Abstract
The sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae), is a pest of many economically important agricultural crops and a vector of plant viruses. Bemisia tabaci harbors facultative endosymbiont species that have been implicated in pest status, including tolerance to insecticides, virus transmission efficiency and tolerance to high-temperatures. The facultative endosymbionts reported in B. tabaci include Arsenophonus, Hamiltonella, Wolbachia, Cardinium, Fritschea and Rickettsia. We collected whitefly populations from weed and crop hosts in south Florida and identified the whitefly species as well as the facultative endosymbionts present in these populations by molecular analysis. In addition, a phylogenetic analysis of whiteflies and their endosymbionts was performed. The only facultative endosymbionts found among the B. tabaci populations collected in Florida were Hamiltonella and Rickettsia. The phylogenetic analysis revealed the low genetic diversity of whiteflies and their endosymbionts. Additionally, the phylogenetic tree clustered Rickettsia from Florida in the R1 genetic group. The results will aid to understand the role of the bacterial endosymbionts in the whitefly host.
Keywords: MEAM1, Rickettsia, Hamiltonella, facultative
1. Introduction
The whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae), is a pest of many agricultural crops and a successful vector of over 300 plant virus species including Begomovirus, Crinivirus, Carlavirus, Torradovirus and Ipomovirus [1,2]. Currently, B. tabaci is considered a species complex based on the mitochondrial cytochrome oxidase I (mtCOI) gene [3]. The different genetic groups are indistinguishable morphologically but several other characteristics differ among them [4,5] such as plant host preference, fecundity, ability to transmit viruses [6,7], dispersal and resistance to insecticides [8]. In the United States, three species of the B. tabaci complex have been reported: the Middle East-Asia Minor 1 (MEAM1), also known as B biotype; the Mediterranean (MED), also named as Q biotype; and the indigenous species New World 1 (NW1) or A biotype [9]. Bemisia tabaci MEAM1 is the primary pest of tomato (Solanum lycopersicum L.) in Florida and recent surveys indicate that MED species has been detected on landscape plants in residential areas [10]. Florida is among the main producers of fresh market tomatoes in the United States, with over 27,000 acres harvested in 2018 at a value of $344 million [11]. In addition to tomatoes, B. tabaci is a major pest of other important vegetable crops in Florida such as peppers, squash, cucumber, beans, eggplant, watermelon and cabbage. It can also heavily attack potato, peanut, soybean and cotton and ornamentals including poinsettia, hibiscus and chrysanthemum [12]. The B. tabaci species complex carries an obligatory primary endosymbiont called Candidatus Portiera aleyrodidarum [13]. In addition, whiteflies may harbor facultative endosymbiont species that have been implicated in pest status, including tolerance to insecticides [14], virus transmission efficiency [15,16] and tolerance to high-temperatures [17]. The facultative endosymbionts reported in B. tabaci include Hamiltonella and Arsenophonus [13], Wolbachia [18], Cardinium [19], Fritschea [20] and Rickettsia [21]. Previous studies have revealed a complex genetic divergence of the different endosymbionts within B. tabaci [9]. The infection dynamics of facultative endosymbionts in B. tabaci are usually associated with whitefly species, gender, host plants and geographical locations [22]. However, there is a lack of information about the facultative endosymbiont diversity in whitefly populations of Florida. In this study, we collected whiteflies from weed and crop hosts in south Florida and identified the whitefly species as well as the facultative endosymbionts by molecular analysis. Additionally, a phylogenetic analysis of the resulted sequenced data was performed. The goal of this study was to report the presence and to infer the diversity among the whitefly facultative endosymbionts in populations of Florida. Knowledge regarding the facultative endosymbionts can contribute to understanding epidemics of whitefly-transmitted viruses and be helpful in resistance monitoring studies. The results obtained will supplement further studies to determine relations between facultative endosymbionts and pest status.
2. Material and Methods
2.1. Field Collection
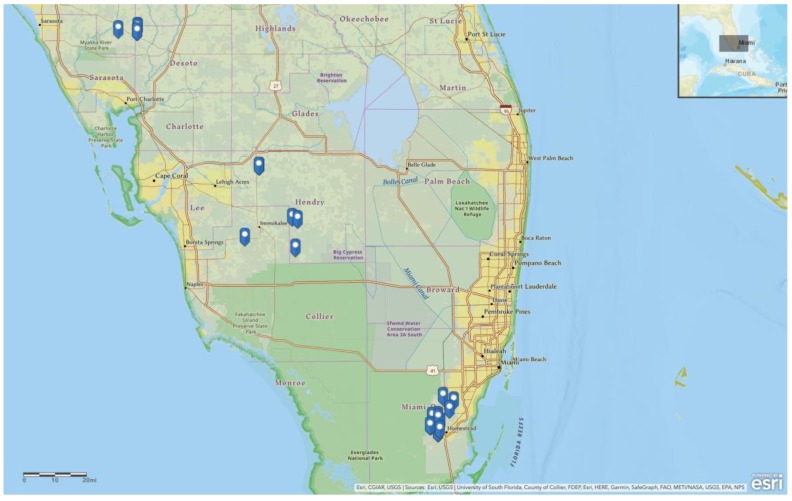
Populations of B. tabaci were collected from five south Florida counties in 2019: Hendry, Collier, Manatee, DeSoto and Miami-Dade (Figure 1). Whitefly adults from different host plants, including tomato, eggplant, watermelon, sweetpotato, hibiscus, Parthenium sp., Euphorbia sp., Amaranthus sp. and Bidens sp., were aspirated into vials using a hand-held aspirator and then transferred to cloth covered cages with cotton (Gossypium hirsutum L) seedlings. The cages were brought back to the University of Florida’s Gulf Coast Research and Education Center in Balm, FL, and maintained on cotton plants in growth rooms at 27 °C (±2 °C), 50% to 75% RH and 14:10 (L:D) photoperiod. Ten whiteflies per population were aspirated and transferred to vials containing 95% EtOH and stored at −20 °C for molecular analysis.
Figure 1.
Blue icons represent the collection sites of the 27 Bemisia tabaci field populations surveyed in the main tomato growing areas of Florida during 2019. The map was generated by the ArcGIS mapping software. Map sources: Esri, CGIAR, USGS, University of South Florida, County of Collier, FDEP, HERE, Garmin, SafeGraph, FAO, METI/NASA, EPA, NPS.
Two laboratory colonies, one of B. tabaci MEAM1, the other of B. tabaci MED, were analyzed in addition to the field populations collected in 2019. The MEAM1 colony was established in the 1990s from whiteflies collected near Bradenton, FL. The MED colony was established in July 2017 from whiteflies collected from hibiscus (Hibiscus rosa-sinensis L) in Palm Beach County, FL. Each colony was maintained in a separate growth room under the same conditions as the field populations.
2.2. Whitefly Identification
Total nucleic acids were extracted from each individual whitefly, following a modified Chelex protocol [23]. Ten insects for each population were individually tested. Bemisia tabaci adults were homogenized in 40 μL of 5% Chelex solution in a 1.5 mL tube. The tube was vortexed for a few seconds and then incubated at 56 °C for 15 min and at 99 °C for 8 min. After centrifugation at 13,000 rpm for 5 min, the supernatant was collected and used as a template for the PCR amplification. The PCR to differentiative MEAM1 from MED was carried out using the primer pair Bem23F (5’-CGGAGCTTGCGCCTTAGTC-3’) and Bem23R (5’-CGGCTTTATCATAGCTCTCGT-3’), which amplifies a microsatellite locus of about 200 bp and 400 bp for MEAM1 and MED [24,25,26], respectively. The PCR product was visualized by electrophoresis in 2% agarose gel stained with Gel Red. Later, at least one sample for each population was used for a PCR with the generic insect primers C1-J-2195 and TL2-N-3014 that amplify a fragment of the mtCOI [26] followed by PCR purification and Sanger sequencing of the fragment (GENEWIZ, South Plainfield, NJ, USA). The nucleotide sequences were analyzed and compared with those present in the GenBank database using BLAST tools (http://blast.ncbi.nlm.nih.gov/Blast).
2.3. Facultative Endosymbionts Identification
The identification of facultative endosymbionts was performed using the same DNA that was used previously for the whitefly species identification. The PCR was carried out with previously described genus-specific primers targeting the 16S or the 23S genes for the identification of Hamiltonella [18], Rickettsia [21], Wolbachia [27], Cardinium [19], Arsenophonus [28] and Fritschea [20]. Later, amplicons of some representative populations were selected for DNA sequencing (GENEWIZ, South Plainfield, NJ, USA). The nucleotide sequences were analyzed and compared with those present in the GenBank database using BLAST tools (http://blast.ncbi.nlm.nih.gov/Blast).
2.4. Phylogenetic Analysis
The Bayesian analyses were conducted using Mr. Bayes v. 3.2.278. They were run for 30 million generations with sampling every 1000 generations. Each analysis consisted of four independent runs, each utilizing four coupled Markov chains. The run convergence was monitored by finding the plateau in the likelihood scores (standard deviation of split frequencies <0.0015). It was discarded in the first 25% of each run. The resulted trees were edited and rooted using FigTree v1.4.2.
The whitefly phylogenetic analysis was performed using the 15 B. tabaci mtCOI sequences obtained in this study added to a global B. tabaci mtCOI dataset [29] totalizing 774 sequences. In addition, phylogenetic analyses of the Partial 16S rDNA gene were carried out for the facultative endosymbionts Hamiltonella and Rickettsia. Multiple sequence alignments were prepared using MAFFT77 within the Geneious 9.1.5 software.
3. Results
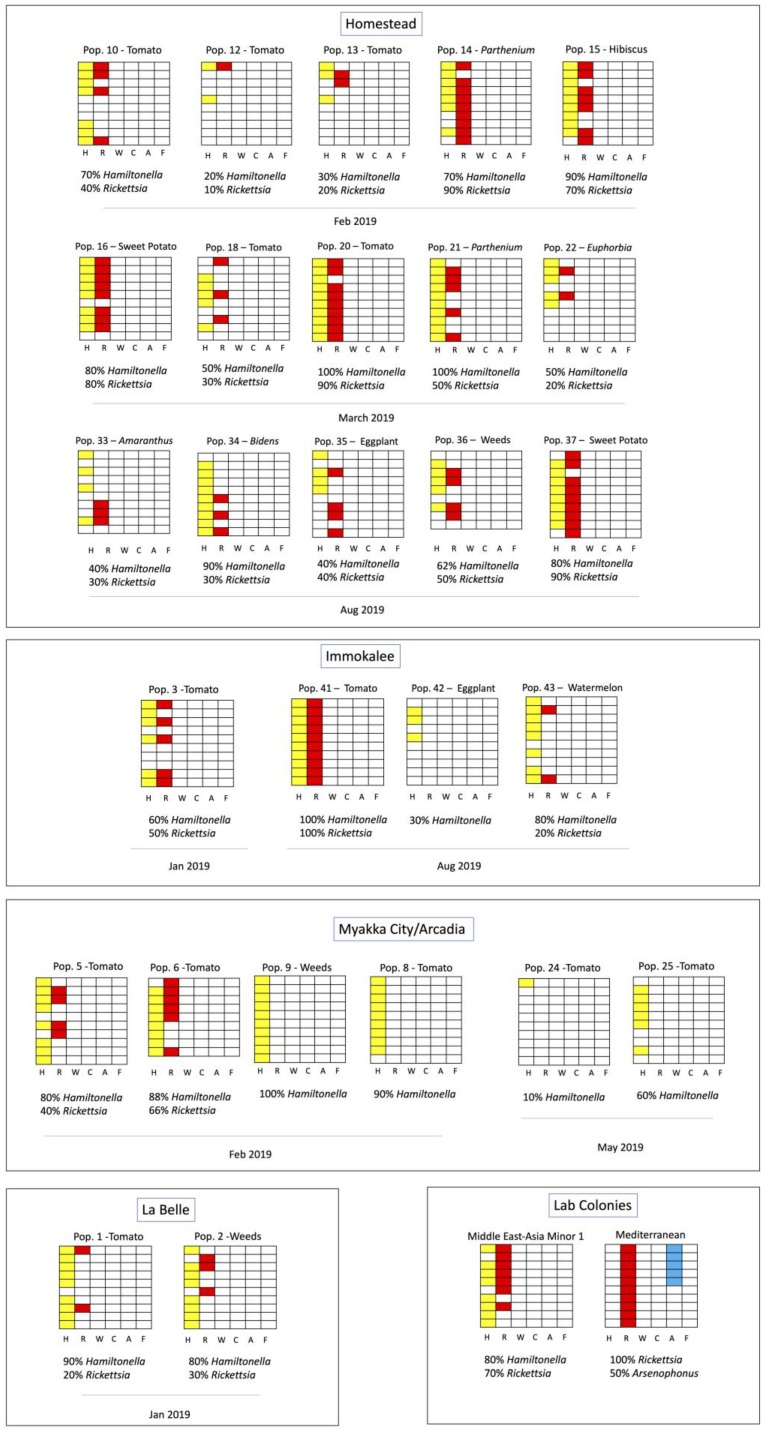
The prevalent facultative endosymbionts pattern observed throughout Florida was Rickettsia in co-infection with Hamiltonella (Figure 2).
Figure 2.
Set of facultative endosymbionts from Bemisia tabaci populations collected in Florida. (H) Hamiltonella, (R) Rickettsia, (W) Wolbachia, (C) Cardinium, (A) Arsenophonus, (F) Fritschea. Vertical columns represent the different symbionts tested as indicated by the letters at the bottom of each column, and each horizontal column represents one individual tested for the presence of the six facultative endosymbionts. Positive infections are represented by color filled columns, yellow (Hamiltonella), red (Rickettsia) and blue (Arsenophonus). Collection site, date, plant host and population IDs are indicated.
This pattern was found in all the regions collected and was the most common pattern in Homestead. In the Myakka City/Arcadia area, most of the specimens found were only harboring Hamiltonella. We also identified the set of endosymbionts for the laboratory colonies, MEAM1 (80% Hamiltonella + 70% Rickettsia) and MED (100% Rickettsia + 50% Arsenophonus), as references. The MED colony was the only population harboring the facultative endosymbionts Arsenophonus.
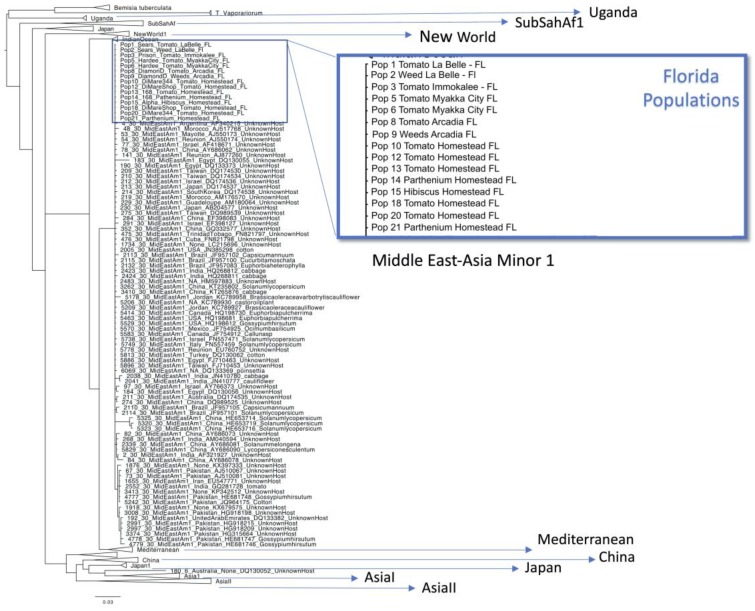
The B. tabaci mtCOI sequence alignment was 625 bp in length. The Hamiltonella alignment consisted of a total of 44 sequences and 746 bp in length. The Rickettsia alignment consisted of a total of 22 sequences and 628 bp in length. The mtCOI phylogenetic analysis grouped all the B. tabaci field populations from Florida in the MEAM1 clade. No variability was observed among the populations collected (Figure 3).
Figure 3.
Phylogenetic analysis of the mtCOI gene from Bemisia tabaci populations collected in Florida.
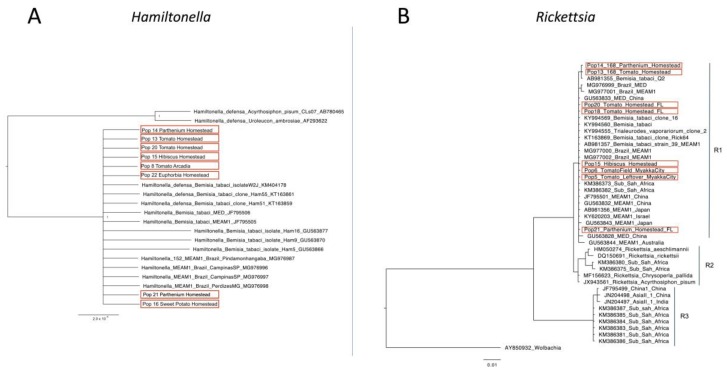
Additionally, low diversity was observed for the 16S gene rDNA of Hamiltonella as all the nucleotide sequences were grouped into one major cluster (Figure 4A). The phylogenetic analysis of the 16S gene rDNA of Rickettsia clustered the Florida populations in the R1 genetic group (Figure 4B).
Figure 4.
Phylogenetic analysis of the 16S gene rDNA of the facultative endosymbionts Hamiltonella (A) and Rickettsia (B) from Bemisia tabaci populations. The samples from Florida, USA, obtained in this study are highlighted in red boxes.
4. Discussion
The survey was conducted in five different counties, and we analyzed 27 field populations from at least seven different host plants, including the cultivated crops tomato, sweetpotato and hibiscus, as well as weeds, including Parthenium sp., Euphorbia sp. and Amaranthus sp. (Table 1). Among the field populations, all the specimens were identified as B. tabaci MEAM1 species and the only facultative endosymbionts found were Hamiltonella and Rickettsia (Figure 1).
Table 1.
Populations of Bemisia tabaci collected in Florida and their set of endosymbionts.
| Pop. ID | Collection Date | Town | Host Crop | Species | Hamiltonella GenBank Accession | Rickettsia GenBank Accession | mtCOI GenBank Accession | Endosymbionts | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H | R | W | C | A | F | ||||||||
| B Colony | 1990s | Bradenton | Gossypium hirsutum | MEAM1 | - | - | - | 8/10 | 7/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| Q Colony | Jul 2017 | Palm Beach | G. hirsutum * | MED | - | - | - | 0/10 | 10/10 | 0/10 | 0/10 | 5/10 | 0/10 |
| 1 | Jan 2019 | LaBelle | Solanum lycopersicum | MEAM1 | - | - | MN972494 | 9/10 | 2/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 2 | Jan 2019 | LaBelle | Weeds | MEAM1 | - | - | MN972495 | 8/10 | 3/10 | 0/6 | 0/10 | 0/10 | 0/10 |
| 3 | Jan 2019 | Immokalee | S. lycopersicum | MEAM1 | - | - | MN972496 | 6/10 | 4/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 5 | Jan 2019 | Myakka City | S. lycopersicum | MEAM1 | - | MN973880 | MN972497 | 8/10 | 4/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 6 | Feb 2019 | Myakka City | S. lycopersicum | MEAM1 | - | MN973881 | MN972498 | 8/9 | 6/8 | 0/9 | 0/10 | 0/10 | 0/10 |
| 8 | Feb 2019 | Arcadia | S. lycopersicum | MEAM1 | MN973892 | - | MN972508 | 9/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 9 | Feb 2019 | Arcadia | Weeds | MEAM1 | - | - | MN972499 | 10/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 10 | Feb 2019 | Homestead | S. lycopersicum | MEAM1 | - | - | MN972500 | 7/10 | 4/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 12 | Feb 2019 | Homestead | S. lycopersicum | MEAM1 | - | - | MN972501 | 2/7 | 1/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 13 | Feb 2019 | Homestead | S. lycopersicum | MEAM1 | MN973889 | MN973886 | MN972502 | 3/10 | 2/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 14 | Feb 2019 | Homestead | Parthenium sp. | MEAM1 | MN973888 | MN973887 | MN972503 | 7/10 | 9/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 15 | Feb 2019 | Homestead | Hibiscus sp. | MEAM1 | MN973891 | MN973882 | MN972504 | 9/10 | 7/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 16 | Mar 2019 | Homestead | Ipomoea batatas | MEAM1 | MN973895 | - | - | 8/10 | 8/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 18 | Mar 2019 | Homestead | S. lycopersicum | MEAM1 | - | MN973883 | MN972505 | 5/10 | 3/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 20 | Mar 2019 | Homestead | S. lycopersicum | MEAM1 | MN973890 | MN973884 | MN972506 | 10/10 | 9/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 21 | Mar 2019 | Homestead | Parthenium sp. | MEAM1 | MN973894 | MN973885 | MN972507 | 10/10 | 5/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 22 | Mar 2019 | Homestead | Euphorbia SP. | MEAM1 | MN973893 | - | - | 5/10 | 2/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 24 | May 2019 | Myakka City | S. lycopersicum | MEAM1 | - | - | - | 1/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 25 | May 2019 | Myakka City | S. lycopersicum | MEAM1 | - | - | - | 6/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 33 | Aug 2019 | Homestead | Amaranthus sp. | MEAM1 | - | - | - | 4/10 | 3/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 34 | Aug 2019 | Homestead | Bidens sp. | MEAM1 | - | - | - | 9/10 | 3/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 35 | Aug 2019 | Homestead | Solanum melongena | MEAM1 | - | - | - | 4/10 | 4/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 36 | Aug 2019 | Homestead | Weeds | MEAM1 | - | - | - | 5/8 | 4/8 | 0/10 | 0/10 | 0/10 | 0/10 |
| 37 | Aug 2019 | Homestead | I. batatas | MEAM1 | - | - | - | 8/10 | 9/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 41 | Aug 2019 | Immokalee | S. lycopersicum | MEAM1 | - | - | - | 10/10 | 10/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 42 | Aug 2019 | Immokalee | S. melongena | MEAM1 | - | - | - | 3/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 43 | Aug 2019 | Immokalee | Citrullus lanatus | MEAM1 | - | - | - | 8/10 | 2/10 | 0/10 | 0/10 | 0/10 | 0/10 |
* Originally collected from hibiscus in the field but has been maintained on cotton since July 2017.
The high rates of Hamiltonella in some populations collected from tomatoes is a concern as this endosymbiont has a role in the transmission of the begomovirus, Tomato yellow leaf curl virus (TYLCV), which causes yellow leaf curl disease in tomato. Hamiltonella encodes a GroEL chaperonin homologue protein that safeguards the begomovirus particles in the hemolymph of the whitefly vector [15]. This disease is a major problem in Florida and is estimated to cause tens of millions of dollars in losses for the tomato crop [30].
The facultative endosymbionts Rickettsia was also found in different rates across Florida. Several populations were found with rates lower than 30% (i.e., Populations #8, #9, #12, #13, #18, #22, #24, #25, #33, #34, #42, #43). A previous study has associated the presence of Rickettsia to increased insecticide susceptibility to acetamiprid, thiamethoxam, spiromesifen and pyriproxyfen in whiteflies [14]. Thus, populations with low incidences of Rickettsia might be a factor to be observed in a resistance monitoring study.
The first collection in the Myakka City/Arcadia area was carried out in February 2019, and a different set of endosymbionts was found in two populations from tomato fields located less than 3 km apart. Population #6 harbored Rickettsia (88%) + Hamiltonella (66%). Population #8 harbored only Hamiltonella (90%). Three months later, we sampled the same tomato field where Population #6 was found and the facultative endosymbionts Rickettsia was no longer detected. The new sampling was identified as Population #24 (10% Hamiltonella). A shift in the dynamics of facultative endosymbionts in whiteflies has been reported before in several studies. In Arizona, USA, a decline was reported in Rickettsia infection from ~95% in 2011 to 36% in 2017 in a six-year survey of B. tabaci MEAM1 in cotton [31]. In Brazil, the frequencies of Hamiltonella and Rickettsia remained very similar in B. tabaci MEAM1 species from 2015 to 2017. However, the same study reported the increasing of infection of Hamiltonella, Rickettsia and Wolbachia in MED species individuals from 2015 to 2017 [32]. In laboratory conditions, Rickettsia is vertically transmitted by female whiteflies at high rates ranging from 98%–99% [31,33]. Therefore, some unknown factors in the field must be responsible for the decline in Rickettsia infection. There are evidences that the decline might be driven by changes in context-dependent fitness effects of hosting the bacteria [31]. In contrast to other studies, the dynamics of Rickettsia in this study were observed in a shorter period of time and could have been driven by specific environmental factors.
The facultative endosymbionts’ phylogenetic trees revealed a very homogeneous genetic background for both Hamiltonella and Rickettsia in Florida (Figure 3). It is already known that Hamiltonella sequences of whiteflies from different parts of the world have low genetic diversity [9] and our data shows that populations from Florida are no exception. This study also classified Rickettsia strains from different parts of the world into three main groups [9]. The Rickettsia strain found in Florida belongs to the clade R1, which consists of invasive and indigenous members from Australia, China, Japan, Israel and Sub-Saharan African countries. A recent study suggested that the majority of the endosymbionts were acquired before the start of B. tabaci complex speciation and their spread occurred after this speciation [9]. The characterization of the Rickettsia strain found in Florida is helpful in a global context for understanding the complexity and diversity of the facultative endosymbionts in B. tabaci.
5. Conclusions
The only facultative endosymbionts found among the B. tabaci populations collected in Florida were Hamiltonella and Rickettsia. The phylogenetic analysis revealed low genetic diversity of whiteflies and their endosymbionts. Further studies are being carried out to verify if there is any evidence of the association between the set of facultative endosymbionts in whiteflies and pest status in the field, such as insecticide resistance.
Author Contributions
Conceptualization, B.R.D.M. and H.A.S.; methodology, B.R.D.M.; software, B.R.D.M.; investigation, B.R.D.M. and H.A.S.; writing—original draft preparation, B.R.D.M.; funding acquisition, H.A.S.; writing—review and editing, B.R.D.M. and H.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded with support from the USDA NIFA SCRI program, grant number 2018-51181-28420.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gilbertson R.L., Batuman O., Webster C.G., Adkins S. Role of the Insect Supervectors Bemisia tabaci and Frankliniella occidentalis in the Emergence and Global Spread of Plant Viruses. Annu. Rev. Virol. 2015;2:67–93. doi: 10.1146/annurev-virology-031413-085410. [DOI] [PubMed] [Google Scholar]
- 2.Navas-Castillo J., Fiallo-Olivé E., Sánchez-Campos S. Emerging Virus Diseases Transmitted by Whiteflies. Annu. Rev. Phytopathol. 2011;49:219–248. doi: 10.1146/annurev-phyto-072910-095235. [DOI] [PubMed] [Google Scholar]
- 3.De Barro P.J., Liu S.-S., Boykin L.M., Dinsdale A.B. Bemisia tabaci: A Statement of Species Status. Annu. Rev. Entomol. 2011;56:1–19. doi: 10.1146/annurev-ento-112408-085504. [DOI] [PubMed] [Google Scholar]
- 4.Brown J.K., Frohlich D.R., Rosell R.C. The Sweetpotato or Silverleaf Whiteflies: Biotypes of Bemisia tabaci or a Species Complex? Annu. Rev. Entomol. 1995;40:511–534. doi: 10.1146/annurev.en.40.010195.002455. [DOI] [Google Scholar]
- 5.Perring T.M. The Bemisia tabaci species complex. Crop Prot. 2001;20:725–737. doi: 10.1016/S0261-2194(01)00109-0. [DOI] [Google Scholar]
- 6.Rossitto De Marchi B., Massaharu Marubayashi J., Madoglio Favara G., Atsushi Yuki V., Watanabe L.F.L.F.M., Barbosa L.F.L.F., Pavan M.A.M.A., Krause-Sakate R. Comparative transmission of five viruses by Bemisia tabaci NW2 and MEAM1. Trop. Plant Pathol. 2017;1 [Google Scholar]
- 7.Polston J.E., De Barro P., Boykin L.M. Transmission specificities of plant viruses with the newly identified species of the Bemisia tabaci species complex. Pest Manag. Sci. 2014;70:1547–1552. doi: 10.1002/ps.3738. [DOI] [PubMed] [Google Scholar]
- 8.Horowitz A.R., Kontsedalov S., Khasdan V., Ishaaya I. Biotypes B and Q of Bemisia tabaci and their relevance to neonicotinoid and pyriproxyfen resistance. Arch. Insect Biochem. 2005;58:216–225. doi: 10.1002/arch.20044. [DOI] [PubMed] [Google Scholar]
- 9.Kanakala S., Ghanim M. Global genetic diversity and geographical distribution of Bemisia tabaci and its bacterial endosymbionts. PLoS ONE. 2019;14:e0213946. doi: 10.1371/journal.pone.0213946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mckenzie C.L., Hodges G., Osborne L.S., Byrne F.J., Shatters R.G. Distribution of Bemisia tabaci (Hemiptera: Aleyrodidae) Biotypes in Florida-Investigating the Q Invasion. J. Econ. Entomol. 2009;102:670–676. doi: 10.1603/029.102.0227. [DOI] [PubMed] [Google Scholar]
- 11.Vegetables Annual Summary. [(accessed on 19 February 2020)]; Available online: http://usda.mannlib.cornell.edu/MannUsda/viewDocumentInfo.do?documentID=1183files/97/viewDocumentInfo.html.
- 12.Bellows T.S., Perring T.M., Gill R.J., Headrick D.H. Description of a Species of Bemisia (Homoptera: Aleyrodidae) Ann. Entomol. Soc. Am. 1994;87:195–206. doi: 10.1093/aesa/87.2.195. [DOI] [Google Scholar]
- 13.Sloan D.B., Moran N.A. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol. Lett. 2012;8:986–989. doi: 10.1098/rsbl.2012.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kontsedalov S., Zchori-Fein E., Chiel E., Gottlieb Y., Inbar M., Ghanim M. The presence of Rickettsia is associated with increased susceptibility of Bemisia tabaci (Homoptera: Aleyrodidae) to insecticides. Pest Manag. Sci. 2008;64:789–792. doi: 10.1002/ps.1595. [DOI] [PubMed] [Google Scholar]
- 15.Morin S., Ghanim M., Sobol I., Czosnek H. The GroEL Protein of the Whitefly Bemisia tabaci Interacts with the Coat Protein of Transmissible and Nontransmissible Begomoviruses in the Yeast Two-Hybrid System. Virology. 2000;276:404–416. doi: 10.1006/viro.2000.0549. [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb Y., Zchori-Fein E., Mozes-Daube N., Kontsedalov S., Skaljac M., Brumin M., Sobol I., Czosnek H., Vavre F., Fleury F., et al. The transmission efficiency of tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J. Virol. 2010;84:9310–9317. doi: 10.1128/JVI.00423-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brumin M., Kontsedalov S., Ghanim M. Rickettsia influences thermotolerance in the whitefly Bemisia tabaci B biotype. Insect Sci. 2011;18:57–66. doi: 10.1111/j.1744-7917.2010.01396.x. [DOI] [Google Scholar]
- 18.Zchori-Fein E., Brown J.K. Diversity of Prokaryotes Associated with Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) Ann. Entomol. Soc. Am. 2002;95:711–718. doi: 10.1603/0013-8746(2002)095[0711:DOPAWB]2.0.CO;2. [DOI] [Google Scholar]
- 19.Weeks A.R., Velten R., Stouthamer R. Incidence of a new sex-ratio-distorting endosymbiotic bacterium among arthropods. Proceedings. Biol. Sci. 2003;270:1857–1865. doi: 10.1098/rspb.2003.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett K.D.E., Thao M., Horn M., Dyszynski G.E., Baumann P. Novel chlamydiae in whiteflies and scale insects: Endosymbionts “Candidatus Fritschea bemisiae” strain Falk and “Candidatus Fritschea eriococci” strain Elm. Int. J. Syst. Evol. Microbiol. 2005;55:1581–1587. doi: 10.1099/ijs.0.63454-0. [DOI] [PubMed] [Google Scholar]
- 21.Gottlieb Y., Ghanim M., Chiel E., Gerling D., Portnoy V., Steinberg S., Tzuri G., Horowitz A.R., Belausov E., Mozes-Daube N., et al. Identification and Localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae) Appl. Environ. Microbiol. 2006;72:3646–3652. doi: 10.1128/AEM.72.5.3646-3652.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan H., Li X., Ge D., Wang S., Wu Q., Xie W., Jiao X., Chu D., Liu B., Xu B., et al. Factors Affecting Population Dynamics of Maternally Transmitted Endosymbionts in Bemisia tabaci. PLoS ONE. 2012;7:e30760. doi: 10.1371/journal.pone.0030760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh P.S., Metzger D.A., Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. doi: 10.2144/000114018. [DOI] [PubMed] [Google Scholar]
- 24.Kontsedalov S., Abu-Moch F., Lebedev G., Czosnek H., Horowitz A.R., Ghanim M. Bemisia tabaci Biotype Dynamics and Resistance to Insecticides in Israel During the Years 2008-2010. J. Integr. Agric. 2012;11:312–320. doi: 10.1016/S2095-3119(12)60015-X. [DOI] [Google Scholar]
- 25.De Barro P.J., Scott K.D., Graham G.C., Lange C.L., Schutze M.K. Isolation and characterization of microsatellite loci in Bemisia tabaci. Mol. Ecol. Notes. 2002;3:40–43. doi: 10.1046/j.1471-8286.2003.00344.x. [DOI] [Google Scholar]
- 26.Frohlich D.R., Torres-Jerez I., Bedford I.D., Markham P.G., Brown J.K. A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol. Ecol. 1999;8:1683–1691. doi: 10.1046/j.1365-294x.1999.00754.x. [DOI] [PubMed] [Google Scholar]
- 27.Heddi A., Grenier A.M., Khatchadourian C., Charles H., Nardon P. Four intracellular genomes direct weevil biology: Nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc. Natl. Acad. Sci. USA. 1999;96:6814–6819. doi: 10.1073/pnas.96.12.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thao M.L., Baumann P. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl. Environ. Microbiol. 2004;70:3401–3406. doi: 10.1128/AEM.70.6.3401-3406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boykin L.M., De Barro P.J. A practical guide to identifying members of the Bemisia tabaci species complex: And other morphologically identical species. Front. Ecol. Evol. 2014;2:45. doi: 10.3389/fevo.2014.00045. [DOI] [Google Scholar]
- 30.Fonsah E.G., Chen Y., Diffie S., Srinivansan R., Riley D. Economic Productivity and Profitability Analysis for Whiteflies and Tomato Yellow Leaf Curl Virus (TYLCV) Management Options. J. Agric. Environ. Sci. 2018;7:1–9. [Google Scholar]
- 31.Bockoven A.A., Bondy E.C., Flores M.J., Kelly S.E., Ravenscraft A.M., Hunter M.S. What Goes Up Might Come Down: The Spectacular Spread of an Endosymbiont Is Followed by Its Decline a Decade Later. Microb. Ecol. 2019:1–13. doi: 10.1007/s00248-019-01417-4. [DOI] [PubMed] [Google Scholar]
- 32.de Moraes L.A., Muller C., de Freitas Bueno R.C.O., Santos A., Bello V.H., De Marchi B.R., Watanabe L.F.M., Marubayashi J.M., Santos B.R., Yuki V.A., et al. Distribution and phylogenetics of whiteflies and their endosymbiont relationships after the Mediterranean species invasion in Brazil. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-32913-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Himler A.G., Adachi-Hagimori T., Bergen J.E., Kozuch A., Kelly S.E., Tabashnik B.E., Chiel E., Duckworth V.E., Dennehy T.J., Zchori-Fein E., et al. Rapid Spread of a Bacterial Symbiont in an Invasive Whitefly Is Driven by Fitness Benefits and Female Bias. Science. 2011;332:254–256. doi: 10.1126/science.1199410. [DOI] [PubMed] [Google Scholar]