Abstract
Porphyromonas gingivalis is regarded as a “keystone pathogen” in periodontitis. The fimbria assists in the initial attachment, biofilm organization, and bacterial adhesion leading to the invasion and colonization of host epithelial cells. The present study aimed to investigate the occurrence of fimA genotypes in patients with chronic periodontitis and healthy individuals in the Indian population, and to study their association with the number of P. gingivalis cells obtained in subgingival plaque samples of these subjects. The study comprised 95 samples from the chronic periodontitis (CP) group and 35 samples from the healthy (H) group, which were detected positive for P. gingivalis in our previous study. Fimbrial genotyping was done by PCR and PCR-restriction fragment length polymorphism (RFLP). The fimA type II was more prevalent in the CP group (55.89%), followed by type IV (30.52%), whereas in the H group, type I was the most prevalent fimbria (51.42%). The quantity of P. gingivalis cells increased with the presence of fimA types II and III. Our results suggest a strong relationship between fimA types II and IV and periodontitis, and between type I and the healthy condition. The colonization of organisms was increased with the occurrence of type II in deep periodontal sites, which could play an important role in the progression of the disease.
Keywords: fimbriae, polymerase chain reaction (PCR), periodontitis, Porphyromonas gingivalis, virulence
1. Introduction
Porphyromonas gingivalis is a Gram-negative, black-pigmented, anerobic bacterium responsible for causing chronic periodontitis by establishing itself in the deep periodontal pockets of the oral cavity [1]. P. gingivalis is regarded as a “keystone pathogen” in periodontitis and plays an essential role in its pathogenesis [2]. Apart from playing a significant role in periodontitis, P. gingivalis is also known to be a risk factor for cardiovascular disease, type 2 diabetes mellitus, and Alzheimer’s disease through immune modification mechanisms [3,4,5]. It is reported that P. gingivalis is not only abundantly present in chronic periodontitis patients, but is also known to occupy the oral cavity of healthy individuals. The pathogenicity of P. gingivalis is ascribed to diverse virulence factors, such as fimbriae, lipopolysaccharides, membrane proteins, capsules, proteases, endotoxins, cysteine proteases, etc. [6]. Fimbriae are arranged on the cell surface of the pathogen and play a vital role in initial attachment and biofilm organization. The bacteria adhere to gingival tissue leading to an invasion and colonization of host epithelial cells [7]. These structures are also known to induce cellular activation and cytokine release, thereby producing an inflammatory response at the infected site [8]. Based on the nucleotide variation, fimbriae are classified into six genotypes (type I, Ib, II, III, IV and V), and are encoded by the fimA gene [9]. Studies have been carried out to examine the presence of these genotypes in different geographical locations and ethnic groups to investigate the relationship between the fimA genotype and disease severity [9,10,11,12,13,14,15]. In Japanese adults, types II and IV have been found to be more prevalent in periodontitis patients, whereas type I has been associated with healthy individuals [9]. In Caucasians, types II and I are more prevalent in periodontitis patients, whereas in Brazilian periodontitis patients, types II and Ib were found to be more prevalent [11,15]. These findings suggest that variability in the frequency of fimbriae could be related to geographical location, ethnic group, and periodontal health status.
Until now, there have been no reports on the complete genotyping of the fimbriae of P. gingivalis in Indian subjects. The present study was aimed to investigate the prevalence of fimA genotypes in patients with chronic periodontitis and healthy individuals, in relation to clinical parameters, in the Indian population. We also aimed to study the association of fimA genotype with the number of P. gingivalis cells obtained in the subgingival plaque samples of these subjects.
2. Materials and Methods
The study comprised 95 samples from the chronic periodontitis (CP) group and 35 samples from the healthy (H) group, which were detected positive for P. gingivalis among 120 samples from each group in our previous study [16]. The subjects were separated into the CP and H groups as per the American Association of Periodontology guidelines [17]. Subjects with diabetes, HIV infection, pregnant women, and those on any medication or antibiotic therapy were excluded from the study. Subjects with at least 20 teeth with gingival inflammation, bleeding on probing, probing depth of ≥5 mm, and clinical attachment loss of ≥3 mm in at least four siteswere included in the CP group. Subjects with no signs of gingival inflammation, absence of bleeding on probing, probing depth of ≤3 mm, and no clinical attachment loss in all the sites were included in the H group.
2.1. Sample Collection
Subgingival plaque samples were collected and pooled using a dental curette as described in our previous study [18]. Briefly, six sites having the deepest probing depths were selected. Sites were isolated with sterile cotton rolls and air dried, and the supragingival plaque was removed. The subgingival sample was collected and pooled in a vial containing Tris EDTA buffer using a dental curette. Clinical parameters such as gingival index (GI) [19], plaque index (PI) [20], probing depth (PD), and clinical attachments loss (CAL) were averaged for each individual in both the healthy and CP groups.
2.2. Polymerase Chain Reaction (PCR)
DNA was extracted from subgingival plaque samples by the “modified proteinase K” method as described previously [21]. A real time polymerase chain reaction (RT-PCR) was carried out for amplification of the 16S rRNA species-specific gene of P. gingivalis in our previous study [16]. Samples found to be positive for P. gingivalis were further processed for the detection of fimA genotypes I to V by polymerase chain reaction (PCR), as described previously [22,23]. The primers specific to each of the fimA types used in the study were as mentioned in Table 1. A reaction mixture with a total volume of 25 µL was prepared by using Ampliqon red 2X mastermix (Ampliqon, Odense, Denmark), which contains Tris-HCL pH 8.5, (NH4)2SO4, 3 mM MgCl2, 0.2% Tween 20, 0.4 mM of each dNTP, 0.2 units/µL Ampliqon Taq DNA polymerase, and inert red dye and stabilizer. The primers were used at 0.5 µM concentration, and 3 µL of the DNA template at approximately 100ng concentration was added to the reaction mixture. The thermal cycling conditions were performed in a veriti 96-well thermal cycler (Applied Biosystems, California, CA, USA). An initial denaturation was done at 95 °C for 5 min, followed by 35 cycles of 95 °C, 58 °C and 72 °C for 30 s each. The final extension was carried out at 72 °C for 5 min.
Table 1.
PCR primers used in the study.
| fimA Type | Primer Sequence 5′-3′ |
Amplification Length (bp) | Reference |
|---|---|---|---|
| I | CTG TGT GTT TAT GGC AAA CTT C AAC CCC GCT CCC TGT ATT CCG A |
392 | [22] |
| Ib | CAG CAG AGC CAA AAA CAA TCG TGT CAG ATA ATT AGC GTC TGC |
271 | [10] |
| II | ACA ACT ATA CTT ATG ACA ATG G AAC CCC GCT CCC TGT ATT CCG A |
257 | [22] |
| III | ATT ACA CCT ACA CAG GTG AGG C AAC CCC GCT CCC TGT ATT CCG A |
247 | [22] |
| IV | CTA TTC AGG TGC TAT TAC CCA A AAC CCC GCT CCC TGT ATT CCG A |
251 | [22] |
| V | AAC AAC AGT CTC CTT GAC AGT G TAT TGG GGG TCG AAC GTT ACT GTC |
462 | [23] |
2.3. Restriction Fragment Length Polymorphism (RFLP)
The samples which were found positive for both type I and type II concurrently were further processed for restriction fragment length polymorphism. The primer pair Ib was used for PCR amplification, and the amplified products were digested with 10 u/µL of FastDigest RsaI enzyme. The reaction mixture was incubated at 37 °C for 5 min [10].
PCR amplified products and restriction digested products were subjected to 2% agarose gel electrophoresis using Tris-acetate EDTA buffer at 80 volts for 2 h. The gel was stained with 0.5 µg/mL of ethidium bromide and observed under a UV gel documentation system (Major Science, Saratoga, CA, USA). The specific fimA type was identified by comparing the band size of each sample with a 100 bp DNA ladder.
2.4. Statistical Analysis
Statistical analysis was carried out by using GraphPad Prism 5.1 (GraphPad Software, Inc., San Diego California, CA, USA). The frequency of fimA genotypes and their statistical association with chronic periodontitis was evaluated by using Fisher’s exact test. Association of fimA genotypes with clinical parameters and the quantity of P. gingivalis was analyzed by the unpaired t-test and the Mann–Whitney U test, respectively. p < 0.05 was considered as statistically significant.
3. Results
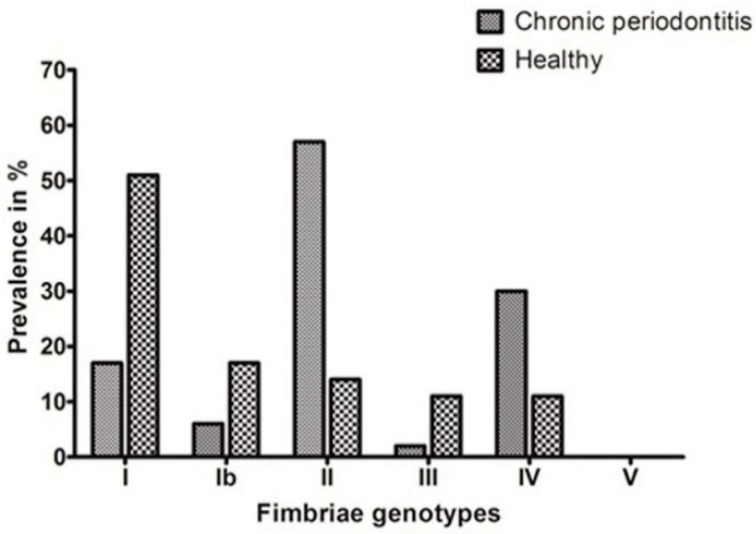
This is a continuation study, which we initiated on 120 samples each from the CP group and H group, for quantitative detection of P. gingivalis in subgingival plaque samples by real time PCR in Indian subjects [16]. In the present study, 95 samples positive for P. gingivalis in the CP group and 35 samples positive in the H group were subjected to fimbrial genotyping by the PCR method. The detection of specific amplified products of fimA genotypes is as shown in Figure 1. The frequency of distribution of these fimA genotypes in the CP and H groups is as shown in Table 2 and Figure 2. Among these genotypes, type II was the most prevalent type in the CP group, followed by type IV, with prevalence rates of 55.89% and 30.52%, respectively. The fimA types I, Ib, and III were at lesser frequencies of 17.89%, 6.31% and 2.1%, respectively. Type V was not detected in the CP group. There was also a co-existence of two fimA types concurrently, with type II and type IV being present in most of the cases. Type II and type IV were significantly associated with the CP group (OR 8.250 and 3.405, respectively) with a p-value < 0.0001 and 0.0393, respectively.
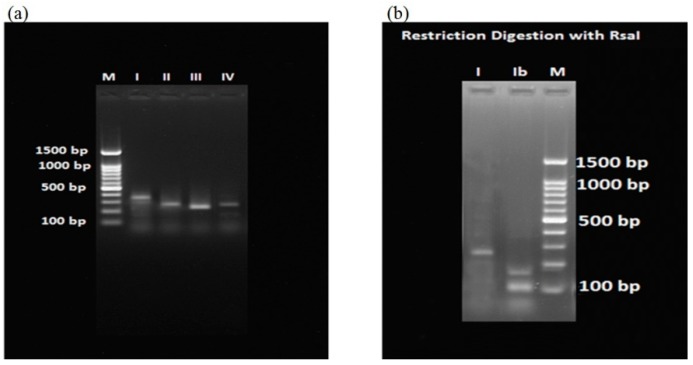
Figure 1.
Agarose gel images showing the amplification of fimA genotypes: (a) amplification of fimA type I (392 bp), type II (257 bp), type III (247 bp), and type IV (251 bp); (b) restriction digestion with the RsaI enzyme yielded type I (271 bp) and type Ib (169 bp, 109 bp).
Table 2.
Frequency distribution of fimA genotypes in subjects detected positive for P. gingivalis in the chronic periodontitis and healthy groups.
| Factor | CP Group | H Group | Odds Ratio | 95% Confidence Interval | p-Value | ||
|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||
| I | 17 | 17.89 | 18 | 51.42 | 0.20 | 0.0884 to 0.4794 | 0.0003 * |
| Ib | 6 | 6.31 | 6 | 17.14 | 0.3258 | 0.09746 to 1.089 | 0.08 |
| II | 55 | 57.89 | 5 | 14.28 | 8.250 | 2.943 to 23.13 | <0.0001 * |
| III | 2 | 2.1 | 4 | 11.4 | 0.1667 | 0.02908 to 0.9551 | 0.0446 * |
| IV | 29 | 30.52 | 4 | 11.4 | 3.405 | 1.101 to 10.54 | 0.0393 * |
| V | 0 | 0 | 0 | 0 | - | - | - |
| I, II | 5 | 5.26 | 1 | 2.8 | 1.889 | 0.2128 to 16.77 | 1.0000 |
| Ib, II | 3 | 3.1 | 0 | 0 | - | - | - |
| II, IV | 4 | 4.2 | 0 | 0 | - | - | - |
| I, IV | 2 | 2.1 | 1 | 2.8 | 0.7234 | 0.06350 to 8.241 | 1.0000 |
CP: chronic periodontitis, H: healthy individuals. * p < 0.05.
Figure 2.
Bar chart showing the frequency distribution of fimA genotypes in subjects detected positive for P. gingivalis in the chronic periodontitis and healthy groups.
In healthy individuals, type I was the most prevalent fimbria, found in 51.42% of cases. Type Ib and type II were found in 17.14% and 14.28% of P. gingivalis positive cases. Type III and type IV were found at a rate of 11.4%, and type V was not found in any of the samples. Co-existence of types I and II was found in 2.8% of cases, and types I and IV were found at the same rates. Type I was found to be significantly associated with the H group (p-value = 0.0003).
The association of these fimA genotypes with clinical parameters in CP is as shown in Table 3. When the comparison was made between the groups with the presence of different genotypes, the mean probing depth was found to be higher (5.76 ± 0.62) for fimA type II when compared to other types, except for type III (6.55 ± 1.06), which was detected in just two samples. The clinical attachment level was also found to be marginally increased (5.40 ± 1.04) with the presence of fimA type II, whereas the probing index (2.52 ± 0.19) and gingival index (2.57 ± 0.21) were found to be highest with the presence of fimA type IV.When the comparison was made within the groups for the presence and absence of different genotypes, probing depth was found to be significantly associated with the presence of type III fimA (p-value = 0.03), whereas the clinical attachment level was significantly associated with the presence of type II fimA (p-value = 0.04).
Table 3.
Association of fimA genotypes with clinical parameters in patients with chronic periodontitis.
| fimA Genotype | Present/Absent | PD | CAL | PI | GI | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | p-Value | Mean ± SD | p-Value | Mean ± SD | p-Value | Mean ± SD | p-Value | ||
| I | Present | 5.54 ± 1.14 | 0.18 | 5.22 ± 1.06 | 0.96 | 2.45 ± 0.21 | 0.2 | 2.46 ± 0.26 | 0.21 |
| Absent | 5.75 ± 0.66 | 5.21 ± 1.08 | 2.52 ± 0.19 | 2.54 ± 0.24 | |||||
| Ib | Present | 5.56 ± 0.27 | 0.51 | 4.98 ± 0.54 | 0.6 | 2.46 ± 0.22 | 0.53 | 2.51 ± 0.22 | 0.85 |
| Absent | 5.72 ± 0.58 | 5.22 ± 1.09 | 2.51 ± 0.19 | 2.53 ± 0.25 | |||||
| II | Present | 5.76 ± 0.62 | 0.32 | 5.40 ± 1.04 | 0.04 * | 2.50 ± 0.19 | 0.73 | 2.50 ± 0.27 | 0.18 |
| Absent | 5.64 ± 0.49 | 5.02 ± 1.03 | 2.52 ± 0.19 | 2.56 ± 0.21 | |||||
| III | Present | 6.55 ± 1.06 | 0.03 * | 6.1 ± 0.42 | 0.24 | 2.45 ± 0.07 | 0.64 | 2.55 ± 0.35 | 0.92 |
| Absent | 5.69 ± 0.55 | 5.19 ± 1.07 | 2.51 ± 0.20 | 2.53 ± 0.24 | |||||
| IV | Present | 5.56 ± 0.39 | 0.08 | 5.15 ± 1.07 | 0.75 | 2.52 ± 0.19 | 0.86 | 2.57 ± 0.21 | 0.36 |
| Absent | 5.78 ± 0.62 | 5.23 ± 1.07 | 2.51 ± 0.20 | 2.51 ± 0.26 | |||||
PD: probing depth, CAL: clinical attachment level, PI: plaque index, GI: gingival index, SD: standard deviation, unpaired t-test.* p < 0.05.
The association of fimA genotypes with the quantities of P. gingivalis is as shown in Table 4. The Mann–Whitney U test was performed to report the difference in the quantities of P. gingivalis obtained in the presence or absence of a specific genotype. The quantity of P. gingivalis was significantly higher in the fimA type II (2.09 × 108) positive samples than in the negative samples (9.52 × 107). The difference was statistically significant with a p-value < 0.001. We also found an increased mean cell count of P. gingivalis in the positive samples of fimA type III (2.77 × 109) compared to the type III negative samples (1.03 × 108). The difference was statistically significant with the p-value = 0.0385.
Table 4.
Association of fimA genotypes with the quantity of P. gingivalis in patients with chronic periodontitis.
| fimA Types | Positive/ Negative |
N | Mean | ± SEM | Median | Interquartile Range (IQR) | p-Value |
|---|---|---|---|---|---|---|---|
| I | Positive | 17 | 2.35 × 107 | 1.30 × 107 | 4.53 × 106 | 1.13 × 107 | 0.1185 |
| Negative | 103 | 1.68 × 108 | 7.45 × 107 | 1.58 × 106 | 9.74 × 106 | ||
| Ib | Positive | 6 | 6.27 × 106 | 1.61 × 106 | 6.56 × 106 | 8.12 × 106 | 0.1894 |
| Negative | 114 | 1.55 × 108 | 6.74 × 107 | 1.35 × 106 | 1.02 × 107 | ||
| II | Positive | 55 | 2.09 × 108 | 9.76 × 107 | 4.61 × 106 | 1.26 × 107 | <0.0001 * |
| Negative | 65 | 9.52 × 107 | 8.49 × 107 | 9.17 × 104 | 5.79 × 106 | ||
| III | Positive | 2 | 2.77 × 109 | 2.75 × 109 | 2.77 × 109 | 5.50 × 109 | 0.0385 * |
| Negative | 118 | 1.03 × 108 | 4.62 × 107 | 1.59 × 106 | 9.84 × 106 | ||
| IV | Positive | 29 | 1.49 × 108 | 1.39 × 108 | 2.08 × 106 | 6.98 × 106 | 0.273 |
| Negative | 91 | 1.47 × 108 | 7.25 × 107 | 1.52 × 106 | 1.04 × 107 |
N: number, SEM: standard error of the mean.* p < 0.05.
4. Discussion
In this study, we have analyzed the association of the presence of fimbrial genotypes with chronic periodontitis and periodontal health status in Indian subjects. In healthy individuals, type I was found to be more prevalent, followed by types Ib and II.The prevalence of fimA types II and IV was found to be higher in the CP group, followed by type I. The prevalence rate of fimA types II and IV in the CP group from our study was in accordance with another previous report carried out on Japanese and Chinese patients [9,13]. This was also in agreement with other studies carried out on Caucasian and Swedish periodontitis patients [15,24]. We also detected fimA type I in greater frequency in periodontitis cases, which is well in accordance with Beikler et al., rather than with other previous reports. Interestingly, Missailidis CG et al. carried out a study in Brazilian patients and found a higher prevalence of type Ib than type IV [11]. The difference in the detection frequencies could be due to the different geographical location, ethnic source, and technical variability.
A study conducted by Zhao et al. showed that genotypes II and IV were more frequently detected in 4–6 mm and >7 mm probing depth groups among patients with periodontitis [13]. Our report also showed that the mean probing depth and clinical attachment loss were marginally higher with the presence of type II fimbriae, whereas the probing index and gingival index were found to be higher with the presence of type IV fimbriae. This suggests the possible role of type II and type IV fimbriae in the initiation and progression of periodontitis in Indian subjects. It could be interesting to see the association of the fimbrial genotype with the different age groups, which we could not do in our study due to the small sample size. Previous reports have suggested that a certain type of relationship does exists between the fimA genotype and age [9,11,15,25].
The prevalence of the fimA genotype was also correlated with the quantity of P. gingivalis obtained by the real time PCR technique. When the comparison was made between genotypes, the cell count of P. gingivalis was greater with the presence of fimA type III, but this type was detected in just two samples. Meanwhile, within the groups, the level of P. gingivalis was found to be significantly higher in the presence of fimA types II and III (p-value < 0.001 and 0.0385, respectively). Although a greater number of samples in fimA type III cases would have provided a clearer idea on its role in the pathogenesis, nevertheless, our study underlines the significant role of fimA type II in the colonization of P. gingivalis in the oral cavity. Miura et al. found that a P. gingivalis isolate with type I fimA had a higher Arg- and Lysine-gingipain activity, suggesting dual-action attachment to gingival tissue by a fimbrial mechanismand destruction by proteinase enzyme activity [12]. This relationship can be further investigated by studying other fimbrial types with other possible virulent mechanisms.
There are two limitations of this study. Small numbers of samples were detected for fimA genotypes (N = 2 for fimA type III, and N = 6 for fimA type Ib). A higher number of samples in the original study itself probably would have provided more numbers of samples for fimA genotypic analysis, in order to draw a fair conclusion for all the genotypes simultaneously. Another limitation was that pooled samples were collected, and therefore the co-existence of different fimA genotypes in the same ecological niche could not be analyzed. In order to relate colonization to a particular ecological niche by a certain genotype, the individual site needs to be assessed.
With the dominance of a specific fimbrial type in the healthy and periodontitis groups, it is evident that variability in the presence of the fimbrial genotype could possibly provide variable pathogenic ability to different P. gingivalis clonal types across all geographic locations. Also, along with the fimbriae, there are other virulence factors, such as outer membrane proteins, capsules, proteases, endotoxin lipopolysaccharides, cytotoxic metabolites, etc. which may act concomitantly to provide differential pathogenic abilities to the P. gingivalis organism. Recently, studies on periodontitis have been focused on the inhibition of the virulence potential of P. gingivalis by different mechanisms to provide health benefits in these subjects [26,27].The variable pathogenic ability due to fimbrial occurrences could also lead us to organize primary and secondary prevention systems and suggests the use of fimbrial proteins, such as fimbrillins in immunoprophylaxis or immunotherapeutic strategies, to eliminate this pathogen from the subgingival microbiota.
5. Conclusions
Our results clearly suggest that there is strong relationship between fimA types II and IV and periodontitis, and that type I is associated with the healthy condition. The study also demonstrated that with the presence of fimA type II, there was increased colonization of P. gingivalis organisms in deep periodontal sites, which could play an important role in the progression of the disease.
Acknowledgments
The authors would like to thank Sanjivini Patil, technical assistant at the Central Research Laboratory, Maratha Mandal’s NGH Institute of Dental Sciences & Research Centre, Belagavi, Karnataka, India for her assistance in the microbiological culture.
Author Contributions
Conceptualization of the study was provided by U.M. and K.B.; design of experiments, methodology, and writing of the paper was accomplished by M.K.; review and editing of the paper was accomplished by V.J., M.K., M.M. and K.P.; statistical analysis was performed by M.R.P. and V.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Darveau R.P., Hajishengallis G., Curtis M.A. Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 2012;91:816–820. doi: 10.1177/0022034512453589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hajishengallis G., Darveau R.P., Curtis M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu C., Fratiglioni L. A major role for cardiovascular burden in age-related cognitive decline. Nat. Rev. Cardiol. 2015;12:267. doi: 10.1038/nrcardio.2014.223. [DOI] [PubMed] [Google Scholar]
- 4.Padmalatha G.V., Bavle R.M., Satyakiran G.V., Paremala K., Sudhakara M., Makarla S. Quantification of Porphyromonasgingivalis in chronic periodontitis patients associated with diabetes mellitus using real-time polymerase chain reaction. J. Oral. Maxillofac. Pathol. 2016;20:413. doi: 10.4103/0973-029X.190933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silvestre F.J., Lauritano D., Carinci F., Silvestre-Rangil J., Martinez-Herrera M., Del Olmo A. Neuroinflammation, Alzheimers disease and periodontal disease: Is there an association between the two processes. J. Biol. Regul. Homeost. Agents. 2017;31:189–196. [PubMed] [Google Scholar]
- 6.Holt S.C., Kesavalu L., Walker S., Genco C.A. Virulence factors of Porphyromonas gingivalis. Periodontology. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 7.Lin X., Wu J., Xie H. Porphyromonas gingivalis minor fimbriae are required for cell-cell interactions. Infect. Immun. 2006;74:6011–6015. doi: 10.1128/IAI.00797-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khlgatian M., Nassar H., Chou H.H., Gibson F.C., Genco C.A. Fimbria-dependent activation of cell adhesion molecule expression in Porphyromonas gingivalis-infected endothelial cells. Infect. Immun. 2002;70:257–267. doi: 10.1128/IAI.70.1.257-267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amano A., Kuboniwa A.M., Nakagawa I., Akiyama S., Morisaki I., Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J. Dent. Res. 2000;79:1664–1668. doi: 10.1177/00220345000790090501. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa. I., Amano A., Ohara-Nemoto Y., Endoh N., Morisaki I., Kimura S., Kawabata S., Hamada S. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J. Periodontal. Res. 2002;37:425–432. doi: 10.1034/j.1600-0765.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- 11.Missailidis C.G., Umeda J.E., Ota-Tsuzuki C., Anzai D., Mayer M.P. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral. Microbiol. Immunol. 2004;19:224–229. doi: 10.1111/j.1399-302X.2004.00140.x. [DOI] [PubMed] [Google Scholar]
- 12.Miura M., Hamachi T., Fujise O., Maeda K. The prevalence and pathogenic differences of Porphyromonas gingivalis fimA genotypes in patients with aggressive periodontitis. J. Periodontal. Res. 2005;40:147–152. doi: 10.1111/j.1600-0765.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhao L., Wu Y.F., Meng S., Yang H., Ou Yang Y.L., Zhou X.D. Prevalence of fimA genotypes of Porphyromonas gingivalis and periodontal health status in Chinese adults. J. Periodontal. Res. 2007;42:511–517. doi: 10.1111/j.1600-0765.2007.00975.x. [DOI] [PubMed] [Google Scholar]
- 14.Fabrizi S., Leon R., Blanc V., Herrera D., Sanz M. Variability of the fimA gene in Porphyromonas gingivalis isolated from periodontitis and non-periodontitis patients. Med. Oral. Patol. Oral. Cir. Bucal. 2013;18:e100–e105. doi: 10.4317/medoral.18042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beikler T., Peters U., Prajaneh S., Prior K., Ehmke B., Flemmig T.F. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur. J. Oral. Sci. 2003;111:390–394. doi: 10.1034/j.1600-0722.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- 16.Kugaji M.S., Muddapur U.M., Bhat K.G., Joshi V.M., Kumbar V.M., Peram M.R. Quantitative evaluation of Porphyromonas gingivalis in Indian subjects with chronic periodontitis by Real-Time Polymerase Chain Reaction. J. Adv. Oral. Res. 2019;10:137–144. doi: 10.1177/2320206819863952. [DOI] [Google Scholar]
- 17.Armitage G.C. Development of a classification system for periodontal diseases and conditions. Ann. Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Joshi V.M., Bhat K.G., Kugaji M.S., Shirahatti R. Characterization and serotype distribution of Aggregatibacteractinomycetemcomitans: Relationship of serotypes to herpesvirus and periodontal status in Indian subjects. Microb. Pathog. 2017;110:189–195. doi: 10.1016/j.micpath.2017.06.041. [DOI] [PubMed] [Google Scholar]
- 19.Silness J., Loe H. Periodontal disease in pregnancy. II. Correlation between oral hygiene and periodontal condition. Acta. Odontol. Scand. 1964;22:121–135. doi: 10.3109/00016356408993968. [DOI] [PubMed] [Google Scholar]
- 20.Loe H. The Gingival Index, the plaque index and the retention index systems. J. Periodontol. 1967;38:610–616. doi: 10.1902/jop.1967.38.6_part2.610. [DOI] [PubMed] [Google Scholar]
- 21.Kugaji M.S., Bhat K.G., Joshi V.M., Pujar M., Mavani P.T. Simplified method of detection of Dialisterinvisus and Olsenellauli in oral cavity samples by Polymerase Chain Reaction. J. Adv. Oral. Res. 2017;8:47–52. doi: 10.1177/2229411217729105. [DOI] [Google Scholar]
- 22.Amano A., Nakagawa I., Kataoka K., Morisaki I., Hamada S. Distribution of Porphyromonas gingivalis strains with fimA genotypes in Periodontitis Patients. J. Clin. Microbiol. 1999;37:1426–1430. doi: 10.1128/JCM.37.5.1426-1430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa I., Amano A., Kimura R.K., Nakamura T., Kawabata S., Hamada S. Distribution and Molecular Characterization of Porphyromonas gingivalis carrying a new type of fimA gene. J. Clin. Microbiol. 2000;38:1909–1914. doi: 10.1128/JCM.38.5.1909-1914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshino T., Laine M.L., Van Winkelhoff A.J., Dahlén G. Genotype variation and capsular serotypes of Porphyromonas gingivalis from chronic periodontitis and periodontal abscesses. FEMS Microbiol. Lett. 2007;270:75–81. doi: 10.1111/j.1574-6968.2007.00651.x. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K., Nakano K., Nomura R., Miyake S., Nakagawa I., Amano A., Ooshima T. Distribution of Porphyromonas gingivalis fimA genotypes in Japanese children and adolescents. J. Periodontol. 2005;76:674–679. doi: 10.1902/jop.2005.76.5.674. [DOI] [PubMed] [Google Scholar]
- 26.Grenier D., Morin M.P., Fournier-Larente J., Chen H. Vitamin D inhibits the growth of and virulence factor gene expression by Porphyromonas gingivalis and blocks activation of the nuclear factor kappa B transcription factor in monocytes. J. Periodontal. Res. 2016;51:359–365. doi: 10.1111/jre.12315. [DOI] [PubMed] [Google Scholar]
- 27.Kugaji M.S., Kumbar V.M., Peram M.R., Patil S., Bhat K.G., Diwan P.V. Effect of Resveratrol on biofilm formation and virulence factor gene expression of Porphyromonas gingivalis in periodontal disease. APMIS. 2019;127:187–195. doi: 10.1111/apm.12930. [DOI] [PubMed] [Google Scholar]