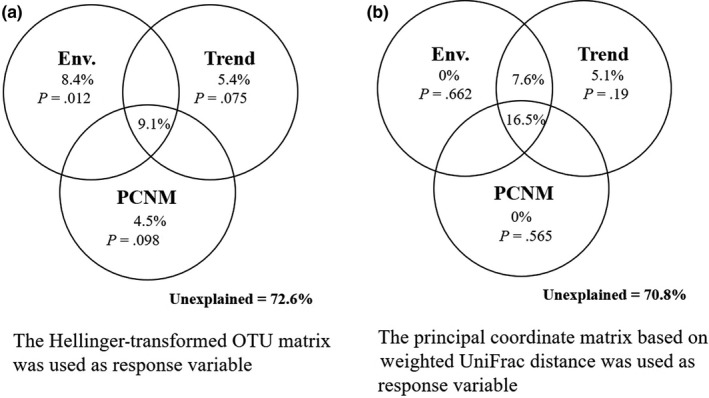
Variance partitioning analysis of bacterioplankton community in Bohai Bay according to seawater environmental factors and geospatial factors. The spatial factors including linear trend (Trend, top right) and PCNM variables (bottom). Forward selection procedures were used to select the best subset of environmental, trend, and PCNM variables explaining community variation, respectively. The community variation was calculated on the Hellinger‐transformed OTU matrix (a) and weighted UniFrac distance matrix (b), respectively. Monte Carlo permutation test was performed on each set without the effect of the other by permuting samples freely (999 permutations)
4. DISCUSSION
4.1. The distribution of bacterial taxa indicating the pollution in different areas of Bohai Bay
In the local areas of Bohai Bay, there were distinct patterns among different regions. At the phylum level, Cyanobacteria were obviously higher in TJ14 than in other sites and the most abundant genus of it was Synechococcus, and it was reported that Synechococcus was more strongly associated with carbon export in the subtropical, nutrient‐depleted, oligotrophic ocean than any other bacterioplankton lineage (Guidi et al., 2016). Among all of the sampling stations, TJ14 was the most coastal and had the lowest water temperature (24°C) and maximum transparency (0.3 m) with relatively lower nutrient concentrations. The light intensities, temperature, and mild oligotrophic conditions may provide the optimal growth conditions for Synechococcus in the studied area. The distribution trend of Synechococcus in the coastal region was in agreement with several other studies in coastal region, given that it was a good indicator for distal sampling stations in the South Sea of Korea (Seo et al., 2017), and in Tillamook Bay, they were dominant in marine samples, while members of Gammaproteobacteria and Bacteroidetes were dominant in freshwater samples (Bernhard, Colbert, Mcmanus, & Field, 2005). At OUT level, the marine Actinobacteria clade OCS155 (OTU104369) showed relatively higher abundance in the group 3 region which located far off the shore and with lower nutrient concentrations. This clade was first described by Fuhrman et al. in oligotrophic “blue‐water” regions from both Pacific and Atlantic Ocean samples (Fuhrman, McCallum, & Davis, 1993), with low levels of particulate organic C and N. In this study, the OCS155 clade was enriched in the offshore area which may indicate that it tends to stay in the oligotrophic areas. Furthermore, members of Flavobacteriaceae (OTU5954, OTU30515, OTU92726, OTU66225) and Rhodobacteraceae (OTU73029, OTU110096, OTU132038) distributed distinctly in different regions of Bohai Bay with strong correlation with nitrogen nutrients. Flavobacteriaceae was found conducting nitrogen fixation in association with plants (Giri & Pati, 2004) and closely related to phytoplankton, mostly present in the microenvironment of the algal sphere (Gomez‐Pereira et al., 2010). It was often reported as particle‐attached bacteria in marine environments (Crump, Armbrust, & Baross, 1999) and correlated with a high nutrient content (Pommier et al., 2007). The nutrient‐content varied coast of Bohai Bay may provide different proper environment for the enrichment or decrement of several members of it. Members of Rhodobacteraceae are purple, photosynthetic bacteria that have the ability of nitrogen fixation and denitrification (Zumft, 1997) and were abundant across most samples as they are the pioneering and dominant microorganisms on submerged surfaces and organic particles, acting as primary colonizers in aquatic environments (Dang & Lovell, 2002, 2015; Elifantz, Horn, Ayon, Cohen, & Minz, 2013). Their significant correlations with nitrogen nutrients or SRP indicate that they contribute to the N or P cycling in the studied area of Bohai Bay. In addition, members of Kiloniellales, Phyllobacteriaceae, and Methylophilaceae all showed significant correlations with nitrogen nutrients and different distribution patterns in the three groups, which all had been reported to participate in nitrogen cycling (Imhoff & Wiese, 2014; Kalyuhznaya et al., 2009; Willems, 2014).
Members of Bacteroidaceae, Porphyromonadaceae, Prevotellaceae, Sphingobacteriaceae, Enterococcaceae, Ochrobactrum, Achromobacter, Arcobacter, and Enterobacteriaceae which showed significant correlations with AN or SRP were only exhibited with higher abundance in TJ02. However, these families contain many sewage‐associated bacteria or pathogens (Finkmann, Altendorf, Stackebrandt, & Lipski, 2000; Pitout & Laupland, 2008; Su & Liu, 2007), although they are reported to have a nitrogen fixation or denitrification ability (Youatt, 1954; Zumft, 1997) which may be responsible for their correlation with the nutrients, it is more likely that they are indicators of anthropogenic contamination in the studied area. Not unexpectedly, a sewage outlet was found near the TJ02 station, which explained that the TJ02‐exclusive taxa were most likely originated from a point source pollution.
As described above, several taxa showed distinct pattern or sensitivity to the different pollution conditions of different regions in Bohai Bay, and these taxa may be good indicator species for environmental monitoring with their abundances significantly correlated with the pollution parameters. For taxa which did not show obviously regular distribution pattern along with the variation of pollution parameter, evidences of human interference may explain for the uncertainty.
4.2. Bacterial alpha diversity was mainly affected by AN in Bohai Bay
The OTU richness and Shannon index were significantly negatively correlated with AN. However, in other studies of similar environments, higher richness was often observed in coastal rather than offshore areas (He et al., 2017; Richa et al., 2017; Shan et al., 2015). Nevertheless, in Guanabara Bay lower richness was observed in the more polluted inner bay water (Vieira et al., 2008), and in the coastal region of Zhejiang area, a negative correlation between AN and bacterial alpha diversity was also detected, although it was not significant (Wang et al., 2015). We compared the AN contents in the listed studies and found that it was highest in Guanabara Bay (0–96.5 μM) (Vieira et al., 2008) and lowest in the Gulf of Naples (0.45–8.22 μM) (Richa et al., 2017), while the AN contents in our study ranged from 0.35–13.53 μM, which was relatively higher than that in the Gulf of Naples. It stands to reason that there may be a positive relationship between bacterial alpha diversity and AN within a certain range, but when the threshold is exceeded the relationship would be changed. Of course, much more surveys need to be done with sampling on a broader scale and perhaps also laboratory experiments. The PLS‐PM results suggested a weak influence of geospatial or physico‐chemical environmental factors on the alpha diversity but the results showed a strong effect of geographic location on the environmental factors, which further illustrated the close relationship between spatial and environmental variables in Bohai Bay.
4.3. The role of environmental and spatial factors in structuring bacterial communities
There was a significant distance–decay relationship in the studied region. A comprehensive review highlighted that environmental selection and dispersal limitation were major processes leading to a distance–decay relationship (Hanson et al., 2012). Distance–decay in similarity of microbial communities in aquatic ecosystems occurred across various spatial scales ranging from 20 m (Lear et al., 2014) to intercontinental scale (Hanson et al., 2012). Bohai Bay with a regional size of about 100 km can be classified as an intermediate scale, for which both environmental and spatial factors were recognized as important for community variation (Martiny et al., 2006). In this study, stronger effects of geographic distance than environmental factor on the variation of beta diversity were detected through the partial Mantel test. It seemed that the dispersal limitation accounted more for the beta‐diversity variation than the environmental heterogeneity. In addition, the VPA results showed that the share of spatial (trend and PCNM) and environmental factors both accounted for the maximum proportion in the explained part under the two matrices (Bray–Curtis and weighted UniFrac), demonstrating the strong influence of both spatial and environmental variables on the BCC variation. Considering the significantly increased environmental variability with geographic distance, it can be speculated that it is the joint effects of environmental and spatial factors that contributed to the explained variation of BCC in the studied area.
The explained fraction compared favorably with other studies in aquatic systems and was almost equal with the explanation of bacterial community variation in Quebec lakes (Beisner et al., 2006) and rock pools at the Baltic Sea coast (Langenheder & Ragnarsson, 2007). In these two studies, environmental factors both contributed more to the BCC variation than the spatial variables, and the environmental factors accounted for most of the explained variation. However, in our study spatial and environmental factors accounted for the largest part of the explanation. A similar study in the coastal region of Zhejiang, East China Sea, achieved a total explanation of 80%, with spatial factors contributing more to the BCC variation than environmental factors (Wang et al., 2015). In said study, the area crossed eight coastal regions including several bay areas, and compared with it, the low explanation in our study may be caused by the small sampling range. However, we offer an analysis of an important local bay that had never been reported.
The low explanation and unexplained part might be attributed to the following reasons: Firstly, dispersal limitation is stronger in isolated habitats and solid substrates but weaker in the pelagic marine environment where ocean currents facilitate microbial dispersal (Hanson et al., 2012). Meanwhile, the sampling time of this study was during the rainfall periods which may contribute to water inputs (the Jiyun river and Haihe river pour into the Bohai Bay), potentially yielding more similarly composed bacterial communities due to the movement of water masses (Langenheder & Ragnarsson, 2007). Secondly, unmeasured variables may contribute for the unexplained BCC variation, which may include abiotic environmental variables such as heavy metals, which constitute a known ecological risk in Bohai Bay (Zhu et al., 2017), as well as TOC (Zheng, Wang, & Liu, 2014) and DOM (Judd, Crump, & Kling, 2006), or biological factors such as chlorophyll a (Qiao et al., 2017), macrophytes, and phytoplankton composition (Niu et al., 2011; Wu et al., 2007). Nonetheless, the main factors which were reported as significant determinants (temperature, DO, salinity, pH, COD, nitrite, nitrate, AN, SRSi, and SRP) were mostly included in this study. In the simple Mantel test, the conductivity, transparency, and nitrogen salts (DIN, AN, nitrate) were detected correlated with beta diversity significantly, which were reported as important factors in structuring the bacterial community (Merlo et al., 2017; Wang et al., 2015; Zhang et al., 2018). However, factors such as DO (Wright, Konwar, & Hallam, 2012), pH (Lindh et al., 2013), and salinity (Lozupone & Knight, 2007) which had been extensively reported as major determinants in microbial community composition in marine environments were not identified as top factors shaping the BCC in this study. The low explanation may therefore be caused by the narrow scale of these environmental factors among the collected samples or may be their effects were masked by the complex physical features in Bohai Bay at the time of sampling. Finally, it has been argued that microbial communities are largely regulated by stochastic events resulting in random community assembly (Sloan et al., 2006). The Mantel correlogram indicated a patchy distribution of communities across most of the spatial scales, suggesting a somewhat random distribution of bacterial communities with geographic distance. The random anthropogenic activities, such as the draining of industrial wastewater and/or domestic sewage, may have caused the microbial community to enter a state of unstable nutrition which subsequently affect the total BCC transiently (He et al., 2017), as evidenced by the relatively high abundance of TJ02‐exclusive taxa. It is therefore possible that we can only explain 27.4%–29.2% of the total variation in BCC among the stations because the communities are truly randomly assembled to a large extent.
Nevertheless, in view of the complex interactions of multitudinous factors that form the basis of the community composition and distribution in coastal areas and adjoining harbors (He et al., 2017), more elaborate work is still needed to elucidate the formation mechanism of microbial communities in the natural environment. This is the key to understanding the mechanisms that generate and maintain microbial biodiversity and predicting ecosystem responses to future environmental changes (Martiny et al., 2011).
5. CONCLUSIONS
This study contributes the first overview of bacterioplankton community distribution and environmental/spatial factors that affect its variation in Bohai Bay using 16S rDNA targeted amplicon sequencing. We found that the environmental factors exhibit spatial autocorrelation in the studied area, and the samples could be classified into three groups according to the environmental and spatial factors. We also identified the bacterioplankton taxa with significant differences among the three groups, which may indicate the effects of the pollution gradient and of dispersal limitation. However, the alpha and beta diversity of the bacterioplankton among all the sites did not exhibit an obvious consistent variation trend with the segmentation of the environmental and spatial factors. A distance–decay relationship was found between the bacterioplankton community similarity and geographic distance. The partial Mantel test showed a stronger effect of geographic distance than the environmental factors on the beta diversity. The variance partitioning analysis showed that environmental and spatial factors contributed most to the explained proportion. We therefore speculated that it was the joint effects of environmental and spatial factors that contributed to the explained variation of BCCs in the studied area of Bohai Bay. The large unexplained part may be caused by the unmeasured biotic/abiotic factors, stochastic factors, and anthropogenic disturbances. This study of planktonic bacterial community in Bohai Bay provides new insights into the microbial ecology of a semi‐enclosed bay.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Wei Zhao: Conceptualization‐Lead, Data curation‐Lead, Formal analysis‐Lead, Funding acquisition‐Lead, Investigation‐Lead, Methodology‐Lead, Project administration‐Lead, Resources‐Lead, Software‐Lead, Supervision‐Lead, Validation‐Lead, Visualization‐Lead, Writing‐original draft‐Lead, Writing‐review & editing‐Lead. Jingjing Wang: Funding acquisition‐Supporting, Writing‐review & editing‐Supporting. Song Xu: Methodology‐Supporting, Resources‐Supporting. Yu Lei: Methodology‐Supporting, Resources‐Supporting. Rong Yang: Funding acquisition‐Supporting. Liuyang Shi: Resources‐Supporting. Xingbiao Wang: Funding acquisition‐Supporting. Zhiyong Huang: Funding acquisition‐Equal, Project administration‐Equal, Supervision‐Equal, Writing‐review & editing‐Equal.
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
We would like to thank the staff of the Tianjin Survey and Design Institute for Water Transport Engineering and Tianjin Key Laboratory of Marine Resources and Chemistry, Tianjin University of Science and Technology, for supporting our sampling. This work was financially supported by the Science and Technology Support Program of Tianjin Municipality under contracts NO. 18ZXSZSF00100, 18YFZCNC01180, and 16YFXTSY00580. Financial support was also provided by the Key Programs of Chinese Academy of Sciences (KFZD‐SW‐219).
APPENDIX 1.
Table A1.
The ANOVA test of environmental variables among different regions (Group1 represents the north region; group2 represents the south region; group3 represents the east region)
| Variables | Unit | North (Group1) | South (Group2) | East (Group3) |
|---|---|---|---|---|
| Longitude | E | 117.85 ± 0.08b | 117.73 ± 0.05b | 118.28 ± 0.30a |
| Latitude | N | 39.05 ± 0.06a | 38.74 ± 0.08b | 38.75 ± 0.10b |
| Temperature | °C | 26.36 ± 1.0b | 30.65 ± 3.26a | 25.51 ± 1.41b |
| Humidity | % | 59.62 ± 5.05ab | 51 ± 10.67b | 67.03 ± 5.46a |
| DO | mg/L | 5.57 ± 0.38b | 6.53 ± 0.77a | 6.69 ± 0.59a |
| Air pressure | kpa | 1,013.80 ± 0.40a | 1,012.88 ± 0.93a | 1,011.83 ± 0.69b |
| Depth | m | 3.28 ± 1.35b | 5.38 ± 2.69b | 14.82 ± 4.94a |
| Water temperature | °C | 27.00 ± 0.23a | 27.91 ± 0.77a | 25.38 ± 1.50b |
| Salinity | ‰ | 27.60 ± 0.92a | 29.30 ± 0.24a | 28.47 ± 2.23a |
| Transparency | cm | 64.00 ± 34.41b | 152.50 ± 61.80a | 190.00 ± 60.55a |
| Conductivity | ms/cm | 42.22 ± 1.62b | 45.18 ± 0.12a | 45.42 ± 0.31a |
| SRP | mg/L | 0.02 ± 0.01a | 0.01 ± 0.00b | 0.008 ± 0.00b |
| Nitrite | mg/L | 0.10 ± 0.05a | 0.10 ± 0.04a | 0.03 ± 0.02b |
| Nitrate | mg/L | 0.47 ± 0.05a | 0.34 ± 0.03b | 0.13 ± 0.03c |
| Ammonia | mg/L | 0.17 ± 0.03a | 0.06 ± 0.07b | 0.06 ± 0.04b |
| DIN | mg/L | 0.74 ± 0.08a | 0.50 ± 0.06b | 0.22 ± 0.05c |
| SRSi | mg/L | 0.88 ± 0.09a | 0.86 ± 0.09a | 0.80 ± 0.10a |
| pH | 7.99 ± 0.04b | 8.15 ± 0.08a | 8.12 ± 0.04a | |
| COD | mg/L | 0.64 ± 0.18b | 1.15 ± 0.39a | 0.82 ± 0.19ab |
Data in the table are the Means ± SD; The different normal letters in the same row indicate significant difference among sites at 0.05 level (n > 3) under Duncan test. Letters in bold means the significant difference of DIN concentrations in each group. Refer to Table 2 for variable abbreviations.
Table A2.
Water quality level of DIN and SRP concentrations
| Water quality levela | II | III | IV | V |
|---|---|---|---|---|
| DIN (mg/L) | 0.2 | 0.3 | 0.4 | 0.5 |
| SRP (mg/L) | 0.015 | 0.03 | 0.03 | 0.045 |
Water quality levels II, III, IV and V are corresponding to relatively clean water, slightly polluted water, medium polluted water and heavily polluted water, respectively.
Table A3.
Spearman's rank correlations between variation of each water chemical parameters (Euclidean distance) and geographic distance between sites
| Variables | r | p | N |
|---|---|---|---|
| Environment | .6357 | .001 | 19 |
| Air temperature | .141 | .162 | 19 |
| Humidity | .2269 | .07 | 19 |
| DO | .17 | .1 | 19 |
| Air pressure | .2564 | .008 | 19 |
| Depth | .1412 | .133 | 19 |
| Water temperature | .4783 | .003 | 19 |
| Salinity | .1455 | .167 | 19 |
| Transparency | .3201 | .004 | 19 |
| Conductivity | .3461 | .018 | 19 |
| SRP | .1064 | .239 | 19 |
| Nitrite | .3389 | .011 | 19 |
| Nitrate | .5266 | .001 | 19 |
| AN | .19 | .054 | 19 |
| IN | .5399 | .001 | 19 |
| SRSi | .0284 | .373 | 19 |
| pH | .1085 | .197 | 19 |
| COD | .1811 | .091 | 19 |
r, correlation coefficients between pairwise distance of each water chemical variables and geographic distance derived from Mantel test with 999 permutations. Data in bold indicate significant correlations (p < .05). Refer to Table 2 for variable abbreviations.
Table A4.
The ANOVA test of OTUs with relative abundance significantly varied among the three clustered regions in the studied area of Bohai Bay (The difference significance test for OTU32430 and OTU132038 were tested through Kruskal‐Wallis test since the test of their variance homogeneity was not significant)
| Group1 | Group2 | Group3 | |
|---|---|---|---|
| OTU104369 | 0.0009 ± 0.001b | 0.0006 ± 0.0006b | 0.0086 ± 0.0107a |
| OTU65886 | 0.0038 ± 0.0051a | 0.0004 ± 0.0004b | 0.0003 ± 0.0003b |
| OTU3224 | 0.001 ± 0.0006a | 0.009 ± 0.0067b | 0.0035 ± 0.0019ab |
| OTU126053 | 0.0004 ± 0.0002b | 0.0113 ± 0.0099a | 0.0009 ± 0.0008b |
| OTU5954 | 0.0149 ± 0.0101a | 0.0044 ± 0.0029b | 0.0027 ± 0.0027b |
| OTU30515 | 0.0001 ± 0b | 0.0003 ± 0.0003b | 0.0099 ± 0.0122a |
| OTU85516 | 0.0004 ± 0.0003b | 0.0067 ± 0.0059a | 0.0014 ± 0.0009b |
| OTU92726 | 0.0032 ± 0.0023a | 0.0139 ± 0.0094b | 0.0174 ± 0.0086b |
| OTU66225 | 0.0008 ± 0.001a | 0.0034 ± 0.0022ab | 0.0057 ± 0.0033b |
| OTU32430 | 0.0025 ± 0.0023a | 0.0062 ± 0.0059ab | 0.0091 ± 0.0038b |
| OTU82462 | 0.0006 ± 0.0004b | 0.0002 ± 0.0001b | 0.0052 ± 0.0058a |
| OTU73029 | 0.0132 ± 0.0101b | 0.0136 ± 0.0097b | 0.0685 ± 0.0566a |
| OTU132038 | 0.0582 ± 0.027b | 0.1289 ± 0.0427a | 0.0639 ± 0.0381b |
| OTU12256 | 0.0011 ± 0.0009b | 0.0013 ± 0.0006b | 0.0059 ± 0.0056a |
| OTU7191 | 0.0025 ± 0.0018b | 0.0066 ± 0.0041a | 0.0028 ± 0.0019b |
| OTU84157 | 0.003 ± 0.0031b | 0.012 ± 0.0084a | 0.0034 ± 0.0026b |
Data in the table are the Means ± SD; The different normal letters in the same row indicate significant difference among groups at 0.05 level (n > 3) under Duncan test except for OTU OTU32430 and OTU132038).
Table A5.
The ANOVA test of bacterial alpha‐diversity among different groups (regions)
| North (Group1) | South (Group2) | East (Group3) | |
|---|---|---|---|
| OTU richness | 662.62 ± 123.99a | 663.66 ± 92.73a | 701.9 ± 89.82a |
| PD | 66.46 ± 11.53a | 64.13 ± 9.53a | 67.45 ± 3.72a |
| Shannon | 4.82 ± 0.60a | 4.24 ± 1.81a | 5.03 ± 0.25a |
Data in the table are the Means ± SD; The different normal letters in the same row indicate significant difference among sites at 0.05 level (n > 3) under Duncan test.
Details of the PLS‐PM
During the PLS‐PM analysis, parameters with a relatively lower loading coefficient of 0.7 were excluded (although the longitude with a loading coefficient of 0.628 was preserved) and at last 13 indicators were selected to establish the model. The LV of geographic location encompassed the MVs of latitude and longitude, physical environmental variables encompassed transparency and air pressure, the chemical environmental variables encompassed conductivity, DO, SRP, nitrate, AN and DIN, and the bacterial alpha diversity was reflected by OTU richness, PD and the Shannon index. The R‐squared values for the correlations between geographic location and chemical or physical environmental variables were both above 0.6, which was considered as indicating a highly credible simple model.
Table A6.
Direct and indirect effects between different latent variables
| Relationships | Direct | Indirect | Total | |
|---|---|---|---|---|
| 1 | Geographic_location → Seawater_environment | 0.798 | 0 | 0.798 |
| 2 | Geographic_location → Nutrient_contents | 0.744 | 0.0806 | 0.824 |
| 3 | Geographic_location → Microbial_alpha_diversity | 0.205 | −0.4323 | −0.227 |
| 4 | Seawater_environment → Nutrient_contents | 0.101 | 0 | 0.101 |
| 5 | Seawater_environment → Microbial_alpha_diversity | −0.419 | −0.0120 | −0.431 |
| 6 | Nutrient_contents → Microbial_alpha_diversity | −0.119 | 0 | −0.119 |
This table was the effects part of PLS‐PM result. This table contains the effects that each construct (latent variables) has on the rest of constructs (latent variables) by taking into consideration the total number of connections in the inner model. The direct effects are given by the path coefficients (showed in Figure 4). But there are also the indirect effects and the total effects. An indirect effect is the influence of one construct on another construct by taking an indirect path. The total effects are the sum of both the direct and indirect effects. The indirect effects are obtained as the product of the path coefficients by taking an indirect path. For instance, consider the impact of Geographic_location on Nutrient_contents: there is an indirect path from Geographic_location to Nutrient_contents that goes through Seawater_environment. If you multiply the path coefficient of Geographic_location on Seawater_environment (0.7984) with the path coefficient of Seawater_environment on Nutrient_contents (0.1009), you get the indirect effect of Geographic_location on Nutrient_contents: 0.0806 = 0.7984 × 0.1009.
Figure A1.
Location of Bohai Bay (area in the rectangular box)
Figure A2.
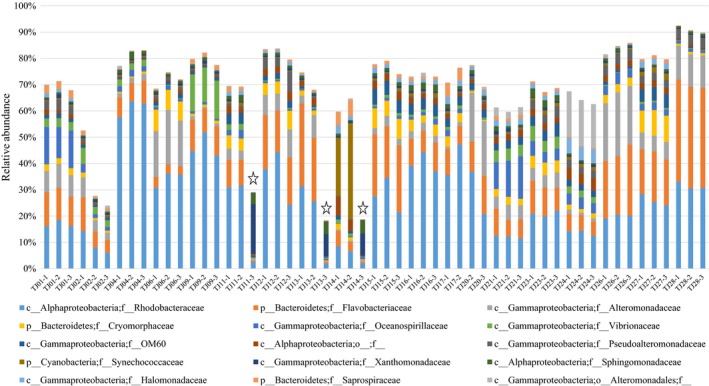
The top 15 taxa (at family level) in samples from the 19 sites in Bohai Bay. Samples signed with star means one replicate which was distinctly biased from the other two replicates of the same sites and these samples were removed before further analysis
Figure A3.
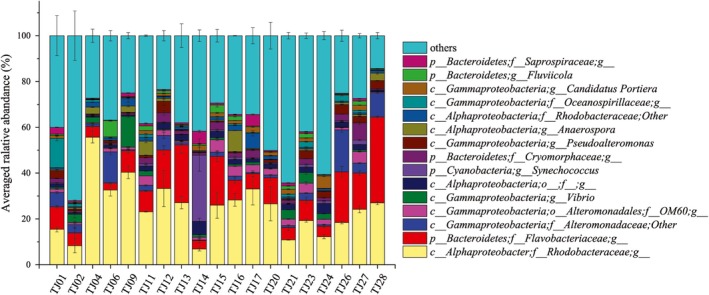
Relative abundances of the dominant taxa in 19 seawater samples of Bohai Bay at genus level. Relative abundances of the phylotypes in the top15 were presented and others represented the sum of the rest phylotypes
Figure A4.
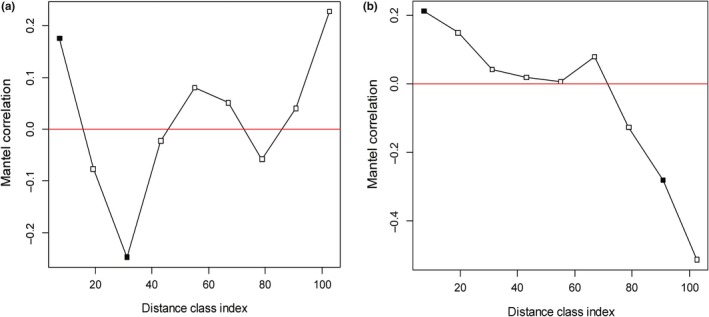
Mantel correlogram demonstrating the significance of distance effect on bacterial beta‐diversity across a subset of different geographic distance scales (classes) among the sampling stations. Pearson correlations between the pairwise matrix of Bray‐Curtis distance (a) or weighted Unifrac distance (b) and geographic distance between stations within each geographic distance class were generated by Mantel tests with 999 permutations. Significant correlations (p < .05, solid squares with positive values) indicate spatial correlation in beta‐diversity, observed at short geographic distance scales among the sampling station
Zhao W, Wang J, Xu S, et al. Bacterioplankton community variation in Bohai Bay (China) is explained by joint effects of environmental and spatial factors. MicrobiologyOpen. 2020;9:e997 10.1002/mbo3.997
DATA AVAILABILITY STATEMENT
All data generated and analyzed in this study are provided in the article. The raw sequencing data are deposited to the NCBI sequence read archive (SRA) under the accession number SRP118494 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA408271). The supplementary material (dataset S1 and dataset S2) is available in the Zenodo repository at https://doi.org/10.5281/zenodo.3581592.
REFERENCES
- Anderson, M. J. (2001). A new method for non‐parametric multivariate analysis of variance. Austral Ecology, 26, 32–46. [Google Scholar]
- AQSIQ . (2007). The specification for marine monitoring of China‐Part 4: Seawater analysis (GB 17378.4‐2007) In General administration of quality supervision, inspection and quarantine (AQSIQ) of the People's Republic of China (in Chinese). [Google Scholar]
- Beisner, B. E. , Peres‐Neto, P. R. , Lindstrom, E. S. , Barnett, A. , & Longhi, M. L. (2006). The role of environmental and spatial processes in structuring lake communities from bacteria to fish. Ecology, 87, 2985–2991. 10.1890/0012-9658(2006)87[2985:TROEAS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bernhard, A. E. , Colbert, D. , Mcmanus, J. , & Field, K. G. (2005). Microbial community dynamics based on 16S rRNA gene profiles in a Pacific Northwest estuary and its tributaries. FEMS Microbiology Ecology, 52, 115–128. 10.1016/j.femsec.2004.10.016 [DOI] [PubMed] [Google Scholar]
- Bokulich, N. A. , Thorngate, J. H. , Richardson, P. M. , & Mills, D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proceedings of the National Academy of Sciences of the United States of America, 111, E139–E148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcard, D. , Gillet, F. , & Legendre, P. (2011). Numerical ecology with R. Matisse, 77, 332–334. [Google Scholar]
- Borcard, D. , Legendre, P. , & Drapeau, P. (1992). Partialling out the spatial component of ecological variation. Ecology, 73, 1045–1055. 10.2307/1940179 [DOI] [Google Scholar]
- Caddy, J. F. (2000). Marine catchment basin effects versus impacts of fisheries on semi‐enclosed seas. ICES Journal of Marine Science, 57, 628–640. 10.1006/jmsc.2000.0739 [DOI] [Google Scholar]
- Caporaso, J. G. , Bittinger, K. , Bushman, F. D. , Desantis, T. Z. , Andersen, G. L. , & Knight, R. (2010). PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics, 26, 266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F. D. , Costello, E. K. , … Knight, R. (2010). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G. , Lauber, C. L. , Walters, W. A. , Berglyons, D. , Lozupone, C. A. , Turnbaugh, P. J. , … Knight, R. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America, 108, 4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, J. H. (1965). The Chesapeake Bay institute technique for the Winkler dissolved oxygen method. Limnology and Oceanography, 10, 141–143. 10.4319/lo.1965.10.1.0141 [DOI] [Google Scholar]
- Chen, Y. H. , Gao, Y. H. , Chen, C. P. , Liang, J. R. , Sun, L. , Zhen, Y. , & Qiao, L. (2016). Seasonal variations of phytoplankton assemblages and its relation to environmental variables in a scallop culture sea area of Bohai Bay, China. Marine Pollution Bulletin, 113, 362–370. 10.1016/j.marpolbul.2016.10.025 [DOI] [PubMed] [Google Scholar]
- Clarke, K. R. (1993). Non‐parametric multivariate analysis of change in community structure. Austral Ecology, 18, 117–143. [Google Scholar]
- Crump, B. C. , Armbrust, E. V. , & Baross, J. A. (1999). Phylogenetic analysis of particle‐attached and free‐living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Applied and Environmental Microbiology, 65, 3192–3204. 10.1128/AEM.65.7.3192-3204.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, H. , & Lovell, C. R. (2002). Numerical dominance and phylotype diversity of marine rhodobacter species during early colonization of submerged surfaces in coastal marine waters as determined by 16S ribosomal DNA sequence analysis and fluorescence in situ hybridization. Applied and Environmental Microbiology, 68, 496–504. 10.1128/AEM.68.2.496-504.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, H. , & Lovell, C. R. (2015). Microbial surface colonization and biofilm development in marine environments. Microbiology and Molecular Biology Reviews, 80, 91–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delong, E. F. , & Karl, D. M. (2005). Genomic perspective in microbial oceanography. Nature, 437, 336–342. [DOI] [PubMed] [Google Scholar]
- Desantis, T. Z. , Hugenholtz, P. , Larsen, N. , Rojas, M. , Brodie, E. L. , Keller, K. , … Andersen, G. L. (2006). Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 72, 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, J. , Xiao, K. , Li, L. , Ding, X. , Liu, H. , Lu, Y. , & Zhou, S. (2013). Temporal and spatial diversity of bacterial communities in coastal waters of the South China sea. PLoS ONE, 8, e66968 10.1371/journal.pone.0066968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. , Haas, B. J. , Clemente, J. C. , Quince, C. , & Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27, 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elifantz, H. , Horn, G. , Ayon, M. , Cohen, Y. , & Minz, D. (2013). Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiology Ecology, 85, 348–357. [DOI] [PubMed] [Google Scholar]
- Farooq, A. , & Francesca, M. (2007). Microbial structuring of marine ecosystems. Nature Reviews Microbiology, 5, 782–791. 10.1038/nrmicro1747 [DOI] [PubMed] [Google Scholar]
- Finkmann, W. , Altendorf, K. , Stackebrandt, E. , & Lipski, A. (2000). Characterization of N2O‐producing Xanthomonas‐like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov. and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. International Journal of Systematic and Evolutionary Microbiology, 50(1), 273–282. 10.1099/00207713-50-1-273 [DOI] [PubMed] [Google Scholar]
- Flynn, D. F. , Mirotchnick, N. , Jain, M. , Palmer, M. I. , & Naeem, S. (2011). Functional and phylogenetic diversity as predictors of biodiversity–ecosystem‐function relationships. Ecology, 92, 1573–1581. 10.1890/10-1245.1 [DOI] [PubMed] [Google Scholar]
- Fuhrman, J. A. (2009). Microbial community structure and its functional implications. Nature, 459, 193–199. 10.1038/nature08058 [DOI] [PubMed] [Google Scholar]
- Fuhrman, J. A. , McCallum, K. , & Davis, A. A. (1993). Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Applied and Environmental Microbiology, 59, 1294–1302. 10.1128/AEM.59.5.1294-1302.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman, J. A. , Steele, J. A. , Hewson, I. , Schwalbach, M. S. , Brown, M. V. , Green, J. L. , & Brown, J. H. (2008). A latitudinal diversity gradient in planktonic marine bacteria. Proceedings of the National Academy of Sciences of the United States of America, 105, 7774–7778. 10.1073/pnas.0803070105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, Y. , He, J. Z. , Zhu, Y. G. , Zhang, J. B. , Xu, Z. , Zhang, L. M. , & Zheng, Y. M. (2008). Differences in soil bacterial diversity: Driven by contemporary disturbances or historical contingencies? The ISME Journal, 2, 254–264. 10.1038/ismej.2008.2 [DOI] [PubMed] [Google Scholar]
- Giri, S. , & Pati, B. R. (2004). A comparative study on phyllosphere nitrogen fixation by newly isolated Corynebacterium sp. & Flavobacterium sp. and their potentialities as biofertilizer. Acta Microbiologica Immunologica Hungarica, 51, 47–56. [DOI] [PubMed] [Google Scholar]
- Gomez‐Pereira, P. R. , Fuchs, B. M. , Alonso, C. , Oliver, M. J. , Van Beusekom, J. E. , & Amann, R. (2010). Distinct flavobacterial communities in contrasting water masses of the north Atlantic Ocean. The ISME Journal, 4, 472–487. 10.1038/ismej.2009.142 [DOI] [PubMed] [Google Scholar]
- Griffith, D. A. , & Peres‐Neto, P. R. (2006). Spatial modeling in ecology: The flexibility of eigenfunction spatial analyses. Ecology, 87, 2603–2613. 10.1890/0012-9658(2006)87[2603:SMIETF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Guidi, L. , Chaffron, S. , Bittner, L. , Eveillard, D. , Larhlimi, A. , Roux, S. , … Gorsky, G. (2016). Plankton networks driving carbon export in the oligotrophic ocean. Nature, 532, 465–470. 10.1038/nature16942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S. , Li, Y. , & Zhang, C. (2014). Phytoplankton community in the Bohai Sea and its relationship with environmental factors. Marine Science Bulletin, 33, 95–105. [Google Scholar]
- Hair, J. F. , Sarstedt, M. , Pieper, T. M. , & Ringle, C. M. (2012). Applications of partial least squares path modeling in management journals: A review of past practices and recommendations for future applications. Long Range Planning, 45, 320–340. [Google Scholar]
- Hanson, C. , Fuhrman, J. , Horner‐Devine, C. , & Martiny, J. (2012). Beyond biogeographic patterns: Processes shaping the microbial landscape. Nature Reviews Microbiology, 10, 497–506. 10.1038/nrmicro2795 [DOI] [PubMed] [Google Scholar]
- He, Y. , Sen, B. , Zhou, S. , Xie, N. , Zhang, Y. , Zhang, J. , & Wang, G. (2017). Distinct seasonal patterns of bacterioplankton abundance and dominance of phyla alpha‐proteobacteria and cyanobacteria in Qinhuangdao coastal waters off the Bohai Sea. Frontiers in Microbiology, 8, 1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlemann, D. P. , Labrenz, M. , Jürgens, K. , Bertilsson, S. , Waniek, J. J. , & Andersson, A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME Journal, 5, 1571–1579. 10.1038/ismej.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhoff, J. F. , & Wiese, J. (2014). The Order Kiloniellales[M]. Berlin Heidelberg, Germany: Springer. [Google Scholar]
- Judd, K. E. , Crump, B. C. , & Kling, G. W. (2006). Variation in dissolved organic matter controls bacterial production and community composition. Ecology, 87, 2068–2079. 10.1890/0012-9658(2006)87[2068:VIDOMC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kalyuhznaya, M. G. , Martens‐Habbena, W. , Wang, T. , Hackett, M. , Stolyar, S. M. , Stahl, D. , … Chistoserdova, L. (2009). Methylophilaceae link methanol oxidation to denitrification in freshwater lake sediment as suggested by stable isotope probing and pure culture analysis. Environmental Microbiology Reports, 1(5), 385–392. 10.1111/j.1758-2229.2009.00046.x [DOI] [PubMed] [Google Scholar]
- Langenheder, S. , & Ragnarsson, H. (2007). The role of environmental and spatial factors for the composition of aquatic bacterial communities. Ecology, 88, 2154–2161. 10.1890/06-2098.1 [DOI] [PubMed] [Google Scholar]
- Lear, G. , Bellamy, J. , Case, B. S. , Lee, J. E. , & Buckley, H. L. (2014). Fine‐scale spatial patterns in bacterial community composition and function within freshwater ponds. The ISME Journal, 8, 1715–1726. 10.1038/ismej.2014.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre, P. , & Gallagher, E. D. (2001). Ecologically meaningful transformations for ordination of species data. Oecologia, 129, 271–280. 10.1007/s004420100716 [DOI] [PubMed] [Google Scholar]
- Lindh, M. V. , Riemann, L. , Baltar, F. , Romerooliva, C. , Salomon, P. S. , Granéli, E. , & Pinhassi, J. (2013). Consequences of increased temperature and acidification on bacterioplankton community composition during a mesocosm spring bloom in the Baltic Sea. Environmental Microbiology Reports, 5, 252–262. 10.1111/1758-2229.12009 [DOI] [PubMed] [Google Scholar]
- Lindström, E. S. , & Langenheder, S. (2012). Local and regional factors influencing bacterial community assembly. Environmental Microbiology Reports, 4, 1–9. 10.1111/j.1758-2229.2011.00257.x [DOI] [PubMed] [Google Scholar]
- Lozupone, C. A. , & Knight, R. (2007). Global patterns in bacterial diversity. Proceedings of the National Academy of Sciences of the United States of America, 104, 11436–11440. 10.1073/pnas.0611525104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc, T. , & Salzberg, S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27, 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny, J. B. , Bohannan, B. J. , Brown, J. H. , Colwell, R. K. , Fuhrman, J. A. , Green, J. L. , … Staley, J. T. (2006). Microbial biogeography: Putting microorganisms on the map. Nature Reviews Microbiology, 4, 102–112. 10.1038/nrmicro1341 [DOI] [PubMed] [Google Scholar]
- Martiny, J. B. , Eisen, J. A. , Penn, K. , Allison, S. D. , & Horner‐Devine, M. C. (2011). Drivers of bacterial beta‐diversity depend on spatial scale. Proceedings of the National Academy of Sciences of the United States of America, 108, 7850–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEP (Ministry of Environmental Protection of the People's Republic of China) . (1997). Sea Water Quality Standard (GB 3097–1997). Retrieved from http://english.mep.gov.cn/standardsreports/standards/water_environment/quality_standard/200710/t20071024111791.htm [Google Scholar]
- Merlo, C. , Amé, M. V. , Bertrand, L. , Abril, A. , Merlo, C. , Amé, M. V. , … Amé, M. V. (2017). Changes in the bacterial community composition of different habitats along a polluted river (Suquía River, Cordoba, Argentina). Ecologia Austral, 27, 72–84. [Google Scholar]
- Niu, Y. , Shen, H. , Chen, J. , Xie, P. , Yang, X. , Tao, M. , … Qi, M. (2011). Phytoplankton community succession shaping bacterioplankton community composition in Lake Taihu, China. Water Research, 45, 4169–4182. 10.1016/j.watres.2011.05.022 [DOI] [PubMed] [Google Scholar]
- Oksanen, J. , Blanchet, F. G. , Friendly, M. , Kindt, R. , Legendre, P. , McGlinn, D. , … Wagner, H. (2019). vegan: Community Ecology Package. R package version 2.5‐6. Retrieved from https://CRAN.R-project.org/package=vegan [Google Scholar]
- Patel, R. K. , & Jain, M. (2012). NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE, 7, e30619 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, S. , Qin, X. , Shi, H. , Zhou, R. , Dai, M. , & Ding, D. (2012). Distribution and controlling factors of phytoplankton assemblages in a semi‐enclosed bay during spring and summer. Marine Pollution Bulletin, 64, 941–948. 10.1016/j.marpolbul.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Peres‐Neto, P. R. , Legendre, P. , Dray, S. , & Borcard, D. (2006). Variation partitioning of species data matrices: Estimation and comparison of fractions. Ecology, 87, 2614–2625. 10.1890/0012-9658(2006)87[2614:VPOSDM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Pitout, J. D. , & Laupland, K. B. (2008). Extended‐spectrum β‐lactamase‐producing Enterobacteriaceae: An emerging public‐health concern. The Lancet Infectious Diseases, 8, 159–166. 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- Pommier, T. , Canback, B. , Riemann, L. , Bostrom, K. H. , Simu, K. , Lundberg, P. , … Hagstrom, A. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Molecular Ecology, 16, 867–880. 10.1111/j.1365-294X.2006.03189.x [DOI] [PubMed] [Google Scholar]
- Pommier, T. , Neal, P. R. , Gasol, J. M. , Coll, M. , Acinas, S. G. , & Pedrós‐Alió, C. (2010). Spatial patterns of bacterial richness and evenness in the NW Mediterranean Sea explored by pyrosequencing of the 16S rRNA. Aquatic Microbial Ecology, 61, 221–233. 10.3354/ame01484 [DOI] [Google Scholar]
- Qiao, Y. , Feng, J. , Cui, S. , & Zhu, L. (2017). Long‐term changes in nutrients, chlorophyll a and their relationships in a semi‐enclosed eutrophic ecosystem, Bohai Bay, China. Marine Pollution Bulletin, 117, 222 10.1016/j.marpolbul.2017.02.002 [DOI] [PubMed] [Google Scholar]
- Richa, K. , Balestra, C. , Piredda, R. , Benes, V. , Borra, M. , Passarelli, A. , … Casotti, R. (2017). Distribution, community composition, and potential metabolic activity of bacterioplankton in an urbanized Mediterranean Sea coastal zone. Applied and Environmental Microbiology, 83, e00497–e517. 10.1128/AEM.00494-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, J. H. (2019). geosphere: Spherical Trigonometry. R package version 1.5‐10. Retrieved from https://CRAN.R-project.org/package=geosphere [Google Scholar]
- Sanchez, G. (2013). PLS Path Modeling with R, Berkeley. Retrieved from http://www.gastonsanchez.com/PLSPathModelingwithR.pdf [Google Scholar]
- Seo, J. H. , Kang, I. , Yang, S. J. , & Cho, J. C. (2017). Characterization of spatial distribution of the bacterial community in the South Sea of Korea. PLoS ONE, 12, e0174159 10.1371/journal.pone.0174159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, D. , Wei, G. , Li, M. , Wang, W. , Li, X. , Gao, Z. , & Shao, Z. (2015). Distribution and diversity of bacterioplankton communities in subtropical seawater around Xiamen Island, China. Microbiological Research, 175, 16–23. 10.1016/j.micres.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Shi, J. , Li, G. , & Wang, P. (2011). Anthropogenic influences on the tidal prism and water exchanges in Jiaozhou Bay, Qingdao, China. Journal of Coastal Research, 27, 57–72. 10.2112/JCOASTRES-D-09-00011.1 [DOI] [Google Scholar]
- Sloan, W. T. , Lunn, M. , Woodcock, S. , Head, I. M. , Nee, S. , & Curtis, T. P. (2006). Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environmental Microbiology, 8, 732–740. 10.1111/j.1462-2920.2005.00956.x [DOI] [PubMed] [Google Scholar]
- Su, Y. C. , & Liu, C. C. (2007). Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiology, 24, 549–558. [DOI] [PubMed] [Google Scholar]
- Tenenhaus, M. , Vinzi, V. E. , Chatelin, Y. M. , & Lauro, C. (2005). PLS path modeling. Computational Statistics and Data Analysis, 48, 159–205. 10.1016/j.csda.2004.03.005 [DOI] [Google Scholar]
- Van der Gucht, K. , Cottenie, K. , Muylaert, K. , Vloemans, N. , Cousin, S. , Declerck, S. , … De Meester, L. (2007). The power of species sorting: Local factors drive bacterial community composition over a wide range of spatial scales. Proceedings of the National Academy of Sciences of the United States of America, 104, 20404–20409. 10.1073/pnas.0707200104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, R. P. , Gonzalez, A. M. , Cardoso, A. M. , Oliveira, D. N. , Albano, R. M. , Clementino, M. M. , … Paranhos, R. (2008). Relationships between bacterial diversity and environmental variables in a tropical marine environment, Rio de Janeiro. Environmental Microbiology, 10, 189–199. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Ye, X. S. , Chen, H. P. , Zhao, Q. F. , Hu, C. J. , He, J. Y. , … Zhang, D. M. (2015). Bacterial biogeography in the coastal waters of northern Zhejiang, East China Sea is highly controlled by spatially structured environmental gradients. Environmental Microbiology, 17, 3898 10.1111/1462-2920.12884 [DOI] [PubMed] [Google Scholar]
- Wang, K. , Ye, X. S. , Zhang, H. J. , Chen, H. P. , Zhang, D. M. , & Liu, L. (2016). Regional variations in the diversity and predicted metabolic potential of benthic prokaryotes in coastal northern Zhejiang, East China Sea. Scientific Reports, 6, 38709 10.1038/srep38709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems, A. (2014). Family Phyllobacteriaceae In Rosenberg E., DeLong E. F., Lory S., Stackebrandt E. & Thompson F. (Eds.), The Prokaryotes (pp. 355-418). Berlin, Heidelberg: Springer. [Google Scholar]
- Wright, J. J. , Konwar, K. M. , & Hallam, S. J. (2012). Microbial ecology of expanding oxygen minimum zones. Nature Reviews Microbiology, 10, 381 10.1038/nrmicro2778 [DOI] [PubMed] [Google Scholar]
- Wu, Q. L. , Zwart, G. , Wu, J. , Kamst‐Van Agterveld, M. P. , Liu, S. , & Hahn, M. W. (2007). Submersed macrophytes play a key role in structuring bacterioplankton community composition in the large, shallow, subtropical Taihu Lake, China. Environmental Microbiology, 9, 2765–2774. 10.1111/j.1462-2920.2007.01388.x [DOI] [PubMed] [Google Scholar]
- Yang, L. , Liu, J. , Zhang, J. , Wang, X. L. , Xu, Y. , Li, X. , & He, L. (2018). Zooplankton community variation and its relationship with environmental variables in Bohai Bay. Journal of Marine Sciences, 36(1), 93–101. 10.3969/j.issn.1001-909X.2018.01.010 [DOI] [Google Scholar]
- Youatt, J. B. (1954). Denitrification of nitrite by a species of Achromobacter. Nature, 173, 826–827. 10.1038/173826a0 [DOI] [PubMed] [Google Scholar]
- Zhang, D. , Wang, X. , Xiong, J. , Zhu, J. , Wang, Y. , Zhao, Q. , … Dai, H. (2014). Bacterioplankton assemblages as biological indicators of shrimp health status. Ecological Indicators, 38, 218–224. 10.1016/j.ecolind.2013.11.002 [DOI] [Google Scholar]
- Zhang, H. , Jia, J. , Chen, S. , Huang, T. , Wang, Y. , Zhao, Z. , … Ma, X. (2018). Dynamics of bacterial and fungal communities during the outbreak and decline of an algal bloom in a drinking water reservoir. International Journal of Environmental Research and Public Health, 15, 361 10.3390/ijerph15020361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhao, Z. , Dai, M. , Jiao, N. , & Herndl, G. J. (2014). Drivers shaping the diversity and biogeography of total and active bacterial communities in the South China Sea. Molecular Ecology, 23, 2260 10.1111/mec.12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, B. , Wang, L. , & Liu, L. (2014). Bacterial community structure and its regulating factors in the intertidal sediment along the Liaodong Bay of Bohai Sea, China. Microbiological Research, 169, 585–592. 10.1016/j.micres.2013.09.019 [DOI] [PubMed] [Google Scholar]
- Zhu, G. , Xie, Z. , Li, T. , Ma, Z. , & Xu, X. (2017). Assessment ecological risk of heavy metal caused by high‐intensity land reclamation in Bohai Bay, China. PLoS ONE, 12, e0175627 10.1371/journal.pone.0175627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft, W. G. (1997). Cell biology and molecular basis of denitrification. Microbiology and Molecular Biology Reviews, 61, 533–616. 10.1128/.61.4.533-616.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and analyzed in this study are provided in the article. The raw sequencing data are deposited to the NCBI sequence read archive (SRA) under the accession number SRP118494 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA408271). The supplementary material (dataset S1 and dataset S2) is available in the Zenodo repository at https://doi.org/10.5281/zenodo.3581592.


