Abstract
TonB-dependent transport system plays a critical role in the transport of nutrients across the energy-deprived outer membrane of Gram-negative bacteria. It contains a specialized outer membrane TonB-dependent transporter (TBDT) and energy generating (ExbB/ExbD) and transducing (TonB) inner membrane multi-protein complex, called TonB complex. Very few TonB complex protein-coding sequences exist in the genomes of Gram-negative bacteria. Interestingly, the TBDT coding alleles are phenomenally high, especially in the genomes of bacteria surviving in complex and stressful environments. Sphingomonads are known to survive in highly polluted environments using rare, recalcitrant, and toxic substances as their sole source of carbon. Naturally, they also contain a huge number of TBDTs in the outer membrane. Out of them, only a few align with the well-characterized TBDTs. The functions of the remaining TBDTs are not known. Predictions made based on genome context and expression pattern suggest their involvement in the transport of xenobiotic compounds across the outer membrane.
Keywords: TonB-dependent transporter (TBDT), sphingomonads, xenobiotics
1. Introduction
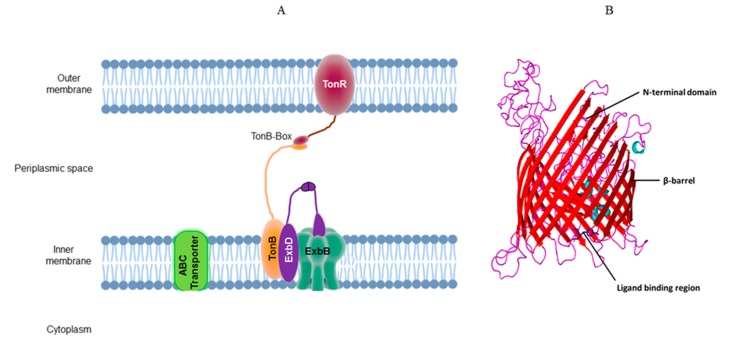
The outer membrane of Gram-negative bacteria performs several important functions. It acts as a barrier to prevent the entry of antibiotics and other toxic chemicals and protects the cell wall by denying access to cell wall degrading enzymes. However, existence of an energy-deprived outer membrane is a hurdle for the uptake of nutrients in Gram-negative bacteria [1]. A majority of nutrients gains entry into periplasmic space by diffusing through the outer membrane via a pore-like structure formed in outer membrane-associated β-barrel containing proteins, otherwise known as porins [2]. However, certain scarcely available nutrients depend on active transport to cross the energy-deprived outer membrane. The active transport mechanism of the outer membrane is known as TonB-dependent transport system. The system contains two components, the inner membrane-associated TonB complex and an outer membrane-associated TonB-dependent transporter (TBDT). The TonB complex contains proton motif force (PMF) components, ExbB/ExbD, and energy transducer TonB in a ratio of 7:2:1 [3]. The TonB complex has unique role in outer membrane transport. The PMF components ExbB/ExbD generate energy by pumping protons across the inner membrane, while TonB transduces this energy to the outer membrane-localized TBDT. The TonB protein contains three domains: the N-terminal transmembrane helix, C-terminal domain, and a proline-rich rigid central domain. The N-terminal region is embedded in the inner membrane and is associated with one of the transmembrane domains of ExbB [4]. The longer C-terminal region extends into periplasmic space and specifically interacts with TonB-box of the outer membrane-localized TBDT. These interactions of TonB and TBDT facilitate the transduction of energy required to transport scarcely available nutrients across the outer membrane. The TBDT possesses unique structural features and exhibits a two-domain structure. The C-terminal domain embedded in the outer membrane contains one of the largest 22-stranded β-barrel with extracellular loops. The N-terminal globular domain, through its unique structural features, establishes cross-talk with the inner membrane-localized TonB complex. An energy coupling consensus pentapeptide motif (ETVIV) designated as “TonB-box” physically interacts with the C-terminal domain of inner membrane-localized, periplasmically exposed TonB [5]. The TBDT undergoes conformational changes upon substrate binding. This conformational change induces structural transition from a state of order to disorder in the TonB-box motif [6]. This disordered state of TonB-box of TBDT is recognized by TonB. The TBDT and TonB interactions are transient, the disordered state of TonB-box returns to an ordered state after completion of substrate transport [7]. TonB plays a critical role in supplying energy required for the structural transition of TBDT. TonB harvests energy generated by PMF components ExbB/ExbD and transduces it to TBDT (Figure 1).
Figure 1.
(A) Schematic diagram of TonB-dependent transport system. (B) Typical structural features of an outer membrane transporter (TBDT), 22 β-barrel structure, N-terminal plug domain, and substrate binding motif are indicated with arrows.
The TonB complex of TBDT is highly conserved among Gram-negative bacteria. The genome sequences of Gram-negative bacteria contain a limited number of alleles to code for TonB complex proteins, TonB and ExbB/ExbD. Though the overall structural features of TBDT are conserved, there exist substantial differences in the residues of ligand binding sites. Such diversity in the residues of ligand binding sites suggests the existence of specialized TBDTs for transport of a variety of scarcely available nutrients in the environment. The copy number of TBDTs shows a very unique pattern among Gram-negative bacteria. Genomes isolated from the cells grown in less stressful environments show existence of a smaller number of TBDT coding alleles. Their numbers in such strains does not exceed four to five alleles per genome. However, the number of alleles coding TBDTs ie more in the genomes of cells isolated from harsh environments [8]. The gut microbiome sequences show an unusually high number of TBDT coding sequences [9]. Similarly, in the genomes of sphingomonads, namely Sphingobium japonicum, Sphingobium indicum, Sphingobium fuliginis, which live in harsh climates, a very high number of TBDT coding alleles are identified. In fact, the number of TBDTs appears to proportionately increase with the complexity of the environment [9]. However, there exists no study to link the increased number of TBDTs and complexity of the habitat. Since sphingomonads survive in habitats polluted with a variety of xenobiotics, in this study we attempt to examine if these TBDTs have a role in the transport of xenobiotic compounds by examining the genome information of certain Sphingomonadaceae members.
2. The TonB-Dependent Transport System
TonB-dependent transport system derived its name due to the fact that phage T1 failed to infect the null mutants of tonA and tonB in Esterichia coli [10]. Subsequent studies performed to understand this unusual observation have identified the physiological role of these two genes. The roles of these two genes are now well established. The tonA codes for TBDT designated as FhuA and is involved in the transport of ferrichrome [10]. Initial studies gave an impression that TonB-dependent transport system, consisting of FhuA, serves exclusively for iron-siderophore complex uptake. However, recent studies have dismantled this myth and showed the involvement of FhuA in the transport of antibiotics. In addition to ferrichrome, FhuA successfully transported siderophore structural mimic Albomycin and Rifamycin CGP 4832, which have no structural similarity with siderophores [11]. Similarly, the outer membrane transporter, BtuB, of E. coli was found to be a member of the TBDT family and interacts with the inner membrane-associated TonB complex to facilitate the active transport of vitamin B12 across the outer membrane [12]. Likewise, a wide variety of TBDTs was identified in many pathogenic and non-pathogenic bacteria to translocate a variety of substrates across the outer membrane via TonB-dependent transport system [13]. Transport of nickel complexes in human pathogen Helicobacter pylori, maltodextrins in the environmental bacteria Caulobacter crescentus, and sucrose in plant pathogen Xanthomonas campestris, pv. campestris are certain classical examples that show involvement of the TonB-dependent transport system in the transport of substrates other than iron. Table 1 gives an exhaustive list of substrates predicted to be transported through TonB-dependent transport system.
Table 1.
Physiological role of TonB-dependent receptors (TBDTs).
| Substrates | TBDTs | Genomes | Evidence | References |
|---|---|---|---|---|
| Iron-siderophore complex | FauA | Bordetella pertussis | Ex | [41] |
| FecA | Escherichia coli | Ex | [42] | |
| FepA | Escherichia coli | Ex | [43] | |
| FhuA | Escherichia coli | Ex | [44] | |
| FptA | Pseudomonas aeruginosa | Ex | [45] | |
| FpvA | Pseudomonas aeruginosa | Ex | [46] | |
| FrpB | Neisseria meningitidis | Ex | [47] | |
| FyuA | Yersenia pestis | Ex | [48] | |
| HemR | Yersenia enterocolitica | Ex | [49] | |
| TbpA | Neisseria meningitidis | Ex | [47] | |
| Cobalamins | BtuB | Escherichia coli | Ex | [50] |
| BPSL0976 | Burkholderia pseudomallei | Pr | [51] | |
| PA1271 | Pseudomonas aeruginosa | Pr | [51] | |
| PA2911 | Pseudomonas aeruginosa | Pr | [51] | |
| RS02718 | Ralstonia solanacearum | Pr | [51] | |
| RSP_2402 | Rhodbacter sphaeroides | Pr | [51] | |
| Sucrose | SuxA | Xanthomonas campestris | Ex | [8] |
| Sfri_3988 | Shewanella frigidimarina | Pr | [8] | |
| Maltose | MalA | Caulobacter vibrioides | Ex | [52] |
| Chitin | XCC2469 | Xanthomonas campestris | Pr | [53] |
| XCC2944 | Xanthomonas campestris | Pr | [53] | |
| CPS_1021 | Colwellia psychrerythraea | Pr | [53] | |
| Sden_2708 | Shewanella denitrificans | Pr | [53] | |
| Xylan | XCC4120 | Xanthomonas campestris | Pr | [8] |
| Copper | NosA | Pseudomonas stutzeri | Pr | [54,55] |
| OprC | Pseudomonas aeruginosa | Pr | [54,55] | |
| NosA | Pseudomonas putida | Ex | [54,55] | |
| OprC | Pseudomonas putida | Ex | [54,55] | |
| Nickel | FrpB4 | Helicobacter pylori | Ex | [13] |
| Daro_3944 | Dechloromonas aromatica | Pr | [56] | |
| BLL6948 | Bradyrhizobium diazoefficiens | Pr | [56] | |
| IL54_0463 | Sphingobium sp. ba1 | Tr | [16] | |
| IL54_3057 | Sphingobium sp. ba1 | Tr | [16] | |
| Thiamin | BF0615 | Bacteroides fragilis | Pr | [57] |
| CPS_0067 | Colwellia psychrerythraea | Pr | [57] | |
| Sputw3181_2365 | Shewanella | Pr | [57] | |
| GOX1347 | Gluconobacter oxydans | Pr | [57] | |
| Cobalt | Daro_1684 | Dechloromonas aromatica | Pr | [56] |
| Pectin | XCC0120 | Xanthomonas campestris | Pr | [8] |
| Colicin | CirA | Escherichia coli | Ex | [58] |
| Heme | HasR | Serratia marcescens | Ex | [49] |
| ShuA | Shigella dysenteriae | Ex | [59] | |
| Sulfanilic acid | BES08_08830 | Novosphingobium resinovorans SA1 | Tr | [36] |
| BES08_18055 | Novosphingobium resinovorans SA1 | Tr | [36] | |
| BES08_18430 | Novosphingobium resinovorans SA1 | Tr | [36] | |
| BES08_18580 | Novosphingobium resinovorans SA1 | Tr | [36] | |
| BES08_22625 | Novosphingobium resinovorans SA1 | Tr | [36] | |
| BES08_23675 | Novosphingobium resinovorans SA1 | Tr | [36] | |
| BES08_26825 | Novosphingobium resinovorans SA1 | Tr | [36] | |
| Various substrates under nutrient limitation | Sala_0027 | Sphingopyxis alaskensis RB2256 | Tr | [14] |
| Sala_0914 | Sphingopyxis alaskensis RB2256 | Tr | [14] | |
| Sala_1228 | Sphingopyxis alaskensis RB2256 | Tr | [14] | |
| Sala_1913 | Sphingopyxis alaskensis RB2256 | Tr | [38] | |
| Sala_3029 | Sphingopyxis alaskensis RB2256 | Tr | [14] | |
| Sala_3108 | Sphingopyxis alaskensis RB2256 | Tr | [14] | |
| Saro_0168 | Novosphingobium aromaticivorans DSM 12444 | Tr | [14] | |
| Saro_1603 | Novosphingobium aromaticivorans DSM 12444 | Tr | [14] | |
| SKA58_00625 | Sphingomonas sp. SKA58 | Tr | [14] | |
| SKA58_14617 | Sphingomonas sp. SKA58 | Tr | [14] | |
| SKA58_18137 | Sphingomonas sp. SKA58 | Tr | [14] | |
| Dibenzo-p-dioxin | Swit_0277 | Sphingomonas wittichii RW1 | Tr | [33] |
| Swit_1066 | Sphingomonas wittichii RW1 | Tr | [33] | |
| Swit_3263 | Sphingomonas wittichii RW1 | Tr | [33] | |
| Swit_4088 | Sphingomonas wittichii RW1 | Tr | [33] | |
| Swit_4197 | Sphingomonas wittichii RW1 | Tr | [33] | |
| Dibenzofuran | Swit_0687 | Sphingomonas wittichii RW1 | Tr | [33] |
| Swit_2477 | Sphingomonas wittichii RW1 | Tr | [33] | |
| Swit_3189 | Sphingomonas wittichii RW1 | Tr | [33] | |
| Swit_3918 | Sphingomonas wittichii RW1 | Tr | [33] | |
| Swit_4025 | Sphingomonas wittichii RW1 | Tr | [33] | |
| Swit_4696 | Sphingomonas wittichii RW1 | Tr | [33] | |
| Swit_4781 | Sphingomonas wittichii RW1 | Tr | [33] |
Ex: experimentally validated, Pr: predicted, Tr: transcriptome and proteome analysis.
3. TonB-Dependent Transporters (TBDTs) and Environmental Adaptation
Sphingomonads survive in various stressful environments. They survive in a nutrient-poor phyllosphere [14], extremely cold marine waters [15] and highly toxic and polluted environments with metals [16], phenanthrene [17,18,19,20], polyethylene glycol [21], alkylphenols [22], dioxins [23], naphthalene [20], diphenyl ethers [24], organophosphates [25,26] and organochlorides [27,28,29]. Their survival under these stressful conditions depends on their ability to use these unusual substrates, hitherto unknown to natural habitats, as a carbon source (Table 2). Such a task can be accomplished with an efficient catabolic repertoire, an effective transport system. The genome sequences of these strains indeed reveal the existence of a novel catabolic repertoire [26]. Interestingly their genomes also contain an unusually high number of TBDTs.
Table 2.
Distribution of TBDTs in sphingomonads.
| Name of the Strain | Seq ID | Genome Size (kb) | Phenotype | No. of TBDTs | Refs |
|---|---|---|---|---|---|
| Sphingomonas wittichii RW1 | NC_009511.1 | 5.38 | Dibenzo-p-dioxin | 153 | [23] |
| Sphingobium sp. YBL2 | NZ_CP010954.1 | 4.77 | Phenylurea | 83 | [60] |
| Sphingopyxis sp. MG | NZ_CP026381.1 | 4.15 | Organo-phosphate | 76 | [26] |
| Sphingobium fuliginis ATCC 27551 |
NZ_CP041016.1, NZ_CP041017.1 |
5.05 | Organo-phosphate | 102 | [25] |
| Sphingobium indicum B90A | NZ_CP013070.1 | 3.65 | Hexachloro-cyclohexane | 45 | [61] |
| Sphingobium japonicum UT26S | NC_014006.1, NC_014013.1 | 4.19 | Hexachloro-cyclohexane | 66 | [62] |
| Novosphingobium sp. PP1Y | NC_015580.1 | 3.9 | Fuel oils | 48 | [63] |
| Novosphingobium aromaticivorans DSM 12444 | NC_007794.1 | 3.5 | Phenanthrene | 55 | [64] |
| Sphingobium sp. SYK-6 | NC_015976.1 | 4.2 | Lignin | 75 | [65] |
| Sphingobium chlorophenolicum L-1 | NC_015593.1, NC_015594.1 | 4.45 | Pentachlorophenol | 99 | [66] |
| Sphingobium sp. 22B | GCA_001580035.1 | 5.36 | Polycyclic aromatic hydrocarbons (PAH) | 85 | [67] |
| Sphingobium sp. AM | GCA_001550165.1 | 5.31 | PAH | 86 | [68] |
| Sphingobium sp. ba1 | GCA_000743655.1 | 4.45 | Growth in high nickel ion concentration | 75 | [16] |
| Novosphingobium resinovorans SA1 | NZ_CP017075.1 | 6.91 | Sulfanilic acid | 148 | [36] |
| Sphingopyxis alaskensis RB2256 | NC_008048.1 | 3.35 | Cold marine water | 39 | [38] |
| Sphingomonas wittichii DC-6 | NZ_CP021181.1 | 5.92 | Dibenzo-p-dioxin | 141 | [69] |
| Acinetobacter baumannii AYE | CU459141.1 | 3.96 | Multi-drug resistance | 06 | [70] |
| Acinetobacter baumannii DS002 | CP027704.1 | 3.43 | Organo-phosphate | 05 | [32] |
| Pseudomonas putida strain JBC17 | CP029693.1 | 6.84 | Dichloro-benzene | 14 | [71] |
4. Unique TonB Complex in Sphingobium fuliginis
Organophosphate (OP)-degrading sphingomonads contain phosphotriesterases (PTE), also known as organophosphate hydrolase (OPH), capable of degrading the third ester linkage found in OP insecticides and nerve agents [30]. The membrane-associated PTE target the membrane in a pre-folded conformation following twin-arginine transport (Tat) pathway. The Tat pathway inserts PTE into the inner membrane-facing periplasmic space of the cell. Recent studies have shown PTE as part of a multiprotein membrane-associated TonB complex. Interestingly, the TonB complex components were co-purified along with PTE. PTE are shown to interact physically with TonB complex components ExbB/ExbD and TonB, showing the existence of a unique four-component TonB complex in S. fuliginis [31]. Co-purification of TonB complex components along with PTE and the inability of pte null mutants of Sphingopyxis wildii to grow in a medium with OP insecticide methyl parathion as a source of phosphate suggest the involvement of a novel TonB-dependent transport system in transport of OP insecticides.
The sphingomonads survive using a variety of organic compounds as a source of carbon and energy [14]. They also contain a rather unusually high number of putative TBDTs when compared to other Gram-negative bacteria surviving in relatively stress-free habitats [32]. Some of these TBDTs are induced when they are grown in the presence of these xenobiotic compounds. In Sphingomonas alaskensis, a threefold increase is noticed in the expression of TBDTs to facilitate transport of nutrients in increased viscous water [15]. Sphingomonads like Sphongobium sp. BA1, Sphingobium cupriresistence, and Novosphingobium PPIY can also withstand the stress imposed by metal ions like Ni2+, Cu2+, and Pb2+, respectively, due to increased expression of TBDTs [16]. Differential expression of TBDTs was evident in Sphingomonas wittichii RW1 strains grown in the presence of dibenzofuran (DF) and dibenzo-p-dioxin (DD). The substrate-specific induction pattern of TBDTs suggests the existence of substrate-specific TonB-dependent transport systems in sphingomonads [33,34]. Supporting this proposition, the induction of TBDTs involved in transport of alginate was only observed when Sphingomonas sps. A1 cells were grown on alginate. These TBDTs directly incorporated alginate molecules into the cytoplasm without degradation [35]. There are 148 TBDTs in Novosphingobium resinovorans SA1 (Table 2). One of them showed a seven-fold increased expression when the cells were grown in sulfanilic acid. Such an increase in TBDT amounts is believed to facilitate active transport of polar sulfanilic acid across the outer membrane [36]. TonB-dependent transport system appears to be advantageous to bacteria in more than one way. Since they transport large molecules across the membrane, it helps bacteria to utilize complex substrates as a source of carbon. Further, the existence of substrate-specific transporters facilitates adaptation of sphingomonads by scavenging nutrients that occur at a very low concentration.
5. TBDTs of S. fuliginis
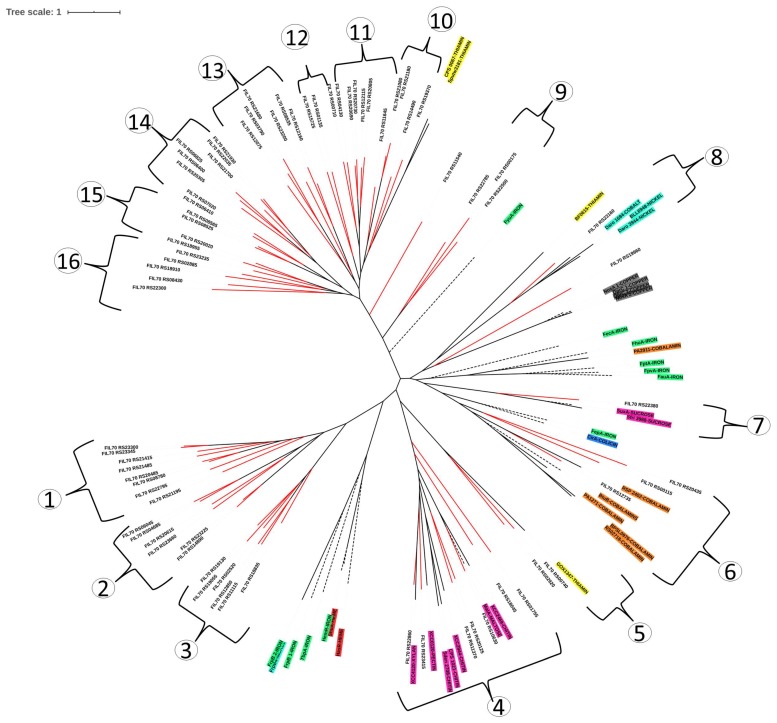
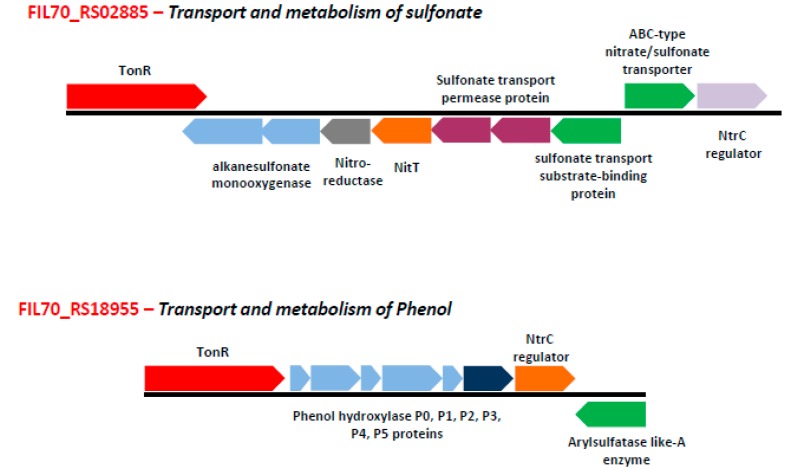
The fully annotated genome sequence of S. fuliginis has shown the existence of more than 100 putative TBDT (SfTBDT) coding sequences. Out of these 100, only 75 SfTBDTs have shown the existence of a 22-stranded β-barrel and N-terminal plug domain, the typical characteristic feature of a TBDT. A phylogenetic tree was constructed by including these uncharacterized SfTBDTs along with TBDTs whose function is either experimentally characterized or predicted based on genome context (Figure 2, Table 1). The phylogenetic tree thus constructed gave 16 TBDT clusters. Of these, only five clusters (cluster numbers 4, 5, 6, 7, and 8) contained TBDTs with known functions. Interestingly, out of 75 putative SfTBDTs only 14 clustered with these five known groups of TBDT sequences. The genes coding transporters often coexist with the genes associated with the metabolism of their cognate substrates. They also contain identical promoters and other regulatory elements to ensure common expression and repression pattern in response to a physiological condition. Therefore, such genomic context of a transporter is taken as an indicator of its function [37]. As expected, the genome context of these 14 TBDTs that clustered with TBDTs of known function supports the results obtained through phylogenetic tree (Figure 3).
Figure 2.
Phylogenetic tree constructed for SfTBDTs. The SfTBDTs clustered with functionally characterized TBDT sequences are present in clusters 4, 5, 6, 7, and 8. Dashed lines of clades indicate experimentally characterized TBDTs. Red lines of the clade indicate uncharacterized TBDTs from Sphingobium fuliginis. TBDTs involved in transport of iron are highlighted with dark green; thiamin in yellow; nickel and cobalt in turquoise; cobalamin in orange; copper in grey; colicin in blue; heme in red; and all carbohydrates in dark pink.
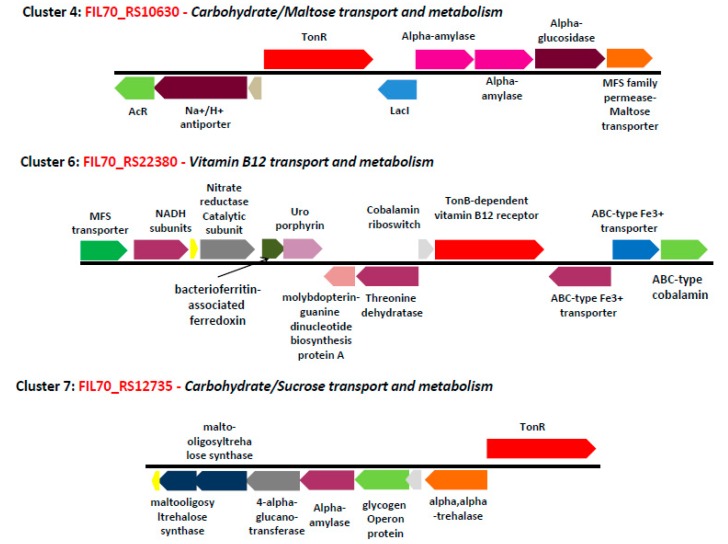
Figure 3.
Genome-context analysis of Sphingobium fuliginis TBDTs found in clusters 4, 6, and 7.
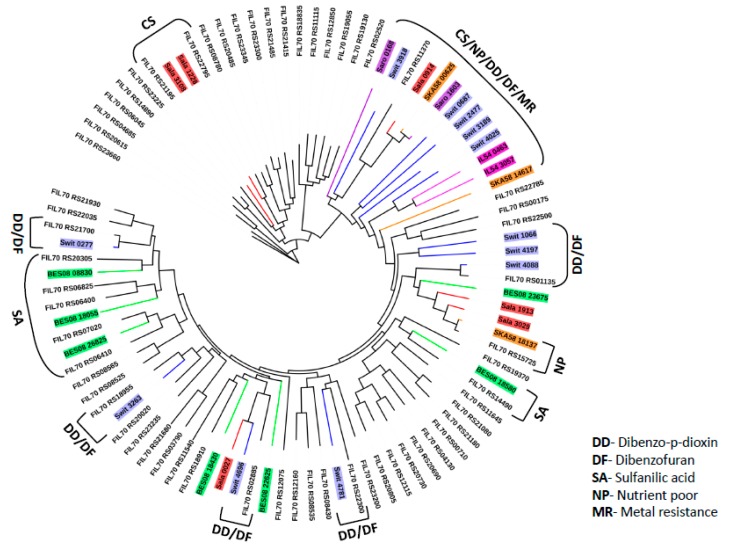
However, the rest of the 61 putative SfTBDTs found in 11 clusters (cluster numbers 1, 2, 3, 9, 10, 11, 12, 13, 14, 15, and 16) of the phylogenetic tree showed no similarity with TBDTs of known function. Transcriptome and proteome analysis was done for certain sphingomonads grown under certain unique physiological conditions. These omics studies showed substrate specific expression patterns of TBDTs [14,16,33,34,36,38,39,40]. About 32 TBDTs showed differential expression in the presence of heavy metals, xenobiotics, temperature stress, and poor nutrient conditions (Table 1). The phylogenetic tree, constructed by including the 61 uncharacterized SfTBDTs and the differentially expressed TBDTs of sphingomonads, gave interesting insights into the functions of SfTBDTs. The SfTBDT (FIL70_RS22795, FIL70_RS21195, and FIL70_RS 11370) clustered with TBDTs of S. alaskensis (Sala_1228, Sala_3108, and Sala_0914) involved in the transport of nutrients facilitating its survival under cold stress conditions (Figure 4). Similarly, the SfTBDTs (FIL70_RS22300, FIL70_RS01135, FIL70_RS18955, FIL70_RS02885, and FIL70_RS21700) clustered with TBDTs of S. wittichii RW1 (Swit_4781, Swit_4088, Swit_3263, Swit_4696, and Swit_0277) was predicted to be involved in the transport of dibenzo-p-dioxin (DD). These TBDTs of S. wittichii were induced in the presence of DD or dibenzofuran (DF). Clustering of SfTBDTs with the DD/DF responsive TBDT of S. wittichii indicates their role in outer membrane transport of these xenobiotics. Similarly, some SfTBDTs (FIL70_RS20305, FIL70_RS06825, FIL70_RS06400, FIL70_RS07020, FIL70_RS06410, and FIL70_RS14490) clustered with sulfanilic acid-responsive TBDTs of Novosphingobium resinovorans SA1 (BES08_08830, BES08_18055, BES08_26825, and BES08_18580). Therefore, these SfTBDTs are implicated in the transport of sulfonated aromatic amines. Gene context analysis was also carried out for two of the TBDTs (FIL_RS02885 and FIL_RS18955) to examine if these two methods provide identical insights on the functions of TBDTs (Figure 5). These two independent strategies followed indicated substrates only for 29 SfTBDTs. Gene context analysis was performed to provide insight on the functional status of the remaining 46 SfTBDTs (Table 3). Although experimental evidence is missing to assign a physiological role to a majority of TBDTs, existence of an unusually high number of TBDTs suggests the presence of robust TonB-dependent transport systems in sphingomonads. Such unique transport mechanisms, together with a novel catabolic repertoire, appear to contribute to the survival of sphingomonads in harsh environments.
Figure 4.
Expression pattern-based phylogenetic tree. Xenobiotic responsive TBDT sequences of Sphingomonas wittichii RW1 (blue), Sphingobium sp. ba1 (dark pink), Sphingopyxis alaskensis RB2256 (red), Sphingomonas sp. SKA58 (orange), and Novosphingobium aromaticivorans DSM 12444 (purple) are included along with 61 uncharacterized SfTBDTs (black) while constructing the phylogenetic tree.
Figure 5.
Gene-context analysis of two SfTBDT sequences that clustered with xenobiotic responsive TBDTs Swit_4696 and Swit_3263 in an expression pattern-based phylogenetic tree.
Table 3.
Predicted functions of SfTBDTs using gene context analysis.
| TBDTs of S. fuliginis | Function Based on Gene Context Analysis |
|---|---|
| FIL70_RS23660 | Amino acid transport and metabolism |
| FIL70_RS20615 | Carbohydrate transport and metabolism |
| FIL70_RS04685 | Amino acid transport and metabolism and inorganic ion transport and metabolism (iron) |
| FIL70_RS06045 | Inorganic ion transport and metabolism (iron) |
| FIL70_RS14890 | Nucleotide transport and metabolism |
| FIL70_RS23225 | Lipid transport and metabolism |
| FIL70_RS08780 | Carbohydrate transport and metabolism (xylan) |
| FIL70_RS20485 | Lipid/carbohydrate transport and metabolism |
| FIL70_RS23345 | Carbohydrate/xylulose/xylan transport and metabolism |
| FIL70_RS21485 | Carbohydrate transport and metabolism |
| FIL70_RS21415 | Amino acid/carbohydrate transport and metabolism |
| FIL70_RS19055 | Inorganic ion transport and metabolism (sulfonate) |
| FIL70_RS18835 | Amino acid transport and metabolism/coenzyme transport and metabolism |
| FIL70_RS11115 | Carbohydrate transport and metabolism |
| FIL70_RS12850 | Amino acid transport and metabolism |
| FIL70_RS19130 | Inorganic ion transport and metabolism (iron)/secondary metabolites biosynthesis, transport, and catabolism (sulfonate) |
| FIL70_RS02520 | Nucleotide transport and metabolism |
| FIL70_RS22785 | Inorganic ion transport and metabolism (nickel) |
| FIL70_RS00175 | Inorganic ion transport and metabolism |
| FIL70_RS22500 | Cobalamin transport and metabolism and inorganic ion transport and metabolism |
| FIL70_RS23300 | Lipid transport and metabolism, secondary metabolites biosynthesis, transport and catabolism, inorganic ion transport and metabolism (zinc) |
| FIL70_RS08535 | Lipid transport and metabolism |
| FIL70_RS12160 | Carbohydrate transport and metabolism |
| FIL70_RS11645 | Lipid transport and metabolism |
| FIL70_RS21180 | Lipid transport and metabolism |
| FIL70_RS21080 | Lipid transport and metabolism |
| FIL70_RS04130 | Benzoate transport |
| FIL70_RS00710 | Inorganic ion transport and metabolism and flavin transport and metabolism |
| FIL70_RS20730 | Coenzyme transport and metabolism (flavin transport and metabolism) |
| FIL70_RS20690 | Coenzyme transport and metabolism (flavin transport and metabolism) |
| FIL70_RS20805 | Flavin/secondary metabolite transport and metabolism |
| FIL70_RS12115 | Coenzyme transport and metabolism/secondary metabolite transport and metabolism |
| FIL70_RS23235 | Inorganic ion transport and metabolism |
| FIL70_RS21680 | Lipid transport and metabolism |
| FIL70_RS03790 | Lipid transport and metabolism |
| FIL70_RS12075 | Lipid transport and metabolism |
| FIL70_RS11540 | Lipid transport and metabolism |
| FIL70_RS08565 | Carbohydrate transport and metabolism |
| FIL70_RS08525 | Carbohydrate transport and metabolism |
| FIL70_RS21930 | Lipid transport and metabolism |
| FIL70_RS22035 | Lipid transport and metabolism |
| FIL70_RS23200 | Carbohydrate transport and metabolism |
| FIL70_RS20020 | Inorganic ion transport and metabolism/secondary metabolites biosynthesis, transport, and catabolism |
| FIL70_RS08430 | Coenzyme transport and metabolism/lipid transport and metabolism |
| FIL70_RS18910 | Inorganic ion transport and metabolism |
| FIL70_RS19370 | Lipid transport and metabolism |
Funding
This research was funded by Council of Scientific and Industrial Research (CSIR), New Delhi, India, grant number 37(1684)17/EMR-II, and Department of Science and Technology (DST), New Delhi, India, grant number EMR/2016/001644. APC was funded by lab fundings.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Zgurskaya H.I., Lopez C.A., Gnanakaran S. Permeability Barrier of Gram-Negative Cell Envelopes and Approaches to Bypass It. ACS Infect. Dis. 2015;1:512–522. doi: 10.1021/acsinfecdis.5b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lepore B.W., Indic M., Pham H., Hearn E.M., Patel D.R., van den Berg B. Ligand-gated diffusion across the bacterial outer membrane. Proc. Natl. Acad. Sci. USA. 2011;108:10121–10126. doi: 10.1073/pnas.1018532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgs P.I., Larsen R.A., Postle K. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol. Microbiol. 2002;44:271–281. doi: 10.1046/j.1365-2958.2002.02880.x. [DOI] [PubMed] [Google Scholar]
- 4.Larsen R.A., Thomas M.G., Wood G.E., Postle K. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (delta V17) by a missense mutation in ExbB. Mol. Microbiol. 1994;13:627–640. doi: 10.1111/j.1365-2958.1994.tb00457.x. [DOI] [PubMed] [Google Scholar]
- 5.Tuckman M., Osburne M.S. In Vivo inhibition of TonB-dependent processes by a TonB box consensus pentapeptide. J. Bacteriol. 1992;174:320–323. doi: 10.1128/JB.174.1.320-323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiener M.C. TonB-dependent outer membrane transport: Going for Baroque? Curr. Opin. Struct. Biol. 2005;15:394–400. doi: 10.1016/j.sbi.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Kadner R.J. Vitamin B12 transport in Escherichia coli: Energy coupling between membranes. Mol. Microbiol. 1990;4:2027–2033. doi: 10.1111/j.1365-2958.1990.tb00562.x. [DOI] [PubMed] [Google Scholar]
- 8.Blanvillain S., Meyer D., Boulanger A., Lautier M., Guynet C., Denance N., Vasse J., Lauber E., Arlat M. Plant carbohydrate scavenging through tonB-dependent receptors: A feature shared by phytopathogenic and aquatic bacteria. PLoS ONE. 2007;2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schauer K., Rodionov D.A., de Reuse H. New substrates for TonB-dependent transport: Do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 2008;33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Killmann H., Videnov G., Jung G., Schwarz H., Braun V. Identification of receptor binding sites by competitive peptide mapping: Phages T1, T5, and phi 80 and colicin M bind to the gating loop of FhuA. J. Bacteriol. 1995;177:694–698. doi: 10.1128/JB.177.3.694-698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun V., Braun M. Iron transport and signaling in Escherichia coli. FEBS Lett. 2002;529:78–85. doi: 10.1016/S0014-5793(02)03185-X. [DOI] [PubMed] [Google Scholar]
- 12.Chimento D.P., Mohanty A.K., Kadner R.J., Wiener M.C. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 2003;10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 13.Schauer K., Gouget B., Carriere M., Labigne A., de Reuse H. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 2007;63:1054–1068. doi: 10.1111/j.1365-2958.2006.05578.x. [DOI] [PubMed] [Google Scholar]
- 14.Delmotte N., Knief C., Chaffron S., Innerebner G., Roschitzki B., Schlapbach R., von Mering C., Vorholt J.A. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. USA. 2009;106:16428–16433. doi: 10.1073/pnas.0905240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ting L., Williams T.J., Cowley M.J., Lauro F.M., Guilhaus M., Raftery M.J., Cavicchioli R. Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environ. Microbiol. 2010;12:2658–2676. doi: 10.1111/j.1462-2920.2010.02235.x. [DOI] [PubMed] [Google Scholar]
- 16.Volpicella M., Leoni C., Manzari C., Chiara M., Picardi E., Piancone E., Italiano F., D’Erchia A., Trotta M., Horner D.S., et al. Transcriptomic analysis of nickel exposure in Sphingobium sp. ba1 cells using RNA-seq. Sci. Rep. 2017;7:8262. doi: 10.1038/s41598-017-08934-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabate J., Vinas M., Bayona J.M., Solanas A.M. Isolation and taxonomic and catabolic characterization of a 3,6-dimethylphenanthrene-utilizing strain of Sphingomonas sp. Can. J. Microbiol. 2003;49:120–129. doi: 10.1139/w03-018. [DOI] [PubMed] [Google Scholar]
- 18.van Herwijnen R., Wattiau P., Bastiaens L., Daal L., Jonker L., Springael D., Govers H.A., Parsons J.R. Elucidation of the metabolic pathway of fluorene and cometabolic pathways of phenanthrene, fluoranthene, anthracene and dibenzothiophene by Sphingomonas sp. LB126. Res. Microbiol. 2003;154:199–206. doi: 10.1016/S0923-2508(03)00039-1. [DOI] [PubMed] [Google Scholar]
- 19.Story S.P., Parker S.H., Hayasaka S.S., Riley M.B., Kline E.L. Convergent and divergent points in catabolic pathways involved in utilization of fluoranthene, naphthalene, anthracene, and phenanthrene by Sphingomonas paucimobilis var. EPA505. J. Ind. Microbiol. Biotechnol. 2001;26:369–382. doi: 10.1038/sj.jim.7000149. [DOI] [PubMed] [Google Scholar]
- 20.Balkwill D.L., Drake G.R., Reeves R.H., Fredrickson J.K., White D.C., Ringelberg D.B., Chandler D.P., Romine M.F., Kennedy D.W., Spadoni C.M. Taxonomic study of aromatic-degrading bacteria from deep-terrestrial-subsurface sediments and description of Sphingomonas aromaticivorans sp. nov., Sphingomonas subterranea sp. nov., and Sphingomonas stygia sp. nov. Int. J. Syst. Bacteriol. 1997;47:191–201. doi: 10.1099/00207713-47-1-191. [DOI] [PubMed] [Google Scholar]
- 21.Charoenpanich J., Tani A., Moriwaki N., Kimbara K., Kawai F. Dual regulation of a polyethylene glycol degradative operon by AraC-type and GalR-type regulators in Sphingopyxis macrogoltabida strain 103. Microbiology. 2006;152:3025–3034. doi: 10.1099/mic.0.29127-0. [DOI] [PubMed] [Google Scholar]
- 22.Kolvenbach B.A., Corvini P.F. The degradation of alkylphenols by Sphingomonas sp. strain TTNP3—A review on seven years of research. New Biotechnol. 2012;30:88–95. doi: 10.1016/j.nbt.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Hong H.B., Chang Y.S., Nam I.H., Fortnagel P., Schmidt S. Biotransformation of 2,7-dichloro- and 1,2,3,4-tetrachlorodibenzo-p-dioxin by Sphingomonas wittichii RW1. Appl. Environ. Microbiol. 2002;68:2584–2588. doi: 10.1128/AEM.68.5.2584-2588.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt S., Wittich R.M., Erdmann D., Wilkes H., Francke W., Fortnagel P. Biodegradation of diphenyl ether and its monohalogenated derivatives by Sphingomonas sp. strain SS3. Appl. Environ. Microbiol. 1992;58:2744–2750. doi: 10.1128/AEM.58.9.2744-2750.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azam S., Parthasarathy S., Singh C., Kumar S., Siddavattam D. Genome Organization and Adaptive Potential of Archetypal Organophosphate Degrading Sphingobium fuliginis ATCC 27551. Genome Biol. Evol. 2019;11:2557–2562. doi: 10.1093/gbe/evz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parthasarathy S., Azam S., Lakshman Sagar A., Narasimha Rao V., Gudla R., Parapatla H., Yakkala H., Ghanta Vemuri S., Siddavattam D. Genome-Guided Insights Reveal Organophosphate-Degrading Brevundimonas diminuta as Sphingopyxis wildii and Define Its Versatile Metabolic Capabilities and Environmental Adaptations. Genome Biol. Evol. 2017;9:77–81. doi: 10.1093/gbe/evw275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verma H., Kumar R., Oldach P., Sangwan N., Khurana J.P., Gilbert J.A., Lal R. Comparative genomic analysis of nine Sphingobium strains: Insights into their evolution and hexachlorocyclohexane (HCH) degradation pathways. BMC Genom. 2014;15:1014. doi: 10.1186/1471-2164-15-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Godoy F.A., Bunster M., Matus V., Aranda C., Gonzalez B., Martinez M.A. Poly-beta-hydroxyalkanoates consumption during degradation of 2,4,6-trichlorophenol by Sphingopyxis chilensis S37. Lett. Appl. Microbiol. 2003;36:315–320. doi: 10.1046/j.1472-765X.2003.01315.x. [DOI] [PubMed] [Google Scholar]
- 29.Cai M., Xun L. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 2002;184:4672–4680. doi: 10.1128/JB.184.17.4672-4680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh B.K., Walker A. Microbial degradation of organophosphorus compounds. FEMS Microbiol. Rev. 2006;30:428–471. doi: 10.1111/j.1574-6976.2006.00018.x. [DOI] [PubMed] [Google Scholar]
- 31.Gudla R., Konduru G.V., Nagarajaram H.A., Siddavattam D. Organophosphate hydrolase interacts with Ton components and is targeted to the membrane only in the presence of the ExbB/ExbD complex. FEBS Lett. 2019;593:581–593. doi: 10.1002/1873-3468.13345. [DOI] [PubMed] [Google Scholar]
- 32.Yakkala H., Samantarrai D., Gribskov M., Siddavattam D. Comparative genome analysis reveals niche-specific genome expansion in Acinetobacter baumannii strains. PLoS ONE. 2019;14:e0218204. doi: 10.1371/journal.pone.0218204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chai B., Tsoi T.V., Iwai S., Liu C., Fish J.A., Gu C., Johnson T.A., Zylstra G., Teppen B.J., Li H., et al. Sphingomonas wittichii Strain RW1 Genome-Wide Gene Expression Shifts in Response to Dioxins and Clay. PLoS ONE. 2016;11:e0157008. doi: 10.1371/journal.pone.0157008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno-Forero S.K., van der Meer J.R. Genome-wide analysis of Sphingomonas wittichii RW1 behaviour during inoculation and growth in contaminated sand. ISME J. 2015;9:150–165. doi: 10.1038/ismej.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashimoto W., Kawai S., Murata K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng. Bugs. 2010;1:97–109. doi: 10.4161/bbug.1.2.10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hegedus B., Kos P.B., Bende G., Bounedjoum N., Maroti G., Laczi K., Szuhaj M., Perei K., Rakhely G. Starvation- and xenobiotic-related transcriptomic responses of the sulfanilic acid-degrading bacterium, Novosphingobium resinovorum SA1. Appl. Microbiol. Biotechnol. 2018;102:305–318. doi: 10.1007/s00253-017-8553-5. [DOI] [PubMed] [Google Scholar]
- 37.Heddle J., Scott D.J., Unzai S., Park S.Y., Tame J.R. Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli. J. Biol. Chem. 2003;278:50322–50329. doi: 10.1074/jbc.M307941200. [DOI] [PubMed] [Google Scholar]
- 38.Colquhoun D.R., Hartmann E.M., Halden R.U. Proteomic profiling of the dioxin-degrading bacterium Sphingomonas wittichii RW1. J. Biomed. Biotechnol. 2012;2012 doi: 10.1155/2012/408690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vancanneyt M., Schut F., Snauwaert C., Goris J., Swings J., Gottschal J.C. Sphingomonas alaskensis sp. nov., a dominant bacterium from a marine oligotrophic environment. Int. J. Syst. Evol. Microbiol. 2001;51:73–79. doi: 10.1099/00207713-51-1-73. [DOI] [PubMed] [Google Scholar]
- 40.Schirmer T., Evans P.R. Structural basis of the allosteric behaviour of phosphofructokinase. Nature. 1990;343:140–145. doi: 10.1038/343140a0. [DOI] [PubMed] [Google Scholar]
- 41.Brillet K., Meksem A., Lauber E., Reimmann C., Cobessi D. Use of an in-house approach to study the three-dimensional structures of various outer membrane proteins: Structure of the alcaligin outer membrane transporter FauA from Bordetella pertussis. Acta Crystallogr. D Biol. Crystallogr. 2009;65:326–331. doi: 10.1107/S0907444909002200. [DOI] [PubMed] [Google Scholar]
- 42.Yue W.W., Grizot S., Buchanan S.K. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J. Mol. Biol. 2003;332:353–368. doi: 10.1016/S0022-2836(03)00855-6. [DOI] [PubMed] [Google Scholar]
- 43.Buchanan S.K., Smith B.S., Venkatramani L., Xia D., Esser L., Palnitkar M., Chakraborty R., van der Helm D., Deisenhofer J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 44.Ferguson A.D., Braun V., Fiedler H.P., Coulton J.W., Diederichs K., Welte W. Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci. 2000;9:956–963. doi: 10.1110/ps.9.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cobessi D., Celia H., Pattus F. Crystal structure at high resolution of ferric-pyochelin and its membrane receptor FptA from Pseudomonas aeruginosa. J. Mol. Biol. 2005;352:893–904. doi: 10.1016/j.jmb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Cobessi D., Celia H., Folschweiller N., Schalk I.J., Abdallah M.A., Pattus F. The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 angstroms resolution. J. Mol. Biol. 2005;347:121–134. doi: 10.1016/j.jmb.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 47.Perkins-Balding D., Ratliff-Griffin M., Stojiljkovic I. Iron transport systems in Neisseria meningitidis. Microbiol. Mol. Biol. Rev. 2004;68:154–171. doi: 10.1128/MMBR.68.1.154-171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noinaj N., Guillier M., Barnard T.J., Buchanan S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cescau S., Cwerman H., Letoffe S., Delepelaire P., Wandersman C., Biville F. Heme acquisition by hemophores. Biometals. 2007;20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- 50.Shultis D.D., Purdy M.D., Banchs C.N., Wiener M.C. Outer membrane active transport: Structure of the BtuB: TonB complex. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 51.Rodionov D.A., Vitreschak A.G., Mironov A.A., Gelfand M.S. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- 52.Neugebauer H., Herrmann C., Kammer W., Schwarz G., Nordheim A., Braun V. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J. Bacteriol. 2005;187:8300–8311. doi: 10.1128/JB.187.24.8300-8311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang C., Rodionov D.A., Li X., Laikova O.N., Gelfand M.S., Zagnitko O.P., Romine M.F., Obraztsova A.Y., Nealson K.H., Osterman A.L. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J. Biol. Chem. 2006;281:29872–29885. doi: 10.1074/jbc.M605052200. [DOI] [PubMed] [Google Scholar]
- 54.Yoneyama H., Nakae T. Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Pt 8Microbiology. 1996;142:2137–2144. doi: 10.1099/13500872-142-8-2137. [DOI] [PubMed] [Google Scholar]
- 55.Lee H.S., Abdelal A.H., Clark M.A., Ingraham J.L. Molecular characterization of nosA, a Pseudomonas stutzeri gene encoding an outer membrane protein required to make copper-containing N2O reductase. J. Bacteriol. 1991;173:5406–5413. doi: 10.1128/JB.173.17.5406-5413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodionov D.A., Hebbeln P., Gelfand M.S., Eitinger T. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: Evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 2006;188:317–327. doi: 10.1128/JB.188.1.317-327.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodionov D.A., Vitreschak A.G., Mironov A.A., Gelfand M.S. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 58.Buchanan S.K., Lukacik P., Grizot S., Ghirlando R., Ali M.M., Barnard T.J., Jakes K.S., Kienker P.K., Esser L. Structure of colicin I receptor bound to the R-domain of colicin Ia: Implications for protein import. EMBO J. 2007;26:2594–2604. doi: 10.1038/sj.emboj.7601693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cobessi D., Meksem A., Brillet K. Structure of the heme/hemoglobin outer membrane receptor ShuA from Shigella dysenteriae: Heme binding by an induced fit mechanism. Proteins. 2010;78:286–294. doi: 10.1002/prot.22539. [DOI] [PubMed] [Google Scholar]
- 60.Gu T., Zhou C., Sorensen S.R., Zhang J., He J., Yu P., Yan X., Li S. The novel bacterial N-demethylase PdmAB is responsible for the initial step of N, N-dimethyl-substituted phenylurea herbicide degradation. Appl. Environ. Microbiol. 2013;79:7846–7856. doi: 10.1128/AEM.02478-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anand S., Sangwan N., Lata P., Kaur J., Dua A., Singh A.K., Verma M., Kaur J., Khurana J.P., Khurana P., et al. Genome sequence of Sphingobium indicum B90A, a hexachlorocyclohexane-degrading bacterium. J. Bacteriol. 2012;194:4471–4472. doi: 10.1128/JB.00901-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagata Y., Ohtsubo Y., Endo R., Ichikawa N., Ankai A., Oguchi A., Fukui S., Fujita N., Tsuda M. Complete genome sequence of the representative gamma-hexachlorocyclohexane-degrading bacterium Sphingobium japonicum UT26. J. Bacteriol. 2010;192:5852–5853. doi: 10.1128/JB.00961-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D’Argenio V., Petrillo M., Cantiello P., Naso B., Cozzuto L., Notomista E., Paolella G., Di Donato A., Salvatore F. De novo sequencing and assembly of the whole genome of Novosphingobium sp. strain PP1Y. J. Bacteriol. 2011;193:4296. doi: 10.1128/JB.05349-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bell S.G., Yang W., Yorke J.A., Zhou W., Wang H., Harmer J., Copley R., Zhang A., Zhou R., Bartlam M., et al. Structure and function of CYP108D1 from Novosphingobium aromaticivorans DSM12444: An aromatic hydrocarbon-binding P450 enzyme. Acta Crystallogr. D Biol. Crystallogr. 2012;68:277–291. doi: 10.1107/S090744491200145X. [DOI] [PubMed] [Google Scholar]
- 65.Barry K.P., Taylor E.A. Characterizing the promiscuity of LigAB, a lignin catabolite degrading extradiol dioxygenase from Sphingomonas paucimobilis SYK-6. Biochemistry. 2013;52:6724–6736. doi: 10.1021/bi400665t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Copley S.D., Rokicki J., Turner P., Daligault H., Nolan M., Land M. The whole genome sequence of Sphingobium chlorophenolicum L-1: Insights into the evolution of the pentachlorophenol degradation pathway. Genome Biol. Evol. 2012;4:184–198. doi: 10.1093/gbe/evr137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Madueno L., Macchi M., Morelli I.S., Coppotelli B.M. Draft Whole-Genome Sequence of Sphingobium sp. 22B, a Polycyclic Aromatic Hydrocarbon-Degrading Bacterium from Semiarid Patagonia, Argentina. Genome Announc. 2016;4 doi: 10.1128/genomeA.00488-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Festa S., Coppotelli B.M., Madueno L., Loviso C.L., Macchi M., Neme Tauil R.M., Valacco M.P., Morelli I.S. Assigning ecological roles to the populations belonging to a phenanthrene-degrading bacterial consortium using omic approaches. PLoS ONE. 2017;12:e0184505. doi: 10.1371/journal.pone.0184505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng M., Yan X., He J., Qiu J., Chen Q. Comparative genome analysis reveals the evolution of chloroacetanilide herbicide mineralization in Sphingomonas wittichii DC-6. Arch Microbiol. 2019;201:907–918. doi: 10.1007/s00203-019-01660-w. [DOI] [PubMed] [Google Scholar]
- 70.Vallenet D., Nordmann P., Barbe V., Poirel L., Mangenot S., Bataille E., Dossat C., Gas S., Kreimeyer A., Lenoble P., et al. Comparative analysis of Acinetobacters: Three genomes for three lifestyles. PLoS ONE. 2008;3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu S.M., Lee Y.H. Genes involved in nutrient competition by Pseudomonas putida JBC17 to suppress green mold in postharvest satsuma mandarin. J. Basic Microbiol. 2015;55:898–906. doi: 10.1002/jobm.201400792. [DOI] [PubMed] [Google Scholar]